13.3
Impact Factor
Theranostics 2024; 14(10):4161-4183. doi:10.7150/thno.95885 This issue Cite
Review
From biogenesis to aptasensors: advancements in analysis for tumor-derived extracellular vesicles research
1. State Key Laboratory of Digital Medical Engineering, School of Biological Science and Medical Engineering, Southeast University, Nanjing 210096, China.
2. China Hunan Key Laboratory of Biomedical Nanomaterials and Devices, Hunan University of Technology, Zhuzhou 412007, PR China.
3. Department of Clinical Laboratory, the Affiliated Drum Tower Hospital of Nanjing University Medical School, Nanjing 210008, China.
4. Institute of Cytology and Genetics, School of Basic Medical Sciences, Hengyang Medical School, University of South China, Hengyang 421001, China.
5. Institute for Future Sciences, University of South China, Changsha Hunan 410000, China.
Received 2024-3-3; Accepted 2024-6-20; Published 2024-7-2
Abstract
Extracellular vesicles (EVs) are enclosed by a nanoscale phospholipid bilayer membrane and typically range in size from 30 to 200 nm. They contain a high concentration of specific proteins, nucleic acids, and lipids, reflecting but not identical to the composition of the parent cell. The inherent characteristics and variety of EVs give them extensive and unique advantages in the field of cancer identification and treatment. Recently, EVs have been recognized as potential tumor markers for the detection of cancer. Aptamers, which are molecules of single-stranded DNA or RNA, demonstrate remarkable specificity and affinity for their targets by adopting distinct tertiary structures. Aptamers offer various advantages over their protein counterparts, such as reduced immunogenicity, the ability for convenient large-scale synthesis, and straightforward chemical modification. In this review, we summarized EVs biogenesis, sample collection, isolation, storage and characterization, and finally provided a comprehensive survey of analysis techniques for EVs detection that are based on aptamers.
Keywords: Extracellular Vesicles, Tumor Markers, Aptamers, Detection
Introduction
Cells release extracellular vesicles (EVs) into the extracellular space; these are small, membrane-bound structures composed of a lipid bilayer. EVs facilitate the transfer of various molecules, including proteins, nucleic acids, and lipids, between cells [1, 2]. Based on their origin and size, EVs typically range from 30 to 200 nm [3, 4]. These vesicles provide critical insights into the health of donor cells and are involved in a wide array of physiological and pathological processes, such as tumor development, immune responses, signal transduction, and antigen presentation [5]. Furthermore, EVs are pivotal for the diagnosis and monitoring of diseases, being detectable in bodily fluids like blood, urine, saliva, and cerebrospinal fluid, which highlights their potential as noninvasive biomarkers [6-8]. Analyzing EVs is challenging due to their small size; however, employing antibodies that target specific protein markers on the EV surface facilitates their visualization and quantification. This approach allows for both qualitative and quantitative assessments of vesicular protein markers [9-11]. Nevertheless, the glycosylation of proteins on EVs can impede the binding of fluorescently labeled antibodies, thus reducing detection sensitivity [12].
Aptamers are short, single-stranded nucleic acids (DNA or RNA) that can bind to specific target molecules with high affinity and specificity. Often referred to as the nucleic acid counterparts of antibodies, aptamers are synthesized through a method known as Systematic Evolution of Ligands by Exponential enrichment (SELEX). This process involves iterative cycles of selection and amplification to isolate aptamers that exhibit the desired binding properties [13]. Aptamers offer numerous benefits over conventional antibodies, including their smaller size, ease of synthesis, and chemical stability [14, 15]. In the context of EVs, aptamers can be tailored to selectively recognize and bind to exosomal proteins or other surface markers, facilitating the development of aptasensors. These biosensors employ aptamers as the recognition element and enable the construction of compact, on-site devices for the detection of EVs [16]. Aptamers are characterized by their high specificity, ease of synthesis & modification, excellent stability, low immunogenicity, and minimal toxicity. They can also be seamlessly integrated into various sensor systems, enhancing the development of portable devices for immediate EVs identification.
In this review, we aim to provide a comprehensive analysis of recent progress in the methodologies for studying EVs. The review explores advancements in several domains, including the production of EVs, sample collection, separation, preservation, and analysis, as illustrated in Figure 1. Additionally, we examine a broad spectrum of aptamer-based detection techniques that employ diverse signaling modalities, such as optical, electrical, and so on.
From Biogenesis to Characterization of Extracellular Vesicles
Biogenesis of EVs
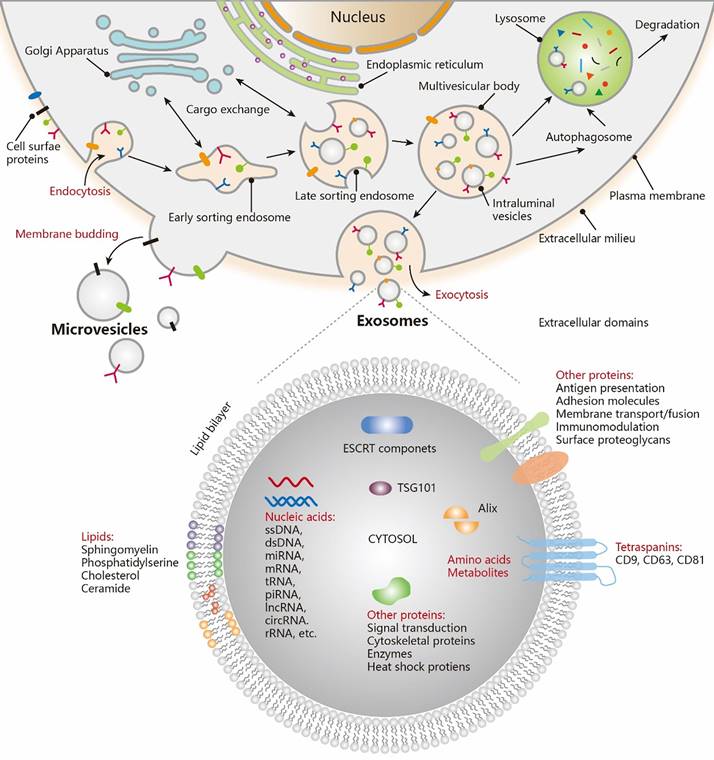
The existence of membrane-structured microvesicles, later named "exosomes," was first demonstrated in maturing erythrocytes in 1983 [17], and the term "exosome" was officially adopted in 1987 [18]. Exosomes, a distinct subtype of EVs, are typically small in size. Numerous studies have shown that cellular communication can occur through the release of EVs [19-21]. EVs are produced through the cellular endocytosis process, carrying intracellular substances, and are prevalent in various body fluids. They play a pivotal role in facilitating intercellular communication, transporting cargo, and regulating diverse pathophysiological processes. The traditional view holds that endocytosis leads to the formation of intraluminal vesicles (ILVs), which merge with the endosome membrane. These endosomes then evolve into multivesicular bodies (MVBs), incorporating "cargo" such as RNA, proteins, and lipids from the cytoplasm (Figure 2 [22]). EVs are expelled through the inward budding of the limiting membrane of MVBs as early endosomes mature into late endosomes [23-26]. However, an alternative theory suggests that the plasma membrane can also directly release EVs through vesicle budding or gradually release them into the extracellular matrix via budding at intracellular plasma membrane-connected compartments [27-29]. Both theories agree that vesicles (ranging from 30-200 nm) are produced from endosomes and plasma membranes through budding. The ILVs are released into the extracellular environment upon the fusion of MVBs with cellular membranes. The primary distinction lies in whether vesicles derived from the plasma membrane should be classified as EVs.
Schematic illustration of analysis for tumor-derived extracellular vesicles, from biogenesis to the development of aptasensors.
Sample collection and handling
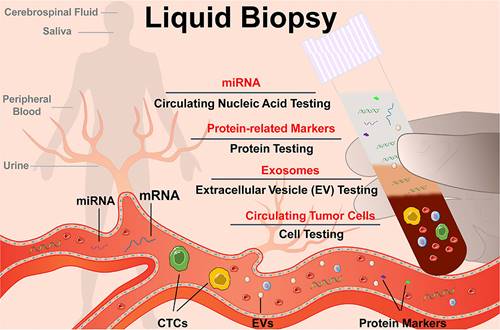
Research has focused on developing novel techniques for the preparation and processing of EVs samples. A variety of physical principles, functional compounds, and engineering methods have been employed to improve the characterization of EVs and associated biomolecules. However, several challenges still need to be addressed in the future. One such challenge is the requirement for distinct processing procedures for EV samples derived from different body fluids (Figure 3) [30]. EVs are demonstrated that presented in almost all the body fluids including cell culture-conditioned media [31-35], blood/serum/plasma [36-40], pleural effusions and ascites [41-44], saliva [45-48], urine [49-55], tear fluid [56-59], and cerebrospinal fluid (CSF) [60-63]. The concentration, composition, and bioactivities of EVs can vary significantly depending on the type of body fluid, collection strategies, storage conditions, and pretreatment methods [64, 65]. Several biological sources of EVs are commonly investigated. A summary of sample collection and handling is presented in Table 1.
Isolation of EVs
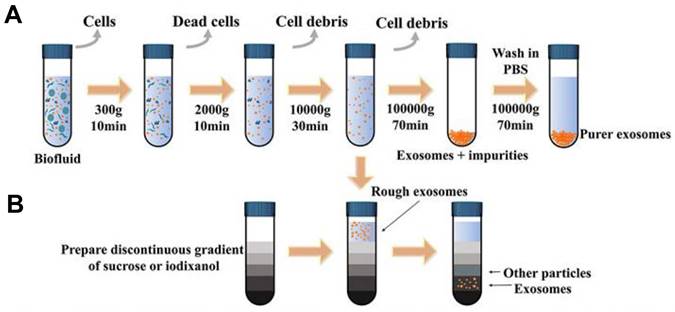
EVs play a crucial role in facilitating information and content exchange between different cells, which significantly impacts the early diagnosis and treatment of tumors. High recovery and purity in separation are essential first steps for EVs -related detection and analysis [66-68]. Various isolation techniques were classified on the basis of the methodology and properties of EVs: (1) ultracentrifugation-based isolation strategies (Figure 4) [69, 70], (2) size-based isolation strategies [71, 72], (3) precipitate and phase separation [73], (4) membrane-based isolation [74], (5) microfluidic analysis [75]. A summary of EVs isolation strategies is presented in Table 2.
Biogenesis and contents of EVs. Exosomes are generated via the double invagination of the PM, including the formation of early sorting endosomes (ESEs) and late sorting endosomes (LSEs), the generation of ILVs within MVBs, the transportation of MVBs to cytoplasmic membrane, and the fusion of MVBs membrane with the cell membrane. Cargo exchange occurs between ESEs (LSEs) and the trans-Golgi network. MVBs can also fuse with lysosomes or autophagosomes for the degradation and recycling of cellular contents. Microvesicles are derived from the direct outward budding of cellular PM. EVs, including exosomes, transport a vast repertoire of different types of proteins, lipids, NAs, and other small molecules. Adapted with permission from [22], copyright 2021 American Chemical Society.
Summary of sample collection and handling
| Sample types | Invasive collection | Storage | Cautions | Reference |
|---|---|---|---|---|
| Cell culture | No | -20°C (temporarily storage) or -80 °C (long-term storage) | • Using exosome-depleted serum or serum-free media in cell culture helps eliminate interference from exogenous EVs. • Eliminate microbial or viral infections before isolation | [31-35] |
| Blood/Serum/Plasma | Yes | -80 °C or in liquid nitrogen | • Use a large diameter needle (such as a 21G needle) to minimize platelet derived EVs • Choose the appropriate anticoagulant (such as sodium citrate or EDTA) • Discard the first 1-2 mL of blood following the puncture • Thawing should be done at 37 °C or room temperature | [36-40] |
| Pleural effusions and ascites | Yes | -80 °C | • Maintain a sterile environment to prevent contamination • Collection site is usually between the ribs in the lower back or side of the chest (pleural effusion) or in the lower abdomen (sites) | [41-44] |
| Saliva | No | -80 °C | • Abstain from consuming food, beverages, and tobacco for a minimum of one hour before sample collection • Centrifuge or filter to remove bacteria, squamous epithelial cells and food debris before isolation | [45-48] |
| Urine | Optional | -80 °C | • Internal reference (such as uromodulin) can be selected to normalize the concentration of EVs in urine • Regulate the diet to avoid the composition of urine can be affected by certain food • It is advised to utilize appropriate protease inhibitors during sample preparation to avoid the deterioration of exosomal proteins while isolation | [49-55] |
| Tear fluid | No | -80 °C | • Ensure a clean and sterile environment • Avoid touching the eyelids or ocular surface • Use appropriate collection methods (such as Schirmer test) • Minimize patient discomfort | [56-59] |
| Cerebrospinal fluid (CSF) | Yes | -80 °C | • CSF can be collected through ventricular/lumbar drain placement or cisternal aspiration during the craniotomy • Filter the CSF samples using a 0.8 μm filter to remove cellular debris after collection | [60-63] |
Schematic diagram of liquid biopsy. Adapted with permission from [30], copyright 2021 American Chemical Society.
Preservation strategies of EVs
It is imperative to conduct the subsequent identification, detection, and analysis of EVs promptly due to their short half-life, which can lead to rapid and irreversible degradation [76-79]. Preservation strategies for EVs are designed to maintain their bioactivities and facilitate transportation. Techniques such as cryopreservation [80-83], freeze-drying [84-86] and spray-drying [87, 88] have been widely applied in various studies and clinical procedures recently. A summary of EVs preservation strategies is revealed in Table 3.
Characterization of EVs
The properties of EVs including size, morphology, and components (proteins, RNA and lipid bilayer) are able to distinguish EVs from others EVs, which is the basis of various EVs characterization methods. In this section, as shown in Table 4, we will discuss various qualitative and quantitative methods commonly used for EVs characterization: (1) western blot (WB) [89], (2) microscopy-based characterization [90-92], (3) nanoparticle tracking analysis (NTA) [93] and dynamic light scattering (DLS) [94, 95], (4) flow cytometry (FCM) [96].
Schematic representation of common exosomal separation techniques. (A) Ultracentrifugation, (B) Density gradient centrifugation. Adapted with permission from [69], copyright 2022 Frontiers.
Summary of EVs isolation strategies
| Isolation techniques | Principle | Advantages | Disadvantages | Reference |
|---|---|---|---|---|
| Differential ultracentrifugation | Distinguishing EVs from other components in the sample by their variations in density and size. | • Widely accepted gold standard • Medium purity separation • Cost-effective • Application of various biological samples types and capacity • Compatibility with downstream analysis | • Time-consuming • Low yield • Co-isolation of different EVs subtypes with similar size and density • Equipment and expertise requirements | [69] |
| Density gradient ultracentrifugation | Distinction in density between EVs and other particles found in a sample. | • High purity separation • Effectively isolation of individual EVs • Preservation of integrity and biological activity | • Time-consuming • Low yield • Complex procedure • Costly • Potential damage to Evs | [70] |
| Ultrafiltration (UF) | EVs in the sample are filtered through the ultrafiltration membrane, with only EVs larger than the membrane's pore size being trapped on the membrane and collected as the retentate. | • Simplicity and ease of use • Time efficiency • Preservation of biological activity • Versatility in EVs size selection • Cost-effectiveness | • Low separation efficiency • Limited purity • Membrane lifespan reduction due to blockage | [71] |
| Size exclusion chromatography (SEC) | Molecules or particles are separated according to their size as they pass through a column filled with porous polymer microspheres. | • Preservation of biophysical and functional properties • Application of various biological samples types and capacity • Compatibility with downstream analysis | • Low yield of EVs • Time-consuming process • Potential for sample dilution • Limited scalability | [72] |
| Polymer precipitation | Utilization of polymers to generate a hydrophobic setting, inducing the less-soluble constituents of EVs to cluster together and settle out of the solution. | • No specialized equipment required • High Extraction Efficiency • Compatibility with Various Sample Types • Cost-Effective | • Low purity • Low recovery • Uneven particle sizes • Damage to exosomes • Hard-to-remove polymers | [73] |
| Immunoaffinity capture | Antibodies specifically recognize the EVs surface markers (CD9, CD63, and CD81) are immobilized onto a solid support (magnetic beads or microfluidic chips). | • High specificity • High purity • Fast and efficient • Compatibility with downstream analysis • Potential for clinical applications | • Limited capture efficiency • Cost and availability of antibodies • Labor-intensive and time-consuming | [74] |
| Microfluidic analysis | Based on the principles of microfluidics and the physical properties of EVs, such as size, immunoaffinity, electrical and acoustic properties. | • Beneficial to limited or precious sample • High efficiency • Easily automated • Integration of multiple functions • Cost-effective | • Sample volume limitations • Device complexity | [75] |
Summary of EVs preservation strategies
| Preservation strategies | Advantages | Disadvantages | Reference |
|---|---|---|---|
| Cryopreservation | • Convenient and inexpensive operation • It is suitable for long-term storage • Easy and quick reconstitution | • Could not maintain the integrated structure of EVs (cryoinjury) • Cryoprotectants components may affect biological properties of EVs • Cold-chain transportation is expensive, result in the cryopreservation was not very appropriate for EVs transportation | [80-83] |
| Freeze-drying | • Lyophilized EVs is acceptable for preservation and transportation under room temperature • The shelf life of lyophilized EVs can be extend to several years • Reduce the risk of contamination during storage | • Molecular structure may be destroyed by the freezing and dehydration pressures generation • Complex and time-consuming process, requires specialized equipment and expertise • Equipment and materials can be expensive | [84-86] |
| Spray-drying | • No freezing procedure • It is available for rapid conversion of EVs solution into a dry powder • It allows for accurate manipulation of the aerodynamic particle size and properties | • The heated gas may lead to damage or disruption of the EVs membrane • The high temperatures and shear forces may lead to aggregation of EVs, resulting in a lower recovery rate | [87, 88] |
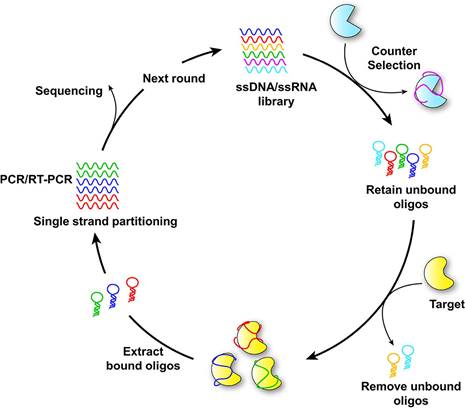
Schematic diagram of SELEX for aptamer selection. Adapted with permission from [30], copyright 2021 American Chemical Society.
Advantages of aptamers as detection probes
Aptamers, brief single-stranded DNA or RNA molecules, can bind to specific target molecules with high affinity and specificity. Due to their ability to recognize and bind to a variety of targets, including proteins, peptides, small molecules, and even entire cells, they are often referred to as "chemical antibodies" [97]. The SELEX process generates aptamers through iterative rounds of selection and amplification, aiming to identify those with high affinity for specific target molecules. Techniques such as surface plasmon resonance (SPR), fluorescence-based assays, and isothermal titration calorimetry (ITC) are utilized to evaluate the binding affinity, specificity, and stability of selected aptamers. Based on the results of analysis and characterization, aptamers can be further refined through rational design or mutagenesis to enhance their binding properties, stability, or other desired characteristics. The principle of aptamer selection by SELEX is illustrated in Figure 5 [30].
In diagnostics, aptamers serve as tools for the detection and quantification of target molecules, including cancer biomarkers, pathogens, and toxins. In therapeutics, aptamers are developed as targeted drug delivery vehicles or as therapeutic agents themselves, capable of inhibiting or modulating the activity of specific proteins [64, 98]. Aptamers have the potential to revolutionize the field of molecular recognition and offer new opportunities for the development of innovative analytical technologies [99]. Aptamers offer several advantages when used as probes in EVs detection: (1) High affinity and specificity: SELEX facilitates the selection of aptamers with high affinity and specificity for targets, enabling accurate and reliable detection of target molecules [15]. (2) Versatility: Aptamers can target a diverse array of entities, including small compounds, proteins, EVs, and cells. This adaptability makes them suitable for detecting various substances across different biological specimens [100]. (3) Batch-to-batch consistency: Aptamers can be synthesized with high reproducibility, ensuring consistent performance across different batches, crucial for diagnostic applications where reliable results are paramount [101]. (4) Ease of modification: Aptamers can be readily modified with various fluorescent labels or other signaling molecules, facilitating the integration of diverse detection methods. This flexibility allows for the development of sensitive assays capable of detecting multiple targets [102]. (5) Enhanced penetration and sensitivity: Due to their smaller size compared to antibodies, aptamers achieve better tissue and cellular penetration, improving detection sensitivity, particularly when the target molecule is in hard-to-reach areas [103]. (6) Reusability: Unlike antibodies, aptamers can be regenerated and reused multiple times without diminishing their binding affinity or specificity, making them ideal for continuous monitoring or repeated measurements [104]. (7) Lower immunogenicity: As synthetic molecules, aptamers do not provoke the same immune response as antibodies, minimizing the risk of adverse immune reactions or interference in diagnostic assays [105]. Overall, the unique properties of aptamers position them as valuable tools for developing precise and accurate detection systems in cancer diagnosis and other biomedical applications.
Summary of current EVs characterization methods
| Methods | Purpose | Advantages | Disadvantages | Reference |
|---|---|---|---|---|
| Western blot (WB) | Specific proteins associated with EVs detection and analysis | • High specificity for proteins • Compatibility with large-scale analysis • Established technique | • Large sample volumes requirement • Lengthy workflow • Lack of specificity for Evs • Inability to quantify EVs concentration • Dependence on antibody availability | [89] |
| Transmission electron microscopy (TEM) | Observation of the internal structures of EVs | • High resolution • Direct measurement • Surface topography and internal structure information • Avoidance of sample distortion (Cryo-EM) | • Extensive and time-consuming sample preparation procedures • Vacuum requirement • Low throughput • Cost and equipment Requirements • Lack of proteins Characterization | [90] |
| Cryo-electron microscopy (Cryo-TEM) | Imaging EVs in natural state | • Preservation of sample integrity • High-resolution • Visualization without staining • Minimal sample distortion | • Expensive equipment • Requirement of cryo-fixation and cryo-sectioning for sample preparation • Ice crystal formation during the freezing process may cause artifacts | [91] |
| Atomic-force microscopy (AFM) | Nanoscale imaging and characterization of the morphology, mechanical properties, and biomolecular components of individual EVs | • High-resolution • Native sample conditions • Mechanical properties (stiffness or elasticity) characterization • Real-time imaging | • Sample deposition may result in the loss or alteration of EVs • Time-consuming • Lack of proteins Characterization • Cost and equipment requirements | [92] |
| Nanoparticle tracking analysis (NTA) | Determination of the size distribution and concentration of EVs | • Size and concentration determination • Real-time observation • Fast detection speed • High sensitivity | • Difficulty in distinguishing contaminated proteins • Influence of camera levels and detection thresholds • Sample preparation may introduce variability and potential loss of EVs | [93] |
| Dynamic light scattering (DLS) | Measurement of hydrodynamic diameter of EVs | • Wide size range measurement • No requirement of labeling or modification of EVs • Rapid analysis • Cost-Effective | • Not suitable for accurately measuring the size distribution of polydisperse vesicles • Difficulty in distinguishing contaminated proteins • Influence of camera levels and detection thresholds | [94, 95] |
| Flow cytometry (FCM) | Analysis and characterization of EVs based on physical and biochemical properties | • High-throughput analysis • Single-particle analysis • Multiparametric analysis • Quantitative analysis • Compatibility with other techniques | • Insufficient detection sensitivity and resolution • May not be able to accurately distinguish between individual EVs and aggregates • Time-consuming sample preparation • Background noise • Instrumentation requirements | [96] |
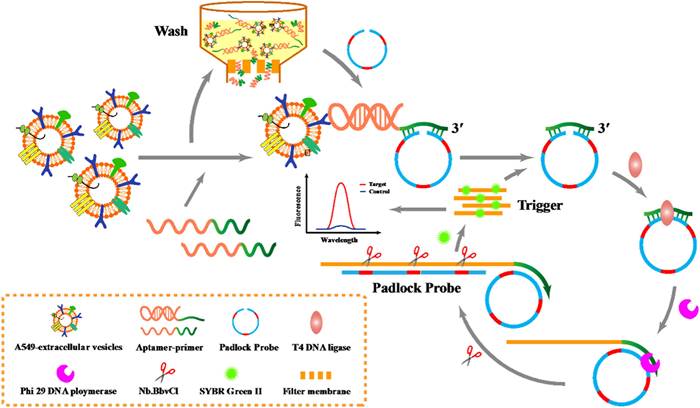
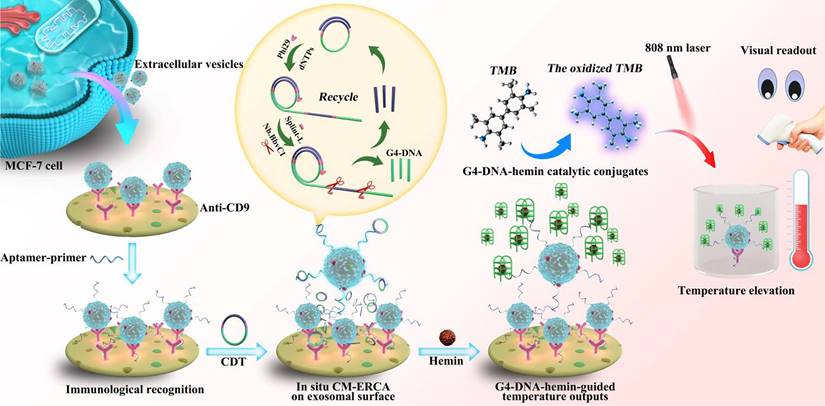
Schematic diagram for P-ERCA. Adapted with permission from [106], copyright 2021 Elsevier.
Detection of EVs
The critical role of EVs is their capacity to facilitate intercellular communication by transporting various biomolecules, such as proteins, nucleic acids, and lipids, between cells. The detection and characterization of EVs have emerged as areas of increasing interest, given their potential as invaluable diagnostic and therapeutic tools. However, the small size and low abundance of EVs in biological fluids present significant challenges for their detection. These challenges have been addressed through recent advancements in analytical technologies, including: (1) fluorescence detection, (2) luminescence detection, (3) electrochemical detection, (4) colorimetric detection, (5) microfluidics detection, and so on. An evaluation of EVs detection methods is summarized in Table 5.
Fluorescence detection
The process of fluorescence detection employs fluorescent molecules or dyes that specifically bind to EVs or their surface markers. These fluorescent probes emit light upon stimulation by a specific wavelength, enabling the visualization and quantification of EVs. In this technique, EVs are typically labeled with fluorescent antibodies or aptamers that selectively bind to surface proteins on the EVs. The resulting fluorescent signal from the labeled EVs can be detected using fluorescence microscopy or flow cytometry. The development of fluorescence detection primarily incorporates four strategies: nucleic acid-assisted amplification, enzyme-assisted amplification, nanomaterials-assisted amplification, and direct fluorescence detection.
Nucleic acid-assisted amplification detection
Nucleic acid-assisted amplification aims to overcome the limitations of low fluorescence intensity exhibited by directly stained EVs and enhances the detection of exosomal protein markers with improved sensitivity. This facilitates more accurate and reliable analysis of EVs in various biological samples. Techniques such as rolling circle amplification (RCA), strand displacement reaction (SDR), hybridization chain reaction (HCR), and catalytic hairpin assembly (CHA) are employed in these processes.
RCA involves amplifying circular DNA templates using a DNA polymerase with strand displacement activity. The RCA process begins with the hybridization of a circular DNA probe to a specific nucleic acid sequence within the EVs. This probe contains a recognition sequence for DNA polymerase and a primer binding site. Upon hybridization to the target sequence, the DNA polymerase initiates the synthesis of new DNA strands, displacing existing strands and generating a long single-stranded DNA molecule. He et al. utilized a padlock probe-based exponential rolling circle amplification (P-ERCA) assay to detect EVs. This method involves conjugating an aptamer targeting the vesicles to a primer sequence, which initiates linear rolling circle amplification in the presence of target EVs [106]. Multiple nicking reactions generate trigger DNA, leading to further circle amplification, and the resulting P-ERCA product exhibits an exponential fluorescence signal as shown in Figure 6. This method successfully discriminates lung cancer EVs in real human serum samples. Additionally, a highly specific technique that utilizes branched rolling circle amplification and a padlock probe demonstrated a shorter reaction time (less than 3 h) and a detection limit of 42.7 particles μL-1 [107]. Similarly, RCA products that employ a CD63 aptamer and DNAzyme achieve targeted identification and amplification of signals within 60 min [108]. For clinical diagnosis of urinary diseases, Wu et al. described a diagnostic protocol that integrates a ratiometric 3D DNA machine with machine learning techniques, achieving a limit of detection (LOD) of 9.9 particles per microliter and a linear range of 104 to 108 particles μL-1 for urinary EVs [109]. Moreover, the classification of disease types exhibits accuracies of 94.7% and 89.5%, respectively.
Summary of EVs detection evaluation
| Detection Type | Method | Target | Linear Range (particles μL-1) | LOD (particles μL-1) | Reference |
|---|---|---|---|---|---|
| Fluorescence detection | RCA | CD9, CD63, Calnexin | 50 - 1.2×103 | 42.22 | [106] |
| MUC1 | 1×102 - 1×106 | 42.7 | [107] | ||
| CD63 | 0.1 - 1×103 | 40 | [108] | ||
| CD63, PSA, EpCAM, MUC1, CEA | 1×10 - 1×105 | 9.9 | [109] | ||
| EpCAM | 2.5×105 to 1×107 | 2.5×105 | [168] | ||
| SDR | CD63, MUC1 | 1.6×102 - 3.2×106, 1×102 - 6.4×105 | 67, 37 | [110] | |
| CD63 | 1×102 -1×106 | 1×102 | [111] | ||
| PTK7 | 5×105 - 7.5×107 | 3.4×105 | [112] | ||
| HCR | CD63, PTK-7 | 5×102 - 1×107 | 4.47×102 | [113] | |
| CD63, EpCAM or CD63, AFP | 1.7×104 - 1.7×1010 | 1.8×102 | [114] | ||
| Nucleolin, PD-L1 | 64 - 106 | 1×102 | [115] | ||
| PTK7 | 1×103 - 1×107 | 4×103 | [116] | ||
| PD-L1 | 1 - 1×106 | 5 | [169] | ||
| CHA | CD63 | 44 - 8.75×105 | 26 | [117] | |
| EpCAM, CD63 | 8.4 - 8.4×105 for EXO-MCF-7, 9.8 - 9.8×104 for EXO-PANC-1 | 0.5 for EXO-MCF-7, 0.1 for EXO-PANC-1 | [118] | ||
| Enzymes-assisted | PD-L1 | 1. ×10-2 pga - 1 pg | 5.21×10-3 pg | [119] | |
| CD9, CD63, CD81, EpCAM, PD-L1 | 6×102 - 6×106 | 72 | [120] | ||
| CD63 | 1×102 - 1×106 | 1×102 | [121] | ||
| CD109, EGFR | 0.1 - 1×105 | 1×102 | [122] | ||
| CD63 | 3.6×102 - 7.19×106 | 360 | [123] | ||
| PD-L1 | 2.5×105 - 1×108 | 9.4×104 | [124] | ||
| CD63, VEGF | 1.75 - 3.5×105 | 1.02 | [170] | ||
| Nano-materials-assisted | CD63, EpCAM, CEA, PTK-7, AFP, PSMA, PDGF | 1.6×102 - 1.6×106 | 1.6×102 | [125] | |
| EpCAM | 5×102-2×104 | 1.86×102 | [126] | ||
| CD63 | 7.5×104 - 1.5×107 | 5×104 | [127] | ||
| MUC1 | 0 - 200 | 8.56 | [128] | ||
| PD-L1 | 0.07 - 0.7 nga | 2.4×10-2 nga | [171] | ||
| Direct fluorescence detection | CD63 | 5×102 - 5×105 | 500 | [131] | |
| CD63, EpCAM | 4.9×104 - 4.9×106 | 4.9×104 | [132] | ||
| Luminescence detection | ECL | EpCAM | 5×102 - 5×106 | 125 | [133] |
| CD63 | 1×102 - 1×105 | 30 | [134] | ||
| CD63, MUC1 | 60 - 1×105 | 36 | [135] | ||
| CD63 | 3.4×102 - 1.7×105 | 74.1 | [136] | ||
| CD63 | 0.95 - 9.5×104 | 0.641 | [172] | ||
| miR21 | 100 aMc - 1 nMc | 4.27 aMc | [173] | ||
| LRET | CD63 | 1×104 - 1×108 | 1.1×103 | [137] | |
| EpCAM | NAb | 80 | [138] | ||
| Electrochemical detection | Direct immobilization | EpCAM | 1×103 - 1×107 | 3.96×102 | [139] |
| miR-21 | NA | 67 aMc | [140] | ||
| CD63, EpCAM | 50 - 1×107 | 13 | [141] | ||
| EpCAM, TLS11a | 60 - 6×106 | 84 | [142] | ||
| EpCAM, CD63 | 1×102 - 1×107 | 58 | [143] | ||
| CD63 | 0.1 - 104 | 42 | [174] | ||
| Label-free | PTK7 | 2.375×103 - 9.5×105 | 6.607×102 | [144] | |
| CD63 | 3.4×102 - 3.4×105 | 5 | [145] | ||
| CD63 | 50 - 1×105 | 16 | [146] | ||
| Colorimetric detection | H2O2 oxidation | EpCAM | 5×103-106 | 1×103 | [147] |
| CD63, HER2, integrin αvβ6 | 10 - 1×108 | 7.77 | [148] | ||
| CD63 | 1×102 - 1×107 | 42 | [149] | ||
| MUC1 | 3.8×103 - 1.2×105 | 1.51×102 | [150] | ||
| CD63 | 1.6×102 - 1.6×103 | 78 | [151] | ||
| MUC1 | 8.3×102 - 5.3×104 | 3.94×102 | [152] | ||
| CD63 | 1×103 - 1×107 | 267 | [153] | ||
| CD9, CD63 | 65-1.3×105 | 2.7 | [154] | ||
| CD63 | 1.9×106 - 3.38×107 | 13.52×105 | [155] | ||
| Nano-materials-assisted | CD63 | 1.4×103 - 2.8×105 | 1.6×102 | [156] | |
| CD63 | 1×103 - 1×107 | 104 | [175] | ||
| Microfluidics integrated detection | EGFR, EpCAM, CD81, CD63 | 1×10 - 1×106 | 83 | [157] | |
| CD63, PD-L1, nucleolin, EpCAM, PTK-7, PSMA | 8.74 - 1.37×105 | 8.74 | [158] | ||
| PD-L1, CA125, CD63, CEA, EpCAM | NAb | 1.58×102 | [159] | ||
| HER2, EpCAM | NAb | Individual EVs | [160] | ||
| PD-L1 | 1×104 - 1×108 | 3.2×103 | [176] | ||
| Others | FRET | CD63 | 10 - 1×106 | 1.4 | [161] |
| CD63, CEA, PD-L1, EpCAM, CA125 | 67.6 - 5.28×106 | 1.95 | [162] | ||
| SERS | CD63 | 32 - 2×106 for SKBR3, 203 - 2×106 for LNCaP, 73 - 2×106 for T84 | 32 for SKBR3, 203 for LNCaP, 73 for T84 | [163] | |
| PSMA | 1.2×102 to 2.4×103 | 19 | [164] | ||
| SPR | CD63 | NA | 5 | [165] | |
| AIE | NAb | 790 - 7.9×103 | 1.3×103 | [166] | |
| ddPCR | PD-L1 | NA | 7.35×10-5 pg | [167] | |
a: the unit is pg when detecting proteins.
b: NA, not found.
c: the unit is aM or nM when detecting miRNA.
Schematic representation of the detection of surface proteins of sEVs using a dual color DNA nanodevice based on an enzyme-free signal amplification and synchronous fluorescence technique. Adapted with permission from [110], copyright 2022 American Chemical Society.
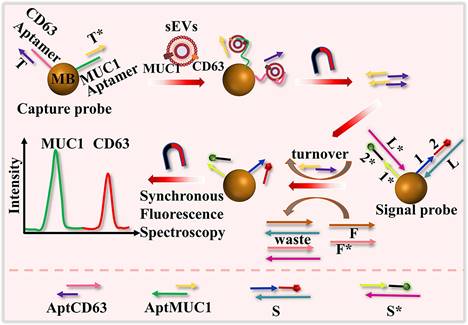
SDR is a branch of nucleic acid amplification methods that leverages the ability of DNA or RNA strands to displace each other from a target sequence through hybridization and enzymatic activity. This method employs nucleic acid probes that specifically recognize and bind to target molecules on EVs. A DNA nanodevice with dual colors was developed to simultaneously analyze surface proteins (CD63 and MUC1) of EVs using toehold-mediated DNA strand displacement signal amplification and synchronous fluorescence techniques, as illustrated in Figure 7 [110]. This assay requires 60 min to complete. For more efficient detection, Zhang introduced a fluorescence-based method for analyzing the surface protein of EVs, utilizing an isothermal cascade nucleic acid amplification approach [111]. This technique transforms signals from the surface membrane proteins of EVs into nucleic acid signals using a carefully designed capture probe that includes a protein aptamer and a partially complementary blocker. DNA polymerase and Nb.BbvCI nicking enzyme are employed to amplify the signal, allowing the detection process to be completed in just 30 min with a linear range of 102 to 106 particles μL-1. Additionally, Chen et al. developed a label-free biosensor, selecting N-methylmesoporphyrin IX as the signal reporter. This compound is capable of inserting into the G-quadruplex structure, thereby generating a significant increase in fluorescence [112]. SDR exhibits exceptional specificity, as the probe is specifically designed to match the target sequence. Moreover, it demonstrates remarkable sensitivity due to the amplification process, enabling the detection of target sequences present in low quantities. Furthermore, SDR can be conducted in a homogeneous solution, eliminating the need for complex sample preparation procedures.
HCR is a technique for amplifying signals through the combination of two hairpin DNA probes, leading to the formation of an extensive DNA chain. This chain undergoes further amplification by continuously opening and extending the hairpin structures, thus significantly enhancing the fluorescence signal. Zhu et al. described a cascade bHCR amplification activated by co-marker recognition, which generates multiple G-quadruplex structures combined with fluorescent dyes to facilitate signal transduction. This method significantly improves accurate detection by effectively minimizing false positives caused by interfering proteins or individual protein markers [113]. Additionally, various advanced approaches have been reported, such as employing DNA nanostructures as "nanoweights" to enrich cancerous EVs [114], and utilizing CRISPR-Cas12a assisted HCR [115]. For clinical applications, He et al. developed a highly sensitive assay that enables direct visualization and measurement of single vesicles in 1 μL plasma, as illustrated in Figure 8 [116]. This assay depends on the specific interaction between activatable aptamer probes and EVs, which are captured by EVs-specific antibodies affixed to the flow cell's surface, leading to the generation of activated fluorescence. The assay operates within a linear range of 103 to 107 particles μL-1.
A novel and effective microchip electrophoresis (MCE)-based method has been developed for detecting MCF-7 EVs, utilizing a triple amplification strategy. This approach combines a cholesterol probe (Chol-probe) with displacement amplification-CHA (SDA-CHA) [117]. The immobilization of CD63 aptamers on avidin magnetic beads facilitates the specific capture of EVs. Following magnetic separation, the Chol-probe complements ssDNA and initiates SDA, leading to the production of multiple DNA sequences (Ta) that trigger CHA, resulting in SDA-CHA amplification. The LOD for MCF-7 EVs was achieved at an impressive level of 26 particles μL-1. Additionally, Zhou et al. proposed a robust fluorescence assay based on aptamer-initiated CHA, achieving an LOD of 0.5 particles μL-1 for MCF-7 cell-derived EVs and 0.1 particles μL-1 for PANC-1 cell-derived EVs, as depicted in Figure 9 [118]. The assay also demonstrated excellent performance in serum samples, with a recovery rate ranging from 95.45% to 106.2%.
Enzymes-assisted amplification detection
DNAzymes are synthetic DNA molecules with enzymatic activity that catalyze specific reactions and can be employed as fluorescence probes for the sensitive and specific identification of EVs. Hao et al. developed a DNAzyme assembly featuring an aptamer-bivalent-cholesterol-anchor, which facilitates straightforward identification of exosomal PD-L1 with a LOD of 5.21 pg mL-1 [119]. This assay is completed in approximately 130 min. Additionally, a sensitive strategy was developed to selectively detect EVs in just 10 min, using multi-component nuclease (MNAzyme) mediated recognition of EpCAM and CD63 proteins on the EV membrane. This strategy enables highly sensitive and specific quantification of EVs in complex biological samples, with an LOD of 72 particles μL-1 [120].
Illustration of the ABDN-based TIRF assay for single-vesicle imaging and detection of circulating tumor-specific Exos in plasma. Adapted with permission from [116], copyright 2019 American Chemical Society.
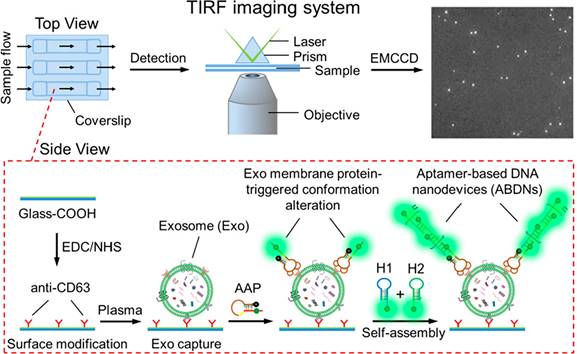
Principle of the aptamer-initiated CHA (AICHA) signal amplification strategy for exosome detection. H1 was modified with a FAM fluorophore and BHQ2 quencher. SA-MB: streptavidin-modified magnetic beads. Biotin-aptamer: biotin modified aptamer. Adapted with permission from [118], copyright 2022 American Chemical Society.
The CRISPR/Cas12a system, a molecular scissor, utilizes the Cas12a protein (also known as Cpf1) and a guide RNA (gRNA) to precisely edit targeted DNA sequences by cleaving both strands at specific locations determined by the gRNA. Zhao's group introduced an allosteric probe-initiated dual cycle amplification-assisted CRISPR-Cas12a, demonstrating a direct correlation with isolated EVs across a concentration range of 102 to 106 particles μL-1 [121]. This approach combines aptamers, PCR-based exponential amplification, and real-time DNA detection using CRISPR/Cas12a. Analysis of clinical serum samples indicated that measuring levels of serum CD109+ and EGFR+ tumor-derived EVs together resulted in high diagnostic accuracy, with an area under the curve (AUC) of 0.934 (95% CI 0.868-1.000), a sensitivity of 84.1%, and a specificity of 85.0% [122]. Additionally, biosensors mediated by terminal deoxynucleotidyl transferase (TdT) [123] and restriction endonucleases (Nt. BstNBI) [124] have also been applied for EV detection.
Nanomaterials-assisted amplification detection
Nano-materials are engineered to enhance the sensitivity and specificity of detection methods, thereby facilitating the precise determination of EVs levels in biological specimens. Jin et al. introduced an EVs -targeted, aptamer nanoprobe-based profiling technique that combines graphene oxide (GO) with aptamers specific to targeted surface proteins, enabling the characterization and quantification of cancerous EVs in a straightforward mix-and-detect format [125]. This assay successfully identified the EVs surface protein PSMA in blood samples from prostate cancer patients, demonstrating its potential for clinical diagnostic applications. In a separate study, Xia et al. employed the BP@Mn2+/DNA hybrid nanosensor for the direct detection of exosomal miRNAs and cancer-specific EVs in clinical plasma samples, aiming to diagnose cancer [126]. Alternatively, signal amplification approaches mediated by copper [127] and gold nanoparticles (AuNPs) [128] utilize a sandwich format to enhance sensitivity, specificity, multiplexing capability, non-destructive detection, compatibility with various sample types, and rapid detection. These features make them invaluable tools for EVs analysis in diverse research and clinical contexts.
Direct fluorescence detection
Traditional techniques for detecting EVs through fluorescence typically require prior purification and signal amplification steps. However, several studies have demonstrated the feasibility of signal amplification-independent fluorescence detection of EVs with exceptional sensitivity [129, 130]. A fluorescence polarization assay utilizing aptamers has been developed, enabling direct quantification of EVs in human plasma without requiring separation or amplification [131]. Additionally, to overcome challenges associated with the sensitive and direct detection of EVs from plasma, Wang et al. introduced a Single Microbead-based Fluorescent Aptasensor (SMFA). This method combines direct magnetic isolation with in situ fluorescence imaging [132]. In this technique, a single microbead (MB) modified with aptamers acts as the reaction carrier, allowing specific EVs with a fluorescent anchor to be highly enriched on the MB. This approach achieves sensitive detection of EVs without requiring signal amplification, by utilizing in situ fluorescence imaging to monitor the signals on the single MB.
Luminescence detection
Luminescent techniques are employed for the detection and quantification of EVs through light emission. These methods confer advantages such as enhanced sensitivity, specificity, and the capability for real-time analysis. In aptamer-based EVs detection, luminescence is primarily generated through two mechanisms: electrochemiluminescence (ECL) and luminescence resonance energy transfer (LRET).
Electrochemiluminescence detection
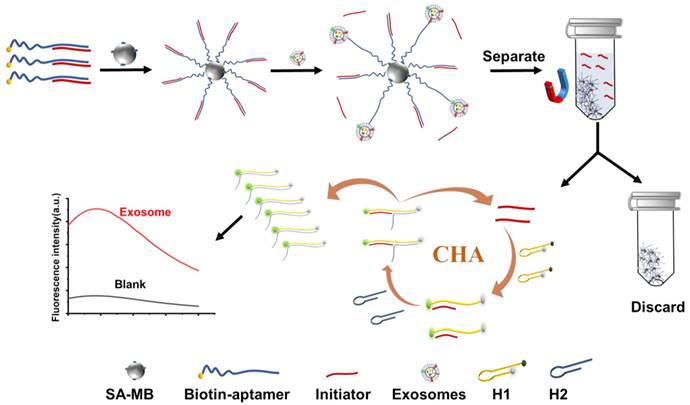
ECL merges the advantages of chemiluminescence and electrochemistry, transforming electrical energy into light emission. This technique provides benefits including low background noise, convenience, rapid response, and high sensitivity. There are two primary strategies for ECL-based EVs detection: signal-enhancement and signal-decrease. The signal-enhancement strategy can be realized through the use of nanomaterials that amplify the ECL signal. Zhang's group developed an ECL biosensor using aptamer-modified Ti3C2 MXenes nanosheets as the ECL nanoprobe [133]. This modification enables efficient capture of EVs on the electrode surface, while the ECL nanoprobe boosts the luminol signals, thereby enhancing EVs detection. Subsequently, they created a highly sensitive ECL biosensor employing a hybrid material composed of AuNPs-decorated Ti3C2 MXenes with aptamer modifications, as shown in Figure 10 [134]. This biosensor achieved a detection limit for HeLa cell-derived EVs of 30 particles μL-1, significantly surpassing the sensitivity of traditional ELISA methods by over a thousandfold. More recently, Nie et al. constructed a self-luminous Faraday cage-type sensing system using a nanoarchitecture that includes a Ti3C2Tx-Bi2S3-x heterostructure and an engineered lipid layer [135]. This sensor detects tumor EVs in gastric cancer patient ascites without additional purification. Conversely, the signal-decrease strategy involves inhibiting electron transfer, leading to a decrease in ECL signal proportional to the concentration of EVs on the electrode, thus enabling quantitative detection. Qiao et al. introduced an ECL aptasensor utilizing mercaptopropionic acid (MPA)-modified Eu3+-doped CdS nanocrystals as emitters [136]. The CD63 aptamer captures EVs, which then transform into a G-quadruplex/hemin DNAzyme. This DNAzyme accelerates the decomposition of H2O2, consequently reducing the ECL signal.
Luminescence resonance energy transfer detection
LRET is a process where energy is non-radiatively transferred from an excited donor molecule to an acceptor molecule via dipole-dipole interactions across significant distances. This method facilitates the detection of EVs in complex biological matrices while effectively reducing background signal interference. A straightforward aptasensor employing the LRET mechanism between upconversion nanoparticles (UCNPs) and gold nanorods (Au NRs) was developed [137]. The extent of luminescence quenching in UCNPs is directly proportional to the EV concentration, which ranges from 1×104 to 1×108 particles μL-1, enabling both detection and quantification of EVs. Additionally, Wang et al. introduced an aptasensor that operates without washing, using LRET between rare-earth-doped UCNPs as the emitter and tetramethyl rhodamine (TAMRA) as the acceptor [138]. In the presence of EVs, TAMRA emits yellow fluorescence at 585 nm due to LRET when the UCNPs-TAMRA system is excited by near-infrared light at 980 nm. Utilizing UCNPs as the energy donor, this method achieves a LOD of 80 particles μL-1, significantly minimizing background interference.
Electrochemical detection
The electrochemical detection method is well-suited for analyzing biological samples due to its high sensitivity, specific target recognition, ease of use, and cost-effectiveness. This approach assesses the properties of targets by monitoring changes in voltage or current. In this process, capture molecules such as antibodies or aptamers are used to bind to the target EVs, while a detection antibody connected to an electroactive signal transducer is utilized to identify the targets and generate electrical signals. The development of electrochemical detection for tumor EVs using aptasensors primarily involves direct immobilization-based detection and label-free detection.
Direct immobilization-based detection
Conductive materials are commonly used to modify electrode surfaces, enhancing electron transfer between the electrode and captured EVs. Various techniques are employed to immobilize aptamers onto these modified surfaces. The electrochemical signals generated by the captured EVs can be detected and quantified, providing valuable insights into the presence and concentration of EVs in the sample. Wang et al. employed advanced aptamer technology, DNA-based nanostructures, and portable electrochemical devices to develop an aptasensor assisted by nanotetrahedra [139]. The precise immobilization of aptamers significantly increased the availability of aptamers containing synthetic nucleobases to bind with suspended EVs, resulting in a 100-fold increase in sensitivity compared to aptasensors modified with individual aptamers. Besides, DNA walkers enhance electrochemical detection by incorporating amplification strategies that increase sensitivity, enabling the detection of EVs at low concentrations. A ratiometric electrochemical biosensor was developed for the direct detection of breast cancer exosomal miR-21, utilizing bipedal DNA walkers powered by locked nucleic acid (LNA) modified toehold mediate strand displacement reaction (TMSDR) [140]. The bipedal walker, with its dual pedals, demonstrates superior signal amplification efficiency compared to a single pedal DNA walker, which is prone to drifting away from tracks. Similarly, Zhao et al. described a highly sensitive detection method utilizing a target-triggered three-dimensional DNA walking machine and exonuclease III-assisted electrochemical proportional biosensing for the detection of exosomes, achieving a LOD of 13 particles μL-1 [141]. An exonuclease III-assisted electrochemical ratiometric sensor was employed to further enhance signal amplification. Besides the aforementioned amplification strategies, signal amplification methods based on nanomaterials have also been widely applied. Wu et al. described the creation of a nanosensor using a dual-aptamer modified reduced graphene oxide (RGO) field-effect transistor (GFET) platform, adorned with AuNPs [142]. This nanosensor achieved a detection limit of 84 particles μL-1 for HepG2-MVs. Furthermore, a novel electrochemical biosensor was developed by integrating hierarchical gold nanoarrays onto two-dimensional Ti2CTx MXene membranes [143]. This biosensor demonstrated a LOD of 58 particles μL-1 within a linear range of 1×102 to 1×107 particles μL-1.
Principle of the ECL biosensor for exosomes detection based on in situ formation of gold nanoparticles decorated Ti3C2 MXenes nanoprobes. Adapted with permission from [134], copyright 2020 American Chemical Society.
Label-free detection
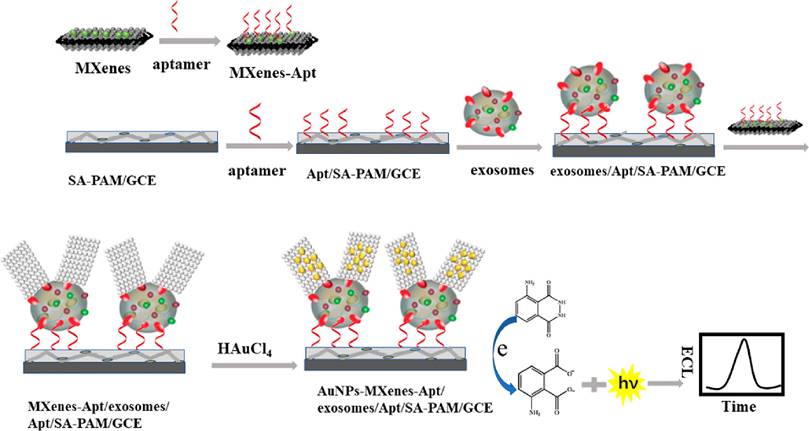
Label-free EC detection facilitates the sensitive detection of EVs in samples without requiring electrode biofunctionalization or the labeling of signal tags on aptamers, thereby circumventing complex and costly multistep procedures. Zhang et al. developed a straightforward electrochemical aptasensor that operates without labeling, utilizing target-induced proximity hybridization of divided aptamers [144]. This sensor captures PTK7-containing EVs using oligonucleotide probes that target PTK7 fragments. In the presence of target EVs, proximity hybridization occurs, leading to the formation of a DNA duplex on the electrode. This duplex subsequently attracts a higher concentration of Ru(NH3)63+, resulting in an enhanced cathodic current signal. The aptasensor effectively detected tumor-derived EVs with a LOD of approximately 660 particles μL-1. In a similar vein, Liu et al. engineered a simple electrochemical biosensor that operates without labels by integrating a paper coated with a metal-organic framework (MOF) onto a screen-printed electrode (SPE) [145]. This biosensor detects EVs using Zr-MOFs and aptamers, initiating a HCR that generates a DNAzyme for signal amplification, as illustrated in Figure 11. This method surpasses many previously documented techniques, achieving a minimal LOD of 5 particles μL-1. A reusable electrochemical biosensor has been developed, which integrates a DNA walker activated by dual-recognition proximity binding using an on-off-on approach [146]. The hairpin DNA (H) underwent a conformational change upon hybridizing with intermediate DNA, significantly increasing the distance between the electroactive label and the electrode surface, known as the sensor's "off" state. Exposure to exonuclease led to the degradation of the intermediate DNA and the restoration of the initial hairpin configuration, reactivating the sensor.
Principle of the paper-based biosensor for exosome assay. Adapted with permission from [145], 2021 American Chemical Society.
Colorimetric detection
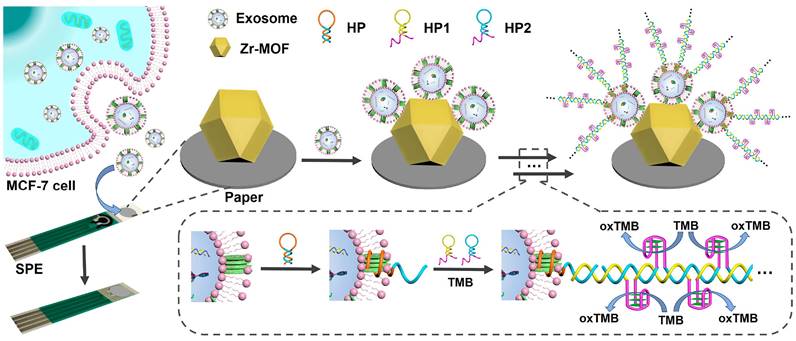
Colorimetric methods for EVs detection have garnered significant attention due to their simplicity, cost-effectiveness, and suitability for point-of-care (POC) applications. These methods depend on the color change in a solution or substrate when EVs are present, enabling visual detection without complex instrumentation. The H₂O₂ oxidation reaction, catalyzed by enzymes such as horseradish peroxidase (HRP), involves the oxidation of a chromogenic substrate in the presence of H₂O₂, resulting in a colored product, typically blue or yellow. This product can be quantified using a spectrophotometer or visually compared to a color chart or standard samples. Zou et al. reported a colorimetric biosensor for point-of-care testing (POCT) that employs RCA and the H₂O₂ oxidation reaction to detect cancerous EVs [147]. In the presence of target EVs, RCA is activated, producing multiple units that bind to DNA probes modified with HRP. This modification causes the transparent ABTS to oxidize, exhibiting strong absorbance in the near-infrared range and enabling both colorimetric and photothermal detection of EVs. Additionally, another colorimetric detection method achieved results within 10 min by catalyzing the transformation of the colorless substrate dopamine into dark-colored polydopamine using HRP in a specially prepared H₂O₂ solution [148]. The LOD was significantly improved to 7.7 particle μL-1, surpassing conventional dot-blot methods by three to five orders of magnitude. Recently, Wang et al. utilized the biotin-streptavidin system combined with MXene nanomaterials to enhance the loading of aptamers and biotin through a dual-effect amplification method, leveraging their substantial specific surface area [149]. Other innovative colorimetric biosensors include those based on enzymatic catalysis by glucose oxidase (GOx) [150, 151], G-quadruplex DNAzyme [152-154], and nano-enzymes [155]. For instance, Ding et al. devised a straightforward platform that uses a DNA-driven photothermal amplification transducer and a simple household thermometer, eliminating the need for sophisticated instruments or labeling processes, as shown in Figure 12 [154]. The LOD was determined to be 2.7 particle μL-1, demonstrating a 103-fold increase in sensitivity compared to fluorescence-based ELISA techniques. Zhang et al. developed a highly responsive and versatile visual technique for detecting EVs, which involves silver deposition on Au NRs induced by enzymatic activity and signal amplification via HCR [156]. CD63 aptamer-labeled magnetic beads were employed to capture EVs, and the exosomal membrane was modified by integrating cholesterol-modified DNA probes. These probes triggered HCR, leading to increased loading of alkaline phosphatase and the production of ascorbic acid. Subsequently, the ascorbic acid reduced silver ions, resulting in the formation of silver shells on the Au NRs, which shifted the plasmon resonance peak towards the blue spectrum. The method achieved a LOD of 1.6×102 particles μL-1 using UV-visible spectroscopy, while visual observation identified limits of 9×103 particles μL-1. However, colorimetric methods may exhibit a limited dynamic range, potentially restricting their capacity to accurately quantify a broad spectrum of EVs concentrations, particularly in samples with significant variability in EVs abundance. Overall, colorimetric methods for EVs detection provide a straightforward and rapid means of analysis, though further research and development are necessary to enhance the performance and reliability of these assays.
Schematic illustration of the DNA-driven photothermal amplification transducer for highly sensitive visual determination of EVs. Adapted with permission from [154], copyright 2023 American Chemical Society.
Microfluidics integrated detection
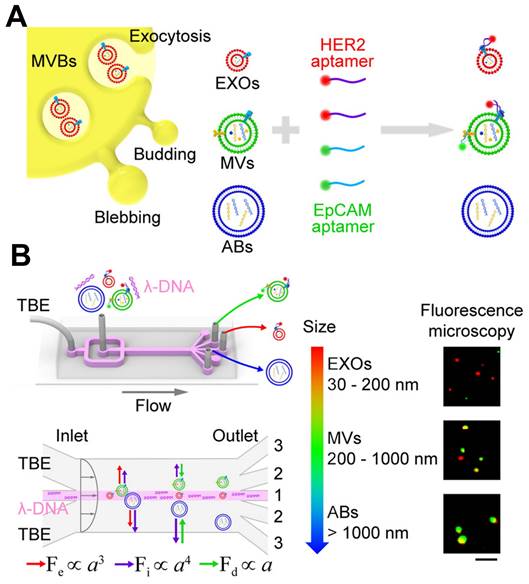
The integration of aptamers with microfluidic devices enables researchers to establish a platform for the capture, isolation, and detection of EVs. Aptamers, immobilized on the surfaces of microfluidic channels or functionalized magnetic beads, facilitate the specific capture of EVs from complex biological samples. Subsequently, the captured EVs can undergo various detection methods for analysis. This integrated approach provides several advantages for EVs detection, including enhanced sensitivity, rapid analysis, and the capacity to analyze small sample volumes. Additionally, it allows for the simultaneous detection of different exosomal markers, enabling the characterization of EVs with specific cargo molecules. The detection of various exosomal miRNAs has been successfully achieved using CHA, which utilizes a hairpin DNA probe and a target RNA molecule to initiate a cascade of signal amplification [157]. Exosomal surface proteins and miRNAs were simultaneously detected in situ using a microcolumn array within a ship-shaped microfluidic device. To minimize signal interference, the identification of exosomal surface proteins and miRNAs was conducted in four separate channels using aptamers labeled with Cy5 and a signal amplification technique based on CHA. Zheng et al. introduced a novel PTCDI-aptamer signal switch strategy for sensitive quantification, combined with a microfluidic chip for specific isolation of EVs [158]. The platform exhibits a wide linear range across six orders of magnitude and a low detection limit of 8.69 particles μL-1 for HepG2 EVs, attributed to the effective on-chip enrichment using Tim4-modified magnetic beads. Zhao et al. reported a novel method that integrates a functionalized membrane with an automated centrifugal microfluidic disc system to isolate and enrich EVs for clinical applications [159]. In clinical sample trials, this approach achieved a diagnostic accuracy of 91% (95% CI 79%-96%), outperforming EVs ELISA for lung cancer detection (area under the curve 0.9378 versus 0.8733, 30 patients). Liu et al. conducted an analysis focused on individual EVs, employing λ-DNA and aptamers for the concurrent isolation of distinct EVs according to their size and the examination of surface proteins, as depicted in Figure 13 [160]. Utilizing a machine learning algorithm, they demonstrated that the isolated microvesicles were superior in differentiating between breast cell lines and Stage II breast cancer patients with varying levels of HER2 expression, through the identification of EVs signatures. In cancer diagnosis, microfluidics offers a promising method for identifying EVs. It provides precise control over fluid flow, enables the integration of various detection strategies, and offers potential for high-throughput analysis. However, further research is necessary to establish standardized detection criteria, enhance machining accuracy, and develop more precise analysis methods for EVs detection based on microfluidics.
Schematic of λ-DNA-mediated sorting of EV subpopulations and aptamer-based analysis of individual EVs. (A) Labeling of cell-originating EVs including exosomes (EXOs, red), microvesicles (MVs, green), and apoptotic bodies (ABs, blue) with fluorescent HER2 and EpCAM aptamers. (B) Size-selective separation of EV subpopulations by λ-DNA mediated viscoelastic microfluidics. Fluorescence microscopy images showed HER2 (red) and EpCAM (green) expression of isolated individual EVs. Scale bar, 5 μm. Adapted with permission from [160], copyright 2019 American Chemical Society.
Others
Fluorescence resonance energy transfer (FRET) is a technique used to detect molecular interactions and conformational changes in biological systems, involving energy transfer between a donor and an acceptor fluorophore. The donor fluorophore is attached to an aptamer or antibody that binds to EVs surface markers, while the acceptor fluorophore is linked to a complementary aptamer or antibody. FRET occurs when EVs bind to these labeled probes, inducing a measurable change in the fluorescence signal. Zhang et al. reported an innovative self-standard ratiometric FRET nanoprobe, consisting of a Cy3-CD63 aptamer and Ti3C2 MXene nanosheets [161]. The Cy3-CD63 aptamer specifically binds to the Ti3C2 MXene nanosheets through hydrogen bonds and metal chelate interactions, leading to the quenching of Cy3 fluorescence. The introduction of EVs disrupts this interaction, resulting in the restoration of Cy3 fluorescence. The detection limit for EVs was established at 1.4 particles μL-1, which is a thousandfold improvement over traditional ELISA techniques. Recently, Xiao et al. introduced a system for detecting extracellular vesicle membrane proteins using FRET signals between aptamer quantum dots and AIEgen dye, eliminating the need for EVs extraction and purification [162]. This system demonstrated significantly higher accuracy (100% versus 65%) and sensitivity (100% versus 55%) in lung cancer diagnosis compared to ELISA. Additionally, by incorporating machine learning analysis, the accuracy of early screening was improved from 96.4% to 100% through the profiling of five biomarkers. FRET allows for real-time monitoring of EVs binding, enabling dynamic studies of interactions and uptake, and provides quantitative data on EVs concentration. Furthermore, FRET assays can be integrated with microfluidic devices or biosensors for enhanced high-throughput and multiplexed EVs analysis.
SERS is an optical analytical method that significantly amplifies the Raman scattering signal of molecules adsorbed onto rough metal surfaces or metal nanostructures. This technique has the potential to advance EVs-related diagnostics and treatments in various fields, including cancer research and precision healthcare. Wang et al. explored the use of magnetic substrates and SERS probes to detect and characterize EVs [163]. They combined capturing substrates with three distinct types of SERS probes, each based on different SERS reporters. The detection of target EVs was indicated by the formation of an apta-immunocomplex, which corresponded to a decrease in the SERS signal in the supernatant. More recently, Cun et al. developed a HCR-amplified strategy featuring an Ag-shell nanostructure induced by alkaline phosphatase for the SERS-based assay of EVs [164]. Prostate cancer-derived EVs were isolated using magnetic beads modified with prostate-specific membrane antigen aptamers. Following this, the release of the HCR-amplified chain incorporated multiple functional moieties, enhancing the signal. The use of magnetic materials simplified the traditional immunoassay steps, achieving a LOD of 19 particles μL-1 within 40 min.
Additionally, Wang et al. demonstrated an aptasensor based on SPR facilitated by dual AuNPs [165]. Electronic coupling between the Au film and AuNPs was achieved through controlled hybridization, leading to significant signal amplification. This technique also observed coupling effects in plasmonic nanostructures. By employing this method, undesirable binding of AuNPs to the SPR chip surface was effectively prevented, and the SPR sensor's functionality was restored by applying a layer of 11-Mercapto-1-undecanol to the Au film surface.
The phenomenon of Aggregation Induced Emission (AIE) is characterized by the enhanced emission intensity of certain fluorescent molecules during aggregation. This property has been extensively utilized across a variety of sensing and imaging applications. Within the field of EVs detection, AIE-based techniques have been refined to improve the sensitivity and specificity of assays. Utilizing the self-assembly capabilities of imprinted polymer technology, EVs serve as templates to construct EVs-specific three-dimensional cavities on the surfaces of magnetic particles [166]. These cavities, formed by the template-eluted imprinted polymers, selectively capture analytes in serum. Then, aptamer-mediated AIE has been employed to selectively activate these captured targets. The effectiveness of this method was evaluated using lysozyme and fuzzy EVs as targets, demonstrating a linear relationship between fluorescence intensity and target concentration in serum, with a notable recovery rate of lysozyme at 107%.
Harnessing the high specificity and sensitivity of droplet digital PCR (ddPCR), Lin et al. introduced a novel approach that combines dual-target-specific aptamer recognition, activated upon connection with the EVs membrane, with ddPCR for the precise quantification of tumor-derived exosomal PD-L1 [167]. This method holds significant potential for transforming EVs into reliable clinical biomarkers and facilitating the exploration of their biological roles.
Conclusions and future perspectives
We have provided an overview of recent advancements in EVs research, encompassing sample collection, isolation, storage, characterization, analysis, and applications in cancer detection. EVs contain a wealth of biomolecules and cellular information, emerging as promising biomarkers for monitoring various physiological processes and diseases. A significant difference in EVs levels between cancer patients and healthy individuals has been documented. Advances in EVs research have enhanced our understanding of their structural features, fundamental constituents, formation processes, physical attributes, and cellular roles. However, the isolation and preservation of EVs present challenges due to their complex formation, variability, and limited abundance. Although multiple isolation techniques exist, none achieve perfect isolation, with each method having limitations in yield, purity, and specificity. To improve specificity and purity, combining various techniques may be necessary, which could introduce variations in EVs concentration and size. Preservation of EVs generally avoids freezing. The significance of aptasensors in diagnostics and detection technologies is growing, playing an essential role. Several physical principles, including acoustics, thermodynamics, electrodynamics, hydrodynamics, and optics, in conjunction with functional molecules such as aptamers and molecular beacons, and engineering techniques like microfluidics, are utilized to augment the analysis of EVs and their associated biomolecules. Nevertheless, the lack of standardized protocols for the isolation, characterization, and analysis of EVs hinders the comparability of results among various studies. Establishing standardized procedures and reference materials is crucial for progressing EVs research and facilitating its clinical translation.
Aptamer-based EVs detection platforms can be tailored for in-situ detection, enhancing the study of EVs dynamics and interactions within complex biological systems. This capability offers significant opportunities for comprehensive profiling of EVs contents and their associations with disease states. Additionally, aptamers can be engineered to bind specifically to target nucleic acids or proteins, enabling the simultaneous detection of multiple analytes within EVs, which is vital for a range of biological and clinical applications. Despite these benefits, challenges remain that must be overcome to facilitate the widespread adoption of aptamer-based EVs detection. These challenges include the heterogeneous nature and small size of EVs, as well as the complex physical and chemical properties of bodily fluids. Improvements in the efficiency of selective separation-capture and overall analytical capabilities are crucial. Furthermore, identifying markers specific to EVs sources and developing high-performance aptamers for novel and personalized EVs targets are imperative. In contrast, aptamer-based detection methods offer potential for high sensitivity, specificity, accessibility, cost-effectiveness, and user-friendliness. With continuous advancements in research and technology, aptasensors are positioned to significantly contribute to the precise identification, analysis, and clinical application of EVs in precision medicine.
Abbreviations
ABTS: 2,2'-azinobis-(3-ethylbenzthiazoline-6-sulphonate); AFM: atomic force microscopy; AuNPs: gold nanoparticles; Au NRs: gold nanorods; CHA: catalytic hairpin assembly; CRISPR: clustered regularly interspaced short palindromic repeats; CSF: cerebrospinal fluid; ddPCR: droplet digital polymerase chain reaction; DLS: dynamic light scattering; DNA: deoxyribonucleic acid; EC: electrochemical; ECL: electrochemiluminescence; EDTA: ethylene diamine tetraacetic acid; ELISA: enzyme-linked immunosorbent assays; EVs: extracellular vesicles; FBS: fetal bovine serum; FCM: flow cytometry; FRET: fluorescence resonance energy transfer; GFET: field-effect transistor; GO: graphene oxide; GOx: glucose oxidase; HCR: hybridization chain reaction; HRP: horseradish peroxidase; ILVs: intraluminal vesicles; ITC: isothermal titration calorimetry; LOD: limit of detection; LRET: luminescence resonance energy transfer; MOF: metal-organic framework; MPA: mercaptopropionic acid; MVBs: multivesicular bodies; MVs: micro vesicles; NTA: nanoparticle tracking analysis; PEG: polyethylene glycol; P-ERCA: probe-based exponential rolling circle amplification; POCT: point-of-care testing; PSMA: prostate-specific membrane antigen; RCA: rolling cycle amplification; RGO: reduced graphene oxide; RNA: ribonucleic acid; SDR: strand displacement reaction; SEC: size exclusion chromatography; SELEX: systematic evolution of ligands by exponential enrichment; SERS: surface-enhanced Raman scattering; SPE: screen-printed electrode; SPR: surface plasmon resonance; TAMRA: tetramethyl rhodamine; TEM: transmission electron microscopy; UCNPs: upconversion nanoparticles; UF: ultrafiltration; UV: ultraviolet; WB: western blot.
Acknowledgements
This work was supported by the NSFC (Nos. 62071119 and 62075098) the National Key Research and Development Program of China (2017YFA0205301 and 2018YFC1602905).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Huang R, He L, Jin L, Li Z, He N, Miao W. Recent advancements in DNA nanotechnology-enabled extracellular vesicles detection and diagnosis: A mini review. Chin Chem Lett. 2023;34:107926
2. György B, Szabó TG, Pásztói M, Pál Z, Misják P, Aradi B. et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667-88
3. Wang T, Fu Y, Sun S, Huang C, Yi Y, Wang J. et al. Exosome-based drug delivery systems in cancer therapy. Chin Chem Lett. 2023;34:107508
4. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487-514
5. Zhang Y, Liu Y, Liu H, Tang WH. Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 2019;9:19
6. Arraud N, Linares R, Tan S, Gounou C, Pasquet JM, Mornet S. et al. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J Thromb Haemost. 2014;12:614-27
7. Zheng L, Hu B, Zhao D, Liu W, Liu Q, Huang Y. et al. Recent progresses of exosome-liposome fusions in drug delivery. Chin Chem Lett. 2024;35:108647
8. Fang Z, Zhang X, Huang H, Wu J. Exosome based miRNA delivery strategy for disease treatment. Chin Chem Lett. 2022;33:1693-704
9. Zhu F, Ji Y, Deng J, Li L, Bai X, Liu X. et al. Microfluidics-based technologies for the analysis of extracellular vesicles at the single-cell level and single-vesicle level. Chin Chem Lett. 2022;33:2893-900
10. Hu S, Hao R, Yu Z, Zhang H, Yang H. Highly efficient labelling of extracellular vesicles for enhanced detection on a microfluidic platform. Chin Chem Lett. 2024;35:108534
11. Li M, Fang F, Sun M, Zhang Y, Hu M, Zhang J. Extracellular vesicles as bioactive nanotherapeutics: An emerging paradigm for regenerative medicine. Theranostics. 2022;12:4879-903
12. Song J, Song B, Yuan L, Yang G. Multiplexed strategies toward clinical translation of extracellular vesicles. Theranostics. 2022;12:6740-61
13. Guo Z, Jin B, Fang Y, Deng Y, Chen Z, Chen H. et al. Selected aptamer specially combing 5-8F cells based on automatic screening instrument. Chin Chem Lett. 2022;33:4208-212
14. Liu Y, Li T, Yang G, Deng Y, Mou X, He N. A simple AuNPs-based colorimetric aptasensor for chlorpyrifos detection. Chin Chem Lett. 2022;33:1913-916
15. Guo Z, Jin B, Fang Y, Jin L, Li S, Deng Y. et al. Automated screening of primary cell-based aptamers for targeting and therapy of pancreatic cancer. Chin Chem Lett. 2024;35:108528
16. Li Y, Wu J, Liu R, Zhang Y, Li X. Extracellular vesicles: catching the light of intercellular communication in fibrotic liver diseases. Theranostics. 2022;12:6955-71
17. Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97:329-39
18. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987;262:9412-20
19. Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ. et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161-72
20. Théry C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P. et al. Molecular characterization of dendritic cell-derived exosomes: selective accumulation of the heat shock protein Hsc73. J Cell Biol. 1999;147:599-610
21. Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA. et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498-503
22. Yan H, Li Y, Cheng S, Zeng Y. Advances in analytical technologies for extracellular vesicles. Anal Chem. 2021;93:4739-74
23. Wang H, Liang P, Zhang L, Shi L, Ge Y, Zhang L. et al. A soluble pH-responsive host-guest-based nanosystem for homogeneous exosomes capture with high-efficiency. Chin Chem Lett. 2023;34:108129
24. Liu J, Zhang L, Zeng W, Zhang L, He N, Lu Z. High-throughput quantitative detection of triple-negative breast cancer-associated expressed miRNAs by rolling circle amplification on fluorescence-encoded microspheres. Chin Chem Lett. 2023;34:108141
25. Dyball LE, Smales CM. Exosomes: Biogenesis, targeting, characterization and their potential as 'Plug & Play' vaccine platforms. Biotechnol J. 2022;17:2100646
26. Xu M, Ji J, Jin D, Wu Y, Wu T, Lin R. et al. The biogenesis and secretion of exosomes and multivesicular bodies (MVBs): Intercellular shuttles and implications in human diseases. Genes Dis. 2023;10:1894-907
27. Yang J, Xu Y. Nanofluidics for sub-single cellular studies: Nascent progress, critical technologies, and future perspectives. Chin Chem Lett. 2022;33:2799-806
28. Beatriz M, Rodrigues RJ, Vilaça R, Egas C, Pinheiro PS, Daley GQ. et al. Extracellular vesicles improve GABAergic transmission in Huntington's disease iPSC-derived neurons. Theranostics. 2023;13:3707-24
29. Xu Y, Feng K, Zhao H, Di L, Wang L, Wang R. Tumor-derived extracellular vesicles as messengers of natural products in cancer treatment. Theranostics. 2022;12:1683-714
30. Wu L, Wang Y, Xu X, Liu Y, Lin B, Zhang M. et al. Aptamer-based detection of circulating targets for precision medicine. Chem Rev. 2021;121:12035-105
31. Mitchell JP, Court J, Mason MD, Tabi Z, Clayton A. Increased exosome production from tumour cell cultures using the Integra CELLine Culture System. J Immunol Methods. 2008;335:98-105
32. Sakakura H, Mii S, Hagiwara S, Kato T, Yamamoto N, Hibi H. et al. CD109 is a component of exosome secreted from cultured cells. Biochem Biophys Res Commun. 2016;469:816-22
33. Driedonks TAP, Nijen Twilhaar MK, Nolte-'t Hoen ENM. Technical approaches to reduce interference of Fetal calf serum derived RNA in the analysis of extracellular vesicle RNA from cultured cells. J Extracell Vesicles. 2019;8:1552059
34. Guo Y, He Z, Chen Z, Chen F, Wang C, Zhou W. et al. Inhibition of Th17 cells by donepezil ameliorates experimental lung fibrosis and pulmonary hypertension. Theranostics. 2023;13:1826-42
35. Lehrich BM, Liang Y, Khosravi P, Federoff HJ, Fiandaca MS. Fetal bovine serum-derived extracellular vesicles persist within vesicle-depleted culture media. Int J Mol Sci. 2018;19:3538
36. Zhang P, Zhou X, He M, Shang Y, Tetlow AL, Godwin AK. et al. Ultrasensitive detection of circulating exosomes with a 3D-nanopatterned microfluidic chip. Nat Biomed Eng. 2019;3:438-51
37. Zhao H, Su E, Huang L, Zai Y, Liu Y, Chen Z. et al. Washing-free chemiluminescence immunoassay for rapid detection of cardiac troponin I in whole blood samples. Chin Chem Lett. 2022;33:743-46
38. Klingeborn M, Skiba NP, Stamer WD, Bowes Rickman C. Isolation of retinal exosome biomarkers from blood by targeted immunocapture. In: Bowes Rickman C, Grimm C, Anderson RE, Ash JD, LaVail MM, Hollyfield JG, Ed. Retinal Degenerative Diseases. Cham: Springer International Publishing. 2019:21-5
39. Jayachandran M, Miller VM, Heit JA, Owen WG. Methodology for isolation, identification and characterization of microvesicles in peripheral blood. J Immunol Methods. 2012;375:207-14
40. Bæk R, Søndergaard EKL, Varming K, Jørgensen MM. The impact of various preanalytical treatments on the phenotype of small extracellular vesicles in blood analyzed by protein microarray. J Immunol Methods. 2016;438:11-20
41. Tian S, Cheng SB, Guo YY, Xie M, Zhan N, Zeng Z. et al. High efficient isolation of tumor cells by a three dimensional scaffold chip for diagnosis of malignant effusions. ACS Appl Bio Mater. 2020;3:2177-84
42. Roca E, Lacroix R, Judicone C, Laroumagne S, Robert S, Cointe S. et al. Detection of EpCAM-positive microparticles in pleural fluid: A new approach to mini-invasively identify patients with malignant pleural effusions. Oncotarget. 2015;7:3347-56
43. Shao MM, Pei XB, Chen QY, Wang F, Wang Z, Zhai K. Macrophage-derived exosome promotes regulatory T cell differentiation in malignant pleural effusion. Front Immunol. 2023;14:1161375
44. Hiroyuki T, Akihisa M, Akira S, Taro I, Minako S, Hideaki I. et al. Exosomal microRNA expression profiling in patients with lung adenocarcinoma-associated malignant pleural effusion. Anticancer Res. 2018;38:6707
45. Ogawa Y, Miura Y, Harazono A, Kanai-Azuma M, Akimoto Y, Kawakami H. et al. Proteomic analysis of two types of exosomes in human whole saliva. Biol Pharm Bull. 2011;34:13-23
46. Hyun KA, Gwak H, Lee J, Kwak B, Jung HI. Salivary exosome and cell-free DNA for cancer detection. Micromachines (Basel). 2018;9:340
47. Gallo A, Tandon M, Alevizos I, Illei GG. The Majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679
48. Ogawa Y, Kanai-Azuma M, Akimoto Y, Kawakami H, Yanoshita R. Exosome-like vesicles with dipeptidyl peptidase IV in human saliva. Biol Pharm Bull. 2008;31:1059-62
49. Zhou H, Yuen PST, Pisitkun T, Gonzales PA, Yasuda H, Dear JW. et al. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69:1471-6
50. Wang D, Sun W. Urinary extracellular microvesicles: Isolation methods and prospects for urinary proteome. Proteomics. 2014;14:1922-32
51. Liu J, Zhang Y, Tian Y, Huang W, Tong N, Fu X. Integrative biology of extracellular vesicles in diabetes mellitus and diabetic complications. Theranostics. 2022;12:1342-72
52. Huang G, Cheng Z, Hildebrand A, Wang C, Cimini M, Roy R. et al. Diabetes impairs cardioprotective function of endothelial progenitor cell-derived extracellular vesicles via H3K9Ac inhibition. Theranostics. 2022;12:4415-30
53. Cho S, Yang HC, Rhee WJ. Development and comparative analysis of human urine exosome isolation strategies. Process Biochem. 2020;88:197-203
54. Zhang N, Sun N, Deng C. Rapid isolation and proteome analysis of urinary exosome based on double interactions of Fe3O4@TiO2-DNA aptamer. Talanta. 2021;221:121571
55. Benito-Martin A, Ucero AC, Zubiri I, Posada-Ayala M, Fernandez-Fernandez B, Cannata-Ortiz P. et al. Osteoprotegerin in exosome-like vesicles from human cultured tubular cells and urine. PLoS One. 2013;8:e72387
56. Li S, Zhu Y, Haghniaz R, Kawakita S, Guan S, Chen J. et al. A microchambers containing contact lens for the noninvasive detection of tear exosomes. Adv Funct Mater. 2022;32:2206620
57. Sachiko I, Hiroki K, Sachiko M, Tomonari K, Motoi B, Yukiya K. et al. Oncogenic miRNAs identified in tear exosomes from metastatic breast cancer patients. Anticancer Res. 2020;40:3091
58. Hefley BS, Deighan C, Vasini B, Khan A, Hjortdal J, Riaz KM. et al. Revealing the presence of tear extracellular vesicles in Keratoconus. Exp Eye Res. 2022;224:109242
59. Shi TT, Zhao RX, Xin Z, Hou ZJ, Wang H, Xie RR. et al. Tear-derived exosomal biomarkers of Graves' ophthalmopathy. Front Immunol. 2022;13:1088606
60. Saugstad JA, Lusardi TA, Van Keuren-Jensen KR, Phillips JI, Lind B, Harrington CA. et al. Analysis of extracellular RNA in cerebrospinal fluid. J Extracell Vesicles. 2017;6:1317577
61. Pan Y, Long W, Liu Q. Current advances and future perspectives of cerebrospinal fluid biopsy in midline brain malignancies. Curr Treat Options Oncol. 2019;20:88
62. Akers JC, Ramakrishnan V, Nolan JP, Duggan E, Fu C-C, Hochberg FH. et al. Comparative analysis of technologies for quantifying extracellular vesicles (EVs) in clinical cerebrospinal fluids (CSF). PLoS One. 2016;11:e0149866
63. Kangas P, Nyman TA, Metsähonkala L, Burns C, Tempest R, Williams T. et al. Towards optimised extracellular vesicle proteomics from cerebrospinal fluid. Sci Rep. 2023;13:9564
64. Słomka A, Wang B, Mocan T, Horhat A, Willms AG, Schmidt-Wolf IGH. et al. Extracellular vesicles and circulating tumour cells-complementary liquid biopsies or standalone concepts? Theranostics. 2022;12:5836-55
65. Yang Z, Ji P, Li Z, Zhang R, Wei M, Yang Y. et al. Improved extracellular vesicle-based mRNA delivery for familial hypercholesterolemia treatment. Theranostics. 2023;13:3467-79
66. Benedikter BJ, Bouwman FG, Vajen T, Heinzmann ACA, Grauls G, Mariman EC. et al. Ultrafiltration combined with size exclusion chromatography efficiently isolates extracellular vesicles from cell culture media for compositional and functional studies. Sci Rep. 2017;7:15297
67. Han G, Li D, Lin Q, Yi J, Lyu Q, Ma Q. et al. Exponential isothermal amplification coupled MALDI-TOF MS for microRNAs detection. Chin Chem Lett. 2023;34:107421
68. Jia Y, Yu L, Ma T, Xu W, Qian H, Sun Y. et al. Small extracellular vesicles isolation and separation: Current techniques, pending questions and clinical applications. Theranostics. 2022;12:6548-75
69. Chen J, Li P, Zhang T, Xu Z, Huang X, Wang R. et al. Review on strategies and technologies for exosome isolation and purification. Front Bioeng Biotechnol. 2022;9:811971
70. Lobb RJ, Becker M, Wen SW, Wong CSF, Wiegmans AP, Leimgruber A. et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015;4:27031
71. Ko M, Kim HJ, Park J, Lee H, Lee KN, Kim K. et al. Isolation of bovine milk exosome using electrophoretic oscillation assisted tangential flow filtration with antifouling of micro-ultrafiltration membrane filters. ACS Appl Bio Mater. 2023;15:26069-80
72. Brooks DE, Haynes CA, Hritcu D, Steels BM, Müller W. Size exclusion chromatography does not require pores. Proc Natl Acad Sci U S A. 2000;97:7064-67
73. Gallart-Palau X, Serra A, Sze SK. Enrichment of extracellular vesicles from tissues of the central nervous system by PROSPR. Mol Neurodegener. 2016;11:41
74. Sharma P, Ludwig S, Muller L, Hong CS, Kirkwood JM, Ferrone S. et al. Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma. J Extracell Vesicles. 2018;7:1435138
75. Wang J, Ma P, Kim DH, Liu BF, Demirci U. Towards microfluidic-based exosome isolation and detection for tumor therapy. Nano Today. 2021;37:101066
76. Yuan F, Li YM, Wang Z. Preserving extracellular vesicles for biomedical applications: consideration of storage stability before and after isolation. Drug Deliv. 2021;28:1501-9
77. Fang Y, Wang Y, Zhu L, Liu H, Su X, Liu Y. et al. A novel cartridge for nucleic acid extraction, amplification and detection of infectious disease pathogens with the help of magnetic nanoparticles. Chin Chem Lett. 2023;34:108092
78. Liu C, Su C. Design strategies and application progress of therapeutic exosomes. Theranostics. 2019;9:1015-28
79. Zhang L, Xu L, Wang Y, Liu J, Tan G, Huang F. et al. A novel therapeutic vaccine based on graphene oxide nanocomposite for tumor immunotherapy. Chin Chem Lett. 2022;33:4089-95
80. Bahr MM, Amer MS, Abo-El-Sooud K, Abdallah AN, El-Tookhy OS. Preservation techniques of stem cells extracellular vesicles: a gate for manufacturing of clinical grade therapeutic extracellular vesicles and long-term clinical trials. Int J Vet Sci Med. 2020;8:1-8
81. Ashwood-Smith M. Mechanisms of cryoprotectant action. Symp Soc Exp Biol. 1987;41:395-406
82. Wu Y, Deng W, Klinke Ii DJ. Exosomes: improved methods to characterize their morphology, RNA content, and surface protein biomarkers. Analyst. 2015;140:6631-42
83. Davis JM, Rowley SD, Braine HG, Piantadosi S, Santos GW. Clinical toxicity of cryopreserved bone marrow graft infusion. Blood. 1990;75:781-6
84. Charoenviriyakul C, Takahashi Y, Nishikawa M, Takakura Y. Preservation of exosomes at room temperature using lyophilization. Int J Pharm. 2018;553:1-7
85. El Baradie KBY, Nouh M, O'Brien Iii F, Liu Y, Fulzele S, Eroglu A. et al. Freeze-dried extracellular vesicles from adipose-derived stem cells prevent hypoxia-induced muscle cell injury. Front Cell Dev Biol. 2020;8:181
86. Guarro M, Suñer F, Lecina M, Borrós S, Fornaguera C. Efficient extracellular vesicles freeze-dry method for direct formulations preparation and use. Colloids Surf B Biointerfaces. 2022;218:112745
87. Gauvin WH, Katta S. Basic concepts of spray dryer design. AICHE J. 1976;22:713-24
88. Kusuma GD, Barabadi M, Tan JL, Morton DAV, Frith JE, Lim R. To protect and to preserve: Novel preservation strategies for extracellular vesicles. Front Pharmacol. 2018;9:1199
89. Trevisan França de Lima L, Müller Bark J, Rasheduzzaman M, Weeramange CE, Punyadeera C. Isolation and characterization of salivary exosomes for cancer biomarker discovery. In: Federico M, Ridolfi B, Ed. Extracellular Vesicles in Diagnosis and Therapy. New York, NY: Springer US. 2022:101-12
90. van der Pol E, Coumans FAW, Grootemaat AE, Gardiner C, Sargent IL, Harrison P. et al. Particle size distribution of exosomes and microvesicles determined by transmission electron microscopy, flow cytometry, nanoparticle tracking analysis, and resistive pulse sensing. J Thromb Haemost. 2014;12:1182-92
91. Tian Y, Ma L, Gong M, Su G, Zhu S, Zhang W. et al. Protein profiling and sizing of extracellular vesicles from colorectal cancer patients via flow cytometry. ACS Nano. 2018;12:671-80
92. Zhang Y, Ju T, Gao M, Song Z, Xu H, Wang Z. et al. Electrical characterization of tumor-derived exosomes by conductive atomic force microscopy. Nanotechnology. 2022;33:295103
93. Soo CY, Song Y, Zheng Y, Campbell EC, Riches AC, Gunn-Moore F. et al. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136:192-7
94. Petersen KE, Manangon E, Hood JL, Wickline SA, Fernandez DP, Johnson WP. et al. A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLS-TEM. Anal Bioanal Chem. 2014;406:7855-66
95. Woo H-K, Cho YK, Lee CY, Lee H, Castro CM, Lee H. Characterization and modulation of surface charges to enhance extracellular vesicle isolation in plasma. Theranostics. 2022;12:1988-98
96. Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D. et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J Extracell Vesicles. 2015;4:25530
97. Lee S, Kang J, Ren S, Laurell T, Kim S, Jeong OC. A cross-contamination-free SELEX platform for a multi-target selection strategy. BioChip J. 2013;7:38-45
98. Ahn JY, Jo M, Dua P, Lee Dk, Kim S. A sol-gel-based microfluidics system enhances the efficiency of RNA aptamer selection. Oligonucleotides. 2011;21:93-100
99. Zhang WY, Zhang W, Liu Z, Li C, Zhu Z, Yang CJ. Highly parallel single-molecule amplification approach based on agarose droplet polymerase chain reaction for efficient and cost-effective aptamer selection. Anal Chem. 2012;84:350-5
100. Saito S. SELEX-based DNA aptamer selection: A perspective from the advancement of separation techniques. Anal Sci. 2021;37:17-26
101. Yu H, Zhu J, Shen G, Deng Y, Geng X, Wang L. Improving aptamer performance: key factors and strategies. Microchim Acta. 2023;190:255
102. Donovan MJ, Meng L, Chen T, Zhang Y, Sefah K, Tan W. Aptamer-drug conjugation for targeted tumor cell therapy. In: Goodchild J, Ed. Therapeutic Oligonucleotides: Methods and Protocols. Totowa, NJ: Humana Press. 2011:141-52
103. Qiao N, Li J, Wu X, Diao D, Zhao J, Li J. et al. Speeding up in vitro discovery of structure-switching aptamers via magnetic cross-linking precipitation. Anal Chem. 2019;91:13383-9
104. Chatterjee B, Kalyani N, Anand A, Khan E, Das S, Bansal V. et al. GOLD SELEX: a novel SELEX approach for the development of high-affinity aptamers against small molecules without residual activity. Microchim Acta. 2020;187:618
105. Narayan C, Veeramani S, Thiel WH. Optimization of RNA aptamer SELEX methods: improved aptamer transcript 3′-end homogeneity, PAGE purification yield, and target-bound aptamer RNA recovery. Nucleic Acid Ther. 2021;32:74-80
106. He L, Yu X, Huang R, Jin L, Liu Y, Deng Y. et al. A novel specific and ultrasensitive method detecting extracellular vesicles secreted from lung cancer by padlock probe-based exponential rolling circle amplification. Nano Today. 2022;42:101334
107. Huang R, He L, Li S, Liu H, Jin L, Chen Z. et al. A simple fluorescence aptasensor for gastric cancer exosome detection based on branched rolling circle amplification. Nanoscale. 2020;12:2445-51
108. Yang Z, She D, Sun C, Gong M, Rong Y. Dumbbell structure probe-triggered rolling circle amplification (RCA)-based detection scaffold for sensitive and specific neonatal infection-related small extracellular vesicle (sEV) detection. Anal Methods. 2022;14:1534-9
109. Wu N, Zhang XY, Xia J, Li X, Yang T, Wang JH. Ratiometric 3D DNA machine combined with machine learning algorithm for ultrasensitive and high-precision screening of early urinary diseases. ACS Nano. 2021;15:19522-34
110. Cheng S, Kong Q, Hu X, Zhang C, Xian Y. An ultrasensitive strand displacement signal amplification-assisted synchronous fluorescence assay for surface proteins of small extracellular vesicle analysis and cancer identification. Anal Chem. 2022;94:1085-91
111. Zhang G. Early-diagnosis of glioma through fluorescence detection of sEVs surface protein via isothermal cascade amplification strategy. Anal Biochem. 2021;633:114370
112. Chen J, Meng HM, An Y, Geng X, Zhao K, Qu L. et al. Structure-switching aptamer triggering hybridization displacement reaction for label-free detection of exosomes. Talanta. 2020;209:120510
113. Zhu J, Sun W, Yao Y, Guo Z, Li Q, Li Z. et al. Combination of specific proteins as markers for accurate detection of extracellular vesicles using proximity ligation-mediated bHCR amplification. Anal Chim Acta. 2023;1267:341322
114. Lu Z, Shi Y, Ma Y, Jia B, Li X, Guan X. et al. Fast and specific enrichment and quantification of cancer-related exosomes by DNA-nanoweight-assisted centrifugation. Anal Chem. 2022;94:9466-71
115. Xing S, Lu Z, Huang Q, Li H, Wang Y, Lai Y. et al. An ultrasensitive hybridization chain reaction-amplified CRISPR-Cas12a aptasensor for extracellular vesicle surface protein quantification. Theranostics. 2020;10:10262-73
116. He D, Ho S-L, Chan H-N, Wang H, Hai L, He X. et al. Molecular-recognition-based DNA nanodevices for enhancing the direct visualization and quantification of single vesicles of tumor Exosomes in plasma microsamples. Anal Chem. 2019;91:2768-75
117. Chen J, Zhang J, Xie Q, Chu Z, Zhang F, Wang Q. Ultrasensitive detection of exosomes by microchip electrophoresis combining with triple amplification strategies. Talanta. 2023;265:124930
118. Zhou J, Lin Q, Huang Z, Xiong H, Yang B, Chen H. et al. Aptamer-initiated catalytic hairpin assembly fluorescence assay for universal, sensitive exosome detection. Anal Chem. 2022;94:5723-8
119. Hao J, Wang J, Dong Y, Yang J, Wang Z, Zhao X. et al. Homogeneous, simple, and direct analysis of exosomal PD-L1 via aptamer-bivalent-cholesterol-anchor assembly of DNAzyme (ABCzyme) for tumor immunotherapy. Anal Chem. 2023;95:6854-62
120. Niu R, Chen X, Sun Z, Wang L, Wang Z, Zhang C. et al. A smart TESTER for reliable discrimination of cancer-derived small extracellular vesicles. Anal Chim Acta. 2023;1276:341636
121. Zhao X, Zeng L, Mei Q, Luo Y. Allosteric probe-Initiated wash-Free method for sensitive extracellular vesicle detection through dual cycle-assisted CRISPR-Cas12a. ACS Sens. 2020;5:2239-46
122. Li H, Xing S, Xu J, He Y, Lai Y, Wang Y. et al. Aptamer-based CRISPR/Cas12a assay for the ultrasensitive detection of extracellular vesicle proteins. Talanta. 2021;221:121670
123. Wang L, Pan Y, Liu Y, Sun Z, Huang Y, Li J. et al. Fabrication of an aptamer-coated liposome complex for the detection and profiling of exosomes based on terminal deoxynucleotidyl transferase-mediated signal amplification. ACS Appl Bio Mater. 2020;12:322-9
124. Feng J, Jia L, Pan W, Fan Y, Guo J, Luo T. et al. Rapid and efficient fluorescent aptasensor for PD-L1 positive extracellular vesicles isolation and analysis: EV-ANCHOR. Chem Eng J. 2023;465:142811
125. Jin D, Yang F, Zhang Y, Liu L, Zhou Y, Wang F. et al. ExoAPP: exosome-oriented, aptamer nanoprobe-enabled surface proteins profiling and detection. Anal Chem. 2018;90:14402-11
126. Xia Q, Zheng J, Bu J, Li R, Li X, Fan S. et al. Mn2+-modified black phosphorus nanosensor for detection of exosomal microRNAs and exosomes. Microchimic Acta. 2023;190:295
127. He F, Wang J, Yin BC, Ye BC. Quantification of exosome based on a copper-mediated signal amplification strategy. Anal Chem. 2018;90:8072-79
128. Liu Z, Zhang W, Zhang X, Wang S, Xia Z, Guo X. et al. Microstructured optical fiber-enhanced light-matter interaction enables highly sensitive exosome-based liquid biopsy of breast cancer. Anal Chem. 2023;95:1095-105
129. Chen H, Ma X, Zhang X, Hu G, Deng Y, Li S. et al. Novel aerosol detection platform for SARS-CoV-2: Based on specific magnetic nanoparticles adsorption sampling and digital droplet PCR detection. Chin Chem Lett. 2023;34:107701
130. Hussain M, Liu X, Zou J, Yang J, Ali Z, Rehman HU. et al. On-chip classification of micro-particles using laser light scattering and machine learning. Chin Chem Lett. 2022;33:1885-88
131. Zhang Z, Tang C, Zhao L, Xu L, Zhou W, Dong Z. et al. Aptamer-based fluorescence polarization assay for separation-free exosome quantification. Nanoscale. 2019;11:10106-13
132. Wang F, Zhang Y, Chen D, Zhang Z, Li Z. Single microbead-based fluorescent aptasensor (SMFA) for direct isolation and in situ quantification of exosomes from plasma. Analyst. 2021;146:3346-51
133. Zhang H, Wang Z, Zhang Q, Wang F, Liu Y. Ti3C2 MXenes nanosheets catalyzed highly efficient electrogenerated chemiluminescence biosensor for the detection of exosomes. Biosens Bioelectron. 2019;124-125:184-90
134. Zhang H, Wang Z, Wang F, Zhang Y, Wang H, Liu Y. In Situ Formation of gold nanoparticles decorated Ti3C2 MXenes nanoprobe for highly sensitive electrogenerated chemiluminescence detection of exosomes and their surface proteins. Anal Chem. 2020;92:5546-53
135. Nie Y, Wang P, Wang S, Ma Q, Su X. Accurate capture and identification of exosomes: nanoarchitecture of the MXene heterostructure/engineered lipid layer. ACS Sens. 2023;8:1850-7
136. Qiao B, Guo Q, Jiang J, Qi Y, Zhang H, He B. et al. An electrochemiluminescent aptasensor for amplified detection of exosomes from breast tumor cells (MCF-7 cells) based on G-quadruplex/hemin DNAzymes. Analyst. 2019;144:3668-75
137. Chen X, Lan J, Liu Y, Li L, Yan L, Xia Y. et al. A paper-supported aptasensor based on upconversion luminescence resonance energy transfer for the accessible determination of exosomes. Biosens Bioelectron. 2018;102:582-8
138. Wang Y, Luo D, Fang Y, Wu W, Wang Y, Xia Y. et al. An aptasensor based on upconversion nanoparticles as LRET donors for the detection of exosomes. Sens Actuators B Chem. 2019;298:126900
139. Wang S, Zhang L, Wan S, Cansiz S, Cui C, Liu Y. et al. Aptasensor with expanded nucleotide using DNA nanotetrahedra for electrochemical detection of cancerous exosomes. ACS Nano. 2017;11:3943-9
140. Zhang J, Wang LL, Hou MF, Xia YK, He WH, Yan A. et al. A ratiometric electrochemical biosensor for the exosomal microRNAs detection based on bipedal DNA walkers propelled by locked nucleic acid modified toehold mediate strand displacement reaction. Biosens Bioelectron. 2018;102:33-40
141. Zhao L, Sun R, He P, Zhang X. Ultrasensitive detection of exosomes by target-triggered three-dimensional DNA walking machine and exonuclease III-assisted electrochemical ratiometric biosensing. Anal Chem. 2019;91:14773-9
142. Wu D, Yu Y, Jin D, Xiao MM, Zhang ZY, Zhang GJ. Dual-aptamer modified graphene field-effect transistor nanosensor for label-free and specific detection of hepatocellular carcinoma-derived microvesicles. Anal Chem. 2020;92:4006-15
143. You Q, Zhuang L, Chang Z, Ge M, Mei Q, Yang L. et al. Hierarchical Au nanoarrays functionalized 2D Ti2CTx MXene membranes for the detection of exosomes isolated from human lung carcinoma cells. Biosens Bioelectron. 2022;216:114647
144. Zhang H, Qiao B, Guo Q, Jiang J, Cai C, Shen J. A facile and label-free electrochemical aptasensor for tumour-derived extracellular vesicle detection based on the target-induced proximity hybridization of split aptamers. Analyst. 2020;145:3557-63
145. Liu X, Gao X, Yang L, Zhao Y, Li F. Metal-organic framework-functionalized paper-based electrochemical biosensor for ultrasensitive exosome assay. Anal Chem. 2021;93:11792-9
146. Guo Y, Liu S, Yang H, Wang P, Feng Q. Regenerable electrochemical biosensor for exosomes detection based on the dual-recognition proximity binding-induced DNA walker. Sens Actuators B Chem. 2021;349:130765
147. Zou L, Liu X, Zhou Y, Mei W, Wang Q, Yang X. et al. Optical fiber amplifier and thermometer assisted point-of-care biosensor for detection of cancerous exosomes. Sens Actuators B Chem. 2022;351:130893
148. Xu L, Chopdat R, Li D, Al-Jamal KT. Development of a simple, sensitive and selective colorimetric aptasensor for the detection of cancer-derived exosomes. Biosens Bioelectron. 2020;169:112576
149. Wang Z, Zhang Z, Pan H, Chang D. An ultrasensitive aptasensor for exosomes detection based on biotin-streptavidin and MXenes. Anal Biochem. 2023;676:115233
150. Xu H, Zheng L, Zhou Y, Ye B-C. An artificial enzyme cascade amplification strategy for highly sensitive and specific detection of breast cancer-derived exosomes. Analyst. 2021;146:5542-9
151. Jiang Q, Xiao Y, Hong AN, Gao Z, Shen Y, Fan Q. et al. Bimetallic metal-organic framework Fe/Co-MIL-88(NH2) exhibiting high peroxidase-like activity and its application in detection of extracellular vesicles. ACS Appl Bio Mater. 2022;14:41800-8
152. Zhou Y, Xu H, Wang H, Ye BC. Detection of breast cancer-derived exosomes using the horseradish peroxidase-mimicking DNAzyme as an aptasensor. Analyst. 2020;145:107-14
153. Ning L, Zhou Y, Xie Y, Duan Z, Yu F, Cheng H. Ultrasensitive and colorimetric small extracellular vesicles (sEVs) analysis via dual-cycle signal tool. J Anal Sci Technol. 2023;14:5
154. Ding Z, Wei Y, Han F, Zhang X, Xu Z. DNA-driven photothermal amplification transducer for highly sensitive visual determination of extracellular vesicles. ACS Sens. 2023;8:2282-9
155. Wang Y-M, Liu J-W, Adkins GB, Shen W, Trinh MP, Duan L-Y. et al. Enhancement of the intrinsic peroxidase-like activity of graphitic carbon nitride nanosheets by ssDNAs and its application for detection of exosomes. Anal Chem. 2017;89:12327-33
156. Zhang Y, Wang D, Yue S, Lu Y, Yang C, Fang J. et al. Sensitive multicolor visual detection of exosomes via dual signal amplification strategy of enzyme-catalyzed metallization of Au nanorods and hybridization chain reaction. ACS Sens. 2019;4:3210-8
157. Chen Y, Gao D, Zhu Q, Chu B, Peng J, Wang J. et al. Rapid exosome isolation and in situ multiplexed detection of exosomal surface proteins and microRNAs on microfluidic platform. Analyst. 2023;148:2387-94
158. Zheng L, Wang H, Zuo P, Liu Y, Xu H, Ye B-C. Rapid on-chip isolation of cancer-associated exosomes andcombined analysis of exosomes and exosomal proteins. Anal Chem. 2022;94:7703-12
159. Zhao L, Wang H, Fu J, Wu X, Liang X, Liu X. et al. Microfluidic-based exosome isolation and highly sensitive aptamer exosome membrane protein detection for lung cancer diagnosis. Biosens Bioelectron. 2022;214:114487
160. Liu C, Zhao J, Tian F, Chang J, Zhang W, Sun J. λ-DNA- and aptamer-mediated sorting and analysis of extracellular vesicles. J Am Chem Soc. 2019;141:3817-21
161. Zhang Q, Wang F, Zhang H, Zhang Y, Liu M, Liu Y. Universal Ti3C2 MXenes based self-standard ratiometric fluorescence resonance energy transfer platform for highly sensitive detection of exosomes. Anal Chem. 2018;90:12737-44
162. Xiao S, Yao Y, Liao S, Xu B, Li X, Zhang Y. et al. Accurate and convenient lung cancer diagnosis through detection of extracellular vesicle membrane proteins via förster resonance energy transfer. Nano Lett. 2023;23:8115-25
163. Wang Z, Zong S, Wang Y, Li N, Li L, Lu J. et al. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale. 2018;10:9053-62
164. Cun F, Huang Z, Lin Q, Yu G, Chen H, Kong J. et al. Hybridized chain reaction-amplified alkaline phosphatase-induced Ag-shell nanostructure for the sensitive and rapid surface-enhanced Raman scattering immunoassay of exosomes. Anal Chem. 2023;95:10025-33
165. Wang Q, Zou L, Yang X, Liu X, Nie W, Zheng Y. et al. Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens Bioelectron. 2019;135:129-36
166. Liao Z, Peng J, Chen S, Zhang P, Chen H, Feng D. et al. Sensitive fluorescent sensor for the fuzzy exosomes in serum based on the exosome imprinted polymer sandwiched with aggregation induced emission. Sens Actuators B Chem. 2022;358:131182
167. Lin B, Tian T, Lu Y, Liu D, Huang M, Zhu L. et al. Tracing tumor-derived exosomal PD-L1 by dual-aptamer activated proximity-induced droplet digital PCR. Angew Chem Int Ed Engl. 2021;60:7582-6
168. Ren Y, Ge K, Lu W, Xie X, Lu Y, Wang M. et al. Multivalent DNA flowers for high-performance isolation, detection, and release of tumor-derived extracellular vesicles. ACS Appl Mater Interfaces. 2023;15:55358-68
169. Ye M, Niu Q, Yan L, Feng J, Wu L, Hu X. et al. Paper-based lipid bilayer magnetic interface for ultrasensitive assay of tumor-derived extracellular vesicles PD-L1. Sens Actuators B Chem. 2024;399:134819
170. Tan W, Zhang C, Cheng S, Hu X, Wang M, Xian Y. DNA gate-based CRISPR-Cas exponential amplification system for ultrasensitive small extracellular vesicle detection to enhance breast cancer diagnosis. Anal Chem. 2024;96:1328-35
171. Zhang YP, Chen HJ, Hu Y, Lin L, Wen HY, Pang DW. et al. Accurate cancer screening and prediction of PD-L1-guided immunotherapy efficacy using quantum dot nanosphere self-assembly and machine learning. Nano Lett. 2024;24:1816-24
172. Jiang Z, Luo K, Yang G, Li Y, Li L, Wang G. et al. An electrochemiluminescent sensor based on glycosyl imprinting and aptamer for the detection of cancer-related extracellular vesicles. Anal Chem. 2024;96:2550-8
173. Shi L, Cai H, Wang H, Wang Q, Shi L, Li T. Proximity-enhanced electrochemiluminescence sensing platform for effective capturing of exosomes and probing internal microRNAs involved in cancer cell apoptosis. Anal Chem. 2023;95:17662-9
174. Jiang B, Zhang T, Liu S, Sheng Y, Hu J. Polydopamine-assisted aptamer-carrying tetrahedral DNA microelectrode sensor for ultrasensitive electrochemical detection of exosomes. J Nanobiotechnology. 2024;22:55
175. Luo J, Wang B, Tang X, Huang P, Yang S, Zhao S. et al. CRISPR/Cas12a-loaded intelligent DNA hydrogel for universal and ultrasensitive exosome assay. View. 2024;5:20230086
176. Cong XZ, Feng J, Zhang HJ, Zhang LZ, Lin TY, Chen G. et al. Microfluidic device-based in vivo detection of PD-L1-positive small extracellular vesicles and its application for tumor monitoring. Anal Chem. 2024;96:2658-65
Author contact
![]() Corresponding authors: Yuan Liu, E-mail: gxuliuyuancom; Yan Deng, E-mail: hndengyancom; Yile Fang, E-mail: fang1lecom; Nongyue He, E-mail: nyhe1958com.
Corresponding authors: Yuan Liu, E-mail: gxuliuyuancom; Yan Deng, E-mail: hndengyancom; Yile Fang, E-mail: fang1lecom; Nongyue He, E-mail: nyhe1958com.
 Global reach, higher impact
Global reach, higher impact