13.3
Impact Factor
Theranostics 2020; 10(22):10326-10340. doi:10.7150/thno.45924 This issue Cite
Research Paper
PML-RARα interaction with TRIB3 impedes PPARγ/RXR function and triggers dyslipidemia in acute promyelocytic leukemia
1. National Clinical Research Center for Metabolic Disease, Department of Metabolism and Endocrinology, the Second Xiangya Hospital, Central South University, Changsha, Hunan, 410011, China.
2. Department of Hematology & Institute of Hematology, Zhejiang Province Key Laboratory of Hematology Oncology Diagnosis and Treatment, The First Affiliated Hospital, Zhejiang University, Hangzhou, Zhejiang, 310058, China.
3. Immunology and Cancer Pharmacology Group, State Key Laboratory of Bioactive Substance and Function of Natural Medicines, Institute of Materia Medica, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, 100050, China.
4. NHC Key Laboratory of Biotechnology of Antibiotics, Institute of Medicinal Biotechnology, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, 100050, China.
5. Department of Hematology, Affiliated Hospital of Xuzhou Medical University, Xuzhou, Jiangsu, 230031 China.
#These authors contributed equally to this work.
Abstract
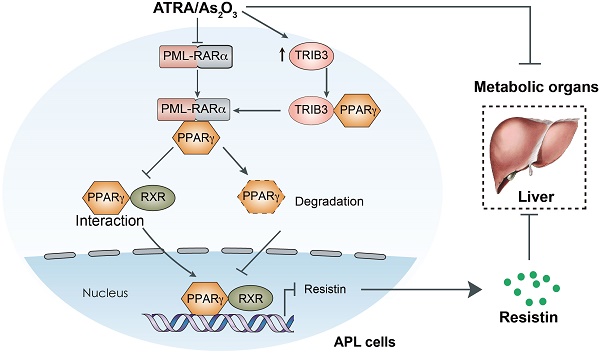
Although dyslipidemia commonly occurs in patients with acute promyelocytic leukemia (APL) in response to anti-APL therapy, the underlying mechanism and the lipid statuses of patients with newly diagnosed APL remain to be addressed.
Methods: We conducted a retrospective study to investigate the lipid profiles of APL patients. PML-RARα transgenic mice and APL cells-transplanted mice were used to assess the effects of APL cells on the blood/liver lipid levels. Subsequently, gene set enrichment analysis, western blot and dual luciferase reporter assay were performed to examine the role and mechanism of PML-RARα and TRIB3 in lipid metabolism regulation in APL patients at pretreatment and after induction therapy.
Results: APL patients exhibited a higher prevalence of dyslipidemia before anti-APL therapy based on a retrospective study. Furthermore, APL cells caused secretion of triglycerides, cholesterol, and PCSK9 from hepatocytes and degradation of low-density lipoprotein receptors in hepatocytes, which elevated the lipid levels in APL cell-transplanted mice and Pml-Rarα transgenic mice. Mechanistically, pseudokinase TRIB3 interacted with PML-RARα to inhibit PPARγ activity by interfering with the interaction of PPARγ and RXR and promoting PPARγ degradation. Thus, reduced PPARγ activity in APL cells decreased leptin but increased resistin expression, causing lipid metabolism disorder in hepatocytes and subsequent dyslipidemia in mice. Although arsenic/ATRA therapy degraded PML-RARα and restored PPARγ expression, it exacerbated dyslipidemia in APL patients. The elevated TRIB3 expression in response to arsenic/ATRA therapy suppressed PPARγ activity by disrupting the PPARγ/RXR dimer, which resulted in dyslipidemia in APL patients undergoing therapy. Indeed, the PPAR activator not only enhanced the anti-APL effects of arsenic/ATRA by suppressing TRIB3 expression but also reduced therapy-induced dyslipidemia in APL patients.
Conclusion: Our work reveals the critical role of the PML-RARα/PPARγ/TRIB3 axis in the development of dyslipidemia in APL patients, potentially conferring a rationale for combining ATRA/arsenic with the PPAR activator for APL treatment.
Keywords: AML, Cancer, leukemia, lipid metabolism, tribbles
 Global reach, higher impact
Global reach, higher impact