13.3
Impact Factor
Theranostics 2022; 12(14):6160-6178. doi:10.7150/thno.70814 This issue Cite
Research Paper
The metabolic genomic atlas reveals potential drivers and clinically relevant insights into the etiology of esophageal squamous cell carcinoma
1. Department of Biochemistry and Biophysics, School of Basic Medical Sciences, Peking University Health Science Center, Beijing, China.
2. Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education/Beijing), Laboratory of Molecular Oncology, Peking University Cancer Hospital & Institute, Beijing 100142, China.
3. Institute of Cancer Research, Shenzhen Bay Laboratory, Shenzhen 518107, China.
4. Research Unit of Molecular Cancer Research, Chinese Academy of Medical Sciences, Beijing, China.
5. Department of Oncology, Cancer Institute, Peking University Shenzhen Hospital, Shenzhen Peking University-Hong Kong University of Science and Technology (PKU-HKUST) Medical Center, Shenzhen 518035, China.
6. Department of Medical Oncology, Sun Yat-Sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou, Guangdong, 510060, P. R. China.
7. BGI-Qingdao, BGI-Shenzhen, Qingdao, 266555, China.
8. Institute of Systems Biomedicine, School of Basic Medical Sciences, Peking University Health Science Center, Beijing, 100191, China.
9. SJTU-BGI Innovation Research Center, Shanghai, China. National Research Center for Translational Medicine, National Key Scientific Infrastructure for Translational Medicine (Shanghai), Shanghai Jiao Tong University, Shanghai, China.
#These authors contributed equally to this work.
Abstract
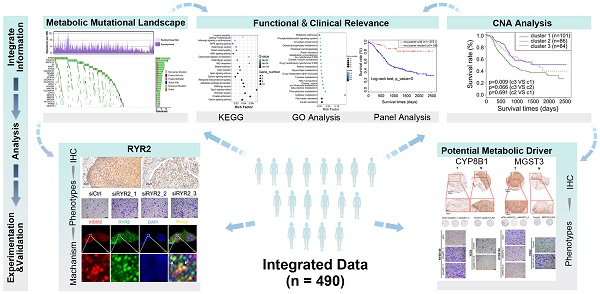
Background: Esophageal squamous cell carcinoma (ESCC) is one of the most common cancers globally, with a poor prognosis and ambiguous therapy target. As a hallmark of cancer, metabolism reprogramming plays a critical role in the development of ESCC; however, the genomic alterations underlying this reconfiguration are still largely unknown.
Methods: We have comprehensively studied the metabolic genomic variations in an integrated ESCC cohort of 490 patients and characterized the somatic alterations associated with various metabolic pathways.
Results: The somatic mutations and copy number alterations (CNAs) occurred heterogeneously in all patients. Using CNA-based clustering, we stratified patients into three clusters and Cluster3 with more deletions marked for worse prognosis. Our findings revealed detailed genetic alterations in components of metabolic pathways and highlighted the role of metal ion channel transporters and non-neuronal/neuronal synapse systems in the development of ESCC. We found a subset of potential metabolic drivers and functionally validated RYR2, MGST3, and CYP8B1 involved in the ESCC-associated malignancy. Another key finding was that we identified 27 metabolic genes with genomic alterations that could serve as independent prognostic factors and figured out two genetic panels that could stratify patients into distinct prognostic groups.
Conclusion: Collectively, our study provided a deep insight into the metabolic landscape in ESCC, extending our understanding of the metabolic reconfiguration underlying the genomic basis of ESCC. Furthermore, our findings revealed potential prognostic factors of ESCC, which are expected to contribute to the accurate determination of the prognosis in the clinic.
Keywords: ESCC, metabolic genomic atlas, somatic mutations, copy number alterations (CNAs), clinical implications
 Global reach, higher impact
Global reach, higher impact