13.3
Impact Factor
Theranostics 2025; 15(8):3289-3315. doi:10.7150/thno.103983 This issue Cite
Review
Engineered Probiotic-Based Biomaterials for Inflammatory Bowel Disease Treatment
1. Department of Inorganic Chemistry, School of Pharmacy, Naval Medical University, Shanghai, 200433, P. R. China.
2. School of Medicine, Shanghai University, Shanghai, 200444, P. R. China.
3. Materdicine lab, School of Life Sciences, Shanghai University, Shanghai, 200444, P. R. China.
#These authors contributed equally.
Received 2024-9-21; Accepted 2024-12-9; Published 2025-2-18
Abstract
Inflammatory bowel disease (IBD) is a chronic condition affecting the intestines, marked by immune-mediated inflammation. This disease is known for its recurrent nature and the challenges it presents in treatment. Recently, probiotic have gained attention as a promising alternative to traditional small molecular drugs and monoclonal antibody chemotherapies for IBD. Probiotic, recognized as a “living” therapeutic agent, offers targeted treatment with minimal side effects and the flexibility for biological modifications, making them highly effective for IBD management. This comprehensive review presents the latest advancements in engineering probiotic-based materials, ranging from basic treatment mechanisms to the modification techniques used in IBD management. It delves deep into how probiotic produces therapeutic effects in the intestinal environment and discusses various strategies to enhance probiotic's efficacy, including genetic modifications and formulation improvements. Additionally, the review addresses the challenges, practical application conditions, and future research directions of probiotic-based therapies in IBD treatment, providing insights into their feasibility and potential clinical implications.
Keywords: engineered probiotic-based materials, living materials, inflammatory bowel disease, probiotics
1. Introduction
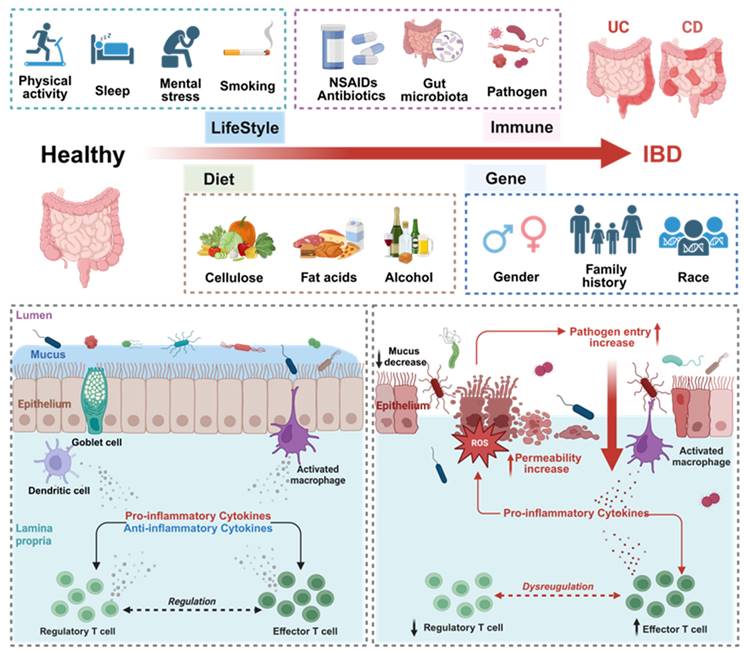
Inflammatory bowel disease (IBD) is an immune-mediated and chronic intestinal disorder, encompassing conditions such as Crohn's disease (CD) and ulcerative colitis (UC), both of which have similar clinical manifestations, mainly including abdominal pain, diarrhea, bloody stools, weight loss, anemia, and low-grade fever. Currently, the exact cause of IBD is still unclear, but it is commonly associated with a combination of lifestyle, behavioral habits, immune system functionality, and environmental and genetic factors (Figure 1) [1]. The current mainstream pharmacological interventions of IBD treatment include non-steroidal anti-inflammatory drugs (5-aminosalicylic acid), corticosteroids (budesonide, prednisone and prednisolone), monoclonal antibodies (adalimumab, infliximab), Janus kinase inhibitors (tofacitinib, upadacitinib), immunosuppressants (methotrexate, cyclosporine, tacrolimus). As the emerging evidence highlights a strong correlation between gut dysbiosis and IBD, antibiotics are now being employed to reestablish a balanced gut microbiome environment [2]. However, existing drug therapies fall short of fully curing IBD, and prolonged usage often results in significant adverse reactions [3,4]. In addition, while fecal microbiota transplantation is a burgeoning therapeutic technique, our grasp of its potential remains nascent. The evolution of individualized intestinal microbiota may offer a more hopeful avenue in the future [5]. Recent studies on the influence of intestinal microorganisms on the dynamic balance mechanism of intestinal mucosa is gradually increasing. The dysbiosis of intestinal microbiota is closely associated with the normal function of the superficial barrier of the colon and the occurrence and development of IBD. Therefore, the potential of probiotics to modulate gut microbiota, bolster the immune system, and rejuvenate epithelium barrier function points towards novel avenues in the diagnosis and treatment of IBD [6].
Comparison of intestinal homeostasis in healthy and IBD conditions and possible pathogenic factors. The current research results on the pathogenesis of IBD are still unclear, and it is generally believed to be related to individual lifestyle habits, immune status, dietary habits, and genetic background. In healthy organisms, the intestinal epithelium maintains a state of immune tolerance. However, upon exposure to inflammatory stimuli, the protective mucus layer and the epithelial cell barrier can become compromised. This leads to abnormal stimulation of immune cells by immunogenic substances, triggering a local immune response. If the antigens are not promptly eliminated, inflammation ensues, further damaging the intestinal barrier. This creates a vicious cycle, perpetuating long-term intestinal inflammation. Created with https://BioRender.com.
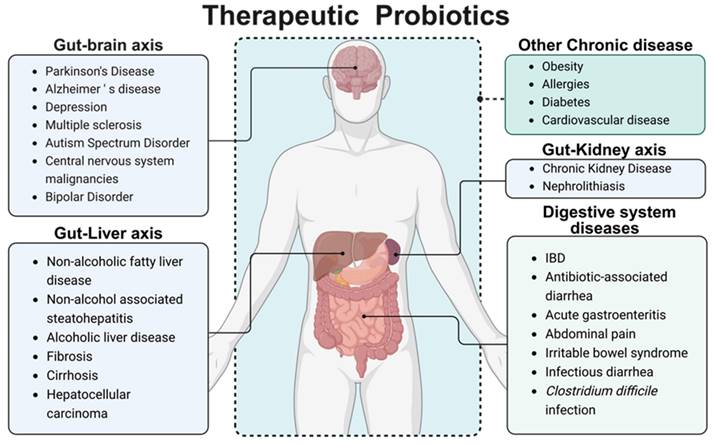
Probiotics are defined as live microorganisms that are beneficial to the human body when consumed in sufficient amounts [7]. Probiotics mainly include two categories, Lactobacillus and Bifidobacterium. In addition, based on their benefits to the human body, some Gram-positive bacteria (such as Lactococcus lactis, Streptococcus thermophilus, Enterococcus faecalis and Bacillus licheniformis) and yeasts (such as Saccharomyces boulardii) are also classified as probiotics. The benefits provided by different probiotic strains vary significantly, with distinct mechanisms underlying each strain's action. Given the variability in gut microbiota composition between individuals, the efficacy of a particular probiotic strain can differ substantially. Current research suggests that probiotics residing in the human gut exert critical regulatory effects on immune function, the gut microbiota environment, intestinal barrier integrity, and even the function of other organs. These effects are mediated through probiotic-host interactions and the secretion of enzymes, organic acids, and small bioactive molecules [8]. It has been widely used in the treatment of abdominal pain, diarrhea, hypertension, diabetes, and other diseases. Given the unique intestinal colonization characteristics of certain bacteria, utilizing specific probiotic strains as biological drug delivery carriers can partially circumvent the limitations associated with traditional drug delivery methodologies in IBD treatment, such as systemic adverse effects and poor drug targeting [9], thereby demonstrating substantial therapeutic potential for the healing of intestinal disease [10]. Subsequent research has illuminated probiotics' capacity to modulate immune responses, maintain intestinal flora equilibrium, and yield pronounced effects in treating acute gastroenteritis [11], IBD [12], antibiotic-associated diarrhea [13], and diarrhea in children with abdominal pain [14], as well as irritable bowel syndrome [15] and Clostridium difficile infection [16]. Moreover, probiotics' therapeutic potential extends to some central nervous system diseases, liver and kidney diseases through the gut-organ axis pathway [17-19]; meanwhile, probiotic have certain therapeutic effects for other common chronic disease like obesity [20], allergies [21], diabetes [22], and cardiovascular disease (Figure 2) [23].
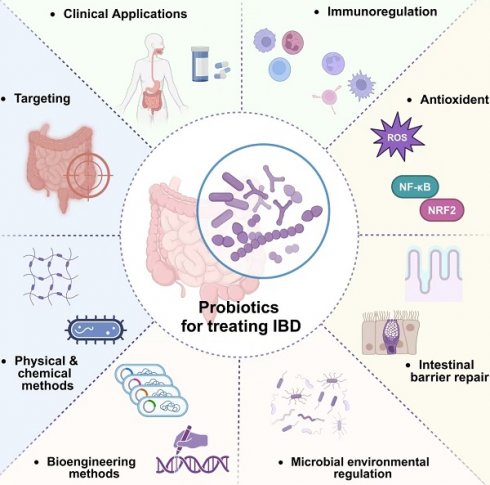
In this review, we delve into the cellular-level pathological alterations of IBD and discuss factors that could influence the disease. Our primary focus lies on probiotics, exploring their therapeutic mechanism in IBD treatment. The intricate interplay between the host organism and probiotic is discussed, with a focus on immunomodulation, antioxidants, intestinal barrier repair, and microbial environmental regulation. Further, we discuss the prevailing targeting strategies and standard methods for the multifunctional transformation of probiotic-based materials. Additionally, we present an in-depth analysis of two well-researched commercial probiotic formulations: VSL#3® and LGG®. The review culminates with an exposition of recent applications of probiotics and prospective trajectories for probiotics within the domain of IBD treatment and beyond, casting a scientific lens on future directions and potential innovations.
2. Treatment mechanism
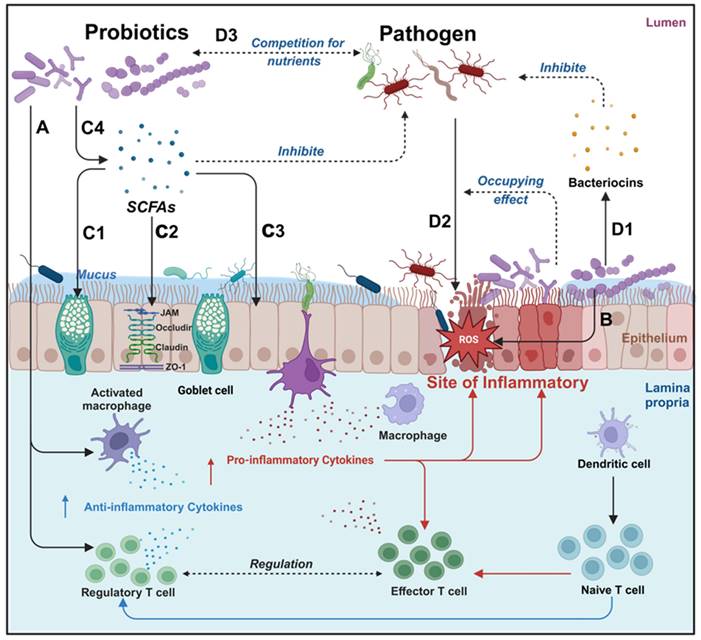
Probiotics manifest a spectrum of health benefits, contingent upon specific strains and the prevailing conditions within the gut microbiota. Probiotics can affect the human body through various mechanisms, including enhancement of digestion, nutrient secretion, modulation of neurotransmitter release, attenuation of pathogenic bacteria virulence, reduction in intestinal adhesion of pathogenic bacteria, synthesis of anti-inflammatory factors, neutralization of pro-inflammatory factors, facilitation of intestinal barrier restoration and modulation of intestinal flora [24]. As shown in Figure 3, probiotics have demonstrated substantial therapeutic potentials in mitigating intestinal inflammation, exhibiting antioxidants, promoting intestinal barrier restoration, and regulating intestinal flora (Table 1).
Therapeutic probiotics. Intestinal microbiota mainly regulates other organs and systems of the body through metabolic pathways (such as secretion of SCFAs, bacteriocins, etc.), immune pathways (stimulating the intestinal tract to maintain normal immune homeostasis and regulate immune factor levels), and neural pathways (affecting neurotransmitter levels). Multiple pathways are mixed with each other. Dietary intervention or exogenous supplementation of probiotics, prebiotics, or synbiotics can help to achieve adjuvant therapeutic effects on various diseases. Created with https://BioRender.com.
Overview of the main mechanism of probiotics in the treatment of IBD. Probiotics have various therapeutic effects on IBD, different probiotics may have different degrees of therapeutic effects, it can be roughly divided into immune regulation, antioxidant, anti-inflammatory, repairing intestinal barriers, and helping the body resist pathogenic microorganisms. A: Immunomodulation: promotion of anti-inflammatory factor expression and inhibition of pro-inflammatory factor expression; B: Neutralization of ROS; C: Probiotics can upregulate mucus protein secretion by goblet cells (C1), enhance tight junction protein function (C2), and intestinal epithelial cell function (C3) by secreting SCFAs (C4); D: Probiotics can inhibit pathogenic bacterium growth by the Secretion of bacteriocins (D1) and reduce pathogenic bacteria adhesion through occupancy effect (D2). Created with https://BioRender.com.
2.1. Immunoregulation
Contemporary research posits a connection between both innate immune deficiencies and aberrations in adaptive immunity with IBD, albeit with the specifics remaining elusive. A plethora of immune alterations in IBD patients defy clear categorization as causes or effects of the disease [25,26]. An upregulation of toll-like receptor 2 (TLR2) and TLR4 expression has been documented in active IBD cases [27], correlating with intestinal inflammatory infiltrates associated with helper T cells (Th cells), and regulatory T cells (Treg cells) [28]. Furthermore, an elevation in antimicrobial antibodies in IBD patients indicates the stimulating effect of gut microbiota on the immune system and its associated pathologies [29]. Inflammatory mediators hold a pivotal role in the progression of IBD.
Several studies conducted on probiotics have demonstrated their capacity to modulate TLR signaling and Th cell differentiation to Th1, Th2, and Th17 effector cells. This modulation results in a downregulation of pro-inflammatory factors [30], concurrently uplifting the levels of anti-inflammatory factors, thereby ameliorating symptoms of intestinal inflammation [31]. The current research results indicate that probiotics can exert various regulatory effects on the immune system, mainly by regulating immune cells, inflammatory pathways, and immune mediators. Depending on the difference of strain, probiotic strains [32-35], surface layer proteins (Slp) [36-39], secretions [40], and extracellular vesicles [41,42] have all shown instances of immune regulation. Moreover, probiotics have been developed as efficient platforms for genetic engineering [43]. Recombinant strains have been engineered to synthesize and secrete a range of anti-inflammatory factors, as well as neutralizing antibodies targeting pro-inflammatory factors [30,44-46], highlighting their significant potential in the treatment of IBD. Dou et al. demonstrated that oral administration of the Lactobacillus casei ATCC 393 strain and its metabolites significantly alleviated dextran sulfate sodium (DSS)-induced UC symptoms in mice, concurrently reducing inflammatory cytokine levels and immune cell infiltration [47]. Hao et al. revealed that the anti-inflammatory effect of Lactobacillus plantarum Q7 is achieved through its secreted extracellular vesicles. After oral administration of extracellular vesicles, the pro-inflammatory factors in the serum were significantly reduced [42]. Chandhni et al. extracted surface proteins from Lactobacillus plantarum MTCC 5690, Lactobacillus fermentum MTCC 568, and Lactobacillus acidophilus NCFM, revealing their respective degrees of anti-inflammatory effects [37]. Luerche et al. emphasized the ability of Lactococcus lactis NCDO 2118 to increase the number of Treg cells and anti-inflammatory dendritic cells (DCs), inhibit interleukin-1β (IL-1β)-induced secretion of interleukin-8 (IL-8) by caco-2 cells, and demonstrate its anti-inflammatory activity in vitro cultured intestinal epithelial cells and DSS-induced UC mice models [48].
Summary of probiotics for treating IBD.
| Strains | Therapeutic agents | Model | Anti-inflammatory | Antioxident | Mucus/Goblet cells | Epithelial cells/TJs | SCFAs | Microbial regulation | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| L. acidophilus;L. helveticus;L. plantarum; | strain | DSS-C57BL/6 | + | + | + | + | + | + | [30] |
| L. plantarum 22A-3 | strain | DSS-C57BL/6 | + | [31] | |||||
| L. paracasei BNCC345679 | strain | DSS-C57BL/6 | + | + | + | + | [32] | ||
| L. plantarum MKMB01/02 | strain | HT-29, caco-2 and THP-1 cells | + | + | + | [33] | |||
| L. acidophilus LA1 | strain | DSS-C57BL/6 | + | + | + | [34] | |||
| B. breve FPHC4024;L. reuteri FPHC2951; | strain | DSS-C57BL/6J | + | + | + | [35] | |||
| L. acidophilus NCFM | Slp | LPS-RAW264.7 cells | + | + | [36] | ||||
| L. acidophilus NCFM | Slp | DSS-C57BL/6;TNBS-C57BL/6 | + | [37] | |||||
| A. muciniphila | Slp | HFD-C57BL/6 | + | [38] | |||||
| A. muciniphila | Slp | CTL and CT26 Cells;DSS-C57BL/6J | + | [39] | |||||
| L. plantarum AKU1009a | derivant | ICR | + | + | [40] | ||||
| L. plantarum BMCM12 | derivant | Type II mucin | + | [41] | |||||
| Lactobacillus plantarum Q7 | Extracellular vesicles | DSS-C57BL/6J | + | + | + | + | [42] | ||
| L. lactis NCDO 2118 | strain | DSS-C57BL/6;Colonic cells | + | + | [48] | ||||
| L. casei Shirota | strain | 2,2′-Azobis (2-amidinopropane) dihydrochloride-Caco-2/TC7 cells | + | + | + | [55] | |||
| L. plantarum ZS62 | strain | DSS-C57BL/6 | + | + | [58] | ||||
| L. plantarum FC225 | strain | ICR | + | [65] | |||||
| B. bifidum | strain | caco-2 cells;DSS-C57BL/6 | + | + | + | [75] | |||
| B. pseudocatenulatum | strain | DSS-C57BL/6J | + | + | + | + | [76] | ||
| B. bifidum | strain | caco-2 cells;DSS-C57BL/6 | + | + | + | + | [77] | ||
| L. gasseri ATCC33323 | strain | DSS-C57BL/6 | + | + | + | [78] | |||
| L. acidophilus CMUL067 | strain | TNBS-BALB/c ByJ | + | + | [79] | ||||
| L. Plantarum | Micro integral membrane protein | DSS-C57BL/6 | + | + | + | + | [80] | ||
| L. plantarum HNU082 | strain | DSS-C57BL/6 | + | + | + | + | + | [95] | |
| L. casei;L. plantarum;L. rhamnosus; | strain | HT-29 cells | + | [109] |
Schematic illustration of complex oxidant interaction mechanism between human cell and probiotics, highlighting the intricate interplay involving ROS. An increase in ROS can directly oxidize Keap1 protein, resulting in the escape of NRF2 and NF-κB from the inhibition of Keap1, entering the nucleus, and initiating an antioxidant stress response. Within the nuclear domain, NRF2 binds to ARE, which can start the transcription of a series of antioxidant enzymes and detoxification enzymes. This transcriptional activity culminates in a reduced production of ROS and mitigation of oxidative damage, thus inhibiting activation of NF-κB and production of inflammatory factors. Conversely, NF-κB can enters the nucleus and binds to the NRF2 promoter region, inhibiting the transcription of NRF2. Upon interaction with ARE, NF-κB will activate the transcription of inflammatory factors and immune related genes, thereby triggering inflammation and immune responses. Secretions from specific strain promote NRF2 pathway expression, while their secreted antioxidant enzymes synergistically enhance cellular antioxidant capacity. Created with https://BioRender.com.
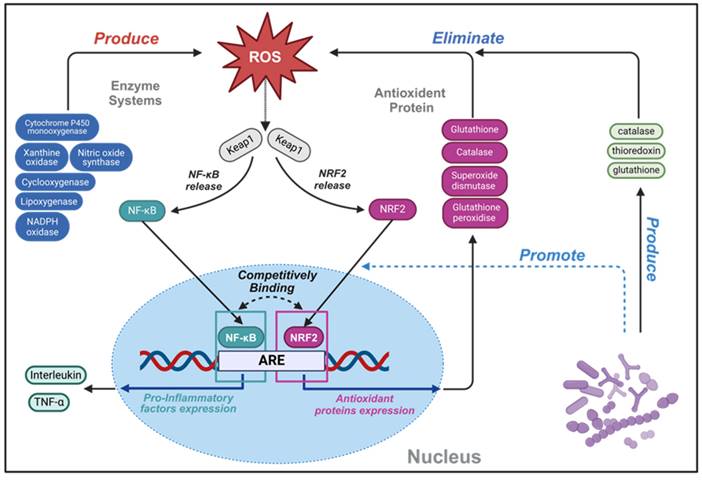
2.2. Antioxidant
Reactive oxygen species (ROS), predominantly arising as metabolic byproducts from mitochondria, peroxisomes and xanthine oxidase, consisting of radicals and nonradical derivatives of oxygen [49]. Although ROS are crucial at moderate levels for maintaining immune function, defending against infection, and synthesizing thyroid hormone, a delicate balance is required to sustain normal cellular functions. Disruptions in ROS homeostasis may cause cellular injury, potentially culminating in disease and caducity [50]. In the intestinal environment, an optimal concentration of ROS is pivotal for stimulating the proliferation and renewal of intestinal epithelial cells, maintain normal intestinal barrier function. Phagocytes, mediated by formyl peptide receptors (FPRs), internally produce high levels of ROS, crucial for the eradication of pathogenic microorganisms [51]. ROS have been implicated in the etiology of inflammation, numbers of factors including intestinal epithelial cells alteration, genetic susceptibility, unregulated mucosal immune system, intestinal microflora dysbiosis, environmental factors, contribute to the development of IBD by influencing the levels of ROS. Furthermore, prolonged oxidative stress exposure can lead to sustained high-level DNA damage, which probably plays a key role in the pathogenesis of IBD [52]. During the inflammatory progress, contact with gut immunogen stimulates intestinal epithelial cells, neutrophils and macrophages can produce ROS, inflicting damage on the intestine barrier. This disruption facilitates further antigen interactions with gut-associated lymphoid tissue and resident immune cells, escalating the secretion of pro-inflammatory cytokines and recruiting additional immune cells. Without timely intestinal barrier restoration, escalating ROS levels perpetuate cellular damage, fueling a vicious inflammatory cycle. Typically, the antioxidant mechanism of probiotic can be broadly categorized into two classes: the transcriptional upregulation of antioxidant proteins and the modulation of inflammatory signaling pathways. These multifaceted strategies underscore the therapeutic potential of probiotics in mitigating oxidative stress and interrupting the inflammatory cascade characteristic of IBD (Figure 4).
2.2.1. Transcription and expression of antioxidant proteins
The antioxidant defense mechanism within probiotics encompasses three predominant enzyme-related antioxidant systems: catalase (CAT), thioredoxin-dependent and glutathione-dependent antioxidant systems [53]. Depend on their specific living environment, various strains have evolved many antioxidant proteins to combat oxidative stress. These mainly include thioredoxin, thioredoxin reductase, CAT, heme-dependent CAT [54], which provide extra competitive advantage compared to other bacteria. In addition to the intrinsic antioxidant capabilities of bacteria, they are known to promote the host expression level of antioxidant protein-related genes [55]. On the other hand, the introduction of exogenous genes coding for antioxidant enzymes into strains can also enhance the antioxidant capacity of bacteria. Carmen et al. demonstrated that oral administration of recombinant superoxide dismutase (SOD) and CAT-producing Streptococcus thermophilus CRL 807 exhibit enhanced anti-inflammatory activities on trinitrobenzene sulfonic acid (TNBS)-induced colonic tissue injury mouse model [56]. Watterlot et al. revealed that intragastric administration of a SOD-producing recombinant Lactobacillus casei BL23 performed better anti-inflammation effects compared to both the standard BL23 strain and SOD treatment alone in DSS-induced colitis mouse model [57]. Moreover, research conducted by Pan et al. indicated that Lactobacillus plantarum ZS62 was capable of upregulating the expression levels of SOD and CAT, thereby preventing and alleviating the symptoms of DSS-induced IBD in mouse model [58].
2.2.2. NRF2 and NF-κB signal pathway
Probiotics employ additional antioxidant mechanisms through modulating the ROS signaling pathways nuclear factor erythroid 2-related factor 2 (NRF2) and nuclear factor kappa-B (NF-κB), both of which play critical roles in the emergence, development, and resolution of ROS-induced inflammation, as depicted in Figure 4 (which delineates the interplay between ROS, NRF2, and NF-κB signaling pathways) [59]. In the nucleus, NRF2 binds to antioxidant response element (ARE) sequences, thereby promoting the transcription of genes, including those encoding the antioxidative enzymes previously described. The NRF2 pathway encompasses a comprehensive set of genes related to ROS detoxification and pro-restitutive function and is recognized as the major regulatory mechanism of organisms to resist environmental oxidative stresses [59,60]. Notably, NRF2-deficient mice exhibit heightened sensitivity to DSS, illustrating the pathway's protective role in colitis models [61].
Conversely, NF-κB represents a pivotal mediator in regulating immune and inflammatory processes, with its activation by ROS leading to the production of pro-inflammatory factors. The abnormal regulation of NF-κB can result in immune dysfunction and inflammatory disease [62]. NF-κB played a protective role in maintaining the integrity of the intestinal barrier and immune homeostasis in IBD patients [63]. The complex relationship of the crosstalk NRF2 and NF-κB signal pathway is highlighted by studies reporting both synergistic and antagonistic interactions between them [64]. Furumoto et al. identified that 10-Oxo-trans-11-octadecenoic acid produced by Lactobacillus plantarum AKU1009a was able to significantly increase the level of NRF2 protein in HepG2 cells and promote the gene expression of a series of related antioxidant enzymes, a phenomenon also observed in mouse organs treated with oral administration of this substance [40]. Another strain, Lactobacillus plantarum FC225, isolated from fermented cabbages, stimulated antioxidative enzyme gene expression in high-fat diet mice via NRF2-dependent transcriptional activation of ARE sites, enhancing the enzyme activities of SOD and glutathione peroxidase (GSH-px) [65].
The Slp of Lactobacillus acidophilus NCFM has been shown to inhibit lipopolysaccharide-induced inflammation through mitogen-activated protein kinase (MAPK) and NF-κB signaling pathways in RAW264.7 cells [36]. Additionally, Lactobacillus casei Shirota has demonstrated the ability to adhere to differentiated intestinal cell-like caco-2 cells, reducing cell damage induced by oxidation of 2,2'-azobis (2-aminopropane) dihydrochloride and inflammatory stress by inhibiting the NF-κB inflammatory pathway [55].
2.3. Intestinal barrier repair
The intestinal barrier of the gastrointestinal tract demarcates human body from external environment, establishing a crucial platform for normal substance exchange between the body and the outside world. This barrier is comprised of mucus, epithelial cells, lamina propria. The development of IBD is often accompanied by a continuous impairment of intestinal barrier function and an increase in intestinal permeability [66]. Pathogens can secrete enzymes to decompose tight junction (TJ) proteins between cells to increase gut barrier permeability [67]. Furthermore, certain inflammatory cytokines also disrupt TJs, undermining the integrity of the gut barrier and facilitating the deeper tissue invasion by pathogenic bacteria.
2.3.1. Mucus
Occupying the outermost layer of the intestinal barrier, mucus is primarily made up of water (90%-95%), electrolytes, lipids (1%-2%), proteins and others [68] formed by the substance secreted from epithelial cells, mainly goblet cells. It is constituted by 21 distinct types of mucins (MUC), denoted as MUC 1 to MUC 21 [69]. The core component of mucus is MUC 2, secreted by goblet cells, acting as a biological lubricant between epithelial cells and luminal microorganisms. It mitigates the direct interaction of gut cells with toxic substances and pathogens, thereby safeguarding them from infection and the impact of digestive juices. While gut microbiota can degenerate MUC proteins in mucus for their physiological activities, adhere and colonize to the epithelial surface, probiotics can increase mucus thickness and occupy the mucus binding sites. On the contrary, pathogens decompose mucin extensively, diminishing mucus thickness. It is noteworthy that various factors, including short chain fatty acids (SCFAs), bacterial components (flagellin, lipoteichoic acid, lipopolysaccharides), can influence MUC expression, stimulating goblet cells to secrete more mucus [68].
It is reported that Goblet cell depletion was observed in patient with IBD frequently, combined with mucus layer deficiency [70]. Due to the amounts of MUC-degrading bacteria increase, the secretions of MUC 2 decrease and the levels of MUC 2 O-glycans change [71]. These perturbations allow more microbiota to reach enterocytes, provoking immune responses, further increasing intestinal barrier permeability and exacerbating inflammation [72]. Akkermansia, a current research hotspot, has shown a unique impact on mucus production compared to other probiotics. Although classified as a mucin-degrading bacterium capable of breaking down proteins, studies indicate that the Amuc_1100 protein expressed on its surface can stimulate goblet cell proliferation, enhance mucin secretion, and reinforce the barrier function of the intestinal epithelium [38].
2.3.2. Epithelial cells and tight junction protein
Situated beneath the mucus layer, intestinal epithelial cells create a boundary between the external environment and the lamina propria. Enterocytes, hyperpolarized epithelial cells constituting the majority of the epithelial cell population, are interconnected by TJ proteins, crucial for maintaining the structural integrity of the intestine. These proteins include occludin, claudins, zonula occludens, tricellulin, cingulin, and junctional adhesion molecules, play an indispensable role in maintaining the integrity of the intestinal barrier and maintaining a reasonable direction of intestinal permeability disruptions in TJ protein expression and apoptosis of intestinal epithelial cells are commonplace in IBD, often culminating in altered intestinal permeability [60,73,74]. Current research has shown that probiotics can upregulate the expression of TJ protein in various ways to protect epithelial cells, and reduce intestinal permeability. Al Sadi et al. found that Bifidobacterium bifidum enhances the TJ barrier function of the mouse gut by activating the P38 kinase pathway and TLR2 dependent pathway [75]. Another strain, Bifidobacterium pseudocatenulatum, protects the intestinal barrier by inhibiting the TLR4/NF-κB pathway, slowing down intestinal inflammation symptoms, and upregulating TJ proteins and mucus expression levels [76]. Hsieh et al. found that the SCFAs metabolites of the Bifidobacterium W1U2 strain can reduce the damage caused by tumor necrosis factor-α (TNF-α) to intestinal epithelial cells and help maintain the integrity of epithelial cells [77]. In another study, Lactobacillus gasseri ATCC33323 regulated the intestinal barrier through constitutive androstane receptor-NR1I3, maintaining the localization of the E-cadherin/β-catenin complex and the E-cadherin/p120 catenin complex, reducing intestinal wall permeability, when E-cadherin was semiknocked out in the mouse intestine, the regulatory ability of Lactobacillus gasseri ATCC33323 was significantly reduced [78]. Mazen et al. collected 11 Lactobacillus and bifidobacteria strains, discovering that 5 strains significantly ameliorated intestinal barrier function and restored TJ protein levels to varying degrees in TNBS-induced mouse model [79]. The presence of micro-integrins on the cell wall surface of Lactobacillus plantarum CGMCC 1258 could repair TJ damage by increasing the expression of TJ proteins such as JAM-1, occludin and claudin-1 [80].
2.3.3. SCFAs
Fatty acids can be classified by the numbers of carbons in the main chain, as short-chain (<6 carbons), medium-chain (6-12 carbons), long-chain (12-21 carbons), or very long chain (>22 carbons) fatty acids. Most SCFAs, including acetate (C2), propionate (C3) and butyrate (C4), in the human intestine arise from bacterial metabolism of dietary fibers, acting as prebiotics, with each prebiotic potentially influencing gut microbiota differently [81]. The concentration of SCFAs at molar ratio in the human gut tract is approximately 60:20:20 for acetate: propionate: butyrate [82]. It has been reported that SCFAs have an important influence on maintaining normal function of the gut barrier. For instance, acetate has demonstrated the ability to suppress TNF-α secretion in lipopolysaccharide-induced mouse-derived blood cells through the g protein-coupled receptor 43 (GPR43) pathway [83] and to activate Lactobacillus bacteriocin synthesis by controlling quorum sensing [84]; butyrate can induce zonula occludens-1 (ZO-1) and occluding assemble through the adenosine monophosphate-activated protein kinase dependent pathway [85]; also butyrate is the main energy source for the colon epithelial cells [86]. Furthermore, SCFAs also associate with body immune system [87,88] and other organ out of gut through gut-organ axis pathway beyond the improvement of intestinal barrier function.
Several studies indicate fecal SCFAs levels reduce in active IBD patients [89,90], with higher levels observed in patients in remission periods compared to those in active phases of the disease [91]. Geirnaert et al. collected six strains of butyrate-producing bacteria, supplemented in vitro to CD patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity [92]. Clinical experiment shows SCFAs enemas induce remission in specific subsets of UC patients [93], whereas histological improvement was observed in UC patients [94]. However, current research on how SCFAs administration affects gut microbiome and immune system in IBD patients remains elusive, and more research needs to be done to explain the causal relationship between IBD and SCFAs.
As SCFAs-producing gut bacteria, Lactobacillus and Bifidobacterium generally produce lactic and acetic acids as primary end products of carbohydrate metabolism. Wu et al. revealed the intake of Lactobacillus plantarum HNU082 upregulated the level of SCFAs, which is strongly negatively correlated with the proinflammatory factors, but strongly positively correlated with the inflammatory suppressor, the relative abundance of SCFAs-producing bacteria is also increased (Figure 5) [95].
The important role of SCFAs in alleviation of DSS-induced UC. A: Gut microbial metabolic pathways associated with SCFAs. B: SCFAs contents. C: Relationship between SCFAs and gut microbiota. D: Relationship between SCFAs and inflammatory cytokines. LP082: L. plantarum HNU082 treated group; SASP: salazosulfasalazine treated group; DSS: colitis group. Reproduced with permission from [95], copyright 2022, ASM Journals.
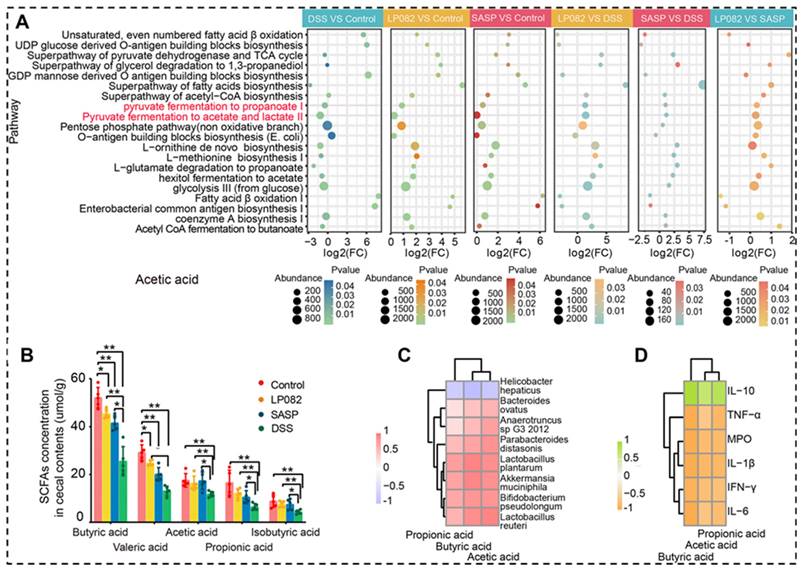
2.4. Microbial environmental regulation
Mounting evidence suggests a close association between gut microbiota and the progression of IBD [96]. The pathogenesis of IBD cannot be attributed to a single intestinal microorganism; instead, it is characterized by a gradual decrease in the overall diversity of intestinal flora as the disease progresses [97]. An increased abundance of Escherichia coli, Vibrio desulfuricans, Clostridium perfringens, and Enterococcus faecalis have been found to be associated with high activity of IBD [98]. A key question that is still waiting for an answer is the causal relationship between dysbiosis and IBD. Additionally, it should be pointed out that other members of the gut microbiome except bacteria, for instance, fungi [99], bacteriophage [100] and archaea [101], also contribute to the disease, though further research is required to elucidate their specific roles. The main mechanism of probiotic antagonistic effect on pathogenic bacteria includes producing bacteriocins, organic acids, competition for nutrients, and occupying effect.
2.4.1. Bacteriocins
Bacteriocins, as peptides or proteins synthesized by bacteria exhibiting the ability to inhibit the growth of specific bacteria strains, are generally classified into two groups: peptides that undergo significant post-translational modifications (class I) and unmodified peptides (class II) [102]. Due to their inhibitory effects on pathogens and low oral toxicity to their host, bacteriocins have potential as therapeutic agents against various infectious diseases. Detailed mechanisms of action have been extensively reviewed elsewhere [102]. Given the crucial role of gut microbiota in IBD progression, antibiotics have been widely used in clinical settings and have proven to be effective [2]. However, bacterial resistance and adverse effects pose limitations to antibiotic use, making bacteriocins a promising alternative.
2.4.2. Competing and occupying effect
The human digestive tract offers a vast surface area for bacterial colonization, the number of bacteria is about 107 in the jejunum, 1011 in the ileum, and 1014 in the colon. Due to differences in pH value and oxygen concentration, the composition of bacteria in different parts of the digestive tract varies, while the colon is the main contributor to the total population of bacteria in the alimentary tract [103]. To avoid mechanical clearance effects by digestive fluid and food, bacteria have evolved many strategies to adhere to the target cell in the gut. Thus, adhesion to host cells is a prerequisite for bacterial survival and proliferation within the host.
Pili, which widely exists on the surface of most Gram-negative bacteria and some Gram- positive bacteria, plays an important role in the adhesion process between bacteria and host cells. Composed of thousands of pili proteins, the tip of it confers varying degrees of adhesion ability to other cells [104]. Some bacteria also possess surface factors that can recognize various classes of host surface cell molecules, including components of the extracellular matrix, transmembrane proteins, exhibiting adhesion characteristics. The mutual binding process between bacteria and host cells is the critical preliminary step for bacteria invasion [105].
Compared to out-cell adhesion, survival within cells means the bacteria can partially avoid the competition from other members of the microbiome and gain more nutrients. Consequently, numbers of different strategies have been evolved to invade host cells by pathogens. These include bacterial surface proteins binding to host cell molecules, leading to bacterial engulfment, exemplified by Listeria monocytogenes [106] and the activation of cell signaling pathways by bacteria, causing cytoskeletal rearrangement of the cell membrane and engulfment of nearby bacteria, as seen in Salmonella [107]. By entering M cells and DCs, pathogens can penetrate the epithelium barrier to facilitate its dissemination in the host [108].
Probiotics can prevent the colonization of pathogenic microorganisms by attaching to the surface of intestinal epithelial cells, thereby physically blocking them through occupying effect [102] competition for nutrients. Abdi et al. isolated 323 strains of Lactobacillus from healthy human breast milk, with 71.8% of the strains showing strong adhesion to intestinal epithelial cells [109]. Another strains Lactobacillus plantarum BMCM12 can secrete extracellular proteins to reduce the adhesion capacity of pathogenic bacteria significantly and protect the intestinal barrier [41].
3. Strategies for engineering probiotic-based materials
In recent years, the field of bacterial modification has witnessed significant advancements with the development of a plethora of physicochemical and biological methods. Modified bacteria can be increasingly utilized as drug transporters or expression carriers for therapeutic interventions. However, the challenging conditions within the gastrointestinal tract, characterized by a harsh pH environment and the presence of various digestive fluids such as pepsin, pancreatic enzymes, and bile acids, pose substantial obstacles [110]. Although specific strains are able to resist bile by producing bile acid salt hydrolase [111], the activity of most of the bacteria will still be greatly affected [112]. At this stage, the main objectives of the modification of bacteria can be summarized as follows: Enhancing the gastrointestinal stability of the bacteria and augmenting their activity [112,113]; Improving the targeting efficacy of the bacteria to ensure colonize and proliferate at specific sites [114]; Endowing bacteria with the capability to express specific genes, facilitating the production of the required substances [115]; Enabling the bacteria to bind to specific drugs, using their targeting properties for the targeted release of drugs [116,117]. It is crucial to acknowledge that relying solely on the modification of the bacterial shell without editing its genes may lead to the dilution of the shell of the modified bacteria as the bacteria continue to divide and multiply, affecting the subsequent therapeutic effect.
In this section, we will delve into colon targeting strategies and bacterial modification methods, categorizing them into physicochemical and biological methods based on the materials and technologies employed. The classification criterion hinges on whether the method endows the bacteria with the ability to autonomously produce substances related to the modification target.
3.1. Engineering approaches for probiotic materials in targeting IBD
The inflammatory sites of IBD patients present significant different in their inflamed tissue sites compared to healthy individuals. These alterations encompass a lower pH value of the inflamed site (2.3~5.5) [118], a higher ROS and inflammatory mediator levels, higher degree of inflammatory cell infiltration and permeability of the colonic epithelium [119], prolonged transit time, reduced level of TJ protein and mucus secretion [120]. Additionally, there is a variation in the distribution of intestinal bacterial flora [121]. Leveraging these differences in pH, transit time, specific inflammatory cells, microbial environment, and ROS levels can facilitate the design of targeted therapy strategies for IBD patients [122,123].
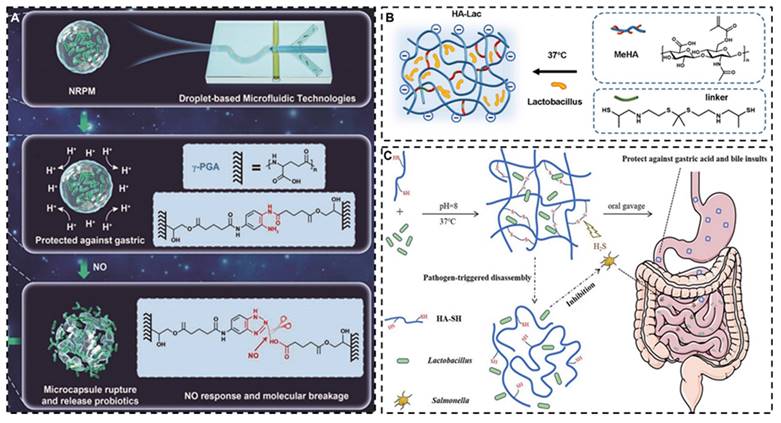
Although the intestinal transit time and pH environment of IBD patients are significantly different from those of the healthy population, considerable inter-patient variability exists. Furthermore, the diarrhea induced by intestinal inflammation can unpredictably alter drug transit times, challenging the achievement of desired therapeutic outcomes with traditional targeting methods. Consequently, it is comparatively more rational to devise targeting strategies based on the distinct characteristics of inflammatory sites, such as the ROS environment [124], inflammatory cell infiltration, and specific microbial milieu, which starkly differ from normal intestinal tissues. Wang et al. designed an NO-sensitive gather γ- glutamate microgel for encapsulating Lactobacillus, which endowed the modified strain with good stability to gastric acid and a good targeting effect on NO gas secreted by intestinal inflammatory tissues [125]. Huang et al. developed a ROS-responsive hyaluronic acid (HA) hydrogel based on physiologically crosslinked methacrylated HA and thiolated thioketal, in which ROS selectively cleaved thioketal linkages to release Lactobacillus reuteri in the inflamed colon tissues [126]. Xiao et al. use HA hydrogel wrap Lactobacillus rhamnoses, which can rapidly drop the drug locally upon contact with H2S produced by the metabolism of pathogenic bacteria, achieving a targeted therapeutic effect (Figure 6) [127]. However, current research on bacterial modification predominantly focuses on non-targeted areas, with studies on targeted modification remaining scarce.
3.2. Physicochemical methods for engineering probiotic-based materials
The bacterial cell wall, directly interacting with the human immune system, has emerged as a focal point in bacterial modification research. Structurally, bacteria cell wall consists of two main components: the cell wall and the protoplasm. The cell wall mainly consists of a glycan skeleton (composed of N-acetylglucosamine and N-acetylcytidylic acid), tetrapeptide side chains and pentapeptide cross-linked bridges [128]. The cell wall's surface, enriched with phosphopeptidic acid and peptidoglycan, is negatively charged and adorned with various functional groups. Through tailored physicochemical methods, chemical bonds can be formed between exogenous substances and cell wall functional groups, thus achieving surface modification that can achieve a variety of functions. The most prevalent mechanisms of bacterial physicochemical modification include electrostatic adsorption, wrapping, and the establishment of various covalent or non-covalent bonds.
Beyond the modification of the bacterial strain itself, adept formulation and encapsulation technologies can enhance the digestive stability of the strain, mitigating the impacts of temperature, oxygen exposure, contact with encapsulation materials, and pressure during formulation, transportation and storage processes. Oral administration stands as the most common delivery route for probiotic products, which are conventionally formulated into tablets, granules, capsules, and other dosage forms. The main research directions include forming physical barriers to isolate the external environment, combining with prebiotics to improve strain viability, and combining with digestive enzyme hydrolases to improve strain digestive tract stability. The commonly used natural materials usually refer to natural polysaccharides and proteins, including soy protein [113], alginate [129], pectin [130-132], chitosan [133], whey protein [134], starch [135], silk sericin [136] and silk fibroin [137]. Synthetic materials include the Eudragit® series [138], hydroxypropyl methyl cellulose phthalate, cellulose acetate and other synthetic polymers (Figure 7) [139,140]. Probiotic encapsulation technology can be categorized based on the relationship between the encapsulated bacteria and the encapsulation mechanism, falling into either block-based bulk encapsulation and single bacterial encapsulation.
Schematic illustration of encapsulated probiotics based on different targeting strategies. A: NO-responsive poly-γ-glutamic acid hydrogel microcapsule. Reproduced with permission from [125], copyright 2022, John Wiley and Sons. B: ROS-responsive HA hydrogel. Reproduced with permission from [126], copyright 2022, Elsevier. C: H2S-triggered HA hydrogel. Reproduced with permission from [127], copyright 2020, America Chemical Society.
3.2.1. Bulk encapsulation
The bulk encapsulation is characterized by the embedding of a substantial number of bacteria within a protective matrix or their adherence to the substrate's surface, as discernible via electron microscopy. At the microstructural level, it can be divided into two forms, nanospheres and nanofibers. As shown in Figure 7, predominantly, the encapsulation techniques encompass include spray-drying [113,141], extraction [129,134], freeze-drying [130], emulsification [135], electrospinning [142,143], 3D printing [144]. The chosen encapsulation method and substrate crucially influence the final encapsulation outcome. Figure 8 exhibits images of bulk encapsulated probiotics with different materials and methods. Within nanospheres, bacteria and matrix are uniformly integrated, though chemical bond forces are not conspicuously dominant. The main advantages of bulk bacterial encapsulation technology are its capacity for encapsulating a large bacterial quantity, low production cost, expedited experimentation, and the simultaneous analysis of multiple bacterial responses, its production process is easier to control. Bulk encapsule technology is usually aimed at improving the shelf life, digestive stability, and storage activity of probiotics, rendering it a promising avenue for industrial application.
3.2.2. Single bacterial encapsulation
In contrast, single bacterial encapsulation typically involves surface coating of protective matrices on bacteria, with an association to the substrate via chemical bond forces or interaction forces, including covalent bonds, diverse electrical properties, hydrophobic interactions, Van der Waals forces, and intermolecular hydrogen bonding (Figure 9). This technology offers precision in bacterial encapsulation, fostering advancements in bacterial-based drug delivery, and providing the bacteria with complex functionalities such as drug delivery [145], multi-antibiotic resistance [146], transcending traditional roles in gastrointestinal tract protection and viability enhancement.
Illustration of molecular interaction, encapsulate methods and materials of engineered probiotics. Different encapsulation materials are combined with strains through various forces, and according to the different preparation methods, the strains exhibit various improvements in physical and chemical properties. Created with https://BioRender.com.
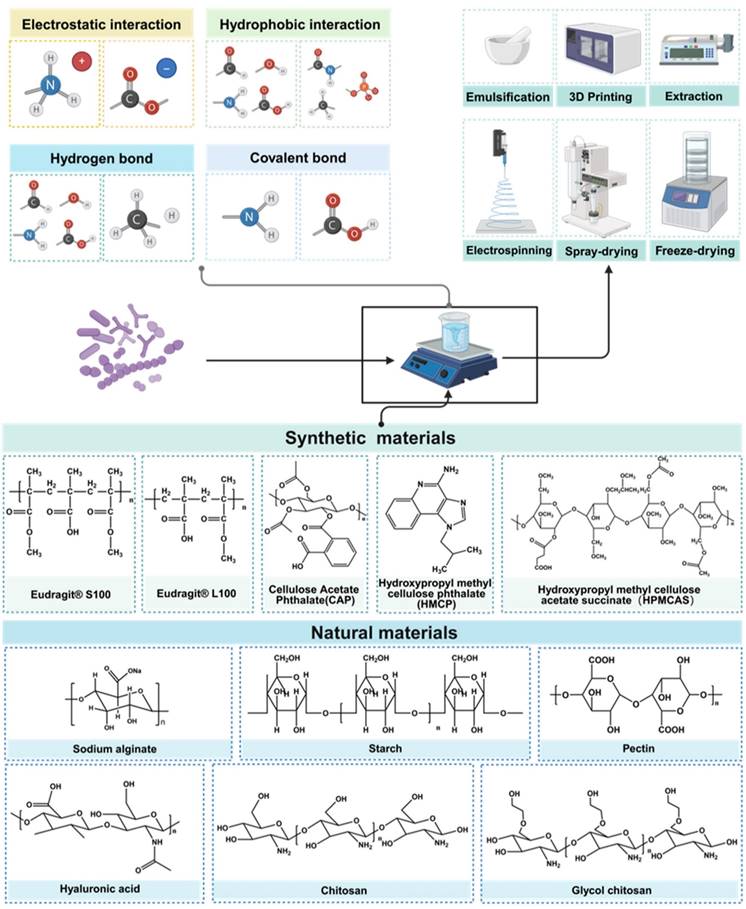
In this section, we broaden our exploration to specific single bacterial encapsulation technologies based on the solution self-assemble (SA) method categorized by different driven forces. Single bacterial encapsulation techniques utilize a variety of principles to construct molecular layered structures with differences primarily in encapsulation mechanisms and layer quantity. Characterized by its spontaneously ordered nanoparticle structuring in solution without external interference, SA relies on interactions such as electrostatic interaction [147,148], hydrogen bond [149], covalent bond [145,150]. The SA method is a natural and spontaneous process that leads to the organized structuring of nanoparticles in a solution without the need for external intervention. Throughout this process, nanoparticles autonomously arrange themselves into ordered structures, facilitated by the interplay of hydrophobic forces, Van der Waals interactions, and electrostatic forces. The simplicity and ease of implementation of SA technology, along with its rapid and straightforward preparation process, result in structures that exhibit commendable stability. A distinct advantage of SA is its frequent application in decorating bacteria with biofilm and biomimetic materials, such as lipid [151], tumor cell membrane [152], which gives the bacteria many unique biological characteristics. However, the controllability and stability of SA technology require improvement. It necessitates the careful selection of appropriate conditions and molecules to regulate its assembly structure and properties.
Images of bulk encapsulated probiotics with different materials and methods. SEM images of alginate-silk sericin-maltitol co-encapsulated L. casei TISTR 1463 with (A1) and without (A2) silk sericin coating. Reproduced with permission from [136], copyright 2022, Elsevier. SEM images of calcium pectin beads surface morphology (B1) and the distribution (B2) of L. paraplantarum L-ZS9 within it. Reproduced with permission from [132], copyright 2022, Elsevier. SEM image of encapsulated L. rhamnosus based on robocasting 3D-printing technology (C). Reproduced with permission from [144], copyright 2023, Elsevier. SEM image (D) of the surface of L. casei 01 beads coated with alginate plus hi-maize starch. Reproduced with permission from [135], copyright 2016, Elsevier. SEM images of hydrogel beads encapsulated with L. rhamnosus GG (LGG®) after freeze-dry: surface view (E1, E3) and fractural section (E2, E4). Reproduced with permission from [130], copyright 2023, Elsevier. SEM images of HA hydrogel-encapsulated L. reuteri (F1, F2). Reproduced with permission from [126], copyright 2022, Elsevier. SEM images (G) of electrospun LGG incorporated in fibrous mats from calcium caseinate-pullulan-LGG fibers. Reproduced with permission from [142], copyright 2022, Elsevier. SEM images (H1) and confocal microscopy images (H2) of L. plantarum-loaded poly(ethylene oxide) nanofibers. Reproduced with permission from [143], copyright 2019, Elsevier. SEM images of calcium-alginate (I1) and calcium-alginate-sucrose (I2) encapsulated LGG®. Reproduced with permission from [141], copyright 2022, Elsevier. UHR FE-SEM images of pectin resistant starch-pectic oligosaccharide hydrogel beads encapsulated L. bulgaricus (J). Reproduced with permission from [131], copyright 2023, Elsevier.
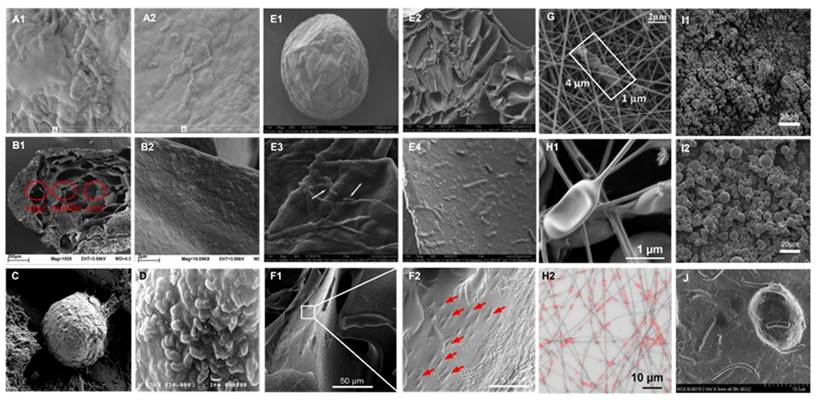
Layer-by-Layer (LBL) method is one specific SA method driven primarily by Van der Waals interactions which provides researchers with the capability to precisely control the thickness of each layer, encapsulating bacteria with various materials and layer counts to create complex bacterial shells. During the LBL process, each layer requires chemical modification of the surface charge before deposition onto the substrate surface. The subsequent mutual adsorption between each layer through electrostatic attraction allows for precise control and adjustment. However, this also implies that the encapsulation procedure is more intricate and time-consuming, necessitating consideration of numerous control conditions, the mastery of advanced laboratory skills by researchers, and presenting challenges in large-scale production.
3.3. Chemical methods for engineering probiotic-based materials
The bacterial cell wall is rich in functional groups, enabling the chemical conjugation of small molecules or other materials to the bacterial surface through a variety of bonding techniques. Such chemical modifications can be used to alter the surface properties of bacteria, similar to physical modification, which plays a crucial role in enhancing the ability of probiotics to target tumor and inflammatory sites, increasing bacterial resilience against external stressors, improving therapeutic efficacy, and augmenting intestinal colonization. The primary advantage of chemical modification lies in its ability to precisely introduce complex and diverse functional groups to the bacterial surface, thereby enabling a wider range of functionalities. Recent advances in click chemistry and bioorthogonal technologies have provided highly efficient and sophisticated tools for bacterial surface engineering (Figure 10). For example, Liu et al. attached PEG to the surface of bacteria by in situ self-polymerization of dopamine, amine-terminated polyethylene glycol (m-PEG-NH2) linked with self-polymerized polydopamine on the surface of bacteria significantly enhancing their ability to penetrate and retention at the intestinal mucus layer, as well as their mobility. Bacteria modified by this approach effectively inhibited pathogen invasion by occupying available niches [153]. In another study from the same group, bacteria were surface-modified with thiol groups, allowing them to form covalent bonds with disulfide-rich mucins in the intestinal environment, leading to a remarkable 170-fold increase in mucosal adhesion [154].
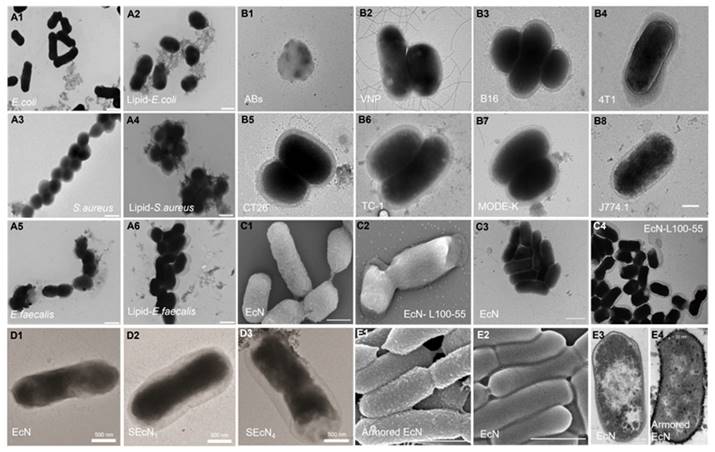
Images of single encapsulated bacteria with different materials and methods. TEM images of uncoated (A1, A3, A5) and Lipid coated bacteria (A2, A4, A6). Reproduced with permission from [151], copyright 2019, Springer Nature. TEM images of apoptotic bodies (B1, Abs), naked Salmonella Typhimurium VNP 20009 (B2, VNP) and VNP coated with different tumor cell membranes (B3-B8). Reproduced with permission from [152], copyright 2022, Elsevier. SEM images of Escherichia coli Nissle 1917 (EcN) (C1) and EcN- Eudragit® L100-55 (C2), TEM images of EcN (C3) and EcN-Eudragit® L100-55 (C4). Reproduced with permission from [138], copyright 2020, John Wiley and Sons. TEM images of EcN (D1), ECN with 1 layer (D2, SEcN1) and 4 layers (D3, SEcN4) of silk fibroin. Reproduced with permission from [137], copyright 2021, John Wiley and Sons. SEM images of armored (E1) and naive EcN (E2), cross-sectional TEM images of naive (E3) or armored EcN (E4). Reproduced with permission from [146], copyright 2022, Springer Nature.
In subsequent studies, researchers have combined chemical and biological techniques, employing plasmid expression to display specific molecules on the bacterial surface, followed by attachment of metal complexes and indocyanine green to enhance photothermal effects [155,156]. Cao et al. synthesized SOD/CAT mimic antioxidant enzymes and combined them with the surface of bacteria through click chemistry reactions, a metal-organic-framework encapsuled iron single-atom SOD/CAT mimic catalyst was linked to the surface of bacteria through Click reaction with boronic acid-poly(ethylene glycol) (C18-PEG-B), providing the bacteria with additional antioxidant capacity, protecting them from inflammatory damage at the inflammatory site, and demonstrating significant therapeutic effects in mouse and beagle dog IBD models [157]. Peng et al. used a click chemistry reaction of azide acetylene to bind HA to bacterial surfaces, the amino groups of the Escherichia coli Nissle 1917 cell wall were diazotization modification, and the diazo groups on the Escherichia coli Nissle 1917 surface are linked to dibenzocyclooctyne (DBCO) functionalized HA through a bioorthogonal reaction, then L100-55 were coated on the outer side, they demonstrated significant immunomodulatory and gut microbiota regulatory effects in mice model [158]. Song et al. developed a modification strategy using bioorthogonal reactions, which modifies DBCO group onto the surface of probiotics and expresses azide groups on the surface of gut microbiota by using metabolic engineering methods. DBCO-functionalized probiotics can undergo bioorthogonal reactions with the gut microbiota, improve their intestinal colonization ability, enhance bacterial delivery efficiency. This technology may have enormous clinical translational potential [159].
3.4. Bioengineering methods for engineering probiotic-based materials
Bioengineering methodologies facilitate the genetic manipulation of probiotics empowering them to express, synthesize, and secrete various specific therapeutic substances in situ. This approach circumvents the loss and some adverse effects associated with the traditional oral administration of drugs through the digestive tract, as illustrated in Figure 11, which delineates the primary mechanisms of action of engineered probiotics in disease management. However, it is critical to acknowledge that many strains suitable for genetic modification are not endogenous to the human gut microbiota. The competitive survival dynamics within the human gastrointestinal ecosystem can impact the colonization efficiency of engineered probiotics, subsequently diminishing their therapeutic potential [160]. Thus, selecting appropriate strains as platforms for genetic modifications becomes imperative [161].
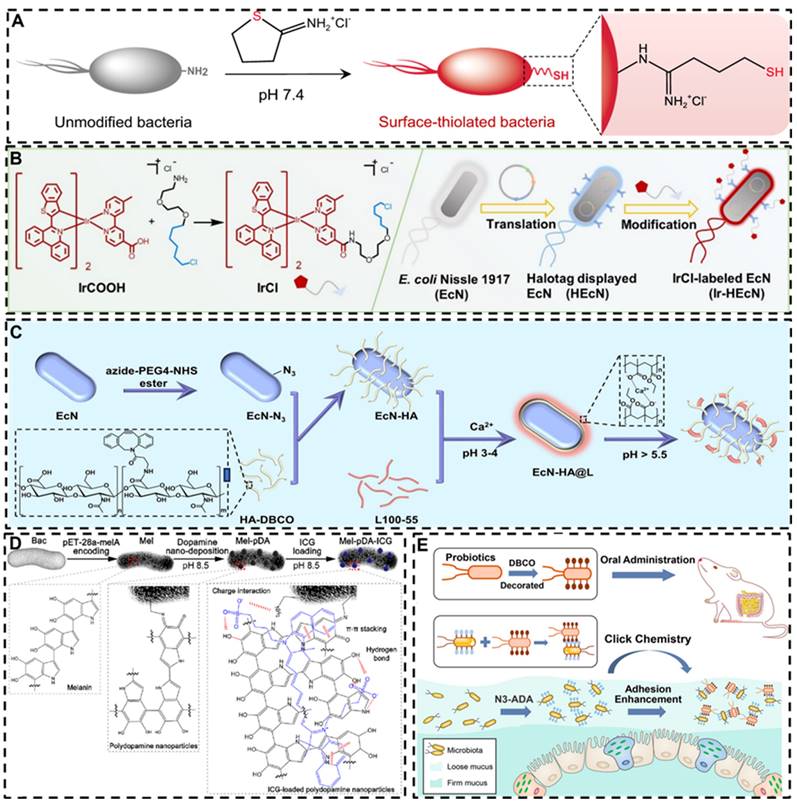
Schematic illustrations of chemically modified probiotics using different strategies and molecules. A: Surface-thiolated ECN. Reproduced with permission from [154], copyright 2022, Springer Nature. B: Iridium (III) photosensitizer-bacteria hybrid. Reproduced with permission from. Reproduced with permission from [155], copyright 2020, John Wiley and Sons. C: Bioorthogonal functionalized EcN. Reproduced with permission from [158], copyright 2024, Elsevier. D: Ternary photosensitive bacteria. Reproduced with permission from [156], copyright 2023, America Chemical Society. E: DBCO and Azido group modified bacteria. Reproduced with permission from [159], copyright 2022, America Chemical Society.
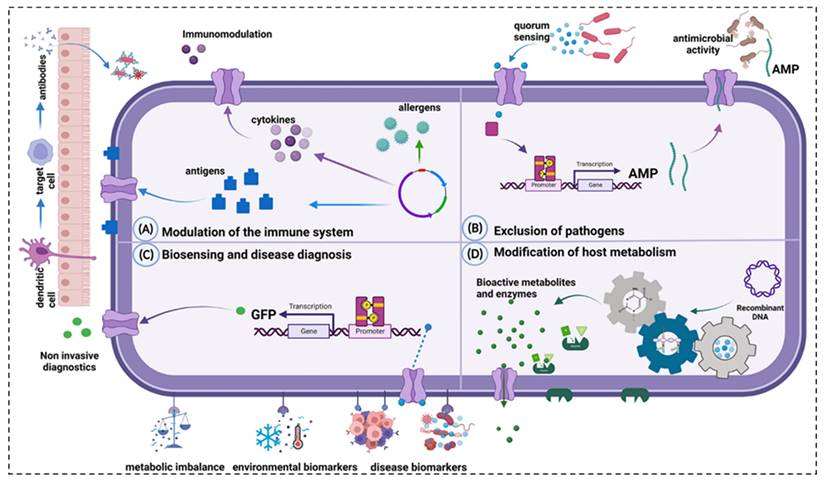
Main mechanisms of action of engineered probiotics. A: Modulation of the immune system. B: Exclusion of pathogens. C: Biosensing and disease diagnosis. D: Modification of host metabolism. Reproduced with permission from [161], copyright 2023, Portland Press, Ltd.
Gene editing techniques have been widely used in the gene transformation of bacteria, for instance, using plasmids and phages as gene carriers to import exogenous genes [162]. CRISPR/CAS gene editing technology has also been used to achieve this goal, which can express a variety of products [163,164]. While plasmid-based methods offer simplicity and a broad selection of genes for introduction [56], there are also risks of instability of introduced genes, possible loss of target genes after several generations, and a high rate of false positives [165]. On the other hand, the CRISPR/CAS gene editing tool offers improved genetic stability by integrating target genes directly into the bacterial genome, facilitating the expression of target proteins [166]. The bacterial genome can be edited to reduce toxicity [167], antigenicity, infectivity and increase the targeting colonizing ability of bacteria [168]. Conventional physical and chemical methods, such as electro-transformation, also play a role in transfecting exogenous genes into probiotic cells [169].
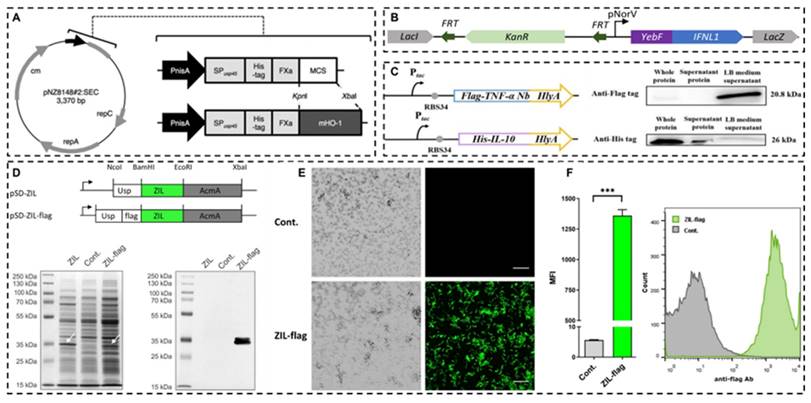
The gene expression induction and regulation system of engineering bacteria developed in recent years can improve the expression efficiency of target genes by adding specific promoter sequences before target genes, including the chlorine induction system [170], lactic P170 induction system [171], phosphate induction system [172], peptide (nisin) regulatory expression system [171,173], sugar-induced expression systems, etc., which can switch the expression of target genes by adding various sugars to the diet of model animals in vitro [174,175]. Shigemori et al. constructed a recombinant Lactococcus lactis NZ9000 capable of secreting the recombinant mouse heme oxygenase-1. The gene expression was controlled by the nisin-inducible promoter, and the plasmid was introduced into the strain using electroporation. Enzyme linked immunosorbent assay (ELISA) results showed that the cell extract of the strain contained a concentration of mouse heme oxygenase-1 of approximately 5μg/mL. Compared with the control strain, the recombinant strain could significantly increase the production of anti-inflammatory mediator IL-10 and reduce the expression of pro-inflammatory mediators in the colon [173]. Chua et al. used Escherichia coli Nissle 1917 as the chassis, constructed an interferon λ1 (IFN L1) producing recombinant strain, and validated the therapeutic effect in the Caco-2/Jurkat T cell co-culture model and scaffold-based 3D co-cultured IBD model. The results showed that the engineered strains could protect the integrity of epithelial cells, increase TJ protein expression, reduce intestinal barrier permeability [176]. Wu et al. constructed a dual bacterial system, the recombinant Escherichia coli Nissle 1917 can express and secrete anti-TNF-α nanobodies and IL-10, respectively, to simultaneously neutralize pro-inflammatory factors to enhance anti-inflammatory effects, regulate gut microbiota, and effectively inhibit colitis induced by DSS [177]. Zahirović et al. constructed recombinant Lactococcus lactis NZ9000, which can express interleukin-6 (IL-6)-binding affibody on the surface of the strain to achieve IL-6 targeting function (Figure 12) [178]. Comprehensive reviews on recent advancements in utilizing lactic acid bacteria as delivery vectors and related biological tools are available in the literature [161,179,180].
4. Clinical applications of probiotics for bowel disease
Current evidence suggests that probiotics may improve symptoms in patients with mild to moderate UC, though their efficacy in CD appears limited. It is important to acknowledge that existing clinical trial results are often inconsistent, and the lack of standardized experimental protocols contributes to this variability. Factors such as strain selection, dosage, treatment duration, as well as differences in patient characteristics and disease severity, complicate the ability to objectively evaluate the therapeutic potential of probiotics in IBD. Moreover, although both UC and CD fall under the category of IBD, significant differences exist in their underlying pathophysiology and clinical manifestations [25,26], which may also explain the differential responses to probiotic interventions.
Another crucial aspect of probiotic therapy is the potential risk of introducing pathogenic microorganisms. Given that IBD patients often exhibit an aberrant immune state within the intestines, and some may concurrently present with Clostridium difficile infections, the introduction of pathogenic bacteria could exacerbate the condition. Therefore, the choice of probiotic strains must take into consideration the patient's immune status, prioritizing strains with established safety profiles and suitable usage contexts. Even strains that are generally considered safe could have deleterious effects in immunocompromised individuals. Additionally, strict precautions should be taken during the production, transportation, storage, and administration of probiotics to prevent contamination with pathogenic microorganisms. Compliance with stringent quality control standards, along with proper transport and storage conditions, is essential, as is educating patients on appropriate usage.
At present, multiple probiotic strains have been approved for the treatment of IBD and have entered the clinical trial stage, including Lactobacillus rhamnosus, Lactobacillus casei, Escherichia coli Nissle 1917, VSL#3®, etc. Here we list the clinical trial records registered on the clinical trials website in Table 2. Considering the progress of clinical trials and the number of preclinical basic studies, we will briefly introduce VSL#3® and LGG® below.
Recombinant probiotics for expressing anti-inflammatory mediators. A: A vector map of the lactococcal secretion vector and schematic representations of gene maps of the vector. Reproduced with permission from [173], copyright 2015, Springer Nature. B: Schematics of the IFN L1 production-secretion cassette for genomic integration. Reproduced with permission from [176], copyright 2023, America Chemical Society. C: Gene route design and Western blot detection results of constitutive engineered bacteria EcN-TNF-α Nanobodies (Nb, top) and EcN-IL10 (bottom). Reproduced with permission from [177], copyright 2024, Springer Nature. D: Gene constructs for expression of IL-6 binding affibody ZIL on the surface of L. lactis NZ9000, and SDS-PAGE, Western blot analysis of whole lysates. E: confocal immunofluorescence microscopy images. F: flow cytometry analysis. Reproduced with permission from [178], copyright 2022, Springer Nature.
Clinical trials of probiotic for IBD therapies [Data from ClinicalTrials.gov, accessed on 6 June 2024.
| NCT Number | Strain | Study Status | Conditions | Sponsor | Phases |
|---|---|---|---|---|---|
| NCT00114465 | Mixed strains (VSL#3®) | COMPLETED | CD | Orphan Australia | PHASE4 |
| NCT00175292 | Mixed strains (VSL#3®) | COMPLETED | CD | University of Alberta | PHASE3 |
| NCT00268164 | L. acidophilus LA5; B. animalis BB12 | TERMINATED | UC | Hvidovre University Hospital | PHASE2 |
| NCT00367705 | Mixed strains (VSL#3®) | UNKNOWN | UC | Hadassah Medical Organization | PHASE4 |
| NCT00305409 | B. longum | COMPLETED | CD | University of Dundee | NA |
| NCT00374374 | L. acidophilus;L. rhamnosus | COMPLETED | CD | Odense University Hospital | NA |
| NCT00374725 | L. acidophilus;L. rhamnosus | COMPLETED | UC | Odense University Hospital | NA |
| NCT00510978 | B. Infantis 35624; | UNKNOWN | UC|CD | University College Cork | PHASE2/3 |
| NCT00510978 | L. Salivarius UCC118 | ||||
| NCT00578799 | Mixed strains (Kyo-Dophilus®) | WITHDRAWN | UC | University of California, Irvine | PHASE1 |
| NCT00803829 | Not Given | COMPLETED | UC | University of Dundee | NA |
| NCT00895336 | L. Rhamnosus GG | WITHDRAWN | UC | Children's Hospital Medical Center, Cincinnati | PHASE2 |
| NCT00944736 | Mixed strains (VSL#3®) | COMPLETED | CD | Children's Mercy Hospital Kansas City | PHASE3 |
| NCT00951548 | Mixed strains (VSL#3®) | COMPLETED | UC | VSL Pharmaceuticals | NA |
| NCT01078935 | Mixed strains | UNKNOWN | CD|UC | The Baruch Padeh Medical Center, Poriya | PHASE4 |
| NCT01173588 | Bifidobacterium | COMPLETED | IBD | Instituto Lala | PHASE3 |
| NCT01193894 | L. plantarum 299V | COMPLETED | UC | Nordisk Rebalance A/S | PHASE2|PHASE3 |
| NCT01479660 | Mixed strains (VSL#3®) | UNKNOWN | UC | Post Graduate Institute of Medical Education and Research, Chandigarh | PHASE4 |
| NCT01548014 | Mixed strains (VSL#3®) | UNKNOWN | CD | Samsung Medical Center | PHASE3 |
| NCT01632462 | Mixed strains (VSL#3®) | UNKNOWN | CD | Federico II University | PHASE4 |
| NCT01698970 | Not Given | COMPLETED | CD | Danone Research | NA |
| NCT01765439 | Mixed strains (VSL#3®) | ACTIVE_NOT_RECRUITING | CD|UC | Charles University, Czech Republic | NA |
| NCT01765998 | Not Given | UNKNOWN | CD | The Baruch Padeh Medical Center, Poriya | PHASE4 |
| NCT01772615 | E. coli Nissle 1917 | COMPLETED | UC | Hvidovre University Hospital | PHASE4 |
| NCT02361957 | Mixed strains (Ecologic®825) | SUSPENDED | UC | Wageningen University | NA |
| NCT02488954 | P. freudenreichii | TERMINATED | UC | Rennes University Hospital | NA |
| NCT03266484 | Not Given | ACTIVE_NOT_RECRUITING | CD|UC | Massachusetts General Hospital | NA |
| NCT03415711 | Mixed strains (VSL#3®) | TERMINATED | UC | VSL Pharmaceuticals | NA |
| NCT03798210 | L. reuteri 4659 | UNKNOWN | UC Flare | Uppsala University | PHASE2 |
| NCT04006977 | Mixed strains | UNKNOWN | UC | Xijing Hospital of Digestive Diseases | NA |
| NCT04102852 | L. Rhamnosus GG | COMPLETED | UC|UC | San Giovanni Addolorata Hospital | PHASE1/2 |
| NCT04223479 | Not Given | COMPLETED | UC | University of Jordan | PHASE2/3 |
| NCT04241029 | Mixed strains (IDOFORM TRAVEL®) | COMPLETED | UC | Oslo University Hospital | NA |
| NCT04305535 | Not Given | UNKNOWN | CD | Instituto de Investigación Sanitaria de la Fundación Jiménez Díaz | NA |
| NCT04804046 | Not Given | TERMINATED | CD | University of Alberta | NA |
| NCT04842149 | B. Breve Bif195 | ACTIVE_NOT_RECRUITING | CD | Hvidovre University Hospital | NA |
| NCT04969679 | E. coli Nissle 1917 (Mutaflor®) | COMPLETED | UC | Kangbuk Samsung Hospital | PHASE4 |
| NCT05118919 | L. Reuteri BGP-014 | RECRUITING | UC | BioGaia Pharma AB | PHASE1/2 |
| NCT05652621 | Not Given | RECRUITING | UC|IBS | The First Affiliated Hospital of Xinxiang Medical College | NA |
| NCT05666960 | Not Given | RECRUITING | UC | Rise Therapeutics LLC | PHASE1 |
| NCT05906043 | Not Given | RECRUITING | IBD | University College Dublin | NA |
| NCT06392061 | Mixed strains (Trilac®) | RECRUITING | IBD | Lebanese University | NA |
| NCT06595719 | Mixed strains | RECRUITING | UC | National University of Malaysia | NA |
| NCT06609447 | Mixed strains (VSL#3®) | RECRUITING | UC | Second Affiliated Hospital, School of Medicine, Zhejiang University | PHASE4 |
| NCT06637683 | B. subtilis;B. clausii (LiveSpo COLON®) | COMPLETED | IBD | Anabio R&D | NA |
| NCT06642883 | Mixed strains (ProLife®) | RECRUITING | UC | University of Padova | NA |
4.1. VSL#3®
VSL#3® (now branded as Visbiome® in the U.S. and Vivomixx® in Europe since January 31, 2016) is a commercially available probiotic blend comprising multiple probiotic strains [181], Visbiome® is a mixed probiotic preparation consisting of eight live lyophilized probiotics, including four strains of Lactobacillus (Lactobacillus casei, Lactobacillus plantarum, Lactobacillus acidophilus, and Lactobacillus delbrueckii), three strains of Bifidobacterium (Bifidobacterium longum, Bifidobacterium shortum, and Bifidobacterium infantis), and one strain of Streptococcus thermophilus. These strains collectively exhibit the capacity to stably colonize the intestinal tract, reduce fecal pH value, locally augment FoxP3 expression in the intestine, and stimulate Treg cells, thereby modulating intestinal immunity upon sustained administration [182]. It has been found that the IL10-induced CD4+ latency-associated peptide (LAP)+Treg pathway may be the main mechanism for its intestinal protective effect [183]. A meta-analysis conducted in 2019 showed that Visbiome® was significantly effective in relieving the symptoms of active UC without an increased risk of adverse effects [184]. Moreover, in pediatric IBD patients, Visbiome® significantly outperformed placebo controls, reducing abdominal pain and rectal bleeding [185,186].
4.2. LGG®
LGG® is another extensively investigated commercial probiotic product, comprising the Lactobacillus rhamnoses GG. This strain demonstrates robust tolerance to gastric and bile acids and has the capacity to secrete proteins that mitigate cytokine-induced apoptosis of epithelial cells [187,188]. Evidence corroborates that a one-week administration of LGG® facilitates colonization in the human colon [189]. The secreted soluble protein HM0539 from LGG® is documented to fortify intestinal integrity by modulating TJ protein expression and enhancing mucus secretion [190]. Additionally, it attenuates Th17 cell activity, reduces IL-17 levels [191] and protects the intestinal epithelium through mucus-binding protein mediated bacteria-body interaction, displaying potent anti-inflammatory effects in vitro [192]. LGG®-derived proteins P40 and P75 have been proven to be able to protect intestinal epithelial cells from apoptosis, promote proliferation, and activate Akt in a Phosphoinositide 3-Kinase-dependent manner [193]. Specifically, P40 has been shown to enhance intestinal cell maturation and differentiation in newborn mice through the EGFR pathway [194]. However, the results of several human clinical trials are conflicting and cannot confirm whether the therapeutic effect is superior to conventional 5-aminosalicylic acid therapy [195]. This necessitates further clinical investigations to rigorously evaluate the therapeutic potential of LGG® in IBD management.
5. Summary and outlook
In this review, we have navigated through the intricate landscape of probiotic and their therapeutic implications in the realm of IBD. We extensively explored the diverse mechanisms by which probiotic exerts its influence, ranging from immunomodulation and antioxidant activities to fortifying the intestinal barriers and reshaping the gut microbiome. The narrative progressed to illuminate potential avenues for targeted therapy, spotlighting current innovations in bacterial modification techniques, and culminating with a critical appraisal of two leading probiotic products under extensive research scrutiny. Through promising, numerous hurdles persist within this domain, necessitating comprehensive and innovative solutions (Figure 13) to advance the field and enhance patient outcomes.
5.1. Strain-specific research
A deep dive into understanding the unique characteristics of different probiotic strains, ensuring a more tailored approach to their application in treating various forms of IBD. Various probiotics are playing an increasingly important role in the treatment of IBD due to their good safety and unique intestinal regulating function. However, the landscape is marred by the limited number of probiotic strains that have successfully transitioned into clinical trial phases with proven efficacy. The existing clinical studies, encompassing diverse patient demographics, geographical locations, and disease variants, further complicate the panorama due to variations in strain selection, dosage, and treatment duration. These factors, coupled with individual lifestyle choices and disease states, contribute to the considerable heterogeneity in clinical outcomes. As delineated by the 2023 ESPEN guideline on Clinical Nutrition in IBD, while probiotics show potential benefits for mild to moderate UC patients, their application in CD remains contentious and unsupported by robust clinical evidence [196].
5.2. Enhanced clinical trials with interdisciplinary collaboration
Rigorous, large-scale, multi-center clinical trials with standardized protocols should be conducted to validate the efficacy of specific strains in diverse patient populations. Looking ahead, the development of probiotic formulations comprising a consortium of strains holds promise for transcending the current limitations imposed by individual variability. This necessitates the initiation of meticulously designed, multi-center, and large-scale clinical trials, devoid of commercial bias, to unravel the intricate patient-probiotic interplay. In tandem, there is a pressing need for collaborative synergy across academia, clinical settings, and industry, aiming to translate scientific discoveries into tangible clinical solutions.
Schematic illustration of the future development direction of probiotics. Created with https://BioRender.com.
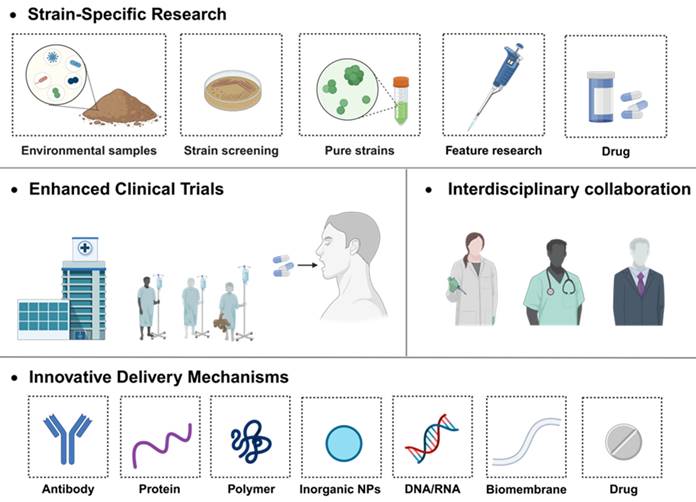
Advancing comprehensive therapies that incorporate bacterial metabolites and probiotic omics represents a promising approach to addressing the inconsistent clinical efficacy of probiotics. A substantial component of host-microbe interactions occurs through bacterial metabolites produced from the fermentation of intestinal contents. Metabolites such as short-chain fatty acids (SCFAs) exert significant regulatory effects on host physiology. Leveraging these metabolites, either as direct therapeutic agents or in conjunction with in vivo microbial interventions, may facilitate more consistent therapeutic outcomes. Furthermore, the development of personalized probiotic formulations, composed of diverse strains, their corresponding metabolites, and prebiotics tailored to the unique characteristics of individual patients, may yield superior clinical benefits compared to the use of single strains. Such combination therapies could lead to enhanced modulation of host microbiota and improved patient outcomes. Nevertheless, these approaches must undergo rigorous efficacy evaluation and be administered by healthcare professionals based on robust clinical evidence. Emerging research has demonstrated that these multi-component probiotic therapies exhibit enhanced efficacy in managing conditions such as inflammatory bowel disease and other gastrointestinal disorders. [197].
5.3. Innovative delivery mechanisms
Traditional chemical drugs and biological antibody therapies remain the primary treatment options for patients with IBD, with probiotics commonly used as adjunctive therapies. Existing research indicates that probiotics can offer therapeutic benefits, particularly in UC, where they have been shown to alleviate symptoms and reduce disease severity. Furthermore, probiotics generally exhibit fewer side effects and a higher safety profile, making them better tolerated by patients. When used in combination with conventional medications, probiotics can help mitigate drug-related side effects and potentially enhance overall treatment efficacy.
However, probiotics are not native to the human intestinal microbiota, and the gut environment is often not conducive to their colonization. To exert therapeutic effects, probiotics must withstand the harsh conditions of the digestive tract and reach the intestinal lumen. Additionally, individual variations in gut microbiota pose significant challenges to the consistent efficacy of probiotics. As a result, the impact of probiotics on treatment outcomes remains limited, and their use in conjunction with traditional therapies is currently a common strategy. Achieving an optimal balance between probiotics and pharmaceutical treatments is an area that warrants further investigation to maximize therapeutic benefits.
As we pivot to the product landscape, oral administration remains the predominant route for strain delivery. Nevertheless, this necessitates innovative approaches to enhance the strains' activity and colonization efficacy within the gastrointestinal milieu. Strategies such as prebiotic supplementation and advanced surface modification technologies have emerged, yet the field is ripe for further innovation, particularly in targeted modification methods and controlled release technologies, tailored specifically for IBD. Furthermore, ensuring prolonged strain viability outside laboratory settings remains a critical consideration for ensuring therapeutic efficacy upon patient administration.
In conclusion, as we grapple with the multifaceted effects of probiotics on human health, a paradigm shift towards a more diversified and nuanced evaluation of therapeutic efficacy is imperative. Future endeavors should be channeled towards establishing a robust classification system, capturing the spectrum of probiotics' effects across diverse patient populations. This, in turn, would facilitate more accurate and personalized evaluations, empowering healthcare practitioners, patients, and researchers alike, and ushering in a new era of optimized, patient-centric probiotic-based therapies.
Abbreviations
IBD: inflammatory bowel disease; CD: Crohn's disease; UC: ulcerative colitis; TLR: toll-like receptor; Th cells: helper t cells; Treg cells: regulatory T cells; Slp: surface layer protein; DSS: dextran sulfate sodium; DCs: dendritic cells; IL-1β: interleukin-1β; IL-8: interleukin-8; ROS: reactive oxygen species; FPRs: formyl peptide receptors; CAT: catalase; SOD: superoxide dismutase; TNBS: trinitrobenzene sulfonic acid; NRF2: nuclear factor erythroid 2-related factor 2; NF-κB: nuclear factor kappa-B; ARE: antioxidant response element; GSH-Px: glutathione peroxidase; MAPK: mitogen-activated protein kinase; TJ: tight junction; MUC: mucins; SCFAs: short chain fatty acids; TNF-α: tumor necrosis factor-α; GPR43: g protein-coupled receptor 43; ZO-1: zonula occludens-1; HA: hyaluronic acid; SA: self-assemble; LBL: layer-by-layer; DBCO: dibenzocyclooctyne; CRISPR/CAS: clustered regularly interspaced short palindromic repeats/ crispr-associated systems; ELISA: Enzyme linked immunosorbent assay; IFN L1: interferon λ1; IL-6: interleukin-6; LAP: latency-associated peptide; ESPEN: the European Society for Clinical Nutrition and Metabolism; TEM: transmission electron microscopy; SEM: scanning electron microscope; UHR FE-SEM: ultra-high resolution field emission scanning electron microscope.
Acknowledgements
This work was financially supported by National Natural Science Foundation of China (Grant No. 82473891), Basic Medicine School of Naval Medical University (JCKFKT-ZD-001), National Natural Science Foundation of China (Grant No. 22205133), “Shuguang Program” supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission (Grant No. 21SG39), Key Science and Technology Research and Development Projects of Jiangxi Province (Grant No. 20223BBH80014) and Wenzhou Basic Scientific Research Project (Grant No. Y20220138).
Author contributions
All authors have given approval to the final version of the manuscript.
Author contributions
Guangze Sang: Writing - original draft & Visualization.
Bingkai Wang: Writing - review & editing.
Yujie Xie: Writing - review & editing; Funding acquisition; Visualization.
Yu Chen: Writing - review & editing.
Feng Yang: Writing - review & editing; Conceptualization.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ananthakrishnan AN, Kaplan GG, Bernstein CN, Burke KE, Lochhead PJ, Sasson AN. et al. Lifestyle, behaviour, and environmental modification for the management of patients with inflammatory bowel diseases: An international organization for study of inflammatory bowel diseases consensus. Lancet Gastroenterol Hepatol. 2022;7:666-78
2. Ledder O, Turner D. Antibiotics in IBD: Still a role in the biological era? Inflamm Bowel Dis. 2018;24:1676-88
3. Núñez F P, Quera R, Bay C, Castro F, Mezzano G. Drug-induced liver injury used in the treatment of inflammatory bowel disease. J Crohns Colitis. 2022;16:1168-76
4. Núñez P, Quera R, Yarur AJ. Safety of janus kinase inhibitors in inflammatory bowel diseases. Drugs. 2023;83:299-314
5. Kaakoush NO. Fecal transplants as a microbiome-based therapeutic. Curr Opin Microbiol. 2020;56:16-23
6. Sasson AN, Ingram RJM, Zhang Z, Taylor LM, Ananthakrishnan AN, Kaplan GG. et al. The role of precision nutrition in the modulation of microbial composition and function in people with inflammatory bowel disease. Lancet Gastroenterol Hepatol. 2021;6:754-69
7. Binda S, Hill C, Johansen E, Obis D, Pot B, Sanders ME. et al. Criteria to qualify microorganisms as “probiotic” in foods and dietary supplements. Front Microbiol. 2020;11:1662
8. Sanders ME, Merenstein DJ, Reid G, Gibson GR, Rastall RA. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat Rev Gastroenterol Hepatol. 2019;16:605-16
9. Wu F, Liu J. Decorated bacteria and the application in drug delivery. Adv Drug Deliv Rev. 2022;188:114443
10. Gareau MG, Sherman PM, Walker WA. Probiotics and the gut microbiota in intestinal health and disease. Nat Rev Gastroenterol Hepatol. 2010;7:503-14
11. Szajewska H, Kołodziej M, Gieruszczak-Białek D, Skórka A, Ruszczyński M, Shamir R. Systematic review with meta-analysis: lactobacillus rhamnosus GG for treating acute gastroenteritis in children - a 2019 update. Aliment Pharmacol Ther. 2019;49:1376-84
12. Sales-Campos H, Soares SC, Freire Oliveira CJ. An introduction of the role of probiotics in human infections and autoimmune diseases. Crit Rev Microbiol. 2019;45:413-32
13. Goodman C, Keating G, Georgousopoulou E, Hespe C, Levett K. Probiotics for the prevention of antibiotic-associated diarrhoea: A systematic review and meta-analysis. BMJ Open. 2021;11:e043054
14. Francavilla R, Miniello V, Magistà AM, De Canio A, Bucci N, Gagliardi F. et al. A randomized controlled trial of lactobacillus GG in children with functional abdominal pain. Pediatrics. 2010;126:e1445-52
15. Simon E, Călinoiu LF, Mitrea L, Vodnar DC. Probiotics, prebiotics, and synbiotics: Implications and beneficial effects against irritable bowel syndrome. Nutrients. 2021;13:2112
16. Goldenberg JZ, Yap C, Lytvyn L, Lo CK-F, Beardsley J, Mertz D. et al. Probiotics for the prevention of clostridium difficile-associated diarrhea in adults and children. Cochrane IBD Group, Ed. Cochrane Database Syst Rev. 2017;2017:CD006095
17. Liu L, Huh JR, Shah K. Microbiota and the gut-brain-axis: Implications for new therapeutic design in the CNS. eBioMedicine. 2022;77:103908
18. Jeong J-J, Park HJ, Cha MG, Park E, Won S-M, Ganesan R. et al. The lactobacillus as a probiotic: Focusing on liver diseases. Microorganisms. 2022;10:288
19. Milosevic I, Vujovic A, Barac A, Djelic M, Korac M, Radovanovic Spurnic A. et al. Gut-liver axis, gut microbiota, and its modulation in the management of liver diseases: A review of the literature. Int J Mol Sci. 2019;20:395
20. Abenavoli L, Scarpellini E, Colica C, Boccuto L, Salehi B, Sharifi-Rad J. et al. Gut microbiota and obesity: A role for probiotics. Nutrients. 2019;11:2690
21. Dargahi N, Johnson J, Donkor O, Vasiljevic T, Apostolopoulos V. Immunomodulatory effects of probiotics: Can they be used to treat allergies and autoimmune diseases? Maturitas. 2019;119:25-38
22. Wang G, Liu J, Xia Y, Ai L. Probiotics-based interventions for diabetes mellitus: A review. Food Biosci. 2021;43:101172
23. March DS, Jones AW, Bishop NC, Burton JO. The efficacy of prebiotic, probiotic, and synbiotic supplementation in modulating gut-derived circulatory particles associated with cardiovascular disease in individuals receiving dialysis: A systematic review and meta-analysis of randomized controlled trials. J Ren Nutr. 2020;30:347-59
24. Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. 2019;25:716-29
25. Torres J, Mehandru S, Colombel J-F, Peyrin-Biroulet L. Crohn's disease. The Lancet. 2017;389:1741-55
26. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel J-F. Ulcerative colitis. The Lancet. 2017;389:1756-70
27. Hausmann M, Kiessling S, Mestermann S, Webb G, Spöttl T, Andus T. et al. Toll-like receptors 2 and 4 are up-regulated during intestinal inflammation. Gastroenterology. 2002;122:1987-2000
28. Hirahara K, Nakayama T. CD4 + T-cell subsets in inflammatory diseases: Beyond the T h 1/T h 2 paradigm. Int Immunol. 2016;28:163-71
29. Rengarajan S, Vivio EE, Parkes M, Peterson DA, Roberson EDO, Newberry RD. et al. Dynamic immunoglobulin responses to gut bacteria during inflammatory bowel disease. Gut Microbes. 2020;11:405
30. Shi J, Xie Q, Yue Y, Chen Q, Zhao L, Evivie SE. et al. Gut microbiota modulation and anti-inflammatory properties of mixed lactobacilli in dextran sodium sulfate-induced colitis in mice. FOOD Funct. 2021;12:5130-43
31. Lamubol J, Ohto N, Kuwahara H, Mizuno M. Lactiplantibacillus plantarum 22A-3-induced TGF-β1 secretion from intestinal epithelial cells stimulated CD103 + DC and Foxp3 + treg differentiation and amelioration of colitis in mice. Food Funct. 2021;12:8044-55
32. Ahmad W, Din AU, Khan TM, Rehman MU, Hassan A, Aziz T. et al. Lacticaseibacillusparacasei BNCC345679 revolutionizes DSS-induced colitis and modulates gut microbiota. Front Microbiol. 2024;15:1343891
33. Devi MB, Bhattacharya A, Kumar A, Singh CT, Das S, Sarma HK. et al. Potential probiotic lactiplantibacillus plantarum strains alleviate TNF-α by regulating ADAM17 protein and ameliorate gut integrity through tight junction protein expression in in vitro model. Cell Commun Signal. 2024;22:1-20
34. Haque M, Kaminsky L, Abdulqadir R, Engers J, Kovtunov E, Rawat M. et al. Lactobacillus acidophilus inhibits the TNF-α-induced increase in intestinal epithelial tight junction permeability via a TLR-2 and PI3K-dependent inhibition of NF-κB activation. Front Immunol. 2024;15:1348010
35. Huang Z, Liu B, Xiao L, Liao M, Huang L, Zhao X. et al. Effects of breast-fed infants-derived limosilactobacillus reuteri and bifidobacterium breve ameliorate DSS-induced colitis in mice. iScience. 2024;27:110902
36. Wang H, Zhang L, Xu S, Pan J, Zhang Q, Lu R. Surface-layer protein from lactobacillus acidophilus NCFM inhibits lipopolysaccharide-induced inflammation through MAPK and NF-κB signaling pathways in RAW264.7 cells. J Agric Food Chem. 2018;66:7655-62
37. Chandhni PR, Pradhan D, Sowmya K, Gupta S, Kadyan S, Choudhary R. et al. Ameliorative effect of surface proteins of probiotic lactobacilli in colitis mouse models. Front Microbiol. 2021;12:679773
38. Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L. et al. A purified membrane protein from akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23:107-13
39. Wang L, Tang L, Feng Y, Zhao S, Han M, Zhang C. et al. A purified membrane protein from akkermansia muciniphila or the pasteurised bacterium blunts colitis associated tumourigenesis by modulation of CD8+ T cells in mice. Gut. 2020;69:1988-97
40. Furumoto H, Nanthirudjanar T, Kume T, Izumi Y, Park S-B, Kitamura N. et al. 10-oxo-trans-11-octadecenoic acid generated from linoleic acid by a gut lactic acid bacterium lactobacillus plantarum is cytoprotective against oxidative stress. Toxicol Appl Pharmacol. 2016;296:1-9
41. Sánchez B, Urdaci MC. Extracellular proteins from lactobacillus plantarum BMCM12 prevent adhesion of enteropathogens to mucin. Curr Microbiol. 2012;64:592-6
42. Hao H, Zhang X, Tong L, Liu Q, Liang X, Bu Y. et al. Effect of extracellular vesicles derived from lactobacillus plantarum Q7 on gut microbiota and ulcerative colitis in mice. Front Immunol. 2021;12:777147
43. Wyszyńska A, Kobierecka P, Bardowski J, Jagusztyn-Krynicka EK. Lactic acid bacteria—20 years exploring their potential as live vectors for mucosal vaccination. Appl Microbiol Biotechnol. 2015;99:2967-77
44. Bermúdez-Humarán LG, Motta J-P, Aubry C, Kharrat P, Rous-Martin L, Sallenave J-M. et al. Serine protease inhibitors protect better than IL-10 and TGF-β anti-inflammatory cytokines against mouse colitis when delivered by recombinant lactococci. Microb Cell Factories. 2015;14:26
45. Schotte L, Steidler L, Vandekerckhove J, Remaut E. Secretion of biologically active murine interleukin-10 by lactococcus lactis. Enzyme Microb Technol. 2000;27:761-5
46. Steidler L, Neirynck S, Huyghebaert N, Snoeck V, Vermeire A, Goddeeris B. et al. Biological containment of genetically modified lactococcus lactis for intestinal delivery of human interleukin 10. Nat Biotechnol. 2003;21:785-9
47. Dou X, Qiao L, Chang J, Yan S, Song X, Chen Y. et al. Lactobacillus casei ATCC 393 and it's metabolites alleviate dextran sulphate sodium-induced ulcerative colitis in mice through the NLRP3-(caspase-1)/IL-1β pathway. Food Funct. 2021;12:12022-35
48. Luerce TD, Gomes-Santos AC, Rocha CS, Moreira TG, Cruz DN, Lemos L. et al. Anti-inflammatory effects of lactococcus lactis NCDO 2118 during the remission period of chemically induced colitis. GUT Pathog. 2014;6:33
49. Holmström KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411-21
50. Yang S, Lian G. ROS and diseases: Role in metabolism and energy supply. Mol Cell Biochem. 2020;467:1-12
51. Neish AS. Redox signaling mediated by the gut microbiota. Free Radic Res. 2013;47:950-7
52. Pereira C, Grácio D, Teixeira JP, Magro F. Oxidative stress and DNA damage: Implications in inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:2403-17
53. Lu J, Holmgren A. The thioredoxin antioxidant system. Free Radic Biol Med. 2014;66:75-87
54. Kong Y, Olejar KJ, On SLW, Chelikani V. The potential of lactobacillus spp. for modulating oxidative stress in the gastrointestinal tract. Antioxidants. 2020;9:610
55. Finamore A, Ambra R, Nobili F, Garaguso I, Raguzzini A, Serafini M. Redox role of lactobacillus casei shirota against the cellular damage induced by 2,2′-azobis (2-amidinopropane) dihydrochloride-induced oxidative and inflammatory stress in enterocytes-like epithelial cells. Front Immunol. 2018;9:1131
56. del Carmen S, de Moreno de LeBlanc A, Martin R, Chain F, Langella P, Bermúdez-Humarán LG. et al. Genetically engineered immunomodulatory streptococcus thermophilus strains producing antioxidant enzymes exhibit enhanced anti-inflammatory activities. Appl Environ Microbiol. 2014;80:869-77
57. Watterlot L, Rochat T, Sokol H, Cherbuy C, Bouloufa I, Lefèvre F. et al. Intragastric administration of a superoxide dismutase-producing recombinant lactobacillus casei BL23 strain attenuates DSS colitis in mice. Int J Food Microbiol. 2010;144:35-41
58. Pan Y, Ning Y, Hu J, Wang Z, Chen X, Zhao X. The preventive effect of lactobacillus plantarum ZS62 on DSS-induced IBD by regulating oxidative stress and the immune response. Oxid Med Cell Longev. 2021;2021:9416794
59. Ahmed SMU, Luo L, Namani A, Wang XJ, Tang X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim Biophys Acta BBA - Mol Basis Dis. 2017;1863:585-97
60. Jin T, Lu H, Zhou Q, Chen D, Zeng Y, Shi J. et al. H2S-releasing versatile montmorillonite nanoformulation trilogically renovates the gut microenvironment for inflammatory bowel disease modulation. Adv Sci. 2024;11:2308092
61. Khor TO, Huang M-T, Kwon KH, Chan JY, Reddy BS, Kong A-N. Nrf2-deficient mice have an increased susceptibility to dextran sulfate sodium-induced colitis. Cancer Res. 2006;66:11580-4
62. Liu T, Zhang L, Joo D, Sun S-C. NF-κB signaling in inflammation. Signal Transduct Target Ther. 2017;2:17023
63. Nenci A, Becker C, Wullaert A, Gareus R, Van Loo G, Danese S. et al. Epithelial NEMO links innate immunity to chronic intestinal inflammation. Nature. 2007;446:557-61
64. Wakabayashi N, Slocum SL, Skoko JJ, Shin S, Kensler TW. When NRF2 Talks, Who's Listening? Antioxid Redox Signal. 2010;13:1649-63
65. Gao D, Gao Z, Zhu G. Antioxidant effects of lactobacillus plantarum via activation of transcription factor Nrf2. Food Funct. 2013;4:982
66. Neurath MF. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat Immunol. 2019;20:970-9
67. Gessain G, Tsai Y-H, Travier L, Bonazzi M, Grayo S, Cossart P. et al. PI3-kinase activation is critical for host barrier permissiveness to listeria monocytogenes. J Exp Med. 2015;212:165-83
68. Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut. 2020;69:2232-43
69. Corfield AP. The interaction of the gut microbiota with the mucus barrier in health and disease in human. Microorganisms. 2018;6:78
70. Gersemann M, Becker S, Kübler I, Koslowski M, Wang G, Herrlinger KR. et al. Differences in goblet cell differentiation between crohn's disease and ulcerative colitis. Differentiation. 2009;77:84-94
71. Cornick S, Tawiah A, Chadee K. Roles and regulation of the mucus barrier in the gut. Tissue Barriers. 2015;3:e982426
72. Gustafsson JK, Johansson MEV. The role of goblet cells and mucus in intestinal homeostasis. Nat Rev Gastroenterol Hepatol. 2022;19:785-803
73. Zeissig S, Burgel N, Gunzel D, Richter J, Mankertz J, Wahnschaffe U. et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active crohn's disease. Gut. 2007;56:61-72
74. Zhang C, Li Q, Xing J, Yang Y, Zhu M, Lin L. et al. Tannic acid and zinc ion coordination of nanase for the treatment of inflammatory bowel disease by promoting mucosal repair and removing reactive oxygen and nitrogen species. Acta Biomater. 2024;177:347-60
75. Al-Sadi R, Dharmaprakash V, Nighot P, Guo S, Nighot M, Do T. et al. Bifidobacterium bifidum enhances the intestinal epithelial tight junction barrier and protects against intestinal inflammation by targeting the toll-like receptor-2 pathway in an NF-κB-independent manner. Int J Mol Sci. 2021;22:8070
76. Chen Y, Yang B, Stanton C, Ross RP, Zhao J, Zhang H. et al. Bifidobacterium pseudocatenulatum ameliorates DSS-induced colitis by maintaining intestinal mechanical barrier, blocking proinflammatory cytokines, inhibiting TLR4/NF-κB signaling, and altering gut microbiota. J Agric Food Chem. 2021;69:1496-512
77. Hsieh C-Y, Osaka T, Moriyama E, Date Y, Kikuchi J, Tsuneda S. Strengthening of the intestinal epithelial tight junction by bifidobacterium bifidum. Physiol Rep. 2015;3:e12327
78. Qian G, Zang H, Tang J, Zhang H, Yu J, Jia H. et al. Lactobacillus gasseri ATCC33323 affects the intestinal mucosal barrier to ameliorate DSS-induced colitis through the NR1I3-mediated regulation of E-cadherin. McLoughlin RM, Ed. PLOS Pathog. 2024;20:e1012541
79. Zaylaa M, Al Kassaa I, Alard J, Peucelle V, Boutillier D, Desramaut J. et al. Probiotics in IBD: Combining in vitro and in vivo models for selecting strains with both anti-inflammatory potential as well as a capacity to restore the gut check for epithelial barrier. J Funct FOODS. 2018;47:304-15
80. Yin M, Yan X, Weng W, Yang Y, Gao R, Liu M. et al. Micro integral membrane protein (MIMP), a newly discovered anti-inflammatory protein of lactobacillus plantarum, enhances the gut barrier and modulates microbiota and inflammatory cytokines. Cell Physiol Biochem. 2018;45:474-90
81. Deehan EC, Yang C, Perez-Muñoz ME, Nguyen NK, Cheng CC, Triador L. et al. Precision microbiome modulation with discrete dietary fiber structures directs short-chain fatty acid production. Cell Host Microbe. 2020;27:389-404.e6
82. Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221-7
83. Masui R, Sasaki M, Funaki Y, Ogasawara N, Mizuno M, Iida A. et al. G protein-coupled receptor 43 moderates gut inflammation through cytokine regulation from mononuclear cells. Inflamm Bowel Dis. 2013;19:2848-56
84. Meng F, Zhao H, Nie T, Lu F, Zhang C, Lu Y. et al. Acetate activates lactobacillus bacteriocin synthesis by controlling quorum sensing. Dozois CM, Ed. Appl Environ Microbiol. 2021;87:e00720-21
85. Peng L, Li Z-R, Green RS, Holzmanr IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in caco-2 cell monolayers. J Nutr. 2009;139:1619-25
86. Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front Microbiol. 2016;7:979
87. Kim CH. Complex regulatory effects of gut microbial short-chain fatty acids on immune tolerance and autoimmunity. Cell Mol Immunol. 2023;20:341-50
88. Bachem A, Makhlouf C, Binger KJ, de Souza DP, Tull D, Hochheiser K. et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity. 2019;51:285-297.e5
89. Huda-Faujan N, Abdulamir AS, Fatimah AB, Anas OM, Shuhaimi M, Yazid AM. et al. The impact of the level of the intestinal short chain fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem J. 2010;4:53-8
90. Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, Eeckhaut V. et al. A decrease of the butyrate-producing species roseburia hominis and faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275-83
91. Kumari R. Fluctuations in butyrate-producing bacteria in ulcerative colitis patients of north India. World J Gastroenterol. 2013;19:3404
92. Geirnaert A, Calatayud M, Grootaert C, Laukens D, Devriese S, Smagghe G. et al. Butyrate-producing bacteria supplemented in vitro to crohn's disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci Rep. 2017;7:11450
93. Breuer RI, Soergel KH, Lashner BA, Christ ML, Hanauer SB, Vanagunas A. et al. Short chain fatty acid rectal irrigation for left-sided ulcerative colitis: A randomised, placebo controlled trial. Gut. 1997;40:485-91
94. Lührs H, Gerke T, Müller JG, Melcher R, Schauber J, Boxberger F. et al. Butyrate inhibits NF-κB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol. 2002;37:458-66
95. Wu Y, Jha R, Li A, Liu H, Zhang Z, Zhang C. et al. Probiotics (lactobacillus plantarum HNU082) supplementation relieves ulcerative colitis by affecting intestinal barrier functions, immunity-related gene expression, gut microbiota, and metabolic pathways in mice. Tang X, Ed. Microbiol Spectr. 2022;10:e01651-22
96. Lee M, Chang EB. Inflammatory bowel diseases (IBD) and the microbiome—searching the crime scene for clues. Gastroenterology. 2021;160:524-37
97. Scanlan PD, Marchesi JR. Micro-eukaryotic diversity of the human distal gut microbiota: Qualitative assessment using culture-dependent and -independent analysis of faeces. ISME J. 2008;2:1183-93
98. Fite A, Macfarlane S, Furrie E, Bahrami B, Cummings JH, Steinke DT. et al. Longitudinal analyses of gut mucosal microbiotas in ulcerative colitis in relation to patient age and disease severity and duration. J Clin Microbiol. 2013;51:849-56
99. Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C. et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66:1039-48
100. Duerkop BA, Kleiner M, Paez-Espino D, Zhu W, Bushnell B, Hassell B. et al. Murine colitis reveals a disease-associated bacteriophage community. Nat Microbiol. 2018;3:1023-31
101. Scanlan PD, Shanahan F, Marchesi JR. Human methanogen diversity and incidence in healthy and diseased colonic groups using mcrA gene analysis. BMC Microbiol. 2008;8:79
102. Heilbronner S, Krismer B, Brötz-Oesterhelt H, Peschel A. The microbiome-shaping roles of bacteriocins. Nat Rev Microbiol. 2021;19:726-39
103. De Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: Mechanistic insights. Gut. 2022;71:1020-32
104. Kline KA, Fälker S, Dahlberg S, Normark S, Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5:580-92
105. Cossart P, Roy CR. Manipulation of host membrane machinery by bacterial pathogens. Curr Opin Cell Biol. 2010;22:547-54
106. Drolia R, Amalaradjou MAR, Ryan V, Tenguria S, Liu D, Bai X. et al. Receptor-targeted engineered probiotics mitigate lethal listeria infection. Nat Commun. 2020;11:6344
107. Misselwitz B, Barrett N, Kreibich S, Vonaesch P, Andritschke D, Rout S. et al. Near surface swimming of salmonella typhimurium explains target-site selection and cooperative invasion. Galán JE, Ed. PLoS Pathog. 2012;8:e1002810
108. Dillon A, Lo DD. M cells: Intelligent engineering of mucosal immune surveillance. Front Immunol. 2019;10:1499
109. Abdi M, Lohrasbi V, Asadi A, Esghaei M, Jazi FM, Rohani M. et al. Interesting probiotic traits of mother's milk lactobacillus isolates; from bacteriocin to inflammatory bowel disease improvement. Microb Pathog. 2021;158:104998
110. Sarao LK, Arora M. Probiotics, prebiotics, and microencapsulation: A review. Crit Rev Food Sci Nutr. 2017;57:344-71
111. Yao M, Li B, Ye H, Huang W, Luo Q, Xiao H. et al. Enhanced viability of probiotics (pediococcus pentosaceus Li05) by encapsulation in microgels doped with inorganic nanoparticles. Food Hydrocoll. 2018;83:246-52
112. Dodoo CC, Wang J, Basit AW, Stapleton P, Gaisford S. Targeted delivery of probiotics to enhance gastrointestinal stability and intestinal colonisation. Int J Pharm. 2017;530:224-9
113. González-Ferrero C, Irache JM, González-Navarro CJ. Soybean protein-based microparticles for oral delivery of probiotics with improved stability during storage and gut resistance. Food Chem. 2018;239:879-88
114. Culligan EP, Hill C, Sleator RD. Probiotics and gastrointestinal disease: successes, problems and future prospects. Gut Pathog. 2009;1:19
115. Steidler L, Hans W, Schotte L, Neirynck S, Obermeier F, Falk W. et al. Treatment of murine colitis by lactococcus lactis secreting interleukin-10. Science. 2000;289:1352-5
116. Centurion F, Merhebi S, Baharfar M, Abbasi R, Zhang C, Mousavi M. et al. Cell-mediated biointerfacial phenolic assembly for probiotic nano encapsulation. Adv Funct Mater. 2022;32:2200775
117. Cao F, Jin L, Zhang C, Gao Y, Qian Z, Wen H. et al. Engineering clinically relevant probiotics with switchable “nano-promoter” and “nano-effector” for precision tumor therapy. Adv Mater. 2024;36:2304257
118. Press AG, Hauptmann IA, Hauptmann L, Fuchs B, Fuchs M, Ewe K. et al. Gastrointestinal pH profiles in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 1998;12:673-8
119. Martini E, Krug SM, Siegmund B, Neurath MF, Becker C. Mend your fences: The epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2017;4:33-46
120. Fischer M, Siva S, Wo JM, Fadda HM. Assessment of small intestinal transit times in ulcerative colitis and crohn's disease patients with different disease activity using video capsule endoscopy. AAPS PharmSciTech. 2017;18:404-9
121. Niu W, Yang F, Fu Z, Dong Y, Zhang Z, Ju J. The role of enteric dysbacteriosis and modulation of gut microbiota in the treatment of inflammatory bowel disease. Microb Pathog. 2022;165:105381
122. Awad A, Madla CM, McCoubrey LE, Ferraro F, Gavins FKH, Buanz A. et al. Clinical translation of advanced colonic drug delivery technologies. Adv Drug Deliv Rev. 2022;181:114076
123. Fan X, Zhang Z, Gao W, Pan Q, Luo K, He B. et al. An engineered butyrate-derived polymer nanoplatform as a mucosa-healing enhancer potentiates the therapeutic effect of magnolol in inflammatory bowel disease. ACS Nano. 2024;18:229-44
124. Zeng Y, Fan M, Zhou Q, Chen D, Jin T, Mu Z. et al. Reactive oxygen species-activated CO versatile nanomedicine with innate gut immune and microbiome remodeling effects for treating inflammatory bowel disease. Adv Funct Mater. 2023;33:2304381
125. Wang R, Guo K, Zhang W, He Y, Yang K, Chen Q. et al. Poly-γ-glutamic acid microgel-encapsulated probiotics with gastric acid resistance and smart inflammatory factor targeted delivery performance to ameliorate colitis. Adv Funct Mater. 2022;32:2113034
126. Huang L, Wang J, Kong L, Wang X, Li Q, Zhang L. et al. ROS-responsive hyaluronic acid hydrogel for targeted delivery of probiotics to relieve colitis. Int J Biol Macromol. 2022;222:1476-86
127. Xiao Y, Lu C, Liu Y, Kong L, Bai H, Mu H. et al. Encapsulation of lactobacillus rhamnosus in hyaluronic acid-based hydrogel for pathogen-targeted delivery to ameliorate enteritis. ACS Appl Mater Interfaces. 2020;12:36967-77
128. Beveridge TJ, Graham LL. Surface layers of bacteria. Microbiol Rev. 1991;55:684-705
129. Yeung TW, Arroyo-Maya IJ, McClements DJ, Sela DA. Microencapsulation of probiotics in hydrogel particles: Enhancing lactococcus lactis subsp. cremoris LM0230 viability using calcium alginate beads. Food Funct. 2016;7:1797-804
130. Li R, Zhang Y, Polk DB, Tomasula PM, Yan F, Liu L. Preserving viability of lactobacillus rhamnosus GG in vitro and in vivo by a new encapsulation system. J Control Release Off J Control Release Soc. 2016;230:79-87
131. Lee Y, Kang Y-R, Chang YH. Effect of pectic oligosaccharide on probiotic survival and physicochemical properties of hydrogel beads for synbiotic encapsulation of lactobacillus bulgaricus. Food Biosci. 2023;51:102260
132. Liu L, Deng J, Guo S, Li Y, Wang L, Liu Y. et al. Stress tolerance and characterization of biofilm-like probiotic bacteria encapsulated in calcium pectin beads. LWT. 2023;184:115072
133. Cook MT, Tzortzis G, Khutoryanskiy VV, Charalampopoulos D. Layer-by-layer coating of alginate matrices with chitosan-alginate for the improved survival and targeted delivery of probiotic bacteria after oral administration. J Mater Chem B. 2012;1:52-60
134. Etchepare M, Nunes G, Nicoloso B, Barin J, Flores E, Mello R. et al. Improvement of the viability of encapsulated probiotics using whey proteins. LWT. 2019;117:108601
135. Pankasemsuk T, Apichartsrangkoon A, Worametrachanon S, Techarang J. Encapsulation of lactobacillus casei 01 by alginate along with hi-maize starch for exposure to a simulated gut model. Food Biosci. 2016;16:32-6
136. Apiwattanasiri P, Charoen R, Rittisak S, Phattayakorn K, Jantrasee S, Savedboworn W. Co-encapsulation efficiency of silk sericin-alginate-prebiotics and the effectiveness of silk sericin coating layer on the survival of probiotic lactobacillus casei. Food Biosci. 2022;46:101576
137. Hou W, Li J, Cao Z, Lin S, Pan C, Pang Y. et al. Decorating bacteria with a therapeutic nanocoating for synergistically enhanced biotherapy. Small. 2021;17:2101810
138. Feng P, Cao Z, Wang X, Li J, Liu J. On-demand bacterial reactivation by restraining within a triggerable nanocoating. Adv Mater. 2020;32:2002406
139. Asgari S, Pourjavadi A, Licht TR, Boisen A, Ajalloueian F. Polymeric carriers for enhanced delivery of probiotics. Adv Drug Deliv Rev. 2020;161-162:1-21
140. Pu Y, Fan X, Zhang Z, Guo Z, Pan Q, Gao W. et al. Harnessing polymer-derived drug delivery systems for combating inflammatory bowel disease. J Controlled Release. 2023;354:1-18
141. Tan LL, Mahotra M, Chan SY, Loo SCJ. In situ alginate crosslinking during spray-drying of lactobacilli probiotics promotes gastrointestinal-targeted delivery. Carbohydr Polym. 2022;286:119279
142. Akkurt S. Encapsulation of lactobacillus rhamnosus GG in edible electrospun mats from calcium and sodium caseinates with pullulan blends. JDS Commun. 2022;2:381-6
143. Škrlec K, Zupančič Š, Prpar Mihevc S, Kocbek P, Kristl J, Berlec A. Development of electrospun nanofibers that enable high loading and long-term viability of probiotics. Eur J Pharm Biopharm. 2019;136:108-19
144. Rosas-Val P, Adhami M, Brotons-Canto A, Gamazo C, Irache JM, Larrañeta E. 3D printing of microencapsulated lactobacillus rhamnosus for oral delivery. Int J Pharm. 2023;641:123058
145. Geng Z, Cao Z, Liu R, Liu K, Liu J, Tan W. Aptamer-assisted tumor localization of bacteria for enhanced biotherapy. Nat Commun. 2021;12:6584
146. Pan J, Gong G, Wang Q, Shang J, He Y, Catania C. et al. A single-cell nanocoating of probiotics for enhanced amelioration of antibiotic-associated diarrhea. Nat Commun. 2022;13:2117
147. Anselmo AC, McHugh KJ, Webster J, Langer R, Jaklenec A. Layer-by-layer encapsulation of probiotics for delivery to the microbiome. Adv Mater. 2016;28:9486-90
148. Liu B, Hu J, Yao H, Zhang L, Liu H. Improved viability of probiotics encapsulated by layer-by-layer assembly using zein nanoparticles and pectin. Food Hydrocoll. 2023;143:108899
149. Pan C, Li J, Hou W, Lin S, Wang L, Pang Y. et al. Polymerization-mediated multifunctionalization of living cells for enhanced cell-based therapy. Adv Mater. 2021;33:2007379
150. Yin L, Meng Z, Zhang Y, Hu K, Chen W, Han K. et al. Bacillus spore-based oral carriers loading curcumin for the therapy of colon cancer. J Controlled Release. 2018;271:31-44
151. Cao Z, Wang X, Pang Y, Cheng S, Liu J. Biointerfacial self-assembly generates lipid membrane coated bacteria for enhanced oral delivery and treatment. Nat Commun. 2019;10:5783
152. Liu R, Cao Z, Wang L, Wang X, Lin S, Wu F. et al. Multimodal oncolytic bacteria by coating with tumor cell derived nanoshells. Nano Today. 2022;45:101537
153. Chen Y, Lin S, Wang L, Zhang Y, Chen H, Fu Z. et al. Reinforcement of the intestinal mucosal barrier via mucus-penetrating PEGylated bacteria. Nat Biomed Eng. 2024;8:823-41
154. Luo H, Chen Y, Kuang X, Wang X, Yang F, Cao Z. et al. Chemical reaction-mediated covalent localization of bacteria. Nat Commun. 2022;13:7808
155. Lin W, Liu Y, Wang J, Zhao Z, Lu K, Meng H. et al. Engineered bacteria labeled with iridium(III) photosensitizers for enhanced photodynamic immunotherapy of solid tumors. 2023; 62: e202310158.
156. Guo H, Cao Z, Li J, Fu Z, Lin S, Wang L. et al. Integrating bacteria with a ternary combination of photosensitizers for monochromatic irradiation-mediated photoacoustic imaging-guided synergistic photothermal therapy. ACS Nano. 2023;17:5059-71
157. Cao F, Jin L, Gao Y, Ding Y, Wen H, Qian Z. et al. Artificial-enzymes-armed bifidobacterium longum probiotics for alleviating intestinal inflammation and microbiota dysbiosis. Nat Nanotechnol. 2023;18:617-27
158. Peng P, Feng T, Yang X, Ding R, Wang J, Chen P. et al. Bioorthogonal conjugation and responsive nanocoating of probiotics for inflammatory bowel disease. J Controlled Release. 2024;374:538-49
159. Song W-F, Yao W-Q, Chen Q-W, Zheng D, Han Z-Y, Zhang X-Z. In situ bioorthogonal conjugation of delivered bacteria with gut inhabitants for enhancing probiotics colonization. ACS Cent Sci. 2022;8:1306-17
160. Charbonneau MR, Isabella VM, Li N, Kurtz CB. Developing a new class of engineered live bacterial therapeutics to treat human diseases. Nat Commun. 2020;11:1738
161. Mugwanda K, Hamese S, Van Zyl WF, Prinsloo E, Du Plessis M, Dicks LMT. et al. Recent advances in genetic tools for engineering probiotic lactic acid bacteria. Biosci Rep. 2023;43:BSR20211299
162. Yu D, Ellis HM, Lee E-C, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in escherichia coli. Proc Natl Acad Sci. 2000;97:5978-83
163. LeBlanc JG, Levit R, Savoy de Giori G, de Moreno de LeBlanc A. Application of vitamin-producing lactic acid bacteria to treat intestinal inflammatory diseases. Appl Microbiol Biotechnol. 2020;104:3331-7
164. Belo GA, Cordeiro BF, Oliveira ER, Braga MP, da Silva SH, Costa BG. et al. SlpB protein enhances the probiotic potential of L. lactis NCDO 2118 in colitis mice model. Front Pharmacol. 2021;12:755825
165. Wang T, Li Y, Li J, Zhang D, Cai N, Zhao G. et al. An update of the suicide plasmid-mediated genome editing system in corynebacterium glutamicum. Microb Biotechnol. 2019;12:907-19
166. Luo ML, Mullis AS, Leenay RT, Beisel CL. Repurposing endogenous type I CRISPR-cas systems for programmable gene repression. Nucleic Acids Res. 2015;43:674-81
167. Lowe DC, Savidge TC, Pickard D, Eckmann L, Kagnoff MF, Dougan G. et al. Characterization of candidate live oral salmonella typhi vaccine strains harboring defined mutations inaroA, aroC, and htrA. Infect Immun. 1999;67:700-7
168. Chen S, Yao Y, Zhang Y, Fan G. CRISPR system: Discovery, development and off-target detection. Cell Signal. 2020;70:109577
169. Wang C, Cui Y, Qu X. Optimization of electrotransformation (ETF) conditions in lactic acid bacteria (LAB). J Microbiol Methods. 2020;174:105944
170. Sanders JW, Venema G, Kok J. A chloride-inducible gene expression cassette and its use in induced lysis of lactococcus lactis. Appl Environ Microbiol. 1997;63:4877-82
171. de Ruyter PG, Kuipers OP, de Vos WM. Controlled gene expression systems for lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662-7
172. Sirén N, Salonen K, Leisola M, Nyyssölä A. A new and efficient phosphate starvation inducible expression system for lactococcus lactis. Appl Microbiol Biotechnol. 2008;79:803-10
173. Shigemori S, Watanabe T, Kudoh K, Ihara M, Nigar S, Yamamoto Y. et al. Oral delivery of lactococcus lactis that secretes bioactive heme oxygenase-1 alleviates development of acute colitis in mice. Microb Cell Factories. 2015;14:189
174. Carmen S del, LeBlanc A de M de, Perdigon G, Pereira VB, Miyoshi A, Azevedo V. et al. Evaluation of the anti-inflammatory effect of milk fermented by a strain of IL-10-producing lactococcus lactis using a murine model of crohn's disease. Microb Physiol. 2011;21:138-46
175. Gomes-Santos AC, Oliveira RP de, Moreira TG, Castro-Junior AB, Horta BC, Lemos L. et al. Hsp65-producing lactococcus lactis prevents inflammatory intestinal disease in mice by IL-10- and TLR2-dependent pathways. Front Immunol. 2017;8:30
176. Chua KJ, Ling H, Hwang IY, Lee HL, March JC, Lee YS. et al. An engineered probiotic produces a type III interferon IFNL1 and reduces inflammations in in vitro inflammatory bowel disease models. ACS Biomater Sci Eng. 2023;9:5123-35
177. Wu Y-Q, Zou Z-P, Zhou Y, Ye B-C. Dual engineered bacteria improve inflammatory bowel disease in mice. Appl Microbiol Biotechnol. 2024;108:333
178. Zahirović A, Berlec A. Targeting IL-6 by engineered lactococcus lactis via surface-displayed affibody. Microb Cell Factories. 2022;21:143
179. Levit R, Cortes-Perez NG, de Moreno de Leblanc A, Loiseau J, Aucouturier A, Langella P. et al. Use of genetically modified lactic acid bacteria and bifidobacteria as live delivery vectors for human and animal health. Gut Microbes. 2022;14:2110821
180. Plavec TV, Berlec A. Engineering of lactic acid bacteria for delivery of therapeutic proteins and peptides. Appl Microbiol Biotechnol. 2019;103:2053-66
181. Cheng F-S, Pan D, Chang B, Jiang M, Sang L-X. Probiotic mixture VSL#3: An overview of basic and clinical studies in chronic diseases. World J Clin Cases. 2020;8:1361-84
182. Pronio A, Montesani C, Butteroni C, Vecchione S, Mumolo G, Vestri A. et al. Probiotic administration in patients with ileal pouch-anal anastomosis for ulcerative colitis is associated with expansion of mucosal regulatory cells. Inflamm Bowel Dis. 2008;14:662-8
183. Di Giacinto C, Marinaro M, Sanchez M, Strober W, Boirivant M. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-β-bearing regulatory cells. J Immunol. 2005;174:3237
184. Dang X, Xu M, Liu D, Zhou D, Yang W. Assessing the efficacy and safety of fecal microbiota transplantation and probiotic VSL#3 for active ulcerative colitis: A systematic review and meta-analysis. Cheungpasitporn W, Ed. PLOS ONE. 2020;15:e0228846
185. Huynh HQ, deBruyn J, Guan L, Diaz H, Li M, Girgis S. et al. Probiotic preparation VSL#3 induces remission in children with mild to moderate acute ulcerative colitis: A pilot study. Inflamm Bowel Dis. 2009;15:760-8
186. Guandalini S, Magazzù G, Chiaro A, La Balestra V, Di Nardo G, Gopalan S. et al. VSL#3 improves symptoms in children with irritable bowel syndrome: A multicenter, randomized, placebo-controlled, double-blind, crossover study. J Pediatr Gastroenterol Nutr. 2010;51:24-30
187. Yan F, Polk DB. Characterization of a probiotic-derived soluble protein which reveals a mechanism of preventive and treatment effects of probiotics on intestinal inflammatory diseases. Gut Microbes. 2012;3:25-8
188. Yan F, Cao H, Cover TL, Washington MK, Shi Y, Liu L. et al. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J Clin Invest. 2011;121:2242-53
189. Pagnini C, Corleto VD, Martorelli M, Lanini C, D'Ambra G, Giulio ED. et al. Mucosal adhesion and anti-inflammatory effects of lactobacillus rhamnosus GG in the human colonic mucosa: A proof-of-concept study. World J Gastroenterol. 2018;24:4652-62
190. Gao J, Li Y, Wan Y, Hu T, Liu L, Yang S. et al. A novel postbiotic from lactobacillus rhamnosus GG with a beneficial effect on intestinal barrier function. Front Microbiol. 2019;10:477
191. Francavilla R, Miniello V, Magistà AM, De Canio A, Bucci N, Gagliardi F. et al. A randomized controlled trial of lactobacillus GG in children with functional abdominal pain. Pediatrics. 2010;126:e1445-1452
192. Stöber H, Maier E, Schmidt H. Protective effects of lactobacilli, bifidobacteria and staphylococci on the infection of cultured HT29 cells with different enterohemorrhagic escherichia coli serotypes are strain-specific. Int J Food Microbiol. 2010;144:133-40
193. Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132:562-75
194. Shen X, Liu L, Peek RM, Acra SA, Moore DJ, Wilson KT. et al. Supplementation of p40, a lactobacillus rhamnosus GG-derived protein, in early life promotes epidermal growth factor receptor-dependent intestinal development and long-term health outcomes. Mucosal Immunol. 2018;11:1316-28
195. Capurso L. Thirty years of lactobacillus rhamnosus GG: A review. J Clin Gastroenterol. 2019;53:S1-41
196. Bischoff SC, Bager P, Escher J, Forbes A, Hébuterne X, Hvas CL. et al. ESPEN guideline on clinical nutrition in inflammatory bowel disease. Clin Nutr. 2023;42:352-79
197. Huang J, Yang Z, Li Y, Chai X, Liang Y, Lin B. et al. Lactobacillus paracasei R3 protects against dextran sulfate sodium (DSS)-induced colitis in mice via regulating Th17/treg cell balance. J Transl Med. 2021;19:356
Author biographies
 Guangze Sang received his B.M. degree in 2016. He is currently pursuing his M.S. degree under the guidance of Prof. Feng Yang at the department of Inorganic Chemistry, School of Pharmacy, Naval Medical University, Shanghai. His research interests focus on targeted drug delivery and bacterial modification.
Guangze Sang received his B.M. degree in 2016. He is currently pursuing his M.S. degree under the guidance of Prof. Feng Yang at the department of Inorganic Chemistry, School of Pharmacy, Naval Medical University, Shanghai. His research interests focus on targeted drug delivery and bacterial modification.
 Dr. Bingkai Wang received his Ph.D. degree from Lanzhou University. He is now a lecturer in Department of Pharmacy, Naval Medical University. His current research interests focused on the design of probiotics-based biomedical materials for anti-inflammatory applications.
Dr. Bingkai Wang received his Ph.D. degree from Lanzhou University. He is now a lecturer in Department of Pharmacy, Naval Medical University. His current research interests focused on the design of probiotics-based biomedical materials for anti-inflammatory applications.
 A/Prof. Yujie Xie obtained her B.Sc. and M.Sc. in Chemistry from the Lanzhou University. She received her PhD from the University of Warwick and then worked as a postdoc at the University of Birmingham. Currently, she is an associate professor in School of Medicine, Shanghai University. Her current research interests focused on the design of polymer-based biomedical materials for anti-tumor applications.
A/Prof. Yujie Xie obtained her B.Sc. and M.Sc. in Chemistry from the Lanzhou University. She received her PhD from the University of Warwick and then worked as a postdoc at the University of Birmingham. Currently, she is an associate professor in School of Medicine, Shanghai University. Her current research interests focused on the design of polymer-based biomedical materials for anti-tumor applications.
 Prof. Yu Chen received his Ph.D. degree at Shanghai Institute of Ceramics, Chinese Academy of Sciences (SICCAS). He is now the full professor in Shanghai University. His research focuses on materdicine, nanomedicine and nanobiotechnology, which involve the design, fabrication and biomedical applications of mesoporous nanoparticles, catalytic biomaterials, two-dimensional nanosheets and 3D-printing bioscaffolds. The focused biomedical applications include drug/gene delivery, molecular imaging, catalytic medicine, catalytic biology, theragenerative biomaterials and in-situ localized disease therapy. He has published more than 300 scientific papers in nanomedicine field with a total citation of more than 48000 times (h-index: 115). He is 2018-2024 Highly Cited Researcher by Clarivate Analytics, Web of Science.
Prof. Yu Chen received his Ph.D. degree at Shanghai Institute of Ceramics, Chinese Academy of Sciences (SICCAS). He is now the full professor in Shanghai University. His research focuses on materdicine, nanomedicine and nanobiotechnology, which involve the design, fabrication and biomedical applications of mesoporous nanoparticles, catalytic biomaterials, two-dimensional nanosheets and 3D-printing bioscaffolds. The focused biomedical applications include drug/gene delivery, molecular imaging, catalytic medicine, catalytic biology, theragenerative biomaterials and in-situ localized disease therapy. He has published more than 300 scientific papers in nanomedicine field with a total citation of more than 48000 times (h-index: 115). He is 2018-2024 Highly Cited Researcher by Clarivate Analytics, Web of Science.
 Prof. Feng Yang obtained his B.S degree in Chemistry from the Lanzhou University, then he received his M.M. and Ph.D. in Naval Medical University, shanghai. He is now the full professor in Naval Medical University, director of the Department of Inorganic Chemistry. His research focuses on targeted and controlled release nanodrug delivery.
Prof. Feng Yang obtained his B.S degree in Chemistry from the Lanzhou University, then he received his M.M. and Ph.D. in Naval Medical University, shanghai. He is now the full professor in Naval Medical University, director of the Department of Inorganic Chemistry. His research focuses on targeted and controlled release nanodrug delivery.
![]() Corresponding authors: wangbkedu.cn; yangfeng1008com; xieyjedu.cn; chenyueduedu.cn.
Corresponding authors: wangbkedu.cn; yangfeng1008com; xieyjedu.cn; chenyueduedu.cn.
 Global reach, higher impact
Global reach, higher impact