13.3
Impact Factor
Theranostics 2022; 12(9):4163-4180. doi:10.7150/thno.71873 This issue Cite
Research Paper
Spatial maps of hepatocellular carcinoma transcriptomes reveal spatial expression patterns in tumor immune microenvironment
1. The department of Medical Genetics, Naval Medical University, Shanghai, 200433, China.
2. The Third Department of Hepatic Surgery, Eastern Hepatobiliary Surgery Hospital Affiliated to Naval Medical University, Shanghai, 200438, China.
3. Department of Pathology, Changhai Hospital Affiliated to Navy Medical University, Shanghai, 200433, China.
4. Translational Medicine Research Center, Medical Innovation Research Division and Fourth Medical Center of the Chinese PLA General Hospital, Beijing, 100048, China.
* Equal contribution
Abstract
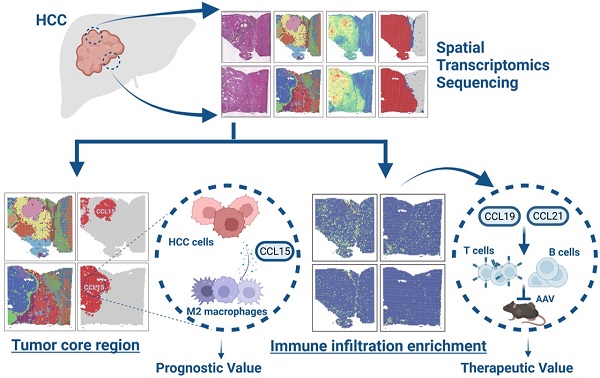
Rationale: Hepatocellular carcinoma (HCC) is a highly heterogeneous and malignant disease with the complex immune microenvironment, which ultimately influence clinic outcomes of patients. However, the spatial expression patterns of diverse immune cells among tumor microenvironment remain to be further deciphered.
Methods: Spatial transcriptomics sequencing (ST) was implemented on two portions of HCC specimens. Differentially expressed genes, cell cycle phases, epithelial-mesenchymal features, pseudo-time and immune infiltration analysis were applied to demonstrate the intratumor heterogeneity and define the specific immune-related regions, and the results were further validated by a second analysis on another ST study. In vitro and in vivo experiments were conducted to confirm the functional mechanisms of key molecules such as CCL15, CCL19 and CCL21. Clinical tissue samples were used to assess their potential prognostic and therapeutic values.
Results: Totally, 7553 spots were categorized into 15 subsets by hierarchical clustering, and malignant subsets with intratumor heterogeneity phenotypes were identified. Spatial heterogeneity from distinct sectors highlights specific chemokines: CCL15 is remarkable in the core region of the carcinoma sector and facilitates the immunosuppressive microenvironment by recruiting and polarizing M2-like macrophages in vitro and in vivo; High expression of CCL15 and CD163 respectively predicts poor prognosis of HCC patients, and the combined application of them has better predictive value. CCL19 and CCL21, sharing similar spatial expression patterns, are highly-correlated and prominent in the immune infiltration enrichment and recruit CD3+ T cells and CD20+ B cells to inhibit the growth of HCC, indicating a good prognosis of HCC patients.
Conclusions: Taken together, our studies preliminarily reveal intratumor heterogeneity of HCC based on ST techniques and unravel the previously unexplored spatial expression patterns in the immune microenvironment. We also highlight the clinical significance and spatial discrepancy of key molecules, providing novel insight for further developing therapeutic strategies in HCC.
Keywords: hepatocellular carcinoma, spatial transcriptomics, heterogeneity, immune microenvironment
 Global reach, higher impact
Global reach, higher impact