13.3
Impact Factor
Theranostics 2022; 12(16):6848-6864. doi:10.7150/thno.73078 This issue Cite
Research Paper
Addressing antimicrobial resistance with the IDentif.AI platform: Rapidly optimizing clinically actionable combination therapy regimens against nontuberculous mycobacteria
1. Department of Pharmacy, Faculty of Science, National University of Singapore, Singapore 117545.
2. The Institute for Digital Medicine (WisDM), Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117456
3. The N.1 Institute for Health (N.1), National University of Singapore, Singapore 117456
4. Department of Biomedical Engineering, College of Design and Engineering, National University of Singapore, Singapore 117583
5. Cancer Science Institute of Singapore, National University of Singapore, Singapore 117599
6. Department of Pharmacology, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117600.
7. NUS Centre for Cancer Research (N2CR), Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117599
8. Bia-Echo Asia Centre for Reproductive Longevity and Equality (ACRLE), Yong Loo Lin School of Medicine, National University of Singapore, Singapore 117600
*Co-First Authors
Abstract
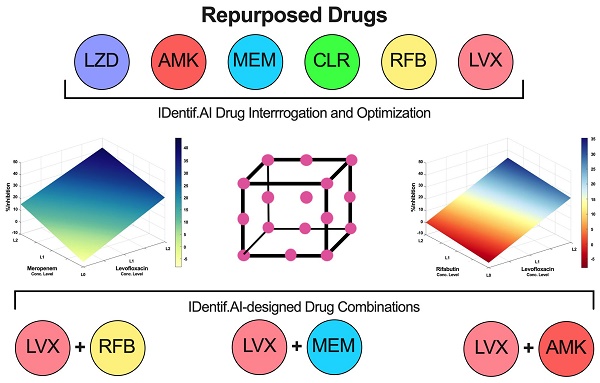
Background: Current standard of care (SOC) regimens against nontuberculous mycobacteria (NTM) usually result in unsatisfactory therapeutic responses, primarily due to multi-drug resistance and antibiotic susceptibility-guided therapies. In the midst of rising incidences in NTM infections, strategies to develop NTM-specific treatments have been explored and validated.
Methods: To provide an alternative approach to address NTM-specific treatment, IDentif.AI was harnessed to rapidly optimize and design effective combination therapy regimens against Mycobacterium abscessus (M. abscessus), the highly resistant and rapid growth species of NTM. IDentif.AI interrogated the drug interaction space from a pool of 6 antibiotics, and pinpointed multiple clinically actionable drug combinations. IDentif.AI-pinpointed actionable combinations were experimentally validated and their interactions were assessed using Bliss independence model and diagonal measurement of n-way drug interactions.
Results: Notably, IDentfi.AI-designed 3- and 4-drug combinations demonstrated greater %Inhibition efficacy than the SOC regimens. The platform also pinpointed two unique drug interactions (Levofloxacin (LVX)/Rifabutin (RFB) and LVX/Meropenem (MEM)) that may serve as the backbone of potential 3- and 4-drug combinations like LVX/MEM/RFB, which exhibited 58.33±4.99 %Inhibition efficacy against M. abscessus. Further analysis of LVX/RFB via Bliss independence model pointed to dose-dependent synergistic interactions in clinically actionable concentrations.
Conclusions: IDentif.AI-designed combinations may provide alternative regimen options to current SOC combinations that are often administered with Amikacin, which has been known to induce ototoxicity in patients. Furthermore, IDentif.AI pinpointed 2-drug interactions may also serve as the backbone for the development of other effective 3- and 4-drug combination therapies. The findings in this study suggest that this platform may contribute to NTM-specific drug development.
Keywords: Combination Therapy, Nontuberculous Mycobacteria, Mycobacterium abscessus, Artificial Intelligence, Infectious Disease
 Global reach, higher impact
Global reach, higher impact