13.3
Impact Factor
Theranostics 2020; 10(23):10548-10562. doi:10.7150/thno.46826 This issue Cite
Research Paper
E. coli nitroreductase NfsA is a reporter gene for non-invasive PET imaging in cancer gene therapy applications
1. Auckland Cancer Society Research Centre, University of Auckland, Auckland, New Zealand.
2. Maurice Wilkins Centre for Molecular Biodiscovery, School of Biological Sciences, University of Auckland, Auckland, New Zealand.
3. School of Biological Sciences, Victoria University of Wellington, Wellington 6012, New Zealand.
4. The M-Lab, Department of Precision Medicine, University of Maastricht, Maastricht, The Netherlands.
5. Current address: The M-Lab, Department of Precision Medicine, University of Maastricht, Maastricht, The Netherlands.
6. Current address: Michael Smith Laboratories, University of British Columbia, Vancouver, BC, V6T 1Z4, Canada.
7. Current address: Walter and Eliza Hall Institute of Medical Research, 4 Research Avenue, Bundoora, Victoria, 3083, Australia.
*Equal contributions to this work.
#Equal contributions to this work.
Abstract
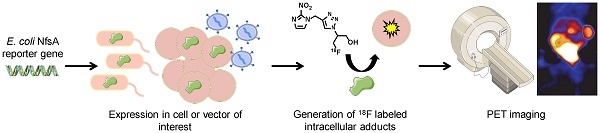
The use of reporter genes to non-invasively image molecular processes inside cells has significant translational potential, particularly in the context of systemically administered gene therapy vectors and adoptively administered cells such as immune or stem cell based therapies. Bacterial nitroreductase enzymes possess ideal properties for reporter gene imaging applications, being of non-human origin and possessing the ability to metabolize a range of clinically relevant nitro(hetero)cyclic substrates.
Methods: A library of eleven Escherichia coli nitroreductase candidates were screened for the ability to efficiently metabolize 2-nitroimidazole based positron emission tomography (PET) probes originally developed as radiotracers for hypoxic cell imaging. Several complementary methods were utilized to detect formation of cell-entrapped metabolites, including various in vitro and in vivo models to establish the capacity of the 2-nitroimidazole PET agent EF5 to quantify expression of a nitroreductase candidate. Proof-of-principle PET imaging studies were successfully conducted using 18F-HX4.
Results: Recombinant enzyme kinetics, bacterial SOS reporter assays, anti-proliferative assays and flow cytometry approaches collectively identified the major oxygen-insensitive nitroreductase NfsA from E. coli (NfsA_Ec) as the most promising nitroreductase reporter gene. Cells expressing NfsA_Ec were demonstrably labelled with the imaging agent EF5 in a manner that was quantitatively superior to hypoxia, in monolayers (2D), multicellular layers (3D), and in human tumor xenograft models. EF5 retention correlated with NfsA_Ec positive cell density over a range of EF5 concentrations in 3D in vitro models and in xenografts in vivo and was predictive of in vivo anti-tumor activity of the cytotoxic prodrug PR-104. Following PET imaging with 18F-HX4, a significantly higher tumor-to-blood ratio was observed in two xenograft models for NfsA_Ec expressing tumors compared to the parental tumors thereof, providing verification of this reporter gene imaging approach.
Conclusion: This study establishes that the bacterial nitroreductase NfsA_Ec can be utilized as an imaging capable reporter gene, with the ability to metabolize and trap 2-nitroimidazole PET imaging agents for non-invasive imaging of gene expression.
Keywords: reporter gene imaging, PET imaging, nitroreductase, gene therapy, drug repurposing
 Global reach, higher impact
Global reach, higher impact