13.3
Impact Factor
Theranostics 2022; 12(17):7586-7602. doi:10.7150/thno.78616 This issue Cite
Research Paper
TIMM44 is a potential therapeutic target of human glioma
1. Institute of Neuroscience, Soochow University, Suzhou, and Department of Neurosurgery, The affiliated Changzhou No. 2 People's Hospital of Nanjing Medical University, Changzhou, China.
2. Department of Neurosurgery, the First Affiliated Hospital of Soochow University, Suzhou, China.
3. Department of Oncology, Dushu Lake Hospital Affiliated of Soochow University, Suzhou, China.
4. Department of Radiotherapy and Oncology, Affiliated Kunshan Hospital of Jiangsu University, Kunshan, China.
5. The Fourth School of Clinical Medicine, The Affiliated Eye Hospital, Nanjing Medical University, Nanjing, China.
6. Jiangsu Key Laboratory of Neuropsychiatric Diseases, Clinical Research Center of Neurological Disease, The Second Affiliated Hospital of Soochow University, Suzhou, China.
#Co-first authors.
Received 2022-9-3; Accepted 2022-10-23; Published 2022-10-31
Abstract
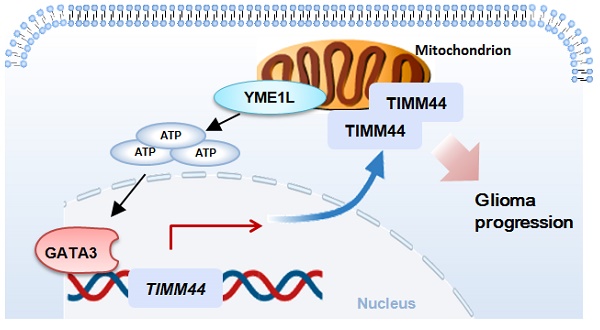
TIMM44 (translocase of inner mitochondrial membrane 44) is essential for the maintenance of mitochondrial functions. Bioinformatics studies and results from the local high-grade glioma tissues showed that TIMM44 mRNA and protein levels are elevated in glioma, correlating with poor overall survival. Mitochondrial TIMM44 upregulation was also detected in patient-derived primary glioma cells and immortalized cell line. In primary and established glioma cells, TIMM44 depletion, using the lentiviral shRNA strategy or the CRISPR/Cas9 knockout (KO) method, robustly inhibited cell viability, proliferation and migration. Moreover, TIMM44 silencing/KO resulted in mitochondrial complex I inhibition, ATP depletion, mitochondrial membrane potential reduction, oxidative stress and DNA damage, and eventually provoked apoptosis. Conversely, ectopic overexpression of TIMM44 augmented glioma cell proliferation and migration. TIMM44 upregulation in glioma is possibly due to increased TIMM44 transcriptional machinery by the transcription factor GATA3 in a YME1L (YME1 Like 1 ATPase)-dependent manner. In vivo, the growth of subcutaneous glioma xenografts was suppressed after intratumoral injection of TIMM44 shRNA adeno-associated virus (AAV). TIMM44 depletion, ATP reduction, oxidative injury and apoptosis were detected in TIMM44 shRNA AAV-injected glioma xenografts. Moreover, the intracranial growth of TIMM44 KO glioma cells in the mouse brain was largely inhibited. Together, overexpressed TIMM44 could be a novel and promising therapeutic target of human glioma.
Keywords: Glioma, TIMM44, Mitochondria, Therapeutic target
Introduction
In central nerve system (CNS), glioma with astrocytic or oligodendroglial origin is the most common malignancy [1, 2]. The current clinical treatments for glioma include tumor resection, radiation therapy and temozolomide-based chemotherapy [1, 2]. The prognosis of glioblastoma (GBM), anaplastic astrocytoma and other high-grade gliomas is extremely poor, leading to significant human mortalities each year [3, 4]. Gliomas often contain heterogeneous mixture of cells exhibiting different cellular and nuclear polymorphism [5-10]. In addition, dysregulation of multiple signaling cascades is essential for the uncontrolled growth and progression of glioma cells [3, 4, 11]. Therefore, identifying novel glioma-driven signaling molecules and exploring the corresponding molecularly-targeted therapies are essential for better and efficient glioma therapy [3, 4, 11, 12]. It has been the research focus of our group [13-15].
Dysfunctional mitochondria are commonly detected in human glioma, participating in tumorigenesis and cancer progression [16-18]. In particular, aberrant energy metabolism, mitochondrial dysfunction, mitochondrial structural abnormalities and dysregulated mitochondrial apoptotic signalling cascade have been observed in malignant gliomas [16-18]. Mitochondrial proteomics abnormalities could provide novel therapeutic targets for glioma [16-18]. Our recent study has shown that overexpression of YME1L (YME1 Like 1 ATPase) [19-23], a mitochondrial inner membrane protease, exerts significant pro-tumorigenic activity in glioma [24].
TIMM44 (translocase of inner mitochondrial membrane 44) locates in the inner mitochondrial membrane [25, 26]. In a ATP-dependent manner, TIMM44 anchors mitochondrial heat shock protein 70 to the translocase of the mitochondrial inner membrane 23 (TIMM23) complex [27-29]. It promotes the import of mitochondrial preproteins to the mitochondrial matrix [27, 28]. This process is dependent on inner membrane potential and ATP hydrolysis on ATPase domain of mtHsp70 [27, 28]. Wang et al., have established an aP2-promoter driven TIMM44 transgenic mouse. The transgenic mice were protected from type 2 diabetes [25]. Different mitochondrial fusion genes, including mitofusin-1 (Mfn1), mitofusin-2 (Mfn2), and optic atrophy 1 (Opa1), were upregulated in TIMM44 transgenic mice [25]. Overexpression of TIMM44 attenuated reactive oxygen species (ROS) production and increased ATP production, promoting human aortic smooth muscle cell proliferation [30].
Yu et al., have shown that human antigen R directly associated with TIMM44 in ovarian cancer cells, vital for the TIMM44 mRNA stability and ovarian cancer cell growth. Conversely, siRNA-induced silencing of TIMM44 inhibited ovarian cancer cell growth [31]. Bonora et al., have identified two novel variants in exon 9 and exon 13 of TIMM44, which could be associated with the development of oncocytic thyroid carcinomas [26]. TIMM44 expression and potential functions in glioma have not been examined thus far. We here will show that TIMM44 overexpression exerts significant pro-tumorigenic activity in glioma.
Results
TIMM44 is overexpressed in GBM, correlating with poor survival
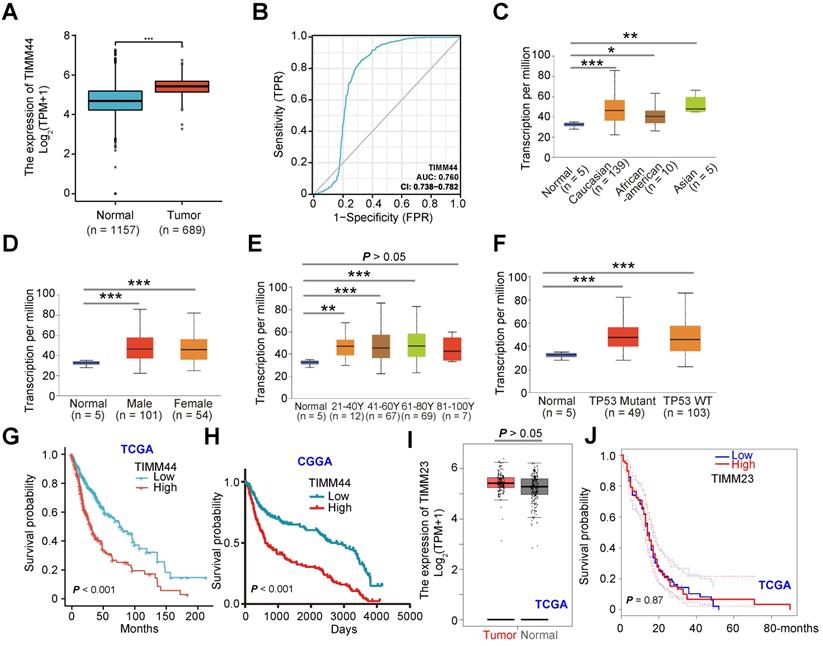
We first analyzed TIMM44 mRNA expression from The Cancer Genome Atlas (TCGA) database combining with the Genotype-Tissue Expression (GTEx) database. A total of 1846 human tissue samples were retrieved, of which 689 were GBM (“Tumor”), 1152 were normal brain tissues, and five were adjacent to cancer. The number of TIMM44 transcripts in GBM tissues was significantly higher than that in the normal brain tissues (“Normal”) (Figure 1A). A receiver operating characteristic (ROC) curve is a plot of the sensitivity versus specificity for a diagnostic test. The different points on the curve indicate the different cut-points determining whether the test results are significant/positive [32]. Area under curve (AUC) is recognized an effective method to summarize the overall diagnostic accuracy of the test. The ROC curve in Figure 1B showed the potential diagnostic value of TIMM44 for glioma. An AUC of 0.760 indicated an acceptable value of TIMM44 overexpression for potential diagnosis of glioma.
By consulting the UALCAN database (http://ualcan.path.uab.edu), the relationship between each subtype grouping and TIMM44 expression of 156 GBM tissues and five para-cancerous tissues was analyzed. TIMM44 was highly expressed in different races (Figure 1C), both genders (Figure 1D) and different age groups (Figure 1E, except for the 81-100Y age). The high expression of TIMM44 was however not correlated with p53 mutation status (Figure 1F). In both TCGA (Figure 1G) and Chinese Glioma Genome Atlas (CGGA) (Figure 1H), patients with high TIMM44 expression in gliomas have worse prognosis and lower survival (Figure 1G and H). Whereas the patients with low TIMM44 expression have better survival (Figure 1G and H). The bioinformatics results showed that TIMM44 is overexpressed in GBM, correlating with poor survival.
TIMM23 is another key component in the mitochondrial protein import machinery, binding to TIMM44 [27-29]. Interestingly, TCGA database results found that the number of TIMM23 transcripts in GBM tissues was not upregulated when compared to that in the normal brain tissues (Figure 1I). Moreover, the overall survivals of high-TIMM23 GBM patients and the low-TIMM23 patients were equivalent (P = 0.87, Figure 1J).
Mitochondrial TIMM44 overexpression in local glioma tissues and cells
We also examined TIMM44 expression in local glioma tissues. A total of sixteen (n = 16) pairs of high-grade (grade III-IV) glioma tissues (“T”) and matched adjacent normal brain tissues (“N”), as reported in our previous studies [13, 15, 24], were analyzed. qRT-PCR results, Figure 2A, demonstrated that TIMM44 mRNA expression in the glioma tissues was significantly higher than that in the normal brain tissues. Western blotting data of four representative patients, Patient-1 to Patient-4, confirmed TIMM44 protein upregulation in glioma tissues (Figure 2B). Combining all 16 pairs of blotting data showed that TIMM44 protein upregulation in glioma tissues was significant (P < 0.05 vs. “N” tissues) (Figure 2C).
The tissue immuno-fluorescence images in Figure 2D demonstrated that TIMM44 protein (labeled in the green fluorescence) was co-localized with the MitoTracker (red fluorescence) in both glioma slides and adjacent normal brain slides of two representative patients (Patient-1# and Patient-2#). TIMM44 green fluorescence intensity in glioma slides was higher than that in the matched normal brain slides (Figure 2D). TIMM44 green fluorescence intensity was recorded from 10 random microscopy views (containing 14-15 nuclei per view) of each slide, and intensity normalized to MitoTracker red fluorescence intensity. As shown, the normalized TIMM44 green fluorescence intensity in the glioma slides was dramatically higher than that in the matched normal brain slides (Figure 2D, the right panel).
Furthermore, the mitochondrial fractions of human tissues of the two representative patients (Patient-1# and Patient-2#) were isolated and tissue lysates were analyzed. TIMM44 protein was only enriched in the mitochondrial lysates (Figure 2E), as indicated by the mitochondrial marker protein VDAC1 (voltage-dependent anion-selective channel 1) (Figure 2E). Both the nuclear marker protein Lamin-B1 and the cytosol marker protein α-Tubulin were not present in the mitochondrial fraction lysates (Figure 2E). Notably, the mitochondrial TIMM44 expression was higher in the glioma tissues (Figure 2E). TIMM23 protein levels were indifferent between glioma tissues and normal brain tissues (Figure 2E). TIMM44, TIMM23 and VDAC1 proteins were not detected in the mitochondria-null lysates of the human tissues (Figure 2F), whereas Lamin-B1 and α-Tubulin were present (Figure 2F). Thus, mitochondria-localized TIMM44 is upregulated in human glioma tissues.
TIMM44 is overexpressed in GBM, correlating with poor survival. TCGA plus GTEx database shows TIMM44 expression (RNA-Seq) in glioma tissues (“Tumor”, n = 689) and in the normal brain tissues (“Normal”, n = 1157) (A). The receiver operating characteristic (ROC) curve results demonstrated the relationship between TIMM44 overexpression and the potential predictive effect on glioma patients' survival (B). The subgroup analyses of TIMM44 mRNA expression and the listed clinical characteristics of glioma patients were shown (C-F). The Kaplan Meier Survival curves of TIMM44-low (in blue) and TIMM44-high (in red) GBM patients from The Cancer Genome Atlas (TCGA) (G) and Chinese Glioma Genome Atlas (CGGA) (H) databases were shown. TCGA database shows TIMM23 expression (RNA-Seq) in glioma tissues (“Tumor”) and in the normal brain tissues (“Normal”) (I). The Kaplan Meier Survival curves of TIMM23-low (in blue) and TIMM23-high (in red) GBM patients from TCGA (J). “TPM” stands for transcripts per million. * P < 0.05. ** P < 0.01. *** P < 0.001.
Mitochondrial TIMM44 overexpression in local glioma tissues and cells. The human glioma tissues (“T”) and the paired adjacent normal brain tissues (“N”), derived from a total of sixteen (16) HGG patients, were homogenized, TIMM44 mRNA and protein expression in tissue lysates was examined by qRT-PCR (A) and Western blotting (B and C) assays, respectively. The human tissue immuno-fluorescence images showed TIMM44 protein (green fluorescence), MitoTracker (red fluorescence), and DAPI (in the red fluorescence) in glioma tissue slides (“T1/T2”) and the adjacent normal brain tissue slides (“N1/N2”) of two representative glioma patients (Patient-1# and Patient-2#) (D). TIMM44 green fluorescence intensity was recorded from 10 random microscopy views of each slide, and its intensity was normalized to MitoTracker red fluorescence intensity (D, the right panel). Mitochondrial lysates and mitochondria-null lysates of the two representative glioma patients (Patient-1# and Patient-2#) were observed and listed proteins were tested (E and F). TIMM44 mRNA (G) and listed proteins (in mitochondrial and mitochondrial-null lysates, H) expression in the primary human astrocytes (“Astrocytes1/2”, derived from two patients), the immortalized (A172) and primary (P1, P2 and P3) glioma cells was shown. The data were presented as mean ± standard deviation (SD).* P < 0.05 vs.“N”/“Astrocytes”.
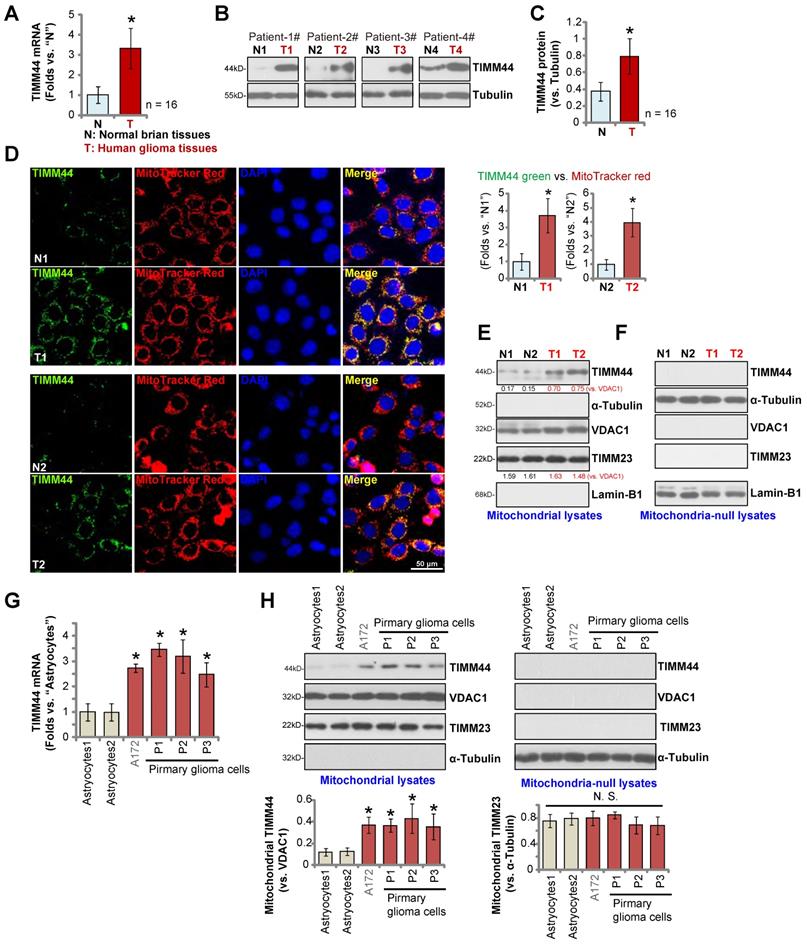
We also examined TIMM44 expression in human glioma cells. Patient-derived primary glioma cells “P1”, “P2” and “P3” (derived from three patients [13]) and the immortalized cell line (A172) were examined. Results showed that TIMM44 mRNA expression in the primary and established glioma cells was higher than that in the primary human astrocytes (“Astrocytes1/2”, derived from two patients, Figure 2G). Moreover, TIMM44 protein upregulation was observed (Figure 2H). TIMM23 and VDAC1 protein expression was equivalent between astrocytes and glioma cells (Figure 2H). The relative expression of mitochondrial TIMM44 protein, after normalization to the mitochondrial proteinVDAC1, was again increased in different glioma cells (Figure 2H). These results confirmed mitochondrial TIMM44 overexpression in local glioma tissues and cells.
TIMM44 depletion induces significant anti-glioma cell activity
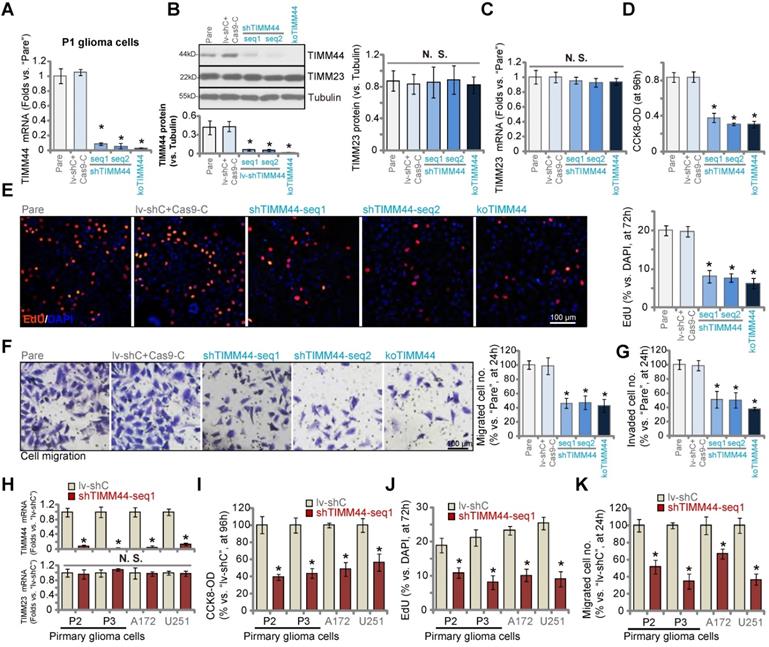
Using the previously described genetic methods [13, 15, 24], we depleted TIMM44 in human glioma cells. Specifically, the lentivirus encoding the TIMM44 shRNA (shTIMM44-seq1/shTIMM44-seq2, with non-overlapping sequences) was added to P1 primary human glioma cells [13, 15, 24]. Stable cells were formed following selection through puromycin-containing medium. Alternatively, a CRISPR/Cas9-TIMM44-KO vector was established and transduced to the Cas9-expressing P1 glioma cells [24]. The transfected cells were thereafter distributed into 96-well plates. Through TIMM44 KO screening, stable TIMM44-KO cells were formed. These cells were named as “koTIMM44” cells. As shown TIMM44 mRNA levels were dramatically decreased in shTIMM44 and ko-TIMM44 P1 glioma cells (Figure 3A). The applied shRNAs and KO construct resulted in TIMM44 protein depletion as well in P1 glioma cells (Figure 3B). TIMM23 mRNA and protein expression was unchanged (Figure 3B and C).
TIMM44 shRNA or KO significantly decreased CCK-8 viability in P1 primary glioma cells (Figure 3D). Moreover, the percentage of EdU-positive nuclei, an indicator of cell proliferation, was dramatically decreased in shTIMM44-expressing P1 glioma cells and koTIMM44 cells (Figure 3E). Moreover, TIMM44 shRNA or KO largely suppressed P1 glioma cell in vitro migration (Figure 3F) and invasion (Figure 3G), tested by “Transwell” and “Matrigel Transwell” assays, respectively. The control P1 glioma cells were with the lentiviral scramble shRNA plus the CRSIPR/Cas9 empty vector (“lv-shC+Cas9-C”), which failed to alter TIMM44-TIMM23 expression (Figure 3A-C) and glioma cell functions (Figure 3D-G).
MB-10 (“MitoBloCK-10”) binds to a specific pocket in the C-terminal domain of TIMM44, thereby inhibiting mitochondrial protein import [33]. Here we found that treatment with MB-10 in P1 primary human glioma cells robustly inhibited CCK-8 cell viability (Figure S1A) and proliferation (EdU-nuclei ratio reduction, Figure S1B). P1 cell migration (Figure S1C) and invasion (Figure S1D) were largely slowed following MB-10 treatment as well.
Other primary human glioma cells (“P2” and “P3”, derived from other patients [13, 15]) and established cell lines (A172 and U251) were infected with shTIMM44-seq1-expressing lentivirus, and puromycin was added to select stable cells. As compared to control glioma cells with the lentiviral scramble shRNA (lv-shC), TIMM44 expression was robustly decreased in shTIMM44-seq1-expressing glioma cells (Figure 3H). The viability of the primary and established glioma cells was decreased following TIMM44 silencing (Figure 3I), which also exerted anti-proliferative activity (Figure 3J) and hindered in vitro migration (Figure 3K) of the glioma cells.
The primary human astrocytes, Astrocytes1 and Astrocyte2, were stably infected with the shTIMM44-seq1-expressing lentivirus, leading to downregulation of TIMM44 mRNA (Figure S2A). TIMM23 mRNA expression was unchanged (Figure S2B). However, in the astrocytes, TIMM44 silencing failed to significantly inhibit cell viability (Figure S2C) and proliferation (Figure S2D).
The mitochondrial functions are disrupted with TIMM44 depletion in glioma cells
Studies have shown that increasing TIMM44 expression can restore mitochondrial functions [25]. We therefore analyzed whether TIMM44 depletion could result in mitochondrial dysfunctions in human glioma cells. As shown, in P1 glioma cells TIMM44 shRNA or KO (see Figure 3) induced conversion of JC-1 aggregates (in red fluorescence) to the JC-1 green monomers (Figure 4A), causing mitochondrial depolarization. Moreover, the mitochondrial complex I activity and the cellular ATP contents were significantly decreased following TIMM44 silencing or depletion in P1 glioma cells (Figure 4B). Further studies showed that cellular ROS contents were significantly increased in P1 cells with TIMM4 shRNA or KO, and the DCF-DA green intensity (Figure 4C) and the CellROX red intensity (Figure 4D) were both increased. Supporting increased lipid peroxidation, we found that the TBAR activity was augmented in TIMM44-silenced or TIMM44-KO P1 glioma cells (Figure 4E). TIMM44 shRNA (by shTIMM44-seq1) or KO robustly inhibited the import of Su9-DHFR fusion protein into the mitochondria of P1 glioma cells (Figure S4A), and about 60-70% reduction of mitochondrial protein import was achieved with TIMM44 silencing/KO (Figure S4A).
TIMM44 depletion induces significant anti-glioma cell activity. The primary human glioma cells, P1, stably expressing the listed TIMM44 shRNA (shTIMM44-seq1/shTIMM44-seq2), the Cas9 construct plus the CRISPR/Cas9-TIMM44-KO construct (“koTIMM44”), or the lentiviral scramble control shRNA plus the Cas9 empty vector (“lv-shC+Cas9-C”), were formed and were tested for the listed genes and proteins (A-C). Alternatively, cells were further cultivated for designated hours, and cellular functions, including cell viability (D), proliferation (E), migration (F) and invasion (G) were examined. Expression of listed mRNAs in the primary glioma cells (“P2” and “P3”) or established lines (A172 and U251) with the TIMM44 shRNA (shTIMM44-seq1) or the lentiviral scramble control shRNA (“lv-shC”) was shown (H). The glioma cells were further cultivated for designated hours, and cell viability (I), proliferation (J) and migration (K) were tested, with the results quantified. “Pare” stands for the parental control glioma cells. The data were presented as mean ± standard deviation (SD). * P < 0.05 vs. “Pare”/“lv-shC”. “N. S.” stands for non-statistical difference (P > 0.05). The in vitro experiments were repeated five times with similar results obtained. Scale bar = 100 µm.
Treatment with the mitochondrial protein import blocker MB-10 induced mitochondrial depolarization (JC-1 green monomers accumulation, Figure S1E) and ROS production (CellROX intensity increasing, Figure S1F). Significant apoptosis activation, evidenced by TUNEL-positive nuclei ratio increasing, was observed as well in MB-10-treated P1 glioma cells (Figure S1G).
mRNA expression of TIMM44-dependent mitochondrial genes, including Opa1, Mfn1 and Mfn2 [25], were dramatically decreased with TIMM44 silencing or KO in P1 glioma cells (Figure 4F). TIMM44 shRNA or KO also decreased the expression of OPA1 protein (the long and un-cleaved form, Figure 4G), but did not alter OPA1 protein cleavage, as the short and cleaved form of OPA1 was unchanged (Figure 4G). Cancer cells have increased mitochondrial fission to achieve proliferative and survival advantages [34-36]. The high-resolution mitochondrial fluorescence images clearly showed that the majority of mitochondria in the parental control P1 glioma cells were small-fragment fission mitochondria (Figure 4H). The average mitochondrial length was quantified from 25 mitochondria per cell of three different cells for each condition. We found that the average mitochondrial length was similar between control glioma cells and TIMM44-silenced/KO glioma cells (Figure 4H). On the contrast, a significant number of fused mitochondria (long fragment) were detected in the non-cancerous primary human astrocytes (“Astrocytes1/2”) (Figure 4H), showing longer average mitochondrial length (Figure 4H).
The mitochondrial functions are disrupted with TIMM44 depletion in glioma cells. P1 glioma cells with applied genetic modifications were cultivated for designated hours, mitochondrial membrane potential (A), the relative mitochondrial complex I activity and cellular ATP contents (B), ROS intensity (DCF-DA/CellROX OD, C and D) and lipid peroxidation (TBAR activity, E) were examined by the assays mentioned. mRNA and listed proteins were tested (F and G). The high-resolution fluorescence images showing mitochondrial fusion/fission morphology in the P1 glioma cells and the primary astrocytes were presented as well (H), and average mitochondrial length was quantified (H). Cell apoptosis (by testing TUNEL-nuclei ratio, I) and relative Caspase-3 activity (J) were examined. “Pare” stands for the parental control glioma cells. The data were presented as mean ± standard deviation (SD). * P < 0.05 vs. “Pare”. “N. S.” stands for non-statistical difference (P > 0.05). The in vitro experiments were repeated five times with similar results obtained. Scale bar = 10 µm /100 µm.
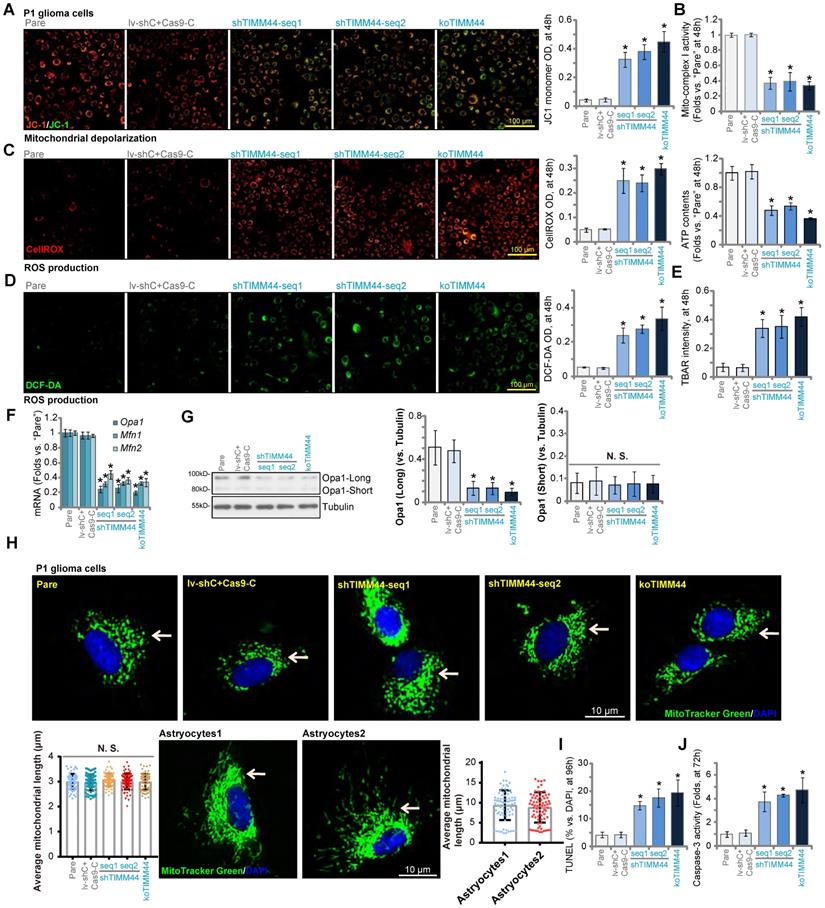
With mitochondrial dysfunction, apoptosis was induced in TIMM44-depeleted glioma cells. In TIMM44-silenced or TIMM44-KO P1 glioma cells, TUNEL-positive nuclei percentage was significantly increased (Figure 4I). Moreover, the relative Caspase-3 activity was augmented (Figure 4J). Supplement ATP or adding the antioxidant NAC robustly ameliorated TIMM44 silencing (by shTIMM44-seq1)-induced viability (CCK-8 OD) reduction and cell death (Trypan blue staining) in P1 glioma cells (Figure S3A and B), suggesting that mitochondrial dysfunction should be the primary cause of TIMM44-depeletion-induced cytotoxicity in glioma cells.
Similar to our results in P1 glioma cells, we found that TIMM44 shRNA decreased mitochondrial complex I activity (Figure S3C) and ATP contents (Figure S3D), while increasing the ROS levels (CellROX intensity increase, Figure S3E) in P2/ P3 primary glioma cells and established lines (A172 and U251). TIMM44 shRNA also increased the number of TUNEL-positive nuclei in the glioma cells (Figure S3F).
In the primary human astrocytes, Astrocytes1 and Astrocyte2, TIMM44 shRNA failed to significantly alter ATP contents (Figure S4B) and ROS levels (CellROX intensity, Figure S4C). Moreover, it failed to increase the Caspase-3 activity (Figure S4D) and provoke apoptosis (Figure S4E).
The pro-cancerous activity by ectopic TIMM44 expression in glioma cells
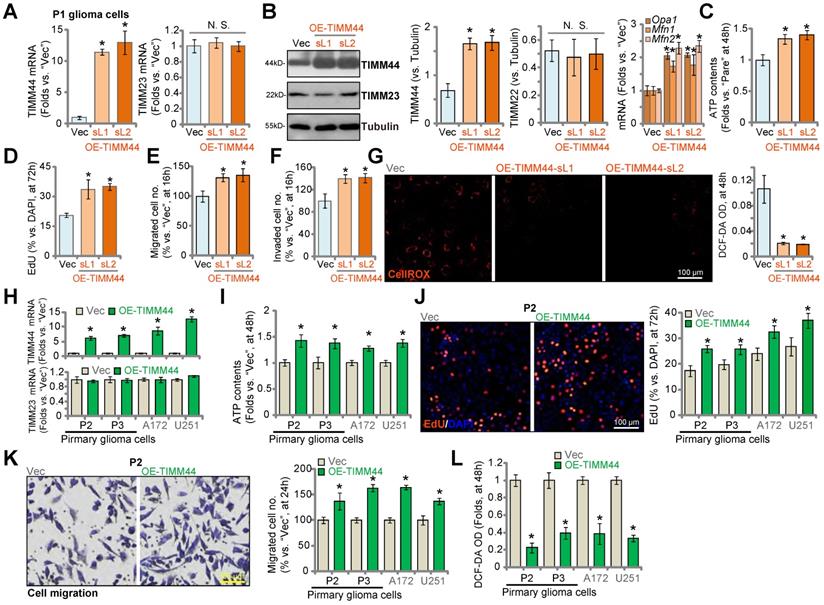
Thus far, we found that genetic depletion of TIMM44 inhibited cell proliferation and migration, disrupted mitochondrial functions and induced apoptosis in primary and established glioma cells. We therefore tested whether ectopic overexpression of TIMM44 could exert pro-glioma cell activity. To this purpose, the lentiviral TIMM44-expressing vector were transduced to P1 primary glioma cells, and puromycin was utilized to select stable cells: OE-TIMM44-sL1 and OE-TIMM44-sL2 (representing two different stable selections). In OE-TIMM44 P1 glioma cells TIMM44 mRNA and protein levels were indeed dramatically increased (Figure 5A and B) and TIMM23 expression was unchanged (Figure 5A and B). Opa1, Mfn1 and Mfn2, TIMM44-dependent mitochondrial genes, were increased in TIMM44-overexpressed P1 glioma cells (Figure 5B). In OE-TIMM44 cells, the cellular ATP contents were significantly increased (Figure 5C). Ectopic overexpression of TIMM44 promoted P1 glioma cell proliferation, which was evidenced by the increased EdU-positive nuclei percentage (Figure 5D). Moreover, enhanced numbers of migrated cells (Figure 5E) and invaded cells (Figure 5F) were observed after TIMM44 overexpression. On the contrast, DCF-DA intensity was decreased (Figure 5G), suggesting decreased ROS contents following TIMM44 overexpression in primary glioma cells.
To P2 and P3 primary glioma cells and established cell lines (A172 and U251), the TIMM44-expressing lentivirus was added, and stable cells were formed (“OE-TIMM44”). As compared to the vector control cells (“Vec”), TIMM44 mRNA, but not TIMM23 mRNA, was increased in OE-TIMM44 glioma cells (Figure 5H). The cellular ATP contents were augmented in the TIMM44-overexpressed glioma cells (Figure 5I). TIMM44 overexpression potentiated cell proliferation (increasing EdU-positive nuclei percentage, Figure 5J) and migration (Figure 5K) in the glioma cells. While the cellular ROS contents, or the DCF-DA intensity, were decreased (Figure 5L). These results further supported the essential role of TIMM44 in the progression of glioma cells.
The lentiviral TIMM44-expressing vector was also transduced to the primary human astrocytes (Astrocytes1 and Astrocyte2), resulting in significant TIMM44 upregulation (“OE-TIMM44”, Figure S5A). TIMM23 mRNA levels were unchanged (Figure S5B). OE-TIMM44 had no significant effect on cell viability (CCK-8 OD, Figure S5C) and proliferation (by measuring EdU-positive nuclei ratio, Figure S5D) in human astrocytes. These results again supported the cancer cell-specific effect by TIMM44.
YME1L promotes GATA3-dependent TIMM44 transcription in glioma cells
Our previous study has shown that YME1L (YME1 Like 1 ATPase), a mitochondrial inner membrane protease, is upregulated in human glioma, and it is essential for glioma cell growth in vitro and in vivo [24]. YME1L shRNA or KO inhibited glioma cell viability, proliferation and migration, and induced apoptosis activation [24]. TCGA database retrieving differentially expressed genes (DEGs) with YME1L in human glioma tissues and the following Pearson Correlation Coefficient analyses showed that TIMM44 is one topYME1L's DEG with the second highest correlation score [24].
Next, YMEL1 shRNA-expressing lentivirus was transfected to P1 primary human glioma cells and stable cells (“lv-shYME1L”) were formed by selection. Alternatively, a lenti-CRSIPR/Cas9-YME1L-knockout (KO)-puro construct was transduced to Cas9-expressing P1 glioma cells, and single stable cells (“koYME1L”) were established following selection and YMEL1 KO screening. As shown YME1L shRNA and KO resulted in significant TIMM44 mRNA (Figure 6A) and protein (Figure 6B) downregulation in P1 glioma cells. YME1L protein was depleted as well (Figure 6B). Expression of TIMM23 mRNA was unchanged (Figure 6C). YME1L silencing or KO resulted in reduction of TIMM44-dependent mitochondrial genes, including Opa1, Mfn1 and Mfn2, in P1 glioma cells (Figure S6A).
The pro-cancerous activity by ectopic TIMM44 expression in glioma cells. The primary human glioma cells, P1, stably expressing the lentiviral TIMM44-expressing vector (OE-TIMM44-sL1/OE-TIMM44-sL2, representing two stable selections) or the empty vector (“Vec”), were formed and tested for the listed genes and proteins (A and B). The glioma cells were further cultivated for designated hours, ATP contents were measured (C); EdU-positive nuclei ratio (D), migrated cell number (E), invaded cell number (F) and CellROX intensify (G) were measured, with results quantified. Expression of listed mRNAs in the primary glioma cells (“P2” and “P3”) or established lines (A172 and U251) with the lentiviral TIMM44-expressing vector (“OE-TIMM44”) or the empty vector (“Vec”) was shown (H). The glioma cells were further cultivated for designated hours, and ATP contents (I), proliferation (EdU-positive nuclei ratio, J), migration (K) and CellROX intensity (L) were tested. The data were presented as mean ± standard deviation (SD). * P < 0.05 vs. “Vec”. “N. S.” stands for non-statistical difference (P > 0.05). The in vitro experiments were repeated five times with similar results obtained. Scale bar = 100 µm.
In contrast, the lentiviral YME1L-expressing construct was transduced to P1 glioma cells. Two stable selections, OE-YME1L-sL1 and OE-YME1L-sL2 [24], were formed after selection using puromycin. We found that TIMM44 mRNA (Figure 6D) and protein (Figure 6E) as well as the YME1L protein (Figure 6E) levels were significantly increased in OE-YME1L glioma cells, whereas TIMM23 mRNA was unchanged (Figure 6F). Opa1, Mfn1 and Mfn2 mRNA levels were increased as well in YEM1L-overexpressed cells (Figure S6B). These results indicated that YME1L is important for TIMM44 expression in primary glioma cells.
Since YME1L is an i-AAA protease but not a transcriptional regulator, it is unlikely that YME1L can directly regulate TIMM44 at mRNA level. We therefore explored the transcriptional mechanism of TIMM44 expression in glioma cells. The tfbind database (https://tfbind.hgc.jp/) predicted transcription factors with the highest binding affinity to TIMM44 promoter. siRNAs specifically targeting these transcription factors were individually transfected to P1 primary glioma cells. Among the tested siRNAs, only GATA3 (GATA binding protein 3) siRNA resulted in robust TIMM44 mRNA downregulation. Importantly, GATA3 chromosome immunoprecipitation (ChIP) results demonstrated that GATA3-TIMM44 promoter binding in P1 glioma cells was robustly decreased after YME1L silencing or depletion (Figure 6G). Whereas overexpression of YME1L increased GATA3-TIMM44 promoter binding (Figure 6H).
shRNA-induced silencing of GATA3 significantly decreased TIMM44 mRNA and protein expression in P1 glioma cells (Figure 6I and J), and YME1L expression was unchanged (Figure 6J). Moreover, in human glioma tissues of four representative patients, GATA3 binding to the TIMM44 promoter was significantly higher than that in the matched surrounding normal brain tissues (Figure 6K). In addition, GATA3-TIMM44 promoter binding was augmented in different glioma cells (Figure 6L). Therefore, YME1L promoted GATA3-dependent TIMM44 transcription in glioma cells. ChIP results demonstrated that the YME1L shRNA-induced decrease of GATA3-TIMM44 promoter binding was attenuated by exogenously adding ATP in P1 glioma cells (Figure S6C). ATP also partially restored TIMM44 mRNA (Figure S6D) and protein (Figure S6E) expression in YME1L-silenced glioma cells.
YME1L promotes GATA3-dependent TIMM44 transcription in glioma cells. P1 primary human glioma cells, stably expressing the YME1L shRNA (lv-shYME1L), the lenti-CRSIPR/Cas9-YME1L-KO-puro construct (“koYME1L”) or the lentiviral scramble shRNA plus the CRSIPR/Cas9 empty vector (“lv-shC+Cas9-C”), were established, expression of listed genes and proteins (in total cell lysates and nuclear lysates) was shown (A-C). P1 glioma cells with the YME1L-expressing construct (“OE-YME1L-sL1 or OE-YME1L-sL2”, two stable selections) or the empty vector (“Vec”) were established, and the expression of listed genes and proteins was shown (D-F). Chromosome IP (ChIP) showed the relative levels of the proposed TIMM44 promoter binding to GATA3 in the P1 glioma cells with genetic modifications (G and H), or in the listed human tissue lysates (K) and astrocytes/glioma cells (L). P1 glioma cells expressing the scramble control shRNA (lv-shC), the lentiviral GATA3 shRNA (“sh-GATA3-seq1/2”, two different sequences) were established, expression of listed genes and protein was shown (I and J). The lv-shYME1L P1 glioma cells were further transduced with or without the lentiviral TIMM44-expressing vector (“OE-TIMM44”), expression of the listed proteins was shown (M). Cells were further cultivated for the indicated time periods, cell proliferation, migration and apoptosis were tested by EdU staining (N), “Transwell” (O), and TUNEL staining (P) assays, respectively. The data were presented as mean ± standard deviation (SD). * P < 0.05 vs. “Pare”/“Vec”/“lv-shC”/“N” tissues/“Astryocytes1”. # P < 0.05. “N. S.” stands for non-statistical difference (P > 0.05). The in vitro experiments were repeated five times with similar results obtained. Scale bar = 100 µm.
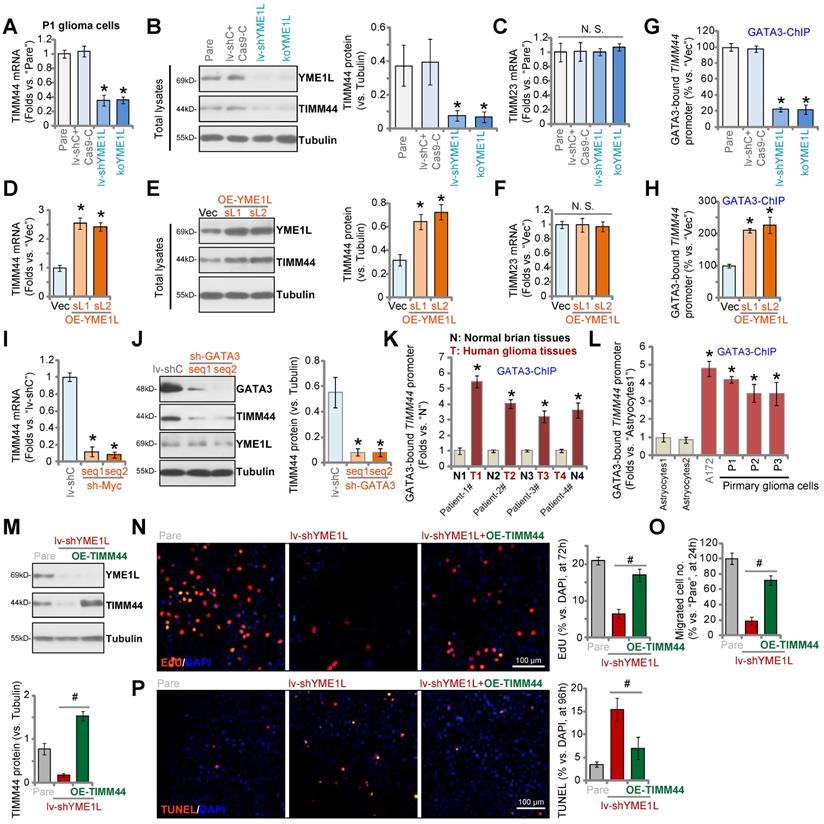
Moreover, overexpression of YME1L, by the lentiviral YME1L-expressing construct (“+OE-YME1L”), failed to restore TIMM44 mRNA expression in GATA3-silenced P1 glioma cells (Figure S6F). Therefore, TIMM44 downregulation in YME1L-deficient glioma cells should be caused by shutdown of GATA3-dependent TIMM44 transcriptional machinery following mitochondrial stress.
Next, the lentiviral TIMM44-expressing construct (“OE-TIMM44”) was transduced to the lv-shYME1L P1 glioma cells and restored TIMM44 expression (Figure 6M). YME1L silencing inhibited P1 glioma cell proliferation (Figure 6N) and migration (Figure 6O), and provoked apoptosis (Figure 6P). Remarkably, OE-TIMM44 inhibited shYME1L-induced anti-glioma cell activity (Figure 6N-P), partially restoring cell proliferation (Figure 6N) and migration (Figure 6O), and alleviating cell apoptosis (Figure 6P).
TIMM44 depletion inhibits subcutaneous and intracranial glioma xenograft growth in vivo
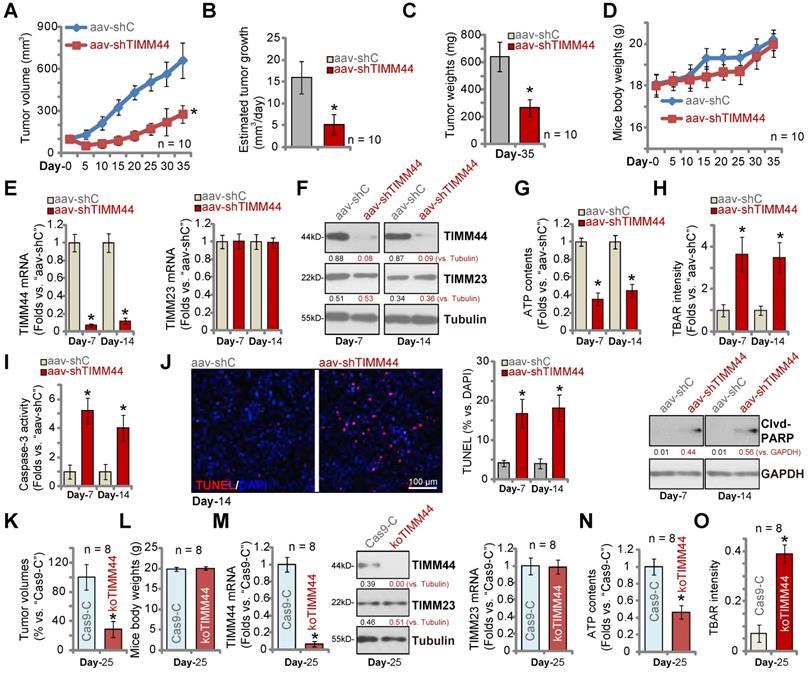
At last we tested the potential effect of TIMM44 on glioma cell growth in vivo. P1 glioma cells were inoculated and subcutaneously (s.c.) injected to the flanks of the nude mice. The subcutaneous P1 glioma xenografts were formed three weeks after cell injection with tumor volumes close to 100 mm3 (“Day-0”). The xenograft-bearing mice were then randomly separated into two different groups. The treatment group received intratumoral injection of TIMM44-shRNA AAV (“aav-shTIMM44”, 10 μL virus per tumor, 1×109 PFU), and the control group received intratumoral injection of the same amount of scramble control shRNA AAV (“aav-shC”). In each group there were ten mice (n = 10), and the AAV was injected every 24h for ten rounds. The xenograft tumor volumes were recorded every five days, from “Day-0” to “Day-35” (seven rounds).
As demonstrated, intratumoral injection of TIMM44-shRNA AAV robustly hindered P1 glioma growth in nude mice, and volumes of aav-shTIMM44 P1 glioma xenografts were significantly lower than the aav-shC xenografts (Figure 7A). A described formula [37] was utilized to calculate the estimated daily tumor growth (in mm3 per day), and results demonstrated that aav-shTIMM44 potently inhibited P1 glioma xenograft growth (Figure 7B). At Day-35, all subcutaneous glioma xenografts of the two groups were isolated and weighted (Figure 7C). As shown aav-shTIMM44-injected tumors were much lighter than aav-shC tumors (Figure 7C). There was however no significant difference in the mice body weights between the two groups (Figure 7D). These results showed that intratumoral injection of TIMM44-shRNA AAV potently inhibited P1 glioma xenograft growth in nude mice.
To examine signaling changes in vivo, at Day-7 and Day-14 we separated one glioma xenograft per group. Total four xenografts were obtained. Each xenograft was cut into five pieces and were analyzed separately. As shown, TIMM44 mRNA and protein expression was robustly decreased in aav-shTIMM44-injected xenograft tumors (Figure 7E and F), whereas TIMM23 mRNA and protein expression was unchanged (Figure 7E and F).
In TIMM44-silenced P1 glioma xenograft tissues, ATP contents were significantly decreased (Figure 7G). Whereas the TBAR activity (Figure 7H) was increased, indicating lipid peroxidation was increased after TIMM44 silencing in vivo. Moreover, the relative Caspase-3 activity (Figure 7I), TUNEL-positive staining and the cleaved-PARP level (Figure 7J) were all significantly increased in aav-shTIMM44 xenograft tissues, supporting apoptosis induction. Therefore, in line with the in vitro findings, ATP reduction, oxidative injury and apoptosis were detected in TIMM44-silenced subcutaneous P1 glioma xenografts.
Alternatively, using the described protocol [13, 15, 24], P1 glioma cells with the CRISPR/Cas9-TIMM44-KO construct (“koTIMM44”) or the control construct (“Cas9-C”) were intracranially injected into the mouse brains. The patient-derived glioma xenografts (PDX) were established. Twenty-five days later (“Day-25”), the first mouse in the Cas9-C group exhibited obvious symptoms, and all mice were sacrificed and PDX tumors were isolated [15]. As shown, compared to the Cas9-C intracranial P1 gliomas, the volumes of koTIMM44 tumors were significantly lower (Figure 7K). The mice body weights were indifferent (Figure 7L). Analyzing tumor tissue lysates, we found that TIMM44 mRNA and protein expression in intracranial koTIMM44 tumors was depleted (Figure 7M). TIMM23 expression was unaffected (Figure 7M). ATP levels were decreased in koTIMM44 tumors (Figure 7N), and the TBAR intensity was increased (Figure 7O). Thus, TIMM44 KO largely inhibited intracranial P1 glioma xenograft growth in mouse brain.
Discussion
Glioblastoma and other high grade gliomas have one of the worst prognosis among all malignancies [11, 38, 39]. The current clinical treatments, including tumor resection, radiation and temozolomide-based chemotherapies, are not able to significantly improve the overall survival and prognosis for the patients [40-42]. It is therefore extremely important to identify novel biomarkers and therapeutic targets of glioma. Increased mitochondrial biogenesis, energy metabolism and oxidative phosphorylation are vital for the tumorigenesis and progression of human glioma [16-18].
Glioma, like other malignant tumors, has aberrant energy metabolism and favors aerobic glycolysis to generate ATP [18]. Dysfunctional mitochondria are often observed in glioma [18]. Moreover, mitochondrial ultrastructure and proteomics are also dysregulated in glioma [18]. These mitochondrial changes play an important role in tumorigenesis and progression of glioma. However, the underlying mechanisms are poorly understood [18]. Sun et al., discovered that transferring normal mitochondria of human astrocytes, through endocytosis, could rescue aerobic respiration and enhance radio-sensitivity into glioma cells, and inhibit glioma xenograft growth in nude mice [43].
TIMM44 is a mitochondrial inner membrane translocase protein essential for the import of the nuclear-encoded proteins to the mitochondria [25, 26, 44]. Mitochondrial TIMM44 dysfunction, due to genetic variants, could be associated with thyroid carcinoma development [26]. Huang et al., has identified TIMM44 as a potential oncogenic gene in colorectal cancer [45]. IR-58, a mitochondria-targeting near-infrared autophagy-enhancer, inhibited TIMM44-SOD2 pathway, causing ROS production, oxidative injury, and colorectal cancer cell apoptosis [45].
TIMM44 depletion inhibits subcutaneous and intracranial glioma xenograft growth in vivo. The subcutaneous P1 glioma xenograft-bearing mice were subject to intratumoral injection of TIMM44-shRNA AAV (“aav-shTIMM44”) or the scramble control shRNA AAV (“aav-shC”). Virus was injected daily for 10 days. The xenograft tumor volumes (A) and mice body weights (D) were recorded every five days. The estimated daily tumor growth (in mm3 per day) (B) was calculated. At Day-35 all tumors were isolated and weighted (C). The listed tumors were homogenized, and expression of listed genes and proteins was tested (E and F). ATP contents (G) and TBAR activity (H) were analyzed in the tissue lysates. Apoptosis intensity and expression of listed proteins were analyzed by the listed assays (I and J). P1 primary human glioma cells (5 × 105 cells of each mouse), with the lenti-CRSIPR/Cas9-TIMM44-KO-puro construct (koTIMM44) or control construct (“Cas9-C”), were intracranially injected to brains of nude mice; After 25 days (“Day-25”), animals were decapitated and tumors were isolated, the tumor volumes (K) and mice body weights were recorded (L). Expression of listed genes and proteins was tested by qRT-PCR and Western blotting assays (M); ATP contents (N) and the TBAR activity (O) were measured as well. The data were presented as mean ± standard deviation (SD). * P < 0.05 versus “aav-shC”/“Cas9-C” groups. Scale bar=100 µm (J).
The results of the present study supported that the mitochondrial protein TIMM44 is an important cancer-promoting molecular for glioma progression. Bioinformatics results and local human glioma tissue data confirmed that TIMM44 mRNA and protein expression is upregulated in glioma. TIMM44 overexpression in glioma correlated with poor overall survival of the patients. In patient-derived primary glioma cells and immortalized cell lines, TIMM44 shRNA or KO (by CRISPR/Cas9 method) potently inhibited cell viability, proliferation and migration. Conversely, ectopic overexpression of TIMM44 by a lentiviral construct augmented cell proliferation and migration. In vivo, the growth of subcutaneous P1 glioma xenografts was suppressed after intratumoral injection of TIMM44 shRNA AAV. Moreover, the growth of intracranial glioma xenografts in nude mice was largely inhibited after TIMM44 KO. These results implied that overexpressed TIMM44 could be a promising and valuable therapeutic target of glioma.
MB-10, through binding to C-terminal region of TIMM44, inhibited mitochondrial protein input, causing viability reduction and apoptosis in cervical cancer HeLa cells [33]. Polson et al., showed that KHS101, a synthetic small-molecule, induced glioma cell death by disrupting the mitochondrial chaperone heat shock protein family D member 1 (HSPD1) [46]. KHS101 disrupted mitochondrial bioenergetic capacity and glycolytic activity, causing energy crisis in glioma cells [46]. Cannabidiol inhibited mitochondrial channel VDAC1 and induced swollen mitochondria, causing apoptosis in glioma cells [47].
We found that overexpressed TIMM44 is essential for maintaining mitochondrial functions in glioma cells. TIMM44 silencing/KO largely inhibited mitochondrial protein input and mitochondrial complex I, causing ATP depletion, mitochondrial membrane potential reduction, oxidative stress and DNA damage, and eventually provoked apoptosis. ATP supplement and the antioxidant NAC can inhibit TIMM44-silencing-induced anti-glioma cell activity. ATP reduction, lipid peroxidation and apoptosis activation were detected as well in TIMM44-depleted subcutaneous and intracranial glioma xenografts. Conversely, TIMM44 overexpression increased cellular ATP contents and alleviated oxidative injury in primary glioma cells. Thus, TIMM44-silencing-induced anti-glioma cell activity was possibly through disrupting mitochondrial functions.
Our previous study has identified an important cancer-promoting function of the mitochondrial protein YME1L in human glioma. YME1L is extremely important for maintaining mitochondrial integrity, morphology, function and plasticity [21, 48, 49]. We previously found that YME1L expression is elevated in human glioma tissues and various glioma cells. Our preliminary results found that depletion of YME1L-targeting microRNAs (miRNAs) could be the primary cause of YME1L elevation in human glioma. YME1L depletion, by shRNA or CRISPR/Cas9 KO, potently inhibited glioma proliferation and migration, and induced mitochondrial dysfunctions and apoptosis. On the other hand, overexpression of YME1L augmented glioma cell proliferation and migration. The orthotopic growth of primary glioma xenografts in nude mice was largely inhibited by YME1L depletion.
In the present study, we found that YME1L could be an important upstream protein for TIMM44 in human glioma cells. TCGA database found that TIMM44 is one of the YME1L's top DEGs. TIMM44 expression in primary human glioma cells was decreased following YME1L silencing or KO, but was elevated after ectopic YME1L overexpression. Significantly, YME1L shRNA-induced anti-glioma cell activity was ameliorated after restoring TIMM44 expression using the TIMM44-expressing construct.
Moreover, we found that YEM1L-promoted TIMM44 expression was possibly through promoting GATA3-dependent TIMM44 transcription. GATA3-TIMM44 promoter binding in glioma cells was robustly decreased after YME1L silencing or depletion, but was increased after YME1L overexpression. GATA3 silencing significantly decreased TIMM44 expression. Moreover, GATA3 binding to the TIMM44 promoter was significantly increased in both human glioma tissues and difference glioma cells. Importantly, YME1L shRNA-induced decrease of GATA3-TIMM44 promoter binding and TIMM44 downregulation were largely attenuated by exogenously adding ATP in P1 glioma cells. These results supported that TIMM44 downregulation in YME1L deficient glioma cells could be caused by shutdown of GATA3-dependent TIMM44 transcriptional machinery (possibly due to mitochondrial stress after YME1L depletion). Upregulation of TIMM44 in glioma could be due to increased TIMM44 transcriptional machinery through GATA3.
Conclusion
Overexpressed TIMM44 could be a novel and promising therapeutic target of human glioma.
Materials and methods
Reagents, chemicals and antibodies
Polybrene, the antioxidant n-acetylcysteine (NAC), ATP, antibiotics, puromycin and medium were provided by Sigma-Aldrich (St. Louis, MO). The antibodies in this study were from Cell Signaling Technology (Beverly, MA) and Abcam (Cambridge, UK). Fluorescence probes, including TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling), MitoTracker Red/MitoTracker Green, tetraethylbenzimidazolylcarbocyanine iodide (JC-1), DAPI (4',6-diamidino-2-phenylindole), EdU (5-ethynyl-20-deoxyuridine), CellROX, dichlorodihydrofluorescein diacetate (DCF-DA), were purchased from Thermo-Fisher Invitrogen (Shanghai, China). MB-10 (“MitoBloCK-10”) [33] was synthesized and verified by Shanghai RuiLu Biotech (Shanghai, China).
Human tissues and cells
The high grade glioma (HGG, grade III-IV) tissues and matched adjacent brain tissues, derived from sixteen (16) written-informed consent primary patients were reported in our previous studies [13, 15]. Fresh tissue lysates were tested by Western blotting and qRT-PCR assays [13, 15]. For the immunofluorescence studies, human glioma and xenograft glioma tissue sections (4 µm in thickness) were incubated with anti-TIMM44 antibody overnight and the green fluorescence secondary antibody (for 1h). The mitochondria were co-stained with the mitochondrial marker MitoTracker Red (Invitrogen) and the nuclear marker DAPI (Invitrogen), the fluorescence signals were observed under confocal fluorescence microscope (Zeiss). The primary human glioma cells derived from three written informed-consent glioma patients (“P1”, “P2” and “P3”, as reported early [13, 15, 24]), the primary human astrocytes (“Astrocytes1/2”, derived from two patients P1 and P2), immortalized glioma cell lines (A172 and U251MG) were reported in our previous studies [13-15, 50]. The protocols of using primary human tissues and cells were according to Declaration of Helsinki, with approval from the Ethics Board of Soochow University.
shRNA
Two TIMM44 shRNA sequences, two different GATA3 shRNA sequences, and one YME1L shRNA sequence were verified and synthesized by Genechem (Shanghai, China), and were individually sub-cloned into a GV369 construct (Genepharm). The construct along with the lentivirus package constructs (Genechem) were co-transfected to HEK-293 cells, thereby generating shRNA lentivirus, which was filtered, enriched (at MOI=15) and added to glioma cells or astrocytes. For in vivo studies, TIMM44 shRNA (-seq1) or the scramble control shRNA was cloned into an adeno-associated virus (AAV) construct (AAV9, Genechem). The shRNA AAV was then generated.
CRISPR/Cas9
The sequence encoding small-guide (sgRNA) targeting human TIMM44 (Target DNA Sequence: CATCTGATAGGTCGACCCAT, PAM Sequence: GGG) or YEM1L [24] was individually inserted into the lenti-CRISPR/Cas9-KO-puro construct (Genechem). The KO construct was then transduced to HEK-293 cells to generate lentivirus. The primary human glioma cells were seeded into six-well plates at 50-60% confluence and transfected with the Cas9-expressing construct (Genechem, Shanghai, China). Stable Cas9-expressing cells were then established after puromycin selection. Cells were then transfected with the lenti-CRISPR/Cas-9 TIMM44 KO construct lentivirus, and puromycin-containing medium was added to select stable cells. TIMM44 KO screening was thereafter performed and the single stable TIMM44 KO cells were established.
TIMM44/YME1L overexpression
The recombinant GV369 lentiviral construct containing the full-length TIMM44 cDNA (NM_006351.4) or YMEL1 cDNA (NM_139312.2) was transfected to cultured glioma cells. The transfected glioma cells were cultured in puromycin-containing medium and thereafter stable cells were formed. The qRT-PCR and Western blotting assays were carried out to verify TIMM44 overexpression. The control glioma cells were transfected with the empty vector.
Cellular fluorescence dye assays
As described [51], cells were cultivated at 50-60% confluence and were stained with the designated fluorescence dyes (including JC-1, DCF-DA, CellROX, TUNEL, EdU and DAPI) and further cultured for applied time periods. Fluorescence images were captured under a fluorescence microscopy (Leica). It intensity was recorded from five random views of each treatment.
Cell migration and invasion
The described cells (at 5 × 104 cells per chamber, in serum-free medium) were initially seeded onto the upper surface of the “Transwell” chamber (BD Biosciences). To the lower compartment, the complete medium with serum was added. After 24h, the migrated cells passing through were fixed and stained [51]. For in vitro cell invasion assays, the chamber surface was coated with Matrigel (Sigma), and other steps were the same.
TBAR assay
Lipid peroxidation in cell and tissue lysates (30 µg per sample) was tested by a thiobarbituric acid reactive substance (TBAR) kit (Cayman Chemical, MI) based on the attached protocol. The kit reagent colorimetrically quantified lipid peroxidation and measured malondialdehyde (MDA). The optical density of TBAR was tested at 545 nm with the reference wavelength at 600 nm at room temperature. It was expressed in nmol per mg of total protein and was always normalized to that of control.
Mitochondrial protein import assays
The detailed protocols were described elsewhere [33]. In brief, prior to import into purified mitochondria, [35S]-methionine and cysteine labeled Su9-DHFR (dihydrofolate reductase) fusion protein (based on the described protocol [33, 52, 53], provided by Shanghai Genechem Co.) was generated with the TNT Quick Coupled Transcription/Translation kit (Promega, Shanghai, China). The isolated mitochondria (200 µg/mL) of human glioma cells was dissolved to the described import buffer [33] and import reaction was started by the addition of translation mix. After 15 min, the import was terminated by trypsin on ice and soybean trypsin inhibitor was thereafter added. Mitochondria were pelleted by centrifugation, disrupted in Laemmli sample buffer and analyzed by SDS-PAGE and autoradiography.
Other assays
The quantitative real-time PCR (qRT-PCR), Western blotting, cell counting kit-8 (CCK-8), Trypan blue staining of cell death, the Caspase-3 activity assay, single strand DNA (ssDNA) ELISA were described in our previous studies [13-15, 50, 54, 55]. ATP contents in cellular and tissue lysates were measured as previously described [56]. The mitochondrial complex I activity in glioma cells with applied treatments was analyzed by a commercial kit (Abcam) according to the attached protocols.
Chromatin immunoprecipitation (ChIP) assay
As described [55] cell lysates were treated by a MisonixSonicator 3000 Homogenizer [57] to achieve fragmented genomic DNA. Lysates were diluted with ChIP dilution buffer and were immunoprecipitated with the anti-GATA3 (ab199428, Abcam) antibody. GATA3-bound DNA was eluted from the protein A/G agarose, and NaCl was included. DNA containing the proposed conserved TIMM44 promoter site (GAGATAGGG) was analyzed via quantitative PCR (qPCR).
Xenograft studies
The athymic nude mice, half male, half female, 6-7 week old, 18.2-19.5 g weight, were purchased from the Shanghai Laboratory Animal Center (SLAC, Shanghai, China) and were maintained under standard conditions. The primary human glioma cells P1 were trypsinized, washed and re-suspended. Cells (in 200 µL of Matrigel basic medium) were subcutaneously injected into the right flanks of the mice. Within three weeks of cell inoculation, the P1 glioma xenografts were established and tumor volume close to 100 mm3. The mice were intratumorally injected with adeno-associated virus (aav)-packed shRNA (1×109 PFU, 10 µL). The mice body weights and the tumor volumes ([15]) were recorded every six days. For intracranial glioma xenograft experiments, P1 glioma cells were intracranially injected as described [58]. Magnetic resonance imaging (MRI) was utilized to visualize the intracranial xenografts. On Day-25, all mice were sacrificed and xenografts were isolated. Tumor volumes were calculated by the described formula [15]. The animal studies were approved by Institutional Animal Care and Use Committee and Ethics Committee of Soochow University.
Statistical analyses
Data in this study were always with normal distribution and were expressed as means ± standard deviation (SD). To examine statistical differences between three and more groups, one-way ANOVA and a Scheffe's f-test (SPSS 23.0, SPSS Co., Chicago, CA) were utilized. The two-tailed unpaired t test (Excel 2007) was utilized to examine significance between two groups. P values < 0.05 were considered statistically significant.
Abbreviations
AAV: Adeno-associated virus; CNS: central nerve system; DAPI: 4',6-diamidino-2-phenylindole; EdU: 5-ethynyl-20-deoxyuridine; CPT1A: carnitine palmitoyltransferase 1A; DHFR: dihydrofolate reductase; DCF-DA: dichlorodihydrofluorescein diacetate; GTEx: Genotype-Tissue Expression; GBM: glioblastoma; KLF4: Krüppel-like factor 4; IHC: immunohistochemistry; KO: knockout; Mfn1: mitofusin-1; Mfn2: mitofusin-2; Opa1: optic atrophy 1; ROS: reactive oxygen species; ROC: receiver operating characteristic; SD: standard deviation; TIMM44: translocase of inner mitochondrial membrane 44; ssDNA: single strand DNA; YME1L: YME1 Like 1 ATPase; TUNEL: terminal deoxynucleotidyl transferase dUTP nick end labeling; JC-1: tetraethylbenzimidazolylcarbocyanine iodide; TBAR: thiobarbituric acid reactive substance.
Supplementary Material
Supplementary figures.
Acknowledgements
Funding
This work was generously supported by Key Research and Development Program of Jiangsu Province (No. BE2019652), Changzhou international cooperation program (CZ20200039), Development Program of Changzhou City (CE20205024) and National Natural Science Foundation of China (81922025, 81802511, 81870679, 82070983, 82171461, 81771457). Suzhou Science and Technology Planning Project (SZM2021025), A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Ethical Approval and Consent to participate
This study was approved by the Ethics Committee of Soochow University.
Availability of data and material
All data generated during this study are included in this published article. Data will be made available upon request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7-30
2. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67:7-30
3. Reardon DA, Wen PY. Glioma in 2014: unravelling tumour heterogeneity-implications for therapy. Nat Rev Clin Oncol. 2015;12:69-70
4. Wen PY, Reardon DA. Neuro-oncology in 2015: Progress in glioma diagnosis, classification and treatment. Nat Rev Neurol. 2016;12:69-70
5. Ellis HP, Greenslade M, Powell B, Spiteri I, Sottoriva A, Kurian KM. Current Challenges in Glioblastoma: Intratumour Heterogeneity, Residual Disease, and Models to Predict Disease Recurrence. Front Oncol. 2015;5:251
6. Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: a social network. Cancer Res. 2011;71:4055-60
7. Little SE, Popov S, Jury A, Bax DA, Doey L, Al-Sarraj S. et al. Receptor tyrosine kinase genes amplified in glioblastoma exhibit a mutual exclusivity in variable proportions reflective of individual tumor heterogeneity. Cancer Res. 2012;72:1614-20
8. Cheng HS, Marvalim C, Zhu P, Law CLD, Low ZYJ, Chong YK. et al. Kinomic profile in patient-derived glioma cells during hypoxia reveals c-MET-PI3K dependency for adaptation. Theranostics. 2021;11:5127-42
9. Zhong C, Tao B, Tang F, Yang X, Peng T, You J. et al. Remodeling cancer stemness by collagen/fibronectin via the AKT and CDC42 signaling pathway crosstalk in glioma. Theranostics. 2021;11:1991-2005
10. Boyd NH, Tran AN, Bernstock JD, Etminan T, Jones AB, Gillespie GY. et al. Glioma stem cells and their roles within the hypoxic tumor microenvironment. Theranostics. 2021;11:665-83
11. Westphal M, Lamszus K. The neurobiology of gliomas: from cell biology to the development of therapeutic approaches. Nat Rev Neurosci. 2011;12:495-508
12. Kwiatkowska A, Symons M. Signaling Determinants of Glioma Cell Invasion. Adv Exp Med Biol. 2020;1202:129-49
13. Wang Y, Liu YY, Chen MB, Cheng KW, Qi LN, Zhang ZQ. et al. Neuronal-driven glioma growth requires Galphai1 and Galphai3. Theranostics. 2021;11:8535-49
14. Shao NY, Wang DX, Wang Y, Li Y, Zhang ZQ, Jiang Q. et al. MicroRNA-29a-3p Downregulation Causes Gab1 Upregulation to Promote Glioma Cell Proliferation. Cell Physiol Biochem. 2018;48:450-60
15. Liu YY, Chen MB, Cheng L, Zhang ZQ, Yu ZQ, Jiang Q. et al. microRNA-200a downregulation in human glioma leads to Galphai1 over-expression, Akt activation, and cell proliferation. Oncogene. 2018;37:2890-902
16. Guntuku L, Naidu VG, Yerra VG. Mitochondrial Dysfunction in Gliomas: Pharmacotherapeutic Potential of Natural Compounds. Curr Neuropharmacol. 2016;14:567-83
17. Griguer CE, Oliva CR. Bioenergetics pathways and therapeutic resistance in gliomas: emerging role of mitochondria. Curr Pharm Des. 2011;17:2421-7
18. Ordys BB, Launay S, Deighton RF, McCulloch J, Whittle IR. The role of mitochondria in glioma pathophysiology. Mol Neurobiol. 2010;42:64-75
19. Hartmann B, Wai T, Hu H, MacVicar T, Musante L, Fischer-Zirnsak B. et al. Homozygous YME1L1 mutation causes mitochondriopathy with optic atrophy and mitochondrial network fragmentation. Elife. 2016 5
20. Rainbolt TK, Saunders JM, Wiseman RL. YME1L degradation reduces mitochondrial proteolytic capacity during oxidative stress. EMBO Rep. 2015;16:97-106
21. Anand R, Wai T, Baker MJ, Kladt N, Schauss AC, Rugarli E. et al. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J Cell Biol. 2014;204:919-29
22. Stiburek L, Cesnekova J, Kostkova O, Fornuskova D, Vinsova K, Wenchich L. et al. YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol Biol Cell. 2012;23:1010-23
23. Coppola M, Pizzigoni A, Banfi S, Bassi MT, Casari G, Incerti B. Identification and characterization of YME1L1, a novel paraplegin-related gene. Genomics. 2000;66:48-54
24. Liu F, Chen G, Zhou L, Wang Y, Zhang Z, Qin X. et al. YME1L overexpression exerts pro-tumorigenic activity in glioma by promoting Gαi1 expression and Akt activation. Protein Cell. 2022 10.1093/procel/pwac011
25. Wang Y, Katayama A, Terami T, Han X, Nunoue T, Zhang D. et al. Translocase of inner mitochondrial membrane 44 alters the mitochondrial fusion and fission dynamics and protects from type 2 diabetes. Metabolism. 2015;64:677-88
26. Bonora E, Evangelisti C, Bonichon F, Tallini G, Romeo G. Novel germline variants identified in the inner mitochondrial membrane transporter TIMM44 and their role in predisposition to oncocytic thyroid carcinomas. Br J Cancer. 2006;95:1529-36
27. Moro F, Okamoto K, Donzeau M, Neupert W, Brunner M. Mitochondrial protein import: molecular basis of the ATP-dependent interaction of MtHsp70 with Tim44. J Biol Chem. 2002;277:6874-80
28. Voos W, von Ahsen O, Muller H, Guiard B, Rassow J, Pfanner N. Differential requirement for the mitochondrial Hsp70-Tim44 complex in unfolding and translocation of preproteins. EMBO J. 1996;15:2668-77
29. Strub A, Rottgers K, Voos W. The Hsp70 peptide-binding domain determines the interaction of the ATPase domain with Tim44 in mitochondria. EMBO J. 2002;21:2626-35
30. Matsuoka T, Wada J, Hashimoto I, Zhang Y, Eguchi J, Ogawa N. et al. Gene delivery of Tim44 reduces mitochondrial superoxide production and ameliorates neointimal proliferation of injured carotid artery in diabetic rats. Diabetes. 2005;54:2882-90
31. Yu X, Li Y, Ding Y, Zhang H, Ding N, Lu M. HuR Promotes Ovarian Cancer Cell Proliferation by Regulating TIMM44 mRNA Stability. Cell Biochem Biophys. 2020;78:447-53
32. Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol. 2010;5:1315-6
33. Miyata N, Tang Z, Conti MA, Johnson ME, Douglas CJ, Hasson SA. et al. Adaptation of a Genetic Screen Reveals an Inhibitor for Mitochondrial Protein Import Component Tim44. J Biol Chem. 2017;292:5429-42
34. Rodrigues T, Ferraz LS. Therapeutic potential of targeting mitochondrial dynamics in cancer. Biochem Pharmacol. 2020;182:114282
35. Yu Y, Peng XD, Qian XJ, Zhang KM, Huang X, Chen YH. et al. Fis1 phosphorylation by Met promotes mitochondrial fission and hepatocellular carcinoma metastasis. Signal Transduct Target Ther. 2021;6:401
36. Tang M, Yang M, Wu G, Mo S, Wu X, Zhang S. et al. Epigenetic Induction of Mitochondrial Fission Is Required for Maintenance of Liver Cancer-Initiating Cells. Cancer Res. 2021;81:3835-48
37. Lv Y, Wang Y, Song Y, Wang SS, Cheng KW, Zhang ZQ. et al. LncRNA PINK1-AS promotes G alpha i1-driven gastric cancer tumorigenesis by sponging microRNA-200a. Oncogene. 2021;40:3826-44
38. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10-29
39. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9-29
40. Khasraw M, Lassman AB. Neuro-oncology: late neurocognitive decline after radiotherapy for low-grade glioma. Nat Rev Neurol. 2009;5:646-7
41. Pollack IF. Neuro-oncology: Therapeutic benefits of reirradiation for recurrent brain tumors. Nat Rev Neurol. 2010;6:533-5
42. Wang Y, Jiang T. Understanding high grade glioma: molecular mechanism, therapy and comprehensive management. Cancer Lett. 2013;331:139-46
43. Sun C, Liu X, Wang B, Wang Z, Liu Y, Di C. et al. Endocytosis-mediated mitochondrial transplantation: Transferring normal human astrocytic mitochondria into glioma cells rescues aerobic respiration and enhances radiosensitivity. Theranostics. 2019;9:3595-607
44. Lin BC, Phung TH, Higgins NR, Greenslade JE, Prado MA, Finley D. et al. ALS/FTD mutations in UBQLN2 are linked to mitochondrial dysfunction through loss-of-function in mitochondrial protein import. Hum Mol Genet. 2021;30:1230-46
45. Huang Y, Zhou J, Luo S, Wang Y, He J, Luo P. et al. Identification of a fluorescent small-molecule enhancer for therapeutic autophagy in colorectal cancer by targeting mitochondrial protein translocase TIM44. Gut. 2018;67:307-19
46. Polson ES, Kuchler VB, Abbosh C, Ross EM, Mathew RK, Beard HA. et al. KHS101 disrupts energy metabolism in human glioblastoma cells and reduces tumor growth in mice. Sci Transl Med. 2018 10
47. Gross C, Ramirez DA, McGrath S, Gustafson DL. Cannabidiol Induces Apoptosis and Perturbs Mitochondrial Function in Human and Canine Glioma Cells. Front Pharmacol. 2021;12:725136
48. Ohba Y, MacVicar T, Langer T. Regulation of mitochondrial plasticity by the i-AAA protease YME1L. Biol Chem. 2020;401:877-90
49. MacVicar T, Ohba Y, Nolte H, Mayer FC, Tatsuta T, Sprenger HG. et al. Lipid signalling drives proteolytic rewiring of mitochondria by YME1L. Nature. 2019;575:361-5
50. Cai S, Li Y, Bai JY, Zhang ZQ, Wang Y, Qiao YB. et al. Galphai3 nuclear translocation causes irradiation resistance in human glioma cells. Oncotarget. 2017;8:35061-8
51. Gao YY, Ling ZY, Zhu YR, Shi C, Wang Y, Zhang XY. et al. The histone acetyltransferase HBO1 functions as a novel oncogenic gene in osteosarcoma. Theranostics. 2021;11:4599-615
52. Iwata K, Nakai M. Interaction between mitochondrial precursor proteins and cytosolic soluble domains of mitochondrial import receptors, Tom20 and Tom70, measured by surface plasmon resonance. Biochem Biophys Res Commun. 1998;253:648-52
53. Maarse AC, Blom J, Grivell LA, Meijer M. MPI1, an essential gene encoding a mitochondrial membrane protein, is possibly involved in protein import into yeast mitochondria. EMBO J. 1992;11:3619-28
54. Sun J, Huang W, Yang SF, Zhang XP, Yu Q, Zhang ZQ. et al. Galphai1 and Galphai3mediate VEGF-induced VEGFR2 endocytosis, signaling and angiogenesis. Theranostics. 2018;8:4695-709
55. Yao J, Wu XY, Yu Q, Yang SF, Yuan J, Zhang ZQ. et al. The requirement of phosphoenolpyruvate carboxykinase 1 for angiogenesis in vitro and in vivo. Sci Adv. 2022;8:eabn6928
56. Yin DP, Zheng YF, Sun P, Yao MY, Xie LX, Dou XW. et al. The pro-tumorigenic activity of p38gamma overexpression in nasopharyngeal carcinoma. Cell Death Dis. 2022;13:210
57. He L, Fan X, Li Y, Chen M, Cui B, Chen G. et al. Overexpression of zinc finger protein 384 (ZNF 384), a poor prognostic predictor, promotes cell growth by upregulating the expression of Cyclin D1 in Hepatocellular carcinoma. Cell Death Dis. 2019;10:444
58. Agnihotri S, Gajadhar AS, Ternamian C, Gorlia T, Diefes KL, Mischel PS. et al. Alkylpurine-DNA-N-glycosylase confers resistance to temozolomide in xenograft models of glioblastoma multiforme and is associated with poor survival in patients. J Clin Invest. 2012;122:253-66
Author contact
![]() Corresponding author: Prof. Fang Liu (czdoctorliucom), Prof. Cong Cao, caocongedu.cn.
Corresponding author: Prof. Fang Liu (czdoctorliucom), Prof. Cong Cao, caocongedu.cn.
 Global reach, higher impact
Global reach, higher impact