13.3
Impact Factor
Theranostics 2018; 8(10):2722-2738. doi:10.7150/thno.21358 This issue Cite
Research Paper
Molecular Detection and Analysis of Exosomes Using Surface-Enhanced Raman Scattering Gold Nanorods and a Miniaturized Device
1. Department of Chemistry, The University of Memphis, Memphis, TN 38152
2. Diagnostics Imaging Department, St Jude Children's Research Hospital, Memphis, TN 38105
3. Department of Computational Biology, St Jude Children's Research Hospital, Memphis, TN 38105
*These authors contributed equally to this work
Abstract
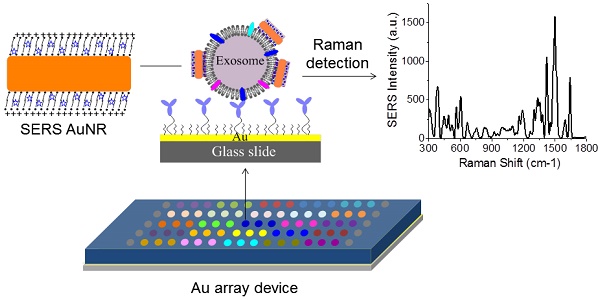
Exosomes are a potential source of cancer biomarkers. Probing tumor-derived exosomes can offer a potential non-invasive way to diagnose cancer, assess cancer progression, and monitor treatment responses. Novel molecular methods would facilitate exosome analysis and accelerate basic and clinical exosome research.
Methods: A standard gold-coated glass microscopy slide was used to develop a miniaturized affinity-based device to capture exosomes in a target-specific manner with the assistance of low-cost 3-D printing technology. Gold nanorods coated with QSY21 Raman reporters were used as the label agent to quantitatively detect the target proteins based on surface enhanced Raman scattering spectroscopy. The expressions of several surface protein markers on exosomes from conditioned culture media of breast cancer cells and from HER2-positive breast cancer patients were quantitatively measured. The data was statistically analyzed and compared with healthy controls.
Results: A miniaturized 17 × 5 Au array device with 2-mm well size was fabricated to capture exosomes in a target-specific manner and detect the target proteins on exosomes with surface enhanced Raman scattering gold nanorods. This assay can specifically detect exosomes with a limit of detection of 2×106 exosomes/mL and analyze over 80 purified samples on a single device within 2 h. Using the assay, we have showed that exosomes derived from MDA-MB-231, MDA-MB-468, and SKBR3 breast cancer cells give distinct protein profiles compared to exosomes derived from MCF12A normal breast cells. We have also showed that exosomes in the plasma from HER2-positive breast cancer patients exhibit significantly (P ≤ 0.01) higher level of HER2 and EpCAM than those from healthy donors.
Conclusion: We have developed a simple, inexpensive, highly efficient, and portable Raman exosome assay for detection and protein profiling of exosomes. Using the assay and model exosomes from breast cancer cells, we have showed that exosomes exhibit diagnostic surface protein markers, reflecting the protein profile of their donor cells. Through proof-of-concept studies, we have identified HER2 and EpCAM biomarkers on exosomes in plasma from HER2-positive breast cancer patients, suggesting the diagnostic potential of these markers for breast cancer diagnostics. This assay would accelerate exosome research and pave a way to the development of novel cancer liquid biopsy for cancer detection and monitoring.
Keywords: Exosome, detection, molecular profiling, cancer, surface enhanced Raman scattering, gold nanorod
 Global reach, higher impact
Global reach, higher impact