13.3
Impact Factor
Theranostics 2019; 9(4):1200-1214. doi:10.7150/thno.31991 This issue Cite
Research Paper
Circulating proteomic panels for diagnosis and risk stratification of acute-on-chronic liver failure in patients with viral hepatitis B
1. State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, 310003 Hangzhou, People's Republic of China
2. Department of Infectious Diseases, Zhejiang University International Hospital, 310004 Hangzhou, People's Republic of China
3. Bio-Macromolecules Analysis Laboratory of Analysis Center of Agrobiology and Environmental Sciences, Zhejiang University, 310058 Hangzhou, People's Republic of China
4. Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, The First Affiliated Hospital, College of Medicine, Zhejiang University, 310003 Hangzhou, People's Republic of China
5. Key Laboratory of Combined Multi-Organ Transplantation, Ministry of Public Health, The First Affiliated Hospital, College of Medicine, Zhejiang University, 310003 Hangzhou, People's Republic of China
6. Key Laboratory of Organ Transplantation, The First Affiliated Hospital, College of Medicine, Zhejiang University, 310003 Hangzhou, People's Republic of China
*These authors contributed equally: Zeyu Sun, Xiaoli Liu, Daxian Wu.
Received 2018-12-4; Accepted 2019-1-17; Published 2019-1-30
Abstract
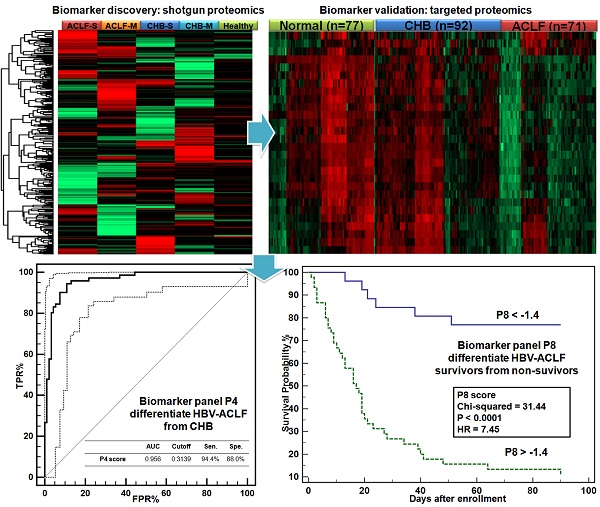
Chronic HBV infection (CHB) can lead to acute-on-chronic liver failure (HBV-ACLF) characterized by high mortality. This study aimed to reveal ACLF-related proteomic alterations, from which protein based diagnostic and prognostic scores for HBV-ACLF were developed.
Methods: Ten healthy controls, 16 CHB, and 19 HBV-ACLF according to COSSH (Chinese group on the study of severe hepatitis B) criteria were enrolled to obtain the comprehensive proteomic portrait related to HBV-ACLF initiation and progression. Potential markers of HBV-ACLF were further selected based on organ specificity and functionality. An additional cohort included 77 healthy controls, 92 CHB and 71 HBV-ACLF was used to validate the proteomic signatures via targeted proteomic assays.
Results: Significant losses of plasma proteins related to multiple functional clusters, including fatty acid metabolism/transport, immuno-response, complement and coagulation systems, were observed in ACLF patients. In the validation study, 28 proteins were confirmed able to separate ACLF, CHB patients. A diagnostic classifier P4 (APOC3, HRG, TF, KLKB1) was built to differentiate ACLF from CHB with high accuracy (auROC = 0.956). A prognostic model P8 (GC, HRG, HPR, SERPINA6, age, NEU, INR and total protein) was built to distinguish survivors from non-survivors in 28 and 90-days follow-up (auROC = 0.882, 0.871), and to stratify ACLF patients into risk subgroups showing significant difference in 28 and 90-days mortality (HR=7.77, 7.45, both P<0.0001). In addition, P8 score correlated with ACLF grades and numbers of extra-hepatic organ failures in ACLF patients, and was able to predict ACLF-associated coagulation and brain failure within 90 days (auROC = 0.815, 0.842).
Conclusions: Proteomic signatures developed in this study reflected the deficiency of key hematological functions in HBV-ACLF patients, and show potential for HBV-ACLF diagnosis and risk prediction in complementary to current clinical based parameters.
Keywords: HBV-ACLF, biomarkers, mass spectrometry, proteomics
Introduction
With more than 257 million population affected, and approximately 887000 deaths caused, hepatitis B virus (HBV) infection poses a global health challenge particularly to developing countries [1-3]. So far there is no definitive cure available for HBV infection [1-3]. Chronic hepatitis B (CHB) patients are at high risk of developing end-stage liver diseases including the increasingly recognized acute-on-chronic liver failure (ACLF) [4-10]. Compare to other CHB sequelae, ACLF is characterized by unpredictable and sudden deterioration of hepatic function with substantial liver damages and ominous short-term prognosis, therefore constitutes a medical emergency [1, 4-6, 9, 10].
Despite discrepancies in recognizing ACLF with different underlying etiologies, hepatologists around world are approaching to the consensus that ACLF is a life-threatening syndrome that needs to be managed aggressively. Current working definitions of ACLF proposed by ESAL[11], APASL[4, 12] and COSSH[10] studies relied heavily on clinical manifestation of symptoms (ascites, jaundice) or presence of organ failures, plus few routine biochemical indicators, reflecting a lack of understanding of ACLF pathogenesis. Although accumulated evidence has suggested the contributions of both substantial hepatic necroinflammation and unregulated immune responses, more investigations are required to define ACLF in pathophysiological and molecular levels, therefore to develop more accurate diagnostic or prognostic criteria to guide interventions. Regarding HBV related ACLF, it is still challenging to unambiguously differentiate ACLF patients from CHB patients with severe HBV flare according to the two working definitions proposed by APASL [4, 12] and COSSH [10]. Must be stressed that ACLF is reversible if interventions are provided promptly, otherwise, patients only receive limited benefits from costly treatments [13, 14]. Given the importance of early intervention, a sensitive and objective diagnosis/prognosis guideline is desperately needed to lower the ACLF-related mortality and the exacerbated economic burdens associated with managing it [4, 9, 14-20]. An unbiased risk assessment system can also aid to select ACLF patients with high urgency for earlier referral, ICU admission, organ supports or liver transplantation (LT). However, current scoring models including the Child-Turcott-Pugh score (CTP), Model for End-Stage Liver Disease (MELD) score [21] and the recently launched CLIF-C ACLF [11] and COSSH score [10] are mostly based on combinations of few clinical indicators and/or subjective judgment from physicians, and have not fully proven to fulfill the challenge [14, 22-24]. In addition, liver biopsy for histological evaluation is considered too risky for coagulopathic ACLF patients. Consequently, there is an unmet need for noninvasive clinical tests to identify ACLF with high confidence and to predict the ACLF-related outcomes.
The emerging paradigm of precision medicine underscores the importance of quantitative portray of molecular features during disease onset and progression. We argue that dynamic markers, such as circulating proteins, may help to capture the disease trajectory and to offer precise prognostication for ACLF patients. Worthy of note, liver is a prominent contributor to the blood proteome, of which a large portion are highly abundant liver-specific species. They include, but not limited to, albumins (HSA, AFP, GC), lipoproteins (APOs), and a collection of coagulation factors and complement components. We therefore hypothesize that the hematological or immunological importance of those circulatory proteins might make them preferable to gauge ACLF progression. In fact, circulating markers had been previously sought to assess liver malfunction and to foresee liver disease outcomes [25-28]. Though significant loss of circulatory proteins was commonly observed in ACLF patients, little efforts have been attempted to search ACLF- specific protein signatures.
Other than being used for biomarker purposes, circulatory proteins also hold the key to develop therapeutic strategy for ACLF patients. Drastic alteration of blood constituents during liver failure has been proposed to cause systematic complications, such as coagulopathy, jaundice, hemodynamic instability, hepatic encephalopathy (HE), hepato-renal syndrome (HRS) and sepsis which eventually lead to extra-hepatic organ failures and death [4-6, 9]. Therefore, hopes have merged to implement extracorporeal artificial liver support systems (ALSS), including a combination of hemofiltration, hemodialysis, plasma exchange/perfusion or albumin dialysis approaches to lower either the mortality or the occurrence of ACLF-related complications [29, 30]. All current ALSS intervention approaches aimed, to a certain extent, to change the constituents of circulatory system. Yet it is frustrating that large-scale clinical studies of those ALSS therapies conducted so far have not shown survival benefits for ACLF patients [29, 30]. We believe that quantitative and comprehensive investigation of blood constituents changes, including its proteome, is vital to the design rationale of novel hematological therapies or to the optimization of current ALSS systems and their implementation.
In this study, we first investigated ACLF-related molecular portrait using quantitative shotgun proteomic analyses, from which ACLF-related functional modulation was revealed. Liver-specific members of deregulated functional groups were validated via targeted proteomics approach in a larger-scale independent sample set, from which two proteomic classifiers were developed to differentiate HBV-ACLF from CHB patients and to predict HBV-ACLF related outcomes in 28 and 90 days after admission. We hope that these circulating proteomic signatures not only reflect the liver function alteration, but also form unbiased criteria to enable individualized medicine and equitable allocation of precious medical resources for HBV-ACLF management, and eventually help to improve current therapeutic strategies to tackle ACLF.
Methods
Study design and patients' enrollment. A total of 244 HBV-infected patients were enrolled from the Department of Infectious Diseases of the First Affiliated Hospital, Zhejiang University from January 2014 to May 2016. Medical history, physical examination and biochemical data were obtained at the enrollment. Survival data was obtained through medical records or by direct contact with the patients or their families at 3-month following enrollment, with death or LT as a composite endpoint. CHB was diagnosed as previously [12], while HBV-ACLF was defined as acute hepatic insult manifesting severe jaundice (Tbil ≥ 12 mg/dL) and coagulopathy (INR ≥ 1.5) on the basis of CHB per COSSH criteria [10]. Detailed inclusion and exclusion criteria, as well as treatment scheme for all patients can be found in the supplementary materials. The research protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki was approved by the Human Ethics Committee of the First Affiliated Hospital, Zhejiang University. All enrolled participants signed a written informed consent to participate in the study.
Eventually, 90 HBV-related ACLF and 108 CHB patients were qualified, together with 87 healthy controls. Subjects were then divided into discovery study groups (10 healthy controls, 16 CHB and 19 ACLF) and validation study groups (77 healthy controls, 92 CHB and 71 ACLF) as schematically summarized in Figure 1.
Collection of plasma samples. Blood samples from all patients were obtained before breakfast on the second day after hospitalization. The samples were immediately processed according to the standardized protocol recommended by the HUPO Plasma Proteome Project [31]. Briefly, blood was collected into plastic K2EDTA tubes (BD), manually inverted for 10 times and centrifuged at 1300 xg for 10 min. Plasma sample was aliquoted and stored at -80 °C until proteomic analysis.
Overall experimental design for biomarker panel development. ACLF patients were identified per COSSH criteria. Large scale shotgun proteomics were used to select ACLF-related marker candidates, which were then validated on separate cohorts using targeted proteomic approach.
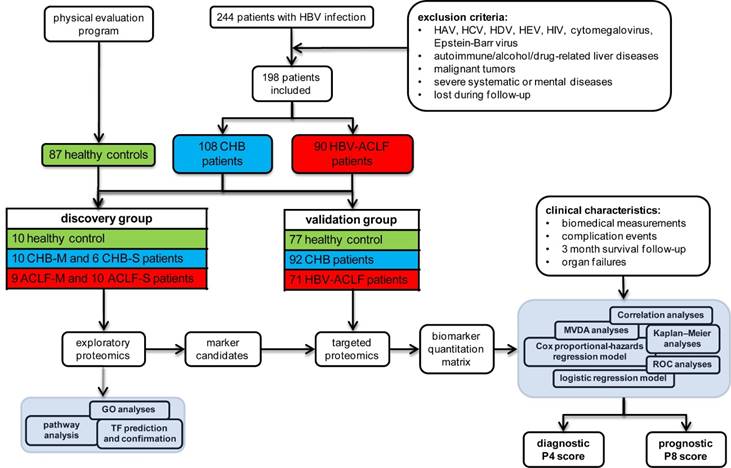
Discovery comparative proteomic analyses. To elucidate the incremental changes in circulation proteome related to CHB and HBV-ACLF progression, patients in two sub-clinical phases of CHB, the mild CHB (CHB-M with increase of ALT but normal Tbil level) and severe CHB (CHB-S with significant increase of ALT and mild jaundice, Tbil level ≥ 2 mg/dL, INR≤1.5), and two sub-clinical phases of HBV-ACLF (ACLF-M with 2>INR≥1.5, and ACLF-S with INR≥2) were included for high-throughput comparative proteomic analyses (Table S1).
Plasma samples from patients within the same group were pooled before proteomic analysis. The pooled plasma was divided into high and low abundant protein fractions by combinatorial peptide ligands library (CPLL) method with a modified protocol described previously [32].
Four parallel comparative proteomics experiments that employed triplex-2MEGA labeling and 2D-RPLC prefractionation coupled with high-throughput MS/MS experiment to compare protein abundances within each group. Two parallel shotgun proteomic experiments were performed to compare low abundant proteins (LAP) from all 5 groups. In the first experiment, LAP peptides from pooled CHB-M (n=10) and CHB-S (n=6) samples were tagged by medium and heavy dimethyl label, respectively, using a 3-plex N-terminal dimethylation after lysine guanidination (2MEGA) protocol described in the supplementary materials. In the second experiment, LAP peptides from ACLF-M (n=9) and ACLF-S (n=10) samples were tagged by medium and heavy dimethyl label, respectively. The control samples (n=10) were pooled and tagged by light dimethyl label and used as a common reference in both experiments. Analogously, another two parallel shotgun proteomic experiments were performed to compare high abundant proteins (HAP) from 5 groups with the identical labeling scheme. The overall design of all four 3-plex 2MEGA-2DLC-MS/MS experiments was illustrated in Figure S1. Detail procedures can be found in the supplementary materials.
2DLC-MS/MS and data analysis. The labeled peptides within each experiment were then mixed and further fractionated with high pH reverse phase chromatography. Peptide fractions were subsequently analyzed by LC-MS/MS. Proteomic data analyses were performed with TransProteomic Pipeline environment (TPP, v.4.8) implementing consensus database search using Comet and MSGF+ against Uniprot human proteome sequences. Protein quantitation was performed with ASAPratio module of TPP extracting isotopic 2MEGA peak clusters from MS1 raw spectra. Detail procedures can be found in the supplementary materials.
Functional clustering analysis. To unveil underlying biological functions associated with proteins significantly changed in plasma from ACLF patients, DEPs from both LAP and HAP fractions were submitted to FunRich v.3 [33] for GO functional clustering using default settings. Key transcription factors predicted by FunRich were validated by Western blotting. Molecular interactions between DEPs were annotated by STRING database [34] and visualized by Cytoscape v3.2.1. In addition, the top enriched pathways were visualized according to Wikipathways annotation.
Western Blot Analysis. Expression of HNF1α and HNF4α were evaluated by western blot using biopsy specimens obtained from CHB patients, HBV-ACLF patients following total hepatectomy, healthy liver transplantation donor. Detailed procedures can be found in the supplementary materials.
Validation study using targeted proteomics. For the validation study, 30 µL plasma collected as described previously was treated by ProteoPrep blue albumin and IgG depletion kit (PROTBA, Sigma) according to the manufacturer's instruction, followed by FASP digestion as described in discovery study, except a 1:20 enzyme to protein ratio was used for tryptic digestion. All samples were analyzed with LC-MS operated under the parallel-reaction monitoring (PRM) acquisition scheme as detailed in the supplementary materials. Eventually, a list of 222 peptides derived from 42 protein markers (Table S7) was generated for the targeted proteomic survey in an independent sample set of 92 CHB and 71 HBV-ACLF patients plus 77 healthy controls (Table 1).
Data processing and multivariate modeling for targeted proteomics data. Acquired PRM data were analyzed by Skyline for transitions identification and peak area integration. Protein intensities were Log2 transformed. Proteins with missing value in more than 60% samples (~146 samples) were excluded. The remaining missing values were considered as low abundance values due to limited MS sensitivity, therefore were replaced by random numbers drawn from a normal distribution with a mean value 1.8x lower and a standard deviation 0.3x of the original data. The final protein relative quantification results were summarized in Table S8 and were used for principal component analysis (PCA) in SIMCA 14.1 (Umetrics).
ACLF diagnostic and prognostic scores development. Proteins quantified by targeted proteomics were subject to ROC analyses individually to evaluate their sensitivity and specificity to discriminate HBV-ACLF from CHB and to predict 3-month endpoint event. A logistic regression model was constructed for HBV-ACLF diagnosis score. In addition, another protein based logistic regression model was built to differentiate CHB-S and CHB-M patients. ROC analyses were used to estimate the performance of diagnostic score.
For HBV-ACLF prognosis, multivariate Cox proportional hazard regression models associated with 3-month mortality caused by HBV-ACLF were used to select protein and clinical parameters for HBV-ACLF mortality risk assessment. To avoid overfitting, multivariate Cox proportional-hazard regression models were built using 8 proteins at a time, and the model with highest prognostic value was then simplified by removing parameters at a time. Non-contributing markers were discarded if there was insignificant loss of prognostic performance of the model. Clinical variables with prognostic value were also selected using the same strategy. Refining steps to select biomarker candidates for downstream validation were summarized in the Figure 2B and Supplementary Materials.
Patient characteristics in the validation set.
| Characteristic | Health Controls (n=77) | CHB (n=92) | HBV-ACLF (n=71) |
|---|---|---|---|
| Age (yrs.) | 43.64±13.43 | 38.83±10.78* | 48.34±12.00§ |
| Gender (female/male) | 23/54 | 17/75 | 2/69§ |
| 3 month survival/death/LT | 77/0/0 | 88/2/2 | 26/28/17§ |
| 28 days survival/death/LT | 77/0/0 | 89/2/1 | 35/22/14§ |
| Laboratory parameters | |||
| Albumin (g/L) | 48.75±2.51 | 38.29±5.79* | 31.83±4.04§ |
| Alanine aminotransferase (IU/L) | 25.43±13.33 | 399.78±368.01* | 376.63±347.95 |
| Aspartate aminotransferase (IU/L) | 18.25±5.39 | 207.02±226.61* | 277.62±286.52§ |
| Total bilirubin (mg/dL) | 0.80±0.34 | 4.66±6.92* | 24.30±8.23§ |
| Creatinine (mg/dL) | 0.863±0.209 | 0.97±1.63* | 0.82±0.27 |
| Prothrombin time (s) | N/A | 13.41±3.37 | 25.45±7.93§ |
| International Normalized Ratio | N/A | 1.17±0.32 | 2.28±0.71§ |
| LogHBV-DNA | 0 | 5.36±2.61* | 4.41±2.72 |
| Ferritin (µg/L) | N/A | 1073.35±1460.51 | 4608.48±5049.88§ |
| Immunoglobulin G (g/L) | 25.66±4.03 | 27.05±4.50* | 26.69±7.60 |
| C reactive protein (mg/L) | N/A | 5.15±6.92 | 14.21±8.33§ |
| White blood cell (109/L) | N/A | 5.24±1.36 | 8.14±3.79§ |
| Neutrophil (109/L) | N/A | 2.90±1.12 | 5.92±3.31§ |
| Lymphocytes (109/L) | N/A | 1.72±0.56 | 1.37±0.72§ |
| Red blood cell (1012/L) | N/A | 4.66±0.61 | 4.13±0.81§ |
| Platelet (109/L) | N/A | 168.87±52.93 | 106.45±43.14§ |
| Total Protein (mg/dL) | N/A | 65.33±6.48 | 58.54±7.05§ |
| K (mM/L) | N/A | 4.19±0.45 | 4.34±0.65 |
| Na (mM/L) | N/A | 139.99±2.28 | 135. 873±3.86§ |
| MELD score | N/A | 8.89±6.86 | 24.85±4.22§ |
| COSSH-ACLF | N/A | 4.10±0.49 | 5.53±0.92§ |
| CLIF-OF | N/A | 6.42±0.91 | 9.66±1.52§ |
| CLIF-C-ACLF | N/A | 26.96±5.48 | 43.80±7.20§ |
| Complication diagnosed at enrollment | |||
| Jaundice (n=111, 46.3%) | 0 (0%) | 40(43.5%)* | 71(100%)§ |
| Hepatic encephalopathy (n=4, 1.7%) | 0 (0%) | 0 (0%) | 4 (5.6%)§ |
| Ascites (n=37, 15.4%) | 0 (0%) | 6(6.5%) | 31(43.7%)§ |
| Alimentary tract hemorrhage (n=1, 0.4%) | 0 (0%) | 0 (0%) | 1 (1.4%) |
| Hepatorenal syndrome (n=3, 1.3%) | 0 (0%) | 1(1%) | 2 (2.8%) |
| Spontaneous Bacterial Peritonitis (n=5, 2.1%) | 0 (0%) | 0 (0%) | 5 (7.0%)§ |
| Comorbidities diagnosed at enrollment | |||
| Liver cirrhosis (n=47, 19.6%) | 0 (0%) | 7(7.6%) | 40(56.3%)§ |
| Diabetes (n=8, 3.3%) | 0 (0%) | 0 (0%) | 8 (11.3%)§ |
| Hypertension (n=12, 5%) | 0 (0%) | 3 (3.3%) | 9(12.7%)§ |
| Precipitation events | |||
| HBV reactivation (n=137, 57.1%) | 0 (0%) | 81(88.0%) | 56(78.9%) |
| Others (Upper GI bleeding, SBP, n=6, 2.5%) | 0 (0%) | 0 (0%) | 6 (8.4%)§ |
| Unknown (n=26, 10.8%) | 0 (0%) | 11 (12.0%) | 15 (21.1%) |
| More than one PEs (n=6, 2.5%) | 0 (0%) | 0 (0%) | 6 (8.4%)§ |
p value <0.05 for comparisons between CHB and healthy controls*, and between HBV-ACLF and CHB§.
Statistical analysis. Mann-Whitney U test and Kruskal-Wallis test were used to analyze quantitative and categorical traits. Differences were considered significant for p-values <0.05, or as otherwise indicated. All statistical analyses were performed in R (v3.2.0) or otherwise specified. Area under the receiver operating curve (auROC) was calculated and compared by Z test (Delong's method). Kaplan-Meier method was used to compare the survival functions in patients partitioned by the prognostic score.
Results
Overall design and clinical synopsis
For the discovery study, we further divided the CHB and HBV-ACLF patients into 4 sub-clinical groups according to the disease progression (Table S1): CHB-M with mild hepatic damage without jaundice (n=10, ALT: 104.9±52.4 U/L, Tbil: 1.0±0.4 mg/dL), and CHB-S who developed jaundice with significant liver damage but not qualified as ACLF at the admission (n=6, ALT: 469.3±401.8 U/L, Tbil: 7.8±4.8 mg/dL); ACLF-M patients who developed both jaundice and coagulopathy (n=9, Tbil: 20.1±9.4 mg/dL, PT: 19.5±2.7 sec) and ACLF-S with severe coagulopathy (n=10, Tbil: 24.3±6.0 mg/dL, PT: 39.3±8.8 sec).
In the validation set (Table 1), all HBV-ACLF patients were diagnosed with jaundice as compared to CHB (43.5%). Ascites were found in almost half of ACLF patients (43.7%) but in only 6 CHB cases (6.5%) at admission. Coagulopathy was presented in all HBV-ACLF patients (INR: 2.28±0.71) but only in two CHB patients (INR: 1.17±0.32). Other complications including HE, GI hemorrhage, HRS and spontaneous bacterial peritonitis (SBP) were occasionally reported at admission in HBV-ACLF patients but not in CHB patients. The MELD scores were 8.89±6.86 and 24.85±4.22, respectively for CHB and HBV-ACLF patients. Mortality (3 month) of HBV-ACLF patients (63.4%) was in accordance with previous reports [4, 11, 35, 36]. Other than significantly elevated level of bilirubin and aspartate aminotransferase, HBV-ACLF patients have no significant difference in serum ALT, HBV-DNA and creatinine level as compared to CHB patients. Total proteins were decreased in HBV-ACLF patients (58.54±7.05 g/L) as compared to CHB (65.33±6.48 g/L). According to the CLIF-OF system, all 71 ACLF patients developed OFs within 90 days (Table S2), including 17 (23.94%) cases with one OF, 54 (76.06%) cases with multiple OFs. As expected, all ACLF patients developed liver failure, while the second most common OF was coagulation failure (34 cases, 47.89%), followed by respiratory failure (32 cases, 45.07%) and brain failure (18 cases, 25.35%). Among 46 ACLF cases with endpoint events (Table S3), 18 cases received LT (39%), while the death causes for the remaining cases were mainly due to MOFs without septic shock or cerebral edema (14 cases, 30%) and severe encephalopathy or cerebral edema that eventually leaded to respiratory arrest (13 cases, 28%).
Summary of high-throughput proteomics analysis of CHB and ACLF in the discovery study. (A) Heatmap representation of abundance profile of all proteins in 5 groups. The 2MEGA quantitation data in Log2 scale were transformed into Z-score by rows. The rows were clustered using the K-mean algorithm. (B) Steps in refining the biomarker candidates down to multi-protein classifier panels. HAP: high abundant protein; LAP: low abundant protein; MVDA: multivariate data analyses. Summary of functional annotation of ACLF by GO biological processes (C), cellular components (D), pathways (E) and predicted TFs (F) related to the ACLF-associated differentially expressed proteins (DEPs). Length of bars represent percentage of DEPs associated with each term. Significance of enrichment (p-value) was noted next to the bar for each term. HDLP, high-density lipoprotein particle; VDLP: Very-low-density lipoprotein particle; ExCel, Extracellular; LPS, Lipopolysaccharide; LBP, Lipopolysaccharide binding protein. Molecular interaction maps between DEPs were annotated by STRING database (G). Color shades (red for upregulation, green for downregulation, colorless for insignificant changes, grey for data not available) represent expression ratio compared with normal condition from all DEPs pooled from LAP and HAP fractions. For protein presented in both HAP and LAP fractions, expression level from HAP was used as final input for network visualization.
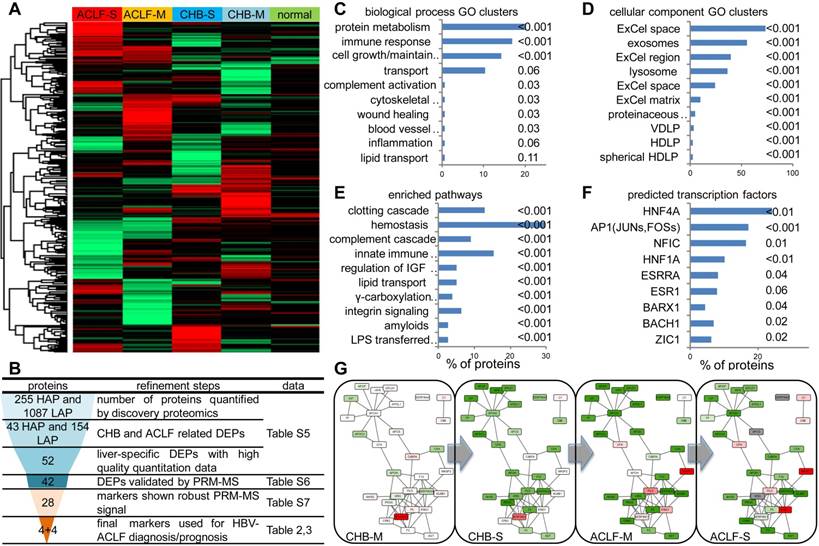
Summary of the proteomic discovery and functional alterations related to HBV-ACLF. To obtain a comprehensive view of proteomic changes related to HBV-ACLF pathogenesis, a high-throughput workflow (Figure S1) employing CPLL techniques was first applied to partition the plasma proteome into high abundant protein fraction (HAPs) and low abundant protein fraction (LAPs). Identification results of all LC-MSMS runs were summarized in Table S5. In total, 255 HAPs and 1087 LAPs throughout all MDLC-MSMS experiments were used for quantification analysis (Figure 2A), from which 43 HAPs and 154 LAPs were considered as differential expression proteins (DEPs, p-value < 0.05 by ASAPratio) related to CHB-ACLF progression (Table S6) and were subject to the following biomarker selection using a multistage refinement workflow summarized in Figure 2B. As shown in Figure 2C-E, biological process GO terms highly enriched were Protein Metabolism (APCS, PLG, KNG1, KLKB1, PROC, PROZ, CAPN1, MMP2, SERPINA4/A5/B6/C1/F2/F2/G1, MASP1/2, UBE2N, HGFAC, HABP2, ITIH1/3/4, ADAMTS13, PCSK9, CPB2, CNDP1, EIF3L), Cell Growth/Maintenance (MEGF6, EFEMP1/2, TIMP1, PFN1, COL6A3, VCL, MSN, CORO1A, CDH5, FBN1, COMP, HSPG2, DPT, FHL1, PCOLCE, ECM1, PRG4, FBLN5), Immune Response (CD5L, ATRN, HP, HPR, IGJ, PIGR, C1QC, ORM1, C4BPA, C7/8B/8G, CD14, SRGN, CLU, IGLL1, LBP, AZGP1, CFP, CAMP, DCD, CFHR2/4/5) and Transport (APOA1/A5/C3/F/H/L1, TF, LTF, HPX, SHBG, MGP, SAA4, HBB, UNC13D, NUP93), while cellular component GO terms highly enriched were Extracellular Space/Matrix, Exosomes, and Lysosome. As expected, vital hematological pathways such as Clotting Cascade (F2/5/12/13A1/13B, SERPINC1/G1, KNG1, KLKB1, PROC), Hemostasis (PLG, TF, HRG, PFN1, SRC, VCL, PTPN6, CDC42, YWHAZ, HBB, EHD3), Complement Cascade (MASP1/2,C1QC,C7/8B/8G) and Innate Immune System (MASP1/2, C1QC, C7/8B/8G, APP, CD14, LBP, UBE2N, PCBP2) were significantly enriched among others. Transcriptional factors (TFs), including HNF-4α, AP1, NFIC and HNF-1α (p<0.01) were predicted as upstream regulators of those differentially expressed proteins during ACLF development (Figure 2F). Validation on liver tissue specimens by WB confirmed significant downregulation of HNF-1α and HNF-4α, both were known for hepatocyte development and liver function maintenance, in ACLF compared to CHB (Figure S3). In addition, interactome analysis (Figure 2G) indicated two highly connected clusters involving lipid transportation (APOA1, APOC3, APOL1, APOF, APOH, HP, HPR, GPLD1, APCS) and coagulation/kinin system (F2, F5, F12, PROC, HRG, PLG, KNG1, KLKB1, SERPING6, SERPINC1). Moderate down-regulation of majority of proteins in these two clusters can be seen in CHB-S, and this trend was more profound in ACLF patients.
Biomarker validation by targeted proteomics and MVDA analysis. Based on the ACLF-related proteomic and functional alteration revealed in the discovery study, we then sought to develop protein marker panels that reflect the ACLF onset and progression with high accuracy. To this aim, we focused on the liver-specific members of deregulated functional groups based on tissue specificity annotation provided by ProteinAtlas and GeneCards databases in addition to data quality criteria detailed in the supplementary materials. A final list of 42 liver specific DEPs with confident 2MEGA quantitation data were included for validation in an additional cohort comprised of 71 HBV-ACLF and 92 CHB patients plus 77 healthy controls (Table 1). Considering the challenge of quantifying dozens of protein candidates in parallel, we employed a median-throughput mass spectrometry based approach, the parallel reaction monitoring (PRM) as the method of choice to measure 222 tryptic peptides containing unique sequences from 42 protein candidates (Table S7). This targeted proteomic assay eventually measured 28 protein candidates with robust signal across the validation sample set (Table S8).
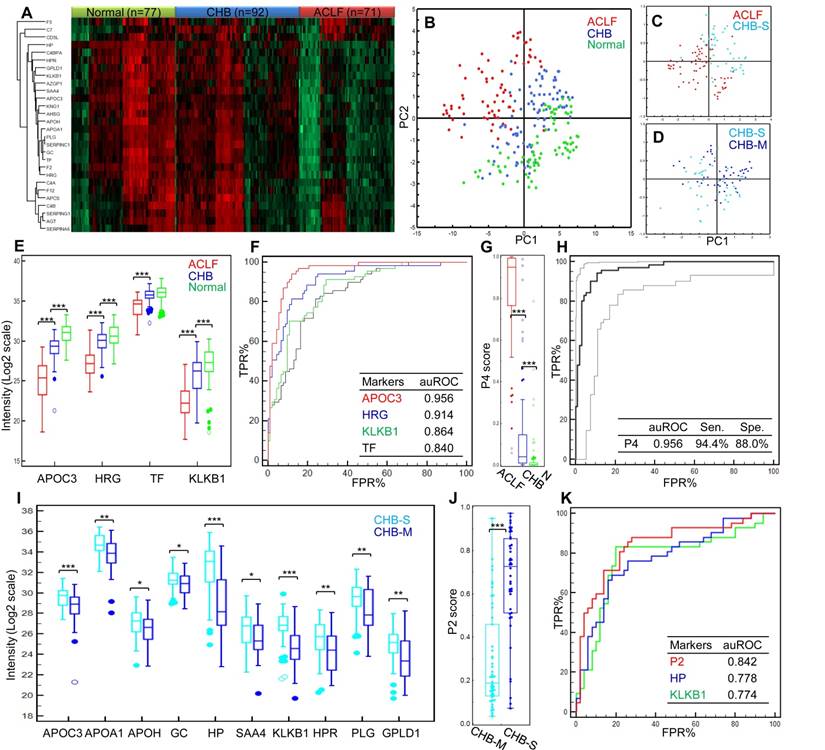
The expression profiles of these markers clearly showed intergroup differences (Figure 3A). The trends of marked down-regulation of most proteins in ACLF samples were in line with the results from the discovery study. The overall PCA model based on these 28 markers demonstrated clear separation of healthy controls, CHB and HBV-ACLF patients (R2X= 0.84, Q2=0.78, Figure 3B). Moreover, the CHB is known for its long course of disease progression, and heterogeneous patient characteristics. We were particularly interested to see if HBV-ACLF can be separated from severe-CHB, i.e. pre-ACLF patients developed mild jaundice (Tbil level ≥ 2 mg/dL, but did not reach ACLF criteria). To this end, the second PCA model (R2X= 0.75, Q2=0.70, Figure 3C) was built showing a clear partition of HBV-ACLF (n=71) and severe-CHB (n=34) patients, who could also be distinguished from the remaining mild-CHB patients (n=58) in the third PCA model (R2X= 0.77, Q2=0.63, Figure 3D). The overall correlation between proteomic signature and the clinical course of HBV-ACLF has encouraged us to further search for specific protein combinations that can be used as ACLF diagnostic and prognostic panels.
Develop diagnosis panel for HBV-ACLF. Proteins shown significant changes in ACLF patients (Bonferroni-adjusted P<0.0001 of log2Intensity, >2 fold-change in raw ACLF/CHB intensity ratio) were subject to ROC analysis individually to evaluate their sensitivity and specificity to discriminate HBV-ACLF from CHB. At this step, 17 proteins with potential ACLF diagnosis value (auROC> 0.8) were found. They were involved with lipid metabolism (APOA1, APOH, APOC3), iron metabolism (HP, HPR, TF), coagulation system (PLG, SERPINC1, F2), complement/kinin system (KNG1, KLKB1, C4BPA), acute-phase response (AHSG, SAA4), vitamin transporter (GC), pathogen clearance (HRG) and GPI-anchor release pathway (GPLD1). To avoid functional redundancy, we selected one candidate at a time from the abovementioned function groups to construct logistic regression models using the stepwise method (variable was entered if p<0.05 and removed if p>0.1). Models were then compared by ROC curves, and the most simplified model (with least parameters) without significant loss of auROC was adopted. This process resulted in a model comprised of 4 protein markers (APOC3, HRG, TF, KLKB1, Table 2), which were all drastically down-regulated in the ACLF samples (Figure 3E). Being the most depleted one in ACLF patients, APOC3 (ACLF/CHB = 0.11) showed the highest sensitivity (94.4%) and specificity (88.0%) with the highest auROC of 0.956 to differentiate ACLF from CHB patients (Figure 3F). The 4-protein based logistic model generated a dichotomous score, dubbed diagnostic P4, to classify each sample. Relation between the probability score of a sample being positively diagnosed as ACLF and the log2 intensity value of each protein marker can be defined as follows:
Ln(P4/(1-P4)) = 17.0668 - 0.92085*APOC3 - 0.75754*HRG + 1.20517*TF - 0.52709*KLKB1
The P4 score was significantly higher in ACLF patients than in CHB and healthy control groups (both p<0.001, Figure 3G). At the threshold point of 0.3139, the sensitivity and specificity of P4 for ACLF diagnosis were 94.4% and 89.1% with an auROC of 0.961 (Figure 3H). Highest Youden index (0.835), i.e. the ability to correctly diagnose true HBV-ACLF and true CHB patients was achieved at the cut-point.
Summary of targeted proteomics analysis of CHB and ACLF in the validation study. (A) Heatmap representation of overall abundance profile of all 28 robustly detected proteins in 3 groups. The protein quantitation data in Log2 scale were transformed into Z-score by rows. The rows were clustered using the K-mean algorithm. To highlight the intergroup differences, samples (columns) were clustered within each group first before making the heatmap. (B) PCA score plot of all samples showing clear separation of ACLF from other groups. (C) PCA plot showed clear separation of ACLF and preclinical severe-CHB (ACLF vs. CHB-S) samples. (D) PCA plot showed no clear separation of mild- and severe-CHB (CHB-M vs. CHB-S) samples. Relative quantities of 4 proteins (E) and the P4 score (G) between HBV-ACLF (red), CHB (blue) and healthy controls (green) were shown. ROC analyses of all 4 diagnostic markers (F) and the P4 diagnostic score (H) to differentiate HBV-ACLF from CHB patients in the validation sample set. Dash lines represent 95% confidence intervals. We also found ten proteins shown decreasing level in CHB-S as compared to CHB-M (I). HP and KLKB1 both showed high diagnostic value (auROC > 0.75) to differentiate CHB-M and CHB-S, the logistic model built based on HP and KLKB1 showed significant difference between two CHB groups (J) and achieved auROC of 0.84 (K). Note: *,**,*** represent p-value <0.05, <0.01, <0.001, respectively. NS: non-significant.
The 4-protein logistic regression classifier (P4) for HBV-ACLF diagnosis.
| Gene | Protein | ACLF/CHB Ratio* | Function | ROC analysis | Logistic regression | ||||
|---|---|---|---|---|---|---|---|---|---|
| auROC | Sen. | Spe. | Coef. | P-value | |||||
| APOC3 | Apolipoprotein C-III | 0.11 | Lipid/cholesterol transport and metabolism (VLDL components) | 0.956 | 0.944 | 0.88 | -0.92085 | 0.0003 | |
| HRG | Histidine-rich glycoprotein | 0.18 | immune complex and pathogen clearance, cell chemotaxis, cell adhesion, angiogenesis, coagulation and fibrinolysis | 0.914 | 0.817 | 0.88 | -0.75754 | 0.0236 | |
| TF | Serotransferrin (Beta-1 metal-binding globulin, Siderophilin) | 0.42 | iron transport/storage | 0.84 | 0.817 | 0.761 | 1.20517 | 0.0154 | |
| KLKB1 | Kallikrein | 0.12 | coagulation regulation, Kinin-renin-angiotensin system | 0.864 | 0.915 | 0.707 | -0.52709 | 0.0030 | |
| Constant | - | - | - | - | - | - | 17.0668 | - | |
| Summary of diagnosis performance of P8 logistic regression classifier | |||||||||
| Cut-off value | ROC-AUC | Sen. | Spe. | J | |||||
| 0.3139 | 0.961 | 0.9437 | 0.8913 | 0.835 | |||||
*all markers shown t-test Bonferroni-adjusted P<0.0001
In addition, the same model building strategy was used to form the logistic model using 10 markers (APOA1, APOC3, APOH, GC, HP, SAA4, KLKB1, HPR, PLG and GPLD1) shown differential abundance between CHB-S and CHB-M (all p<0.05, Figure 3I). The final logistic model, dubbed P2 comprised of HP and KLKB1 (both auROC>0.75), can be expressed as follows:
Ln(P2/(1-P2)) = 20.6397 - 0.31803*HP - 0.43152*KLKB1
Using this P2 score (Figure 3J), CHB-S samples can be distinguished from CHB-M samples with high sensitivity (88.1%) and specificity (72.0%), and the auROC reached 0.842 in our data set (Figure 3K).
Develop prognosis panel for HBV-ACLF. Acute liver failure in patients of chronic liver diseases such as CHB is considered to be a complex and challenging condition that portends ominous prognosis. We hereby aimed to test if protein candidates discovered in this study can be used to gauge the severity of ACLF and used in complementary to clinical prognostic variables to predict short-turn mortality (28 and 90 days post admission) for ACLF.
Multivariable Cox proportional-hazard regression modeling revealed 4 proteins (GC, HRG, HPR, SERPINA6) and 4 clinical variables (INR, Age, NEU, total protein level) associated significantly (all p<0.05) with 3-month mortality in HBV-ACLF patients (Table 3). Relation between the relative hazard of an ACLF patient reaching endpoint and 8 parameters can be defined as follows:
Ln(Ht/Ht0) = 0.4903*GC + 0.3335*HPR - 0.639*HRG - 0.2866*SERPINA6 + 1.4392*INR + 0.05412*Age + 0.2145*NEU - 0.09234*TotalProtein
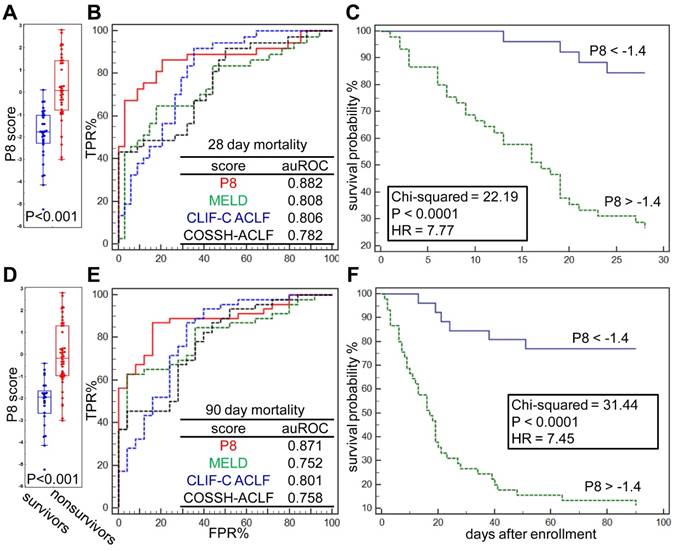
The differences of prognostic P8 score between survivors and nonsurvivors at 28 and 90 days post admission were significant (both p<0.001, Figure 4A, D). At the threshold point of -1.4, the sensitivity and specificity of P8 to predict 28-day endpoints were 86.96% and 84.00%, respectively, and reached 86.49% and 79.41%, respectively, for 90-day endpoints (Figure 4B, E). As summarized in Table 4, the P8 (auROC = 0.882) score showed better prediction power for 28-day endpoints than MELD (auROC =0.808), COOSH (auROC =0.782) or CLIF-C ACLF (auROC =0.806). Similar high prediction power was achieved for 90-day endpoint (auROC = 0.871) by P8 than MELD (auROC =0.752), COOSH (auROC =0.758) or CLIF-C ACLF (auROC =0.801). As shown in Figure 4C, 4F, higher prognostic P8 score was strongly correlated with poor outcome in 28 and 90 days (log-rank test, HR=7.77, and 7.45, respectively, both P<0.0001). Higher P8 score also correlated with higher ACLF grades per COSSH criteria (Figure S5A), as higher ACLF grades typically portend ominous outcomes.
Development of P8 score for HBV-ACLF prognosis.
| Multivariate Cox proportional-hazards regression | ||||
|---|---|---|---|---|
| Covariate | b | P-value | RH | 95% CI of RH |
| GC | 0.4903 | 0.0047 | 1.6328 | 1.1646 to 2.2893 |
| HRG | -0.6390 | <0.0001 | 0.5278 | 0.4003 to 0.6960 |
| HPR | 0.3335 | 0.0005 | 1.3958 | 1.1579 to 1.6827 |
| SERPINA6 | -0.2866 | 0.0002 | 0.7508 | 0.6454 to 0.8733 |
| INR | 1.4392 | <0.0001 | 4.2174 | 2.5552 to 6.9608 |
| Age | 0.05412 | 0.0002 | 1.0556 | 1.0265 to 1.0856 |
| NEU | 0.2145 | <0.0001 | 1.2392 | 1.1180 to 1.3736 |
| Total protein | -0.09234 | 0.0004 | 0.9118 | 0.8667 to 0.9593 |
| Protein markers description | ||||
| Gene | Protein | Function | ||
| GC | Vitamin D-binding protein | vitamin D transport, enhancement of the chemotactic activity for neutrophil in inflammation, macrophage activation | ||
| HRG | Histidine-rich glycoprotein | immune complex and pathogen clearance, cell chemotaxis, cell adhesion, angiogenesis, coagulation and fibrinolysis | ||
| HPR | Haptoglobin-related protein | HDL particle associated, immune response, iron/heme metabolism | ||
| SERPINA6 | Corticosteroid binding globulin (Transcortin) | glucocorticoids transport | ||
Model summary: Chi-squared=60.851, P < 0.0001.
It is believed that the critical therapeutic "golden window" exists between the development of liver failure and extra-hepatic OFs, after which mortality and the cost of treatment increase dramatically [15, 22, 36]. Regarding common OF events, we found that ACLF patients who developed multiple OFs had significant higher P8 scores than those with extra-hepatic 0-1 OF (p <0.001). However, P8 displayed no differences between patients with 2, 3 or 4 OFs (Figure S5B). Compared to other ACLF prognostic scores, we found the P8 score showed higher predictive power for coagulation and brain failure (auROC = 0.815, 0.842, respectively), while none of current scores reached auROC of 0.8 for these two OF types. However, we found the P8 showed limited value to predict respiratory failure (Figure S5C-H), while the COSSH score outperformed all other score by the auROC of 0.764.
Discussion
Liver failure was considered as one of the most challenging and intractable condition in the fields of hepatology and critical care today. Particularly for patients with chronic etiologies, such as viral and alcoholic hepatitis, liver failure comes in the form of ACLF, which is considered as more ominous than ALF. Extensive research efforts are still required to improve and standardize the definition and diagnosis frame for this complicated life-threatening syndrome. This study mainly focused on ACLF patients based on CHB background which was described systematically by the recent COSSH study [10] that shown cirrhosis-free HBV-ACLF have comparably high mortality as to cirrhosis-based ACLF described by the CLIF-C ACLF [11]. Regardless of etiology, the underlying pathophysiology of ACLF presumed to be the results of severe hepatic necroinflammation and uncoordinated immune responses, and the molecular events that lead to these events remain to be elucidated.
Comparison of scores for HBV-ACLF prognosis.
| Score | auROC | Sensitivity | Specificity |
|---|---|---|---|
| For 28 days mortality prediction | |||
| P8 | 0.882 | 86.96% | 84.00% |
| MELD | 0.808 | 63.04% | 96.00% |
| CLIF-C ACLF | 0.806 | 86.96% | 68.00% |
| COSSH-ACLF | 0.782 | 86.96% | 56.00% |
| For 90 days mortality prediction | |||
| P8 | 0.871 | 86.49% | 79.41% |
| MELD | 0.752 | 64.86% | 82.35% |
| CLIF-C ACLF | 0.801 | 91.89% | 64.71% |
| COSSH-ACLF | 0.758 | 43.24% | 100% |
Prognostic performance of the P8 score. (A) Difference of Prognostic P8 between survivors and nonsurvivors at 28 days post admission. (B) Comparison of Prognostic P8, MELD, CLIF-C ACLF and COSSH-ACLF score in prediction of 28-day endpoint event by ROC plots. ROC characteristics of 4 scores were summarized in the table. (C) Kaplan-Meier analysis shown the difference of 28-day survival probability in HBV-ACLF subgroups partitioned by Prognostic P8 score at its threshold derived via ROC analysis. The same analyses were also performed for 90-days survival (D, E, F).
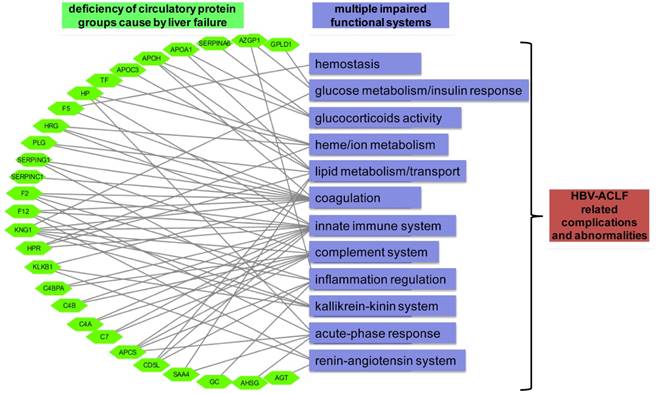
Graphical paradigm for proposed roles of deficient plasma proteins in ACLF. Each quantified circulatory protein (green hexagon) by targeted proteomics was linked to their documented hematological functions (purple rectangular) that may related to the development of HBV-ACLF related complications.
Considering the rapid progression and high mortality of ACLF, it is critical to implement aggressive treatments at the early stage of ACLF with the help of sensitive and objective diagnostic measurements. Score systems based on biochemical and clinical indicators including APACHE-II, CTP and MELD and several variations thereof for liver disease severity assessment have not been thoroughly validated in HBV-ACLF cohorts [11, 21, 37-41]. Arguably, both CTP and MELD were initially developed to monitor the progression of chronic liver diseases, mainly cirrhosis. Their performance to portray acute hepatic deterioration, such as ACLF, requires prudent validation. Specifically, the narrow score range based on subjective judgments makes the CTP a less precise tool to estimate liver damage [14]. One should also be cautious to use MELD on HBV-ACLF patients among whom exacerbated renal damage is uncommon [42]. Alternative to current clinical indicators, molecular indicators including blood markers allow quantitative assessment of the disease progression. Unfortunately, conventional markers such as ALT, AST have limited value even as diagnostic or prognostic adjuncts for ACLF [14]. Regarding this, we sought to provide a comprehensive view of molecular continuum from CHB to ACLF via a hypothesis-free proteomic approach, based on which biomarkers can be derived.
One of the key observations of our initial proteomic survey was the concerted decreasing level of many secretory proteins, involved with protein metabolism, immune response, cell growth, transport and complement system during ACLF pathogenesis (Figure 2C). Notably, decreasing level of proteins in two prominent functional blocks, the fatty acid metabolism/transport and the complement/coagulation cascades, was evident at CHB-S stage and became more pronounced in ACLF stages (Figure S4). Disrupted expression of hepatic lipoproteins had been reported previously to portend a pessimistic ACLF outcome [43, 44]. Other than lipid metabolism, heme metabolism (TF, HP and HPR) was also inhibited in our ACLF patients in accordance with previous reports [45]. Coagulation impairment is also a hallmark of ACLF [11, 42]. Our data suggested the prolonged INR in ACLF patients can be explained by the exacerbated deficiency of a series of coagulation factors (F2, F5, F12, SERPINC1 and CPB2). The complement-kinin-coagulation crosstalk was previously known to contribute to the development of life-threatening thromboinflammation [46, 47]. Regarding this, our data revealed significant loss of complement-kinin players (C4A,C4B, C4BPA, CD5L, C7, PLG, MASP1, KLKB1 and KNG1) in ACLF. It is interesting that recent studies have point out that thrombosis is more common and dangerous condition than spontaneous bleeding in patients with liver failure despite prolonged INR [48-50]. We also need to mention that low level of plasma kallikrein (KLKB1) and angiotensinogen (AGT) suggesting the dysregulation of renin-angiotensin system (RAS) in ACLF cases. Deregulated RAS had been well-documented in heart, renal and cerebral failures, and inhibited hepatic synthesis of AGT had been reported to correlate with the histologic abnormality and disease severity in ALF patients previously [51]. Depletion of key members in those hematological groups was further verified by targeted proteomics in a larger sample set (Figure 3A). Based on these observations, we propose that the loss of liver secreted functional proteins may contribute to the development of complications and abnormalities associated with ACLF as summarized in Figure 5.
The observed extensive proteome alteration and its hypothetic link to ACLF-related complications may help to explain the ineffectiveness of many none-LT therapeutic options, including conventional internal medicine, albumin infusion or ALSS interventions, which do not have the ability to correct such drastic change of blood proteome. We believe future development of ACLF therapies should focus on the upstream molecular events before the occurrence of such large-scale proteomic disturbance. Despite its complicated pathology, HBV-ACLF is essentially initiated by liver failure, which was reflected by the altered proteome of large portion being liver secreted species. We inferred that the marked downregulation of these functional plasma constituents can be ascribed to the profound defect of hepatic protein generation in HBV-ACLF patients. Such dramatic “shutdown” of liver synthesis function in ACLF was also evidenced by the down-regulation of two key hepatic function maintainers, HNF-1α and HNF-4α [52-54] in the liver of ACLF patients (Figure S3). The down regulation of these two hepatic nuclear factors also indicates impaired hepatocellular regeneration [53, 55], which was augmented by our data showing lower levels of liver regeneration related proteins in the ACLF patients (MST1, F2, APOA1/5, KLKB1, CPB2, PON3, ANGFTL6,Table S6). Targeting these upstream pathways orchestrating hepatic functions and regeneration may offer new path towards effective therapy for ACLF.
Given that a wealth of liver-specific and ACLF associated proteins was discovered in the first step, this study then aimed to develop early diagnostic and risk stratification marker panel for HBV-ACLF management. The challenge of verifying multitude of candidates in large sample sets prompted us to resort to targeted proteomics technology with high multiplexability, which has emerged as a powerful analytical tool for biomarker validation in recent years [56]. Compared to conventional antibody-based method such as ELISA, mass spectrometry based PRM approach can efficiently capture the multifactorial nature of the disease course and is particularly useful to develop biomarker panels. In this study, 28 DEPs verified by PRM assays showed clear ability to differentiate CHB and ACLF patients (Figure 3A,B). Among those candidates, 4 markers (APOC3, HRG, TF, KNG1, Table 2) were eventually incorporated into the final discriminative P4 score. Using P4, each sample can be assigned with a probability score from 0-1 reflecting the likelihood of that patient being HBV-ACLF with high accuracy (auROC=0.961). In addition to their ability to discriminate samples accurately, the final resulting diagnostic markers combination also reflects the loss of multiple key hepatic and hematological functions in the midst of liver failure, including lipid transportation and metabolism (APOC3), iron transportation and metabolism (TF), coagulation/complement system (KLKB1) and inflammation/immune-response (HRG). Therefore, we believe judgment based on all proteins collectively in this panel is more informative than markers used individually. Other than focusing on separating ACLF and CHB, we also argued that recognizing CHB-S as early as possible may help to prevent the ACLF onset. Since there was almost separation between CHB-S and CHB-M (Figure 3D), we did capture a subset of 10 markers correlated with the aggravation of CHB (Figure 3I) which might be valuable to capture the subtle molecular disturbance priori to ACLF onset.
One of the mysteries of ACLF is that different patients may progress to divergent outcomes [42]. We therefore investigated the ability of all proteins to predict HBV-ACLF related mortality. In this study, 4 proteins associated with poor prognosis were identified. It is interesting that 3 out of 4 prognostic proteins differed from the 4 diagnostic proteins, hence suggesting there were unique biological processes contributed to the ACLF deterioration after onset. The only protein included in both panel was histidine-rich glycoprotein (HRG) which reflected the weakened innate immunity in ACLF patients. Intriguingly, recent reports have shown the decreasing level of HRG can lead to excessive systemic inflammation and immunothrombosis related to septic lethality and MOFs [57], both were known to signify worst ACLF outcome [8, 36]. Another member of innate immune system with ACLF prognostic value was haptoglobin-related protein (HPR) which was also involved with heme/iron metabolism. However, no report so far has investigated HPR in liver disease yet, despite its high liver specificity. The prognostic significance of inadequate immune system in ACLF was also evidenced by the third prognostic marker, the corticosteroid binding globulin (CBG or SERPINA6). As adrenal corticosteroids is essential for normal immune functions, deficiency of major corticoids transporters like CBG and HSA indicates crippled glucocorticoid activity in ACLF patients. Such changes are the basis of hepato-adrenal syndrome which has been described in patients with acute liver failure and end-stage cirrhosis, possibly due to low HDL level and low steroidogenesis [58-60].The fourth prognostic maker, vitamin D-binding protein (or actin-free Gc globulin) has long been connected to a list of liver diseases, and was viewed as a valuable prognostic marker to signify MOFs and sepsis events during acute liver failure [61-63]. The implication of vitamin D deficiency on inflammation has long been proposed but without a clear explanation [64]. Given its relevance, future studies are needed to clarify whether vitamin D deficiency merely reflects the impaired hepatic function or if it plays critical role in progression of ACLF. Since there were already known clinical parameters for ACLF prognosis, and protein markers developed in this study shown low degree of negative correlation with them (Figure S6), we further formulated a refined model, i.e. Prognostic P8 based on 4 proteins in conjunction with 4 clinical risk factors: INR, NEU, age and total protein level. As expected, higher INR, age and decreased total protein level were widely documented in ACLF patients [10, 11, 36]. The P8 panel also included neutrophil which has been reported as prognostic factor in ACLF [65, 66]. The prognostic P8 score selected ACLF subgroups with high likelihood of reaching short-term endpoints. Moreover, as previous CONONIC and COSSH cohort studies have both concluded that ACLF patients developing multiple extra-hepatic OFs have significant higher mortality, our study shows the P8 score has such risk stratification ability to predict multiple extra-hepatic OFs, and specifically to predict coagulation and brain failures, both of which were the most common extra-hepatic OF types in HBV-ACLF. Another interesting finding of this study is that liver cirrhosis status seems to have limited impact on the circulatory proteome in ACLF patients, as neither P4 or P8 score or the levels of 28 proteins differed between subgroups with or without cirrhosis (all p>0.05). This data may add to the current argument and different perspectives from Asian and European hepatologists on whether pre-existing cirrhosis is a sine qua non criteria for ACLF diagnosis [19, 22, 67]. Our data indicated, at least on the plasma proteome level, ACLF patients regardless having cirrhosis or not, may result in a molecular homogeneity (Figure S7) which needs to be confirmed in future studies. Of note, COSSH ACLF cohorts were mostly insulted by hepatic events while CONONIC ACLF cohorts mostly insulted by extra-hepatic events, but we observed no proteomic or P4, P8 score differences regarding precipitating insult types suggesting that the proteomic signature developed in this study were associated with the inherent pathological characteristics of ACLF. In summary, we believe this prognostic score is meaningful to guide aggressive therapeutic intervention, and to maximize the power of future clinical studies.
It should be noted that the current outcome prediction was given by the proteomic signature measured in patients at admission. However, the conditions of ACLF patients are often unstable, and the evolving differences between survivors and nonsurvivors increases as time to death decreases. Recent study showed serial evaluations in the time frame of 3-7 day after admission correlated more to the final outcome of ACLF patients [68]. Given the correlation of protein signature alteration and the pathological stages illustrated in our study, we believe the sequential measurements of these protein markers in ACLF patients will provide more precise portrait of molecular trajectory toward convalescence or deterioration, thus to improve prognostic performance of the current admission-based model. We also want to stress that this study has produced a rich pool of markers that highly associated with ACLF progress, despite only few were integrated into the final P4 or P8 score. However, one should not underestimate the potential value and biological roles of the rest candidates which were not included in our final panels. The depletion of these 28 markers and their associated clinical consequences still need to be explored to develop corresponding diagnosis, prognosis or even intervention strategy.
Abbreviations
2DLC-MS/MS: 2 dimensional liquid chromatography-tandem mass spectrometry; 2MEGA: dimethylation after lysine guanidination; ACLF: acute-on-chronic liver failure; ACN: acetonitrile; AGT: angiotensinogen; ALB: albumin; ALF: acute liver failure; ALT: alanine aminotransferase; APOA1: apolipoprotein A-I; AST: aspartate amino transferase; CHB: chronic hepatitis B; CI: confidence interval; CPLL: combinatorial peptide ligand library; CTP: Child-Turcott-Pugh score; Cr: creatinine; DDA: data-dependent acquisition; FA: formic acid; FASP: filter-aided sample preparation; FDR: false discovery rate; GC: vitamin D binding protein; GGT: gamma-glutamyl transferase; HAP: high-abundance proteins; HBV: hepatitis B virus; HCD: higher-energy collision dissociation; HE: hepatic encephalopathy; HNF-1a: hepatocyte nuclear factor 1a; HNF-4a: hepatocyte nuclear factor 4a; HP: haptoglobin; HR: hazard ratio; HRG: histidine-rich glycoprotein; INR: international normalized ratio; KNG1: kininogen-1; LAP: low-abundance proteins; LC: liver cirrhosis; LT: liver transplantation; MELD: model of end-stage liver disease; MOF: multi-organ failure; MWCO: molecular weight cut-off; PRM: parallel reaction monitoring; RAS: renin-angiotensin system; PSMs: peptide-spectra matches; ROC: receiver operating characteristic; SAA4: serum amyloid A-4; SBP: spontaneous bacterial peritonitis; Tbil: total bilirubin; TEAB: triethylammonium bicarbonate; TF: serotransferrin; TPP: transproteomic pipeline environment.
Supplementary Material
Supplementary materials and methods, figures, tables, references.
Table S6.
Table S7.
Table S8.
Acknowledgements
The study was funded by The National Key Research and Development Program (2017YFC1200200), National Natural Science Foundation of China (81400589), Foundation for Innovative Research Groups of the National Natural Science Foundation of China (81121002), Independent Project Fund of the State Key Laboratory for Diagnosis and Treatment of Infectious Disease, Chinese High-Tech Research and Development Programs (2012AA020204), National S&T Major Project (2012ZX10002004), and Zhejiang Provincial Medicine and Health Science and Technology Project (2016KYA083, 2016147735). We want to thank all patients contributed plasma samples for this study, and the clinical team at the Department of Infectious Diseases at the First Affiliated Hospital, College of Medicine, Zhejiang University.
Author contributions
Z.S., X.L., and D.W. contributed equally to this work. L.L., Z.S. and X.L. conceived and designed the study; L.L., X.X., Z.S. and X.L. obtained funding, contributed reagents and analytic tools; Z.S., D.W., H.G., Y.Y., J.J., J.W. and X.X. coordinated samples and clinical data collection; Z.S., J.J., Q.G., D.W.,J.W. and Y.X. performed sample preparation and mass spectrometry analyses; Z.S., D.W., H.Z. and J.W. processed mass spectrometry data and performed multivariate modeling; Z.S., D.W., X.L. and Z.J wrote the manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Dienstag JL. Hepatitis B virus infection. N Engl J Med. 2008;359:1486-500
2. Mortality GBD, Causes of Death C. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117-71
3. MacLachlan JH, Locarnini S, Cowie BC. Estimating the global prevalence of hepatitis B. Lancet. 2015;386:1515-7
4. Sarin SK, Kedarisetty CK, Abbas Z, Amarapurkar D, Bihari C, Chan AC. et al. Acute-on-chronic liver failure: consensus recommendations of the Asian Pacific Association for the Study of the Liver (APASL) 2014. Hepatol Int. 2014;8:453-71
5. Asrani SK, O'Leary JG. Acute-on-chronic liver failure. Clin Liver Dis. 2014;18:561-74
6. Bernal W, Jalan R, Quaglia A, Simpson K, Wendon J, Burroughs A. Acute-on-chronic liver failure. Lancet. 2015;386:1576-87
7. Jalan R, Yurdaydin C, Bajaj JS, Acharya SK, Arroyo V, Lin HC. et al. Toward an improved definition of acute-on-chronic liver failure. Gastroenterology. 2014;147:4-10
8. Moreau R, Jalan R, Arroyo V. Acute-on-Chronic Liver Failure: Recent Concepts. J Clin Exp Hepatol. 2015;5:81-5
9. Sarin SK, Choudhury A. Acute-on-chronic liver failure: terminology, mechanisms and management. Nat Rev Gastroenterol Hepatol. 2016;13:131-49
10. Wu T, Li J, Shao L, Xin J, Jiang L, Zhou Q. et al. Development of diagnostic criteria and a prognostic score for hepatitis B virus-related acute-on-chronic liver failure. Gut. 2017
11. Jalan R, Saliba F, Pavesi M, Amoros A, Moreau R, Gines P. et al. Development and validation of a prognostic score to predict mortality in patients with acute-on-chronic liver failure. J Hepatol. 2014;61:1038-47
12. Sarin SK, Kumar M, Lau GK, Abbas Z, Chan HL, Chen CJ. et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int. 2016;10:1-98
13. Graziadei IW. The clinical challenges of acute on chronic liver failure. Liver Int. 2011;31(Suppl 3):24-6
14. Chen EQ, Zeng F, Zhou LY, Tang H. Early warning and clinical outcome prediction of acute-on-chronic hepatitis B liver failure. World J Gastroenterol. 2015;21:11964-73
15. Jindal A, Rastogi A, Sarin SK. Reviewing the diagnostic criteria for acute-on-chronic liver failure. Expert Rev Gastroenterol Hepatol. 2016;10:1385-95
16. Sun LJ, Yu JW, Zhao YH, Kang P, Li SC. Influential factors of prognosis in lamivudine treatment for patients with acute-on-chronic hepatitis B liver failure. J Gastroenterol Hepatol. 2010;25:583-90
17. Garg H, Sarin SK, Kumar M, Garg V, Sharma BC, Kumar A. Tenofovir improves the outcome in patients with spontaneous reactivation of hepatitis B presenting as acute-on-chronic liver failure. Hepatology. 2011;53:774-80
18. Jindal A, Kumar M, Sarin SK. Management of acute hepatitis B and reactivation of hepatitis B. Liver Int. 2013;33(Suppl 1):164-75
19. Jalan R, Stadlbauer V, Sen S, Cheshire L, Chang YM, Mookerjee RP. Role of predisposition, injury, response and organ failure in the prognosis of patients with acute-on-chronic liver failure: a prospective cohort study. Crit Care. 2012;16:R227
20. Sen S, Williams R, Jalan R. The pathophysiological basis of acute-on-chronic liver failure. Liver. 2002;22(Suppl 2):5-13
21. Kamath PS, Wiesner RH, Malinchoc M, Kremers W, Therneau TM, Kosberg CL. et al. A model to predict survival in patients with end-stage liver disease. Hepatology. 2001;33:464-70
22. Zhang Q, Li Y, Han T, Nie C, Cai J, Liu H. et al. Comparison of current diagnostic criteria for acute-on-chronic liver failure. PLoS One. 2015;10:e0122158
23. Wang X, Sarin SK, Ning Q. Definition of ACLF and inclusion criteria for extra-hepatic organ failure. Hepatol Int. 2015;9:360-5
24. Wu D, Sun Z, Liu X, Rao Q, Chen W, Wang J. et al. HINT: a novel prognostic model for patients with hepatitis B virus-related acute-on-chronic liver failure. Aliment Pharmacol Ther. 2018;48:750-60
25. Talens S, Hoekstra J, Dirkx SP, Darwish Murad S, Trebicka J, Elias E. et al. Proteomic analysis reveals that apolipoprotein A1 levels are decreased in patients with Budd-Chiari syndrome. J Hepatol. 2011;54:908-14
26. Shang S, Plymoth A, Ge S, Feng Z, Rosen HR, Sangrajrang S. et al. Identification of osteopontin as a novel marker for early hepatocellular carcinoma. Hepatology. 2012;55:483-90
27. Molleken C, Sitek B, Henkel C, Poschmann G, Sipos B, Wiese S. et al. Detection of novel biomarkers of liver cirrhosis by proteomic analysis. Hepatology. 2009;49:1257-66
28. Wang J, Sun Z, Jiang J, Wu D, Liu X, Xie Z. et al. Proteomic signature of acute liver failure: from discovery and verification in a pig model to confirmation in humans. Mol Cell Proteomics. 2017
29. Rademacher S, Oppert M, Jorres A. Artificial extracorporeal liver support therapy in patients with severe liver failure. Expert Rev Gastroenterol Hepatol. 2011;5:591-9
30. Maiwall R, Maras JS, Nayak SL, Sarin SK. Liver dialysis in acute-on-chronic liver failure: current and future perspectives. Hepatol Int. 2014;8(Suppl 2):505-13
31. Rai AJ, Gelfand CA, Haywood BC, Warunek DJ, Yi J, Schuchard MD. et al. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005;5:3262-77
32. Tu C, Li J, Young R, Page BJ, Engler F, Halfon MS. et al. Combinatorial peptide ligand library treatment followed by a dual-enzyme, dual-activation approach on a nanoflow liquid chromatography/orbitrap/electron transfer dissociation system for comprehensive analysis of swine plasma proteome. Anal Chem. 2011;83:4802-13
33. Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA. et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15:2597-601
34. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J. et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447-52
35. Sun QF, Ding JG, Xu DZ, Chen YP, Hong L, Ye ZY. et al. Prediction of the prognosis of patients with acute-on-chronic hepatitis B liver failure using the model for end-stage liver disease scoring system and a novel logistic regression model. J Viral Hepat. 2009;16:464-70
36. Moreau R, Jalan R, Gines P, Pavesi M, Angeli P, Cordoba J. et al. Acute-on-chronic liver failure is a distinct syndrome that develops in patients with acute decompensation of cirrhosis. Gastroenterology. 2013;144:1426-37 1437 e1-9
37. Biggins SW, Kim WR, Terrault NA, Saab S, Balan V, Schiano T. et al. Evidence-based incorporation of serum sodium concentration into MELD. Gastroenterology. 2006;130:1652-60
38. Luca A, Angermayr B, Bertolini G, Koenig F, Vizzini G, Ploner M. et al. An integrated MELD model including serum sodium and age improves the prediction of early mortality in patients with cirrhosis. Liver Transpl. 2007;13:1174-80
39. Shen Y, Liu YM, Wang B, Zhu YG, Wang YY, Wang XL. et al. External validation and comparison of six prognostic models in a prospective cohort of HBV-ACLF in China. Ann Hepatol. 2016;15:236-45
40. Yan H, Wu W, Yang Y, Wu Y, Yang Q, Shi Y. A novel integrated Model for End-Stage Liver Disease model predicts short-term prognosis of hepatitis B virus-related acute-on-chronic liver failure patients. Hepatol Res. 2015;45:405-14
41. Duseja A, Choudhary NS, Gupta S, Dhiman RK, Chawla Y. APACHE II score is superior to SOFA, CTP and MELD in predicting the short-term mortality in patients with acute-on-chronic liver failure (ACLF). J Dig Dis. 2013;14:484-90
42. Li H, Chen LY, Zhang NN, Li ST, Zeng B, Pavesi M. et al. Characteristics, Diagnosis and Prognosis of Acute-on-Chronic Liver Failure in Cirrhosis Associated to Hepatitis B. Sci Rep. 2016;6:25487
43. Etogo-Asse FE, Vincent RP, Hughes SA, Auzinger G, Le Roux CW, Wendon J. et al. High density lipoprotein in patients with liver failure; relation to sepsis, adrenal function and outcome of illness. Liver Int. 2012;32:128-36
44. Reyes-Soffer G, Moon B, Hernandez-Ono A, Dionizovik-Dimanovski M, Jimenez J, Obunike J. et al. Complex effects of inhibiting hepatic apolipoprotein B100 synthesis in humans. Sci Transl Med. 2016;8:323ra12
45. Maras JS, Maiwall R, Harsha HC, Das S, Hussain MS, Kumar C. et al. Dysregulated iron homeostasis is strongly associated with multiorgan failure and early mortality in acute-on-chronic liver failure. Hepatology. 2015;61:1306-20
46. Amara U, Flierl MA, Rittirsch D, Klos A, Chen H, Acker B. et al. Molecular intercommunication between the complement and coagulation systems. J Immunol. 2010;185:5628-36
47. Ekdahl KN, Teramura Y, Hamad OA, Asif S, Duehrkop C, Fromell K. et al. Dangerous liaisons: complement, coagulation, and kallikrein/kinin cross-talk act as a linchpin in the events leading to thromboinflammation. Immunol Rev. 2016;274:245-69
48. Agarwal B, Wright G, Gatt A, Riddell A, Vemala V, Mallett S. et al. Evaluation of coagulation abnormalities in acute liver failure. J Hepatol. 2012;57:780-6
49. Stravitz RT, Ellerbe C, Durkalski V, Schilsky M, Fontana RJ, Peterseim C. et al. Bleeding complications in acute liver failure. Hepatology. 2018;67:1931-42
50. Stravitz RT, Lisman T, Luketic VA, Sterling RK, Puri P, Fuchs M. et al. Minimal effects of acute liver injury/acute liver failure on hemostasis as assessed by thromboelastography. J Hepatol. 2012;56:129-36
51. Colmenero J, Bataller R, Sancho-Bru P, Bellot P, Miquel R, Moreno M. et al. Hepatic expression of candidate genes in patients with alcoholic hepatitis: correlation with disease severity. Gastroenterology. 2007;132:687-97
52. Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL. et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378-81
53. Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000;14:464-74
54. Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol. 2001;21:1393-403
55. Costa RH, Kalinichenko VV, Holterman AX, Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331-47
56. Meyer JG, Schilling B. Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert Rev Proteomics. 2017;14:419-29
57. Wake H, Mori S, Liu K, Morioka Y, Teshigawara K, Sakaguchi M. et al. Histidine-Rich Glycoprotein Prevents Septic Lethality through Regulation of Immunothrombosis and Inflammation. EBioMedicine. 2016;9:180-94
58. Fernandez J, Escorsell A, Zabalza M, Felipe V, Navasa M, Mas A. et al. Adrenal insufficiency in patients with cirrhosis and septic shock: Effect of treatment with hydrocortisone on survival. Hepatology. 2006;44:1288-95
59. Harry R, Auzinger G, Wendon J. The clinical importance of adrenal insufficiency in acute hepatic dysfunction. Hepatology. 2002;36:395-402
60. Tsai MH, Peng YS, Chen YC, Liu NJ, Ho YP, Fang JT. et al. Adrenal insufficiency in patients with cirrhosis, severe sepsis and septic shock. Hepatology. 2006;43:673-81
61. Antoniades CG, Berry PA, Bruce M, Cross TJ, Portal AJ, Hussain MJ. et al. Actin-free Gc globulin: a rapidly assessed biomarker of organ dysfunction in acute liver failure and cirrhosis. Liver Transpl. 2007;13:1254-61
62. Bagchi A, Kumar S, Ray PC, Das BC, Gumma PK, Kar P. Predictive value of serum actin-free Gc-globulin for complications and outcome in acute liver failure. J Viral Hepat. 2015;22:192-200
63. Schiodt FV, Bangert K, Shakil AO, McCashland T, Murray N, Hay JE. et al. Predictive value of actin-free Gc-globulin in acute liver failure. Liver Transpl. 2007;13:1324-9
64. Mangin M, Sinha R, Fincher K. Inflammation and vitamin D: the infection connection. Inflamm Res. 2014;63:803-19
65. Moreau N, Wittebole X, Fleury Y, Forget P, Laterre PF, Castanares-Zapatero D. Neutrophil-to-Lymphocyte Ratio Predicts Death in Acute-on-Chronic Liver Failure Patients Admitted to the Intensive Care Unit: A Retrospective Cohort Study. Shock. 2018;49:385-92
66. Agiasotelli D, Alexopoulou A, Vasilieva L, Kalpakou G, Papadaki S, Dourakis SP. Evaluation of neutrophil/leukocyte ratio and organ failure score as predictors of reversibility and survival following an acute-on-chronic liver failure event. Hepatol Res. 2016;46:514-20
67. Olson JC, Kamath PS. Acute-on-chronic liver failure: concept, natural history, and prognosis. Curr Opin Crit Care. 2011;17:165-9
68. Gustot T, Fernandez J, Garcia E, Morando F, Caraceni P, Alessandria C. et al. Clinical Course of acute-on-chronic liver failure syndrome and effects on prognosis. Hepatology. 2015;62:243-52
Author contact
![]() Corresponding author: Lanjuan Li, State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, School of Medicine, Zhejiang University. #79 Qingchun Road, Hangzhou, China. 310003. Tel: 86-571-87236758 Fax: 86-571-87236459, E-mail: ljliedu.cn
Corresponding author: Lanjuan Li, State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, School of Medicine, Zhejiang University. #79 Qingchun Road, Hangzhou, China. 310003. Tel: 86-571-87236758 Fax: 86-571-87236459, E-mail: ljliedu.cn
 Global reach, higher impact
Global reach, higher impact