13.3
Impact Factor
Theranostics 2025; 15(11):5277-5311. doi:10.7150/thno.113650 This issue Cite
Review
Exosomes: innovative biomarkers leading the charge in non-invasive cancer diagnostics
1. Department of Neurosurgery, Haikou Affiliated Hospital of Central South University Xiangya School of Medicine, Haikou, China, 570208.
2. School of Basic Medicine and Clinical Pharmacy, China Pharmaceutical University, Nanjing, China, 211198.
3. Department of Neurosurgery, Central Hospital of Zhuzhou, Zhuzhou, Hunan, China, 412000.
4. Department of Neurosurgery, Affiliated Nanhua Hospital, University of South China, Hengyang, Hunan, China, 533000.
5. Department of Neurosurgery Second Branche, Hunan Provincial People's Hospital (The First Affiliated Hospital of Hunan Normal University), Changsha, Hunan, China, 410005.
6. Department of Radiation Oncology, The First Affiliated Hospital Zhejiang University, Hangzhou, China, 310009.
*Co-first author: Jiale Li, Ailin Wang.
Received 2025-3-12; Accepted 2025-4-6; Published 2025-4-13
Abstract
Exosomes, nanoscale extracellular vesicles secreted by diverse cell types, have emerged as promising biomarkers for non-invasive tumor diagnostics, offering significant advantages over traditional methods. These vesicles, typically ranging from 30 to 150 nanometers in size, carry a diverse cargo of proteins, lipids, RNA, and microRNAs, which reflect the molecular alterations occurring within their parent cells. Notably, exosomes can be isolated from easily accessible biofluids such as blood, urine, and saliva, making them ideal candidates for liquid biopsy applications. This review explores the transformative potential of exosome-based biomarkers in the early detection and monitoring of cancers across diverse organ systems, including respiratory, digestive, hematological, neurological, endocrine malignancies and so on. Special emphasis is placed on their application in clinical trials, where exosome-based diagnostics have demonstrated promising results in detecting tumors at early stages and monitoring treatment responses, offering a less invasive and more accessible alternative to traditional biopsies. While recent advancements in exosome isolation and characterization technologies have significantly improved the sensitivity and specificity of these diagnostics, challenges such as biological heterogeneity, lack of standardization, and regulatory hurdles remain. Nevertheless, exosome-based diagnostics hold the promise of providing real-time, dynamic insights into tumor progression, enhancing personalized medicine. The integration of exosomes into clinical practice could revolutionize cancer diagnostics and therapy, improving patient outcomes. Further research and large-scale clinical validation are essential to fully realize the clinical potential of exosome-based biomarker applications in routine clinical settings.
1. Introduction
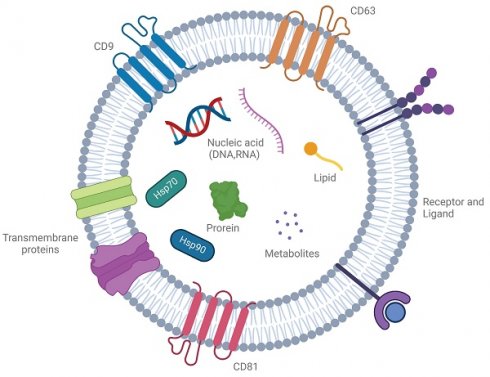
Exosomes are nanosized, membrane-bound vesicles ranging from 30 to 150 nm, produced by many cell types, including cancer cells, immune cells, and neurons. These vesicles play a crucial role in intercellular communication by transferring bioactive molecules such as proteins, lipids, RNAs, and metabolites [1] (Figure 1). Known for protecting their cargo from degradation, exosomes are key messengers in both normal and disease-related processes, reflecting the molecular profile of their parent cells. Particularly, exosomes derived from diseased cells, such as cancer or infected cells, carry disease-specific signatures, making them promising candidates for non-invasive diagnostic applications [2].
Traditionally, disease diagnosis has relied on invasive techniques like tissue biopsies. Although these are valuable, they are limited by sampling biases, the need for complex surgical procedures, and the inability to capture the dynamic progression of diseases. This is especially true for cancers, where the molecular variation across tumor regions complicates diagnostic accuracy [3]. As an alternative, liquid biopsy--a non-invasive approach analyzing biofluids such as blood, urine, and saliva for disease-related biomarkers--has gained attention. Exosomes, due to their stable presence in these biofluids, have become ideal candidates for liquid biopsy. Their molecular content mirrors the alterations in disease states, offering real-time insights into disease detection, monitoring, and progression [4]. Recent advancements in exosome isolation and characterization techniques, such as ultracentrifugation, immunoaffinity capture, and microfluidics, have further boosted the potential of exosome-based diagnostics [5]. These technologies allow for the extraction and analysis of exosomal proteins, RNAs, and lipids, which have shown promise as biomarkers for early disease detection and monitoring. For example, exosomal miRNAs and proteins have been identified as biomarkers for early-stage cancers, while exosomal RNAs have been used to detect infectious diseases and monitor neurodegenerative conditions [6].
However, several challenges must be addressed to fully realize the diagnostic potential of exosomes. These include the need for standardized, scalable isolation methods, the identification of specific and sensitive biomarkers, and the clinical validation of exosome-based diagnostics across diverse patient populations. Additionally, the heterogeneity of exosomes, both within and between diseases, presents another challenge for their widespread use [7]. This review aims to provide a comprehensive overview of exosomal biomarkers and their transformative potential in non-invasive diagnostics. By examining the current state of exosome-based diagnostic technologies, the molecular potential of exosomal content, and the clinical feasibility of their application, we seek to highlight impact of exosomes on revolutionizing early disease detection, monitoring disease progression, and facilitating personalized medicine. Ultimately, the integration of exosome-based diagnostics into clinical practice holds the promise of improving patient outcomes by offering a more precise, less invasive, and accessible approach to disease management.
A cell-to-cell transit system in the human body with pleiotropic functions. Exosomes are extracellular vesicles generated by all cells and they carry nucleic acids, proteins, lipids, and metabolites. They are mediators of near and long-distance intercellular communication in health and disease and affect various aspects of cell biology. Created with BioRender.com.
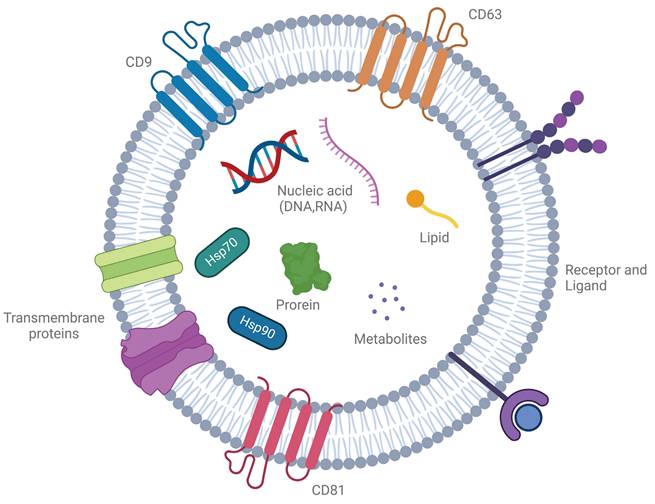
Overview of the process for exosome biogenesis. Exosome biogenesis is primarily centered around the formation of multivesicular bodies (MVBs). These structures are typically derived from the process of endocytosis, during which various mechanisms facilitate the inward budding of the plasma membrane, leading to the creation of early endosomes. As these endosomes mature, they undergo further processes that result in the formation of MVBs, which ultimately give rise to exosomes. Created with BioRender.com.
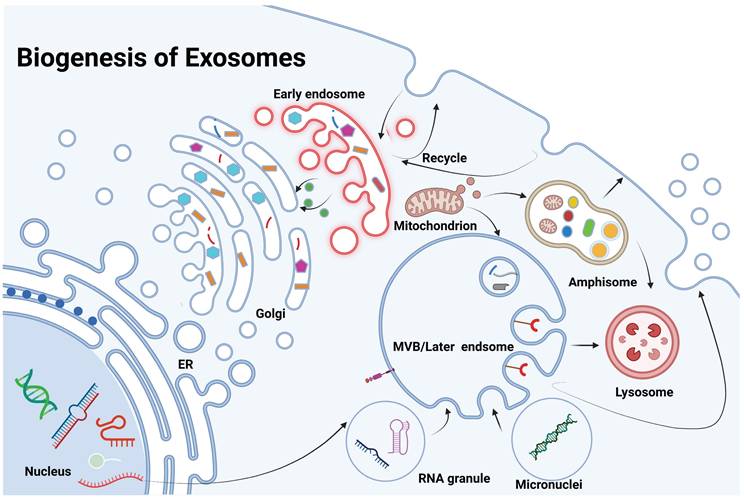
2. Exosome biogenesis and role in tumors
2.1. Biogenesis of exosomes
Exosome biogenesis is a highly regulated, multi-step process involving the formation, maturation, and release of multivesicular bodies (MVBs), which are key intermediates in exosome production. This process begins with the invagination of the plasma membrane to form early endosomes that sort cellular cargo. As these early endosomes mature into late endosomes or MVBs, further inward budding generates intraluminal vesicles (ILVs), which become exosomes upon release. These ILVs selectively encapsulate lipids, proteins, and nucleic acids, reflecting the physiological or pathological state of the source cell [8] (Figure 2). The release of exosomes is regulated by the fate of MVBs, which may either fuse with lysosomes for degradation or with the plasma membrane to release exosomes. This process is controlled by Rab GTPases, such as Rab27a and Rab27b, ensuring proper timing and spatial regulation of exosome release [9]. There are two primary pathways for ILV formation and cargo sorting: the ESCRT-dependent and ESCRT-independent pathways. The ESCRT-dependent pathway involves the endosomal sorting complex required for transport (ESCRT), which mediates the formation of ILVs through a series of sequential complexes (ESCRT-0, -I, -II, and -III). ESCRT-0 recognizes ubiquitinated proteins, while ESCRT-I and -II facilitate membrane deformation, and ESCRT-III completes vesicle scission. Accessory proteins like Alix and TSG101 assist in this process, with VPS4 providing energy for complex disassembly. The ESCRT-independent pathway relies on lipid and protein-based mechanisms. Ceramide, generated by neutral sphingomyelinase 2 (nSMase2), induces membrane curvature, and tetraspanins like CD9, CD63, and CD81 organize the membrane and aid in cargo sorting [10]. Cargo loading into ILVs is highly selective and involves various molecular players. Heat shock proteins (e.g., HSP90, HSP70) incorporate functional proteins, while RNA-binding proteins like hnRNPA2B1 and YBX1 selectively package microRNAs and other non-coding RNAs into exosomes. Lipids such as cholesterol and phosphatidylserine stabilize the vesicles and facilitate membrane fusion during release [11].
The above-explained mechanisms of exosome biogenesis are closely linked to tumor initiation and progression. In cancer, dysregulated exosome biogenesis leads to the secretion of exosomes that carry oncogenic cargo, influencing the tumor microenvironment (TME) and promoting tumor progression. These exosomes, enriched with growth factors, metalloproteinases, and microRNAs, reprogram normal cells into malignant phenotypes and contribute to clonal expansion and intratumoral heterogeneity. Exosomes also facilitate immune evasion by carrying immunosuppressive molecules such as PD-L1, TGF-β, and IL-10, while promoting metastasis through the transfer of matrix metalloproteinases (MMPs). Furthermore, exosomes play a key role in drug resistance by transferring resistance factors, including drug efflux pumps and mutant proteins, along with miRNAs that affect apoptosis and drug metabolism, complicating treatment strategies [12].
The dynamic interplay between exosome biogenesis and cancer progression highlights the importance of exosomes as not only vehicles for intercellular communication but also active mediators of tumor progression, immune modulation, and treatment resistance. Given their central role in these processes, exosomes present an intriguing therapeutic target for modulating cancer progression, as well as a non-invasive biomarker for early detection, monitoring disease progression, and evaluating therapeutic responses. Understanding the precise molecular mechanisms underlying exosome biogenesis and cargo selection in cancer cells is essential for the development of exosome-based therapies and diagnostic tools [13].
2.2. The role of exosomes in tumor progression
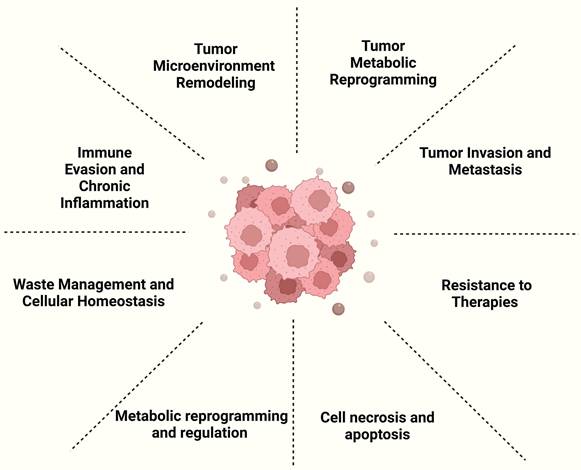
Exosomes serve as a key factor in cancer progression by remodeling the tumor microenvironment and influencing key processes such as tumor cell growth, migration, immune evasion, and therapy resistance. They facilitate tumor growth by transporting molecules, including non-coding RNAs, that regulate cancer cell behavior and promote angiogenesis. Exosomes also trigger macrophage polarization and chronic inflammation, enhancing tumor survival and dissemination. Furthermore, exosomes have been identified as key mediators of resistance to cancer therapies, as they carry molecules associated with drug resistance, diminishing the efficacy of therapies including chemotherapy and radiotherapy. Due to their presence in body fluids, exosomes hold significant potential as non-invasive biomarkers for cancer detection and prognosis [14] (Figure 3). Below, we provide a selection of representative examples that, while not exhaustive, highlight their significance.
2.2.1. Tumor Microenvironment (TME) remodeling
Exosomes are pivotal mediators of intercellular communication within the TME, a sophisticated ecosystem consisting of tumor cells, stromal cells (including cancer-associated fibroblasts or CAFs), immune cells, blood vessels, and the extracellular matrix (ECM) [15]. These nanoscale vesicles, containing bioactive molecules such as proteins, RNAs (including miRNAs, mRNAs, and lncRNAs), lipids, and metabolites, profoundly influence the TME's dynamics, facilitating tumor progression, metastasis, and immune evasion. Tumor-derived exosomes alter the behavior of surrounding non-tumor cells by transferring tumor-specific molecules, such as oncogenic miRNAs, which induce CAFs to adopt pro-tumor phenotypes, thereby promoting tumor growth, invasion, and metastatic potential [16]. Additionally, exosomes facilitate immune evasion by delivering immunosuppressive molecules, including PD-L1 and TGF-β, which dampen anti-tumor immune responses and reprogram immune cells, including macrophages, towards an immunosuppressive M2 phenotype. Furthermore, exosomes are crucial for tumor angiogenesis by transferring angiogenic factors, such as VEGF, to endothelial cells, stimulating new blood vessel formation and increasing endothelial barrier permeability. This facilitates tumor cell extravasation and metastasis. Exosomes also mediate ECM remodeling by carrying matrix metalloproteinases (MMPs) and other proteases that degrade ECM components, including fibronectin and collagen, thereby enhancing tumor cell invasion and facilitating metastatic spread. Through these mechanisms, exosomes serve as critical modulators of the TME, promoting tumor progression, immune escape, and metastasis, and therefore represent promising targets for therapeutic intervention [17].
2.2.2. Tumor metabolic reprogramming
Tumor cells undergo significant metabolic reprogramming, which is considered one of the hallmarks of cancer. This adaptation enables them to meet the increased energy demands required for rapid proliferation and survival in hostile microenvironments. A hallmark of this metabolic shift is the Warburg effect, where cancer cells mainly depend on glycolysis for ATP generation, even under normoxic conditions. This metabolic reprogramming facilitates cancer cells to efficiently produce energy and biosynthetic essential intermediates for growth. In addition to glucose metabolism, altered lipid and amino acid metabolism are also essential for sustaining cancer cell growth and metastasis [18]. Exosomes, small vesicles secreted by tumor cells, serve as key mediators of metabolic reprogramming within the TME. These extracellular vesicles carry a diverse array of molecules that allow exosomes to facilitate communication between surrounding stromal cells and cancer cells. Exosomes from tumor cells transfer metabolic signals, such as lactate, fatty acids, and miRNAs, to neighboring cells, including CAFs, endothelial cells, and immune cells, thereby promoting the metabolic adaptation necessary for tumor growth [19]. One of the central processes driven by exosomes is the "reverse Warburg effect." Tumor cells export lactate via exosomes, which is then taken up by CAFs. This lactate is converted back into pyruvate, which fuels oxidative phosphorylation (OXPHOS) in CAFs, providing them with the energy needed to support tumor cells. This metabolic symbiosis between cancer cells and stromal cells facilitates the continuous growth and invasion of tumors. Additionally, exosomes enable metabolic reprogramming of immune cells within the TME. For instance, exosome-mediated transfer of metabolic molecules can alter the function of macrophages, promoting their polarization toward an immunosuppressive M2 phenotype that supports tumor progression and immune evasion [20]. Exosomes are also key contributors to the regulation of amino acid and lipid metabolism within the TME. Exosome-mediated transfer of essential metabolic intermediates enhances tumor cell proliferation and survival. By reprogramming the metabolic landscape of stromal and immune cells, exosomes create a supportive environment that favors tumor progression, metastasis, and resistance to therapeutic interventions [21].
The role of exosomes in tumor progression. Created with BioRender.com.
2.2.3. Immune evasion
Immune evasion is a fundamental mechanism that enables tumor cells to escape recognition and destruction by the host immune system, contributing significantly to cancer progression, metastasis, and therapeutic resistance. Tumors exploit a variety of strategies to subvert immune surveillance, ensuring their survival and growth within the hostile TME. These mechanisms include immune suppression, alteration of immune cell function, and modulation of immune checkpoints, which collectively enable tumor cells to evade both adaptive and innate immune responses [22]. One of the primary immune evasion mechanisms is the reprogramming of tumor-associated immune cells, such as tumor-associated macrophages (TAMs), dendritic cells, and regulatory T cells (Tregs), into immunosuppressive phenotypes. For example, TAMs are frequently polarized toward an M2-like phenotype within the TME, promoting immune suppression and enhancing tumor progression. Tumor cells can also induce Tregs, which suppress the effectiveness of cytotoxic T cells and natural killer (NK) cells, further shielding the cancer from immune attack [23]. Moreover, tumor cells frequently exploit immune checkpoint pathways, such as the PD-1/PD-L1 axis, to inhibit T cell activation. PD-L1 is often upregulated on the surface of tumor cells and immune cells within the TME, binding to the PD-1 receptor on T cells and suppressing their ability to mount an effective anti-tumor response. This mechanism not only prevents T cells from attacking the tumor but also promotes tumor cell survival by dampening the immune response [24]. Other immune checkpoints, such as CTLA-4, are similarly exploited by tumors to impair immune activation and promote immune tolerance [25]. Exosomes, small vesicles secreted by tumor cells, play a key role in immune evasion by transferring immunosuppressive molecules, including PD-L1, TGF-β, and immunomodulatory miRNAs, to immune cells. Through this mechanism, exosomes facilitate the polarization of macrophages into an M2 phenotype and promote Treg expansion, further suppressing anti-tumor immunity. Additionally, exosomes can transfer tumor antigens to dendritic cells, altering their function and impairing their ability to initiate a robust anti-tumor immune response [26]. The metabolic reprogramming of the TME also contributes to immune evasion. Tumor cells often induce a hypoxic, acidic, and nutrient-deprived environment that inhibits the function of immune cells. For instance, low oxygen levels and high lactate concentrations within the TME can impair the cytotoxic activity of NK cells and CD8+ T cells, further facilitating immune escape. Tumor-derived exosomes, which carry metabolic molecules such as lactate, exacerbate this immunosuppressive microenvironment by influencing the metabolism of both immune and stromal cells [27].
2.2.4. Tumor invasion and metastasis
Cancer progression through invasion and metastasis is a principal cause of morbidity and mortality, and the capacity of tumor cells to infiltrate local tissues and metastasize is essential for cancer progression [28]. Exosomes, small vesicles secreted by tumor cells, act as a key factor in mediating the complex processes of invasion and metastasis. These nanoscale vesicles carry a range of bioactive molecules which facilitate communication between tumor cells and the surrounding TME, as well as distant organs involved in metastasis. Exosomes contribute to tumor invasion by promoting the degradation of the ECM, a crucial barrier that must be disrupted for tumor cells to invade surrounding tissues. Tumor-derived exosomes carry MMPs and urokinase-type plasminogen activator (uPA), both of which are enzymes that degrade ECM components such as collagen, fibronectin, and laminins. By facilitating ECM degradation, exosomes enable tumor cells to migrate and invade neighboring tissues, a critical step in the metastatic cascade [29]. Furthermore, exosomes can modulate the phenotype of stromal cells, such as fibroblasts, endothelial cells, and immune cells, to create a permissive environment for invasion. For example, exosomes derived from tumor cells can induce CAFs to promote ECM remodeling and secretion of pro-inflammatory cytokines, which further support tumor cell invasion [30]. In the context of metastasis, exosomes play a key role in establishing the pre-metastatic niche (PMN), a microenvironment in distant organs that is primed to support the survival and colonization of circulating tumor cells. Tumor-derived exosomes can transfer a variety of factors that alter the behavior of stromal cells, endothelial cells, and immune cells in distant organs, creating a favorable environment for metastasis [31]. For instance, exosomes can recruit immune cells such as myeloid-derived suppressor cells (MDSCs) and macrophages to distant organs, where they facilitate immune suppression and support the metastatic process. Exosomes can also transfer factors such as vascular endothelial growth factor (VEGF), which promote angiogenesis and increase vascular permeability, allowing tumor cells to more easily extravasate from the bloodstream and colonize secondary tissues [32]. Moreover, exosomes mediate signaling between tumor cells and distant metastatic sites by modulating the metabolic and immune landscape of the TME. Exosomes carry metabolic signals, including lactate and lipids, which influence the metabolism of stromal and immune cells in the metastatic microenvironment. These metabolic changes promote tumor cell survival and immune evasion at metastatic sites. Additionally, exosomes can modulate immune cell function by transferring immunosuppressive molecules such as PD-L1, TGF-β, and miRNAs, which help tumor cells evade immune detection and destruction in distant organs [33].
2.2.5. Resistance to therapies
Drug-resistant tumors pose a significant therapeutic challenge in cancer treatment, contributing to therapy failure and poor patient outcomes [34]. Exosomes, small vesicles secreted by tumor cells, are now pivotal in mediating the mechanisms underlying drug resistance [35]. These extracellular vesicles carry a range of bioactive molecules that facilitate intercellular communication between tumor cells, stromal cells, and immune cells within the TME. One of the primary mechanisms by which exosomes mediate drug resistance is through the transfer of molecules that modulate drug efflux. Tumor cells often upregulate ATP-binding cassette (ABC) transporters, such as P-glycoprotein, which actively expel chemotherapy drugs from the cell, causing reduced intracellular drug levels and compromised therapy outcomes [36]. Exosomes play a critical role in this process by transferring these drug efflux pumps to neighboring cells, including CAFs and endothelial cells, thereby facilitating the spread of resistance across the tumor. This intercellular exchange of drug resistance markers contributes to the heterogeneous nature of resistance within a tumor, complicating treatment strategies [37]. Exosomes also play a role in drug resistance by mediating changes in the tumor microenvironment that promote cellular survival and evade drug-induced apoptosis. For instance, exosomes can carry and transfer survival factors including epidermal growth factor receptor (EGFR) ligands, VEGF, and anti-apoptotic proteins like Bcl-2. By transferring these factors, exosomes enhance the proliferative and survival capabilities of recipient cells, making them less susceptible to chemotherapy and targeted therapies [38]. Moreover, exosomes can carry miRNAs and lncRNAs that regulate apoptosis-related pathways, further contributing to the resistance phenotype. For example, exosome-mediated delivery of miR-21, which targets pro-apoptotic genes, has been demonstrated to enhance resistance to various chemotherapeutic agents [39]. Additionally, exosomes can influence the immune response to cancer therapies, promoting immune evasion and resistance to immunotherapies. Tumor-derived exosomes often carry immunosuppressive molecules including PD-L1, TGF-β, and FasL, which can suppress the activity of cytotoxic T cells, NK cells, and other immune effector cells. By modulating the immune landscape of the TME, exosomes create an environment that supports tumor survival and enhances resistance to immune checkpoint inhibitors and other immunotherapies [40]. The metabolic reprogramming of the TME also acts as a major factor in drug resistance, and exosomes are involved in this process. Tumor cells and their microenvironment often undergo metabolic changes, such as increased glycolysis, to support rapid growth and survival. Exosomes can carry metabolic molecules, including lactate and lipids, to neighboring cells, promoting a shift in the metabolic profile of the TME that supports drug resistance. For instance, exosome-mediated transfer of lactate to nearby cells has been shown to induce a metabolic shift that enables cells to resist oxidative stress and survive under low oxygen conditions, enhancing their resistance to chemotherapy and radiation therapy [41].
3. Conventional cancer diagnostic methods vs Exosome-based cancer diagnostics
3.1. Conventional cancer diagnostic methods
Conventional cancer diagnostic methods, including imaging techniques, histological examination, blood and biomarker testing, and molecular biological tests, are essential for early-stage detection, cancer staging, and prognosis evaluation. However, each method has limitations in sensitivity, specificity, and invasiveness [42]. Imaging techniques, such as CT, MRI, PET, and ultrasound, provide critical information on tumor size, location, and metabolic activity, but may struggle with detecting early-stage cancer or distinguishing benign from malignant lesions. It is worth noting that, with the continuous advancement of diagnostics, the integration of radiomics and artificial intelligence, particularly PSMA-PET/CT imaging, can significantly enhance the diagnostic accuracy, staging, treatment planning, and outcome prediction for certain cancers [43]. Histological examination offers reliable tumor classification and grading but lacks molecular insights. Blood-based tests, like liquid biopsies, offer non-invasive alternatives for detecting cancer-related biomarkers but can face challenges with false positives and negatives. Molecular biological tests, including NGS and PCR, provide detailed genetic insights, aiding in personalized treatment plans, but can be expensive and technically complex. Despite their advantages, these methods do not fully capture the tumor's molecular heterogeneity or microenvironment, limiting their ability to assess treatment responses and resistance [44].
3.2. Advantages of exosome-based diagnostics
Exosome biomarkers offer significant advantages over traditional diagnostic methods in cancer, providing a promising avenue for non-invasive, highly sensitive, and real-time monitoring of tumor dynamics. Exosomes are nanoscale extracellular vesicles secreted by tumor cells into the bloodstream and other bodily fluids, carrying a wealth of bioactive molecules, which reflect the genetic and phenotypic characteristics of the originating tumor. Unlike other biomarkers, exosomes are stable in circulation, making them reliable candidates for liquid biopsy applications, particularly for early cancer detection, monitoring treatment response, and detecting minimal residual disease [45]. One of the primary benefits of exosome-based diagnostics is their ability to capture the heterogeneity of tumors. Tumors are often composed of genetically diverse cell populations, and exosomes provide a snapshot of this diversity by carrying molecular information from different tumor subclones. This feature allows exosomes to offer a more comprehensive understanding of tumor biology compared to traditional methods, which may fail to fully represent tumor heterogeneity. Moreover, exosomes can be derived from various biofluids, including blood, urine, and saliva, enabling minimally invasive sampling that provides an accessible and convenient alternative to tissue biopsy, which can be invasive and challenging to perform repeatedly [46]. Exosomes also outperform conventional biomarkers in their ability to detect early-stage cancers and monitor real-time treatment response. Because exosomes contain tumor-specific molecules, their presence and composition change with tumor progression, offering valuable insights into disease status before the emergence of clinically detectable tumors. Additionally, exosome-derived biomarkers can be used to assess therapeutic efficacy, providing a dynamic picture of tumor evolution and resistance mechanisms. For example, the detection of exosome-mediated changes in protein expression or RNA levels can signal the development of drug resistance or immune evasion, facilitating early intervention and adjustment of therapeutic strategies [47]. Furthermore, exosomes facilitate the detection of molecular signatures linked to immunotherapy, targeted therapy, and chemotherapy. Their ability to convey genetic, proteomic, and lipidomic information makes them powerful tools for personalized cancer care, facilitating the selection of the most appropriate treatment for individual patients. This molecular profiling capacity offers a level of precision that traditional biomarkers, such as serum protein markers, cannot match [48].
3.3. Limitations of exosome-based diagnostics
Exosome-based cancer diagnostics hold significant promise but face several challenges that hinder their clinical implementation. One major limitation lies in the purification and detection methods used to isolate and analyze exosomes. Common purification techniques, such as ultracentrifugation, size-exclusion chromatography (SEC), immunocapture, and precipitation kits, each come with their own advantages and limitations, especially in terms of yield, purity, and processing efficiency. These methods can produce exosome preparations of varying quality, which affects the consistency of biomarker identification [49]. In addition, the sensitivity, specificity, and throughput of detection techniques can vary widely. Methods such as fluorescence-based techniques, nanoparticle tracking analysis (NTA), Western blotting, enzyme-linked immunosorbent assays (ELISA), and newer approaches like surface-enhanced Raman spectroscopy (SERS) and electrochemical sensors all offer unique capabilities, including label-free detection and multiplexing. However, these methods are not without their challenges, and their application depends on the clinical needs and the disease-specific characteristics of the exosomes. For instance, detecting low-abundance biomarkers in bodily fluids like blood requires highly sensitive techniques, which can be costly and time-consuming [50] (Table 1).
Methods for exosome-based diagnostic purification and detection
| Method | Type | Strengths | Limitations |
|---|---|---|---|
| Ultracentrifugation | Purification | Cost-effective, widely used | Time-consuming, low yield, may co-isolate contaminants |
| Size Exclusion Chromatography | Purification | High purity, gentle process | Lower throughput, requires specialized equipment |
| Immunocapture | Purification | High specificity, rapid isolation | Potential bias toward targeted subpopulations |
| Precipitation Kits | Purification | Fast, cost-effective | Lower purity, co-precipitation of contaminants |
| Fluorescence-Based Techniques | Detection | High sensitivity, suitable for real-time analysis | Requires fluorescent labels, complex assay setup |
| Nanoparticle Tracking Analysis | Detection | Quantifies size and concentration | Lacks specificity for surface markers |
| Western Blotting | Detection | Identifies specific proteins or biomarkers | Requires large sample volumes, time-consuming |
| Enzyme-Linked Immunosorbent Assay | Detection | High sensitivity for protein biomarkers | Time-consuming, requires prior exosome isolation |
| Surface-Enhanced Raman Spectroscopy | Detection | High sensitivity, label-free, multiplexing capabilities | Complex sample preparation, requires specialized equipment |
| Electrochemical Sensors | Detection | Low-cost, sensitive, suitable for point-of-care testing | Requires electrode-based setups, limited to specific biomarkers |
The inherent heterogeneity of exosomes further complicates their diagnostic application. Exosome content can differ significantly depending on the cancer type, tumor microenvironment, and even between individual patients, making it difficult to identify reliable, universal biomarkers for broad cancer detection. This variability, combined with a lack of standardized protocols for exosome isolation and characterization, leads to inconsistent results and hampers the reproducibility of exosome-based diagnostic tests [51]. Finally, while exosome-based diagnostics are non-invasive, their utility is still constrained by the absence of large-scale studies that can validate their diagnostic accuracy and sensitivity across diverse patient populations. These challenges highlight the need for further research into standardized methodologies, improved detection sensitivity, and extensive clinical validation to fully harness the potential of exosome-based diagnostics in cancer detection [52].
4. Exosomal biomarkers in tumor diagnostics
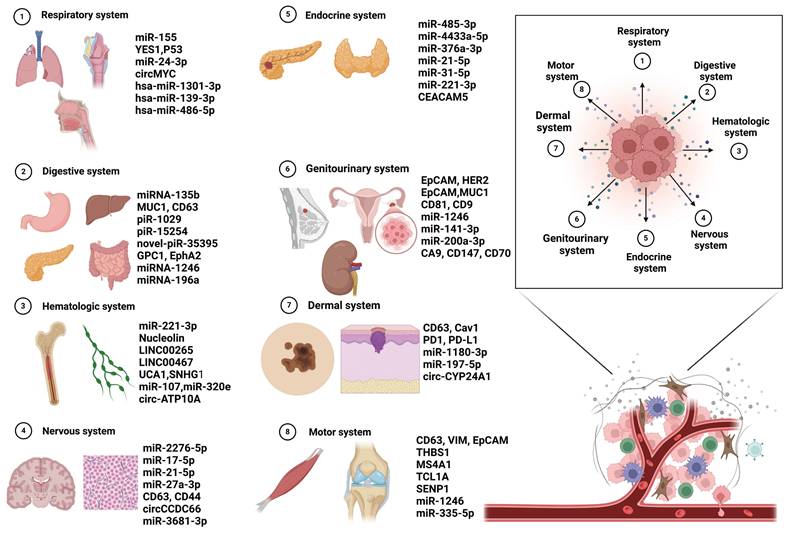
Cancer continues to be a major global health issue, where early detection is key to enhancing patient survival and optimizing treatment outcomes. Fueled by rapid advancements in molecular biology and nanotechnology, the scientific community has progressively unraveled the complexities of intercellular communication networks. In this landscape, exosomes--nanoscale vesicles secreted by cells--have emerged as powerful diagnostic tools due to their rich content of nucleic acids, proteins, and other biomolecules. Exosomal biomarkers offer exceptional sensitivity and specificity, and their utilization in non-invasive liquid biopsy techniques enables dynamic monitoring. This breakthrough paves the way for a new era in the early detection of cancer, promising more accurate and timely diagnoses [53] (Figure 4).
4.1. Respiratory system
Lung cancer is the most common and deadly respiratory malignancy, accounting for the highest incidence and mortality rates globally. It is mainly divided into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC), with NSCLC making up around 85% of cases [54]. Lung cancer's high prevalence and poor prognosis make it a major challenge in respiratory oncology. Its development involves a complex process driven by genetic and environmental factors, as well as changes in molecular pathways [55]. Advances in molecular biology have helped identify genetic mutations and disrupted signaling pathways, enabling targeted therapies and precision medicine. Despite these advancements, survival rates for advanced lung cancer remain low [56]. Current diagnostic limitations include the absence of early symptoms, leading to diagnoses at advanced stages, often missing the optimal treatment window. Imaging tools like chest X-rays and CT scans have limited sensitivity and specificity, leading to false results and affecting diagnostic accuracy. Diagnosis often relies on invasive tissue biopsies, which can be complex and risky, discouraging some patients. While biomarker detection holds promise for classification and treatment guidance, their clinical utility is still limited [57]. Furthermore, the high cost and limited access to molecular diagnostics and personalized therapies restrict their widespread use. Uneven distribution of medical resources can also delay diagnoses, further impacting prognosis. Thus, improving early screening, non-invasive diagnostic methods, biomarker development, and resource allocation is essential to enhance diagnosis and treatment outcomes [58]. Exosomes, as biomarkers, provide a more convenient and effective option for the early diagnosis of lung cancer.
Yunpeng Fan et al. developed the Integrated Concentration and Determination System of Exosomes (ICDSE), which combines engineered red blood cells (RBCs) and a plasmonic sensor for efficient screening of exosomal miR-155 in NSCLC patients. The RBCs, functionalized with CD63-specific aptamers, capture exosomes, which are then isolated using simple centrifugation. A plasmonic sensor amplifies the miRNA signal with a detection limit of 2.03 fM, offering a fast, cost-effective, and highly sensitive approach for exosome-based liquid biopsies and early cancer diagnosis [59]. Esther Redin et al. identified YES1 as a novel oncogenic target and biomarker in SCLC. YES1, detectable in exosomes, correlates with tumor characteristics and prognosis, with high expression linked to shorter progression-free survival (PFS) and overall survival (OS). Its role in predicting sensitivity to therapies like CH6953755 and dasatinib enhances its clinical utility, offering a real-time disease monitoring tool and personalized treatment strategy [60]. Li Ming's team showed that seven autoantibodies detected on plasma-derived small extracellular vesicles (sEVs) outperform serum autoantibodies in early lung cancer diagnosis, with improved sensitivity and specificity. These sEV-associated autoantibodies are enriched by tumor-associated antigens (TAAs) and may aid immune evasion in lung cancer. This highlights the potential of sEVs as non-invasive biomarkers for early lung cancer screening and their role in immune modulation [61]. Another study combined Exosome-SERS with Artificial Intelligence (AI) to detect early-stage cancers, including lung, breast, and colon cancers, with high sensitivity (90.2%) and specificity (94.4%). The system also identified the tissue of origin with an AUC of 0.945, offering a rapid, label-free, and cost-effective diagnostic tool for clinical use, supporting precision medicine in exosome-based cancer research [62].
Exosomal biomarkers of systemic tumors. Application in tumors of different systems and some examples of biomarkers. Created with BioRender.com.
Nasopharyngeal carcinoma (NPC) is a malignancy of the nasopharynx, often linked to Epstein-Barr virus (EBV) infection. It is most common in Southeast Asia, southern China, and North Africa, with risk factors such as genetic predisposition, diet, and environmental exposures. Most cases are undifferentiated carcinoma, with symptoms like nasal obstruction, nosebleeds, hearing loss, and enlarged cervical lymph nodes. Diagnosis involves endoscopy, imaging, biopsy, and plasma EBV DNA as a biomarker. Treatment typically involves radiation and chemotherapy, with advancements like IMRT improving outcomes [63]. Despite advances in diagnostic methods, early detection of NPC remains challenging. The nasopharynx's anatomical location complicates detection during routine exams, often leading to delayed diagnosis until symptoms become apparent. Initial symptoms such as nasal congestion or mild hearing loss are commonly mistaken for conditions like sinusitis. Although MRI and CT scans are sensitive, they may not reliably differentiate early-stage NPC from benign lesions [64]. Biopsy, the gold standard, is invasive and may not be performed quickly, particularly in asymptomatic cases. Biomarkers like EBV DNA improve diagnostic accuracy but remain limited by availability and cost. These challenges underscore the need for better screening, more specific biomarkers, and greater public awareness for earlier detection and improved patient outcomes [65]. Exosomal biomarkers offer a promising new avenue for the early diagnosis of NPC.
Chuanben Chen et al. identify potential plasma biomarkers for early-stage NPC by analyzing exosomal miRNAs via RNA sequencing, revealing 31 differentially expressed miRNAs (21 upregulated, 10 downregulated). Notably, hsa-miR-1301-3p is significantly upregulated and validated as a promising diagnostic biomarker. Bioinformatics analysis indicates that the target genes of these miRNAs are enriched in cancer-related pathways, such as PI3K-Akt and MAPK signaling. These findings suggest that exosomal miRNAs, particularly hsa-miR-1301-3p, could serve as non-invasive biomarkers for early NPC diagnosis [66]. Jiang Li et al. show that exosomal miR-24-3p, enriched in NPC-derived exosomes, suppresses T-cell proliferation and differentiation by targeting FGF11 and modulating ERK and STAT pathways. Hypoxia enhances miR-24-3p expression, amplifying its immunosuppressive effects.
Exosomal biomarkers in respiratory system cancers
| Cancer type | Biomarkers | Source | Analytical Technique | Expression | Effectiveness | Reference |
|---|---|---|---|---|---|---|
| miR-155 | Plasma | Integrated concentration and determination system of exosomes (ICDSE) | Upregulated | LOD: 2.03 fM; Specific & Cost-effective | [59] | |
| Lung cancer | YES1 | Plasma | Western blot, IHC, FISH | Upregulated | Independent predictor of poor prognosis, correlates with reduced PFS and OS | [60] |
| Autoantibody panel on sEVs (P53, PGP9.5, SOX2, GAGE7, GBU4-5, MAGEA1, CAGE) | Plasma | ELISA, AUC analysis | Upregulated | Sensitivity: 65.56%, Specificity: 96.88%, AUC: 0.8007 | [61] | |
| Multi-cancer types (lung, breast, colon, liver, pancreas, stomach) | Raman signal patterns of exosomes | Plasma | Surface-enhanced Raman spectroscopy (SERS) with AI | Differential patterns of Raman signals | AUC: 0.970; Sensitivity: 90.2%; Specificity: 94.4%; Tissue of origin (TOO) accuracy: 72% | [62] |
| Nasopharyngeal carcinoma | hsa-miR-1301-3p | Plasma | RNA sequencing, qRT-PCR, GO/KEGG analysis | Upregulated | Differential expression confirmed (Fold Change > 2, P < 0.05); GO/KEGG pathways indicate roles in PI3K-Akt and MAPK pathways | [66] |
| miR-24-3p | Serum | RT-qPCR, Luciferase reporter assay, FACS analysis | Upregulated | Elevated serum miR-24-3p levels link to poor DFS (p<0.05) and immune regulation. | [67] | |
| circMYC | Serum | qRT-PCR, AUROC analysis, siRNA knockdown studies | Upregulated in radioresistant NPC | AUROC: 0.945 Sensitivity: 90.24%, Specificity: 94.51% | [68] |
Elevated serum miR-24-3p levels correlate with worse disease-free survival, while higher FGF11 expression associates with improved prognosis and greater densities of tumor-infiltrating lymphocytes. These findings suggest miR-24-3p and FGF11 as potential prognostic biomarkers and therapeutic targets for NPC [67]. Rong Gui et al. reveal exosomal circMYC as a potential biomarker and therapeutic target for radioresistant NPC. CircMYC is upregulated in the serum exosomes of NPC patients, especially those with radioresistance, and correlates with larger tumor size, lymph node metastasis, advanced TNM stage, poor survival outcomes, and higher recurrence rates. Functional studies show that circMYC promotes proliferation and reduces radiosensitivity by sponging tumor-suppressive miRNAs, influencing AGO1 and CRY2 pathways. ROC analysis confirms circMYC's high sensitivity and specificity in distinguishing radioresistant from radiosensitive NPC patients, highlighting its clinical potential [68].
Exosomal biomarkers are revolutionizing the early diagnosis, prognosis, and therapeutic monitoring of respiratory tumors, including lung cancer, NPC, and LSCC (Table 2). These vesicles, enriched with miRNAs (e.g., miR-21), lncRNAs (e.g., HOTAIR), circRNAs (e.g., circMYC), and proteins (e.g., IGFBP7), offer high stability and specificity for liquid biopsies. With diagnostic accuracies often exceeding AUC 0.95, they provide insights into tumor aggressiveness, metastasis, and survival. Functionally, they affect key pathways like PI3K-Akt and MAPK, driving tumor progression and therapy resistance. Emerging technologies like Exosome-SERS-AI enable multi-cancer detection. Despite challenges in standardizing methods, exosomal biomarkers hold great promise for improving early detection, risk stratification, and personalized treatment in respiratory tumors.
4.2. Digestive system
Gastric cancer (GC) ranks as the fifth most prevalent type of cancer and is responsible for the third highest number of cancer-related deaths worldwide. Over 80% of GC patients are diagnosed at a late stage [69]. The late diagnosis of GC significantly contributes to its high mortality rate. Currently, GC detection relies on endoscopy and histological biopsy, which are invasive, time-consuming, and expensive. The delay in diagnosis and limited screening among high-risk populations have drawn considerable attention. Consequently, developing rapid and early diagnostic methods is crucial for improving GC patient survival [70]. In recent times, liquid biopsy using specific gastric biomarkers has gained recognition as a less invasive alternative to traditional biopsies. Body fluids offer a range of potential biomarkers, including proteins, DNAs, RNAs, and extracellular vesicles, for GC detection. The characteristics of exosomes render them a highly potential option for liquid biopsy applications [71].
Qiang Ma et al. developed a novel electrochemiluminescence (ECL) sensor using MoS2 QDs-MXene heterostructures and Au NPs@biomimetic lipid layers to detect miRNA-135b, a gastric cancer biomarker in exosomes. The MoS2 QDs-MXene heterostructure amplifies the signal, while the Au NPs@biomimetic lipid layer provides a specific, antifouling platform for miRNA capture. The sensor achieves highly sensitive detection with a limit of 10 fM, offering a rapid, non-invasive method for gastric cancer diagnosis and monitoring [72]. Additionally, Qiang Ma et al. developed a magnetoplasmonic metasurface-modulated ECL sensor for ultrasensitive detection of gastric cancer-derived extracellular vesicles (EVs). Using Fe3O4@Au yolk-shell nanoparticles and MUC1 aptamers for recognition, this system enhances ECL signals and achieves high sensitivity (detection limit of 200 particles/mL). This approach provides a powerful tool for early gastric cancer diagnosis and monitoring, particularly for peritoneal metastasis [73]. Z. Li et al. introduced a droplet digital branched rolling circle amplification (ddBRCA) biosensor for ultrasensitive detection of gastric cancer-derived EVs. The platform uses stem-loop hairpin DNA (APP) with MUC1-specific aptamers to recognize target EVs, triggering a BRCA reaction that amplifies fluorescence signals. This method allows single-EV analysis with a detection limit of 12 particles/mL and provides a rapid, highly sensitive, and clinically validated tool for non-invasive gastric cancer diagnostics [74].
Hepatocellular carcinoma (HCC), which accounts for 75% to 85% of primary liver cancer cases, is the most common form of liver cancer and ranks as the third leading cause of cancer-related mortality globally. Diagnosis typically involves imaging and histopathological biopsies. However, up to 30% of HCC patients show negative alpha-fetoprotein (AFP) results, limiting AFP's sensitivity and specificity for screening [75]. Imaging has high specificity but relatively low sensitivity, making detection of small tumors difficult. Histopathological biopsies, although informative, are invasive and prone to false negatives. As early symptoms and specific biomarkers are often absent, most HCC patients are diagnosed at advanced stages, limiting treatment efficacy. Early detection is crucial for improving patient outcomes [76]. The occurrence and progression of liver cancer is complex, and its molecular mechanisms remain unclear. While viral infections, alcohol, and non-alcoholic hepatotoxicity are major contributors to HCC, the full pathogenesis is not completely understood. Recent in vivo and in vitro studies suggest that exosomes play a critical role in the initiation, progression, diagnosis, and treatment of HCC, positioning them as promising candidates for novel biomarkers [77].
Xiaobing Zhang et al. discovered that serum exosome-derived PIWI-interacting RNAs (piRNAs) are promising biomarkers for HCC diagnosis. Among 253 differentially expressed piRNAs, five (piR-1029, piR-15254, novel-piR-35395, novel-piR-43597, and novel-piR-32132) were highly upregulated in HCC patients, showing superior diagnostic accuracy (AUROC up to 0.986) compared to AFP, even in low tumor burden cases. Unique base distribution patterns in HCC-derived piRNAs may help differentiate HCC from non-tumor donors. This highlights piRNAs' potential as stable and specific biomarkers for non-invasive early HCC detection [78]. Minqiang Lu et al. identified the exosomal lncRNA THEMIS2-211 as a tumor-derived biomarker significantly upregulated in HCC, with diagnostic and prognostic value. THEMIS2-211 acts as a competing endogenous RNA (ceRNA), sponging miR-940 to upregulate SPOCK1, driving tumor proliferation, invasion, and EMT. Its exosomal origin ensures stability, outperforming AFP in early-stage HCC diagnosis. Targeting the THEMIS2-211/miR-940/SPOCK1 axis could offer new therapeutic strategies for HCC [79]. Fubing Wang et al. developed a SiO₂-coated 3D hierarchical porous chip (SiO₂-chip) for efficient exosome enrichment, identifying lncRNAs LUCAT-1 and EGFR-AS1 as specific biomarkers for early HCC detection and monitoring. The SiO₂-chip captures exosomes using a 3D porous scaffold, enabling rapid isolation from minimal plasma samples. These lncRNAs show superior diagnostic accuracy (AUC: 0.947) and prognostic value, correlating with tumor progression and poor survival. The platform offers high sensitivity (detection limit: 10,000 particles/mL), speed (under 10 minutes), and robustness, making it a powerful tool for non-invasive HCC diagnostics and treatment monitoring [80].
Pancreatic cancer, particularly pancreatic ductal adenocarcinoma (PDAC), is a highly aggressive malignancy with one of the poorest prognoses. It is often asymptomatic in its early stages, leading to late-stage diagnosis when the disease is typically advanced or metastatic. Major risk factors include obesity, smoking, chronic pancreatitis, diabetes, and a family history of the disease [81]. Despite advances in surgery, chemotherapy, and targeted therapies, the five-year survival rate remains low due to late detection. Diagnosis of pancreatic cancer is challenging due to the asymptomatic nature of early stages and the lack of specific biomarkers. Current diagnostic methods, such as imaging (CT, MRI, and endoscopic ultrasound) and serum biomarkers like CA19-9, have limitations. Imaging may fail to detect small or early-stage tumors, and CA19-9 lacks sensitivity and specificity, as its levels can be elevated in benign conditions like pancreatitis. Invasive biopsies are not always feasible due to the tumor's location [82]. These limitations lead to delayed diagnosis, emphasizing the need for more accurate, non-invasive diagnostic tools for early detection and improved outcomes in this challenging cancer.
Jianlin Shi et al. developed a nanoliquid biopsy (nLB) assay using dual biomarkers, GPC1 and EphA2, to detect pancreatic cancer tumor exosomes (T-Exos). With magnetic nanoparticles for isolation and gold nanoparticles for signal amplification, the assay achieves high sensitivity (78 pg/mL detection limit) and 100% specificity. It provides precise early-stage diagnosis (AUC: 1.0) and reliable tumor monitoring, making it a promising non-invasive tool for clinical use [83]. Lingling Wu et al. created a multiplex detection strategy for tumor-derived extracellular vesicle microRNAs (tEV-miRNAs) using encoded-targeted fusion beads (ETFBs) and aptamer-modified supported lipid bilayers (SLBs). This system captures and profiles six key miRNAs from plasma without exosome isolation, achieving 98% diagnostic accuracy and rapid processing (within 2 hours), making it a non-invasive and efficient tool for early pancreatic cancer detection [84]. L. James Lee et al. introduced an Immune Lipoplex Nanoparticle (ILN) biochip for detecting GPC1 mRNA in exosomes and GPC1 protein in tumor-associated microvesicles (tMVs). This dual biomarker approach achieves high diagnostic accuracy (AUC: 0.960) and provides prognostic value, correlating low GPC1 levels with prolonged survival in late-stage PDAC patients. It offers high sensitivity and rapid processing, making it a powerful tool for PDAC diagnosis and prognosis [85].
Colorectal cancer (CRC) is a common and life-threatening malignancy that typically starts as benign polyps, which can become cancerous over time if not removed. Risk factors include age, high consumption of processed or red meats, low fiber intake, obesity, smoking, excessive alcohol, inflammatory bowel disease, and a family history of CRC [86]. It is worth noting that although older patients with pT4 disease are more prone to severe postoperative complications, there is no consensus on whether age affects survival outcomes. The prognosis of older patients may be confounded by differences in stage at presentation, tumor location, preexisting comorbidities, and the type of treatment received [87]. Symptoms include changes in bowel habits, abdominal pain, blood in the stool, fatigue, and weight loss. Regular screening, such as colonoscopy and stool-based tests, is essential for early detection and prevention. Current diagnostic methods for CRC have limitations. Colonoscopy is the gold standard but is invasive, time-consuming, and requires bowel preparation, deterring some patients [88]. Non-invasive stool-based tests, such as fecal immunochemical tests (FIT) and stool DNA tests, have limited sensitivity and may miss early-stage lesions. Imaging techniques like CT colonography are less invasive but have reduced accuracy for detecting small or flat polyps, often requiring follow-up procedures. Additionally, the lack of early symptoms often leads to delayed diagnoses and poorer outcomes. These limitations highlight the need for more accurate, non-invasive, and patient-friendly diagnostic tools for early CRC detection [89]. Exosomes, secreted by cancer cells, play a crucial role in early CRC diagnosis as carriers of tumor-specific biomarkers. These nanosized vesicles can be detected in accessible body fluids like blood and stool, enabling non-invasive liquid biopsy approaches. Exosome-based diagnostics offer high sensitivity and specificity, allowing for the detection of CRC at early stages, even before clinical symptoms arise. This makes exosomes a promising tool for early detection, treatment guidance, and improved patient outcome [90].
Meilin Wang et al. identify exosomal circLPAR1 as a stable and specific biomarker for CRC diagnosis and progression. circLPAR1 is significantly downregulated in CRC, with plasma exosomal levels showing an AUC of 0.875, achieving 87.3% sensitivity and 76.3% specificity when combined with traditional biomarkers like CEA and CA19-9. Functionally, circLPAR1 acts as a tumor suppressor by disrupting the METTL3-eIF3h interaction, thereby inhibiting BRD4 translation, a key oncogene in CRC. Its levels correlate with disease progression and decrease after surgical resection, offering both diagnostic and prognostic value. Advantages include its high stability in exosomes, non-invasive detectability in plasma, and its ability to distinguish CRC from other cancers, making it a powerful tool for early detection and treatment monitoring [91]. Chu Wang et al. developed a rapid and cost-effective EV-based proteomics strategy for early CRC diagnosis. By using DSPE-functionalized beads, EVs were efficiently isolated from plasma within 10 minutes, followed by proteomic analysis using SP3 technology and DIA-MS. A machine learning model identified 10 key EV protein biomarkers, achieving 89.3% diagnostic accuracy and an AUC of over 0.94. The mechanism leverages hydrophobic interactions for specific EV capture and integrates mass spectrometry data with machine learning for precise biomarker identification. This approach is non-invasive, scalable, and highly accurate, offering significant potential for early CRC detection and broader clinical applications [92]. Shutian Zhang et al. identified a 10-gene sEV-RNA signature for the early detection of T1a stage CRC and advanced adenoma (AA) through whole-transcriptomic profiling of circulating sEV-derived RNAs. This signature, enriched in cancer-related pathways, achieved high diagnostic accuracy (AUC: 0.94, 99.0% sensitivity, 79.3% specificity) and was validated in independent cohorts. The mechanism leverages sEVs' ability to protect RNAs from degradation, enabling robust liquid biopsy-based detection. Its advantages include early detection of precancerous and early-stage CRC lesions, non-invasiveness, high stability of sEV-RNAs, and scalability for large-scale clinical applications, addressing critical gaps in CRC screening [93].
Exosomal biomarkers are transforming the non-invasive diagnosis of digestive system tumors (Table 3). These biomarkers, such as serum exosome-derived piRNAs and dual markers like GPC1 and EphA2, enable early detection with high sensitivity and specificity, outperforming traditional methods like CA19-9 and AFP. Their integration into liquid biopsy platforms offers a patient-friendly and minimally invasive alternative to conventional diagnostics, allowing for routine screening, instantaneous tracking of treatment response, and early detection of recurrence. With their stability, scalability, and ability to provide biological insights, exosomal biomarkers are poised to revolutionize cancer diagnosis and management, improving outcomes for patients with digestive system tumors.
Exosomal biomarkers in digestive system cancers
| Cancer type | Biomarkers | Source | Analytical Technique | Expression | Effectiveness | Reference |
|---|---|---|---|---|---|---|
| miRNA-135b | Ascites | MoS2 QDs-MXene-based ECL sensor, qRT-PCR | Upregulated | LOD: 10 fM; Linear range: 30 fM-20 nM; High specificity and sensitivity for GC diagnosis | [72] | |
| Gastric cancer | MUC1 | Ascites | Magnetoplasmonic metasurface-modulated ECL sensor | Upregulated | Detection range: 5.8×102 to 5.8×107 particles/mL; LOD: 200 particles/mL; High specificity and stability | [73] |
| MUC1, CD63 | Serum | Droplet Digital Branched Rolling Circle Amplification (ddBRCA) | Upregulated | Detection range: 2×102 to 3.2×103 particles/mL; LOD: 12 particles/mL; AUC (MUC1): 0.8086; AUC (CD63): 0.7832; Combined AUC: 0.8705; Sensitivity: 82.35%; Specificity: 66.67% | [74] | |
| piR-1029, piR-15254, novel-piR-35395, novel-piR-32132, novel-piR-43597 | Serum | sRNA-seq, qRT-PCR, AUROC analysis | Upregulated | AUROC: 0.986 (combined); High sensitivity (82.35%) and specificity (66.67%); Superior to AFP | [78] | |
| Hepatocellular carcinoma | lncRNA THEMIS2-211, PRKACA-202, H19-204 | Plasma | qRT-PCR, AUROC analysis analysis | Upregulated | AUROC: THEMIS2-211 (0.832), PRKACA-202 (0.804), H19-204 (0.701); Combined AUROC: 0.879; Superior to AFP for early-stage HCC | [79] |
| lncRNA LUCAT-1, EGFR-AS1 | Plasma | SiO₂-chip for exosome enrichment, qRT-PCR, AUROC analysis | Upregulated | Combined AUROC: 0.897 (LUCAT-1 + EGFR-AS1); Improved diagnostic accuracy when combined with AFP and DCP | [80] | |
| GPC1, EphA2 | Plasma | Nanoliquid Biopsy (nLB) with ICP-MS quantification | Upregulated | AUROC: 1.0 (Stage I/II PDAC vs NC); LOD: 78 pg/mL GPC1; Sensitivity and Specificity: 100% | [83] | |
| Pancreatic cancer | miRNA-21, miRNA-16, miRNA-10b, miRNA-155, miRNA-1246, miRNA-196a | Plasma | Encoded fusion-mediated miRNA profiling with flow cytometry | Upregulated | AUROC: 0.98 (combined miRNAs for PDAC vs healthy donors); Sensitivity: 98%; Process time: 2 hours for multiplex detection | [84] |
| GPC1 mRNA, GPC1 protein | Serum | Immune Lipoplex Nanoparticle (ILN) biochip, qRT-PCR, Western blot, TEM | Upregulated | AUROC (Stage I/II): 0.960 (dual-biomarker); AUROC (Stage III/IV): 0.973; Improved sensitivity and specificity compared to CA19-9 | [85] | |
| Colorectal cancer | circLPAR1 | Plasma | RNA sequencing, FISH, qRT-PCR, RNA pull-down, RIP assay | Downregulated in CRC plasma and tissues; Upregulated after surgery | AUROC: 0.875 (combined with CEA and CA19-9); Sensitivity: 87.3%; Specificity: 76.3%; Correlates with tumor suppression and improved survival | [91] |
| Panel of 10 protein markers (e.g., APOA4, GPX3, SNCA, THBS4, etc.) | Plasma | DSPE-functionalized bead isolation, SP3, DIA-MS, Machine Learning | Dysregulated proteins in early CRC and polyp patients | AUROC: 1.0 (SVM model); Accuracy: 89.3%; Sensitivity and specificity improved significantly | [92] | |
| Colorectal cancer and advanced adenoma | 10 RNA markers, including miR-425-5p, let-7f-5p, C19orf43, TOP1, PPDPF, lnc-MKRN2-42:1, HIST2H2AA4, and LNC-EV-9572 | Plasma | RNA sequencing, qRT-PCR, t-SNE clustering, machine learning (SVM, Lasso regression) | Biomarkers showed distinct profiles in T1a CRC/AA patients compared to normal controls | AUROC: 0.88 for AA and 0.80 for CRC using multi-RNA signatures; Sensitivity: 99.0%, Specificity: 79.3% with RT-qPCR | [93] |
4.3. Hematologic system
Leukemia is a type of hematologic malignancies marked by the uncontrolled growth of abnormal white blood cells in the bone marrow and blood. It disrupts the normal production of blood cells, leading to symptoms such as anemia, infections, bleeding tendencies, fatigue, and bone pain. Leukemia is broadly classified into acute and chronic forms, with further subtypes such as acute lymphoblastic leukemia (ALL), acute myeloid leukemia (AML), chronic lymphocytic leukemia (CLL), and chronic myeloid leukemia (CML) [94]. Risk factors include genetic predispositions, exposure to radiation or certain chemicals, and prior chemotherapy. Diagnosis involves blood tests, bone marrow examination, and molecular studies. Advances in targeted therapies, immunotherapies, and stem cell transplantation have significantly improved outcomes, especially when leukemia is detected early and treated promptly. The diagnosis of leukemia faces several limitations despite advances in diagnostic tools. While blood tests and bone marrow examinations are essential for identifying abnormal cells, these methods are invasive and can be uncomfortable for patients [95]. Molecular and cytogenetic analyses provide critical insights but are time-consuming and require specialized equipment, limiting their accessibility in resource-constrained settings. Additionally, early-stage or indolent forms of leukemia, such as CLL, may be asymptomatic and go undetected during routine screenings. The lack of specific biomarkers for certain subtypes further complicates early diagnosis and differentiation between similar hematologic disorders. These challenges underscore the need for more precise, non-invasive, and rapid diagnostic approaches to improve leukemia detection and patient outcomes [96].
Hui Cheng et al. discovered that sEVs derived from AML cells promote leukemogenesis by transferring miR-221-3p, a key microRNA. miR-221-3p enhances AML cell proliferation and survival by targeting Gbp2, thereby activating the PI3K/AKT signaling pathway, while also impairing the function and differentiation of hematopoietic stem and progenitor cells (HSPCs). The findings highlight the role of AML-sEVs in creating a leukemia-permissive bone marrow niche. This mechanism not only offers insights into AML progression but also positions miR-221-3p as a potential biomarker for diagnosis and a therapeutic target to disrupt the leukemogenic microenvironment, providing a novel avenue for AML management [97]. Lin Huang et al. developed a highly sensitive colorimetric biosensor for detecting leukemia-derived exosomes by targeting nucleolin, a protein enriched on exosome surfaces. The mechanism combines rolling circle amplification (RCA) with dual signal amplification using gold nanoparticles and horseradish peroxidase (GNPs-HRP) to produce a visible colorimetric response. This biosensor achieves a detection limit as low as 100 particles/μL and requires minimal sample volume (40 μL). Its advantages include high specificity, non-invasiveness, cost-effectiveness, and simplicity, as it allows visual detection without complex instruments. These features make it a promising tool for early leukemia diagnosis and treatment monitoring [98]. Ling Zhang et al. identified four plasma exosomal lncRNAs--UCA1, LINC00467, LINC00265 and SNHG1--as potential biomarkers for AML diagnosis and treatment monitoring. These lncRNAs, with distinct expression patterns in AML patients, allow differentiation from healthy controls and tracking of treatment response, particularly during chemotherapy or stem cell transplantation. The mechanism involves the stable encapsulation of lncRNAs within exosomes, protecting them from degradation and enabling reliable detection. Advantages include non-invasive sampling, high stability, real-time monitoring of disease progression, and improved diagnostic accuracy when combining multiple lncRNAs, making them a promising tool for AML management [99].
Multiple myeloma (MM) is a malignancy of plasma cell disorder characterized by the clonal proliferation of abnormal plasma cells in the bone marrow. These cells produce excessive monoclonal immunoglobulins (M-proteins), leading to various complications such as bone lesions, anemia, renal dysfunction, hypercalcemia, and an increased risk of infections. MM typically develops in older adults and is often preceded by precursor conditions like monoclonal gammopathy of undetermined significance (MGUS) or smoldering myeloma. Diagnosis involves blood and urine tests (for M-proteins), bone marrow biopsy, and imaging studies to detect bone damage [100]. Advances in treatments, including proteasome inhibitors, immunomodulatory drugs, monoclonal antibodies, and autologous stem cell transplantation, have significantly improved outcomes, but MM remains incurable, with relapses common. Early detection and tailored therapeutic strategies are essential for managing the disease and enhancing patient survival and quality of life.
Chen Kuisheng et al. discovered that cancer-associated fibroblast (CAF)-derived exosomal miR-21 plays a critical role in promoting angiogenesis in MM by enhancing endothelial cell proliferation, migration, and tube formation. The mechanism involves miR-21 activating pro-angiogenic pathways in MM endothelial cells and facilitating the transformation of normal fibroblasts into CAFs, which further secrete pro-angiogenic exosomes. This exosomal miR-21 is stable, specific to the tumor microenvironment, and detectable in circulation, making it a promising biomarker for MM progression and a therapeutic target to suppress tumor angiogenesis, offering non-invasive diagnostic and treatment potential [101]. Xing Cui et al. identify circ-ATP10A, an exosome-enriched circular RNA, as a novel biomarker for MM. circ-ATP10A promotes angiogenesis by acting as a miRNA sponge, sequestering miRNAs like hsa-miR-6758-3p and modulating the expression of angiogenesis-related genes such as VEGFB and HIF1A. It is significantly upregulated in MM patients and correlates with increased bone marrow microvessel density and poor survival outcomes. Advantages of circ-ATP10A include its high diagnostic accuracy (AUC: 0.854), non-invasive detection via serum exosomes, stability in circulation, and prognostic value, making it a promising tool for MM diagnosis, prognosis, and as a therapeutic target to inhibit angiogenesis-driven tumor progression [102].Yanwei Luo et al. identified LRG1, a protein enriched in platelet-derived exosomes, as a key promoter of MM progression. LRG1 interacts with OLFM4 to activate EMT and angiogenesis, enhancing MM cell proliferation, tumor invasiveness, and vascular formation. Elevated levels of exosomal LRG1 in MM patients correlate with advanced disease stages and poor survival outcomes, highlighting its role as a biomarker and therapeutic target. Advantages of exosomal LRG1 include its high specificity to MM, non-invasive detection through liquid biopsy, and potential for targeted therapies to disrupt its tumor-promoting effects, offering new strategies for MM diagnosis and treatment [103].
Lymphoma is a form of cancer that originates in the lymphatic system, which plays a crucial role in immune defense. It occurs when lymphocytes, a type of white blood cell, grow uncontrollably and form tumors in lymph nodes, spleen, bone marrow, or other organs. Lymphoma is broadly classified into Hodgkin lymphoma (HL) and non-Hodgkin lymphoma (NHL), with NHL being more common and diverse. Risk factors include weakened immunity, certain infections (e.g., Epstein-Barr virus or HIV), and exposure to radiation or chemicals. Symptoms often include fatigue, fever, painless swelling of lymph nodes, night sweats, and unexplained weight loss [104]. Diagnosis typically involves imaging, blood tests, and lymph node biopsy. Advances in targeted therapies, immunotherapy, and stem cell transplantation have significantly improved outcomes, with many forms of lymphoma being treatable or even curable, especially when detected early. The diagnosis of lymphoma faces several limitations despite advances in medical technology. Traditional methods like lymph node biopsy, while accurate, are invasive and can be uncomfortable for patients. Imaging techniques such as CT, MRI, or PET scans are useful but may fail to detect very small lesions or distinguish lymphoma from benign conditions [105]. Blood tests, while supportive, lack the sensitivity and specificity to definitively diagnose lymphoma. Furthermore, the diverse subtypes of lymphoma, particularly in NHL, complicate diagnosis and require advanced immunophenotyping or molecular testing, which may not be readily available in all clinical settings. These challenges highlight the need for non-invasive, specific, and accessible diagnostic tools to improve early detection and accurate classification of lymphoma subtypes [106].
Seok Jin Kim et al. identify serum-derived exosomal miR-320e, miR-21-5p and miR-4454 as key prognostic biomarkers for extranodal natural killer/T-cell lymphoma (ENKTL). These miRNAs are upregulated in advanced disease stages, associated with poor survival, and linked to treatment resistance, particularly to etoposide. Mechanistically, they promote tumor progression by enhancing inflammatory cytokine secretion, inducing M2-like macrophage polarization, and creating an immune-suppressive tumor microenvironment. Advantages include their stability within exosomes, non-invasive detectability via liquid biopsy, strong correlation with prognosis and treatment response, and potential as therapeutic targets to overcome resistance and improve patient outcomes [107]. Xin Wang et al. identified exosomal miR-107 as a tumor suppressor in diffuse large B-cell lymphoma (DLBCL), with significantly reduced levels in patients correlating with advanced disease and poor survival. Mechanistically, miR-107 targets and downregulates the oncogenic protein 14-3-3η, disrupting PI3K/Akt signaling and cell cycle regulation, thereby inhibiting tumor cell proliferation, invasion, and promoting apoptosis. Advantages of miR-107 include its high diagnostic accuracy (AUC: 0.854), non-invasive detectability via plasma exosomes, stability in circulation, prognostic relevance, and therapeutic potential as a target to suppress tumor progression and overcome resistance in DLBCL [108].
The findings highlight the transformative potential of exosomal biomarkers in diagnosing and managing hematologic malignancies, including AML, MM, ENKTL, and DLBCL (Table 4). These studies demonstrate the utility of exosomal microRNAs (e.g., miR-221-3p, miR-107) and long non-coding RNAs (e.g., LINC00265) for early detection, prognostic stratification, and treatment monitoring with high sensitivity and specificity. Exosome-based liquid biopsies provide a non-invasive, stable, and reliable alternative to traditional diagnostic methods like bone marrow biopsies. Additionally, these biomarkers reveal tumor-promoting mechanisms, such as angiogenesis and immune modulation, offering therapeutic insights. Collectively, these advancements pave the way for precision medicine and improved outcomes in hematologic oncology.
Exosomal biomarkers in hematologic system cancers
| Cancer type | Biomarkers | Source | Analytical Technique | Expression | Effectiveness | Reference |
|---|---|---|---|---|---|---|
| miR-221-3p | Bone marrow and plasma | RNA sequencing, qRT-PCR, flow cytometry, Western blot, TEM | Upregulated | Promotes leukemogenesis by inhibiting apoptosis and facilitating cell cycle entry; Targeting Gbp2 to regulate PI3K/AKT pathway | [97] | |
| Leukemia | Nucleolin | Leukemia-derived exosomes | Rolling Circle Amplification (RCA) with dual signal amplification (colorimetric biosensor) | Upregulated | LOD: 100 particles/μL; High specificity; Successfully distinguishes leukemia patients from healthy individuals | [98] |
| Exosomal lncRNAs (LINC00265, LINC00467, UCA1, SNHG1) | Plasma | qRT-PCR, AUROC analysis, TEM, NTA, Western blot | LINC00265, LINC00467, UCA1 downregulated; SNHG1 upregulated in AML patients compared to healthy donors | AUROC for combined biomarkers: 0.8685; sensitivity: 85%-100%; specificity: 50%-65%; effective in diagnosis and treatment monitoring | [99] | |
| miR-21 | Bone marrow | qRT-PCR, Western blot, TEM, NTA | Upregulated | miR-21 promotes angiogenesis in MM by enhancing MMEC proliferation, migration, and tube formation; transforms NFs into CAFs | [101] | |
| Multiple Myelom | circ-ATP10A | Serum | RNA-seq, qRT-PCR, TEM, NTA, Immunohistochemistry | Upregulated | AUROC: 0.854; Sensitivity: 87.5%; Specificity: 75%; Prognostic biomarker for angiogenesis and MM progression | [102] |
| LRG1 | Platelet | Proteomics, ELISA, qRT-PCR, Western blot, TEM, NTA | Upregulated | Promotes MM progression via EMT activation and angiogenesis; High LRG1 correlates with poor prognosis; | [103] | |
| miR-21-5p, miR-320e, miR-4454 | Serum | Nanostring nCounter miRNA array, qRT-PCR, TEM, NTA, KEGG analysis | Upregulated | Predicts poor overall survival (OS); Associated with treatment failure; miR-21-5p (OS: p < 0.001); miR-4454 (OS: p < 0.001) | [107] | |
| Lymphoma | miR-107 | Plasma | qRT-PCR, RNA-seq, Western blot, TEM, NTA | Downregulated | AUROC: 0.854 for diagnostic power; inhibits cell proliferation and invasion by targeting 14-3-3η; strong prognostic relevance | [108] |
4.4. Nervous system
Gliomas are the most common primary brain tumors, originating from neural stem or progenitor cells that acquire tumor-initiating genetic mutations. These tumors are classified under the World Health Organization (WHO) grading system, ranging from Grade 1 (benign) to Grade 4 (highly malignant). Adult diffuse gliomas, such as isocitrate dehydrogenase (IDH)-mutant astrocytomas and oligodendrogliomas, often have better prognoses, while IDH-wild-type glioblastomas are highly aggressive and carry poor survival outcomes. Pediatric gliomas include both low-grade and high-grade forms, with distinct molecular alterations, such as histone mutations in midline gliomas, leading to their aggressive behavior [109]. The development of gliomas involves genetic and epigenetic changes, including mutations in IDH, TP53, and ATRX, alongside alterations in pathways like PI3K/AKT and MAPK, contributing to tumor growth and resistance to therapy. Gliomas are challenging to treat due to their invasive nature, resistance mechanisms, and immunosuppressive microenvironment. Standard therapies include surgery, radiotherapy, and chemotherapy, but emerging molecularly targeted therapies offer hope for improved outcomes [110]. Advancements in molecular profiling and biomarker discovery have enhanced diagnostic accuracy, enabling personalized treatment approaches. In particular, the total number of lncRNAs has been steadily increasing, thanks to more sensitive detection methods. Today, the number of lncRNAs surpasses that of all protein-coding genes. Primarily transcribed by RNA polymerase II, lncRNAs undergo various post-transcriptional modifications. They are found in multiple cellular compartments, including the nucleus, nucleolus, cytoplasm, and mitochondria. Growing evidence suggests a mechanistic link between lncRNA dysregulation and numerous human diseases, including cancer, positioning lncRNAs as promising therapeutic targets and biomarkers [111]. However, challenges in early detection, the complexity of molecular subtypes, and limited treatment efficacy for high-grade gliomas underscore the need for continued research and innovation in glioma management [112]. The emergence of exosomes as biomarkers has provided new possibilities for the early detection of glioma.
Shiguang Zhao et al. identified exosomal miR-2276-5p as a promising glioma biomarker, with low expression in glioma patients, particularly high-grade ones, and poor survival correlation. It targets RAB13, impacting tumor progression and the JAK/STAT3 pathway, and shows strong diagnostic accuracy (AUC: 0.8107). miR-2276-5p's non-invasive detection via plasma exosomes highlights its potential as both a biomarker and therapeutic target [113]. Huilin Shao et al. introduced EZ-READ, a blood-based glioblastoma (GB) diagnostic platform, detecting mRNA and miRNA signatures from EVs with high sensitivity (detection limit: 9 RNA copies) and accuracy (AUC: 0.897). EZ-READ offers a rapid, non-invasive alternative to PCR, ideal for real-time monitoring and personalized medicine [114]. Yanlin Song et al. developed a multiplex optical biochip for exosome detection in glioblastoma, using nanochains to capture exosomes and amplify signals. It can simultaneously detect multiple biomarkers (e.g., CD44, CD133) within 30 minutes, providing a cost-effective, non-invasive approach for early diagnosis and personalized treatment [115]. Guan Sun et al. identified circBTG2, enriched in exosomes from RBP-J-overexpressed macrophages, as a glioma biomarker. It regulates key oncogenic pathways through the circBTG2/miR-25-3p/PTEN axis, offering both diagnostic and therapeutic potential for liquid biopsy applications and personalized treatment [116].
Pituitary adenomas (PAs) are typically benign tumors of the pituitary gland, classified by size into microadenomas (<10 mm) and macroadenomas (>10 mm), and by hormone activity into functional (hormone-secreting) and non-functional (non-secreting) types. Functional adenomas cause hormone-related syndromes, such as prolactinomas (hyperprolactinemia), acromegaly (excess growth hormone), or Cushing's disease (excess cortisol), while non-functional pituitary adenomas (NFPAs) often present with symptoms like headaches or visual disturbances due to compression of surrounding structures [117]. Diagnosis involves hormonal tests, MRI imaging, and sometimes visual field assessments. Treatment options include medical therapy (e.g., dopamine agonists for prolactinomas), surgery (typically via a transsphenoidal approach), and, in some cases, radiation therapy. While benign, pituitary adenomas can cause significant health issues, underscoring the importance of early diagnosis and treatment. The diagnosis of PAs faces challenges due to their diverse presentations and subtle early symptoms. Functional adenomas may cause hormonal syndromes like Cushing's disease or acromegaly, but these symptoms often develop gradually and overlap with other conditions, delaying recognition. NFPAs are typically asymptomatic until they grow large enough to cause headaches or visual disturbances. Small microadenomas can be difficult to detect on imaging, and hormonal tests may yield false positives or negatives due to confounding factors. Differentiating benign adenomas from malignant tumors is also challenging without invasive procedures. These issues emphasize the need for better imaging techniques, specific biomarkers, and improved clinical awareness [118]. Exosomes, as biomarkers, provide a convenient and effective approach for the diagnosis of PAs.
Peizhi Zhou et al. highlight the potential of exosomal miRNA profiling for non-invasive diagnosis and prognosis of non-functioning pituitary adenomas (NFPAs). hsa-miR-486-5p was identified as a promising biomarker with high diagnostic accuracy and strong predictive value for tumor recurrence or progression. Next-generation sequencing revealed 54 differentially expressed miRNAs, with bioinformatics suggesting that hsa-miR-486-5p regulates tumor progression via the MAPK signaling pathway, DNA repair, and epigenetic mechanisms. Higher levels of hsa-miR-486-5p were associated with worse progression-free survival, supporting its use for personalized treatment strategies. Further validation in larger cohorts is needed [119]. Weiping Liu et al. identified serum exosomal circCCDC66 as a promising biomarker for the diagnosis and prognosis of pituitary adenomas (PAs). circCCDC66 was significantly upregulated in PA patients, showing strong correlation between exosomal and tissue levels. It demonstrated high diagnostic accuracy (AUC: 0.8719) with 80% sensitivity and 84% specificity. Post-surgical levels decreased but increased at recurrence, aiding in tumor monitoring. Lower circCCDC66 expression correlated with longer progression-free survival, suggesting its role in early detection and recurrence prediction. Further large-scale validation studies are needed [120]. Guojun Zhang et al. emphasized the potential of EMT-related markers in serum exosomes as non-invasive biomarkers for diagnosing and monitoring invasive pituitary adenomas (IPAs). Invasive PAs showed higher mesenchymal marker expression (N-cadherin, vimentin) and lower epithelial marker expression (E-cadherin, Epcam) in serum exosomes and tumor tissues. The N-cadherin/Epcam ratio was significantly elevated in invasive cases, aiding differentiation from non-invasive adenomas. The TGF-β/Smad signaling pathway was also found to negatively regulate EMT in pituitary adenomas, with therapeutic implications. These findings suggest that EMT-related exosomal markers could be valuable for non-invasive diagnosis and monitoring of IPA progression, though larger studies are needed for validation [121].
The studies highlight the potential of exosomal biomarkers for diagnosing, prognosing, and monitoring nervous system tumors, including gliomas, glioblastomas, and PCNSL (Table 5). Exosomal miRNAs like miR-2276-5p and circBTG2 are promising for glioma diagnosis and progression, while platforms like EZ-READ and biochips enable non-invasive glioblastoma subtyping and monitoring. These biomarkers allow non-invasive sampling, improved sensitivity, and real-time monitoring, though more clinical validation is needed.
4.5. Endocrine system
Thyroid cancer, the most common endocrine malignancy, includes four types: papillary thyroid cancer (PTC), follicular thyroid cancer (FTC), medullary thyroid cancer (MTC), and anaplastic thyroid cancer (ATC). Symptoms include a painless neck lump, hoarseness, and difficulty swallowing or breathing. Diagnosis involves imaging, biopsy, and thyroid function tests, with treatment typically including surgery, radioactive iodine, and sometimes chemotherapy for advanced cases [122]. PTC and FTC generally have an excellent prognosis with early detection. Despite advances in diagnostic techniques, thyroid cancer diagnosis faces challenges. Early-stage thyroid cancer is often asymptomatic, making detection difficult without routine screening [123]. Thyroid ultrasound, while essential for evaluating nodules, often cannot distinguish benign from malignant ones, leading to unnecessary biopsies and increased costs [124]. FNAB can yield indeterminate cytology (e.g., Bethesda categories III and IV), creating uncertainty about the next steps [125]. Molecular testing improves accuracy but is limited by high costs and inconsistent availability [126]. Diagnosing aggressive forms like ATC or small metastases remains difficult, as current imaging may miss microscopic disease. These issues highlight the need for better diagnostic tools, including precise imaging, advanced molecular biomarkers, and risk stratification systems.
Yongsheng Zhao et al. identified plasma exosomal miR-485-3p and miR-4433a-5p as non-invasive biomarkers for diagnosing and stratifying papillary thyroid cancer (PTC). MiR-485-3p showed the highest diagnostic accuracy (AUC: 0.8581, sensitivity: 85.42%, specificity: 73.33%) and was linked to high-risk features like larger tumor size, advanced stage, BRAF mutation, and lymph node metastasis, making these miRNAs valuable for distinguishing PTC and identifying aggressive disease [127]. Pei-Jie Huang et al. identified urinary exosomal TIMP and Angiopoietin-1 as biomarkers for predicting lymph node metastasis (LNM) in well-differentiated thyroid cancer (WDTC). Higher TIMP levels were associated with LNM, and these biomarkers could complement existing tools like MACIS for surgical planning and follow-up [128]. Diego Russo et al. identified deregulated exosomal miRNAs (miR-31-5p, miR-222-3p, let-7i-3p) as key players in aggressive PTC subtypes. These miRNAs, upregulated in tumor-derived exosomes, are linked to tumorigenic pathways and could serve as biomarkers and therapeutic targets for advanced PTC [129].
Pancreatic neuroendocrine tumors (pNETs) are rare neoplasms, making up 1-2% of pancreatic tumors [130]. They are classified into functional and non-functional types. Functional pNETs produce hormones, leading to conditions like hypoglycemia in insulinomas, ulcers in gastrinomas, and watery diarrhea in VIPomas. Non-functional pNETs, which are more common, present with abdominal pain, jaundice, or weight loss as the tumor grows or metastasizes [131]. Diagnosis involves imaging (CT, MRI, Ga-68 DOTATATE PET/CT) and biomarkers like chromogranin A. Treatment includes surgery for localized tumors, somatostatin analogs for functional tumors, and targeted therapies or peptide receptor radionuclide therapy (PRRT) for advanced cases. Prognosis varies, emphasizing the need for early detection and personalized treatment [132]. Diagnosing pNETs is challenging due to their rarity, heterogeneity, and subtle symptoms. Functional pNETs have distinct syndromes but can overlap with common conditions, causing delays in diagnosis. Non-functional pNETs often remain asymptomatic until advanced stages or metastasis, complicating early detection. Imaging techniques, such as CT, MRI, and Ga-68 DOTATATE PET/CT, are essential but may miss small or poorly differentiated tumors. Biochemical markers like chromogranin A lack specificity, with non-tumor factors (e.g., renal impairment, proton pump inhibitors) affecting results [133]. Additionally, distinguishing pNETs from other pancreatic cancers and assessing tumor aggressiveness remains difficult. These challenges highlight the need for more sensitive biomarkers, advanced imaging, and integrated diagnostic approaches for better early detection and accurate classification of pNETs.
Exosomal biomarkers in nervous system cancers
| Cancer type | Biomarkers | Source | Analytical Technique | Expression | Effectiveness | Reference |
|---|---|---|---|---|---|---|
| miR-2276-5p | Plasma | qRT-PCR, AUROC analysis, TEM, NTA, western blot | Downregulated | AUROC: 0.8107; Prognostic biomarker for survival; Lower expression associated with poor survival rates | [113] | |
| Glioma | miR-17-5p, miR-21-5p, miR-27a-3p, LOX, SLCO3A1 | Plasma | EZ-READ platform, qRT-PCR, RNA sequencing, ROC analysis, TEM, NTA | Upregulated | AUROC for diagnosis: 0.854; AUROC for subtyping: 0.897; enables non-invasive subtype classification and prognosis | [114] |
| CD63, CD44, CD133 | Plasma | Multiplex optical biochip with nanochain patterns | Upregulated | Detects GBM-specific exosomes with high sensitivity (LOD: 6×10⁷ particles/mL); rapid detection (~30 min) | [115] | |
| circRNA BTG2, miR-25-3p, PTEN | Exosomes from RBP-J overexpressed macrophages | qRT-PCR, RNA-seq, TEM, NTA, Luciferase assay, Transwell | circRNA BTG2 upregulated; miR-25-3p downregulated; PTEN upregulated | Inhibits glioma progression; reduces proliferation and invasion via the circBTG2/miR-25-3p/PTEN pathway NFs into CAFs | [116] | |
| hsa-miR-486-5p, hsa-miR-151a-5p, hsa-miR-652-3p_R+1, hsa-miR-1180-3p | Serum | NGS, qRT-PCR, microarray, AUROC analysis, TEM, NTA, western blot | Upregulated | AUROC: 0.943 (hsa-miR-486-5p); potential biomarker for NFPA diagnosis, progression, and relapse prediction | [119] | |
| Pituitary adenoma | circCCDC66 | Serum | qRT-PCR, AUROC analysis, TEM, NTA, western blot | Upregulated | AUROC: 0.872; Sensitivity: 80%; Specificity: 84%; prognostic marker for tumor recurrence and progression | [120] |
| N-cadherin, E-cadherin, Epcam, Vimentin | Serum | qRT-PCR, western blot, TEM, NTA, immunohistochemistry | N-cadherin and vimentin upregulated; E-cadherin and Epcam downregulated in PA | AUROC for N-cadherin/Epcam ratio: 0.83; effective in assessing tumor invasiveness and predicting recurrence | [121] |
Exosomal biomarkers in endocrine system cancers
| Cancer type | Biomarkers | Source | Analytical Technique | Expression | Effectiveness | Reference |
|---|---|---|---|---|---|---|
| miR-485-3p, miR-4433a-5p, miR-376a-3p | Plasma | Small RNA sequencing, qRT-PCR, AUROC analysis, TEM, NTA, Western blot | Upregulated | AUROC: miR-485-3p (0.858), miR-4433a-5p (0.812); serve as diagnostic and prognostic biomarkers for PTC | [127] | |
| Thyroid cancer | Angiopoietin-1, TIMP | Urinary | LC-MRM/MS, qRT-PCR, AUROC analysis, TEM, NTA | Upregulated | AUROC: Angiopoietin-1 (0.857), TIMP (0.892); potential biomarkers for preoperative screening and prognosis | [128] |
| miR-21-5p, miR-31-5p, miR-221-3p, miR-222-3p, let-7i-3p | Exosomes from thyroid cancer cell lines | qRT-PCR, RNA-seq, AUROC analysis, TEM, NTA, Western blot | Upregulated | AUROC: miR-31-5p (approximately 0.9), miR-222-3p (approximately 0.9); miRNAs linked to invasion, angiogenesis, and tumor progression | [129] | |
| Pancreatic Neuroendocrine Neoplasm | miR-4488 | Hypoxic tumor-derived exosomes | RNA-seq, qRT-PCR, TEM, NTA, WB, Luciferase assay | Upregulated | miR-4488 facilitates M2 macrophage polarization, promotes metastasis via RTN3/FABP5-mediated fatty acid oxidation and PI3K/AKT/mTOR pathway | [134] |
| CEACAM5 | Hypoxic tumor-derived exosomes | qRT-PCR, Western blot, ChIP, TEM, NTA, Transwell assay, Flow cytometry | Upregulated | AUROC: 0.873 for differentiating metastatic and non-metastatic pNETs; promotes metastasis via MAPK pathway and MMP9 upregulation | [135] |
Qiyun Tang et al. reveal that hypoxic pNENts-derived exosomal miR-4488 promotes liver metastasis by reprogramming tumor-associated macrophages (TAMs) into an M2-like phenotype. miR-4488 downregulates RTN3 in macrophages, enhancing fatty acid oxidation (FAO) via FABP5 and activating the PI3K/AKT/mTOR pathway, which drives M2 polarization. These M2 macrophages secrete MMP2, facilitating tumor migration and liver metastasis. In vivo models confirmed the metastatic role of hypoxic exosomes, while targeting miR-4488, RTN3, or MMP2 significantly reduced metastasis. These findings suggest that miR-4488 and its downstream targets (RTN3, FAO, MMP2) could serve as promising biomarkers and therapeutic targets to prevent or treat pNEt metastasis [134]. The research team also conducted another related study. The study identifies hypoxia-induced exosomal CEACAM5 as a key driver of pancreatic neuroendocrine tumor (pNET) metastasis by promoting tumor-associated macrophage (TAM) M2 polarization and enhancing MMP9 secretion. CEACAM5, upregulated under hypoxic conditions via HIF1α, activates the MAPK/ERK pathway in TAMs, leading to M2 polarization and increased extracellular matrix degradation through MMP9, which facilitates tumor cell migration and invasion. In vivo models confirmed that CEACAM5-rich exosomes significantly increased metastatic burden, while knockdown of CEACAM5 or inhibition of exosome secretion and MMP9 reduced metastasis. These findings suggest CEACAM5 as a promising biomarker and therapeutic target for metastatic pNETs, offering new strategies to disrupt tumor progression in this aggressive cancer type [135].
The studies highlight the potential of exosomal biomarkers in diagnosing and managing endocrine tumors (Table 6). Exosomal miRNAs like miR-485-3p and miR-4433a-5p provide strong diagnostic accuracy for PTC, while urinary exosomal proteins (TIMP, Angiopoietin-1) aid in lymph node metastasis assessment for WDTC. In pNETs, exosomal components like miR-4488 and CEACAM5 promote macrophage M2 polarization and metastasis, offering diagnostic and therapeutic potential. These findings emphasize the role of exosomal biomarkers in early detection, risk assessment, and treatment monitoring.
4.6. Genitourinary system
Breast cancer (BC) is one of the most common malignancies and the leading cause of cancer-related deaths among women worldwide. It arises from the abnormal growth of cells in the breast tissue, often originating in the ducts (ductal carcinoma) or lobules (lobular carcinoma). Key risk factors are genetic predisposition (e.g., BRCA1/2 mutations), hormonal factors, lifestyle influences such as obesity and alcohol consumption, and a family history of the disease [136]. Symptoms may include a lump in the breast, changes in breast shape or size, skin dimpling, or nipple discharge. Diagnosis typically involves imaging techniques like mammography and ultrasound, followed by biopsy for histopathological and molecular analysis. Advances in treatment, including surgery, radiotherapy, chemotherapy, targeted therapies, and immunotherapy, have significantly improved survival rates. Early detection through screening programs remains crucial for improving outcomes, as breast cancer is most treatable in its early stages [137]. Despite advancements in BC diagnostics, several limitations remain. Imaging techniques like mammography, while effective for early detection, often struggle to distinguish between benign and malignant lesions, leading to false positives and unnecessary biopsies. Additionally, dense breast tissue can obscure tumors, reducing diagnostic accuracy. Biopsy, the gold standard for confirmation, is invasive and may cause patient discomfort. Furthermore, molecular and genetic testing, although valuable for identifying subtypes and guiding treatment, is expensive and not universally accessible. Early-stage cancers or those in non-palpable areas may still go undetected, emphasizing the need for more sensitive, non-invasive, and cost-effective diagnostic tools to address these challenges [138]. The emergence of exosomal biomarkers offers promising solutions to these challenges in BC diagnosis.
Jinfeng Zhu et al. developed aptamer-based plasmonic metasurfaces (APM) for the label-free detection of breast tumor-derived exosomes, enabling highly sensitive and cost-effective BC diagnosis and molecular subtyping. The mechanism leverages optimized near-field plasmonic effects using thiol-modified aptamers, achieving an improved detection limit of 1.04 × 10⁴ particles/mL and a diagnostic accuracy (AUC: 99.3%) superior to traditional biomarkers like CA153. Advantages include high sensitivity, non-invasive serum-based detection, cost-effectiveness due to aptamer use, simplified workflows without signal amplification, and the ability to classify molecular subtypes such as HER2-positive BC, making it a valuable tool for early detection and personalized treatment strategies [139]. Shuqing Sun et al, developed the CRISPR/Cas12a and Aptamer-Chemiluminescence-Based Analysis (CACBA) platform for BC diagnosis by detecting tumor-related proteins (EpCAM and MUC1) on exosome surfaces. The mechanism combines CRISPR/Cas12a for total exosome quantification with aptamer-chemiluminescence assays to calculate the relative abundance of tumor-specific exosomal proteins. This approach achieved exceptional diagnostic accuracy (AUC = 1.00) and sensitivity, with detection limits as low as 1.45 × 10² particles/μl. Advantages include its non-invasive nature, high specificity, robustness against sample variability, cost-effectiveness due to aptamer use, and adaptability for detecting other tumor-related biomarkers, making it a versatile and reliable tool for clinical cancer diagnostics [140]. Genxi Li et al. developed a biomimetic vesicle system that utilizes cancer cell membrane camouflage to selectively bind and fuse with homologous BC exosomes, enabling precise molecular subtyping of BC. The system detects endogenous exosomal RNA biomarkers (e.g., miR-375 for ER-positive and PD-L1 mRNA for triple-negative subtypes) to trigger catalytic DNA assembly, amplifying electrochemical signals for highly sensitive detection with a limit of 557 particles/mL. Key benefits also encompass high specificity and sensitivity, the option for non-invasive detection through liquid biopsy, the ability to monitor disease according to its stage, and versatility for multiple cancer types, positioning it as a powerful solution for BC diagnosis and personalized treatment planning [141].
Ovarian cancer (OC) is a leading cause of gynecological cancer-related deaths, often asymptomatic in early stages, earning it the nickname "silent killer." It primarily originates from epithelial cells but can also arise from germ cells or stromal tissues. Symptoms like abdominal bloating and pelvic pain are nonspecific, leading to delayed diagnosis. Risk factors include genetic predisposition (e.g., BRCA1/2 mutations), family history, age, and hormonal influences. Despite treatment advances, prognosis for advanced-stage cases remains poor due to high recurrence rates [142]. Current diagnostic tools, including imaging, CA-125 and HE4 blood tests, are limited by low sensitivity and specificity, often resulting in false positives or negatives, especially for early-stage detection [143]. CA125 and HE4 are the only approved biomarkers for use in epithelial OC; however, they are not sufficient for early detection. Multivariate index (MVI) assays have been developed to mitigate the limitations of single serum biomarkers in epithelial OC, especially during the pre-surgical evaluation of adnexal masses. The Risk of malignancy algorithm (ROMA) integrates menopausal status, CA125 and HE4 concentrations to diagnose women with a pelvic mass. miRNAs may have remarkable potential in various aspects of epithelial OC prediction. However, further work is needed regarding its characterization as a biomarker. In particular, the steps involved in processing samples need to be standardized and the platforms for detecting miRNA in tumours and blood need to be refined [144]. Invasive procedures like biopsies are costly and risky. Exosomal biomarkers offer a promising non-invasive alternative for early OC detection [145].
Yanlong Xing et al. developed a SERS-microfluidic platform (S-MMEV) for non-invasive phenotyping of sEVs in serum, enabling early-stage OC diagnosis. The system combines SERS with microfluidics to detect multiple biomarkers, such as EpCAM, CD24, and CA125, with a detection limit as low as 10 particles/mL. By capturing sEVs with anti-CD63 antibodies and using SERS nanoprobes for biomarker labeling, it achieves high sensitivity and specificity (AUC: 0.9467 for early-stage detection). Its advantages include rapid turnaround (within one hour), the ability to detect multiple biomarkers simultaneously, non-invasive sample collection via liquid biopsy, and potential adaptability to various cancer types, making it a valuable tool for early detection and treatment monitoring [146]. Genxi Li et al. developed an electrochemical biosensor based on entropy-driven autocatalytic DNA circuits (EADC) for the ultra-sensitive detection of ovarian cancer-derived exosomes. The biosensor captures exosomes expressing MUC1 glycoprotein using aptamer-functionalized probes, triggering an autocatalytic DNAzyme reaction that amplifies electrochemical signals. It achieved an impressive detection limit of 30 particles/μL and demonstrated high specificity in distinguishing OC exosomes from normal cell-derived exosomes. Strengths include its high sensitivity, non-invasive detection via serum samples, cost-effectiveness due to its enzyme-free design, adaptability for other biomarkers, and potential for portable, real-time cancer diagnostics, making it a powerful tool for early ovarian cancer detection [147]. Zhiyong Liang et al. developed the sEVmiR-EOC model, a non-invasive diagnostic and prognostic tool for epithelial ovarian carcinoma (EOC) based on seven serum-derived sEV microRNAs (e.g., miR-141-3p, miR-200c-3p). This model achieved superior diagnostic accuracy (AUC: 0.913-0.973) compared to CA125, particularly in detecting early-stage EOC (AUC: 0.903 vs. 0.694). It also demonstrated prognostic utility by correlating miRNA expression changes with patient outcomes. The approach provides high sensitivity, specificity, and the capability to track disease progression, making it a promising method for early detection, treatment monitoring, and personalized management of OC, with potential adaptability for other types of cancer [148] (Figure 5).
Renal cell carcinoma (RCC) is the most common kidney cancer, accounting for about 90% of adult cases. It originates in the renal tubules and is known for its aggressive nature and resistance to traditional therapies like chemotherapy and radiotherapy [149]. Early-stage RCC is often asymptomatic, while advanced stages may present with hematuria, flank pain, and an abdominal mass. Risk factors include smoking, obesity, hypertension, and genetic factors like mutations in the VHL tumor suppressor gene. The loss of functional VHL protein is considered a defining event in the development of clear-cell RCC, and the principal downstream oncogenic mechanism appears to be HIF-2α accumulation and constitutive HIF transcription factor activity. Belzutifan is a potent small-molecule inhibitor of HIF-2α that prevents heterodimerization with HIF-1β into an active transcription factor and has shown activity in clear-cell RCC. A recently published phase III study showed a significant benefit of belzutifan over everolimus concerning progression-free survival and objective response in participants with advanced clear-cell RCC who had previously received immune checkpoint and antiangiogenic therapies [150]. Diagnosis faces challenges, as RCC is frequently detected incidentally and current imaging methods (CT, MRI, ultrasound) may not distinguish benign from malignant lesions, often requiring invasive biopsies for confirmation [151]. The heterogeneity of RCC further complicates diagnosis, emphasizing the need for advanced diagnostic tools, including molecular biomarkers and improved imaging methods for early, non-invasive detection. The cargo in tumor-derived exosomes, such as the range of miRNAs, can serve as biomarkers for clear cell RCC in the serum and urine of patients, offering valuable targets for early detection and monitoring of the disease [152].
Fubo Wang et al. identified five exosomal mRNA biomarkers (CUL9, KMT2D, PBRM1, PREX2, SETD2) for early detection and differential diagnosis of clear cell renal cell carcinoma (ccRCC). The diagnostic signature with KMT2D and PREX2 achieved high accuracy (AUC: 0.836-0.830), and the differential signature with CUL9, KMT2D, and PREX2 distinguished ccRCC from benign renal masses (AUC: 0.816). This approach, validated across multiple cohorts, offers exceptional sensitivity, specificity, and non-invasive detection using blood-derived exosomes, making it a valuable tool for early ccRCC detection and personalized treatment [153]. Maria Jesus Alvarez-Cubero et al. identified exosomal mitochondrial DNA (mtDNA) as a non-invasive biomarker for RCC diagnosis and aggressiveness. mtDNA genes like HV1 and CYB showed significant differences between RCC patients and healthy controls, correlating with tumor stage and metastatic potential. Using plasma-derived exosomes and digital PCR, the study achieved high diagnostic accuracy (AUC: 0.833 for HV1), demonstrating the stability of exosomal mtDNA in circulation and its value for monitoring RCC progression and guiding personalized treatments [154]. Kerstin Junker et al. identified CD147, CA9, and CD70 as specific tumor markers on exosomes from ccRCC, distinguishing them from normal tissue exosomes. These markers, validated in ccRCC cell lines, primary tumors, and patient samples, reflect key tumorigenic processes like matrix degradation, angiogenesis, metabolism, and immune evasion. The study introduced a refined EV isolation protocol for high-purity EV recovery, showing that these markers outperform EpCAM in diagnostic performance and have potential for non-invasive early diagnosis and therapeutic monitoring of ccRCC [155].
The studies highlight significant advancements in non-invasive diagnostics for urogenital cancers (Table 7). Exosomal biomarkers such as miR-2276-5p (OC), miR-141-3p (RCC), and tumor-specific surface markers (e.g., CD147, CA9, CD70) are emerging as key diagnostic tools. Advanced detection technologies, including SERS-based biosensors, CRISPR/Cas12a assays, and autocatalytic DNA circuits, enable highly sensitive and specific detection of these biomarkers in liquid biopsy samples. With diagnostic accuracies (AUC > 0.9), these platforms allow early cancer detection, differentiation of malignant and benign conditions, and precise molecular subtyping. They also support disease monitoring and prognostic assessments, crucial for personalized treatment. Novel isolation and amplification methods ensure reproducibility, making these approaches clinically feasible. These innovations represent a major shift in urogenital cancer diagnostics, improving early detection, prognosis, and treatment outcomes.
4.7. Dermal system
Malignant melanoma (MM) is an aggressive skin cancer originating from melanocytes, responsible for most skin cancer-related deaths due to its rapid metastasis. Risk factors include UV exposure, fair skin, sunburns, and genetic mutations (e.g., BRAF, NRAS) [156]. Early symptoms involve changes in moles, following the ABCDE rule. Diagnosis faces challenges as early-stage melanomas resemble benign lesions, and visual assessments may miss atypical cases. Dermoscopy and biopsy are the gold standards but are invasive and require expertise. Advanced melanomas often metastasize early, and the lack of specific biomarkers complicates non-invasive screening [157]. Myeloid-derived suppressor cells, Treg cells and tumour-associated macrophages constitute immunosuppressive cells present within the tumour microenvironment, which release ROS amongst other factors, effectively inhibiting NK cell response. Higher levels of fibroblasts secrete more metalloproteinases, resulting in further shedding of ligands that could link to NK cells. Fibroblasts even have a more direct impact on NK cells by preventing cytokine induced activating receptor upregulation. NK cell function is also impaired by the presence of melanoma derived exosomes. This finding highlights the potential of exosomal biomarkers in diagnostics [158].
Schematic representation of the sEVmiR-EOC model for noninvasive diagnostic and prognostic predictions of EOC. The changes in the sEVmiR-EOC RiskScore between preoperative and postoperative blood samples were analyzed, and we found that the sEVmiR-EOC model could predict the prognosis of EOC patients. The study indicates that serum sEV miRNAs could be promising noninvasive biomarkers for the diagnosis and prognosis of EOC. Reproduced from reference [148] Copyright 2023, with permission from American Chemical Society.
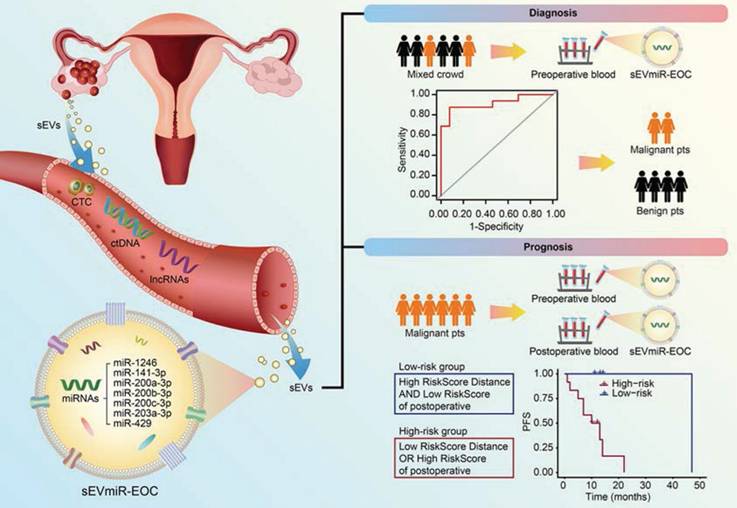
Exosomal biomarkers in genitourinary system cancers
| Cancer type | Biomarkers | Source | Analytical Technique | Expression | Effectiveness | Reference |
|---|---|---|---|---|---|---|
| EpCAM, HER2 | Serum | Plasmonic metasurface (APM), qRT-PCR, TEM, NTA, SEM, AUROC analysis | Upregulated | AUROC: 99.3% for EpCAM; APM biosensors distinguish BC from controls and classify HER2 levels with P < 0.0001 | [139] | |
| Breast cancer | EpCAM, MUC1 | Plasma | CRISPR/Cas12a, Aptamer-chemiluminescence (ACL), TEM, NTA, Western blot, Flow Cytometry | Upregulated | AUROC: EpCAM (+) % = 0.9889, MUC1(+) % = 0.9630; High sensitivity and specificity in differentiating breast cancer from healthy controls | [140] |
| miR-375, PD-L1 mRNA | Serum | Electrochemical analysis, qRT-PCR, TEM, NTA, Western blot | Upregulated | Detection limit: 557 particles/mL for miR-375; Reliable subtype differentiation with homotypic recognition; Quantitative analysis supports diagnosis and monitoring | [141] | |
| Ovarian cancer | CD81, CD9, EpCAM, EGFR, CD24, CA125 | Serum | SERS-Microfluidic platform, TEM, NTA, Western blot, AUROC analysis | Upregulated | AUROC: 0.9467 for early-stage OC (stage I-II); 0.9538 for all OC patients. High sensitivity (1.00) and specificity (0.95) in early diagnosis. | [146] |
| MUC1 | Serum | Electrochemical biosensor with EADC, TEM, NTA, SWV | Upregulated | LOD: 30 particles/µL; Linear range: 79 to 315,000 particles/µL; Differentiates OC exosomes with high specificity and sensitivity | [147] | |
| miR-1246, miR-141-3p, miR-200a-3p, miR-200b-3p, miR-200c-3p, miR-203a-3p, miR-429 | Serum | RNA sequencing, qRT-PCR, TEM, NTA, LASSO regression, AUROC analysis | Upregulated | AUROC: 0.973 (testing cohort), 0.924 (validation cohort); Higher sensitivity and specificity than CA125 for early-stage OC detection | [148] | |
| Renal cell carcinoma | KMT2D, PREX2 | Serum | RNA sequencing, qRT-PCR, Logistic regression, AUROC analysis | Upregulated | AUROC: 0.836 (training cohort), 0.830 (validation cohort) for ccRCC vs. healthy; AUROC: 0.816 for ccRCC vs. benign renal masses | [153] |
| HV1 (Hypervariable Region 1), CYB (Cytochrome B) | Plasma | qPCR, dPCR, NGS, AUROC analysis | Upregulated | AUROC: 0.833 (HV1 long), 0.810 (CYB long); Effective in differentiating RCC cases and assessing aggressiveness | [154] | |
| CA9, CD147, CD70 | Serum | Western blot, TEM, NTA, IHC, qRT-PCR | Upregulated | Potential to enhance tumor-specific EV isolation; diagnostic markers for ccRCC with high specificity | [155] |
Se-Hwan Paek et al. developed a calcium switch-based immuno-isolation method to enrich CD63-positive exosomes for early melanoma detection. This approach increased diagnostic sensitivity (87.5%) compared to bulk exosome analysis (68%) and accurately distinguished melanoma patients from healthy controls (specificity: 85.7%). The method offers a non-invasive, reliable alternative for cancer detection and can be applied to other cancers [159]. Amalia Azzariti et al. identified PD1+ and PD-L1+ EVs as biomarkers for predicting response to immune checkpoint inhibitors (ICIs) in metastatic melanoma. High levels of these EVs correlated with poorer prognosis and contributed to resistance by neutralizing therapeutic antibodies and suppressing T-cell activity. Liquid biopsy of circulating EVs offers a predictive, minimally invasive method for monitoring ICI response [160]. Cong Peng et al. found that plasma exosomal miR-1180-3p is a promising non-invasive biomarker for cutaneous melanoma, with an AUC of 0.729. miR-1180-3p was downregulated in melanoma patients and suppressed tumor cell proliferation and migration by targeting key genes involved in melanoma progression. This highlights its potential for early detection and personalized treatment [161].
Non-melanoma skin cancer (NMSC), the most common cancer worldwide, primarily includes basal cell carcinoma (BCC) and squamous cell carcinoma (SCC) [162]. BCC is generally slow-growing with minimal metastatic risk, while SCC has a higher potential for invasion and metastasis. Key risk factors include chronic UV radiation exposure, fair skin, immunosuppression, and genetic predispositions like xeroderma pigmentosum [163]. NMSC typically manifests as non-healing lesions or erythematous plaques on sun-exposed areas, with BCC appearing as pearly nodules and SCC as scaly, erythematous lesions [164]. Diagnosis of non-melanoma skin cancer (NMSC) relies on clinical examination and biopsy, with treatments ranging from surgery (e.g., Mohs) to cryotherapy, radiation, and topical therapies. Advanced cases may require systemic immunotherapy, such as cemiplimab. Early intervention offers an excellent prognosis, while UV protection and regular skin checks are key to managing the increasing global incidence of NMSC. Diagnosis faces limitations, especially in early detection and differentiation from benign conditions. Clinical examination and dermoscopy depend on clinician experience and may miss subtle presentations, particularly in early-stage tumors or patients with darker skin. Biopsy is invasive and prone to sampling errors. Emerging imaging techniques, like ultrasound and reflectance confocal microscopy, are not widely available. There is also a lack of reliable non-invasive biomarkers for distinguishing aggressive subtypes. These challenges can lead to delayed or misdirected treatment. Advancements in non-invasive diagnostics, such as liquid biopsy, molecular profiling, and AI-assisted imaging, are needed to improve early detection and accuracy [165].
A.L.S. Chang et al. identify serum exosomal miR-197-5p as a key regulator of metastasis in BCC, highlighting its role in modulating the tumor microenvironment. RNA sequencing and qPCR revealed that miR-197-5p, among other miRNAs, was significantly upregulated in serum exosomes from metastatic BCC (MBCC) patients compared to non-metastatic cases. Functional studies demonstrated that MBCC-derived exosomes enhanced fibroblast proliferation, migration, and invasion, with miR-197-5p specifically promoting fibroblast activation. Inhibition of miR-197-5p significantly reduced fibroblast migration and metabolic activity, underscoring its role in metastasis. These findings imply that miR-197-5p could serve as a non-invasive biomarker for metastatic BCC and a potential therapeutic target, pending further validation in larger clinical studies [166]. Jiuhong Li et al. identify exosomal circ-CYP24A1 as a critical driver of cutaneous squamous cell carcinoma (cSCC) progression and a potential diagnostic biomarker and therapeutic target. circ-CYP24A1 was significantly overexpressed in plasma-derived exosomes from cSCC patients and correlated with tumor size, thickness, and serum SCC-Ag levels. Functional studies revealed that circ-CYP24A1 promotes cSCC cell proliferation, migration, and invasion while inhibiting apoptosis, partially through the regulation of downstream targets such as CDS2, MAVS, and SOGA1. Pathway analysis linked circ-CYP24A1 to immune and cell cycle regulatory pathways, highlighting its role in tumor-stromal communication. Targeting circ-CYP24A1 in exosomes significantly reduced tumorigenic behaviors, underscoring its potential as a non-invasive biomarker for cSCC diagnosis and a novel therapeutic target for limiting tumor progression [167].
Exosomes are emerging as highly valuable tools in the diagnosis and monitoring of skin cancers, including basal cell carcinoma, malignant melanoma, and squamous cell carcinoma (Table 8). By carrying tumor-specific biomarkers such as CD63, Cav1, circ-CYP24A1, and miR-1180-3p, exosomes enable highly sensitive and specific detection of early-stage cancers and offer insights into disease progression. Moreover, exosomal molecules like PD1 and PD-L1 are closely linked to immunotherapy resistance and poor prognosis in metastatic melanoma, making them effective for predicting therapeutic outcomes and enabling real-time monitoring of treatment efficacy. Additionally, exosomal miRNAs such as miR-197-5p play a role in metastatic progression, highlighting their potential as therapeutic targets. With their high stability in circulation, non-invasive collection from bodily fluids, and ability to provide dynamic molecular insights, exosomes represent an ideal platform for liquid biopsy, offering transformative opportunities for early detection, personalized treatment, and improved patient management in skin cancers.
4.8. Motor system
Osteosarcoma, the most common primary malignant bone tumor, primarily affects adolescents and young adults. It arises from mesenchymal cells and commonly occurs in the long bones, especially near the metaphysis of the distal femur, proximal tibia, and humerus [168]. Risk factors include genetic mutations (e.g., RB1, TP53), Paget's disease, and radiation exposure. Symptoms, such as localized pain and swelling, often worsen over time. Treatment involves chemotherapy and surgical resection, with limb-sparing surgeries improving quality of life. However, metastasis and recurrence make it a challenging disease [169]. Early diagnosis is difficult, as initial symptoms are often nonspecific and can be mistaken for benign conditions. Current imaging techniques, while useful, cannot distinguish between benign and malignant lesions without biopsies, which are invasive and carry risks. Additionally, there are no established blood-based biomarkers for osteosarcoma, limiting early detection. These diagnostic challenges often lead to advanced-stage diagnoses when metastases are more common, highlighting the need for innovative, non-invasive diagnostic methods to improve early detection and outcomes.
Liang Qiao et al. developed a microfluidic-SERS platform for non-invasive osteosarcoma diagnosis by profiling plasma-derived exosomal biomarkers (CD63, VIM, EpCAM). The system combines tangential flow microfluidics with SERS nanotags, achieving high sensitivity (2 exosomes/μL) and diagnostic accuracy (sensitivity: 100%, specificity: 90%, overall accuracy: 95%). Requiring minimal plasma volume (50 µL) and delivering results in 5 hours, the platform is faster, more sensitive, and cost-effective compared to traditional methods. Its ability to detect multiple biomarkers simultaneously offers significant potential for early osteosarcoma detection and personalized treatment planning [170]. Amos HP Loh et al. identified exosomal mRNA biomarkers (THBS1, MS4A1, TCL1A) from peripheral blood as non-invasive indicators of tumor burden, immune response, and minimal residual disease (MRD) in pediatric osteosarcoma. These biomarkers correlate with disease progression and treatment response, with exosomal mRNA outperforming circulating tumor cells (CTCs) in reliability and RNA integrity for monitoring disease dynamics. This advancement offers potential for real-time, minimally invasive monitoring and personalized treatment strategies [171]. Jinbo Liu et al. identified plasma exosome-derived SENP1 as a highly accurate biomarker for prognosis and monitoring of osteosarcoma. SENP1 levels were elevated in osteosarcoma patients and correlated with clinical features like tumor size, stage, and metastasis. The study showed high prognostic accuracy with AUCs of 0.96 for 3-year DFS and OS. SENP1 levels also decreased following treatment, emphasizing its role in real-time monitoring. These findings highlight the potential of plasma exosome-derived SENP1 for early intervention, personalized treatment, and improved prognostic accuracy in osteosarcoma [172].
Exosome biomarkers like SENP1, miR-335-5p, miR-1246, and CD147 are valuable tools for diagnosing, prognosing, and managing musculoskeletal tumors (Table 9). Found in plasma-derived exosomes, they offer high sensitivity and specificity for tumor detection. SENP1 shows strong prognostic value in osteosarcoma (AUROC: 0.90 for 1-year, 0.96 for 3-year survival). Exosomal biomarkers enable non-invasive early detection, monitoring, and provide insights into tumor biology, such as promoting metastasis and tumor-stroma interaction. They hold promise for personalized oncology, improving diagnostics, prognosis, and therapy.
5. Clinical feasibility and challenges
Exosomes have emerged as promising biomarkers for diagnosis, garnering significant attention in recent years. Considerable progress has been made through ongoing clinical trials. Studies increasingly demonstrate the unique advantages of exosomes in early detection, disease monitoring, and treatment evaluation, positioning them as strong candidates for non-invasive diagnostic tools. Currently, numerous clinical trials are underway to validate the efficacy and reliability of exosome-based diagnostics across various tumors [173]. We have compiled clinical trial data in Table 10, covering biomarkers (protein and RNA, etc.) for detecting various cancers such as gastric, thyroid, colorectal, lung, bladder, kidney, lymphoma, pancreatic, ovarian, sarcoma, osteosarcoma, cholangiocarcinoma, melanoma, and so on. Several of these clinical trials have already shown the effectiveness of sEVs or exosomes as biomarkers.
Exosomal biomarkers in dermal system cancers
| Cancer type | Biomarkers | Source | Analytical Technique | Expression | Effectiveness | Reference |
|---|---|---|---|---|---|---|
| CD63, Cav1 | Serum | ELISA, Immuno-magnetic separation, TEM, SEM, DLS | Upregulated | Sensitivity: 87.5% for early-stage melanoma; Enhanced Cav1/CD9 ratio by 7.7-11.3 times compared to bulk exosomes | [159] | |
| Malignant melanoma | PD1, PD-L1 | Plasma | TEM, DLS, NTA, Flow Cytometry | Upregulated | AUROC: 0.86 (PFS, PD1), 0.975 (OS, PD1); independent predictors for resistance to ICI therapy | [160] |
| miR-1180-3p | Plasma | RNA sequencing, qRT-PCR, AUROC analysis, TEM | Downregulated | AUROC: 0.729 for melanoma detection; Associated with tumor proliferation, migration, and invasion | [161] | |
| Non-melanoma skin cancer | miR-197-5p | Serum | RNA sequencing, qRT-PCR, TEM, NTA | Upregulated | miR-197-5p promotes fibroblast migration and invasion; potential biomarker for MBCC progression and metastasis | [166] |
| circ-CYP24A1 | Plasma | RNA sequencing, qRT-PCR, NTA, TEM, Western blot | Upregulated | circ-CYP24A1 promotes cSCC proliferation, migration, and invasion; potential therapeutic target upregulation | [167] |
Exosomal biomarkers in motor system cancers
| Cancer type | Biomarkers | Source | Analytical Technique | Expression | Effectiveness | Reference |
|---|---|---|---|---|---|---|
| CD63, VIM, EpCAM | Plasma | Microfluidic-SERS, TEM, NTA, Western blot | Upregulated | AUROC: 0.971 (training set); Sensitivity: 100%, Specificity: 90%, Accuracy: 95% | [170] | |
| Osteosarcoma | THBS1, MS4A1, TCL1A | Plasma | NanoString, qRT-PCR, TEM, NTA | Upregulated | THBS1 associated with poor response to chemotherapy; potential biomarkers for minimal residual disease (MRD) | [171] |
| SENP1 (Sentrin SUMO-Specific Protease 1) | Plasma | ELISA, TEM, NTA, Western Blot | Upregulated | AUROC: 0.90 (1-year DFS), 0.96 (3-year DFS); predictive of poor prognosis and survival outcomes | [172] |
Clinical trials of cancer exosome biomarkers
| NCT number | Biomarkers | Origin | Tumor | Number of included subjects | Status | Start time | Investigators or contacts |
|---|---|---|---|---|---|---|---|
| NCT06707961 | PCA3 mRNA | Abdominal cavity drainage fluid | Prostate cancer | 100 | Not yet recruiting | 2024 | Huaqi Zhan et al. |
| NCT05854030 | miRNA | Blood | Lung squamous carcinoma | 60 | Recruiting | 2022 | Richeng Jiang et al. |
| NCT06342427 | miRNA | Blood | Gastric cancer | 809 | Completed | 2023 | Ajay Goel et al. |
| NCT03738319 | miRNA/ lncRNA | Blood | Epithelia ovarian cancer | 160 | Unknown status | 2018 | Lei Li et al. |
| NCT03108677 | RNA | Blood | Osteosarcoma | 90 | Active, not recruiting | 2017 | Yuhui Shen et al. |
| NCT03874559 | Uncertain | Blood | Rectal cancer | 30 | Recruiting | 2018 | Andrew Hoover et al. |
| NCT03102268 | ncRNA | Bile | Cholangiocarcinoma | 80 | Unknown status | 2017 | Lin Miao et al. |
| NCT06342414 | exo-miRNA | Blood | Primary liver cancer | 400 | Recruiting | 2024 | Ajay Goel et al. |
| NCT04530890 | ctDNA | Blood | Breast Cancer | 1000 | Recruiting | 2021 | Camille Evrard et al. |
| NCT06278064 | Protein | Blood | Upper Gastrointestinal Cancer | 562 | Recruiting | 2024 | Li Min et al. |
| NCT04499794 | EML4-ALK | Blood | Non-small cell lung cancer | 75 | Recruiting | 2020 | Yutao Liu et al. |
| NCT02890849 | PD-L1 mRNA | Blood | Non-small cell lung cancer | 60 | Completed | 2016 | Jianguo Sun et al. |
| NCT06654622 | miRNA | Blood | Colorectal cancer | 200 | Recruiting | 2023 | Ajay Goel et al. |
| NCT05270174 | lncRNA-ELNAT1 | Urine | Bladder cancer | 74 | Not yet recruiting | 2023 | Changhao Chen et al. |
| NCT02147418 | Protein | Oropharyngeal rinse | Human Papillomavirus-Positive Oropharyngeal Squamous Cell Carcinoma | 30 | Recruiting | 2015 | Andrew Cowan et al. |
| NCT03800121 | Uncertain | Blood | Sarcoma | 34 | Recruiting | 2018 | Alice HERVIEU et al. |
A study developed a liquid biopsy method using the lncRNA GClnc1 derived from exosomes for early gastric cancer (EGC) diagnosis. Involving 2,141 participants, GClnc1 was significantly upregulated in gastric cancer tissues and circulating exosomes, outperforming traditional biomarkers (CEA, CA72-4, CA19-9) with high sensitivity (>85%) and specificity (>78%) in validation cohorts. It distinguished precancerous lesions from gastric cancer and identified patients negative for traditional biomarkers. Its levels decreased post-surgery, confirming specificity, and it was stable under conditions like room temperature and freeze-thaw cycles, making it a reliable tool for early detection and curative surgery (NCT05397548) [174]. Further advancements in cancer detection include the DESTINEX trial, focused on exosome-based diagnostics for gastric cancer, which was completed (NCT06342427). This study aims to develop a cost-effective blood test for early gastric cancer detection by combining genetic markers, such as cell-free and exosomal microRNAs, to enhance accuracy. This approach could significantly reduce gastric cancer mortality and introduce new screening methods [175]. A study developed the ExoVita Pancreas blood-based EV classifier for early detection of pancreatic ductal adenocarcinoma (PDAC). Analyzing seven protein biomarkers in EVs isolated from plasma, the classifier achieved high sensitivity (93.3%) and specificity (91.0%) in training and 90.0% sensitivity and 92.8% specificity in independent validation. It outperformed traditional markers like CA19-9, demonstrating its potential for early-stage PDAC detection and high-risk population screening. Ongoing investigations include the ExoLuminate Study (NCT0562552) to evaluate this method in high-risk PDAC patients [176]. Lin Miao et al. identified bile-derived exosomal miR-483-5p and miR-126-3p as biomarkers for distinguishing malignant from benign biliary obstructions. RNA sequencing in 82 patients showed significant elevation of both miRNAs in malignant cases. miR-483-5p had an AUC of 0.81 (81.1% sensitivity and specificity), while miR-126-3p had an AUC of 0.74 (73.0% sensitivity, 86.5% specificity), outperforming CA19-9. These findings highlight the potential of miR-483-5p and miR-126-3p as effective, non-invasive diagnostic tools for malignant biliary obstructions (NCT03102268) [177]. The GeparSixto clinical trial demonstrated the potential of exosomal miRNAs, particularly miR-155 and miR-301, as non-invasive biomarkers for tumor diagnostics. Specifically, miR-155 levels were significantly associated with pathological complete response (pCR) in both HER2-positive and triple-negative breast cancer (TNBC) patients, with a p-value of 0.002 in univariate and 0.003 in multivariate models. Similarly, miR-301 showed a strong correlation with pCR, with p-values of 0.002 in univariate and 0.001 in multivariate analyses. The study found that these biomarkers could potentially predict the therapeutic response to neoadjuvant therapy, particularly in the carboplatin treatment arm. However, further studies are required to validate these biomarkers in larger cohorts and to improve exosome isolation methods, as current exosome populations are heterogeneous, containing both cancerous and normal exosomes [178]. One study on exosomal HOTTIP as a potential diagnostic and prognostic biomarker for GC found that exosomal HOTTIP levels were significantly elevated in GC patients compared to healthy controls, with a p-value of <0.001. The diagnostic capability of exosomal HOTTIP was superior to traditional markers such as CEA, CA 19-9, and CA 72-4, with an AUC of 0.827, compared to 0.653, 0.685, and 0.639 for the latter, respectively. Additionally, high levels of exosomal HOTTIP were correlated with worse OS, with Kaplan-Meier analysis indicating a significant association (logrank P < 0.001). Multivariate Cox regression confirmed that exosomal HOTTIP was an independent prognostic factor for poor OS in GC patients (P = 0.027). These findings suggest that exosomal HOTTIP could serve as a valuable non-invasive biomarker for both diagnosing and predicting the prognosis of gastric cancer [179].
Although exosomal biomarkers have shown promising results in clinical trials, several challenges still hinder their further development and application. Firstly, the isolation and purification of exosomes remain challenging, as current methods often lack specificity and scalability, leading to contaminants and inconsistent results [180]. Secondly, the absence of standardized protocols for isolation, quantification, and characterization creates reproducibility issues across studies. Additionally, the heterogeneity of exosomes complicates biomarker identification, while existing detection methods lack the sensitivity required for low-abundance biomarkers [181]. Furthermore, the limited understanding of exosomal biogenesis and cargo selection mechanisms hinders data interpretation [182]. Moreover, scalability issues for clinical applications and regulatory hurdles for biomarker validation pose significant barriers. Finally, biological variability among patients and limited access to large clinical sample cohorts further complicate biomarker discovery and validation. Addressing these challenges requires integrated advancements in technology, biology, and clinical frameworks to fully realize the potential of exosomal biomarkers [183].
6. Future directions and opportunities
The future of exosome-based diagnostics is poised for significant advancement, with several promising directions paving the way for its development. The integration of AI and machine learning (ML) offers the potential to enhance data analysis, enabling greater diagnostic accuracy, predictive modeling, and the ability to identify novel biomarker patterns from large, complex datasets [184]. Exosomal biomarkers hold significant promise in personalized medicine and precision diagnostics, providing opportunities for tailored disease management, early intervention, and improved patient outcomes. Beyond diagnostics, exosome research is expanding into therapeutic applications, including drug delivery systems and targeted gene therapies, further highlighting their clinical versatility. To fully realize their clinical utility, large-scale, multicenter studies are essential for validating these biomarkers across diverse populations, ensuring reproducibility, and establishing standardized protocols for exosome isolation, characterization, and biomarker quantification [185]. Fubo Wang et al. discovered that by analyzing tumor RNA derived from exosomes, a set of blood-based biomarkers for multi-cancer early detection was identified. These biomarkers were validated through a multi-phase, multi-center study and used to develop a machine learning-based diagnostic platform (ETR.sig). This platform efficiently distinguishes cancer from healthy controls, with an AUC of 0.915, and accurately classifies multiple cancer types, with a multi-class model achieving an AUC of 0.983. The study also demonstrated that these exosomal RNA biomarkers are closely associated with tumor progression and prognosis, with higher ETR.sig scores correlating with advanced cancer stages and poorer survival outcomes [186]. Furthermore, leveraging advances in microfluidics and biosensor technology could streamline exosome analysis, making it faster, more cost-effective, and accessible for routine clinical use [187]. The iExoDisc, developed by Xudong Zhao and colleagues, is an automated centrifugal microfluidic platform for efficiently isolating exosomes from blood samples and performing glycan analysis. The benefits of this platform include: completing exosome isolation within 45 minutes, saving significant time compared to traditional methods like ultracentrifugation; improving exosome purity by 3 to 6 times and achieving a recovery rate of 74.7%; additionally, iExoDisc can identify potential diagnostic markers, such as galactosylation and sialylation, from plasma samples of triple-negative breast cancer (TNBC) patients. This technology provides a new solution for early cancer diagnosis and liquid biopsy [188]. Exploring their application in new disease areas, such as cardiovascular, metabolic, autoimmune disorders, and even rare genetic diseases, could vastly expand their impact and provide novel insights into disease pathogenesis [189]. Additionally, future research should focus on exploring multi-biomarker analysis and combined diagnostic strategies to enhance both the sensitivity and specificity of diagnostics. The performance of a single biomarker may be limited, but integrating various exosomal RNAs, proteins, and lipids can provide a more comprehensive view of the multifaceted nature of disease states. Such multi-biomarker combinations are expected to offer more reliable tools for risk stratification and disease classification, ultimately enabling more precise disease management. Moreover, developing multimodal diagnostic approaches that combine exosomal biomarkers with other diagnostic technologies, such as imaging and immunoassays, could further improve diagnostic comprehensiveness and accuracy [190]. Technological advancements will continue to drive the evolution of the exosome field. Emerging nanotechnologies, enhanced isolation techniques, and high-throughput analytical platforms are poised to significantly reduce the time and cost of exosome detection. At the same time, the development of miniaturized, portable diagnostic devices--such as microchips for bedside testing--could bring exosome diagnostics directly into clinical practice, particularly in resource-limited settings. Furthermore, optimizing the engineering of exosomes could lead to the production of specific exosome mimics or "smart exosomes" that more precisely deliver diagnostic and therapeutic information, thereby providing more targeted solutions for future personalized medicine [191]. From a regulatory and policy perspective, the future of exosome diagnostics and therapeutics will benefit from more clearly defined regulatory frameworks and industry guidelines. Establishing well-defined regulations and industry standards can reduce uncertainties in the development process, accelerate the approval of new products, and build trust among the public and medical community. Collaboration between industry and regulatory agencies will be critical for developing unified quality benchmarks, validation standards, and safety evaluation protocols [5, 192]. Finally, patient engagement and education will be crucial to advancing exosome research and clinical applications. Increasing public awareness of exosome technologies and their potential not only enhances patient acceptance of exosome-based diagnostics and therapies, but also encourages greater participation in clinical trials, providing richer datasets for research. Strengthening medical education, including helping clinicians understand how to interpret exosomal biomarkers and integrate them into existing clinical pathways, will also facilitate the widespread adoption and application of this technology in routine clinical practice.
Addressing these opportunities will help overcome current challenges, such as variability in isolation techniques, limited scalability, and a lack of universally accepted biomarkers. By resolving these hurdles and fostering interdisciplinary collaboration, the transformative potential of exosomal biomarkers in advancing diagnostics, therapeutic monitoring, and patient care can be fully unlocked, marking a new era in precision medicine.
7. Conclusion
Exosomal biomarkers have the potential to fundamentally transform the landscape of non-invasive cancer diagnostics, offering a highly promising alternative to traditional invasive methods. These nanoscale vesicles, carrying a rich cargo of proteins, lipids, RNA, and microRNAs, provide a snapshot of the molecular state of their parent cells, reflecting critical changes that occur during disease progression. As such, exosomes represent an unparalleled tool for detecting cancer at early stages, monitoring tumor dynamics, and assessing treatment responses in real time. One of the most compelling advantages of exosome-based diagnostics is their ability to be extracted from easily accessible biofluids such as blood, saliva, and urine, positioning them as ideal candidates for liquid biopsy technologies. The ability to perform repeated, non-invasive sampling makes exosomes particularly attractive for continuous monitoring of tumor progression and therapeutic efficacy, significantly improving patient outcomes. Recent advancements in exosome isolation and characterization techniques have substantially enhanced their sensitivity and specificity, allowing for the detection of tumor-specific biomarkers even at low concentrations in biofluids. Clinical trials incorporating exosome-based diagnostics have demonstrated their utility across various cancer types, from respiratory and digestive system cancers to hematological and neurological malignancies. These trials have showcased exosomes as reliable biomarkers for early cancer detection, offering diagnostic accuracy on par with or exceeding traditional methods such as tissue biopsies and imaging. Additionally, exosome-based diagnostics offer significant advantages in terms of their ability to capture tumor heterogeneity, providing a comprehensive picture of disease status, which is often challenging to achieve through conventional approaches. Despite these exciting developments, several challenges remain in fully realizing the potential of exosome-based diagnostics. The biological heterogeneity of exosomes, both within and between different diseases, poses a significant obstacle in identifying universal biomarkers with sufficient specificity. Furthermore, the deficiency of standardized guidelines for exosome isolation, characterization, and analysis continues to hinder the widespread implementation of these technologies in clinical settings. Regulatory hurdles, such as the need for clinical validation and approval by regulatory bodies, also represent significant barriers to the clinical adoption of exosome-based diagnostics. Nevertheless, the future of exosome-based diagnostics remains incredibly promising. With continued advancements in exosome isolation methods, the development of more refined and standardized analytical techniques, and the successful completion of large-scale clinical trials, exosomes are poised to revolutionize the field of personalized medicine. Their ability to provide dynamic, real-time insights into tumor behavior and response to treatment could significantly enhance the precision and effectiveness of cancer therapy. As exosome-based technologies mature, they are expected to play a central role in early cancer detection, improving patient prognosis through earlier interventions, better monitoring of disease progression, and more tailored therapeutic strategies.
Ultimately, integrating exosomal biomarkers into routine clinical practice will offer a paradigm shift in cancer diagnostics and management. It will empower clinicians with powerful tools for personalized treatment, enable more effective and less invasive monitoring, and enhance patient outcomes by allowing for timely intervention. With the field undergoing further evolution, exosomes are positioned to play a transformative role in cancer care, providing promise for enhanced accuracy, cost-effective, and accessible diagnostic solutions.
Abbreviations
MVBs: multivesicular bodies;
ILVs: intraluminal vesicles;
DC: dendritic cell;
NK: natural killer;
TDEs: tumor-derived exosomes;
MSC: mesenchymal stem cell;
Tregs: regulatory T cells;
MDSCs: myeloid-derived suppressor cells;
LOX: lysyl oxidase;
EMT: epithelial-mesenchymal transition;
ABC: ATP-binding cassette;
BBB: blood-brain barrier;
NSCLC: non-small cell lung cancer;
SCLC: small cell lung cancer;
ICDSE: Integrated Concentration and Determination System of Exosomes;
PFS: progression-free survival;
OS: overall survival;
sEVs: small extracellular vesicles;
TAAs: tumor-associated antigens;
SERS: Surface-enhanced Raman Spectroscopy;
AI: Artificial Intelligence;
NPC: nasopharyngeal carcinoma;
EBV: Epstein-Barr virus;
LSCC: laryngeal squamous cell carcinoma;
AUC: Area Under the Curve;
GC: gastric cancer;
HCC: hepatocellular carcinoma;
PDAC: pancreatic ductal adenocarcinoma;
CRC: colorectal cancer;
ALL: acute lymphoblastic leukemia;
AML: acute myeloid leukemia;
CLL: chronic lymphocytic leukemia;
CML: chronic myeloid leukemia;
MM: multiple myeloma;
HL: Hodgkin lymphoma;
NHL: non-Hodgkin lymphoma;
PAs: pituitary adenomas;
PCNSL: primary central nervous system lymphoma;
PTC: papillary thyroid cancer;
MTC: medullary thyroid cancer;
FTC: follicular thyroid cancer;
ATC: anaplastic thyroid cancer;
PNETs: pancreatic neuroendocrine tumors;
BC: breast cancer;
OC: ovarian cancer;
RCC: renal cell carcinoma;
MM: malignant melanoma;
NMSC: non-melanoma skin cancer;
RMS: rhabdomyosarcoma.
Acknowledgements
We express our sincere thanks to all authors of this study. This work was supported by National Natural Science Foundation of China (grant/award number: 82460479, 82160492); Supported by the specific research fund of The Innovation Platform for Academicians of Hainan Province (YSPTZX202033); Hainan Province Clinical Medical Research Center (grant no. LCYX202410); Hainan Province Clinical Medical Center (grant no. 0202068); Natural Science Foundation of Hunan Province (No. 2024JJ5363, 2024JJ9290); Changsha Natural Science Foundation (grant no. kq2403145).
Authors contributions
Jiale Li and Zigui Chen wrote the manuscript.
Jiale Li, Zigui Chen, and Jiachong Wang designed the study and provided funding support.
Jiale Li drew the figures and tables.
Ailin Wang, Haijun Guo, Wei Zheng, Rui Chen, Changfeng Miao, Dandan Zheng, Jun Peng, Jiachong Wang, and Zigui Chen revised the manuscript. All of the authors have read the manuscript and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Nail HM, Chiu CC, Leung CH, Ahmed MMM, Wang HD. Exosomal miRNA-mediated intercellular communications and immunomodulatory effects in tumor microenvironments. J Biomed Sci. 2023;30:69
2. Araujo-Abad S, Berna JM, Lloret-Lopez E, López-Cortés A, Saceda M, de Juan Romero C. Exosomes: from basic research to clinical diagnostic and therapeutic applications in cancer. Cell Oncol (Dordr). 2024
3. Ilié M, Hofman P. Pros: Can tissue biopsy be replaced by liquid biopsy? Transl Lung Cancer Res. 2016;5:420-3
4. Tang H, Yu D, Zhang J, Wang M, Fu M, Qian Y. et al. The new advance of exosome-based liquid biopsy for cancer diagnosis. J Nanobiotechnology. 2024;22:610
5. Lai JJ, Chau ZL, Chen SY, Hill JJ, Korpany KV, Liang NW. et al. Exosome Processing and Characterization Approaches for Research and Technology Development. Adv Sci (Weinh). 2022;9:e2103222
6. Tao H, Gao B. Exosomes for neurodegenerative diseases: diagnosis and targeted therapy. J Neurol. 2024;271:3050-62
7. Lin X, Zhu J, Shen J, Zhang Y, Zhu J. Advances in exosome plasmonic sensing: Device integration strategies and AI-aided diagnosis. Biosens Bioelectron. 2024;266:116718
8. Han QF, Li WJ, Hu KS, Gao J, Zhai WL, Yang JH, Zhang SJ. Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer. Mol Cancer. 2022;21:207
9. Krylova SV, Feng D. The Machinery of Exosomes: Biogenesis, Release, and Uptake. Int J Mol Sci. 2023;24:1337
10. Arya SB, Collie SP, Parent CA. The ins-and-outs of exosome biogenesis, secretion, and internalization. Trends Cell Biol. 2024;34:90-108
11. Lee YJ, Shin KJ, Chae YC. Regulation of cargo selection in exosome biogenesis and its biomedical applications in cancer. Exp Mol Med. 2024;56:877-89
12. McAndrews KM, Kalluri R. Mechanisms associated with biogenesis of exosomes in cancer. Mol Cancer. 2019;18:52
13. Li MY, Liu LZ, Dong M. Progress on pivotal role and application of exosome in lung cancer carcinogenesis, diagnosis, therapy and prognosis. Mol Cancer. 2021;20:22
14. Paskeh MDA, Entezari M, Mirzaei S, Zabolian A, Saleki H, Naghdi MJ. et al. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J Hematol Oncol. 2022;15:83
15. Anderson NM, Simon MC. The tumor microenvironment. Curr Biol. 2020;30:R921-r5
16. Maia J, Caja S, Strano Moraes MC, Couto N, Costa-Silva B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front Cell Dev Biol. 2018;6:18
17. Huang Y, Kanada M, Ye J, Deng Y, He Q, Lei Z. et al. Exosome-mediated remodeling of the tumor microenvironment: From local to distant intercellular communication. Cancer Lett. 2022;543:215796
18. Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science. 2020;368:eaaw5473
19. Tan S, Yang Y, Yang W, Han Y, Huang L, Yang R. et al. Exosomal cargos-mediated metabolic reprogramming in tumor microenvironment. J Exp Clin Cancer Res. 2023;42:59
20. Liang L, Li W, Li X, Jin X, Liao Q, Li Y, Zhou Y. 'Reverse Warburg effect' of cancer-associated fibroblasts (Review). Int J Oncol. 2022;60:67
21. Bajaj N, Sharma D. Uncovering metabolic signatures in cancer-derived exosomes: LC-MS/MS and NMR profiling. Nanoscale. 2025;17:287-303
22. Galassi C, Chan TA, Vitale I, Galluzzi L. The hallmarks of cancer immune evasion. Cancer Cell. 2024;42:1825-63
23. Zhang H, Li S, Wang D, Liu S, Xiao T, Gu W. et al. Metabolic reprogramming and immune evasion: the interplay in the tumor microenvironment. Biomark Res. 2024;12:96
24. Sun C, Mezzadra R, Schumacher TN. Regulation and Function of the PD-L1 Checkpoint. Immunity. 2018;48:434-52
25. Zhang H, Dai Z, Wu W, Wang Z, Zhang N, Zhang L. et al. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J Exp Clin Cancer Res. 2021;40:184
26. Hosseini R, Hosseinzadeh N, Asef-Kabiri L, Akbari A, Ghezelbash B, Sarvnaz H, Akbari ME. Small extracellular vesicle TGF-β in cancer progression and immune evasion. Cancer Gene Ther. 2023;30:1309-22
27. Arner EN, Rathmell JC. Metabolic programming and immune suppression in the tumor microenvironment. Cancer cell. 2023;41:421-33
28. Sulekha Suresh D, Guruvayoorappan C. Molecular principles of tissue invasion and metastasis. Am J Physiol Cell Physiol. 2023;324:C971-c91
29. Quesnel A, Broughton A, Karagiannis GS, Filippou PS. Message in the bottle: regulation of the tumor microenvironment via exosome-driven proteolysis. Cancer Metastasis Rev. 2022;41:789-801
30. Li C, Teixeira AF, Zhu HJ, Ten Dijke P. Cancer associated-fibroblast-derived exosomes in cancer progression. Mol Cancer. 2021;20:154
31. Yang X, Zhang Y, Zhang Y, Zhang S, Qiu L, Zhuang Z. et al. The Key Role of Exosomes on the Pre-metastatic Niche Formation in Tumors. Front Mol Biosci. 2021;8:703640
32. Wang Y, Jia J, Wang F, Fang Y, Yang Y, Zhou Q. et al. Pre-metastatic niche: formation, characteristics and therapeutic implication. Signal Transduct Target Ther. 2024;9:236
33. Kugeratski FG, Kalluri R. Exosomes as mediators of immune regulation and immunotherapy in cancer. FEBS J. 2021;288:10-35
34. Wang X, Zhang H, Chen X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019;2:141-60
35. Dong X, Bai X, Ni J, Zhang H, Duan W, Graham P, Li Y. Exosomes and breast cancer drug resistance. Cell Death Dis. 2020;11:987
36. Cunha A, Silva PM, Sarmento B, Queirós O. Targeting glucose metabolism in cancer cells as an approach to overcoming drug resistance. Pharmaceutics. 2023;15:2610
37. Yang Q, Xu J, Gu J, Shi H, Zhang J, Zhang J. et al. Extracellular Vesicles in Cancer Drug Resistance: Roles, Mechanisms, and Implications. Adv Sci (Weinh). 2022;9:e2201609
38. Easwaran VB, Pai KMS, Pai KSR. Mesenchymal Stem Cell-Derived Exosomes in Cancer Resistance Against Therapeutics. Cancers (Basel). 2025;17:831
39. Akhtarkhavari T, Bahrami AR, Matin MM. Downregulation of miR-21 as a promising strategy to overcome drug resistance in cancer. Eur J Pharmacol. 2022;932:175233
40. Xie F, Xu M, Lu J, Mao L, Wang S. The role of exosomal PD-L1 in tumor progression and immunotherapy. Mol Cancer. 2019;18:146
41. Polónia B, Xavier CPR, Kopecka J, Riganti C, Vasconcelos MH. The role of Extracellular Vesicles in glycolytic and lipid metabolic reprogramming of cancer cells: Consequences for drug resistance. Cytokine Growth Factor Rev. 2023;73:150-62
42. Crosby D, Bhatia S, Brindle KM, Coussens LM, Dive C, Emberton M. et al. Early detection of cancer. Science. 2022;375:eaay9040
43. Tapper W, Carneiro G, Mikropoulos C, Thomas SA, Evans PM, Boussios S. The Application of Radiomics and AI to Molecular Imaging for Prostate Cancer. J Pers Med. 2024;14:287
44. Vanderpoel J, Stevens AL, Emond B, Lafeuille MH, Hilts A, Lefebvre P, Morrison L. Total cost of testing for genomic alterations associated with next-generation sequencing versus polymerase chain reaction testing strategies among patients with metastatic non-small cell lung cancer. J Med Econ. 2022;25:457-68
45. Preethi KA, Selvakumar SC, Ross K, Jayaraman S, Tusubira D, Sekar D. Liquid biopsy: Exosomal microRNAs as novel diagnostic and prognostic biomarkers in cancer. Mol Cancer. 2022;21:54
46. Yu D, Li Y, Wang M, Gu J, Xu W, Cai H. et al. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer. 2022;21:56
47. Zhang Y, Chen L, Ye X, Wu Z, Zhang Z, Sun B. et al. Expression and mechanism of exosome-mediated A FOXM1 related long noncoding RNA in gastric cancer. J Nanobiotechnology. 2021;19:133
48. Batista IA, Machado JC, Melo SA. Advances in exosomes utilization for clinical applications in cancer. Trends Cancer. 2024;10:947-68
49. Li J, Wang J, Chen Z. Emerging role of exosomes in cancer therapy: progress and challenges. Mol Cancer. 2025;24:13
50. Xiong H, Huang Z, Yang Z, Lin Q, Yang B, Fang X. et al. Recent Progress in Detection and Profiling of Cancer Cell-Derived Exosomes. Small. 2021;17:e2007971
51. Peng X-X, Qin X, Qin Y, Xiang Y, Zhang G-J, Yang F. Bioprobes-regulated precision biosensing of exosomes: From the nanovesicle surface to the inside. Coord Chem Rev. 2022;463:214538
52. Wang Z, Wang Q, Qin F, Chen J. Exosomes: a promising avenue for cancer diagnosis beyond treatment. Front Cell Dev Biol. 2024;12:1344705
53. Yasamineh S, Nikben N, Hamed Ahmed M, Abdul Kareem R, Kadhim Al-Aridhy A, Hosseini Hooshiar M. Increasing the sensitivity and accuracy of detecting exosomes as biomarkers for cancer monitoring using optical nanobiosensors. Cancer Cell Int. 2024;24:189
54. Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398:535-54
55. Leiter A, Veluswamy RR, Wisnivesky JP. The global burden of lung cancer: current status and future trends. Nat Rev Clin Oncol. 2023;20:624-39
56. Li Y, Yan B, He S. Advances and challenges in the treatment of lung cancer. Biomed Pharmacother. 2023;169:115891
57. Dantas E, Murthy A, Ahmed T, Ahmed M, Ramsamooj S, Hurd MA. et al. TIMP1 is an early biomarker for detection and prognosis of lung cancer. Clin Transl Med. 2023;13:e1391
58. Chen P, Liu Y, Wen Y, Zhou C. Non-small cell lung cancer in China. Cancer Commun (Lond). 2022;42:937-70
59. Fan Y, Wu W, Xie N, Huang Y, Wu H, Zhang J. et al. Biocompatible engineered erythrocytes as plasmonic sensor initiators for high-sensitive screening of non-small cell lung cancer-derived exosomal miRNA in an integrated system. Biosens Bioelectron. 2023;219:114802
60. Redin E, Garrido-Martin EM, Valencia K, Redrado M, Solorzano JL, Carias R. et al. YES1 Is a Druggable Oncogenic Target in SCLC. J Thorac Oncol. 2022;17:1387-403
61. Hua Y, Dai C, He Q, Cai X, Li M. Autoantibody panel on small extracellular vesicles for the early detection of lung cancer. Clin Immunol. 2022;245:109175
62. Shin H, Choi BH, Shim O, Kim J, Park Y, Cho SK. et al. Single test-based diagnosis of multiple cancer types using Exosome-SERS-AI for early stage cancers. Nat Commun. 2023;14:1644
63. Wong KCW, Hui EP, Lo KW, Lam WKJ, Johnson D, Li L. et al. Nasopharyngeal carcinoma: an evolving paradigm. Nat Rev Clin Oncol. 2021;18:679-95
64. Wang Z, Fang M, Zhang J, Tang L, Zhong L, Li H. et al. Radiomics and Deep Learning in Nasopharyngeal Carcinoma: A Review. IEEE Rev Biomed Eng. 2024;17:118-35
65. Liu W, Chen G, Gong X, Wang Y, Zheng Y, Liao X. et al. The diagnostic value of EBV-DNA and EBV-related antibodies detection for nasopharyngeal carcinoma: a meta-analysis. Cancer Cell Int. 2021;21:164
66. Zheng W, Ye W, Wu Z, Huang X, Xu Y, Chen Q. et al. Identification of potential plasma biomarkers in early-stage nasopharyngeal carcinoma-derived exosomes based on RNA sequencing. Cancer Cell Int. 2021;21:185
67. Ye SB, Zhang H, Cai TT, Liu YN, Ni JJ, He J. et al. Exosomal miR-24-3p impedes T-cell function by targeting FGF11 and serves as a potential prognostic biomarker for nasopharyngeal carcinoma. J Pathol. 2016;240:329-40
68. Luo Y, Ma J, Liu F, Guo J, Gui R. Diagnostic value of exosomal circMYC in radioresistant nasopharyngeal carcinoma. Head Neck. 2020;42:3702-11
69. Ajani JA, D'Amico TA, Bentrem DJ, Chao J, Cooke D, Corvera C. et al. Gastric Cancer, Version 2.2022, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2022;20:167-92
70. Conti CB, Agnesi S, Scaravaglio M, Masseria P, Dinelli ME, Oldani M, Uggeri F. Early Gastric Cancer: Update on Prevention, Diagnosis and Treatment. Int J Environ Res Public Health. 2023;20:2149
71. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977
72. Guo Y, Nie Y, Wang P, Li Z, Ma Q. MoS(2) QDs-MXene heterostructure-based ECL sensor for the detection of miRNA-135b in gastric cancer exosomes. Talanta. 2023;259:124559
73. Liang Z, Wang P, Li Z, Wang D, Ma Q. Magnetoplasmonic Metasurface-Modulated Electrochemiluminescence Strategy for Extracellular Vesicle Detection. Anal Chem. 2024;96:4909-17
74. Huang R, Di K, Adeel K, Fan B, Gu X, Xu H. et al. The exploration of droplet digital branched rolling circle amplification based ultrasensitive biosensor for gastric cancer cell-derived extracellular vesicles detection. Mater Today Adv. 2022;16:100296
75. Tayob N, Kanwal F, Alsarraj A, Hernaez R, El-Serag HB. The Performance of AFP, AFP-3, DCP as Biomarkers for Detection of Hepatocellular Carcinoma (HCC): A Phase 3 Biomarker Study in the United States. Clin Gastroenterol Hepatol. 2023;21:415-23.e4
76. Attia AM, Rezaee-Zavareh MS, Hwang SY, Kim N, Adetyan H, Yalda T. et al. Novel Biomarkers for Early Detection of Hepatocellular Carcinoma. Diagnostics (Basel). 2024;14:2278
77. Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y. et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer. 2023;22:55
78. Rui T, Wang K, Xiang A, Guo J, Tang N, Jin X. et al. Serum Exosome-Derived piRNAs Could Be Promising Biomarkers for HCC Diagnosis. Int J Nanomedicine. 2023;18:1989-2001
79. Yao J, Hua X, Shi J, Hu X, Lui K, He K. et al. LncRNA THEMIS2-211, a tumor-originated circulating exosomal biomarker, promotes the growth and metastasis of hepatocellular carcinoma by functioning as a competing endogenous RNA. FASEB J. 2022;36:e22238
80. Yi K, Wang Y, Rong Y, Bao Y, Liang Y, Chen Y. et al. Transcriptomic Signature of 3D Hierarchical Porous Chip Enriched Exosomes for Early Detection and Progression Monitoring of Hepatocellular Carcinoma. Adv Sci (Weinh). 2024;11:e2305204
81. Huang J, Lok V, Ngai CH, Zhang L, Yuan J, Lao XQ. et al. Worldwide Burden of, Risk Factors for, and Trends in Pancreatic Cancer. Gastroenterology. 2021;160:744-54
82. Zhang L, Sanagapalli S, Stoita A. Challenges in diagnosis of pancreatic cancer. World J Gastroenterol. 2018;24:2047-60
83. Yu Z, Yang Y, Fang W, Hu P, Liu Y, Shi J. Dual Tumor Exosome Biomarker Co-recognitions Based Nanoliquid Biopsy for the Accurate Early Diagnosis of Pancreatic Cancer. ACS Nano. 2023;17:11384-95
84. Feng J, Shu Y, An Y, Niu Q, Fan Q, Lei Y. et al. Encoded Fusion-Mediated MicroRNA Signature Profiling of Tumor-Derived Extracellular Vesicles for Pancreatic Cancer Diagnosis. Anal Chem. 2023;95:7743-52
85. Li H, Chiang CL, Kwak KJ, Wang X, Doddi S, Ramanathan LV. et al. Extracellular Vesicular Analysis of Glypican 1 mRNA and Protein for Pancreatic Cancer Diagnosis and Prognosis. Adv Sci (Weinh). 2024;11:e2306373
86. Sullivan BA, Noujaim M, Roper J. Cause, Epidemiology, and Histology of Polyps and Pathways to Colorectal Cancer. Gastrointest Endosc Clin N Am. 2022;32:177-94
87. Osseis M, Nehmeh WA, Rassy N, Derienne J, Noun R, Salloum C. et al. Surgery for T4 Colorectal Cancer in Older Patients: Determinants of Outcomes. J Pers Med. 2022;12:1534
88. Fasulo E, D'Amico F, Zilli A, Furfaro F, Cicerone C, Parigi TL. et al. Advancing Colorectal Cancer Prevention in Inflammatory Bowel Disease (IBD): Challenges and Innovations in Endoscopic Surveillance. Cancers (Basel). 2025;17:60
89. Kamel F, Eltarhoni K, Nisar P, Soloviev M. Colorectal Cancer Diagnosis: The Obstacles We Face in Determining a Non-Invasive Test and Current Advances in Biomarker Detection. Cancers (Basel). 2022;14:1889
90. Titu S, Gata VA, Decea RM, Mocan T, Dina C, Irimie A, Lisencu CI. Exosomes in Colorectal Cancer: From Physiology to Clinical Applications. Int J Mol Sci. 2023;24:4382
91. Zheng R, Zhang K, Tan S, Gao F, Zhang Y, Xu W. et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol Cancer. 2022;21:49
92. Zhang J, Gao Z, Xiao W, Jin N, Zeng J, Wang F. et al. A simplified and efficient extracellular vesicle-based proteomics strategy for early diagnosis of colorectal cancer. Chem Sci. 2024;15:18419-30
93. Min L, Bu F, Meng J, Liu X, Guo Q, Zhao L. et al. Circulating small extracellular vesicle RNA profiling for the detection of T1a stage colorectal cancer and precancerous advanced adenoma. Elife. 2024;12:RP88675
94. Whiteley AE, Price TT, Cantelli G, Sipkins DA. Leukaemia: a model metastatic disease. Nat Rev Cancer. 2021;21:461-75
95. Percival ME, Lai C, Estey E, Hourigan CS. Bone marrow evaluation for diagnosis and monitoring of acute myeloid leukemia. Blood Rev. 2017;31:185-92
96. Shadman M. Diagnosis and Treatment of Chronic Lymphocytic Leukemia: A Review. JAMA. 2023;329:918-32
97. Li M, Sun G, Zhao J, Pu S, Lv Y, Wang Y. et al. Small extracellular vesicles derived from acute myeloid leukemia cells promote leukemogenesis by transferring miR-221-3p. Haematologica. 2024;109:3209-21
98. Li C, Zhou M, Wang H, Wang J, Huang L. Rolling circle amplification assisted dual signal amplification colorimetric biosensor for ultrasensitive detection of leukemia-derived exosomes. Talanta. 2022;245:123444
99. Xiao Q, Lin C, Peng M, Ren J, Jing Y, Lei L. et al. Circulating plasma exosomal long non-coding RNAs LINC00265, LINC00467, UCA1, and SNHG1 as biomarkers for diagnosis and treatment monitoring of acute myeloid leukemia. Front Oncol. 2022;12:1033143
100. Rajkumar SV. Multiple myeloma: 2024 update on diagnosis, risk-stratification, and management. Am J Hematol. 2024;99:1802-24
101. Miaomiao S, Xiaoqian W, Yuwei S, Chao C, Chenbo Y, Yinghao L. et al. Cancer-associated fibroblast-derived exosome microRNA-21 promotes angiogenesis in multiple myeloma. Sci Rep. 2023;13:9671
102. Yu M, Yu J, Zhang Y, Sun X, Sun R, Xia M. et al. A novel circRNA-miRNA-mRNA network revealed exosomal circ-ATP10A as a biomarker for multiple myeloma angiogenesis. Bioengineered. 2022;13:667-83
103. Gao M, Dong H, Jiang S, Chen F, Fu Y, Luo Y. Activated platelet-derived exosomal LRG1 promotes multiple myeloma cell growth. Oncogenesis. 2024;13:21
104. Armitage JO, Gascoyne RD, Lunning MA, Cavalli F. Non-Hodgkin lymphoma. Lancet. 2017;390:298-310
105. Chiavazza C, Pellerino A, Ferrio F, Cistaro A, Soffietti R, Rudà R. Primary CNS Lymphomas: Challenges in Diagnosis and Monitoring. Biomed Res Int. 2018;2018:3606970
106. Lewis WD, Lilly S, Jones KL. Lymphoma: Diagnosis and Treatment. Am Fam Physician. 2020;101:34-41
107. Ryu KJ, Lee JY, Choi ME, Yoon SE, Cho J, Ko YH. et al. Serum-Derived Exosomal MicroRNA Profiles Can Predict Poor Survival Outcomes in Patients with Extranodal Natural Killer/T-Cell Lymphoma. Cancers (Basel). 2020;12:3548
108. Liu J, Han Y, Hu S, Cai Y, Yang J, Ren S. et al. Circulating Exosomal MiR-107 Restrains Tumorigenesis in Diffuse Large B-Cell Lymphoma by Targeting 14-3-3η. Front Cell Dev Biol. 2021;9:667800
109. Weller M, Wen PY, Chang SM, Dirven L, Lim M, Monje M, Reifenberger G. Glioma. Nat Rev Dis Primers. 2024;10:33
110. Regmi M, Wang Y, Liu W, Dai Y, Liu S, Ma K. et al. From glioma gloom to immune bloom: unveiling novel immunotherapeutic paradigms-a review. J Exp Clin Cancer Res. 2024;43:47
111. Pokorná M, Černá M, Boussios S, Ovsepian SV, O'Leary VB. lncRNA Biomarkers of Glioblastoma Multiforme. Biomedicines. 2024;12:932
112. Xu C, Hou P, Li X, Xiao M, Zhang Z, Li Z. et al. Comprehensive understanding of glioblastoma molecular phenotypes: classification, characteristics, and transition. Cancer Biol Med. 2024;21:363-81
113. Sun J, Sun Z, Gareev I, Yan T, Chen X, Ahmad A. et al. Exosomal miR-2276-5p in Plasma Is a Potential Diagnostic and Prognostic Biomarker in Glioma. Front Cell Dev Biol. 2021;9:671202
114. Zhang Y, Wong CY, Lim CZJ, Chen Q, Yu Z, Natalia A. et al. Multiplexed RNA profiling by regenerative catalysis enables blood-based subtyping of brain tumors. Nat Commun. 2023;14:4278
115. Yang X, Zhang Z, Wu Y, Wang H, Yun Y, Sun Y. et al. Printed Divisional Optical Biochip for Multiplex Visualizable Exosome Analysis at Point-of-Care. Adv Mater. 2024;36:e2304935
116. Shi L, Cao Y, Yuan W, Guo J, Sun G. Exosomal circRNA BTG2 derived from RBP-J overexpressed-macrophages inhibits glioma progression via miR-25-3p/PTEN. Cell Death Dis. 2022;13:506
117. Singh AR, Kumar P. Comparative study on clinicopathological characteristics of functional and non-functional subtypes in pituitary adenomas. Bull Natl Res Cent. 2024;48:64
118. Tritos NA, Miller KK. Diagnosis and Management of Pituitary Adenomas: A Review. JAMA. 2023;329:1386-98
119. Lyu L, Li H, Chen C, Yu Y, Wang L, Yin S. et al. Exosomal miRNA Profiling is a Potential Screening Route for Non-Functional Pituitary Adenoma. Front Cell Dev Biol. 2021;9:771354
120. Yue X, Lan F, Liu W. Serum exosomal circCCDC66 as a potential diagnostic and prognostic biomarker for pituitary adenomas. Front Oncol. 2023;13:1268778
121. Chen K, Li G, Kang X, Liu P, Qian L, Shi Y. et al. EMT-Related Markers in Serum Exosomes are Potential Diagnostic Biomarkers for Invasive Pituitary Adenomas. Neuropsychiatr Dis Treat. 2021;17:3769-80
122. Nabhan F, Dedhia PH, Ringel MD. Thyroid cancer, recent advances in diagnosis and therapy. Int J Cancer. 2021;149:984-92
123. Drake T, Caicedo-Granados E. Most Thyroid Surgeries Start With an Asymptomatic Patient. JAMA Otolaryngol Head Neck Surg. 2022;148:818-9
124. Ghai S, Goldstein DP, Sawka AM. Ultrasound Imaging in Active Surveillance of Small, Low-Risk Papillary Thyroid Cancer. Korean J Radiol. 2024;25:749-55
125. Houdek D, Cooke-Hubley S, Puttagunta L, Morrish D. Factors affecting thyroid nodule fine needle aspiration non-diagnostic rates: a retrospective association study of 1975 thyroid biopsies. Thyroid Res. 2021;14:2
126. Livhits MJ, Zhu CY, Kuo EJ, Nguyen DT, Kim J, Tseng CH. et al. Effectiveness of Molecular Testing Techniques for Diagnosis of Indeterminate Thyroid Nodules: A Randomized Clinical Trial. JAMA Oncol. 2021;7:70-7
127. Dai D, Tan Y, Guo L, Tang A, Zhao Y. Identification of exosomal miRNA biomarkers for diagnosis of papillary thyroid cancer by small RNA sequencing. Eur J Endocrinol. 2020;182:111-21
128. Wang C-Y, Shih S-R, Chen K-Y, Huang P-J. Urinary Exosomal Tissue TIMP and Angiopoietin-1 Are Preoperative Novel Biomarkers of Well-Differentiated Thyroid Cancer. Biomedicines. 2023;11:24
129. Maggisano V, Capriglione F, Verrienti A, Celano M, Gagliardi A, Bulotta S. et al. Identification of Exosomal microRNAs and Their Targets in Papillary Thyroid Cancer Cells. Biomedicines. 2022;10:961
130. Yao JC, Shah MH, Ito T, Bohas CL, Wolin EM, Van Cutsem E. et al. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med. 2011;364:514-23
131. Falconi M, Bartsch DK, Eriksson B, Klöppel G, Lopes JM, O'Connor JM. et al. ENETS Consensus Guidelines for the management of patients with digestive neuroendocrine neoplasms of the digestive system: well-differentiated pancreatic non-functioning tumors. Neuroendocrinology. 2012;95:120-34
132. Chang A, Sherman SK, Howe JR, Sahai V. Progress in the Management of Pancreatic Neuroendocrine Tumors. Annu Rev Med. 2022;73:213-29
133. Konukiewitz B, Jesinghaus M, Kasajima A, Klöppel G. Neuroendocrine neoplasms of the pancreas: diagnosis and pitfalls. Virchows Archiv. 2022;480:247-57
134. Lu F, Ye M, Shen Y, Xu Y, Hu C, Chen J. et al. Hypoxic tumor-derived exosomal miR-4488 induces macrophage M2 polarization to promote liver metastasis of pancreatic neuroendocrine neoplasm through RTN3/FABP5 mediated fatty acid oxidation. Int J Biol Sci. 2024;20:3201-18
135. Ye M, Lu F, Gu D, Xue B, Xu L, Hu C. et al. Hypoxia exosome derived CEACAM5 promotes tumor-associated macrophages M2 polarization to accelerate pancreatic neuroendocrine tumors metastasis via MMP9. FASEB J. 2024;38:e23762
136. Smolarz B, Nowak AZ, Romanowicz H. Breast Cancer-Epidemiology, Classification, Pathogenesis and Treatment (Review of Literature). Cancers. 2022;14:2569
137. Hong R, Xu B. Breast cancer: an up-to-date review and future perspectives. Cancer Commun (Lond). 2022;42:913-36
138. Bhushan A, Gonsalves A, Menon JU. Current State of Breast Cancer Diagnosis, Treatment, and Theranostics. Pharmaceutics. 2021;13:723
139. Hu R, Lin S, Li F, Shen J, Xie Y, Deng B. et al. Exploring aptamer-based metasurfaces for label-free plasmonic biosensing of breast tumor-derived exosomes. Adv Opt Mater. 2024;12:2401180
140. Guan X, Zhao J, Sha Z, Liang Y, Huang J, Zhang J, Sun S. CRISPR/Cas12a and aptamer-chemiluminescence based analysis for the relative abundance determination of tumor-related protein positive exosomes for breast cancer diagnosis. Biosens Bioelectron. 2024;259:116380
141. Cao Y, Yu X, Zeng T, Fu Z, Zhao Y, Nie B. et al. Molecular Characterization of Exosomes for Subtype-Based Diagnosis of Breast Cancer. J Am Chem Soc. 2022;144:13475-86
142. Matulonis UA, Sood AK, Fallowfield L, Howitt BE, Sehouli J, Karlan BY. Ovarian cancer. Nat Rev Dis Primers. 2016;2:16061
143. Charkhchi P, Cybulski C, Gronwald J, Wong FO, Narod SA, Akbari MR. CA125 and Ovarian Cancer: A Comprehensive Review. Cancers (Basel). 2020;12:3730
144. Ghose A, McCann L, Makker S, Mukherjee U, Gullapalli SVN, Erekkath J. et al. Diagnostic biomarkers in ovarian cancer: advances beyond CA125 and HE4. Ther Adv Med Oncol. 2024;16:17588359241233225
145. Liberto JM, Chen S-Y, Shih I-M, Wang T-H, Wang T-L, Pisanic TR. Current and Emerging Methods for Ovarian Cancer Screening and Diagnostics: A Comprehensive Review. Cancers (Basel). 2022;14:2885
146. Chen X, Tang J, Zhao Y, Wang R, Sang S, Yu F, Xing Y. Sensitive phenotyping of serum extracellular vesicles on a SERS-microfluidic platform for early-stage clinical diagnosis of ovarian carcinoma. Biosens Bioelectron. 2025;267:116724
147. Deng Y, Zhou T, Hu K, Peng Y, Jia X, Yang J, Li G. An electrochemical biosensor designed with entropy-driven autocatalytic DNA circuits for sensitive detection of ovarian cancer-derived exosomes. Biosens Bioelectron. 2024;250:116060
148. Li L, Zhang F, Zhang J, Shi X, Wu H, Chao X. et al. Identifying Serum Small Extracellular Vesicle MicroRNA as a Noninvasive Diagnostic and Prognostic Biomarker for Ovarian Cancer. ACS Nano. 2023;17:19197-210
149. Ljungberg B, Albiges L, Abu-Ghanem Y, Bedke J, Capitanio U, Dabestani S. et al. European Association of Urology Guidelines on Renal Cell Carcinoma: The 2022 Update. Eur Urol. 2022;82:399-410
150. Choueiri TK, Powles T, Peltola K, de Velasco G, Burotto M, Suarez C. et al. Belzutifan versus Everolimus for Advanced Renal-Cell Carcinoma. N Engl J Med. 2024;391:710-21
151. Bhatt JR, Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat Rev Urol. 2014;11:517-25
152. Boussios S, Devo P, Goodall ICA, Sirlantzis K, Ghose A, Shinde SD. et al. Exosomes in the Diagnosis and Treatment of Renal Cell Cancer. Int J Mol Sci. 2023;24:14356
153. He X, Tian F, Guo F, Zhang F, Zhang H, Ji J. et al. Circulating exosomal mRNA signatures for the early diagnosis of clear cell renal cell carcinoma. BMC Med. 2022;20:270
154. Arance E, Ramírez V, Rubio-Roldan A, Ocaña-Peinado FM, Romero-Cachinero C, Jódar-Reyes AB. et al. Determination of Exosome Mitochondrial DNA as a Biomarker of Renal Cancer Aggressiveness. Cancers (Basel). 2022;14:199
155. Himbert D, Zeuschner P, Ayoubian H, Heinzelmann J, Stöckle M, Junker K. Characterization of CD147, CA9, and CD70 as Tumor-Specific Markers on Extracellular Vesicles in Clear Cell Renal Cell Carcinoma. Diagnostics (Basel). 2020;10:1034
156. Schadendorf D, Fisher DE, Garbe C, Gershenwald JE, Grob JJ, Halpern A. et al. Melanoma. Nat Rev Dis Primers. 2015;1:15003
157. Long GV, Swetter SM, Menzies AM, Gershenwald JE, Scolyer RA. Cutaneous melanoma. Lancet. 2023;402:485-502
158. Das A, Ghose A, Naicker K, Sanchez E, Chargari C, Rassy E, Boussios S. Advances in adoptive T-cell therapy for metastatic melanoma. Curr Res Transl Med. 2023;71:103404
159. Choi DY, Park JN, Paek SH, Choi SC, Paek SH. Detecting early-stage malignant melanoma using a calcium switch-enriched exosome subpopulation containing tumor markers as a sample. Biosens Bioelectron. 2022;198:113828
160. Serratì S, Guida M, Di Fonte R, De Summa S, Strippoli S, Iacobazzi RM. et al. Circulating extracellular vesicles expressing PD1 and PD-L1 predict response and mediate resistance to checkpoint inhibitors immunotherapy in metastatic melanoma. Mol Cancer. 2022;21:20
161. Guo Y, Zhang X, Wang L, Li M, Shen M, Zhou Z. et al. The plasma exosomal miR-1180-3p serves as a novel potential diagnostic marker for cutaneous melanoma. Cancer Cell Int. 2021;21:487
162. Sol S, Boncimino F, Todorova K, Waszyn SE, Mandinova A. Therapeutic Approaches for Non-Melanoma Skin Cancer: Standard of Care and Emerging Modalities. Int J Mol Sci. 2024;25:7056
163. Didona D, Paolino G, Bottoni U, Cantisani C. Non Melanoma Skin Cancer Pathogenesis Overview. Biomedicines. 2018;6:6
164. Cives M, Mannavola F, Lospalluti L, Sergi MC, Cazzato G, Filoni E. et al. Non-Melanoma Skin Cancers: Biological and Clinical Features. Int J Mol Sci. 2020;21:5394
165. Durante G, Broseghini E, Comito F, Naddeo M, Milani M, Salamon I. et al. Circulating microRNA biomarkers in melanoma and non-melanoma skin cancer. Expert Rev Mol Diagn. 2022;22:305-18
166. Chang J, Tran DC, Zhu GA, Li R, Whitson R, Kim YH. et al. Initial in vitro functional characterization of serum exosomal microRNAs from patients with metastatic basal cell carcinoma. Br J Dermatol. 2017;177:e187-e90
167. Zhang Z, Guo H, Yang W, Li J. Exosomal Circular RNA RNA-seq Profiling and the Carcinogenic Role of Exosomal circ-CYP24A1 in Cutaneous Squamous Cell Carcinoma. Front Med (Lausanne). 2021;8:675842
168. Beird HC, Bielack SS, Flanagan AM, Gill J, Heymann D, Janeway KA. et al. Osteosarcoma. Nat Rev Dis Primers. 2022;8:77
169. Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320-5
170. Han Z, Peng X, Yang Y, Yi J, Zhao D, Bao Q. et al. Integrated microfluidic-SERS for exosome biomarker profiling and osteosarcoma diagnosis. Biosens Bioelectron. 2022;217:114709
171. Ong JLK, Jalaludin NFF, Wong MK, Tan SH, Angelina C, Sukhatme SA. et al. Exosomal mRNA Cargo are biomarkers of tumor and immune cell populations in pediatric osteosarcoma. Transl Oncol. 2024;46:102008
172. Wang L, Wu J, Song S, Chen H, Hu Y, Xu B, Liu J. Plasma Exosome-Derived Sentrin SUMO-Specific Protease 1: A Prognostic Biomarker in Patients With Osteosarcoma. Front Oncol. 2021;11:625109
173. Rezaie J, Feghhi M, Etemadi T. A review on exosomes application in clinical trials: perspective, questions, and challenges. Cell Commun Signal. 2022;20:145
174. Guo X, Peng Y, Song Q, Wei J, Wang X, Ru Y. et al. A Liquid Biopsy Signature for the Early Detection of Gastric Cancer in Patients. Gastroenterology. 2023;165:402-13.e13
175. Xiao Q, Tan M, Yan G, Peng L. Revolutionizing lung cancer treatment: harnessing exosomes as early diagnostic biomarkers, therapeutics and nano-delivery platforms. J Nanobiotechnology. 2025;23:232
176. Hinestrosa JP, Sears RC, Dhani H, Lewis JM, Schroeder G, Balcer HI. et al. Development of a blood-based extracellular vesicle classifier for detection of early-stage pancreatic ductal adenocarcinoma. Commun Med (Lond). 2023;3:146
177. Ge X, Tang L, Wang Y, Wang N, Zhou J, Deng X. et al. The diagnostic value of exosomal miRNAs in human bile of malignant biliary obstructions. Dig Liver Dis. 2021;53:760-5
178. Stevic I, Müller V, Weber K, Fasching PA, Karn T, Marmé F. et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 2018;16:179
179. Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X. et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17:68
180. Shirejini SZ, Inci F. The Yin and Yang of exosome isolation methods: conventional practice, microfluidics, and commercial kits. Biotechnol Adv. 2022;54:107814
181. Manno M, Bongiovanni A, Margolis L, Bergese P, Arosio P. The physico-chemical landscape of extracellular vesicles. Nat Rev Bioeng. 2025;3:68-82
182. Li R, Hinnant B, Li Z. Regulation and Mechanisms of Exosome Cargo Selection and Secretion. In: Li Z, editor. Extracellular Vesicle: Biology and Translational Application. Singapore: Springer Nature Singapore. 2024 p. 9-27
183. Patel K, El-Khoury JM, Aarsand AK, Badrick T, Jones GRD, Sikaris K, Parnas ML. Current Utility and Reliability of Biological Variability. Clin Chem. 2021;67:1050-5
184. Shin H, Kang Y, Choi KW, Kim S, Ham BJ, Choi Y. Artificial Intelligence-Based Major Depressive Disorder (MDD) Diagnosis Using Raman Spectroscopic Features of Plasma Exosomes. Anal Chem. 2023;95:6410-6
185. Greening DW, Xu R, Ji H, Tauro BJ, Simpson RJ. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol Biol. 2015;1295:179-209
186. Wang F, Wang C, Chen S, Wei C, Ji J, Liu Y. et al. Identification of blood-derived exosomal tumor RNA signatures as noninvasive diagnostic biomarkers for multi-cancer: a multi-phase, multi-center study. Mol Cancer. 2025;24:60
187. Shen J, Ma Z, Xu J, Xue T, Lv X, Zhu G, Huang B. Exosome Isolation and Detection: From Microfluidic Chips to Nanoplasmonic Biosensor. ACS Appl Mater Interfaces. 2024
188. Zhao X, Liu X, Chen T, Xie H, Li S, Zhang Y. et al. Fully Integrated Centrifugal Microfluidics for Rapid Exosome Isolation, Glycan Analysis, and Point-of-Care Diagnosis. ACS Nano. 2025;19:8948-65
189. Wang J, Yue BL, Huang YZ, Lan XY, Liu WJ, Chen H. Exosomal RNAs: Novel Potential Biomarkers for Diseases-A Review. Int J Mol Sci. 2022;23:2461
190. Shende P, Augustine S, Prabhakar B, Gaud RS. Advanced multimodal diagnostic approaches for detection of lung cancer. Expert Rev Mol Diagn. 2019;19:409-17
191. Surappa S, Multani P, Parlatan U, Sinawang PD, Kaifi J, Akin D, Demirci U. Integrated "lab-on-a-chip" microfluidic systems for isolation, enrichment, and analysis of cancer biomarkers. Lab Chip. 2023;23:2942-58
192. Wang CK, Tsai TH, Lee CH. Regulation of exosomes as biologic medicines: Regulatory challenges faced in exosome development and manufacturing processes. Clin Transl Sci. 2024;17:e13904
Author contact
![]() Corresponding authors: Jiachong Wang, Department of Neurosurgery, Haikou Affiliated Hospital of Central South University Xiangya School of Medicine, Haikou, China, 570208, Email: wjc1972com. Zigui Chen, Department of Neurosurgery, Haikou Affiliated Hospital of Central South University Xiangya School of Medicine, Haikou, China, 570208, Email: chenzigui_001com.
Corresponding authors: Jiachong Wang, Department of Neurosurgery, Haikou Affiliated Hospital of Central South University Xiangya School of Medicine, Haikou, China, 570208, Email: wjc1972com. Zigui Chen, Department of Neurosurgery, Haikou Affiliated Hospital of Central South University Xiangya School of Medicine, Haikou, China, 570208, Email: chenzigui_001com.
 Global reach, higher impact
Global reach, higher impact