13.3
Impact Factor
Theranostics 2025; 15(1):245-257. doi:10.7150/thno.102650 This issue Cite
Review
tRNA-derived small RNAs in disease immunity
1. Department of Radiation Oncology, Sichuan Clinical Research Center for Cancer, Sichuan Cancer Hospital & Institute, Sichuan Cancer Center, Affiliated Cancer Hospital of University of Electronic Science and Technology of China, Chengdu, China.
2. Department of Respiratory and Critical Care, Chengdu Third People's Hospital, Chengdu, China.
* These authors contributed equally to this work.
Received 2024-8-22; Accepted 2024-11-8; Published 2025-1-1
Abstract
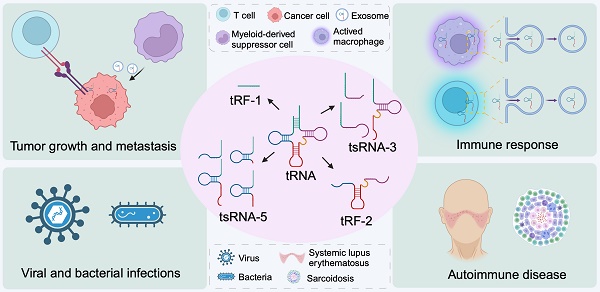
Recently, members of a unique species of non-coding RNA, known as transfer RNA-derived small RNAs (tsRNAs) have been reported to serve multiple molecular functions, including in cells that mediate immunity. Because of their low molecular weights, tsRNAs were previously difficult to detect and were thus overlooked, until now. In this review, we delve into the biogenesis of tsRNAs and their diverse biological functions, ranging from transcriptional regulation to modulation of mRNA translation. We highlight the current evidence demonstrating their involvement in the immune response, as well as how tsRNAs modulate immunity to influence tumor growth and spread, autoimmune disease pathology and infection by pathogens. We surmise that tsRNAs are likely informative as diagnostic markers of cellular homeostasis and disease, and that therapeutic targeting of tsRNAs could be beneficial for a range of human diseases. Improved knowledge on the functions for tsRNAs in the mammalian immune system will enable us to leverage tsRNAs for their effective clinical use as treatments for human health challenges.
Keywords: tRNA-derived small RNA, non-coding RNA, biogenesis, adaptive immunity, tumor immunology, autoimmune diseases
Introduction
Transfer RNAs (tRNAs) are indispensable molecular components that comprise the cellular protein synthesis machinery of cells, serving to direct the accurate assembly of polypeptides [1, 2]. Recently, the traditional view of tRNAs as mere translators of genetic information has been challenged by the discovery of tRNA-derived small RNAs (tsRNAs) which, remarkably, have revealed further regulatory complexity for tRNAs and their functional products within cells. Indeed, tsRNAs, generated through alternative processing pathways, have been found to influence gene expression and mediate cellular responses to stress [3-7]. Studies on how tsRNAs exert their molecular functions in cells of the immune system has begun to reveal how they influence signaling pathways. For example, tsRNAs have been shown to play a role in the activation of immune cells and the production of cytokines, which are crucial for mounting an effective immune response against a pathogen challenge [8, 9]. Moreover, tsRNAs have also been postulated to function as modulators of the immune responses in cancer, particularly in tumor growth and immune evasion [10]. Furthermore, tsRNAs have also been suggested to play a critical role in autoimmune disorders [11]. Here, we will review our current understanding of the role of tsRNAs in cells of the immune system and how they are crucial to health and disease.
Biogenesis and classification of tsRNA
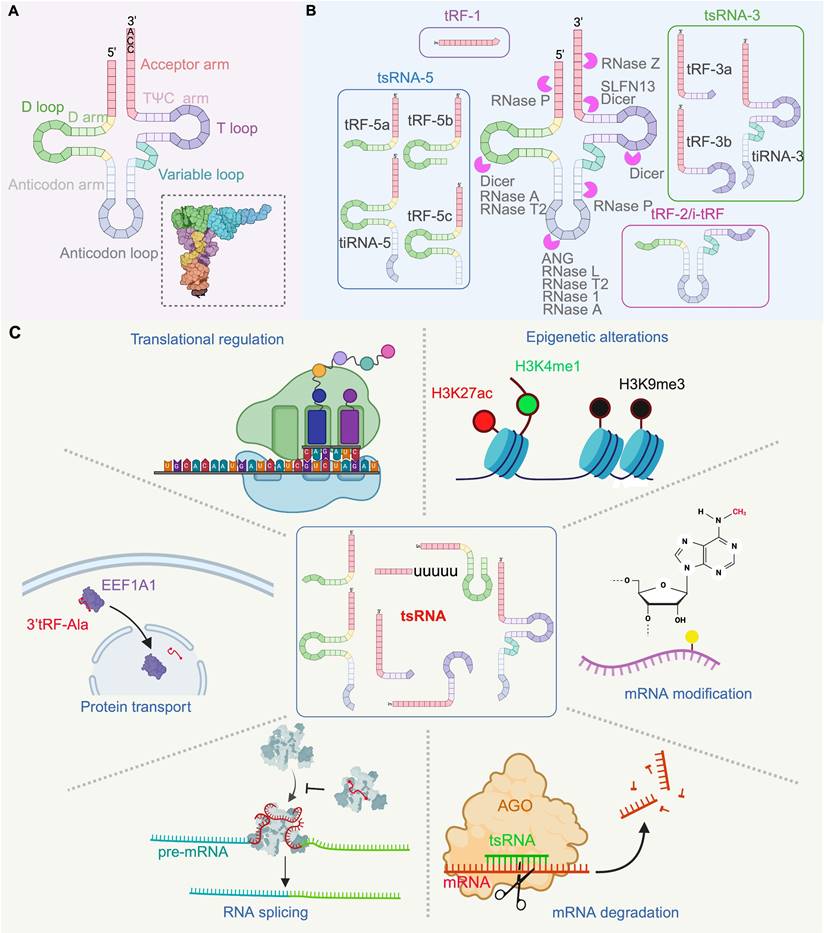
The biogenesis of tsRNA involves the transcription of tRNA genes by RNA polymerase III, followed by the subsequent maturation of pre-tRNAs into mature tRNAs with a “cloverleaf” structure which, in turn, folds further into an L-shaped conformation (Figure 1A) [1, 12-15]. This molecular assembly process includes the removal of 5' and 3' sequences by RNase P and RNase Z or ELAC2, respectively, as well as the addition of a 5'-CCA-3' tail by a nucleotidyl transferase. Mature tRNAs fold into an L-shaped conformation with exposed regions that are susceptible to cleavage, often at the anticodon or loop regions, by various enzymes including RNase P, Dicer, and ANG [3, 16-22]. Also, tRNA cleavage is known to occur outside of the loop regions. Other enzymes that can cleave tRNA to generate tsRNA include RNase 1, RNase Z, and members of the RNase A family like ANG [23-25], RNase T2 [26, 27], and certain interferon-stimulated genes, such as RNase L, Schlafen 11 (SLFN11) and Schlafen 13 (SLFN13) [28-30]. However, the exact cleavage mechanisms and sites for some of these enzymes, remain under investigation. Notably, the presence of tsRNAs appears to be closely linked to chemical modifications of tRNAs, with over 170 distinct modifications identified [31-36]. Such modifications, including cytosine-C5 methylation by DNMT2 and NSUN2, enhance the stability of tRNAs as well as translational decoding [37, 38]. Also, it has been found that tRNAs that are deficient in chemical modifications are more likely to be cleaved into tsRNA. For instance, the m5C38 modification of target tRNAs by DNMT2 can reduce the generation of 5' tRNA halves under stress [39, 40]. Furthermore, reduced levels of m5C48/49N modification arising from a deficiency in the modifier known as SUN2 can lead to increased levels of 5' tRNA halves [41]. The absence of certain modification-related enzymes such as PUS7, TRMT10A, ALKBH3, and BCDIN3D can also alter the relative composition of tsRNA species and their abundance within cells [22, 42-45]. Collectively, these observations underscore the impact of tRNA modifications on tsRNA biogenesis and in the event that there is no significant change to the quantity of parental tRNA species from which these are derived. What remains unclear is the entire lexicon of sequence motifs recognised by RNA-bodifying enzymes, the full complexity of modification states of tsRNAs, as well as how targeting enzymes facilitate tsRNA biogenesis. Moreover, whether modification-dependent protein interactions or tRNA folding dynamics regulate the susceptibility of tRNA to ribonuclease activity, all also remain to be clarified.
Currently, tsRNAs can be classified along two main groups based on size and origin: tRNA halves (tiRNAs) and tRNA-derived fragments (tRFs) (Figure 1B). For tiRNAs, these arise from the cleavage at the anticodon loop of mature tRNAs and are further divided into 5' tiRNAs (30-35 nt) and 3' tiRNAs (40-50 nt) [46, 47]. In contrast, tRFs are shorter in length, ranging from 13 to 30 nt, and include specific types such as 3U-tRF (tRF-1), tRF-3, tRF-5, and internal tRF (i-tRF or tRF-2), while tRF-1 originates from the 3' end of pre-tRNAs and varies in length. Of note, tRF-3, derived from the TΨC loop of mature tRNAs, is divided into tRF-3a (~18 nt) and tRF-3b (~22 nt), while tRF-5 extends from the tRNA terminus, and is subclassified further as tRF-5a (14-16 nt), tRF-5b (22-24 nt), and tRF-5c (27-30 nt). The i-tRFs are derived from anticodon loops of mature tRNAs.
Functional mechanisms of tsRNAs
Transcriptional regulation
Increasingly, tsRNAs have emerged as significant modulators of transcription through epigenetic mechanisms, that is, through gene expression changes without DNA sequence alterations [48, 49]. Indeed, tsRNAs have been described in influence DNA methylation, histone modification, and non-coding RNA regulation, all of which control gene expression at the transcriptional level to mediate cellular function in health and disease [50]. In the context of non-small cell lung cancer (NSCLC), it has been reported that upregulation of tsRNA AS-tDR-007333 in NSCLC cells could promote cell proliferation and migration by influencing histone modifications at the MED29 promoter [51]. In another example, the tsRNA known as tRF-GG has been found to influence the production of non-coding RNAs, a function that is influenced by the stability and activity of Cajal bodies within cells. Indeed, tRF-GG was reported to modulate the transcriptional repression of MERVL elements by heterochromatin through U7 snRNA regulation which, in turn, impacted histone protein availability and led to gene regulation effects [52]. Furthermore, tsRNAs have been found to interact with PIWI proteins, suggesting they could be relevant to gene silencing. For example, a tRF-5c species derived from tRNA-Glu has been reported to interact with Piwi-like protein 4, so as to recruit chromatin-modifying enzymes to the CD1A promoter which, in turn, enhances H3K9 methylation, so as to suppress CD1A gene transcription [53]. Other tsRNAs have also been identified in complexes with Piwi-like protein 2, suggestive of their influence kin downstream gene transcription [54]. Further research is needed to understand these mechanisms fully.
Post-transcriptional regulation
The roles for tsRNAs in post-transcriptional regulatory functions can, in part, be attributable to their document interaction with Argonaute (AGO) proteins [55]. Indeed, tsRNAs are capable of forming RNA-induced silencing complexes (RISC) with AGO proteins to regulate target RNA expression through base-pairing, a process that is thought to involve Dicer [56-59]. For example, tRF3008A from tRNA-Val interacts with AGO proteins to destabilize the FOXK1 transcript in colorectal cancer (CRC) cells, and this leads to reduced proliferation [60]. Notably, tsRNAs interact with specific AGO proteins, with some studies showing tsRNAs binding to AGO1, 3, and 4 [61]; with others studies showing interactions with AGO2 [58, 62]. Furthermore, tsRNAs are involved in the regulation of m6A methylation, an epigenetic modification that modulates target gene expression. For example, tRF-22 interacts with the 3' UTR of METTL3 mRNA in an AGO2-dependent manner, so as to modulate m6A methylation activity which, in turn, affects the expression of Axin1 and Arid1b in cells [63].
Structure, classification and function of tRNA-derived small RNA (tsRNA). (A) tRNA structure: The tRNA mocular consists of four arms (acceptor arm, D arm, TΨC arm and anticodon arm), three loops (D loop, TΨC loop and anticodon loop) and a variable loop structure. The L-shaped tertiary structure model of tRNA is depicted in the lower right corner. (B) Classification and Biogenesisof tsRNA: tsRNA is primarily generated by specific nucleases (such as ANG, Dicer, SLFN13, RNase 1, RNase P, RNase Z, RNase T2, RNaseA, RNase L, etc.) that cleave mature tRNAs or during the processing of tRNA precursors. (C) Function of tsRNA: tsRNA participates in the regulation of numerous of biological processes, including epigenetic alterations, mRNA modifications, gene sciencing, RNA splicing, translational regulation and protein transport. For example, the 3'tRF-Val directly binds to the chaperone molecule EEF1A1, an interaction that does not affect the levels of EEF1A1 mRNA and protein but mediates its transport to the nucleus. This figure was created with BioRender.com.
Beyond their roles in RISC complex assembly and function, tsRNAs also interact with RNA-binding proteins (RBPs) to sequester them from interacting with other RNAs, leading to changes in the landscape for mRNA stability within cells [64-67]. Indeed, as an example, i-tRFs from multiple tRNAs can interact with YBX1, leading to the destabilization of oncogenic transcripts in breast cancer (BC) cells [64]. Similarly, tRF-5 from tRNA-Gln interacts with IGF2BP1 to displace IGF2BP2-bound c-MYC RNA, leading to reduced mRNA stability of c-MYC transcripts [68, 69]. Moreover, tsRNAs also regulate protein transport, such as in the case of 3'tRF-Val which has been shown to bind EEF1A1 and mediate the nuclear transport of RBPs and modulating p53 signaling pathways [70]. In another such example, tRF-29-79 facilitates the cytoplasmic transport of the RNA-binding protein PTBP1 which, in turn, affects alternative splicing of SLC1A5 [71]. In addition to protein transport, tsRNAs have also been found to modulate target protein stability, such as in the case of tiRNA-Gly that interacts with RBM17 so as to enhance its prevalence within cells through inhibiton of proteasomal degradation [72], as well as in the case of tiRNA-Val-CAC-2 which stabilizes FUBP1 protein within cells while concomitantly promoting c-MYC transcription [73].
Translational regulation
Current evidence indicates that tsRNAs play a significant role in the translational regulation of protein synthesis, a critical process that includes transcription, mRNA processing, and ribosomal translation [74, 75]. Indeed, various reports have indicated that tsRNAs modulate the translation of proteins [24, 76-78], such as when under stress conditions like hypoxia or nutrient deprivation, where cells repress translation to conserve on intracellular resources. In that scenario, tsRNAs such as 5'tiRNAs, have been shown to interact with proteins like YBX1, a member of the cold shock family of proteins and an essential regulator of transcription and translation [79], which can inhibit global protein translation by promoting stress granule formation [80]. Interestingly, such tiRNAs contain a TOG motif that forms G-quadruplex structures, important for translational repression [81, 82]. Expanding on this notion, Lyons et al. discovered that G4-tiRNAs target the HEAT1 domain of eIF4G, leading to impaired ribosome scanning on mRNAs and the formation of stress granules [83]. Moreover, tRF-5 can inhibit translation by sequestering PABPC1, in the context of PUS7-dependent ψ8 modification [42], while a tRF-1, tRF-Gln-CTG-026, can repress global protein synthesis by affecting the association between TSR1 and the pre-40S ribosome [76]. In addition to nuclear genome-derived tsRNA species, mitochondrial tsRNAs have also been reported to participate in translational regulation, such as in the case of 5'tsRNA-Glu-CTC, a mitochondrial tsRNA that disrupts mt-tRNA-Leu aminoacylation as well as the translation of mitochondria-encoded proteins [84]. In contrast to the tsRNAs that inhibit translation, some tsRNAs, such as 3'tsRNA-Leu-CAG, can enigmatically enhance translation and promote pre-18S ribosomal RNA processing, as well as increase the availability of 40S ribosomal subunits within cells [77, 85]. These lines of evidence speak to the versatile roles of tsRNAs in controlling protein synthesis in homeostasis and stress.
A summary illustration for how tsRNAs contribute to gene expression, is provided in Figure 1C.
The role of tsRNAs in immunity
The immune system of mammals encompassing a variety of immune cells and organs, altogether representing an intricate defense mechanism that protects against infection and illness [86, 87]. Comprising both innate and adaptive immunity, it is responsible for the identification and elimination of foreign pathogens such as bacteria, viruses, and parasites, while also monitoring and clearing abnormal cells within the body [86, 87]. Recent studies have revealed the presence of tsRNAs within immune cells [88], including candidate tsRNAs that influence myeloid cell differentiation. For example, the tsRNA known as 5'tiRNA-Pro-CGG-1, found in extracellular vesicles from osteoblasts, has been shown to enhance protein translation, cell proliferation, and myeloid differentiation when transferred to granulocyte-monocyte progenitors [89]. This suggests a potential role for tsRNA in immune modulation.
Functions for tsRNAs in innate immunity
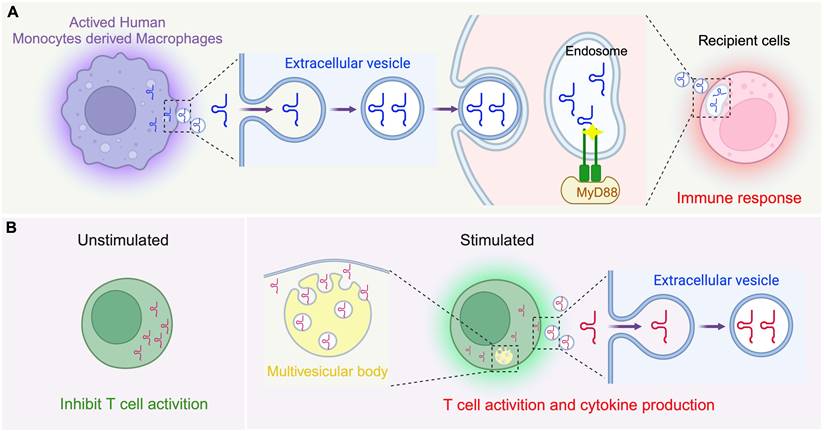
The innate immune system acts as the frontline defense of the body against invading pathogens, utilizing both physical barriers like the skin and mucous membranes, and cellular components such as leukocytes to initiate a swift defense [90, 91]. A critical element of this system is the Toll-like receptor (TLR) family, with TLR7 being particularly significant for its ability to detect single-stranded RNAs from foreign pathogens. Our understanding of the specific endogenous RNAs that activate TLR7 has remained poor until recent findings, as follows. Pawar et al. identified that 5'-tRNA half molecules are key activators of TLR7 [92], such that, during mycobacterial infections, TLR7 activation on the cell surface leads to an upregulation of 5'-tRNA half molecules in human monocyte-derived macrophages (HMDMs), leading to their accumulation within extracellular vesicles (EVs), with specific tsRNA species such as EV-5'-tRNA-His-GUG showing higher concentrations than the most prevalent EV-microRNAs (Figure 2A) [92]. Notably, the mechanism by which these molecules are transferred to recipient cells so as to activate TLR7 within endosomes has been experimentally confirmed as a new pathway for tRNA in mediating innate immunity, and 5'-tRNA half molecules are now recognizes as “immune activators”. In another example, the 5'-tRNA-Val-CAC/AAC half, prevalent in macrophage EVs, has been demonstrated to robustly activate TLR7 as part of a molecular mechanism that mediates bacterial clearance, and this function relies on the terminal GUUU sequence of this tsRNA [93]. Thus, these findings have expanded our understanding of tsRNA as intrinsic ligands for TLR7 and their active participation in signalling the innate immune response.
Functions for tsRNAs in adaptive immunity
Adaptive immunity provides specific responses through B and T cells, offering long-term protection and forming immunological memory to rapidly respond to the same pathogens upon re-exposure [94, 95]. Recent studies have suggested that tsRNAs have the potential to modulate T cell activation. Chiou et al. discovered a significant enrichment of tsRFs in EVs released by T cells, with 45% of tsRNAs being more abundant than miRNAs [96]. Upon activation, T cells induced the release of a distinct subset of tsRNAs derived from specific regions within their parental tRNAs, excluding variable loops, via EVs. Disruption of EV biogenesis pathways results in the intracellular retention of these tRFs within multivesicular bodies (MVBs). The use of antisense oligonucleotides to target these tsRNAs has been shown to enhance T cell activation, indicating a selective release mechanism of tRFs into EVs via MVBs, potentially removing suppressive factors within the immune response (Figure 2B) [96]. While these insights have broadened our understanding of tsRNAs in the adaptive immune response, how tsRNAs mediate the complex dynamics of immune cell functions remains to be better clarified.
Regulation of immune cell function by tsRNA. (A) In innate immunity, activated human monocyte-derived macrophages secrete extracellular vesicles (EVs) containing tsRNA, particularly EV-5'-tRNA-His-GUG half molecules, to activate the immune response. The 5'-tRNA-Val-CAC/AAC half-molecule effectively targets and eliminates intracellular pathogens by activating TLR7. (B) In adaptive immunity, T cell activation triggers the formation of multivesicular body (MVB) and the release of a specific set of tRFs via EVs derived from the 5' end and 3' inner region of trRNA without variable loops. This figure was created with BioRender.com.
The roles for tsRNAs in mediating immunity in disease
The roles for tsRNAs in tumor immunology
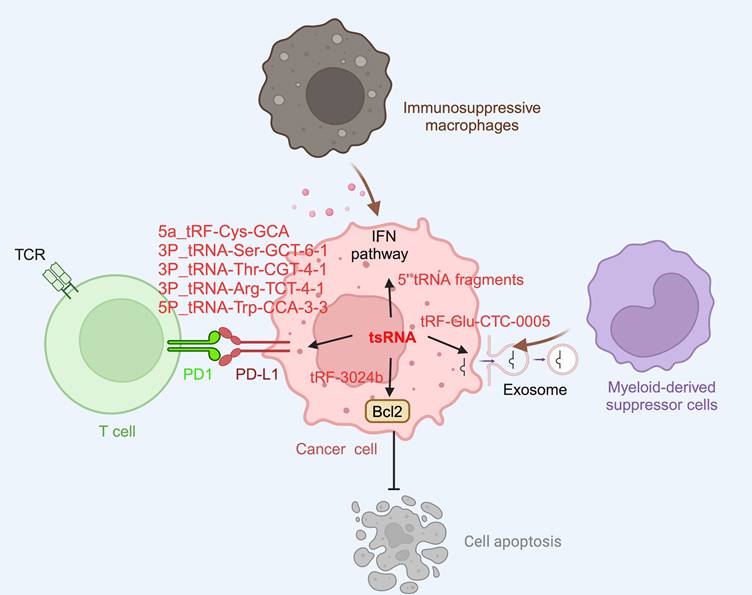
Cancer is a leading cause of mortality and a significant factor in reducing life expectancy globally. In 2020, it was estimated that there were 19.3 million new cancer cases worldwide, excluding non-melanoma skin cancer, and nearly 10.0 million cancer-related deaths [97]. Understanding the pathogenesis of cancer and developing better diagnostics and effective treatment strategies for this condition is an urgent priority worldwide. It has been recognized that the avoidance of immune destruction is one of the 14 hallmarks of cancer, and this underscores the indispensable role of the immune system in cancer development and progression [98]. Recent studies have shed light on the regulatory role of tsRNAs in tumor proliferation [70, 99, 100], apoptosis [66, 70], and tumor metastasis [72, 101-103], all features of which influence cancer development and severity. Moreover, the association of tsRNAs with tumor immunity has also been reported (Figure 3). For instance, a study by Shan et al. demonstrated a significant link between T cell activation and the abundance of tsRNAs such as ts-34 and ts-49, in breast cancer patients [104]. In that study, it was reported subjects with T cell exhaustion had low levels of ts-34 or high levels of ts-49 which were associated with enhanced survival rates [104]. In lung cancer, tsRNAs such as 5a_tRF-Cys-GCA, 3P_tRNA-Ser-GCT-6-1, 3P_tRNA-Thr-CGT-4-1, 3P_tRNA-Arg-TCT-4-1, and 5P_tRNA-Trp-CCA-3-3 were correlated with the PD-L1 immune checkpoint and PD-L1 signaling pathway-related genes [105]. These findings indicate that the presence of tsRNAs are relevant to cancer and, arguably, targeting such tsRNAs might have potential therapeutic significance when treating this condition.
tsRNA in tumor immunology. tsRNA within tumor cells contribute to immune evasion by enhancing the expression of PD-L1. Specific tsRNA species, including 5a_tRF-Cys-GCA, 3P_tRNA-Ser-GCT-6-1, 3P_tRNA-Thr-CGT-4-1, 3P_tRNA-Arg-TCT-4-1, and 5P_tRNA-Trp-CCA-3-3, have been identified to play a role in this process. However, the precise molecular mechanisms remain unclear. Additionally, tsRNA can recruit immunosuppressive macrophagesand myeloid-derived suppressor cells by modulating the interferon pathway, as exemplified by 5'-tRNA fragments. They also contribute to the formation of an immunosuppressive microenvironment by secreting exosomes containing tsRNAs, such as tRF-Glu-CTC-0005. Furthermore, tsRNAs can impede the effectiveness of immunotherapy by inhibiting apoptosis. For instance, tRF-3024b reduces tumor cell apoptosis by promoting BCL-2 expression, thereby inhibiting the therapeutic effects of immune checkpoint inhibitors. This figure was created with BioRender.com.
Recent studies have shed light into the mechanisms through which tsRNA regulates tumor immunity. In prostate cancer, METTL1 depletion results in the emergence of a novel class of 5' tsRNAs, which modulate translation control to favor the synthesis of key regulators of tumor growth suppression, interferon pathways, and immune effectors [106]. In esophageal squamous cell carcinoma (ESCC), tRF-3024b, which upregultes in ESCC cells that survived co-culture with cytotoxic T lymphocytes (CTLs), has been shown to reduce tumor cell apoptosis by sequestering miR-192-5p and promoting BCL-2 expression, thereby enhancing the protective effects of BCL-2 [107]. This finding is suggestive of an approach through which modulation of key tsRNAs could enhance the response of ESCC to CTLs as an strategy to improve the effectiveness of immunotherapy for ESCC. In another example, 3'tRF-Ala-AGC is upregulated in BC specimens as well as in adriamycin-resistant cancer cells, and this tsRNA has been linked to the promotion of malignant cell activity and the facilitation of M2 macrophage polarization via interacting with the Type 1-associated death domain protein (TRADD). Moreover, it was found that overexpression of 3'tRF-Ala-AGC in M2 macrophages can activate the NF-κB signaling pathway in BC cells [9]. Furthermore, the association of tsRNA with exosomes has been implicated in tumor immunity, with tRF-GluCTC-0005 in pancreatic cancer-derived exosomes being shown to be important for recruiting myeloid-derived suppressor cells to a tumor site, leading to the formation of an immunosuppressive microenvironment [108]. The molecular mechanism for the actions of tRF-GluCTC-0005 in this cellular context has been found to involve its binding to the 3' untranslated region of WDR1 mRNA in hepatic stellate cells, leading to stabilization of this mRNA, as well as the activity of YAP, a modulator of cellular gene expression [108].
The roles for tsRNA in autoimmune disorders
Systemic lupus erythematosus (SLE) is an autoimmune disorder that is associated with a loss of nuclear and cytoplasmic self-antigens, leading to the production of autoantibodies and the formation of immune complexes [109, 110]. These factors contribute to inflammation that affects multiple organ systems. Recent research has found that tsRNAs may serve as diagnostic biomarkers for SLE that are detected in the sera, particularly when changes to levels of tRF-His-GTG-1 and levels of anti-dsDNA are observed [111]. Moreover, serum tsRNAs have been shown to directly target signaling molecules that play a pivotal role in the regulation of the immune system [111]. Furthermore, tsRNAs are differentially expressed in peripheral blood mononuclear cells from SLE patients compared to healthy controls. Bioinformatic analysis of these differences suggests that the altered target genes are prominently enriched in T cell receptor signaling pathways, Th1 and Th2 cell differentiation, and primary immunodeficiency [112]. Another study of tsRNAs in SLE found that, mesenchymal stem cell exosomal tsRNA-21109 could ameliorate SLE by inhibiting macrophage M1 polarization [11, 113]. In addition, research by Geng et al. demonstrated that tsRNAs, such as tRF-3009, were involved in the modulation of IFN-α-induced oxidative phosphorylation in CD4+ T cells of lupus patients. In vitro analysis of CD4+ T cells overexpressing tRF-3009 revealed a correlation between this product and type I IFN (interferon) as well as oxidative phosphorylation pathways. Notably, IFN-α demonstrated the capacity to stimulate the generation of reactive oxygen species (ROS) and ATP in CD4+ T cells. Conversely, the knockdown of tRF-3009 reversed these effects. Overexpression of tRF-3009 alone in CD4+ T cells was sufficient to enhance oxygen consumption rate, ROS production, and ATP generation [114].
Sarcoidosis is a systemic autoimmune disease characterized by the formation of non-caseating granulomas, a pathological hallmark of the condition as noted in recent studies [115]. Notably, three specific tsRNAs, namely tiRNA-Glu-TTC-001, tiRNA-Lys-CTT-003, and tRF-Ser-TGA-007, were reported to be significantly dysregulated in sarcoidosis, and suggest that these tsRNAs mediate the pathophysiology of this disease [116]. A further bioinformatics study revealed the possible involvement of such tsRNAs in mediating chemokine signaling, cAMP- and cGMP-PKG pathways, retrograde endorphin signaling, and the FoxO pathway [116]. Despite these advances in knowledge, the precise mechanisms by which these tsRNAs modulate the immune response in sarcoidosis remain to be defined.
The involvement of tsRNAs in cellular infections
Recent studies has illuminated the involvement of tsRNAs in viral infections [59, 117], as evidenced by the promotion of RSV replication by 5'-tRF-Gly-CCC and 5'-tRF-Lys-CTT [118]. In patients diagnosed with COVID-19, caused an infection by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), tsRNAs, particularly from the tRF-5, have been detected in nasopharyngeal swabs of affected individuals [119]. Intriguingly, post-SARS-CoV-2 infection, an upregulation of specific tsRNAs has been observed in the blood, with higher levels correlating with the severity of COVID-19 symptoms. Specifically, the 3'CCA tsRNAs derived from tRNA-Gly have been found to be significantly associated with the inflammatory marker C-reactive protein, and could therefore present as a possible target for therapeutic modulation [120]. In bovine studies, dysregulation of five tsRNAs (tRF-36-8JZ8RN58X2NF79E, tRF-20-0PF05B2I, tRF-27-W4R951KHZKK, tRF-22-S3M8309NF, and tRF-26-M87SFR2W9J0) in calves infected with bovine leukemia virus is indicative of their association with such an infection [121]. In addition, tsRNAs have been implicated in bacterial infections. For example, Gumas et al. reported a global and substantial upregulation of plasma small non-coding RNAs (sncRNAs) in patients infected with mycobacterium tuberculosis (MTB), with tsRNAs being the most significantly elevated class [122]. These sncRNAs, which are notably abundant in MTB-infected patients, potently activate human macrophages via TLR7, triggering cytokine production [122]. These findings collectively offer insight into how tsRNAs might influence the immune response when challenged by an infectious disease, and reveals putative tsRNA candidates that could be modulated for therapeutic impact.
The roles for tsRNAs in other health conditions
The modulation of the immune system by tsRNAs has been observed in a spectrum of health challenges. For example, in hypertrophic scarring, the overexpression of tsRNA-14783 is linked to macrophage polarization towards the M2 phenotype, and this, in turn, has been reported to influence scar formation through the upregulation of M2 macrophage markers such as TGF-β, IL-10, and CD206, and downregulation of M1 macrophage markers like IL-1 and NOS2 [123, 124]. In the context of diabetes, it has been reported that alterations to tsRNAs profiles within the islets of NOD mice are observed during the early stages of type 1 diabetes. A subset of these tsRNAs, including Gly-GCC-5'H and Leu-CCA-I that are enriched in EVs from CD4+/CD25- T cells, impacts beta cell gene expression and immune regulation, and predisposing them to apoptosis, and this suggests a role for tsRNAs in diabetes pathogenesis [125]. In another scenario, high-throughput sequencing studies have identified differential tsRNA expression (tDR-006826, tDR-006049, tDR-001271 and tDR-001276) in bone marrow mesenchymal stem cells of patients with fibrous dysplasia (FD), a disease characterized by abnormal bone tissue replacement. These tsRNAs were associated with immune response regulation, as revealed by GO and KEGG pathway analysis [126]. In renal ischemia-reperfusion injury, differential tsRNA expression, including tiRNA-Gly-GC-003, tiRNA-Lys-CTT-003, and tiRNA-His-GTG-002, has been associated with natural killer cell-mediated cytotoxicity pathways, and this could suggest that tsRNAs could influence acute kidney injury progression following ischemia-reperfusion [127]. In pigs, intrauterine growth restriction is marked by differential tsRNA expression, with tRF-5c predominating and originating mostly from tRNA-Gly-GCC. Other significant tsRNAs, including tiRNA-Ser-TGA-001 and tRF-Val-AAC-034, have also been implicated in immune system processes, with predicted target genes including TNF, TLR4, CD44, MAPK1, and STAT1, potentially regulating T cell receptor and toll-like receptor signaling pathways [128]. In the context of post-muscle injury, 5'tiRNA-Gly is upregulated and correlates with inflammation, promoting pro-inflammatory cytokines (IL-1β, IL-6) and M1 macrophage markers (TNF-α, CD80, MCP-1), increasing the percentage of CD86+ macrophages and inhibiting the percentage of CD206+ macrophages. Notably the mechanism of action for 5'tiRNA-Gly appears to involve modulation of the gene Tgfbr1 in an AGO1- and AGO3-dependent manner. Through modulation of the TGF-β signaling pathway, 5'tiRNA-Gly regulates the expression of downstream genes related to inflammation, satellite cell activation, and myoblast differentiation. These findings indicate that 5'tiRNA-Gly may modulate skeletal muscle regeneration through TGF-β-mediated inflammation, and this, in turn, suggests that 5'tiRNA-Gly may be a viable target for the treatment of skeletal muscle regeneration [129].
A comprehensive overview of the functions of tsRNAs in across multiple diseases and in the context of immunity is presented in Table 1, and a schematic representation of the role for tsRNAs in disease immunity is provided in Figure 4.
Conclusion and future perspectives
Here, we have reviewed the evidence for tsRNAs as pivotal regulators in a spectrum of biological processes, including in immune responses and in the pathogenesis of a wide range of diseases. The intricate biogenesis of tsRNAs and their diverse functionalities, particularly in immunological contexts, are increasingly being recognized. Despite these advancements, there is a sense that we are only beginning to understand how tsRNAs carry out its cellular functions to health and disease. Notably, the accurate detection of tsRNAs remains a significant challenge due to their small size, while the presence of modifications on tsRNAs can interfere with RNA-seq approaches for detecting these non-coding RNAs. However, with the recognition of the pivotal roles that small RNAs play in both physiological and pathological processes, there has recently been a surge in the development of improved sequencing technologies that have been specifically designed to detect small RNAs with high sensitivity and specificity. Such approaches include CPA-seq [130], PANDORA-seq [16] and AQRNA-seq [131], all of which hold promise to improve detection of tsRNAs. Additionally, post-sequencing, it is imperative to verify whether these tsRNAs are mere degradation products or possess functional roles.
The clinical application of tsRNAs in immunity shows promise, with evidence that tsRNAs could be targeted as biomarkers or as therapeutic targets. For example, tsRNAs identified in conditions such as cancer and autoimmune disorders could be targets for restoring immune balance. In cancer, where immune evasion is a critical hallmark, tsRNAs could be leveraged to enhance the efficacy of immunotherapies, whereby immunosuppressive tsRNAs could be inhibited, while the activity of tsRNAs that promote anti-tumor immunity could be boosted. In the context of autoimmune diseases, tsRNAs could be used to suppress an overactive immune response.
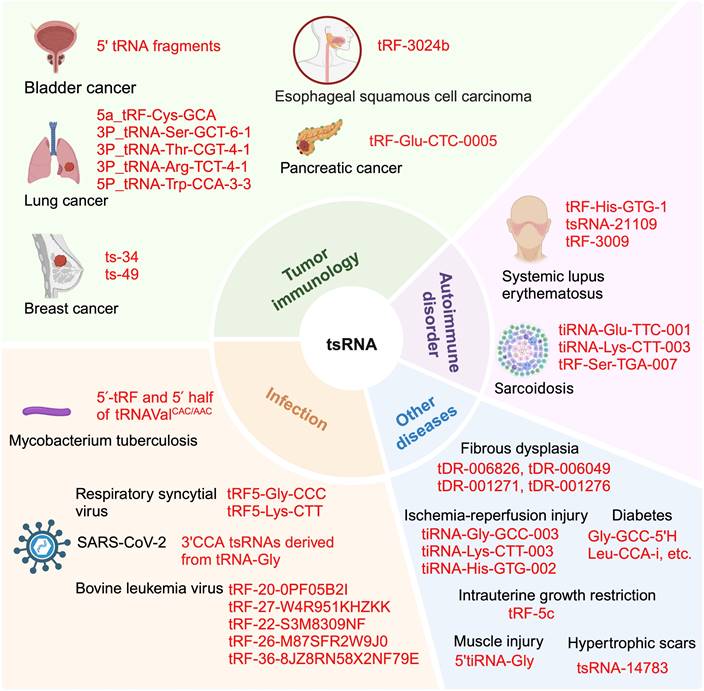
tsRNA's involvement in diverse immune-related diseases. tsRNA plays a role in various immune-related conditions, including different types of cancer such as bladder cancer, lung cancer, braest cancer, esophageal squamous cell carcinoma and pancreatic cancer. It is also implicated in autoimmune diseases like systemic lupus erythematosus and and sarcoidosis. Additionally, tsRNA is associated with bacterial and viral infections, including respiratory syncytial, SARS-Cov-2 and bovine leukemia virus. Other conditions where tsRNA is involved encompass hypertrophic scars, intrauterin growth restriction, diabetes, ischemia-reperfusion injury, muscle injury and fibrous dysplasia. This figure was created with BioRender.com.
The role of tsRNAs in disease immunity
| Disease | tsRNA | Function | Mechanism | Reference |
|---|---|---|---|---|
| Breast cancer | ts-34, ts-49 | Relates to T cell activation status. | \ | [104] |
| 3'tRF-Ala-AGC | Promotes cell malignant activity and facilitates M2 polarization of macrophages. | Binds Type 1-associated death domain protein, and its overexpression in M2 macrophages activates NF-κB signaling pathway in BC cells. | [9] | |
| Lung cancer | 5a_tRF-Cys-GCA, 3P_tRNA-Ser-GCT-6-1, 3P_tRNA-Thr-CGT-4-1, 3P_tRNA-Arg-TCT-4-1, 5P_tRNA-Trp-CCA-3-3 | Positively associates with PD-L1 immune checkpoint and correlates with genes that target in PD-L1 checkpoint signaling pathway. | \ | [105] |
| Prostate cancer | 5' tsRNAs due to METTL1 depletion | Suppresses prostate tumour growth, and enhances the response to immune checkpoint blockade therapy. | Represses translation initiation, and activates IFN signalling pathway. | [106] |
| Esophageal squamous cell carcinoma | tRF-3024b | Reduces the apoptosis of tumor cells when co-cultured with cytotoxic T lymphocytes. | Promotes the expression of B-cell lymphoma-2 by sequestering miR-192-5p, a microRNA that would normally inhibit BCL-2 expression. | [107] |
| Pancreatic cancer | tRF-GluCTC-0005 | tRF-GluCTC-0005 in pancreatic cancer-derived exosomes recruits myeloid-derived suppressor cells in liver, and creates an immunosuppressive microenvironment, as well as advances liver metastasis from pancreatic cancer. | Binds to the 3' untranslated region of WDR1 mRNA in hepatic stellate cells, stabilizing the mRNA and affecting the YAP protein's activity. | [108] |
| Systemic lupus erythematosus | Differentially expressed tsRNAs | Enriched in T cell receptor signaling pathways, Th1 and Th2 cell differentiation, and primary immunodeficiency. | \ | [112] |
| tsRNA-21109 | tsRNA-21109-privative MSC-exo upregulates M1 markers, downregulates M2 markers, and increases levels of TNF-α and IL-1β in macrophages. | \ | [11] | |
| tRF-3009 | Positively correlates with SLE disease activity index, active lupus nephritis and serum IFN-α levels. | Upregulates IFN-α-induced ROS and ATP production in CD4+ T cells. | [114] | |
| Sarcoidosis | tiRNA-Glu-TTC-001, tiRNA-Lys-CTT-003, tRF-Ser-TGA-007 | May play roles in chemokine, cAMP, cGMP-PKG, retrograde endorphin, and FoxO signalling pathways by bioinformatics analyses. | \ | [116] |
| post-SARS-CoV-2 infection | 3'CCA tsRNAs derived from tRNA-Gly | Associates with the inflammatory marker C-reactive protein. | \ | [120] |
| MTB infection | 5'tsRNAs derived from tRNA-His-GUG, tRNA-Glu-CUC, tRNA-Val-CAC/AAC | upregulated in patients infected with MTB, and could be as potent activators of macrophage TLR7. | \ | [122] |
| Hypertrophic scarring | tsRNA-14783 | Promotes scar formation by regulating macrophage polarization towards the M2 phenotype. | \ | [123, 124] |
| Diabetes | Gly-GCC-5'H, Leu-CCA-i, etc. | Impacts beta cell gene expression and immune regulation, predisposing them to apoptosis | These tsRNAs in EVs from CD4+/CD25- T cells move into beta cells through adoptive transfer. | [125] |
| Fibrous dysplasia | tDR-006826, tDR-006049, tDR-001271, tDR-001276 | Involved in immune response regulation, as revealed using bioinformatics analyses. | \ | [126] |
| Renal ischemia-reperfusion injury | tiRNA-Gly-GC-003, tiRNA-Lys-CTT-003, tiRNA-His-GTG-002 | Associates with natural killer cell-mediated cytotoxicity pathways, as reveaed using bioinformatics analyses. | \ | [127] |
| Intrauterine growth restriction | tiRNA-Ser-TGA-001, tRF-Val-AAC-034 | Mediates the immunocompromise caused by intrauterine growth restriction. | May target TNF, TLR4, CD44, MAPK1, and STAT1, potentially regulate T cell receptor and toll-like receptor signaling pathways | [128] |
| Muscle injury | 5'tiRNA-Gly | Upregulated after muscle injury and positively correlated with inflammation; promotes expression of proinflammatory cytokines, the M1 markers of macrophages, the percentages of CD86+macrophages, and myogenic differentiation marker genes of myoblast. | Targets TGFBR1 in a AGO1- and AGO3-dependet manner to regulate the TGF-β signaling pathway. | [129] |
“\” indicates that the molecular mechanism is currently unknown
Moreover, when considering infectious diseases, tsRNAs could be leveraged to enhance the immune defense. We surmise that potential clinical applications of tsRNAs in the context of disease immunity can be summarized as follows: firstly, as immune-modulators, certain 5'-tRNA halves from tRNA-His-GUG that activate TLR7, could be key as immune activators [92, 93]. Secondly, the levels of tsRNAs could be predictive of the immune status of patients and possible be prognostic for disease severity, such as for BC, where T-cell activation is significantly associated with ts-34 and ts-49 [104]. Thirdly, tsRNAs could be investigated as therapeutic targets. For example, knockdown of tRF-3009 in lupus patients' CD4+ T cells reverses IFN-α-induced oxidative phosphorylation and this approach could be effective as a strategy to develop this tsRNA as therapeutic targets for SLE [114]. Fourthly, tsRNA agonists may be developed as an exosome-delivered therapeutic, since it has been shown that exosomal tsRNA-21109 from mesenchymal stem cells can SLE by inhibiting macrophage M1 polarization [11]. Finally, the design of inhibitors that target tsRNAs could enhance the efficacy of immunotherapies. For example, in ESCC, tRF-3024b enhances the tolerance of ESCC to CTLs, suggesting that inhibitors could be designed to increase tumor cell sensitivity to CTLs [107].
In conclusion, we find that tsRNAs are relevant to disease immunity, and there remains a clear need for clarification of the mechanistic functions of tsRNAs in health and disease, particularly in the context of immune system interactions. Future research is also needed to corroborate bioinformatics predictions and explore the translational potential of tsRNAs within clinical settings. Moreover, the stability of tsRNAs within the immune microenvironment and their potential for targeted delivery as therapeutic agents are also important areas for research. As our understanding of tsRNA biology expands, so too does the opportunity to harness their potential in managing tissue homeostasis, as well as immune-related diseases and other health challenges.
Acknowledgements
We thank Julian Heng (ORCID: 0000-0002-0378-7078) for providing professional English-language editing (Certificate No. 0Hy4Ah4Y).
Funding
This work was supported by the Sichuan Cancer Hospital Outstanding Youth Funding (Grant No. YB2021031).
Author Contributions
Conceptualization, H,J.; writing-original draft preparation, H,J., L.Z.; writing-review and editing, H,J., L.Z.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Orellana EA, Siegal E, Gregory RI. tRNA dysregulation and disease. Nat Rev Genet. 2022;23:651-64
2. Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamecnik PC. A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem. 1958;231:241-57
3. Chen Q, Zhang X, Shi J, Yan M, Zhou T. Origins and evolving functionalities of tRNA-derived small RNAs. Trends Biochem Sci. 2021;46:790-804
4. Yu M, Lu B, Zhang J, Ding J, Liu P, Lu Y. tRNA-derived RNA fragments in cancer: current status and future perspectives. J Hematol Oncol. 2020;13:121
5. Muthukumar S, Li CT, Liu RJ, Bellodi C. Roles and regulation of tRNA-derived small RNAs in animals. Nat Rev Mol Cell Biol. 2024;25:359-78
6. Li G, Manning AC, Bagi A, Yang X, Gokulnath P, Spanos M. et al. Distinct Stress-Dependent Signatures of Cellular and Extracellular tRNA-Derived Small RNAs. Adv Sci (Weinh). 2022;9:e2200829
7. Ren B, Wang X, Duan J, Ma J. Rhizobial. tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science. 2019;365:919-22
8. Lu S, Wei X, Tao L, Dong D, Hu W, Zhang Q. et al. A novel tRNA-derived fragment tRF-3022b modulates cell apoptosis and M2 macrophage polarization via binding to cytokines in colorectal cancer. J Hematol Oncol. 2022;15:176
9. Mo D, Tang X, Ma Y, Chen D, Xu W, Jiang N. et al. tRNA-derived fragment 3'tRF-AlaAGC modulates cell chemoresistance and M2 macrophage polarization via binding to TRADD in breast cancer. J Transl Med. 2024;22:706
10. Yang M, Mo Y, Ren D, Liu S, Zeng Z, Xiong W. Transfer RNA-derived small RNAs in tumor microenvironment. Mol Cancer. 2023;22:32
11. Dou R, Zhang X, Xu X, Wang P, Yan B. Mesenchymal stem cell exosomal tsRNA-21109 alleviate systemic lupus erythematosus by inhibiting macrophage M1 polarization. Mol Immunol. 2021;139:106-14
12. Turowski TW, Tollervey D. Transcription by RNA polymerase III: insights into mechanism and regulation. Biochem Soc Trans. 2016;44:1367-75
13. Frank DN, Pace NR. Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu Rev Biochem. 1998;67:153-80
14. Maraia RJ, Lamichhane TN. 3' processing of eukaryotic precursor tRNAs. Wiley Interdiscip Rev RNA. 2011;2:362-75
15. Suzuki T. The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol. 2021;22:375-92
16. Shi J, Zhang Y, Tan D, Zhang X, Yan M, Zhang Y. et al. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat Cell Biol. 2021;23:424-36
17. Kumar P, Mudunuri SB, Anaya J, Dutta A. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015;43:D141-5
18. Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W. et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22:1609-12
19. Kikuchi Y, Sasaki N, Ando-Yamagami Y. Cleavage of tRNA within the mature tRNA sequence by the catalytic RNA of RNase P: implication for the formation of the primer tRNA fragment for reverse transcription in copia retrovirus-like particles. Proc Natl Acad Sci U S A. 1990;87:8105-9
20. Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW. et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147-60
21. Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673-95
22. Reinsborough CW, Ipas H, Abell NS, Nottingham RM, Yao J, Devanathan SK. et al. BCDIN3D regulates tRNAHis 3' fragment processing. PLoS Genet. 2019;15:e1008273
23. Nechooshtan G, Yunusov D, Chang K, Gingeras TR. Processing by RNase 1 forms tRNA halves and distinct Y RNA fragments in the extracellular environment. Nucleic Acids Res. 2020;48:8035-49
24. Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35-42
25. Zhu B, Lee SJ, Tan M, Wang ED, Richardson CC. Gene 5.5 protein of bacteriophage T7 in complex with Escherichia coli nucleoid protein H-NS and transfer RNA masks transfer RNA priming in T7 DNA replication. Proc Natl Acad Sci U S A. 2012;109:8050-5
26. Megel C, Hummel G, Lalande S, Ubrig E, Cognat V, Morelle G. et al. Plant RNases T2, but not Dicer-like proteins, are major players of tRNA-derived fragments biogenesis. Nucleic Acids Res. 2019;47:941-52
27. Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J Cell Biol. 2009;185:43-50
28. Donovan J, Rath S, Kolet-Mandrikov D, Korennykh A. Rapid RNase L-driven arrest of protein synthesis in the dsRNA response without degradation of translation machinery. RNA. 2017;23:1660-71
29. Li M, Kao E, Gao X, Sandig H, Limmer K, Pavon-Eternod M. et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature. 2012;491:125-8
30. Li M, Kao E, Malone D, Gao X, Wang JYJ, David M. DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat Struct Mol Biol. 2018;25:1047-58
31. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A. 1974;71:3971-5
32. Oberbauer V, Schaefer MR. tRNA-Derived Small RNAs: Biogenesis, Modification, Function and Potential Impact on Human Disease Development. Genes (Basel). 2018;9:607
33. Pan T. Modifications and functional genomics of human transfer RNA. Cell Res. 2018;28:395-404
34. Phizicky EM, Hopper AK. tRNA processing, modification, and subcellular dynamics: past, present, and future. RNA. 2015;21:483-5
35. Wang X, Matuszek Z, Huang Y, Parisien M, Dai Q, Clark W. et al. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA. 2018;24:1305-13
36. Pereira M, Ribeiro DR, Pinheiro MM, Ferreira M, Kellner S, Soares AR. m(5)U54 tRNA Hypomodification by Lack of TRMT2A Drives the Generation of tRNA-Derived Small RNAs. Int J Mol Sci. 2021;22:2941
37. Durdevic Z, Mobin MB, Hanna K, Lyko F, Schaefer M. The RNA methyltransferase Dnmt2 is required for efficient Dicer-2-dependent siRNA pathway activity in Drosophila. Cell Rep. 2013;4:931-7
38. Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M. et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590-5
39. Tuorto F, Herbst F, Alerasool N, Bender S, Popp O, Federico G. et al. The tRNA methyltransferase Dnmt2 is required for accurate polypeptide synthesis during haematopoiesis. EMBO J. 2015;34:2350-62
40. Zhang Y, Zhang X, Shi J, Tuorto F, Li X, Liu Y. et al. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol. 2018;20:535-40
41. Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P. et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020-39
42. Guzzi N, Ciesla M, Ngoc PCT, Lang S, Arora S, Dimitriou M. et al. Pseudouridylation of tRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell. 2018;173:1204-16 e26
43. Cosentino C, Toivonen S, Diaz Villamil E, Atta M, Ravanat JL, Demine S. et al. Pancreatic beta-cell tRNA hypomethylation and fragmentation link TRMT10A deficiency with diabetes. Nucleic Acids Res. 2018;46:10302-18
44. Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J. et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533-45
45. Martinez A, Yamashita S, Nagaike T, Sakaguchi Y, Suzuki T, Tomita K. Human BCDIN3D monomethylates cytoplasmic histidine transfer RNA. Nucleic Acids Res. 2017;45:5423-36
46. Su Z, Wilson B, Kumar P, Dutta A. Noncanonical Roles of tRNAs: tRNA Fragments and Beyond. Annu Rev Genet. 2020;54:47-69
47. Kumar P, Kuscu C, Dutta A. Biogenesis and Function of Transfer RNA-Related Fragments (tRFs). Trends Biochem Sci. 2016;41:679-89
48. Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12-27
49. Recillas-Targa F. Cancer Epigenetics: An Overview. Arch Med Res. 2022;53:732-40
50. Bheda P, Schneider R. Epigenetics reloaded: the single-cell revolution. Trends Cell Biol. 2014;24:712-23
51. Yang W, Gao K, Qian Y, Huang Y, Xiang Q, Chen C. et al. A novel tRNA-derived fragment AS-tDR-007333 promotes the malignancy of NSCLC via the HSPB1/MED29 and ELK4/MED29 axes. J Hematol Oncol. 2022;15:53
52. Boskovic A, Bing XY, Kaymak E, Rando OJ. Control of noncoding RNA production and histone levels by a 5' tRNA fragment. Genes Dev. 2020;34:118-31
53. Zhang X, He X, Liu C, Liu J, Hu Q, Pan T. et al. IL-4 Inhibits the Biogenesis of an Epigenetically Suppressive PIWI-Interacting RNA To Upregulate CD1a Molecules on Monocytes/Dendritic Cells. J Immunol. 2016;196:1591-603
54. Pekarsky Y, Balatti V, Palamarchuk A, Rizzotto L, Veneziano D, Nigita G. et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci U S A. 2016;113:5071-6
55. Martinez G, Choudury SG, Slotkin RK. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res. 2017;45:5142-52
56. Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA. 2018;24:1093-105
57. Su Z, Kuscu C, Malik A, Shibata E, Dutta A. Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J Biol Chem. 2019;294:16930-41
58. Di Fazio A, Schlackow M, Pong SK, Alagia A, Gullerova M. Dicer dependent tRNA derived small RNAs promote nascent RNA silencing. Nucleic Acids Res. 2022;50:1734-52
59. Choi EJ, Ren J, Zhang K, Wu W, Lee YS, Lee I. et al. The Importance of AGO 1 and 4 in Post-Transcriptional Gene Regulatory Function of tRF5-GluCTC, an Respiratory Syncytial Virus-Induced tRNA-Derived RNA Fragment. Int J Mol Sci. 2020;21:8766
60. Han Y, Peng Y, Liu S, Wang X, Cai C, Guo C. et al. tRF3008A suppresses the progression and metastasis of colorectal cancer by destabilizing FOXK1 in an AGO-dependent manner. J Exp Clin Cancer Res. 2022;41:32
61. Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78
62. Green JA, Ansari MY, Ball HC, Haqqi TM. tRNA-derived fragments (tRFs) regulate post-transcriptional gene expression via AGO-dependent mechanism in IL-1beta stimulated chondrocytes. Osteoarthritis Cartilage. 2020;28:1102-10
63. Liu C, Li M, Shen Y, Han X, Wei R, Wang Y. et al. Targeting choroidal vasculopathy via up-regulation of tRNA-derived fragment tRF-22 expression for controlling progression of myopia. J Transl Med. 2023;21:412
64. Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-Derived Fragments Suppress Breast Cancer Progression via YBX1 Displacement. Cell. 2015;161:790-802
65. Cho H, Lee W, Kim GW, Lee SH, Moon JS, Kim M. et al. Regulation of La/SSB-dependent viral gene expression by pre-tRNA 3' trailer-derived tRNA fragments. Nucleic Acids Res. 2019;47:9888-901
66. Yu M, Yi J, Qiu Q, Yao D, Li J, Yang J. et al. Pan-cancer tRNA-derived fragment CAT1 coordinates RBPMS to stabilize NOTCH2 mRNA to promote tumorigenesis. Cell Rep. 2023;42:113408
67. Li Y, Gao J, Wang Y, Cai J, Wu D, Wang L. et al. The functions of a 5' tRNA-Ala-derived fragment in gene expression. Plant Physiol. 2023;193:1126-41
68. Krishna S, Yim DG, Lakshmanan V, Tirumalai V, Koh JL, Park JE. et al. Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 2019;20:e47789
69. Weidensdorfer D, Stohr N, Baude A, Lederer M, Kohn M, Schierhorn A. et al. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA. 2009;15:104-15
70. Cui H, Li H, Wu H, Du F, Xie X, Zeng S. et al. A novel 3'tRNA-derived fragment tRF-Val promotes proliferation and inhibits apoptosis by targeting EEF1A1 in gastric cancer. Cell Death Dis. 2022;13:471
71. Shi Y, Pan Z, Feng Y, Zhou Q, Wang Q, Wang H. et al. tRF-29-79 regulates lung adenocarcinoma progression through mediating glutamine transporter SLC1A5. Carcinogenesis. 2024;45:409-23
72. Han L, Lai H, Yang Y, Hu J, Li Z, Ma B. et al. A 5'-tRNA halve, tiRNA-Gly promotes cell proliferation and migration via binding to RBM17 and inducing alternative splicing in papillary thyroid cancer. J Exp Clin Cancer Res. 2021;40:222
73. Xiong Q, Zhang Y, Xu Y, Yang Y, Zhang Z, Zhou Y. et al. tiRNA-Val-CAC-2 interacts with FUBP1 to promote pancreatic cancer metastasis by activating c-MYC transcription. Oncogene. 2024;43:1274-1287
74. Merrick WC, Pavitt GD. Protein Synthesis Initiation in Eukaryotic Cells. Cold Spring Harb Perspect Biol. 2018;10:a033092
75. Haselkorn R, Rothman-Denes LB. Protein synthesis. Annu Rev Biochem. 1973;42:397-438
76. Ying S, Li P, Wang J, Chen K, Zou Y, Dai M. et al. tRF-Gln-CTG-026 ameliorates liver injury by alleviating global protein synthesis. Signal Transduct Target Ther. 2023;8:144
77. Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H. et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57-62
78. Sobala A, Hutvagner G. Small RNAs derived from the 5' end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10:553-63
79. Bates M, Boland A, McDermott N, Marignol L. YB-1: The key to personalised prostate cancer management? Cancer Lett. 2020;490:66-75
80. Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43:613-23
81. Ivanov P, O'Day E, Emara MM, Wagner G, Lieberman J, Anderson P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci U S A. 2014;111:18201-6
82. Lyons SM, Gudanis D, Coyne SM, Gdaniec Z, Ivanov P. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat Commun. 2017;8:1127
83. Lyons SM, Kharel P, Akiyama Y, Ojha S, Dave D, Tsvetkov V. et al. eIF4G has intrinsic G-quadruplex binding activity that is required for tiRNA function. Nucleic Acids Res. 2020;48:6223-33
84. Li D, Gao X, Ma X, Wang M, Cheng C, Xue T. et al. Aging-induced tRNA(Glu)-derived fragment impairs glutamate biosynthesis by targeting mitochondrial translation-dependent cristae organization. Cell Metab. 2024;36:1059-75 e9
85. Kim HK, Xu J, Chu K, Park H, Jang H, Li P. et al. A tRNA-Derived Small RNA Regulates Ribosomal Protein S28 Protein Levels after Translation Initiation in Humans and Mice. Cell Rep. 2019;29:3816-24 e4
86. Parkin J, Cohen B. An overview of the immune system. Lancet. 2001;357:1777-89
87. Huse M. Mechanical forces in the immune system. Nat Rev Immunol. 2017;17:679-90
88. Lu D, Yamawaki T, Zhou H, Chou WY, Chhoa M, Lamas E. et al. Limited differential expression of miRNAs and other small RNAs in LPS-stimulated human monocytes. PLoS One. 2019;14:e0214296
89. Kfoury YS, Ji F, Mazzola M, Sykes DB, Scherer AK, Anselmo A. et al. tiRNA signaling via stress-regulated vesicle transfer in the hematopoietic niche. Cell Stem Cell. 2021;28:2090-103 e9
90. Mantovani A, Garlanda C. Humoral Innate Immunity and Acute-Phase Proteins. N Engl J Med. 2023;388:439-52
91. Pradeu T, Thomma B, Girardin SE, Lemaitre B. The conceptual foundations of innate immunity: Taking stock 30 years later. Immunity. 2024;57:613-31
92. Pawar K, Shigematsu M, Sharbati S, Kirino Y. Infection-induced 5'-half molecules of tRNAHisGUG activate Toll-like receptor 7. PLoS Biol. 2020;18:e3000982
93. Pawar K, Kawamura T, Kirino Y. The tRNA(Val) half: A strong endogenous Toll-like receptor 7 ligand with a 5'-terminal universal sequence signature. Proc Natl Acad Sci U S A. 2024;121:e2319569121
94. Netea MG, Schlitzer A, Placek K, Joosten LAB, Schultze JL. Innate and Adaptive Immune Memory: an Evolutionary Continuum in the Host's Response to Pathogens. Cell Host Microbe. 2019;25:13-26
95. Carroll SL, Pasare C, Barton GM. Control of adaptive immunity by pattern recognition receptors. Immunity. 2024;57:632-48
96. Chiou NT, Kageyama R, Ansel KM. Selective Export into Extracellular Vesicles and Function of tRNA Fragments during T Cell Activation. Cell Rep. 2018;25:3356-70 e4
97. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-49
98. Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31-46
99. Zou L, Yang Y, Zhou B, Li W, Liu K, Li G. et al. tRF-3013b inhibits gallbladder cancer proliferation by targeting TPRG1L. Cell Mol Biol Lett. 2022;27:99
100. Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K. et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404-9
101. Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W. et al. tRF/miR-1280 Suppresses Stem Cell-like Cells and Metastasis in Colorectal Cancer. Cancer Res. 2017;77:3194-206
102. Chen F, Song C, Meng F, Zhu Y, Chen X, Fang X. et al. 5'-tRF-GlyGCC promotes breast cancer metastasis by increasing fat mass and obesity-associated protein demethylase activity. Int J Biol Macromol. 2023;226:397-409
103. Xiong Q, Zhang Y, Xu Y, Yang Y, Zhang Z, Zhou Y. et al. tiRNA-Val-CAC-2 interacts with FUBP1 to promote pancreatic cancer metastasis by activating c-MYC transcription. Oncogene. 2024;43:1274-87
104. Shan N, Li N, Dai Q, Hou L, Yan X, Amei A. et al. Interplay of tRNA-Derived Fragments and T Cell Activation in Breast Cancer Patient Survival. Cancers (Basel). 2020;12:2230
105. Gao Z, Jijiwa M, Nasu M, Borgard H, Gong T, Xu J. et al. Comprehensive landscape of tRNA-derived fragments in lung cancer. Mol Ther Oncolytics. 2022;26:207-25
106. Garcia-Vilchez R, Anazco-Guenkova AM, Dietmann S, Lopez J, Moron-Calvente V, D'Ambrosi S. et al. METTL1 promotes tumorigenesis through tRNA-derived fragment biogenesis in prostate cancer. Mol Cancer. 2023;22:119
107. Wang L, Peng B, Yan Y, Liu G, Yang D, Wang Q. et al. The tRF-3024b hijacks miR-192-5p to increase BCL-2-mediated resistance to cytotoxic T lymphocytes in Esophageal Squamous Cell Carcinoma. Int Immunopharmacol. 2024;126:111135
108. Chen W, Peng W, Wang R, Bai S, Cao M, Xiong S. et al. Exosome-derived tRNA fragments tRF-GluCTC-0005 promotes pancreatic cancer liver metastasis by activating hepatic stellate cells. Cell Death Dis. 2024;15:102
109. Durcan L, O'Dwyer T, Petri M. Management strategies and future directions for systemic lupus erythematosus in adults. Lancet. 2019;393:2332-43
110. Kiriakidou M, Ching CL. Systemic Lupus Erythematosus. Ann Intern Med. 2020;172:ITC81-ITC96
111. Yang P, Zhang X, Chen S, Tao Y, Ning M, Zhu Y. et al. A Novel Serum tsRNA for Diagnosis and Prediction of Nephritis in SLE. Front Immunol. 2021;12:735105
112. Xu H, Chen W, Zheng F, Tang D, Dai W, Huang S. et al. The potential role of tRNAs and small RNAs derived from tRNAs in the occurrence and development of systemic lupus erythematosus. Biochem Biophys Res Commun. 2020;527:561-7
113. Pencheva T, Gindeva R, Strashimirov D. [Dipsogenic and pressor responses after intraventricular administration of angiotensin II in rats treated neonatally with the hormone]. Eksp Med Morfol. 1987;26:14-8
114. Geng G, Wang H, Xin W, Liu Z, Chen J, Danting Z. et al. tRNA derived fragment (tRF)-3009 participates in modulation of IFN-alpha-induced CD4(+) T cell oxidative phosphorylation in lupus patients. J Transl Med. 2021;19:305
115. Drent M, Crouser ED, Grunewald J. Challenges of Sarcoidosis and Its Management. N Engl J Med. 2021;385:1018-32
116. Zhao M, Tian C, Di X, Cong S, Cao Y, Zhou X. et al. Systematic and Comprehensive Analysis of tRNA-Derived Small RNAs Reveals Their Potential Regulatory Roles and Clinical Relevance in Sarcoidosis. J Inflamm Res. 2023;16:2357-74
117. Visser M, Maree HJ, Rees DJ, Burger JT. High-throughput sequencing reveals small RNAs involved in ASGV infection. BMC Genomics. 2014;15:568
118. Zhou J, Liu S, Chen Y, Fu Y, Silver AJ, Hill MS. et al. Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J Gen Virol. 2017;98:1600-10
119. Wu W, Choi EJ, Wang B, Zhang K, Adam A, Huang G. et al. Changes of Small Non-coding RNAs by Severe Acute Respiratory Syndrome Coronavirus 2 Infection. Front Mol Biosci. 2022;9:821137
120. Liu X, Wen YZ, Huang ZL, Shen X, Wang JH, Luo YH. et al. SARS-CoV-2 causes a significant stress response mediated by small RNAs in the blood of COVID-19 patients. Mol Ther Nucleic Acids. 2022;27:751-62
121. Goldkamp AK, Lahuis CH, Hagen DE, Taxis TM. Influence of Maternal BLV Infection on miRNA and tRF Expression in Calves. Pathogens. 2023;12:1312
122. Gumas J, Kawamura T, Shigematsu M, Kirino Y. Immunostimulatory short non-coding RNAs in the circulation of patients with tuberculosis infection. Mol Ther Nucleic Acids. 2024;35:102156
123. Ogawa R. The Most Current Algorithms for the Treatment and Prevention of Hypertrophic Scars and Keloids: A 2020 Update of the Algorithms Published 10 Years Ago. Plast Reconstr Surg. 2022;149:79e-94e
124. Wang X, Hu Z. tRNA derived fragment tsRNA-14783 promotes M2 polarization of macrophages in keloid. Biochem Biophys Res Commun. 2022;636:119-27
125. Brozzi F, Jacovetti C, Cosentino C, Menoud V, Wu K, Bayazit MB. et al. tRNA-derived fragments in T lymphocyte-beta cell crosstalk and in type 1 diabetes pathogenesis in NOD mice. Diabetologia. 2024;67:2260-2274
126. Ling Z, Xiao N, Li Y, Xie H, Xiao T, Jiang H. et al. Differential expression profiles and function prediction of tRNA-derived fragments in fibrous dysplasia. Arch Oral Biol. 2022;135:105347
127. Li D, Zhang H, Wu X, Dai Q, Tang S, Liu Y. et al. Role of tRNA derived fragments in renal ischemia-reperfusion injury. Ren Fail. 2022;44:815-25
128. Ma J, Gan M, Chen J, Chen L, Zhao Y, Zhu Y. et al. Characteristics of tRNA-Derived Small RNAs and microRNAs Associated with Immunocompromise in an Intrauterine Growth-Restricted Pig Model. Animals (Basel). 2022;12:2102
129. Shen L, Liao T, Chen Q, Lei Y, Wang L, Gu H. et al. tRNA-derived small RNA, 5'tiRNA-Gly-CCC, promotes skeletal muscle regeneration through the inflammatory response. J Cachexia Sarcopenia Muscle. 2023;14:1033-45
130. Wang H, Huang R, Li L, Zhu J, Li Z, Peng C. et al. CPA-seq reveals small ncRNAs with methylated nucleosides and diverse termini. Cell Discov. 2021;7:25
131. Hu JF, Yim D, Ma D, Huber SM, Davis N, Bacusmo JM. et al. Quantitative mapping of the cellular small RNA landscape with AQRNA-seq. Nat Biotechnol. 2021;39:978-88
Author contact
![]() Corresponding author: Hongyuan Jia (Email: jiahongyuanorg.cn).
Corresponding author: Hongyuan Jia (Email: jiahongyuanorg.cn).
 Global reach, higher impact
Global reach, higher impact