13.3
Impact Factor
Theranostics 2022; 12(3):1074-1096. doi:10.7150/thno.65694 This issue Cite
Research Paper
Single-cell RNA landscape of the osteoimmunology microenvironment in periodontitis
1. Department of Basic Science of Stomatology, The Affiliated Stomatological Hospital of Nanjing Medical University, Nanjing, China; Jiangsu Province Key Laboratory of Oral Diseases, Nanjing, China; Jiangsu Province Engineering Research Center of Stomatological Translational Medicine, Nanjing, China.
2. Department of Pathology and Laboratory Medicine and Center for Musculoskeletal Research, University of Rochester Medical Center, Rochester, NY, USA.
3. State Key Laboratory of Reproductive Medicine, The Research Center for Bone and Stem Cells, Department of Anatomy, Histology and Embryology, Nanjing Medical University, Nanjing, China.
*These authors contributed equally to this work.
Abstract
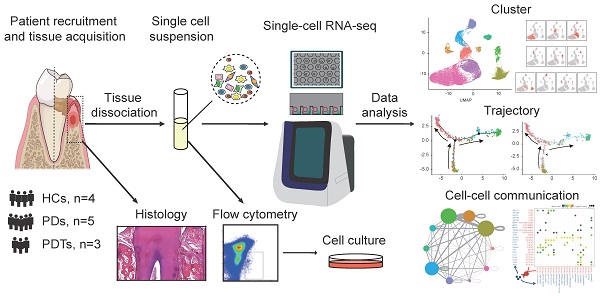
Single-cell RNA sequencing (scRNA-seq) enables specific profiling of cell populations at single-cell resolution. The osteoimmunology microenvironment in the occurrence and development of periodontitis remains poorly understood at the single-cell level. In this study, we used single-cell transcriptomics to comprehensively reveal the complexities of the molecular components and differences with counterparts residing in periodontal tissues.
Methods: We performed scRNA-seq to identify 51248 single cells from healthy controls (n=4), patients with severe chronic periodontitis (n=5), and patients with severe chronic periodontitis after initial periodontal therapy within 1 month (n=3). Uniform manifold approximation and projection (UMAP) were further conducted to explore the cellular composition of periodontal tissues. Pseudotime cell trajectory and RNA velocity analysis, combined with gene enrichment analysis were used to reveal the molecular pathways underlying cell fate decisions. CellPhoneDB were performed to identify ligand-receptor pairs among the major cell types in the osteoimmunology microenvironment of periodontal tissues.
Results: A cell atlas of the osteoimmunology microenvironment in periodontal tissues was characterized and included ten major cell types, such as fibroblasts, monocytic cells, endothelial cells, and T and B cells. The enrichment of TNFRSF21+ fibroblasts with high expression of CXCL1, CXCL2, CXCL5, CXCL6, CXCL13, and IL24 was detected in patients with periodontitis compared to healthy individuals. The fractions of CD55+ mesenchymal stem cells (MSCs), APOE+ pre-osteoblasts (pre-OBs), and IBSP+ osteoblasts decreased significantly in response to initial periodontal therapy. In addition, CXCL12+ MSC-like pericytes could convert their identity into a pre-OB state during inflammatory responses even after initial periodontal therapy confirmed by single-cell trajectory. Moreover, we portrayed the distinct subtypes of monocytic cells and abundant endothelial cells significantly involved in the immune response. The heterogeneity of T and B cells in periodontal tissues was characterized. Finally, we mapped osteoblast/osteoclast differentiation mediators to their source cell populations by identifying ligand-receptor pairs and highlighted the effects of Ephrin-Eph signaling on bone regeneration after initial periodontal therapy.
Conclusions: Our analyses uncovered striking spatiotemporal dynamics in gene expression, population composition, and cell-cell interactions during periodontitis progression. These findings provide insights into the cellular and molecular underpinning of periodontal bone regeneration.
Keywords: Osteoimmunology, Periodontitis, Single-cell RNA-seq, Mesenchymal stem cells, alveolar bone
 Global reach, higher impact
Global reach, higher impact