13.3
Impact Factor
Theranostics 2025; 15(5):1987-2005. doi:10.7150/thno.98199 This issue Cite
Research Paper
Stress granule formation is regulated by signaling machinery involving Sch9/Ypk1, sphingolipids, and Ubi4
1. Department of Chemistry and Molecular Biology, University of Gothenburg, S-413 90, Göteborg, Sweden.
2. Guangzhou National Laboratory, Guangzhou, Guangdong, China.
3. School of Biomedical Engineering, Guangzhou Medical University, Guangzhou, Guangdong, China.
4. State Key Laboratory of Subtropical Silviculture, School of Forestry and Biotechnology, Zhejiang A&F University, Lin'an, Hangzhou, 311300, China.
5. EATRIS Center for Large-scale cell-based screening, Department of Chemistry and Molecular Biology, University of Gothenburg, S-413 90, Göteborg, Sweden.
† These authors contributed equally to this work.
Received 2024-5-7; Accepted 2024-11-18; Published 2025-1-6
Abstract

Rationale: Stress granules (SGs) are membraneless organelles that are formed in response to various stresses. Multiple cellular processes have been reported to be involved in SG formation. However, the signaling cascades that coordinate SG formation remain to be elucidated.
Methods: By performing two high-content imaging-based phenomic screens, we identified multiple signaling components that form a possible signal transduction pathway that regulates SG formation.
Results: We found that Sch9 and Ypk1 function in an early step of SG formation, leading to a decrease in intermediate long-chain base sphingolipids (LCBs). This further downregulates the polyubiquitin precursor protein Ubi4 through upregulating the deubiquitinase Ubp3. Decreased levels of cellular free ubiquitin may subsequently facilitate Lsm7 phase separation and thus trigger SG formation.
Conclusion: The signaling pathway identified in this work, together with its conserved components, provides valuable clues for understanding the mechanisms underlying SG formation and SG-associated human diseases.
Keywords: stress granules, mammalian target of rapamycin (mTOR), ubiquitination, sphingolipids, phase separation
Introduction
Cytoplasmic stress granules (SGs) are membraneless organelles that temporarily assemble to facilitate rapid cessation of translation in response to stress conditions [1-9] or normal cellular processes such as G protein activation [10]. SGs typically include a plethora of translation initiation components and nontranslating mRNAs, in addition to proteins that regulate mRNA function [1, 3, 11, 12]. Accumulating evidence has shown that ribonucleoprotein particle (RNP) granules, including SGs, form through a process called liquid-liquid phase separation (LLPS). These highly conserved subcellular structures are tightly regulated and thought to form via LLPS of low-complexity protein domains [13-16]. Moreover, RNAs can impact the phase separation of various RNA-binding proteins (RBPs) [17-19]. In addition, cellular signaling and catalytic proteins have been shown to be sequestered in SGs [5, 20, 21]. Dysregulation of SGs is connected to many pathologies, ranging from neurodegenerative diseases to cancers [22-28].
The formation of SGs affects cellular signaling pathways, thereby controlling cell fate or function [21, 24, 29-33]. Accordingly, SG formation affects target of rapamycin (TOR) signaling, which coordinates cellular responses to stressful conditions such as nutritional deficiencies. A key component of the yeast TOR1 complex, Kog1 (raptor in mammalian cells), is transiently recruited to SGs upon heat shock or oxidative stress, thereby delaying the reactivation of TORC1 signaling or preventing mTORC1 hyperactivation-induced apoptosis [24, 29]. However, whether TORC1/2 or any of their downstream components have active signaling roles in regulating SG formation is unclear.
The sphingolipid metabolism pathway has important roles in signal transmission and cell recognition, and disorders in this pathway affect neural tissues [34-36]. The composition of sphingolipids may be altered due to changes under physiological conditions. For example, when yeast cells enter the stationary phase, there is a large increase in total free and sphingolipid-bound long chain bases, which are regulated mainly by intrinsic factors, such as Orm1 and Orm2, and serine palmitoyltransferase [37]. In contrast, under glucose restriction, the amount of free long chain bases decreases [37]. These results highlight the physiological role of long chain bases in stress resistance and survival. Furthermore, sphingolipid have been reported to mediate processing body (PB, another type of RNA granule) formation and translation initiation during heat stress [34]. However, whether there is a link between the physiological role of sphingolipids and SG formation has not been determined.
Recent progress has highlighted the ubiquitin-based machinery that regulates SG formation. It has been shown that free ubiquitin, rather than ubiquitin-conjugated proteins, colocalizes to SGs and that free ubiquitin may alter SG protein interactions [38]. In accordance, specific noncovalent ubiquitin binding may disrupt the LLPS of SG-residing proteins [39]. The cellular ubiquitin pool comprises free ubiquitin and ubiquitin conjugates. Under normal conditions, the bulk of ubiquitin is controlled by three ubiquitin-fused ribosomal proteins, RPL40A (UBI1), RPL40B (UBI2), and RPS31 (UBI3). In yeast, an additional gene, UBI4, accounts for the main source of de novo ubiquitin synthesis under various stresses, including heat stress, oxidative stress, starvation, and the stationary phase [40, 41], while the expression of the other three ubiquitin genes is suppressed during stress conditions [42]. In addition to Ubi4, the yeast deubiquitinase Ubp3 (human USP10) has been found to be a constituent and regulator of SGs [43-45].
In this work, we report a signaling pathway that controls the formation of SGs under 2-deoxyglucose (2-DG) treatment. Using an imaging-based phenomic screen, we found that Sch9 and Ypk1 are downstream effectors of TORC1/2 complexes involved in the regulation of SG formation. The TORC1/2-Sch9/Ypk1 signaling cascade further decreases the levels of major long-chain base sphingolipids (LCBs) in the cell. This subsequently downregulates the expression of the ubiquitin-proteasome system (UPS) component Ubi4, in part by upregulating the deubiquitinase Ubp3. In addition, we found that a SG initiation factor - Lsm7 - is located downstream of the Ubi4-Ubp3 signaling. Ubp3 positively regulates Lsm7 phase separation, thereby triggering SG formation.
Results
Essential components involved in SG formation
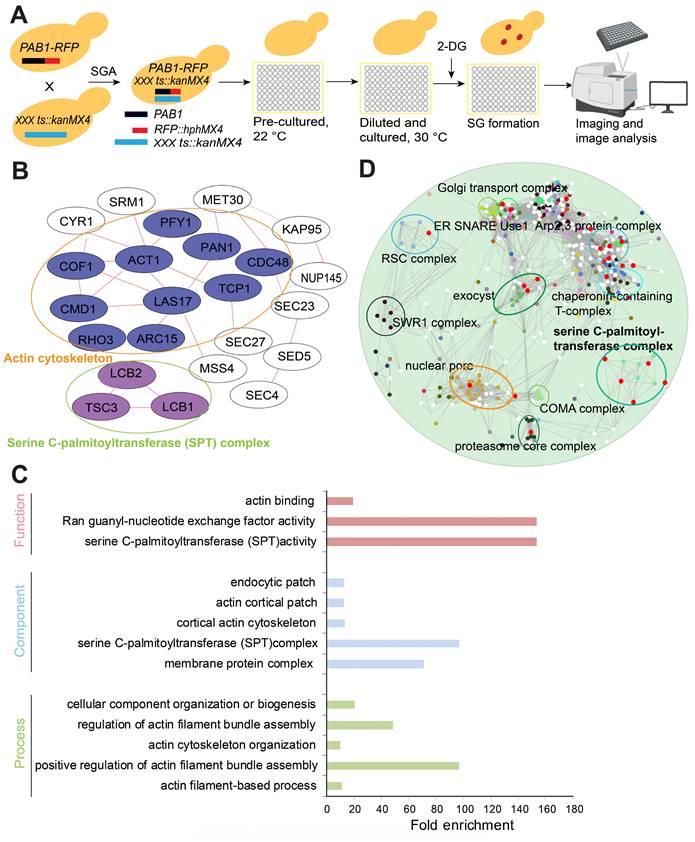
To identify essential SG signaling components, we systematically quantified SG formation using the yeast essential gene temperature sensitive (ts) allele collection [46]. We chose 400 mM 2-DG as the SG inducer since 2-DG can induce robust and steady SG formation [47]. The library for the SG formation screen was constructed by crossing a query strain (Y7092) bearing a red fluorescent protein-tagged Pab1 (Pab1-RFP) with the ts allele collection by using the SGA method [48] (Figure 1A). After manual confirmation, we obtained 31 mutants with significantly increased SG formation. These confirmed hits were analyzed for physical interaction networks using the GeneMANIA plug-in for Cytoscape (Cytoscape v3.2.1) [49]. The network analysis indicated that the genes of the mutants with increased SG formation were enriched for products that physically interact with each other. The interacting proteins included those with cellular functions such as (1) serine C-palmitoyltransferase activity (sphingolipid biosynthesis) and (2) actin cytoskeleton/binding (Figure 1B).
The top hits were also analyzed using the Gene Ontology Term Finder [50] for enrichment of Gene Ontology (GO) biological processes, molecular functions, and cellular component categories via comparisons with a background set of genes (TS-V5) [46]. The enriched cellular functions were enriched for (1) serine C-palmitoyltransferase activity (sphingolipid biosynthesis), (2) actin binding, and (3) Ran guanyl-nucleotide exchange factor activity; the enriched cellular components were (1) serine C-palmitoyltransferase complex, (2) endocytic patch, (3) actin cortical patch, (4) cortical actin cytoskeleton, and (5) membrane protein complex; and the enriched biological processes were (1) cellular component organization or biogenesis, (2) regulation of actin filament bundle assembly, (3) positive regulation of actin filament bundle assembly, (4) actin cytoskeleton organization, and (5) actin filament-based process (Figure 1C).
Furthermore, we performed a genetic interaction profile similarity network analysis (annotated with protein complex) for the ts alleles that displayed increased SG formation. The results showed that these ts alleles were enriched in the following complexes: serine C-palmitoyltransferase complex, Arp2,3 protein complex, exocyst, proteasome core complex, nuclear pore, and Golgi transport complex; or interacted with these complexes: chaperonin-containing T-complex, COMA complex, SWR1 complex, RSC complex, nuclear pore, and ER SNARE Use1 (Figure 1D). From these analyses, ts mutants of the serine palmitoyltransferase complex (lcb1-4, lcb2-16 and tsc3-2) were identified as a consistent significantly enriched functional group from our screen.
A high-content screening-based study identified the serine C-palmitoyltransferase complex as a major regulator of SG formation. A, The workflow for screening SG formation (Pab1-RFP foci are represented by red dots) in response to temperature-sensitive (ts) alleles of essential genes under 2-DG treatment. B, Network analysis of the genes whose ts alleles showed increased SG formation. The genes were grouped into modules based on their known physical interactions (red lines) and published cellular component information. C, Functional enrichment analysis of the ts alleles associated with increased SG formation. Mutants with confirmed SG phenotypes were analyzed for enrichment of the GO biological process, function and component categories. Enriched groups were scored by comparison to the essential gene temperature-sensitive allele collection using a cutoff of P < 0.05. D, A genetic interaction profile similarity network analysis (annotated with protein complex) for the ts alleles that showed increased SG formation.
From the screen, we also identified 47 ts mutants with significantly decreased SG formation. Gene Ontology (GO) analysis indicated that gene mutants associated with decreased SG formation were significantly enriched for the following cellular components: (1) TOR signaling, (2) proteasomal core complex, and (3) DNA polymerase complex (Figure S1A). The functional enrichment analysis of the ts alleles associated with decreased SG formation revealed enrichment of “cellular function” GO terms (1) helicase activity, (2) protein kinase activator activity, and (3) kinase activator activity; “component” GO terms (1) delta DNA polymerase complex, (2) proteasome core complex, (3) cytosolic proteasome complex, (4) proteasome core complex, beta-subunit complex, and (5) TOR complex; and “process” GO terms (1) regulation of cell growth, (2) nucleocytoplasmic transport, (3) establishment or maintenance of actin cytoskeleton polarity, (4) ribonucleoprotein complex biogenesis, (5) proteasomal ubiquitin-independent protein catabolic process, and (5) TOR signaling (Figure S1B).
LCBs as regulators of SG formation
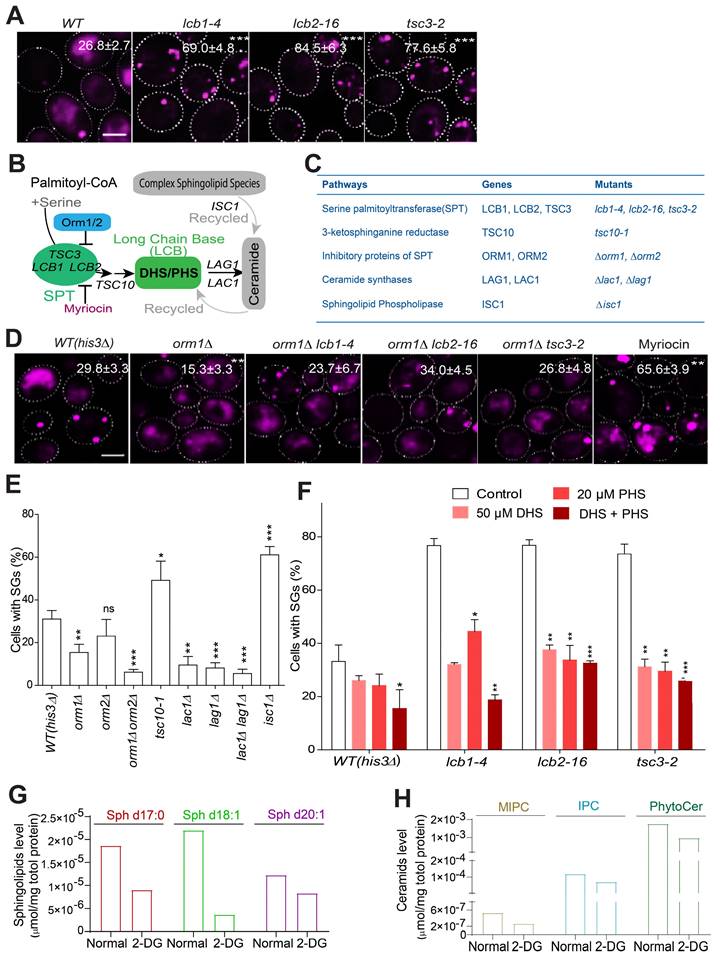
Among the increased SG formation hits, our screen identified three ts mutants of the serine palmitoyltransferase complex (lcb1-4, lcb2-16 and tsc3-2), which formed a consistently enriched group according to different analyses (Figure 1B-D). The increased SG-formation phenotype of these mutants was also among the top groups of hits (Table S1). This complex is highly conserved across eukaryotic cells and is the first rate-limiting step during sphingolipid biosynthesis. Therefore, we hypothesized that the sphingolipid metabolism pathway may play a role in the regulation of SG formation. We first manually confirmed the increased SG phenotype of these mutants (Figure 2A), and showed that their increase in SG formation was not due to enhanced expression of SG marker Pab1-RFP (Figure S2A).
The sphingolipid synthesis pathway is known to be regulated by multiple cellular components, including Orm1/2 and Lac1/Lag1, and the recycling pathway coordinated by Isc1 (Figure 2B) [51, 52]. To better understand how sphingolipid synthesis pathway components influence SG formation, we tested available mutants of these regulators (Figure 2C). First, we found that the derepression of serine palmitoyltransferase (SPT) by the deletion of ORM1 decreased SG formation compared to that in the control strain (his3∆) (Figure 2D), while the time SGs took to fuse was not changed between the two strains (Figure S2B). And this deletion further dampened SG induction in the lcb1-4, lcb2-1, and tsc3-2 mutants (Figure 2D). Since ORM1 has the paralog ORM2, we tested the ORM2 single mutant, in addition to the orm1∆ orm2∆ double mutant, for its SG phenotype. The phenotype of the orm2∆ single mutant was not significant (although a trend toward a decrease in SG formation was observed), but the orm1∆ orm2∆ double mutant exhibited a significant additive effect on decreasing SG formation (Figure 2E). Accordingly, myriocin, a drug that primarily inhibits the SPT complex and leads to the depletion of sphingolipids such as dihydrosphingosine (DHS) [53], greatly increased SG formation (Figure 2D). Third, we found that the tsc10-1 mutant (Tsc10 is a component that catalyzes the second step in phytosphingosine synthesis) exhibited a similar phenotype to that of the SPT mutants, and SG formation was decreased in the lac1∆, lag1∆, and lac1∆lag1∆ mutants but strongly increased in the isc1∆ mutant (Figure 2E). Previous research has demonstrated that phytosphingosine (PHS) and DHS are increased in lac1∆ and lag1∆ cells [54] . Moreover, in the isc1∆ mutant, the levels of DHS and PHS might decrease because Isc1 is responsible for the recycling of ceramides to generate DHS and PHS [51, 55]. We then hypothesized that DHS and PHS may be important signaling components that negatively regulate SG formation. Indeed, we found that exogenous DHS, PHS, or their combination could suppress the increase in SG formation in the SPT ts mutants (Figure 2F). In support of this, the major LCBs and ceramides were decreased in the wild-type strain upon 2-DG treatment (Figure 2G-H), and the protein expression level of the SPT component Tsc3 decreased upon 2-DG treatment (Figure S2C). Meanwhile, we used another SG marker eIF4G1 [56, 57] to further verify our findings. The data showed that the foci formation of eIF4G1 was similar to that of Pab1 under myriocin or DHS + PHS treatment (Figure S2D).
To explore if the signaling cascade here reported is a common regulator of SGs formation in yeasts, we applied other stress inducers, including glucose depletion and oxidative stress. It was found that the SPT ts alleles had similar SGs phenotype under glucose depletion (Figure S2E) and arsenite treatment (Figure S2F) as 2-DG treatment, which suggest the signaling cascade we elucidated can be conserved in yeast cells.
These data suggested that DHS and PHS function as signaling components that negatively regulate SG formation and further motivated us to search for upstream regulators and downstream effectors of DHS and PHS in the SG formation process (Figure 3A).
LCBs as regulators of SG formation. A, Mutants of the serine palmitoyltransferase (SPT) complex (LCB1, LCB2, and TSC3) significantly increase SG formation. The values represent the percentage of cells with SGs and are shown as the means ± S.D.s. Six clones were examined for each strain, and >300 cells were analyzed for each colony. The scale bars indicate 2 µm, * P < 0.05; ** P < 0.01; *** P < 0.001 (unpaired two-tailed Student's t test). B, A schematic diagram showing how the key sphingolipid metabolism pathway intermediates, LCB sphingolipids, are regulated by multiple genes/pathways. C, The genes related to sphingolipid metabolism pathway and the corresponding mutants examined in this study were shown. D, Deletion of ORM1 decreased SG formation in the WT strain; suppressed SG induction in the lcb1-4, lcb2-1, and tsc3-2 mutants; and the inhibition of the SPT complex by myriocin increased SG formation. The values represent the percentage of cells with SGs and are shown as the means ± S.D.s. Six clones were examined for each strain, and >300 cells were analyzed for each colony. The scale bars indicate 2 µm, * P < 0.05; ** P < 0.01; *** P < 0.001 (one-way ANOVA followed by Dunnett's test). E, SG formation was increased in the tsc10-1 and isc1∆ mutants and was decreased in the orm1∆, orm1∆orm2, ∆lac1∆, lag1∆, and lac1∆lag1∆ mutants. Same as in (D). F, Exogenous DHS, PHS, and their combination (50 μM DHS + 20 μM PHS) can restore the increased SG formation in the SPT ts mutants. G-H, The major long chain-base sphingolipids (dihydrosphingosine (C17 base), sphingosine (d18:1), sphingosine (d20:1)) and ceramides (MIPC, IPC, PhytoCer) decreased in WT (his3Δ) cells after 2-DG treatment (2 h). 7 OD WT cells were collected after 2 h of 400 mM 2-DG treatment in SD medium and subject to lipidomics analysis. One representative experiment was shown.
Sch9 and Ypk1 function upstream of LCBs
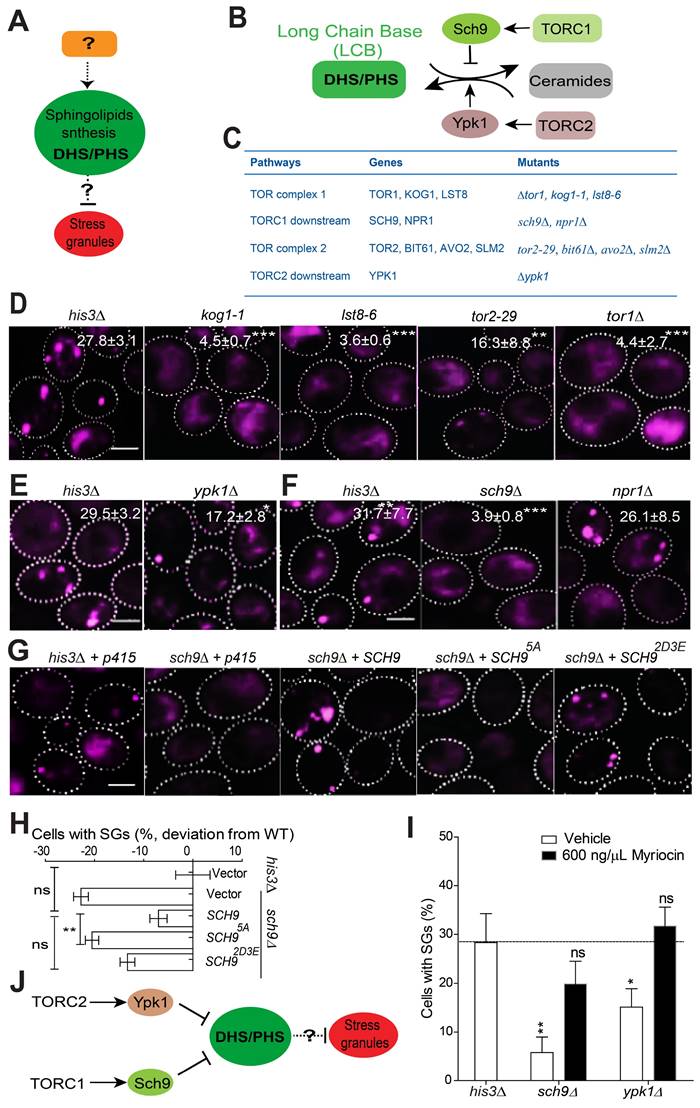
Sch9 and Ypk1 are known to be the main components that regulate sphingolipid metabolism (Figure 3B-C; summarized from Kitagaki et al. and Muir et al.) [51, 52]. Ypk1 and Sch9 function as key regulators of sphingolipid metabolism and maintain low levels of LCBs and LCB phosphates (LCBPs) and high levels of ceramides [51, 55]. Given that Sch9 and Ypk1 are known downstream effectors of TORC1 and TORC2, respectively, we hypothesized that Sch9 and Ypk1 may serve as upstream regulators of LCBs to govern SG formation together with the TORC1/2 complex.
The TORC1 complex has been implicated in SG formation. In mammalian cells, under heat shock or oxidative stress, the components of mTORC1 are recruited into SGs by astrin [24, 29]. Moreover, the mTOR effector kinases S6 kinase 1 (S6K1) and S6 kinase 2 (S6K2) also localize to SGs in human cells under mild oxidative stress [58]. TORC2 mediates the heat stress response in Drosophila by promoting the autonomous formation of SGs [59]. We first verified whether the TORC1/2 complex plays an active role in regulating SG formation under 2DG-induced stress conditions. Our screening data and further manual confirmation showed that the TORC1/2-related mutants kog1-1, lst8-6, tor2-29 and tor1∆ exhibited significant defects in SG formation (Figure S1A-B and Figure 3D) without altering the expression of the SG marker Pab1-RFP (Figure S3A). These findings indicated that TOR complexes play a functional role in regulating SG formation under 2-DG-induced stress conditions. We then evaluated the SG phenotype in the absence of other available TORC1/2 core components, including Avo2, Bit61, and Slm2. All these mutants exhibited a substantial decrease in SG formation (Figure S3B), and their SG-defective phenotypes were not due to a decreased expression level of the SG marker Pab1-RFP (Figure S3C). Next, we found that the Kog1-GFP, Lst8-GFP, and Bit61-GFP signals did not co-localize with SGs (Figure S3D). This indicated that their SG phenotypes do not result from a reduction in their gene products as SG components.
TORC2 is known to control sphingolipid homeostasis through Ypk1 [51, 60]. We found that SG formation decreased in ypk1∆ strains (Figure 3E). These data, together with the findings that SG formation is decreased in other core TORC2 component mutants (tor2-29, avo2∆, bit61∆, and slm2∆) (Figure S3B-C), indicate that Ypk1 may work downstream of TORC2 in the signal transduction process during SG formation. However, given that the SG phenotype of ypk1∆ strains is weaker than that of avo2∆, bit61∆, and slm2∆ strains, this indicates that TORC2 signaling may involve other effectors in addition to YPK1 that regulate 2-DG-induced SG formation.
Yeast TORC1 functions mainly through Npr1 and Sch9 [61, 62]. We found that sch9∆ cells displayed an SG formation defect, while the npr1∆ mutation had no clear effect (Figure 3F). We further found that the inactive form SCH95A failed to complement the SG phenotype of the sch9∆ mutant but that the constitutively active mutant SCH92D3E could (Figure 3G-H).
To clarify the link between SCH9/YPK1 and LCBs, we added myriocin to the sch9∆ and ypk1∆ mutants to determine whether inhibiting LCB synthesis could reverse the SG phenotype. As shown in Figure 3I, the addition of myriocin reversed the defects in SG formation; this result indicates a connection between SCH9/YPK1 and LCBs in the regulation of SG formation. Again, we showed that the SG phenotype of sch9∆ and ypk1∆ mutants is similar under arsenite (Figure S3E) as under 2-DG treatment, suggesting that the signaling cascade is a common regulator of SG formation in yeast cells.
Taken together, these results suggest that Sch9 and Ypk1 work upstream of LCB sphingolipids to regulate SG formation (Figure 3J). However, the factors located downstream of LCBs in the SG formation control pathway remain to be determined.
Reduced LCBs decrease Ubi4 expression levels and thereby enable SG formation
To search for components that work downstream of the sphingolipid pathway in regulating SG formation, we isolated all currently known interactors of the sphingolipid metabolism-related genes that were identified from our screen. We found that the sphingolipid-related gene LCB1 has a genetic interaction with the UPS gene UBI4 [63, 64] (Figure 4A). Additionally, the ORM2 gene, which inhibits LCB1/2, also genetically interacts with UBI4 [65], and the protein products of ORM2 and UBP3 physically interact [66] (Figure 4A). Furthermore, UBI4 is known to be induced by starvation and other stressors [40].
TORC1-Sch9 and TORC2-Ypk1 function upstream of LCB sphingolipids. A, A schematic diagram showing that LCBs regulate SG formation; however, the upstream regulators and downstream effectors involved are unknown. B, A schematic diagram showing the regulation of the sphingolipid metabolism pathway by the TORC1/2 signaling components identified from our screen in budding yeast. C, The genes related to TORC1/2 signaling pathway and the corresponding mutants examined in this study were shown. D, TORC1/2 ts mutants (kog1-1, lst8‑6, tor2-29) and the null mutant tor1∆ exhibit significant defects in SG formation. The values represent the percentage of cells with SGs and are shown as the means ± S.D.s. Six clones were examined for each strain, and >300 cells were analyzed for each colony. The scale bars indicate 2 µm, ** P < 0.01; *** P < 0.001 (unpaired two-tailed Student's t test). E, Deletion of the TORC2 downstream effector YPK1 results in a significant defect in SG formation. As in (C), * P < 0.05 (unpaired two-tailed Student's t test). F, Deletion of SCH9 results in a clear defect in SG formation, while deletion of NPR1 has no effects. As in (D), *** P < 0.001 (unpaired two-tailed Student's t test). G-H, The inactive form of SCH95A fails to complement the SG phenotype of the sch9∆ mutant, but the constitutively active mutant SCH92D3E can restore SG formation. The values represent the percentage of cells with SGs and are shown as the means ± S.D.s. Six clones were examined for each strain, and >300 cells were analyzed for each colony. The scale bar indicates 2 µm, ** P < 0.01 (sch9∆ + SCH95A vs. sch9∆ + SCH9); ns, not significant (WT + vector vs. sch9∆ + SCH9; sch9∆ + SCH9 vs. sch9∆ + SCH92D3E) (one-way ANOVA followed by Tukey's test). I, The addition of myriocin reversed the defects in SG formation in the sch9∆ and ypk1∆ mutants. Same as in (G). J, A diagram summarizing the current data and suggesting that TORC1-Sch9 and TORC2-Ypk1 function upstream of LCB sphingolipids in the regulation of SG formation.
Ubi4 suppresses SG formation. A, Interaction network analysis of the SPT complex genes LCB1, LCB2, and TSC3 (dark lines indicate a reduction in SG formation in the null mutants, and red lines indicate the induction of SG formation in the null mutants). The green lines indicate physical interactions, and the purple lines indicate genetic interactions. B, Deletion of UBI4 triggers SG formation. Eight clones were examined for each strain, and >300 cells were analyzed for each colony. The scale bar indicates 2 µm, *** P < 0.001 (unpaired two-tailed Student's t test). C, Deletion of UBI4 increases the number of SGs in the cell. Six clones were examined for each strain, and ~90 SG-containing cells were analyzed for each colony. One representative result is shown. The values are the numbers of SGs, and the P values are from unpaired two-tailed Student's t tests. D, Overexpression of UBI4 decreases SG formation. Six clones were examined for each strain, and ~90 SG-containing cells were analyzed for each colony. One representative result is shown, and the scale bar indicates 2 µm. The values are the numbers of SGs, and the P values are from unpaired two-tailed Student's t tests. E-F, Ubi4-GFP (C-terminally GFP-tagged) expression is downregulated in lcb1-4 and lcb2-16. One representative result with three different clones for each strain is shown. The values are presented as the means of arbitrary units (the intensity of the target bands was normalized to the Pgk1 level) for each clone. G-H, Ubi4-GFP expression was markedly increased by exogenous LCBs (25 µM DHS + 10 µM PHS or DHS + PHS). Same as in (D-E).
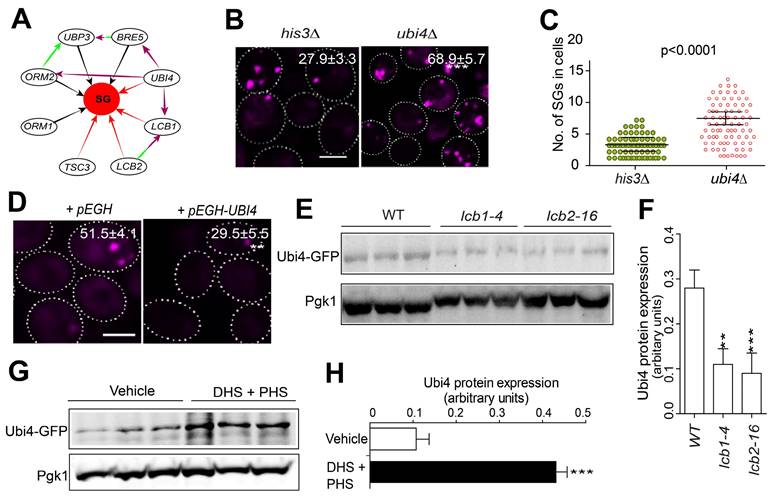
Following these clues, we wondered whether Ubi4 might act downstream of the sphingolipid pathway in SG formation signaling. We found that the deletion of UBI4 triggered SG formation (Figure 4B-C). Consistently, overexpression of UBI4 strongly suppressed SG formation (Figure 4D). Meanwhile, the Pab1-RFP protein expression was not changed in the Ubi4 deletion or overexpression strains (Figure S4). We further evaluated the relationship between SPT components and UBI4 by investigating the Ubi4 protein expression level in the WT and the SPT ts mutants. We found that Ubi4 expression was significantly downregulated in lcb1-4 and tsc3-2 (Figure 4E-F). This finding suggested that UBI4 could work downstream of the SPT complex in SG formation signaling. However, further evidence is needed to clarify how the SPT complex influences UBI4 during this process. To address this question, we investigated the expression level of Ubi4 in cells exposed to the two main products of the sphingolipid synthesis pathway, DHS and PHS, and found that the level of Ubi4 was significantly increased (Figure 4G-H). This result showed that the SPT complex potentially regulates UBI4 through influencing its downstream products DHS and PHS, and the cellular levels of DHS and PHS positively regulate the expression level of Ubi4. Taken together, these results suggest that Ubi4 may function as a downstream target of LCBs in the regulation of SG formation.
LCBs enhance Ubi4 expression through suppressing Ubp3
The next question is how LCBs regulate Ubi4. To explore possible intermediators that signal from LCBs to Ubi4, we first surveyed published databases and found that Ubp3 can genetically interact with Ubi4 [67, 68]. The yeast Ubp3 is an mRNA-binding ubiquitin-specific protease that cleaves ubiquitin fusions [69], supported by its cofactor Bre5. The catalytic activity of Ubp3 is essential for SG assembly in yeast [45]. Moreover, USP10, the mammalian homolog of Ubp3, has been found to be a constituent and regulator of SGs [43, 44]. These findings motivated us to explore the relationship between Ubp3 and Ubi4 in the context of SG formation. First, we examined whether Ubp3 could also be regulated by LCBs. Upon the addition of exogenous DHS + PHS, Ubp3 protein expression was significantly downregulated (Figure 5A-B). Second, myriocin could not induce SG formation once UBP3 was deleted (Figure S5A), and the SG phenotypes of the SPT ts alleles (lcb1-4, lcb2-16 and tsc3-2) and isc1 null mutant strains were abolished by UBP3 deletion (Figure S5B). These data suggested that Ubp3 works downstream of the sphingolipid metabolism pathway in SG formation regulation and that the expression level of Ubp3 was negatively regulated by the level of DHS and PHS in the cell.
Subsequently, we determined whether Ubp3 could directly influence the expression of Ubi4. As shown in Figure 5C-D, the deletion of UBP3 robustly induced Ubi4 expression under both normal conditions and after 2-DG treatment. These results indicate that Ubi4 acts downstream of Ubp3, and that Ubp3 negatively regulates the level of Ubi4. Moreover, we found that Ubi4 protein expression was not affected by LCBs in the absence of Ubp3 (Figure 5E-F), suggesting an indispensable role for Ubp3 in the regulation of Ubi4 by LCBs. Finally, the deletion of UBI4 in the ubp3Δ mutant resulted in a normal SG formation level compared to that in the WT (Figure 5G). Also, the Pab1 expression was not affected in the Ubp3 deletion strain (Figure S5C). Moreover, the changes in SG formation of ubi4∆ and ubp3∆ mutants are independent of PBs (Figure S5D-E). Last, we showed that the SG phenotype of ubi4∆ and ubp3∆ mutants is similar under arsenite (Figure S5F) as under 2-DG treatment, suggesting that the Ubi4 and Ubp3 may commonly regulate SG formation under stressful conditions in yeast cells.
Taken together, the above results suggest that, in the process of SG formation, LCBs function through suppressing Ubp3 expression to upregulate the level of Ubi4.
Ubp3 and Ubi4 regulate Lsm7 foci formation
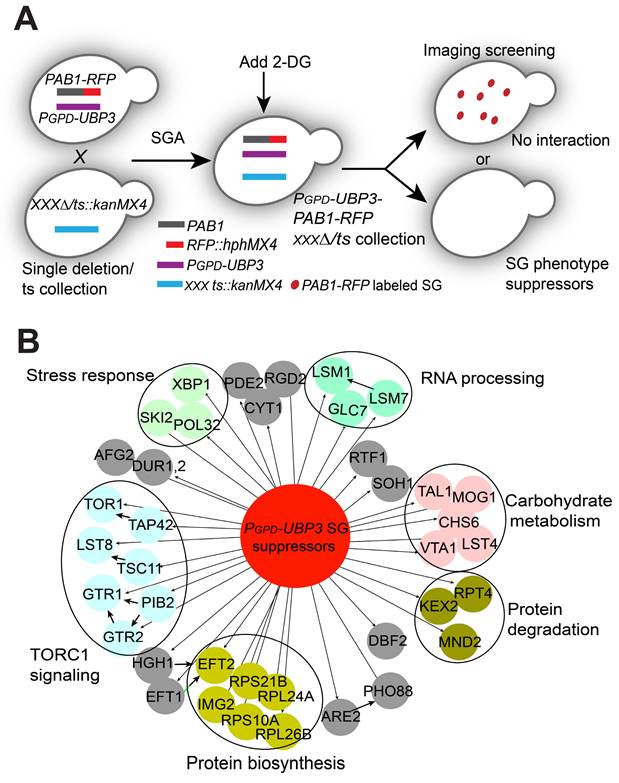
Next, we aimed to explore the possible downstream targets/substrates of Ubi4 and/or Ubp3 in SG formation signaling. Since we found that cells with endogenous overexpression of UBP3 had an increased SG formation phenotype (Figure S6), we decided to use the UBP3 overexpressor to perform another imaging-based genome-wide screen to search for further downstream factors. We reasoned that the downstream targets of Ubi4 and Ubp3 should fulfill two criteria: 1) their null mutations should directly abolish/suppress the increase in SG formation induced by UBP3 overproduction, and 2) they should belong to or be associated with critical SG structural components. Therefore, we performed a genome-wide phenotypic suppressor screen (Figure 6A) to identify the components that can suppress the increase in SG formation caused by Ubp3 overexpression (PGPD-UBP3). We isolated 45 suppressors that were enriched for processes related to (1) TOR complex 1 signaling, (2) RNA processing, (3) stress response, (4) protein degradation, and (5) carbohydrate metabolism (Figure 6B). The RNA processing group is of particular interest since one important protein, Lsm7, was recently reported to be a SG component and a key early phase separation factor that promotes the initiation of SGs under 2-DG treatment [70]. A manual test confirmed the phenotype of the knockout of LSM7 almost completely suppressing the increase in SG formation in the UBP3 overexpression strains (Figure 7A-B). Therefore, we wanted to investigate whether and how Lsm7 was affected by Ubi4/Ubp3.
As reported, Lsm7 can undergo phase separation under 2-DG treatment, thereby promoting SG formation [70]. We thus determined whether Lsm7 foci (phase-separated droplets) could be regulated by Ubp3 and Ubi4. We found that the number of Lsm7-GFP foci was significantly decreased in the UBP3 deletion strain and increased in the UBP3 overproducer strain (Figure 7C). These data suggest that Ubp3 may play a functional role in regulating Lsm7 phase separation. To test this, we first assessed the effect of Ubp3 on the solubility of Lsm7. We found that the deletion of UBP3 significantly reduced the proportion of insoluble Lsm7 protein, while UBP3 overproduction dramatically increased this proportion; and that the total level of Lsm7 was not altered (Figure 7D). Reciprocally, the deletion of Ubi4 enhanced the number of Lsm7 foci and the insoluble Lsm7 fraction (Figure 7E-F). Taken together, these results indicate that Ubp3 positively regulates the phase separation of Lsm7 by suppressing Ubi4, thereby promoting SG formation under 2-DG treatment.
Sphingolipids metabolism pathway regulates SG formation in mammalian cells
To further test whether our findings in yeast cells could be transferred to mammalian cells, we used wild-type and SPTLC2 (the human LCB2-like isoform) knockout HEK293t cells and G3BP1 as a marker for SGs. We found significantly increased SG formation under 2-DG treatment in wild-type cells, and SPTLC2 knockout enhanced this effect (Figure 8A). Furthermore, in both wild-type and SPTLC2 knockout HEK293t cells treated with 2-DG, myriocin significantly increased SG formation, whereas DHS suppressed SG formation (Figure 8B). These data demonstrated that the sphingolipid pathway regulates SGs formation in both yeast and mammalian cells.
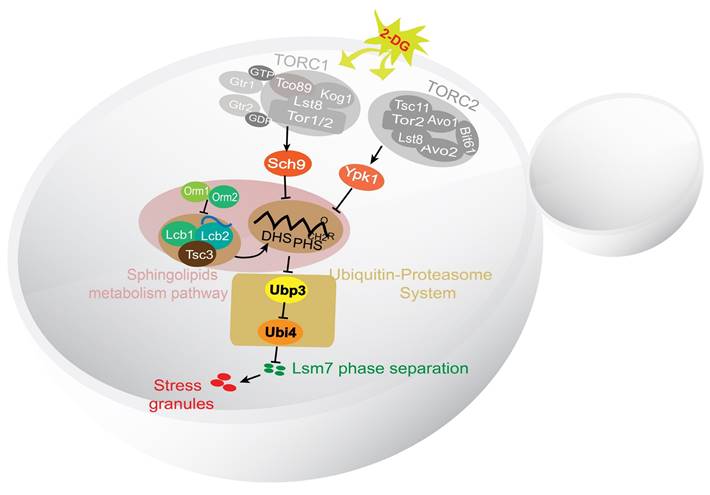
In summary, we have identified the major components of a potential signaling pathway controlling SG formation in yeast (Figure 9). Sch9 and Ypk1 were found to be involved in this pathway downstream of TORC1/2. The stress signal was transduced through Sch9 and Ypk1 to result in a decreased level of LCBs in the cell. The decreased LCBs caused an increase in the level of Ubp3, and the higher Ubp3 expression further represses the expression of Ubi4, the lowered Ubi4 level stimulates the phase separation of Lsm7, which in turn seeds the formation of SGs.
LCBs upregulate Ubi4 expression through suppressing Ubp3. A-B, Ubp3-GFP (C-terminally tagged with GFP) expression was downregulated by exogenous LCBs (25 µM DHS + 10 µM PHS or DHS + PHS). One representative result with three different clones for each strain is shown. The values are the means of the arbitrary units in (B) (the intensity of the target bands was normalized to the Pgk1 level) for each clone. *** P < 0.001 (unpaired two-tailed Student's t test). C-D, The Ubi4-GFP expression level increased when Ubp3 was deleted. One representative result with two different clones for each strain is shown. Same as in (A-B). ** P < 0.01 (unpaired two-tailed Student's t test). NG, normal glucose; DG, 400 mM 2-DG. E-F, The Ubi4-GFP expression level was not changed by exogenous LCBs (25 µM DHS + 10 µM PHS or DHS + PHS) when UBP3 was deleted. One representative result with two different clones for each strain is shown. Same as in (A-B). ns, not significant (unpaired two-tailed Student's t test). G, Deletion of UBI4 did not enhance SG formation in the absence of Ubp3. The values represent the percentage of cells with SGs and are shown as the means ± S.D.s. Six clones were examined for each strain, and >300 cells were analyzed for each colony; the scale bar indicates 2 µm. *** P < 0.001 (ubp3∆ vs WT); ns, not significant (ubp3∆ubi4∆ vs WT) (one-way ANOVA followed by Tukey's test).
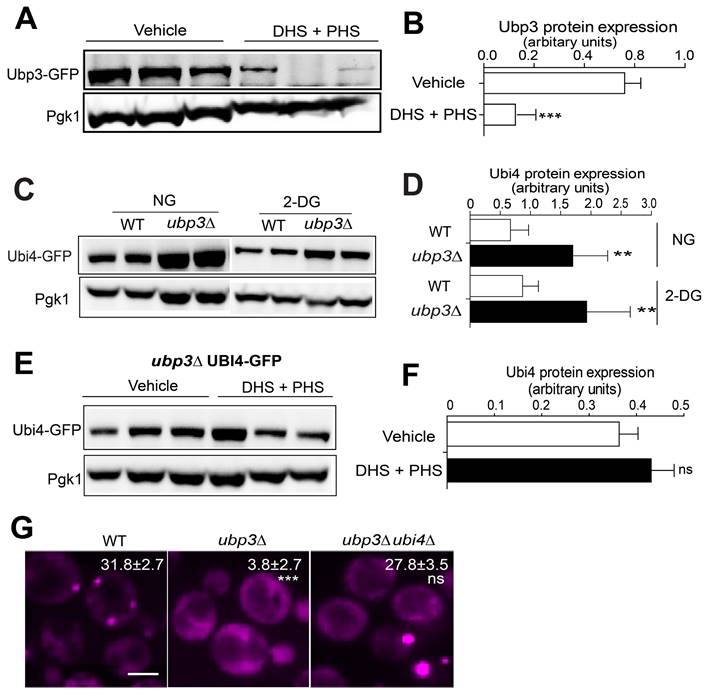
A high-content screening-based study identified genes that suppress UBP3 overexpression-induced SG formation. A, The workflow for screening PGPD-UBP3-mediated SG formation suppression. B, Network and functional analyses of the 45 PGPD-UBP3 SG formation suppressors. Hits showing SG formation-suppressing effects (nodes linked with dark arrows) were grouped into modules based on their known physical interactions (bold dark arrows) and published cellular processing information. The functional groups are shown in different colored nodes. Dark circles indicate subunits or complexes within groups.
Discussion
In this study, we have identified the major components of a signaling pathway controlling the formation of SGs in yeast under 2-DG treatment (Figure 9). We show that the TORC1/2-Sch9/Ypk1 signaling cascade regulates SG formation mainly through modulating the major long-chain base sphingolipids (LCBs) in the cell. Further we demonstrate that the ubiquitin-proteasome system components Ubi4 and Ubp3 initiate the phase separation of Lsm7, which later triggers SG formation.
Sphingolipids have been linked to stress responses because they mediate transcriptional reprogramming, translation of heat-shock proteins, and protein quality control [36, 63]. Our observation of the suppression of SG formation by LCBs could explain the proapoptotic properties of these factors since SG formation prevents cell apoptosis not only by inhibiting translation initiation but also by sequestering certain apoptotic factors [71]. Therefore, the inhibition of SG formation may release apoptotic regulatory factors that trigger apoptosis or result in the inability to restrict the accumulation of ROS, which is a key inducer of apoptosis [43]. These reports, together with our results, suggest that sphingolipids play a wide role in stress responses and cell fate by modulating SG formation.
Data from this work suggest that the LCB sphingolipids may serve as messengers to regulate Ubi4 expression via Ubp3 under stress. These results may help us understand the mechanism by which the depletion of ubiquitin results in neuronal cell death and further neurodegenerative diseases. The ubiquitin pathway is highly active in synapses [72], and neurons are susceptible to ubiquitin depletion under stress [73, 74]. Furthermore, the depletion of ubiquitin may lead to persistent SG formation under stress, which can sequester some functional proteins or impair ribostasis and cause other pathological changes in neurons [23, 75]. Additionally, a recent study has shown that the number of ubiquitin units encoded by UBI4 influences proteostasis and stress survival and that the optimal UBI4 repeats vary under different stress conditions [76], suggesting an important role for ubiquitin during eukaryotic stress adaptation. However, whether SG formation is involved in this evolutionary alternative has yet to be determined.
Ubp3 and Ubi4 regulate Lsm7 phase separation. A-B, Knockout of LSM7 almost completely suppressed the increase in SG formation in the PGPD-UBP3 strain. The bars in (A) show the means ± S.E.Ms. (n = 6). The circle represents the mean of individual clones. Six clones were examined for each strain, and >300 cells were analyzed for each clone. The scale bars indicate 2 µm, ** P < 0.01; *** P < 0.001 (one-way ANOVA followed by Tukey's test (B)). C, Lsm7-GFP aggregates were significantly decreased in the ubp3∆ strain and increased in the PGPD-UBP3 strain (upper panel, SGs; values represent the percentage of cells with SGs; lower panel, Lsm7 aggregates; values represent the percentage of cells with Lsm7 aggregates). D, Deletion of UBP3 significantly reduced the insoluble form (P) and increased the soluble form (S) of the Lsm7 protein, while UBP3 overproduction dramatically increased the insoluble form (P) and decreased the soluble form (S). The data are representative of three independent experiments. The values are presented as the means± SDs of arbitrary units (the intensities of the target bands were normalized to the Pgk1 levels in the corresponding fractions; for the pellet fractions, the values were normalized to the total Pgk1 level). * P < 0.05; (pellet fraction, one-way ANOVA followed by Dunnett's test). # P < 0.05 (soluble fractions, one-way ANOVA followed by Dunnett's test). E, Lsm7-GFP aggregates were significantly increased in the ubi4∆ strain (upper panel, SGs; values represent the percentage of cells with SGs; lower panel, Lsm7 aggregates; values represent the percentage of cells with Lsm7 aggregates). (C, E) Four clones were tested for each strain, and 300 cells were analyzed for each clone. The results are presented as the means ± S.D.s. Scale bars indicate 2 µm. ** P < 0.01; *** P < 0.001 (one-way ANOVA followed by Dunnett's test). F, Deletion of UBI4 increased the insoluble form (P) and decreased the soluble form (S). The data are representative of three independent experiments. The values are shown in F. * P < 0.05; (pellet fraction, one-way ANOVA followed by Dunnett's test).
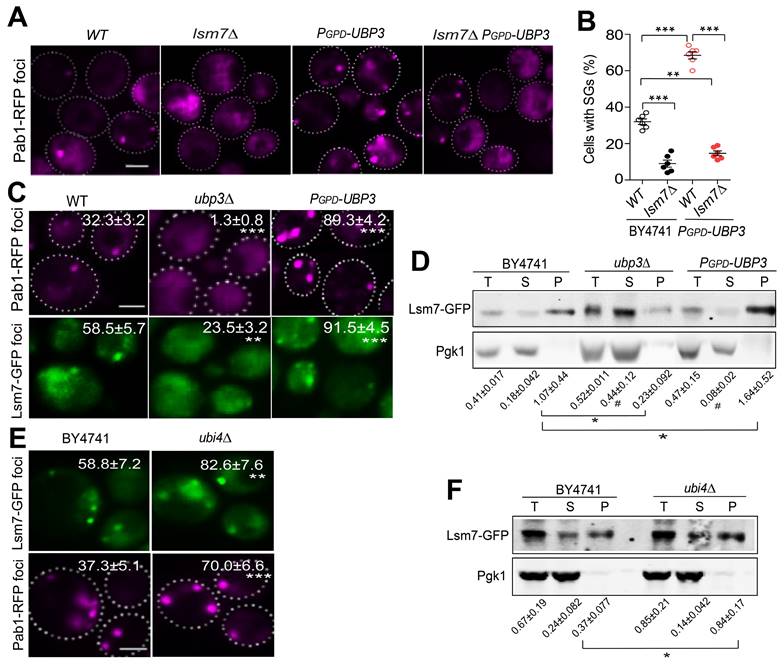
Sphingolipids metabolism pathway regulates SG formation in mammalian cells. A, SPTLC2 knockout HEK293t cells had increased SG formation (G3BP1 foci) under 2-DG treatment. Scale bars indicate 20 μm. Values represent the percentage of cells with SGs (G3BP1 foci), >300 cells were analyzed for each sample. Data were analyzed using the two-way ANOVA, *** P < 0.001 (2-DG vs Control); ### P < 0.001(SPTLC2 KO vs WT under 2-DG treatment). B, Myriocin induced, while DHS suppressed SG formation in the WT or SPTLC2 knockout HEK293t cells under 2-DG treatment. Scale bars indicate 20 μm. Values represent the percentage of cells with SGs (G3BP1 foci), >300 cells were analyzed for each sample. Data were analyzed using the two-way ANOVA, ** P < 0.01, *** P < 0.001 (vs WT treated with 2DG + Vehicle); # P < 0.05, ### P < 0.001 (vs SPTLC2 KO treated with 2-DG + Vehicle).
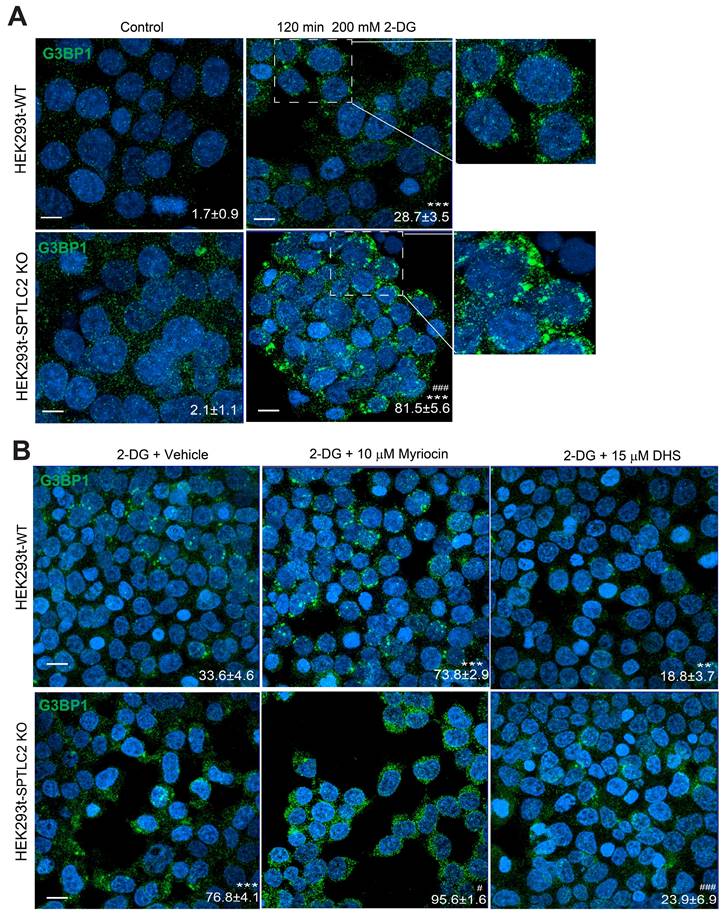
A working model for the signaling pathway controlling the formation of SGs in yeast under 2-DG treatment. A working model demonstrating the SG formation signaling pathway identified in this work. TORC1/2 was determined to be involved in an early step in this pathway. The signal was further transduced through its downstream effectors Sch9 and Ypk1 to influence the level of LCBs. Downstream of the LCBs, Ubi4 and Ubp3 together modulate Lsm7 phase separation and thereby regulate SG formation.
SGs colocalize with free ubiquitin but not polyubiquitinated proteins [38], which may suggest a potential role of free ubiquitin in the regulation of SG assembly. Moreover, SGs are formed through LLPS of certain proteins and RNA, where ubiquitin and polyubiquitin have been shown to eliminate the phase separation behavior of SG components [39]. This may further suggest that free ubiquitin could affect SG formation by influencing the LLPS of SG constituents. Accordingly, a recent study reported that Lsm7, a component of SGs, undergoes phase separation upon 2-DG treatment, which subsequently induces SG formation [70]. Our observations further provide evidence that Ubp3 positively regulates the phase separation of Lsm7 via the suppression of Ubi4 expression, thereby promoting SG formation under 2-DG treatment. Nevertheless, the existence and possible functions of other components that participate in this interaction between Ubp3, Ubi4, and Lsm7 need to be elucidated.
Here, we report key components in a possible pathway that controls SG formation under 2-DG- and arsentie-induced stress conditions. This pathway might represent a mechanism for SG formation under energy/nutrient depletion and oxidative stress conditions, where much of the microbial biomass in the world is believed to exist [77]. Furthermore, we have shown that the pathway regulating SG formation is also conserved in mammalian cells. However, the functions of these key SG regulators need to be further investigated in mammalian model systems. Such studies will improve our understanding of the mechanisms underlying SG-induced drug resistance and the relationship between SGs and age-related diseases such as cancer and neurodegeneration.
Methods
Cell culture
All the strains used in this study were from the BY4741/4742 or SGA (S288C) background (the strains are listed in Table S2) and were grown at 30 °C or at the indicated temperatures. Yeast rich medium (YP) containing 1% Bacto yeast extract and 2% Bacto peptone was supplemented with 2% glucose (YPD). Yeast minimal medium (YNBD) contained 0.67% Difco yeast nitrogen base without amino acids and 2% glucose. Supplements essential for auxotrophic strains were added to 20 mg/L for bases and amino acids (complete) except for leucine (SC-Leu), histidine (SC-His) or uracil (SC-Ura). In this study, the his3∆ strain was used as the wild-type control strain for the generation of mutants via the SGA approach.
Yeast strain construction
Single, double, or triple null mutants were constructed by PCR amplification and insertion with selective markers, including LEU2, natMX4, and kanMX4 (primers are listed in Table S3). The strains used for protein expression and localization analyses were picked directly from the yeast GFP collection or constructed by using standard PCR to integrate an Aequorea victoria GFP (S65T) tag into the yeast chromosome (C-terminus of the ORF) through homologous recombination, after which the constructs were expressed using endogenous promoters [78]. The UBP3 overexpression strain PGPD-UBP3 was constructed by amplifying the natNT2-containing cassette from the pYM-N15 plasmid using specific primers (Table S3) and then inserting it into the upstream promoter region of UBP3 by homologous recombination.
Screening for SG formation in temperature-sensitive (ts) alleles
To efficiently incorporate the SG marker (Pab1-RFP) into the yeast ts collection (TS-V5), the yeast synthetic genetic array (SGA) methodology was applied to combine the Pab1-RFP marker into a single haploid cell through standard mating and meiotic recombination via a robotic SGA procedure using the Singer RoToR HDA system (Singer Instruments) [46, 48]. The screening and manual confirmation were performed as described previously, with some modifications [47]. Briefly, the ts mutants were precultured at 22 °C for 2-3 days, diluted to an OD600 of 0.2 in SD media and grown with shaking at 30 °C until an OD600 of 0.6 was reached. The cells were then treated with 400 mM 2-DG for 2 h, fixed and washed. Images were acquired in an automated cellular imaging and analysis system (ImageXpress MICRO (MDC)). The SG formation in the mutants was quantified by MetaXpress (version 3.1) software by using the MetaXpress subprogram. For manual quantification, the cells were precultured at 22 °C for 2 days, diluted and grown at 30 °C to an OD600 of 0.5 before the application of 2-DG, formaldehyde fixed and washed as described above. The SG phenotype was microscopically studied (Zeiss AxioObserver. Z1, Germany) and quantified (the ratio of cells with Pab1-RFP foci) using ImageJ software with the cell-counter plug-in.
Genome-wide high-content screening for PGPD-UBP3 suppressors
To construct the null/ts allele collection with UBP3 overexpression, PGPD-UBP3 and Pab1-RFP were both introduced into the yeast query strain Y7039 (MATα, can1∆::STE2pr-Sp_Ura3 lyp1∆, his3∆, leu2∆, ura3∆, met15∆). The collection was then constructed using the SGA method described above. The SG formation in this collection was then screened and manually confirmed as described above with some modifications (precultured in SGA final media [SD-Leu/Arg/Lys + S-AEC, Canavanine, and Hygromycin B] at 30 °C for 2 days).
Functional enrichment and interaction network analysis
The functional enrichment of the confirmed hits from the SG formation screen (ts alleles library) was tested using the BiNGO plug-in for Cytoscape (Cytoscape v3.2.1) [49]. The Gene Ontology biological process term with the highest enrichment in a particular cluster was used to label the cluster on the network. The top hits were also analyzed by using the Gene Ontology Term Finder [50] for enrichment of Gene Ontology (GO) biological processes, molecular functions, and cellular component categories by comparison with a background set of genes (the top 60% of the ts alleles [TS-V5] that show an effect on growth rate at 30 °C; listed in Li et al. [46]). A cutoff of P < 0.05 was used. For the screens for SG components, the interaction network diagram of the hits was extracted from the interaction analysis using Osprey 1.2.0, and the physical interactions among confirmed hits were added according to the BioGRID interaction database.
For the screens for SG components and for PGPD-UBP3 SG suppressors, the interaction network diagram of the hits was constructed via interaction analysis using Osprey 1.2.0, and the physical interactions among the confirmed hits were added to the BioGRID interaction database. The top hits from the PGPD-UBP3 SG suppressor screen were also analyzed using the Gene Ontology Term Finder for enrichment of Gene Ontology (GO) biological processes by comparison with a background set list representing TS-V5 plus SGA-V2 and an array of slow-growing mutants. A cutoff of P < 0.05 was used.
SG induction with 2-DG
For the two screens described above, cells were grown in 96-well plates with a starting OD600 of 0.2. When the OD600 reached 0.6, the cells were treated with 2-DG for 2 h at a final concentration of 400 mM to induce SG formation.
Treatment with myriocin, DL-dihydrosphingosine and phytosphingosine hydrochloride
To study the SG phenotype resulting from SPT complex inhibition, cells were grown with a starting OD600 of 0.1 in the presence of a vehicle (methanol) or 600 ng/mL myriocin (Sigma, M1177). The cells were treated with 400 mM 2-DG for 2 h until the OD600 reached 0.5 (5 h were needed with methanol alone but 9 h were needed with myriocin), after which the cells were fixed with 3.7% formaldehyde and washed twice with PBS. To study the sphingolipid synthesis pathway, DL-dihydrosphingosine (DHS; Sigma, D6783) and phytosphingosine hydrochloride (PHS; Sigma, P2795) were used to treat cells as described above for myriocin treatment.
Fluorescence microscopy
The cells were grown to an OD600 of 0.5 with or without treatment as described above. The cells were then fixed with 3.7% formaldehyde for 30 min and washed twice with PBS. A Zeiss Axiovert 200 M fluorescence microscope was used to obtain images using the GFP and RFP channels.
Treatment with arsenite
The cells were cultured to an OD600 of 0.5. Sodium arsenite (NaAsO2; Sigma-Aldrich, St. Louise, MO, USA) was added to cell cultures at the concentration of 2 mM for 3 h. The cells were then fixed with 3.7% formaldehyde for 30 min and washed twice with PBS. A Zeiss Axiovert 200 M fluorescence microscope was used to obtain images using the RFP channel.
Time-lapse imaging assay
The cells were incubated to an OD600 of 0.5. The observation was started right after when the cells were treated with 400 mM 2-DG and transferred to 0.25 mg/mL concanavalin A coated 96-well plate. Images were acquired in an automated cellular imaging and analysis system (ImageXpress MICRO (MDC)) using the RFP channel with an interval of 3 min for 6 h. The SGs fusion time was studied by ImageJ software. 100 SGs fusion cases were measured in each strain. The fusion time was calculated as the duration between the two SGs observed until they fuse into one stable SG.
Lipidomics analysis
Indicated yeast cells were cultured in SD medium to log phase, and then treated with or without 400 mM 2-DG for another 2 h. About 7 OD cells were collected to 1.5 mL centrifuge tube and were immediately snap-frozen in liquid nitrogen and transported on dry ice for lipidomic analysis.
The yeast lipidomics analysis was conducted at LipidALL Technologies as described previously[79]. Briefly, lipid extraction was performed by homogenizing yeast cells in 750 µL of chloroform:methanol 1:2 (v/v) containing 10% deionized water with glass beads on an automated bead shaker. The homogenate was then incubated at 4 °C for 1 h, followed by the addition of 350 µL deionized water and 250 µL chloroform. The lower organic phase containing lipids was extracted by centrifugation and was further dried in the SpeedVac under OH mode. Samples were stored at -80 °C until further analysis.
Lipidomics methodology was reported according to standard guidelines. Polar lipids PC, LPC, Sph, and PhytoCer were analyzed in the ESI positive ion mode on Agilent 1260 HPLC coupled to Sciex 5500 QTRAP, with source parameters CUR 10, TEM 400 °C, GS1 30, GS2 30. Spiked internal standards, including d9-PC32:0 (16:0/16:0), d7-PE33:1 (15:0/18:1), d31-PS, d7-PG33:1 (15:0/18:1), d7-PI33:1 (15:0/18:1), d7PA33:1 (15:0/18:1), Cer (d18:1-d7/15:0), d7-LPC18:1, d7-LPE18:1, C17:0-LPA, C17:1-LPI, C17:1-LPS, d17:1-Sph (Avanti Polar Lipids. Inc.) were used to quantify individual lipid species.
Western blotting
The total yeast protein extraction method [80] was used with modifications. Approximately 1.0 unit OD600 of yeast cells was harvested and incubated in 1 mL of 0.2 M NaOH for 20 min on ice. The mixture was subsequently resuspended in 50 µL of HU sample buffer (8 M urea, 0.2 M Tris-HCl (pH 6.8), 1 mM EDTA, 5% SDS, 1% β-mercaptoethanol, and 0.0025% bromophenol blue) and heated for 10 min at 70 °C. All samples were electrophoresed on 10% Tris-HCl/SDS-polyacrylamide gels (Bio-Rad) and transferred onto polyvinylidene difluoride membranes (Merck Millipore). The membranes were then incubated with primary antibodies (mouse anti-Pgk1 (Invitrogen, #459250), rabbit anti-GFP (Abcam, ab6556), or anti-RFP (Abcam, ab62341)) followed by secondary antibodies (goat anti-mouse IRDye® 680RD (LICOR, IR Dye 926-68180) or goat anti-rabbit IRDye® 800CW (LICOR, IR Dye 926-32210)). Relative protein expression was analyzed by an Odyssey® imaging system (LICOR) and was normalized to that of Pgk1.
Protein aggregation assay
Soluble and insoluble (aggregate) proteins were extracted as described previously [81]. Yeast cells were grown to an OD600 of 0.6-0.8 and harvested by centrifugation. Approximately 10 units of OD600 cells were resuspended in 300 μL of 50 mM Tris-HCl (pH 8.5)/500 mM NaCl/1 mM phenylmethylsulfonyl fluoride (PMSF)/EDTA-free protease inhibitors (Roche). The cells were pulverized with a vortex mixer and glass beads (diameter 0.5 mm; Biospec) and were precleared by centrifugation at 3,000 × g for 5 min. A 200-μL volume of the supernatant was taken, and 90 μL was stored as total protein. The remaining 110 μL, which was used for fractionation of soluble and insoluble proteins, was centrifuged at 16,000 × g for 20 min. The resulting supernatant contained the soluble fraction. The pellet, which contained the insoluble fraction, was washed in 110 μL of 50 mM Tris-HCl (pH 8.5)/150 mM NaCl/1 mM PMSF/EDTA-free protease inhibitors (Roche), centrifuged at 16,000 × g for 20 min, and resuspended in 110 μL of 8 M urea/2% SDS/50 mM Tris (pH 8.5)/150 mM NaCl/1 mM PMSF/2 mM dithiothreitol/EDTA-free protease inhibitors (Roche). The same volumes of soluble and insoluble fractions were used for quantitative western blotting. To calculate the soluble or insoluble fraction of a protein, the signal of the supernatant or pellet was divided by the total signal of the supernatant and pellet of the same sample.
SG formation examination in mammalian cells
The SPTLC2 knockout cell line (HEK293t) was constructed as described previously [36]. Cells were cultured in Dulbecco's modified Eagle's medium (Gibco, c11995500bt) supplemented with 10% fetal bovine serum (FBS, ExcellBio) and 1% penicillin-streptomycin (HycloneTM, SV30010) in a humidified incubator containing 5% CO2 at 37 °C.
For SG formation examination studies, cells were seeded onto confocal imaging plates (Nest, 801002) and treated with 10 μM myriocin or 15 μM DHS when the cells had reached 90% confluency. 200 mM 2-DG was added to induce SG formation (2 h). Cells were fixed with 4% paraformaldehyde for 15 min and rinsed in PBS for further use.
For immunofluorescent studies, fixed cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min, and blocked with 10% donkey serum for 1 hour. Samples were further incubated with G3BP1 (Proteintech 13057-2-AP) in blocking buffer at 4 °C overnight. Samples were washed three times with PBS and incubated with goat anti rabbit Alexa Fluor 488 (Thermo Fisher Scientific) for 2 h at room temperature, followed by DAPI (Thermo Fisher Scientific P36931) staining for 15 min. Images were captured and visualized using a Leica DMI4000 fluorescence microscope.
Statistical analysis
No statistical methods were used to predetermine the sample size. Appropriate statistical analyses were performed depending on the comparisons made in the text and figure legends. One-way or two-way ANOVA following Dunnett's test, Tukey's test, or two-tailed unpaired Student's t test was performed using Prism version 8 (GraphPad, Inc.). P values are designated *P < 0.05, **P < 0.01, and ***P < 0.001. All the graphs show the mean and error bars, which represent the standard error of the deviation (S.D.) or the standard error of the mean (S.E.M.).
Supplementary Material
Supplementary figures.
Supplementary table.
Acknowledgements
The authors would like to thank Charles Boone for the generous contribution of the Moby plasmids and Joris Winderickx for kindly sharing the strains and plasmids. We also thank Simon Alberti, Marc Pilon, and Ralf Braun for their comments and suggestions on this project. We acknowledge the EATRIS Center for Large-scale Cell-based Screening at the University of Gothenburg for support in performing the high-content genome-wide screening, as well as the Centre for Cellular Imaging at the University of Gothenburg for microscopy support. This work was supported by grants from the Swedish Cancer Society - Cancerfonden (CAN 2017/643, 19/0069 and 23 2769 Pj to B.L.; 19/0133 and 22/2014 to P.S.), the Swedish Natural Research Council (VR 2019-03604 and 2023-03560 to B.L.), the Guangdong Basic and Applied Basic Research Foundation (2023A1515010483 to L.C.), the Young Scientists Program of Guangzhou National Laboratory (Grant No. QNPG23-02 to L.C.), and the Major Project of Guangzhou National Laboratory (Grant No. GZNL2023A02008).
Author contributions
B.L. conceived the project, and B.L., P.S., T. N., L.C., and H.L. supervised the research. L.C., Y.G., X.H., X.Y., M.L., S.J., and X.C. performed the experiments. L.C. and B.L. analyzed the data and wrote the manuscript, which was further edited by the other coauthors.
Data availability
- The high-content screening data and a full list of hits related to Figure 1A-C and Figure 6B are included in Table S1.
- Raw data related to the core figures and supplementary figures are provided as a source data file.
- All the other raw data are high-content screening-based images or images acquired by conventional microscopy. Due to the large size (55 GB in total), these original images are available from the corresponding author upon request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803-8
2. Protter DSW, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26:668-79
3. Jain S, Wheeler Joshua R, Walters Robert W, Agrawal A, Barsic A, Parker R. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell. 2016;164:487-98
4. Wheeler JR, Matheny T, Jain S, Abrisch R, Parker R. Distinct stages in stress granule assembly and disassembly. eLife. 2016;5:e18413
5. Buchan JR. mRNP granules: Assembly, function, and connections with disease. RNA Biol. 2014;11:1019-30
6. Buchan JR, Kolaitis R-M, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461-74
7. Grousl T, Vojtova J, Hasek J, Vomastek T. Yeast stress granules at a glance. Yeast. 2022;39:247-61
8. Hofmann S, Kedersha N, Anderson P, Ivanov P. Molecular mechanisms of stress granule assembly and disassembly. Biochim Biophys Acta Mol Cell Res. 2021;1868:118876
9. Zhou H, Luo J, Mou K, Peng L, Li X, Lei Y. et al. Stress granules: functions and mechanisms in cancer. Cell Biosci. 2023;13:86
10. Qifti A, Jackson L, Singla A, Garwain O, Scarlata S. Stimulation of phospholipase Cβ1 by Gαq promotes the assembly of stress granule proteins. Sci Signal. 2021;14:eaav1012
11. Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431-42
12. Youn JY, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI. et al. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell. 2018;69:517-32.e11
13. Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J. et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729-32
14. Riback JA, Katanski CD, Kear-Scott JL, Pilipenko EV, Rojek AE, Sosnick TR. et al. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 2017;168:1028-40
15. Li P, Banjade S, Cheng H-C, Kim S, Chen B, Guo L. et al. Phase transitions in the assembly of multi-valent signaling proteins. Nature. 2012;483:336-40
16. Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30:39-58
17. Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA. et al. RNA controls polyQ protein phase transitions. Mol Cell. 2015;60:220-30
18. Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj Anderson P, Kim Hong J. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123-33
19. Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I. et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360:918-21
20. McDonald KK, Aulas A, Destroismaisons L, Pickles S, Beleac E, Camu W. et al. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Gen. 2011;20:1400-10
21. Somasekharan SP, El-Naggar A, Leprivier G, Cheng H, Hajee S, G.P GT. et al. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol. 2015;208:913-29
22. Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. BBA Gene Regul Mech. 2015;1849:861-70
23. Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154:727-36
24. Thedieck K, Holzwarth B, Prentzell MT, Boehlke C, Kläsener K, Ruf S. et al. Inhibition of mTORC1 by astrin and stress granules prevents apoptosis in cancer cells. Cell. 2013;154:859-74
25. Fang MY, Markmiller S, Vu AQ, Javaherian A, Dowdle WE, Jolivet P. et al. Small-molecule modulation of TDP-43 recruitment to stress granules prevents persistent TDP-43 accumulation in ALS/FTD. Neuron. 2019;103:802-19.e11
26. Attwood KM, Robichaud A, Westhaver LP, Castle EL, Brandman DM, Balgi AD. et al. Raloxifene prevents stress granule dissolution, impairs translational control and promotes cell death during hypoxia in glioblastoma cells. Cell Death Dis. 2020;11:989
27. Gupta N, Badeaux M, Liu Y, Naxerova K, Sgroi D, Munn LL. et al. Stress granule-associated protein G3BP2 regulates breast tumor initiation. Proc Natl Acad Sci U S A. 2017;114:1033-8
28. Fonteneau G, Redding A, Hoag-Lee H, Sim ES, Heinrich S, Gaida MM. et al. Stress granules determine the development of obesity-associated pancreatic cancer. Cancer Discov. 2022;12:1984-2005
29. Takahara T, Maeda T. Transient sequestration of TORC1 into stress granules during heat stress. Mol Cell. 2012;47:242-52
30. Arimoto K, Fukuda H, Imajoh-Ohmi S, Saito H, Takekawa M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat Cell Biol. 2008;10:1324-32
31. Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430-6
32. Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener. 2012;7:56
33. Eiermann N, Haneke K, Sun Z, Stoecklin G, Ruggieri A. Dance with the devil: stress granules and signaling in antiviral responses. Viruses. 2020;12:984
34. Cowart LA, Gandy Jason L, Tholanikunnel B, Hannun Yusuf A. Sphingolipids mediate formation of mRNA processing bodies during the heat-stress response of Saccharomyces cerevisiae. Biochem J. 2010;431:31-8
35. Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18:33-50
36. Cao X, Wu X, Zhao L, Zheng J, Jin X, Hao X. et al. Maturation and detoxification of synphilin-1 inclusion bodies regulated by sphingolipids. eLife. 2024 p. RP92180
37. Lester RL, Withers BR, Schultz MA, Dickson RC. Iron, glucose and intrinsic factors alter sphingolipid composition as yeast cells enter stationary phase. Biochim Biophys Acta. 2013;1831:726-36
38. Markmiller S, Fulzele A, Higgins R, Leonard M, Yeo GW, Bennett EJ. Active protein neddylation or ubiquitylation is dispensable for stress granule dynamics. Cell Rep. 2019;27:1356-63.e3
39. Dao TP, Kolaitis R-M, Kim HJ, O'Donovan K, Martyniak B, Colicino E. et al. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol Cell. 2018;69:965-78.e6
40. Finley D, Özkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035-46
41. Cheng L, Watt R, Piper PW. Polyubiquitin gene expression contributes to oxidative stress resistance in respiratory yeast (Saccharomyces cerevisiae). Mol Gen Genet. 1994;243:358-62
42. Walters RW, Muhlrad D, Garcia J, Parker R. Differential effects of Ydj1 and Sis1 on Hsp70-mediated clearance of stress granules in Saccharomyces cerevisiae. RNA. 2015;21:1660-71
43. Takahashi M, Higuchi M, Matsuki H, Yoshita M, Ohsawa T, Oie M. et al. Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol Cell Biol. 2013;33:815-29
44. Kedersha N, Panas MD, Achorn CA, Lyons S, Tisdale S, Hickman T. et al. G3BP-Caprin1-USP10 complexes mediate stress granule condensation and associate with 40S subunits. J Cell Biol. 2016;212:845-60
45. Nostramo R, Varia SN, Zhang B, Emerson MM, Herman PK. The catalytic activity of the Ubp3 deubiquitinating protease is required for efficient stress granule assembly in Saccharomyces cerevisiae. Mol Cell Biol. 2016;36:173-83
46. Li Z, Vizeacoumar FJ, Bahr S, Li J, Warringer J, Vizeacoumar FS. et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotech. 2011;29:361-7
47. Yang X, Shen Y, Garre E, Hao X, Krumlinde D, Cvijović M. et al. Stress granule-defective mutants deregulate stress responsive transcripts. PLoS Genet. 2014;10:e1004763
48. Baryshnikova A, Costanzo M, Dixon S, Vizeacoumar FJ, Myers CL, Andrews B. et al. Synthetic genetic array (SGA) analysis in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Methods Enzymol: Academic Press. 2010 p. 145-79
49. Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448-9
50. Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM. et al. GO::TermFinder-open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710-5
51. Muir A, Ramachandran S, Roelants FM, Timmons G, Thorner J. TORC2-dependent protein kinase Ypk1 phosphorylates ceramide synthase to stimulate synthesis of complex sphingolipids. eLife. 2014;3:e03779
52. Kitagaki H, Cowart LA, Matmati N, Vaena de Avalos S, Novgorodov SA, Zeidan YH. et al. Isc1 regulates sphingolipid metabolism in yeast mitochondria. Biochim Biophys Acta. 2007;1768:2849-61
53. Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem Biophys Res Commun. 1995;211:396-403
54. Schorling S, Vallée B, Barz WP, Riezman H, Oesterhelt D. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisae. Mol Biol Cell. 2001;12:3417-27
55. Swinnen E, Wilms T, Idkowiak-Baldys J, Smets B, De Snijder P, Accardo S. et al. The protein kinase Sch9 is a key regulator of sphingolipid metabolism in Saccharomyces cerevisiae. Mol Biol Cell. 2014;25:196-211
56. Hoyle NP, Castelli LM, Campbell SG, Holmes LE, Ashe MP. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J Cell Biol. 2007;179:65-74
57. Begovich K, Wilhelm JE. An in vitro assembly system identifies roles for RNA nucleation and ATP in yeast stress granule formation. Mol Cell. 2020;79:991-1007
58. Sfakianos AP, Mellor LE, Pang YF, Kritsiligkou P, Needs H, Abou-Hamdan H. et al. The mTOR-S6 kinase pathway promotes stress granule assembly. Cell Death Differ. 2018;25:1766-80
59. Jevtov I, Zacharogianni M, van Oorschot MM, van Zadelhoff G, Aguilera-Gomez A, Vuillez I. et al. TORC2 mediates the heat stress response in Drosophila by promoting the formation of stress granules. J Cell Sci. 2015;128:2497-508
60. Roelants FM, Breslow DK, Muir A, Weissman JS, Thorner J. Protein kinase Ypk1 phosphorylates regulatory proteins Orm1 and Orm2 to control sphingolipid homeostasis in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2011;108:19222-7
61. MacGurn JA, Hsu P-C, Smolka MB, Emr SD. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell. 2011;147:1104-17
62. Urban J, Soulard A, Huber A, Lippman S, Mukhopadhyay D, Deloche O. et al. Sch9 Is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007;26:663-74
63. Meier KD, Deloche O, Kajiwara K, Funato K, Riezman H. Sphingoid base is required for translation initiation during heat stress in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1164-75
64. Friant S, Meier KD, Riezman H. Increased ubiquitin-dependent degradation can replace the essential requirement for heat shock protein induction. EMBO J. 2003;22:3783-91
65. Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G. et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921-6
66. Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS. et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806-10
67. Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G. et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420
68. Mayor T, Lipford JR, Graumann J, Smith GT, Deshaies RJ. Analysis of polyubiquitin conjugates reveals that the Rpn10 substrate receptor contributes to the turnover of multiple proteasome targets. Mol Cell Proteomics. 2005;4:741-51
69. Baker RT, Tobias JW, Varshavsky A. Ubiquitin-specific proteases of Saccharomyces cerevisiae-cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem. 1992;267:23364-75
70. Lindström M, Chen L, Jiang S, Zhang D, Gao Y, Zheng J. et al. Lsm7 phase-separated condensates trigger stress granule formation. Nat Commun. 2022;13:3701
71. Kedersha N, Ivanov P, Anderson P. Stress granules and cell signaling: more than just a passing phase? Trends Biochem Sci. 2013;38:494-506
72. Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231
73. Tan Z, Qu W, Tu W, Liu W, Baudry M, Schreiber SS. p53 accumulation due to down-regulation of ubiquitin: relevance for neuronal apoptosis. Cell Death Differ. 2000;7:675-81
74. Tan Z, Tu W, Schreiber SS. Downregulation of free ubiquitin: a novel mechanism of p53 stabilization and neuronal cell death. Brain Res Mol Brain Res. 2001;91:179-88
75. Chen L, Liu B. Relationships between stress granules, oxidative stress, and neurodegenerative diseases. Oxid Med Cell Longev. 2017;2017:1809592
76. Gemayel R, Yang Y, Dzialo MC, Kominek J, Vowinckel J, Saels V. et al. Variable repeats in the eukaryotic polyubiquitin gene ubi4 modulate proteostasis and stress survival. Nat Commun. 2017;8:397
77. Lewis D, Gattie D. The ecology of quiescent microbes. ASM News. 1991;57:27-32
78. Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS. et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686-91
79. Liu S, Chen M, Wang Y, Lei Y, Huang T, Zhang Y. et al. The ER calcium channel Csg2 integrates sphingolipid metabolism with autophagy. Nat Commun. 2023;14:3725
80. Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857-60
81. Cherkasov V, Hofmann S, Druffel-Augustin S, Mogk A, Tyedmers J, Stoecklin G. et al. Coordination of translational control and protein homeostasis during severe heat stress. Curr Biol. 2013;23:2452-62
Author contact
![]() Corresponding authors: beidong.liugu.se; chen_lihuaac.cn.
Corresponding authors: beidong.liugu.se; chen_lihuaac.cn.
 Global reach, higher impact
Global reach, higher impact