13.3
Impact Factor
Theranostics 2023; 13(10):3290-3309. doi:10.7150/thno.84978 This issue Cite
Research Paper
Comprehensive machine learning-generated classifier identifies pro-metastatic characteristics and predicts individual treatment in pancreatic cancer: A multicenter cohort study based on super-enhancer profiling
1. Department of General Surgery, Pancreatic Disease Center, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, P.R. China.
2. Research Institute of Pancreatic Diseases, Shanghai Key Laboratory of Translational Research for Pancreatic Neoplasms, Shanghai Jiao Tong University School of Medicine, Shanghai, P.R. China.
3. State Key Laboratory of Oncogenes and Related Genes, Institute of Translational Medicine, Shanghai Jiao Tong University, Shanghai, P.R. China.
4. Department of General Surgery, Taiyuan Central Hospital, Shanxi, P.R. China.
† These authors have contributed equally to this work and share the first authorship.
Abstract
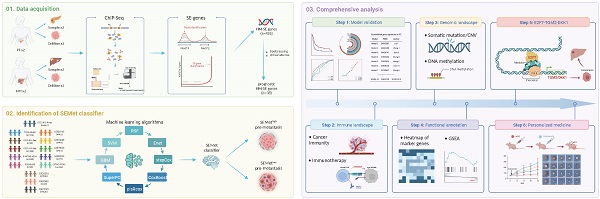
Rationale: Accumulating evidence illustrated that the reprogramming of the super-enhancers (SEs) landscape could promote the acquisition of metastatic features in pancreatic cancer (PC). Given the anatomy-based TNM staging is limited by the heterogeneous clinical outcomes in treatment, it is of great clinical significance to tailor individual stratification and to develop alternative therapeutic strategies for metastatic PC patients based on SEs.
Methods: In our study, ChIP-Seq analysis for H3K27ac was performed in primary pancreatic tumors (PTs) and hepatic metastases (HMs). Bootstrapping and univariate Cox analysis were implemented to screen prognostic HM-acquired, SE-associated genes (HM-SE genes). Then, based on 1705 PC patients from 14 multicenter cohorts, 188 machine-learning (ML) algorithm integrations were utilized to develop a comprehensive super-enhancer-related metastatic (SEMet) classifier.
Results: We established a novel SEMet classifier based on 38 prognostic HM-SE genes. Compared to other clinical traits and 33 published signatures, the SEMet classifier possessed robust and powerful performance in predicting prognosis. In addition, patients in the SEMetlow subgroup owned dismal survival rates, more frequent genomic alterations, and more activated cancer immunity cycle as well as better benefits in immunotherapy. Remarkably, there existed a tight correlation between the SEMetlow subgroup and metastatic phenotypes of PC. Among 18 SEMet genes, we demonstrated that E2F7 may promote PC metastasis through the upregulation of TGM2 and DKK1. Finally, after in silico screening of potential compounds targeted SEMet classifier, results revealed that flumethasone could enhance the sensitivity of metastatic PC to routine gemcitabine chemotherapy.
Conclusion: Overall, our study provided new insights into personalized treatment approaches in the clinical management of metastatic PC patients.
Keywords: super-enhancer, machine-learning algorithm, personalized treatment, multicenter study, metastatic pancreatic cancer
 Global reach, higher impact
Global reach, higher impact