13.3
Impact Factor
Theranostics 2025; 15(11):5238-5257. doi:10.7150/thno.113312 This issue Cite
Review
Old players and new insights: unraveling the role of RNA-binding proteins in brain tumors
1. Department of Neurosurgery, Beijing Tiantan Hospital, Capital Medical University, Beijing, China.
2. China National Clinical Research Center for Neurological Diseases, Fengtai, Beijing, China.
3. Department of Neurosurgery, Qilu Hospital, Cheeloo College of Medicine and Institute of Brain and Brain-Inspired Science, Shandong University, Jinan, 250012, China.
4. Jinan Microecological Biomedicine Shandong Laboratory, Jinan, 250117, China and Shandong Key Laboratory of Brain Health and Function Remodeling, Jinan 250012, China.
* The authors have made equal contributions to this work as corresponding authors.
Received 2025-3-6; Accepted 2025-3-27; Published 2025-4-13
Abstract
The human genome harbors >1,600 evolutionarily conserved RNA-binding proteins (RBPs), with extensive multi-omics investigations documenting their pervasive dysregulation in malignancies ranging from glioblastoma to melanoma. These RBPs are integral to the complex regulatory networks governing hallmark cancer processes. Recent studies have investigated the multifaceted contributions of RBPs to tumorigenesis, tumor metabolism, the tumor-immune microenvironment, and resistance to therapy. This complexity is further compounded by the intricate regulation of RNA function at various levels by RBPs, as well as the post-translational modifications of RBPs, which improve their functional capacity. Moreover, numerous RBP-based therapeutics have emerged, each underpinned by distinct molecular mechanisms that extend from genomic analysis to the interference of RBPs' function. This review aims to provide a comprehensive overview of the recent progress in the meticulous roles of RBPs in brain tumors and to explore potential therapeutic interventions targeting these RBPs, complemented by a discussion of innovative techniques emerging in this research field. Advances in deciphering RNA-RBP interactomes and refining targeted therapeutic strategies are revealing the transformative potential of RBP-centric approaches in brain tumor treatment, establishing them as pivotal agents for overcoming current clinical challenges.
Keywords: RNA-binding proteins, Brain tumors, Glioblastoma, Targeted therapy, Inhibitor.
Introduction
Tumorigenesis, a complex and progressive process, poses a significant threat to human health, and brain tumors are among the most formidable types. It starts with the transformation of a benign cell into a precursor cell with tumorigenic potential, driven by genetic mutations that disrupt the balance of pro-oncogenic and anti-oncogenic signaling. In the context of brain tumors, the dysregulation of RNA-binding proteins (RBPs) is a crucial factor contributing to this imbalance [1,2]. Current high-throughput screening has effectively discovered more than 1600 RBPs in the human genome, representing around 7.5% of all proteins [3]. RBPs play a vital role in maintaining cellular homeostasis and are involved in various stages of the gene expression network [4]. They are key regulators of multiple processes in the RNA life cycle, such as splicing, polyadenylation, nucleotide editing, stability, cellular localization, and translation [5]. In brain cells, the proper function of RBPs is essential for normalphysiological functions, and any alterations in their expression or function can have a significant impact on the development of brain tumors.
Despite the efforts made in the treatment of brain tumors, the outcomes remain unsatisfactory. Current therapeutic interventions, including surgical resection, radiation, and chemotherapy, have not been able to effectively combat the high incidence and poor prognosis of brain tumors. This highlights the urgent need for new therapeutic strategies. The increasing evidence of the important roles of RBPs in brain tumors, including glioblastoma, meningiomas, medulloblastoma, pituitary adenoma, and brain metastases, offers a new hope [6]. Nearly two-thirds of adults with glioblastoma, the most aggressive form of brain tumors, die within two years of diagnosis [7]. By modulating RNA-RBP binding, we may be able to target the key molecular mechanisms that drive the growth and progression of brain tumors, and develop more effective and precise therapeutic approaches [8].
The molecular subtyping and gene profiles of brain tumors have provided us with a deeper understanding of their heterogeneity [9]. Global changes in post-transcriptional regulation, often mediated by RBPs, are an important contributing factor to the differences in these molecular features among different subtypes of brain tumors [10]. RBPs are involved in every step of RNA processing, including splicing, modification, and translation and responsible for the production of more than 74% of human genes in both normal and malignant tissues [11]. In the context of brain tumors, the dysregulation of RBPs can lead to significant consequences, making them a highly promising target forprecise treatment.
This review aims to provide a comprehensive overview of the recent advancements in exploring the molecular mechanism of RBPs in brain tumors. It summarizes the potential of RBPs as novel therapeutic targets and the application of RBP-based therapies. By highlighting the importance of RBPs in the development and progression of brain tumors, this review hopes to inspire further research and development in this field, and contribute to the improvement of the treatment outcomes of brain tumors, bringing new hope to patients.
1. RBPs' general structure and functional complexes
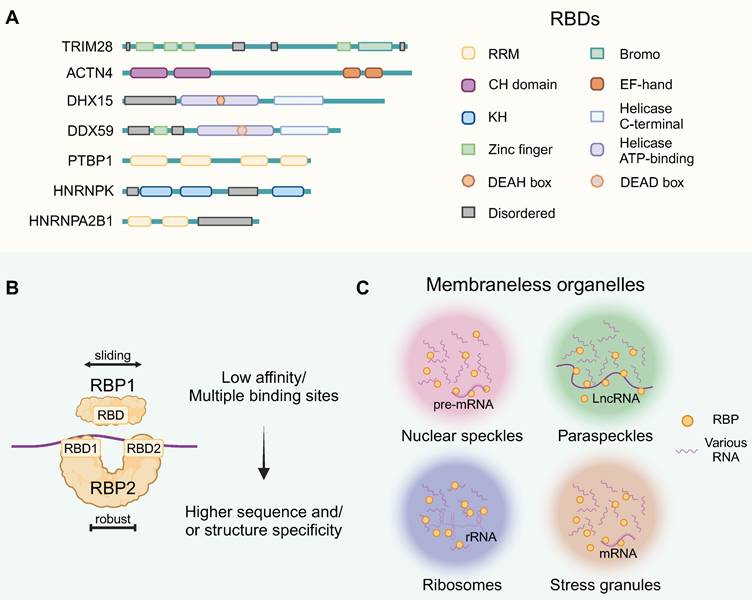
RBPs interact with RNA molecules by recognizing specific sequences or structures through their RNA-binding domain (RBD) configurations. Structurally, these well-defined RBDs include the RNA-recognition motif (RRM), human heterogeneous nuclear ribonucleoprotein (hnRNP) K-homology domain (KH), zinc-finger domains, and the DEAD-box helicase domain, among others (Figure 1A). These abundant mRNA-binding domain classes recognize 4-6 nucleotide segments, usually in combinations or repeats, enhancing RBP binding strength [12]. In the meanwhile, nearly half of RBPs lack typical RBDs due to atypical interactions with RNA [13]. RBPs are crucial for assembling functional ribonucleoprotein particles (RNPs), primarily through interactions with both coding and noncoding RNAs. The RNP complexes composed of single or tandem RBD-containing proteins bind to their RNA targets, and multiple RBDs could provide higher sequence or structural specificity (Figure 1B).
RNP granules constituted membraneless organelles (MLOs), formed via complex RNA-RNA, RNA-protein, and protein-protein interactions [14]. They are notable for their distinct size, concentration and multifaceted interactions (Figure 1C). MLOs can be formed through liquid-liquid phase separation (LLPS) and are thought to be associated with the disordered regions of RBPs in some contexts. This figure highlights the contextual versatility of RBPs, providing a foundation for elucidating the functional mechanisms of RBPs.
Among membraneless organelles, nuclear speckles are found in the nucleus and involved in the storage and assembly of pre-mRNA splicing factors. LLPS of RBP CLK2 initiate the reorganization of nuclear speckles, restricting mRNA splicing and hampering the maintenance of glioblastoma stem cells (GSCs) [15]. Paraspeckles contain an architectural lncRNA that attracts proteins to initiate the LLPS process. In GBM, elevated paraspeckle RBP NONO are associated with worse prognosis through abnormal LLPS [16]. The ribosome, the cellular machinery for protein synthesis, contains a large amount of rRNA and ribosomal proteins. Based on ribosome-associated RBP signiture, it is possible to establish a prognosis prediction system for GBM [17]. Moreover, the absence of RBPs DDX56 and WDR75 disrupts ribosomal protein regulation, inducing nucleolar stress that reduces proliferation and triggers apoptosis in glioma cells through caspase activation [18,19]. Stress granules sequester and store untranslated mRNAs during stress conditions, protecting mRNAs from degradation. RBP MSI1 and eIF2a play a leading role in stress granule formation that grants GSC with stemness properties and chemoresistance [20]. These are several examples of the oncogenic functions of RBPs within cellular organelles. These highlight the contextual versatility of RBPs, providing a foundation for elucidating the functional mechanisms of RBPs.
2. RBPs achieve their function via gene expression regulation
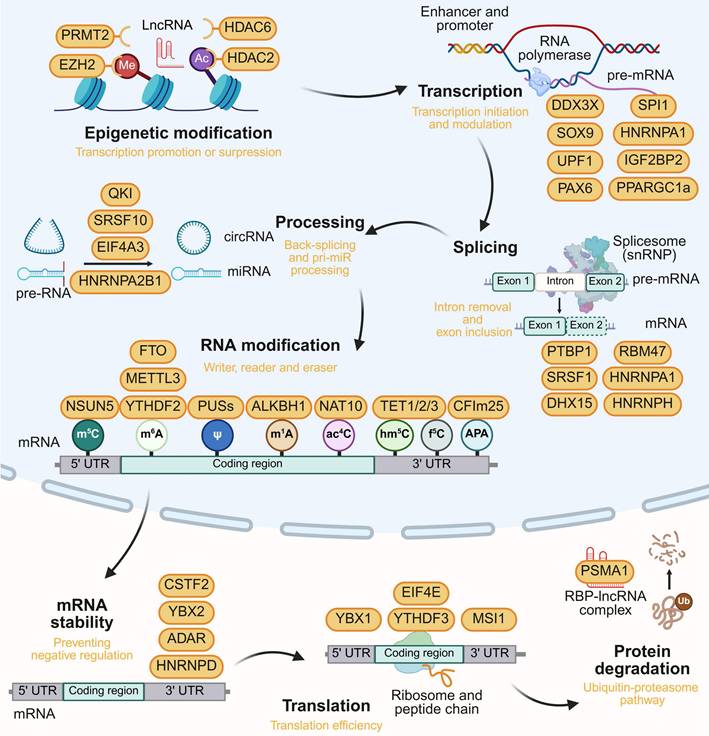
This part outlines the complex interactions of RBPs with regulatory noncoding RNAs (ncRNAs) and mRNA in cancer biology, emphasizing RBP roles in gene expression and biological function. It's clear that RBP-RNA complexes are diverse in brain tumors and forming regulatory networks. The relationship between RBP dysregulation and tumor behavior is summarized in Figure 2.
2.1 RBPs in epigenetic regulation
Epigenetic changes result in inheritable gene expression alterations without DNA sequence alteration. In brain tumors, RBPs can exert epigenetic regulation on their downstream target genes.
Mechanistic studies of the long noncoding RNA (lncRNA) HOTAIR have shown that its 5' region binds PRC2 for H3K27 trimethylation, while the 3' region binds LSD1/CoREST/REST for H3K4 demethylation [21]. HOTAIR modulates cell cycle progression in GBM via its interaction through RBP EZH2 and PRC2 complex [22]. Histone H2B acetylation is partially driven by RBP EP300 in classical-like and immune-low GBM, as determined by stratification based on snRNA-seq [23]. Histone deacetylases (HDACs) play significant roles in gliomas by regulating gene expression, with their inhibitors showing potential therapeutic value. Lnc00461 stability is maintained through interactions with HDAC6 and RBPs like CNOT6 and FUS. Targeting Lnc00461 in GBM can decrease cell division-related proteins and subsequently induces cell cycle arrest [24]. RBP HDAC2 promotes the binding of c-Myc to the CCL1 promoter, thereby activating tumor immunosuppression in GBM [25]. Diminished RBP PRMT1 and H4R3me2 levels are observed in IDH1R132H gliomas. The PRMT1-PTX3 axis is essential for controlling ferritin genes/iron storage, and its inhibition triggers ferritinophagic flux [26].
Structural and functional diversity of RBPs in brain tumors. (A) Schematic presentation of the well-defined functional RBDs of RBPs, including domains and motifs. And the intrinsically disordered region was annotated. (B) Protein-RNA Interaction Mechanisms. Individual RNA binding domains typically exhibit lower affinities compared to proteins possessing multiple such domains, with most RBPs featuring multiple RBDs for enhanced specificity in sequence and/or structural recognition. (C) RBP-RNA complexes form MLOs, which are characterized by distinct size, concentration, interactions, and functions. The RNA components within these MLOs include rRNA, snRNA, mRNA, lncRNA, and others. This figure was created with Biorender.
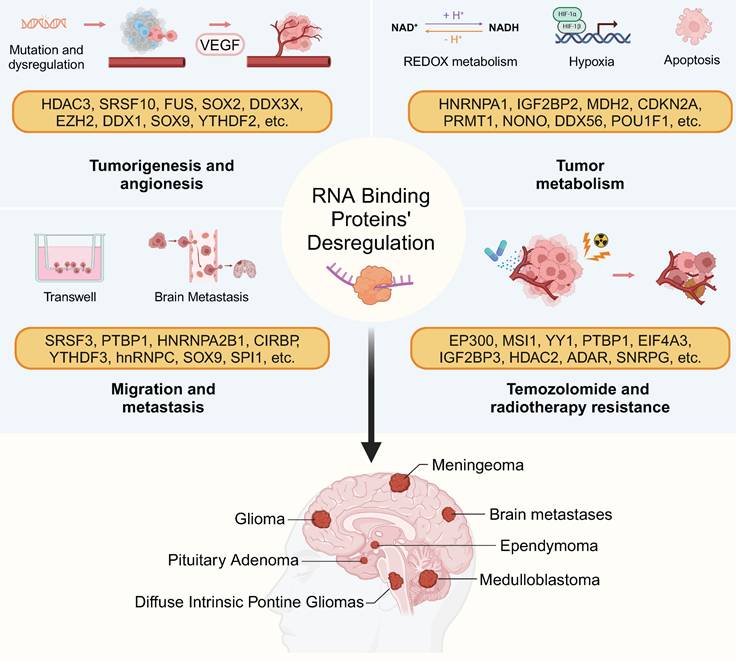
RBP-driven hallmarks of brain tumor pathogenesis. RBPs govern tumor angiogenesis, metabolic reprogramming, therapy resistance, and immune evasion through context-dependent RNA regulation. This figure was created with Biorender.
2.2 RBPs in transcription
RBPs also regulate tumorigenesis through transcription independently of histone modifications. RBP Ddx3x governs hindbrain development and prevents medulloblastoma by modulating Hox genes. It senses oncogenic stress in mice prone to Wnt- or Shh-driven medulloblastoma, suppressing tumorigenesis [27]. RBP SOX9, together with FOXG1, reshapes the enhancer activation landscape, driving EGFRvIII-dependent tumorigenesis in GBM [28]. Furthermore, RBP UPF1 binds to circRPPH1, maintaining its stability. This interaction forms a feedback loop with transcription factor ATF3, contributing to a stable stream that enhances tumorigenesis in GSCs [29]. p53 maintains quiescence by promoting fatty acid oxidation in neural stem cells (NSCs) and suppressing tumor initiation. The upregulation of transcriptional induction of RBP PPARGC1a activates PPARα, leading to the upregulation of fatty acid oxidation genes [30]. Previous research has pinpointed multiple transcription factors involved in pituitary development and tumor formation. Both POU1F1 (also known as PIT-1) involved in somatotroph, lactotroph, and thyrotroph adenoma, and NR5A1 (also known as SF-1) involved in gonadotroph adenoma, are RNA-binding proteins [31,32].
RBPs influence tumor metabolism and invasion through transcriptional regulation. Hypoxia, characteristic of glioblastoma microenvironment, elevates miR-1246 in glioma-derived exosomes through increased transcription and selective packaging, assisted by RBPs such as POU5F1 and hnRNPA1 [33]. Moreover, the hypoxia-inducible lncRNA HIF1A-AS2, specific to the mesenchymal subtype, is upregulated in GSCs. It interacts with RBPs IGF2BP2 and DHX9 to sustain HMGA1 expression [34]. The RBP SPI1 plays a complex role in transcriptional modulation during GBM progression. SPI1 induced MIR222HG expression in MES-GBM tissues and inhibited the transcription of FTO in GBM [35,36]. RBP Pax6 severely impairs tumor propagation by downregulating Olig2 in PDGF-induced oligodendroglioma and is proposed as a positive prognostic marker for gliomas [37,38].
2.3 RBPs in pre-mRNA splicing and processing
RBPs play a crucial role in RNA splicing by modulating spliceosome assembly and splice site selection. Glioblastomas can be distinguished from healthy brain samples by the altered expression of key spliceosome components and splicing factors, including SRSF family, PTBP1 [39]. Overexpression of circSMARCA5 boosts the inclusion of exon 4 in the SRSF3 RNA isoform, typically skipped by SRSF1, creating an nonsense-mediated decay (NMD) substrate that promotes glioma cell migration [40]. The lncRNA LINREP regulates PTBP1-mediated AS, notably skipping RTN4 exon 3 to produce diverse splice variants [41]. The splicing of a brain-enriched cassette exon in the tumor suppressor ANXA7 is mediated by PTBP1, which subsequently enhances EGFR signaling during glioma progression [42].
In another research, 58 consistently upregulated RBPs in GSCs were identified by measuring the expression levels of 1542 human RBPs in GBM and glioma stem cell samples [43]. The EGFRvIII upregulates the splicing factor hnRNPA1 through mTOR and MYC, leading to hnRNPA1-dependent splicing of the protein Max. This results in the promotion of glycolytic gene expression and significantly shorter survival in glioma patients [44]. The splicing factor hnRNPH regulates the aberrant splice event IG20, creating an anti-apoptotic isoform that redirects TNF-α/TRAIL-induced death signals to promote survival [45]. RBM47 promotes the inclusion of exon 20 of TJP1, and the resulting TJP1 isoform improves the assembly of actin stress fibers, thereby promoting EMT in neuroblastoma [46]. The RBP DHX15 acts as a tumor suppressor gene in glioma, suppressing the expression of NF-κB downstream target genes involved in splicing [47]. In summary, the splicing variants mediated by RBPs in brain tumors are highly diverse and still requeired further exploration.
In addition to splicing, RBPs directly bind to RNA to regulate RNA processing. RBP QKI has been identified as the primary regulator of the biogenesis of circular RNAs during EMT, indicating its involvement in promoting brain tumor invasion [48]. SRSF10 enhances circ-ATXN1 production in glioma by binding its 5' and 3' ends. Circ-ATXN1, within an RNA-induced silencing complex, sequesters miR-526b-3p to modulate glioma angiogenesis [49]. EIF4A3-induced circRNA ASAP1 expression, along with flanking regions, promotes tumorigenesis and temozolomide resistance in glioblastoma [50]. Additionally, eIF4A3 induced cyclization of circMMP9, resulting in increased circMMP9 expression in GBM by binding to the MMP9 mRNA transcript [51]. The m6A modification on pri-miR-10a is recognized by HNRNPA2B1, which then recruits processor DGCR8 to facilitate pri-miR-10a processing [36].
2.4 RBPs in mRNA modification
N6-methyladenosine (m6A), the most prevalent mRNA modification, is abundant in coding sequences and 3'-UTRs, especially in long internal exons and near stop codons. m6A proteins--methyltransferases (METTL3/METTL14), demethylases (FTO/ALKBH5), and binding proteins--play key roles in mRNA processing, particularly in stability and translation regulation [52]. Human antigen R (HuR) recruitment was essential for the stabilization of lncRNA LINREP via m6A formation, which implicated in the regulation of PTBP1-induced AS [53]. YTHDF2 enhances UBXN1 mRNA decay through METTL3-catalyzed m6A methylation, which boosts NF-κB activity. Elevated UBXN1 levels counteract the tumorigenic impact of YTHDF2 overexpression and correlate with improved GBM patient survival [54].
As for other types of RNA modifications, 5-Methylcytosine (m5C) occurs not only in tRNAs and rRNAs but also in mRNA [55]. NSUN5, which introduces m5C at the C3782 site in human 28S rRNA, induces a general reduction in protein synthesis, facilitating an adaptive translational response for glioma cell survival under stress [56]. TET proteins (TET1/2/3) catalyze the oxidative conversion of DNA and RNA m5C to hm5C and f5C, respectively, and dysregulation of TET family proteins has been implicated in glioma [57]. As for N1-methyladenosine (m1A) modification and pseudouridine (ψ) modification, the m1A eraser ALKBH1 and pseudouridine synthases (PUSs, ψ writers) also function as RBPs and play crucial roles in GBM [58,59]. The upregulation of NAT10, an enzyme responsible for N4-acetylcytidine (ac4C) modification, promotes the acetylation of PARP1, thereby inducing cell death in GBM cells [60]. CFIm25 modulates proximal poly(A) site selection through alternative polyadenylation (APA). Knockdown of CFIm25 results in the shortening of 3'UTRs in over 1,400 genes, subsequently leading to elevated expression of oncogenes in GBM [61]. However, these mRNA modifications in brain tumors still require further research.
2.5 RBP in translational and post-translational regulation
The translation process is facilitated by RBPs from the Superfamily II DNA and RNA helicase family within the ribosome, and the efficiency of translation is closely related to mRNA stability. The lncRNA ATXN8OS stabilizes GLS2 mRNA by recruiting RBP ADAR, thereby limiting TMZ resistance in glioma [62]. Additionally, CSTF2 interacts with the mRNA of the BAD to truncate its 3'-UTR, adversely affecting BAD-mediated apoptosis and promoting the survival of GBM cells [63]. YBX2 binds and stabilizes HNF4G mRNA to regulate blood-tumor barrier (BTB) permeability [64]. There is a potential mechanism of APA in GBM involving the influence of multiple RBPs on the stability and translation of APA isoforms [65]. It is noteworthy that there are also examples of RBPs negatively regulating RNA stability. The RBP HNRNPD can decrease the stability of ZHX2 mRNA and promote the formation of vasculogenic mimicry in GBM [66].
Elevated YTHDF3 RBP levels facilitats blood-brain barrier (BBB) penetration and angiogenesis. YTHDF3 boosts translation of m6A-rich ST6GALNAC5, GJA1, and EGFR transcripts, which are involved in brain metastasis [67]. The Msi1 mRNA has an extensive 3'-UTR that contains several AU- and U-rich sequences, often targeted by HuR. The HuR binds to the Msi1 3'-UTR, improving MSI1 mRNA stability and translation in GBM [68]. RBP YBX1 binds to the 5'-UTR of CCT4 mRNA to promote the translation of CCT4, which activates the mTOR signaling pathway [69]. Previous research has shown that global eIF4E-mediated translation is inhibited in glioma [70]. However, stable silencing of DDX28 in hypoxic human glioblastoma cells increased eIF4E2 binding to the m7GTP cap structure and translation of eIF4E2 target mRNAs [71].
In post-translational regulation, suppression of RBP SNRPG facilitates the cytosolic translocation of MYC and nuclear translocation of p53. This enhances the inhibitory effect of TMZ and reduces the mismatch repair protein MLH1 in GBM [72]. LncRNA MDHDH serves as a molecular scaffold, binding RBP PSMA1 and MDH2, to promote MDH2 degradation, affecting mitochondrial membrane potential and the NAD+/NADH balance, ultimately impeding GBM glycolysis [73]. The role of RBPs in brain tumors is listed in Table 1 and the model is summarized in Figure 3. This hierarchy emphasizes the potential of stage-specific RBP targeting to disrupt tumorigenic networks.
The role of RNA binding protein in brain tumor.
| RBP | Cancer role | Cancer type | Cellular mechanisms | Ref. | |
|---|---|---|---|---|---|
| Histone modification | EZH2 | Proto-oncogenic | Glioblastoma | Promote H3K27 trimethylation | [22] |
| EP300 | Proto-oncogenic | Ependymoma | Promote H2B acetylation | [23] | |
| HDAC2 | Proto-oncogenic | Glioblastoma | Deacetylation of histones and non-histones | [25,120] | |
| PRMT1 | Proto-oncogenic | Glioma | Promote H4R3me2 of PTX3 promoter | [26] | |
| Transcriptional regulation | DDX3X | Tumor-suppressing | Medulloblastoma | Modulating Hox expression and suppresses medulloblastoma formation | [27] |
| PPARGC1a | Proto-oncogenic | Glioblastoma | Activates PPARα expression in GSCs | [30] | |
| POU1F1 and NR5A1 | Proto-oncogenic | Pituitary adenoma | Pituitary development and tumorigenesis through transcription | [31,32] | |
| IGF2BP2 and DHX9 | Proto-oncogenic | Glioblastoma | Sustain HMGA1 expression in mesenchymal subtype GBM | [34] | |
| Pax6 | Tumor-suppressing | Oligodendroglioma and glioma | Surpressing Olig2 expression | [37,38] | |
| Splicing and processing | SRSF1 | Tumor-suppressing | Glioma | Inclusion of exon 4 in the SRSF3 | [40] |
| PTBP1 | Proto-oncogenic | Glioblastoma | Involved in exon skipping of RTN4 and ANXA7 | [41,42] | |
| hnRNPA1 | Proto-oncogenic | Glioma | Splicing of the Max mRNA | [44] | |
| RBM47 | Proto-oncogenic | Neuroblastoma | Promoted the inclusion of exon 20 of TJP1 | [46] | |
| DHX15 | Tumor-suppressing | Glioma | Alterantive splicing of glioma | [47] | |
| SRSF10 | Proto-oncogenic | Glioma | Enhances circ-ATXN1 production by binding its 5' and 3' ends | [49] | |
| EIF4A3 | Proto-oncogenic | Glioblastoma | Promoted circular RNA biogenesis and cyclization | [50,51] | |
| RNA modification | YTHDF2 | Proto-oncogenic | Glioblastoma | Enhances UBXN1 mRNA decay through m6A methylation | [54] |
| NSUN5 | Proto-oncogenic | Glioma | Introduces m5C at the C3782 site in 28S rRNA | [56] | |
| NAT10 | Proto-oncogenic | Glioblastoma | Promotes the acetylation of PARP1 mRNA | [60] | |
| CFIm25 | Proto-oncogenic | Glioblastoma | Introduces APA in mRNA 3'UTRs | [61] | |
| Translational and post-translational regulation | ADAR | Tumor-suppressing | Glioma | Stabilizes GLS2 mRNA with lncRNA ATXN8OS | [62] |
| HNRNPD | Proto-oncogenic | Glioblastoma | Decrease the stability of ZHX2 mRNA | [66] | |
| YTHDF3 | Proto-oncogenic | Brain metastasis | Enhances translation of m6A-enriched transcripts | [67] | |
| YBX1 | Proto-oncogenic | Glioblastoma | Bounds the 5′UTR of CCT4 to promote the translation | [69] | |
| EIF4E2 | Proto-oncogenic | Glioblastoma | Binding the m7GTP cap structure to promote translation | [71] | |
| SNRPG | Proto-oncogenic | Glioblastoma | Facilitates the cytosolic translocalization of MYC | [72] | |
| DDX31 | Proto-oncogenic | Medulloblastoma | Blocking interaction between the E3 ubiquitin ligase and p53 | [91] | |
| Multiple pathway | SOX9 | Proto-oncogenic | Medulloblastoma, Glioblastoma | Gene transcription and cell transdifferentiation | [28,92,97,98] |
| SOX2 | Proto-oncogenic | Glioblastoma | Maintaining the self-renewal of GSC | [80,88] | |
| STAT3 | Proto-oncogenic | Glioblastoma | Astrogliogenesis and cell proliferation | [84,125,126] | |
| QKI | Proto-oncogenic | Glioblastoma, brain metastasis | Regulator of biogenesis of circular RNAs and exerting functions within EVs | [48,108,109] | |
| HuR | Proto-oncogenic | Glioblastoma | mRNA stabilization through m6A modification and 3'-UTR binding | [53,68] | |
| HNRNPA2B1 | Proto-oncogenic | Glioblastoma | Processing pri-miR and exerting functions within EVs | [36,110,111] |
Multi-layered regulatory roles of RBPs in gene expression. RBPs modulate epigenetic modification, transcription, splicing, RNA processing RNA modification, mRNA stability, translation and protein degradation, with representative oncogenic/tumor-suppressive RBPs annotated at each regulatory layer. This figure was created with Biorender.
3. RBPs' expression and function regulation
3.1 RBPs' expression regulation
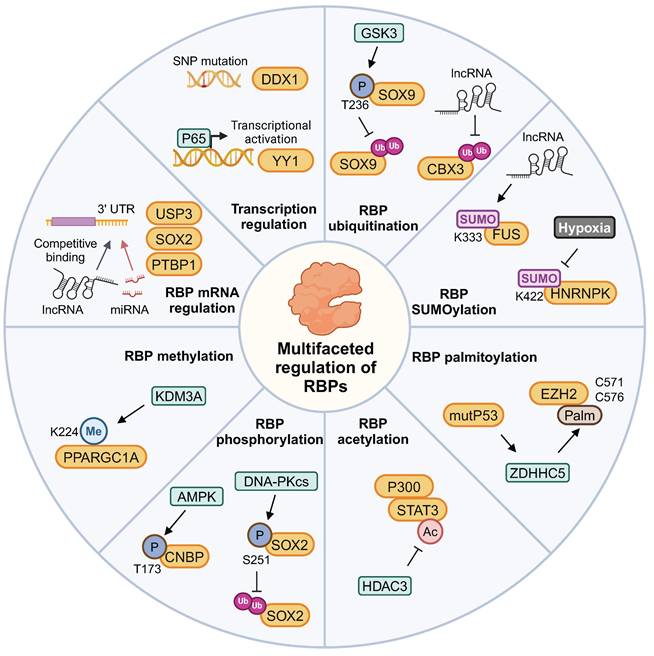
This complex interaction network between RBP and ncRNA has been gradually revealed in the context of brain tumors. The SNP rs72780850 variant in the RBP DDX1 promoter shows enhanced transcription factor binding and is linked to increased DDX1 expression, potentially elevating neuroblastoma risk [74]. Continuous radiation stimulates the nuclear translocation of p65. Consequently, there is increased expression of RBP YY1, which induces radioresistance in glioma cells [75]. CDKN2A expression, a RBP biomarker for aggressive meningiomas, is regulated by genetic aberrations and miRNAs in meningiomas and pilocytic astrocytomas [76,77].
The lncRNA HOXA-AS3 functions as a mir-455-5p sponge, upregulating RBP USP3 expression to facilitate GBM progression [78]. PTB-AS significantly increases PTBP1 level by directly binding to its 3'-UTR, and masking the miR-9 binding site in the PTBP1-3'-UTR to prevent PTBP1 degradation [79]. lncRNA NEAT1 indirectly regulates RBP SOX2 expression by targeting miR-132 in glioma [80]. miR-146a diminishes the expression of RBP hnRNPC, which then inhibits the migratory and invasive activity of brain metastatic cells by downregulating the expression of MMP-1, uPA, and uPAR [81]. LINC00470 lncRNA dynamically regulates RBP subcellular localization by forming a ternary complex with FUS and AKT in the cytoplasm, which activates AKT, prevents nuclear translocation, and fosters glioblastoma tumorigenesis [82].
3.2 RBPs' function regulation by protein modification
Protein modifications play a significant role in the regulation of brain tumors by RBPs, as previously reported. The PPARGC1A K224R mutant expression significantly increased mitochondrial biogenesis under hypoxic conditions. This effect is attributed to the loss of PPARGC1A K224 monomethylation, which is regulated by oxygen availability-dependent KDM3A [83]. HDAC3 modulates the acetylation state of STAT3 and competes with STAT3 for P300 binding, thereby antagonizing astrogliogenesis. This regulation influences phenotypic commitment during oligodendrocyte-astrocytic fate decision [84]. Mutant p53 increases ZDHHC5 and NF-Y expression, enhancing GSCs via EZH2 palmitoylation and phosphorylation modifications [85].
The cyclic adenosine 3′,5′-monophosphate pathway activation promoted RBP FLNA phosphorylation in GH-secreting tumoral pituitary cells, abolishing antitumor effects on pituitary cells [86]. Cerebellar granule cell precursor proliferation is governed by Hedgehog signaling. AMP-activated protein kinase phosphorylates RBP CNBP at threonine 173 upon Hedgehog signaling, enhancing medulloblastoma cell proliferation [87]. Additionally, DNA-PKcs phosphorylates RBP SOX2 at S251, stabilizing SOX2 by preventing WWP2-mediated ubiquitination. Consequently, this stabilization promotes the maintenance of GSCs [88].
RBP HNRNPL prevents ACTN4 from being degraded by the proteasome, a process involving ubiquitination. This protection enhances NF-κB subunit's nuclear accumulation and contributs to GBM progression [89]. Moreover, LINC00998 stabilizes RBP CBX3, prevents its ubiquitination degradation, and regulates the c-Met/Akt/mTOR signaling pathway in GBM [90]. RBP DDX31 binds to NPM, blocking the interaction between E3 ubiquitin ligase and p53, leading to p53 stabilization in medulloblastoma tumorigenesis [91]. Increased levels of RBP SOX9 correlate with medulloblastoma spread and unfavorable outcomes. The phosphorylation of SOX9 at T236 competes with its ubiquitination by E3 ligase FBW7 [92].
SUMO (Small Ubiquitin-like Modifier) is a small ubiquitin-like protein that participates in post-translational modification of proteins, with particularly high levels of SUMOylated proteins observed in GBM. Hypoxia enhances the cytoplasmic translocation of hnRNPA2B1 by enhancing its SUMOylation, a process that can be inhibited by the SUMOylation inhibitor TAK-981 [93]. The lncRNA RMST enhances RBP FUS SUMOylation, particularly promoting SUMO1 attachment at K333. This modification strengthens FUS-hnRNPD interaction, stabilizing these proteins and inducing mitophagy [94]. GSCs preferentially exhibit high levels of SUMO1-modified SUMOylation. The SUMO1 SUMOylation inhibitor juglone significantly abrogated GSC maintenance and GSC-driven tumor growth [95]. The expression of HNRNPK-SUMO1 is primarily localized within the infiltration regions of GBM. The SUMOylation of the K422 residue on HNRNPK impairs its capacity to bind DNA, consequently disrupting subsequent transcriptional processes [96]. The multifaceted regulatory patterns of RBPs are summarized in Figure 4. Focusing on these modifications provides a strategic approach to indirectly modulate RBP function within the context of tumor biochemistry.
The regulation of gene expression and post-translational modifications of RBPs in brain tumors. The post-translational modifications of RBPs include methylation, phosphorylation, acetylation, palmitoylation, ubiquitination, and SUMOylation. This figure was created with Biorender.
4. RBP influences on brain tumor microenvironment
4.1 RBP functions in cellular microenvironment
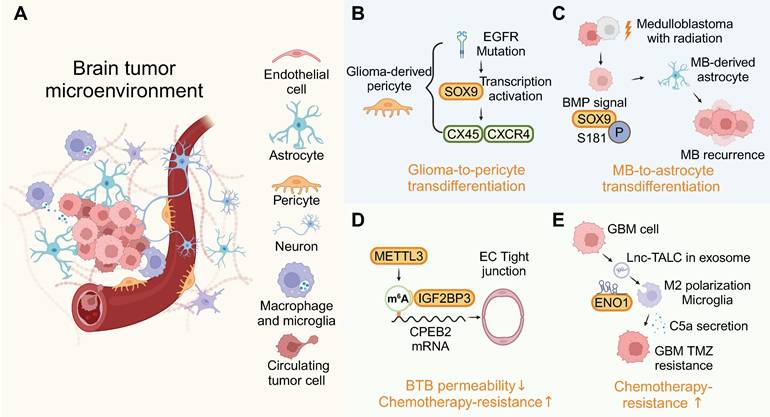
While the BBB is widely recognized as a major obstacle in the treatment of brain tumors, the role of RBPs within the BBB remains poorly understood. RBPs are involved in BBB-mediated brain tumor progression through various cellular microenvironments (Figure 5A).
Dysregulation of RBPs can notably affect cell phenotypes in the brain's microenvironment. EGFR mutations prompt glioma cells to act as pericytes, dependent on BMX/SOX9, thereby stabilizing blood vessels and attracting immune cells via a glioma-to-pericyte transition [97] (Figure 5B). Astrocytes are derived from the transdifferentiation of tumor cells in relapsed medulloblastoma. Treatment with BMP proteins significantly induced astrocytogenesis and phosphorylation of the RBP SOX9, effectively stimulating tumor cell transdifferentiation into astrocytes (Figure 5C) [98]. The accumulation of microglia in the brain tumor microenvironment is associated with malignancy and a poor prognosis for these tumors. Activation of TLR2 along with other TLRs transforms microglia into a glioma-supportive phenotype [99]. RNP complex LOC-DHX15 promote infiltration of immunosuppressive glioma-associated microglia and macrophages [100].
TARBP2 is an RBP bound to SNHG7, leading to an extended half-life of SNHG7 and, consequently, an increased permeability of BBB [101]. Upregulation of RBP METTL3 and IGF2BP3 in glioma microvascular endothelial cells (ECs) increased the stability of CPEB2 mRNA through m6A methylation, thereby regulating BTB permeability (Figure 5D) [102]. The expression of RBP YBX2 could influence BTB permeability through YBX2-mediated transcriptional activities [64]. Recent GBM research underscores the reciprocal signaling between tumors and neurons, perpetuating a cycle of increased proliferation, synaptic incorporation, and heightened brain activity [103]. Furthermore, RBP nucleolin plays a crucial role in the axonal trafficking of mRNA and influences neuronal growth, highlighting the multifaceted functions of RBPs in both the tumor and its microenvironment [104]. By analyzing patient samples and patient-derived xenograft (PDX) models that recapitulate the invasive pattern, it is suggested that overexpressed RBP CIRBP is essential for brain metastasis [105].
4.2 RBP functions in extracellular vesicles
Extracellular vesicles (EVs), including exosomes, are cellular messengers that transport various molecules including RNA. Recent studies have revealed that RBPs are present within EVs, with some RBPs playing a role in the selective packaging of ncRNAs into exosomes. RBP hnRNPA2B1, MSI, AGO2, and YBX1 have been implicated in the selective sorting of specific RNAs into exosomes [106]. Moreover, the RBP-RNA interaction was primarily detected through their binding site in EVs. For example, RBPs could be detected in EVs through inferred RNA targets, or EV-transcripts were found to harbor sequence motifs mirroring the activity of RBPs [107].
RNA within EVs has multiple roles, and the RBP QKI, found in EVs, is linked to primary brain tumors and breast cancer tissues. Elevated QKI levels are associated with poorer survival, indicating its potential as a prognostic marker for glioblastoma and brain metastasis [108,109]. Furthermore, RBP-ncRNA-exosome mechanisms are involved in establishing an immunosuppressive microenvironment in brain tumors. circNEIL3, encapsulated by RBP hnRNPA2B1 into exosomes, is transferred to tumor-associated macrophages, conferring them immunosuppressive capabilities via IGF2BP3 stabilization [110]. The binding of miR-30b-3p to hnRNPA2B1 facilitates its transfer into EVs. The EV-packaged miR-30b-3p decreased apoptosis and increased the proliferation of glioma [111]. RBP hnRNPA1 facilitates the inclusion of exo-miR-1246, which drives monocyte polarization towards myeloid-derived suppressor cells (MDSCs). These MDSCs inhibit CD8+ T cell proliferation, aiding in the creation of an immunosuppressive microenvironment [33]. As a result of lnc-TALC binding to RBP ENO1, Lnc-TALC could be incorporated into exosomes and transmitted to microglia and promote M2 polarization (Figure 5E) [112]. These findings all indicate that the role of EV-associated RBPs in brain tumors can no longer be ignored. The above insights highlight the potential of microenvironment-focused RBP therapeutics as a complementary approach to traditional tumor cell-targeted strategies.
RBP-mediated crosstalk in the tumor microenvironment. (A) RBPs drive cellular plasticity in endothelial cells, astrocytes, and immune cells to foster a pro-tumorigenic niche. (B-E) Mechanistic case studies illustrate RBP roles in transdifferentiation, and chemotherapy resistance. This figure was created with Biorender.
5. Targeting RBPs in therapeutic strategy
5.1 Engineered particles
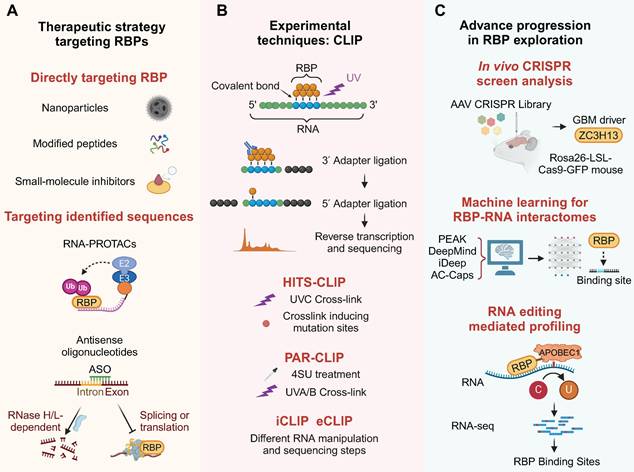
Various approaches, such as small-molecule inhibitors, peptides and nanomedicines, are employed to modulate RBPs for therapeutic purposes in cancer treatment, aiming to control gene expression and biological functions.
Engineered particles were designed to target tumor-associated RBPs. Considering the existence of the BBB, corresponding delivery strategies have also been developed for nanomedicines [113]. A β-cyclodextrin nanoparticle formulation encapsulating the RBP TLR7/8 agonist R848 was developed to target blood-borne macrophages, bypassing the need for adaptive immunity in the glioma microenvironment [114]. Immunotherapy using a TLR9 agonist CpG ODN offers a novel strategy. CpG is efficiently loaded into apolipoprotein E peptide-directed polymersomes, forming a CpG nano-immunoadjuvant that can cross the BBB and target glioma and cervical lymph nodes [115]. The RBP WT1-mediated upregulation of CD97 promotes cellular invasiveness [116]. Based on this target, a modified 9-mer WT1 peptide was tested in a Phase II clinical trial, indicating the potential application of RBP-related vaccines in GBM [117]. The Iodine-123 Meitner-Auger PARP1 inhibitor (123I-MAPi), an iodine-123 labeled PARP1 inhibitor, is a targeted therapy for GBM that utilizes Auger electrons for precise apoptosis induction [118].
5.2 Small molecule inhibitors
Developing RBP-targeted therapies involves screening RNAi oligos or small molecules that decrease RBP-RNA binding and modify target RNA levels. Using high-throughput methods to identify RBP-bound RNAs and analyzing large datasets could lead to new cancer treatments or prognostic markers [119].
RBP HDAC2 affects several physiological processes through the deacetylation of histones and non-histones [120]. PCI-24781, identified through the Connectivity Map bioinformatic tool, is a novel HDAC2 inhibitor specific to the GBM signature. It works synergistically with TMZ to improve its efficacy and GBM survival [121]. The EP300 inhibitor CPI-1612 suppressed H3K27ac and the associated transcription complex and synergistically sensitized TMZ in vitro by brain penetration [122]. The epigenetic modifier KDM1A inhibitor NCD38 can cross the BBB, and KDM1A inhibition increases the efficacy of TMZ for GBM [123]. The palmitoylation of OCT4 is indispensable for protecting it from lysosomal degradation and maintaining GSCs. Its competitive palmitoylation inhibitor, CPP-S1, inhibits the self-renewal ability and tumorigenicity of GSCs [124].
STAT3 is traditionally regarded as a transcription factor. However, it is now widely accepted STAT3 is also a highly versatile RBP [125]. The continuous activation of STAT3 in a significant proportion of GBM, which positions STAT3 as a promising therapeutic target and a prognostic marker for GBM [126]. The compound SS-4, developed through computational modeling for STAT3 binding, specifically targets STAT3 Y-705 phosphorylation in GBM [127]. Treatment with WP1066, a STAT3 inhibitor currently used in clinical settings for pediatric brain tumors, inhibits tumor growth in H3K27M-mutant diffuse midline gliomas [128] and high-grade gliomas with arginine or valine substitutions in the histone H3.3 glycine 34 residue [129]. In the case of HuR, the focus is primarily on three ongoing targeted strategies. These include inhibiting HuR translocation from the nucleus to the cytoplasm, inhibiting the ability of HuR to bind to target RNA, and silencing HuR expression. The corresponding inhibitor, MS-444, was verified in GBM [130].
Auranofin was used to disrupt the binding and splicing of the RBP NONO, thereby inhibiting the malignancy of GBM [131]. The RBP CDK9 inhibitors, including SNS032, LY2857785, AZD4573, NVP229, and JSH15030, significantly reduced the viability and self-renewal of GBM cells [132]. The sensitivity of PLX-4720 is associated with the expression of RBP SPI1 and improves the radiosensitivity of mesenchymal GSCs [35]. Inhibition of the RBP SPI1 using DB2313 restored downstream FTO expression and alleviated the tumor burden in GBM [36]. Treatment with DHODH inhibitors like brequinar or ML390 depletes pyrimidine levels in glioblastoma and causes nucleolar stress. This stress causes mislocalization of the RBP NPM1 and promotes p53 transcription factor stabilization [133]. The PARP-1 inhibitor PJ-34 suppresses the migration of brain microvascular endothelial cells, thereby influencing the growth and progression of brain tumors [134]. The selective nanobody NB237 against RBP TRIM28 inhibits the invasiveness and spread of GSCs in vivo [135].
METTL3 facilitates medulloblastoma growth through the enhancement of Sonic Hedgehog signaling, and its inhibition by STM2457 impedes tumor advancement [136]. The LIN28B inhibitor 1632 reduced medulloblastoma cell growth and provided preliminary preclinical results for drugs for medulloblastoma [137]. HB007, identified through a cancer cell-based drug screen, selectively degraded SUMO1 in GBM PDX model [138]. Compound 25, discovered via SPR screening, potently binds hnRNPK and disrupts its interaction with the c-myc promoter, offering a selective approach to modulate oncogene transcription without targeting DNA secondary structures [139]. Furthermore, TERT and LRP1, both recognized as RBPs, have small-molecule drugs that have progressed to clinical trial stages [140,141].
Finally, considering that most RBPs are difficult to target with drugs, RNA-PROTACs represent a novel approach for targeting RBPs. These chimeric structures employ small RNA mimics as targeting groups that dock the RNA-binding site of the RBP, whereupon a conjugated E3-recruiting peptide directs the RBP for proteasomal degradation [142]. This targeting approach has been proven to be effective against the glioma oncogene LIN28.
5.3 Antisense oligonucleotides (ASO)
ASOs are short, single-stranded nucleic acids (DNA or RNA) ranging from 12 to 24 nucleotides in length. They are designed to bind to specific complementary mRNA targets through base pairing, thereby modulating gene expression. ASO are categorized into RNase H/L-dependent ASOs that facilitate RNA cleavage and steric blockade ASOs that hinder RBP binding [143]. Modifications enhance ASO stability, cellular uptake, and bioavailability for diverse applications [144].
An ASO targeting LINC02283 and the concomitant RBP PDGFRA successfully inhibited GBM tumorigenesis [145]. The selected ASOs specifically delay the growth of patient-derived H3.3K27M cells grown by mediating the depletion of the H3-3A protein in diffuse intrinsic pontine gliomas (DIPG) [146].
In addition, a decoy oligonucleotide similar to ASO, composed of several repeats of an RNA motif, can specifically bind to splicing factors such as SRSF1. This binding inhibits their splicing activity and suppresses the growth of gliomas [147]. This type of RBP decoy also hinders the translation of mRNA mediated by YBX1 [69]. And an Aptamer-RIBOTAC designed based on ASO can activate endogenous RNase L to precisely degrade oncogenic miRNAs in gliomas, offering another promising therapeutic approach [148]. The strategies targeting RBPs have been summarized in Figure 6A. This panel resonates with the emphasis on innovation in precision medicine, highlighting tools that bridge the gap between mechanistic discovery and clinical translation. The various approaches for targeting RBPs and their specific applications are comprehensively summarized in Table 2.
Technological advances in RBP research. (A) Therapeutic modalities targeting RBPs, including nanoparticles, peptides, inhibitors, PROTACs, and ASOs. (B) Evolution of CLIP-based methods for high-resolution RBP-RNA interactome mapping. (C) CRISPR screening and machine learning enable systematic discovery of clinically actionable RBP targets. This figure was created with Biorender.
The RBP associated treatment entering clinical trials and in preliminary test.
| RBP | Therapeutic agent | Drug Types | Cancer | Phase | NCT number | Mechanism | Ref. |
|---|---|---|---|---|---|---|---|
| WT1 | WT1 peptide | Vaccine | Recurrent GBM | II | Not provide | Inducing an antitumor immune response in brain | [117] |
| HDAC2 | PCI-24781 | Inhibitor | Glioblastoma | I | NCT05698524 | Enhance sensitivity to temozolomide | [121] |
| STAT3 | WP1066 | Inhibitor | Medulloblastoma, malignant glioma | I | NCT04334863 | Target the STAT3 pathway in GBM and activate the immune system | [128,129] |
| STAT3 | WP1066 | Inhibitor | Recurrent Glioblastoma, Metastatic Melanoma | I | NCT01904123 | Target the STAT3 pathway in GBM and activate the immune system | [128,129] |
| STAT3 | WP1066 | Inhibitor | Glioblastoma | II | NCT05879250 | Target the STAT3 pathway in GBM and activate the immune system | [128,129] |
| TERT | GRN163L | Inhibitor | Glioblastoma, Astrocytoma, Oligodendroglioma | II | NCT01836549 | Inhibition of telomerase activity | [140] |
| LRP1 | ANG1005 | Inhibitor | Recurrent High-Grade Glioma | II | NCT01967810 | Penetrate malignant cells via LRP1 transport system | [141] |
| TLR7/8 | CDNP-R848 | Agonist | Glioblastoma | / | / | Reshapes the immunosuppressive tumor microenvironment to a proinflammatory state | [114] |
| TLR9 | t-NanoCpG | Agonist | Glioblastoma | / | / | Stimulates the maturation of dendritic cells and production of proinflammatory cytokines | [115] |
| EP300 | CPI-1612 | Inhibitor | Glioblastoma | / | / | SuppressedH3K27ac and the associated transcription complex | [122] |
| KDM1A | NCD38 | Inhibitor | Glioblastoma | / | / | Increased sensitivity to temozolomide | [123] |
| OCT4 | CPP-S1 | Inhibitor | GSCs | / | / | Inhibitthe palmitoylation of OCT4 | [124] |
| STAT3 | SS-4 | Inhibitor | Glioblastoma | / | / | Selectivelyinhibited STAT3 tyrosine (Y)-705 phosphorylation | [127] |
| HuR | MS-444 | Inhibitor | Glioblastoma | / | / | Inhibition of HuR dimerization | [130] |
| NONO | Auranofin | Inhibitor | Glioblastoma | / | / | Disturb the binding and splicing of NONO | [131] |
| NPM1 | Brequinar, ML390 | Inhibitor | Glioblastoma | / | / | Aberrant redistribution of the NPM1 | [133] |
| METTL3 | STM2457 | Inhibitor | SHH-medulloblastoma | / | / | Inhibit catalytic function | [136] |
| TRIM28 | NB237 | Inhibitor | GSCs | / | / | Inhibition of GSCs invasiveness and spread | [135] |
| SPI1 | PLX-4720 | Inhibitor | GSCs | / | / | Enhance radiosensitivity through SPI1 | [35] |
| SPI1 | DB2313 | Inhibitor | Glioblastoma | / | / | Restore downstream FTO expression | [36] |
| CDK9 | SNS032, LY2857785, AZD4573, NVP229, JSH15030 | Inhibitor | Glioblastoma | / | / | Reduce the cell viability and self-renewal | [132] |
| PARP1 | PJ-34 | Inhibitor | Glioma | / | / | Suppress the brain microvascular endothelial cells migration | [134] |
| LIN28B | Inhibitor 1632 | Inhibitor | Medulloblastoma | / | / | Inhibit LIN28B-let-7-PBK axis | [137] |
| SUMO1 | HB007 | Inhibitor | Brain tumor | / | / | Selectively degraded SUMO1 through ubiquitination | [138] |
| hnRNPK | compound 25 | Inhibitor | Glioblastoma | / | / | Disrupt the interaction between hnRNPK and c-myc promoter i-motif | [139] |
| PDGFRA | ASO | ASO | Glioblastoma | / | / | ASO targeting lncRNA to block the LINC02283-PDGFRA interaction | [145] |
| H3-3A | ASO | ASO | DIPG | / | / | Direct RNase H-mediated knockdown of H3-3A mRNA | [146] |
| SRSF1 | Decoy oligonucleotides | ASO-like drugs | Glioblastoma | / | / | Inhibit splicing and biological activities of splicing factors | [147] |
6. Advancements and progressions in RBP networks
6.1 Advancements in technologies for RBP-RNA interaction
The information regarding which RNA sequence or structure is bound by the RBPs is crucial. Over the past decades, various methods have been developed to determine this, including RNA immunoprecipitation (RIP) [149], targets of RNA-binding proteins identified by editing (TRIBE) [150], and RNA tagging [151]. These techniques have significantly improved our understanding of RNA-protein interactions and their roles in cellular processes. Each method has its strengths and limitations, and the choice of method often depends on the specific research question and available resources.
Crosslinking and immunoprecipitation (CLIP) is a widely used technique for studying protein-RNA interactions in vivo on a transcriptome-wide scale, and its applications have expanded over time (Figure 6B). CLIP utilizes ultraviolet (UV) light to create covalent bonds between RNA and attached proteins, followed by immunoprecipitation with specific RBP antibodies to isolate RBP-RNA complexes, initially used to map direct interactions in the brain's RNA network [152].
CLIP's evolution has spawned variants that boost efficiency across steps like RNA fragmentation and RBP purification. HITS-CLIP, which integrates high-throughput sequencing, offers a comprehensive view of RBP-RNA interactions at nucleotide resolution, enabling the study of RNA-binding sites in distinct brain regions [153]. Individual-nucleotide CLIP (iCLIP) is an advanced protocol that uses leftover amino acids post-crosslinking to stop reverse transcriptase, allowing for precise mapping of RBP-RNA interactions in GBM. It incorporates a second adaptor with a unique sequencing barcode during circularization for detailed interaction site localization [154]. Enhanced CLIP (eCLIP) refines the iCLIP process by incorporating a size-matched control during RNA-protein purification, facilitating a more accurate assessment of the specificity in RNA-PTBP1/IGF2BP2 interactions with HOTAIRM1 [155]. Photoactivatable ribonucleotide-enhanced CLIP (PAR-CLIP) enhances the mapping of RBP-RNA interactions by incorporating 4-thiouridine or 6-thioguanosine into nascent RNAs, followed by UV-A light-induced crosslinking to RBPs, facilitating precise interaction site identification through RNA-seq analysis in brain. The integration of RNA-Seq data with the application of PAR-CLIP for YBX1 revealed direct post-transcriptional control in medulloblastomas [156].
Nevertheless, the CLIP technique remains dependent on substantial sample sizes and highly efficient RBP antibodies, and it is limited to the analysis of a single RBP at a time. More powerful and precise methods for exploring RBPs are continuously being updated, although they have not yet been applied in the field of brain tumors. A novel approach combining quantitative proteomics with photoreactive nucleotide-enhanced UV crosslinking and oligo (dT) purification has been established to identify mRNA-bound proteins [157]. The RNA interactome capture (RIC) technique employs magnetic beads to enrich and analyze the mRNA interactome, thereby overcoming the limitation of relying on highly efficient antibodies [158,159]. "cat RNA-Protein Interaction Design" utilizes RNA-binding motifs to forecast protein-RNA interaction probabilities. Its sequence fragmentation method facilitates the analysis of both long linear and circular RNAs [160]. LACE-seq, designed to reveal RBP-regulated RNA networks in single oocytes, surpasses various CLIP-based methods in sensitivity, accuracy, precision, and efficiency, potentially offering a clearer view of brain tumorigenesis and the microenvironment at the single-cell level [161]. Beyond genomic and transcriptomic analyses, protein-protein interaction networks and dynamic subcellular localization provide a holistic view of the RBPs' mutational landscapes [162,163]. It is anticipated that these methods will also be applied to the exploration of brain tumors.
6.2 CRISPR screening
CRISPR-Cas9 technology has emerged as a new tool for RBP screening and regulation. Advancements in delivery systems for CRISPR-Cas9 have been crucial for RBP-focused cancer therapies, facilitating the discovery and targeting of cancer-related RBPs. Effective delivery of CRISPR-Cas9 is key to deepening our understanding of RBPs in cancer and their translation into clinical therapeutics [164]. Adeno-associated virus-based CRISPR screening in glioblastoma has been used to create tumor models that mimic human disease. This approach involves injecting a library of viruses targeting common cancer genes into the brains of mice with conditional Cas9, leading to the formation of tumors. This method has identified co-occurring driver combinations like RBP Zc3h13-Rb1 and provided insights into the functional suppressors of gliomagenesis. The CRISPR-Cas9 system's efficient delivery is crucial for understanding and advancing RBPs as therapeutic targets in cancer [165].
6.3 Multimodal integration and machine learning
With the growing volume of RBP data, it's essential to understand how motif enrichment influences RBP-RNA binding profiles in cells. The ENCODE consortium has generated extensive CLIP data using the eCLIP method on 150 RBPs, providing a rich resource for this research [166]. New computational methods such as PEKA have been developed to identify motifs in CLIP data, enabling comprehensive analysis of RBP binding preferences [167]. The refinement of CLIP methodologies has significantly accelerated the identification of RBP-RNA interactomes.
The HARD-AP method retrieves RBPs and tightly associated RNA regulatory complexes, and systematically maps RNA-binding sites across primary organs using machine learning-based modeling [168]. In addition, other notable deep learning approaches for predicting RBP binding sites include DeepMind and iDeep [169,170]. An attention-based capsule network (AC-Caps method) can reliably process large-scale RBP binding site data on lncRNA [171]. This integration has, in turn, enabled the construction of predictive models for regulatory networks involved in RBP-RNA interactions within brain.
6.4 RNA editing applications
Comprehending RBP functions requires identifying their in vivo targets, yet CLIP and its adaptations may face challenges like a lack of suitable antibodies or inefficiency in small cell populations. Several recently designed genetic approaches were developed to identify RBP targets under such circumstances.
TRIBE and similar editing-based profiling techniques are innovative genetic tools for investigating post-transcriptional regulation and RBP detection [172]. A novel approach involves modifying sequences near methylation sites, which could enable m6A detection by standard RNA-seq, overcoming limitations of current antibody-dependent methods. The RBP APOBEC1, a cytidine deaminase that targets both DNA and RNA, induces cytidine to uridine (C to U) editing [173]. Originally identified for editing ApoB mRNA, APOBEC1 has been repurposed in CRISPR/Cas9 systems for inducing C to U transitions at specific single-stranded DNA locations [174]. Fusing APOBEC1 to the m6A-binding YTH domain could enable targeted editing of cytidines near m6A sites in RNAs. Detecting these edits with RNA-Seq may provide a clearer picture of the m6A landscape in brain tumors [67,175]. This innovative approach provides a promising avenue for advancing our understanding of the complex dynamics of protein-RNA interactions and their implications in brain tumor pathology. These advanced methods for exploring RBP networks are summarized in Figure 6C.
7. Conclusions and perspectives
The past two decades have witnessed a surge in mechanistic studies that have significantly expanded our understanding of RBPs within the RNA regulation network. In addition to their conventional intracellular locations, RBPs can localize to the cell membrane surface, where they facilitate the cell-penetrating peptide entry and engage in communication within the cellular microenvironment [176]. Moreover, RBPs not only facilitate the transdifferentiation of cells within the brain microenvironment, as previously mentioned, but also serve as independent prognostic risk factors for brain tumors. They have the potential to predict patient outcomes due to their association with immune checkpoint responses and chemotherapeutic efficacy [177,178]. The role of the IGFBP family is of particular interest, as it significantly contributes to the interferon pathway and the activation of T cells and NK cells in various brain tumor types [179,180]. These findings indicate that the functions of RBPs in brain tumors are not yet fully understood and continue to possess significant potential value.
With the advent of novel methodologies, a genome-wide perspective on the RNA structure-dependent interactions of RBPs under physiological and pathological states is now possible. This method not only improves our knowledge but also facilitates the discovery of RBP-RNA interactions with therapeutic potential. The field of compound screening is rapidly advancing, with innovative techniques being developed to modulate RBP interactomes, thereby offering new therapeutic avenues. Despite this progress, targeting RBPs presents several challenges, particularly regarding delivery methods and translation of the RBP-RNA network's potential in clinical applications for brain tumor treatment.
The involvement of RBPs in brain tumor pathogenesis has sparked interest in their modulation as a novel therapeutic strategy. Approaches targeting the RNA-RBP network include RNA-based therapies, such as ASOs, siRNAs, RNA aptamers, and mRNAs, which are now part of clinical medicine. Moreover, small molecules that target RNA structures are under development, using strategies like conjugating RNA recognition elements to ribonuclease-activating compounds [181]. Compound screening is crucial for identifying protein-based inhibitors that can block RNA-protein interactions, with several small molecules already identified that bind to RBPs and inactivate specific RBDs [182]. Additionally, compounds that disrupt the nucleocytoplasmic shuttling of RBPs, such as the YBX1 inhibitor HSc025, offer alternative strategies for modulating RBP functions [183].
The intracellular delivery of therapeutic RNA without inducing immune responses is essential for effective RNA-based RBP therapy. The delivery systems are categorized into polymer-based, lipid-based, and conjugate-based systems, with the ability to target specific cells either passively or actively [184]. Active targeting uses specific ligands to bind molecular targets, such as the clinically established GalNAc-linked siRNA and ASO conjugates [185]. Biomimetic nanovesicles, including endogenous EVs and artificial nanovesicles, can cross the BBB, making them ideal for brain-targeted drug delivery [186]. EVs can serve as delivery platforms for RNAs like miRNAs and siRNAs in cancer therapy [187]. The secretory autophagy pathway facilitates the extracellular release of RBPs and RNA cargo via EVs using the LC3-conjugation machinery [188]. Exosome targeting can be improved by adding peptides to their surface and loading ncRNAs for therapeutic efficacy, effectively combining targeting and therapeutic capabilities. Overall, for clinical applications of exosome-based therapies, the production and quality control of exosomes must be meticulously addressed [186].
The RNA-RBP regulatory network has emerged as a pivotal axis for advancing brain tumor therapy. Integrating RBP profiles into molecular diagnostics could enable personalized therapies by defining actionable targets across tumor subtypes. Clinically, RBP-targeting strategies (e.g., ASOs and PROTACs) show potential to reprogram oncogenic RNA networks with high specificity. However, challenges remain in delivering these agents across the blood-brain barrier and addressing tumor heterogeneity. Advances in nanovesicles and exosome-based delivery systems may overcome these limitations, while liquid biopsies monitoring exosomal RBPs could refine treatment response assessment [189]. Moving forward, efforts should focus on multi-omics mapping of RBP interactions, validating biomarkers in clinical cohorts, and combining RBP inhibitors with immunotherapy or radiation to counter resistance. By aligning mechanistic insights with scalable therapeutic platforms, RBP research embodies the theranostic vision of bridging molecular discovery to patient care.
Abbreviations
RBPs: RNA-binding proteins; CNS: Central Nervous System; 3'-UTRs: 3' untranslated regions; GBM: glioblastoma; RBDs: RNA-binding domains; RRM: RNA-recognition motif; hnRNP: human heterogeneous nuclear ribonucleoprotein; KH: K-homology domain; RNPs: ribonucleoprotein particles; MLOs: membraneless organelles; ncRNAs: noncoding RNAs; lncRNA: long noncoding RNA; PRC2: polycomb repressive complex 2; H3K27: histone H3 lysine 27; HDAC: histone deacetylase; TMZ: temozolomide; GSCs: glioblastoma stem cells; NMD: nonsense-mediated decay; m6A: N6-methyladenosine; rRNAs: ribosomal RNAs; BBB: blood-brain barrier; EMT: epithelial-mesenchymal transition; IDH: isocitrate dehydrogenase; NSCs: neural stem cells; m5C: 5-Methylcytosine; hm5C: 5-hydroxymethylcytidine; f5C: 5-formyl-cytidine; m1A: N1-methyladenosine; PUSs: pseudouridine synthases; HuR: human antigen R; APA: alternative polyadenylation; SUMO: small ubiquitin-like modifier; LLPS: liquid-liquid phase separation; ECs: endothelial cells; BTB: blood-tumor barrier; EVs: extracellular vesicles; MDSCs: myeloid-derived suppressor cells; RNAi: RNA interference; siRNA: small interfering RNA; 123I-MAPi: Iodine-123 Meitner-Auger PARP1 inhibitor; ASO: antisense oligonucleotides; DIPG: diffuse intrinsic pontine gliomas; RIP: RNA immunoprecipitation; TRIBE: targets of RNA-binding proteins identified by editing; CLIP: crosslinking and immunoprecipitation; UV: ultraviolet; HITS-CLIP: high-throughput sequencing CLIP; iCLIP: individual-nucleotide CLIP; eCLIP: enhanced CLIP; PAR-CLIP: photoactivatable ribonucleotide-enhanced CLIP; PEKA: Positionally Enriched K-mer Analysis; RIC: RNA interactome capture.
Acknowledgements
The authors acknowledge the BioRender (www.biorender.com), as figures in this review were created with BioRender platform.
Funding
This work was supported by the National Natural Science Foundation of China (82472932, 82071996, 82403089 and 82003075), the Beijing Natural Science Foundation (7212008), Beijing Hospitals Authority Youth Program (QML20210502), Beijing Postdoctoral Research Foundation, "Tiantan-CIBR" Joint Project, the Special Foundation for Taishan Scholars (tsqn201909173), the Department of Science & Technology of Shandong Province (ZR2022ZD36), and the Research Project of Jinan Microecological Biomedicine Shandong Laboratory (JNL-2023007C and SYS202202).
Author contributions
A.C., W.J. and X.W.: Conceptualization, Formal analysis, Funding acquisition, and Writing - original draft, review & editing; J.L. and C.Z.: Investigation and Writing - review & editing. X.G. and X.L.: Funding acquisition and Writing - review & editing. All authors reviewed the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Gebauer F, Schwarzl T, Valcárcel J, Hentze MW. RNA-binding proteins in human genetic disease. Nat Rev Genet. 2021;22:185-98
2. Hashimoto S, Kishimoto T. Roles of RNA-binding proteins in immune diseases and cancer. Semin Cancer Biol. 2022;86:310-24
3. Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829-45
4. Qin H, Ni H, Liu Y, Yuan Y, Xi T, Li X. et al. RNA-binding proteins in tumor progression. J Hematol Oncol. 2020;13:90
5. He S, Valkov E, Cheloufi S, Murn J. The nexus between RNA-binding proteins and their effectors. Nat Rev Genet. 2023;24:276-94
6. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D. et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro-Oncology. 2021;23:1231-51
7. Tykocki T, Eltayeb M. Ten-year survival in glioblastoma. A systematic review. J Clin Neurosci. 2018;54:7-13
8. Arvanitis CD, Ferraro GB, Jain RK. The blood-brain barrier and blood-tumour barrier in brain tumours and metastases. Nat Rev Cancer. 2020;20:26-41
9. Yang K, Wu Z, Zhang H, Zhang N, Wu W, Wang Z. et al. Glioma targeted therapy: insight into future of molecular approaches. Mol Cancer. 2022;21:39
10. Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Xiao R. et al. A large-scale binding and functional map of human RNA-binding proteins. Nature. 2020;583:711-9
11. Zhao Y, Mir C, Garcia-Mayea Y, Paciucci R, Kondoh H, LLeonart ME. RNA-binding proteins: Underestimated contributors in tumorigenesis. Semin Cancer Biol. 2022;86:431-44
12. Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479-90
13. Beckmann BM, Castello A, Medenbach J. The expanding universe of ribonucleoproteins: of novel RNA-binding proteins and unconventional interactions. Pflügers Archiv - European Journal of Physiology. 2016;468:1029-40
14. Li Y, Liu Y, Yu X-Y, Xu Y, Pan X, Sun Y. et al. Membraneless organelles in health and disease: exploring the molecular basis, physiological roles and pathological implications. Sig Transduct Target Ther. 2024;9:305
15. Wang B, Li J, Song Y, Qin X, Lu X, Huang W. et al. CLK2 Condensates Reorganize Nuclear Speckles and Induce Intron Retention. Advanced Science. 2024;11:2309588
16. Wei Y, Luo H, Yee PP, Zhang L, Liu Z, Zheng H. et al. Paraspeckle Protein NONO Promotes TAZ Phase Separation in the Nucleus to Drive the Oncogenic Transcriptional Program. Adv Sci. 2021;8:2102653
17. Chen Z, Wu H, Yang H, Fan Y, Zhao S, Zhang M. Identification and validation of RNA-binding protein-related gene signature revealed potential associations with immunosuppression and drug sensitivity in glioma. Cancer Medicine. 2021;10:7418-39
18. Pryszlak M, Wiggans M, Chen X, Jaramillo JE, Burns SE, Richards LM. et al. The DEAD-box helicase DDX56 is a conserved stemness regulator in normal and cancer stem cells. Cell Reports. 2021;34:108903
19. Moudry P, Chroma K, Bursac S, Volarevic S, Bartek J. RNA-interference screen for p53 regulators unveils a role of WDR75 in ribosome biogenesis. Cell Death Differ. 2022;29:687-96
20. Chen H-Y, Lin L-T, Wang M-L, Tsai K-L, Huang P-I, Yang Y-P. et al. Musashi-1 promotes chemoresistant granule formation by PKR/eIF2α signalling cascade in refractory glioblastoma. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 2018;1864:1850-61
21. Zheng Y-C, Ma J, Wang Z, Li J, Jiang B, Zhou W. et al. A Systematic Review of Histone Lysine-Specific Demethylase 1 and Its Inhibitors: LSD1 AND ITS INHIBITORS. Med Res Rev. 2015;35:1032-71
22. Zhao J, Jin W, Yi K, Wang Q, Zhou J, Tan Y. et al. Combination LSD1 and HOTAIR-EZH2 inhibition disrupts cell cycle processes and induces apoptosis in glioblastoma cells. Pharmacol Res. 2021;171:105764
23. Wang L-B, Karpova A, Gritsenko MA, Kyle JE, Cao S, Li Y. et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021;39:509-528.e20
24. Wu A-C, Yang W-B, Chang K-Y, Lee J-S, Liou J-P, Su R-Y. et al. HDAC6 involves in regulating the lncRNA-microRNA-mRNA network to promote the proliferation of glioblastoma cells. J Exp Clin Canc Res. 2022;41:47
25. Sun T, Liu B, Cai L, Zhou Y, Yang W, Li Y. Suberanilohydroxamic acid (SAHA), a HDAC inhibitor, suppresses the effect of Treg cells by targeting the c-Myc/CCL1 pathway in glioma stem cells and improves PD-L1 blockade therapy. J Neurooncol. 2024;168:457-71
26. Lathoria K, Gowda P, Umdor SB, Patrick S, Suri V, Sen E. PRMT1 driven PTX3 regulates ferritinophagy in glioma. Autophagy. 2023;19:1997-2014
27. Patmore DM, Jassim A, Nathan E, Gilbertson RJ, Tahan D, Hoffmann N. et al. DDX3X Suppresses the Susceptibility of Hindbrain Lineages to Medulloblastoma. Dev Cell. 2020;54:455-470.e5
28. Liu F, Hon GC, Villa GR, Turner KM, Ikegami S, Yang H. et al. EGFR Mutation Promotes Glioblastoma through Epigenome and Transcription Factor Network Remodeling. Mol Cell. 2015;60:307-18
29. Xu J, Zhang G, Hu J, Li H, Zhao J, Zong S. et al. UPF1/circRPPH1/ATF3 feedback loop promotes the malignant phenotype and stemness of GSCs. Cell Death Dis. 2022;13:645
30. Amodeo V, Davies T, Martinez-Segura A, Clements MP, Ragdale HS, Bailey A. et al. Diet suppresses glioblastoma initiation in mice by maintaining quiescence of mutation-bearing neural stem cells. Dev Cell. 2023;58:836-846.e6
31. Zhang S, Cui Y, Ma X, Yong J, Yan L, Yang M. et al. Single-cell transcriptomics identifies divergent developmental lineage trajectories during human pituitary development. Nat Commun. 2020;11:5275
32. Zhang Q, Yao B, Long X, Chen Z, He M, Wu Y. et al. Single-cell sequencing identifies differentiation-related markers for molecular classification and recurrence prediction of PitNET. Cell Reports Medicine. 2023;4:100934
33. Qiu W, Guo X, Li B, Wang J, Qi Y, Chen Z. et al. Exosomal miR-1246 from glioma patient body fluids drives the differentiation and activation of myeloid-derived suppressor cells. Molecular Therapy. 2021;29:3449-64
34. Mineo M, Ricklefs F, Rooj AK, Lyons SM, Ivanov P, Ansari KI. et al. The Long Non-coding RNA HIF1A-AS2 Facilitates the Maintenance of Mesenchymal Glioblastoma Stem-like Cells in Hypoxic Niches. Cell Reports. 2016;15:2500-9
35. Fan Y, Gao Z, Xu J, Wang H, Guo Q, Li B. et al. SPI1-mediated MIR222HG transcription promotes proneural-to-mesenchymal transition of glioma stem cells and immunosuppressive polarization of macrophages. Theranostics. 2023;13:3310-29
36. Zhang S, Zhao S, Qi Y, Li B, Wang H, Pan Z. et al. SPI1-induced downregulation of FTO promotes GBM progression by regulating pri-miR-10a processing in an m6A-dependent manner. Molecular Therapy - Nucleic Acids. 2022;27:699-717
37. Appolloni I, Calzolari F, Barilari M, Terrile M, Daga A, Malatesta P. Antagonistic modulation of gliomagenesis by Pax6 and Olig2 in PDGF-induced oligodendroglioma. Int J Cancer. 2012;131:E1078-87
38. Zhou Y-H, Wu X, Tan F, Shi Y-X, Glass T, Liu TJ. et al. PAX6 suppresses growth of human glioblastoma cells. J Neurooncol. 2005;71:223-9
39. Fuentes-Fayos AC, Vázquez-Borrego MC, Jiménez-Vacas JM, Bejarano L, Pedraza-Arévalo S, L.-López F. et al. Splicing machinery dysregulation drives glioblastoma development/aggressiveness: oncogenic role of SRSF3. Brain. 2020;143:3273-93
40. Barbagallo D, Caponnetto A, Cirnigliaro M, Brex D, Barbagallo C, D'Angeli F. et al. CircSMARCA5 Inhibits Migration of Glioblastoma Multiforme Cells by Regulating a Molecular Axis Involving Splicing Factors SRSF1/SRSF3/PTB. International Journal of Molecular Sciences. 2018;19:480
41. Ji X, Liu Z, Gao J, Bing X, He D, Liu W. et al. N6-Methyladenosine-modified lncRNA LINREP promotes Glioblastoma progression by recruiting the PTBP1/HuR complex. Cell Death & Differentiation. 2023;30:54-68
42. Ferrarese R, Harsh GR, Yadav AK, Bug E, Maticzka D, Reichardt W. et al. Lineage-specific splicing of a brain-enriched alternative exon promotes glioblastoma progression. J Clin Invest. 2014;124:2861-76
43. Correa BR, De Araujo PR, Qiao M, Burns SC, Chen C, Schlegel R. et al. Functional genomics analyses of RNA-binding proteins reveal the splicing regulator SNRPB as an oncogenic candidate in glioblastoma. Genome Biol. 2016;17:125
44. Babic I, Anderson ES, Tanaka K, Guo D, Masui K, Li B. et al. EGFR Mutation-Induced Alternative Splicing of Max Contributes to Growth of Glycolytic Tumors in Brain Cancer. Cell Metab. 2013;17:1000-8
45. LeFave CV, Squatrito M, Vorlova S, Rocco GL, Brennan CW, Holland EC. et al. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas: hnRNPH-dependent splicing switch in gliomas. The EMBO Journal. 2011;30:4084-97
46. Kim Y-E, Won M, Lee S-G, Park C, Song C-H, Kim KK. RBM47-regulated alternative splicing of TJP1 promotes actin stress fiber assembly during epithelial-to-mesenchymal transition. Oncogene. 2019;38:6521-36
47. Ito S, Koso H, Sakamoto K, Watanabe S. RNA helicase DHX15 acts as a tumour suppressor in glioma. Br J Cancer. 2017;117:1349-59
48. Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA. et al. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell. 2015;160:1125-34
49. Liu X, Shen S, Zhu L, Su R, Zheng J, Ruan X. et al. SRSF10 inhibits biogenesis of circ-ATXN1 to regulate glioma angiogenesis via miR-526b-3p/MMP2 pathway. J Exp Clin Cancer Res. 2020;39:121
50. Wei Y, Lu C, Zhou P, Zhao L, Lyu X, Yin J. et al. EIF4A3-induced circular RNA ASAP1 promotes tumorigenesis and temozolomide resistance of glioblastoma via NRAS/MEK1/ERK1-2 signaling. Neuro-Oncology. 2021;23:611-24
51. Wang R, Zhang S, Chen X, Li N, Li J, Jia R. et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17:166
52. Wang T, Kong S, Tao M, Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol Cancer. 2020;19:88
53. Ji X, Liu Z, Gao J, Bing X, He D, Liu W. et al. N6-Methyladenosine-modified lncRNA LINREP promotes Glioblastoma progression by recruiting the PTBP1/HuR complex. Cell Death Differ. 2023;30:54-68
54. Chai R-C, Chang Y-Z, Chang X, Pang B, An SY, Zhang K-N. et al. YTHDF2 facilitates UBXN1 mRNA decay by recognizing METTL3-mediated m6A modification to activate NF-κB and promote the malignant progression of glioma. J Hematol Oncol. 2021;14:109
55. Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ. et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023-33
56. Janin M, Ortiz-Barahona V, De Moura MC, Martínez-Cardús A, Llinàs-Arias P, Soler M. et al. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 2019;138:1053-74
57. Carella A, Tejedor JR, García MG, Urdinguio RG, Bayón GF, Sierra M. et al. Epigenetic downregulation of TET3 reduces genome-wide 5hmC levels and promotes glioblastoma tumorigenesis. Int J Cancer. 2020;146:373-87
58. Xie Q, Wu TP, Gimple RC, Li Z, Prager BC, Wu Q. et al. N-methyladenine DNA Modification in Glioblastoma. Cell. 2018;175:1228-1243.e20
59. Mongiardi MP, Savino M, Falchetti ML, Illi B, Bozzo F, Valle C. et al. c-MYC inhibition impairs hypoxia response in glioblastoma multiforme. Oncotarget. 2016;7:33257-71
60. Liang S, Wang X, Piao M, Chen X, Wang Z, Li C. et al. Activated SIRT1 contributes to DPT-induced glioma cell parthanatos by upregulation of NOX2 and NAT10. Acta Pharmacol Sin. 2023;44:2125-38
61. Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu A-B. et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412-6
62. Luo J, Bai R, Liu Y, Bi H, Shi X, Qu C. Long non-coding RNA ATXN8OS promotes ferroptosis and inhibits the temozolomide-resistance of gliomas through the ADAR/GLS2 pathway. Brain Res Bull. 2022;186:27-37
63. Xu Y, Yuan F, Sun Q, Zhao L, Hong Y, Tong S. et al. The RNA-binding protein CSTF2 regulates BAD to inhibit apoptosis in glioblastoma. Int J Biol Macromol. 2023;226:915-26
64. Ding Y, Liu X, Yang C, Ruan X, Wang D, Liu Y. et al. Pseudogene RPL32P3 regulates the blood-tumor barrier permeability via the YBX2/HNF4G axis. Cell Death Discovery. 2021;7:367
65. Shao J, Zhang J, Zhang Z, Jiang H, Lou X, Huang B. et al. Alternative Polyadenylation in Glioblastoma Multiforme and Changes in Predicted RNA Binding Protein Profiles. OMICS: A Journal of Integrative Biology. 2013;17:136-49
66. Yu S, Ruan X, Liu X, Zhang F, Wang D, Liu Y. et al. HNRNPD interacts with ZHX2 regulating the vasculogenic mimicry formation of glioma cells via linc00707/miR-651-3p/SP2 axis. Cell Death Dis. 2021;12:153
67. Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S. et al. YTHDF3 Induces the Translation of m6A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell. 2020;38:857-871.e7
68. Vo DT, Abdelmohsen K, Martindale JL, Qiao M, Tominaga K, Burton TL. et al. The Oncogenic RNA-Binding Protein Musashi1 Is Regulated by HuR via mRNA Translation and Stability in Glioblastoma Cells. Mol Cancer Res. 2012;10:143-55
69. Wang J-Z, Zhu H, You P, Liu H, Wang W-K, Fan X. et al. Upregulated YB-1 protein promotes glioblastoma growth through a YB-1/CCT4/mLST8/mTOR pathway. J Clin Invest. 2022;132:e146536
70. Gera JF, Mellinghoff IK, Shi Y, Rettig MB, Tran C, Hsu J. et al. AKT Activity Determines Sensitivity to Mammalian Target of Rapamycin (mTOR) Inhibitors by Regulating Cyclin D1 and c-myc Expression. Journal of Biological Chemistry. 2004;279:2737-46
71. Evagelou SL, Bebenek O, Specker EJ, Uniacke J. DEAD Box Protein Family Member DDX28 Is a Negative Regulator of Hypoxia-Inducible Factor 2α- and Eukaryotic Initiation Factor 4E2-Directed Hypoxic Translation. Molecular and Cellular Biology. 2020;40:e00610-19
72. Lan Y, Lou J, Hu J, Yu Z, Lyu W, Zhang B. Downregulation of SNRPG induces cell cycle arrest and sensitizes human glioblastoma cells to temozolomide by targeting Myc through a p53-dependent signaling pathway. Cancer Biology and Medicine. 2020;17:112-31
73. He D, Xin T, Pang B, Sun J, Liu ZH, Qin Z. et al. A novel lncRNA MDHDH suppresses glioblastoma multiforme by acting as a scaffold for MDH2 and PSMA1 to regulate NAD+ metabolism and autophagy. J Exp Clin Canc Res. 2022;41:349
74. Jin Y, Shi J, Wang H, Lu J, Chen C, Yu Y. et al. MYC-associated protein X binding with the variant rs72780850 in RNA helicase DEAD box 1 for susceptibility to neuroblastoma. Science China Life Sciences. 2021;64:991-9
75. Gu J, Mu N, Jia B, Guo Q, Pan L, Zhu M. et al. Targeting radiation-tolerant persister cells as a strategy for inhibiting radioresistance and recurrence in glioblastoma. Neuro-Oncology. 2022;24:1056-70
76. Wang JZ, Patil V, Liu J, Dogan H, Tabatabai G, Yefet LS. et al. Increased mRNA expression of CDKN2A is a transcriptomic marker of clinically aggressive meningiomas. Acta Neuropathol (berl). 2023;146:145-62
77. Jones TA, Jeyapalan JN, Forshew T, Tatevossian RG, Lawson ARJ, Patel SN. et al. Molecular analysis of pediatric brain tumors identifies microRNAs in pilocytic astrocytomas that target the MAPK and NF-κB pathways. Acta Neuropathologica Communications. 2015;3:86
78. Chen W, Li Q, Zhang G, Wang H, Zhu Z, Chen L. LncRNA HOXA-AS3 promotes the malignancy of glioblastoma through regulating miR-455-5p/USP3 axis. J Cell Mol Med. 2020;24:11755-67
79. Zhu L, Wei Q, Qi Y, Ruan X, Wu F, Li L. et al. PTB-AS, a Novel Natural Antisense Transcript, Promotes Glioma Progression by Improving PTBP1 mRNA Stability with SND1. Mol Ther. 2019;27:1621-37
80. Zhou K, Zhang C, Yao H, Zhang X, Zhou Y, Che Y. et al. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol Cancer. 2018;17:105
81. Hwang SJ, Seol HJ, Park YM, Kim KH, Gorospe M, Nam D-H. et al. MicroRNA-146a suppresses metastatic activity in brain metastasis. Mol Cells. 2012;34:329-34
82. Liu C, Zhang Y, She X, Fan L, Li P, Feng J. et al. A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J Hematol Oncol. 2018;11:77
83. Qian X, Li X, Shi Z, Bai X, Xia Y, Zheng Y. et al. KDM3A Senses Oxygen Availability to Regulate PGC-1α-Mediated Mitochondrial Biogenesis. Mol Cell. 2019;76:885-895.e7
84. Zhang L, He X, Liu L, Jiang M, Zhao C, Wang H. et al. Hdac3 Interaction with p300 Histone Acetyltransferase Regulates the Oligodendrocyte and Astrocyte Lineage Fate Switch. Dev Cell. 2016;36:316-30
85. Chen X, Ma H, Wang Z, Zhang S, Yang H, Fang Z. EZH2 Palmitoylation Mediated by ZDHHC5 in p53-Mutant Glioma Drives Malignant Development and Progression. Cancer Res. 2017;77:4998-5010
86. Peverelli E, Giardino E, Mangili F, Treppiedi D, Catalano R, Ferrante E. et al. cAMP/PKA-induced filamin A (FLNA) phosphorylation inhibits SST2 signal transduction in GH-secreting pituitary tumor cells. Cancer Lett. 2018;435:101-9
87. D'Amico D, Antonucci L, Di Magno L, Coni S, Sdruscia G, Macone A. et al. Non-canonical Hedgehog/AMPK-Mediated Control of Polyamine Metabolism Supports Neuronal and Medulloblastoma Cell Growth. Dev Cell. 2015;35:21-35
88. Fang X, Huang Z, Zhai K, Huang Q, Tao W, Kim L. et al. Inhibiting DNA-PK induces glioma stem cell differentiation and sensitizes glioblastoma to radiation in mice. Sci Transl Med. 2021;13:eabc7275
89. Ji J, Xu R, Ding K, Bao G, Zhang X, Huang B. et al. Long Noncoding RNA SChLAP1 Forms a Growth-Promoting Complex with HNRNPL in Human Glioblastoma through Stabilization of ACTN4 and Activation of NF-κB Signaling. Clin Cancer Res. 2019;25:6868-81
90. Cai H, Yu Y, Ni X, Li C, Hu Y, Wang J. et al. LncRNA LINC00998 inhibits the malignant glioma phenotype via the CBX3-mediated c-Met/Akt/mTOR axis. Cell Death Dis. 2020;11:1032
91. Bish R, Vogel C. RNA Binding Protein-Mediated Post-Transcriptional Gene Regulation in Medulloblastoma. Mol Cells. 2014;37:357-64
92. Suryo Rahmanto A, Savov V, Brunner A, Bolin S, Weishaupt H, Malyukova A. et al. FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. Embo J. 2016;35:2192-212
93. Guo Q, Fan Y, Wang Q, Li B, Qiu W, Qi Y. et al. Glioblastoma upregulates SUMOylation of hnRNP A2/B1 to eliminate the tumor suppressor miR-204-3p, accelerating angiogenesis under hypoxia. Cell Death Dis. 2023;14:147
94. Liu C, Peng Z, Li P, Fu H, Feng J, Zhang Y. et al. lncRNA RMST Suppressed GBM Cell Mitophagy through Enhancing FUS SUMOylation. Molecular Therapy - Nucleic Acids. 2020;19:1198-208
95. Zhang A, Tao W, Zhai K, Fang X, Huang Z, Yu JS. et al. Protein sumoylation with SUMO1 promoted by Pin1 in glioma stem cells augments glioblastoma malignancy. Neuro-Oncology. 2020;22:1809-21
96. Zhao W, Wang J, Zhao F, Li Y, Li Z, Li X. et al. SUMOylation modification of HNRNPK at the K422 site promotes invasion in glioblastoma. Int J Biol Sci. 2024;20:5715-30
97. Segura-Collar B, Garranzo-Asensio M, Herranz B, Hernández-SanMiguel E, Cejalvo T, Casas BS. et al. Tumor-Derived Pericytes Driven by EGFR Mutations Govern the Vascular and Immune Microenvironment of Gliomas. Cancer Res. 2021;81:2142-56
98. Guo D, Wang Y, Cheng Y, Liao S, Hu J, Du F. et al. Tumor cells generate astrocyte-like cells that contribute to SHH-driven medulloblastoma relapse. J Exp Med. 2021;218:e20202350
99. Vinnakota K, Hu F, Ku M-C, Georgieva PB, Szulzewsky F, Pohlmann A. et al. Toll-like receptor 2 mediates microglia/brain macrophage MT1-MMP expression and glioma expansion. Neuro-Oncology. 2013;15:1457-68
100. Wu L, Zhao Z, Shin YJ, Yin Y, Raju A, Vaiyapuri TS. et al. Tumour microenvironment programming by an RNA-RNA-binding protein complex creates a druggable vulnerability in IDH-wild-type glioblastoma. Nat Cell Biol. 2024;26:1003-18
101. Ning H, Zhang L, Zhu B, Zhou X, Zhang T, Ma T. TARBP2-stablized SNHG7 regulates blood-brain barrier permeability by acting as a competing endogenous RNA to miR-17-5p/NFATC3 in Aβ-microenvironment. Cell Death Dis. 2022;13:457
102. Zhang M, Yang C, Ruan X, Liu X, Wang D, Liu L. et al. CPEB2 m6A methylation regulates blood-tumor barrier permeability by regulating splicing factor SRSF5 stability. Communications Biology. 2022;5:908
103. Venkatesh HS, Morishita W, Geraghty AC, Silverbush D, Gillespie SM, Arzt M. et al. Electrical and synaptic integration of glioma into neural circuits. Nature. 2019;573:539-45
104. Doron-Mandel E, Koppel I, Abraham O, Rishal I, Smith TP, Buchanan CN. et al. The glycine arginine-rich domain of the RNA-binding protein nucleolin regulates its subcellular localization. Embo J. 2021;40:e107158
105. Dankner M, Caron M, Al-Saadi T, Yu W, Ouellet V, Ezzeddine R. et al. Invasive growth associated with cold-inducible RNA-binding protein expression drives recurrence of surgically resected brain metastases. Neuro-Oncology. 2021;23:1470-80
106. Wang T, Zhang H. Exploring the roles and molecular mechanisms of RNA binding proteins in the sorting of noncoding RNAs into exosomes during tumor progression. J Adv Res. 2024;65:105-123
107. Fabbiano F, Corsi J, Gurrieri E, Trevisan C, Notarangelo M, D'Agostino VG. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? Journal of Extracellular Vesicles. 2020;10:e12043
108. Chen A-J, Paik J-H, Zhang H, Shukla SA, Mortensen R, Hu J. et al. STAR RNA-binding protein Quaking suppresses cancer via stabilization of specific miRNA. Genes Dev. 2012;26:1459-72
109. Li ZZ, Kondo T, Murata T, Ebersole TA, Nishi T, Tada K. et al. Expression of Hqk Encoding a KH RNA Binding Protein Is Altered in Human Glioma. Jpn J Cancer Res. 2002;93:167-77
110. Pan Z, Zhao R, Li B, Qi Y, Qiu W, Guo Q. et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol Cancer. 2022;21:16
111. Yin J, Ge X, Shi Z, Yu C, Lu C, Wei Y. et al. Extracellular vesicles derived from hypoxic glioma stem-like cells confer temozolomide resistance on glioblastoma by delivering miR-30b-3p. Theranostics. 2021;11:1763-79
112. Li Z, Meng X, Wu P, Zha C, Han B, Li L. et al. Glioblastoma Cell-Derived lncRNA-Containing Exosomes Induce Microglia to Produce Complement C5, Promoting Chemotherapy Resistance. Cancer Immunology Research. 2021;9:1383-99
113. Liu H-J, Xu P. Strategies to overcome/penetrate the BBB for systemic nanoparticle delivery to the brain/brain tumor. Advanced Drug Delivery Reviews. 2022;191:114619
114. Turco V, Pfleiderer K, Hunger J, Horvat NK, Karimian-Jazi K, Schregel K. et al. T cell-independent eradication of experimental glioma by intravenous TLR7/8-agonist-loaded nanoparticles. Nat Commun. 2023;14:771
115. Wei J, Wu D, Zhao S, Shao Y, Xia Y, Ni D. et al. Immunotherapy of Malignant Glioma by Noninvasive Administration of TLR9 Agonist CpG Nano-Immunoadjuvant. Advanced Science. 2022;9:2103689
116. Chidambaram A, Fillmore HL, Van Meter TE, Dumur CI, Broaddus WC. Novel report of expression and function of CD97 in malignant gliomas: correlation with Wilms tumor 1 expression and glioma cell invasiveness: Laboratory investigation. JNS. 2012;116:843-53
117. Izumoto S, Tsuboi A, Oka Y, Suzuki T, Hashiba T, Kagawa N. et al. Phase II clinical trial of Wilms tumor 1 peptide vaccination for patients with recurrent glioblastoma multiforme. JNS. 2008;108:963-71
118. Pirovano G, Jannetti SA, Carter LM, Sadique A, Kossatz S, Guru N. et al. Targeted Brain Tumor Radiotherapy Using an Auger Emitter. Clin Cancer Res. 2020;26:2871-81
119. Hong S. RNA Binding Protein as an Emerging Therapeutic Target for Cancer Prevention and Treatment. Journal of Cancer Prevention. 2017;22:203-10
120. Krämer OH. HDAC2: a critical factor in health and disease. Trends Pharmacol Sci. 2009;30:647-55
121. Vengoji R, Atri P, Macha MA, Seshacharyulu P, Perumal N, Mallya K. et al. Differential gene expression-based connectivity mapping identified novel drug candidate and improved Temozolomide efficacy for Glioblastoma. J Exp Clin Canc Res. 2021;40:335
122. Mladek AC, Yan H, Tian S, Decker PA, Burgenske DM, Bakken K. et al. RBBP4-p300 axis modulates expression of genes essential for cell survival and is a potential target for therapy in glioblastoma. Neuro-Oncology. 2022;24:1261-72
123. Alejo S, Palacios BE, Venkata PP, He Y, Li W, Johnson JD. et al. Lysine-specific histone demethylase 1A (KDM1A/LSD1) inhibition attenuates DNA double-strand break repair and augments the efficacy of temozolomide in glioblastoma. Neuro Oncol. 2023;25:1249-61
124. Chen X, Niu W, Fan X, Yang H, Zhao C, Fan J. et al. Oct4A palmitoylation modulates tumorigenicity and stemness in human glioblastoma cells. Neuro-Oncology. 2023;25:82-96
125. Argoetti A, Shalev D, Polyak G, Shima N, Biran H, Lahav T. et al. lncRNA NORAD modulates STAT3/STAT1 balance and innate immune responses in human cells via interaction with STAT3. Nat Commun. 2025;16:571
126. Chang N, Ahn SH, Kong D-S, Lee HW, Nam D-H. The role of STAT3 in glioblastoma progression through dual influences on tumor cells and the immune microenvironment. Mol Cell Endocrinol. 2017;451:53-65
127. Wang Y, Yang C, Sims MM, Sacher JR, Raje M, Deokar H. et al. SS-4 is a highly selective small molecule inhibitor of STAT3 tyrosine phosphorylation that potently inhibits GBM tumorigenesis in vitro and in vivo. Cancer Lett. 2022;533:215614
128. Zhang L, Nesvick CL, Day CA, Choi J, Lu VM, Peterson T. et al. STAT3 is a biologically relevant therapeutic target in H3K27M-mutant diffuse midline glioma. Neuro-Oncology. 2022;24:1700-11
129. Sweha SR, Chung C, Natarajan SK, Panwalkar P, Pun M, Ghali A. et al. Epigenetically defined therapeutic targeting in H3.3G34R/V high-grade gliomas. Sci Transl Med. 2021;13:eabf7860
130. Wang J, Hjelmeland AB, Nabors LB, King PH. Anti-cancer effects of the HuR inhibitor, MS-444, in malignant glioma cells. Cancer Biol Ther. 2019;20:979-88
131. Wang X, Han M, Wang S, Sun Y, Zhao W, Xue Z. et al. Targeting the splicing factor NONO inhibits GBM progression through GPX1 intron retention. Theranostics. 2022;12:5451-69
132. Bhutada I, Khambati F, Cheng S-Y, Tiek DM, Duckett D, Lawrence H. et al. CDK7 and CDK9 inhibition interferes with transcription, translation, and stemness, and induces cytotoxicity in GBM irrespective of temozolomide sensitivity. Neuro-Oncology. 2023 noad143
133. Lafita-Navarro MC, Venkateswaran N, Kilgore JA, Kanji S, Han J, Barnes S. et al. Inhibition of the de novo pyrimidine biosynthesis pathway limits ribosomal RNA transcription causing nucleolar stress in glioblastoma cells. Khagi S, Ed. Plos Genet. 2020;16:e1009117
134. Motta C, D'Angeli F, Scalia M, Satriano C, Barbagallo D, Naletova I. et al. PJ-34 inhibits PARP-1 expression and ERK phosphorylation in glioma-conditioned brain microvascular endothelial cells. Eur J Pharmacol. 2015;761:55-64
135. Porčnik A, Novak M, Breznik B, Majc B, Hrastar B, Šamec N. et al. TRIM28 Selective Nanobody Reduces Glioblastoma Stem Cell Invasion. Molecules. 2021;26:5141
136. Zhang Z-W, Teng X, Zhao F, Ma C, Zhang J, Xiao L-F. et al. METTL3 regulates m6A methylation of PTCH1 and GLI2 in Sonic hedgehog signaling to promote tumor progression in SHH-medulloblastoma. Cell Reports. 2022;41:111530
137. Shahab SW, Roggeveen CM, Sun J, Kunhiraman H, McSwain LF, Juraschka K. et al. The LIN28B -let-7- PBK pathway is essential for group 3 medulloblastoma tumor growth and survival. Mol Oncol. 2023;17:1784-802
138. Bellail AC, Jin HR, Lo H-Y, Jung SH, Hamdouchi C, Kim D. et al. Ubiquitination and degradation of SUMO1 by small-molecule degraders extends survival of mice with patient-derived tumors. Sci Transl Med. 2021;13:eabh1486
139. Shu B, Zeng P, Kang S, Li P-H, Hu D, Kuang G. et al. Syntheses and evaluation of new Quinoline derivatives for inhibition of hnRNP K in regulating oncogene c-myc transcription. Bioorg Chem. 2019;85:1-17
140. Marian CO, Cho SK, Mcellin BM, Maher EA, Hatanpaa KJ, Madden CJ. et al. The Telomerase Antagonist, Imetelstat, Efficiently Targets Glioblastoma Tumor-Initiating Cells Leading to Decreased Proliferation and Tumor Growth. Clinical Cancer Research. 2010;16:154-63
141. Bertrand Y, Currie J-C, Poirier J, Demeule M, Abulrob A, Fatehi D. et al. Influence of glioma tumour microenvironment on the transport of ANG1005 via low-density lipoprotein receptor-related protein 1. Br J Cancer. 2011;105:1697-707
142. Ghidini A, Cléry A, Halloy F, Allain FHT, Hall J. RNA-PROTACs: Degraders of RNA-Binding Proteins. Angew Chem Int Ed. 2021;60:3163-9
143. Kumar S, Fry LE, Wang J-H, Martin KR, Hewitt AW, Chen FK. et al. RNA-targeting strategies as a platform for ocular gene therapy. Prog Retin Eye Res. 2023;92:101110
144. Crooke ST, Liang X, Crooke RM, Baker BF, Geary RS. Antisense drug discovery and development technology considered in a pharmacological context. Biochem Pharmacol. 2021;189:114196
145. Goenka A, Song X, Tiek D, Iglesia RP, Lu M, Zeng C. et al. Oncogenic long noncoding RNA LINC02283 enhances PDGF receptor A-mediated signaling and drives glioblastoma tumorigenesis. Neuro-Oncology. 2023 noad065
146. Zhang Q, Yang L, Liu YH, Wilkinson JE, Krainer AR. Antisense oligonucleotide therapy for H3.3K27M diffuse midline glioma. Sci Transl Med. 2023;15:eadd8280
147. Denichenko P, Mogilevsky M, Cléry A, Welte T, Biran J, Shimshon O. et al. Specific inhibition of splicing factor activity by decoy RNA oligonucleotides. Nat Commun. 2019;10:1590
148. Fang Y, Wu Q, Wang F, Liu Y, Zhang H, Yang C. et al. Aptamer-RIBOTAC Strategy Enabling Tumor-Specific Targeted Degradation of MicroRNA for Precise Cancer Therapy. Small Methods. 2024 2400349
149. Brooks SA. Characterization of the mRNA ligands bound by the RNA binding protein hnRNP A2 utilizing a novel in vivo technique. Nucleic Acids Research. 2000;28:49e-49
150. Lambert N, Robertson A, Jangi M, McGeary S, Sharp PA, Burge CB. RNA Bind-n-Seq: Quantitative Assessment of the Sequence and Structural Binding Specificity of RNA Binding Proteins. Molecular Cell. 2014;54:887-900
151. Lapointe CP, Wilinski D, Saunders HAJ, Wickens M. Protein-RNA networks revealed through covalent RNA marks. Nat Methods. 2015;12:1163-70
152. Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP Identifies Nova-Regulated RNA Networks in the Brain. Science. 2003;302:1212-5
153. Maurin T, Lebrigand K, Castagnola S, Paquet A, Jarjat M, Popa A. et al. HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res. 2018;46:6344-55
154. Wu S-L, Fu X, Huang J, Jia T-T, Zong F-Y, Mu S-R. et al. Genome-wide analysis of YB-1-RNA interactions reveals a novel role of YB-1 in miRNA processing in glioblastoma multiforme. Nucleic Acids Res. 2015;43:8516-28
155. Han W, Wang S, Qi Y, Wu F, Tian N, Qiang B. et al. Targeting HOTAIRM1 ameliorates glioblastoma by disrupting mitochondrial oxidative phosphorylation and serine metabolism. iScience. 2022;25:104823
156. Kloetgen A, Duggimpudi S, Schuschel K, Hezaveh K, Picard D, Schaal H. et al. YBX1 Indirectly Targets Heterochromatin-Repressed Inflammatory Response-Related Apoptosis Genes through Regulating CBX5 mRNA. International Journal of Molecular Sciences. 2020;21:4453
157. Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M. et al. The mRNA-Bound Proteome and Its Global Occupancy Profile on Protein-Coding Transcripts. Mol Cell. 2012;46:674-90
158. Castello A, Horos R, Strein C, Fischer B, Eichelbaum K, Steinmetz LM. et al. System-wide identification of RNA-binding proteins by interactome capture. Nat Protoc. 2013;8:491-500
159. Perez-Perri JI, Noerenberg M, Kamel W, Lenz CE, Mohammed S, Hentze MW. et al. Global analysis of RNA-binding protein dynamics by comparative and enhanced RNA interactome capture. Nat Protoc. 2021;16:27-60
160. Armaos A, Colantoni A, Proietti G, Rupert J, Tartaglia GG. cat RAPID omics v2.0 : going deeper and wider in the prediction of protein-RNA interactions. Nucleic Acids Res. 2021;49:W72-9
161. Su R, Fan L-H, Cao C, Wang L, Du Z, Cai Z. et al. Global profiling of RNA-binding protein target sites by LACE-seq. Nat Cell Biol. 2021;23:664-75
162. Nag S, Goswami B, Das Mandal S, Ray PS. Cooperation and competition by RNA-binding proteins in cancer. Semin Cancer Biol. 2022;86:286-97
163. Ziff OJ, Harley J, Wang Y, Neeves J, Tyzack G, Ibrahim F. et al. Nucleocytoplasmic mRNA redistribution accompanies RNA binding protein mislocalization in ALS motor neurons and is restored by VCP ATPase inhibition. Neuron. 2023;111:3011-3027.e7
164. Yan J, Kang DD, Turnbull G, Dong Y. Delivery of CRISPR-Cas9 system for screening and editing RNA binding proteins in cancer. Adv Drug Delivery Rev. 2022;180:114042
165. Chow RD, Guzman CD, Wang G, Schmidt F, Youngblood MW, Ye L. et al. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat Neurosci. 2017;20:1329-41
166. Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I. et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46:D794-801
167. Kuret K, Amalietti AG, Jones DM, Capitanchik C, Ule J. Positional motif analysis reveals the extent of specificity of protein-RNA interactions observed by CLIP. Genome Biol. 2022;23:191
168. Ren Y, Liao H, Yan J, Lu H, Mao X, Wang C. et al. Capture of RNA-binding proteins across mouse tissues using HARD-AP. Nat Commun. 2024;15:8421
169. Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol. 2015;33:831-8
170. Pan X, Rijnbeek P, Yan J, Shen H-B. Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks. BMC Genomics. 2018;19:511
171. Song J, Tian S, Yu L, Xing Y, Yang Q, Duan X. et al. AC-Caps: Attention Based Capsule Network for Predicting RBP Binding Sites of LncRNA. Interdiscip Sci Comput Life Sci. 2020;12:414-23
172. Brannan KW, Chaim IA, Marina RJ, Yee BA, Kofman ER, Lorenz DA. et al. Robust single-cell discovery of RNA targets of RNA-binding proteins and ribosomes. Nat Methods. 2021;18:507-19
173. Navaratnam N, Morrison JR, Bhattacharya S, Patel D, Funahashi T, Giannoni F. et al. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J Biol Chem. 1993;268:20709-12
174. Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420-4
175. Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q. et al. The RNA m6A Reader YTHDF2 Maintains Oncogene Expression and Is a Targetable Dependency in Glioblastoma Stem Cells. Cancer Discov. 2021;11:480-99
176. Perr J, Langen A, Almahayni K, Nestola G, Chai P, Lebedenko CG. et al. RNA-binding proteins and glycoRNAs form domains on the cell surface for cell-penetrating peptide entry. Cell. 2025 S0092867425001096
177. Tian R, Li Y, Liu Q, Shu M. Identification and Validation of an Immune-Associated RNA-Binding Proteins Signature to Predict Clinical Outcomes and Therapeutic Responses in Glioma Patients. Cancers. 2021;13:1730
178. Wei L, Zou C, Chen L, Lin Y, Liang L, Hu B. et al. Molecular Insights and Prognosis Associated With RBM8A in Glioblastoma. Front Mol Biosci. 2022;9:876603
179. Elcheva IA, Gowda CP, Bogush D, Gornostaeva S, Fakhardo A, Sheth N. et al. IGF2BP family of RNA-binding proteins regulate innate and adaptive immune responses in cancer cells and tumor microenvironment. Front Immunol. 2023;14:1224516
180. Shao W, Zhao H, Zhang S, Ding Q, Guo Y, Hou K. et al. A pan-cancer landscape of IGF2BPs and their association with prognosis, stemness and tumor immune microenvironment. Front Oncol. 2023;12:1049183
181. Tong Y, Lee Y, Liu X, Childs-Disney JL, Suresh BM, Benhamou RI. et al. Programming inactive RNA-binding small molecules into bioactive degraders. Nature. 2023;618:169-79
182. Julio AR, Backus KM. New approaches to target RNA binding proteins. Curr Opin Chem Biol. 2021;62:13-23
183. Hasegawa M, Matsushita Y, Horikawa M, Higashi K, Tomigahara Y, Kaneko H. et al. A novel inhibitor of Smad-dependent transcriptional activation suppresses tissue fibrosis in mouse models of systemic sclerosis. Arthritis & Rheumatism. 2009;60:3465-75
184. Paunovska K, Loughrey D, Dahlman JE. Drug delivery systems for RNA therapeutics. Nat Rev Genet. 2022;23:265-80
185. Garrelfs SF, Frishberg Y, Hulton SA, Koren MJ, O'Riordan WD, Cochat P. et al. Lumasiran, an RNAi Therapeutic for Primary Hyperoxaluria Type 1. N Engl J Med. 2021;384:1216-26
186. You Q, Liang F, Wu G, Cao F, Liu J, He Z. et al. The Landscape of Biomimetic Nanovesicles in Brain Diseases. Adv Mater. 2024;36:2306583
187. Sun W, Cui H, Xu T, Yue J, Liang J, You W. et al. RNA binding proteins in extracellular vesicles and their potential value for cancer diagnosis and treatment (Review). Int J Oncol. 2023;63:114
188. Leidal AM, Huang HH, Marsh T, Solvik T, Zhang D, Ye J. et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat Cell Biol. 2020;22:187-99
189. Zhao J, Li Q, Hu J, Yu H, Shen Y, Lai H. et al. Circular RNA landscape in extracellular vesicles from human biofluids. Genome Med. 2024;16:126
Author contact
![]() Corresponding authors: Department of Neurosurgery, Qilu Hospital, Cheeloo College of Medicine and Institute of Brain and Brain-Inspired Science, Shandong University, Jinan, China. Email: Dr. Anjing Chen: chenajedu.cn; Department of Neurosurgery, Beijing Tiantan Hospital, Capital Medical University, Beijing, China. Email: Dr. Wang Jia: jwttyycom.
Corresponding authors: Department of Neurosurgery, Qilu Hospital, Cheeloo College of Medicine and Institute of Brain and Brain-Inspired Science, Shandong University, Jinan, China. Email: Dr. Anjing Chen: chenajedu.cn; Department of Neurosurgery, Beijing Tiantan Hospital, Capital Medical University, Beijing, China. Email: Dr. Wang Jia: jwttyycom.
 Global reach, higher impact
Global reach, higher impact