13.3
Impact Factor
Theranostics 2021; 11(16):8027-8042. doi:10.7150/thno.61207 This issue Cite
Review
Radiomics in prostate cancer imaging for a personalized treatment approach - current aspects of methodology and a systematic review on validated studies
1. Department of Radiation Oncology, Medical Center - University of Freiburg, Faculty of Medicine. University of Freiburg, Germany
2. German Cancer Consortium (DKTK). Partner Site Freiburg, Germany
3. Department of Radiology, Medical Center - University of Freiburg, Faculty of Medicine. University of Freiburg, Germany
4. Department of Nuclear Medicine, Medical Center - University of Freiburg, Faculty of Medicine. University of Freiburg, Germany
5. Department of Radiation Oncology - Division of Medical Physics, Medical Center - University of Freiburg, Faculty of Medicine. University of Freiburg, Germany
6. Radiotherapy and Radiation Oncology, Faculty of Medicine and University Hospital Carl Gustav Carus, Technische Universität Dresden.
7. OncoRay-National Center for Radiation Research in Oncology, Faculty of Medicine and University Hospital Carl Gustav Carus, Technische Universität Dresden, Helmholtz-Zentrum Dresden-Rossendorf, Dresden, Germany
8. Institute of Computer Engineering, Vienne University of Technology, Vienna, Austria
9. Department of Biomedical Sciences, Humanitas University, Via Rita Levi Montalcini 4, 20090 Pieve Emanuele - Milan, Italy
10. IRCCS Humanitas Research Hospital, Via Manzoni 56, 20089 Rozzano - Milan, Italy
11. Berta-Ottenstein-Programme, Faculty of Medicine, University of Freiburg, Germany
12. German Oncology Center, European University of Cyprus, Limassol, Cyprus
* shared authorship
Received 2021-4-2; Accepted 2021-6-17; Published 2021-7-6
Abstract
Prostate cancer (PCa) is one of the most frequently diagnosed malignancies of men in the world. Due to a variety of treatment options in different risk groups, proper diagnostic and risk stratification is pivotal in treatment of PCa. The development of precise medical imaging procedures simultaneously to improvements in big data analysis has led to the establishment of radiomics - a computer-based method of extracting and analyzing image features quantitatively. This approach bears the potential to assess and improve PCa detection, tissue characterization and clinical outcome prediction. This article gives an overview on the current aspects of methodology and systematically reviews available literature on radiomics in PCa patients, showing its potential for personalized therapy approaches. The qualitative synthesis includes all imaging modalities and focuses on validated studies, putting forward future directions.
Introduction
In global cancer statistics of men prostate cancer (PCa) is the most frequently diagnosed malignancies in the world and the fifth leading cause of death worldwide [1,2]. Therefore, the development of accurate diagnostic tools is of great importance. Many modern imaging modalities provide a great value in screening, diagnosis, treatment response measurement and prognosis evaluation of PCa patients. A suspicious digital rectal examination and/or an elevation of prostate specific antigen (PSA) in blood serum lead to transrectal ultrasound (TRUS) guided biopsy for histopathologic verification of PCa [3]. In recent years an augmented approach of this strategy, including magnetic resonance imaging (MRI), has gained traction in clinical application and was incorporated into guidelines [4]. MRI is not only employed prior to biopsy, but for local staging and follow up [5]. Nevertheless, diagnostic accuracy is still hampered by inter-observer variability and exactness of lesion detection does not seem to be warranted [6-8]. In an attempt to improve interpretation, reporting and acquisition standards for global harmonization “Prostate Imaging - Reporting and Data System Version 2.1” (PI-RADSv2.1) were established [9]. Bone scans and computer tomography (CT) used to be standard of care (SoC) for staging and re-staging. As of late prostate specific membrane antigen positron emission tomography (PSMA-PET) has been implemented into clinical practice as recommended in current guidelines for staging and restaging [10-13]. Additionally, growing evidence proclaims the use of PSMA-PET for intraprostatic lesion detection and segmentation [14-18]. In addition to an accurate diagnosis, proper and decisive risk stratification is crucial, due to a variety of treatment options in different clinical scenarios. However, recommended models for risk classification [5,19,20] might not always predict the final outcome in every disease stage of PCa [21,22]. Thus, new concepts for adequate detection and risk stratification towards precision medicine and personalized treatment are required. With the rise of big data analysis, the computer-based extraction of pre-defined image features in terms of “hand-crafted” radiomic features (RF) is an emerging field in research that might satisfy this need. It is hypothesized that medical images contain more information than discernible visually by trained professionals. Simplified these RF might provide more information about a tumor or other tissues facilitating diagnosis, risk stratification and therapeutic outcome. The advantage of radiomics is the utilization of SoC images without additional required effort and the abundance of medical images available, which can be utilized for longitudinal monitoring. Another benefit is that radiomics examines whole tumors as opposed to biopsy schedules which are prone to sampling errors due to intratumoral heterogeneity [23,24]. Thereby, radiomics offer great potential for personalization of therapeutic approaches, in particular for image-based disciplines such as radiation oncology. This review gives an overview of methodological aspects of radiomics firstly, followed by the methodology of our literature search and a qualitative synthesis of radiomics in prostate cancer subdivided by imaging modalities and based on a systematic search. There have been reviews on this topic but mostly focusing on MRI whereas our review includes all imaging modalities and concentrates on papers with internal or external validation [25-29].
Current Methodological Aspects of Radiomic Feature Extraction
Radiomics Pipeline
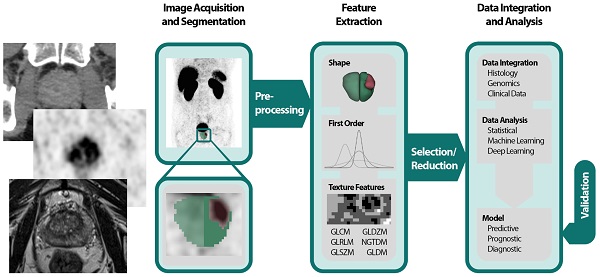
The Radiomics Pipeline (Figure 1) is the entire sequence of data processing from imaging to a diagnostic, predictive or prognostic model based on RF. It is subdivided into three major operations.
(1) Image acquisition and preprocessing
(2) High-throughput feature extraction
(3) Data integration and data analysis
The Image Biomarker Standardization Initiative (IBSI) published a reference manual to harmonize the feature extraction by providing (i) definitions, (ii) a standardization of the radiomics pipeline, (iii) reference datasets and (iv) a reporting scheme [30].
(1) Image acquisition and preprocessing
All imaging modalities mentioned in the introduction can be utilized for PCa radiomics: TRUS, MRI, PSMA-PET, CT and bone scan. It is important to mention that heterogeneities in image acquisition und image reconstruction algorithms due to different local standards are culpable for missing repeatability and reproducibility of RF [31-34]. Prospective trails with fixed imaging protocols could aim to ensure that a scan yields similar results in the same patient when repeated on the same system i.e. repeatability as well as on different systems and institutes i.e. reproducibility [33]. After image acquisition the volume of interest (VOI) is delineated manually, semi-automatically or fully automatically. If manual segmentation is performed a sophisticated protocol should be used throughout the whole dataset to minimize inter-observer variability [35]. Subsequently and before feature extraction images should be pre-processed, e.g., by intensity inhomogeneity correction or noise filtering for MR-images [32]. It is known that preprocessing sequences can also have a significant impact on the robustness and reproducibility of RF and identification of generalizable and consistent preprocessing algorithms is a pivotal step [36].
(2) High-throughput feature extraction
The spatial and gray level information of the segmented voxels is used in numerous mathematical calculations to extract pre-defined “hand-crafted” RF. They can be computed with various open-source packages like PyRadiomics [37], IBEX [38], RaCat [39], QIFE [40], MaZda [41], CERR [42] or LIFEx [43] as well as commercial products [44]. Additionally, radiomic codes implemented in MatLab ® are commonly used. It is very important to validate the used software tools, especially homemade software, with datasets provided by the IBSI [30], to increase reproducibility, robustness and comparability across different platforms. Current versions of PyRadiomics, LIFEx, RaCat, CERR and adapted version of IBEX comply with IBSI. Besides the extraction of hand-crafted RF, convolutional neuronal networks as a subfield of machine learning (ML) can be used for pattern recognition and image feature analyses by applying the actual images [45]. This can be done in combination with predefined “hand-crafted” features [46,47], but mostly ML algorithms engineer models, based on large amounts of data, autonomously [48].
Radiomics pipeline depicts the data processing and operations to build a radiomics model with validation. First an image is acquired and segmented manually, semiautomatically or fully automatically. Then feature extraction is performed after preprocessing. Feature classes are shape features, first order features and texture features. Due to the abundance of RF a selection or reduction should be performed before or while integrating with histology, genomics or clinical data. Data analysis can be performed by using classical statistical models, with machine learning or deep learning. A predictive, prognostic or diagnostic model is built and should be internally or externally validated. Abbreviations: GLCM=gray level co-occurrence matrix; GLDZM = gray level distance zone matrix; GLRLM = gray level run length matrix; GLSZM = gray level size zone matrix; NGTDM = neighboring gray tone difference matrix; NGLDM = neighboring gray level dependence matrix; RF= Radiomic features.
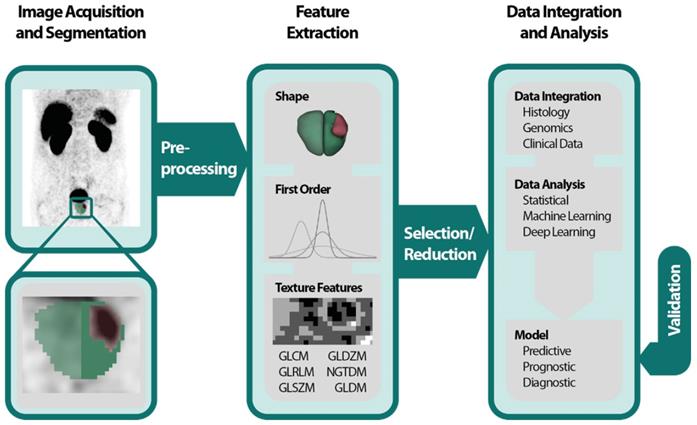
(3) Data integration and data analysis
Often a vast number of RF are computed, and the abundance of RF demands feature selection and/or reduction to avoid overfitting and to exclude not relevant or redundant features. Many features are correlated with each other; these redundant features might be depicted with heatmaps and should be omitted [49,50]. Additionally, ML algorithms like minimum redundancy and maximum relevance or fisher score can be used to assess the correlation between RF [51,52]. Other options for feature reduction are prioritizing robust features [34]. An overview of feature reduction steps including quantitative comparisons of performance is given by Leger et al [53] and Parmar and colleagues [54]. The analysis of the remaining features can be conducted by using the RF alone or in combination with other clinical parameters by applying classical statistical methods or ML for data integration and modelling. Examples of ML classifiers are random forests, support vector machines and nearest neighbors for instance [51,55]. To avoid overfitting, it is recommended to control false positive results by correcting for multiple testing when the data analysis is based on classical statistical methods [56,57]. After the generation of a model based on RF, validation should be executed to evaluate its performance and to assess generalizability [58]. In recent years ever more emphasis has been laid on this last step [33-35]. During an internal validation, the data is usually split in 3 datasets: (i) one training dataset for optimizing the parameters of a model, (ii) one validation dataset for hyperparameter optimization e.g., the depth of a tree or a deep-learning architecture and (iii) one test set for the final assessment. The latter might be used independently for validation. During cross-validation (CV) usually small datasets are divided accordingly in an iterative process. K-fold CV partitions the dataset in k subsets using one as a validation and the rest for training. This process is repeated for each subset. Leave-one-out CV operates similarly but leaves one patient for validation while using the rest for training CV should be used with caution especially with leave-one-out CV tending to be overly optimistic [59,60]. External validation based on independent datasets from different institutes enables the highest quality of validation [58].
Hand-crafted Radiomic Features
“Hand-crafted” RF [48] can be grouped in shape descriptors, 1st order features and texture features. Shape features describe the morphology of the VOI for instance the size, volume or diameter. 1st Order features are based on an intensity histogram derived of the segmented voxels [24]. Texture features are more advanced and do not only rely on voxel intensities i.e., gray levels but on spatial information as well. First introduced by Haralick et al. the gray level co-occurrence matrix (GLCM) assesses the gray levels of pairs of neighboring voxels [61]. Others like the gray level size zone matrix (GLSZM) [62] and the gray level run length matrix (GLRLM) [63] analyze groups of consecutive voxels, zones, or runs of connected voxels in one direction, respectively. For a more complete description of features, texture matrices and their mathematical calculations we recommend the IBSI and their documentation [30]. High-order features are calculated on filter transformed images like wavelets or gaussian bandpass filer [24].
Methodology
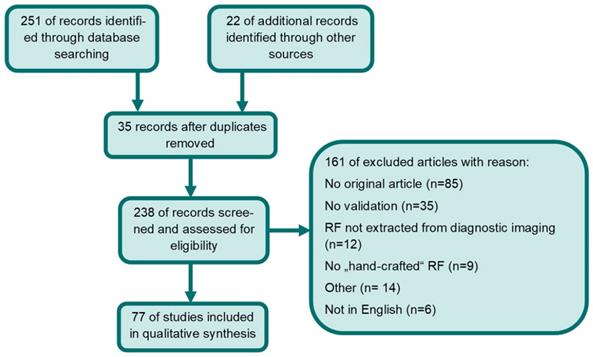
Studies eligible for inclusion complied with the following criteria: articles had to be on PCa radiomics with predefined “hand-crafted” features derived from MRI, TRUS, CT, Choline- or PSMA-PET and needed to apply internal or external validation. Excluded were papers not written in English and non-original articles. Two of the co-authors (SKBS and ASB) performed independently a PubMed/Medline, EMBASE and Cochrane Library database search for the terms: (cancer of prostate[MeSH Terms]) AND ((texture features) OR (radiomics)). If the two independent readers included or excluded studies differently a third reader (CZ) decided on eligibility. This was performed in 11 cases. The time period considered in this literature review was from 1st of January 2014 [64] to 1st of January 2021. 251 articles were located. Additionally, 22 manuscripts were identified through other sources (e.g., google scholar or references in screened manuscripts). 35 duplicates were removed. Only articles that met inclusion criteria were included. Finally, 77 studies were included in the qualitative synthesis. Please see Figure 2 for a detailed description of the performed literature search according to PRISMA [65]. Due to heterogeneity of imaging modalities, in the applied Radiomics pipeline and the analyzed endpoints no quantitative analysis was performed. Additionally, we assessed whether the utilized software complied with IBSI.
Furthermore, ongoing clinical trials were screened on “clinicaltrials.gov”. Studies eligible for inclusion fulfilled the following criteria: ongoing trials on PCa RFs with “hand-crafted” features derived of MRI, TRUS, CT or PSMA-PET. Trials with unknown status were excluded. CZ performed the search for the terms (“Condition or disease: prostate cancer” AND “radiomicsl” OR “texture features”). Six trials were located, and one trial (NCT03294122) was excluded due to unknown trial status.
Flow diagram of systematic database search and records excluded. Abbreviations: RF= radiomic features
Results
I MRI
Literature research revealed 57 original papers computing RF on multiparametric magnetic resonance tomography (mpMRI) imaging (see Table 1). The most common segmented VOI was intraprostatic tumor (n = 48) [66-114], of which five studies focused on tumor location in transitional zone (TZ) [72,73,81,95,112] and five studies in peripheral zone (PZ) [68,71,76,82,95]. 14 studies selected the prostate as VOI [69,71,73,78,82,87,96,97,104,110,112, 115-118] of which one study analyzed the prostate excluding urethra and intraprostatic dominant lesions [97] and one study differentiated between different prostate zones [115]. Four studies considered PCa localization in fused histopathologic information as VOI [75,119-121] and one study the area of biopsy including the surrounding 15 mm [114]. One study included peritumoral areas [96]. The rectal wall was delineated as VOI in one study [122]. RFs were extracted from T2-weighted images (T2w, n = 52) [66-76,78-91,93-107,109-113,116-122], apparent diffusion coefficient (ADC, n = 38) [67,70-72,74,77-81,83-85,87,89-96,98-100,103-105,108-113,115,116,120,122], diffusion weighted imaging (DWI, n = 21) [66,69-73,76,77,80,82,86,88,90,94,98,105,107,112,115-117], dynamic contrast enhanced (DCE, n = 13) [66,73, 79,80,82,89,91,104-107,114,120], computed high-b DWI (CHB-DWI) (n = 5) [69,74,75,100,115], T1-weighted sequences (T1w, n = 3) [72,106,117] and diffusion tensor imaging (n = 1) [76], respectively. PCa detection (n = 19) [66-82,114,115], Gleason Score (GS) discrimination or upgrading from biopsy to prostatectomy (n = 22) [83-100,116,119-121], extracapsular extension (n = 5) [90,103-105,120], biochemical recurrence (n=4) [108-110,117], segmentation (n = 3) [112, 113, 118], bone metastasis (n = 2) [106,107], treatment response (n = 1) [111], and rectal toxicity (n = 1) were investigated as primary endpoints [122]. Most of the studies adopted internal validation (n = 50) [66,67,69-72,74-95,97,99,100,103-107,109,111-117,119-122] utilizing ML algorithms like CV (n = 26) and/or leave-one-patient-out CV (n = 10) [66,67,69,72,74,75,81,83,84,86,88,90-94,97,99,103,104,111,113-115,119, 122], as well as independent internal validation cohorts (n = 24) [70,71,76-80,82,85,87,89,90, 95,100,105-107,109,112,116,117,120,121]. External validation was performed in seven studies [68,73,96, 98,108,110,118]. Eleven studies complied with IBSI [71,78-80,94,97,103,105,108,109,118].
List of included articles on RFs derived of MRI. In the second column # are the number of patients enrolled retrospectively (R) or prospectively (P). In the fourth column the volume of interest (VOI) is presented accompanied by the type of segmentation in brackets M = manual, SA = semiautomatic and A = fully automatic. The last column contains information on validation. “e” stands for external validation and “I” for internal. The number stands for the number of cohorts used. 2 means one for development and one for testing.
| Prostate cancer detection | ||||||
|---|---|---|---|---|---|---|
| Study | # | Imaging Modality | VOI (Segmentation) | Endpoint(s) | Results | Validation |
| Cameron et al. [67] | 5 (R) | T2w, ADC | PCa (A +M) | PCa detection | RF model outperformed conventional mpMRI feature models. | i (LOO) |
| Cameron et al. [66] | 13 (R) | T2w, DWI, DCE | PCa (A) | Classifiers for PCa detection | RF model outperformed conventional mpMRI feature models. | i (CV, LOO) |
| Viswanath et al. [68] | 85 (R) | T2w | PCa, PZ, central gland (M) | Classifier for voxel-wise PCa detection | Boosted Decision Tree classifier has the highest ROC-AUC for detecting PCa., Boosted Quadratic-Discriminant Analysis is the most accurate and robust in detection of PCa extent across three sites. The ground truth was established by whole mount histology. | e (CV with external centers) |
| Khalvati et al. [69] | 20 (R ) | T2w, DWI, CDI, CHB-DWI | PCa, prostate (M) | Classifier for PCa detection | Support vector machine classifier improved PCa auto-detection. | i (LOO) |
| Xu et al. [70] | 331 (R) | T2w, DWI, ADC | PCa | Benign vs. malignant lesions | BpMRI improved discrimination between benign and malignant lesions. | i (2) |
| Bonekamp et al. [71] | 316 (R) | T2w, DWI and ADC | PCa, PZ, prostate (M) | PCa ISUP ≥2 | Quantitative ADC measurement improves differentiation of benign vs malignant lesions, ML comparable, performance of zone-specific models was lower. | i (2+CV) |
| Sidhu et al. [72] | 76 (R) | T1w, T2w, DWI, ADC | PCa in TZ (M) | PCa detection in TZ | TZ derived RF can discriminate TZ-PCa. | i (LOO) |
| Ginsburg et al. [73] | 80 (R) | T2w, DWI, DCE | PCa, TZ, prostate (M) | PCa detection in TZ and PZ | TZ-specific classifier significantly improves accuracy of PZ-PCa detection. | e (3 institutions) |
| Parra et al.[114] | 52 (R) | DCE | Habitat = biopsy +15 mm (M) | PCa detection (significant) | Habitats from DCE predict clinically significant PCa well. | i (LOO) |
| Khalvati et al. [74] | 30 (R) | T2w, ADC, CHB-DW, CDI | PCa (A) | Framework for PCa detection | Proposed framework (MPCaD can be utilized to detect and localize PCa. | i (LOO) |
| Wang et al. [75] | 54 (R) | T2w, CHB-DW | PCa, histological-radiological correlation (M) | Classifier for PCa detection (significant) | SVM classifier improves performance of PI-RADS v2 for clinically relevant PCa. | i (LOO) |
| Gholizadeh et al.[76] | 16 (P) | T2WI, DWI, DTI | PCa in PZ(M) | Differentiation pf PCa and non-PCa | Voxel‐based supervised machine learning models generated a binary classification of cancer probability maps. | i (2+LOO) |
| Hu et al. [77] | 136 (P) | DWI, ADC | PCa (M) | PCa detection | A mixed model based on the clinically independent risk factors and mp-MRI radiomics score showed the best performance. | i (2) |
| Woźnicki et al.[78] | 191 (R) | T2w, ADC | PCa + Prostate (M) | PCa detection and clinical significance | An ensemble machine learning model combining radiomics, PI-RADS, prostate specific antigen density and digital rectal examination resulted in a good predictive performance. | i (2+CV) |
| Qi et al. [79] | 199 (R) | T2w, ADC, DCE | PCa (M) | PCa prediction on patients with PSA level of 4-10ng/ml | The combined model incorporating all sequences, age, PSA density and the PI‐RADS v2 score yielded good performance for prediction of PCa. | i (2) |
| Dulhanty et al. [115] | 101 (R) | ADC, CHB-DWI | Prostate zones (M) | PCa detection based on 10 anatomical zones | Zone-level radiomic sequences distinguish between positive and negative zones. | i (CV) |
| Bleker et al. [80] | 206 (P) | T2w, DWI, ADC, DCE | PCa (SA) | csPCa in PZ | Addition of DCE-RFs does not improve performance of T2w- and DWI-RF based models. Multivariate RF selection with extreme gradient boosting outperformed univariate selection. | i (2) |
| Wu et al.[81] | 90 (R) | T2w, ADC | PCa in TZ (M) | Differentiation of PCa inTZ | Proposed models using quantitative ADC, shape and texture features, show good performance for TZ PCa detection and remained accurate when comparing TZ PCa with stromal BPH and in smaller lesions. | i (CV) |
| Kwon et al. [82] | 344 (R | T2w, DCE, DWI, proton density-weighted | Prostate, PZ, PCa (M) | Detection of csPCA Classification methods | Random forest classification showed the highest AUC. | i (2) |
| Gleason score | ||||||
| Hectors et al.[83] | 64 (R) | T2w, ADC, diffusion kurtosis imaging maps | PCa (unknown) | Aggressiveness (GS, Gene expression, Decipher) | 14 RF with significant correlation to GS, 40 DWI features with significant correlation to Gene expression, ML models with excellent performance to predict Decipher score ≥ 6. | i (CV) |
| Chaddad et al. [85] | 99 (R) | T2w, ADC | PCa (M) | GS grouping (6/3+4/4+3) | Joint Intensity Matrix-derived RF (n=5) are independent predictors of GS. | i (2) |
| Chaddad et al. [84] | 99 (R) | T2w, ADC | PCa (A) | GS grouping (6/3+4/4+3) | T2w and ADC derived RF can predict GS. | i (CV) |
| Sun et al. [119] | 30 (R) | T2w | PCa on histology (M) | GS, Risk groups | ADC, GLCM and GLRLM discriminate between high grade and low grade PCa. The combination further improved AUC. | i (CV) |
| Jensen et al. [86] | 112 (R) | T2w, DWI | PCa (M) | GS, risk group | Zonal-specific DWI and T2w derived RF differentiate between PCa lesions of all GS. | i (LOO + CV) |
| Chen et al. [87] | 381 (R) | ADC, T2w | PCa, prostate(M) | PCa/non-PCa, high grade GS /low grade GS 6 compared to PI-RADSv2 | T2w and ADC RF show high efficacy in distinguishing PCa vs non-PCa and high-grade vs low-grade PCa. | i (2) |
| Toivonen et al. [88] | 62 (R) | T2w, DWI, T2-mapping | PCa | GS | T2w and DWI derived RF show good classification performance for GS of PCa. | i (LPOCV + CV) |
| Zhang et al. [89] | 166 (R) | T2w, ADC, DCE | PCa (M) | PCa upgrading | T2w, ADC and DCE derived RF can predict GS upgrading from biopsy to radical prostatectomy. | i (2) |
| Min et al. [90] | 280 (R) | T2w, DWI, ADC | PCa (M) | PCa detection (significant) | MpMRI derived RF discriminate between GS 3+4 or lower. | I (CV) |
| Li et al. [91] | 63 (R) | T2w, ADC, DCE | PCa (M) | GS in CG PCa | Support vector machine classification achieves accurate GS classification of PCa in central gland. | i (CV) |
| Rozenberg et al. [92] | 54 (R) | ADC | PCa (M) | Prediction of GS upgrading and Differentiation of GS 3+4 and 4+3 | ADC derived texture features are not predictive of GS upgrading after radical prostatectomy. | i (CV) |
| McGarry et al. [120] | 48 (P) | T2w, ADC, DCE | PCa on histology (M) | Gleason probability maps | RF based mapping successfully stratifies high- and low-risk PCa. | i (2) |
| Penzias et al. [121] | 36 (R) | T2w | PCa on histology (M) | GS, risk group, correlation with QH | RF and quantitative histomorphometry features correlated with these RF are predictive for of GS. | i (2) |
| Fehr et al. [93] | 217 (R) | T2w, ADC | PCa (M) | GS risk group differentiation | Automatic classifiers achieve accurate classification of GS. | i (CV) |
| Hou et al. [94] | 263 (R) | T2w, DWI, ADC | PCa, (M) | Clincially significant PCa (GS≥7) in PIRADS 3 lesions | Radiomics ML model of all sequences has potential to predict csPCa in PIRADS 3 lesions to guide biopsy. | i (CV) |
| Li et al. [95] | 381 (R) | T2w, ADC | PCa in TZ and PZ (M) | Clincically significant PCa | Radiomics model can predict csPCa with high accuracy (AUC ≥-98). | i (2) |
| Gong et al.[116] | 489 (R) | T2w, DWI, ADC | Prostate | Identification of high grade PCa (>GS7) | DWI RF-model and combination of T2w and DWI achieved high accuracy in prediction of GS >7. | i (2, CV) |
| Algohary et al. [96] | 231 (R) | T2w, ADC | PCa lesion, peritumoral area (M) | Differentiation of PCa Risk Groups according to D'Amico | Combination of peritumoral and intratumoral RFs improved the risk stratification results by 3-6% compared to intra-tumoral features alone. | e (2) |
| Gugliandolo et al. [97] | 65 (R) | T2w | Prostate excluding urethra and dominant intraprostatic lesions (M) | Prediction of GS, PIRADS v2 Score and Risk Group | Radiomic signature consisting of the combination of 3D GLCM and intensity domain category features were able to discriminate between low- and intermediate-grade malignancy. | i (CV, LOO) |
| Zhang et al. [98] | 159 (R) | T2w, DWI, ADC | PCa (M) | Discrimintation of csPCa and clincially insignificant PCa | A radiomic signature of 10 features, was significantly associated with csPCa. A nomogram of this signature and ADC values showed even better AUCs. | e (2, CV) |
| Algohary et al. [99] | 56 (R) | T2w, ADC | PCa (M) | Prediction of csPCa in active surveillance patients | 7 T2w-based and 3 ADC-based RF exhibited statistically significant differences between malignant and normal regions in the training groups. The 3 constructed ML models yielded good accuracy | i (CV) |
| Abraham et al. [100] | 162 (R) | T2w, ADC, high B-Value Diffusion-Weighted (BVAL) | PCa (A) | Classification of Grade Groups | The novel method using texture features and stacked sparse autoencoder was able to classify PCa grade groups moderately. | i (2, CV) |
| Extracapsular extension | ||||||
| Ma et al. [120] | 119 (R) | T2w | PCa (M) | ECE of PCa | T2w derived RF predict side specific ECE. | i (2) |
| Ma et al. [90] | 210 (R) | T2w | PCa (M) | ECE prior to RP | T2w derived RF outperformed radiologist in predicting ECE. | i (2) |
| Stanzione et al. [103] | 39(R) | T2w, AdC | PCa index Lesions (M) | Classifier for ECE prediction | Bayesian Network was the best classifier for ECE prediction. | i (CV) |
| Losnegard et al. [104] | 228 (R) | T2w, ADC, DCE | Prostate, PCa (M+A) | ECE Prediction in high and unfav. Intermediate risk PCa | 12 RF extracted from manual segmentation combined with a Random Forest classifier can predict ECE with an AUC of 0.74. Features from T2W and ADC showed a good performance. A combined model performed even better. | i (CV) |
| Xu et al. [105] | 95 (R) | T2w, DWI, ADC, DCE | PCa (M) | ECE | 8 RF were used to build a radiomics model with an AUC of 0.92. A radiomics nomogram with clinical features yielded similar results. | i (2) |
| Bone metastasis | ||||||
| Wang et al. [106] | 176 (R) | T2w, DCE T1w | PCa (M) | Bone metastasis prediction | T2w and DCE derived RF were predictors for BM. | i (2) |
| Zhang et al. [107] | 116 (R) | T2, DWI, DCE | PCa (M) | Prediction of bone metastasis in newly diagnosed PCa | The radiomics nomogram based on 11 RFs and clinical risk factors, showed good performance to promote individualized prediction of bone metastasis. | i (2) |
| Biochemical recurrence | ||||||
| Bourbonne et al. [109] | 107 (R) | T2w, ADC | PCa (SA) | Prediction of BCR and biochemical relapse free survival after RP in high risk PCa | One ADC derived RF (SZEGLSZM) was predictive for BCR and bRFS (AUC 0.76). | i (2) |
| Bourbonne et al. [108] | 195 (R) | ADC | PCa (SA) | BCR | External validation of the identified ADC derived RF (SZEGLSZM) for BCR and bRFS prediction after RP. | e (2) |
| Shiradka et al. [110] | 120 (R) | T2w and ADc | PCa, prostate (M) | BCR after RP or RT | BpMRI RF-trained machine learning classifier can be predictive of BCR. | e (2) |
| Zhong et al.[117] | 91 (R) | T1w, T2w, DWI | Prostate (M) | BRC of localized PCa after RT and neoadjuvant endocrine therapy. | MRI derived RFs can predict BCR after RT with good performance. | i (2, CV) |
| Treatment response | ||||||
| Abdollahi et al. [111] | 33 (P) | T2w, ADC, pre- and post IMRT | PCa (M) | Therapy response (RT), GS, T-stage | T2w and ADC derived RF and ML correlate with IMRT response. | i (CV) |
| Toxicity | ||||||
| Abdollahi et al. [122] | 33 (P) | T2w, ADC | Rectal wall (M) | Rectal toxicity | Pre-IMRT MRI RF predict rectal toxicity. | i (CV) |
| Segmentation | ||||||
| Sunoqrot et al. [118] | 635 (R) | T2w | Prostate gland (M) | Quality System for automated prostate segmentation | Proposal of a quality check for automated segmentation of the prostate in T2W MR image. | e (2, CV) |
| Lay et al. [112] | 224 (R) | T2w, ADC, DWI | PCa (M) Prostate and TZ (A) | PCa segmentation | Random forest sampling strategy and instance-level weighting improve PCa detection performance compared to support vector machine. | i (2, CV) |
| Giannini et al. [113] | 58 (R) | T2w, ADC | PCa (M) | PCa segmentation | Proposed method with GLCM texture features computed on ADC and T2w images reduced the number of false positives and increased the precision of PCa detection. | i (CV) |
Abbreviations: ADC=Apparent diffusion coefficient, BCR=biochemical recurrence, bpMRI=biparametric magnetic resonance imaging, bRFS=biochemical recurrence free survival, CDI=current density imaging, csPCa= clinically significant prostate cancer, CV=cross validation, DCE=dynamic contrast enhanced, DTI= diffusion. tensor imaging, DWI=diffusion weighted imaging, GLCM= gray level co-occurrence matrix, GLRLM=grey-level run length matrix, GS=Gleason score, IMRT=intensity modulated radiotherapy, LOO=leave one out, LPOCV=leave-pair-out cross-validation, M=manual confirmation, ML=machine learning, mpMRI=multiparametric magnetic resonance imaging, PCa=Prostate cancer; PZ=peripheral zone, RF=radiomic feature, ROC-AUC=are under the receiver operating characteristics curve, RP=radical prostatectomy, T1w= T1-weighted imaging, T2w=T2-weighted imaging, TZ= transitional zone.
In two preliminary studies, Cameron et al. developed a model based on mpMRI RF for PCa detection implementing a comprehensible identification scheme by grouping features into the categories morphology, asymmetry, physiology and size (MAPS) [66, 67]. The model had an accuracy of 87% and outperformed models based on conventional mpMRI features [66]. Furthermore Khalvati et al. proposed a RF based framework for PCa detection and localization [74]. Additional studies, among which is the externally validated study by Viswanath et al. (ROC-AUC 0.683-0.768 across 3 sites), showed good area under the receiver operating characteristics curve (ROC-AUC) for PCa detection [68-70,75]. Five studies considered the tumor location (TZ vs. PZ) for RF based PCa detection [71-73,76,81]. Two of these demonstrated improved PZ and TZ lesion classification with ADC based RF [71] and a high ROC-AUC value of 0.86 [72]. Gholizadeh et al. developed a framework of combined T2w, DWI and DTI features for differentiation of PCa and non-PCa voxels [76]. Bleker et al. demonstrated that the addition of DCE-RFs does not improve performance of T2w and DWI-RF based models to detect clinically significant PCa in the PZ [80]. A multi-institutional and externally validated study by Ginsburg et al. showed lower results for a PZ specific classifier for PCa detection with ROC-AUC of 0.6-0.71 [73].
GS prediction and discrimination were assessed in 22 studies [83-100,116,119-121]. Most of these studies showed that RF models can differentiate between GS groups (low-, intermediate- and high risk), to predict GS or to predict GS upgrading between the biopsy and radical prostatectomy [83-91,93,114,119,120]. Chaddad et al. introduced novel RF based on Joint Intensity Matrix to predict GS (ROC-AUC 0.64-0.82 depending on GS groups) [84]. A study from Rozenberg et al. however, could not show that ADC features were predictive for GS upgrading in intermediate-risk prostate cancer [92]. Penzias et al. demonstrated that RF and quantitative histomorphometry correlate and are predictive for GS [121]. Hou et al. investigated the prediction of clinically significant PCa (GS≥7) in PIRADS 3 lesions, which could be a useful tool for biopsy guidance [94]. Zhang and colleagues showed that a radiomic signature, consisting of 10 features, identified clinically significantly PCa (GS ≥ 3+4) with AUC values of 0.95 (training), 0.86 (internal validation), and 0.81 (external validation) [84]. Algohary et al. reported that RFs from T2w and DWI sequences are associated with clinically significant PCa, being even more relevant than PIRADSv2 evaluation in some patients [99]. In an external validated study, a combination of intra- and peritumoral RF resulted in AUCs of 0.87 and 0.75 for the differentiation of low risk PCa versus high-risk PCa or intermediate- and high risk PCa defined by D'Amico Risk Classification [82].
Five studies investigated RF for the prediction of extracapsular extension and reported high AUC values between 0.80-0.90 for radiomic signatures based on T2w and ADC sequences [90,103,120] that outperformed clinical or nomogram models [104,105]. Two studies from Wang et al. and Zhang et al. showed that mpMRI derived RFs show good performance for bone metastasis prediction in untreated PCa with an ROC-AUC up to 0.92 [85,107]. Six studies analyzed the performance of RF in terms of outcome [108-111,117,122]. Bourbonne et al. externally validated an ADC based RF (SZEGLSZM), which was identified in a previous study [94] for biochemical recurrence (BCR) prediction after surgery with an accuracy of 0.76 [108,109]. Shiradkar et al. demonstrated a ML classifier derived from T2w and ADC RF with good prediction of BCR after surgery or RT. which was externally validated with a AUC of 0.73 [110]. Another RF model by Zhong et al. showed good performance for BCR prediction after RT of localized PCa [117]. Abdollahi and colleagues indicated that RF from pre- and post-treatment ADC images are predictive in terms of treatment response after primary external beam radiotherapy [111]. Another study from this group demonstrated that RF of pre-radiotherapy images provided good ROC-AUC values of up to 0.81 for rectal toxicity prediction [122]. One study by Sunoqrot et al. elaborated a quality system to asses automated prostate segmentations with external validation [118] and two studies from Lay et al. and Giannini et al. addressed RF-based PCa segmentation [112,113].
II PSMA-PET
Literature research revealed five original papers [123-127] using PET images to extract RF (see Table 2). Four studies used intraprostatic tumor as VOI for RF extraction [124-127] and one study non-PCa tissue in PET [123]. One study performed external validation of their results, the remaining studies were internally validated by CV [124,125] or two independent cohorts [126,127]. Four of these studies complied with IBSI [123-126].
Three studies aimed for GS discrimination [124-126] and demonstrated excellent ROC-AUC values between 0.81-0.91. Two studies chose intraprostatic tumor detection as study endpoint [123,126]. A study by Zambolgou et al. reported two distinct RFs (SAE, local binary pattern small-area emphasis; SZNUN, local binary pattern size-zone non-uniformity normalized) with good performance to detect significant PCa lesions not visible in PSMA-PET/CT. This result was externally validated by an independent cohort [123]. Cysouw et al. demonstrated a RF based machine learning model to predict lymph node involvement, presence of metastases, GS prediction (≥8) and presence of extracapsular extension [125].
III Other imaging modalities
Literature research revealed six original papers using CT scans [128-133] and four using TRUS imaging (n = 4) [47,134-136] to extract RF (see Table 3). The respective VOI for RF extraction were prostatic gland (n = 6) [47, 128, 129, 133-135], intraprostatic tumor (n=5) [136], bone metastases (n = 1) [132], lymph node metastasis (n=1) [131] and rectal/bladder wall (n = 1) [130]. All studies were internally validated by CV (n = 8) [128-134,136] or two independent cohorts (n = 3) [47,131,135]. Only one study complied with IBSI [131].
GS discrimination by RF was the aim of four studies using TRUS [134], CT [128,133] or CBCT images [129] and reported excellent ROC-AUC values between 0.77-0.98 including one or multiple RF for modeling. Three studies defined intraprostatic tumor detection in TRUS images [47,134,136] as a study endpoint. Again, the implementations of one or multiple RF led to very promising results in PCa detection. The study of Wu et al. implemented RF for automatic prostate gland delineation in TRUS images [135] and observed similar results compared to manual delineation by experts. One study [130] implemented RF to predict bladder and bowel toxicity after radiotherapy of PCa patients and reported ROC-AUCs of up to 0.77 by integrating clinical information with RF. The study of Osman et al. suggested that RF derived from CT images might enhance interpretation of treatment response of bone metastases [133] and Acar et al. demonstrated that RFs derived from CT images of PSMA-PET/CT scans could accurately distinguish between metastatic lesions and sclerotic area [132]. The RF model in a study by Peeken et al. outperformed conventional measures for detection of lymph nodes metastases [131].
IV Ongoing trials
In total, 5 studies were identified using mpMR imaging (n = 4), PET (n = 1), CT (n = 1) and bone scans (n = 1) to extract RF (see Table 4). Four studies evaluate RF for outcome prediction during or after several treatment approaches: active surveillance, surgery, radiotherapy, or radionuclide therapy in addition to chemotherapy. Two of those four studies integrate RF with molecular markers for modelling. One study evaluates whether RF extracted from lesions describe histologic characteristics, lymph node involvement and extension.
List of included articles on RFs derived from PSMA-PET images. In the second column # are the number of patients enrolled retrospectively (R) or prospectively (P). In the fourth column the volume of interest (VOI) is presented accompanied by the type of segmentation in brackets M = manual, SA = semiautomatic and A = fully automatic. The last column contains information on validation. The number stands for the number of cohorts used. 2 means one for development and one for testing.
| Study | # | Imaging Modality | VOI (Segmentation) | Endpoint(s) | Results | Validation |
|---|---|---|---|---|---|---|
| Zamboglou et al. [123] | 20 (P) 52 (R) | [68Ga]Ga-PSMA-11 PET | Non-PCa tissue | Visually not-detected lesions | 2 distinct RF with good performance (SAE, SZNUN) | e (2) |
| Papp et al.[124] | 52 (P) | [18F]FMC/ [68Ga]Ga-PSMA-11 PET/MRI | PCa (M) | Risk group discrimination, BCR | Machine learning RF based models. | i (CV) |
| Cysouw et al.[125] | 76 (P) | [18F]DCFPyL PET | PCa (SA) | Lymph node metastasis, metastasis, GS≥ 8, extracapsular extension | Radiomics-based machine learning models. | i (CV) |
| Zamboglou et al. [126] | 60 (R) | PSMA-PET | PCa (M) on PET images and on co-registered histology | PCa detection, GS, pN1 status | QSZHGE: quantization algorithm + short zones high gray-level emphasis. | i (2) |
| Alongi et al. [127] | 46 (R) | 18F-Choline PET | PCa (unknown) | PCa patients' outcome | 13 selected RF. | i (2) |
Abbreviations: CV= cross-validation; PCa = prostate cancer; GS = Gleason score, SAE, local binary pattern small-area emphasis; SZNUN, local binary pattern size-zone non-uniformity QSZHGE= quantization algorithm + short zones high gray-level emphasis.
List of included articles on RFs derived from other imaging modalities than MRI. In the second column # are the number of patients enrolled retrospectively (R) or prospectively (P). In the fourth column the volume of interest (VOI) is presented accompanied by the type of segmentation in brackets M = manual, SA = semiautomatic and A = fully automatic. The last column contains information on validation. “e” stands for external validation and “I” for internal. The number stands for the number of cohorts used. 2 means one cohort for development and one for testing.
| Study | # | Imaging Modality | VOI (Segmentation) | Endpoint(s) | Results | Validation |
|---|---|---|---|---|---|---|
| Prostate | ||||||
| Zhang et al. [47] | 113 (R) | TRUS: B-mode, Sono- elastography | Prostate (M) | PCa detection | Multimodal feature (4 RFs) learning. | i (2) |
| Wildboer et al. [134]. | 50 (R) | TRUS: B-mode, contrast enhanced US | Prostate (A) | PCa detection, GS | Multiparametric classifier (n=14). | i (CV) |
| Wu et al. [135] | 132 & 5 videos (R) | TRUS: B-mode | Prostate (A&M) | Prostate segmentation | Prostate segmentation framework utilizing speckle-induced texture features. | i (2) |
| Huang et al. [136] | 342 (R) | TRUS (M) | Rectangle around the biopsy core | PCa detection | RF for a support vector machine classifier. | i (CV) |
| Osman et al. [133] | 342 (R) | CT | Prostate (M) | GS, risk group discrimination | Radiomics classifier. | i (2, CV) |
| Tanadini-Lang et al. [128] | 41 (R) | CT perfusion | Prostate (M) | GS, risk group discrimination | Single and combined use of RF and conventional CT perfusion parameters. | i (CV) |
| Bosetti et al. [129] | 31 (R) | Cone-beam CT | Prostate (M) | Tumor stage, GS, PSA level, risk group discrimination, BCR | Histogram-based Energy and Kurtosis and a shape-based feature predict BCR and high risk. | i (CV) |
| Toxicity | ||||||
| Mostafei et al. [130] | 64 (P) | CT | Pre-treatment Rectal-& Bladder wall (M) | RT toxicity GI/GU ≥ grade 1 CTCAEv4.03 | Cystitis: clinical-radiomics (n=4) model. Proctitis: radiomics (n=3) model. | i (CV) |
| Lymph nodes | ||||||
| Peeken et al. [131] | 80 (R) | Contrast-enhanced CT from PSMA PET/CT scans | Lymph nodes (M) | Lymph node metastasis | Radiomics model significantly outperformed all conventional CT parameters. | i (CV, 2) |
| Bone metastases | ||||||
| Acar et al. [132] | 75 (R) | CT from PSMA PET/CT | Bone metastases | Discrimination of bone metastases that responded after treatment | Weighted k-nearest neighborhood algorithm. | i (CV) |
Abbreviations: CT = computed tomography, CV= cross-validation, GI=gastrointestinal, GS = Gleason score, GU=genitourinary; PCa = prostate cancer; QSZHGE= quantization algorithm + short zones high gray-level emphasis, TRUS = Transrectal Ultrasound
List of identified ongoing trials to extract radiomic features. Only aims concerning radiomics are mentioned above. In the second column # are the number of patients enrolled retrospectively (R) or prospectively (P). The third column displays the imaging modality (mpMRI=multiparametric magnetic resonance imaging, PSMA/FDG-PET=prostate specific membrane antigen fluorodeoxyglucose positron emission tomography, CT=computer tomography). The fourth column gives an overview of the study's aim(s).
| Study | # | Imaging Modality | Aim(s) |
|---|---|---|---|
| NCT03979573 | 90 (P) | mpMRI | Identification and monitoring of patients with RF in combination with clinical and molecular markers during active surveillance of PCa to reduce discontinuation. |
| NCT02242773 | 207 (P) | mpMRI | Correlation of RF with progression during active surveillance and with genomic signatures and other biomarkers. |
| NCT03180398 | 20 (P) | mpMRI | Extracted RF are used to identify dominant lesions within the prostate. These RF are monitored longitudinally to analyze their correlation with the local control. |
| NCT04219059 | 200 (R) | mpMRI | Evaluates if RF on primitive prostate lesions can describe histological characteristics, lymph node involvement and disease extension. |
| NCT04343885 | 140 (P) | PSMA/FDG-PET, CT, bone scans | Prognostic and predictive value RF from PET, CT or bone scans after Lutetium-177 PSMA radionuclide treatment and/ or chemotherapy. |
Discussion
PCa radiomics is an emerging research field with a high potential to offer non-invasive and longitudinal biomarkers for personalized medicine. In our review based on a qualitative synthesis of 77 studies, most papers address MRI based RFs, which is not surprising since MRI is the actual SoC for primary PCa staging. Other imaging modalities such as CT, PSMA-PET, TRUS and bone scan are less commonly used, but their application has improved in the recent years. This trend might proceed with the increased usage of PSMA PET/CT for staging of primary, recurrent, and metastasized PCa patients. One major focus of the included papers was PCa detection. Keeping in mind that image interpretation and segmentation is hampered by interobserver variability [6,137] implementation of RF might enhance diagnostic performance. Advances in automated segmentation of intraprostatic tumor lesions, for example by deep learning-based approaches such as convolutional neural networks, might overcome this limitation [138].
The other focus is GS discrimination, reflecting the need for improvements in risk stratification. It is not surprising that most of the studies chose GS discrimination, since GS is the most established histologic biomarker. In clinical routine, the GS before primary PCa therapy is evaluated in tissue cores obtained by biopsy. However, due to intratumoral heterogeneity the GS in biopsy cores and prostatectomy specimen is discordant in 20-60% of the patients [139,140]. Nevertheless, the bioptic GS has a significant impact on clinical management as it defines the patient's risk group influencing for example the duration of androgen deprivation therapy or the dose to the prostate during radiation therapy [141]. RF-based GS prediction might account for intratumoral heterogeneity leading to over- or underestimation of the GS in biopsy specimen. For instance, Zamboglou et al. demonstrated that a PSMA PET-derived RF (QSZHGE) may outperform biopsy mapping for GS 7 vs ≥8 discrimination [126]. Recently, Chu et al. examined the PSMA expression in a combined cohort of more than 18 000 radical prostatectomy specimens and observed a correlation between PSMA expression and the GS [142]. This finding provides a strong biological rationale for non-invasive GS prediction based on RF extracted from PSMA PET images.
However, several studies proposed that a thorough analysis of PCa tissue characteristics (e.g. by genomic analyses) might outperform GS for risk prediction [143]. Radiogenomics combines RF analysis with genomic information thus linking both research fields. Our literature search revealed five studies but none of the studies were internally or externally validated and thus excluded. Nevertheless, they should be mentioned, highlighting this modern and innovative approach [25,144-147]. A pilot study by Sun et al. showed weak correlations between RF and hypoxia gene expressions, providing an opportunity to assess the hypoxia status in PCa [146]. Two studies by McCann et al. and Switlyk et al. demonstrated an association between RF and the genetic marker phosphatase and tensin homolog [144,147]. Stoyanova et al. identified radiomic signatures which reflected genes that are over- and underexpressed in aggressive prostate cancer [25]. Additionally, another study with a small patient cohort by Kesch et al. suggests that RF signatures could distinguish between lesions of different aggressiveness [145].
Direct prediction of treatment outcome with RF is investigated in ongoing clinical trials especially. A possible explanation for this finding is the long follow-up time needed to provide reliable clinical information of treatment outcomes in PCa patients. Just a few manuscripts (n = 24) address extraprostatic extension (n = 6), BCR (n = 6), segmentation (n = 4), bone metastasis (n = 3), lymph node detection (n = 3) and radiotherapy toxicity (n = 2). Considering that most PCa patients are long-term survivors after treatment a reliable prediction of toxicity is warranted. Due to the lack of predictive models for toxicity prediction, we consider this field of major interest for future studies. Some of the excluded studies featured interesting concepts for the use of radiomics and treatment associated toxicity in PCa patients. Radiotherapy toxicity prediction was investigated for femoral head fractures [148] and urethral strictures after high-dose rate brachytherapy [149]. One paper used RF for response assessment of PCa bone lesions derived of and ADC maps [150]. Rossi et al. did not compute RF on imaging but on rectum and bladder 3D dose-volume histogram distributions. This add-on improved the prediction of late toxicities after radiotherapy [151]. These extensive fields of application demonstrate the great potential of radiomics and its clinical implementation from diagnosis to outcome and toxicity prediction in an era of big data and individualized medicine.
Overall, most of the included studies presented good to high AUC values. However, these findings need to be considered diligently regarding publication bias and the variability observed in RF. As illustrated above the radiomic pipeline is a sequence of operations and each operation can be modified [31]. RF and models are sensitive to those modifications and consequently, investigations on RF variability, robustness and reproducibility are demanded [31].
Texture features are increasingly sensitive to acquisition parameters with growing spatial resolution [152] as well as reconstruction algorithms [153]. Yang et al proposed a simulation framework to asses robustness and accuracy of radiomic textural features with different MRI acquisition parameters and reconstruction algorithms [153]. Recently Rai et al. developed a 3D printable phantom to measure repeatability and reproducibility of MRI-based radiomic features which could facilitate multi-center studies to harmonize image protocols and thereby tackling some of these challenges [160].
Multiple segmentations can reduce variability and bias in RF extraction of manually, semiautomatically or automatically segmented VOIs [154]. To increase robustness of segmentation manual methods should be avoided. In PET images, Bashir et al demonstrated that semiautomatic threshold-based methods yield superior interobserver reproducibility [155]. Additionally, CNN based segmentation methods showed good performance [156].
Isaakson et al investigated normalization techniques to enhance comparability across different subjects and visits [158]. Scalco et al. investigated different generally adopted image intensity normalization techniques for T2w-MRI images and demonstrated a relevant impact on reproducibility of RFs [154].
Schwier et al. investigated the variability of RF in MRI by using different filters, normalization, and image discretization techniques and observed that RF were sensitive to these pre-processing procedures. Hence, they recommended detailed reporting of the pre-processing steps and the use of open-source software [29]. Orlhac et al. reported that ComBat harmonization is efficient and enables MRI data pooling from different scanners and centers [155].
Two studies investigated repeatability of MRI-derived RFs and concluded that repeatability of many RFs is moderate and that a set of reproducible image features is desirable [156, 157]. Delgadillo et al. investigated repeatability of RF derived from CBCTs and reported that only five radiomic features were repeatable in < 97% of the reconstruction and preprocessing methods [159]. Bologna et al. proposed an approach to assess RF stability without multiple acquisitions and segmentations that could be used for preliminary RF selection. In addition, the authors advocated that RF derived of ADC maps behave differently based on the region extracted e.g. RF derived from head and neck tumors are less stable than those derived of sarcomas [161]. Pfaehler et al. recommends to investigate the repeatability of RF for every tumor type as well and for every PET-Tracer [30].
These papers demonstrate the fragility of RFs and the need of reproducible RF sets in order to enable a broad clinical application.
Consequentially, more research on prostate MRI and PSMA-PET RF robustness should be performed. Other approaches to tackle RF variability is the standardization of RF definitions and calculations which IBSI tries to promote [28]. The radiomics quality score, a tool to evaluate methodologic quality of radiomic studies, could also be used [32]. With higher quality, evidence on RF robustness like the recent metanalysis of Zwanenburg et al. pitfalls could be uncovered and described [33]. These methodological aspects seem all the more important, since only a few studies identified in this review are explicitly IBSI compliant and future work needs to focus on this issue. We furthermore encountered problems to validate the studies IBSI compliance, since most studies don't give sufficient information about the used software and calculations of RF. We therefore plead for uniform and detailed specifications.
Nevertheless, validation is pivotal considering the variability of RF. 35 of 238 articles were excluded due to missing validation. In internal validation different types might be utilized like the aforementioned ML algorithms, k-fold CV or leave-one-out CV, as well as independent datasets for model development and validation. A proper methodology and the separation of training and validation dataset is demanded at all times [157]. Our synthesis detected 64 articles with internal validation (k-fold CV n = 36; leave-one-out CV n = 11, two cohorts n = 29). 14 studies used more than one validation type. External validation is the gold standard and was performed in eight of the identified articles. Only one manuscript reported about external validation of an already published model [108]. These findings put ever more emphasis on the validation of radiomics models especially externally and from already published models [58].
Many studies used ML for model building and verification. ML and deep learning as a subfield are emerging and harbor great potential [48]. Li et al. used deep learning in combination with “hand-crafted” features and has successfully applied it in differentiating unilateral breast cancer from low-risk patients [46]. Segmentation of PCa lesions by deep learning networks is explored without “hand-crafted” features [158].
This review focusses on the clinical aspects of RF demonstrating its great potential to affect management of PCa. However, some technical aspects have not been further investigated: information on the used algorithms for RF extraction or ML approaches were not provided. Additionally, we did not state whether the published models or the parameters are publicly available.
In conclusion, most research in PCa radiomics focuses on PCa detection and GS discrimination. MRI as SoC is the most used imaging modality for RF computation for now, but PSMA-PET is gaining evidence in a wide variety of clinical settings. Most of the results suggest good to high performance of radiomics models but should be considered carefully due to RF variability. Further research is demanded on RF sensitivity and robustness especially on RF extracted of prostate MRI and PSMA-PET.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries
3. Mottet N, Bellmunt J, Bolla M, Briers E, Cumberbatch MG, De Santis M. et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2017;71(4):618-29
4. Kasivisvanathan V, Rannikko AS, Borghi M, Panebianco V, Mynderse LA, Vaarala MH. et al. MRI-targeted orstandard biopsy for prostate-cancer diagnosis. N Engl J Med. 2018 10;378(19):1767-1777
5. Van der Poel H, van der Kwast T, Aben K, Mottet N, Mason N. Imaging and T category for prostate cancer in the 8th edition of the union for international cancer control TNM classification. Eur Urol Oncol. 2020;3(5):563-564
6. Steenbergen P, Haustermans K, Lerut E, Oyen R, De Wever L, Van den Bergh L. et al. Prostate tumor delineation using multiparametric magnetic resonance imaging: Inter-observer variability and pathology validation. Radiother Oncol. 2015;115(2):186-90
7. Panebianco V, Giganti F, Kitzing YX, Cornud F, Campa R, De Rubeis G. et al. An update of pitfalls in prostate mpMRI: a practical approach through the lens of PI-RADS v. 2 guidelines. Insights Imaging. 2018;9(1):87-101
8. Johnson DC, Raman SS, Mirak SA, Kwan L, Bajgiran AM, Hsu W. et al. Detection of individual prostate cancer foci via multiparametric magnetic resonance imaging. Eur Urol. 2019;75(5):712-20
9. Turkbey B, Rosenkrantz AB, Haider MA, Padhani AR, Villeirs G, Macura KJ. et al. Prostate imaging reporting and data system version 2.1: 2019 update of prostate imaging reporting and data system version 2. Eur Urol. 2019;76(3):340-51
10. Roach PJ, Francis R, Emmett L, Hsiao E, Kneebone A, Hruby G. et al. The impact of <sup>68</sup> Ga-PSMA PET/CT on management intent in prostate cancer: results of an Australian prospective multicenter study. J Nucl Med. 2018;59(1):82-8
11. Hope TA, Goodman JZ, Allen IE, Calais J, Fendler WP, Carroll PR. Metaanalysis of 68Ga-PSMA-11 PET accuracy for the detection of prostate cancer validated by histopathology. J Nucl Med. 2019;60(6):786-93
12. Yaxley JW, Raveenthiran S, Nouhaud F-X, Samaratunga H, Yaxley WJ, Coughlin G. et al. Risk of metastatic disease on 68gallium-prostate-specific membrane antigen positron emission tomography/computed tomography scan for primary staging of 1253 men at the diagnosis of prostate cancer. BJU Int. 2019;124(3):401-7
13. Hofman MS, Lawrentschuk N, Francis RJ, Tang C, Vela I, Thomas P. et al. Prostate-specific membrane antigen PET-CT in patients with high-risk prostate cancer before curative-intent surgery or radiotherapy (proPSMA): a prospective, randomised, multi-centre study. Lancet. 2020 11;395(10231):1208-1216
14. Chen M, Zhang Q, Zhang C, Zhao X, Marra G, Gao J. et al. Combination of 68 Ga-PSMA PET/CT and multiparameter MRI improves the detection of clinically significant prostate cancer: a lesion by lesion analysis. J Nucl Med. 2018;60(7):944-949
15. Bettermann AS, Zamboglou C, Kiefer S, Jilg CA, Spohn S, Kranz-Rudolph J. et al. [68Ga-]PSMA-11 PET/CT and multiparametric MRI for gross tumor volume delineation in a slice by slice analysis with whole mount histopathology as a reference standard - Implications for focal radiotherapy planning in primary prostate cancer. Radiother Oncol. 2019 141:214-219
16. Donato P, Morton A, Yaxley J, Ranasinghe S, Teloken PE, Kyle S. et al. 68Ga-PSMA PET/CT better characterises localised prostate cancer after MRI and transperineal prostate biopsy: Is 68Ga-PSMA PET/CT guided biopsy the future? Eur J Nucl Med Mol Imaging. 2020;47(8):1843-1851
17. Spohn SKB, Kramer M, Kiefer S, Bronsert P, Sigle A, Schultze-Seemann W. et al. Comparison of Manual and Semi-Automatic [(18)F]PSMA-1007 PET Based Contouring Techniques for Intraprostatic Tumor Delineation in Patients With Primary Prostate Cancer and Validation With Histopathology as Standard of Reference. Front Oncol. 2020;10:600690
18. Zamboglou C, Kramer M, Kiefer S, Bronsert P, Ceci L, Sigle A. et al. The impact of the co-registration technique and analysis methodology in comparison studies between advanced imaging modalities and whole-mount-histology reference in primary prostate cancer. Sci Rep. 2021;11(1):5836
19. D'Amico AV, Whittington R, Malkowicz SB, Schultz D, Blank K, Broderick GA. et al. Biochemical outcome after radical prostatectomy, external beam radiation therapy, or interstitial radiation therapy for clinically localized prostate cancer. JAMA. 1998;280(11):969-74
20. Gleason DF, Mellinger GT. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J Urol. 2002 167(2, Part 2):953-8
21. Williams SG, Duchesne GM, Gogna NK, Millar JL, Pickles T, Pratt GR. et al. An international multicenter study evaluating the impact of an alternative biochemical failure definition on the judgment of prostate cancer risk. Int J Radiat Oncol Biol Phys. 2006;65(2):351-7
22. Schmidt-Hegemann N-S, Stief C, Kim T-H, Eze C, Kirste S, Strouthos I. et al. Outcome after PSMA PET/CT-based salvage radiotherapy in patients with biochemical recurrence after radical prostatectomy: a 2-institution retrospective analysis. J Nucl Med. 2019;60(2):227-33
23. Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501(7467):338-45
24. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563-77
25. Stoyanova R, Pollack A, Takhar M, Lynne C, Parra N, Lam LLC. et al. Association of multiparametric MRI quantitative imaging features with prostate cancer gene expression in MRI-targeted prostate biopsies. Oncotarget. 2016 7(33)
26. Yang F, Ford JC, Dogan N, Padgett KR, Breto AL, Abramowitz MC. et al. Magnetic resonance imaging (MRI)-based radiomics for prostate cancer radiotherapy. Transl Androl Urol. 2018;7(3):445-58
27. Patel N, Henry A, Scarsbrook A. The value of MR textural analysis in prostate cancer. Clin Radiol. 2019;74(11):876-85
28. Sun Y, Reynolds HM, Parameswaran B, Wraith D, Finnegan ME, Williams S. et al. Multiparametric MRI and radiomics in prostate cancer: a review. Australas Phys Eng Sci Med. 2019;42(1):3-25
29. Yao S, Jiang H, Song B. Radiomics in prostate cancer: basic concepts and current state-of-the-art. Chin J Acad Radiol. 2020;2(3):47-55
30. Zwanenburg A, Leger S, Vallières M, Löck S. Image biomarker standardisation initiative. Radiology. 2020:191145.
31. Schwier M, Griethuysen vJ, Vangel MG, Pieper S, Peled S, Tempany C. et al. Repeatability of multiparametric prostate MRI radiomics features. Sci Rep. 2019;9(1):9441
32. Pfaehler E, Beukinga RJ, de Jong JR, Slart RHJA, Slump CH, Dierckx RAJO. et al. Repeatability of 18F-FDG PET radiomic features: A phantom study to explore sensitivity to image reconstruction settings, noise, and delineation method. Med Phys. 2019;46(2):665-78
33. Traverso A, Wee L, Dekker A, Gillies R. Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol Biol Phys. 2018;102(4):1143-58
34. Sanduleanu S, Woodruff HC, de Jong EEC, van Timmeren JE, Jochems A, Dubois L. et al. Tracking tumor biology with radiomics: A systematic review utilizing a radiomics quality score. Radiother Oncol. 2018;127(3):349-60
35. Zwanenburg A. Radiomics in nuclear medicine: robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur J Nucl Med Mol Imaging. 2019;46(13):2638-55
36. Moradmand H, Aghamiri SMR, Ghaderi R. Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J Appl Clin Med Phys. 2020;21(1):179-90
37. van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V. et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77(21):e104-e7
38. Bettinelli A, Branchini M, De Monte F, Scaggion A, Paiusco M. Technical Note: An IBEX adaption toward image biomarker standardization. Med Phys. 2020;47(3):1167-73
39. Pfaehler E, Zwanenburg A, de Jong JR, Boellaard R. RaCaT: An open source and easy to use radiomics calculator tool. PLoS One. 2019;14(2):e0212223-e
40. Echegaray S, Bakr S, Rubin DL, Napel S. Quantitative Image Feature Engine (QIFE): an open-source, modular engine for 3D quantitative feature extraction from volumetric medical images. J Digit Imaging. 2018;31(4):403-14
41. Szczypiński PM, Strzelecki M, Materka A, Klepaczko A. MaZda-a software package for image texture analysis. Comput Methods Programs Biomed. 2009;94(1):66-76
42. Apte AP, Iyer A, Crispin-Ortuzar M, Pandya R, van Dijk LV, Spezi E. et al. Technical Note: Extension of CERR for computational radiomics: A comprehensive MATLAB platform for reproducible radiomics research. Med Phys. 2018;45(8):3713-20
43. Nioche C, Orlhac F, Boughdad S, Reuzé S, Goya-Outi J, Robert C. et al. LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 2018;78(16):4786-4789
44. Larue RTHM, Defraene G, De Ruysscher D, Lambin P, van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol. 2017;90(1070):20160665
45. Mun SK, Wong KH, Lo SB, Li Y, Bayarsaikhan S. Artificial intelligence for the future radiology diagnostic service. Front Mol Biosci. 2020;7:614258
46. Li H, Giger ML, Huynh BQ, Antropova NO. Deep learning in breast cancer risk assessment: evaluation of convolutional neural networks on a clinical dataset of full-field digital mammograms. J Med Imaging (Bellingham). 2017;4(4):041304
47. Zhang Q, Xiong J, Cai Y, Shi J, Xu S, Zhang B. Multimodal feature learning and fusion on B-mode ultrasonography and sonoelastography using point-wise gated deep networks for prostate cancer diagnosis. Biomed Tech (Berl). 2020;65(1):87-98
48. Levine AB, Schlosser C, Grewal J, Coope R, Jones SJM, Yip S. Rise of the machines: advances in deep learning for cancer diagnosis. Trends Cancer. 2019;5(3):157-69
49. Orlhac F, Soussan M, Maisonobe J-A, Garcia CA, Vanderlinden B, Buvat I. Tumor texture analysis in 18F-FDG PET: relationships between texture parameters, histogram Indices, standardized uptake values, metabolic volumes, and total lesion glycolysis. J Nucl Med. 2014;55(3):414-22
50. Guezennec C, Bourhis D, Orlhac F, Robin P, Corre J-B, Delcroix O. et al. Inter-observer and segmentation method variability of textural analysis in pre-therapeutic FDG PET/CT in head and neck cancer. PLoS One. 2019;14(3):e0214299
51. Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH. et al. Robust Radiomics feature quantification using semiautomatic volumetric segmentation. PLoS One. 2014;9(7):e102107
52. Caicedo JC, Cooper S, Heigwer F, Warchal S, Qiu P, Molnar C. et al. Data-analysis strategies for image-based cell profiling. Nat Methods. 2017;14(9):849-63
53. Leger S, Zwanenburg A, Pilz K, Lohaus F, Linge A, Zöphel K. et al. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep. 2017;7(1):13206
54. Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep. 2015;5(1):13087
55. Avanzo M, Wei L, Stancanello J, Vallières M, Rao A, Morin O. et al. Machine and deep learning methods for radiomics. Med Phys. 2020;47(5):e185-e202
56. Benjamini Y, Hochberg Y. Controlling The false discovery rate - A practical and powerful approach to multiple testing. J. R. Statist Soc. B. 1995;57:289-300
57. Chalkidou A, O'Doherty MJ, Marsden PK. False discovery rates in PET and CT studies with texture features: a systematic review. PLoS One. 2015;10(5):e0124165
58. Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ. 2015 350
59. Efron B. Estimating the Error Rate of a Prediction Rule: Improvement on Cross-Validation. J Am Stat Assocn. 1983;78(382):316-31
60. Baumann K. Cross-validation as the objective function for variable-selection techniques. Trends Analyt Chem. 2003;22(6):395-406
61. Haralick RM, Shanmugam K. Textural features for image classification. IEEE Trans Syst Man Cybern Syst. 1973(6):610-21.
62. Thibault G, Angulo J, Meyer F. Advanced statistical matrices for texture characterization: application to cell classification. IEEE Trans Biomed Eng. 2014;61(3):630-7
63. Galloway MM. Texture analysis using gray level run lengths. Computer Graphics and Image Processing. 1975;4(2):172-9
64. Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Carvalho S. et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5(1):4006
65. Moher D. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264
66. Cameron A, Khalvati F, Haider MA, Wong A. MAPS: a quantitative radiomics approach for prostate cancer detection. IEEE Trans Biomed Eng. 2016;63(6):1145-56
67. Cameron A, Modhafar A, Khalvati F, Lui D, Shafiee MJ, Wong A, et al., editors. Multiparametric MRI prostate cancer analysis via a hybrid morphological-textural model. 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. 2014 2014/08/262014
68. Viswanath SE, Chirra PV, Yim MC, Rofsky NM, Purysko AS, Rosen MA. et al. Comparing radiomic classifiers and classifier ensembles for detection of peripheral zone prostate tumors on T2-weighted MRI: a multi-site study. BMC Med Imaging. 2019;19(1):22
69. Khalvati F, Wong A, Haider MA. Automated prostate cancer detection via comprehensive multi-parametric magnetic resonance imaging texture feature models. BMC Med Imaging. 2015;15:27
70. Xu M, Fang M, Zou J, Yang S, Yu D, Zhong L. et al. Using biparametric MRI radiomics signature to differentiate between benign and malignant prostate lesions. Eur J Radiol. 2019;114:38-44
71. Bonekamp D, Kohl S, Wiesenfarth M, Schelb P, Radtke JP, Götz M. et al. Radiomic machine learning for characterization of prostate lesions with MRI: comparison to ADC values. Radiology. 2018;289(1):128-37
72. Sidhu HS, Benigno S, Ganeshan B, Dikaios N, Johnston EW, Allen C. et al. Textural analysis of multiparametric MRI detects transition zone prostate cancer. Eur Radiol. 2017;27(6):2348-58
73. Ginsburg SB, Algohary A, Pahwa S, Gulani V, Ponsky L, Aronen HJ. et al. Radiomic features for prostate cancer detection on MRI differ between the transition and peripheral zones: Preliminary findings from a multi-institutional study. J Magn Reson Imaging. 2017;46(1):184-93
74. Khalvati F, Zhang J, Chung AG, Shafiee MJ, Wong A, Haider MA. MPCaD: a multi-scale radiomics-driven framework for automated prostate cancer localization and detection. BMC Med Imaging. 2018;18(1):16
75. Wang J, Wu C-J, Bao M-L, Zhang J, Wang X-N, Zhang Y-D. Machine learning-based analysis of MR radiomics can help to improve the diagnostic performance of PI-RADS v2 in clinically relevant prostate cancer. Eur Radiol. 2017;27(10):4082-90
76. Gholizadeh N, Simpson J, Ramadan S, Denham J, Lau P, Siddique S. et al. Voxel-based supervised machine learning of peripheral zone prostate cancer using noncontrast multiparametric MRI. J Appl Clin Med Phys. 2020;21(10):179-91
77. Hu L, Zhou DW, Fu CX, Benkert T, Jiang CY, Li RT. et al. Advanced zoomed diffusion-weighted imaging vs. full-field-of-view diffusion-weighted imaging in prostate cancer detection: a radiomic features study. Eur Radiol. 2020;31(3):1760-1769
78. Woźnicki P, Westhoff N, Huber T, Riffel P, Froelich MF, Gresser E. et al. Multiparametric MRI for prostate cancer characterization: combined use of radiomics model with PI-RADS and clinical parameters. Cancers (Basel). 2020 12(7)
79. Qi Y, Zhang S, Wei J, Zhang G, Lei J, Yan W. et al. Multiparametric MRI-based radiomics for prostate cancer screening with PSA in 4-10 ng/mL to reduce unnecessary biopsies. J Magn Reson Imaging. 2020;51(6):1890-9
80. Bleker J, Kwee TC, Dierckx RAJO, de Jong IJ, Huisman H, Yakar D. Multiparametric MRI and auto-fixed volume of interest-based radiomics signature for clinically significant peripheral zone prostate cancer. Eur Radiol. 2020;30(3):1313-24
81. Wu M, Krishna S, Thornhill RE, Flood TA, McInnes MDF, Schieda N. Transition zone prostate cancer: Logistic regression and machine-learning models of quantitative ADC, shape and texture features are highly accurate for diagnosis. J Magn Reson Imaging. 2019;50(3):940-50
82. Kwon D, Reis IM, Breto AL, Tschudi Y, Gautney N, Zavala-Romero O. et al. Classification of suspicious lesions on prostate multiparametric MRI using machine learning. J Med Imaging (Bellingham). 2018;5(3):034502
83. Hectors SJ, Cherny M, Yadav KK, Beksaç AT, Thulasidass H, Lewis S. et al. Radiomics features measured with multiparametric magnetic resonance imaging predict prostate cancer aggressiveness. J Urol. 2019;202(3):498-505
84. Chaddad A, Kucharczyk MJ, Niazi T. Multimodal radiomic features for the predicting Gleason score of prostate cancer. Cancers (Basel). 2018;10(8):249
85. Chaddad A, Niazi T, Probst S, Bladou F, Anidjar M, Bahoric B. Predicting Gleason score of prostate cancer patients using radiomic analysis. Front Oncol. 2018;8:630
86. Jensen C, Carl J, Boesen L, Langkilde NC, Østergaard LR. Assessment of prostate cancer prognostic Gleason grade group using zonal-specific features extracted from biparametric MRI using a KNN classifier. J Appl Clin Med Phys. 2019;20(2):146-53
87. Chen T, Li M, Gu Y, Zhang Y, Yang S, Wei C. et al. Prostate cancer differentiation and aggressiveness: assessment with a radiomic-based model vs. PI-RADS v2. J Magn Reson Imaging. 2019;49(3):875-84
88. Toivonen J, Montoya Perez I, Movahedi P, Merisaari H, Pesola M, Taimen P. et al. Radiomics and machine learning of multisequence multiparametric prostate MRI: Towards improved non-invasive prostate cancer characterization. PLoS One. 2019 14(7)
89. Zhang G-m-y, Han Y-q, Wei J-w, Qi Y-f, Gu D-s, Lei J. et al. Radiomics based on MRI as a biomarker to guide therapy by predicting upgrading of prostate cancer from biopsy to radical prostatectomy. J Magn Reson Imaging. 2020;52(4):1239-1248
90. Min X, Li M, Dong D, Feng Z, Zhang P, Ke Z. et al. Multi-parametric MRI-based radiomics signature for discriminating between clinically significant and insignificant prostate cancer: Cross-validation of a machine learning method. Eur J Radiol. 2019;115:16-21
91. Li J, Weng Z, Xu H, Zhang Z, Miao H, Chen W. et al. Support Vector Machines (SVM) classification of prostate cancer Gleason score in central gland using multiparametric magnetic resonance images: A cross-validated study. Eur J Radiol. 2018;98:61-7
92. Rozenberg R, Thornhill RE, Flood TA, Hakim SW, Lim C, Schieda N. Whole-tumor Qquantitative apparent diffusion coefficient histogram and texture analysis to predict Gleason score upgrading in intermediate-risk 3 + 4 = 7 prostate cancer. AJR Am J Roentgenol. 2016;206(4):775-82
93. Fehr D, Veeraraghavan H, Wibmer A, Gondo T, Matsumoto K, Vargas HA. et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci U S A. 2015;112(46):E6265-E73
94. Hou Y, Bao M-L, Wu C-J, Zhang J, Zhang Y-D, Shi H-B. A radiomics machine learning-based redefining score robustly identifies clinically significant prostate cancer in equivocal PI-RADS score 3 lesions. Abdom Radiol (NY). 2020;45(12):4223-34
95. Li M, Chen T, Zhao W, Wei C, Li X, Duan S. et al. Radiomics prediction model for the improved diagnosis of clinically significant prostate cancer on biparametric MRI. Quant Imaging Med Surg. 2020;10(2):368-79
96. Algohary A, Shiradkar R, Pahwa S, Purysko A, Verma S, Moses D. et al. Combination of peri-tumoral and intra-tumoral radiomic features on bi-parametric MRI accurately stratifies prostate cancer risk: a multi-site study. Cancers (Basel). 2020 12(8)
97. Gugliandolo SG, Pepa M, Isaksson LJ, Marvaso G, Raimondi S, Botta F. et al. MRI-based radiomics signature for localized prostate cancer: a new clinical tool for cancer aggressiveness prediction? Sub-study of prospective phase II trial on ultra-hypofractionated radiotherapy (AIRC IG-13218). Eur Radiol. 2021;31(2):716-28
98. Zhang Y, Chen W, Yue X, Shen J, Gao C, Pang P. et al. Development of a novel, multi-parametric, MRI-based radiomic nomogram for differentiating between clinically significant and insignificant prostate cancer. Front Oncol. 2020;10:888
99. Algohary A, Viswanath S, Shiradkar R, Ghose S, Pahwa S, Moses D. et al. Radiomic features on MRI enable risk categorization of prostate cancer patients on active surveillance: Preliminary findings. J Magn Reson Imaging. 2018 22;10.1002/jmri.25983
100. Abraham B, Nair MS. Computer-aided classification of prostate cancer grade groups from MRI images using texture features and stacked sparse autoencoder. Comput Med Imaging Graph. 2018;69:60-8
101. Ma S, Xie H, Wang H, Han C, Yang J, Lin Z. et al. MRI-based radiomics signature for the preoperative prediction of extracapsular extension of prostate cancer. J Magn Reson Imaging. 2019;50(6):1914-25
102. Ma S, Xie H, Wang H, Yang J, Han C, Wang X. et al. Preoperative prediction of extracapsular extension: radiomics signature based on magnetic resonance imaging to stage prostate cancer. Mol Imaging Biol. 2019;22(3):711-72
103. Stanzione A, Cuocolo R, Cocozza S, Romeo V, Persico F, Fusco F. et al. Detection of extraprostaticextension of cancer on biparametric MRI combining texture analysis and machine learning: preliminary results. Acad Radiol. 2019;26(10):1338-44
104. Losnegård A, Reisæter LAR, Halvorsen OJ, Jurek J, Assmus J, Arnes JB. et al. Magnetic resonance radiomics for prediction of extraprostatic extension in non-favorable intermediate- and high-risk prostate cancer patients. Acta Radiol. 2020;61(11):1570-9
105. Xu L, Zhang G, Zhao L, Mao L, Li X, Yan W. et al. Radiomics based on multiparametric magnetic resonance imaging to predict extraprostatic extension of prostate cancer. Front Oncol. 2020;10:940
106. Wang Y, Yu B, Zhong F, Guo Q, Li K, Hou Y. et al. MRI-based texture analysis of the primary tumor for pre-treatment prediction of bone metastases in prostate cancer. Magn Reson Imaging. 2019;60:76-84
107. Zhang W, Mao N, Wang Y, Xie H, Duan S, Zhang X. et al. A Radiomics nomogram for predicting bone metastasis in newly diagnosed prostate cancer patients. Eur J Radiol. 2020;128:109020
108. Bourbonne V, Fournier G, Vallieres M, Lucia F, Doucet L, Tissot V. et al. External validation of an MRI-derived radiomics model to predict biochemical recurrence after surgery for high-risk prostate cancer. Cancers (Basel). 2020 12(4)
109. Bourbonne V, Vallières M, Lucia F, Doucet L, Visvikis D, Tissot V. et al. MRI-derived radiomics to guide post-operative management for high-risk prostate cancer. Front Oncol. 2019;9:807
110. Shiradkar R, Ghose S, Jambor I, Taimen P, Ettala O, Purysko AS. et al. Radiomic features from pretreatment biparametric MRI predict prostate cancer biochemical recurrence: Preliminary findings. J Magn Reson Imaging. 2018;48(6):1626-36
111. Abdollahi H, Mofid B, Shiri I, Razzaghdoust A, Saadipoor A, Mahdavi A. et al. Machine learning-based radiomic models to predict intensity-modulated radiation therapy response, Gleason score and stage in prostate cancer. Radiol med. 2019;124(6):555-67
112. Lay N, Tsehay Y, Greer MD, Turkbey B, Kwak JT, Choyke PL. et al. Detection of prostate cancer in multiparametric MRI using random forest with instance weighting. J Med Imaging (Bellingham). 2017;4(2):024506
113. Giannini V, Rosati S, Regge D, Balestra G. Specificity improvement of a CAD system for multiparametric MR prostate cancer using texture features and artificial neural networks. Health and Technol (Berl). 2017;7(1):71-80
114. Parra NA, Lu H, Choi J, Gage K, Pow-Sang J, Gillies RJ. et al. Habitats in DCE-MRI to predict clinically significant prostate cancers. Tomography. 2019;5(1):68-76
115. Dulhanty C, Wang L, Cheng M, Gunraj H, Khalvati F, Haider MA. et al. Radiomics driven diffusion weighted imaging sensing strategies for zone-Level Prostate Cancer Sensing. Sensors (Basel). 2020 20(5)
116. Gong L, Xu M, Fang M, Zou J, Yang S, Yu X. et al. Noninvasive prediction of high-grade prostate cancer via biparametric MRI radiomics. J Magn Reson Imaging. 2020;52(4):1102-9
117. Zhong Q-Z, Long L-H, Liu A, Li C-M, Xiu X, Hou X-Y. et al. Radiomics of multiparametric MRI to predict biochemical recurrence of localized prostate cancer after radiation therapy. Front Oncol. 2020;10:731
118. Sunoqrot MRS, Selnæs KM, Sandsmark E, Nketiah GA, Zavala-Romero O, Stoyanova R. et al. A quality control system for automated prostate segmentation on T2-Weighted MRI. Diagnostics (Basel). 2020 10(9)
119. Sun Y, Reynolds HM, Wraith D, Williams S, Finnegan ME, Mitchell C. et al. Automatic stratification of prostate tumour aggressiveness using multiparametric MRI: a horizontal comparison of texture features. Acta Oncol. 2019;58(8):1118-26
120. McGarry SD, Bukowy JD, Iczkowski KA, Unteriner JG, Duvnjak P, Lowman AK. et al. Gleason probability maps: a radiomics tool for mapping prostate cancer likelihood in MRI space. Tomography (Ann Arbor, Mich). 2019;5(1):127-34
121. Penzias G, Singanamalli A, Elliott R, Gollamudi J, Shih N, Feldman M. et al. Identifying the morphologic basis for radiomic features in distinguishing different Gleason grades of prostate cancer on MRI: Preliminary findings. PLoS One. 2018;13(8):e0200730
122. Abdollahi H, Mahdavi SR, Mofid B, Bakhshandeh M, Razzaghdoust A, Saadipoor A. et al. Rectal wall MRI radiomics in prostate cancer patients: prediction of and correlation with early rectal toxicity. Int J Radiat Oncol Biol Phys. 2018;94(9):829-37
123. Zamboglou C, Bettermann AS, Gratzke C, Mix M, Ruf J, Kiefer S. et al. Uncovering the invisible-prevalence, characteristics, and radiomics feature-based detection of visually undetectable intraprostatic tumor lesions in (68)GaPSMA-11 PET images of patients with primary prostate cancer. Eur J Nucl Med Mol Imaging. 2021Jun;48(6):1987-1997
124. Papp L, Spielvogel CP, Grubmüller B, Grahovac M, Krajnc D, Ecsedi B. et al. Supervised machine learning enables non-invasive lesion characterization in primary prostate cancer with [(68)Ga]Ga-PSMA-11 PET/MRI. Eur J Nucl Med Mol Imaging.Jun;48(6):1795-1805.
125. Cysouw MCF, Jansen BHE, van de Brug T, Oprea-Lager DE, Pfaehler E, de Vries BM. et al. Machine learning-based analysis of [(18)F]DCFPyL PET radiomics for risk stratification in primary prostate cancer. Eur J Nucl Med Mol Imaging. 2021;48(2):340-9
126. Zamboglou C, Carles M, Fechter T, Kiefer S, Reichel K, Fassbender TF. et al. Radiomic features from PSMA PET for non-invasive intraprostatic tumor discrimination and characterization in patients with intermediate- and high-risk prostate cancer - a comparison study with histology reference. Theranostics. 2019;9(9):2595-605
127. Alongi P, Laudicella R, Stefano A, Caobelli F, Comelli A, Vento A. et al. Choline PET/CT features to predict survival outcome in high risk prostate cancer restaging: a preliminary machine-learning radiomics study. Q J Nucl Med Mol Imaging. 2020
128. Tanadini-Lang S, Bogowicz M, Veit-Haibach P, Huellner M, Pauli C, Shukla V. et al. Exploratory radiomics in computed tomography perfusion of prostate cancer. Anticancer Res. 2018;38(2):685-90
129. Bosetti DG, Ruinelli L, Piliero MA, van der Gaag LC, Pesce GA, Valli M. et al. Cone-beam computed tomography-based radiomics in prostate cancer: a mono-institutional study. Strahlenther Onkol. 2020;196(10):943-51
130. Mostafaei S, Abdollahi H, Kazempour Dehkordi S, Shiri I, Razzaghdoust A, Zoljalali Moghaddam SH. et al. CT imaging markers to improve radiation toxicity prediction in prostate cancer radiotherapy by stacking regression algorithm. Radiol med. 2020;125(1):87-97
131. Peeken JC, Shouman MA, Kroenke M, Rauscher I, Maurer T, Gschwend JE. et al. A CT-based radiomics model to detect prostate cancer lymph node metastases in PSMA radioguided surgery patients. Eur J Nucl Med Mol Imaging. 2020;47(13):2968-77
132. Acar E, Leblebici A, Ellidokuz BE, Başbınar Y, Kaya GÇ. Machine learning for differentiating metastatic and completely responded sclerotic bone lesion in prostate cancer: a retrospective radiomics study. Br J Radiol. 2019;92(1101):20190286
133. Osman SOS, Leijenaar RTH, Cole AJ, Lyons CA, Hounsell AR, Prise KM. et al. Computed tomography-based radiomics for risk stratification in prostate cancer. Int J Radiat Oncol Biol Phys. 2019;105(2):448-56
134. Wildeboer RR, Mannaerts CK, van Sloun RJG, Budäus L, Tilki D, Wijkstra H. et al. Automated multiparametric localization of prostate cancer based on B-mode, shear-wave elastography, and contrast-enhanced ultrasound radiomics. Eur Radiol. 2020;30(2):806-15
135. Wu P, Liu Y, Li Y, Liu B. Robust prostate segmentation using intrinsic properties of TRUS images. IEEE Trans Med Imaging. 2015;34(6):1321-35
136. Huang X, Chen M, Liu P, Du Y. Texture feature-based classification on transrectal ultrasound image for prostatic cancer detection. Comput Math Methods Med. 2020;2020:7359375
137. Zamboglou C, Fassbender TF, Steffan L, Schiller F, Fechter T, Carles M. et al. Validation of different PSMA-PET/CT-based contouring techniques for intraprostatic tumor definition using histopathology as standard of reference. Radiother Oncol. 2019;141:208-213
138. Kostyszyn D, Fechter T, Bartl N, Grosu AL, Gratzke C, Sigle A. et al. Intraprostatic tumour segmentation on PSMA-PET images in patients with primary prostate cancer with a convolutional neural network. J Nucl Med. 2021 1;62(6):823-828
139. Divrik RT, Eroglu A, Sahin A, Zorlu F, Ozen H. Increasing the number of biopsies increases the concordance of Gleason scores of needle biopsies and prostatectomy specimens. Urol Oncol. 2007;25(5):376-82
140. San Francisco IF, DeWolf WC, Rosen S, Upton M, Olumi AF. Extended prostate needle biopsy improves concordance of Gleason grading between prostate needle biopsy and radical prostatectomy. J Urol. 2003;169(1):136-40
141. Mottet N, Bellmunt J, Bolla M, Briers E, Cumberbatch MG, De Santis M. et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2017;71(4):618-29
142. Chu CE, Alshalalfa M, Sjöström M, Zhao SG, Liu Y, Chou J. et al. Prostate-specific membrane mntigen and fluciclovine transporter genes are associated with variable clinical features and molecular subtypes ofprimary prostate cancer. Eur Urol. 2021;79(6):717-21
143. Spratt DE, Yousefi K, Deheshi S, Ross AE, Den RB, Schaeffer EM. et al. Individual patient-level meta-analysis of the performance of the decipher genomic classifier in high-risk men after prostatectomy to predict development of metastatic disease. J Clin Oncol. 2017;35(18):1991-8
144. McCann SM, Jiang Y, Fan X, Wang J, Antic T, Prior F. et al. Quantitative multiparametric MRI features and PTEN expression of peripheral zone prostate cancer: a pilot study. AJR Am J Roentgenol. 2016;206(3):559-65
145. Kesch C, Radtke J-P, Wintsche A, Wiesenfarth M, Luttje M, Gasch C. et al. Correlation between genomic index lesions and mpMRI and 68Ga-PSMA-PET/CT imaging features in primary prostate cancer. Sci Rep. 2018;8(1):16708
146. Sun Y, Williams S, Byrne D, Keam S, Reynolds HM, Mitchell C. et al. Association analysis between quantitative MRI features and hypoxia-related genetic profiles in prostate cancer: a pilot study. Br J Radiol. 2019;92(1104):20190373
147. Switlyk MD, Salberg UB, Geier OM, Vlatkovic L, Lilleby W, Lyng H. et al. PTEN Expression in Prostate Cancer: Relationship With Clinicopathologic Features and Multiparametric MRI Findings. AJR Am J Roentgenol. 2019:1-9
148. Abdollahi H, Mahdavi SR, Shiri I, Mofid B, Bakhshandeh M, Rahmani K. Magnetic resonance imaging radiomic feature analysis of radiation-induced femoral head changes in prostate cancer radiotherapy. J Cancer Res Ther. 2019;15(Supplement):S11-S9
149. Tsang YM, Vignarajah D, McWilliam A, Tharmalingam H, Lowe G, Choudhury A. et al. A pilot study on dosimetric and radiomics analysis of urethral strictures following HDR brachytherapy as monotherapy for localized prostate cancer. Br J Radiol. 2019;93(1106):20190760
150. Reischauer C, Patzwahl R, Koh D-M, Froehlich JM, Gutzeit A. Texture analysis of apparent diffusion coefficient maps for treatment response assessment in prostate cancer bone metastases-A pilot study. Eur J Radiol. 2018;101:184-90
151. Rossi L, Bijman R, Schillemans W, Aluwini S, Cavedon C, Witte M. et al. Texture analysis of 3D dose distributions for predictive modelling of toxicity rates in radiotherapy. Radiother Oncol. 2018;129(3):548-553
152. Mayerhoefer ME, Szomolanyi P, Jirak D, Materka A, Trattnig S. Effects of MRI acquisition parameter variations and protocol heterogeneity on the results of texture analysis and pattern discrimination: An application-oriented study. Med Phys. 2009;36(4):1236-43
153. Yang F, Dogan N, Stoyanova R, Ford JC. Evaluation of radiomic texture feature error due to MRI acquisition and reconstruction: A simulation study utilizing ground truth. Phys Med. 2018;50:26-36
154. Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J. et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749-62
155. Bashir U, Azad G, Siddique MM, Dhillon S, Patel N, Bassett P. et al. The effects of segmentation algorithms on the measurement of (18)F-FDG PET texture parameters in non-small cell lung cancer. EJNMMI Res. 2017;7(1):60
156. Hatt M, Laurent B, Ouahabi A, Fayad H, Tan S, Li L. et al. The first MICCAI challenge on PET tumor segmentation. Med Image Anal. 2018;44:177-95
157. Zwanenburg A, Löck S. Why validation of prognostic models matters? Radiother Oncol. 2018;127(3):370-3
158. Wang X, Yang W, Weinreb J, Han J, Li Q, Kong X. et al. Searching for prostate cancer by fully automated magnetic resonance imaging classification: deep learning versus non-deep learning. Sci Rep. 2017;7(1):1-8
159. Delgadillo R, Spieler BO, Ford JC, Kwon D, Yang F, Studenski M, Padgett KR, Abramowitz MC, Dal Pra A, Stoyanova R, Pollack A, Dogan N. Repeatability of CBCT radiomic features and their correlation with CT radiomic features for prostate cancer. Med Phys. 2021May;48(5):2386-2399 doi: 10.1002/mp.14787. Epub 2021 Mar 16. PMID: 33598943
160. Rai R, Holloway LC, Brink C, Field M, Christiansen RL, Sun Y, Barton MB, Liney GP. Multicenter evaluation of MRI-based radiomic features: A phantom study. Med Phys. 2020Jul;47(7):3054-3063 doi: 10.1002/mp.14173. Epub 2020 Apr 30. PMID: 32277703
161. Bologna M, Corino VDA, Montin E, Messina A, Calareso G, Greco FG, Sdao S, Mainardi LT. Assessment of Stability and Discrimination Capacity of Radiomic Features on Apparent Diffusion Coefficient Images. J Digit Imaging. 2018Dec;31(6):879-894 doi: 10.1007/s10278-018-0092-9. PMID: 29725965; PMCID: PMC6261192
Author contact
![]() Corresponding author: Simon Spohn, Department of Radiation Oncology, Medical Center - University of Freiburg, Robert-Koch Straße 3, 79106 Freiburg. Telephone: +49 (0)761 270 94010; Email: simon.spohnde
Corresponding author: Simon Spohn, Department of Radiation Oncology, Medical Center - University of Freiburg, Robert-Koch Straße 3, 79106 Freiburg. Telephone: +49 (0)761 270 94010; Email: simon.spohnde
 Global reach, higher impact
Global reach, higher impact