13.3
Impact Factor
Theranostics 2024; 14(12):4683-4700. doi:10.7150/thno.99019 This issue Cite
Review
The role of m6A-associated membraneless organelles in the RNA metabolism processes and human diseases
1. College of Pharmacy and Department of Hepatology, Institute of Hepatology and Metabolic Diseases, the Affiliated Hospital of Hangzhou Normal University, Hangzhou Normal University, Hangzhou, Zhejiang 311121, China.
2. Key Laboratory of Elemene Class Anti-Cancer Chinese Medicines; Engineering Laboratory of Development and Application of Traditional Chinese Medicines; Collaborative Innovation Center of Traditional Chinese Medicines of Zhejiang Province, Hangzhou Normal University, Hangzhou, Zhejiang 311121, China.
3. Department of Gastroenterology, Affiliated Nanjing Jiangbei Hospital of Xinglin College, Nantong University, Nanjing 210044, P. R. China.
4. Laboratory of Cancer Genomics, Division of Cellular and Molecular Research, National Cancer Centre Singapore, 169610, Singapore.
Received 2024-5-29; Accepted 2024-7-25; Published 2024-8-6
Abstract
N6-methyladenosine (m6A) is the most abundant post-transcriptional dynamic RNA modification process in eukaryotes, extensively implicated in cellular growth, embryonic development and immune homeostasis. One of the most profound biological functions of m6A is to regulate RNA metabolism, thereby determining the fate of RNA. Notably, the regulation of m6A-mediated organized RNA metabolism critically relies on the assembly of membraneless organelles (MLOs) in both the nucleus and cytoplasm, such as nuclear speckles, stress granules and processing bodies. In addition, m6A-associated MLOs exert a pivotal role in governing diverse RNA metabolic processes encompassing transcription, splicing, transport, decay and translation. However, emerging evidence suggests that dysregulated m6A levels contribute to the formation of pathological condensates in a range of human diseases, including tumorigenesis, reproductive diseases, neurological diseases and respiratory diseases. To date, the molecular mechanism by which m6A regulates the aggregation of biomolecular condensates associated with RNA metabolism is unclear. In this review, we comprehensively summarize the updated biochemical processes of m6A-associated MLOs, particularly focusing on their impact on RNA metabolism and their pivotal role in disease development and related biological mechanisms. Furthermore, we propose that m6A-associated MLOs could serve as predictive markers for disease progression and potential drug targets in the future.
Keywords: N6-methyladenosine, membraneless organelle, RNA metabolism, liquid-liquid phase separation, human diseases
Introduction
To date, more than one hundred chemical decorations on RNA have been discovered in eukaryotes [1]. N6-methyladenosine (m6A), a methyl modification on adenosine, represents the most prevalent and dynamic post-transcriptional modification in eukaryotic mRNA as well as certain noncoding RNAs (ncRNAs) [2, 3]. Commonly, m6A is installed by methyltransferases (writers), such as METTL3 and METTL16 [4, 5], and removed by demethylases (erasers), such as ALKBH3, ALKBH5 and FTO [6, 7]. Besides, readers preferentially bind to m6A determines the fate of mRNA. Different discovered readers, such as YTH (YT521-B homology) domain proteins [8], IGF2BP proteins [9] and HnRNP family proteins [10], are involved in RNA transcription, splicing, export, degradation and translation. Physiologically, the dynamic m6A modification plays a crucial role in germ cell meiosis [11], embryonic development [12] and immune response [13]. However, destruction of the physiological levels of m6A leads to the emergence and development of various diseases, including infertility [14], neurodegenerative disease [15] and tumorigenesis [16]. These unexpected effects are likely attributed to metabolic disorders of key mRNAs associated with m6A modification. Intriguingly, a substantial pool of mRNAs marked by m6A facilitates the formation of membraneless organelles (MLOs) or biomolecular condensates (BioMCs) formation through interacting with m6A writers, readers or erasers, which represents a crucial process in mRNA metabolism and turnover [16-18]. Liquid-liquid phase separation (LLPS) promotes the assembly and formation of these m6A-assocaited MLOs or -BioMCs [18, 19]. Many reports indicate that multivalent low-affinity interactions, encompassing RNA-RNA, RNA-protein and protein-protein, can trigger LLPS [20]. Importantly, post-transcriptional modification, particularly m6A, provides “available bacon” for different RNA binding proteins containing internally disordered regions (IDRs) during the process of LLPS.
Recently, MLOs or BioMCs driven by m6A-modified RNAs have been proved to participate in transcription [21], alternative splicing (AS) [22], miRNA biogenesis [23], export [24], mRNA decay [18] and translation [25]. These m6A-associated MLOs or BioMCs ensure efficient RNA metabolism under physiological conditions, thereby facilitating robust cellular homeostasis. Nevertheless, maladjustment of m6A modification, such as deficiency of ALKBH5, a crucial m6A demethylase, results in diminished nuclear speckle activity and dysregulated mRNA export and splicing in mammalian cells. This distinctively leads to impaired fertility in male mice although diverse biological functions of ALKBH5 have been revealed successively [6]. Differently, MLOs driven by m6A modifications, such as nuclear YTHDC1-m6A condensates (nYACs), protect oncogenic mRNAs from degradation by polyA tail exosome targeting complex (PAXT), which contributes to the survival of cancer cells in acute myeloid leukemia [26]. Moreover, excessive MLOs formation such as stress granules (SGs) is associated with tauopathy, and hnRNPA2B1 (a reader for m6A) mutation leads to Tau aggregation and sequesters itself in SGs [27]. Although it has been established that m6A-associated MLOs play a regulatory role in various pathological processes, a comprehensive understanding of how these MLOs precisely affect specific biochemical processes, and the precise functional association between MLOs and the emergence and processes of different human diseases remains elusive.
In this review, we will outline the updated biochemical processes of m6A-associated MLOs, especially for RNA metabolism, and explore their implications in disease development and related biological mechanisms. Importantly, we put forward the hypothesis that the number of m6A-associated MLOs may predict the disease progression, and intervening in MLOs production or changing m6A levels will be an effective strategy to alleviate the disease process, especially for cancer cell growth.
The selected biological reaction, substrate and significance of m6A modification
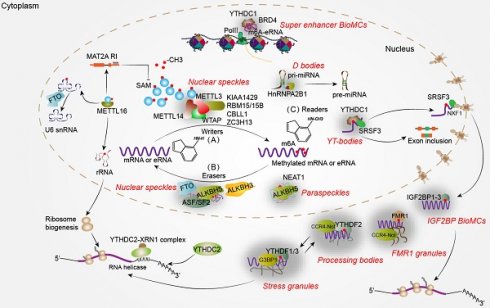
Studies have found that a variety of chemical modifications, such as methylation, acetylation and phosphorylation, occur in DNA, RNA and protein, among which RNA is the most abundant substrate for chemical modification. More than 170 RNA chemical modifications have been identified yet, which are widely distributed in mRNA, tRNA, rRNA, spliceosomal RNA and other ncRNAs [1, 28]. RNA methylation accounted for more than 60% of RNA modification, and the common RNA methylation included 6-methyladenine (m6A), 5-methylcytosine (m5C), 1-methyladenine (m1A), 5-hydroxymethylcytosine (h5mC) and pseudouridine (Ψ) [29]. Among them, m6A is the most abundant type of methylation modification in eukaryotic mRNA, which regulates gene expression post-transcriptionally [2, 3]. Methylation occurs in the N6 of adenine within highly conserved RRACH (R: purine; A: m6A; H: non-guanine) region, that is a dynamic modification regulated by methyltransferases (writers), demethylases (erasers), and m6A-binding proteins (readers) (Figure 1). Generally, the m6A modification frequently occurs near the translation stop codon and in 3'-UTR of mRNA [30]. “Writers” transfer the methyl groups from the donor S-adenosylmethionine (SAM) to the N6 position of adenosine [31]. Current studies shows that m6A is marked by writer complex, which consists of the METTL3-METTL14 heterodimer and many vital adaptor proteins, WTAP [32], KIAA1429 (also known as VIRMA) [33], RBM15, or its paralogue RBM15B [34], CBLL1 and ZC3H13 [35]. METTL3 and METTL14 form a complex that facilitates the transfer of methyl groups to the sixth adenine of mRNA. WTAP plays a regulatory role in the recruitment of METTL3-METTL14 heterodimeric enzyme complex to nuclear speckles (NSs) which are involved in RNA transcription, AS and export, although it has no catalytic activity [32]. In addition, KIAA1429, RBM15 and ZC3H13 are also part of the MTC and collaborate synergistically even though their different roles in mRNA or ncRNA methylation processes [34-36]. METTL16 has been identified as an independent U6 snRNA m6A methyltransferase, distinct from the METTL3-METTL14 complex. Generally, SAM serves as a methyl group donor for cellular methylation reactions. Importantly, METTL16 regulates the SAM cycle through controlling the intron retention of SAM synthetase MAT2A, which necessitates methylation substrate on hairpin1 (hp1) in the 3'-UTR of MAT2A [37]. Moreover, METTL16 in the nucleolus regulates rRNA maturation, ribosome biogenesis and mRNA translation, possibly through engaging in rRNA methylation [38]. In addition, METTL5 and ZCCHC4 have also been identified as the enzyme that methylates rRNA, 18S and 28S, respectively [39, 40]. FTO, ALKBH3 and ALKBH5 are demethylases that catalyze the reversal of m6A modification on different RNA molecules [6, 7, 41]. FTO mediates the demethylation of RNA in both the nucleus and cytoplasm, exhibiting cell type-dependent variations and exerting distinct effects on various RNA substrates, including mRNA, snRNA and tRNA [42]. ALKBH3 mediates the demethylation of m6A in mammalian tRNA, thereby augmenting protein synthesis [41]. ALKBH5 catalyzes m6A elimination on nuclear RNA (mostly mRNA), which is confirmed to colocalize with NSs to regulate nuclear RNA export and alternative splicing [6].
Different types of RNA undergo m6A modification through the regulation of methyltransferase and demethylase despite no sequence changes, thereby dynamically controlling various biological processes. Notably, m6A readers, a class of RNA-binding proteins, recognize and selectively bind to the sites modified by m6A, guiding the fate of target RNA. Currently, known m6A readers mainly include three families, YTH domain-containing proteins (YTHDF1-3, YTHDC1-2), hnRNP protein family (hnRNPA2B1, hnRNPC and hnRNPG) and insulin-like growth factor 2-mRNA binding proteins (IGF2BP1-3) [9, 10, 43]. YTHDF1-3 and YTHDC1-2 are canonical m6A readers that share the highly conserved YTH domain, enabling recognition of m6A-modified sites through formation of an aromatic cage; however, they exhibit distinct subcellular location and perform diverse molecular biological functions [44]. YTHDF1-3 and YTHDC2 are mainly located in the cytoplasm. YTHDF1 facilitates cap-dependent RNA translation through recruiting eukaryotic initiation translation factor (EIF3) and coupling m6A-modified mRNA to ribosomes [45]. Most academic perspectives hold that YTHDF2 accelerates the degradation of m6A-modified mRNA through different pathways. For instance, YTHDF2 can recruit the CCR4-NOT deadenylase complex or HRSP12-RNase P-MRP complex to mediate deadenylation of m6A-modified mRNAs and facilitate their endoribonucleolytic cleavage, respectively [46]. Interestingly, YTHDF3 exerts an impact on the translation and degradation of m6A-modified RNAs by cooperating with YTHDF1 or YTHDF2, thereby indicating the synergistic functional interplay among these three YTHDF proteins inside cells [47]. Nuclear YTHDC1 actively participates in the intricate processes of RNA splicing and transport through recognizing m6A-modified sites and interacting with splicing factor SRSF3 [24, 48]. Increasing studies indicate that cytoplasmic YTHDC2 has an impact on RNA degradation and translation. Unlike other readers, YTHDC2 has been demonstrated to perform RNA helicase activity through binding U-rich motifs in 3'-UTR of mRNAs during the mitotic-meiotic transition of mouse germ cells, independent of its m6A-reading capacity [49]. HnRNP protein families have a wide function in regulating RNA metabolism and have also been identified as “m6A readers”. It is noteworthy that although some of them do not directly bind to m6A-modified sites, they instead interact with RNA regions proximal to m6A modification via a so-called “m6A-switch” mechanism [50, 51]. Through this non-canonical mechanism, m6A induces RNA unfolding and enhances the accessibility of hnRNPC, hnRNPG and hnRNPA2B1 to specific RNA binding sites, thereby contributing to the refinement of their alternative splicing activity [50-52]. Besides, hnRNPA2B1 has been implicated in the processing of primary miRNA (pri-miRNA) through its interaction with m6A-modified pri-miRNA and the microRNA Microprocessor complex protein DGCR8 [23]. Additionally, IGF2BPs are identified as a class of non-canonical m6A readers that augment mRNA stability and translation by specifically binding to m6A-modified sites through their KH domains although the binding affinity of IGF2BPs for m6A-modified RNAs is significantly inferior compared to that exhibited by YTH domains [9, 53]. Besides, fragile X mental-retardation protein (FMR1) [54], EIF3 [55] and proline rich coiled-coil 2 A (PRRC2A) [56] have also been identified as m6A readers that actively participate in RNA metabolism through distinct molecular mechanism. Collectively, different readers are distributed in distinct subcellular locations for binding to m6A-modified RNAs, thereby orchestrating the process of RNA metabolism [57]. Overall, m6A modification plays a pivotal role in regulating the whole processes of RNA metabolism, encompassing transcription, splicing, transport, translation and stability (Figure 1).
Noteworthily, m6A modification and its regulators play a crucial role in a variety of physiological processes, including gametogenesis [58], embryogenesis [59], immune response [60] and bone generation [61]. However, maladjustment of m6A modification leads to the emergence and development of diseases [62], such as embryonic lethality [63], infertility [64], immune system dysfunction [13] and tumorigenesis. Intriguingly, emerging evidence suggests that m6A-modified RNAs are enriched within the MLOs or BioMCs, in which they recruit and juxtapose m6A readers, facilitating them to undergo LLPS and to form RNA-protein droplets [19]. These m6A-associated MLOs or BioMCs have been demonstrated to effectively regulate RNA metabolism in a coordinated manner (Figure 1).
Overview of RNA-associated membraneless organelles
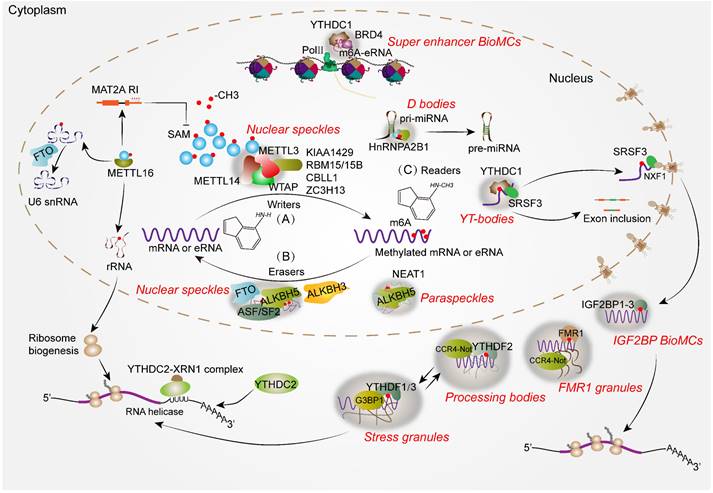
To maintain various biochemical reactions in order, there are subcellular compartments in eukaryotic cells that isolate biomolecules, including canonical membranous organelles, such as mitochondria, Golgi bodies, endoplasmic reticulum and nuclei. Interestingly, some dynamic organelles or apparatus without surrounding membranes, about 0.2~2 μm in size, are also found in the cells, known as MLOs or BioMCs, which exhibit different morphologies and are enriched with abundant proteins and nucleic acids [20]. These MLOs have a liquid-like feature and bear liquid droplets, which are assembled or disassembled depending on the cellular condition and partially external stimuli. Non-covalent interactions between various proteins and nucleic acids serve as driving forces for LLPS, facilitating the formation of MLOs [65]. It is worth noting that MLOs play essential roles in diverse cellular processes (Table 1), such as chromosome maintenance [66], RNA metabolism [20], protein turnover [67], immune signal transduction [68] and cytoskeleton remodeling [69].
m6A-associated MLOs or BioMCs run through the life of RNA. (A) METTL16 controls cellular SAM levels through regulating the intron retention of SAM synthetase, MAT2A. Generally, m6A modifications on mRNA, eRNA or ncRNA are catalyzed by distinct methyltransferases such as METTL16 or “writer complex-MTC” comprising the METTL3-METTL14 heterodimer and essential adaptor proteins including WTAP, KIAA1429, RBM15/15B, CBLL1 and ZC3H13. In this process, METTL3 enhances the assembly of MTC through LLPS, while WTAP recruits METTL3-METTL14 heterodimer to NSs. METTL16 can promote the biogenesis of ribosome through methylating rRNA in the nucleolus, indirectly enhancing mRNA translation. (B) The removal of methyl groups is completed by demethylases (erasers), including FTO, ALKBH5 and ALKBH3. FTO and ALKBH5 are predominantly localized in NSs, where their demethylation activity facilitates RNA nuclear retention or alternative splicing. Besides, ALKBH5 promotes the assembly of PSs through LLPS and demethylating NEAT1, a vital scaffolding lncRNA for PSs formation, thereby enhancing its stability. (C) The fate of m6A-modified RNA largely depends on the formation MLOs or BioMCs mediated by different m6A readers, which serves as an indicator for RNA metabolism direction. YTHDC1 promotes the formation of nuclear MLOs to regulate RNA transcription, splicing and export. YTHDC1 forms YT-Bodies at transcription active sites and adjacent to NSs, defining splicing factor SRSF3 location in the NSs as well as enhancing its RNA-binding affinity, thereby promoting exon inclusion. In addition, the interaction between YTHDC1 and SRSF3 contributes to RNA crowding to NXF1, which is required for nuclear methylated mRNA export. Additionally, YTHDC1 drives the formation of super enhancer BioMCs through interacting m6A-modified eRNAs and BRD4, leading to enhanced RNA transcription. HnRNPA2B1 represents another type of m6A reader that regulates miRNA processing potentially by enhancing D-bodies formation through interactions with DGCR8 and DROSHA. Cytoplasmic m6A readers including YTHDF1-3, IGF2BP1-3 and YTHDC2 are implicated in regulating RNA stabilization, degradation and translation processes. YTHDF1-3 enhances the formation of SGs upon external stimuli. After stimuli removal, certain mRNAs combined with initiation factors will transport into the ribosomes for translation. Compared with YTHDF1/3, YTHDF2 colocalizes with both SGs and PBs, primarily mediating RNA degradation via interacting with CCR4-Not complex. FMR1 promotes maternal RNA degradation by forming FMR1 granules with m6A-modified RNAs and CCR4-Not. IGF2BPs are a group of m6A readers that enhance the stability of m6A-modified mRNA and further promote their translation through forming IGF2BP BioMCs.
The engaged cell process or function of representative MLOs or BioMCs in eukaryotes.
| Localization | Name | Core LLPS protein/RNA | Specific cellular environment | Cellular processes/function | Reference |
|---|---|---|---|---|---|
| Nucleus | Nucleolus | Fibrillarin | No | Regulates rRNA transcription, processing and modification, thereby facilitating the assembly of ribosome | [20, 65] |
| Nuclear speckle | SRRM2, MALAT1 | No | Regulates pre-mRNA synthesis and splicing, mRNP maturation and RNA transport | [20, 22] | |
| Paraspeckle | NEAT1, NONO, SFPQ | No | Regulates RNA retention, transcription and stability | [20, 104] | |
| Cajal body | Coilin, SMN | No | Regulates pre-snRNA transcription, modification and snRNP assembly | [70] | |
| Gem | SMN | No | Promotes maturation of snRNPs | [20, 65] | |
| Nuclear stress body | HSATIII lncRNA, HSF1 | Thermal stress | Regulates pre-mRNA splicing upon thermal stress | [20] | |
| PML nuclear body | PML | No | Regulates chromosome association and transcription | [20, 65] | |
| D body | DCL1 | No | Promotes miRNA processing | [96] | |
| Heterochromatin condensates | HP1, MECP2 | No | Regulates chromosome maintenance, segregation and transcriptional silencing | [66] | |
| YAP condensates | YAP | Hyperosmotic stress | Activates downstream gene transcription involved in cell proliferation | [75] | |
| Super enhancer condensates | BRD4, MED1 | No | Drives robust gene transcription involved in cell identity | [76] | |
| Cytoplasm | Stress granule | G3BP1/2 | Different cell stress | Regulates RNA storage, decay and translational control upon various stress condition | [74] |
| Processing body | DCP1A, LSM14A | No | Regulates RNA storage and decay | [71, 72] | |
| P62 body | P62 | No | Ensures effective autophagic degradation of ubiquitinated proteins and damaged organelle clearance | [67] | |
| cGAS condensates | GAS | Cytoplasmic DNA arising | Activates innate immune signaling | [68] | |
| D-granules | DIAPH3 | Different cell stress | Regulates actin cytoskeletal remodeling upon various cell stress condition | [69] |
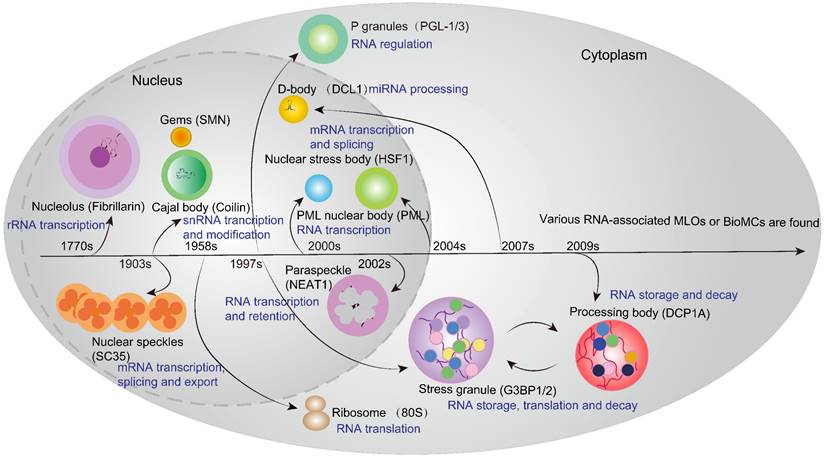
With the discovery of the nucleolus (the first identified MLO) in the 1770s, other RNA-associated MLOs, such as Cajal bodies (CBs), Gems, NSs, paraspeckles, promyelocytic leukemia protein nuclear bodies (PML-NBs) and nuclear stress body in the nucleus, as well as stress granules (SGs), processing bodies (PBs) and P granule (Caenorhabditis elegans) in the cytoplasm, have gradually been identified in eukaryotes [65] (Figure 2). Their different functions in gene regulation, especially in the RNA metabolism process, are profoundly investigated. For example, the nucleolus and CBs are involved in the biogenesis of rRNA and snRNA respectively, which contributes to the production of macromolecular machineries including ribosomes and spliceosomes. Intriguingly, these two MLOs show the close physical and functional relationship in the nucleus. CBs are the center for the maturation of small nuclear ribonucleoproteins (snRNPs), which are essential for spliceosome assembly, snRNA transcription and modification. Moreover, accumulating studies reveal that CBs are also required for the assembly of small nucleolar ribonucleoproteins (snoRNPs) that are vital for rRNA processing within the nucleolus [70]. Notably, PBs in the cytoplasm share some common components with CBs, and communicate and cooperate with each other in the regulation of RNA metabolism. PBs are cytoplasmic MLOs that play an important role in microRNA-induced silencing complex (miRISC) [71], mRNA decay and translation repression [72]. The indirect crosstalk between these two independent MLOs can be well illustrated by an example that alternative mRNA splicing and mRNA decay in normal organisms require the balance of LSM1-7 and LSM2-8 complexes in PBs and CBs, respectively [73]. It is evident that they share some LSM proteins to play a collaborative role in RNA processing and metabolism. In addition, PBs and SGs are two classical cytosolic MLOs that share some RNPs and mRNAs, and they play a crucial role in translation control and RNA storage. However, SGs only become visible under stressed conditions while PBs are constitutively observed by microscopy. Upon stress, cytosolic translation halts and mRNAs that have been bound to ribosomes or other specific proteins converge to form SGs. Meanwhile, translation-stopped mRNAs that lack binding to translation initiation factors enter the PBs [74]. Thus, MLOs may share some components and are assembled when cellular biochemical reaction or condition changes are going to happen. Additionally, it is noteworthy that certain nuclear condensates, such as the nuclear YAP droplet or BRD4 droplet, promote robust downstream gene transcription associated with cellular proliferation and differentiation [75, 76]. In addition to the classical RNA-associated MLOs, essential proteins such as cyclic GMP-AMP synthase (cGAS) and P62 can also drive the formation of MLOs, which function as sensors for cytoplasmic DNAs and ubiquitinated proteins, thereby participating in cellular immune response and autophagy [67, 68]. However, the aberrant formation or disaggregation of MLOs disrupts biochemical reaction processes and gene regulation, which will lead to the emergence of human diseases, such as Reye's syndrome [66] and Parkinson's disease [77]. Moreover, cancer cells employ some MLOs (e.g. SGs) to gain the adaptability in the harsh tumor microenvironment, thereby compromising the efficacy and efficiency of anti-cancer therapies [78]. Therefore, restoring or targeting these MLOs may emerge as a promising therapeutic strategy for human diseases.
Extensive studies have demonstrated that multiple folded domains, short linear motifs (SLiMs), IDRs and post-translational modification (PTM) of certain proteins lead to the vulnerability of LLPS to form MLOs [79-81]. Notably, post-transcriptional modification, particularly m6A modification, has a great impact on the formation of MLOs, which in turn affects RNA metabolism under both physiological and pathological conditions. In this review, we provide a comprehensive summary of recent studies on the regulation of RNA metabolism processes by m6A-associtated MLOs, with a particular emphasis on the role of m6A modification in driving LLPS in human diseases. Furthermore, we propose that m6A-associated MLOs could serve as predictive markers for disease progression and potential drug targets in the future.
M6A-associated MLOs participate in RNA metabolism processes
Approximately 25% of mRNA molecules encompass at least one m6A base [82]. Accumulating studies demonstrate that this post-transcriptional modification of mRNA exerts regulatory control over its fate, particularly during the processes of RNA transcription, splicing, export, translation and decay. How m6A modification guides these processes is poorly understood. Recently, m6A readers, including YTHDF1-3, have been identified as LLPS proteins to undergo phase separation in cells. Both the N-terminal IDR domain and the C-terminal YTH domain responsible for m6A binding are critical for BioMCs formation [17]. Under stress conditions, polymethylated mRNA, not single m6A modified mRNA, enhances phase separation by serving as a multivalent scaffold for binding to YTHDF proteins and juxtaposing their low-complexity domains [82].
Moreover, m6A modification in the nucleus is predominantly mediated by methyltransferase complex (MTC), whose assembly has been reported to be dependent on METTL3 LLPS [83]. These condensates partition into distinct endogenous MLOs, such as PBs, SGs and NSs, thereby governing the regulation of RNA metabolism [17, 82, 83]. In the following, we will provide a summary of recently found MLOs regulated by m6A modification in RNA metabolism processes (Figure 1).
History of RNA-associated MLOs in Eukaryotes. Since the discovery of the nucleolus (the first identified MLO) in the 1770s, other RNA-associated MLOs have gradually been identified in eukaryotes, such as Cajal bodies (CBs), nuclear speckles (NSs), paraspeckles, D-body, promyelocytic leukemia protein nuclear bodies (PML-NBs) and nuclear stress body in the nucleus, as well as translational machinery ribosomes, P granules, stress granules (SGs) and processing bodies (PBs) in the cytoplasm. Different MLOs can be marked by key component proteins, such as coilin for CB, SC35 for NSs, and NEAT1 for paraspeckles. These MLOs play a pivotal role in various RNA metabolism processes encompassing transcription, modification, splicing, storage, decay and translation.
RNA transcription
M6A is an important modification involved in regulating chromatin states and transcription dynamics. For instance, METTL3 labels m6A modifications to chromosome-associated regulatory RNAs (carRNAs), such as those overlapping enhancers or super-enhancers. The nuclear reader YTHDC1 specifically recognizes these m6A-marked carRNAs (mainly LINE1 repeats), and subsequently elicits their degradation through the nuclear exosome mediated pathway. Knockout of METTL3 or YTHDC1 in mouse embryonic stem cells enhances chromatin accessibility and activates transcription [84]. This study suggests that m6A modification on carRNAs exert an influence on the adjacent chromatin state and downstream transcription. Inconsistently, Lee et al. found that the m6A-labeled enhancer RNAs (eRNAs) are pervasively present in human cells. These m6A-modified eRNAs absorb the nuclear m6A reader YTHDC1 to strong enhancers, further stimulating gene activation and transcription [21]. Knockdown of YTHDC1 inhibits enhancer and gene transcription instead of altering eRNA stability. Mechanistically, m6A-eRNAs/YTHDC1 interaction facilitates LLPS and promotes the formation of transcriptional activator BRD4 condensates (Figure 1). Further experiments confirmed that arginine residues enriched in IDR2 at the C-terminal of YTHDC1 are vital for the formation of its condensate, which further enhanced the formation of BRD4 coactivator condensates [21]. This study reveals the critical role of m6A-marked eRNA condensates in crosstalk between epitranscriptome and epigenome, which are reminiscent of nuclear speckles in the nucleus [22]. However, they did not rule out whether YTHDC1 plays a role in activating gene transcription through direct binding to promoters. Besides, these studies did not establish a causal relationship between m6A levels, number of m6A sites, identity of eRNAs with condensation formation. Furthermore, their investigation did not provide a comprehensive definition of the condensates or unveil the physiological and pathological role of these m6A-associated super enhancer condensates.
Pre-mRNA splicing and miRNA processing
AS is an important process mediated by spliceosome that produces different RNA isoforms from the same pre-mRNA, which leads to the diversity of proteins involved in biological processes [85]. Progressive evidence suggests that m6A modification and its regulators play a pivotal role in the assembly of splicing condensates, as well as in the biogenesis and recruitment of splicing factors to precisely orchestrate AS [22, 86]. The study conducted by Dominissini et al. first revealed that m6A affects RNA splicing, as evidenced by differential spliced exons and introns induced by METTL3 knockdown in HepG2 cells, of which most were methylated [87]. Notably, acute deletion of METTL3 coupled with nascent RNA profiling only reveal several dozens of AS events, such as alternative 5′ splice sites (A5SS) and intron retention (RI), instead of widespread splicing defects [88], in line with the findings from recent studies showing that m6A deposition onto RNA is mostly occluded by exon-junction complex [88, 89]. Thus, nuclear splicing inversely governs m6A methylation at the proximal splice site of pre-mRNA. These findings suggest a reciprocal and synergistic relationship between m6A modification and RNA splicing. NSs within the nucleoplasm are vital MLOs that provide the reaction sites for RNA transcription, splicing and modification [22]. Notably, it has been acknowledged that NSs contain MTC core proteins, including METTL3, METTL14 and WTAP, in which splicing factor WTAP is required for recruiting METTL3-METTL14 to NSs [22]. Interestingly, METTL3 has been reported to endogenously colocalize with NSs and exhibit self-interaction capabilities, thereby promoting the formation of condensates in nucleus. The presence of nascent RNAs plays a crucial role in facilitating the opto-condensate formation of METTL3. However, mutation of SAM binding sites or treatment with an inhibitor targeting MAT2A (SAM synthetase) abolished or prevented the formation of METTL3 condensates. This study suggests that the dynamic assembly of MTC is largely dependent on METTL3 phase separation, and WTAP may regulate the size of endogenous METTL3 droplet [83]. In short, m6A modification is essential for NSs formation.
In addition, YTHDC1, a key m6A reader implicated in AS, was also identified as a pivotal component of NSs. In contrast to the NSs-promoting function of METTL3, YTHDC1 forms YT-Bodies at transcriptionally active sites and adjacent to NSs, thereby delineating the localization of splicing factor SRSF3 within NSs and enhancing its RNA-binding affinity. However, it competitively suppresses these abilities of SRSF10, thereby promoting exon inclusion and repressing exon skipping [48, 90]. Moreover, YTHDC1 interacts with three mRNA 3'-end processing factors, namely CPSF6, SRSF3, and SRSF6, to modulate alternative polyadenylation and splicing during mouse oocyte development [91]. It seems that YTHDC1 potentially regulates the recruitment of splicing factors to NSs, thereby manipulating AS. Indeed, m6A-eRNA, m6A binding domain and IDR are essential for the formation of YTHDC1 nuclear condensates [21]. Consequently, it is plausible that YTHDC1 serves as a scaffold protein for NSs formation and further studies are required to confirm this hypothesis.
Moreover, ALKBH5 and FTO, two m6A eraser proteins, are also found to colocalize with NSs [6, 7]. Deficiency of ALKBH5 affects assembly of splicing factor ASF/SF2 and SPRK1 in NSs, causing reduced phosphorylation of ASF/SF2 and disturbed pre-mRNA splicing [6, 11]. Interestingly, disturbance of ALKBH5 led to the disappearance of NSs. It is speculated that ALKBH5 may also be a scaffold protein for NS formation since the critical study has confirmed that ALKBH5 LLPS facilitates the formation of BioMCs. The demethylation activity of FTO in the nucleus has a significant impact on pre-mRNAs processing, including exon inclusion and 3'-end processing [92]. However, knockdown of FTO did not impact the number of NSs, which suggests that FTO may not be essential for NSs formation but rather serves as an adaptor protein for pre-mRNA splicing through demethylating m6A-modified transcripts and cooperating with other splicing factors in NSs. Furthermore, an unprecedented study conducted by Mauer et al. revealed that FTO selectively demethylates the N6,2′-O-dimethyladenosine modification (m6Am) on snRNAs before it happens cap hypermethylation (m7G), thereby exerting further influence on AS [93]. SnRNAs are the core spliceosome components involved in pre-mRNA splicing, necessitating a series of modifications to protect them from degradation before being incorporated into small nuclear ribonucleoproteins (snRNP) [94]. Thus, FTO may also indirectly affect AS through regulating the assembly of spliceosome.
These above-mentioned studies suggest that m6A core proteins not only regulate m6A methylation or demethylation, but also promote the formation of NSs, the assembly of splicing machinery and participate in NSs-mediated pre-mRNA splicing. NSs share some proteins with other MLOs, such as CBs, which is the assembly compartments of spliceosomal snRNPs before their relocation to NSs. Will m6A modification or regulators also impact the assembly of snRNPs in CBs?
Recent studies have also indicated that m6A is enriched in primary microRNA (miRNA) transcripts, which play a pivotal role in controlling miRNA biogenesis within the cellular nucleus [95]. Alarcon et al. found that hnRNPA2B1 is a nuclear “reader” of the m6A mark, mediating miRNA processing and AS. Knockdown of hnRNPA2B1 resulted in similar genome-wide AS events observed with METTL3 deficiency. Notably, depletion of hnRNPA2B1 led to the nuclear accumulation of specific pri-miRNAs, also resembling the effects seen with METTL3 knockdown. Mechanistic studies revealed that hnRNPA2B1 can recruit DGCR8, a component of microprocessor complex, to a subset of precursor miRNAs, thereby promoting pri-miRNA processing [23]. Heuristically, a recent study found that phase separation of SERRATE promotes the assembly of dicing bodies (D-bodies), which are the miRNA processing MLOs in Arabidopsis [96]. The low complexity domains (LCDs) present in multiple RNA-binding proteins (RBPs) promote LLPS. Indeed, hnRNPA2B1 has two RRM regions at the N-terminus and a conserved LCD at the C-terminus. Upon cellular stress conditions, hnRNPA2B1 is recruited to SGs [97]. It is fascinating to assume that hnRNPA2B1 could serve as the key driver protein for LLPS, facilitating recognition of m6A-modified pri-miRNA and interaction with the microprocessor complex, thereby portioning into condensates involved in pri-miRNA processing, such as D-bodies.
RNA export
Accumulating evidence suggests that RNA modification, particularly m6A, is intimately associated with RNA export or nuclear retention [98]. It has been reported that hyperphosphorylated ASF/SF2, a splicing factor highly enriched in multisite phosphorylation of its C-terminal RS domain, regulates pre-mRNA splicing in NSs while hypophosphorylated ASF/SF2 facilitates the association between the TAP-p15 complex and mRNA cargo to enhance mRNA export and further modulates translation initiation of bound mRNA [99-101]. As above mentioned, ALKBH5 regulates pre-mRNA splicing in NSs by promoting assembly of splicing factor ASF/SF2. Interestingly, knockdown of ALKBH5 diminished phosphorylation of ASF/SF2 and promoted the production of methylated transcripts, which facilitates mRNA export to the cytoplasm [6]. This suggests that m6A promotes RNA export and ALKBH5-mediated demethylation activity enables selective entry of pre-mRNA into NSs for processing. Upon viral infection, DDX46 recruited ALKBH5 to demethylate m6A-modified host antiviral transcripts, which consequently enhances their nuclear retention, represses cytoplasmic translation and limits interferon production [102]. Importantly, ALKBH5 has been reported to promote the assembly of paraspeckles (PSs) through LLPS and demethylating NEAT1, a scaffolding lncRNA for PSs formation, and enhancing its stability [103, 104]. Therefore, ALKBH5 may promote the nuclear retention of transcripts through demethylating m6A-modified transcripts and promoting the assembly of nuclear m6A-associated MLOs. On the contrary, YTHDC1 facilitates nuclear RNA export through recognizing or binding to m6A-modified mRNA [24, 105, 106]. Mechanistically, the interaction between YTHDC1 and SRSF3 contributes to RNA crowding to nuclear RNA export factor 1 (NXF1), which is essential for efficient export of methylated mRNAs from the nucleus [24]. This finding suggests that YTHDC1 is important for substrate RNA selection in the mRNA export pathway. In this nuclear-to-cytoplasmic process, SRSF3 functions as an adapter protein facilitating the export of methylated nuclear mRNAs bound by both YTHDC1 and NXF1, rather than acting solely as a splicing factor [24, 107]. Whether this process is carried out in m6A-associated MLOs is unknown. Given the numerous reports highlighting YTHDC1's involvement in nuclear MLOs formation, particularly in RNA transcription and splicing via LLPS, it is intriguing to investigate its potential contribution towards the assembly of RNA transport condensates.
RNA degradation
Recent studies have demonstrated that m6A modification plays a crucial role in governing mRNA degradation [108, 109]. For decades, the complex of YTHDF proteins and m6A-modified mRNAs have been reported to partition into intercellular condensates through LLPS, leading to the formation of cytoplasmic MLOs such as PBs and SGs. PBs are enriched with deadenylase, decapping complex and 5'-3' exoribonuclease, indicating their crucial role in post-transcriptional regulation [72]. Evidence indicates that PBs and SGs present “docked” against each other during the assembly of SGs, despite being considered distinct MLOs. This intriguing presentation is a result of the sharing RBPs and mRNAs shuttling between PBs and SGs [74]. In particular, YTHDF2 presents the versatile ability to partition into different subcellular compartments, such as PBs under normal conditions and SGs during cellular stress [82]. Functionally, YTHDF2 serves as the prototypical m6A reader that actively engages in two canonical mRNA decay pathways. YTHDF2 contains a C-terminal RNA-binding domain known as the YTH domain, for binding to m6A-modified mRNA, and a P/Q/N-rich N terminus (N-YTHDF2), responsible for directing the localization of YTHDF2-mRNA complex to cellular RNA decay sites such as PBs [110]. In detail, YTHDF2 promotes the decay of its mRNA targets through recruiting CCR4-NOT deadenylase complex [111]. Interestingly, HRSP12, a 14.5 kDa translational inhibitor protein, has been recognized as an adaptor to connect YTHDF2 with RNase P/MRP (endoribonucleases), which subsequently elicits the rapid degradation of YTHDF2-bound RNAs. If m6A-modified RNAs possess HRSP12 binding sites upstream and downstream YTHDF2-binding sites, along with RNase P/MRP-directed cleavage sites, these RNAs will undergo preferential degradation through the RNase P/MRP-mediated endoribonucleolytic pathway coupled to the CCR4-NOT complex-mediated deadenylation pathway. Despite lacking HRSP12 binding sites, YTHDF2 may still interact with HRSP12 and trigger both decay pathways less efficiently [46]. Nevertheless, deadenylation is known to precede PB formation, and subsequent 3'-5' exoribonuclease cleavage in exosome is also carried out outside PBs. Thus, the remaining RNA intermediates may be processed by decapping reaction and 5'-3' degradation in PBs. Besides involvement in RNA decay, PBs are also identified as a cytoplasmic hub for untranslated RNA storage [72, 112]. Recently, YTHDFs were found to synergistically destabilize mRNAs, irrespective of their m6A modification. Evidently, the triple knockdown of all three members of YTHDF proteins leads to an increased number of PBs and enhanced mRNA stability, independent of methylation status [113]. Consequently, these proteins also serve a non-redundant role as scaffolds in maintaining RNA granule formation.
Early embryonic development depends on maternal RNAs and proteins since the zygotic genomes are transcriptionally silent [114]. Fragile X mental-retardation protein (FMR1), a ribosome-associated RBP, is present in several cytoplasmic RNP granules, such as PBs, stress granules and neuronal granules, which was found to participate in embryogenesis [54]. In Drosophila, FMR1 exhibits a preferential binding affinity towards m6A-modified mRNAs, which induces FMR1 granule condensation through recruiting numerous unmodified mRNAs. Specifically, the LC domain initiates the self-assembly of FMR1 BioMCs while KH domains enhance affinity for capturing m6A-modified mRNAs. Importantly, these dynamic m6A-associated FMR1 BioMCs facilitate degradation of maternal RNA and contribute to normal development [54].
Collectively, MLOs or BioMCs formed by YTHDFs, FMR1 and m6A-modified transcripts may provide sites for efficient degradation of redundant RNAs or storage of crucial RNAs under physiological conditions. However, cancer cells can benefit from these “RNA shredders” through promoting the decay of tumor suppressor genes or storing oncogenic transcripts.
RNA translation
Accumulating studies suggest that m6A modification on RNA translation is multifaceted and complicated, such as recruiting translation initiation factors and interacting with them, promoting ribosome biogenesis, SGs and PBs formation [38, 115, 116]. For example, METTL16 colocalizes with the nucleolus, a phase-separated nuclear condensate, and regulates rRNA maturation likely through methylating pre-rRNA prior to processing, which is required for ribosome biogenesis (Figure 1). Moreover, METTL16 interacted with specific nucleolar proteins, including NOLC1, TCOF, UBF1/2 and fibrillarin. Knockout of METTL16 resulted in a reduction in the number of nucleoli, indicating its essential role in nucleolus formation [38, 117]. However, it remains unconfirmed whether METTL16 can drive nucleoli formation through LLPS due to its possession of multiple RNA-binding sites and a long IDR.
As abovementioned, YTHDF2 primarily participates in RNA decay mediated by PBs. Interestingly, YTHDF1 and YTHDF3 may selectively promote translation through binding to m6A motif. SGs are a group of RNP granules formed under various stress conditions, such as chemicals, UV light, heat or oxygen radical stimulation, which function in controlling RNA translation and degradation [20]. Recent studies have revealed that m6A-modified RNAs are also enriched in SGs, and m6A-binding YTHDFs enhance the formation of SGs [17]. All three YTHDF proteins are found to colocalize with G3BP1, a classical molecular marker of SGs. However, due to significant differences in their protein sequence within LCDs, YTHDF1/YTHDF3 and YTHDF2 form distinct RNA granules, as evidenced by the co-localization of YTHDF2 with both SGs and PBs. Knockdown of YTHDF1/3, but not YTHDF2, resulted in a substantial reduction in the number of SGs, along with significantly diminished m6A and polyA signals within these granules [17]. This finding suggests that YTHDF1 and YTHDF3 may exert selective control over RNA translation in comparison to YTHDF2. Consistently, growing studies show that YTHDF1 and YTHDF3 facilitate translation of many m6A-modified mRNAs, such as EIF3C, ATG2A, ATG14 and FZD7 during tumorigenesis [118-120]. In the cytosol, mRNAs undergo dynamic categorization into translation and non-translation pools. A recent study conducted by Shan et al. has revealed that m6A modification is an important element switching mRNAs from polysome to PBs, which was mediated by IGF2BP3, a crucial m6A reader in the cytoplasm [25]. They also confirmed that decreased m6A modification has no impact on the number of PBs, but it affects mRNA trafficking into PBs. IGF2BP3 directly interacts with core proteins DDX6 and 4E-T in PBs, as well as with TIAR, a core protein in SGs [25]. Therefore, IGF2BP3 may facilitate the assembly of m6A-assocaited MLOs involved in mRNA storage and stability during cellular stress. When stress is “unlocked”, some mRNAs bound to initiation factors can be transported into ribosomes for translation. In conclusion, m6A-associated SGs can be a vital translation adaptor under cell stress conditions.
The role of m6A-associated MLOs in human diseases
It is widely known that maladjustment of RNA metabolism due to aberrant post-transcriptional modification will lead to the emergence of multiple human diseases, including respiratory, neurological, reproductive diseases and tumorigenesis. As abovementioned, under stable or stressful conditions, m6A-modified mRNA is required for the assembly of biomolecular condensates involved in various RNA metabolism processes. Noteworthily, certain MLOs or BioMCs that are induced by aberrant m6A levels or regulators (writers, readers and erasers) have been progressively recognized as pivotal factors driving diverse diseases (Table 2).
Tumorigenesis
Recently, Cheng et al. found that m6A is critical for YTHDC1 to drive LLPS and promotes the formation of nuclear YTHDC1-m6A condensates (nYACs). Compared to normal hematopoietic stem and progenitor cells, there was a significant upregulation in the number of nYACs observed in acute myeloid leukemia (AML) cells [26]. Functional experiments have confirmed that nYACs are essential for AML cell survival and differentiation control. Further experiments confirmed that both IDR domains and m6A-binding regions of YTHDC1 are vital for nYACs to exert their oncogenic function in AML. Interestingly, 40% and 35% of nYACs were identified as NSs and super-enhancer condensates respectively, which consequently govern RNA abundance and indirect RNA splicing. Mechanistically, nYACs repressed the PAXT-mediated nuclear decay of m6A-modified mRNAs, such as Myc and GINS1 [26], which coincides with the fact that YTHDC1 recognizes m6A-modified RNA transcripts in the nucleus and promote their stability and transport [24]. Thus, these nuclear bodies induced by m6A-modified mRNA and YTHDC1 LLPS are crucial regulators for tumor cell survival and differentiation repression (Figure 3). However, the clinical significance of nYACs in cancer patients remains undisclosed.
In addition, RNA-binding motif protein 15 (RBM15) has been identified as a component of the MTC and functions as a mediator for METTL3 recruitment. It also forms substantial nuclear condensates that exhibit dynamic fusion and fission characteristics [121]. Ectopic expression of RBM15 promotes the proliferation and oncogenic transformation of NIH3T3 cells both in vivo and in vitro. Intriguingly, RBM15 condensates exhibit colocalization with abundant m6A signals, partially interacting with or being embedded into the NSs, while showing no association with other nuclear MLOs, including PML bodies, CBs, paraspeckles, nucleoli and chromatin.
The role and mechanism of m6A-associated MLOs in human diseases. NA represents not available.
| M6A regulator | Protein domain driving LLPS | MLOs | Subcellular location | Disease | Function | RNA metabolism processes | Reference |
|---|---|---|---|---|---|---|---|
| YTHDC1 | N-terminal IDRs and C-terminal YTH domain | YTHDC1-m6A condensates | Nucleus | Acute myeloid leukemia | Regulate cell growth and differentiation control | Protect oncogenic transcripts from decay by the PAXT complex | [26] |
| RBM15 | RRM domain, IDRs at N-and SPOC at C-terminal | RBM15-m6A-condensates | Nucleus | Tumorigenesis | Promote oncogenic transformation of normal cell | Promote the stability of oncogenic transcripts, such as STYK1 | [121] |
| IGF2BP1 | M6A binding domain | HPV E7-IGF2BP1 condensates | Cytoplasm | HPV-associated cancers | Promote cell growth and migration | Stabilize HPV E7 mRNA | [122] |
| IGF2BP1 | KH3/4 domain | RUNX-IT1 -IGF2BP1-GPX4 condensates | Cytoplasm | Breast cancer | Promote tumorigenesis | Stabilize GPX4 mRNA | [124] |
| YTHDF3 | M6A binding domain | SGs | Cytoplasm | Prostate cancer | Promote stress adaptive cell survival | Regulate the translation of AR mRNA | [127] |
| YBX2 | CSD domain and mRNA binding domain | YBX1-YTHDF2 condensates | Cytoplasm | Endometrial cancer | Delay the cell proliferation and may promote chemotherapy resistance | Promote the degradation of HSPA6 mRNA | [128] |
| ALKBH5 | NA | NSs | Nucleus | Men infertility | Deficiency of ALKBH5 leads to the impaired fertility | Promote accurate RNA splicing and impact RNA export | [6, 11] |
| YTHDC1 | NA | Large RNA-containing granules | Cytoplasm | Developmental defect | Deficiency of YTHDC1 leads to impaired gametogenesis and embryogenesis | Regulate alternative polyadenylation and splicing | [91] |
| HnRNPA2B1 | Prion-like domain and m6A-binding region | Stress granules (oTau-hnRNPA2B1-m6A condensate) | Cytoplasm | AD | Promote tau fibrillization, nuclear membrane disruption, and progressive neurodegeneration. | Control RNA translation | [27] |
| TDP43/YTHDF2 | NA | TDP43-YTHDF2-m6A condensates | Cytoplasm | ALS | Promote TDP43-mediated toxicity | Enhance RNA destabilization | [138] |
| TLS/FUS | IDR domain | TLS/FUS condensates | Cytoplasm | ALS | Promote the cell viability | M6A-modified short RNA fragments inhibit the LLPS of TLS/FUS | [141] |
| YTHDF1 | IDR domain and prion-like domain | SGs | Cytoplasm | Asthma | Promote airway inflammatory responses | Enhance the translation of CLOCK mRNA | [144] |
| METTL14 | Thr72 phosphorylation at IDR domain | METTL14 condensates | Nucleus | Tuberculosis | Inhibit intracellular survival of M. tuberculosis | Enhance m6A modification of Nox2 | [142] |
Newly found m6A-associated MLOs or BioMCs during tumorigenesis. Nuclear m6A-assocaited MLOs, including nYACs and RBM15 condensates, as well as cytoplasmic IGF2BP1 condensates and SGs, play an oncogenic role in human cancers. nYACs protect oncogenic transcripts c-Myc and GINS1 from nuclear PAXT-exosome mediated RNA decay, thus promoting cancer cell survival in AML. RBM15 recruits METTL3 to methylation compartments through LLPS to methylate STYK1 and enhance the stability of STYK1, thereby inducing oncogenic transformation of NIH3T3 cells. Cytoplasmic m6A-HPV E7 mRNA-IGF2BP1 condensates enhance the stability of HPV E7 mRNA, promoting cell growth and migration in HPV-associated cancers. In addition, lncRNA RUNX-IT1 can directly bind to IGF2BP1 and promote IGF2BP1 condensates formation, leading to increased occupancy on m6A-modified GPX4 mRNA and enhancing its stability, thereby inhibiting ferroptosis in breast cancer. SGs induced by AR pathway inhibition (ARPI) play a cell-protective role in prostate cancer. Upon ARPI stress, m6A-modified AR mRNA undergoes LLPS with YTHDF3 while unmodified AR mRNA undergoes phase separation with G3BP1.Upon resolution of stress conditions, translation of AR mRNA occurs regulated by YTHDF3.
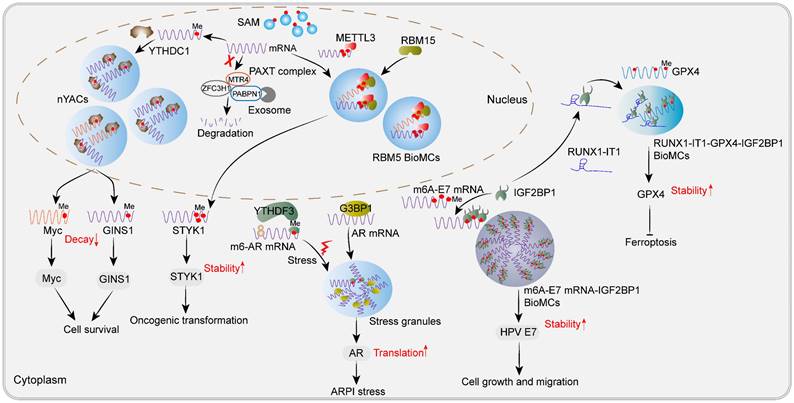
Though RRM domain, IDRs in the N-terminal and SPOC in the C-terminal truncates tend to form smaller droplets, concrete domains responsible for RBM15 LLPS have not been dissected yet. Subsequent proteomics, transcriptomics and RNA methylomics experiments indicated that RBM15 condensates may represent a novel class of subnuclear MLOs involved in facilitating m6A deposition on oncogenic transcripts, such as serine/threonine/tyrosine kinase 1 (STYK1), thereby enhancing their stability [121]. Other known stability enhancers for m6A-modified transcripts, IGF2BP3 and YTHDC1, were also found in RBM15 condensates [121]. Collectively, YTHDC1 or RBM15 condensates dispersed in the nucleus have made great contributions to the stabilization of the oncogenic transcripts, thereby promoting tumorigenesis, and future anti-tumor strategies can inhibit the formation of these condensates.
Excitingly, inhibiting some m6A-associated BioMCs showed good efficacy in cancer cells and mouse allogeneic tumor model [122]. Human papillomavirus (HPV)-induced carcinogenesis relies heavily on virus early protein 7 (E7), which makes E7 a promising drug therapy target [123]. Recently, Wang et al. found that m6A-modified E7 mRNA-IGF2BP1 condensates, which are oncogenic in nature, stabilize E7 mRNA but exhibit thermal instability and form insoluble aggregates. Clearly, m6A modification plays a crucial role in facilitating the stability of E7 mRNA and enhancing the assembly of oncogenic condensates through binding to IGF2BP1, as demonstrated by the negligible impact on heat-induced condensate formation upon deletion of IDR region of IGF2BP1. Upon prolonged heat stress treatment, these condensates undergo a phase transition and form insoluble aggregates, which are subsequently degraded by the ubiquitin-proteasome system. Noteworthily, heat stress selectively destabilizes E7 mRNA and downregulates E7 protein expression without affecting other m6A-modified transcripts such as CREBBP, RAB21, and FBXO41. Significantly, daily heat treatment inhibited the malignancy of HPV16-positive cells both in vitro and in vivo through lowering E7 mRNA expression along with IGF2BP1 [122]. Thus, targeting m6A-E7 mRNA-IGF2BP1 condensates could be a promising therapeutic strategy for HPV-associated cancers, highlighting the clinical potential of anti-tumor therapy by focusing on m6A-associated MLOs. Most recently, Wang et al. found that RUNX1 intronic transcript 1 (RUNX1-IT1), known as a tumor driver lncRNA in breast cancer, directly interacts with IGF2BP1 and partitions into BioMCs, thereby occupying and stabilizing glutathione peroxidase 4 (GPX4) mRNA, and then promoting GPX4 protein expression while repressing ferroptosis [124]. Rapid clustering of IGF2BP1 has been recently identified as an initiator for SGs assembly upon osmotic shock. The KH3/4 di-domain and an IDR of IGF2BP1 have been confirmed to play crucial roles in mediating this clustering process [125]. Deletion of the KH3/4 domain led to the disappearance of these IGF2BP1 condensates [124]. However, the specific binding region between IGF2BP1 and RUNX1-IT1 remains unclear. Overall, it is proposed that IGF2BP1 acts as a phase-separated protein driving the formation of m6A mRNA-stabilized MLOs during tumorigenesis, and future anti-tumor therapy may target these IGF2BP1 condensates through inhibiting IGF2BP1 LLPS or dissolving them. Further studies can isolate these MLOs for more comprehensive RNA and proteomic analysis to identify additional potential targets.
Androgen receptor (AR) pathway inhibition (ARPI) triggers strong and lasting responses in advanced prostate cancer [126]. Recently, Syam et al. found that m6A-modified AR mRNA undergoes liquid-liquid phase separation with YTHDF3, while unmodified AR mRNA undergoes phase separation with G3BP1 in response to ARPI-induced stress in prostate cancer (PCA). These processes facilitate the formation of SGs and drive rapid adaptive changes that confer protection to PCA cells against ARPI-induced stress by regulating the translation of AR mRNA. Inhibiting this adaptive response through knockdown of YTHDF3 or G3BP1 makes PCA cells vulnerable to ARPI stress [127]. This study suggests that inhibiting m6A modification of AR mRNA or YTHDF3 could serve as potential therapeutic strategies for ARPI-based therapies in PCA. In contrast to the translational regulatory role of YTHDF3, Y-box binding protein 2 (YBX2) forms stable cytoplasmic condensates through recruiting YTHDF2 and promotes the degradation of m6A-modified HSPA6 mRNA in endometrial adenocarcinoma-derived Ishikawa cells. The cold-shock domain (CSD) and mRNA-binding domain of YBX2 are responsible for recruiting both YTHDF2 and mRNA to LLPS [128]. These cytoplasmic condensates may protect cancer cells from chemotherapy.
Collectively, m6A-associated MLOs, including nYACs, RBM15 condensates, IGF2BP1 condensates, SGs and YBX2-YTHDF2 condensates, ensure stable oncogenic mRNA metabolism by regulating RNA decay, nuclear export, stability and translation in cancer cells (Figure 3). Some therapeutics, such as targeting the m6A regulator, dissolving the condensates or inhibiting m6A modification of specific mRNA, could be explored as promising anti-cancer strategies.
Reproductive diseases
Recently, an increasing body of literature has emphasized the crucial role of m6A in RNA metabolism of reproductive systems, including embryo implantation [129], spermatogenesis [11] and embryonic development [12]. For example, male mice with ALKBH5 knockout exhibited elevated levels of m6A in mRNA, which was associated with reduced fertility attributed to compromised spermatogenesis. Mechanistically, ALKBH5 regulates mRNA export, stability and the assembly of RNA splicing factor in NSs [6]. The demethylation activity of ALKBH5 and its interaction with splicing factors are crucial determinants of ALKBH5's role in regulating RNA metabolism in NSs. Demethylation activity of ALKBH5 in the NSs of spermatocytes and round spermatids is also necessary for accurate splicing and the production of longer 3'-UTR mRNAs [11]. Overall, we suspect that maintaining physiological level of ALKBH5 is essential for RNA metabolism in germ cells; however, aberrant dysfunction of ALKBH5 may lead to male infertility. Nevertheless, there is currently a lack of clinical evidence supporting this hypothesis and the specific protein regions responsible for positioning ALKBH5 in NSs have not been dissected yet.
The m6A modification and enzymes also play a critical role in normal embryogenesis and maternal RNA degradation. YTHDC1, a well-known protein involved in LLPS, forms nuclear granules to regulate RNA metabolism. Nuclear YTHDC1 has been confirmed to regulate alternative polyadenylation and splicing through interacting with CPSF6, SRSF3 and SRSF7, which is crucial for mouse embryo viability and germline development. However, knockout of YTHDC1 in oocytes leads to the formation of large novel RNA-containing granules that may comprise abnormal processed transcripts [91]. Like PBs, these cytoplasmic granules may serve as a repository for misprocessed RNA transcripts. Indifferently, in Drosophila, loss of maternal FMR1, a novel m6A reader, not YTHDF or YTHDC, resulted in embryonic lethality. Mechanistically, FMR1 could form dynamic condensates with m6A-modified RNA molecules, which controls maternal mRNA decay and translation in an m6A-dependent manner [128]. Thus, these m6A-associated BioMCs play a crucial role in the processes of gametogenesis and embryogenesis, and ALKBH5 and YTHDC1 are indispensable for RNA processing mediated by NSs in mammals.
In summary, m6A-modified mRNA facilitates the formation of MLOs associated with correct RNA metabolism in reproductive organs. However, aberrant expression levels of m6A writer, reader or eraser proteins may affect the formation and function of these m6A-associated MLOs, leading to infertility or embryonic lethality. Thus, modulating m6A levels to restore the integrity of m6A-associted MLOs could represent a promising strategy for addressing infertility or promoting embryo survival. However, the lack of clinical significance makes this hypothesis a long way from being realized.
Neurological diseases
Aberrant expression of YTHDFs and disturbed m6A modification have been documented to contribute to the pathogenesis of different human neurological disorders [130]. Given the pivotal role of YTHDFs in SGs formation, it is meaningful to elucidate whether SGs triggered by a combination of YTHDFs and stress conditions exert an impact on disease processes. It is widely acknowledged that the microtubule-associated protein tau plays a crucial role in maintaining the stability of microtubules in healthy homeostasis. However, tau oligomerization and hyperphosphorylation accumulate within the somato-dendritic arbor upon stress or disease challenging, which accelerates the progression of tauopathy, such as Alzheimer's disease (AD) [131]. Tau oligomerization propagates tau pathology and exhibits extensive co-localization with cytoplasmic TIA1-positive SGs, as evidenced by co-localization with TIA1, PABP and eIF3η [132]. Increasing evidence indicates that LLPS and protein aggregates play pivotal roles in SGs formation, particularly in neurodegenerative diseases like tauopathy. Furthermore, RBPs such as TDP-43 (TAR-binding protein of 43 kDa), TLS/FUS (Translocated in LipoSarcoma/Fused in Sarcoma) and hnRNPA2B1, are known to translocate from the nucleus to the cytoplasm and localize within SGs [133]. Among them, a prion-like domain (LCD) mutation of hnRNPA2B1, an indirect m6A reader, has been reported to enhance aggregation and drive itself to be sequestrated in SGs in multisystem proteinopathy (MSP). Notably, hnRNPA2B1 is merely located in the nucleus in normal muscle but it accumulates at cytoplasmic inclusions in some muscle fibers of MSP patients [134, 135]. Thus, hnRNPA2B1 may participate in SG formation through LLPS in human neurological diseases.
Recently, Jiang et al. found that light-induced tau oligomerization (o-tau) promotes the assembly and neurotoxicity of SGs, concomitant with the cytoplasmic translocation of hnRNPA2B1 in cultured neurons. Further mechanistic experiments confirmed that tau phosphorylation enhances its interaction with hnRNPA2B1 via accelerating oligomerization. HnRNPA2B1 was proved to be the key bridge protein connecting o-tau and m6A modified transcripts. Evidently, knockdown of hnRNPA2B1 drastically attenuated the formation of SGs, as suggested by reduced PABP and EIF3η, and ameliorated tau-mediated neurodegeneration in both neurons and tau mice, as suggested by decreased levels of o-Tau, m6A and cleaved caspase-3. Thus, the oTau-hnRNPA2B1-m6A complex enhances SGs formation and regulates the translational response to stress conditions. Importantly, hnRNPA2B1 was found to co-localize with both o-tau and cytoplasmic m6A in human AD samples, indicating the clinical significance for m6A-associated SGs [27]. However, the specific region within hnRNPA2B1 that drives toxic SG formation remains elusive. The LCD in C-terminal and RNA-binding domain may be responsible for SGs formation in AD. Further investigations are required to validate this hypothesis.
TDP43, an hnRNP, is also a nuclear RNA-binding protein that regulates RNA metabolism. However, cytoplasmic mislocalization and aggregation of TDP43 are frequently presented in neurons of amyotrophic lateral sclerosis (ALS) patients [136, 137]. Recently, Michael McMillan and coworkers found a significant enrichment of m6A-modified transcripts in post-mortem samples obtained from ALS patients. Interestingly, methylated RNAs and YTHDF2 foci are found to be colocalized with TDP43 pathology in the spinal cord and frontal cortex of ALS patient, suggesting that aberrant cytoplasmic aggregates of TDP43 may predominantly influence m6A-marked RNA metabolism. Furthermore, knockout of Ythdf2 alleviated TDP43-related neural toxicity in rodents, whereas overexpression of Ythdf2 led to fatal outcomes [138]. In TDP43-overxpressing human iPSCs-derived neuron, a wide RNA destabilization was observed. Considering the recognition of m6A-labeled RNA in PBs and its subsequent degradation mediated by YTHDF2, it is worth investigating whether TDP43-binding m6A transcripts recognized by YTHDF2 undergo degradation in PBs. Moreover, whether YTHDF2 enhances LLPS of TDP43-m6A binding complex is related to the instability of neuroprotective transcripts requires further clarification.
In addition, aberrant cytoplasmic aggregation of TLS/FUS, a nuclear RNA-binding protein, is also implicated in the pathogenesis of ALS. Previous studies have emphasized that post-translational modifications, such as arginine methylation and phosphorylation of TLS/FUS's IDR region, influence LLPS and subsequent aggregation [139, 140]. Unexpectedly, Ryoma et al. proposed that TLS/FUS might serve as a potential m6A reader candidate even though it does not contain YTH domain. They further found that m6A modified short RNA fragments effectively suppress LLPS of TLS/FUS or its mutants. Additionally, treatment with m6A modified short RNA treatment derived from pncRNA-D obviously attenuated cytoplasmic TLS/FUS foci induced by sorbitol, a type of hyperosmotic stress, in cultured neural cells, thereby promoting cell viability. Moreover, there was a slight reduction in SGs, as TLS/FUS also exhibited colocalization with SGs [141]. This study is reminiscent of the fact that polymethylated mRNA promotes LLPS rather than a single m6A methylated mRNA [82]. Thus, treatment with m6A modified short RNA fragments may be a strategy to inhibit MSP. Further studies are required for investigating the therapeutic efficacy of m6A-modified RNA fragments both in vitro and in vivo.
Respiratory diseases
Accumulating evidence indicates that m6A modification is involved in anti-viral or -bacterial immune response [13, 142, 143]. Notably, RNA metabolism associated with m6A in some respiratory diseases also depends on the formation of MLOs. In allergic airway inflammation, allergens promote the LLPS of YTHDF1 and trigger the formation of a complex comprising dimeric YTHDF1 and CLOCK mRNA, which localizes within SGs. This stress adaptive process induced by YTHDF1 LLPS enhances the translation of CLOCK mRNA, thereby promoting the generation of NLRP3 inflammasome and secretion of IL-1β, leading to airway inflammatory responses [144]. This study suggests that YTHDF1 condensates play a vital role in the pathogenesis of allergic airway inflammation through regulating CLOCK mRNA translation. Moreover, aberrant upregulation of YTHDF1 was observed in asthma patients compared to control subjects [144], therefore targeting YTHDF1 condensates may be a clinical strategy for improving allergic airway inflammation.
Mycobacterium tuberculosis (M. tuberculosis) is a highly successful intracellular pathogen responsible for causing tuberculosis (TB), resulting in 10 million active cases and 1.5 million deaths annually [145]. Upon M. tuberculosis infection, METTL14 LLPS promoted the formation of MTC complex and mediated the methylation of NOX2 mRNA in macrophages. However, EsxB, a M. tuberculosis-specific secretory protein encoded by an RD-1 region, repressed METTL14 LLPS through inhibiting p38-mediated phosphorylation of Thr72 within the IDR domain of METTL14, thereby disrupting the m6A methylation of NOX2 mRNA and its subsequent interaction with IGF2BP1, leading to NOX2 mRNA instability. Consequently, this results in increased intracellular survival ratio of M. tuberculosis by diminishing ROS levels. Importantly, enhanced T72 phosphorylation of METTL14 was clinically associated with Nox2 mRNA expression level and ROS production in primary TB patients [142]. Therefore, METTL14 condensates in macrophages may contribute to the eradication of M. tuberculosis through catalyzing m6A methylation.
Concluding remarks and prospect
Over the past few decades, a growing body of evidence has suggested that m6A modification plays an essential role in the physiological and pathological processes of eukaryotes. Importantly, m6A-modified RNA and its regulators such as METTL3, YTHDF1-3 and YTHDC1 were proven to be enhancers for MLOs formation. These m6A-associated MLOs run through the life of mRNA, engaging in RNA transcription, splicing, export, decay and translation; however, maladjustment of which causes reduced fertility, enhanced cancer cell growth, tauopathy and respiratory disorders. In parallel, disorders of RNA metabolism will follow, encompassing aberrant alternative splicing, subcellular mislocalization of RNAs and augmented stability of oncogenic transcripts. Although many studies have primarily demonstrated that m6A modification in different human diseases, it is imperative to acknowledge that m6A-associated MLOs serve as pivotal biochemical reaction sites and exert substantial regulatory roles in disease progression. Thus, m6A-associated MLOs may be clinical diagnostic markers and attractive drug targets despite the lack of authoritative reports establishing a direct correlation between m6A-associtaed MLOs and disease severity. Mutation of LCD in m6A regulators, such as hnRNPA2B1, results in ectopic production of SGs, which leads to multisystem proteinopathy. It is meaningful to investigate the correlation between the mutation of m6A regulators in the LCD and related human diseases, particularly cancer. Alteration of m6A modification through targeting the LLPS driven region in m6A writer, reader or eraser may be an effective strategy for correcting disorders of RNA metabolism induced by m6A-associated MLOs in human diseases.
Abbreviations
AD: Alzheimer's disease; ALS: amyotrophic lateral sclerosis; AML: Acute myeloid leukemia; AR: Androgen receptor; ARPI: Androgen receptor pathway inhibition; AS: Alternative splicing; BioMCs: Biomolecular condensates; carRNAs: Chromosome-associated regulatory RNAs; CBs: Cajal bodies; CSD: Cold-shock domain; D-bodies: Dicing bodies; E7: Virus early protein 7; EIF3: Initiation translation factor 3; eRNAs: enhancer RNAs; FMR1: Fragile X mental-retardation protein 1; GPX4: Glutathione peroxidase 4; hp1: Hairpin1; HPV: Human papillomavirus; IDRs: Internally disordered regions; LCD: Low complexity domain; LLPS: Liquid-liquid phase separation; M. tuberculosis: Mycobacterium tuberculosis; m6A: N6-methyladenosine; miRISC: microRNA-induced silencing complex; MLOs: Membraneless organelles; MSP: Multisystem proteinopathy; MTC: Methyltransferase complex; ncRNAs: Noncoding RNAs; NSs: Nuclear speckles; NXF1: Nuclear RNA export factor 1; nYACs: Nuclear YTHDC1-m6A condensates; O-tau: Tau oligomerization; PAXT: PolyA tail exosome targeting complex; PBs: Processing bodies; PCA: Prostate cancer; PML-NBs: Promyelocytic leukemia protein nuclear bodies; PTM: Post-translational modification; RBM15: RNA-binding motif protein 15; RBPs: RNA-binding proteins; RUNX1-IT1: RUNX1 intronic transcript 1; SAM: S-adenosylmethionine; SGs: Stress granules; SLiMs: Short linear motifs; snRNPs: Small nuclear ribonucleoproteins; TB: Tuberculosis; TDP-43: TAR-binding protein of 43 kDa; TLS/FUS: Translocated in LipoSarcoma/Fused in Sarcoma; YBX2: Y-box binding protein 2; YTH: YT521-B homology.
Acknowledgements
The present work was supported by grants from "Pioneer" and "Leading Goose" R&D Program of Zhejiang Province (2022C03004); Zhejiang Provincial Natural Science Foundation of China (LQ24H160029 and LR21H160001); National Natural Science Foundation of China (82072646 and 82372664); The joint foundation of National Administration of Traditional Chinese Medicine & Zhejiang Province - major project (GZY-ZJ-KJ-24045); Start-up grant of HZNU (No.4275C50223204036) and National program for the cultivation of academic subjects (No.4275C52023106).
Author contributions
F.T.B drafted and amended the manuscript and tables; H.Y.W contributed to the figure design, literature collection and manuscript checking; C.X, K.L.S, D.Z and L.T.W contributed to the literature collection, writing and checking of manuscript; J.X.C, F.T.B and L.Y conceived and designed the review, and contributed to the manuscript revision. All authors approved the final version of the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E. et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022;50:D231-D235
2. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187-1200
3. Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15:313-326
4. Liu JZ, Yue YN, Han DL, Wang X, Fu Y, Zhang L. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93-95
5. Han L, Dong L, Leung K, Zhao Z, Li Y, Gao L. et al. METTL16 drives leukemogenesis and leukemia stem cell self-renewal by reprogramming BCAA metabolism. Cell Stem Cell. 2023;30:52-68 e13
6. Zheng GQ, Dahl JA, Niu YM, Fedorcsak P, Huang CM, Li CJ. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29
7. Jia GF, Fu Y, Zhao X, Dai Q, Zheng GQ, Yang Y. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-887
8. Sikorski V, Selberg S, Lalowski M, Karelson M, Kankuri E. The structure and function of YTHDF epitranscriptomic m(6)A readers. Trends Pharmacol Sci. 2023;44:335-353
9. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-295
10. Zhao YC, Shi YF, Shen HF, Xie WZ. m6A-binding proteins: the emerging crucial performers in epigenetics. Journal of Hematology & Oncology. 2020;13:35
11. Tang C, Klukovich R, Peng H, Wang Z, Yu T, Zhang Y. et al. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3'-UTR mRNAs in male germ cells. Proc Natl Acad Sci U S A. 2018;115:E325-E333
12. Wei J, Yu X, Yang L, Liu X, Gao B, Huang B. et al. FTO mediates LINE1 m(6)A demethylation and chromatin regulation in mESCs and mouse development. Science. 2022;376:968-973
13. Li N, Hui H, Bray B, Gonzalez GM, Zeller M, Anderson KG. et al. METTL3 regulates viral m6A RNA modification and host cell innate immune responses during SARS-CoV-2 infection. Cell Rep. 2021;35:109091
14. Chen JY, Fang YW, Xu Y, Sun HT. Role of m6A modification in female infertility and reproductive system diseases. Int J Biol Sci. 2022;18:3592-3604
15. Shafik AM, Zhang FR, Guo ZX, Dai Q, Pajdzik K, Li YP. et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer's disease. Genome Biol. 2021;22:17
16. Xu ZJ, Peng B, Cai Y, Wu GT, Huang JZ, Gao M. et al. N6-methyladenosine RNA modification in cancer therapeutic resistance: Current status and perspectives. Biochem Pharmacol. 2020;182:114258
17. Fu Y, Zhuang X. m(6)A-binding YTHDF proteins promote stress granule formation. Nat Chem Biol. 2020;16:955-963
18. Li J, Chen K, Dong X, Xu Y, Sun Q, Wang H. et al. YTHDF1 promotes mRNA degradation via YTHDF1-AGO2 interaction and phase separation. Cell Prolif. 2022;55:e13157
19. Su YF, Maimaitiyiming Y, Wang LF, Cheng XD, Hsu CH. Modulation of phase separation by RNA: A glimpse on N6-methyladenosine modification. Front Cell Dev Biol. 2021;9:786454
20. Hirose T, Ninomiya K, Nakagawa S, Yamazaki T. A guide to membraneless organelles and their various roles in gene regulation. Nat Rev Mol Cell Bio. 2023;24:288-304
21. Lee JH, Wang R, Xiong F, Krakowiak J, Liao Z, Nguyen PT. et al. Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation. Mol Cell. 2021;81:3368-3385
22. Galganski L, Urbanek MO, Krzyzosiak WJ. Nuclear speckles: molecular organization, biological function and role in disease. Nucleic Acids Res. 2017;45:10350-10368
23. Alarcon CR, Goodarzi H, Lee H, Liu XH, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299-1308
24. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y. et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017;6:e31311
25. Shan T, Liu F, Wen M, Chen Z, Li S, Wang Y. et al. m(6)A modification negatively regulates translation by switching mRNA from polysome to P-body via IGF2BP3. Mol Cell. 2023;83:4494-4508
26. Cheng Y, Xie W, Pickering BF, Chu KL, Savino AM, Yang X. et al. N(6)-Methyladenosine on mRNA facilitates a phase-separated nuclear body that suppresses myeloid leukemic differentiation. Cancer Cell. 2021;39:958-972
27. Jiang L, Lin W, Zhang C, Ash PEA, Verma M, Kwan J. et al. Interaction of tau with HNRNPA2B1 and N(6)-methyladenosine RNA mediates the progression of tauopathy. Mol Cell. 2021;81:4209-4227
28. Wiener D, Schwartz S. The epitranscriptome beyond m(6)A. Nat Rev Genet. 2021;22:119-131
29. Zhang M, Zhang X, Ma Y, Yi C. New directions for Psi and m1A decoding in mRNA: deciphering the stoichiometry and function. RNA. 2024;30:537-547
30. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635-1646
31. Deng LJ, Deng WQ, Fan SR, Chen MF, Qi M, Lyu WY. et al. m6A modification: recent advances, anticancer targeted drug discovery and beyond. Mol Cancer. 2022;21:52
32. Scholler E, Weichmann F, Treiber T, Ringle S, Treiber N, Flatley A. et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA. 2018;24:499-512
33. Shan M, Liu D, Sun L, Yang M, He M, Zhang Y. et al. KIAA1429 facilitates metastasis via m6A-YTHDC1-dependent RND3 down-regulation in hepatocellular carcinoma cells. Cancer Lett. 2024;584:216598
34. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M. et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369-373
35. Wen J, Lv R, Ma H, Shen H, He C, Wang J. et al. Zc3h13 Regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol Cell. 2018;69:1028-1038
36. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG. et al. Perturbation of m6A Writers Reveals Two Distinct Classes of mRNA Methylation at Internal and 5′ Sites. Cell Rep. 2014;8:284-296
37. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP. et al. The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169:824-835
38. Xue M, Dong L, Zhang H, Li Y, Qiu K, Zhao Z. et al. METTL16 promotes liver cancer stem cell self-renewal via controlling ribosome biogenesis and mRNA translation. J Hematol Oncol. 2024;17:7
39. Tran NV, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P. et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Research. 2019;47:7719-7733
40. Ma HH, Wang XY, Cai JB, Dai Q, Natchiar SK, Lv RT. et al. Publisher Correction: N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol. 2019;15:549
41. Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K. et al. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep. 2017;7:42271
42. Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC. et al. Differential m(6)A, m(6)A(m), and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell. 2018;71:973-985
43. Shi H, Wei J, He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74:640-650
44. Liao J, Wei Y, Liang J, Wen J, Chen X, Zhang B. et al. Insight into the structure, physiological function, and role in cancer of m6A readers-YTH domain-containing proteins. Cell Death Discov. 2022;8:137
45. Liu Y, Yang D, Liu T, Chen J, Yu J, Yi P. N6-methyladenosine-mediated gene regulation and therapeutic implications. Trends Mol Med. 2023;29:454-467
46. Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK. et al. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol Cell. 2019;74:494-507
47. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ. et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315-328
48. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF. et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61:507-519
49. Li L, Krasnykov K, Homolka D, Gos P, Mendel M, Fish RJ. et al. The XRN1-regulated RNA helicase activity of YTHDC2 ensures mouse fertility independently of m(6)A recognition. Mol Cell. 2022;82:1678-1690
50. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560-564
51. Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR. et al. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun. 2018;9:420
52. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Research. 2017;45:6051-6063
53. Wei G. RNA m6A modification, signals for degradation or stabilisation? Biochem Soc Trans. 2024;52:707-717
54. Zhang GQ, Xu YR, Wang XN, Zhu YX, Wang LL, Zhang WX. et al. Dynamic FMR1 granule phase switch instructed by m6A modification contributes to maternal RNA decay. Nat Commun. 2022;13:859
55. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O. et al. 5' UTR m(6)A promotes Cap-independent translation. Cell. 2015;163:999-1010
56. Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T. et al. A novel m(6)A reader Prrc2a controls oligodendroglial specification and myelination. Cell Res. 2019;29:23-41
57. Zaccara S, Ries RJ, Jaffrey SR. Publisher Correction: Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2023;24:770
58. Fang F, Wang X, Li Z, Ni K, Xiong C. Epigenetic regulation of mRNA N6-methyladenosine modifications in mammalian gametogenesis. Mol Hum Reprod. 2021;27:gaab025
59. Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M. et al. Stem cells. M6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002-1006
60. Aufgebauer CJ, Bland KM, Horner SM. Modifying the antiviral innate immune response by selective writing, erasing, and reading of m(6)A on viral and cellular RNA. Cell Chem Biol. 2024;31:100-109
61. Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang Y. et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. 2018;9:4772
62. Liu S, Chen L, Zhang Y, Zhou Y, He Y, Chen Z. et al. M6AREG: m6A-centered regulation of disease development and drug response. Nucleic Acids Res. 2023;51:D1333-D1344
63. Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z. et al. Ythdf2-mediated m(6)A mRNA clearance modulates neural development in mice. Genome Biol. 2018;19:69
64. Hsu PJ, Zhu YF, Ma HH, Guo YH, Shi XD, Liu YY. et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Research. 2017;27:1115-1127
65. Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Bio. 2017;18:285-298
66. Li CH, Coffey EL, Dall'Agnese A, Hannett NM, Tang X, Henninger JE. et al. MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature. 2020;586:440-444
67. Komatsu M. p62 bodies: Phase separation, NRF2 activation, and selective autophagic degradation. IUBMB Life. 2022;74:1200-1208
68. Du M, Chen ZJ. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704-709
69. Zhang K, Huang M, Li A, Wen J, Yan L, Li Y. et al. DIAPH3 condensates formed by liquid-liquid phase separation act as a regulatory hub for stress-induced actin cytoskeleton remodeling. Cell Rep. 2023;42:111986
70. Trinkle-Mulcahy L, Sleeman JE. The Cajal body and the nucleolus: "In a relationship" or "It's complicated"? Rna Biol. 2017;14:739-751
71. Ding L, Spencer A, Morita K, Han M. The developmental timing regulator AIN-1 interacts with miRISCs and may target the argonaute protein ALG-1 to cytoplasmic P bodies in C. elegans. Mol Cell. 2005;19:437-447
72. Luo Y, Na Z, Slavoff SA. P-Bodies: composition, properties, and functions. Biochemistry. 2018;57:2424-2431
73. Vindry C, Marnef A, Broomhead H, Twyffels L, Ozgur S, Stoecklin G. et al. Dual RNA processing roles of Pat1b via cytoplasmic Lsm1-7 and nuclear Lsm2-8 complexes. Cell Rep. 2017;20:1187-1200
74. Youn JY, Dyakov BJA, Zhang J, Knight JDR, Vernon RM, Forman-Kay JD. et al. Properties of stress granule and P-Body proteomes. Mol Cell. 2019;76:286-294
75. Cai D, Feliciano D, Dong P, Flores E, Gruebele M, Porat-Shliom N. et al. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat Cell Biol. 2019;21:1578-1589
76. Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958
77. Hallacli E, Kayatekin C, Nazeen S, Wang XH, Sheinkopf Z, Sathyakumar S. et al. The Parkinson's disease protein alpha-synuclein is a modulator of processing bodies and mRNA stability. Cell. 2022;185:2035-2065
78. Lavalée M, Curdy N, Laurent C, Fournié JJ, Franchini DM. Cancer cell adaptability: turning ribonucleoprotein granules into targets. Trends Cancer. 2021;7:902-915
79. Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ. et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123-133
80. Li J, Zhang M, Ma W, Yang B, Lu H, Zhou F. et al. Post-translational modifications in liquid-liquid phase separation: a comprehensive review. Mol Biomed. 2022;3:13
81. Saito M, Hess D, Eglinger J, Fritsch AW, Kreysing M, Weinert BT. et al. Acetylation of intrinsically disordered regions regulates phase separation. Nat Chem Biol. 2019;15:51-61
82. Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF. et al. m(6)A enhances the phase separation potential of mRNA. Nature. 2019;571:424-428
83. Han D, Longhini AP, Zhang X, Hoang V, Wilson MZ, Kosik KS. Dynamic assembly of the mRNA m6A methyltransferase complex is regulated by METTL3 phase separation. PLoS Biol. 2022;20:e3001535
84. Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM. et al. N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science. 2020;367:580-586
85. Chang CJ, Rajasekaran M, Qiao YT, Dong H, Wang Y, Xia HP. et al. The aberrant upregulation of exon 10-inclusive SREK1 through SRSF10 acts as an oncogenic driver in human hepatocellular carcinoma. Nat Commun. 2022;13:1363
86. Wu Y, Jin M, Fernandez M, Hart KL, Liao A, Ge X. et al. METTL3-mediated m6A modification controls splicing factor abundance and contributes to aggressive CLL. Blood Cancer Discov. 2023;4:228-245
87. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-206
88. Wei G, Almeida M, Pintacuda G, Coker H, Bowness JS, Ule J. et al. Acute depletion of METTL3 implicates N6-methyladenosine in alternative intron/exon inclusion in the nascent transcriptome. Genome Research. 2021;31:1395-1408
89. Uzonyi A, Dierks D, Nir R, Kwon OS, Toth U, Barbosa I. et al. Exclusion of m6A from splice-site proximal regions by the exon junction complex dictates m6A topologies and mRNA stability. Mol Cell. 2023;83:237-251
90. Widagdo J, Anggono V, Wong JJ. The multifaceted effects of YTHDC1-mediated nuclear m(6)A recognition. Trends Genet. 2022;38:325-332
91. Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD. et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. Plos Genet. 2018;14:e1007412
92. Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3'-end processing. Nucleic Acids Res. 2017;45:11356-11370
93. Mauer J, Sindelar M, Despic V, Guez T, Hawley BR, Vasseur JJ. et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat Chem Biol. 2019;15:340-347
94. Shukla S, Parker R. Quality control of assembly-defective U1 snRNAs by decapping and 5′-to-3′ exonucleolytic digestion. P Natl Acad Sci USA. 2014;111:E3277-E3286
95. Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482-485
96. Xie D, Chen M, Niu J, Wang L, Li Y, Fang X. et al. Phase separation of SERRATE drives dicing body assembly and promotes miRNA processing in Arabidopsis. Nat Cell Biol. 2021;23:32-39
97. Baradaran-Heravi Y, Van Broeckhoven C, van der Zee J. Stress granule mediated protein aggregation and underlying gene defects in the FTD-ALS spectrum. Neurobiol Dis. 2020;134:104639
98. Khan M, Hou S, Chen M, Lei H. Mechanisms of RNA export and nuclear retention. Wiley Interdiscip Rev RNA. 2023;14:e1755
99. Michlewski G, Sanford JR, Cáceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179-189
100. Ma CT, Velazquez-Dones A, Hagopian JC, Ghosh G, Fu XD, Adams JA. Ordered multi-site phosphorylation of the splicing factor ASF/SF2 by SRPK1. J Mol Biol. 2008;376:55-68
101. Lai MC, Tarn WY. Hypophosphorylated ASF/SF2 binds TAP and is present in messenger ribonucleoproteins. J Biol Chem. 2004;279:31745-31749
102. Zheng Q, Hou J, Zhou Y, Li Z, Cao X. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat Immunol. 2017;18:1094-1103
103. Dong F, Qin X, Wang B, Li Q, Hu J, Cheng X. et al. ALKBH5 facilitates hypoxia-induced paraspeckle assembly and IL8 secretion to generate an immunosuppressive tumor microenvironment. Cancer Res. 2021;81:5876-5888
104. Qin X, Long Y, Bai X, Cao L, Yan H, Zhang K. et al. The disordered C terminus of ALKBH5 promotes phase separation and paraspeckles assembly. J Biol Chem. 2023;299:105071
105. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD. et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695
106. Qiao Y, Sun Q, Chen X, He L, Wang D, Su R. et al. Nuclear m6A reader YTHDC1 promotes muscle stem cell activation/proliferation by regulating mRNA splicing and nuclear export. Elife. 2023;12:e82703
107. Ratnadiwakara M, Archer SK, Dent CI, De Los Mozos IR, Beilharz TH, Knaupp AS. et al. SRSF3 promotes pluripotency through mRNA export and coordination of the pluripotency gene expression program. Elife. 2018;7:e37419
108. Lee Y, Choe J, Park OH, Kim YK. Molecular mechanisms driving mRNA degradation by m(6)A modification. Trends Genet. 2020;36:177-188
109. Boo SH, Ha H, Lee Y, Shin MK, Lee S, Kim YK. UPF1 promotes rapid degradation of m6A-containing RNAs. Cell Rep. 2022;39:110861
110. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117-120
111. Du H, Zhao Y, He JQ, Zhang Y, Xi HR, Liu MF. et al. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626
112. Hubstenberger A, Courel M, Benard M, Souquere S, Ernoult-Lange M, Chouaib R. et al. P-Body purification reveals the condensation of repressed mRNA regulons. Mol Cell. 2017;68:144-157
113. Zou Z, Sepich-Poore C, Zhou X, Wei J, He C. The mechanism underlying redundant functions of the YTHDF proteins. Genome Biol. 2023;24:17
114. Despic V, Neugebauer KM. RNA tales - how embryos read and discard messages from mom. J Cell Sci. 2018;131:jcs201996
115. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335-345
116. Su R, Dong L, Li YC, Gao M, He PC, Liu W. et al. METTL16 exerts an m6A-independent function to facilitate translation and tumorigenesis. Nat Cell Biol. 2022;24:205-216
117. Stixová L, Komurková D, Kovaríková AS, Fagherazzi P, Bártová E. Localization of METTL16 at the nuclear periphery and the nucleolus is cell cycle-specific and METTL16 interacts with several nucleolar proteins. Life-Basel. 2021;11:669
118. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y. et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020;48:3816-3831
119. Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang H. et al. HIF-1alpha-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct Target Ther. 2021;6:76
120. Pi J, Wang W, Ji M, Wang X, Wei X, Jin J. et al. YTHDF1 promotes gastric carcinogenesis by controlling translation of FZD7. Cancer Res. 2021;81:2651-2665
121. Jiang A, Zhang SW, Wang XY, Li D. RBM15 condensates modulate m6A modification of STYK1 to promote tumorigenesis. Comput Struct Biotec. 2022;20:4825-4836
122. Wang LF, Zhan GK, Maimaitiyiming Y, Su YF, Lin ST, Liu JF. et al. m(6)A modification confers thermal vulnerability to HPV E7 oncotranscripts via reverse regulation of its reader protein IGF2BP1 upon heat stress. Cell Rep. 2022;41:111546
123. Nagarsheth NB, Norberg SM, Sinkoe AL, Adhikary S, Meyer TJ, Lack JB. et al. TCR-engineered T cells targeting E7 for patients with metastatic HPV-associated epithelial cancers. Nat Med. 2021;27:419-425
124. Wang ST, Wang YF, Li Q, Zeng KX, Li XM, Feng XH. RUNX1-IT1 favors breast cancer carcinogenesis through regulation of IGF2BP1/GPX4 axis. Discov Oncol. 2023;14:42
125. Zeng WJ, Lu CX, Shi YY, Wu CY, Chen XX, Li CM. et al. Initiation of stress granule assembly by rapid clustering of IGF2BP proteins upon osmotic shock. Bba-Mol Cell Res. 2020;1867:118795
126. Wyatt AW, Gleave ME. Targeting the adaptive molecular landscape of castration-resistant prostate cancer. Embo Mol Med. 2015;7:878-894
127. Somasekharan SP, Saxena N, Zhang F, Beraldi E, Huang JN, Gentle C. et al. Regulation of AR mRNA translation in response to acute AR pathway inhibition. Nucleic Acids Res. 2022;50:1069-1091
128. Cai Y, Li N, Li H. YBX2 modulates mRNA stability via interaction with YTHDF2 in endometrial cancer cells. Exp Cell Res. 2023;427:113586
129. Zheng ZH, Zhang GL, Jiang RF, Hong YQ, Zhang QY, He JP. et al. METTL3 is essential for normal progesterone signaling during embryo implantation via m6A-mediated translation control of progesterone receptor. P Natl Acad Sci USA. 2023;120:e2214684120
130. Wan X, Ge Y, Xu S, Feng Y, Zhu Y, Yin L. et al. m(6)A modification and its role in neural development and neurological diseases. Epigenomics. 2023;15:819-833
131. Saha P, Sen N. Tauopathy: A common mechanism for neurodegeneration and brain aging. Mech Ageing Dev. 2019;178:72-79
132. Jiang L, Ash PEA, Maziuk BF, Ballance HI, Boudeau S, Abdullatif AA. et al. TIA1 regulates the generation and response to toxic tau oligomers. Acta Neuropathol. 2019;137:259-277
133. Cruz A, Verma M, Wolozin B. The pathophysiology of Tau and stress granules in disease. Adv Exp Med Biol. 2019;1184:359-372
134. Korb MK, Kimonis VE, Mozaffar T. Multisystem proteinopathy: Where myopathy and motor neuron disease converge. Muscle Nerve. 2021;63:442-454
135. Paul KR, Molliex A, Cascarina S, Boncella AE, Taylor JP, Ross ED. Effects of mutations on the aggregation propensity of the human Prion-like protein hnRNPA2B1. Mol Cell Biol. 2017;37:e00652-16
136. Carter GC, Hsiung CH, Simpson L, Yang H, Zhang X. N-terminal domain of TDP43 enhances liquid-liquid phase separation of globular proteins. J Mol Biol. 2021;433:166948
137. Tziortzouda P, Van Den Bosch L, Hirth F. Triad of TDP43 control in neurodegeneration: autoregulation, localization and aggregation. Nat Rev Neurosci. 2021;22:197-208
138. McMillan M, Gomez N, Hsieh C, Bekier M, Li X, Miguez R. et al. RNA methylation influences TDP43 binding and disease pathogenesis in models of amyotrophic lateral sclerosis and frontotemporal dementia. Mol Cell. 2023;83:219-236
139. Monahan Z, Ryan VH, Janke AM, Burke KA, Rhoads SN, Zerze GH. et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017;36:2951-2967
140. Qamar S, Wang GZ, Randle SJ, Ruggeri FS, Varela JA, Lin JQ. et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell. 2018;173:720-734
141. Yoneda R, Ueda N, Kurokawa R. m(6)A modified short RNA fragments inhibit cytoplasmic TLS/FUS aggregation induced by hyperosmotic stress. Int J Mol Sci. 2021;22:11014
142. Ma M, Duan Y, Peng C, Wu Y, Zhang X, Chang B. et al. Mycobacterium tuberculosis inhibits METTL14-mediated m(6)A methylation of Nox2 mRNA and suppresses anti-TB immunity. Cell Discov. 2024;10:36
143. Wang L, Wen MY, Cao XT. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science. 2019;365:eaav0758
144. Wang J, Zhou Y, Zhang M, Wu Y, Wu Q, Su W. et al. YTHDF1-CLOCK axis contributes to pathogenesis of allergic airway inflammation through LLPS. Cell Rep. 2024;43:113947
145. Chakaya J, Khan M, Ntoumi F, Aklillu E, Fatima R, Mwaba P. et al. Global Tuberculosis report 2020-reflections on the global TB burden, treatment and prevention efforts. Int J Infect Dis. 2021;113(Suppl 1):S7-S12
Author contact
![]() Corresponding authors: Dr. Jie Ying, +86- 025-57068090, Email: yj_8611com; Dr. Jianxiang Chen, +86-0571-28865408, Email: chenjxedu.cn; Dr. Fangtian Bu, +86-0571-28863415, Email: bufangtianedu.cn.
Corresponding authors: Dr. Jie Ying, +86- 025-57068090, Email: yj_8611com; Dr. Jianxiang Chen, +86-0571-28865408, Email: chenjxedu.cn; Dr. Fangtian Bu, +86-0571-28863415, Email: bufangtianedu.cn.
 Global reach, higher impact
Global reach, higher impact