13.3
Impact Factor
Theranostics 2025; 15(1):216-232. doi:10.7150/thno.99664 This issue Cite
Research Paper
Tumor immunomodulation by nanoparticle and focused ultrasound alters gut microbiome in a sexually dimorphic manner
1. Department of Radiation Oncology, UT Southwestern Medical Center, Dallas, TX 75390, USA.
2. Department of Veterinary Pathobiology, Oklahoma State University, Stillwater, OK 74078, USA.
Received 2024-7-1; Accepted 2024-10-21; Published 2025-1-1
Abstract
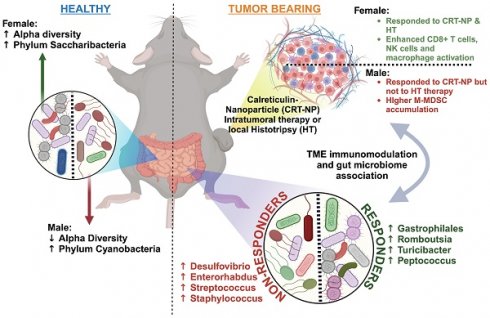
Background: Local immunomodulation with nanoparticles (NPs) and focused ultrasound (FUS) is recognized for triggering anti-tumor immunity. However, the impact of these tumor immunomodulations on sex-specific microbiome diversity at distant sites and their correlation with therapeutic effectiveness remains unknown. Here, we conducted local intratumoral therapy using immunogenic cell death-enhancing Calreticulin-Nanoparticles (CRT-NPs) and FUS in male and female mice. We identified immune-related microbiome populations, aiming to translate our findings into clinical applications.
Methods: CRT-NPs were synthesized by loading CRT-delivering plasmids into cationic liposomes. Local tumor therapy was performed using CRT-NP and FUS-based histotripsy (HT) on poorly immunogenic Mouse Oral Squamous Cell Carcinoma (MOC2) in the flank regions of male and female mice. Fecal samples were collected and analyzed before and three weeks post-treatment. The microbiome features were then correlated with immune cell dynamics within tumors and systemic cytokine responses to identify prognostic biomarkers in both male and female subjects.
Results: Intratumorally administered CRT-NP induced tumor remission and immune cell activation in both male and female mice, whereas HT was ineffective in males and showed efficacy only in females. Turicibacter and Peptococcus inversely correlated with tumor growth, while Enterorhabdus, Subdologranulum, Desulfovibrio, and Aldercreutzia-Asaccharobacter showed direct correlations with tumor growth. HT induced higher levels of Turicibacter in MOC2-bearing females, while males displayed increased Enterorhabdus and Streptococcus populations. Independent of sex, treatments promoting CD4+ T helper cells, functional CD8+ T cells, and total macrophage infiltration correlated with higher levels of Gastrophilales, Romboutsia, Turicibacter, and Peptococcus. Alternatively, Enterorhabdus, Desulfovibrio, Streptococcus, and Staphylococcus corresponded to poor treatment outcomes in both sexes.
Conclusion: An enhanced abundance of Enterorhabdus, Desulfovibrio, Streptococcus, and Staphylococcus in response to immunomodulatory therapies could serve as predictive biomarkers in a sex-independent manner. These findings could also be potentially extended to the realm of personalized interventions through fecal transplantations to reverse immunosuppressive phenotypes in males and improve patient outcomes.
Keywords: Gut Microbiome, Immunomodulation, Nanoparticle, Focused Ultrasound, Sexual dimorphism
Introduction
In recent years, immunotherapy utilizing tumor immunomodulatory nanoparticles (NPs) has garnered significant attention due to their capacity to enhance anti-tumor immune responses [1]. These NPs, encompassing lipid-based NPs (LNPs), polymeric NPs, gold NPs (AuNPs), and mesoporous silica NPs (MSNs), offer unique advantages such as precise targeting, controlled release, and versatility in payload delivery [2-5]. Through diverse mechanisms, including the delivery of immunostimulatory agents, inhibition of immune checkpoints, and modulation of the tumor microenvironment, the NPs can bolster immune-mediated tumor eradication [6]. Many NPs exploit immunogenic cell death (ICD), characterized by the release/expression of damage-associated molecular patterns (DAMPs) from dying tumor cells, thus activating anti-tumor immune responses. We recently developed a lipid-based CRT plasmid encapsulating NP (CRT-NP) that can directly upregulate CRT-based DAMP in tumor cells [7]. Data suggest that upon local intratumor administration, CRT-NP improved immune-mediated tumor regression and survival outcomes in several preclinical models [8].
Like NPs, therapeutic devices, such as focused ultrasound (FUS), radiation, and lasers can treat solid tumors, while also modifying the tumor immune microenvironment. When FUS is applied to ablate tumor tissue, it induces localized cell death within the targeted region. This cell death results in the release of various tumor molecules/antigens from the dying tumor cells into the extracellular space [9]. Specifically, focused ultrasound (FUS)-based histotripsy (HT) generates controlled microscale cavitation bubbles within the targeted tumor microenvironment (TME), resulting in mechanical disruption and fragmentation of the tumor cells for release of DAMPs, proteins, peptides, and other molecules that are recognized as foreign by the immune system [10]. We have shown that FUS-based histotripsy represents a promising approach for not only achieving local tumor control but also for eliciting systemic anti-tumor immune responses, making it a valuable adjunct to cancer immunotherapy strategies [11].
Males and females exhibit distinctive physiological and biological differences, and sex based dysbiosis can significantly influence their anti-tumor immunity [12]. Recent advancements in microbiome research have shown that gut microbiome is a sex dependent biological variable that could impact tumorigenesis [12]. Sexual hormones such as estrogen and androgen could select for different gut microbiota resulting in substantial gut microbiota differences in male vs female [13]. However, very little is known about the role of sexual dimorphism driven gut microbiome compositional differences on therapeutic efficacy of NPs and HT. Understanding the correlation between these sex-specific differences and microbiome plasticity could open new avenues for personalized immunotherapy, thereby advancing the frontiers of sex-personalized therapeutic strategies. In this study, we characterized the distant gut microbiome-local tumor axis in both male and female subjects using a poorly immunogenic MOC2 model following CRT-NP and FUS local treatments. Since both FUS and CRT-NP local treatments work via different mechanisms, we hypothesized that they would induce distinct sex-specific changes in the tumor-microbiome axis. Furthermore, we postulated that, irrespective of tumor types, our data mining would enable the identification of unique sex-independent microbial signatures that could potentially serve as future candidates for fecal transplantations to reverse immunosuppressive phenotypes in such scenarios.
Our data shows that immunomodulatory therapies based on CRT-NPs and HT indeed elicit distinct therapeutic outcomes in male and female mice, which are correlated with their respective microbiome signatures. Notably, our identification of the presence of sex-independent bacterial signatures can personalize such therapies and enhance outcomes in patients.
Materials and Methods
Materials
The lipids DOTAP (1,2-dioleoyl-3-trimethylammonium-propane) (cat#890890) and cholesterol (cat#700000) were purchased from Avanti Polar Lipids, Inc, Alabaster, AL. Nanoplasmid containing CRT gene (NTC 9385R-Calreticulin) controlled by the CMV promoter was obtained from Nature Technology Company, Lincoln. Chloroform (cat#C2432) was purchased from Sigma-Aldrich, St. Louis, MO, USA. HEPES, sterile 1M buffer (cat#J848), and Triton X-100 (cat#0694) were purchased from VWR life science, Radnor, PA, USA. Agarose Precast Gel (3% TBE EtBr Wide Mini Ready - 20-well) (cat#1613030) was purchased from Bio-Rad, Hercules, CA, USA. DMEM (cat#11965092), Fetal bovine serum, FBS (cat#10082147), Penicillin-Streptomycin (cat#15140122), PBS (cat#10010023), Collagenase IV (cat#17104019), were purchased from ThermoFisher/Gibco, Waltham, MA, USA. The following Fluorochrome-conjugated monoclonal antibodies (mAbs) for flow cytometry were purchased from BioLegend, San Diego, CA, USA: APC-Cy7 anti-CD45 (cat#103115), PerCP anti-CD3 epsilon (cat#100325), PE-Cy5 anti-CD4 (cat#100410), PE-Cy7 anti-CD8a (cat#100721), PE anti-Granzyme B (cat#372208), BV510 anti-IFNγ (cat#505841), BV605 anti-PD-1 (cat#135219), BV510 anti-Ly-6G (cat#127633), PE anti-Ly-6C (cat#128007), BV650 anti-CD11b (cat#101239), APC anti-CD206 (141707), BV785 anti-MHC II (cat#107645), BV421 anti-CD11c (cat#117329), AF488 anti-CD86 (cat#105017), PE-Cy7 anti-PDL1 (cat#124314), AF488 anti-FOXP3 (cat#126406) and BV711 anti-NK1.1 (cat#108745). 10X RBC Lysis Buffer (cat# 00-4300-54) was purchased from Invitrogen, Waltham, MA, USA. Mouse OSCC (MOC2) cell line (cat#EWL002-FP) was purchased from Kerafast, Boston, MA, USA. Liberase™ TM Research Grade (cat#5401119001) was procured from Millipore sigma, St. Louis, MO, USA. Transcription factor buffer set (cat#562574) was purchased from BD Biosciences, San Jose, CA, USA.
Synthesis and characterization of CRT-NP
Unilamellar cationic liposomes were formulated by a thin-film hydration method as we previously published [8]. Briefly, DOTAP and cholesterol at a 1:1 molar ratio were dissolved in chloroform, and the solvent was removed using a Rotary Evaporator (Heidolph, Wood Dale, IL, USA). The resulting lipid film was desiccated overnight and then hydrated using 10 mM HEPES buffer (pH 7.4) at 55 °C and sonicated to homogeneity. To ensure stability and uniform size, the liposome formulation was subjected to extrusion five times using an extruder with two stacked Whatman polycarbonate nanopore filters (200 nm pore diameter; GE Healthcare, Chicago, IL, USA). Subsequently, the liposome preparations were gently mixed with pDNA (CRT) solutions (1:20) in a total volume of 100 µl of CRT-NP lipoplex solution, and the mixture was incubated for 30 min at room temperature. Before and after mixing with the pDNA, the size, polydispersity index (PDI), and surface charge were assessed using dynamic light scattering (DLS) and zeta potential measurements with a Brookhaven ZetaPALS instrument (Holtsville, NY, USA). Plasmid encapsulation was determined by gel retardation assay.
In vivo mice study and CRT-NP/FUS treatments
All animal-related procedures were approved and carried out under the guidelines of the Oklahoma State University Animal Care and Use Committee. 7-8 weeks old male and female C57BL/6 mice were purchased from Charles River Laboratories. For tumor inoculation, MOC2 cells derived from female mice at 70-80% confluency were harvested, washed, and diluted with sterile cold PBS to generate a dose of 1.5 × 105 cells in 100 μL per mouse, and transplanted subcutaneously (s.c) into the right flank of 36 mice (18 males and 18 females) mice using a 27-gauge needle (BD, Franklin Lakes, NJ, USA). In our preliminary studies, nanoparticles (NP) with an irrelevant plasmid did not demonstrate efficacy. Therefore, we included untreated and HT as comparative controls for this study. Once the tumors reached a size of 40-60 mm3 (Day 7), male and female mice (n=6 mice/group) were randomly assigned to the following treatment groups: 1) Control, 2) CRT-NP, and 3) Histotripsy (HT) [14-17]. Mice received three intratumoral injections of CRT-NP (20 µg plasmid DNA per injection) over one week, while the HT group received two HT treatments, spaced three days apart [7, 10]. HT treatments covering 10-20% of tumor in anesthetized mice were administered using an Alpinion system (Bothell, WA, USA) with FUS transducer featuring a 1.5 MHz central frequency with following parameters: 5 Hz PRF, 1% duty cycle, 600 W acoustic power, 20 s treatment time per focal point. Tumor growth was monitored and measured daily using serial caliper (General Tools Fraction™, New York, NY, USA) and the tumor volume was calculated based on the formula (Length X Width2)/2, where length was the largest dimension and width was the smallest dimension perpendicular to the length.
Fecal sample collection, storage, and microbiome analysis
Mice were allowed a 2-week acclimatization period and provided with a standardized diet for the study duration. Fecal pellets (4-6 pellets per mouse, n=5 mice per group) for microbiome analysis [15, 17] were obtained from individual mice kept in clean containers before inoculation and on the day of sacrifice and were preserved at -80°C until further processing. Fecal pellets were processed and analyzed for microbiome at ZymoBIOMICS® Targeted Sequencing Service, (Zymo Research Corporation, Irvine, CA, USA). Briefly, DNA extraction was performed using ZymoBIOMICS®-96 MagBead DNA Kit (Zymo Research, Irvine, CA, USA). Bacterial 16S ribosomal RNA gene targeted sequencing was performed using the Quick-16S™ NGS Library Prep Kit (Zymo Research, Irvine, CA, USA) and V3-V4 region of the 16S rRNA gene were amplified using PCR and targeted library sequence was prepared. The final library was sequenced on Illumina® MiSeq™ with a v3 reagent kit (600 cycles). Microbiome data was analyzed primarily using MicrobiomeAnalyst [18]. Briefly, BIOM files were imported to MicrobiomeAnalyst web tool with Qiime v.1.9.1 for community profiling and comparative analysis. Data were filtered to remove low count (<20%) and low variance (<10%), and then scaled and normalized at the total sum scale. Community composition comparisons and Alpha diversity metrics were calculated on filtered reads using “Microbiome R packages” [19]. Beta diversity was performed using ordination-based method followed by calculating the Bray-Curtis Index. Differential abundance analysis (DAA) was performed using DESeq2 [20] and edgeR [21] with default parameters. The resulting FDR-corrected p-values were used for further analysis. Differences in taxonomy abundance were determined using Linear Discriminant Analysis Effect size (LEfSe) [22] employing the Kruskal-Walli's rank sum test. Euclidean distance was used for community clustering with the Ward algorithm. Correlation heatmap analysis was used to describe the relationship between immune cells, cytokines, tumor volume and specific taxa at genus level. The correlation analysis was based on the R stats package (version.4.02), Hmisc package, and heatmap package. Networks were constructed using igraph r package (https://igraph.org) and visualized with the interactive platform Gephi (https://gephi.org/) and Chord plot was generated using circlize R package. Data visualization package ggplot2 (https://github.com/tidyverse/ggplot2) and Microbiome Analyst internal scripts were used for plotting figures.
Immune cell analysis of harvested tumors
Mice were sacrificed on Day 20 post-inoculation, and blood and tumor tissues were collected. Tumors were excised, weighed, and processed on the same day for flow cytometry. Tumors were minced and digested with 200 U per mL Collagenase IV solution followed by filtration through a 70 μm cell strainer (Corning Inc., Corning, NY, USA) to obtain single cell suspensions. Cells were treated with RBS lysis buffer for 20 mins at room temperature and then stained with specific anti-mouse fluorochrome-conjugated antibody combinations for 60 mins on ice in the dark to distinguish immune cell populations. To stain for intracellular markers, IFNγ, Granzyme-B (GZMB), CD206 and FOXp3, cells were washed after surface marker staining, fixed and permeabilized with a transcription factor buffer set (BD Biosciences), and incubated with intracellular antibody for 60 mins in the dark on ice. Stained cells were run in an LSRII flow cytometer (BD Biosciences). Compensations were performed with single-stained cells. FlowJo software v.10.2 (Treestar Inc., Ashland, OR, USA) and NovoExpress software v1.6.2 (Agilent, Santa Clara, CA, USA) were used for data analysis. Gating for different immune cell population were done as follows: CD45+ (Tumor infiltrating leukocytes; TILs), CD45+ CD3+ (Total T-cells), CD45+ CD3+ CD4+ CD8- (TH, CD4+ T helper cells), CD45+ CD3+ CD8- CD4+ FOXP3+ (Treg, Regulatory T-cells), CD45+ CD3+ CD4- CD8+ (TC, CD8+ T-cells), CD45+ CD3+ CD4- CD8+ GZMB+/IFNγ+ (Effector cytotoxic T-cells), CD45+ CD3+ CD4- CD8+ PD1+ (Exhausted T cell), CD45+ CD3- NK 1.1+ (NK cells), CD45+ CD3- NK 1.1+ GZMB+/IFNγ+ (Cytotoxic NK cells), CD45+ CD3- NK 1.1+ PD1+ (Mature NK cells) CD45+ CD11b- CD11c+ (DC, Dendritic cells), CD45+ CD11b- CD11c+ MHC-II+ CD86+ (Activated DCs), CD45+ CD11c- CD11b+ (Total Macrophages), CD45+ CD11c- CD11b+ CD86+ MHC-II+ (Activated M1 Macrophages), CD45+ CD11c- CD11b+ 206+ (M2 Macrophages), CD45+ CD11b+ Ly6CHi Ly6G- (Monocytic-Myeloid derived suppressor cells, M-MDSC) & Ly6CLo Ly6G+ (Polymorphonuclear-MDSC, PMN-MDSC).
Serum cytokine analysis
Murine cytokines, chemokines, and growth factors were determined using the Luminex™ 200 system (Luminex, Austin, TX, USA) by Eve Technologies Corp (Calgary, Alberta) using mice serum. Eve technologies utilize the Mouse Cytokine/Chemokine 32-Plex Discovery Assay® Array (MD32) (MilliporeSigma, Burlington, Massachusetts, USA). The 32 cytokines included Eotaxin, G-CSF, GM-CSF, IFNγ, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-12p40, IL-12p70, IL-13, IL-15, IL-17A, IP-10, KC, LIF, LIX, MCP-1, M-CSF, MIG, MIP-1α, MIP-1β, MIP-2, RANTES, TNFα, and VEGF-A, with assay sensitivity ranging from 3.2 to 21.0 pg/mL. The analysis utilized 2X diluted 50 μL of serum sample per mouse, and the results were presented in picogram per mL concentration.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 10.0 software (GraphPad Software Inc, La Jolla, CA, USA). Data are presented as mean ± SEM unless otherwise indicated. All microbiome-related analyses were carried out in R version 2024.4.2 and Microbiome Analyst in-built tests. Analysis of differences between 2 normally distributed test groups was performed using an unpaired t-test assuming unequal variance. The differences between the treatments compared to the untreated control were analyzed by two-way ANOVA. For analysis of three or more groups, one-way ANOVA with Fisher test was performed. P values less than 0.05 were considered significant and represented as *p<0.05, **p<0.005, ***p<0.0005, ****p<0.0001. Pearson's correlation coefficient and Spearman's rank correlation coefficient were utilized in this study. The co-occurrence event to be a robust correlation if the spearman's correlation coefficient (ρ) was both >0.7 and statistically significant (P-value <0.05).
Results
Healthy C57BL6 male and female mice showed distinct gut microbiome signatures
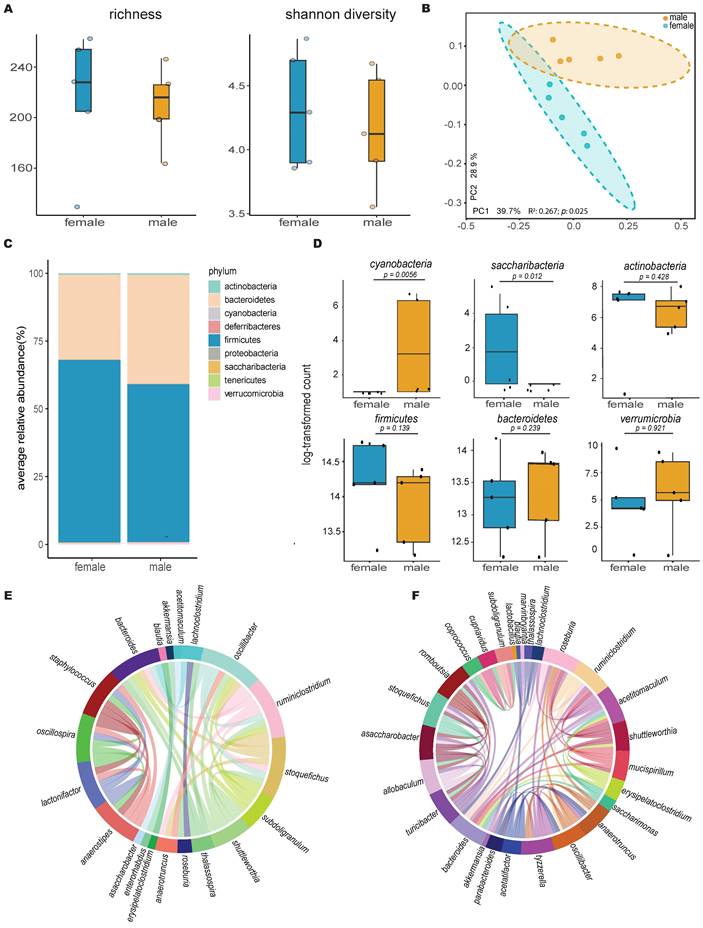
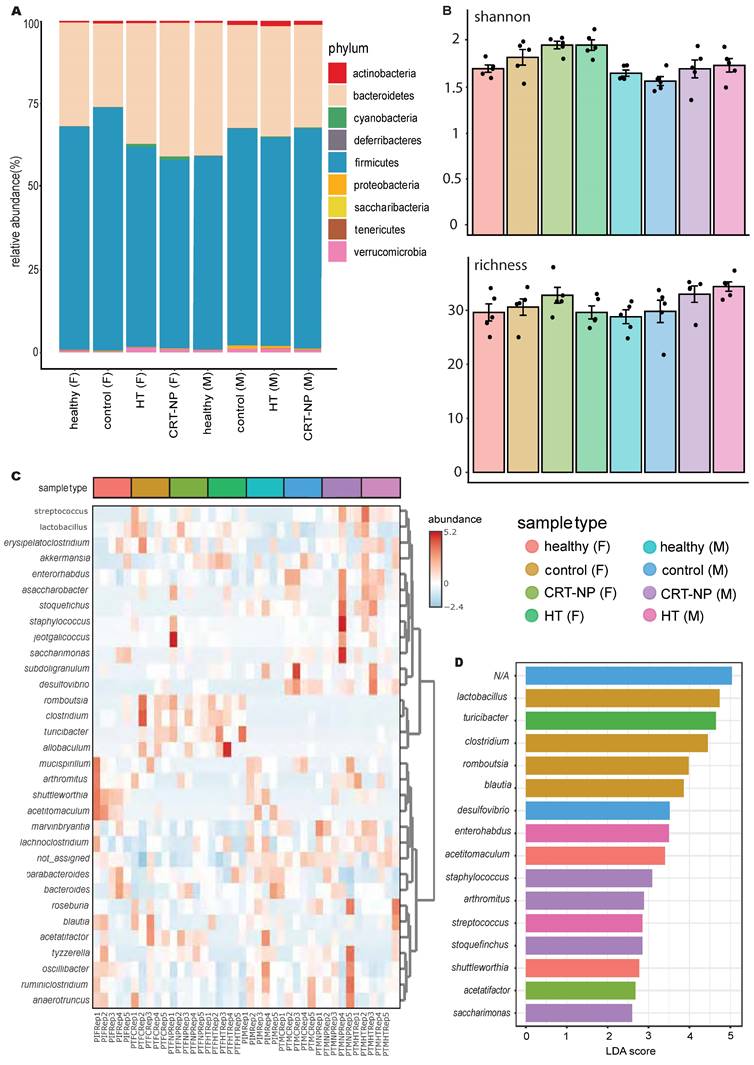
Comparison of the gut microbiome between healthy male and female mice using 16S rRNA gene sequencing revealed higher bacterial diversity in the female group, as measured by two distinct metrics: Chao1 richness and Shannon diversity (Figure 1A). This was further supported by principal coordinates analysis of Bray-Curtis distances, which showed a significant separation between the gut microbiotas of male and female mice (n=5, p=0.025, R²=0.267, PERMANOVA; Figure 1B), with distinct compositional differences between the male and female groups (Figure 1C). Additional quantitative analysis of core microbiome abundance showed an abundance of OTUs in the Verrucomicrobia phylum distinctively in males (Figure S1).
Statistical comparison using the Kruskal-Wallis test of the most abundant phyla revealed that males had a significantly higher abundance of Cyanobacteria, while females had a higher abundance of Saccharibacteria (Figure 1D). Lefse analysis (LDA > 2 & FDR < 0.05) at the genus level identified two distinct populations, Clostridium and Stoquefichus (Figure S2), with healthy females showing a higher abundance of Clostridium (p=0.034) and males having a higher abundance of Stoquefichus (p=0.007).
Further analysis using Chord diagrams depicting genus-level Spearman correlation networks (p < 0.05) within male (Figure 1E) and female (Figure 1F) mice revealed distinct microbiome interaction patterns. In males, notable correlations were observed between several genera, with strong interactions among Oscillibacter, Ruminiclostridium, Stoquefichus, and Bacteroides, indicating a tightly interconnected network. In contrast, the female microbiome network showed a different set of correlations, particularly between Roseburia, Ruminiclostridium, Bacteroides, and Oscillibacter. Unique genera such as Staphylococcus, Lactonifactor, Anaerostipes, and Enterorhabdus were more prevalent in males, while Turicibacter, Tyzzerella, Parabacteroides, Saccharimonas, Mucispirillum, Romboutsia, and Marvinbryantia were predominant in females. Comparing the two networks, clear differences emerged in the central genera within the correlation structures. While some genera, such as Ruminiclostridium and Bacteroides, appeared in both networks, their interaction partners differed, reflecting gender-specific microbial community dynamics.
Male mice displayed significantly faster MOC2 tumor growth compared to females with alterations in gut microbiome profiles
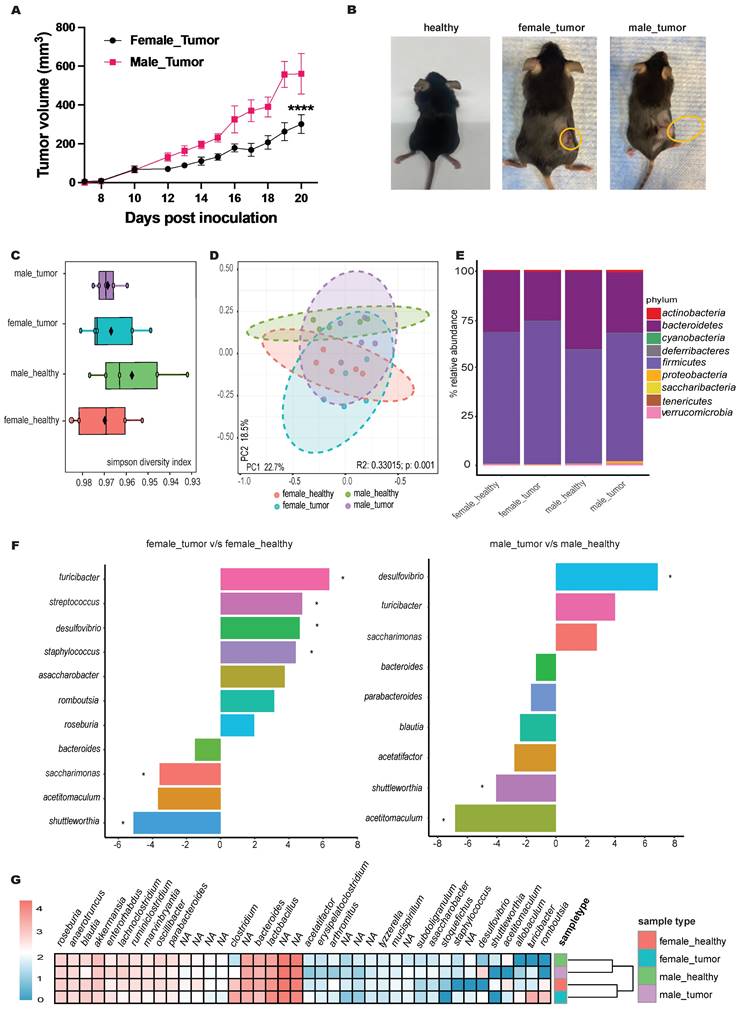
Male mice exhibited approximately 2-fold faster MOC2 tumor growth rates 10 days post-inoculation compared to females (Figure 2A-B). These changes were associated with shifts in microbiome alpha diversity compared to healthy mice (Figure 2C). For instance, the Simpson diversity index revealed lower microbial diversity in tumor-bearing mice, with principal component analysis (PCA) showing distinct clustering of microbial communities between healthy and tumor-bearing mice (p=0.001; Figure 2D).
Qualitative abundance analysis at the phylum level (Figure 2E) in both male and female tumor-bearing mice showed an increased Firmicutes-to-Bacteroidetes ratio, with males uniquely exhibiting elevated levels of Proteobacteria. EdgeR statistical comparison revealed that tumor inoculation caused more significant changes in genera abundance in females than in males (Figure 2F). Specifically, females showed an increase in Turicibacter, Streptococcus, and Staphylococcus, along with a decrease in Saccharimonas. In contrast, males exhibited a significant decrease in Acetitomaculum. Although not statistically significant, the abundance of Saccharimonas increased in males with tumors, opposite to the trend seen in females. Sex-independent changes included an increase in Desulfovibrio and a decrease in Shuttleworthia. To visualize the broader changes in tumor-bearing male and female mice compared to healthy controls, phenotype clustering using DESeq2 grouped the top 40 genera into male- and female-specific clusters (Figure 2G).
In-situ immunomodulation with CRT-NP and HT induced distinct therapeutic responses between male and female mice
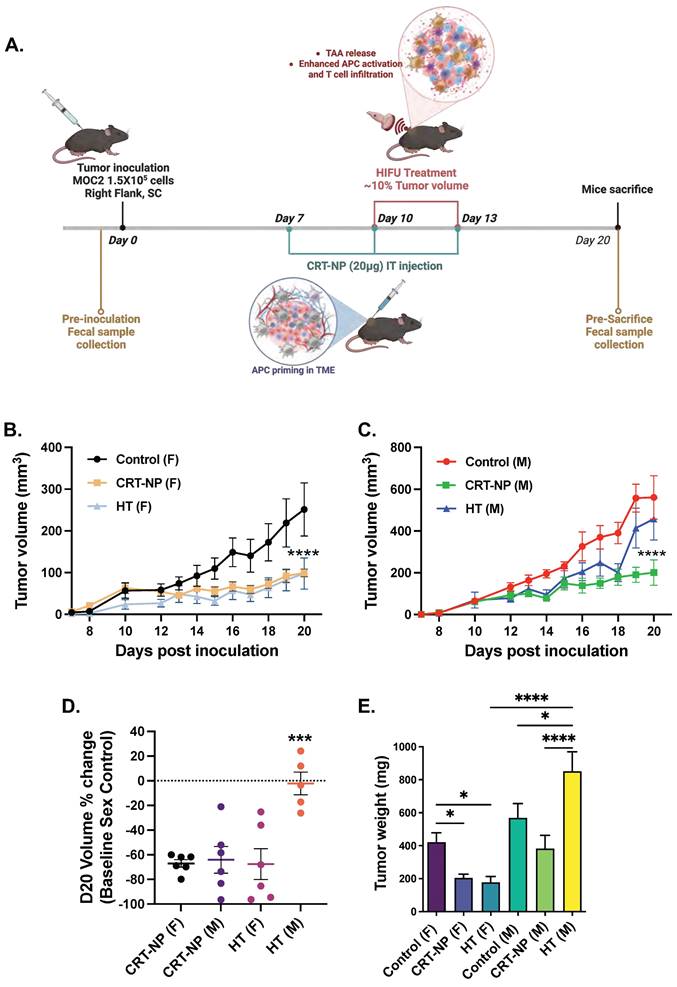
Female mice bearing MOC2 tumors exhibited a significant reduction (~70%) in tumor growth when treated with both CRT-NP and HT therapies (Figure 3B-D). In contrast, among male mice, only CRT-NP displayed substantial therapeutic effectiveness, leading to a notable 60% decrease in both tumor growth and weights, mirroring the response observed in female mice. In contrast, HT treatment was ineffective in male mice (Figure 3C-E).
Distinct sets of immune cells controlled the therapeutic outcomes of CRT-NP and HT therapies
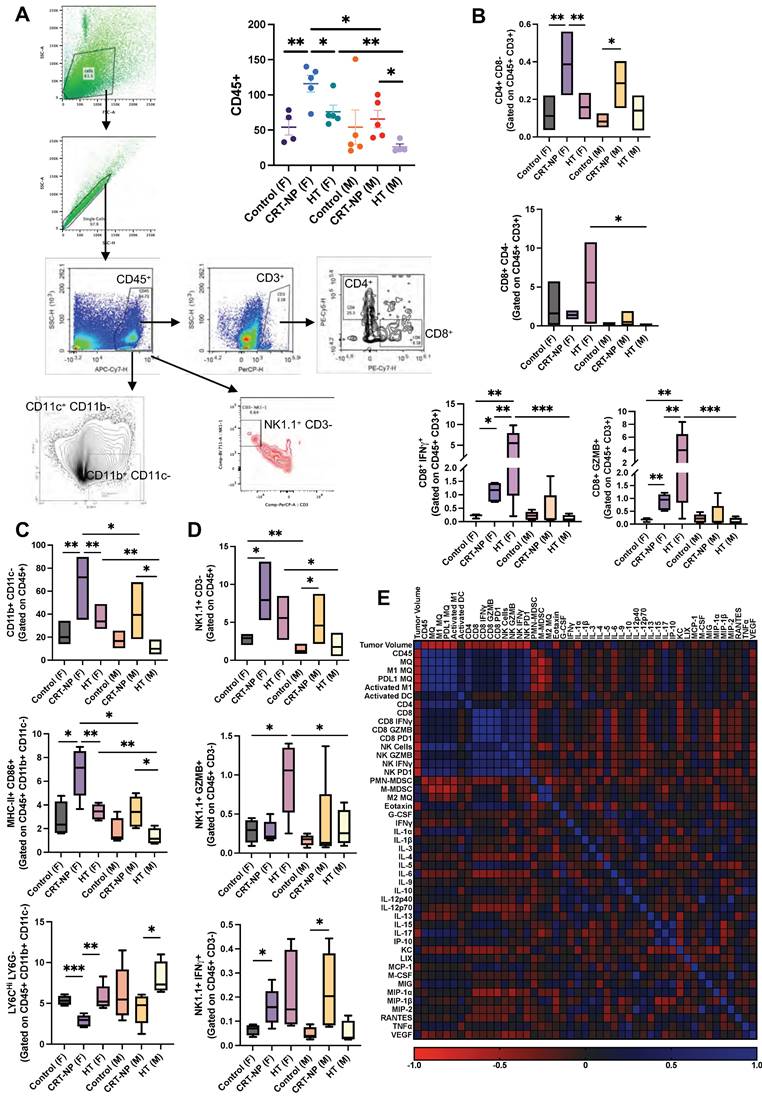
Flow cytometry analysis of immune cell populations in male and female mice showed CRT-NP treatment was associated with an increase in CD45+ hematopoietic cell populations, whereas HT-treated males exhibited a decrease in this population (Figure 4A). Notably, CRT-NP treatment enhanced CD4+ T helper cell populations (3.5-4 folds) in both genders and promoted the infiltration of total macrophages (3.5 folds in female & 2 folds in male) and M1 macrophages (~3.5 folds in female & 2.5 folds in male) with decline (~2 folds in female) in M-MDSC infiltration within the tumor microenvironment (TME; Figure 4B-C). Conversely, HT-treated females exhibited the highest proportion of activated CD8+ cytotoxic T cells (16 folds) and NK cells expressing IFNγ (3 folds) and GZMB (4 folds), a phenomenon not observed in males across treatment groups (Figure 4B-D).
Gut microbiota differs significantly between healthy female and male mice. A) Alpha diversity calculations between healthy female and male mice, with each point representing an individual mouse in the group (n=5); B) Principal coordinate analysis of Bray-Curtis distances showing significant separation between the gender groups (PERMANOVA test values); C) Stacked bar plot illustrating the percent relative abundances of bacterial phyla between healthy female and male mice; D) Log-transformed counts of the most abundant and significant bacterial phyla between healthy female and male mice, with p-values determined using the Kruskal-Wallis test; E & F) Circos plots of Spearman correlations between genera in (E) male and (F) female mice. Each band represents the strength of the monotonic relationship between paired genera, with the Spearman correlation coefficient set at a minimum of 0.7 and the p value < 0.01.
MOC2 tumor growth pattern showed sex-based difference and shift in microbial abundance. A) Line graphs depicting differential MOC2 tumor growth rates in male and female mice (n=6, ****p<0.0001, Two-way ANOVA with Tukey's multiple comparison); B) Representative images of healthy and tumor bearing female and male mice. C) Microbiome alpha diversity calculated using Simpson index (n=5); D) Principal coordinate analysis of Bray-Curtis distances shows significant separation between groups (PERMANOVA test values). Each point represents an individual's gut microbiota based on 16S sequencing; E) Bar plot of the relative genus-level abundance in healthy and tumor-bearing male and female mice; F) Differential abundance analysis of the microbiome between healthy and tumor-bearing male and female mice using edgeR (species with p < 0.05 shown, * significance at FDR < 0.05; tumor-bearing mice as baseline); G) Clustered heatmap of the top 40 genera for four groups of mice, with female and male (healthy and tumor-bearing) forming distinct clusters.
CRT-NP & HT immunotherapies showed superior therapeutic response in female mice compared to male. A) C57BL6 mice bearing unilateral flank tumors were treated seven days post-inoculation (n=6). Three intratumoral injections of CRT-NP at 20 µg CRT plasmid was performed. The HT group received two treatments covering 10-20% of the total tumor volume. Fecal samples were collected from all mice before inoculation and at the time of mice sacrifice (20 days post inoculation). B&C), Line graphs representing MOC2 tumor growth post CRT-NP & HT treatments in female (B) and male (C). D) Median tumor regression percentage with CRT-NP & HT treatments in female and male mice compared to untreated control at the day of mice sacrifice (D20 post-inoculation). E) Tumors weights at the completion of study endpoint. Two-way ANOVA test was used to analyze tumor growth in line graph and one-way ANOVA was used for bar graphs analysis, * p<0.05, *** p<0.0005, **** p<0.0001.
CRT-NP & HT therapies efficacy is driven by distinct set of immune cells. A) Flow cytometry analysis and gating strategy utilized to estimate distinct CD45+ immune cell number infiltrating MOC2 TME. B) Bar graph representing cell counts of CD8- CD4+ T helper cells, CD8+ CD4- cytotoxic T cells, and IFNγ and GranzymeB expressing CD8+ cytotoxic T cells, in TME of untreated, CRT-NP & HT treated tumors. C) Bar graph representing cell counts of CD11b+ CD11c- macrophages, MHC-II+ CD86+ M1 macrophages, Ly6Chi LY6G- M-MDSCs, in TME of untreated, CRT-NP & HT treated tumors of male and female mice. D) Bar graph representing CD3- Natural Killer cell counts, NK1.1+ CD3- NK cells, NK1.1+ GranzymeB+ cytotoxic NK cells, NK1.1+ IFNγ+ activated NK cells, in TME of untreated, CRT-NP & HT treated tumors of male and female mice. E) Pearson R correlation plot representing interaction profile of various TME associated immune cells, serum cytokines and chemokines levels and tumor volume. Immune cell counts are normalized and represented as cell number per mg of tumor. Difference among treatment groups (n=6) were analyzed using One-way ANOVA with Fisher test, * p<0.05, ** p<0.005, *** p<0.0005. P values for Pearson R correlation analysis is provided in Table 1. MQ: Macrophages, Activated M1: MHC-II+ CD86+ macrophages, Activated DC: MHC-II+ CD86+ dendritic cells (CD11c+ CD11b-).
Correlation of immune cells with serum cytokines
Expression of IFNγ and GZMB on CD8 T cells showed significant correlation with Eotaxin, MIP1α, KC, IL-5, IL-6. Whereas GZMB expression on NK cells showed significant correlation with IL-4 and MIP1α (Figure 4E). Pearson r correlation of various immune cells, cytokine and chemokines revealed significant correlation between IFNγ and PD1 expression on NK cells with total and activated macrophage population along with total and cytotoxic CD8+ T cells which also correlated with GZMB expression on NK cells.
Gut microbiome alters with specific immunomodulatory treatment in sex dependent and independent manner
OTU clustering revealed a reduction in the Firmicutes to Bacteroidetes ratio and an increase in Cyanobacteria in females treated with CRT-NP and HT compared to untreated control and healthy mice. In contrast, males did not exhibit noticeable changes with treatment, except for a decrease in Proteobacteria populations in the CRT-NP treated group compared to other groups (Figure 5A). Both male and female mice exhibited increased alpha diversity with CRT-NP and HT treatments employing Shannon, Simpson and Chao1 matrix (Figure 5B). The hierarchical clustering and correlation analysis of OTUs at the genus level revealed distinct population patterns (Figure 5C). Further identification of distinct genera using LefSe analysis identified 32 genera with increasing LDA scores (>2), top 15 represented in Figure 5D. HT treatment in females induced higher accumulation of Turicibacter in females, while in males Enterorhabdus and Streptococcus were increased. With CRT-NP treatment, higher abundance of Acetatifactor was observed in females whereas Staphylococcus, Arthromitus, Stoquefichus and Saccharimonas were elevated.
Specific microbial genera and immune features correlates with immunotherapeutic outcomes
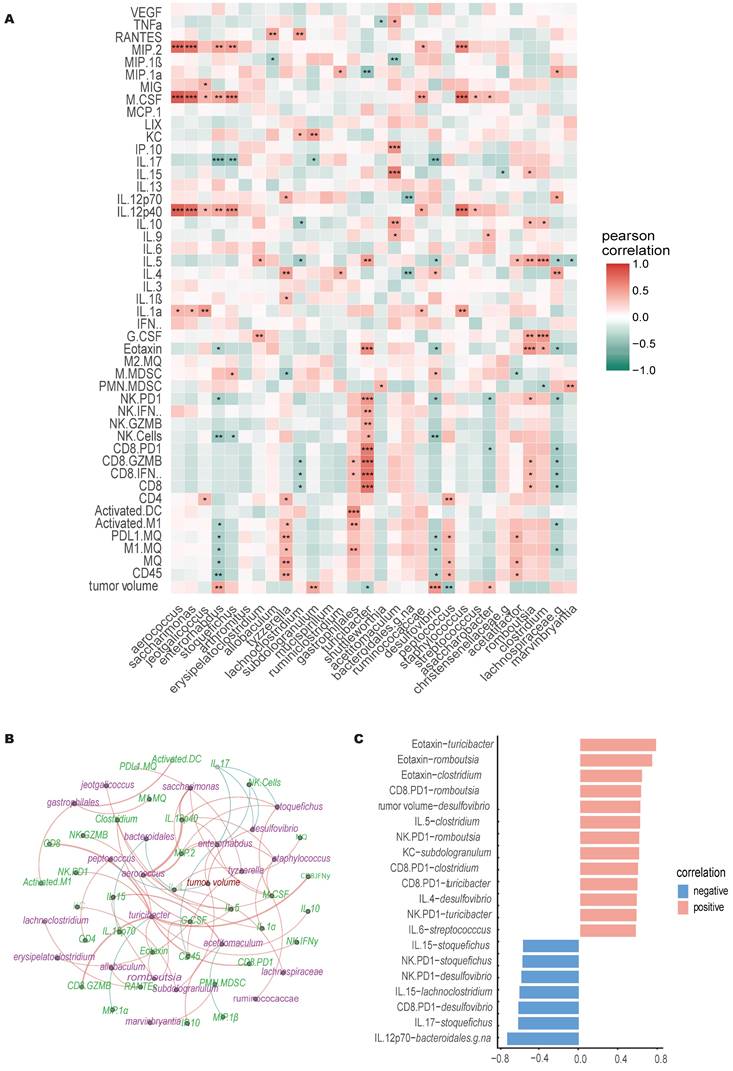
Multifactor LefSe comparison, identified a total of 32 genera that displayed LDA score >3.5. Pearson r correlation analysis conducted on 31 identified genera (excluding non-classified genus, NA) revealed both direct and inverse correlations with various immune cells, cytokines, chemokines, and tumor growth (Figure 6A-C). Notably, Turicibacter and Peptococcus exhibited an inverse relationship with tumor growth, while Enterorhabdus, Subdologranulum, Desulfovibrio, and Adlercreutzia-Assacharobacter were directly correlated with tumor growth.
Correlation with immune cell dynamics indicated that Tyzzerella and Gastrophilales showed positive correlation with M1 macrophage abundance in TME, whereas Lachnospiraceae and Enterorhabdus was inversely correlated. Gastrophilales also correlated positively with activated DC and cytotoxic CD8+ T cells (IFNγ+ & GZMB+) accumulation in TME. Similarly, Turicibacter showed positive correlation with higher CD8+ T cell and NK cell infiltration and activation in tumors. Lachnoclostridium and Lachnospiraceae showed inverse correlation with CD8 T cells cytotoxicity whereas Romboutsia showed positive correlation. Higher levels of Jeotgalicoccus, Tyzzerella and Peptococcus were associated with higher CD4+ T cells infiltration in tumors. Enterorhabdus, Stoquefichus and Desulfovibrio abundance was negatively correlated with NK cell tumor infiltration. Higher M-MDSC accumulation was associated with higher Stoquefichus and Desulfovibrio, and with lower Tyzzerella and Acetatifactor. Additionally, PMN-MDSCs accumulation were negatively correlated with Clostridium and positively with Shuttleworthia and Marvinbryantia.
Acetitomaculum and Romboutsia showed positive correlation with lymphocyte stimulating cytokines whereas Aerococcus, Saccharimonas, Stoquefichus, Enterorhabdus, Ruminicocaccae, Staphylococcus, Streptococcus positively correlated with inflammatory, pro-tumoral cytokines like IL-1, M-CSF, MIP-2, IL12p40 or IL-17, etc. Similar to tumor growth, Desulfovibrio and Turicibacter showed contrasting correlation with IL-5 and Eotaxin serum levels. Desulfovibrio also correlated with significantly with CD4+ T cells associated cytokines, positively with IL-4 levels but inversely with IL-17 levels. On the other hand, higher Turicibacter were associated with lower pro-tumoral MIP-1α levels.
Discussion
Changes in alpha and beta microbiome diversity, are known to be strongly associated with altered anti-tumor immunity [23]. While the influence of sexual hormones on immune responses and microbiome dynamics is recognized [24, 25], the extent to which microbiome signatures induced by specific treatments (such as CRT-NP and HT) consistently appear across genders, or operate independently, remains uncertain. This understanding is crucial for customizing personalized solid tumor immunotherapy regimens.
P value for Pearson R correlation between tumor burden, TME infiltrating immune cells and serum cytokines and chemokines level.
| Variables | Correlation score | P-value | Variables | Correlation score | P-value | Variables | Correlation score | P-value |
|---|---|---|---|---|---|---|---|---|
| Volume-CD45 | -0.52 | 0.00421634 | PDL1 MQ-M-MDSC | -0.61 | 0.00055154 | CD8 PD1-IL-6 | -0.37 | 0.04659335 |
| Volume-MQ | -0.62 | 0.00039106 | PDL1 MQ-M2 MQ | -0.37 | 0.04993837 | CD8 PD1-MCP-1 | 0.44 | 0.01763022 |
| Volume-M1 MQ | -0.58 | 0.00106689 | PDL1 MQ-IL-17 | 0.44 | 0.01849909 | NK cells-NK GZMB | 0.49 | 0.00683037 |
| Volume-PDL1 MQ | -0.66 | 0.00011364 | Activated M1-Activated DC | 0.39 | 0.03919269 | NK cells-NK IFNγ | 0.53 | 0.00357604 |
| Volume-Activated M1 | -0.57 | 0.00144691 | Activated M1-CD4 | 0.67 | 7.1165E-05 | NK cells-NK PD1 | 0.91 | 1.7061E-11 |
| Volume-CD4 | -0.48 | 0.00920969 | Activated M1-NK Cells | 0.68 | 5.9748E-05 | NK cells-M-MDSC | -0.41 | 0.02738338 |
| Volume-CD8 | -0.38 | 0.04219493 | Activated M1-NK IFNγ | 0.47 | 0.01141053 | NK cells-Eotaxin | 0.43 | 0.02103778 |
| Volume-CD8 PD1 | -0.42 | 0.0236728 | Activated M1-NK PD1 | 0.56 | 0.00185548 | NK cells-IL-17 | 0.48 | 0.00939359 |
| Volume-NK Cells | -0.67 | 8.7192E-05 | Activated M1-PMN-MDSC | -0.46 | 0.01318932 | NK cells-VEGF | -0.4 | 0.03228272 |
| Volume-NK GZMB | -0.47 | 0.01109233 | Activated M1-M-MDSC | -0.49 | 0.00743 | NK GZMB-NK IFNγ | 0.64 | 0.0002028 |
| Volume-NK IFNγ | -0.52 | 0.00386049 | Activated M1-IL-1α | 0.47 | 0.01147772 | NK GZMB-NK PD1 | 0.69 | 3.6844E-05 |
| Volume-NK PD1 | -0.64 | 0.0002259 | Activated M1-IL-17 | 0.39 | 0.03591859 | NK GZMB-IL-4 | -0.39 | 0.03697605 |
| Volume-M-MDSC | 0.47 | 0.01051002 | Activated DC-IL-3 | 0.52 | 0.0050792 | NK GZMB-MIP-1α | -0.48 | 0.01225285 |
| Volume-M2 MQ | 0.39 | 0.03869564 | CD4-NK Cells | 0.62 | 0.00043071 | NK IFNγ-NK PD1 | 0.63 | 0.00032468 |
| Volume-Eotaxin | -0.44 | 0.01887789 | CD4-NK IFNγ | 0.43 | 0.02159545 | NK PD1-Eotaxin | 0.5 | 0.00603451 |
| Volume-IL-6 | 0.43 | 0.02113829 | CD4-NK PD1 | 0.43 | 0.02095208 | PMN-MDSC-M2 MQ | -0.52 | 0.00399718 |
| Volume-IL-17 | -0.41 | 0.02676211 | CD4-IL-1α | 0.43 | 0.02050272 | M-MDSC-M2 MQ | 0.8 | 2.3522E-07 |
| Volume-MIP-1β | 0.37 | 0.04801781 | CD8-CD8 IFNγ | 0.99 | 1.4308E-29 | M-MDSC-IL-17 | -0.43 | 0.0216771 |
| CD45-MQ | 0.84 | 2.1345E-08 | CD8-CD8 GZMB | 0.98 | 9.2678E-22 | Eotaxin-IL-1α | 0.4 | 0.03247099 |
| CD45-M1 MQ | 0.81 | 1.2673E-07 | CD8-CD8 PD1 | 0.95 | 1.5493E-15 | Eotaxin-IL-5 | 0.43 | 0.02068092 |
| CD45-PDL1 MQ | 0.79 | 3.6753E-07 | CD8-NK Cells | 0.44 | 0.0184868 | Eotaxin-IL-15 | 0.44 | 0.0167475 |
| CD45-Activated M1 | 0.8 | 2.1758E-07 | CD8-NK GZMB | 0.69 | 4.4474E-05 | Eotaxin-IL-17 | 0.38 | 0.04066252 |
| CD45-CD4 | 0.53 | 0.00352439 | CD8-NK IFNγ | 0.59 | 0.0008205 | Eotaxin-MIP-1β | -0.38 | 0.04307274 |
| CD45-NK Cells | 0.68 | 6.0604E-05 | CD8-NK PD1 | 0.72 | 1.0613E-05 | G-CSF-IL-5 | 0.48 | 0.0096817 |
| CD45-NK PD1 | 0.58 | 0.00098454 | CD8-Eotaxin | 0.42 | 0.02528201 | G-CSF-IL-6 | 0.61 | 0.00055369 |
| CD45-PMN-MDSC | -0.52 | 0.00450039 | CD8-IL-5 | 0.41 | 0.02905943 | G-CSF-IL-10 | 0.47 | 0.01051729 |
| CD45-IL-17 | 0.48 | 0.00864797 | CD8-IL-6 | -0.39 | 0.03618232 | IL-1α-IL-12p40 | 0.46 | 0.01725341 |
| MQ-M1 MQ | 0.93 | 8.0992E-13 | CD8-MIP-1α | -0.43 | 0.02544701 | IL-1α-IL-15 | 0.41 | 0.02979496 |
| MQ-PDL1 MQ | 0.98 | 5.3277E-22 | CD8 IFNγ-CD8 GZMB | 0.99 | 3.5215E-28 | IL-1α-M-CSF | 0.48 | 0.0093922 |
| MQ-Activated M1 | 0.92 | 4.0689E-12 | CD8 IFNγ-CD8 PD1 | 0.93 | 5.0268E-13 | IL-1α-MIG | 0.52 | 0.00438417 |
| MQ-CD4 | 0.7 | 3.0944E-05 | CD8 IFNγ-NK Cells | 0.42 | 0.02283366 | IL-1α-MIP-2 | 0.39 | 0.03521646 |
| MQ-NK Cells | 0.8 | 3.1608E-07 | CD8 IFNγ-NK GZMB | 0.67 | 9.5083E-05 | IL-1β-IL-4 | 0.64 | 0.00022742 |
| MQ-NK IFNγ | 0.45 | 0.01397965 | CD8 IFNγ-NK IFNγ | 0.58 | 0.00102969 | IL-1β-IL-10 | 0.4 | 0.03243689 |
| MQ-NK PD1 | 0.63 | 0.00031533 | CD8 IFNγ-NK PD1 | 0.71 | 1.6229E-05 | IL-1β-IL-12p70 | 0.72 | 3.7999E-05 |
| MQ-M-MDSC | -0.59 | 0.00074024 | CD8 IFNγ-Eotaxin | 0.43 | 0.0220556 | IL-3-MIP-1β | 0.57 | 0.00159166 |
| MQ-IL-1α | 0.37 | 0.04861054 | CD8 IFNγ-IL-5 | 0.41 | 0.02891788 | IL-4-IL-12p70 | 0.69 | 0.0001086 |
| MQ-IL-17 | 0.43 | 0.0206379 | CD8 IFNγ-IL-6 | -0.38 | 0.0404886 | IL-10-TNFα | 0.63 | 0.00028375 |
| M1 MQ-PDL1 MQ | 0.9 | 4.9573E-11 | CD8 IFNγ-MIP-1α | -0.43 | 0.02824918 | IL-10-VEGF | 0.44 | 0.01678669 |
| M1 MQ-Activated M1 | 0.99 | 1.7604E-33 | CD8 GZMB-CD8 PD1 | 0.9 | 7.0141E-11 | IL-12p40-M-CSF | 0.96 | 2.5473E-15 |
| M1 MQ-CD4 | 0.69 | 4.7562E-05 | CD8 GZMB-NK Cells | 0.39 | 0.03736217 | IL-12p40-MIP-2 | 0.86 | 1.2327E-08 |
| M1 MQ-NK Cells | 0.7 | 2.3297E-05 | CD8 GZMB-NK GZMB | 0.62 | 0.00041501 | IL-12p70-MIP-1β | 0.47 | 0.01655269 |
| M1 MQ-NK IFNγ | 0.48 | 0.00897125 | CD8 GZMB-NK IFNγ | 0.56 | 0.00182819 | IL-13-KC | -0.41 | 0.02728385 |
| M1 MQ-NK PD1 | 0.59 | 0.00086081 | CD8 GZMB-NK PD1 | 0.68 | 5.293E-05 | IL-13-VEGF | 0.53 | 0.0031358 |
| M1 MQ-PMN-MDSC | -0.46 | 0.01222074 | CD8 GZMB-Eotaxin | 0.44 | 0.01814214 | IL-15-IP-10 | 0.81 | 1.3313E-07 |
| M1 MQ-M-MDSC | -0.48 | 0.00841607 | CD8 GZMB-IL-5 | 0.42 | 0.02538759 | IL-15-MIG | 0.44 | 0.01703965 |
| M1 MQ-IL-1α | 0.46 | 0.01251544 | CD8 GZMB-IL-6 | -0.38 | 0.04224085 | IL-15-MIP-1β | -0.44 | 0.01629144 |
| M1 MQ-IL-17 | 0.4 | 0.03272108 | CD8 GZMB-MIP-1α | -0.41 | 0.03601448 | IL-17-MIP-1β | -0.4 | 0.03257627 |
| PDL1 MQ-Activated M1 | 0.88 | 2.5978E-10 | CD8 PD1-NK Cells | 0.45 | 0.01491434 | IP-10-MIG | 0.59 | 0.0008301 |
| PDL1 MQ-CD4 | 0.69 | 3.4299E-05 | CD8 PD1-NK GZMB | 0.77 | 1.0194E-06 | KC-MIP-1α | -0.39 | 0.04445848 |
| PDL1 MQ-NK Cells | 0.85 | 7.414E-09 | CD8 PD1-NK IFNγ | 0.63 | 0.00031228 | MCP-1-TNFα | 0.54 | 0.00276259 |
| PDL1 MQ-NK IFNγ | 0.47 | 0.01013809 | CD8 PD1-NK PD1 | 0.72 | 1.3719E-05 | M-CSF-MIP-2 | 0.87 | 1.0176E-09 |
| PDL1 MQ-NK PD1 | 0.69 | 3.9799E-05 | CD8 PD1-IL-5 | 0.38 | 0.0429076 | TNFα-VEGF | 0.42 | 0.0240414 |
Local immunomodulation alters gut microbiome and key genus abundance. A) OTU clustering at the phylum level in healthy and tumor-bearing male and female mice following CRT-NP and HT treatments (n=5). B) Alpha diversity comparison between different groups calculated using Shannon and chao1 alpha diversity index. C) Hierarchical clustering and correlation analysis of OTUs at the genus level in different treatment groups. D) Bar graph representing top 15 populations identified with LEfSe analysis at the genus level across multiple treatments. The cutoff of LDA score in the LEfSe analysis is 2.0.
Correlation of immune cells, cytokines, and gut microbiome. A) Pearson correlation heatmap of genera with tumor burden, TME infiltrating immune cells, serum cytokines and chemokines levels, * significance at P < 0.05, ** significance at P < 0.01, and *** significance at P < 0.001, n=30. B) In the bacteria-immune correlation network for each treatment, only significant correlations were considered. Blue lines represent negative correlations, and orange lines represent positive correlations. The thickness of the lines indicates the strength of the correlation. C) Significant Pearson correlations for bacteria-immune pairs (p<0.001) are shown, with the x-axis representing the correlation coefficient and the y-axis representing the bacteria-immune pairs.
Multifactor LefSe analysis details comprising P values, FDR and LDA score.
| Pvalues | FDR | Female_Pre-Inoculation | Male_Pre-Inoculation | Female_CRT-NP_treated | Female_Control | Male_CRT-NP_treated | Male_ Control | Female_HT_treated | Male_HT_treated | LDA score | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Desulfovibrio | 5.74E-05 | 0.0018381 | 0 | 0 | 8923.2 | 13121 | 23937 | 65572 | 0 | 52876 | 4.52 |
| Clostridium | 0.00024135 | 0.0038616 | 144010 | 2235.3 | 496230 | 565880 | 17645 | 21349 | 520580 | 20501 | 5.45 |
| Romboutsia | 0.000450.00045025 | 0.0048026 | 24719 | 0 | 158840 | 192030 | 984.86 | 0 | 178310 | 0 | 4.98 |
| Shuttleworthia | 0.00096931 | 0.0066677 | 11787 | 5221.1 | 0 | 0 | 2607.7 | 0 | 0 | 2062.6 | 3.77 |
| Streptococcus | 0.0010418 | 0.0066677 | 0 | 0 | 5280.6 | 8284.8 | 11392 | 2751.9 | 3833.4 | 13979 | 3.84 |
| Turicibacter | 0.0022415 | 0.011955 | 4654.3 | 0 | 482360 | 445440 | 6643.5 | 8649.2 | 892120 | 55124 | 5.65 |
| NA | 0.0064335 | 0.02941 | 7217100 | 7732300 | 6081800 | 5922800 | 7172200 | 7935700 | 5763100 | 7419200 | 6.04 |
| *Stoquefichus | 0.0091161 | 0.032931 | 620.57 | 11988 | 0 | 0 | 14034 | 5291.2 | 3409.9 | 13328 | 3.85 |
| Acetitomaculum | 0.0092618 | 0.032931 | 50578 | 29897 | 0 | 5320 | 1679.9 | 0 | 3477.8 | 0 | 4.4 |
| Acetatifactor | 0.032508 | 0.10403 | 2206.8 | 6379.8 | 10709 | 10133 | 1481.7 | 1102.3 | 1425.1 | 2209 | 3.68 |
| Staphylococcus | 0.036809 | 0.10708 | 0 | 0 | 13984 | 6369.6 | 24300 | 4299.5 | 0 | 15509 | 4.08 |
| *Saccharimonas | 0.045966 | 0.14924 | 3567.4 | 0 | 1827.9 | 0 | 7857.8 | 3684.3 | 841.96 | 1785.9 | 3.59 |
| Blautia | 0.075145 | 0.17052 | 100250 | 88686 | 8505.8 | 151680 | 30877 | 22304 | 55417 | 93517 | 4.85 |
| Enterorhabdus | 0.07573 | 0.17052 | 33655 | 35822 | 38280 | 61203 | 70522 | 92715 | 32401 | 95952 | 4.5 |
| Lactobacillus | 0.079934 | 0.17052 | 1277100 | 780290 | 1692900 | 1869000 | 1415600 | 780770 | 1662800 | 1123500 | 5.74 |
| *Arthromitus | 0.091419 | 0.18284 | 15928 | 8667.3 | 6396.2 | 7877.8 | 16403 | 1317.7 | 1003.2 | 5054.9 | 3.89 |
| Marvinbryantia | 0.12256 | 0.21885 | 53581 | 55119 | 33252 | 30160 | 62123 | 61358 | 13895 | 61901 | 4.38 |
| Tyzzerella | 0.12841 | 0.21885 | 6650.5 | 7494.2 | 9120.3 | 4228.4 | 9373.7 | 5807.6 | 0 | 2565.3 | 3.67 |
| Mucispirillum | 0.12994 | 0.21885 | 11109 | 10519 | 0 | 8302.8 | 6782.8 | 7310.1 | 0 | 1727.7 | 3.74 |
| Akkermansia | 0.14236 | 0.22777 | 49596 | 73143 | 100480 | 26760 | 74688 | 120500 | 155060 | 129800 | 4.81 |
| Oscillibacter | 0.15956 | 0.23488 | 74674 | 74724 | 40051 | 41532 | 66715 | 48148 | 25458 | 27517 | 4.39 |
| Jeotgalicoccus | 0.16148 | 0.23488 | 0 | 0 | 41366 | 1698.6 | 24765 | 4616.9 | 5984.3 | 16919 | 4.32 |
| Bacteroides | 0.17131 | 0.23834 | 570990 | 617050 | 478070 | 246170 | 479380 | 400770 | 372240 | 378120 | 5.27 |
| Parabacteroides | 0.20288 | 0.25587 | 66808 | 76631 | 50495 | 40998 | 38219 | 37863 | 53454 | 33063 | 4.34 |
| Erysipelatoclostridium | 0.2051 | 0.25587 | 5843.2 | 2158 | 6741.9 | 8117.6 | 4450.7 | 1752.3 | 5401.9 | 9518.6 | 3.59 |
| Adlercreutzia_Asaccharobacter | 0.2079 | 0.25587 | 0 | 2739.4 | 10338 | 3593.1 | 9115.1 | 8772.8 | 4061.2 | 21234 | 4.03 |
| Anaerotruncus | 0.24641 | 0.28779 | 70138 | 66565 | 37435 | 71962 | 72789 | 81796 | 22867 | 47024 | 4.47 |
| Roseburia | 0.25182 | 0.28779 | 24690 | 77833 | 34194 | 92600 | 83066 | 77479 | 47523 | 115050 | 4.65 |
| Lachnoclostridium | 0.26355 | 0.29082 | 123250 | 164210 | 110920 | 96061 | 171100 | 132620 | 109940 | 183460 | 4.64 |
| Allobaculum | 0.3536 | 0.37717 | 4188.9 | 0 | 16492 | 28145 | 0 | 1865.5 | 43499 | 7153.8 | 4.34 |
| Ruminiclostridium | 0.38684 | 0.39932 | 50676 | 65313 | 23773 | 37873 | 77226 | 53185 | 18436 | 41748 | 4.47 |
| Subdoligranulum | 0.78416 | 0.78416 | 1610.4 | 5052.1 | 1238.7 | 2625 | 2115.9 | 10619 | 3477.6 | 8565.9 | 3.67 |
We first profiled the gut microbiomes of healthy, non-tumor-bearing male and female mice. At the phylum level, females exhibited a higher Firmicutes-to-Bacteroidetes ratio, consistent with previous studies [26]. Additionally, we observed elevated levels of Saccharibacteria in females, which has been associated with higher sugar metabolism [27, 28] and protection from inflammatory damage induced by pathogens in mammalian hosts [29]. In contrast, the gut microbiome of males showed a significantly higher presence of Cyanobacteria, whose derivatives and metabolites are currently being studied for their anti-tumor properties [30]. However, it is worth noting that cyanobacterial blooms, particularly their toxins—such as microcystins, cylindrospermopsin, and lipopolysaccharides—are known to contribute to gastric cancers [31]. Given the contradictory outcomes of HT in males, we hypothesize that its efficacy may be treatment-dependent. Among various phyla, Firmicutes emerged as a key regulator of sex-dependent disparities, as most genera showing differences between males and females belonged to this phylum. For example, Staphylococcus, Lactonifactor, and Anaerostipes were more abundant in males, while Turicibacter, Tyzzerella, Romboutsia, and Marvinbryantia were more prevalent in females.
The MOC2-bearing male mice demonstrated rapid tumor progression consistent with poorly immunogenic profiles, as observed in other models [32, 33]. Since the MOC2 cell lines used in this study were derived from female mice, it was surprising to find that the tumors grew faster in males, as they should ideally have grown slower due to tumor antigen mismatch between sexes. Prior research indicates that oropharyngeal squamous carcinoma with higher estrogen receptor α (ERα) expressions is associated with improved survival among those receiving chemoradiation [34]. While the ERα expression profile of MOC2 is not known, the improved outcome in females vs. males suggests potential hormonal signaling influencing growth patterns. We also examined the tumors of untreated male and female mice and observed moderately higher CD8 T cell and NK cell infiltration in female tumors. These observations led us to hypothesize that microbiome-related factors between sexes could be at-play, which we investigated in this study.
Our data suggested that NK cell infiltration correlated with four bacterial genera, among which Desulfovibrio (DSV) was persistently higher in males. DSV has been shown to be in higher abundance in gastric, colon, rectal, and breast cancer patients [35], inducing colonic damage and pre-metastatic niche in mice liver by increasing IL-1β, TNFα, CXCL12 and various matrix metalloproteases [36]. We found that DSV, similarly, correlated positively with faster tumor growth, higher serum IL-4 and reduced Eotaxin and IL-17 levels (Figure 6). In addition to DSV, three other genera (Adlercreutzia-Asaccharobacter, Subdoligranulum, and Enterorhabdus) showed a positive correlation with tumor growth, while Turicibacter and Peptococcus exhibited a negative correlation. Previous studies have linked a decrease in Peptococcus abundance with tumor growth in esophageal squamous cell carcinoma [37], prostate cancer patients [38], and cachectic cancer patients [39]. Chen et al. demonstrated that higher gut Peptococcus abundance decreases the risk of non-small cell lung cancer via CD4+ T cells [40]. Similarly, we observed a positive correlation between Peptococcus abundance and CD4+ T cell infiltration in MOC2 tumors. Interestingly, Peptococcus also correlated with increased macrophage numbers in the TME and higher PD-L1 expression on macrophages, potentially limiting its use as a predictive marker for successful immunotherapy outcomes.
Finally, we assessed the microbiome dynamics in MOC2-bearing mice treated with CRT-NP and FUS in both sexes. The intratumoral route of CRT-NP administration was selected to explore how localized treatment of a distant tumor affects gut microbiome abundance. This approach contrasts with other studies that have focused on tumor cell and tumor microenvironment (TME) resident microbiome interactions [41-43], or on gut microbiome modulation through systemic treatments [44, 45]. However, the impact of localized tumor treatment on the gut microbiome, particularly in distant tumors, remains underexplored. CRT-NP induced a similar decline in MOC2 tumor growth in both sexes. However, HT failed to control MOC2 tumor growth in males, while efficiently retarding tumor growth in females, mirroring the efficacy of CRT-NP therapy (Figure 3). Unlike caliper measurements of tumor volume, males showed higher tumor weight compared to untreated controls (Figure 3C-E), suggesting a faster inward spread of the tumor toward the peritoneal space by the day of sacrifice, which was not fully captured by caliper measurements. To elucidate the potential immune-mediated mechanisms underlying these outcomes, we examined the tumor immune microenvironment and systemic cytokine profiles. We observed an increase in CD4+ T cells and macrophages in both sexes following CRT-NP treatments, with levels higher in females. Tumor-infiltrating macrophages displayed polarization toward the M1 type, accompanied by a decrease in MDSC population. In contrast, with HT, TME immune cell profiling showed a higher engagement of cytotoxic NK cells and CD8 T cells in HT-treated female tumors compared to all other groups (Figure 4). Pearson r analysis showed strong correlation of cytotoxic T cells and NK cells with Turicibacter abundance (Figure 6C) which increased significantly in HT treated females. In contrast, genera like Streptococcus and Enterorhabdus were highly abundant in HT-treated male mice (non-responders) and showed a positive correlation with serum IL-1, M-CSF, MIP-2, IL12p40 or IL-17, pro-tumoral cytokines, and chemokines. Streptococcus and Staphylococcus species are well-known to promote the development of cancer in various tissues [46, 47]. A retrospective analysis of the intestinal microbiome composition in 44 pancreatic cancer patients demonstrated a significant increase in Streptococcus content [48]. Their higher abundance was also noted in non-responder NSCLC patients receiving anti-PD1 therapy [49]. Enterorhabdus has been associated with ileocecal inflammation [50], and its gut levels negatively correlated with IFNγ serum levels [51]. In addition to protumoral cytokines, we observed Enterorhabdus showed a negative correlation with intratumoral M1 macrophage activation and NK cell numbers (Figure 6). Additionally, a positive correlation of Turicibacter with cytotoxic T cells and NK cell populations in the tumor microenvironment as well as serum levels of Eotaxin and IL-5 indicate a treatment-specific change in the gut microbiome (Figure 6A-C). Recent research has suggested that Turicibacter plays a role in creating a favorable tumor microenvironment by enhancing the production of metabolites, such as short-chain fatty acids (SCFAs), which can improve the recruitment and activation of immune cells like cytotoxic T lymphocytes and natural killer cells [52, 53].
Our study has some limitations. We used a single tumor line in a single mouse strain. Panshak et al. previously demonstrated that females showed a higher response to the B16-F10/BL6 syngeneic mouse model melanoma than males [32]. Thus, as evidenced by our correlative analysis and prior reports, our findings could be generalized based on sex with the added element of microbiome signatures, along with cytokine and immune cell features. Furthermore, the translational impact of our findings would be best demonstrated by microbial transplantation from female FMT into untreated males to show reduced tumor growth. The goal of this study was to highlight and identify sexually dimorphic signatures indicative of microbiome-correlated outcomes. Future studies performing microbial depletion experiments or fecal transplantations to validate these findings would provide more direct evidence of the causal relationship between these microbial species and treatment response. However, those studies were beyond the scope of this paper.
Conclusion
To summarize, our study highlights the potential significance of sex-specific factors and microbiota composition in treatment response. Particularly, higher levels of Enterorhabdus, DSV, Streptococcus and Staphylococcus in fecal samples after NP and HT immunomodulatory treatment can serve as predictive biomarker for treatment response, independent of sex. The differential response observed in male mice with HT underscores the importance of considering sex-specific factors and associated microbiota composition in personalized therapies. Characterizing biomarkers before treatment initiation can aid in the selection and stratification of male and female patients who are expected to respond most favorably to treatments.
Supplementary Material
Supplementary figures.
Acknowledgements
We thank the National Cancer Institute of the National Institutes of Health under Award numbers 5R37CA239150 & RO1CA284741, the Focused Ultrasound Foundation, and Barret Family Professor of Cancer Research at UT Southwestern Medical Center for supporting this research.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Gorbet M-J, Ranjan A. Cancer immunotherapy with immunoadjuvants, nanoparticles, and checkpoint inhibitors: Recent progress and challenges in treatment and tracking response to immunotherapy. Pharmacol Ther. 2020;207:107456 -
2. Zhang Z, Yao S, Hu Y, Zhao X, Lee RJ. Application of lipid-based nanoparticles in cancer immunotherapy. Front Immunol. 2022;13:967505
3. Yu Z, Shen X, Yu H, Tu H, Chittasupho C, Zhao Y. Smart Polymeric Nanoparticles in Cancer Immunotherapy. Pharmaceutics. 2023;15:775
4. He JS, Liu SJ, Zhang YR, Chu XD, Lin ZB, Zhao Z. et al. The Application of and Strategy for Gold Nanoparticles in Cancer Immunotherapy. Front Pharmacol. 2021;12:687399
5. Yu A, Dai X, Wang Z, Chen H, Guo B, Huang L. Recent Advances of Mesoporous Silica as a Platform for Cancer Immunotherapy. Biosensors (Basel). 2022;12:109
6. Gorbet MJ, Singh A, Mao C, Fiering S, Ranjan A. Using nanoparticles for in situ vaccination against cancer: mechanisms and immunotherapy benefits. Int J Hyperthermia. 2020;37:18-33
7. Sethuraman SN, Singh MP, Patil G, Li S, Fiering S, Hoopes PJ. et al. Novel calreticulin-nanoparticle in combination with focused ultrasound induces immunogenic cell death in melanoma to enhance antitumor immunity. Theranostics. 2020;10:3397-412
8. Chandrasekar SV, Singh A, Ranjan A. Overcoming Resistance to Immune Checkpoint Inhibitor Therapy Using Calreticulin-Inducing Nanoparticle. Pharmaceutics. 2023;15:1693
9. Ashar H, Ranjan A. Immunomodulation and targeted drug delivery with high intensity focused ultrasound (HIFU): Principles and mechanisms. Pharmacol Ther. 2023;244:108393
10. Thim EA, Kitelinger LE, Rivera-Escalera F, Mathew AS, Elliott MR, Bullock TNJ. et al. Focused ultrasound ablation of melanoma with boiling histotripsy yields abscopal tumor control and antigen-dependent dendritic cell activation. bioRxiv. 2024
11. Singh MP, Sethuraman SN, Miller C, Malayer J, Ranjan A. Boiling histotripsy and in-situ CD40 stimulation improve the checkpoint blockade therapy of poorly immunogenic tumors. Theranostics. 2021;11:540-54
12. Wang L, Tang L, Zhai D, Song M, Li W, Xu S. et al. The role of the sex hormone-gut microbiome axis in tumor immunotherapy. Gut Microbes. 2023;15:2185035
13. Yoon K, Kim N. Roles of Sex Hormones and Gender in the Gut Microbiota. Journal of neurogastroenterology and motility. 2021;27:314-25
14. Li Y, Tinoco R, Elmen L, Segota I, Xian Y, Fujita Y. et al. Gut microbiota dependent anti-tumor immunity restricts melanoma growth in Rnf5(-/-) mice. Nat Commun. 2019;10:1492
15. Kang J, Li C, Gao X, Liu Z, Chen C, Luo D. Metformin inhibits tumor growth and affects intestinal flora in diabetic tumor-bearing mice. Eur J Pharmacol. 2021;912:174605
16. Shi Q, Wang J, Zhou M, Zheng R, Zhang X, Liu B. Gut Lactobacillus contribute to the progression of breast cancer by affecting the anti-tumor activities of immune cells in the TME of tumor-bearing mice. Int Immunopharmacol. 2023;124:111039
17. Zhang SL, Mao YQ, Zhang ZY, Li ZM, Kong CY, Chen HL. et al. Pectin supplement significantly enhanced the anti-PD-1 efficacy in tumor-bearing mice humanized with gut microbiota from patients with colorectal cancer. Theranostics. 2021;11:4155-70
18. Lu Y, Zhou G, Ewald J, Pang Z, Shiri T, Xia J. MicrobiomeAnalyst 2.0: comprehensive statistical, functional and integrative analysis of microbiome data. Nucleic Acids Research. 2023;51:W310-W8
19. Lahti L SS. Microbiome R package. 2019.
20. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology. 2014;15:550
21. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-40
22. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS. et al. Metagenomic biomarker discovery and explanation. Genome biology. 2011;12:R60
23. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell. 2018;33:570-80
24. Cook MB, Dawsey SM, Freedman ND, Inskip PD, Wichner SM, Quraishi SM. et al. Sex disparities in cancer incidence by period and age. Cancer Epidemiol Biomarkers Prev. 2009;18:1174-82
25. Hankinson SE, Eliassen AH. Endogenous estrogen, testosterone and progesterone levels in relation to breast cancer risk. J Steroid Biochem Mol Biol. 2007;106:24-30
26. Koliada A, Moseiko V, Romanenko M, Lushchak O, Kryzhanovska N, Guryanov V. et al. Sex differences in the phylum-level human gut microbiota composition. BMC Microbiol. 2021;21:131
27. Figueroa-Gonzalez PA, Bornemann TLV, Adam PS, Plewka J, Revesz F, von Hagen CA. et al. Saccharibacteria as Organic Carbon Sinks in Hydrocarbon-Fueled Communities. Front Microbiol. 2020;11:587782
28. Sanchez-Quinto A, Cerqueda-Garcia D, Falcon LI, Gaona O, Martinez-Correa S, Nieto J. et al. Gut Microbiome in Children from Indigenous and Urban Communities in Mexico: Different Subsistence Models, Different Microbiomes. Microorganisms. 2020;8:1592
29. Chipashvili O, Utter DR, Bedree JK, Ma Y, Schulte F, Mascarin G. et al. Episymbiotic Saccharibacteria suppresses gingival inflammation and bone loss in mice through host bacterial modulation. Cell Host Microbe. 2021;29:1649-62 e7
30. Bouyahya A, Bakrim S, Chamkhi I, Taha D, El Omari N, El Mneyiy N. et al. Bioactive substances of cyanobacteria and microalgae: Sources, metabolism, and anticancer mechanism insights. Biomed Pharmacother. 2024;170:115989
31. Kubickova B, Babica P, Hilscherová K, Sindlerová L. Effects of cyanobacterial toxins on the human gastrointestinal tract and the mucosal innate immune system. Environ Sci Eur. 2019;31:31
32. Dakup PP, Porter KI, Little AA, Zhang H, Gaddameedhi S. Sex differences in the association between tumor growth and T cell response in a melanoma mouse model. Cancer Immunol Immunother. 2020;69:2157-62
33. Ray AL, Nofchissey RA, Khan MA, Reidy MA, Lerner MR, Wu X. et al. The role of sex in the innate and adaptive immune environment of metastatic colorectal cancer. Br J Cancer. 2020;123:624-32
34. Koenigs MB, Lefranc-Torres A, Bonilla-Velez J, Patel KB, Hayes DN, Glomski K. et al. Association of Estrogen Receptor Alpha Expression With Survival in Oropharyngeal Cancer Following Chemoradiation Therapy. Journal of the National Cancer Institute. 2019;111:933-42
35. Singh SB, Carroll-Portillo A, Lin HC. Desulfovibrio in the Gut: The Enemy within? Microorganisms. 2023;11:1772
36. Yu Y, Cai Y, Yang B, Xie S, Shen W, Wu Y. et al. High-Fat Diet Enhances the Liver Metastasis Potential of Colorectal Cancer through Microbiota Dysbiosis. Cancers (Basel). 2022;14:2573
37. Chen X, Winckler B, Lu M, Cheng H, Yuan Z, Yang Y. et al. Oral Microbiota and Risk for Esophageal Squamous Cell Carcinoma in a High-Risk Area of China. PLoS One. 2015;10:e0143603
38. Gonçalves MFM, Pina-Vaz T, Fernandes AR, Miranda IM, Silva CM, Rodrigues AG. et al. Microbiota of Urine, Glans and Prostate Biopsies in Patients with Prostate Cancer Reveals a Dysbiosis in the Genitourinary System. Cancers. 2023;15:1423
39. Ubachs J, Ziemons J, Soons Z, Aarnoutse R, Dijk DPJ, Penders J. et al. Gut microbiota and short-chain fatty acid alterations in cachectic cancer patients. J Cachexia Sarcopeni. 2021;12:2007-21
40. Chen J, Yu X, Wu X, Chai K, Wang S. Causal relationships between gut microbiota, immune cell, and Non-small cell lung cancer: a two-step, two-sample Mendelian randomization study. J Cancer. 2024;15:1890-7
41. Jiang J, Mei J, Ma Y, Jiang S, Zhang J, Yi S. et al. Tumor hijacks macrophages and microbiota through extracellular vesicles. Exploration (Beijing). 2022;2:20210144
42. Jiang J, Mei J, Yi S, Feng C, Ma Y, Liu Y. et al. Tumor associated macrophage and microbe: The potential targets of tumor vaccine delivery. Adv Drug Deliv Rev. 2022;180:114046
43. Wong CC, Yu J. Gut microbiota in colorectal cancer development and therapy. Nat Rev Clin Oncol. 2023;20:429-52
44. Li X, Zhang S, Guo G, Han J, Yu J. Gut microbiome in modulating immune checkpoint inhibitors. EBioMedicine. 2022;82:104163
45. Li S, Zhu S, Yu J. The role of gut microbiota and metabolites in cancer chemotherapy. J Adv Res. 2024;64:223-235
46. Zi M, Zhang Y, Hu C, Zhang S, Chen J, Yuan L. et al. A literature review on the potential clinical implications of streptococci in gastric cancer. Front Microbiol. 2022;13:1010465
47. Wei Y, Sandhu E, Yang X, Yang J, Ren Y, Gao X. Bidirectional Functional Effects of Staphylococcus on Carcinogenesis. Microorganisms. 2022;10:2353
48. Yang J, Ma Y, Tan Q, Zhou B, Yu D, Jin M. et al. Gut Streptococcus is a microbial marker for the occurrence and liver metastasis of pancreatic cancer. Front Microbiol. 2023;14:1184869
49. Botticelli A, Vernocchi P, Marini F, Quagliariello A, Cerbelli B, Reddel S. et al. Gut metabolomics profiling of non-small cell lung cancer (NSCLC) patients under immunotherapy treatment. J Transl Med. 2020;18:49
50. Shlisky J, Bloom DE, Beaudreault AR, Tucker KL, Keller HH, Freund-Levi Y. et al. Nutritional Considerations for Healthy Aging and Reduction in Age-Related Chronic Disease. Adv Nutr. 2017;8:17-26
51. Pagliai G, Russo E, Niccolai E, Dinu M, Di Pilato V, Magrini A. et al. Influence of a 3-month low-calorie Mediterranean diet compared to the vegetarian diet on human gut microbiota and SCFA: the CARDIVEG Study. Eur J Nutr. 2020;59:2011-24
52. Hamada K, Isobe J, Hattori K, Hosonuma M, Baba Y, Murayama M. et al. Turicibacter and Acidaminococcus predict immune-related adverse events and efficacy of immune checkpoint inhibitor. Front Immunol. 2023;14:1164724
53. Lynch JB, Gonzalez EL, Choy K, Faull KF, Jewell T, Arellano A. et al. Gut microbiota Turicibacter strains differentially modify bile acids and host lipids. Nat Commun. 2023;14:3669
Author contact
![]() Corresponding author: Ashish Ranjan, B.V.Sc., Ph.D., Barret Family Professor of Cancer Research, Vice Chair: Research Innovations & Comparative Oncology, Director: One Medicine Clinics, Department of Radiation Oncology, UT Southwestern Medical Center, 2280 Inwood Road, Dallas, Texas 75390. Phone: 214-645-7603; ashish.ranjanedu.
Corresponding author: Ashish Ranjan, B.V.Sc., Ph.D., Barret Family Professor of Cancer Research, Vice Chair: Research Innovations & Comparative Oncology, Director: One Medicine Clinics, Department of Radiation Oncology, UT Southwestern Medical Center, 2280 Inwood Road, Dallas, Texas 75390. Phone: 214-645-7603; ashish.ranjanedu.
 Global reach, higher impact
Global reach, higher impact