13.3
Impact Factor
Theranostics 2019; 9(14):4141-4155. doi:10.7150/thno.35033 This issue Cite
Research Paper
Survey of the translation shifts in hepatocellular carcinoma with ribosome profiling
1. MOE Key Laboratory of Bioinformatics, Tsinghua University, Beijing 100084, China
2. Center for Synthetic & Systems Biology, Tsinghua University, Beijing 100084, China
3. School of Life Sciences, Tsinghua University, Beijing 100084, China
4. Joint Graduate Program of Peking-Tsinghua-National Institute of Biological Science, Tsinghua University, Beijing 100084, China.
5. Department of Hepatobiliary Surgery, The General Hospital of Chinese People's Liberation Army, Beijing 100853, China.
6. Department of Liver Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100730, China.
# These authors contributed equally to this work.
Abstract
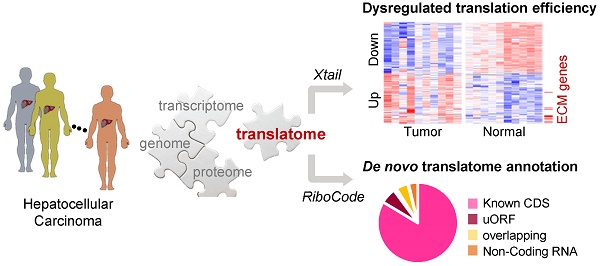
Despite the critical position of translation in the multilevel gene expression regulation program, high-resolution and genome-wide view of the landscape of RNA translation in solid tumors is still limited.
Methods: With a ribosome profiling procedure optimized for solid tissue samples, we profiled the translatomes of liver tumors and their adjacent noncancerous normal liver tissues from 10 patients with hepatocellular carcinoma (HCC). A set of bioinformatics tools was then applied to these data for the mining of novel insights into the translation shifts in HCC.
Results: This is the first translatome data resource for dissecting dysregulated translation in HCC at the sub-codon resolution. Based on our data, quantitative comparisons of mRNA translation rates yielded the genes and processes that were subjected to patient specific or universal dysregulations of translation efficiencies in tumors. For example, multiple proteins involved in extracellular matrix organization exhibited significant translational upregulation in tumors. We then experimentally validated the tumor-promoting functions of two such genes as examples: AGRN and VWA1. In addition, the data was also used for de novo annotation of the translatomes in tumors and normal tissues, including multiple types of novel non-canonical small ORFs, which would be a resource for further functional studies.
Conclusions: The present study generates the first survey of the HCC translatome with ribosome profiling, which is an insightful data resource for dissecting the translatome shift in liver cancer, at sub-codon resolution.
Keywords: hepatocellular carcinoma, ribosome profiling, RNA translation, translatome, translation efficiency
 Global reach, higher impact
Global reach, higher impact