13.3
Impact Factor
Theranostics 2022; 12(11):4980-4992. doi:10.7150/thno.73222 This issue Cite
Research Paper
Single-cell profiling of microenvironment components by spatial localization in pancreatic ductal adenocarcinoma
1. Department of Surgery, College of Medicine; University of Florida, Gainesville, FL 32610.
2. Department of Pathology, College of Medicine; University of Florida, Gainesville, FL 32610.
3. Department of Biostatistics, College of Medicine; University of Florida, Gainesville, FL 32610.
4. Department of Medicine, College of Medicine; University of Florida, Gainesville, FL 32610.
#These authors contributed equally to this work.
Received 2022-3-24; Accepted 2022-6-4; Published 2022-6-27
Abstract
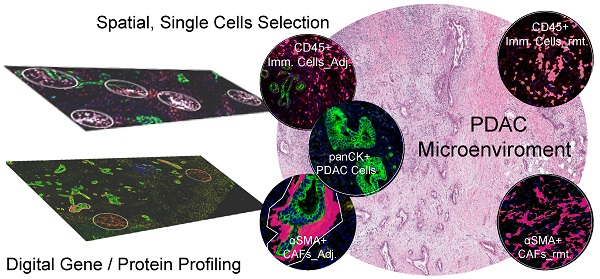
Rationale: The biology of the pancreatic ductal adenocarcinoma (PDAC) is heterogenous, but how heterogenity of the tumor microenvironment contributes to disparate patient outcomes remains essentially unstudied.
Methods: A strategy employing multiplex digital spatial profiling (mplxDSP) technology was employed to evaluate the nature and dynamics of microenvironment components including cancer associated fibroblasts (CAFs) and infiltrating immune cells at the single-cell level based upon their spatial relationship within the tumor.
Results: We report that myofibroblasts directly adjacent to PDAC tumors comparatively overexpress genes (BATF3, IL12B, ITGB8, CD4 and IFNAR1), constructing pathways prone to stimulating an adaptive immune response. Markers of innate immune cells (Natural Killer cells, Dendritic Cells and macrophages) are predominant in CD45+ cells immediately adjacent to PDAC tumor, however, the checkpoint protein CTLA4 is also overwhelmingly expressed, fostering tolerance. Finaly, mRNA profiling of adjacent CAFs identified clusters of genes that correlate with survival.
Conclusion: CAFs and leukocytes in close proximity to PDAC significantly differ from those remote from the tumor, providing insight into microenvironment influence on immune tolerance mediated through relative populations of leukocytes and subsets of CAFs and monocytes. mRNA expression profiling of CAFs adjacent to PDAC cells may hold promise for prognostication.
Keywords: pancreas, digital spatial profiling, tumor microenvironment, stromal interaction, immune tolerance
Introduction
There is a fascinating, relatively unique feature of pancreatic ductal adenocarcinoma (PDAC): its mass is usually comprised of less than 20% epithelial tumor cells, and is mostly comprised of surrounding stroma. This desmoplastic stromal reaction in PDAC is majorly attributed to the activation of quiescent stellate cells becoming alpha-smooth muscle actin (α-SMA) expressing myofibroblasts, often termed as cancer-associated fibroblasts (CAFs) or tumor-associated stromal (TAS) cells. Initial studies of the impact of α-SMA+ cells suggested they contributed to disease progression by promoting cell proliferation and invasion as well as drug resistance [1, 2]. However, this concept has recently been challenged as depletion of α-SMA+ myofibroblasts in genetically-engineered murine models of pancreas cancer accelerates tumor progression [3]; Genetic reduction or pharmacologic inhibition of sonic hedgehog (Shh) pathway resulted in reduced stromal content with more aggressive tumors in KPC mice [4, 5], and a similar phenomenon was simultaneously reported in type I collagen (Col1) deficient tumors [6]. We previously reported TAS cells express and deliver tumor suppressive miRNA-145 to neighboring PDAC cells [7], offering a potential mechanism for these observations. More recent studies identified distinct stromal types and subtypes of α-SMA+ cells suggesting that stromal heterogenicity associates with tumor development and progression [8-10]. For example, Hutton, et al. reported CD105pos pancreatic fibroblasts are typically more abundant in PDAC tumors and tumor permissive, whereas CD105neg fibroblasts suppress tumor growth in a manner dependent on adaptive immunity [11]. The heterogeneity of pancreatic fibroblast has raised attention in recent studies. Several research groups have reported different signature markers that could define pancreatic fibroblasts functions.
Yet, the nature and dynamics of α-SMA+ cells and their controversial role in PDAC tumor microenvironment remain poorly elucidated.
This fibrotic PDAC microenvironment is also highly immunosuppressive, conducive for tumor development and growth. In general, the PDAC microenvironment is equipped with upregulated tumor supporting cells [i.e., myeloid-derived suppressor cells (MDSCs), tumor-associated macrophages (TAMs), regulatory T cells (Tregs), etc.], while tumor destroying cells are downregulated [i.e., nature killer (NK) cells, CD8+ cells, etc.] [12]. Recent work from Carstens, et al. utilized a novel computational imaging technology for spatially-characterized, heterogeneous T lymphocytes subpopulations infiltration in PDAC. The study delivered an important message of utilizing spatial information to improve our understanding of the heterogenous tumor microenvironment with respect to its correlation with patient outcome, specifically for stratifying PDAC patients for effective immunotherapy [13].
In this study, we aimed to investigate how spatial relationships between the three major components of PDAC including PDAC cells, activated myofibroblasts (CAFs), and immune cells contributes to heterogeneity within those compartments using novel multiplex digital spatial profiling (mplxDSP) technology. We further hypothesized that spatial-driven analyses of these contributors to the microenvironment may offer prognostic value.
Methods
Patients
Four Patients of PDAC who underwent surgical resection at the University of Florida (UF) Department of Surgery were enrolled in this study following UF Institutional Review Board (IRB) approved protocols. All patients were white race with no smoking history. Pathological diagnosis was confirmed to be invasive, moderately differentiated PDAC. Main clinical features of these patients are summarized in Supplemental Table 1. Metastasis was absent in all patients at time of resection. None received neoadjuvant chemotherapy or radiation. Patients were selected and grouped based on the clinical outcomes: reported disease associated death within approx. 12 months after surgical resection due to recurrence (Rcr) and patients with prolonged overall survival longer than 36 months after surgical resection with no reported recurrence, grouped as no disease evidence (NDE).
Morphologic markers labeling and regions of interest (ROIs selection)
Formalin-fixed and paraffin-embedded (FFPE) tumor blocks were preserved at the time of surgical resection. One of the 5-µm serial sections from each tissue was stained for histological analysis of hematoxylin and eosin (HE) and used as reference for ROIs selection. DSP morphologic labeling for ROIs selection for three single cell ROIs using fluorescently labeled antibodies specific for total immune cells (anti-CD45), epithelial cell marker (anti-panCK), and (anti-αSMA), along with SYTO 13 DNA dye. A total of 24 ROIs for each cell types were defined and selected, except for tumor protein profiling, which used 12 ROIs. Examples are shown in Figures 1A-E and 3A-F. All ROIs were identified within serial sections from the same tumor core. Quality control was performed within the GeoMx DSP analysis.
mplxDSP transcripts and protein panel
To quantitate gene expression at mRNA level, a minimum of 300 µm surface area of one ROI were selected. The nanoString GeoMx® Immune Pathways RNA panel consists of 78 genes, including 73 target genes and 5 internal reference genes. This gene panel covers a number of cytokines and chemokine signaling molecules, key genes involved in tumorigenesis and tumor immunology (The complete gene list and annotations is listed in Suppl. Table S2, provided by nanoString, Sept 2019 MK1810). For measuring protein expression, we used the nanoString GeoMx® Immune Cell Profiling Panel, which consists of a panel of 21 proteins including 3 housekeeping proteins (Suppl. Table S3). High-resolution multiplex profiling for selected cells followed the manufacturer's instructions. Photocleaved probes were hybridized to nanoString optical barcodes for digital counting and quantified in nCounter platform. Final digital counts were normalized with internal spike-in controls to account for system variation and background was corrected using the negative probe controls.
Enhanced pathway Analysis
The differentially expressed genes identified between groups were input into the online software of Ingenuity Pathway Analysis (IPCA; www.integenuity.com) to curate gene interactions and correlation networks. A core analysis was employed to identify the most significant canonical pathways, molecular functions and network interactions. Pathways with p value <0.05 was considered statistcally significant.
Statistical Analysis
To identify differential expressed genes, we used the linear model followed by the empirical Bayes moderated t-statistics test (two tailed), which are implemented in the R software limma package [14]. We employed the Benjamini-Hochberg correction [15] to adjust for the multiple comparison and the false discovery rate (q-value) was reported. Unsupervised clustering analysis was performed using hierarchical clustering analysis. Dimension reduction was performed using principal component analysis. All statistical analyses were carried out using R (v. 4.1.1) software.
Results
RNA mplxDSP of spatially distributed αSMA+ CAFs in PDAC microenvironment
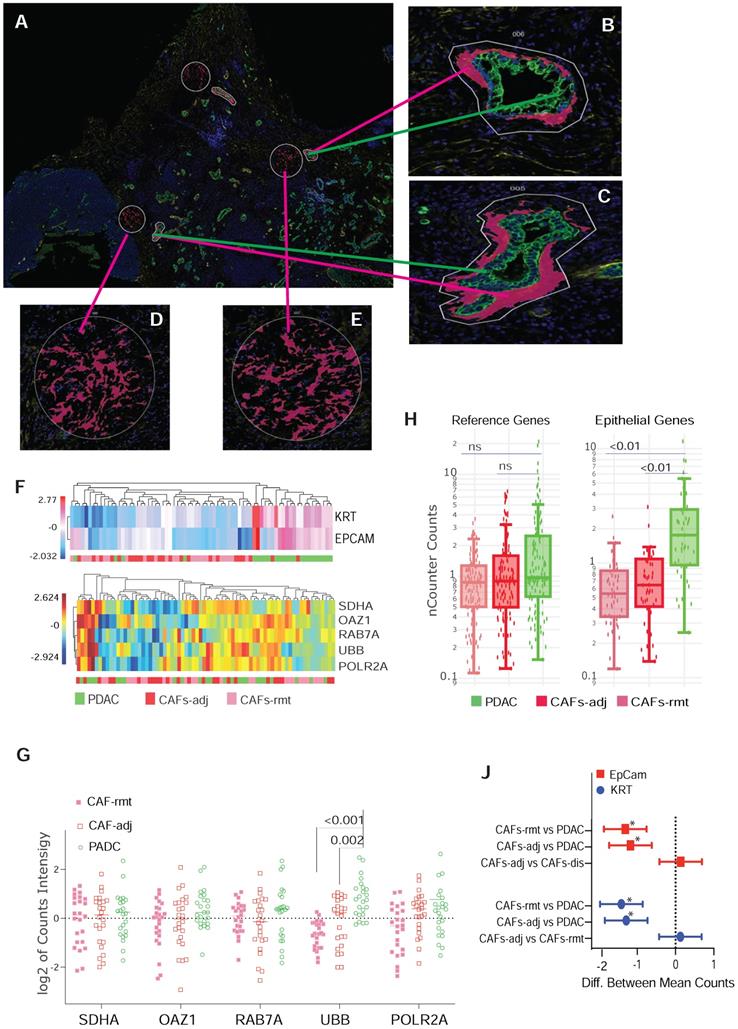
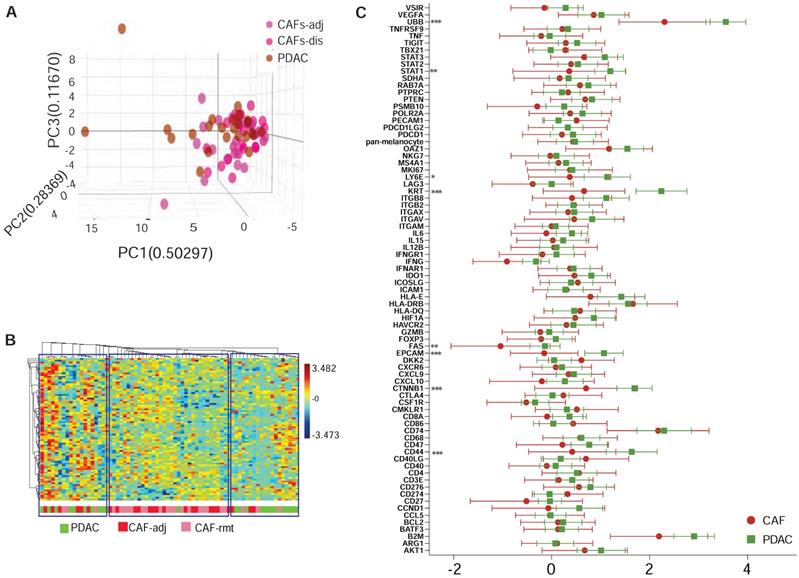
We previously observed that CAFs immediately adjacent to transformed epithelial cells expressed more intensive α-SMA immunostaining [16]. This observation was confirmed by others and contributed to the discovery of sub-populations of CAFs in the PDAC microenvironment by co-culture of murine PSCs and PDAC organoids [9]. Our goal here was to test the capacity of spatially resolved gene profiling for defining heterogeneously distributed CAFs using mplxDSP technology. ROIs of CAF-rmt is chosen to avoid immediately next to PDAC tumor, the mean distance of a cluster of CAFs-rmt to the nearest PDAC epithelial compartment is at approximal of 331.1 µm (SD = 66.03). We profiled and compared 78 immuno-oncogenes from CAFs located immediately adjacent to PDAC epithelial compartment (adjacent CAFs, CAFs-adj) and CAFs located remote from cancer cells (PDAC distal CAFs, CAFs-rmt). Twenty-four individual regions of interest (ROIs) of each CAFs-adj and CAFs-rmt (defined by morphology marker anti-α-SMA antibody labeling) along with 24 ROIs of PDAC cells (defined by morphology marker anti-panCK antibody labeling) were selected for profiling (Figure 1A-E). The 78 RNA multiplex panel consists of 73 target genes and 5 reference genes (Suppl. Table S2). Unsupervised clustering analysis demonstrated the correspondent expression of epithelial genes of EpCam and KRT separating panCK+ PDAC cells from CAFs, despite few PDAC ROIs also expressing low level of epithelial expression (Figure 1F, top panel). As expected, no obvious cluster was assembled by the expression of the panel of 5 reference genes among all samples (Figure 1F, bottom panel). No differential expression of any of these genes was noticed between the two locations of CAFs. However, among the five reference genes (SDHA, OAZ1, RAB7A, UBB and POLR2A, Figure 1G), UBB alone was found differentially expressed with significant higher level in PDAC cells compared to CAFs (mean differences between CAFs-adj and CAFs-rmt to PDAC are -0.957 (p = 0.002) and -1.477 (p < 0.001) respectively). These data are in line with reported UBB upregulation in cancers [17, 18]. We thus excluded UBB from the reference gene panel and the initial data were then normalized with the panel of the remaining four reference genes (of SDHA, OAZ1, RAB7A, and POLR2A) for further analysis. Based on the adjusted normalization, we observed no differential expression levels of total reference genes but distinguishable epithelial gene expressions between PDAC cells to CAFs-adj and CAFs-rmt (Figure 1H-J). Within the 73 target genes and 5 reference genes, 8 genes (approximal 10%) were detected with significantly higher expression levels in PDAC cells than CAFs (Suppl. Table S4 and figure 2C). Despite that this panel of 78 genes was not able to distinguish the two types of cells (Figure 2B), the hierarchical cluster analysis showed three dominate clusters: PDAC, CAFs and a mixture of both.
Protein mplxDSP of spatially distributed CD45+ immune cells in PDAC microenvironment
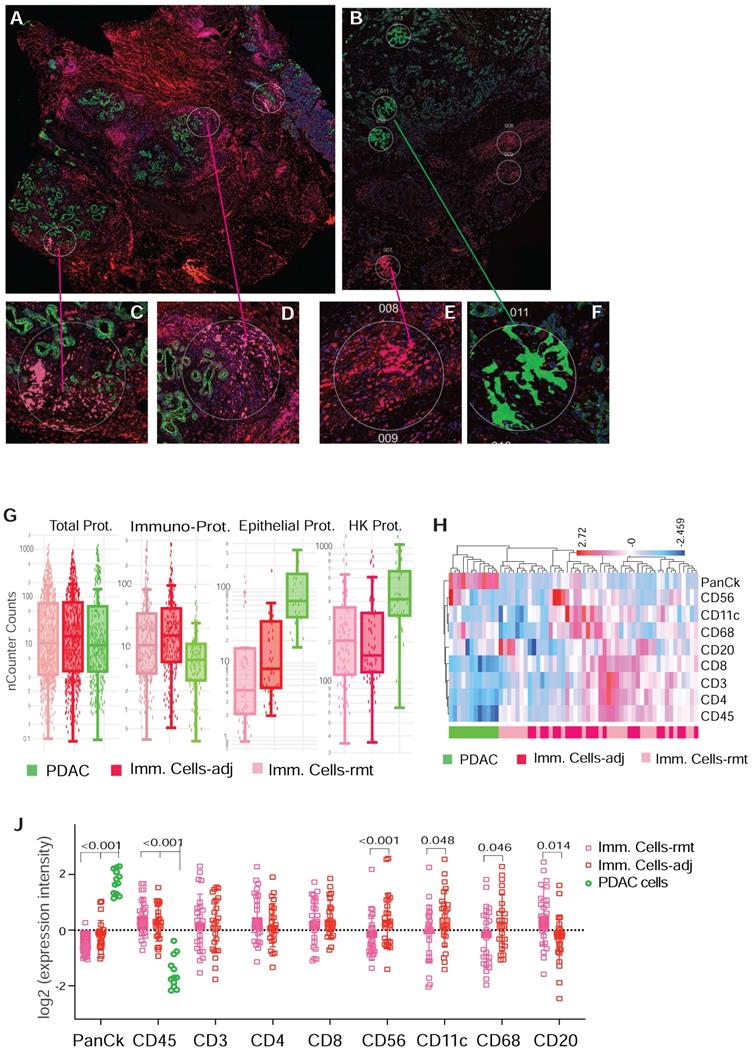
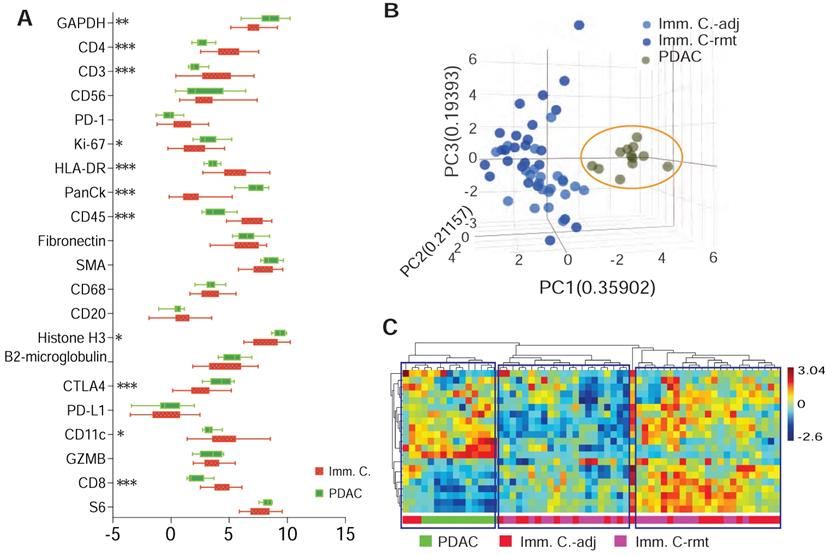
A computational method using immunohistochemistry spatial probing information showed that human PDAC contains a heterogeneous T-cell population [13]. We employed mplxDSP technology to further characterize immune cells in PDAC microenvironment. The mplxDSP immune-protein panel (Suppl. Table S3) includes markers for total immune cells (CD45), subtypes of T-cells (CD3, CD4, CD8), Dendric cells (CD11c), B cells (CD20), Natural Killer cells (CD56) and M2 macrophage cells (CD68). We again divided immune cells into two groups by their localization to tumor lesion: CD45+ immune cells in immediate vicinity of PDAC area (immune cells-adjacent, Imm. Cells-adj) and those with no direct contact with PDAC cells (immune cells-remote, Imm. Cells-rmt). For each subtype of immune cells, 24 ROIs were selected and profiled to determine the relative abundance of these populations of immune cells. PDAC cells as counterpart compartment were also tested from 12 selected ROIs (Figure 3A-F & Suppl. Figure S2). As expected, total nCount counts (expression level) of markers for immune cells are found higher in CD45+ immune cells, both in Imm. Cells-adj and Imm. Cells-rmt in comparison to PDAC cells, and vice versa, the total nCount counts (expression level) of the epithelial pan-CK are high in PDAC cells and low in all CD45+ immune cells (Figure 3G). However, it is noticed that total nCount counts for the three housekeeping proteins (Histone H3, S6 and GAPDH) were also significantly more intensive in PDAC cells in comparison to immune cells (Figure 3G). Given that the total nCount counts of all proteins are even in both PDAC and immune cells, normalization was then performed based on the total protein counts. Demonstrated in Figure 3H, ROI selection successfully clustered two distinct segmentations of PDAC cells from immune cells, yet not between Imm.Cells-adj and Imm. Cells-rmt. As we further compared the subtypes of immune cells by distance to tumor lesion, we also observed the increased protein levels of NK cell marker CD56 (p < 0.001) and decreased B cell marker CD20 (p = 0.014) in immune cells adjacent (Imm.Cells-adj) to PDAC compared to those in distant stroma (Imm. Cells-rmt). DC marker CD11c (p = 0.048) and M2 macrophage marker CD68 (p = 0.046) were also seen slightly increased in Imm.Cells-adj. No differential protein expression of T cell markers, neither CD3, CD4, nor CD8, were observed (Figure 3J). Among the 3 housekeeping proteins and 18 target proteins, over 60% of proteins (11/18) from the panel were differentially expressed between PDAC and immune cells (Figure 4A). A group of PDAC cells was thus clearly separated from immune cells as indicated by PCA (Figure 4B) and hierarchical cluster analysis (Figure 4C). Thus, comparison of the Imm. Cells-adj to Imm. Cells-rmt demonstrated these quantities of these proteins are similar between the two ROIs, despite that some cells appears having more intensive expression than the others (Figure 4C).
Leukopoiesis is enriched in CAFs adjacent to PDAC
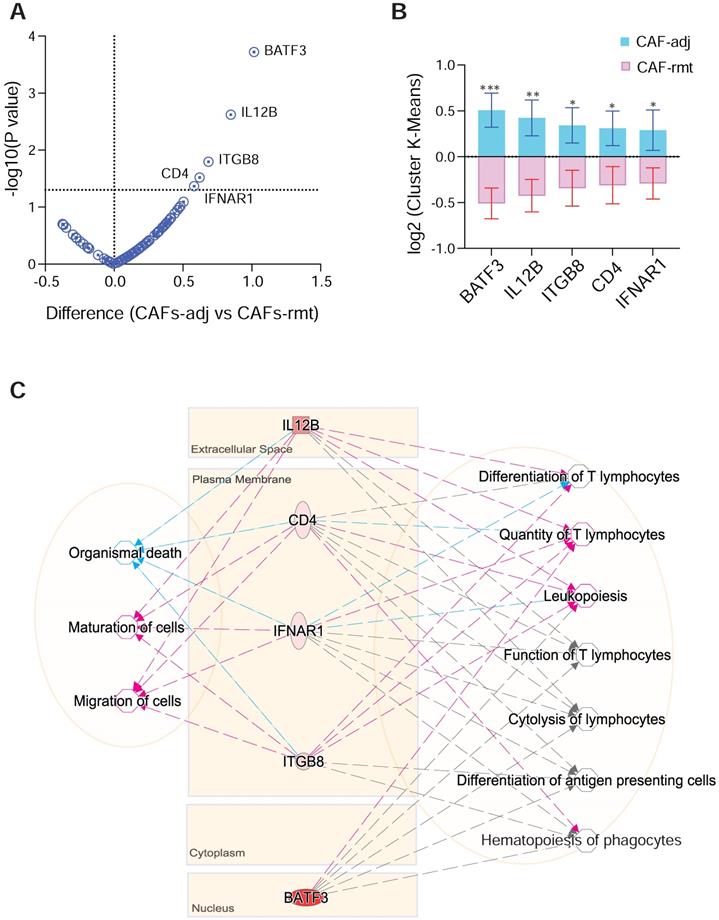
We next compared target genes involved in tumor-immune pathways using mpxDSP mRNA 73-multiplex panel between CAFs in PDAC-adjacent (CAFs-adj) and remote (CAFs-rmt) locations. The key molecules in this panel are involved in tumor-immune biological activities, such as cytokine and chemokine signaling, Wnt signaling, tumor proliferation and apoptosis, especially in immune pathways of antigen presentation and checkpoint, myeloid suppression and activation, T cell activation, as well as immune cell adhesion and migration. Five genes (BATF3, IL12b, ITGB8, CD4 and IFNAR1) were found to have increased expression levels in CAFs-adj comparing to CAFs-rmt (Figure 5A-B). Interestingly, genes of IL12b, ITGB8, CD4 and IFNAR1 all have reportedly similar function to decrease Organismal Death (p value = 3.88E-03) [19-21], but increase Maturation of Cells (p value = 2.94E-06) [22-24], and Migration of Cells (p value = 2.37E-03) [25]. The set of genes are also known affecting immune cells differentiation and function, highlighted as increased Quantify of T lymphocytes (p value = 6.42E-08) [26-29], Differentiation of T lymphocytes (p value = 1.61E-06), and Leukopoiesis (p value = 5.01E-07) [30-33]. Despite known involvement in Hematopoiesis of Phagocytes (p value = 1.42E-07) [32], Function of T lymphocytes (p value = 1.71E-07) [34], Differentiation of Antigen Presenting cells (p value = 1.78E-07) [35] and Cytolysis of lymphocytes (p value = 1.31E-09) [36], their precise role in these functions is yet not clear (Figure 5C).
Checkpoint protein CTLA4 overexpression in immune cells adjacent to PDAC
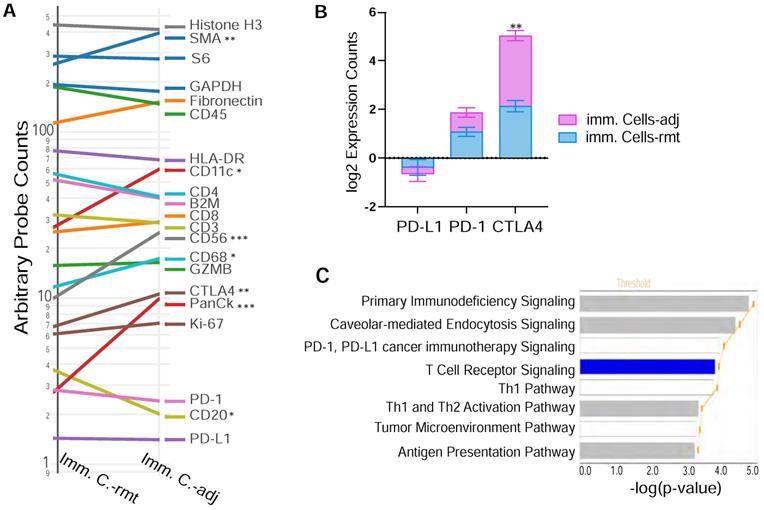
Figure 6A demonstrates the expression of 18 target proteins plus 3 housekeeping proteins in the nanoString™ immuno-oncology proteins panel from both immune cells adjacent and remote to PDAC cells. As shown by the trend comparison, the three housekeeping proteins (Histone H3, GAPDH and S6) together with the 2 stromal proteins (SMA and Fibronectin), as well as total immune cell marker CD45 are strongly expressed in both locations of immune cells adjacent (imm. C.-adj) and remote (imm. C.-rmt), with only SMA highly expressed in PDAC adjacent immune cells (p < 0.01). Level of pan-CK (a cocktail of anti-KRT1, KRT10, KRT2, KRT16, KRT5, KRT6B, KRT19, KRT6A, KRT8, KRT14, KRT3 antibodies) was also significantly higher in immune cells adjacent to PDAC (p < 0.001). As described above in Figure 6A, NK cell marker CD56 is expressed at higher levels in PDAC adjacent immune cells by trend analysis, but B cell marker protein CD20 is found expressed at lower levels in PDAC adjacent immune cells. Noticeably, checkpoint PD-L1 and PD-1 have relatively lower expression levels and are not differentially expressed between PDAC adjacent (imm. C.-adj) and remote (imm. C.-rmt) immune cells. However, checkpoint protein CTLA4 expression was found expressed at moderate levels and considerably higher in PDAC adjacent immune cells (imm. C.-adj) compared to remote immune cells (Figure 6A & 6B), which is also a major contributor to downregulated T cell receptor signaling pathway predicted by IPA (p = 1.29E-04).
ROIs selection and characterization for CAFs by locolization. (A-E) Representative ROIs selection using immunofluorescent staining for morphological markers. (B & C) Enlarged images of PDAC cells with panCK antibody labeling, green-colored and immediate adjacented CAFs (CAF-adj) labeled with α-SMA antibody, red-colored. (D & E) CAFs distant to tumor lesion (CAF-rmt) also labeled with α-SMA antibody, red-colored. (F) Dendrogram of unsupervised hierarchically clustered analysis of the expression of epithelial genes of Epcam and KRT in log2 scale (top panel) and the expression of reference genes SDHA, OAZ1, RAB7A, UBB and POLR2A (bottom panel). (G) Dot plot of the expression level of the 5 reference genes individually by two way ANOVA, y-axil indicates log2 scale of reading intensity; displayed numbers are p-value. (H) Box plot of non-parametric analysis of total nCount counts for the set of four reference genes (SDHA, OAZ1, RAB7A, and POLR2A) and the two epithelial genes (EpCan and panCK); displayed numbers are p-value, ns = not significant. (J) Line plot of 95% Confidence Intervals (CI) of 2-way ANOVA multiple comparison of between PDAC cells, CAF-adj and CAF-rmt for EpCam and KRT; * = p < 0.001.
Multiplex DSP for RNA profiling CAFs vs PDAC cells. (A) Comparison profiling of 78 RNA gene expression of CAF and PDAC cells, * = p < 0.05, ** = p < 0.01, *** = p < 0.001. (B) Tri-dimentional diagam showing the result of principal component analysis (PCA) using the panel of 78 genes, each dot represent one ROI. (C) Dendrogram showing the unsupervised hierarchically cluster analysis of relative gene expression (y-axus) across all ROIs (x-axis).
Multiplex DSP with potential for recurrence prediction
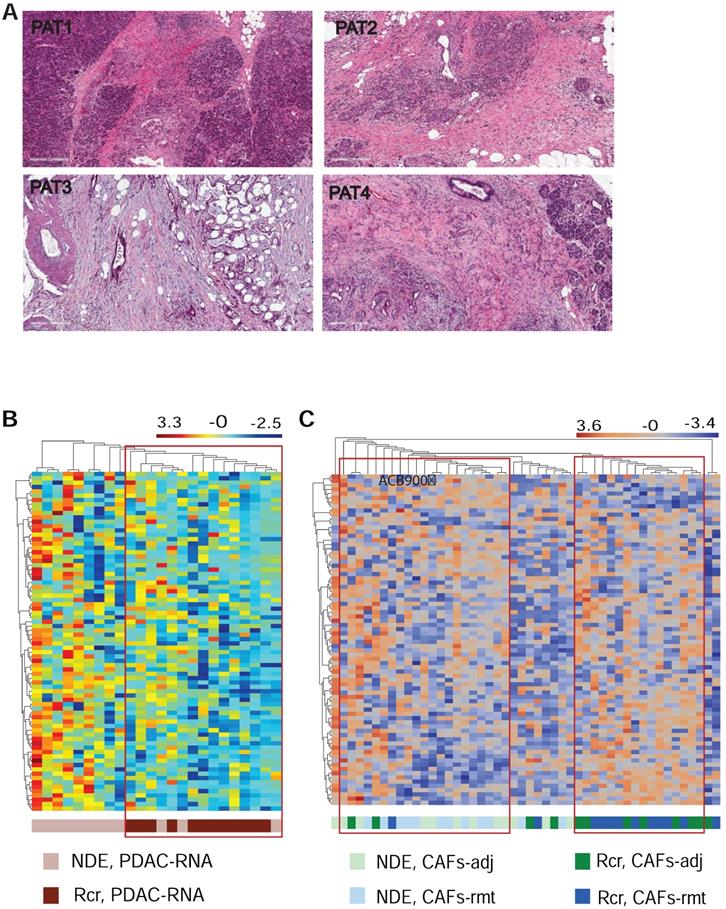
Lastly, we explored whether mplexDSP, with the capability of single cell profiling, can surpass the bulk tissue profiling and provide better insights in predicting recurrence and patient survival. Four patients who underwent pancreaticoduodenectomy (Whipple surgery) were recruited in the study. Pathologic examination confirmed diagnosis of invasive, moderately or moderately to poorly differentiated PDAC (Suppl. Table S1 & Figure 7A). Among them, two patients encountered recurrence and death within 14 months (Rcr), and the other two patients experienced longer survival (over 36 months) with no reported recurrence (termed as no disease evidence, NDE) at the time of last follow up. Using the panel of 81-RNA data from mplexDSP, we observed tentative clusters that potentially distinguish patients with recurrence from patients with no reported recurrence. As suggested by Figure 7B, PDAC cell RNA multiplex showed a clearer cluster consisting all ROIs (12/12, 100%) from recurrent patients with mixture of 20% (3/15) ROIs from patients without recurrence. Profiling this gene set on SMA+ stromal cells also demonstrated tentative separation of recurrence from no disease evidence. As shown in Figure 7C, three tentative clusters are identified: first a Rcr-ROIs formed cluster including 66.7% of ROIs from recurrent tumors (16/24,); a second NDE-ROIs-dominated cluster including 87.5% (18/21) of ROIs from tumors of no disease evidence, with 18 of NDE-ROIs (18/24, 66.7%) and 3 Rcr-ROIs (3/24, 12.5%); lastly, a small cluster of 8 ROIs mingled across both with and without recurrence patient samples. Despite the lack of tightness of perfect clusters, both single cell analysis of PDAC or stromal cells displayed better heatmap construction in comparison to mixed-cell profiling (Suppl. Figure S1).
Representative ROIs of CD45+ immune cells selection in PDAC microenvironment. (A & B) Immunofluorescent staining of morphologic markers on PDAC tumor tissue. (C, D & F) Regions of interest (ROIs) of anti-panCK antibody labeled PDAC cells (green-colored) and immune cells in vicinity (Imm.Cells-adj, red-colored). (E) Regions of interest (ROIs) for selection of immune cells in remote area (Imm. Cells-rmt, red-colored). (G) Box plot of total nCounter Counts of multiplex DSP analysis for the total 18-plex proteins, CD45, panCK, and the three housekeeping proteins (panels from left to right). (H) Dendrogram showing distinct clusters of PDAC cells from immune cells -adj and -rmt based on the expression of markers; and (J) Expression levels of marker proteins determined by DSP from three sorts of ROIs for PDAC cells and immune cells adjacent and remote, displayed numbers are p-value.
DSP defines differential protein expression of PDAC cells from immune cells. (A) Comparison profiling of multiplex DSP panel of 21 protein expression in selected CD45+ immune cells and panCK+ PDAC cells, * = p < 0.05, ** = p < 0.01, *** = p < 0.001. (B) A tri-dimention plot of PCA showing PDAC cells cluster from immune cells; and (C) Dendrogram of heiracherical cluster analysis. Three tantative clusters are boxed. Color bar indicates z-score value.
Discussion
Here we performed a study that simultaneously determines gene and protein expression from single cells of the three major cell types within the pancreatic tumor microenvironment using FFPE tumor sections. Despite the small cohort of patients, our data reveal insightful and informative cues of the tumor-stroma-immune contexture, highlighting the potential power of such approach as a novel tool to better understand PDAC tumor biology and for biomarker discovery.
We, and others, have observed the phenomenon of PDAC tumors defined by stromal cells with varying intensity of αSMA expression depending upon their proximity to the epithelial cancer cells [9, 16]. Using an organoid and PSC co-culture model, Öhlund et al. defined two distinct subtypes of CAFs based on their distance to tumor: the αSMAhigh IL-6low periglandular myofibroblasts (myCAFs) and more distantly distributed αSMAlow IL-6high inflammatory CAFs (iCAFs) [9]. More importantly, the authors reported the two subtypes are reversible, indicating a dynamic feature of CAFs. In this work, using the mplxDSP technology, we were able to perform spatial transcriptional profiles on human tissue using FFPE sections. Accordingly, differentially expressed transcripts are found between adjacent or remote αSMA+ myofibroblasts, supporting the theory that the PDAC microenvironment is comprised of distinct phenotypes of stromal cells. The pathway analysis of our data suggests that the periglandular stromal cells likely participate in recruiting immune cells and stimulating immune cell differentiation and expansion. Despite the reported conflicting function of αSMA+ myofibroblasts, there is increased awareness that stroma demonstrate an interplay of tumor promoting and tumor suppressing effects [37]. It is now rational to believe that this phenomenon could be attributed to spatial distribution of CAFs. This hypothesis is well supported by the recent discovery of subTMEs from the study of Grünwald et al., the existence of “reactive” and “deserted” subTME associated with fibroblast plasticity [38].
DSP defines differential gene expression and pathway activities of CAFs by localisation. (A) Volcano plot and (B) bar graft of statistical significance against cluster differential genes between CAFs-adj and CAFs-rmt; * p < 0.05, ** p < 0.01, *** p < 0.001; and (C) Gene-to-function relationship depicted by IPA. Color of lines and arrows correspondent to the relationship of genes to the annotated functions: red colored lines and arrows indicate increased function, blue colored lines and arrows indicate decreased function, and gray colored lines and arrows indicated that the gene is involved without further details.
In addition to αSMA+ myofibroblasts, we also analyzed the microenvironment immune cell distributions and their associated properties. Representative work by Carstens, et al. used computational, 8-color IHC multispectral imaging to discover that the distribution of the total numbers of T cell and subpopulation of T cells differs by location in relation to tumor, and more importantly, correlates with PDAC patient survival [13]. Our approach of mplxDPS measures the levels of infiltrated immune cells by the transcript expression intensity, including both T cells and innate immune cells. While we observed no significant spatial distribution of T cell subpopulations, we detected an increase in the innate immune compartments, e.g. macrophage, DC and NK cells, in immune cells directly adjacent to PDAC tumor. In the setting of cancer, innate immunity has been documented to both promote and inhibit T cell priming and effector activity [39]. Alas, we are not in a position to test the function of these innate immune cells in this work, however, our data raise support further investigation of how the proximity of innate immune components relative to the tumor may be important to T cells in influencing and shaping the known immunosuppressive PDAC microenvironment.
Therapeutic targeting of immune checkpoint molecules through inhibition of CTLA-4, PD-1 and PD-L1 has led to a paradigm shift in the treatment of many solid tumors [40], but has shown less encouraging outcomes in the treatment of PDAC [41]. There is increased awareness of the impact of the expression profile of immune checkpoints on immune-modulatory therapies to specific individuals. Although a consensus view that successful treatment depends on optimal patient selection, reaching this goal remains a challenge without a guiding biomarker. For example, Steele et al. comprehensively characterized the tumor microenvironment and found that CD8+ T cells expressed markedly immune checkpoint profiles in individual patient and did not cluster by disease stage [42]. Our interesting finding of CTLA-4 overexpression (but not PD1 or PDL1) in immune cells adjacent to PDAC tumor cells, provides compelling evidence that spatial information might be used to guide immunotherapy selection, but this requires additional investigation.
There are still many questions left to answer: The function of αSMA+ myofibroblasts beyond a simple physical barrier as conventionally acknowledged; The involvement of all tumoral and stromal components that contribute to the immunosuppressive PDCA microenvironment; The strategy of spatially defined single cell biomarkers for improving disease prognostication or the application of precision medicine, etc. We demonstrate here an initial effort toward uncovering these answers and also demonstrated the great advantages of mplxDSP technology.
DSP defines differential CTLA4 protein expression in PDAC adjacent immune cells. (A) Trend line plot to compare the eighteen target genes and three housekeeping genes expression between immunce cells adjacent (imm. C.-adj) and remote (imm. C.-rmt) to PDAC area. X-axis is grouped segments of ROIs and y-axis plots for nCount probe counts. * p < 0.05, ** p < 0.01, *** p < 0.001; (B) bar graft of the protein expression for the three checkpoint proteins PD-L1, PD-1 and CTLA4 in both immunce cells -adj and -rmt; ** p < 0.01. (C) The top significant function annotations predicted by IPA show impoverished T Cell Receptor Signalling pathway in immune cells adjacent to PDAC. The significance of canonical pathways was determined with default threshold of -log(p-value) = 1.3 (p-value = 0.05).
Differential gene expression and pathway activation. (A) HE staining of FFPE of the four resected tumors; (B & C) heatmap cluster dendrograms of RNA profiling panCK+ PDAC cells and for αSMA+ CAFs respectively.
Conclusion
Our work has provided a novel insight into the distinguishable phenotypes of non-tumor cells that inscribe the heterogenicity of the pancreatic tumor microenvironment. How to utilize spatial biomarkers for clinical benefit awaits further exploration and validation with larger patient cohorts.
Supplementary Material
Supplementary figures.
Supplementary tables.
Acknowledgements
We thank Gang Su (Department of Surgery, University of Florida) for her assist in FFPE preparation and Katy Richards-Hrdlicka (nanoString) for analysis training.
Funding
This research was funded by National Cancer Institute and National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health under award number 1U01DK108320 (SJH). Authors also receive salary support from the National Human Genome Research Institute of the NIH (T32 HG008958 to K.M.H and A.N.R.).
Author Contributions
Conceptualization, S.H., Z.H., and S.J.H.; methodology, S.H., D.F, G.T, L.M., and G.S.; investigation, S.H, D.F., G.T., and L.M.; clinical resources, K.H., A.N.R., and S.J.H.; writing—original draft preparation, S.H, and G.T.; supervision, S.H., Z.H., and S.J.H.; writing—review and editing, S.H, D.F., G.T., L.M., G.S., K.H., A.N.R., Z.H., T.G., and S.J.H.; All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of University of Florida (IRB201600873).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Hwang RF, Moore T, Arumugam T, Ramachandran V, Amos KD, Rivera A. et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008;68:918-26
2. Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D. et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457-61
3. Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR. et al. Depletion of Carcinoma-Associated Fibroblasts and Fibrosis Induces Immunosuppression and Accelerates Pancreas Cancer with Reduced Survival. Cancer Cell. 2015;28:831-3
4. Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA. et al. Stromal Elements Act to Restrain, Rather Than Support, Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2014;25:735-47
5. Lee JJ, Perera RM, Wang H, Wu DC, Liu XS, Han S. et al. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A. 2014;111:E3091-100
6. Chen Y, Kim J, Yang S, Wang H, Wu CJ, Sugimoto H. et al. Type I collagen deletion in alphaSMA(+) myofibroblasts augments immune suppression and accelerates progression of pancreatic cancer. Cancer Cell. 2021;39:548-65 e6
7. Han S, Gonzalo DH, Feely M, Rinaldi C, Belsare S, Zhai H. et al. Stroma-derived extracellular vesicles deliver tumor-suppressive miRNAs to pancreatic cancer cells. Oncotarget. 2018;9:5764-77
8. Ogawa Y, Masugi Y, Abe T, Yamazaki K, Ueno A, Fujii-Nishimura Y. et al. Three Distinct Stroma Types in Human Pancreatic Cancer Identified by Image Analysis of Fibroblast Subpopulations and Collagen. Clin Cancer Res. 2021;27:107-19
9. Ohlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M. et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214:579-96
10. Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA. et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168-78
11. Hutton C, Heider F, Blanco-Gomez A, Banyard A, Kononov A, Zhang X. et al. Single-cell analysis defines a pancreatic fibroblast lineage that supports anti-tumor immunity. Cancer Cell. 2021;39:1227-44 e20
12. Looi CK, Chung FF, Leong CO, Wong SF, Rosli R, Mai CW. Therapeutic challenges and current immunomodulatory strategies in targeting the immunosuppressive pancreatic tumor microenvironment. J Exp Clin Cancer Res. 2019;38:162
13. Carstens JL, Correa de Sampaio P, Yang D, Barua S, Wang H, Rao A. et al. Spatial computation of intratumoral T cells correlates with survival of patients with pancreatic cancer. Nat Commun. 2017;8:15095
14. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47
15. Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc B. 1995;57:289-300
16. Han S, Underwood P, Hughes SJ. From tumor microenvironment communicants to biomarker discovery: Selectively packaged extracellular vesicular cargoes in pancreatic cancer. Cytokine Growth F R. 2020;51:61-8
17. Tang Y, Geng Y, Luo J, Shen W, Zhu W, Meng C. et al. Downregulation of ubiquitin inhibits the proliferation and radioresistance of non-small cell lung cancer cells in vitro and in vivo. Sci Rep. 2015;5:9476
18. Scarpa ES, Tasini F, Crinelli R, Ceccarini C, Magnani M, Bianchi M. The Ubiquitin Gene Expression Pattern and Sensitivity to UBB and UBC Knockdown Differentiate Primary 23132/87 and Metastatic MKN45 Gastric Cancer Cells. Int J Mol Sci. 2020 21
19. Dillon CP, Weinlich R, Rodriguez DA, Cripps JG, Quarato G, Gurung P. et al. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell. 2014;157:1189-202
20. La Gruta NL, Kedzierska K, Stambas J, Doherty PC. A question of self-preservation: immunopathology in influenza virus infection. Immunol Cell Biol. 2007;85:85-92
21. Nocera DA, Roselli E, Araya P, Nunez NG, Lienenklaus S, Jablonska J. et al. In vivo Visualizing the IFN-beta Response Required for Tumor Growth Control in a Therapeutic Model of Polyadenylic-Polyuridylic Acid Administration. J Immunol. 2016;196:2860-9
22. Honda K, Sakaguchi S, Nakajima C, Watanabe A, Yanai H, Matsumoto M. et al. Selective contribution of IFN-alpha/beta signaling to the maturation of dendritic cells induced by double-stranded RNA or viral infection. Proc Natl Acad Sci U S A. 2003;100:10872-7
23. Uehori J, Fukase K, Akazawa T, Uematsu S, Akira S, Funami K. et al. Dendritic cell maturation induced by muramyl dipeptide (MDP) derivatives: monoacylated MDP confers TLR2/TLR4 activation. J Immunol. 2005;174:7096-103
24. Arnold TD, Lizama CO, Cautivo KM, Santander N, Lin L, Qiu H. et al. Impaired alphaVbeta8 and TGFbeta signaling lead to microglial dysmaturation and neuromotor dysfunction. J Exp Med. 2019;216:900-15
25. Kitching AR, Tipping PG, Kurimoto M, Holdsworth SR. IL-18 has IL-12-independent effects in delayed-type hypersensitivity: Studies in cell-mediated crescentic glomerulonephritis. Journal of Immunology. 2000;165:4649-57
26. Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M. et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097-100
27. Li MO, Flavell RA. TGF-beta: A master of all T cell trades. Cell. 2008;134:392-404
28. Royer DJ, Conrady CD, Carr DJ. Herpesvirus-Associated Lymphadenitis Distorts Fibroblastic Reticular Cell Microarchitecture and Attenuates CD8 T Cell Responses to Neurotropic Infection in Mice Lacking the STING-IFNalpha/beta Defense Pathways. J Immunol. 2016;197:2338-52
29. Chong MMW, Simpson N, Ciofani M, Chen G, Collins A, Littman DR. Epigenetic propagation of CD4 expression is established by the Cd4 proximal enhancer in helper T cells. Gene Dev. 2010;24:659-69
30. Lee SY, Jung YO, Kim DJ, Kang CM, Moon YM, Heo YJ. et al. IL-12p40 Homodimer Ameliorates Experimental Autoimmune Arthritis. J Immunol. 2015;195:3001-10
31. Melton AC, Bailey-Bucktrout SL, Travis MA, Fife BT, Bluestone JA, Sheppard D. Expression of alphavbeta8 integrin on dendritic cells regulates Th17 cell development and experimental autoimmune encephalomyelitis in mice. J Clin Invest. 2010;120:4436-44
32. Zhen A, Krutzik SR, Levin BR, Kasparian S, Zack JA, Kitchen SG. CD4 ligation on human blood monocytes triggers macrophage differentiation and enhances HIV infection. J Virol. 2014;88:9934-46
33. Chen YL, Chen TT, Pai LM, Wesoly J, Bluyssen HA, Lee CK. A type I IFN-Flt3 ligand axis augments plasmacytoid dendritic cell development from common lymphoid progenitors. J Exp Med. 2013;210:2515-22
34. Lin PL, Rutledge T, Green AM, Bigbee M, Fuhrman C, Klein E. et al. CD4 T Cell Depletion Exacerbates Acute Mycobacterium tuberculosis While Reactivation of Latent Infection Is Dependent on Severity of Tissue Depletion in Cynomolgus Macaques. Aids Res Hum Retrov. 2012;28:1693-702
35. Satpathy AT, Briseno CG, Lee JS, Ng D, Manieri NA, Kc W. et al. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat Immunol. 2013;14:937-48
36. Schleicher U, Liese J, Knippertz I, Kurzmann C, Hesse A, Heit A. et al. NK cell activation in visceral leishmaniasis requires TLR9, myeloid DCs, and IL-12, but is independent of plasmacytoid DCs. J Exp Med. 2007;204:893-906
37. Bijlsma MF, van Laarhoven HW. The conflicting roles of tumor stroma in pancreatic cancer and their contribution to the failure of clinical trials: a systematic review and critical appraisal. Cancer Metastasis Rev. 2015;34:97-114
38. Grunwald BT, Devisme A, Andrieux G, Vyas F, Aliar K, McCloskey CW. et al. Spatially confined sub-tumor microenvironments in pancreatic cancer. Cell. 2021;184:5577-92 e18
39. Chang RNB, Beatty GL. The interplay between innate and adaptive immunity in cancer shapes the productivity of cancer immunosurveillance. J Leukocyte Biol. 2020;108:363-76
40. Brower V. Checkpoint blockade immunotherapy for cancer comes of age. J Natl Cancer Inst. 2015 107
41. Schizas D, Charalampakis N, Kole C, Economopoulou P, Koustas E, Gkotsis E. et al. Immunotherapy for pancreatic cancer: A 2020 update. Cancer Treat Rev. 2020;86:102016
42. Steele NG, Carpenter ES, Kemp SB, Sirihorachai VR, The S, Delrosario L. et al. Multimodal Mapping of the Tumor and Peripheral Blood Immune Landscape in Human Pancreatic Cancer. Nat Cancer. 2020;1:1097-112
Author contact
![]() Corresponding author: Song Han, Department of Surgery, University of Florida College of Medicine, 1600 SW Archer Rd, Gainesville, FL 32610. E-mail: song.hanufl.edu.
Corresponding author: Song Han, Department of Surgery, University of Florida College of Medicine, 1600 SW Archer Rd, Gainesville, FL 32610. E-mail: song.hanufl.edu.
 Global reach, higher impact
Global reach, higher impact