13.3
Impact Factor
Theranostics 2022; 12(16):6931-6954. doi:10.7150/thno.77949 This issue Cite
Review
Artificial intelligence in pancreatic cancer
1. Department of General Surgery, State Key Laboratory of Complex Severe and Rare Diseases, Peking Union Medical College Hospital, Chinese Academy of Medical Science and Peking Union Medical College, Beijing 100730, China.
2. School of Medicine, Tsinghua University, Beijing, 100084, China.
*These authors contributed equally to this work.
Received 2022-8-11; Accepted 2022-9-24; Published 2022-10-3
Abstract
Pancreatic cancer is the deadliest disease, with a five-year overall survival rate of just 11%. The pancreatic cancer patients diagnosed with early screening have a median overall survival of nearly ten years, compared with 1.5 years for those not diagnosed with early screening. Therefore, early diagnosis and early treatment of pancreatic cancer are particularly critical. However, as a rare disease, the general screening cost of pancreatic cancer is high, the accuracy of existing tumor markers is not enough, and the efficacy of treatment methods is not exact. In terms of early diagnosis, artificial intelligence technology can quickly locate high-risk groups through medical images, pathological examination, biomarkers, and other aspects, then screening pancreatic cancer lesions early. At the same time, the artificial intelligence algorithm can also be used to predict the survival time, recurrence risk, metastasis, and therapy response which could affect the prognosis. In addition, artificial intelligence is widely used in pancreatic cancer health records, estimating medical imaging parameters, developing computer-aided diagnosis systems, etc. Advances in AI applications for pancreatic cancer will require a concerted effort among clinicians, basic scientists, statisticians, and engineers. Although it has some limitations, it will play an essential role in overcoming pancreatic cancer in the foreseeable future due to its mighty computing power.
Keywords: Artificial intelligence, machine learning, pancreatic cancer, early detection, prognosis prediction
Introduction
Pancreatic cancer (PC) is the deadliest form of all cancer. The five-year relative survival rate for PC is only 11% in the USA, which is the lowest among all cancers [1]. There were 495773 new cases and 466003 deaths from PC worldwide in 2020, accounting for 2.6% of all new cancer diagnoses and 4.7% of all cancer deaths, respectively [2]. In China, the incidence and mortality of PC among tumors are 2.47% and 3.64%, respectively [3]. The main reason for such a poor prognosis of PC is the late diagnosis, with only about 20% of patients being diagnosed at an early stage. Most patients have non-specific first symptoms, such as jaundice, fatigue, change in bowel habits, and indigestion, that make it difficult to distinguish from non-cancer diseases [4]. Most chemotherapy [5-7], targeted therapy [8], and immunotherapy [9-11] are ineffective because most patients are already in the progressive stage with local invasion and distant metastases at the detection time [12,13]. A multicenter study demonstrated that patients with PC detected by screening had a 5-year survival rate of 73.3% and a median survival time of 9.8 years, compared with 1.5 years for patients with PC seen by non-screening [14]. To diagnose early-stage PC accurately is desperately needed [15,16].
Radiographic imaging-based investigations [17-20] are fundamental techniques in PC screening, including endoscopic ultrasound (EUS), computed tomography (CT), magnetic resonance imaging (MRI), etc. Related techniques based on the above methods, such as EUS-guided fine needle aspiration (FNA) and biopsy (FNB), contrast-enhanced EUS (CE-EUS), CT (CE-CT), MRI (CE-MRI), and positron emission tomography-computed tomography (PET/CT), can further improve the accuracy of diagnosis. However, screening in the asymptomatic population is not recommended due to the economic burden and the relatively low incidence of PC in the general population [21,22]. Also, early PC lacks biomarkers. Carbohydrate Antigen 19-9 (CA19-9), the best-validated biomarker in PC, does not have enough accuracy and specificity in screening early PC [23,24]. Thus, many scientists are working to develop new early screening methods. Meanwhile, it is also essential to use better ways to assess treatment efficacy and prognosis, which will facilitate the development of appropriate clinical treatment options and critical drugs [25,26].
Artificial intelligence (AI) is a branch of computer science dedicated to producing a new kind of intelligent machine that can respond similarly to human intelligence [27]. Nowadays, many researchers are attempting to apply AI to the medical field, including healthcare [28], oncology [29], cardiology [30], and more. Compared with traditional biometric methods, AI has greater flexibility and scalability, which allows it to be deployed for many tasks. Another advantage is its ability to integrate a large number of different data types and understand complex relationships between variables in a flexible, trainable manner. As the scale of medical data continues to expand and computer computing power continues to improve, AI is showing more and more advantages in processing big data. In clinical practice, AI can perform routine tasks consistently, freeing up physicians' time to solve more complex clinical problems [27,31]. For PC, AI-assisted diagnostic techniques are also gaining more attention. The 2020 AI and Early Detection of Pancreatic Cancer Virtual Summit discussed and highlighted the potential of AI in the early diagnosis of PC [32]. In the recent meeting of The Alliance of Pancreatic Cancer Consortia, the discussion focused on imaging methods and the use of AI for the early detection of PC [33].
Despite the unparalleled advantages of AI, there are still many concerns about its application in the clinical field. For example, no one model can solve all problems, and all models have their range of adaptation [34]. There is a risk that AI algorithms may ignore specific differences, such as gender and race, which can lead to bias because of the heterogeneity between the training set and other patients [35-37]. AI also faces many ethical issues in clinical practice, such as the need for researchers and healthcare organizations to protect data from hacking for patient privacy. Healthcare systems should strive to ensure that the benefits of AI are passed on to all patients they serve, not just those with access to more resources. Also, it is difficult to assign liability for medical malpractice arising from defects in AI [37,38]. In addition, AI's transparency and interpretability are challenged by factors such as patient privacy, algorithm interpretability, publication bias, etc. [39,40]. Most of the AI devices approved by the FDA have only undergone retrospective studies. The lack of prospective studies may lead to unexpected conditions during the clinical application of AI devices [41,42]. Solving these problems is a key point for the future of AI in clinical applications.
In this review, using “artificial intelligence”, “machine learning”, and “pancreatic cancer” as the keywords, we searched the relevant literature published by July 2022 in PubMed, Embase, Web of Science, and other databases. We summarized the application of AI in several aspects of PC. Compared with the existing studies, our review summarizes more comprehensively [43,44]. We outlined how AI could help in medical image analysis, pathological examination, and biomarkers in the tumor diagnosis process. In prognosis respect, the AI analysis includes survival time, recurrence risk, metastasis, and therapy response. Finally, we summarized the current status of AI in PC and discussed the future challenges and directions for the field.
State-of-the-art AI Algorithms involved in Pancreatic Cancer
Machine Learning
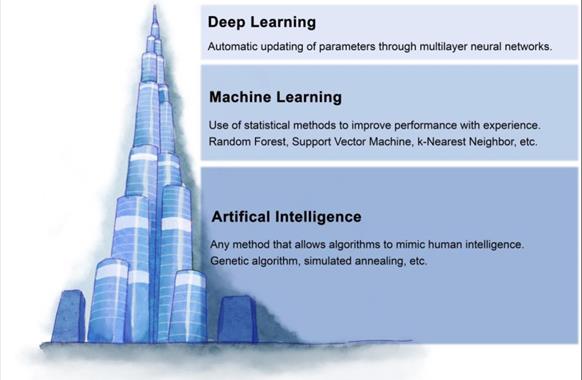
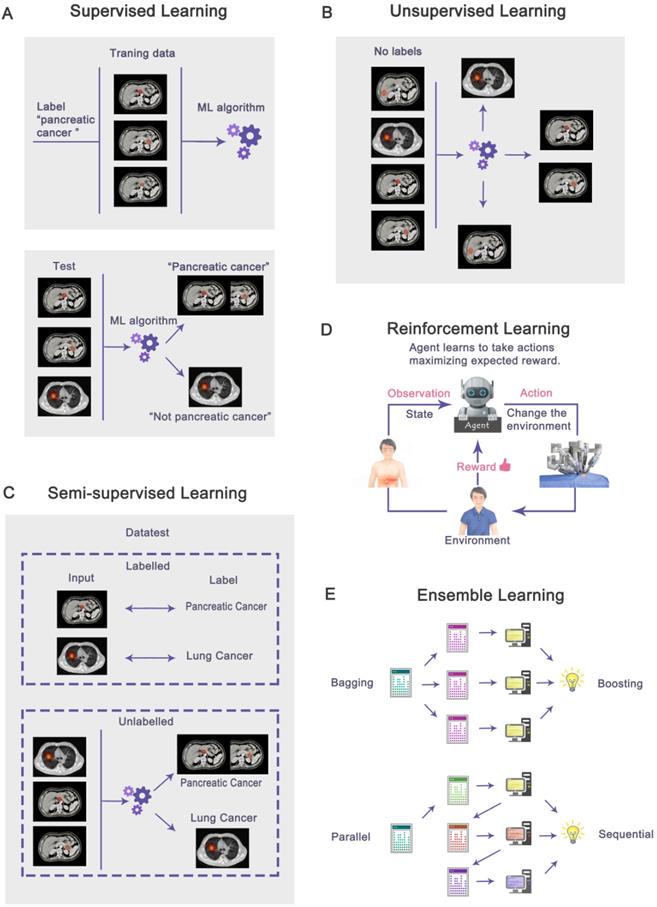
Machine learning (ML) is a subfield of AI that solves the problem of how to build computers that improve automatically through experience (Figure 1). Based on a large amount of feature data, ML can use specific algorithms to learn how to accomplish a task [45]. In today's medical field, there is a massive amount of data generated every day, and it becomes a challenge to integrate this data to make predictions. The most significant advantage of ML is the ability to integrate vast amounts of data and combine the observed and predicted quantities in nonlinear and highly interactive ways [46]. ML techniques can be broadly classified based on the type of labels. Based on labels, machine learning can be classified as supervised, unsupervised, semi-supervised, and reinforcement learning. There is also ensemble learning that integrates multiple algorithms (Figure 2A-2E).
Receiver operating characteristics (ROC) curves help organize ML classifiers and visualize their performance. ROC curve is a line graph plotted with sensitivity as the vertical coordinate and (1-specificity) as the horizontal coordinate. The area under the ROC curve (AUC) is the evaluation metric, and the larger the AUC value, the better the corresponding algorithm performs [47]. Other metrics, including accuracy, sensitivity, specificity, F1-Score, positive predictive value (PPV), and negative predictive value (NPV), are also commonly used to evaluate the result of the ML [48].
Supervised Learning
Supervised learning is constructing a model in which each observation vector has a corresponding response variable. In other words, all data is labeled. By fitting a model that relates responses to predictors, supervised learning can accurately predict future observed responses or better understand the relationship between responses and predictors [49,50]. Examples of such algorithms include Logistic Regressions (LR), Decision Trees (DT), Support Vector Machines (SVM), Naïve Bayes (NB), Artificial Neural Networks (ANN), etc., and the best application scenario for each algorithm varies [51]. In this review, most of the algorithms used in PC are supervised learning. A typical application of supervised learning algorithms is the precise diagnosis, including detection, grading, and differential diagnosis, using radiomics, digital pathology slides, or biomarkers. The prognosis of PC is also widely used to predict survival time, recurrence rate, metastasis, and therapy response.
Unsupervised Learning
Unsupervised learning means we can know the observation vector, not the associated response. In other words, all data is unlabeled. Using the observation vector's data makes it possible to perform clustering, correlation evaluation, dimensionality reduction, etc. [50,52]. Examples of such algorithms include K-mean clustering [53], Principal Component Analysis (PCA) [54], Non-negative Matrix Factorization (NMF) [55], etc. The application of unsupervised learning in PC is relatively rare, but there have been attempts to do so, including classification [56], feature extraction of CT images [57,58] or pathological slides [59], and estimation of medical imaging parameters [60].
Semi-supervised Learning
As its name suggests, semi-supervised learning is somewhere between supervised and unsupervised learning, allowing the use of large amounts of available unlabeled data in combination with small labeled datasets in many use cases. Semi-supervised learning can utilize a small amount of labeled data to obtain better performance than supervised learning while utilizing less labeled data to achieve the same level of performance close to that of supervised learning [61,62]. Examples of such algorithms include generative models, self-training, co-training, graph-based learning, Semi-supervised support vector machines, etc. [61,63].
The application of semi-supervised learning is mostly seen in medical imaging. Supervised learning algorithms may lack annotated data because annotation of medical imaging data is time-consuming and requires a high level of expertise. By using semi-supervised learning algorithms, the task of segmentation or diagnosis using medical images can be accomplished with fewer annotations [64]. For example, CT images of PC can be used for segmentation and diagnosis [65].
The relationship between artificial intelligence, machine learning, and deep learning. Artificial intelligence refers to the use of machines to simulate human intelligence. Machine learning is a subfield of artificial intelligence, which mainly studies how to simulate or realize the learning function in human intelligence. The deep learning model is a subset of machine learning, which is a model combining multi-layer neural networks.
Based on labels, machine learning can be classified as supervised (A), unsupervised (B), semi-supervised (C), reinforcement learning (D), and ensemble learning (E) that integrates multiple algorithms. In supervised learning, all data is labeled, while unsupervised learning is unlabeled. Semi-supervised learning contains a small amount of labeled data and a large amount of unlabeled data. Reinforcement learning is when the agent interacts with the unknown environment and obtains rewards or punishments from the environment. In ensemble learning, multiple algorithms are integrated to solve problems. The algorithms may be parallel (Bagging) or sequential (Boosting).
Reinforcement Learning
The reinforcement learning process is guided by a specific goal. Agents interact with the unknown environment and get reward or punishment feedback from the environment. Then, it uses this feedback to train itself and collect experience and knowledge about the environment to achieve specific goals [66]. Reinforcement learning can also be combined with deep learning to become deep reinforcement learning. It uses dynamic data and labels to bring feedback signals into the learning process rather than constructed, static dataset labels as in traditional machine learning [67].
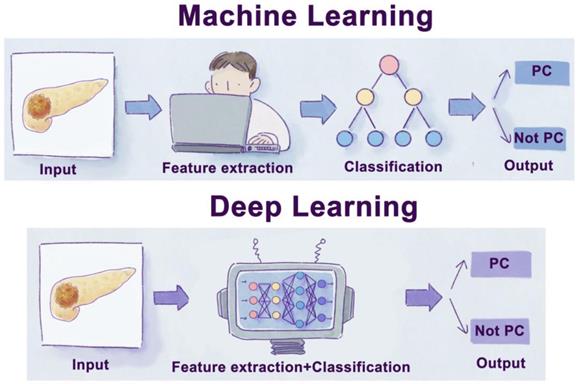
Schematic diagram of machine learning and deep learning process. Traditional machine learning usually needs four steps: input, feature extraction, classification, and output. Moreover, deep learning is a subset of a machine learning algorithm, which can extract labels by itself without manual extraction.
Reinforcement learning algorithms are commonly used in decision-making in the medical field. Due to the heterogeneity of patients' conditions and treatment responses, it is challenging to realize precision medicine. Reinforcement learning can construct dynamic treatment regimens that consider the immediate effect of treatment and the long-term benefit to the patient [68,69]. For PC, reinforcement learning algorithms can generate high-quality treatment plans for pancreas stereotactic body radiation therapy (SBRT) to achieve optimal metering distribution [70].
Ensemble Learning
Rather than a single algorithm, ensemble learning seamlessly integrates various machine learning algorithms into a unified framework, typically for supervised learning. Specifically, ensemble learning samples the data and produces prediction results using multiple learners. The above results are combined, and the errors of individual learners are potentially compensated by other learners, resulting in better prediction performance [71,72]. Depending on whether the different learners are independent of each other, the ensemble approach can be divided into two main frameworks: the dependent and independent [71]. The output of each learner of the dependent framework affects the next learner, which is represented by AdaBoost in the “Boosting” algorithm [73]. In an independent framework, individual learners can output in parallel, which is represented by Random Forest (RF) in the “Bagging” algorithm [74]. Both dependent and independent frameworks have applications in diagnosing [75-77] and prognosis [78-81] PC.
Deep Learning
Deep learning (DL) is a subset of ML algorithms (Figure 1). It allows a machine to feed raw data and automatically build complex concepts. Take image recognition as an example. The mapping from many different pixels to an image is very complex. DL solves this difficulty by decomposing the complex mappings required to recognize an image into a series of simple nested mappings. The algorithm can be divided into one visible layer and several hidden layers. The visible layer is where the image is fed, while hidden layers are where the algorithm gradually extracts the features from the image [82,83]. Compared to shallow ML and traditional data analysis methods, DL models have superior performance in many applications, especially in domains with extensive and high-dimensional data. However, shallow ML performs better for low-dimensional data, especially when a limited training set [84]. With the significant development of computer technology, many DL algorithms, such as Convolutional Neural Networks (CNN), Recurrent Neural Networks (RNN), MultiLayer Perceptron (MLP), Generative Adversarial Networks (GAN), and Deep Belief Networks (DBN), have been widely used in the field of oncology (Figure 3) [85-87].
AI in Tumor Diagnosis Process
Reading medical images to make judgments is essentially a problem of recognizing complex patterns, which computers can be trained using ML models to achieve efficient and repeatable recognition. AI can play a role in several steps in medical image-based PC diagnosis, including image reconstruction, segmentation, and detection, characterization, grading of pancreatic disease based on image features. Using similar techniques, AI can also identify digitized histopathology slides. It can potentially improve the accuracy, reproducibility, and efficiency of diagnosis using histological sections. In addition, the computer can analyze biomarker information with high throughput and accuracy, thus identifying tumor-related biomarkers more efficiently and using this information for diagnosis.
AI in Medical images-based diagnosis
Imaging techniques play an essential role in the diagnosis of PC. Current clinical imaging modalities include EUS, CT, MRI, and PET, with different advantages and disadvantages in clinical applications (Table 1). In the traditional process of medical image analysis, experienced radiologists are required. With AI technology, it is possible to free imaging physicians from tedious and repetitive labor to handle tasks that require more creativity.
Advantages and disadvantages of imaging modalities for pancreatic cancer
| Modality | Advantages | Disadvantages |
|---|---|---|
| EUS | High-resolution; Useful in tissue sampling | Invasive; Operator dependent |
| CT | High spatial resolution; Widely available | Poor contrast resolution |
| MRI | High contrast resolution; High sensitivity to small tumor and metastasis | Limited availability; Image artifacts |
| PET | Provides functional metabolic information | Poor spatial and contrast resolution; Physiological FDG uptake disturbance |
Radiomics refers to the high-throughput extraction of many image features from radiographic images, which may be challenging to recognize or quantify by the human eye. Radiomics can be used to identify lesions, allowing for early detection and diagnosis of disease. Also, radiological features can predict prognoses, such as survival, tumor metastasis, and treatment response, and correlate with genomic, transcriptomic, or proteomic features [88-91]. Conventional workflow in radiomics usually contains four steps: image acquisition, segmentation, feature extraction, and analysis [92]. For image acquisition, standard protocols are needed to minimize confounding variables [93]. Segmentation involves identifying the images' regions of interest (ROIs) and defining the boundaries in the pictures. While this step can be done manually by practiced radiologists, many ML methods have been used for image segmentation [88,94,95]. The dice similarity coefficient (DSC), used to measure the similarity of two sets, is the most used metric in evaluating segmentation performances. In some research, segmentation performances also used Hausdorff distance and intersection over union for evaluation [96]. The next step is extracting radiomics features from ROIs, including histogram-based, texture-based, model-based, transform-based, and shape-based features. Radiomic features are usually numerous, highly correlated, and redundant features that need to be filtered out before they can be used for model building [91,94]. The final step is to build a predictive model using ML and evaluate the model's performance.
Endoscopic Ultrasound
EUS is widely used in diagnosing pancreatic lesions because it provides high-resolution images of the pancreas without being disrupted by gas, bone, or subcutaneous fat. EUS and its related techniques, such as CE-EUS and EUS elastography, show high specificity and sensitivity in diagnosing pancreatic diseases. Furthermore, it is frequently used to identify regional lymph nodes and assess the relationship of tumors to nearby vascular structures [19,97]. In addition, EUS can guide tissue sampling to obtain pathological information about the cancerous tissue [20,98]. The disadvantage is that EUS is an invasive procedure with a risk of pancreatitis or bleeding. The method is also demanding on the operator, and improper handling may reduce the accuracy of the diagnosis [99,100]. As early as 2001, scientists had researched using neural networks to enhance EUS to detect and diagnose PC. Many studies have emerged in recent years (Table 2) [101-112].
AI in EUS is frequently used to aid in the differential diagnosis of PC and other conditions. In our statistics, a few studies applied AI in the differential diagnosis of PC. The overall AUC, accuracy, sensitivity, and specificity were 0.940-0.986, 80%-98.26%, 87.59%-100%, 50%-93.38%, respectively [101-109]. Udriştoiu et al. combined CNN and long short-term memory (LSTM) neural networks to construct an ML algorithm for differential diagnosis of focal pancreatic masses, using multi-sequences EUS (grayscale, color Doppler, arterial and venous phase contrast-enhancement, and elastography). Their model achieved high AUC and accuracy among the studies [105]. Kuwahara et al. used CNN (ResNet-50) in turn to extract image features and distinguish between benign and malignant intraductal papillary mucinous neoplasm (IPMN) [110].
Applying AI based on EUS in the differential diagnosis of pancreatic cancer and other pancreatic tumors
| Reference | Sample size | Data source | Algorithms | Aim | Best result |
|---|---|---|---|---|---|
| Zhu et al. [101] | 388 cases | EUS | SVM | PC vs CP | Accuracy (94.2%), Sensitivity (96.25%), Specificity (93.38%), PPV (92.21%), NPV (96.68%) |
| Udriştoiu et al. [102] | 65 cases | multi-sequences EUS | CNN, LSTM neural network | PDAC vs CPP vs PNET | AUC (0.98), Accuracy (98.26%) |
| Tong et al. [103] | 558 cases | CE-EUS | CNN (ResNet-50) | PDAC vs CP | AUC (0.986) |
| Tonozuka et al. [104] | 1390 images | EUS | CNN | PDAC vs CP | AUC (0.940), Sensitivity (92.4%), Specificity (84.1%), PPV (86.8%), NPV (90.7%) |
| Sǎftoiu et al. [105] | 167 cases | CEH-EUS | ANN | PC vs CP | Specificity (94.44%), Sensitivity (94.64%), PPV (97.24%), NPV (89.47%) |
| Marya et al. [106] | 1174461 images from 583 cases | EUS | CNN | AIP vs PDAC vs CP vs NP | For PDAC: AUC (0.976), Sensitivity (95%), Specificity (91%), PPV (87%), NPV (97%) |
| Sǎftoiu et al. [107] | 68 cases | EUS elastography | NN_MLP | PC vs CP vs NP vs PNET | AUC (0.932), Accuracy (89.7%), Sensitivity (91.4%), Specificity (87.9%), PPV (88.9%), NPV (90.6%) |
| Norton et al. [108] | 35 cases | EUS | NN | PC vs CP | Accuracy (80%), Sensitivity (100%), Specificity (50%) |
| Sǎftoiu et al. [109] | 258 cases | EUS elastography | ANN_MLP | PC vs CP | AUC (0.94), Accuracy (91.14%), Sensitivity (87.59%), Specificity (82.94%), PPV (96.25%), NPV (57.22%) |
| Kuwahara et al. [110] | 3,970 images | EUS | CNN (ResNet-50) | Diagnosis of malignancy in IPMN | AUC (0.98), Accuracy (94.0%), Sensitivity (95.7%), Specificity (92.6%), PPV (91.7%), NPV (96.2%) |
| Iwasa et al. [111] | 100 cases | CE-EUS | U-Net | PC segmentation | Median IoU (0.77) |
| Zhang et al. [112] | 2207+19486 images | EUS | ResNet | Pancreas segmentation; station recognition | Pancreas segmentation: DSC (71.5%); Station recognition: accuracy (82.4%) |
Abbreviations: AIP: autoimmune pancreatitis; ANN: artificial neural network; AUC: area under the curve; CE-EUS: contrast-enhanced EUS; CEH-EUS: contrast-enhanced harmonic EUS; CNN: convolutional neural network; CP: chronic pancreatitis; CPP: chronic pseudotumoral pancreatitis; DSC: dice similarity coefficient; IPMN: intraductal papillary mucinous neoplasm; IoU: intersection over union; LSTM: long short-term memory; MLP: multilayer perceptron; NN: neural network; NP: normal pancreas; NPV: negative predictive value; PDAC: pancreatic ductal adenocarcinoma; PNET: pancreatic neuroendocrine tumor; PPV: positive predictive value; SVM: support vector machine.
The physician can also use AI in EUS for pancreas segmentation. Iwasa et al. analyzed 100 patients with different PC. Segmentation was performed using U-Net with 100 epochs and was evaluated with 4-fold cross-validation. The median intersection over the union of all cases was 0.77 [111]. Zhang et al. developed a station classification model and a pancreas segmentation model for EUS training and quality control. The DSC of the pancreas segmentation model was 71.5%, and the accuracy of the station recognition model reached 82.4% [112].
Computed Tomography
CT is the dominant imaging modality for diagnosing and staging PC, which is more widely available and less expensive than other imaging modalities. Due to the high spatial resolution of CT can be used for diagnosing and staging the tumor, identifying vascular involvement, tumor resectability analysis, etc. However, the tumor may not be visible due to the poor contrast resolution of CT. With the use of multiplanar reformations, 3D techniques, and spatial and temporal resolution improvement, CT has achieved high sensitivity (96%) in tumor identification [18,97]. AI can assist CT-based diagnosis in many ways, including pancreas segmentation, diagnosis and staging of PC, differential diagnosis, and resectability analysis (summarized in Table 3).
In our statistics, several studies focused on PC or PC precursor lesions diagnosis or prediction by AI-assisted CT. Their AUC, accuracy, sensitivity, specificity were 0.79-0.999, 77.66%-99.2%, 76.64%-100%, 85.59%-98.5%, respectively [113-122]. The method of Chu et al. [120] has the highest accuracy (99.2%) among the studies. 190 pancreatic ductal adenocarcinoma (PDAC) patients and 190 healthy control cases with 64-MDCT scans were included, and 0.75-mm slices of venous phase images were chosen for segmentation and radiomics analysis. Images were manually segmented, and their features were extracted by a binary mask and selected by minimum-redundancy maximum-relevancy feature selection. Finally, an RF classifier was constructed to classify PDAC from the normal pancreas. All of the PDAC cases were correctly classified. Only one normal case from a renal donor was classified as PDAC, giving an AUC of 0.999, an accuracy of 99.2%, a sensitivity of 100% and a specificity of 98.5%.
Some studies focused on the differential diagnosis of pancreatic disease. Ikeda et al. investigated a neural network classifier for the differential diagnosis of PDAC and mass-forming pancreatitis, with an AUC of 0.866 [123]. Chen et al. combined imaging features and enhanced CT texture analysis. Then they used LASSO and RFE_LinearSVC algorithms to select features to differentiate pancreatic serous cystadenomas (SCN) from pancreatic mucinous cystadenomas (MCN), with an AUC of 0.932 [124]. Ren et al. extracted 792 radiomics features from the late arterial and portal venous phases of CE-CT. They then used an RF classifier for differential diagnosis between pancreatic adenosquamous carcinoma (PASC) and PDAC and achieved an AUC of 0.98 [125]. Xie et al. extracted and screened ten optimal imaging features and applied an RF algorithm to build a Rad-score to discriminate between MCN and atypical SCN. The method achieved an AUC of 0.97 [126]. Li et al. extracted 1409 radiomics features from the portal phase of multidetector computed tomography (MDCT). After removing irrelevant features and Bonferroni correction, four features by LASSO regression were still significantly associated with focal-type autoimmune pancreatitis (AIP) and PDAC. The LASSO logistic regression formula was used to obtain the rad-score for discriminating focal-type AIP from PDAC (AUC 0.97) [127]. In addition to using traditional algorithms (PyRadiomics), Ziegelmayer et al. used deep CNN for radiomics feature extraction. For the prediction of AIP or PDAC, an extremely randomized tree classifier was fit on the extracted features with an AUC of 0.90 [128]. Yang et al. adopted the RF method to construct a diagnostic prediction model based on textural parameters of CE-CT images to discriminate between SCN and MCN [129].
Application of AI based on CT in the differential diagnosis of pancreatic cancer and other pancreatic tumors
| Reference | Sample size | Data source | Algorithms | Aim | Best result |
|---|---|---|---|---|---|
| Ma et al. [113] | 3494 images from 190 cases | CE-CT | CNN | PC diagnosis | Accuracy (95.47%), Sensitivity (91.58%), Specificity (98.27%) |
| Liu et al. [114] | 338 cases | CE-CT | faster R-CNN | PC diagnosis | AUC (0.9632), Precision (76.64%) |
| Si et al. [115] | 143,945 images from 319 cases | CE-CT | ResNet18, U-net32, ResNet34 | PC diagnosis | AUC (0.871), Accuracy (82.7%), Sensitivity (86.8%), Specificity (69.5%) |
| Qiu et al. [116] | 312 cases | Plain CT | MSTA architecture, SVM | PDAC diagnosis | AUC (0.88), Accuracy (81.19%), Sensitivity (76.64%), Specificity (85.59%) |
| Qureshi et al. [117] | 216 cases | CE-CT | RFE_NB | PDAC prediction | Accuracy (86%) |
| Ebrahimian et al. [118] | 103 cases | DECT | RF | Benign vs Malignant Pancreatic Lesions | AUC (0.94), Accuracy (89%), Sensitivity (90%), Specificity (88%) |
| Chakraborty et al. [119] | 103 cases | CE-CT | RF, SVM | Predict High Risk IPMN | AUC (0.81) |
| Chu et al. [120] | 380 cases | MDCT | RF | PDAC detection | AUC (0.999), Accuracy (99.2%), Sensitivity (100%), Specificity (98.5%) |
| Mukherjee et al. [121] | 420 cases | CE-CT | KNN, SVM, RF, and XGBoost | PDAC detection | AUC (0.98), Accuracy (92.2%), Sensitivity (95.5%), Specificity (90.3%) |
| Polk et al. [122] | 51 cases | CE-CT | LR | IPMN malignancy prediction | AUC (0.93) |
| Ikeda et al. [123] | 71 cases | CE-CT | NN | PDAC vs mass-forming pancreatitis | AUC (0.916) |
| Chen et al. [124] | 100 cases | CE-CT | LASSO, SVM (RFE_LinearSVC) | SCN vs MCN | AUC (0.932), Sensitivity (87.5%), Specificity (82.4%) |
| Ren et al. [125] | 112 cases | CE-CT | RF | PASC vs PDAC | AUC (0.98), Accuracy (94.5%), Sensitivity (98.3%), Specificity (90.1%), PPV (91.9%), NPV (97.8%) |
| Xie et al. [126] | 216 cases | CE-CT | RF | MCN vs atypical SCN | AUC (0.734), Accuracy (72.8%), Sensitivity (74.8%), Specificity (70.5%), PPV (73.2%), NPV (79.8%) |
| Li et al. [127] | 97 cases | MDCT | LASSO regression | focal-type AIP vs PDAC | AUC (0.97), Accuracy (94%), Sensitivity (95%), Specificity (93%) |
| Ziegelmayer et al. [128] | 86 cases | CE-CT | Deep CNN+ Extremely Randomized Trees | AIP vs PDAC | AUC (0.90), Sensitivity (89%), Specificity (83%) |
| Yang et al. [129] | 78 cases | CE-CT | RF+ LASSO | MCN vs SCN | AUC (0.77), Accuracy (85%), Sensitivity (95%), Specificity (83%) |
| Gao et al. [143] | 170 cases | CE-CT | mRMR+ LASSO | MCN vs SCN | AUC (0.91), Accuracy (85%), Sensitivity (92%), Specificity (81%) |
| Panda et al. [130] | 1917 images | Venous phase CT | 3D CNN | Pancreas segmentation | DSC (91%), HD (0.15mm) |
| Mahmoudi et al. [131] | 157 cases | CT | 3D CNN, U-Net, TAU-Net | PDAC segmentation | DSC (60.6%), Precision (57.8%), Recall (78.0%), HD (3.73mm) |
| Huang et al. [132] | 170 cases | CE-CT | U-net | PNET segmentation, grading | DSC (81.8%), AUC (0.87) |
| Lim et al. [133] | 1006 cases | CE-CT | 3D U-Net | Pancreas segmentation and volumetry | DSC (84.2%), Precision (86.9%), Recall (84.2%) |
| Boers et al. [134] | 1995 images | Venous phase CT | 3D U-net | Pancreas segmentation | DSC (78.1%) |
| Xie et al. [135] | 82 cases | CE-CT | RSTN | Pancreas segmentation | DSC (84.53%) |
| Wang et al. [65] | 800 images | Venous phase CT | IGA-Net | PDAC segmentation; NP vs PDAC | DSC (60.29%), Sensitivity (99.75%), Specificity (96.50%) |
| Zhou et al. [136] | 14 cases | 4DCT | ResNet-50, FPN | Tumor positioning | DSC (98%) |
| Abel et al. [137] | 221 images | Venous phase CT | nnU-Net | PCL detection | Sensitivity (78.8%) |
| Lyu et al. [138] | 47 cases | CE-CT | DLIR-H | PC resectability prediction | AUC (0.91), Sensitivity (97%), Specificity (87%) |
| Chang et al. [139] | 401 cases | CE-CT | SVM+LASSO | PDAC grading | AUC (0.961), Accuracy (90.1%), Sensitivity (88.6%), Specificity (91.7%), PPV (92.1%), NPV (88.0%) |
| Luo et al. [140] | 112 cases | CE-CT | CNN | PNET grading | AUC (0.82), Accuracy (82.1%), Sensitivity (88.3%), Specificity (84.6%) |
| Wan et al. [57] | 137 cases | CT | SAE+ mRMR+ SVM | PNET grading | AUC (0.715, SAE-based model), (0.771, hybrid feature-based model) |
| Wan et al. [58] | 114 cases | CE-CT | SAE+ SVM/MLR/ANN | PNET grading | AUC (0.845, SVM) (0.856, MLR) (1.00, ANN) |
Abbreviations: 4DCT: four dimensions CT; AIP: autoimmune pancreatitis; ANN: artificial neural network; AUC: area under the curve; CE-CT: contrast-enhanced CT; CNN: convolutional neural network; CT: computed tomography; DECT: dual energy CT; DLIR: deep learning image reconstruction; DSC: dice similarity coefficient; FPN: feature pyramid network; HD: Hausdorff distance; IGA-Net: Inductive Attention Guidance Network; IPMN: intraductal papillary mucinous neoplasm; KNN: k-nearest neighbor; LASSO: least absolute shrinkage and selection operator; MCN: pancreatic mucinous cystadenoma; MDCT: multidetector CT; MLR: multivariable logistic regression; mRMR: minimum redundancy; MSTA: multiresolution-statistical texture analysis; NB: naïve Bayes; NN: neural network; NP: normal pancreas; PASC: pancreatic adenosquamous carcinoma; PCL: pancreatic cystic lesion; PDAC: pancreatic ductal adenocarcinoma; PNET: pancreatic neuroendocrine tumor; RF: random forest; RFE: recursive feature elimination; RSTN: recursive feature elimination; SAE: sparse autoencoder; SCN: pancreatic serous cystadenoma; SVM: support vector machine.
AI-based on MRI is applied in the differential diagnosis of pancreatic cancer and other pancreatic tumors
| Reference | Sample size | Data source | Algorithms | Aim | Best result |
|---|---|---|---|---|---|
| Li et al. [146] | 267 samples from 4 modalities (T1: 67, T2: 68, DWI: 68, AP: 64) | T1, T2, DWI, AP MRI | UDA+ meta learning+ GCN | PC segmentation | DSC (62.08%, T1), (61.35%, T2), (61.88%, DWI), (60.43%, AP) |
| Chen et al. [147] | 73 cases | multi-sequences MRI | Spiral-ResUNet | PC segmentation | DSC (65.60%), Jaccard index (49.64%), HD (7.27mm), Recall (76.69%), Precision (62.96%) |
| Liang et al. [148] | 56 DCE MRI sets | DCE MRI | CNN (SGDM) | PDAC segmentation | DSC (71%), HD (7.36mm), MSD (1.78mm) |
| Goldenberg et al. [149] | 30 mouse models | T1 relaxation, CEST, and DCE MRI | SVM | PC classification | Accuracy (87.5%, CEST) (85.1%, DCE) |
| Cui et al. [150] | 202 cases | T1-w, T2-w, CET1-w MRI | LASSO | BD-IPMN grading | AUC (0.903), Specificity (94.8%), Sensitivity (73.4%) |
| Corral et al. [151] | 139 cases | multi-sequences MRI | CNN | IPMN classification | AUC (0.783), Sensitivity (75%), Specificity (78%), PPV (73%), NPV (81%) |
| Hussein et al. [56] | 171 cases | T2 MRI | SVM, RF, 3D CNN | IPMN classification | Unsupervised:Accuracy (58.04%), Sensitivity (58.61%), Specificity (41.67%); Supervised: Accuracy (84.22%), Sensitivity (97.2%), Specificity (46.5%) |
| Cheng et al. [152] | 60 cases | CE-CT, T2 MRI | LR, SVM | Malignant IPMN prediction | MRI+SVM: AUC (0.940), Accuracy (86.7%), Sensitivity (95.7%), Specificity (81.1%), PPV (75.9%), NPV (96.8%) CT+SVM: AUC (0.864), Accuracy (83.3%), Sensitivity (78.3%), Specificity (86.5%), PPV (78.3%), NPV (86.5%) |
Abbreviations: AUC: area under the curve; BD-IPMN: branching type IPMN; CEST: chemical exchange saturation transfer; CNN: convolutional neural network; DCE: dynamic contrast enhancement; DSC: dice similarity coefficient; GCN: Graph Convolutional Networks; HD: Hausdorff distance; IPMN: intraductal papillary mucinous neoplasm; LR: logistic regression; MLP: multilayer perceptron; MSD: mean surface distance; NPV: negative predictive value; PC: pancreatic cancer; PDAC: pancreatic ductal adenocarcinoma; PPV: positive predictive value; RF: random forest; SGDM: stochastic gradient descent with momentum; SVM: support vector machine; UDA: unsupervised domain adaptation; AP MRI: atrial phase MRI; DWI: diffusion weighted imaging.
Several studies used AI-assisted CT for pancreas or PC segmentation. Their DSCs ranged from 60.6% to 91% [65,130-135]. Panda et al. developed a two-stage 3D CNN model based on a modified U-net architecture. 1917 portal venous phase CT scans with normal pancreas were used for training, validation, and testing. The mean DSC of their method was 91%, the highest among the studies. The authors also demonstrated that their approach could be applied to CT images containing PC (mean DSC=0.96) [130]. Zhou et al. developed a 4DCT-based method for tumor positioning without pancreas segmentation. Using 4DCT, they built a digitally reconstructed radiograph dataset for each patient, with clinical target volume (CTV) contours labeled. Then the datasets trained the ResNet and FPN algorithm to predict CTV. DSC of their method was 98%, which shows the accuracy of the positioning [136].
For other applications of AI in CT, Abel et al. developed and evaluated an algorithm based on a two-step nnU-Net architecture for automated detection of pancreatic cystic lesions (PCL) in CT [137]. Lyu et al. used high strength levels of the DL image reconstruction (DLIR-H) algorithm to predict the resectability of PC [138]. Chang et al. extracted radiomics features of CE-CT images by the SVM model and generated a radiomics signature by the LASSO model for the preoperative prediction of histological grades of PDAC. The radiomics signature for the training set and external validation data had an AUC of 0.961 and 0.770, respectively [139]. Luo et al. built a CNN-based model to analyze CE-CT images for pancreatic neuroendocrine neoplasms (PNET) grading [140]. Wan et al. built handcrafted, SAE, and hybrid features-based SVM prediction models for PNET grading. Among them, the hybrid feature model performs best (AUC 0.771) [57].
In addition to using CT image features to aid the precise diagnosis of PC, some studies attempted to improve CT image quality. Ohira et al. constructed a deep CNN that generates virtual monochromatic images from single-energy computed tomography (SECT) images for improved PC imaging quality [141]. Noda et al. used a DL image reconstruction algorithm to reconstruct dual-energy computed tomography images, thus assisting PC diagnosis [142].
Magnetic Resonance Imaging
MRI is of great value in diagnosing PC due to its ability to collect many types and superior soft-tissue contrast. The best application circumstances for MRI include: (1) detection of small non-contour-deforming tumors, (2) evaluation of local extension and vascular encasement, and (3) determination of lymph node, liver, and peritoneal metastases [17,144]. However, MRI is usually more expensive than CT. Also, metal implants may cause image artifacts and hinder imaging [145]. The applications of AI in MRI-based diagnosis include pancreas segmentation, PC classification, and grading (summarized in Table 4) [56,146-152].
Li et al. collected four modalities of MRI for PC segmentation. Since MRI image labeling is time-consuming and laborious, they attempted to train the algorithm on labeled MRI images in one modality and test the model's performance in another modality to achieve unsupervised labeling of MRI images. The DSC of their methods on different models are 62.08% (T1), 61.35% (T2), 61.88% (DWI), and 60.43% (AP) [146]. To achieve pancreas segmentation, Chen et al. developed a spiral-transformation algorithm to map 3D images onto a 2D plane. Combined with U-Net, their method has a relatively high mean DSC (65.60%) [147]. Liang et al. trained the CNN with Stochastic Gradient Descent with Momentum algorithm, and their method got 71% DSC [148].
Four teams used AI-assisted MRI to classify or grade PC. Goldenberg et al. built three groups of tumor models with different types of PDAC cells. The method of support vector machine predicted the correct tumor model with 87.5% (CEST MRI) and 85.1% accuracy (DCE MRI) [149]. Cui et al. used multivariate logistic regression to analyze the extracted MRI image features, and the AUC of their method was 0.903 [150]. Corral et al. used a CNN to classify IPMN. Sensitivity and specificity to identify high-grade dysplasia or cancer were 75% and 78%, respectively. Moreover, the AUC was 0.78 [151]. Hussein et al. were among the few teams using an unsupervised learning algorithm. 3D CNN was used to classify IPMN. Their method's accuracy, sensitivity, and specificity were 58.04%, 58.61%, and 41.67% [56].
Cheng et al. compared the predictive value of two medical imaging methods for predicting malignant IPMN. Radiomics features were extracted from arterial and venous phase images of CT and T2-weighted images of MRI, respectively. The LASSO algorithm was used for feature selection, and LR and SVM algorithms were applied to construct radiomics models. The results show that the MRI-based model (AUC 0.940) has a better performance compared to the CT-based model (AUC 0.864) [152].
Positron Emission Tomography
Positron Emission Tomography (PET) is a molecular imaging technique that has a vital role in diagnosing and staging tumors (Table 5). In combination with CT technology, PET/CT can help localize functional abnormalities and provide information on the biological characteristics of the tumors, such as metabolism, hypoxia, and proliferation [153,154]. Fluorine 18-fluorodeoxyglucose (FDG), a glucose analogue, is the most widely used radiotracer in PET. However, glucose metabolism is not specific, and physiological uptake of FDG by inflamed tissues may lead to false-positive results [155]. Also, FDG intake is reduced in patients with hyperglycemia, leading to a false negative result [156]. For PC, FDG-PET is more sensitive than CT for treatment monitoring after radiotherapy and description of recurrence after tumor resection [17].
Li et al. used simple linear iterative clustering on PET/CT pseudo-color images for pancreas segmentation and developed threshold component analysis to select the most beneficial feature combination. Then they designed a hybrid feedback-support vector machine-random forest (HFB-SVM-RF) model to identify normal pancreas or PC. The DSC and Jaccard index for pancreas segmentation is 78.9% and 65.4%, respectively. For PC diagnosis, the accuracy and sensitivity of their method were 96.47% and 97,51%, respectively [75]. Liu et al. extracted 502 radiomics features from dual-time PET/CT and used SVM to build a classifier to distinguish between PDAC and AIP. The AUC, Accuracy, Sensitivity, and Specificity were 0.9668, 89.91%, 85.31%, and 96.04%, respectively [157]. Xing extracted 251 expert-designed features from PET/CT images and combined them into five feature sets according to their modalities and dimensions. Four feature selection strategies (Spearman's rank correlation coefficient, minimum redundancy maximum relevance, support vector machine recursive feature elimination, and no feature selection) and four machine learning classifiers (RF, adaptive boosting, SVM with the Gaussian radial basis function, and SVM with the linear kernel function) were used to found the optimal feature set. Based on the best combination of the feature selection strategy and classifier, the model that differentiates AIP from PDAC was developed and achieve an AUC of 0.93 [158]. Xing et al. extracted radiomics features from PET/CT images using Pyradiomics and used the XGBoost algorithm to build a prediction model for PDAC pathological grade prediction [159].
Applying AI based on PET in the differential diagnosis of pancreatic cancer and other pancreatic tumors
| Reference | Sample Size | Data Source | Algorithm | Aim | Best result |
|---|---|---|---|---|---|
| Li et al. [75] | 80 cases | PET/CT | HFB-SVM-RF | PC diagnosis | Accuracy (96.47%), Sensitivity (95.23%), Specificity (97.51%) |
| Liu et al. [157] | 112 cases | PET/CT | SVM | PDAC vs AIP | AUC (0.9668), Accuracy (89.91%), Sensitivity (85.31%), Specificity (96.04%) |
| Zhang et al. [158] | 111 cases | PET/CT | RF, adaptive boosting, SVM | PDAC vs AIP | AUC (0.93), Accuracy (85%), Sensitivity (86%), Specificity (84%) |
| Xing et al. [159] | 149 cases | PET/CT | XGBoost | PDAC grading | AUC (0.994) |
Abbreviations: AIP: autoimmune pancreatitis; AUC: area under the curve; CT: computed tomography; NP: normal pancreas; PASC: pancreatic adenosquamous carcinoma; PDAC: pancreatic ductal adenocarcinoma; PET: positron emission tomography; RF: random forest; SVM: support vector machine.
Application of AI based on pathological examination in the differential diagnosis of pancreatic cancer and other pancreatic tumors
| Reference | Sample Size | Data Source | Algorithm | Aim | Best result |
|---|---|---|---|---|---|
| Song et al. [163] | 11 images from 7 cases | WSI | DCM+BAYES, KNN, SVM, ANN | SCN vs MCN | Accuracy (90%), Sensitivity (89%), Specificity (91%), PPV (91%), NPV (89%) |
| Song et al. [164] | 240 images | WSI | SVM | PDAC diagnosis and grading | For diagnosis: Accuracy (94.38%), Sensitivity (93.13%), Specificity (95.63%), PPV (95.78%), NPV (93.50%); For grading: Accuracy (77.03%), Sensitivity (74.38%), Specificity (79.69%), PPV (79.65%), NPV (75.40%) |
| Kriegsmann et al. [165] | 201 cases | WSI | CNN | PIN, PDAC identification | Balanced accuracy (73% for non-aggregated; 92% for aggregated) |
| Niazi et al. [166] | 33 cases | Ki67 stained slides | CNN | PNET identification | Accuracy (96.2%), Sensitivity (97.8%), Specificity (88.8%) |
| Vance et al. [76] | 31 cases | WSI and MxIF | RF | PDAC cell populations identification | Accuracy (90.0%) |
| Momeni-Boroujeni et al. [167] | 277images from 75 cases | FNA | MNN | Benign vs malignant pancreatic cytology | Accuracy (100%) For atypical cases: Accuracy (77%), Sensitivity (80%), Specificity (75%) |
| Naito et al. [168] | 532 images | EUS-FNB | CNN | PDAC detection | AUC (0.9836), Accuracy (94.17%), Sensitivity (93.02%), Specificity (97.06%) |
| Kurita et al. [169] | 85 cases | cyst fluid and EUS-FNA | NN | Benign vs malignant PCLs | AUC (0.966), Accuracy (92.9%), Sensitivity (95.7%), Specificity (91.9%), PPV (81.5%), NPV (98.3%) |
Abbreviations: ANN: artificial neural network; AUC: area under the curve; BAYES: Batesian classifier; CNN: convolutional neural network; DCM: direction cumulative map; FNA: fine needle aspiration; FNB: fine needle biopsy; KNN: k-nearest neighbor; MCN: pancreatic mucinous cystadenoma; NPV: negative predictive value; PC: pancreatic cancer; PIN: pancreatic intraepithelial neoplasia; PPV: positive predictive value; MNN: multilayer perceptron neural network; MxIF: cyclic multiplexed-immunofluorescence; NN: neural network; PDAC: pancreatic ductal adenocarcinoma; RF: random forest; PNET: pancreatic neuroendocrine tumor; SCN: pancreatic serous cystadenoma; SVM: support vector machine; WSI: Whole slide imaging.
AI-assisted Pathological Examination
In addition to the above radiographic images, The pathologist can apply AI to Hematoxylin and Eosin (H&E)-stained or immunofluorescent-stained whole slides images (WSI) for PC diagnosis [160]. FNA and FNB are essential diagnostic methods for suspended PC with high accuracy [161,162]. AI can assist in the diagnosis of PC by analyzing cytology and biochemical characteristics of FNA/FNB samples. Here, we summarized the application of AI in pathological examination (Table 6) [76,163-169].
Song et al. constructed a system to automatically segment epithelial cell nuclei on slide images and extract morphological features. They subsequently used multiple classifiers to demonstrate the effectiveness of the designed method in the differential diagnostic of SCN and MCN [163]. Using a similar approach, Song also worked on diagnosing and grading PDAC [164]. Kriegsmann et al. used CNN to construct models for automatic localization and quantification of tissue categories in whole tissue slides, including pancreatic intraepithelial neoplasia and PDAC [165]. Ki67 index has clear guidance for proliferation rate and grading of PNET. However, the non-specificity of Ki67 staining and counterstaining hinders the accurate quantification of the Ki67 index. Therefore, Niazi et al. proposed a DL method based on Ki67-stained biopsy images to distinguish NET from non-tumor areas automatically and achieved 97.8% sensitivity and 88.8% specificity [166]. Vance et al. combined WSI and cyclic multiplexed immunofluorescence to collect 31 markers of PDAC and used an RF algorithm to identify tumor cell populations, achieving an accuracy of 87% [76].
Momeni-Boroujeni et al. used a K-means clustering algorithm to segment cell clusters from FNA-based slides, extracted the morphological features of the cell clusters, and trained a multilayer perceptron neural network (MNN) with these features. Then tested their ability to discriminate between benign and malignant pancreatic cytology (accuracy 100%) [167]. Vance et al. trained the CNN using FNB-based slides to assess PDAC and achieved an AUC of 0.984 [168]. Kurita et al. combined biomarkers in the cyst fluid, cytological features obtained by FNA, and clinical variables to training neural networks for differentiating malignant and benign PCLs [169].
The applications of AI in Biomarkers
Biomarkers have a significant role in diagnosing, staging, and treating PC. The use of appropriate biomarkers for screening in high-risk populations is an essential aspect of the early diagnosis of PC. However, the current PC biomarkers lack sufficient sensitivity and specificity for clinical application [44,161,170]. Liquid biopsies allow a comprehensive cancer profile to be evaluated in a non-invasive and real-time manner and help discover more data for cancer research, including CTCs, cfDNA, exosomes, etc. The biomarkers in these data can be analyzed using AI for their association with diseases. With the expansion of data obtained from liquid biopsies, scientists can better apply AI to biomarker-based early diagnosis of cancers [161,171,172].
As summarized in Table 7, various biomarkers have been used to diagnose or detect PC with the aid of AI, including exosomes [173-175], proteins [176-179], cell-free DNA (cfDNA) [77], circulating microRNA [180], extracellular vesicles long RNA [181], gene expression pattern [182], etc. The above studies mainly contain two categories, 1) biomarkers were known, these data trained ML algorithms to obtain prediction models for PC [173,178,179,181,182], and 2) biomarkers were uncertain, features in the dataset needed to be extracted, then ML models were used to evaluate the value of these features in the diagnosis of PC [77,174-177,180].
Genomics
All cancers occur due to a series of mutations in the cellular genome. The most common driver genes of PDAC are KRAS, CDKN2A, TP53, and SMAD4, and genetic alterations of the SWI/SNF and COMPASS complexes significantly impact PC. The genome can be used as biomarkers for PC diagnosis, and genome studies can also reveal features that make PC therapeutically susceptible [183].
A part of some researchers used AI to analyze the existing genomic data in the database to find their association with PC. Wang et al. used 78 PDAC samples from the GEO database as the training set. By combining Support Vector Machine Recursive Feature Elimination (SVM-RFE) and Large Margin Distribution Machine Recursive Feature Elimination (LDM-RFE) algorithms, they predicted seven differentially expressed genes as specific biomarkers for PC [184]. Ko et al. developed a Gene Vector for Each Sample (GVES) model, which generated vector representations of genes using gene expression and biological network data from the TCGA database. In cases of small sample sizes, GVES had good accuracy for predicting prognostic genes [185]. Cristiano et al. developed an approach to evaluate fragmentation patterns of cfDNA across the genome and constructed a gradient tree boosting (GBM) model to detect cancer. For PC, the AUC, accuracy, and specificity were 0.86, 67%, and 71%, respectively [77].
Applications of artificial intelligence in biomarker-based pancreatic cancer diagnosis
| Reference | Sample Size | Data Source | Algorithm | Aim | Best result |
|---|---|---|---|---|---|
| Chen et al. [173] | 28 samples* | DNA-PAINT (exosomes) | LDA | Cancer detection | Accuracy (100%) |
| Zheng et al. [174] | 220 cases** | MALDI-TOF-MS (exosomes) | ANN | Cancer discrimination | AUC (0.86) |
| Ko et al. [175] | 28 mice + 34 cases | ExoTENPO chip (exosomes) | LDA | PC diagnosis | Accuracy (100%) |
| Gao et al. [176] | 199 cases | SELDI-TOF-MS (proteomes) | SVM, KNN, ANN | PC diagnosis | AUC (0.971), Sensitivity (96.67%), Specificity (100%) |
| Yu et al. [177] | 100 serum samples | SELDI-proteinchip | DT | PC prediction | Sensitivity (88.9%), Specificity (74.1%) |
| Yang et al. [178] | 913 serum samples | Multiple serum tumor markers | ANN, LR | PC diagnosis | AUC (0.905), Accuracy (83.53%), Sensitivity (90.86%), Specificity (67.50%) |
| Qiao et al. [179] | 136 cases | CT images+ serum tumor markers | 2D-3D CNN | Image segmentation; PC vs CP | For image segmentation: DSC (84.32%); For PC vs CP: Accuracy (87.63%), Sensitivity (94.57%), Specificity (93.25%), PPV (84.57%), NPV (90.34%) |
| Cristiano et al. [77] | 34 cases | Cell-free DNA | GBM | Cancer detection | AUC (0.86), Accuracy (67%), Specificity (71%) |
| Alizadeh Savareh et al. [180] | 671 cases | GEO database (circulating microRNA) | PSO-ANN-NCA | PC diagnosis | Accuracy (93%), Sensitivity (93%), Specificity (92%) |
| Yu et al. [181] | 501 cases | exLR | SVM | PDAC detection | AUC (0.960), Accuracy (90.43%), Sensitivity (93.39%), Specificity (85.07%) |
| Almeida et al. [182] | 648 samples | Gene expression microarray | ANN | PDAC prediction | F1-score (0.86), Accuracy (89.66%), Sensitivity (87.6%), Specificity (83.1%) |
| Yang et al. [197] | 204 cases | Liquid biopsy | KNN, SVM, LDA, LR, and Naive Bayes | PC diagnosis and staging | For diagnosis: AUC (0.95), Accuracy (92%), Sensitivity (88%), Specificity (95%); For staging: Accuracy (84%), Sensitivity (78%), Specificity (88%) |
| Sinkala et al. [198] | 185 cases | TCGA database (proteins, mRNAs, miRNAs, and DNA methylation patterns) | NCA, SVM, DT, LR, ET, KNN | PC subtypes differentiation | Accuracy (98.7% for mRNA-based KNN classifier; 97.8% for the DNA methylation-based SVM classifier) |
| Zhang et al. [199] | 1183 cases*** | LDI-MS | SVM | Pan-cancer diagnosis and classification | For PC: Accuracy (100%) |
*Including 9 healthy samples, 10 breast cancer samples, 9 PC samples;
**Including 79 breast cancer cases, 57 PC cases, 84 healthy controls;
***Including 97 PC cases.
Abbreviations: ANN: artificial neural network; AUC: area under the curve; CNN: convolutional neural network; CP: chronic pancreatitis; DT: decision tree; DNA-PAINT: DNA points accumulation for imaging in nanoscale topography; ET: ensemble tree; exLR: extracellular vesicles long RNA; GBM: gradient tree boosting; KNN: k-nearest neighbor; LDA: liner discriminate analysis; LDI-MS: laser desorption/ionization mass spectrometry; LR: logistic regression; MALDI-TOF-MS: matrix-assisted laser desorption/ionization time-of-flight MS; MLP: multilayer perceptron; NCA: neighborhood component analysis; PC: pancreatic cancer; PDAC: pancreatic ductal adenocarcinoma; PPV: positive predict value; SELDI-TOF-MS: surface-enhanced laser desorption/ionization time-offlight mass spectrometry; SVM: support vector machine.
Transcriptomics
Both coding and non-coding RNAs are essential for gene expression. In addition to mRNAs directly related to protein expression, many non-coding RNAs are involved in or reveal tumor progressions, such as miRNA, lncRNA, and circRNA [186]. The study of PC transcriptomics helps to understand the mechanism of tumor progression and provides valuable prognostic markers [187].
Xuan et al. obtained miRNA-disease association data from the human miRNA-disease database and built a network representation learning and CNN-based model to predict disease miRNAs. The AUC of the model in PC miRNA prediction is 0.971 [188]. Some researchers have also identified biomarkers directly from pathological samples. Mori et al. directly sequenced the RNA from PDAC tumor tissues and normal tissues, and DL analyzed the data. The selected genes were all important prognostic factors for PC based on the TCGA database [189]. Alizadeh Savareh et al. identified a series of circulating miRNAs associated with PC by analyzing four GEO microarray datasets. The value of the top miRNAs was then assessed by ML methods (Particle Swarm Optimization (PSO) + ANN and Neighborhood Component Analysis (NCA)). The final model, which consist of five miRNAs (miR-663a, miR-1469, miR-92a-2-5p, miR-125b-1-3p and miR-532-5p), showed good diagnostic results on the investigated cases and validation set (Accuracy: 0.93, Sensitivity: 0.93, Specificity: 0.92) [180]. Yu et al. analyzed the extracellular vesicles' long RNA profile of PDAC, chronic pancreatitis (CP), and healthy plasma samples. The d-signature was identified using an SVM algorithm to detect PDAC (0.960), identify resectable stage I/II cancer (AUC 0.949), and distinguish PDAC from CP (AUC 0.931) [181]. Almeida et al. identify five differentially expressed genes (AHNAK2, KRT19, LAMB3, LAMC2, and S100P) from a gene expression microarray meta-analysis to train an ANN to classify PDAC or healthy samples. The ANN model could classify the test samples with a sensitivity of 87.6% and a specificity of 83.1% [182].
Proteomics
Proteins are the performers of gene-encoded functions. Although proteins are the direct products of mRNA translation, many unexpected associations in the proteome are primarily absent in RNA. Tumor proteome is closely related to epithelial and mesenchymal markers, cell lineage sensitivity, etc. Interventions on the proteome also provide new avenues for tumor treatment [190].
Gao et al. used SELDI-TOF-MS to analyze serum proteomes from PC patients and healthy controls. SVM analysis of the spectra was used to generate a predictive algorithm based on maximally differentially expressed proteins between PC patients and healthy controls. Four significant peaks were used to build a classifier to distinguish PC patients from healthy controls. Combining the SELDI protein peaks and CA19-9, their classifier achieves the AUC of 0.971 [176]. Yu et al. used SDLDI-Proteinchip to analyze serum protein profiling from PC patients and healthy controls. A decision-tree algorithm was trained to separate PC from controls. The sensitivity and specificity of their method were 88.9% and 74.1%, respectively [177]. Yang et al. used three tumor markers (CA19-9, CA125, and CEA) from serum specimens to train the ANN model for PC diagnosis. The AUC, accuracy, sensitivity, and specificity were 0.905, 83.53%, 90.86%, 67.50%, respectively [178].
Exosomes
Exosomes are extracellular vesicles secreted by eukaryotic cells involved in intercellular communication containing nucleic acids, proteins, lipids, and glycoconjugates. It can regulate tumor cell proliferation, metastasis, Epithelial-to-Mesenchymal Transition (EMT), and angiogenesis during tumor development. In clinical practice, it can be used as a biomarker for tumor diagnosis, grading, and prognosis prediction [191,192]. Many studies have used exosomes to diagnose, treat, and monitor treatment response in PC [193].
Chen et al. developed a quantitative analysis platform for continuously quantifying multiple exosomal surface biomarkers from blood samples. Four exosomal surface biomarkers (HER2, GPC-1, EpCAM, EGFR) were immunostained to calculate the number. Linear discriminant analysis was further used to identify exosomes from pancreatic and breast cancer samples. The accuracy of their method was 100% [173]. Zheng et al. used sequential size exclusion chromatography (SSEC) to separate exosomes from human plasma. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) data of samples were collected, and a multi-classifier artificial neural network (denoted as Exo-ANN) model was used to identify pancreatic and breast cancer samples. The AUC of their method for PC was 0.86 [174]. Ko et al. developed a multichannel nanofluidic system to isolate exosomes with an ExoTENPO chip, profiled the RNA cargo inside these exosomes, and applied a linear discriminate analysis (LDA) algorithm to identify samples derived from PC patients. Eight exosomal mRNA biomarkers were identified in their mice studies and used to distinguish PC patients from healthy controls. All samples (N=24) in the blinded test set were classified correctly [175].
Multi-omics
It is also possible to combine multiple types of biomarkers to detect PC. The analysis of multiple biomarkers revealed the complex molecular landscape of PC and offered the possibility of precision medicine [194-196]. Yang et al. constructed a multi-analyte panel, including extracellular vesicle miRNAs and mRNAs, cfDNA, and CA19-9. These data are used in the training of various ML algorithms. When applied to PDAC diagnosis, the model achieved an AUC of 0.95 and an accuracy of 92%. Furthermore, the model achieved an accuracy of 84% for disease staging [197]. Sinkala et al. extracted several types of biomarkers from the TCGA database. Then they used neighborhood component analysis (NCA) to identify biomarker sets. Different biomarkers trained several ML algorithms for PC subtypes differentiation [198]. Zhang et al. reported a laser desorption/ionization (LDI) mass spectrometry-based liquid biopsy for cancer screening and classification. The study included many cancer types, with 100% accuracy for PC detection in an internal validation cohort [199]. Cheng et al. deployed ALICE (Automated Liquid Biopsy Cell Enumerator) to identify and enumerate minute amounts of tumor cell phenotypes bestrewed in massive leukocytes and discovered two subpopulations of circulating hybrid cells from PC patients [200]. Qiao et al. used CT images to train a 2D-3D CNN model for pancreas segmentation and achieved an average DSC of 84.32%. The diagnostic performance (accuracy 87.63%, sensitivity 94.57%, specificity 93.25%, PPV 84.57%, NPV 90.34%) of CT combined with tumor marker (CA-50, CA-199, CA-242) was better than CT or serum tumor markers only [179].
AI in Prognosis
Accurately predicting the prognosis of PC has important implications for clinical decision-making. This information can help clinicians decide on treatment options, analyze the outcome of pancreatectomy, improve the management of patients, etc. However, classical prognostic factors, such as lymph node status and American Joint Committee on Cancer (AJCC) stage, are not entirely relevant in some long-term survivors [201,202]. Also, long-term survivors and general patients did not show significant differences in their mutation profiles [203]. These facts make it challenging to predict the prognosis of PC. Due to its excellent computational power, AI was used to analyze PC prognoses, including survival time [204-221], recurrence risk [78,221-224], metastasis [225-230], therapy response [79-81,231-240], etc.
Survival time
The non-invasive identification of specific imaging features (or signatures) that can predict tumor genomic alterations is termed “radiogenomics,” which integrates radiomics and genomics information. The gene expression profiles obtained in radiogenomics can be used as biomarkers to predict prognosis [241]. With the aid of ML, the radiologist used radiological images (CT, MRI) to detect multiple gene expression profiles in PC, including p53 status and PD-L1 expression [204], FAP expression [205], and ITGAV expression [206]. These genes had been shown to have predictive ability for the prognosis of PC.
Radiomics can also be applied alone for prognosis prediction. Xu et al. used EUS images to predict the prognosis of PC patients undergoing interstitial brachytherapy [207]. By extracting the radiomics features of FDG-PET [208,209] or CT [210-212] images and combining them with ML models, researchers could improve the accuracy of survival prediction for PC patients. In addition to direct extraction of image features, CT images have also been used to analyze patient body compositions [213] and tumor heterogeneity [214] in PC to predict survival.
In addition to the radioactive approach mentioned above, some non-imaging methods can predict PC survival. Walczak et al. and Aronsson et al. combined clinical variables (sex, age, year of diagnosis, tumor stage, etc.) with ANN algorithms to predict survival in PC (91% sensitivity and 38% specificity) [215] and invasive IPMN (82% accuracy and 83% precision) [216], respectively. Using the ML algorithm, Hayward et al. combined clinical variables and treatment records to predict PC clinical performance (patient survival time, quality of life scores, surgical outcomes, and tumor characteristics) [217]. Biomarker analysis is also a common approach in prognosis. Yokoyama et al. evaluated the methylation status of MUC1, MUC2, and MUC4 promoter regions and integrated these results and clinical pathologic features. Then they used SVM-, NN-, and multinomial-based methods to develop a prognostic classifier [218]. Winter et al. used the NetRank algorithm to filtrate marker genes prognostic for the outcome from PC gene expression profiles. Accuracies were assessed using SVM classifiers and Monte Carlo cross-validation [219]. A wavelet-based DL method was proposed by Tang et al. to select variables and predict prognosis for PC by training with multi-omics data (genomic, epigenomic, and clinical cohort information). This method predicts prognosis better than the traditional LASSO model (AUC: 0.937 vs. 0.802) [220]. Beak et al. used multi-omics data to analyze survival and recurrence in PC, with data sources including whole-exome sequencing, RNA sequencing, microRNA sequencing, DNA methylation data, and other clinical data. LR analysis generally revealed the best performance for both disease-free survival (DFS) and overall survival (OS) (accuracy = 0.762 and AUC= 0.795 for DFS; accuracy = 0.776 and AUC = 0.769 for OS) [221].
Recurrence risk
Clinical features, such as CA19-9 level, tumor location, size, stage, and differentiation degree, are of considerable importance for predicting the risk of recurrence. Li et al. collected demographics and various biochemical and pathological variables of PDAC patients from multiple institutions and used six ML algorithms to construct predictive models. SVM and KNN models had the highest accuracy in predicting 1-year and 2-year recurrence (70.9% and 73.4%), respectively [222]. Lee et al. compared the effects of RF and Cox models on the prognosis of PDAC. Training these two models using multiple clinical variables yielded a mean c-index of 0.6805 and 0.7738 for the RF and Cox models, respectively [78].
In combination with clinical features from patients, radiomics features can be used to predict the risk of recurrence of PC. He et al. collected PDAC patients' CT images performed three months after surgery for radiomics analysis. Using clinicoradiological information and radiomics feature jointly or separately, multivariable LR was applied to construct the local recurrences model of PDAC. The combined model achieved an AUC of 0.742 in the validation cohort, which is better than the clinicoradiological-only risk model (AUC 0.533), and the radiomics-only risk model (AUC 0.730) [223]. Li et al. preprocessed CE-CT and extracted and selected optimal radiomics features from intratumoral volume (ITV) and peritumoral volume (PTV). Then, ANN and LR models were employed to develop the ITV model, PTV model, combined model, clinical model, and radiomics-clinical model. Radiomics-clinical model outperformed other models in predicting 1-year recurrence (AUC 0.764 for validation set) and 2-year recurrence (AUC 0.773 for validation set) [224].
Metastasis
The lymph node metastasis status of PDAC significantly impacts the choice of treatment options, the risk of postoperative recurrence, and the overall survival rate of patients [242,243]. Therefore, correct prediction of lymph node metastasis status can enhance patient prognosis. An et al. analyzed preoperative DECT images of regional lymph node dissection in PDAC patients using the Res-Net 18 algorithm to classify lymph node metastasis. The authors compared the prediction effects of virtual monoenergetic images at different voltages. 100 + 150 keV DECT yielded the best predictions (AUC 0.87). If key clinical features (CT-reported T stage, LN status, glutamyl transpeptidase, and glucose) are integrated can further improve the prediction of the model (AUC 0.92) [225]. Some studies employed CT radiomics for PC lymph node metastasis prediction, and they all applied multivariable LR to construct a radiomics-based model with AUCs ranging from 0.75 to 0.912 [226-228]. Shi et al. employed T2-weighted (T2WI) and portal venous phase (PVP) MRI images for lymph node metastasis analysis. Radiomics features were extracted by PHIgo software and selected by a gradient-boosting decision tree. T2WI combined with the PVP model has the best performance. The AUCs were 0.834 and 0.807 in the training and validation cohorts, respectively [229].
Liver metastases are more common after PDAC resection but are unpredictable and lead to a poorer prognosis. Zambirinis et al. performed liver radiomics analysis on preoperative CE-CT scans and developed an LR classifier to predict early liver metastasis. By incorporating preoperative clinicopathological variables with radiomics features, their model achieved an AUC of 0.76 [230].
Therapy response
AI has also been used to predict treatment responses, including chemotherapy, radiotherapy, immunotherapy, and surgery. Wei et al. used a variational autoencoder (VAE) algorithm to extract tumor transcriptome features. Regularized gradient boosted decision trees (XGBoost) were further used to predict chemotherapy drug response for cancer (for PC: AUROC 0.738; AUPRC 0.764) [80]. Cos et al. collected preoperative activity metrics (step count, heart rate, and sleep time series) from patients with the help of wearable devices and built ML models to predict whether the pancreatectomy achieved the desired outcome (the absence of postoperative pancreatic fistulae, etc.). Their model combined clinical characteristics and achieved an AUC of 0.7875 [231]. Facciorusso et al. developed ANN and LR models to predict pain response after repeat echoendoscopic celiac plexus neurolysis. The predictive performance of the ANN model was higher than the LR model (AUC 0.94 vs. 0.85) [232]. Using clinical data and MRI images, Kaissis et al. distinguished two subtypes of PDAC (KRT81+ and KRT81-) by gradient boosted-tree algorithm. Subsequently, they assessed chemotherapy response and survival stratified by subtype and radiographic characteristics. Patients with the KRT81+ subtype responded significantly better to gemcitabine-based chemotherapy than FOLFIRINOX (HR 2.33) [81]. Schperberg et al. combined clinical trials, drug-related biomarkers, and molecular profile information to construct an RF model to predict drug oncologic outcomes in randomized clinical trials. The Spearman correlation (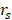 ) between their predicted model's and actual outcomes was statistically significant (progression-free survival (PFS):
) between their predicted model's and actual outcomes was statistically significant (progression-free survival (PFS): 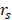 = 0.879, overall survival (OS):
= 0.879, overall survival (OS): 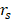 = 0.878, P < .0001) [79]. Nasief et al. collected CT images of patients during chemotherapy and compared the changes in radiomics features therein. Bayesian-regularization-neural-network was used to build a response prediction model with AUC of 0.98 for kurtosis-coarseness-NESTD (normalized-entropy-to-standard-deviation-difference) combination [233].
= 0.878, P < .0001) [79]. Nasief et al. collected CT images of patients during chemotherapy and compared the changes in radiomics features therein. Bayesian-regularization-neural-network was used to build a response prediction model with AUC of 0.98 for kurtosis-coarseness-NESTD (normalized-entropy-to-standard-deviation-difference) combination [233].
Stereotactic body radiotherapy (SBRT) is a therapeutic option in PC care, which permits the precise application of high-dose radiation in 1 to 5 fractions to a limited target volume. It has been proven that SBRT has significantly better outcomes than chemotherapy alone or in combination with conventional external-beam radiotherapy (EBRT) [244]. Several studies with radiomics have emerged to predict the response to SBRT. Based on the radiomics features of CT and clinical characteristics, Gregucci et al. applied a multivariate LR model to predict local response to SBRT for locally advanced PC (AUC 0.851) [234]. Based on CT radiomics features, Parr et al. used the gradient boosting machine model to construct OS (c-index 0.66) and recurrence prediction model (AUC 0.78) for PC following SBRT [235]. Simpson et al. extracted radiomics features from low field strength (0.35 T) MRI for predicting treatment response in PC patients undergoing SBRT. RF algorithm was adopted to construct a prediction model with the AUCs of 0.81 and 0.845 in two similar studies [236,237].
Immunotherapy has shown remarkable efficacy against various tumors, but PC has shown minimal response to immunotherapy. Tumor-infiltrating lymphocytes (TILs) have been proven to be associated with immunotherapy response [245], OS, and PFS [246]. Analysis of TILs may help identify PC patients most likely to respond to immunotherapy. Bian et al. developed an XGBoost-based model for preoperative prediction TILs in PDAC patients with CT radiomics features (AUC 0.79) [238]. Based on MRI radiomics features, Bian et al. also predicted Tumor-infiltrating CD8+ T cell [239] and other TILs [240] in PDAC patients with LDA-based model (AUC 0.76) and XGBoost-based model (AUC 0.79), respectively.
Some researchers have used ML to analyze the recovery condition of long-term survivors of PC. Kong et al. analyzed CT images by ML to determine changes in body composition (skeletal muscle, adipose tissue) in long-term survivors of pancreatic head cancer. They performed a multi-factor LR analysis of the factors affecting the changes in body composition. Their research concluded that long-term survivors of PC did not recover their preoperative body composition status, and preoperative sarcopenia and recurrence influenced body composition changes [247]. The tumor-stroma ratio (TSR) is an independent prognostic factor for solid tumors [248]. Based on MRI radiomics features, Meng et al. developed and validated an XGBoost classifier for evaluating TSR in patients with PDAC for interstitial targeted therapy selection and monitoring (AUC 0.78 in the validation set) [249].
Other applications of AI in PC
In addition to the research mentioned above, AI has also been applied to many aspects of PC, including predicting gene mutation [250,251], nucleus segmentation [252], tumor target localization [253], and predicting the risk of ICU admission for PC patients [254], etc.
Electronic health records (EHR) are also considered to be useful for early screening of PC. In a workshop (Early Detection of Pancreatic Cancer: Opportunities and Challenges in Utilizing Electronic Health Records) held in March 2021, experts from multiple fields assessed the feasibility of AI-based data extraction and modeling applied to EHRs and identified future directions [255]. In another article published the same year, Malhotra et al. used logistic regression on EHRs to screen people at high risk of PC. Their method could indicate cancer risk over a decade before diagnosing PC patients [256]. Roch et al. developed a natural language processing-based pancreatic cyst identification system. It could search and identify keywords of pancreatic cysts based on electronic medical records (EMR), with sensitivity and specificity of 99.9% and 98.8%, respectively. This system could help capture patients at risk of PC [257].
Some studies have also focused on analyzing the omics of PC. Kim et al. classified PC using ML and DL to classify quantitative proteomics data [258]. Song et al. employed AI techniques to deconvolute spatial transcriptomics data to uncover the cell states and subpopulations based on spatial localization [259]. Bagante et al. integrated whole-exome sequencing data with the help of artificial neural networks for cell-of-origin pattern prediction and molecular subtypes classification of hepato-pancreato-biliary cancers. Combining the clinical data and the above information, they also analyzed the prognosis of cancer patients using random survival forest and Cox analysis [260].
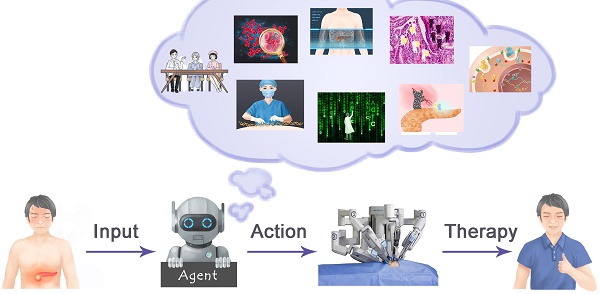
Application of artificial intelligence in multiple related fields of pancreatic cancer. Artificial intelligence can use one type of data alone to make predictions about pancreatic cancer or integrate multi-omics information for analysis.
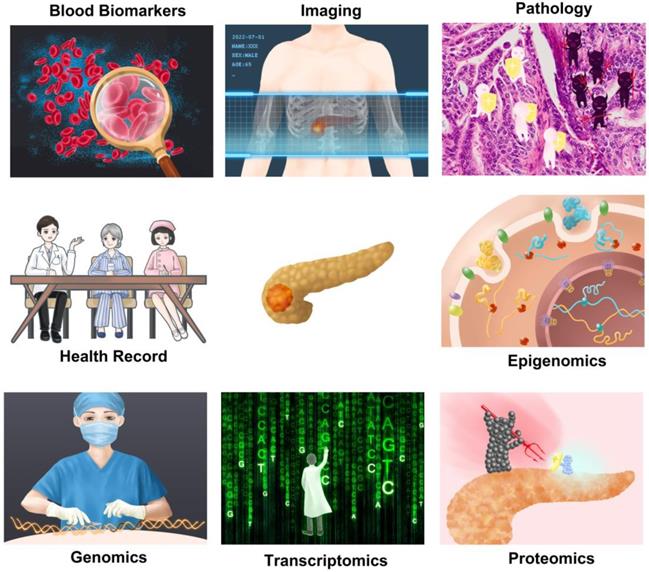
Some studies have attempted to apply DL to estimate medical imaging parameters. Misha et al. present an unsupervised physics-informed DL algorithm of intravoxel incoherent motion (IVIM) model called IVIM-NEToptim to fit diffusion-weighted imaging (DWI)-MRI data. MRI images of 23 PDAC patients showed IVIM-NEToptim superior performance to the least squares and Bayesian approaches at SNRs < 50 [60]. Ottens et al. presented various frameworks, including non-linear least squares (NLLS), Gated Recurrent Unit (GRU), FCN, LSTM, GRU, CNN, and U-Net, that analyze DCE-MRI concentration curves and output extended Tofts-Kety parameter estimates. Testing on 28 PC patients showed that GRU had the best performance [261].
Some researchers have attempted to develop new computer-aided diagnosis (CAD) systems. Li et al. constructed a Raman spectroscopic system using CNN models to efficiently distinguish between cancerous and normal pancreatic tissue. The AUCs of all three Raman image-based (1D, 2D Raman images, and 2D Raman PC1) methods were close to 0.99 [262]. Jadhav et al. developed a 3D virtual pancreatography system using ML algorithms for non-invasive diagnosis and classification of pancreatic lesions, the precursors of PC [263]. Dmitriev et al. developed a CAD system for classifying pancreatic cystic lesions based on RF and CNN. They proposed a visual analytics approach to uncover the system's decision-making process [264]. Combining clinical and radiological characteristics, Kang et al. built LR and ML models to predict the risk of malignant IPMN. After 200 repetitions, the mean AUCs of their ML and LR models were comparable (0.725 vs. 0.725) [265].
Challenges and Future Perspectives
Although AI has many applications on PC, many challenges have remained (Figure 4). Data accessibility and bias may affect the effectiveness of AI predictions. In radiomics, the image's quality may affect the construction of AI models. An assessment of quality gaps in public pancreas imaging datasets found that a substantial proportion of CT images were unsuitable for AI due to biliary stents or other factors [266]. Minorities are often underrepresented in clinical trials, leading to biased results [267,268]. For AI, insufficient data on minorities may result in the inability to adequately assess patient diversity for algorithm development and testing.
Though radiomics is thought to hold promise for addressing many issues in cancer care, there are still some concerns, such as reproducibility. Variations include intra-individual test-retest repeatability, image-acquisition technique, multi-machine reproducibility, segmentation reproducibility, radiomics feature definition, parameter setting, and implementation. All challenge the reproducibility of radiomics [269-271]. To improve the reproducibility of radiomics, scientists have attempted many approaches. The repeatability of radiomics features can be assessed using consistency correlation coefficients (CCCs) [272]. In order to build a more reproducible model, only repeatable features should be retained for subsequent model construction. Many studies reuse the dataset from which the model was developed for validation, lacking validation from external datasets. Using an external dataset for validation can improve the reproducibility of the model. Since there are many mature algorithms and software for radiomics, standardizing the process of radiomics, including image acquisition, segmentation, and feature extraction, can help to improve reproducibility [269,271,273].
Due to the specificity of the medicine, an interpretable algorithm is preferred. A flawed algorithm can lead to terrible consequences. Hundreds of hospitals that used IBM Waston Health's cancer AI algorithm for recommending treatments proved to have some errors in their operation [274]. However, a trade-off exists between performance and explainability at present. The DL models usually perform better, but they are often the least explainable because they are purely data-driven, and the underlying structures are challenging to interpret [32,275]. Three main approaches have attempted to address the interpretability of DL models: (1) proxy models, which use more traditional statistical models to explain the operational properties of DL; (2) visualization, which shows the internal mechanisms of DL models; and (3) internal interpretability approach, where the model can explain by itself which parts are essential [276,277]. This suggests that before CAD systems can be used in the clinic, they must be approved for safety and efficacy to avoid patient harm.
As knowledge of the disease continues to expand, the data collected will gradually increase, and clinical decision-making will become more and more complex. Training AI by a single type of medical image or biomarker is not perfect for the diagnosis and prognosis of PC. It should be noted that the study of multimodal features based on image and multi-omics is a new direction for future research. Despite the challenges of AI in PC, it will eventually emerge in all areas of PC due to its great advantage in integrating complex data. In clinical practice, building viable healthcare AI systems requires the joint work of experts in multiple fields, including clinicians, basic scientists, statisticians, and engineers. As AI technology advances and various experts collaborate, its features will become more powerful and accurate.
Conclusions
Here we summarized the applications of AI on PC. AI-based early screening may be a critical factor in improving the prognosis of PC patients. The combination of medical images and AI may become an essential part of CAD systems to assist physicians in making adequate and accurate diagnoses. In addition, the role of AI in multi-omics and pathology cannot be ignored. With the decrease in computing costs and improved computer technology and biotechnology, AI will make a fantastic process in the medical field. This progress requires a collaborative effort between clinicians, basic scientists, statisticians, and engineers. Despite some limitations, it will still dramatically improve many aspects of PC in the foreseeable future because of its powerful computing capabilities.
Abbreviations
4DCT: four dimensions CT; AIP: autoimmune pancreatitis; ANN: artificial neural network; AUC: area under the curve; BAYES: Batesian classifier; BD-IPMN: branching type IPMN; CE-CT: contrast-enhanced CT; CE-EUS: contrast-enhanced EUS; CEH-EUS: contrast-enhanced harmonic EUS; CNN: convolutional neural network; CEST: chemical exchange saturation transfer; CP: chronic pancreatitis; CPP: chronic pseudotumoral pancreatitis; CT: computed tomography; DCE: dynamic contrast enhancement; DCM: direction cumulative map; DECT: dual energy CT; DLIR: deep learning image reconstruction; DNA-PAINT: DNA points accumulation for imaging in nanoscale topography; DSC: dice similarity coefficient; DT: decision tree; ET: ensemble tree; exLR: extracellular vesicles long RNA; FNA: fine needle aspiration; FNB: fine needle biopsy; FPN: feature pyramid network; GBM: gradient tree boosting; GCN: Graph Convolutional Networks; HD: Hausdorff distance; IGA-Net: Inductive Attention Guidance Network; IPMN: intraductal papillary mucinous neoplasm; IoU: intersection over union; KNN: k-nearest neighbor; LASSO: least absolute shrinkage and selection operator; LDA: liner discriminate analysis; LDI-MS: laser desorption/ionization mass spectrometry; LR: logistic regression; LSTM: long short-term memory; MALDI-TOF-MS: matrix-assisted laser desorption/ionization time-of-flight MS; MCN: pancreatic mucinous cystadenoma; MDCT: multidetector CT; MLP: multilayer perceptron; MLR: multivariable logistic regression; MNN: multilayer perceptron neural network; mRMR: minimum redundancy; MSD: mean surface distance; MSTA: multiresolution-statistical texture analysis; MxIF: cyclic multiplexed-immunofluorescence; NB: naïve Bayes; NCA: neighborhood component analysis; NN: neural network; NP: normal pancreas; NPV: negative predictive value; PASC: pancreatic adenosquamous carcinoma; PC: pancreatic cancer; PCL: pancreatic cystic lesion; PDAC: pancreatic ductal adenocarcinoma; PET: positron emission tomography; PIN: pancreatic intraepithelial neoplasia; PNET: pancreatic neuroendocrine tumor; PPV: positive predictive value; RF: random forest; RFE: recursive feature elimination; RSTN: recursive feature elimination; SAE: sparse autoencoder; SCN: pancreatic serous cystadenoma; SELDI-TOF-MS: surface-enhanced laser desorption/ionization time-offlight mass spectrometry; SGDM: stochastic gradient descent with momentum; SVM: support vector machine; UDA: unsupervised domain adaptation; WSI: Whole slide imaging.
Acknowledgements
This study is supported by the National Natural Science Foundation of China (81972324) and the CAMS Innovation Fund for Medical Sciences (2021-I2M-1-002).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA: A Cancer Journal for Clinicians. 2022;72:7-33
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians. 2021;71:209-49
3. Zheng R, Zhang S, Zeng H, Wang S, Sun K, Chen R. et al. Cancer incidence and mortality in China, 2016. Journal of the National Cancer Center. 2022;2:1-9
4. Walter FM, Mills K, Mendonça SC, Abel GA, Basu B, Carroll N. et al. Symptoms and patient factors associated with diagnostic intervals for pancreatic cancer (SYMPTOM pancreatic study): a prospective cohort study. Lancet Gastroenterol Hepatol. 2016;1:298-306
5. Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y. et al. FOLFIRINOX versus Gemcitabine for Metastatic Pancreatic Cancer. New Engl J Med. 2011;364:1817-25
6. Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M. et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. New Engl J Med. 2013;369:1691-703
7. Schultheis B, Strumberg D, Bergmann L, Graeven U, Hanauske AR, Lipp R. et al. Results of a phase II trial of S-1 as first-line treatment of metastatic pancreatic cancer (CESAR-study group). Invest New Drug. 2012;30:1184-92
8. Qian Y, Gong Y, Fan Z, Luo G, Huang Q, Deng S. et al. Molecular alterations and targeted therapy in pancreatic ductal adenocarcinoma. J Hematol Oncol. 2020;13:130
9. Bear AS, Vonderheide RH, O'Hara MH. Challenges and Opportunities for Pancreatic Cancer Immunotherapy. Cancer Cell. 2020;38:788-802
10. Balachandran VP, Beatty GL, Dougan SK. Broadening the Impact of Immunotherapy to Pancreatic Cancer: Challenges and Opportunities. Gastroenterology. 2019;156:2056-72
11. Schizas D, Charalampakis N, Kole C, Economopoulou P, Koustas E, Gkotsis E. et al. Immunotherapy for pancreatic cancer: A 2020 update. Cancer Treat Rev. 2020;86:102016
12. McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, McCain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroentero. 2018;24:4846-61
13. Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F. et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349-61
14. Dbouk M, Katona BW, Brand RE, Chak A, Syngal S, Farrell JJ. et al. The Multicenter Cancer of Pancreas Screening Study: Impact on Stage and Survival. J Clin Oncol. 2022: O2200298.
15. Khalaf N, El-Serag HB, Abrams HR, Thrift AP. Burden of Pancreatic Cancer: From Epidemiology to Practice. Clin Gastroenterol H. 2021;19:876-84
16. Klein AP. Pancreatic cancer epidemiology: understanding the role of lifestyle and inherited risk factors. Nat Rev Gastro Hepat. 2021;18:493-502
17. Lee ES, Lee JM. Imaging diagnosis of pancreatic cancer: a state-of-the-art review. World J Gastroentero. 2014;20:7864-77
18. Raman SP, Horton KM, Fishman EK. Multimodality imaging of pancreatic cancer-computed tomography, magnetic resonance imaging, and positron emission tomography. The Cancer Journal. 2012;18:511-22
19. Kitano M, Yoshida T, Itonaga M, Tamura T, Hatamaru K, Yamashita Y. Impact of endoscopic ultrasonography on diagnosis of pancreatic cancer. J Gastroenterol. 2019;54:19-32
20. Moutinho-Ribeiro P, Iglesias-Garcia J, Gaspar R, Macedo G. Early pancreatic cancer - The role of endoscopic ultrasound with or without tissue acquisition in diagnosis and staging. Digest Liver Dis. 2019;51:4-9
21. Force UPST. Screening for Pancreatic Cancer: US Preventive Services Task Force Reaffirmation Recommendation Statement. Jama-J Am Med Assoc. 2019;322:438-44
22. Chari ST, Kelly K, Hollingsworth MA, Thayer SP, Ahlquist DA, Andersen DK. et al. Early detection of sporadic pancreatic cancer: summative review. Pancreas. 2015;44:693-712
23. Luo G, Jin K, Deng S, Cheng H, Fan Z, Gong Y. et al. Roles of CA19-9 in pancreatic cancer: Biomarker, predictor and promoter. Bba-Rev Cancer. 2021;1875:188409
24. Xing H, Wang J, Wang Y, Tong M, Hu H, Huang C. et al. Diagnostic Value of CA 19-9 and Carcinoembryonic Antigen for Pancreatic Cancer: A Meta-Analysis. Gastroent Res Pract. 2018;2018:8704751
25. Wang X, Chung WY, Correa E, Zhu Y, Issa E, Dennison AR. The integration of artificial intelligence models to augment imaging modalities in pancreatic cancer. Journal of Pancreatology. 2020;3:173-80
26. Neoptolemos JP, Kleeff J, Michl P, Costello E, Greenhalf W, Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nature reviews. Gastroenterology & hepatology. 2018;15:333-48
27. Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. New Engl J Med. 2019;380:1347-58
28. Yu KH, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat Biomed Eng. 2018;2:719-31
29. Shimizu H, Nakayama KI. Artificial intelligence in oncology. Cancer Sci. 2020;111:1452-60
30. Johnson KW, Torres Soto J, Glicksberg BS, Shameer K, Miotto R, Ali M. et al. Artificial Intelligence in Cardiology. J Am Coll Cardiol. 2018;71:2668-79
31. Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019;20:e262-73
32. Kenner B, Chari ST, Kelsen D, Klimstra DS, Pandol SJ, Rosenthal M. et al. Artificial Intelligence and Early Detection of Pancreatic Cancer: 2020 Summative Review. Pancreas. 2021;50:251-79
33. Young MR, Abrams N, Ghosh S, Rinaudo JAS, Marquez G, Srivastava S. Prediagnostic Image Data, Artificial Intelligence, and Pancreatic Cancer: A Tell-Tale Sign to Early Detection. Pancreas. 2020;49:882-6
34. Gruson D, Helleputte T, Rousseau P, Gruson D. Data science, artificial intelligence, and machine learning: Opportunities for laboratory medicine and the value of positive regulation. Clin Biochem. 2019;69:1-7
35. Cirillo D, Catuara-Solarz S, Morey C, Guney E, Subirats L, Mellino S. et al. Sex and gender differences and biases in artificial intelligence for biomedicine and healthcare. Npj Digit Med. 2020 3
36. Charpignon ML, Celi LA, Samuel MC. Who does the model learn from? Comment. Lancet Digit Health. 2021;3:E275-6
37. Brady AP, Neri E. Artificial Intelligence in Radiology-Ethical Considerations. Diagnostics. 2020 10
38. Safdar NM, Banja JD, Meltzer CC. Ethical considerations in artificial intelligence. Eur J Radiol. 2020 122
39. Stupple A, Singerman D, Celi LA. The reproducibility crisis in the age of digital medicine. Npj Digit Med. 2019 2
40. Haibe-Kains B, Adam GA, Hosny A, Khodakarami F, Shraddha T, Kusko R. et al. Transparency and reproducibility in artificial intelligence. Nature. 2020;586:E14-7
41. Wu E, Wu K, Daneshjou R, Ouyang D, Ho DE, Zou J. How medical AI devices are evaluated: limitations and recommendations from an analysis of FDA approvals. Nat Med. 2021;27:582-4
42. Benjamens S, Dhunnoo P, Mesko B. The state of artificial intelligence-based FDA-approved medical devices and algorithms: an online database. Npj Digit Med. 2020 3
43. Mendoza Ladd A, Diehl DL. Artificial intelligence for early detection of pancreatic adenocarcinoma: The future is promising. World J Gastroentero. 2021;27:1283-95
44. Pereira SP, Oldfield L, Ney A, Hart PA, Keane MG, Pandol SJ. et al. Early detection of pancreatic cancer. Lancet Gastroenterol. 2020;5:698-710
45. Jordan MI, Mitchell TM. Machine learning: Trends, perspectives, and prospects. Science. 2015;349:255-60
46. Obermeyer Z, Emanuel EJ. Predicting the Future - Big Data, Machine Learning, and Clinical Medicine. New Engl J Med. 2016;375:1216-9
47. Fawcett T. An introduction to ROC analysis. Pattern Recogn Lett. 2006;27:861-74
48. Rahmani AM, Yousefpoor E, Yousefpoor MS, Mehmood Z, Haider A, Hosseinzadeh M. et al. Machine Learning (ML) in Medicine: Review, Applications, and Challenges. Mathematics-Basel. 2021 9
49. Jiang T, Gradus JL, Rosellini AJ. Supervised Machine Learning: A Brief Primer. Behav Ther. 2020;51:675-87
50. James G, Witten D, Hastie T, Tibshirani R, James G, Witten D, et al. An Introduction to Statistical Learning with Applications in R Introduction; 2013
51. Caruana R, Niculescu-Mizil A. An Empirical Comparison of Supervised Learning Algorithms. InICML '06. New York, NY, USA. 2006 p. 161-8
52. Berry MW, Mohamed A, Yap BW. Supervised and Unsupervised Learning for Data Science. 2020.
53. Sinaga KP, Yang MS. Unsupervised K-Means Clustering Algorithm. Ieee Access. 2020;8:80716-27
54. Jolliffe IT, Cadima J. Principal component analysis: a review and recent developments. Philos T R Soc a. 2016 374
55. Lee DD, Seung HS. Algorithms for non-negative matrix factorization. In: Leen TK, Dietterich TG, Tresp V,*.^editors. ADVANCES IN NEURAL INFORMATION PROCESSING SYSTEMS 13. 14th Annual Neural Information Processing Systems Conference (NIPS). 2001 p. 556-62
56. Hussein S, Kandel P, Bolan CW, Wallace MB, Bagci U. Lung and Pancreatic Tumor Characterization in the Deep Learning Era: Novel Supervised and Unsupervised Learning Approaches. Ieee T Med Imaging. 2019;38:1777-87
57. Wan YD, Yang PF, Xu L, Yang J, Luo C, Wang J. et al. Radiomics analysis combining unsupervised learning and handcrafted features: A multiple-disease study. Med Phys. 2021;48:7003-15
58. Wan YD, Xu L, Yang PF, Cao ZZ, Luo C, Shen XY. et al. Feasibility of predicting pancreatic neuroendocrine tumor grade using deep features from unsupervised learning. In: Chen PH, Deserno TM,*.^editors. MEDICAL IMAGING 2020: IMAGING INFORMATICS FOR HEALTHCARE, RESEARCH, AND APPLICATIONS. SPIE Conference on Medical Imaging - Imaging Informatics for Healthcare, Research, and Applications. 2020
59. Asano K, Ono N, Iwamoto C, Ohuchida K, Shindo K, Kanaya S. Feature extraction and Cluster analysis of Pancreatic Pathological Image Based on Unsupervised Convolutional Neural Network. In: Zheng H, Callejas Z, Griol D, Wang H, Hu X, Schmidt H, et al. PROCEEDINGS 2018 IEEE INTERNATIONAL CONFERENCE ON BIOINFORMATICS AND BIOMEDICINE (BIBM). IEEE International Conference on Bioinformatics and Biomedicine (BIBM) - Human Genomics. 2018 p. 2738-40
60. Kaandorp M, Barbieri S, Klaassen R, van Laarhoven H, Crezee H, While PT. et al. Improved unsupervised physics-informed deep learning for intravoxel incoherent motion modeling and evaluation in pancreatic cancer patients. Magn Reson Med. 2021;86:2250-65
61. van Engelen JE, Hoos HH. A survey on semi-supervised learning. Mach Learn. 2020;109:373-440
62. Xiaojin Zhu ABG. Introduction To Semi-Supervised learning: Springer Cham; 2009
63. Sheikhpour R, Sarram MA, Gharaghani S, Chahooki MAZ. A Survey on semi-supervised feature selection methods. Pattern Recogn. 2017;64:141-58
64. Cheplygina V, de Bruijne M, Pluim J. Not-so-supervised: A survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med Image Anal. 2019;54:280-96
65. Wang Y, Tang P, Zhou Y, Shen W, Fishman EK, Yuille AL. Learning Inductive Attention Guidance for Partially Supervised Pancreatic Ductal Adenocarcinoma Prediction. Ieee T Med Imaging. 2021;40:2723-35
66. Naeem M, Rizvi S, Coronato A. A Gentle Introduction to Reinforcement Learning and its Application in Different Fields. Ieee Access. 2020;8:209320-44
67. Wang HN, Liu N, Zhang YY, Feng DW, Huang F, Li DS. et al. Deep reinforcement learning: a survey. Front Inform Tech El. 2020;21:1726-44
68. Coronato A, Naeem M, De Pietro G, Paragliola G. Reinforcement learning for intelligent healthcare applications: A survey. Artif Intell Med. 2020 109
69. Jonsson A. Deep Reinforcement Learning in Medicine. Kidney Dis-Basel. 2019;5:18-22
70. Zhang JH, Wang CH, Sheng Y, Palta M, Czito B, Willett C. et al. An Interpretable Planning Bot for Pancreas Stereotactic Body Radiation Therapy. Int J Radiat Oncol. 2021;109:1076-85
71. Sagi O, Rokach L. Ensemble learning: A survey. Wires Data Min Knowl. 2018 8
72. Dong XB, Yu ZW, Cao WM, Shi YF, Ma QL. A survey on ensemble learning. Front Comput Sci-Chi. 2020;14:241-58
73. Bartlett PL, Traskin M. AdaBoost is consistent. J Mach Learn Res. 2007;8:2347-68
74. Breiman L. Random forests. Mach Learn. 2001;45:5-32
75. Li S, Jiang H, Wang Z, Zhang G, Yao Y. An effective computer aided diagnosis model for pancreas cancer on PET/CT images. Comput Meth Prog Bio. 2018;165:205-14
76. Vance K, Alitinok A, Winfree S, Jensen-Smith H, Swanson BJ, Grandgenett PM. et al. Machine learning analyses of highly-multiplexed immunofluorescence identifies distinct tumor and stromal cell populations in primary pancreatic tumors. Cancer Biomark. 2022;33:219-35
77. Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570:385-9
78. Lee K, Jang J, Yu Y, Heo JS, Han H, Yoon Y. et al. Usefulness of artificial intelligence for predicting recurrence following surgery for pancreatic cancer: Retrospective cohort study. Int J Surg. 2021;93:106050
79. Schperberg AV, Boichard A, Tsigelny IF, Richard SB, Kurzrock R. Machine learning model to predict oncologic outcomes for drugs in randomized clinical trials. Int J Cancer. 2020;147:2537-49
80. Wei Q, Ramsey SA. Predicting chemotherapy response using a variational autoencoder approach. Bmc Bioinformatics. 2021;22:453
81. Kaissis G, Ziegelmayer S, Lohöfer F, Steiger K, Algül H, Muckenhuber A. et al. A machine learning algorithm predicts molecular subtypes in pancreatic ductal adenocarcinoma with differential response to gemcitabine-based versus FOLFIRINOX chemotherapy. Plos One. 2019;14:e218642
82. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-44
83. Goodfellow I, Bengio Y, Courville A, Goodfellow I, Bengio Y, Courville A. Deep Learning; 2016
84. Janiesch C, Zschech P, Heinrich K. Machine learning and deep learning. Electron Mark. 2021;31:685-95
85. Tran KA, Kondrashova O, Bradley A, Williams ED, Pearson JV, Waddell N. Deep learning in cancer diagnosis, prognosis and treatment selection. Genome Med. 2021;13:152
86. Echle A, Rindtorff NT, Brinker TJ, Luedde T, Pearson AT, Kather JN. Deep learning in cancer pathology: a new generation of clinical biomarkers. Brit J Cancer. 2021;124:686-96
87. Wang CW, Khalil MA, Firdi NP. A Survey on Deep Learning for Precision Oncology. Diagnostics. 2022 12
88. Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P. et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441-6
89. Gillies RJ, Schabath MB. Radiomics Improves Cancer Screening and Early Detection. Cancer Epidem Biomar. 2020;29:2556-67
90. He M, Xue H, Jin Z. Radiomics in pancreatic ductal adenocarcinoma: a state of art review. Journal of Pancreatology. 2020;3:195-200
91. Mayerhoefer ME, Materka A, Langs G, Haggstrom I, Szczypinski P, Gibbs P. et al. Introduction to Radiomics. J Nucl Med. 2020;61:488-95
92. Thomasian NM, Kamel IR, Bai HX. Machine intelligence in non-invasive endocrine cancer diagnostics. Nat Rev Endocrinol. 2022;18:81-95
93. Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol. 2016;61:R150-66
94. Dalal V, Carmicheal J, Dhaliwal A, Jain M, Kaur S, Batra SK. Radiomics in stratification of pancreatic cystic lesions: Machine learning in action. Cancer Lett. 2020;469:228-37
95. Kumar H, DeSouza SV, Petrov MS. Automated pancreas segmentation from computed tomography and magnetic resonance images: A systematic review. Scientific Reports. 2019;178:319-28
96. Taha AA, Hanbury A. Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. Bmc Med Imaging. 2015;15:29
97. Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395:2008-20
98. Bhutani MS, Koduru P, Joshi V, Saxena P, Suzuki R, Irisawa A. et al. The role of endoscopic ultrasound in pancreatic cancer screening. Endosc Ultrasound. 2016;5:8-16
99. Eloubeidi MA, Decker GA, Chandrasekhara V, Chathadi KV, Early DS, Evans JA. et al. The role of endoscopy in the evaluation and management of patients with solid pancreatic neoplasia. Gastrointest Endosc. 2016;83:17-28
100. Tummers WS, Willmann JK, Bonsing BA, Vahrmeijer AL, Gambhir SS, Swijnenburg RJ. Advances in Diagnostic and Intraoperative Molecular Imaging of Pancreatic Cancer. Pancreas. 2018;47:675-89
101. Zhu M, Xu C, Yu J, Wu Y, Li C, Zhang M. et al. Differentiation of pancreatic cancer and chronic pancreatitis using computer-aided diagnosis of endoscopic ultrasound (EUS) images: a diagnostic test. Plos One. 2013;8:e63820
102. Udriștoiu AL, Cazacu IM, Gruionu LG, Gruionu G, Iacob AV, Burtea DE. et al. Real-time computer-aided diagnosis of focal pancreatic masses from endoscopic ultrasound imaging based on a hybrid convolutional and long short-term memory neural network model. Plos One. 2021;16:e251701
103. Tong T, Gu J, Xu D, Song L, Zhao Q, Cheng F. et al. Deep learning radiomics based on contrast-enhanced ultrasound images for assisted diagnosis of pancreatic ductal adenocarcinoma and chronic pancreatitis. Bmc Med. 2022;20:74
104. Tonozuka R, Itoi T, Nagata N, Kojima H, Sofuni A, Tsuchiya T. et al. Deep learning analysis for the detection of pancreatic cancer on endosonographic images: a pilot study. J Hepato-Bil-Pan Sci. 2021;28:95-104
105. Săftoiu A, Vilmann P, Dietrich CF, Iglesias-Garcia J, Hocke M, Seicean A. et al. Quantitative contrast-enhanced harmonic EUS in differential diagnosis of focal pancreatic masses (with videos). Gastrointest Endosc. 2015;82:59-69
106. Marya NB, Powers PD, Chari ST, Gleeson FC, Leggett CL, Abu Dayyeh BK. et al. Utilisation of artificial intelligence for the development of an EUS-convolutional neural network model trained to enhance the diagnosis of autoimmune pancreatitis. Gut. 2021;70:1335-44
107. Săftoiu A, Vilmann P, Gorunescu F, Gheonea DI, Gorunescu M, Ciurea T. et al. Neural network analysis of dynamic sequences of EUS elastography used for the differential diagnosis of chronic pancreatitis and pancreatic cancer. Gastrointest Endosc. 2008;68:1086-94
108. Norton ID, Zheng Y, Wiersema MS, Greenleaf J, Clain JE, Dimagno EP. Neural network analysis of EUS images to differentiate between pancreatic malignancy and pancreatitis. Gastrointest Endosc. 2001;54:625-9
109. Săftoiu A, Vilmann P, Gorunescu F, Janssen J, Hocke M, Larsen M. et al. Efficacy of an artificial neural network-based approach to endoscopic ultrasound elastography in diagnosis of focal pancreatic masses. Clin Gastroenterol H. 2012;10:84-90
110. Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S, Obata M. et al. Usefulness of Deep Learning Analysis for the Diagnosis of Malignancy in Intraductal Papillary Mucinous Neoplasms of the Pancreas. Clin Transl Gastroen. 2019;10:1-8
111. Iwasa Y, Iwashita T, Takeuchi Y, Ichikawa H, Mita N, Uemura S. et al. Automatic Segmentation of Pancreatic Tumors Using Deep Learning on a Video Image of Contrast-Enhanced Endoscopic Ultrasound. J Clin Med. 2021 10
112. Zhang J, Zhu L, Yao L, Ding X, Chen D, Wu H. et al. Deep learning-based pancreas segmentation and station recognition system in EUS: development and validation of a useful training tool (with video). Gastrointest Endosc. 2020;92:874-85
113. Ma H, Liu ZX, Zhang JJ, Wu FT, Xu CF, Shen Z. et al. Construction of a convolutional neural network classifier developed by computed tomography images for pancreatic cancer diagnosis. World J Gastroentero. 2020;26:5156-68
114. Liu SL, Li S, Guo YT, Zhou YP, Zhang ZD, Li S. et al. Establishment and application of an artificial intelligence diagnosis system for pancreatic cancer with a faster region-based convolutional neural network. Chinese Med J-Peking. 2019;132:2795-803
115. Si K, Xue Y, Yu X, Zhu X, Li Q, Gong W. et al. Fully end-to-end deep-learning-based diagnosis of pancreatic tumors. Theranostics. 2021;11:1982-90
116. Qiu JJ, Yin J, Qian W, Liu JH, Huang ZX, Yu HP. et al. A Novel Multiresolution-Statistical Texture Analysis Architecture: Radiomics-Aided Diagnosis of PDAC Based on Plain CT Images. Ieee T Med Imaging. 2021;40:12-25
117. Qureshi TA, Gaddam S, Wachsman AM, Wang L, Azab L, Asadpour V. et al. Predicting pancreatic ductal adenocarcinoma using artificial intelligence analysis of pre-diagnostic computed tomography images. Cancer Biomark. 2022;33:211-7
118. Ebrahimian S, Singh R, Netaji A, Madhusudhan KS, Homayounieh F, Primak A. et al. Characterization of Benign and Malignant Pancreatic Lesions with DECT Quantitative Metrics and Radiomics. Acad Radiol. 2022;29:705-13
119. Chakraborty J, Midya A, Gazit L, Attiyeh M, Langdon-Embry L, Allen PJ. et al. CT radiomics to predict high-risk intraductal papillary mucinous neoplasms of the pancreas. Med Phys. 2018;45:5019-29
120. Chu LC, Park S, Kawamoto S, Fouladi DF, Shayesteh S, Zinreich ES. et al. Utility of CT Radiomics Features in Differentiation of Pancreatic Ductal Adenocarcinoma From Normal Pancreatic Tissue. Am J Roentgenol. 2019;213:349-57
121. Mukherjee S, Patra A, Khasawneh H, Korfiatis P, Rajamohan N, Suman G. et al. Radiomics-Based Machine-Learning Models Can Detect Pancreatic Cancer on Prediagnostic CTs at a Substantial Lead Time Prior to Clinical Diagnosis. Gastroenterology. 2022
122. Polk SL, Choi JW, McGettigan MJ, Rose T, Ahmed A, Kim J. et al. Multiphase computed tomography radiomics of pancreatic intraductal papillary mucinous neoplasms to predict malignancy. World J Gastroentero. 2020;26:3458-71
123. Ikeda M, Ito S, Ishigaki T, Yamauchi K. Evaluation of a neural network classifier for pancreatic masses based on CT findings. Comput Med Imag Grap. 1997;21:175-83
124. Chen HY, Deng XY, Pan Y, Chen JY, Liu YY, Chen WJ. et al. Pancreatic Serous Cystic Neoplasms and Mucinous Cystic Neoplasms: Differential Diagnosis by Combining Imaging Features and Enhanced CT Texture Analysis. Front Oncol. 2021;11:745001
125. Ren S, Zhao R, Cui W, Qiu W, Guo K, Cao Y. et al. Computed Tomography-Based Radiomics Signature for the Preoperative Differentiation of Pancreatic Adenosquamous Carcinoma From Pancreatic Ductal Adenocarcinoma. Front Oncol. 2020;10:1618
126. Xie T, Wang X, Zhang Z, Zhou Z. CT-Based Radiomics Analysis for Preoperative Diagnosis of Pancreatic Mucinous Cystic Neoplasm and Atypical Serous Cystadenomas. Front Oncol. 2021;11:621520
127. Li J, Liu F, Fang X, Cao K, Meng Y, Zhang H. et al. CT Radiomics Features in Differentiation of Focal-Type Autoimmune Pancreatitis from Pancreatic Ductal Adenocarcinoma: A Propensity Score Analysis. Acad Radiol. 2022;29:358-66
128. Ziegelmayer S, Kaissis G, Harder F, Jungmann F, Müller T, Makowski M. et al. Deep Convolutional Neural Network-Assisted Feature Extraction for Diagnostic Discrimination and Feature Visualization in Pancreatic Ductal Adenocarcinoma (PDAC) versus Autoimmune Pancreatitis (AIP). J Clin Med. 2020 9
129. Yang J, Guo X, Ou X, Zhang W, Ma X. Discrimination of Pancreatic Serous Cystadenomas From Mucinous Cystadenomas With CT Textural Features: Based on Machine Learning. Front Oncol. 2019;9:494
130. Panda A, Korfiatis P, Suman G, Garg SK, Polley EC, Singh DP. et al. Two-stage deep learning model for fully automated pancreas segmentation on computed tomography: Comparison with intra-reader and inter-reader reliability at full and reduced radiation dose on an external dataset. Med Phys. 2021;48:2468-81
131. Mahmoudi T, Kouzahkanan ZM, Radmard AR, Kafieh R, Salehnia A, Davarpanah AH. et al. Segmentation of pancreatic ductal adenocarcinoma (PDAC) and surrounding vessels in CT images using deep convolutional neural networks and texture descriptors. Sci Rep-Uk. 2022;12:3092
132. Huang B, Lin X, Shen J, Chen X, Chen J, Li ZP. et al. Accurate and Feasible Deep Learning Based Semi-Automatic Segmentation in CT for Radiomics Analysis in Pancreatic Neuroendocrine Neoplasms. Ieee J Biomed Health. 2021;25:3498-506
133. Lim SH, Kim YJ, Park YH, Kim D, Kim KG, Lee DH. Automated pancreas segmentation and volumetry using deep neural network on computed tomography. Diagnostics. 2022;12:4075
134. Boers TGW, Hu Y, Gibson E, Barratt DC, Bonmati E, Krdzalic J. et al. Interactive 3D U-net for the segmentation of the pancreas in computed tomography scans. Physics in Medicine & Biology. 2020;65:65002
135. Xie L, Yu Q, Zhou Y, Wang Y, Fishman EK, Yuille AL. Recurrent Saliency Transformation Network for Tiny Target Segmentation in Abdominal CT Scans. Ieee T Med Imaging. 2020;39:514-25
136. Zhou D, Nakamura M, Mukumoto N, Yoshimura M, Mizowaki T. Development of a deep learning-based patient-specific target contour prediction model for markerless tumor positioning. Med Phys. 2022;49:1382-90
137. Abel L, Wasserthal J, Weikert T, Sauter AW, Nesic I, Obradovic M. et al. Automated Detection of Pancreatic Cystic Lesions on CT Using Deep Learning. Plos One. 2021 11
138. Lyu P, Neely B, Solomon J, Rigiroli F, Ding Y, Schwartz FR. et al. Effect of deep learning image reconstruction in the prediction of resectability of pancreatic cancer: Diagnostic performance and reader confidence. Eur J Radiol. 2021;141:109825
139. Chang N, Cui L, Luo Y, Chang Z, Yu B, Liu Z. Development and multicenter validation of a CT-based radiomics signature for discriminating histological grades of pancreatic ductal adenocarcinoma. Quant Imag Med Surg. 2020;10:692-702
140. Luo Y, Chen X, Chen J, Song C, Shen J, Xiao H. et al. Preoperative Prediction of Pancreatic Neuroendocrine Neoplasms Grading Based on Enhanced Computed Tomography Imaging: Validation of Deep Learning with a Convolutional Neural Network. Neuroendocrinology. 2020;110:338-50
141. Ohira S, Koike Y, Akino Y, Kanayama N, Wada K, Ueda Y. et al. Improvement of image quality for pancreatic cancer using deep learning-generated virtual monochromatic images: Comparison with single-energy computed tomography. Physica Medica. 2021;85:8-14
142. Noda Y, Kawai N, Nagata S, Nakamura F, Mori T, Miyoshi T. et al. Deep learning image reconstruction algorithm for pancreatic protocol dual-energy computed tomography: image quality and quantification of iodine concentration. Eur Radiol. 2022;32:384-94
143. Gao J, Han F, Wang X, Duan S, Zhang J. Multi-Phase CT-Based Radiomics Nomogram for Discrimination Between Pancreatic Serous Cystic Neoplasm From Mucinous Cystic Neoplasm. Front Oncol. 2021;11:699812
144. Vachiranubhap B, Kim YH, Balci NC, Semelka RC. Magnetic resonance imaging of adenocarcinoma of the pancreas. Topics in Magnetic Resonance Imaging. 2009;20:3-9
145. Hargreaves BA, Worters PW, Pauly KB, Pauly JM, Koch KM, Gold GE. Metal-induced artifacts in MRI. Am J Roentgenol. 2011;197:547-55
146. Li J, Feng C, Lin X, Qian X. Utilizing GCN and Meta-Learning Strategy in Unsupervised Domain Adaptation for Pancreatic Cancer Segmentation. Ieee J Biomed Health. 2022;26:79-89
147. Chen X, Chen Z, Li J, Zhang YD, Lin X, Qian X. Model-Driven Deep Learning Method for Pancreatic Cancer Segmentation Based on Spiral-Transformation. Ieee T Med Imaging. 2022;41:75-87
148. Liang Y, Schott D, Zhang Y, Wang Z, Nasief H, Paulson E. et al. Auto-segmentation of pancreatic tumor in multi-parametric MRI using deep convolutional neural networks. Radiother Oncol. 2020;145:193-200
149. Goldenberg JM, Cárdenas-Rodríguez J, Pagel MD. Machine learning improves classification of preclinical models of pancreatic cancer with chemical exchange saturation transfer MRI. Magn Reson Med. 2019;81:594-601
150. Cui S, Tang T, Su Q, Wang Y, Shu Z, Yang W. et al. Radiomic nomogram based on MRI to predict grade of branching type intraductal papillary mucinous neoplasms of the pancreas: a multicenter study. Cancer Imaging. 2021;21:26
151. Corral JE, Hussein S, Kandel P, Bolan CW, Bagci U, Wallace MB. Deep Learning to Classify Intraductal Papillary Mucinous Neoplasms Using Magnetic Resonance Imaging. Pancreas. 2019;48:805-10
152. Cheng S, Shi H, Lu M, Wang C, Duan S, Xu Q. et al. Radiomics Analysis for Predicting Malignant Potential of Intraductal Papillary Mucinous Neoplasms of the Pancreas: Comparison of CT and MRI. Acad Radiol. 2022;29:367-75
153. Fonti R, Conson M, Del VS. PET/CT in radiation oncology. Semin Oncol. 2019;46:202-9
154. Blodgett TM, Meltzer CC, Townsend DW. PET/CT: form and function. Radiology. 2007;242:360-85
155. Yokoyama Y, Nagino M, Hiromatsu T, Yuasa N, Oda K, Arai T. et al. Intense PET signal in the degenerative necrosis superimposed on chronic pancreatitis. Pancreas. 2005;31:192-4
156. Feldman MK, Gandhi NS. Imaging Evaluation of Pancreatic Cancer. Surg Clin N Am. 2016;96:1235-56
157. Liu Z, Li M, Zuo C, Yang Z, Yang X, Ren S. et al. Radiomics model of dual-time 2-[(18)F]FDG PET/CT imaging to distinguish between pancreatic ductal adenocarcinoma and autoimmune pancreatitis. Eur Radiol. 2021;31:6983-91
158. Zhang Y, Cheng C, Liu Z, Wang L, Pan G, Sun G. et al. Radiomics analysis for the differentiation of autoimmune pancreatitis and pancreatic ductal adenocarcinoma in (18) F-FDG PET/CT. Med Phys. 2019;46:4520-30
159. Xing H, Hao Z, Zhu W, Sun D, Ding J, Zhang H. et al. Preoperative prediction of pathological grade in pancreatic ductal adenocarcinoma based on (18)F-FDG PET/CT radiomics. Ejnmmi Res. 2021;11:19
160. Elemento O, Leslie C, Lundin J, Tourassi G. Artificial intelligence in cancer research, diagnosis and therapy. Nat Rev Cancer. 2021;21:747-52
161. Yang J, Xu R, Wang C, Qiu J, Ren B, You L. Early screening and diagnosis strategies of pancreatic cancer: a comprehensive review. Cancer Commun. 2021;41:1257-74
162. Nicola M, Onorati M, Albertoni MM, Bianchi CL, De Nucci G, Mandelli ED. et al. Fine Needle Aspiration versus Fine Needle Biopsy of Biliopancreatic Lesions: Are They Really Opposing Techniques or Can They Be Complementary? Our Experience in a Large Cohort of Cases from a Single Institution. Acta Cytol. 2021;65:40-7
163. Song JW, Lee JH, Choi JH, Chun SJ. Automatic differential diagnosis of pancreatic serous and mucinous cystadenomas based on morphological features. Comput Biol Med. 2013;43:1-15
164. Song JW, Lee JH. New morphological features for grading pancreatic ductal adenocarcinomas. Biomed Res Int. 2013;2013:175271
165. Kriegsmann M, Kriegsmann K, Steinbuss G, Zgorzelski C, Kraft A, Gaida MM. Deep Learning in Pancreatic Tissue: Identification of Anatomical Structures, Pancreatic Intraepithelial Neoplasia, and Ductal Adenocarcinoma. International Journal of Molecular Sciences. 2021 22
166. Niazi MKK, Tavolara TE, Arole V, Hartman DJ, Pantanowitz L, Gurcan MN. Identifying tumor in pancreatic neuroendocrine neoplasms from Ki67 images using transfer learning. Plos One. 2018;13:e195621
167. Momeni-Boroujeni A, Yousefi E, Somma J. Computer-assisted cytologic diagnosis in pancreatic FNA: An application of neural networks to image analysis. Cancer Cytopathol. 2017;125:926-33
168. Naito Y, Tsuneki M, Fukushima N, Koga Y, Higashi M, Notohara K. et al. A deep learning model to detect pancreatic ductal adenocarcinoma on endoscopic ultrasound-guided fine-needle biopsy. Sci Rep-Uk. 2021;11:8454
169. Kurita Y, Kuwahara T, Hara K, Mizuno N, Okuno N, Matsumoto S. et al. Diagnostic ability of artificial intelligence using deep learning analysis of cyst fluid in differentiating malignant from benign pancreatic cystic lesions. Sci Rep-Uk. 2019;9:6893
170. Singhi AD, Koay EJ, Chari ST, Maitra A. Early Detection of Pancreatic Cancer: Opportunities and Challenges. Gastroenterology. 2019;156:2024-40
171. Bhinder B, Gilvary C, Madhukar NS, Elemento O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 2021;11:900-15
172. Iovanna J. Implementing biological markers as a tool to guide clinical care of patients with pancreatic cancer. Transl Oncol. 2021;14:100965
173. Chen C, Zong S, Liu Y, Wang Z, Zhang Y, Chen B. et al. Profiling of Exosomal Biomarkers for Accurate Cancer Identification: Combining DNA-PAINT with Machine- Learning-Based Classification. Small. 2019;15:e1901014
174. Zheng H, Zhao J, Wang X, Yan S, Chu H, Gao M. et al. Integrated Pipeline of Rapid Isolation and Analysis of Human Plasma Exosomes for Cancer Discrimination Based on Deep Learning of MALDI-TOF MS Fingerprints. Anal Chem. 2022;94:1831-9
175. Ko J, Bhagwat N, Yee SS, Ortiz N, Sahmoud A, Black T. et al. Combining Machine Learning and Nanofluidic Technology To Diagnose Pancreatic Cancer Using Exosomes. Acs Nano. 2017;11:11182-93
176. Gao H, Zheng Z, Yue Z, Liu F, Zhou L, Zhao X. Evaluation of serum diagnosis of pancreatic cancer by using surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Int J Mol Med. 2012;30:1061-8
177. Yu Y, Chen S, Wang LS, Chen WL, Guo WJ, Yan H. et al. Prediction of pancreatic cancer by serum biomarkers using surface-enhanced laser desorption/ionization-based decision tree classification. Cancer Biomark. 2005;68:79-86
178. Yang Y, Chen H, Wang D, Luo W, Zhu B, Zhang Z. Diagnosis of pancreatic carcinoma based on combined measurement of multiple serum tumor markers using artificial neural network analysis. Chinese Med J-Peking. 2014;127:1891-6
179. Qiao ZM, Ge JL, He WP, Xu XY, He JX. Artificial Intelligence Algorithm-Based Computerized Tomography Image Features Combined with Serum Tumor Markers for Diagnosis of Pancreatic Cancer. Comput Math Method M. 2022. 2022
180. Alizadeh Savareh B, Asadzadeh Aghdaie H, Behmanesh A, Bashiri A, Sadeghi A, Zali M. et al. A machine learning approach identified a diagnostic model for pancreatic cancer through using circulating microRNA signatures. Pancreatology. 2020;20:1195-204
181. Yu S, Li Y, Liao Z, Wang Z, Wang Z, Li Y. et al. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut. 2020;69:540-50
182. Almeida PP, Cardoso CP, de Freitas LM. PDAC-ANN: an artificial neural network to predict pancreatic ductal adenocarcinoma based on gene expression. Bmc Cancer. 2020;20:82
183. Hayashi A, Hong J, Iacobuzio-Donahue CA. The pancreatic cancer genome revisited. Nat Rev Gastro Hepat. 2021;18:469-81
184. Wang Y, Liu K, Ma Q, Tan Y, Du W, Lv Y. et al. Pancreatic cancer biomarker detection by two support vector strategies for recursive feature elimination. Biomark Med. 2019;13:105-21
185. Ko S, Choi J, Ahn J. GVES: machine learning model for identification of prognostic genes with a small dataset. Sci Rep-Uk. 2021;11:439
186. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21:22-36
187. Peng J, Sun BF, Chen CY, Zhou JY, Chen YS, Chen H. et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019;29:725-38
188. Xuan P, Sun H, Wang X, Zhang T, Pan S. Inferring the Disease-Associated miRNAs Based on Network Representation Learning and Convolutional Neural Networks. Int J Mol Sci. 2019 20
189. Mori Y, Yokota H, Hoshino I, Iwatate Y, Wakamatsu K, Uno T. et al. Deep learning-based gene selection in comprehensive gene analysis in pancreatic cancer. Sci Rep-Uk. 2021;11:16521
190. Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER, Kalocsay M. et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell. 2020;180:387-402
191. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487-514
192. Dai J, Su Y, Zhong S, Cong L, Liu B, Yang J. et al. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct Tar. 2020;5:145
193. Ariston GA, Wang F, Jiao Q, Yvette U, Yang X, Al-Ameri SA. et al. The involvement of exosomes in the diagnosis and treatment of pancreatic cancer. Mol Cancer. 2020;19:132
194. Connor AA, Gallinger S. Pancreatic cancer evolution and heterogeneity: integrating omics and clinical data. Nat Rev Cancer. 2022;22:131-42
195. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2017; 32: 185-203.
196. Cao L, Huang C, Cui ZD, Hu Y, Lih TM, Savage SR. et al. Proteogenomic characterization of pancreatic ductal adenocarcinoma. Cell. 2021;184:5031-52
197. Yang Z, LaRiviere MJ, Ko J, Till JE, Christensen T, Yee SS. et al. A Multianalyte Panel Consisting of Extracellular Vesicle miRNAs and mRNAs, cfDNA, and CA19-9 Shows Utility for Diagnosis and Staging of Pancreatic Ductal Adenocarcinoma. Clin Cancer Res. 2020;26:3248-58
198. Sinkala M, Mulder N, Martin D. Machine Learning and Network Analyses Reveal Disease Subtypes of Pancreatic Cancer and their Molecular Characteristics. Sci Rep-Uk. 2020;10:1212
199. Zhang H, Zhao L, Jiang J, Zheng J, Yang L, Li Y. et al. Multiplexed nanomaterial-assisted laser desorption/ionization for pan-cancer diagnosis and classification. Nat Commun. 2022;13:617
200. Cheng KS, Pan R, Pan H, Li B, Meena SS, Xing H. et al. ALICE: a hybrid AI paradigm with enhanced connectivity and cybersecurity for a serendipitous encounter with circulating hybrid cells. Theranostics. 2020;10:11026-48
201. Stark AP, Sacks GD, Rochefort MM, Donahue TR, Reber HA, Tomlinson JS. et al. Long-term survival in patients with pancreatic ductal adenocarcinoma. Surgery. 2016;159:1520-7
202. Hruban RH, Gaida MM, Thompson E, Hong S, Noë M, Brosens LAA. et al. Why is pancreatic cancer so deadly? The pathologist's view. J Pathol. 2019;248:131-41
203. Dal Molin M, Zhang M, de Wilde RF, Ottenhof NA, Rezaee N, Wolfgang CL. et al. Very Long-term Survival Following Resection for Pancreatic Cancer Is Not Explained by Commonly Mutated Genes: Results of Whole-Exome Sequencing Analysis. Clin Cancer Res. 2015;21:1944-50
204. Iwatate Y, Hoshino I, Yokota H, Ishige F, Itami M, Mori Y. et al. Radiogenomics for predicting p53 status, PD-L1 expression, and prognosis with machine learning in pancreatic cancer. Brit J Cancer. 2020;123:1253-61
205. Meng Y, Zhang H, Li Q, Xing P, Liu F, Cao K. et al. Noncontrast Magnetic Resonance Radiomics and Multilayer Perceptron Network Classifier: An approach for Predicting Fibroblast Activation Protein Expression in Patients With Pancreatic Ductal Adenocarcinoma. J Magn Reson Imaging. 2021;54:1432-43
206. Iwatate Y, Yokota H, Hoshino I, Ishige F, Kuwayama N, Itami M. et al. Machine learning with imaging features to predict the expression of ITGAV, which is a poor prognostic factor derived from transcriptome analysis in pancreatic cancer. Int J Oncol. 2022 60
207. Xu W, Liu Y, Lu Z, Jin ZD, Hu YH, Yu JG. et al. A new endoscopic ultrasonography image processing method to evaluate the prognosis for pancreatic cancer treated with interstitial brachytherapy. World J Gastroentero. 2013;19:6479-84
208. Toyama Y, Hotta M, Motoi F, Takanami K, Minamimoto R, Takase K. Prognostic value of FDG-PET radiomics with machine learning in pancreatic cancer. Sci Rep-Uk. 2020;10:17024
209. Lee JW, Park SH, Ahn H, Lee SM, Jang SJ. Predicting Survival in Patients with Pancreatic Cancer by Integrating Bone Marrow FDG Uptake and Radiomic Features of Primary Tumor in PET/CT. Cancers. 2021 13
210. Zhang Y, Lobo-Mueller EM, Karanicolas P, Gallinger S, Haider MA, Khalvati F. Improving prognostic performance in resectable pancreatic ductal adenocarcinoma using radiomics and deep learning features fusion in CT images. Sci Rep-Uk. 2021;11:1378
211. Park S, Sham JG, Kawamoto S, Blair AB, Rozich N, Fouladi DF. et al. CT Radiomics-Based Preoperative Survival Prediction in Patients With Pancreatic Ductal Adenocarcinoma. Am J Roentgenol. 2021;217:1104-12
212. Xie T, Wang X, Li M, Tong T, Yu X, Zhou Z. Pancreatic ductal adenocarcinoma: a radiomics nomogram outperforms clinical model and TNM staging for survival estimation after curative resection. Eur Radiol. 2020;30:2513-24
213. Hsu TH, Schawkat K, Berkowitz SJ, Wei JL, Makoyeva A, Legare K. et al. Artificial intelligence to assess body composition on routine abdominal CT scans and predict mortality in pancreatic cancer- A recipe for your local application. Eur J Radiol. 2021;142:109834
214. Chakraborty J, Langdon-Embry L, Cunanan KM, Escalon JG, Allen PJ, Lowery MA. et al. Preliminary study of tumor heterogeneity in imaging predicts two year survival in pancreatic cancer patients. Plos One. 2017;12:e188022
215. Walczak S, Velanovich V. An Evaluation of Artificial Neural Networks in Predicting Pancreatic Cancer Survival. J Gastrointest Surg. 2017;21:1606-12
216. Aronsson L, Andersson R, Ansari D. Artificial neural networks versus LASSO regression for the prediction of long-term survival after surgery for invasive IPMN of the pancreas. Eur J Cancer. 2021;16:e249206
217. Hayward J, Alvarez SA, Ruiz C, Sullivan M, Tseng J, Whalen G. Machine learning of clinical performance in a pancreatic cancer database. Artif Intell Med. 2010;49:187-95
218. Yokoyama S, Hamada T, Higashi M, Matsuo K, Maemura K, Kurahara H. et al. Predicted Prognosis of Patients with Pancreatic Cancer by Machine Learning. Clin Cancer Res. 2020;26:2411-21
219. Winter C, Kristiansen G, Kersting S, Roy J, Aust D, Knösel T. et al. Google goes cancer: improving outcome prediction for cancer patients by network-based ranking of marker genes. Plos Comput Biol. 2012;8:e1002511
220. Tang B, Chen Y, Wang Y, Nie J. A Wavelet-Based Learning Model Enhances Molecular Prognosis in Pancreatic Adenocarcinoma. Biomed Res Int. 2021;2021:7865856
221. Baek B, Lee H. Prediction of survival and recurrence in patients with pancreatic cancer by integrating multi-omics data. Sci Rep-Uk. 2020;10:18951
222. Li X, Yang L, Yuan Z, Lou J, Fan Y, Shi A. et al. Multi-institutional development and external validation of machine learning-based models to predict relapse risk of pancreatic ductal adenocarcinoma after radical resection. J Transl Med. 2021;19:281
223. He M, Chen X, Wels M, Lades F, Li Y, Liu Z. et al. Computed Tomography-based Radiomics Evaluation of Postoperative Local Recurrence of Pancreatic Ductal Adenocarcinoma. Acad Radiol. 2022
224. Li X, Wan Y, Lou J, Xu L, Shi A, Yang L. et al. Preoperative recurrence prediction in pancreatic ductal adenocarcinoma after radical resection using radiomics of diagnostic computed tomography. Eclinicalmedicine. 2022;43:101215
225. An C, Li D, Li S, Li W, Tong T, Liu L. et al. Deep learning radiomics of dual-energy computed tomography for predicting lymph node metastases of pancreatic ductal adenocarcinoma. Eur J Nucl Med Mol I. 2022;49:1187-99
226. Liang X, Cai W, Liu X, Jin M, Ruan L, Yan S. A radiomics model that predicts lymph node status in pancreatic cancer to guide clinical decision making: A retrospective study. J Cancer. 2021;12:6050-7
227. Li K, Yao Q, Xiao J, Li M, Yang J, Hou W. et al. Contrast-enhanced CT radiomics for predicting lymph node metastasis in pancreatic ductal adenocarcinoma: a pilot study. Cancer Imaging. 2020;20:12
228. Bian Y, Guo S, Jiang H, Gao S, Shao C, Cao K. et al. Radiomics nomogram for the preoperative prediction of lymph node metastasis in pancreatic ductal adenocarcinoma. Cancer Imaging. 2022;22:4
229. Shi L, Wang L, Wu C, Wei Y, Zhang Y, Chen J. Preoperative Prediction of Lymph Node Metastasis of Pancreatic Ductal Adenocarcinoma Based on a Radiomics Nomogram of Dual-Parametric MRI Imaging. Front Oncol. 2022;12:927077
230. Zambirinis CP, Midya A, Chakraborty J, Chou JF, Zheng J, McIntyre CA. et al. Recurrence After Resection of Pancreatic Cancer: Can Radiomics Predict Patients at Greatest Risk of Liver Metastasis? Ann Surg Oncol. 2022;29:4962-74
231. Cos H, Li D, Williams G, Chininis J, Dai R, Zhang J. et al. Predicting Outcomes in Patients Undergoing Pancreatectomy Using Wearable Technology and Machine Learning: Prospective Cohort Study. J Med Internet Res. 2021;23:e23595
232. Facciorusso A, Del Prete V, Antonino M, Buccino VR, Muscatiello N. Response to repeat echoendoscopic celiac plexus neurolysis in pancreatic cancer patients: A machine learning approach. Pancreatology. 2019;19:866-72
233. Nasief H, Zheng C, Schott D, Hall W, Tsai S, Erickson B. et al. A machine learning based delta-radiomics process for early prediction of treatment response of pancreatic cancer. Npj Precis Oncol. 2019;3:25
234. Gregucci F, Fiorentino A, Mazzola R, Ricchetti F, Bonaparte I, Surgo A. et al. Radiomic analysis to predict local response in locally advanced pancreatic cancer treated with stereotactic body radiation therapy. Radiol Med. 2022;127:100-7
235. Parr E, Du Q, Zhang C, Lin C, Kamal A, McAlister J. et al. Radiomics-Based Outcome Prediction for Pancreatic Cancer Following Stereotactic Body Radiotherapy. Cancers. 2020 12
236. Simpson G, Spieler B, Dogan N, Portelance L, Mellon EA, Kwon D. et al. Predictive value of 0.35 T magnetic resonance imaging radiomic features in stereotactic ablative body radiotherapy of pancreatic cancer: A pilot study. Med Phys. 2020;47:3682-90
237. Simpson G, Jin W, Spieler B, Portelance L, Mellon E, Kwon D. et al. Predictive Value of Delta-Radiomics Texture Features in 0.35 Tesla Magnetic Resonance Setup Images Acquired During Stereotactic Ablative Radiotherapy of Pancreatic Cancer. Front Oncol. 2022;12:807725
238. Bian Y, Liu YF, Li J, Liu F, Fang X, Lu J. et al. Machine Learning for Computed Tomography Radiomics: Prediction of Tumor-Infiltrating Lymphocytes in Patients With Pancreatic Ductal Adenocarcinoma. Pancreas. 2022;51:549-58
239. Bian Y, Liu C, Li Q, Meng Y, Liu F, Zhang H. et al. Preoperative Radiomics Approach to Evaluating Tumor-Infiltrating CD8(+) T Cells in Patients With Pancreatic Ductal Adenocarcinoma Using Noncontrast Magnetic Resonance Imaging. J Magn Reson Imaging. 2022;55:803-14
240. Bian Y, Liu YF, Jiang H, Meng Y, Liu F, Cao K. et al. Machine learning for MRI radiomics: a study predicting tumor-infiltrating lymphocytes in patients with pancreatic ductal adenocarcinoma. Abdom Radiol. 2021;46:4800-16
241. Bodalal Z, Trebeschi S, Nguyen-Kim TDL, Schats W, Beets-Tan R. Radiogenomics: bridging imaging and genomics. Abdom Radiol. 2019;44:1960-84
242. Valsangkar NP, Bush DM, Michaelson JS, Ferrone CR, Wargo JA, Lillemoe KD. et al. N0/N1, PNL, or LNR? The effect of lymph node number on accurate survival prediction in pancreatic ductal adenocarcinoma. J Gastrointest Surg. 2013;17:257-66
243. Choi SB, Han HJ, Park P, Kim WB, Song TJ, Choi SY. Systematic review of the clinical significance of lymph node micrometastases of pancreatic adenocarcinoma following surgical resection. Pancreatology. 2017;17:342-9
244. de Geus S, Eskander MF, Kasumova GG, Ng SC, Kent TS, Mancias JD. et al. Stereotactic body radiotherapy for unresected pancreatic cancer: A nationwide review. Cancer-Am Cancer Soc. 2017;123:4158-67
245. Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321-30
246. Jamieson NB, Mohamed M, Oien KA, Foulis AK, Dickson EJ, Imrie CW. et al. The Relationship Between Tumor Inflammatory Cell Infiltrate and Outcome in Patients with Pancreatic Ductal Adenocarcinoma. Ann Surg Oncol. 2012;19:3581-90
247. Kong HH, Kim KW, Ko YS, Kim SC, Lee JH, Song KB. et al. Longitudinal Changes in Body Composition of Long-Term Survivors of Pancreatic Head Cancer and Factors Affecting the Changes. J Clin Med. 2021 10
248. Wu J, Liang C, Chen M, Su W. Association between tumor-stroma ratio and prognosis in solid tumor patients: a systematic review and meta-analysis. Oncotarget. 2016;7:68954-65
249. Meng Y, Zhang H, Li Q, Liu F, Fang X, Li J. et al. Magnetic Resonance Radiomics and Machine-learning Models: An Approach for Evaluating Tumor-stroma Ratio in Patients with Pancreatic Ductal Adenocarcinoma. Acad Radiol. 2022;29:523-35
250. Chen X, Lin X, Shen Q, Qian X. Combined Spiral Transformation and Model-Driven Multi-Modal Deep Learning Scheme for Automatic Prediction of TP53 Mutation in Pancreatic Cancer. Ieee T Med Imaging. 2021;40:735-47
251. Gao J, Chen X, Li X, Miao F, Fang W, Li B. et al. Differentiating TP53 Mutation Status in Pancreatic Ductal Adenocarcinoma Using Multiparametric MRI-Derived Radiomics. Front Oncol. 2021;11:632130
252. Xing F, Xie Y, Yang L. An Automatic Learning-Based Framework for Robust Nucleus Segmentation. Ieee T Med Imaging. 2016;35:550-66
253. Zhao W, Shen L, Han B, Yang Y, Cheng K, Toesca DAS. et al. Markerless Pancreatic Tumor Target Localization Enabled By Deep Learning. International Journal of Radiation Oncology*Biology*Physics. 2019;105:432-9
254. Zhang Y, Zhu S, Yuan Z, Li Q, Ding R, Bao X. et al. Risk factors and socio-economic burden in pancreatic ductal adenocarcinoma operation: a machine learning based analysis. Bmc Cancer. 2020;20:1161
255. Kenner BJ, Abrams ND, Chari ST, Field BF, Goldberg AE, Hoos WA. et al. Early Detection of Pancreatic Cancer: Applying Artificial Intelligence to Electronic Health Records. Pancreas. 2021;50:916-22
256. Malhotra A, Rachet B, Bonaventure A, Pereira SP, Woods LM. Can we screen for pancreatic cancer? Identifying a sub-population of patients at high risk of subsequent diagnosis using machine learning techniques applied to primary care data. Plos One. 2021;16:e251876
257. Roch AM, Mehrabi S, Krishnan A, Schmidt HE, Kesterson J, Beesley C. et al. Automated pancreatic cyst screening using natural language processing: a new tool in the early detection of pancreatic cancer. Hpb. 2015;17:447-53
258. Kim H, Kim Y, Han B, Jang JY, Kim Y. Clinically Applicable Deep Learning Algorithm Using Quantitative Proteomic Data. J Proteome Res. 2019;18:3195-202
259. Song Q, Su J. DSTG: deconvoluting spatial transcriptomics data through graph-based artificial intelligence. Brief Bioinform. 2021 22
260. Bagante F, Spolverato G, Ruzzenente A, Luchini C, Tsilimigras DI, Campagnaro T. et al. Artificial neural networks for multi-omics classifications of hepato-pancreato-biliary cancers: towards the clinical application of genetic data. Phys Medica. 2021;148:348-58
261. Ottens T, Barbieri S, Orton MR, Klaassen R, van Laarhoven H, Crezee H. et al. Deep learning DCE-MRI parameter estimation: Application in pancreatic cancer. Med Image Anal. 2022;80:102512
262. Li Z, Li Z, Chen Q, Ramos A, Zhang J, Boudreaux JP. et al. Detection of pancreatic cancer by convolutional-neural-network-assisted spontaneous Raman spectroscopy with critical feature visualization. Neural Networks. 2021;144:455-64
263. Jadhav S, Dmitriev K, Marino J, Barish M, Kaufman AE. 3D Virtual Pancreatography. Ieee T Vis Comput Gr. 2022;28:1457-68
264. Dmitriev K, Marino J, Baker K, Kaufman AE. Visual Analytics of a Computer-Aided Diagnosis System for Pancreatic Lesions. Ieee T Vis Comput Gr. 2021;27:2174-85
265. Kang JS, Lee C, Song W, Choo W, Lee S, Lee S. et al. Risk prediction for malignant intraductal papillary mucinous neoplasm of the pancreas: logistic regression versus machine learning. Sci Rep-Uk. 2020;10:20140
266. Suman G, Patra A, Korfiatis P, Majumder S, Chari ST, Truty MJ. et al. Quality gaps in public pancreas imaging datasets: Implications & challenges for AI applications. Pancreatology. 2021;21:1001-8
267. Niranjan SJ, Wenzel JA, Martin MY, Fouad MN, Vickers SM, Konety BR. et al. Perceived Institutional Barriers Among Clinical and Research Professionals: Minority Participation in Oncology Clinical Trials. Jco Oncol Pract. 2021;17:e666-75
268. Ma MA, Gutiérrez DE, Frausto JM, Al-Delaimy WK. Minority Representation in Clinical Trials in the United States: Trends Over the Past 25 Years. Mayo Clin Proc. 2021;96:264-6
269. Zhao B. Understanding Sources of Variation to Improve the Reproducibility of Radiomics. Front Oncol. 2021;11:633176
270. Yamashita R, Perrin T, Chakraborty J, Chou JF, Horvat N, Koszalka MA. et al. Radiomic feature reproducibility in contrast-enhanced CT of the pancreas is affected by variabilities in scan parameters and manual segmentation. Eur Radiol. 2020;30:195-205
271. Park JE, Park SY, Kim HJ, Kim HS. Reproducibility and Generalizability in Radiomics Modeling: Possible Strategies in Radiologic and Statistical Perspectives. Korean J Radiol. 2019;20:1124-37
272. Lin LI. A concordance correlation coefficient to evaluate reproducibility. Biometrics. 1989;45:255-68
273. Zwanenburg A. Radiomics in nuclear medicine: robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur J Nucl Med Mol I. 2019;46:2638-55
274. Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25:44-56
275. Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. Bmc Med. 2019;17:195
276. Papadimitroulas P, Brocki L, Chung NC, Marchadour W, Vermet F, Gaubert L. et al. Artificial intelligence: Deep learning in oncological radiomics and challenges of interpretability and data harmonization. Phys Medica. 2021;83:108-21
277. Teng QY, Liu Z, Song YQ, Han K, Lu Y. A survey on the interpretability of deep learning in medical diagnosis. Multimedia Syst. 2022;25:1-21
Author contact
![]() Corresponding author: Junchao Guo, Tel.: +86-010-69152600; Fax: +86-010-69152601; E-mail: guojccn.
Corresponding author: Junchao Guo, Tel.: +86-010-69152600; Fax: +86-010-69152601; E-mail: guojccn.
 Global reach, higher impact
Global reach, higher impact