13.3
Impact Factor
Theranostics 2023; 13(3):1165-1179. doi:10.7150/thno.81583 This issue Cite
Research Paper
Colonization with two different Blastocystis subtypes in DSS-induced colitis mice is associated with strikingly different microbiome and pathological features
1. Laboratory of Molecular and Cellular Parasitology, Department of Microbiology and Immunology, Healthy Longevity Translational Research Programme, Yong Loo Lin School of Medicine, National University of Singapore, 5 Science Drive 2, Singapore 117545, Singapore
2. Department of Microbiology and Immunology, Immunology Translational Research Programme, Yong Loo Lin School of Medicine, National University of Singapore, 5 Science Drive 2, Singapore 117545, Singapore
3. Department of Pharmacy, Faculty of Science, National University of Singapore, 18 Science Drive 4, Singapore 117559, Singapore
4. The Key Laboratory of Animal Disease and Human Health of Sichuan Province, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, Sichuan, 611130, China
Received 2022-12-6; Accepted 2023-1-4; Published 2023-2-5
Abstract
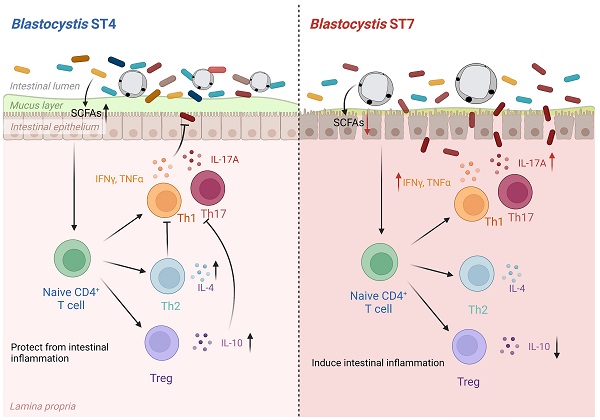
Rationale: The gut microbiota plays a significant role in the pathogenesis of inflammatory bowel disease (IBD). However, the role of Blastocystis infection and Blastocystis-altered gut microbiota in the development of inflammatory diseases and their underlying mechanisms are not well understood.
Methods: We investigated the effect of Blastocystis ST4 and ST7 infection on the intestinal microbiota, metabolism, and host immune responses, and then explored the role of Blastocystis-altered gut microbiome in the development of dextran sulfate sodium (DSS)-induced colitis in mice.
Results: This study showed that prior colonization with ST4 conferred protection from DSS-induced colitis through elevating the abundance of beneficial bacteria, short-chain fatty acid (SCFA) production and the proportion of Foxp3+ and IL-10-producing CD4+ T cells. Conversely, prior ST7 infection exacerbated the severity of colitis by increasing the proportion of pathogenic bacteria and inducing pro-inflammatory IL-17A and TNF-α-producing CD4+ T cells. Furthermore, transplantation of ST4- and ST7-altered microbiota resulted in similar phenotypes.
Conclusions: Our data showed that ST4 and ST7 infection exert strikingly differential effects on the gut microbiota, and these could influence the susceptibility to colitis. ST4 colonization prevented DSS-induced colitis in mice and may be considered as a novel therapeutic strategy against immunological diseases in the future, while ST7 infection is a potential risk factor for the development of experimentally induced colitis that warrants attention.
Keywords: Blastocystis, Gut microbiota, IBD, DSS-induced colitis, Short-chain fatty acids
Introduction
Inflammatory bowel disease (IBD) is a chronic gastrointestinal (GI) disorder, including Crohn's disease (CD) and ulcerative colitis (UC) [1]. Numerous factors are thought to play important roles in the pathogenesis of IBD, including the gut microbiome and immune dysregulation [2, 3]. The gut microbiome is composed of trillions of indigenous bacteria, archaea, fungi, viruses, and protists [4]. Studies have primarily focused on associations between bacteria and IBD, while the influence of gut protists on this disease is largely unexplored [5]. Several protists are typically known for causing dysenteric infections, such as Entamoeba histolytica, and Cryptosporidium spp. [6, 7]. However, the impact of other protists on host health remains debated, particularly for Blastocystis. Blastocystis is the most common gut protist found in humans and animals, and harbors extensive genetic heterogeneity [8]. The relationship between Blastocystis and IBD is controversial. Several studies have shown that Blastocystis prevalence was higher in IBD patients than in healthy individuals [9, 10], while Blastocystis was also considered to be an enteric commensal as it was found to be more abundant in asymptomatic individuals and associated with increased gut bacterial diversity [11].
Blastocystis colonization or infection could modulate the gut microbial composition and may influence the progression of IBD [12]. Experimental colitis induced by dextran sulfate sodium (DSS) in rodents is a common animal model of human IBD research [13]. Our previous study showed that infection with Blastocystis ST7 exacerbated the severity of DSS-induced colitis in a mouse model. This was accompanied with decreased proportions of beneficial bacteria such as Bifidobacterium and Lactobacillus [14]. However, our recent study showed that colonization with Blastocystis ST4 can reduce the severity of colitis through modulating the gut microbiota and activation of T helper-2 (Th2) and regulatory T cell (Treg) immune responses [15]. These earlier studies involved inoculation of Blastocystis after DSS treatment and investigated the effects of Blastocystis infection or colonization in promoting recovery or exacerbating DSS-induced colitis. However, whether prior Blastocystis infection has an impact on subsequent experimentally induced colitis and whether Blastocystis-altered gut microbiota is also able to influence the development of IBD remains unexplored.
Although several studies have shown the associations between Blastocystis colonization and host health, most studies do not demonstrate whether the effect of Blastocystis on host health is a direct result of immune regulation or an indirect effect of microbiome modulation. On another hand, studies looking at the effect of Blastocystis on the gut microbiota employed the use of one subtype, although it has been reported that distinct subtypes interact differently with bacteria. In this study, we compared the effects of colonization by two Blastocystis subtypes, ST4 and ST7, available as axenic cultures (pure cultures grown in the absence of bacteria) that have shown distinct effects on gut epithelial barrier function and immune responses in several in vitro studies [16, 17]. ST4 and ST7 were originally identified in a Wistar rat and a patient with gastrointestinal symptoms, respectively [18, 19]. We observed that prior ST4 colonization can prevent induced colitis when mice were later challenged with DSS, through elevating the proportion of beneficial bacteria, SCFAs, and protective immune responses. In contrast, prior infection with ST7 exacerbated colitis through inducing pro-inflammatory Th1 and Th17 immune responses. These results provide evidence that distinct modifications of gut microbial composition are triggered by specific Blastocystis subtypes, and that Blastocystis-altered gut microbiota play roles in the development of experimental colitis.
Materials and methods
Blastocystis cultures
Axenized cultures of Blastocystis ST4 and ST7 were used in this study. ST4-WR1 was first identified in a healthy Wistar rat in National University of Singapore (NUS), Singapore [19]. ST7-B was originally isolated from a diarrheal patient at the Singapore General Hospital in the early 1990s before the Institutional Review Board was established at the NUS [18]. Blastocystis was maintained in 10 ml of pre-reduced Iscove's modified Dulbecco's medium (IMDM) (Gibco) supplemented with heat-inactivated 10% horse serum (Gibco), and NaHCO3. Blastocystis cells were cultured in an Anaerojar (Oxoid) with Oxoid CO2 Gen Sachet (Thermo Scientific) at 37 °C. A hemocytometer (Kova International) was used to calculate the number of Blastocystis cells.
Mice and treatment
C57BL/6 mice, aged 8-12 weeks and bred in-house, were maintained in the animal facility of the National University of Singapore (NUS). Littermates of the same sex and age were randomly assigned to the different experimental groups. All animal experiments were performed under the Singapore National Advisory Committee for Laboratory Animal Research guidelines and approved by the Institutional Animal Care and Use Committee of NUS (protocol no. R19-1259).
Oral infection of Blastocystis. Mice (n = 6 per group) were orally gavaged with 5 × 107 live Blastocystis ST4 and ST7 cells suspended in sterile phosphate-buffered saline (PBS). The control mice (n = 6) were orally gavaged with equal amounts of PBS at the same time. The mice used in all the experiments were age and sex matched. Each treatment was housed together in two cages (n = 3).
DSS-induced colitis. Mice were exposed to 2% DSS (molecular mass = 36,000-50,000 Da; MP Biomedicals) w/v in drinking water for 7 days. The weight, stool consistency, and presence of fecal blood were recorded each day. The disease activity index (DAI) was used to determine the severity of colitis as previously described [20].
Antibiotic administration. Mice were administrated with a broad-spectrum antibiotic cocktail ampicillin (1 g/L), vancomycin (500 mg/L), neomycin sulfate (1 g/L), and metronidazole (1 g/L) in drinking water for 14 days [21], before the microbiota transplant experiments.
Microbiota transfer
To exclude the effect of Blastocystis on the severity of colitis, feces collected from ST4 and ST7 infected mice and non-infected control mice were frozen at -80°C, and then thawed in pre-reduced PBS. This procedure ruptures Blastocystis cells, and the absence of live Blastocystis cells was confirmed by culturing the feces in Jones' medium. Fecal microbiota transplantation (FMT) was performed as described [22]. In brief, frozen feces were pooled from control, ST4-colonized, and ST7-infected mice, respectively, diluted with pre-reduced PBS (50 mg/ml), and administered to recipient mice by oral gavage (10 mg/mice) three times a week for one week.
Scanning electron microscopy
Scanning electron microscopy (SEM) was performed as previously described [23]. In brief, the cecum/colon tissues were collected and then processed by opening the gut longitudinally without disturbing the intestinal contents. The opened tissues were pinned down to a silicone mat in four corners and fixed in 2.5% glutaraldehyde at 4°C overnight, and then washed twice with PBS. The washed tissues were kept at 4°C until further processing. The tissues were processed by post-fixing in 1% osmium tetroxide for 1 h, and then dehydrated first in 25% ethanol for 5 minutes followed by 50%,75% and 95% ethanol for 10 minutes each. After which, the sample was fixed in 100% ethanol for 10 minutes (3 changes). The dried samples were coated with 25 nm of gold and imaged on a field emission JSM-6701F SEM at a voltage of 10 kV.
DNA extraction and real-time quantitative PCR
Total genomic DNA from feces of control, ST4-colonized, and ST7-infected mice at day 0 (before Blastocystis inoculation), and day 14 (end of experiment) was extracted using QIAamp Fast DNA Stool Mini Kit (Qiagen) according to the manufacturer's protocol. Real-time quantitative PCR (qPCR) was used to estimate the number of Blastocystis cells in fecal samples at the end point as described [24].
Colon histology
Colonic tissues were collected at the end of each experiment and fixed in 4% neutral buffered formalin, and then processed and embedded in paraffin. Sections of 4.5 μm were prepared and stained with hematoxylin and eosin (H&E). Histology scoring was performed in a blinded fashion, whereby changes in intestinal crypt architecture, degree of inflammation, and epithelium damage were scored as previously described [25]. A combined score of crypt architecture, tissue damage, goblet cell loss, and inflammatory cell infiltration was determined as follows: crypt architecture: score 0, normal; 1, irregular; 2, moderate crypt loss (10-50%); 3, severe crypt loss (50-90%); 4, small/medium sized ulcers (<10 crypt widths); 5, large ulcers (>10 crypt widths). Tissue damage: score 0, no damage; 1, discrete lesions; 2, mucosal erosions; 3, extensive mucosal damage. Goblet cell loss: score 0, normal; 1, 10-25% loss; 2, 25-50% loss; 3, = >50% loss. Inflammatory cell infiltration: score 0, occasional infiltration; 1, increasing leukocytes in lamina propria; 2, confluence of leukocytes extending to submucosa; 3, transmural extension of inflammatory infiltrates.
16S rRNA gene sequencing and bioinformatics analysis
Genomic DNA was extracted from fecal samples. The 16S rRNA gene sequencing library was constructed using a MetaVX Library Preparation Kit (South Plainfield, NJ). Briefly, 20-30 ng of DNA was used to generate amplicons that cover V3 and V4 hypervariable regions of the 16S rRNA gene of bacteria. The forward primer contains the sequence 'CCTACGGRRBGCASCAGKVRVGAAT', and the reverse primers contain the sequence 'GGACTACNVGGGTWTCTAATCC'. The 25 µl PCR mixture was prepared with 2.5 µl of TransStart buffer, 2 µl of dNTPs, 1 µl of each primer, 0.5 µl of TransStart Taq DNA polymerase, and 20 ng template DNA. PCR was performed by the following program: 3 min of denaturation at 94℃, 24 cycles of 5 s at 95℃, 90 s of annealing at 57℃, 10 s of elongation at 72℃, and a final extension at 72℃ for 5 min. Indexed adapters were added to the ends of the amplicons by limited cycle PCR. Finally, the library was purified using magnetic beads. DNA concentration was determined with a microplate reader (Tecan, Infinite 200 Pro) and the expected ~600 bp fragment size was determined by 1.5% agarose gel electrophoresis. Next generation sequencing was conducted on an Illumina Novaseq Platform (Illumina, San Diego, USA) at Azenta Inc (USA). Automated cluster generation and 250 paired-end sequencing with dual reads were performed according to the manufacturer's instructions.
Paired-end sequences of positive and negative reads were filtered, denoised, and had chimeras removed to obtain amplicon sequence variants (ASVs) through DADA2 using Quantitative Insights into Microbial Ecology 2 (Qiime2) plugin [26]. The ribosomal database program (RDP) Classifier Bayes algorithm was trained using Silva 138 database to perform a taxonomic analysis [27]. Shannon and Chao1 were used to estimate the bacterial diversity and richness respectively. Beta diversity was assessed by permutational multivariate analysis of variance (PERMANOVA). Principal coordinates analysis (PCoA) plots were constructed based on Bray-Curtis dissimilarity to illustrate the differences in community structure between different groups. Heatmaps were used to show the different taxa between groups. Linear discriminant analysis effect size (LEfSe) analysis was performed to detect bacterial taxa with significantly different abundance among different groups with P-value <0.05 and LDA score >4 (https://huttenhower.sph.harvard.edu/galaxy/).
RNA sequencing and bioinformatics analysis
Colon tissues were collected and washed with PBS to discard the luminal contents, and then dissected into 1 cm pieces for RNA extraction. RNA was extracted using RNAzol RT (Sigma-Aldrich) according to the manufacturer's protocol. One μg total RNA was used for the library preparation. Poly(A) mRNA isolation was performed using Oligo (dT) beads. The mRNA fragmentation was performed using divalent cations and high temperatures. Priming was performed using Random Primers (Invitrogen). First-strand cDNA and the second-strand cDNA were synthesized. The purified double-stranded cDNA was then treated to repair both ends and a dA-tailing in one reaction, followed by a T-A ligation to add adaptors to both ends was performed. Size selection of Adaptor-ligated DNA was then performed using MagMAX™ DNA Multi-Sample Ultra Kit (Applied Biosystems). Each sample was then amplified by PCR using P5 ('AATGATACGGCGACCACCGA') and P7 ('CAAGCAGAAGACGGCATACGAGAT') primers and the PCR products were validated using Agilent 2100. Then libraries with different indexes were multiplexed and loaded on an Illumina Novaseq6000 instrument at Azenta Inc (USA).
To remove adapters, primers, or fragments thereof, and quality of bases lower than 20, pass filter data of fastq format were processed by Cutadapt (V4.1, phred cutoff: 20, error rate: 0.1, adapter overlap: 1bp, min. length: 75, proportion of N: 0.1) to give high-quality clean data. Reference genome sequences and gene model annotation files were downloaded from ENSEMBL (https://asia.ensembl.org/index.html). Hisat2 (v2.1.0) was used to index reference genome sequences [28]. Principal component analysis (PCA) was generated by corrplot package in R language. Differential expression analysis was done using the Limma Bioconductor package [29]. Padj of genes was set at < 0.05 to detect differential expressed genes (DEGs). GOSeq (v1.34.1) was used to identify GO terms that annotate a list of enriched genes with a significant padj less than 0.05. KEGG (Kyoto Encyclopedia of Genes and Genomes) is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances (http://en. wikipedia.org/wiki/KEGG). We used in-house scripts to enrich significant differential expression genes in KEGG pathways. Gene Set Enrichment Analysis (GSEA) was generated in GSEABase package in R [30, 31].
LC/MS/MS assay
Liquid chromatography/tandem mass spectrometry (LC/MS/MS) was carried out for analysis of short-chain fatty acids (SCFAs) in derivatized stool extracts as previously described [32]. Briefly, 500 μl of ice-cold extraction solvent containing 10 μM of d5-benzoic acid as internal standard (IS) was added to 250 mg of fecal sample and subjected to vortex mixing for 5 min at ambient temperature. The suspension was then centrifuged at 18,000g for 10 min at 4 °C. The supernatant was carefully removed and centrifuged again at 18,000g for 10 min at 4 °C. An aliquot of 100 μl was subsequently derivatized using a final concentration of 10 mM aniline and 5 mM EDC for 2 h at 4 °C. The derivatization reaction was quenched using a final concentration of 18 mM succinic acid and 4.6 mM 2-mercaptoethanol for 2 h at 4 °C. All samples were stored at 4 °C until analysis on the same day. Analysis was performed using an Agilent 1290 Infinity LC system (Agilent Technologies) interfaced with an AB Sciex QTRAP 5500 hybrid linear ion-trap quadrupole mass spectrometer equipped with a TurboIonSpray source (Applied Biosystems).
Isolation of colon lamina propria cells
To analyze colonic lymphocytes, the colon tissues were longitudinally opened and washed with ice-cold PBS to remove luminal contents. The tissues were cut into 1 cm pieces and incubated in Roswell Park Memorial Institute (RPMI) 1640 medium (Sigma-Aldrich) containing 1 mM EDTA (Sigma-Aldrich) and 1 mM DTT (Sigma-Aldrich) at room temperature for 20 min under slow rotation and spun down to remove the supernatant. The remaining tissue pieces were incubated in RPMI containing 25% HEPES, 10% fetal calf serum (FCS), 1 mM EDTA, and 1 mM DTT at 37 ℃ for 1 h under slow rotation and then washed by PBS to remove epithelial cells and intraepithelial lymphocytes. Tissue pieces were digested with Liberase (Sigma-Aldrich) at 37 ℃ for 30 min under slow rotation. The digested tissue pieces and supernatants were filtered by 70 μm cell strainer and glass wool separately. After centrifugation, pellets containing the LP lymphocytes were harvested.
Flow cytometry analysis
Lymphocytes were stimulated for 6 h with a cell stimulation cocktail of PMA (50 ng/ml), ionomycin (750 ng/ml), and GolgiStop (monensin, BD Biosciences). Live/dead stain was used to evaluate the viability of the cells using live/dead fixable viability stain kits. For surface staining, stimulated cells were stained with TCR beta monoclonal antibody (BUV510, eBioscience), and anti-CD4 (BUV395; Biolegend). Fixation and permeabilization buffers from Biolegend were used for intracellular cytokine staining. Fixed and permeabilized cells were stained with fluorochrome-conjugated anti-mouse antibodies against interleukin (IL)-4 (BUV421; Biolegend), IL-10 (PE; Biolegend), IL-17A (APC; eBioscience), interferon-gamma (IFN-γ) (BUV711; Biolegend), and tumor necrosis factor (TNF-α) (APC; eBioscience) at 4 ℃ for 10 h. Flow cytometry analysis was performed on Fortessa X-20 (BD biosciences) and the data were analyzed using FlowJo_V10 software.
Statistical analysis
All statistical analysis was performed using Prism 8 (Graphpad Software Inc.). Two independent experiments were performed. For comparisons of two groups, Student's two-tailed t-test was used. When three or more groups were analyzed, one-way or two-way ANOVA with multiple comparisons test was used. Error bars on graphs display the mean and SEM. P values of < 0.05 were considered significant; the following symbols were used to indicate significance levels: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Results
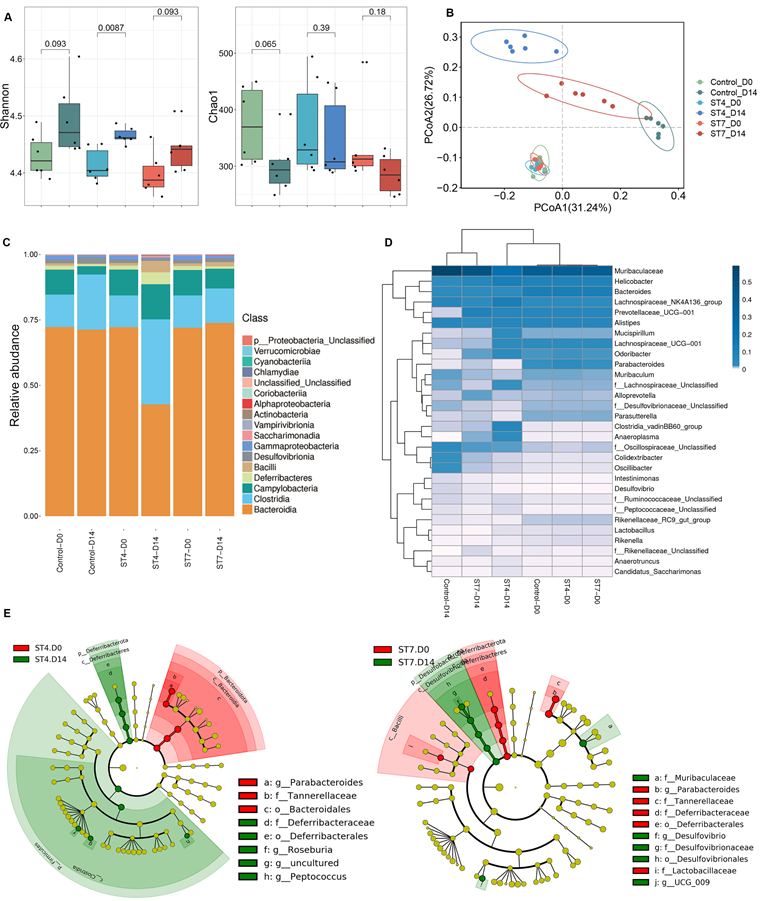
Effects of Blastocystis ST4 and ST7 infection on bacterial compositions
C57BL/6 mice were orally gavaged with Blastocystis ST4 and ST7 three times a week for two weeks (Figure S1A). The number of Blastocystis cells and colonization status was confirmed by qPCR and scanning electron microscopy (SEM) (Figure S1B-C). Although the bacterial richness reflected by Chao1 did not show significant differences among the three groups, the Shannon index, reflecting the bacterial diversity, was significantly increased in the ST4-colonized mice (Figure 1A). ST7 effects on bacterial diversity were comparable to those in fecal samples at day 0 (Figure 1A). We next performed PCoA analysis based on Bray-Curtis dissimilarities to investigate the effect of Blastocystis colonization on microbial community structure (β-diversity). At baseline (day 0), the ST4, ST7, and control mice showed similar microbiota profiles (PERMANOVA p > 0.05; Figure 1B and Table S1), suggesting those group mice showed similar gut microbiota compositions before Blastocystis inoculation. After infection for two weeks, all these three groups showed significant differences compared to the baseline (PERMANOVA p < 0.05; Figure 1B and Table S1). ST4-colonized mice showed increase in the abundance of Clostridia and decrease in the proportion of Bacteroidia (Figure 1C and Figure S1E). At the genus level, the Lachnospiraceae NK4A136 group, and Clostridia vadinBB60 group were enriched in ST4-clonized mice (Figure 1D and Figure S1F). Bacteria from these genera are known to be beneficial bacteria, which can produce SCFAs that are a carbon source for intestinal epithelial cells and induction of Treg cells [33, 34]. In contrast, ST7 infection caused the reduction in beneficial bacteria Lachnospiraceae UCG-001, and Lactobacillus, and increased the sulfate-producing bacteria Desulfovibrio (Figure 1D and Figure S1F). The enrichment of Desulfovibrio was observed in UC patients, suggesting the species of Desulfovibrio genus can be involved in colitis development [35, 36]. Similarly, the proportion of Lachnospiraceae UCG-001, and Lactobacillus declined, and Desulfovibrio was enriched in control mice (Figure S1F).
Linear discriminant analysis effect size (LEfSe) analysis revealed that the Deferribacterales, Deferribacteraceae, Roseburia, and Peptococcus were significantly more abundant in ST4-colonized group compared to the baseline. In contrast, significant increases of Desulfovibrionales, Desulfovibrionaceae, Muribaculaceae, and Desulfovibrio were observed in ST7-infected groups (Figure 1E). In general, ST4 colonization was associated with higher proportion of beneficial bacteria, while ST7 infection decreased the abundance of beneficial bacteria and increased sulfate-producing bacteria, which may predispose the host to colitis.
Blastocystis ST4 and ST7 infection caused differential gut microbial signatures. (A) Alpha diversity was measured by Shannon and Chao1 indices (n = 6). (B) Principal co-ordinates analysis (PCoA) based on Bray-Curtis dissimilarities of fecal gut microbiota derived from control (green dots), ST4 (blue dots), and ST7 (red dots) mice at day 0 and day 14. (C) Relative abundance of the different taxa at the class level. (D) Heatmap showing Blastocystis ST4 and ST7-associated taxonomic markers at day 14. (E) LefSe analysis showed the differentially abundant bacterial taxa between ST4 and ST7 groups relative to the baseline. Data are representative of two independent experiments and shown as the mean ± SEM.
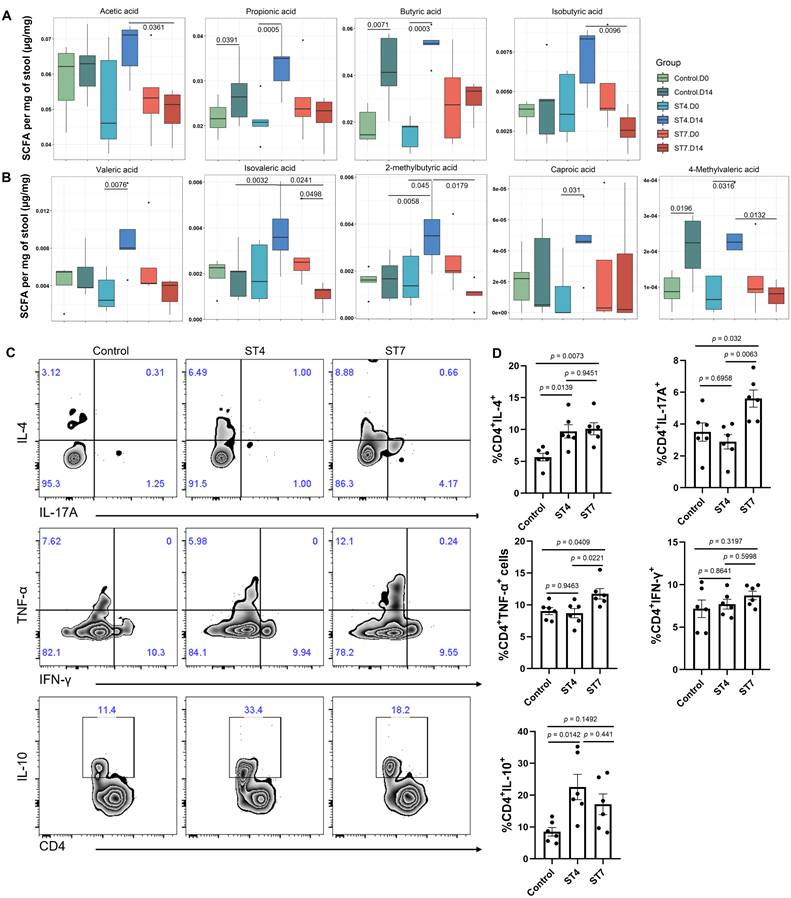
Effects of Blastocystis ST4 and ST7 infection on SCFA production and colonic T helper cells
We next investigated whether Blastocystis infection affected the production of SCFAs, which constitute the primary energy sources for colon epithelium and play a role in maintaining intestinal homeostasis [37]. Interestingly, ST4-colonized mice showed a significant increase in the majority of SCFAs (Figure 2A-B). However, ST7 did not and instead was associated with reduced isovaleric acid. (Figure 2A-B). As microbiota-derived SCFAs can regulate the adaptive immune response [38], we next investigated whether ST4 and ST7 infection could also influence host immunity (Figure S2A). ST4-colonized mice showed a substantial increase in IL-4 and IL-10-producing CD4+ T cells (Figure 2C-D), which is consistent with our previous data [15]. Interestingly, the Th1 compartment (expressing TNF-α) and the Th17 subset (expressing IL-17A) were mildly but significantly increased in ST7-infected mice (Figure 2C-D). The effects of ST4 and ST7 infection on host overall health were also investigated. We observed that both ST4 and ST7-infected mice showed comparable effects on body weight change, DAI, colon length, and histopathology scores when compared to control mice (Figure S2B-C). These data suggest that ST4 and ST7 infection did not cause obvious pathological changes, but elicited different immune responses, which may affect the outcome of the host's response to other stimuli.
Blastocystis ST4 and ST7 infection influence SCFAs production and colonic T helper cells. (A) The concentration of acetic, propionic, butyric, and isobutyric in control, ST4, and ST7 infected mice. (B) The concentration of valeric acid, isovaleric, 2-methylbutyric, and caproic acid, and 4-methylvaleric acid. (C) Zebra plots show staining for IL-4, IL-17A, TNF-α, IFN-γ and IL-10 within CD4+ T cells. (D) Bar charts show the percentage of IL-4, IL-17A, TNF-α, IFN-γ and IL-10 expressing CD4+ T cells. One-way ANOVA p-values adjusted for Tukey's multiple comparisons are shown. Data are representative of two independent experiments and shown as the mean ± SEM.
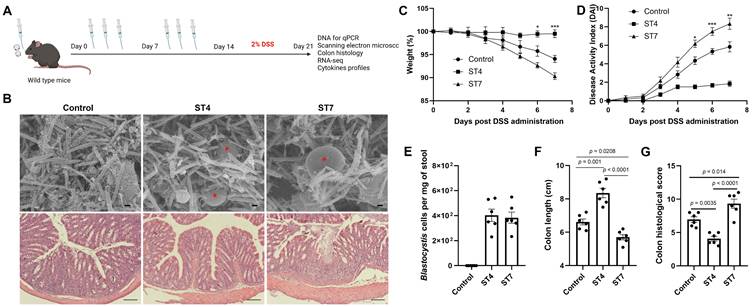
The effects of Blastocystis ST4 and ST7 colonization on subsequent DSS challenge. (A) Study design. (B) SEM of colonic and cecum tissues from control, ST4, and ST7 infected mice (upper panel), and Blastocystis are indicated with red asterisk (Scale bar = 1 μm). H&E-stained colon sections (lower panel) from the three groups (Scale bar = 100 μm). Weight changes (C), and DAI (D) in control, ST4, and ST7 infected mice. (E) The number of Blastocystis cells in ST4 and ST7 infected mice. Colon length (F) and histological scores (G) in the three groups. Data are representative of two independent experiments and shown as the mean ± SEM. Two-way ANOVA with Dunnett's multiple comparison testing were used for multiple comparisons. *p < 0.05; **p < 0.01; ***p < 0.001.
Effects of Blastocystis ST4 and ST7 infection on the severity of experimental colitis
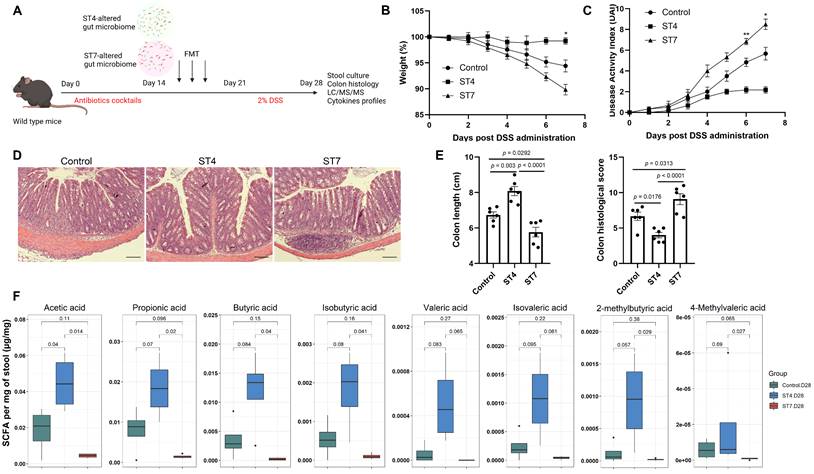
The gut microbiota is implicated in pathogenesis of IBD [39]. Next, we explored the effects of altered gut microbiota caused by Blastocystis in the development of experimentally induced colitis. Mice were infected with Blastocystis and then subjected to induced colitis through oral DSS administration for 7 days (Figure 3A). The Blastocystis colonization status and the number of Blastocystis cells was examined (Figure 3B and 3E). We observed that ST7-infected mice treated with 2% DSS showed progressive weight loss and DAI from day 5 to day 7 (Figure 3C-D). In contrast, mice colonized with ST4 did not develop these symptoms (Figure 3C-D). Additionally, ST7 infected mice exhibited shorter colon lengths when compared to non-infected and ST4 colonized mice (Figure 3F). Histological analysis of colon tissues showed severe inflammation with loss of crypts and infiltration of inflammatory cells in ST7 infected mice, whereas ST4 colonized mice were protected from DSS-induced damage to the colon (Figure 3B and 3G). Altogether, priming mice with ST4 and ST7 showed distinct effects on the severity of DSS-induced colitis.
Effects of Blastocystis ST4 and ST7 infection on mucosal gene expression in DSS-induced colitis mice
To assess the impact of Blastocystis infection on the intestinal epithelium, we analyzed the transcriptional profile of colon tissues from DSS-induced colitis mice. PCA plot showed samples from the same group clustered together (Figure 4A). Pairwise comparisons between the different groups revealed that ST4 colonized mice had 109 differential expressed genes (DEGs) (33 up-regulated and 76 down-regulated) when compared to control mice (Figure 4B). ST7 colonized mice have a far greater number of DEGs with control group than ST4, with 1,142 (391 up-regulated and 751 down-regulated) (Figure 4B). We next analyzed Gene Ontology (GO) terms, which provides a framework and set of concepts for describing the functions of gene products. Interestingly, colonization with ST4 mostly activated humoral immune response and immunoglobulin production pathways (Figure 4C). We observed that ST7 infection was related to the adaptive immune response (Figure 4C). KEGG pathway analysis showed both ST4 and ST7 infection were related to cytokine-cytokine receptor interactions (Figure S3). In addition, other categories of immune-related pathways, such as Th17 cell differentiation and inflammatory bowel diseases, were associated with ST7 infection (Figure S3). To better understand the molecular mechanisms of complex biochemical reactions, we then applied GSEA on the DEGs. We found that ST4 colonization caused a marked reduction in normalized enrichment scores (NESs) for inflammatory bowel disease, T cell receptor signaling pathway, Th1 and Th2 cell differentiation, and Th17 cell differentiation (Figure 4D). In contrast, increased scores for each of these gene sets were found in ST7 infected groups (Figure 4D). In general, the induction seems to be higher for ST7 from the gene ratio which induced more gene profile responses. Mice with prior ST4 colonization are refractory to colitis, associated with protective immune responses, while ST7 infection caused worsening of colitis, related to pro-inflammatory immune responses.
Gene expression in ST4 and ST7-infected colon tissues after DSS treatment. (A) Principal component analysis (PCA) plot comparing mouse transcriptomes among control, ST4, and ST7 infected mice. (B) Volcano plots showing log2 fold change plotted against log of mean normalized expression counts. (C) The top 10 most enriched GO terms found in the analysis of DEGs in ST4 vs. Control group, and ST7 vs. Control group. GO terms were ranked by their significance. (D) GSEA analysis showed the gene sets of inflammatory bowel disease, T cell receptor signaling pathway, Th1 and Th2 cell differentiation, and Th17 cell differentiation. Graphs depict the enrichment score (y axis) with negative values where gene sets are inhibited, and positive values where they are induced. Each vertical bar on the x axis represents an individual gene within the gene set, and its relative ranking against all genes analyzed. P and Padj value is indicated on the graph.
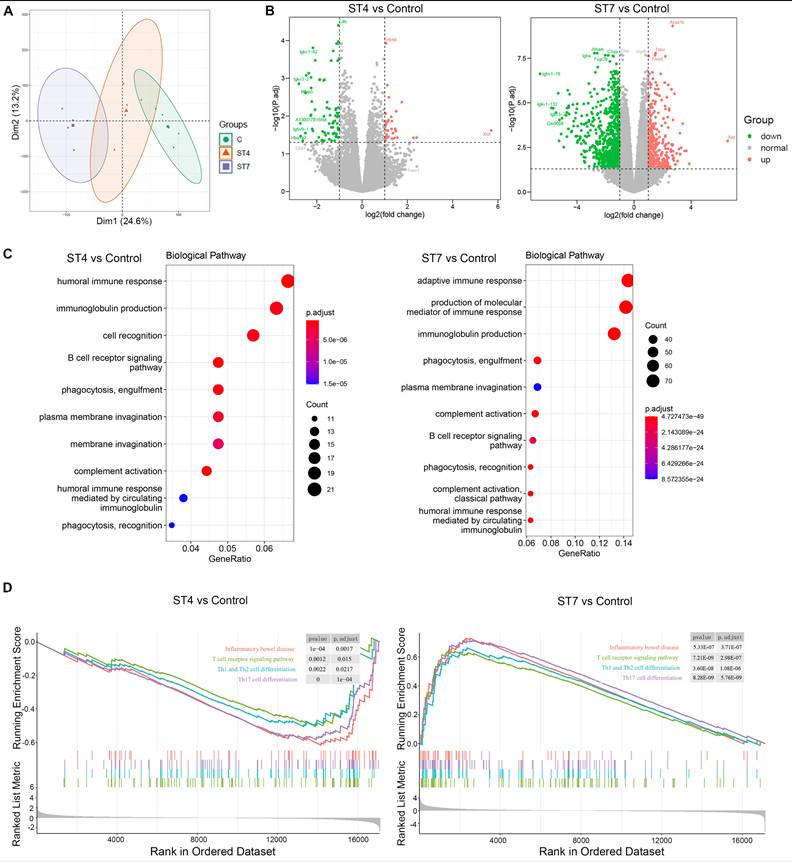
Effects of Blastocystis ST4 and ST7 infection on colonic T helper cells in DSS-induced colitis mice
T helper cells are thought to play a crucial role in the development of IBD [40]. To further investigate whether ST4 and ST7 infection caused changes related to T helper cells, we measured the distribution of Th1, Th2, Th17, and Treg cells within T cells isolated from colonic mucosa. In contrast to ST4 colonized mice, infection with ST7 markedly increased the percentage of pro-inflammatory cytokines IL-17A and TNF-α-producing T cells in the colonic lamina propria (LP) (Figure 5A-B). Concomitantly, the percentage of T cells expressing anti-inflammatory cytokine IL-10 was significantly reduced (Figure 5C). Forkhead box protein 3 (Foxp3) is essential in the development, maintenance, and function of Treg cells, which play a critical role in the maintenance of intestinal homeostasis [41]. ST4 colonized mice showed a substantial accumulation of Foxp3+ Tregs (Figure 5D). Together, these results suggest that increased susceptibility to colitis after ST7 infection is associated with the induction of pro-inflammatory Th1 and Th17 responses, while ST4 colonization confers resistance to colitis related to protective immune responses.
Colonic immune profiles in control, ST4, and ST7 infected mice after treatment with DSS. (A) Zebra plots and bar charts show staining and the percentage of IL-4, and IL-17A within CD4+ T cells. (B) Zebra plots and bar charts show staining and the percentage of TNF-α, and IFN-γ within CD4+ T cells. Zebra plots and bar charts show staining and the percentage of IL-10 (C) and Foxp3 (D) within CD4+ T cells. Data are representative of two independent experiments and shown as the mean ± SEM. One-way ANOVA p-values adjusted for Tukey's multiple comparisons are shown.
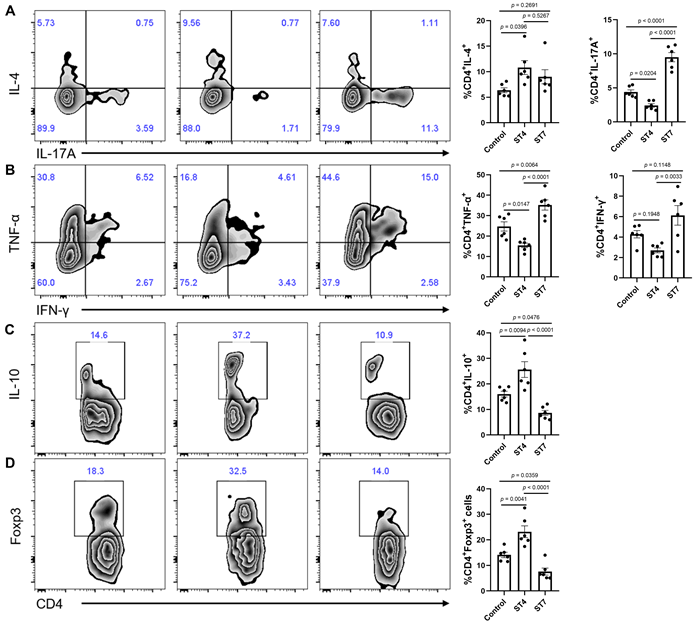
Effects of Blastocystis-altered gut microbiota on the severity of experimental colitis
We next sought to determine whether the Blastocystis-altered microbiota played a direct role in the worsening of, or protection against, DSS colitis. To exclude the effects of Blastocystis on the severity of colitis, feces from Blastocystis infected mice were frozen at -80°C at least one week before starting the FMT. This treatment ruptures and destroys Blastocystis organisms (Figure S4). Recipient mice were treated with antibiotic cocktails to deplete the gut microbiota and then gavaged with ST4 and ST7-altered gut microbiota (Figure 6A). The recipient mice that received FMT from ST4 donor mice had less severe colitis when challenged with DSS, reflected by weight changes and DAI (Figure 6B-C). In contrast, body weight decreased significantly, and DAI tended to be higher in recipients of the ST7-altered microbiota (Figure 6B-C). Additionally, recipients of the ST7-altered microbiota had significantly shorter colon lengths than control recipients, whereas recipients of the ST4-altered microbiota had significantly longer colon lengths (Figure 6E). Histology analysis showed that recipients of the ST4-altered microbiota had significantly lower histological scores than control and ST7-altered microbiota recipients (Figure 6D-E). These findings provide evidence that the Blastocystis-altered microbiota, even without Blastocystis present, was sufficient to worsen or protect the mice from the DSS-induced colitis.
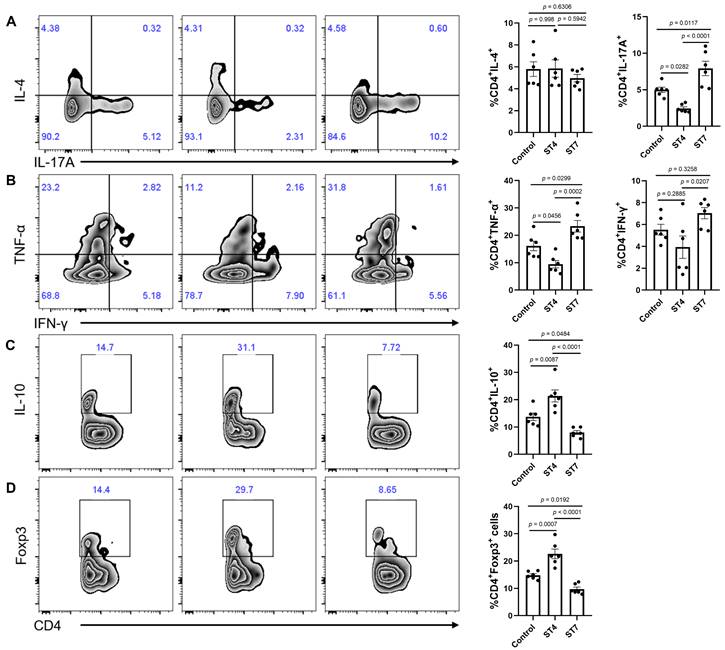
Effects of Blastocystis-altered gut microbiota on SCFA production and colonic T helper cells
To understand mechanistically how ST4 and ST7-altered gut microbiota differentially impact DSS-induced colitis, we measured the concentration of SCFAs among the three groups to determine whether these were associated with the development of colitis. The concentrations of acetic acid, propionic acid, butyric acid, isobutyric acid, 2-methylbutyric acid, and 4-methylvaleric acid in fecal samples of ST4-colonized mice were significantly higher than in ST7-infected mice (Figure 6F). We next sought to investigate whether the Blastocystis altered microbiota can modulate the colonic adaptive immune responses. Although there was no significant difference in the percentage of IL-4-producing Th2 cells among the three groups, we observed a substantial increase in IL-10+ and Foxp3+ Treg cells in the colonic mucosa of ST4-altered microbiota recipients (Figure 7A, C-D). Similarly, we also detected a significant expansion of TNF-α-producing CD4+ Th1 cells and IL-17A-producing CD4+ Th17 cells after receipt of the ST7-altered microbiota (Figure 7A-B). These data showed that Blastocystis-altered gut microbiota play a significant role in the regulation of the pathogenesis of DSS-induced colitis by affecting adaptive immune responses.
Discussion
Growing evidence suggests that parasitic infections can influence the severity of colitis in a mouse model of IBD [42, 43]. Infection with the tapeworm Hymenolepis diminuta suppressed experimentally-induced colitis in mice dependent on gut microbiota, suggesting the gut microbiota may influence the outcome of infection and disease [44]. In this study, two subtypes were used to investigate the effects of Blastocystis on host health. ST4 is more often found in rodents than ST7 which is commonly found in birds [45], suggesting that subtype specificity may play a role in the clinical outcome of Blastocystis infection. We demonstrated that mice colonized with Blastocystis ST4 showed an increase in the beneficial bacteria Lachnospiraceae and Clostridia vadinBB60 group, which protected the mice when challenged with DSS-induced colitis. However, Blastocystis ST7 infection decreased the abundance of beneficial bacteria Lachnospiraceae UCG-001, Lactobacillus, and Roseburia, and increased the level of sulfate-producing bacteria Desulfovibrio, which may have exacerbated the severity of the experimental colitis. These findings suggested that Blastocystis may serve as a critical component of the anti-colitic or pro-colitic effects in an animal model of colitis through regulating the gut microbiota.
Effects of Blastocystis-altered gut microbiota on the severity of experimental colitis. (A) Study design. Weight changes (B) and DAI (C) in control, ST4, and ST7 altered microbiota recipient mice. (D) H&E-stained colon sections from the three groups (Scale bar = 100 μm). (E) colon length and histological scores in the three groups. (F) The concentration of acetic, propionic, butyric, and isobutyric, valeric acid, isovaleric, 2-methylbutyric, and caproic acid, and 4-methylvaleric acid in the recipient mice. Data are representative of two independent experiments and shown as the mean ± SEM. One-way ANOVA p-values adjusted for Tukey's multiple comparisons are shown (E, F). Two-way ANOVA with Dunnett's multiple comparison testing were used for multiple comparisons (B, C). *p < 0.05; **p < 0.01.
Colonic immune profiles from recipient mice. (A) Zebra plots and bar charts show staining and the percentage of IL-4, and IL-17A within CD4+ T cells. (B) Zebra plots and bar charts show staining and the percentage of TNF-α, and IFN-γ within CD4+ T cells. Zebra plots and bar charts show staining and the percentage of IL-10 (C) and Foxp3 (D) within CD4+ T cells. Data are representative of two independent experiments and shown as the mean ± SEM. One-way ANOVA p-values adjusted for Tukey's multiple comparisons are shown.
The associations between Blastocystis and gut microbiota have been explored in several studies in recent years [46, 47]. Although most studies showed Blastocystis-carriers are asymptomatic and have healthy gut microbiota, other studies revealed that Blastocystis infection was associated with decreased executive functions in humans [48], and fecal dysbiosis in a mouse model [49]. It has been suggested that different subtypes exhibited differential effects on the development of the experimental colitis [14, 15, 50]. For example, long-term Blastocystis ST3 colonization promotes a faster recovery from dinitrobenzene sulfonic acid (DNBS) induced colitis through stabilization of the gut microbiota [50]. In contrast, severe colonic damage and ulceration were observed in Blastocystis ST7 infected mice when exposed to DSS accompanied by disrupted gut microbiota [14].
In this study, we observed that Blastocystis ST4 not only increased the well-known SCFA-producing bacteria but increased the concentration of SCFAs, while Blastocystis ST7 did not show such effects, and, to some extent, decreased the level of some SCFAs. SCFAs are the main metabolites produced by the gut microbiota through the anaerobic fermentation of substrates, such as fibers and resistant starch, which play a crucial role in maintaining intestinal homoeostasis [51]. Decreased numbers of SCFA-producing bacteria and lower concentrations of fecal SCFAs were also observed in IBD patients [52, 53]. SCFAs also regulate immune cells, especially Tregs, by inhibiting histone deacetylase (HDAC) activity or through GPRC receptors, exerting anti-inflammatory effects [54].
High Th1 and Th17 cytokine production in colonic mucosa was related to clinical manifestation of colitis [55]. Th2 immune responses can antagonize the production of Th1 cytokines to alleviate colitis [56]. Besides, IL-10-producing Treg cells also can suppress pathogenic Th17 cell responses [57]. Our data support a mechanism whereby colonization with Blastocystis ST4 causes an increased abundance of SCFA-producing bacteria and concentration of SCFAs, which mediates the suppression of colitis by the expansion of Tregs in the colonic mucosa. In contrast, ST7 infection decreased the proportion of beneficial bacteria and SCFAs and increased the sulfate-reducing bacteria, which worsened the severity of colitis through the induction of pro-inflammatory Th1 and Th17 cytokines.
It has been demonstrated that the excretory or secretory products from helminths can inhibit tuft and goblet cells to block the effects of IL-4 and IL-13 in mice [58]. On the other hand, secreted products can induce Foxp3+ Tregs by activating TGF-β signaling [59]. Although live Blastocystis cells were excluded from the Blastocystis-altered microbiota FMT experiments, we could not exclude the influence of excretory/secretory products on the severity of colitis through FMT. Future directions will explore whether Blastocystis can affect gut microbiota composition through secreted products or extracellular vesicles, and whether Blastocystis can also modulate the development and progression of inflammatory diseases through excretory/secretory products and other small molecules, such as bile acids and tryptophan metabolites.
FMT is a common therapeutic strategy for restoring disrupted microbiota [60]. It has been used as an ordinary standard therapy for recurrent C. difficile infection (rCDI) [61], and some trials have reported its potential to alleviate or treat IBD [62]. The present consensus for FMT is to exclude Blastocystis-positive donor samples [61, 63], though the pathogenicity of Blastocystis remains controversial [64]. The presence of Blastocystis-positive (ST1 and ST3) donor samples for FMT in treatment rCDI did not cause any adverse gastrointestinal symptoms [65], suggesting that certain subtypes can be used for FMT. Since ST1, ST2, and ST3 are very common in humans, donors colonized by one of these 3 subtypes are probably acceptable. Our data show that transplantation of ST4-altered gut microbiota protected mice from later DSS-induced colitis, while ST7 caused converse effects, suggesting the necessity for differentiation of Blastocystis-positive donors at the subtype level. Besides, it should be noted that the effects of different strains from the same subtype may also impact intestinal health. For example, the extensive intra-ST7 variability in inducing intestinal barrier dysfunction was observed in our previous study [16], suggesting intra-subtype variability may explain the discrepancy of pathogenicity in Blastocystis.
Conclusions
We report, for the first time, that colonization with Blastocystis ST4 exerted a protective effect against the onset of intestinal inflammation by increasing the proportion of beneficial bacteria and SCFAs, and the regulation of colonic CD4 T cells. ST7 infection caused opposite effects on DSS-induced acute colitis. Moreover, transplantation of ST4 and ST7-altered gut microbiota was sufficient to transfer the protective and inflammatory phenotypes respectively. As the murine model is not exactly reflective of the human gut in the case of Blastocystis infection, it is necessary to perform, in future studies, human gut microbiome studies to reveal the precise roles of ST4 and ST7 on intestinal inflammation.
Abbreviations
IBD: Inflammatory bowel disease; CD: Crohn's disease; UC: ulcerative colitis; Th: T helper; Treg: T regulatory; DSS: Dextran sulfate sodium; IMDM: Iscove's modified Dulbecco's medium; PBS: Phosphate-buffered saline; DAI: Disease activity index; H&E: Hematoxylin and eosin; SEM: Scanning electron microscopy; QIIME: Quantitative Insights into Microbial Ecology; PERMANOVA: Permutational multivariate analysis of variance; PCoA: Principal co-ordinates analysis; PCA: Principal component analysis; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; GSEA: Gene Set Enrichment Analysis; RPMI: Roswell Park Memorial Institute; FCS: Fetal calf serum; FBS: Fetal Bovine Serum; IL: Interleukin; IFN-γ: Interferon gamma; TNF-α: Tumor necrosis factor alpha; LP: Lamina propria; FMT: Fecal microbiota transplantation; SCFAs: Short-chain fatty acids; HDAC: Histone deacetylase.
Supplementary Material
Supplementary figures and table.
Acknowledgements
We would like to thank Chua Yen Leong for assistance in animal work, and Dr. Zhiyuan Lu for the help in histology experiments. We also would like to thank Ms. Geok Choo Ng and Dr. Paul Edward Hutchinson for providing technical support.
Funding
This work was supported by NUHS Seed Grant R-571-000-061-114 to NRJG, R-148-000-277-114 to ECYC and R-571-000-037-114 to KSWT. KSWT also acknowledges the Blastocystis Under One Health Grant (A-8000685-00-00) for supporting this study.
Author contributions
L.D. performed experiments, analyzed and interpreted the data; L.W. and L.D. performed immunological experiments; C.W.P. and Y.Z. performed and interpreted histological analyses; K.Y.Q.D., Y.G. and E.C.Y.C. performed and interpreted LC/MS/MS assays; T.T.A. and B.M. performed the scanning electron microscopy experiments; G.P. and N.R.J.G. provided scientific insights and critically reviewed the manuscript. K.S.W.T. designed, conceived, and supervised the study.
Availability of data and materials
The datasets generated and analyzed in the current study are available in the Sequence Read Archive (SRA) database at NCBI under BioProject ID PRJNA856792 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA856792).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205-17
2. Plichta DR, Graham DB, Subramanian S, Xavier RJ. Therapeutic opportunities in inflammatory bowel disease: mechanistic dissection of host-microbiome relationships. Cell. 2019;178:1041-56
3. de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13:13-27
4. Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8:51
5. Guzzo GL, Andrews JM, Weyrich LS. The neglected gut microbiome: fungi, protozoa, and bacteriophages in inflammatory bowel disease. Inflamm Bowel Dis. 2022 1112
6. Moonah SN, Jiang NM, Petri WA Jr. Host immune response to intestinal amebiasis. PLoS Pathog. 2013;9:e1003489
7. Kotloff KL. The burden and etiology of diarrheal illness in developing countries. Pediatr Clin North Am. 2017;64:799-814
8. Tan KSW. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin Microbiol Rev. 2008;21:639-65
9. Cekin AH, Cekin Y, Adakan Y, Tasdemir E, Koclar FG, Yolcular BO. Blastocystosis in patients with gastrointestinal symptoms: a case-control study. BMC Gastroenterol. 2012;12:122
10. Shirvani G, Fasihi-Harandi M, Raiesi O, Bazargan N, Zahedi MJ, Sharifi I. et al. Prevalence and molecular subtyping of Blastocystis from patients with irritable bowel syndrome, inflammatory bowel disease and chronic urticaria in iran. Acta Parasitol. 2020;65:90-6
11. Stensvold CR, van der Giezen M. Associations between gut microbiota and common luminal intestinal parasites. Trends Parasitol. 2018;34:369-77
12. Deng L, Wojciech L, Gascoigne NRJ, Peng G, Tan KSW. New insights into the interactions between Blastocystis, the gut microbiota, and host immunity. PLoS Pathog. 2021;17:e1009253
13. Solomon L, Mansor S, Mallon P, Donnelly E, Hoper M, Loughrey M, Kirk S, Gardiner K. The dextran sulphate sodium (DSS) model of colitis: an overview. Comparative Clin Pathol. 2010;19:235-9
14. Yason JA, Liang YR, Png CW, Zhang Y, Tan KSW. Interactions between a pathogenic Blastocystis subtype and gut microbiota: in vitro and in vivo studies. Microbiome. 2019;7:30
15. Deng L, Wojciech L, Png CW, Koh EY, Aung TT, Kioh DYQ. et al. Experimental colonization with Blastocystis ST4 is associated with protective immune responses and modulation of gut microbiome in a DSS-induced colitis mouse model. Cellul Mol Life Sci. 2022;79:245
16. Wu Z, Mirza H, Tan KS. Intra-subtype variation in enteroadhesion accounts for differences in epithelial barrier disruption and is associated with metronidazole resistance in Blastocystis subtype-7. PLoS Negl Trop Dis. 2014;8:e2885
17. Lim MX, Png CW, Tay CY, Teo JD, Jiao H, Lehming N, Tan KS, Zhang Y. Differential regulation of proinflammatory cytokine expression by mitogen-activated protein kinases in macrophages in response to intestinal parasite infection. Infect Immun. 2014;82:4789-801
18. Ho LC, Singh M, Suresh G, Ng GC, Yap EH. Axenic culture of Blastocystis hominis in Iscove's modified Dulbecco's medium. Parasitology research. 1993;79:614-6
19. Chen XQ, Singh M, Ho LC, Moe KT, Tan SW, Yap EH. A survey of Blastocystis sp. in rodents. Lab Ani Sci. 1997;47:91-4
20. Cooper HS, Murthy SN, Shah RS, Sedergran DJ. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69:238-49
21. Belda E, Voland L, Tremaroli V, Falony G, Adriouch S, Assmann KE. et al. Impairment of gut microbial biotin metabolism and host biotin status in severe obesity: effect of biotin and prebiotic supplementation on improved metabolism. Gut. 2022;71:2463-80
22. Burrello C, Garavaglia F, Cribiù FM, Ercoli G, Lopez G, Troisi J. et al. Therapeutic faecal microbiota transplantation controls intestinal inflammation through IL10 secretion by immune cells. Nat Comm. 2018;9:5184
23. Mishra A, Lai GC, Yao LJ, Aung TT, Shental N, Rotter-Maskowitz A. et al. Microbial exposure during early human development primes fetal immune cells. Cell. 2021;184:3394-409.e20
24. Poirier P, Wawrzyniak I, Albert A, El Alaoui H, Delbac F, Livrelli V. Development and evaluation of a real-time PCR assay for detection and quantification of Blastocystis parasites in human stool samples: prospective study of patients with hematological malignancies. J Clin Microbiol. 2011;49:975-83
25. Melgar S, Karlsson L, Rehnström E, Karlsson A, Utkovic H, Jansson L, Michaëlsson E. Validation of murine dextran sulfate sodium-induced colitis using four therapeutic agents for human inflammatory bowel disease. Intern Immunopharmacol. 2008;8:836-44
26. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581-3
27. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590-6
28. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357-60
29. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl Aacids Res. 2015;43:e47
30. Subramanian A, Tamayo P, Mootha Vamsi K, Mukherjee S, Ebert Benjamin L, Gillette Michael A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Nal Acad Sci USA. 2005;102:15545-50
31. Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267-73
32. Chan JC, Kioh DY, Yap GC, Lee BW, Chan EC. A novel LCMSMS method for quantitative measurement of short-chain fatty acids in human stool derivatized with (12)C- and (13)C-labelled aniline. J Pharmaceutical Biomedical Anal. 2017;138:43-53
33. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569-73
34. Vital M, Karch A, Pieper DH. Colonic butyrate-producing communities in humans: an overview using omics data. mSystems. 2017;2:e00130-17
35. Kushkevych I, Dordević D, Vítězová M. Possible synergy effect of hydrogen sulfide and acetate produced by sulfate-reducing bacteria on inflammatory bowel disease development. J Adv Res. 2021;27:71-8
36. Rowan F, Docherty NG, Murphy M, Murphy B, Coffey JC, O'Connell PR. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis Colon Rectum. 2010;53:1530-6
37. van der Hee B, Wells JM. Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends Microbiol. 2021;29:700-12
38. Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y. et al. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat Comm. 2020;11:4457
39. Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, Andoh A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018;11:1-10
40. Imam T, Park S, Kaplan MH, Olson MR. Effector T Helper Cell Subsets in Inflammatory Bowel Diseases. Front Immunol. 2018;9:1212
41. Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057-61
42. Lopes F, Matisz C, Reyes JL, Jijon H, Al-Darmaki A, Kaplan GG, McKay DM. Helminth regulation of immunity: a three-pronged approach to treat colitis. Inflamm Bowel Dis. 2016;22:2499-512
43. Smith P, Mangan NE, Walsh CM, Fallon RE, McKenzie ANJ, van Rooijen N, Fallon PG. Infection with a Helminth Parasite Prevents Experimental Colitis via a Macrophage-Mediated Mechanism. J Immunol. 2007;178:4557
44. Shute A, Callejas BE, Li S, Wang A, Jayme TS, Ohland C, Lewis IA, Layden BT, Buret AG, McKay DM. Cooperation between host immunity and the gut bacteria is essential for helminth-evoked suppression of colitis. Microbiome. 2021;9:186
45. Hublin JSY, Maloney JG, Santin M. Blastocystis in domesticated and wild mammals and birds. Research in veterinary science. 2021;135:260-282
46. Even G, Lokmer A, Rodrigues J, Audebert C, Viscogliosi E, Ségurel L, Chabé M. changes in the human gut microbiota associated with colonization by Blastocystis sp. and Entamoeba spp. in non-industrialized populations. Front Cell Infecti Microbiol. 2021;11:533528
47. Stensvold CR, Sørland BA, Berg R, Andersen LO, van der Giezen M, Bowtell JL, El-Badry AA, Belkessa S, Kurt Ö, Nielsen HV. Stool microbiota diversity analysis of Blastocystis-positive and Blastocystis-negative individuals. Microorganisms. 2022;10:326
48. Mayneris-Perxachs J, Arnoriaga-Rodríguez M, Garre-Olmo J, Puig J, Ramos R, Trelis M. et al. Presence of Blastocystis in gut microbiota is associated with cognitive traits and decreased executive function. ISME J. 2022;16:2181-97
49. Defaye M, Nourrisson C, Baudu E, Lashermes A, Meynier M, Meleine M. et al. Fecal dysbiosis associated with colonic hypersensitivity and behavioral alterations in chronically Blastocystis-infected rats. Sci Rep. 2020;10:9146
50. Billy V, Lhotská Z, Jirků M, Kadlecová O, Frgelecová L, Parfrey LW, Pomajbíková KJ. Blastocystis colonization alters the gut microbiome and, in some cases, promotes faster recovery from induced colitis. Front Microbiol. 2021;12:641483
51. Russo E, Giudici F, Fiorindi C, Ficari F, Scaringi S, Amedei A. Immunomodulating activity and therapeutic effects of short chain fatty acids and tryptophan post-biotics in inflammatory bowel disease. Front Immunol. 2019;10:2754
52. Ferrer-Picón E, Dotti I, Corraliza AM, Mayorgas A, Esteller M, Perales JC. et al. intestinal inflammation modulates the epithelial response to butyrate in patients with inflammatory bowel disease. se. Inflamm Bowel Dis. 2020;26:43-55
53. Qiu P, Ishimoto T, Fu L, Zhang J, Zhang Z, Liu Y. The gut microbiota in inflammatory bowel disease. Front Cell Infect Microbiol. 2022;12:733992
54. Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J, Kim CH. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015;8:80-93
55. Harbour Stacey N, Maynard Craig L, Zindl Carlene L, Schoeb Trenton R, Weaver Casey T. Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc Nal Acad Sci USA. 2015;112:7061-6
56. Motomura Y, Wang H, Deng Y, El-Sharkawy RT, Verdu EF, Khan WI. Helminth antigen-based strategy to ameliorate inflammation in an experimental model of colitis. Clin Exp Immunol. 2009;155:88-95
57. Chaudhry A, Samstein RM, Treuting P, Liang Y, Pils MC, Heinrich J-M. et al. Interleukin-10 signaling in regulatory T cells is required for suppression of Th17 cell-mediated inflammation. Immunity. 2011;34:566-78
58. Drurey C, Lindholm HT, Coakley G, Poveda MC, Löser S, Doolan R. et al. Intestinal epithelial tuft cell induction is negated by a murine helminth and its secreted products. J Exp Medi. 2022;219:e20211140
59. Johnston CJC, Smyth DJ, Kodali RB, White MPJ, Harcus Y, Filbey KJ. et al. A structurally distinct TGF-β mimic from an intestinal helminth parasite potently induces regulatory T cells. Nat Comm. 2017;8:1741
60. Zhang F, Cui B, He X, Nie Y, Wu K, Fan D. Microbiota transplantation: concept, methodology and strategy for its modernization. Protein Cell. 2018;9:462-73
61. Terveer EM, van Beurden YH, Goorhuis A, Mulder CJJ, Kuijper EJ, Keller JJ. Faecal microbiota transplantation in clinical practice. Gut. 2018;67:196
62. de Groot PF, Frissen MN, de Clercq NC, Nieuwdorp M. Fecal microbiota transplantation in metabolic syndrome: History, present and future. Gut Microbes. 2017;8:253-67
63. Cammarota G, Ianiro G, Tilg H, Rajilić-Stojanović M, Kump P, Satokari R. et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut. 2017;66:569-80
64. Andersen LO, Stensvold CR. Blastocystis in health and disease: are we moving from a clinical to a public health perspective? J Clin Microbiol. 2016;54:524-8
65. Terveer EM, van Gool T, Ooijevaar RE, Sanders I, Boeije-Koppenol E, Keller JJ. et al. Human transmission of Blastocystis by Fecal Microbiota Transplantation without development of gastrointestinal symptoms in recipients. Clin Infect Dise. 2019;71:2630-6
Author contact
![]() Corresponding author: Prof. Kevin Tan. Email: mictankedu.sg
Corresponding author: Prof. Kevin Tan. Email: mictankedu.sg
 Global reach, higher impact
Global reach, higher impact