13.3
Impact Factor
Theranostics 2018; 8(10):2722-2738. doi:10.7150/thno.21358 This issue Cite
Research Paper
Molecular Detection and Analysis of Exosomes Using Surface-Enhanced Raman Scattering Gold Nanorods and a Miniaturized Device
1. Department of Chemistry, The University of Memphis, Memphis, TN 38152
2. Diagnostics Imaging Department, St Jude Children's Research Hospital, Memphis, TN 38105
3. Department of Computational Biology, St Jude Children's Research Hospital, Memphis, TN 38105
*These authors contributed equally to this work
Received 2017-6-6; Accepted 2018-1-9; Published 2018-4-9
Abstract
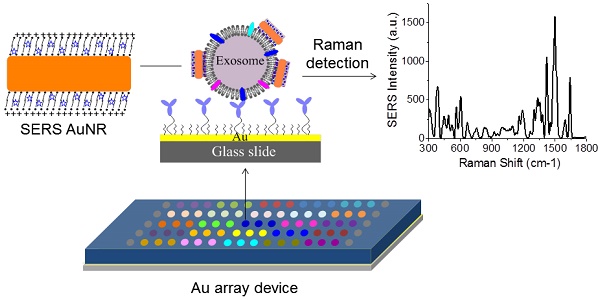
Exosomes are a potential source of cancer biomarkers. Probing tumor-derived exosomes can offer a potential non-invasive way to diagnose cancer, assess cancer progression, and monitor treatment responses. Novel molecular methods would facilitate exosome analysis and accelerate basic and clinical exosome research.
Methods: A standard gold-coated glass microscopy slide was used to develop a miniaturized affinity-based device to capture exosomes in a target-specific manner with the assistance of low-cost 3-D printing technology. Gold nanorods coated with QSY21 Raman reporters were used as the label agent to quantitatively detect the target proteins based on surface enhanced Raman scattering spectroscopy. The expressions of several surface protein markers on exosomes from conditioned culture media of breast cancer cells and from HER2-positive breast cancer patients were quantitatively measured. The data was statistically analyzed and compared with healthy controls.
Results: A miniaturized 17 × 5 Au array device with 2-mm well size was fabricated to capture exosomes in a target-specific manner and detect the target proteins on exosomes with surface enhanced Raman scattering gold nanorods. This assay can specifically detect exosomes with a limit of detection of 2×106 exosomes/mL and analyze over 80 purified samples on a single device within 2 h. Using the assay, we have showed that exosomes derived from MDA-MB-231, MDA-MB-468, and SKBR3 breast cancer cells give distinct protein profiles compared to exosomes derived from MCF12A normal breast cells. We have also showed that exosomes in the plasma from HER2-positive breast cancer patients exhibit significantly (P ≤ 0.01) higher level of HER2 and EpCAM than those from healthy donors.
Conclusion: We have developed a simple, inexpensive, highly efficient, and portable Raman exosome assay for detection and protein profiling of exosomes. Using the assay and model exosomes from breast cancer cells, we have showed that exosomes exhibit diagnostic surface protein markers, reflecting the protein profile of their donor cells. Through proof-of-concept studies, we have identified HER2 and EpCAM biomarkers on exosomes in plasma from HER2-positive breast cancer patients, suggesting the diagnostic potential of these markers for breast cancer diagnostics. This assay would accelerate exosome research and pave a way to the development of novel cancer liquid biopsy for cancer detection and monitoring.
Keywords: Exosome, detection, molecular profiling, cancer, surface enhanced Raman scattering, gold nanorod
Introduction
Extracellular vesicles, especially exosomes, are receiving increasing interest as a resource of biomarkers in medicine [1]. Although they were discovered in the early 1980's, exosomes have only recently moved into intense investigations regarding their biogenesis, composition, and functions [2-5]. It is now believed that exosomes are 40-200 nm membrane-bound vesicles derived from multivesicular bodies and released into the extracellular environment by many cell types [6-8]. They carry molecular constituents of their originating cells including proteins, nucleic acids, and lipids and represent an important mode of intercellular communication by horizontal transfer of their molecular contents between cells [9-14].
Growing evidence suggests that cancer-derived exosomes can transfer oncogenic activity and regulate angiogenesis, immunity, and metastasis to promote tumorigenesis and progression [15-21]. For example, Peinado et al. demonstrated that exosomes from highly metastatic melanoma cells educated bone marrow progenitor cells toward a pro-metastatic phenotype via horizontal transfer of exosomal Met [22]. Zhou et al. showed that exosome-mediated transfer of miR-105 in metastatic breast cancer cells efficiently destroyed vascular endothelial barriers to promote metastasis [23]. Exosomes have been found in various body fluids such as blood, urine, saliva, and cerebrospinal fluid [24-27]. Thus, exosomes are a promising resource of cancer biomarkers to noninvasively screen for cancer, assess cancer progression, and monitor treatment responses [28-36].
Despite their diagnostic and therapeutic potential, the clinical use of exosomes as cancer biomarkers is, however, still very limited. One of the major challenges is molecular detection and analysis due to their small size and complex biological environment. To ensure analytical accuracy, exosomes usually need to be isolated and purified from cell culture supernatant or plasma before analysis. Classical methods for exosome isolation are differential centrifugation, filtration, immunomagnetic separations, and microfluidics [37-40]. Differential centrifugation consists of a series of low, high and ultrahigh speed centrifugations to separate exosomes from cell debris, larger microvesicles, and proteins based on size and density. It is the gold-standard method to purify exosomes. After purification, exosomes have been commonly analyzed for protein compositions using western blot, enzyme-linked immunosorbent assays (ELISA), and mass spectrometry [41-45]. These traditional approaches have greatly helped understand exosome biology, but they are impractical for longitudinal studies and clinical use because they are time-consuming and labor-intensive. Technologies for exosome detection and analysis have been greatly advanced in past years [43, 46]. For example, the nPLEX assay has improved detection sensitivity as high as 1000-fold compared to ELISA [47]. New flow cytometry instrumentation can analyze individual exosomes down to 70-80 nm [48]. In these techniques, exosomes are detected based on fluorescence [48-58], surface plasmon resonance (SPR) [47, 59-61], light scattering plasmon resonance [62], nuclear magnetic resonance [63], electrochemical [64-69], and mechanical approaches [70].
Here we report a new method for exosome detection and protein profiling using surface-enhanced Raman scattering (SERS) nanotags in combination with a miniaturized capture platform. SERS is the enhancement of Raman signals from small molecules that are proximal to a metal surface via electromagnetic and chemical mechanisms [71]. It is an ultrasensitive vibrational spectroscopic technique, with Raman enhancements as high as 1015 for small molecules, such as organic dyes, on plasmonic nanoparticle surfaces [72, 73]. The SERS effect has been previously used to probe exosome composition [74-77], but here we report the use of SERS nanotags for exosome detection and analysis. SERS nanotags, which are plasmonic nanoparticles carrying abundant Raman reporters, such as organic dyes, can provide highly sensitive and specific detection by controlling the surface chemistry, size, and structure of the plasmonic nanoparticles, and the surface density of the Raman reporters [78]. Compared to the classic fluorescence method, SERS gives fingerprinting signals that distinguish interferences from a biological background. The SERS spectrum only requires a simple baseline correction using a multi-segment polynomial fitting to subtract SERS background (broad continuum emission). This baseline correction can be incorporated in the signal correction software and thus the as-acquired spectrum does not need further signal separation processing for quantitative analysis. In addition, signal acquisition is extremely fast when SERS nanotags are used (1 s or faster per spectrum) due to the high sensitivity of SERS nanotags. Due to these attributes, SERS nanotags have emerged as a popular class of biological labels and have been well used for cancer detection, including biomarker detection in body fluids [71, 78-89]. Here we report the first application of SERS nanotags for exosome detection and analysis. We used small gold nanorods (AuNRs) as the SERS substrate. The AuNRs are sufficiently small (~35 nm in the longitudinal dimension) in comparison with the small exosomes. The anisotropic rod structure promotes SERS effects due to the high electromagnetic fields at the ends of the rods [90]. We made use of 3D printing technology to improve analytical efficiency. 3D printers are cheap, portable, and easy-to-use. They are accessible to large populations, especially in resource-limited environments. Using a 3D-printed array template, we made an antibody array to capture exosomes in a target-specific manner on Au-coated standard glass microscopy slides. Combining the advantages of SERS nanotags and 3-D printing technology, this simple and low-cost assay offers dozens of test sites on a single palm-sized chip, provides results within 2 h, and has a microliter sample requirement at femtomolar concentrations. Due to its simplicity, high efficiency, and high sensitivity, this assay has great potential for clinical applications for biomarker discovery and understanding of the role of exosomes in cancer development.
Materials and Methods
Materials
All reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. Antibodies were purchased from Biolegend (San Diego, CA). QSY21 carboxylic acid-succinimidyl ester was purchased from Thermo Fisher Scientific. PE-labeled antibodies were purchased from Miltenyi Biotec (Auburn, CA). All cell lines were purchased from ATCC (Manassas, VA). Cell culture media were purchased from VWR (Radnor, PA) and fetal bovine serum (FBS) was purchased from Fisher Scientific (Waltham, MA).
Synthesis of gold nanorods (AuNRs)
Small gold nanorods (AuNRs) were synthesized by modifying the classic seed-mediated growth method [91, 92]. This method involves two steps: preparation of Au seeds and growth of Au seeds into AuNRs in a growth solution. To make the Au seed solution, 0.5 mL of 1 mM chloroauric acid (HAuCl4) was added to 1.5 mL of 0.2M cetyltrimethylammonium bromide (CTAB) solution with constant stirring. 120 μL of 10 mM ice-cold sodium borohydride (NaBH4) was quickly injected and the solution was stirred for 3 min to form the Au seed solution. The Au seed solution was kept undisturbed for 3 h in a 25 °C water bath before its use. In a different glass vial, 5 mL of 1 mM HAuCl4 was added to 5 mL of 0.2 M CTAB solution followed by addition of 125 μL of 4 mM silver nitrate (AgNO3). After mixing by stirring, 12 μL of Au seed solution was quickly injected into the solution and left undisturbed for 10 min to form small AuNRs. The solution was centrifuged at 16,000 × g for 10 min and the AuNR pellet was resuspended in ultrapure water for further use.
Preparation of SERS AuNRs
100 μL of 100 μM QSY21 carboxylic acid (hydrolyzed from QSY21 carboxylic acid-succinimidyl ester) aqueous solution was added to 1 mL of 2 nM AuNRs and the mixture was stirred for 15 min at RT to allow adsorption of the dye onto the AuNRs. After purification by centrifugation (16,000 × g, 10 min), the QSY21 carboxylic acid-adsorbed AuNRs were resuspended in phosphate buffer solution (PBS) to make 1 nM solution. The solution was aged at room temperature (RT) for 2 h before use.
Au thin film deposition on microscopy glass slides
A standard microscopy glass slide (75 × 25 × 1 mm) was coated with a 10 nm thick Au film by the magnetron sputtering technique using an ORION-AJA system from a 99.99% pure Au target. The deposition of the Au layer was performed on a 4 nm titanium layer previously deposited from a 99.99% pure titanium target on the glass slide. The slide-target distance was kept at 15 cm during the process. The film thickness was controlled by an INFICON SQM-160 quartz crystal monitor/controller equipment. The rotating substrate-holder was kept at 80 rpm. The films were grown in an atmosphere of argon at 3.0 mTorr and a gas flow of 15 sccm, with the DC power supply set to 100 W and the pressure before inserting the argon was 4.0×10-8 Torr. The whole process took 4 h.
Fabrication of array template
Plastic (polylactic acid) array templates with specified well size and inter-well distance were fabricated using a MakerBot Replicator PC 3-D printer. The template was attached to a rubber array via a layer of glue composed of 60% silicone and 40% mineral spirit. This rubber array was made from a 1.6 mm thick rubber sheet with the same dimensions as the template via puncture. The assembled plastic and rubber arrays were used as a template array to make an antibody array on the Au-coated glass slides.
Fabrication of the antibody-based capture array
The template array was attached onto the surface of the Au-coated glass slide using 3/4" wide heavy-duty binder clips. 15 μL of 50 μg/mL target-specific antibody-linked polyethylene glycol thiols (HS-PEG-Ab) in PBS was added into the wells and incubated for 5 h at RT. The HS-PEG-Ab was prepared in advance by reacting antibodies with thiol- polyethylene glycol- N-hydroxysuccinimide esters (HS-PEG-NHS 5000, 1:100) at 4 °C overnight. The free HS-PEG-NHS was separated by membrane filtration with a 10 kD Nanosep filter (PALL Life Sciences). The antibody-treated wells were washed three times with phosphate buffer solution-tween (PBST) (100 mL PBS + 0.5 mL Tween 20 (0.5%)) to get rid of unbound proteins. Then, 15 μL of 0.1 mM 11-mercaptoundecyl tetra(ethylene glycol) (MU-TEG) was added into the wells and incubated for 30 min at RT to saturate the Au surface. The antibody-functionalized wells were washed three times with PBST and stored at 4 °C for further use. Isotype IgG was used as the negative control.
Isolation and characterization of exosomes in culture media
Human breast cancer cells MDA-MB-231 (MM231), MDA-MB-468 (MM468), and SKBR3 were cultured in Dulbecco's Modified Eagle Medium (DMEM) with high glucose (MM231 and MM468) RPMI 1640 medium (SKBR3) with 10% fetal bovine serum at 37 °C under 5% CO2. Human breast normal cells MCF12A (immortalized) were cultured in Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12) medium with 5% fetal horse serum, 1% Pen/Strep (100×), 0.5 mg/mL hydrocortisone, 10 μg/mL bovine insulin, 100 ng/mL cholera toxin, and 20 ng/mL epidermal growth factor (EGF). Cells were grown in conditioned cell culture media (media + 10% exosome-free FBS) for 48 h. The exosome-free FBS was obtained by separating exosomes from FBS with ultracentrifugation (100,000 ×g, 24 h). To collect exosomes, the conditioned cell culture supernatant was collected and centrifuged at 430 ×g at RT for 10 min. The supernatant was collected and centrifuged at 16,500 ×g at 4 °C for 30 min. The supernatant was collected and centrifuged at 100,000 ×g at 4 °C for 70 min. After removing the supernatant, the exosome pellet was resuspended in cold sterile PBS and centrifuged again at 100,000 ×g at 4 °C for 70 min. The exosome pellet was resuspended in cold sterile PBS, filtered with a 0.2 μm PES filter (Agilent Technologies), and stored at -80 °C until use. The concentration and size distribution of exosomes were characterized using nanoparticle tracking analysis (NTA) with a NanoSight LM10 microscope (Malvern Instruments, Inc).
Isolation and characterization of exosomes in plasma samples
Plasma samples from six human epidermal growth factor receptor 2 (HER2) positive breast cancer patients (stage III) and three healthy donors were purchased through the XpressBank from Asterand Bioscience (Detroit, Michigan). The samples were collected in 2016 and 2017 and stored in liquid nitrogen (LN). The samples were available for research uses under IRB exemption through the BioSPOKE™ custom biospecimen procurement service. The identity information of each subject was coded with a unique Donor Identification Number (DIN) and we do not have access to the identifying information. To purify exosomes, the plasma samples were diluted with sterile PBS and centrifuged at 16,500 × g for 30 min at 4 °C. The supernatant was collected and centrifuged at 100,000 × g for 70 min at 4 °C. The exosome pellet was resuspended in cold sterile PBS and centrifuged again at 100,000 × g for 70 min at 4 °C. The pellet was resuspended in cold sterile PBS, filtered with a 0.2 μm PES filter, and characterized with NTA to determine the concentration and size distribution of exosomes. The exosomes were stored at -80 °C until use. Exosomes from healthy donors were obtained from fresh whole blood samples through Analytical Biological Science (Wilmington, DE). The whole blood samples were centrifuged two times at 2,500 ×g for 15 min at 4 °C. Plasma was collected as the supernatant and processed further based on the above procedures to obtain exosomes.
Exosome binding, SERS detection, and fluorescence imaging
15 μL of 6.25×107/mL exosomes was added to the antibody-functionalized Au array wells and incubated for 30 min at RT. After washing the wells three times with PBS, 15 μL of 1 nM SERS AuNRs was added and incubated for 30 min. After washing three times with PBS, 15 μL of PBS was added and exosomes in the wells were detected with a TSI ProRaman spectrometer (λ = 785 nm). The laser beam size at focus was 200 μm. Each spectrum was collected with a laser power of 50 mW and acquisition time of 1 s. Baseline correction using multi-segment polynomial fitting was automatically performed by the signal acquisition software (EZRaman Reader v8.1.8) to subtract SERS background (broad continuum emission). The peak at 1497 cm-1, which is the strongest among all the peaks of the QSY21 SERS spectrum, was used as the representative peak for analysis. To account for variations from instrumentation response and batch-to-batch nanotag preparation, the spectrum of the SERS nanotag solution (0.1 nM) during each experiment was collected and the intensity of the 1497 cm-1 peak was normalized to 2500 a.u., the typical value of a 0.1 nM nanotag solution. This gave a correction factor for each nanotag to correct the signal intensity from exosomes labeled with that nanotag during each experiment. The corrected intensity of the 1497 cm-1 peak was used for analysis. The whole process from exosome binding to signal readout took ~2 h. To confirm the captured exosomes, exosomes were labeled with 1 mM 3,3'-Dioctadecyloxacarbocyanine perchlorate (DiO) in PBS for 15 min at RT. Exosomes were then washed with PBS and examined by a fluorescence microscope (Olympus IX 71) with a Prior Lumen 200 illumination system. The excitation was 482/35 nm and emission was 536/40 nm.
Enzyme-linked immunosorbent assay (ELISA)
50 μL of 6.25×108/mL exosomes were added into a 96-well polystyrene plate (Corning Incorporated) and incubated at 4 °C overnight. The wells were washed three times with Dulbecco's phosphate-buffered saline (DPBS) followed by incubation with 100 μL of blocking solution (DPBS with 4% BSA) at RT for 2 h. After washing three times with DPBS, each well was treated with the following solutions sequentially: 50 μL of 2 μg/mL target-specific antibodies (2 h, RT), 50 μL of horseradish peroxidase (HRP)-conjugated anti-mouse IgG antibody (ThermoFisher, 1:60 dilution in blocking solution; 1 h, RT), and 100 μL of 3,3,5,5-tetramethylbenzidine solution (TMB, Sigma- Aldrich; 30 min, RT). The wells were washed three times with DBPS between steps. After the TMB incubation, 100 μL of 2 M sulfuric acid (H2SO4) was added to stop the reaction. The optical density of each well was measured at 450 nm using a BioTEK ELx800 absorbance microplate reader. Isotype IgG was used as the control.
Statistical analysis
Statistical analysis was performed to compare the expression levels of target proteins across different cell lines using analysis of variance (ANOVA) with post hoc Scheffe method [93]. A p-value ≤ 0.01 was considered significantly different. The mean difference between different groups was considered to be significant if the absolute value was greater than the minimum significant difference derived from the Scheffe method. The marker difference between breast cancer patients and healthy donors was evaluated from generalized estimation equations (geepack v1.2-1 in R) to account for the measurement correlation within each individual. The diagnostic value of identified markers in breast cancer patients was evaluated by receiver operation characteristic (ROC) curve analysis using R packages.
Results and Discussion
Figure 1A shows the schematic design of the Raman exosome assay. It contains three major steps: (1) preparation of the antibody array, (2) labeling of the captured exosomes with SERS AuNRs, (3) detection of exosomes with a portable Raman spectrometer. The antibody array was fabricated on a Au-coated standard glass microscopy slide (75 × 25 × 1 mm) with the assistance of a 3-D printed array template. Exosomes were captured on the Au slide via the target-specific antibodies. To detect the captured exosomes, we made use of the surface properties of exosomes and AuNRs. Exosomes are negatively charged (zeta potential around -10 mV) because of their lipid membrane. AuNRs are positively charged (zeta potential around +35 mV) because of the bilayer CTAB capping agent. The Raman reporter was incorporated in the CTAB bilayer via hydrophobic interactions to give SERS signals for detection. Thus, we hypothesized that captured exosomes via surface proteins could be detected with AuNRs via SERS through electrostatic interactions between the AuNRs and exosomes (Figure 1B-C).
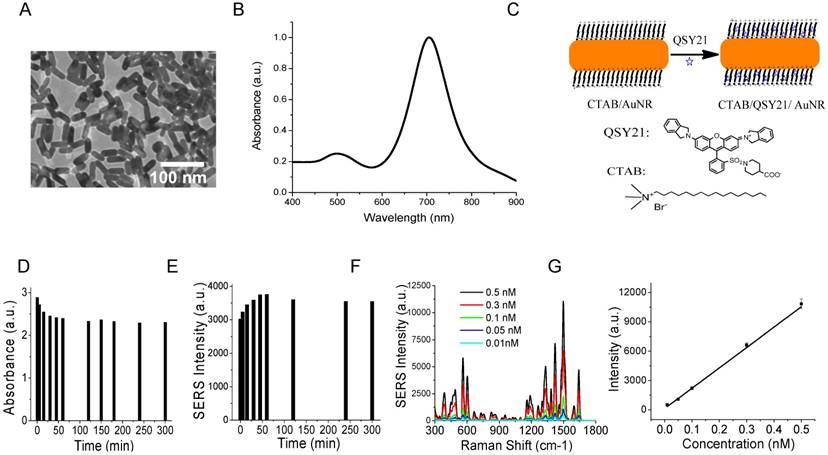
AuNRs were synthesized using the well-established seed-mediated growth method [91, 92]. To ensure efficient binding to the small exosomes, we synthesized small AuNRs by controlling the growth time to ten minutes after seed injection. The AuNRs were 35 nm in length and 12 nm in width on average, with localized surface plasmon resonance (LSPR) at 720 nm (Figure 2A-B). It has been reported that AuNRs with such LSPR properties give stronger SERS activities than those with LSPR at shorter or longer wavelengths due to the competitive effect of SERS enhancement and extinction [94]. We took advantage of the unique surface chemistry of AuNRs for the preparation of SERS AuNRs. The as-prepared Au NRs were stabilized with positively charged CTAB in a bilayer structure [95]. This bilayer of CTAB provides a hydrophobic pocket for loading hydrophobic molecules such as organic dyes via hydrophobic interactions (Figure 2C). Organic dye QSY21 was used as the Raman reporter because it is non-fluorescent and gives fingerprinting signals [86, 96]. To load hydrophobic QSY21 onto aqueous AuNRs, we used the amphiphilic form QSY21 carboxylic acid. The QSY21 carboxylic acid was formed by hydrolyzing QSY21 carboxylic acid-succinimidyl ester in water. QSY21-coated AuNRs were formed by mixing QSY21 carboxylic acid with AuNRs (5000:1) in water with constant mixing for 15 min. Free reporters were separated by centrifugation.
We investigated the stability of the SERS AuNRs in PBS by monitoring their absorption and SERS spectra with time after preparation (Figure S1A-B and Figure 2D-E). Within the 5 h study time, we found that the absorption intensity of the SERS AuNRs gradually decreased by 20% and SERS signal intensity increased by 18% within the first 2 h. Then, the signals did not change within the next 3 h. These results show that the SERS AuNRs are slightly aggregated within 2 h after preparation but then they are stable for hours. For comparison, we investigated the stability of the AuNRs in PBS without QSY21. The results showed that the absorption intensity of the AuNRs decreased by 7% within 2 h and then was constant within the next few hours (Figure S1C). The mechanism for this phenomenon remains to be explored. But, it is not surprising that the SERS AuNRs are stable as CTAB is a known strong capping agent. Based on the stability studies, we thus let the SERS AuNRs age for 2 h before use. The labeling time was only 30 min and the Raman measurement was performed right after labeling. Thus, during our sample processing and signal measurement, the SERS AuNRs were stable and signals from exosomes were reliable. The SERS signals of QSY21-coated AuNRs showed excellent linearity respective to the concentration of the AuNRs, with a correlation coefficient of 0.98 (Figure 2F-G). Thus, the QSY21-coated AuNRs can be used for reliable and quantitative detection and profiling.
Design of the Raman exosome assay. (A) Schematic overview of the Raman exosome assay. A protein array was fabricated on a gold chip (75 × 25 × 1 mm) using a 3-D-printed template array. Exosomes were recognized and immobilized on the Au chip via the target-specific antibodies anchored on the surface of the chip. Immobilized exosomes were detected by surface enhanced Raman scattering gold nanorods that bind to exosomes through electrostatic interactions between cetyltrimethylammonium bromide on gold nanorods and lipid membrane on exosomes. (B) Side view of the interactions of exosome lipid membrane and SERS AuNR. (C) Top view of the interactions of exosome lipid membrane and SERS AuNR.
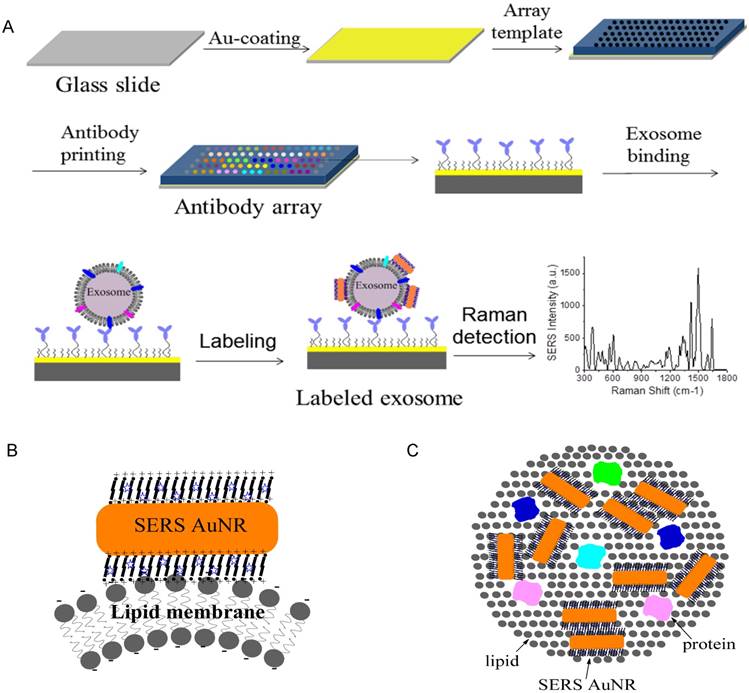
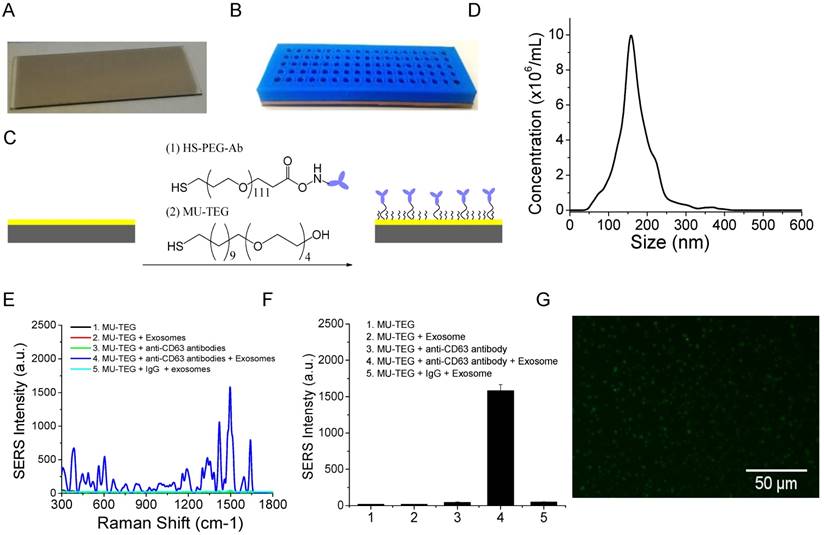
The microscopy glass slide was coated with a 10 nm Au film to facilitate surface chemical modification via high vacuum thin film deposition with an AJA deposition unit. The Au film is optically transparent and thus allows for optical imaging. A photographic picture of the Au-coated glass slide is shown in Figure 3A. To increase sample throughput, we separated the Au slide into an array of wells using a 5 mm-thick 3-D-printed plastic array template (Figure 3B). Suitable arrays should ensure (1) no leaking of the wells and (2) clean manual washing of the samples in the wells. We printed and tested a number of arrays with variable well sizes and inter-well distances. We found that the smallest size of the well was 2 mm in diameter and the smallest distance between neighboring wells was 2 mm. Accordingly, a 17 × 5 array can be made per template. This template provides 85 test sites per slide. Each well can hold a maximum of 15 μL solution. The template was attached on top of the Au chip with a rubber interface array that allowed tight sealing of the plastic array template to the Au chip so that the solution in each well did not leak out. The Au surface of the wells was functionalized with target-specific antibodies to capture exosomes (Figure 3C). This was done by incubation with target-specific HS-PEG-Ab followed by saturation with hydrophilic MU-TEG. HS-PEG-Ab was prepared by reacting HS-PEG-NHS (MW 5000) with antibodies at 4 °C overnight followed by purification with a 10 kD Nanosep filter. The shorter MU-TEG was used to minimize nonspecific interactions of exosomes and SERS AuNRs with the Au slide.
Characterizations of AuNRs and SERS AuNRs. (A) TEM image of AuNRs. (B) Absorption spectrum of AuNRs. (C) Schematic of the preparation of SERS AuNRs using QSY21 as the Raman reporter. (D, E) Stability of CTAB/QSY21/AuNRs in PBS at different time after preparation, which was monitored by absorption (D) and Raman (E) measurements. Absorption in (D) was measured at localized surface plasma resonance of AuNRs. SERS signal intensity in (E) was measured at the 1497 cm-1 peak. (F) SERS signal intensity of QSY21-coated AuNRs at different particle concentrations. (G) The SERS signal intensity of QSY21-coated AuNRs at different AuNR concentrations. Data in (G) were presented as mean values from three replicated experiments with standard deviation.
To examine the specificity of the chemically modified Au slide and the SERS AuNRs for exosome capture and detection, we isolated and purified exosomes from the MM231 model breast cancer cell line. Exosomes were isolated from conditioned culture supernatant using the standard differential method. In this isolation method, cell debris was separated by low-speed centrifugation (430 × g) and microvesicles by medium speed centrifugation (16,5000 × g). Exosome pellet was collected after ultracentrifugation at 100,000 ×g. The exosomes were characterized by NTA to determine their concentration and size distribution. The MM231 exosomes have sizes of 168 ± 49 nm (mean ± standard deviation (SD)) (Figure 3D). Figure 3E shows SERS spectra of CD63-targeted exosomes compared with several controls. The isotype IgG control gave a signal intensity of 48 a.u. at 1497 cm-1. When anti-CD63 antibodies were used, the signal intensity increased to 1582 a.u., which was 33 times stronger than that of IgG control. When exosomes, antibodies, or both exosomes and antibodies were absent, the signals were 44, 17, and 19 a.u., respectively (Figure 3F). These studies demonstrated that the SERS AuNRs and the antibody capture Au slide specifically captured exosomes with targeted surface proteins and detected them without significant nonspecific interference. The captured exosomes with CD63 antibodies were further confirmed using fluorescence imaging with DiO as the membrane labeling agent (Figure 3G). The fluorescence image also shows that exosomes were distributed evenly on the Au surface. In each experiment, SERS spectra from different locations in the Au array wells were collected and the averaged spectrum was used for analysis to account for variation in exosome density at different locations in the well (Figure S2).
Characterizations of the Au array device for target-specific exosome capture and detection. (A) A photograph of the Au-coated microscopy glass slide. (B) A photograph of the Au array device formed using a 3-D-printed template array and the Au-coated microscopy glass slide. The Au array was 17 × 5, with a well size of 2 mm and a gap of 2 mm. (C) Schematic of the chemical surface modification of the Au surface with target-specific antibodies. (D) Size distribution of MM231 exosomes characterized with NTA. (E, F) Evaluation of the specificity of the Raman exosome assay with (E) showing the average SERS spectra (n=3) and (F) showing the mean signal intensity of the 1497 cm-1 peak from different experiments. SERS AuNRs were added in all experiments from 1 to 5. (G) Fluorescence image of MM231 exosomes captured with CD63 antibodies.
To examine the sensitivity of the assay, we did a titration experiment with a series of dilutions using CD63 as the capture antibody. Figure 4A shows the averaged SERS spectra (n=3) from exosomes with different concentrations. The dose (exosome concentration) - response (averaged SERS intensity of the 1497 cm-1 peak with SD from the triplicate experiments) curve based on data in Figure 4A is shown in Figure 4B. The studies showed that the limit of detection (LOD) was 2×106/mL (3.3 fM). The concentration of exosomes in human plasma is >109/mL [97]. Thus, our assay can detect exosomes at a concentration 500 times lower than a typical concentration of exosomes in plasma. This sensitivity was achieved using a 200 μm Raman probe (λ = 785 nm) with low laser power (50 mW). The acquisition time for each spectrum was only 1 s. The as-acquired spectrum was baseline-corrected by the EZRaman Reader V8.1.8 MV signal acquisition software to separate broad emission SERS background. Based on the exosome volume (15 μL), concentration (2×106/mL), the size of the well (2 mm), and the size of the laser spot (200 μm), we calculated that the LOD on the Au slide was 300 exosomes. The SERS signal intensity was linearly proportional to the concentration of exosomes at a range of 106 to 108 exosome/mL, with a correlation coefficient R2 = 0.97 (Figure 4B inset). The working concentration we used was 6.25 × 107 /mL with a working volume of 15 μL. This sample consumption is comparable to that of the most recent high-sensitivity exosome detection technique based on plasmon resonance light scattering properties of gold nanoparticles [62].
To examine the feasibility of the Raman assay for exosome molecular profiling, we analyzed 8 surface proteins from three different categories: epithelial marker EpCAM, breast cancer markers CD44, HER2, EGFR, IGFR, and exosome markers CD81, CD63, CD9 on the model MM231 exosomes. Figure 4C shows the averaged SERS spectrum for each target protein (n=3) and Figure 4D shows the expression profile of all 8 proteins on MM231 exosomes using the data in Figure 4C. Isotype IgG was used as the negative control to examine the nonspecific interactions of the antibodies. The results show that MM231 exosomes have high expression of CD44 and the three exosome markers CD81, CD63, and CD9. They have very low expression of EpCAM and the other three breast cancer markers HER2, EGFR, and IGF1R. These results are consistent with a literature report using a SPR method [59]. MM231 cells are known to overexpress (3+) CD44 with low expression (0-1+) of HER2, EGFR, and EpCAM [98-103], which was further confirmed by flow cytometry analysis (Figure 5). This suggests that exosomes reflect the surface marker expressions on their parental cells and thus can be used as a resource for cancer biomarkers. Our SERS method was further validated using the gold standard ELISA. ELISA was carried out using the indirect approach, in which exosomes were adsorbed onto 96-well plates and then labeled with antibodies targeting each protein. The antibodies were recognized with HRP-conjugated secondary IgG antibody and then detected with the chromogenic substrate TMB. Figure 4E shows the protein profile on MM231 exosomes using ELISA. Similar to the results with the SERS method, the exosomes have high expression of CD44, CD81, CD63, and CD9 and low expression of EpCAM, HER2, EGFR, and IGF1R. A quantitative comparison shows that our Raman assay has a high correlation to ELISA, with a correlation coefficient R2 of 0.97 (Figure 4F). Compared to ELISA, our Raman assay is simpler and faster. With the functionalized Au chips, the sample preparation only requires exosome capture and labeling, which takes about 1.5 h. However, ELISA involves a cumbersome procedure including exosome adsorption, plate blocking, primary antibody binding, secondary antibody binding, and the enzyme-catalyzed signal production, with a total sample processing time > 24 h. In addition, this assay provides point-of-care use because of the portable nature of the Au chip and Raman spectrometer.
Evaluations of the capabilities of the Raman exosome assay for detection and protein profiling of exosomes. (A) Average SERS spectra (n=3) of exosomes at different concentrations captured with CD63 antibodies. (B) Dose (exosome concentration) - response (mean SERS signal intensity of the 1497 cm-1 peak) curve. Linear range is shown in the inset. (C) Average SERS spectra (n=3) from exosomes using different capture antibodies. IgG was used as the control. (D) Expression profile of target proteins on exosomes determined with the Raman exosome assay. Data are presented as the mean intensity of the 1497 cm-1 peak with standard deviation (n=3). (E) Expression profile of target proteins on exosomes determined with ELISA. Data are presented as the mean absorbance (λ = 450 nm) with standard deviation (n=3). (F) Correlation of the Raman exosome assay with ELISA. Exosomes were derived from MM231 cells.
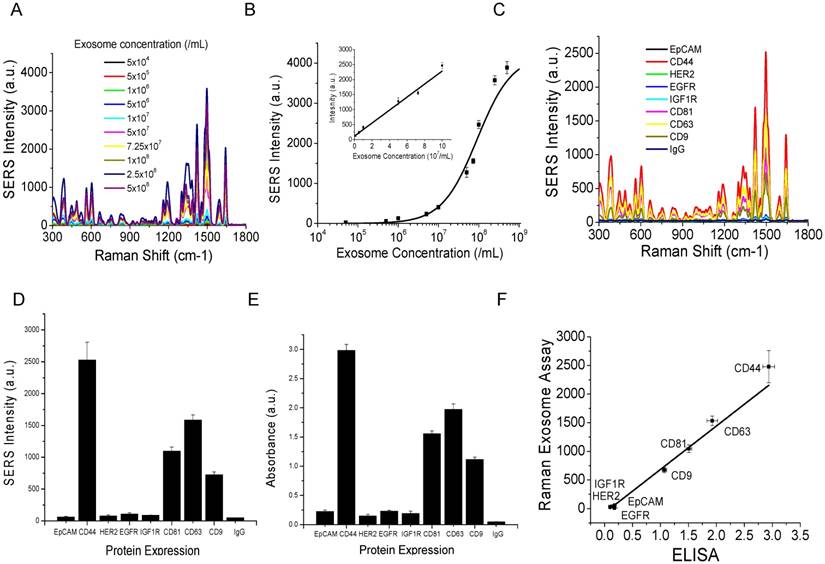
Flow cytometry analyses of the expression of surface protein markers on cancer (MM231, MM468, and SKBR3) and normal (MCF12A) cells. Cells were labeled with PE-conjugated antibodies. PE-conjugated IgG was used as the control.
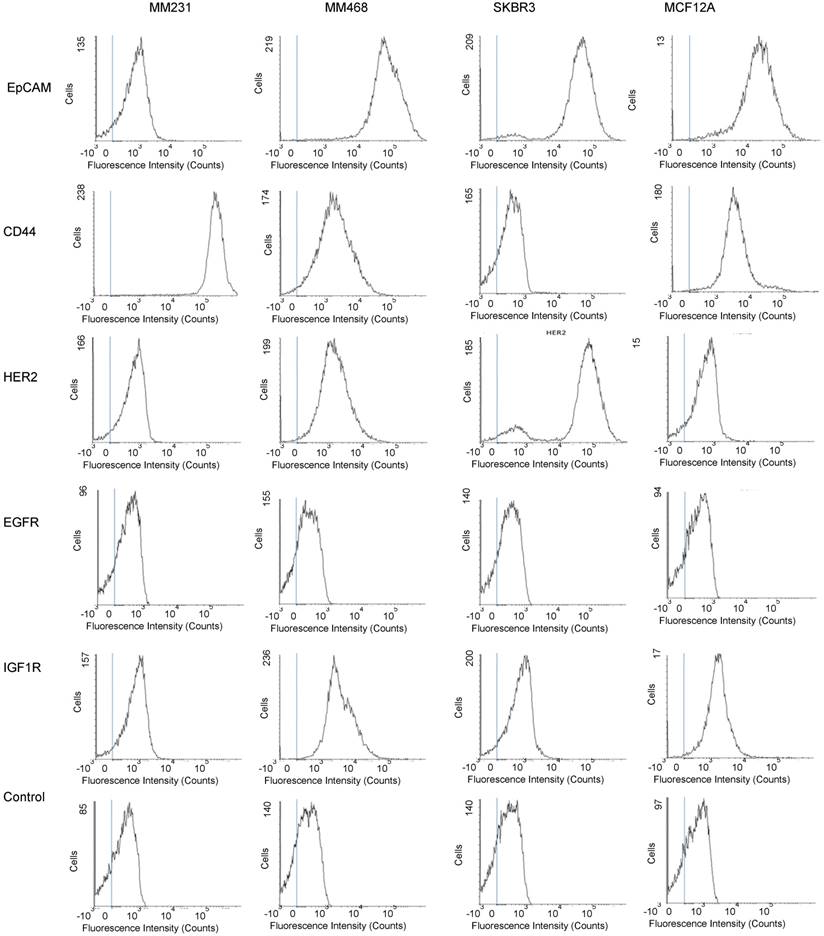
To evaluate whether our method can differentiate exosomes from different cancer cell lines and differentiate cancer cells from normal cells, we profiled exosomes from two additional breast cancer cell lines, MM468 and SKBR3, and one normal (immortalized) breast cell line, MCF12A. Flow cytometry measurements revealed that the surface markers of MM468 cells are EpCAM (strong), HER2 (moderate), and IGFR (moderate) (Figure 5). The surface markers of SKBR3 cells are EpCAM (strong) and HER2 (strong). Both cell lines have different marker expression patterns from the MM231 cancer cells. The MCF12A normal cells are EpCAM positive, but their level is much lower compared to that of MM468 and SKBR3 cells. The size of exosomes derived from MM468, SKBR3, and MCF12A was 170 ± 54, 165 ± 38, 156 ± 32 nm, respectively (Figure S3). Figure 6 shows a comparison of the protein profiles on exosomes from these cancer and normal cells. The exosomes exhibited significantly (p < 0.01) different surface marker profiles across different cell lines. Exosomes from MM468 cells have high expression of EpCAM and moderate expression of HER2 and IGF1R. Exosomes from SKBR3 cells have high expression of EpCAM and HER2. As described above, exosomes from MM231 cells have high expression of CD44 and negligible EpCAM. Exosomes from the MCF12A normal cells have moderate EpCAM expression. Compared to the protein expression on the surface of these cells from flow cytometry analysis, it is clear that exosomes reflect their originating cells' surface protein marker expressions, irrespective of cell lines. Thus, exosomes can be used to identify signature markers for cancer detection by surface protein profiling. All cell lines are positive for the three exosome markers CD81, CD63, and CD9.
Protein profiling of exosomes derived from breast cancer cells (MM231, MM468, and SKBR3) and normal cells (MCF12A). (A-D) Average SERS spectra (n=3) of exosomes targeting different surface proteins. IgG was used as the control. (E) Comparison of protein expressions on cancer and normal cells. Data are presented as the mean intensity of the 1497 cm-1 peak with standard deviation (n=3). (F) Colorimetric comparison of protein expressions on cancer and normal cells based on data in (E). The p-values among the four cell lines for EpCAM, CD44, HER2, EGFR, IGF1R, CD81, CD63, and CD9 are 1.1×10-6, 5.5×10-8, 1.3×10-14, 4.7×10-3, 1.6×10-6, 1.5×10-8, 2.5×10-7, and 1.7×10-8, respectively.
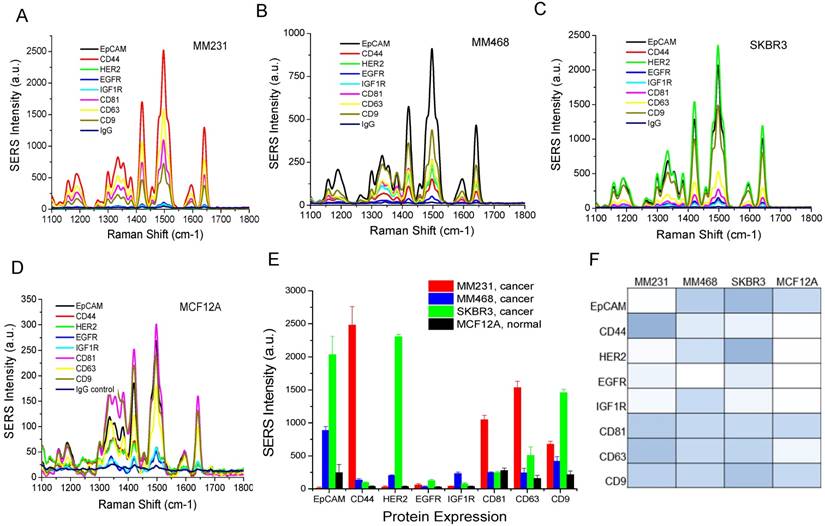
To evaluate the clinical potential, we used the Raman assay to analyze exosomes in breast cancer patients. Due to the heterogeneous breast cancer types, we chose HER2-positive patients (n=10) for a proof-of-concept study. The disease includes invasive lobular carcinoma, infiltrating ductal carcinoma, and adenocarcinoma of the breast in stages I, II, and III. We obtained patient plasma samples from the XpressBank at Asterand Bioscience. Although research has shown that liquid nitrogen-frozen exosomes are stable for over 5 years [104], we used samples collected within the last three years to ensure high quality. To collect plasma samples from healthy donors (n=5), we obtained fresh whole blood and extracted exosomes by differential centrifugation. Using NTA, we determined that exosomes in plasma samples had mean sizes from 120 nm to 170 nm, with concentrations from 1.0×109/mL to 3×1010/mL (Figure 7 and Table 1). There were no significant differences in the sizes between cancer patients and healthy donors. The concentration was heterogeneous in patients and healthy donors. Exosome concentrations in patient 2 and patient 5 were much higher than those in the other patients. Exosome concentrations in healthy donors were higher than those in most patients. The variations in concentration cannot be exclusively ascribed to the age of the samples since all the healthy samples were freshly collected at the same time and newer than all the patient samples. Thus, the concentration depends on the individual, irrespective of disease state.
Size distributions of exosomes in human plasma samples from breast cancer patients and healthy donors characterized with NTA.
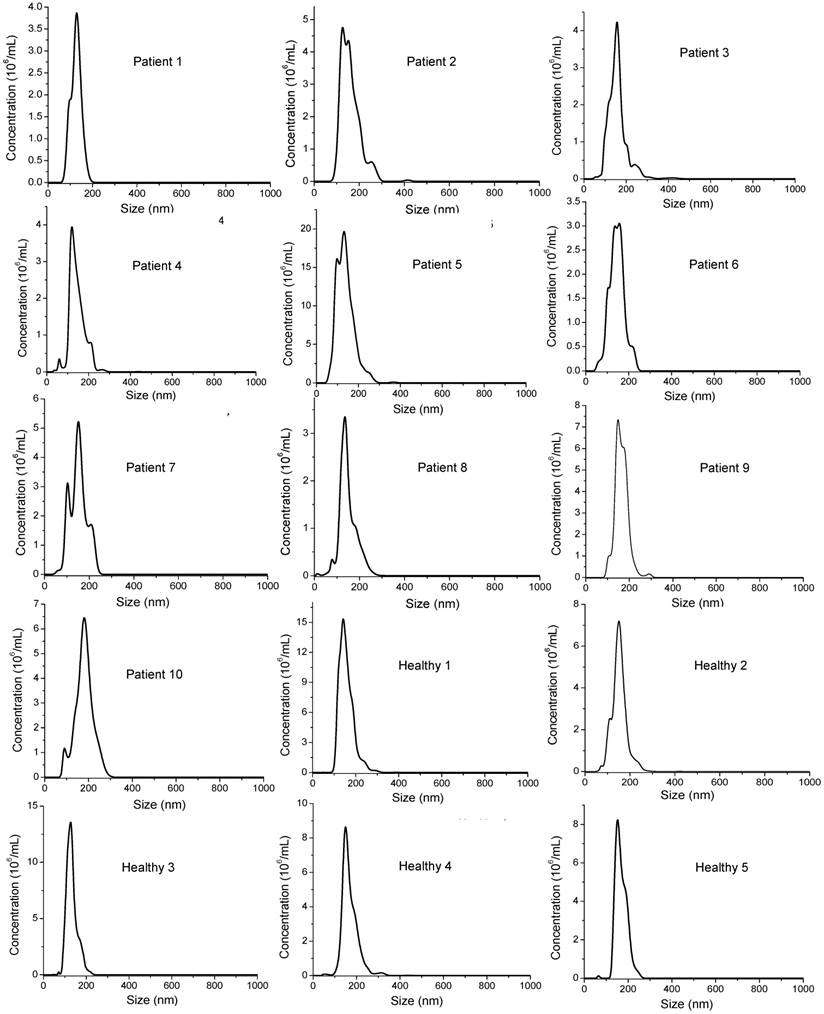
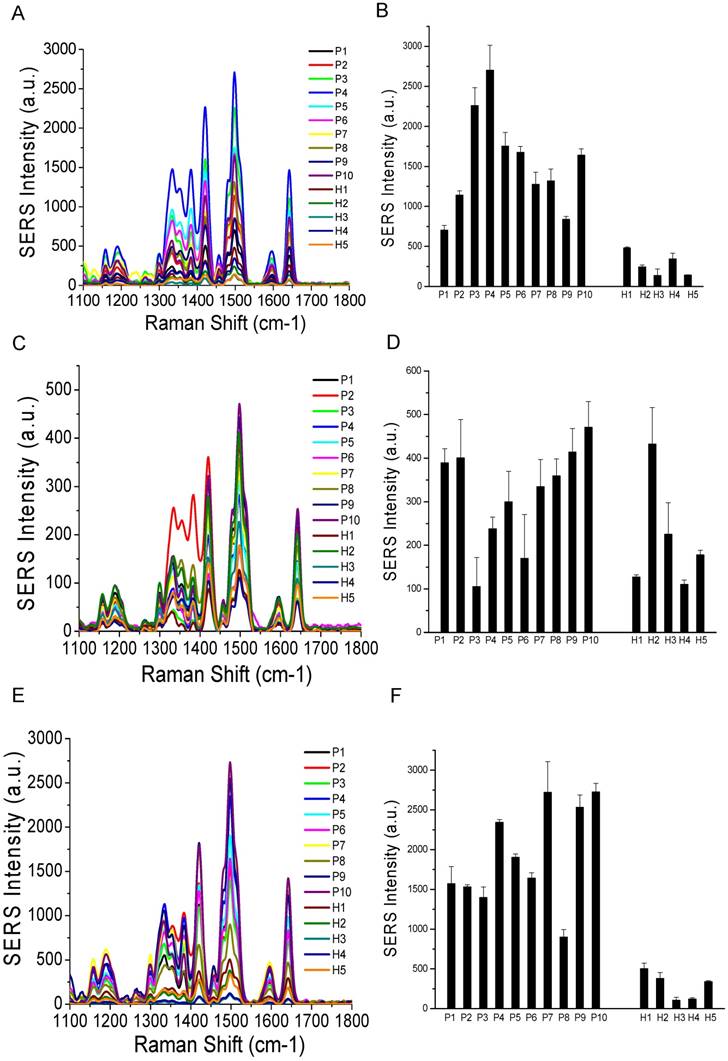
Comparisons of surface marker expressions of EpCAM (A, B), CD44 (C, D), and HER2 (E, F) between cancer patients and healthy donors. (A, C, and E) Average SERS spectra (n=3) from each subject. (B, D, and F) Protein expression profiles based on the data in the SERS spectra. Data are presented as the mean intensity of the 1497 cm-1 peak with standard deviation. The p-value between cancer patients and healthy donors for EpCAM, CD44, and HER2 was 7.4×10-11, 0.097, and < 2.2×10-16, respectively.
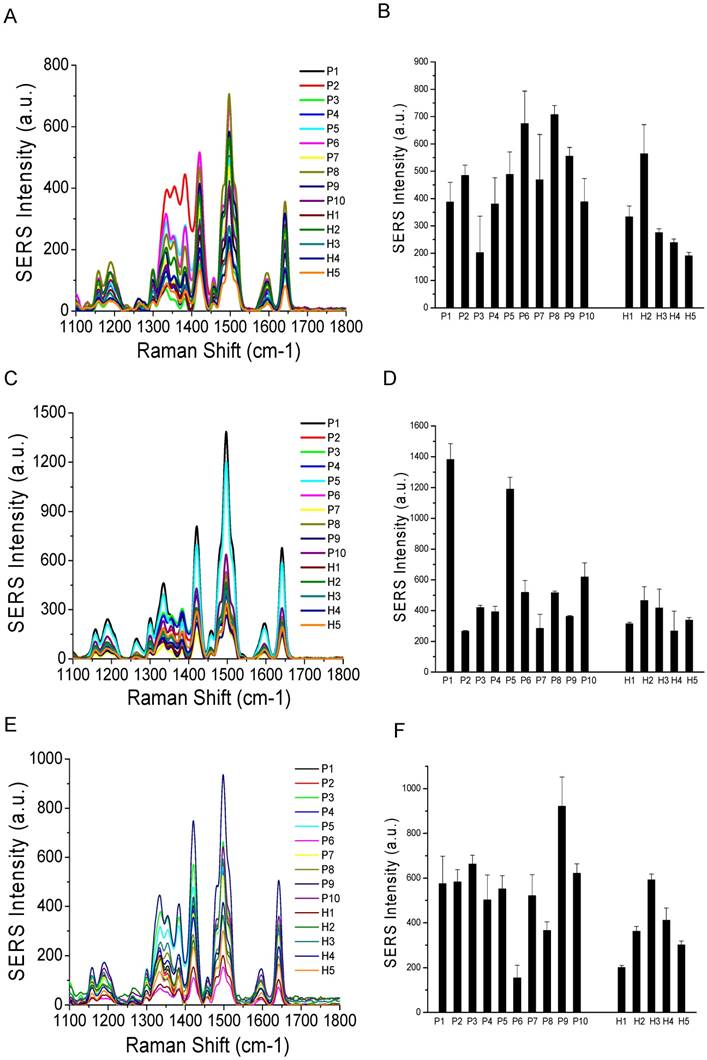
Comparisons of surface marker expressions of CD81 (A, B), CD63 (C, D), and CD9 (E, F) between cancer patients and healthy donors. (A, C, and E) Average SERS spectra (n=3) from each subject. (B, D, and F) Protein expression profiles based on the data in the SERS spectra. Data are presented as the mean intensity of the 1497 cm-1 peak with standard deviation. The p-value between cancer patients and healthy donors for CD81, CD63, and CD9 was 0.037, 0.049, and 0.038, respectively.
Characterization of the size and concentration of exosomes in human plasma samples.
| Sample ID | Clinical diagnosis | Stage | Size (nm) | Concentration (exosomes/mL) |
|---|---|---|---|---|
| Patient 1 | Invasive lobular carcinoma | III | 129 ± 29 | (4.2 ± 0.5) × 109 |
| Patient 2 | Infiltrating ductal carcinoma | III | 161 ± 43 | (1.6 ± 0.1) × 1010 |
| Patient 3 | Adenocarcinoma | III | 158 ± 44 | (2.0 ± 0.3) × 109 |
| Patient 4 | Invasive lobular carcinoma | III | 145 ± 38 | (1.9 ± 0.2) × 109 |
| Patient 5 | Invasive lobular carcinoma | III | 137 ± 42 | (1.3 ± 0.1) × 1010 |
| Patient 6 | Invasive lobular carcinoma | III | 144 ± 33 | (2.2 ± 0.2) × 109 |
| Patient 7 | Infiltrating duct carcinoma | I | 151 ± 38 | (1.4 ± 0.1) × 109 |
| Patient 8 | Infiltrating ductal carcinoma | I | 148 ± 37 | (1.1 ± 0.1) × 109 |
| Patient 9 | Adenocarcinoma | II | 168 ± 36 | (2.1 ± 0.2) × 109 |
| Patient 10 | Infiltrating ductal carcinoma | II | 154 ± 35 | (1.2 ± 0.1) × 109 |
| Healthy 1 | - | - | 157 ± 34 | (9.0 ± 0.3) × 109 |
| Healthy 2 | - | - | 152 ± 33 | (7.5 ± 0.6) × 109 |
| Healthy 3 | - | - | 133 ± 25 | (1.8 ± 0.2) × 109 |
| Healthy 4 | - | - | 169 ± 37 | (8.8 ± 0.5) × 109 |
| Healthy 5 | - | - | 161 ± 32 | (7.6 ± 0.2) × 109 |
Based on our in vitro studies, EpCAM, CD44, and HER2 are biomarkers to distinguish breast cancer exosomes from normal cell-derived exosomes. Thus, we chose these three markers to analyze exosomes from the plasma samples (Figure 8), together with the three exosome markers CD81, CD63, and CD9 (Figure 9). The levels of EpCAM and HER2 were significantly higher in the tested breast cancer patient samples than in the control groups (p < 0.01 for both markers). The levels of CD44 (p = 0.097), CD81 (p = 0.037), CD63 (p = 0.049), and CD9 (p = 0.038) were not significantly different in the patient samples than in the controls. Our finding of HER2 marker (AUC = 1 from ROC curve, Figure S4) on exosomes in the HER2-positive breast cancer patient is consistent with previous studies with a SPR method [60]. In addition, we identified EpCAM as another biomarker to differentiate exosomes from breast cancer patients from normal controls (AUC = 1, Figure S4). EpCAM has been previously identified as an exosome-based biomarker for ovarian cancer in ascites samples [47]. Here we report EpCAM as an exosome-based biomarker for breast cancer. The early promise of these proteins for breast cancer diagnosis, however, requires further validation with larger cohorts.
Conclusion
In conclusion, we have developed a simple, rapid, inexpensive, highly sensitive, and highly specific Raman-based assay for point-of-care detection and molecular profiling of exosomes. Using the assay and model exosomes from breast cancer cells, we showed that exosomes reflect their donor cancer cells' surface biomarker expressions, suggesting the potential of exosomes as biomarkers for cancer detection and investigation. Our assay can be used to differentiate different subtypes of cancer cells and differentiate cancer cells from normal cells. Using the assay, we have identified HER2 and EpCAM biomarkers on exosomes for diagnosis of HER2-positive breast cancer patients. The assay can be widely used for basic and clinical cancer research. Using the classic high speed and high throughput micro drop printing technology, the assay can be readily translated into a microarray platform, offering throughput close to 1,000 test sites on a single Au chip. In addition, the microdrop printing technology can automate the sample processing steps, dramatically enhancing the efficiencies of exosome molecular analysis. This next-generation Raman exosome assay has the potential to revolutionize exosome research and realize a novel cancer liquid biopsy approach for cancer research and early detection. We would like to point out that our method is a detection method that requires initial exosome isolation. Future development should also consider methodology improvement that allows for direct detection of exosomes in plasma.
Abbreviations
SERS: surface-enhanced Raman scattering; gold nanorods: AuNRs; 3-D: 3-dimension; FBS: fetal bovine serum; HAuCl4: chloroauric acid; CTAB: cetyltrimethylammonium bromide; NaBH4: sodium borohydride; AgNO3: silver nitrate; PBS: phosphate buffer solution; RT: room temperature; HS-PEG-Ab: antibody-linked polyethylene glycol thiols; HS-PEG-NHS: thiol- polyethylene glycol- N-hydroxysuccinimide esters; MU-PEG: 11-mercaptoundecyl tetra(ethylene glycol); PBST: phosphate buffer solution-tween; MM231: MDA-MB-231; MM468: MDA-MB-468; DMEM: Dulbecco's Modified Eagle Medium; DMEM/F-12: Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12; EGF: epidermal growth factor; NTA: nanoparticle tracking analysis; HER2: human epidermal growth factor receptor 2; EpCAM: epithelial cell adhesion molecule; EGFR: epidermal growth factor receptor; IGF1R: Insulin-like growth factor 1 receptor; IgG: immunoglobulin; DIN: donor identification number; LN: liquid nitrogen; PVDF: polyvinylidene difluoride; DiO: 3,3'-Dioctadecyloxacarbocyanine perchlorate; ELISA: Enzyme-linked immunosorbent assay; DPBS: Dulbecco's phosphate-buffered saline; TMB: 3,3,5,5-tetramethylbenzidine solution; H2SO4: sulfuric acid; HRP: horseradish peroxidase; ANOVA: analysis of variance; ROC: receiver operation characteristic; LSPR: localized surface plasmon resonance; SD: standard.
Supplementary Material
Supplementary figures.
Acknowledgements
We gratefully acknowledge the support from the Research Investment Fund from the University of Memphis and Institute of Technology Development Grant from The University of Memphis Foundation.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Properzi F, Logozzi M, Fais S. Exosomes: the future of biomarkers in medicine. Biomarkers Med. 2013;7:769-78
2. Trams EG, Lauter CJ, Salem Jr N, Heine U. Exfoliation of membrane ecto-enzymes in the form of microvesicles. BBA Biomemb. 1981;645:63-70
3. Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967-77
4. Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and of the transferrin receptor in rat reticulocytes recycling. J Cell Biol. 1983;97:329-39
5. Harding CV, Heuser JE, Stahl PD. Exosomes: Looking back three decades and into the future. J Cell Biol. 2012;200:367-71
6. Denzer K, Kleijmeer MJ, Heijnen HF, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. J Cell Sci. 2000;113:3365-74
7. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373-83
8. Kowal J, Théry TM. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116-25
9. Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569-79
10. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654-659
11. Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581-93
12. Mathivanan SHJ, Simpson RJ. Exosomes: Extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907-20
13. Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255-89
14. Lo Cicero A, Stahl PD, Raposo G. Extracellular vesicles shuffling intercellular messages: for good or for bad. Curr Opin Cell Biol. 2015;35:69-77
15. Zhang HG, Grizzle WE. A novel pathway of local and distant intercellular communication that facilitates the growth and metastasis of neoplastic lesions. Am J Pathol. 2014;184:28-41
16. Shkarina KA, Cherednyk OV, Voloschenko II, Trembach OM, Trembach IO, Khoruzhenko AI. Exosomes: messengers and mediators of tumor-stromal interactions. Biopoly Cell. 2014;30:426-35
17. Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J hematol Oncol. 2015;8:83-7
18. Brinton LT, Sloane HS, Kester M, Kelly KS. Formation and role of exosomes in cancer. Cell Mol Life Sci. 2015;72:659-71
19. Guo W, Gao Y, Li N, Shao F, Wang C, Wang P, Yang Z, Li R, He J. Exosomes: new players in cancer. Oncol Rep. 2017;38:665-75
20. Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2017;126:1208-15
21. Nawaz M, Camussi G, Valadi H, Nazarenko I, Ekström K, Wang X, Principe S, Shah N, Ashraf NM, Fatima F, Neder L, Kislinger T. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Rev Urol. 2014;11:688-701
22. Peinado H, Aleckovic M, Lavotshkin S. et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883-91
23. Zhou W, Fong MY, Min Y. et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501-15
24. Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA. 2004;101:13368-73
25. Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int. Immunol. 2005;17:879-87
26. Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, Hill AF. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol. 2007;211:582-90
27. Ogawa Y, Miura Y, Harazono A, Kanai-Azuma M, Akimoto Y, Kawakami H, Yamaguchi T, Toda T, Endo T, Tsubuki M, Yanoshita R. Proteomic analysis of two types of exosomes in human whole saliva. Biol Pharm Bull. 2011;34:13-23
28. Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6:267-83
29. Vlassov AV, Magdaleno S, Setterquist R, Conrad R. Exosomes: current knowledge of their composition, biological functions, and diagnostic and therapeutic potentials. Biochim Biophys. 2012;1820:940-8
30. Choi DS, Lee J, Go G, Kim YK, Gho YS. Circulating extracellular vesicles in cancer diagnosis and monitoring: an appraisal of clinical potential. Mol Diagn Ther. 2013;17:265-71
31. Revenfeld ALS, Bæk R, Nielsen MH, Stensballe A, Varming K, Jørgensen M. Diagnostic and prognostic potential of extracellular vesicles in peripheral blood. Clin Ther. 2014;36:830-46
32. D'Souza-Schorey C, Di Vizio D. Biology and proteomics of extracellular vesicles: harnessing their clinical potential. Expert Rev Proteomics. 2014;11:251-3
33. Verma M, Lam TK, Hebert E, Divi RL. Extracellular vesicles: potential applications in cancer diagnosis, prognosis, and epidemiology. BMC Clin Pathol. 2015;15:1-9
34. Munson P, Shukla A. Exosomes: potential in cancer diagnosis and therapy. Medicine. 2015;2:310-27
35. Alderton GK. Diagnosis: fishing for exosomes. Nat Rev Cancer. 2015;15:453-5
36. Soung YH, Ford S, Zhang V, Chung J. Exosomes in cancer diagnostics. Cancers. 2017;9:8
37. Thery C, Amigorena S, Raposo G, Clayton A. Curr Protoc Cell Biol. 2006; 22: Chapter 3(Unit 3).
38. Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RM. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293-304
39. Lobb RJ, Belanie Becker M, Wen SW, Wong CSF, Wiegmans AP, Leimgruber A, Mo¨ ller A. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J Extracell Vesicles. 2015;4:27031
40. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics. 2017;7:789-804
41. Yakimchuk K. Exosomes: isolation and characterization methods and specific markers. Mater Methods. 2015;5:1450-3
42. Peterson MF, Otoc N, Sethi JK, Gupta A, Antes TJ. Integrated systems for exosome investigation. Methods. 2015;87:31-45
43. Khatun Z, Bhat A, Sharma S, Sharma A. Elucidating diversity of exosomes: biophysical and molecular characterization methods. Nanomedicine (Lond). 2016;11:2359-67
44. Szatanek R, Baj-Krzyworzeka M, Zimoch J, Lekka M, Siedlar M, Baran J. The methods of choice for extracellular vesicles (EVs) characterization. Int J Mol Sci. 2017;18:1153-7
45. Yoshioka Y, Konishi Y, Kosaka N, Katsuda T, Kato T, Ochiya T. Comparative marker analysis of extracellular vesicles in different human cancer types. J Extracell Vesicles. 2013;2:20424
46. Sunkara V, Woo HK, Cho YK. Emerging techniques in the isolation and characterization of extracellular vesicles and their roles in cancer diagnostics and prognostics. Analyst. 2016;141:371-81
47. Im H, Shao H, II Park Y, Peterson YM, Castro CM, Weissleder R, Lee H. Label-free detection and molecular profiling of exosomes with a nano-l,,asmonic sensor. Nat Biotechnol. 2014;32:490-5
48. Stoner SA, Duggan E, Condello D, Guerrero A, Turk JR, Narayanan PK, Nolan JP. High sensitivity flow cytometry of membrane vesicles. Cytometry A. 2016;89A:196-206
49. Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip. 2014;14:1891-900
50. He M, Crow J, Roth M, Zeng Y, Godwin AK. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip. 2014;14:3773-80
51. He M, Zeng YJ. Microfluidic exosome analysis toward liquid biopsy for cancer. Lab Autom. 2016;21:599-608
52. Zho Z, Yang Y, Zeng Y, He M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip. 2016;16:489-96
53. Marcoux G, Duchez AC, Cloutier N, Provost P, Nigrovic PA, Boilard E. Revealing the diversity of extracellular vesicles using high-dimensional flow cytometry analyses. Sci Rep. 2016;6:35928
54. Inglis H, Norris P, Danesh A. Techniques for the analysis of extracellular vesicles using flow cytometry. J Vis Exp. 2015;97:e52484
55. Morales-Kastresana A, Telford B, Musich TA. et al. Labeling extracellular vesicles for nanoscale flow cytometry. Sci Rep. 2017;7:1878
56. Pospichalova V, Svoboda J, Dave Z. et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J Extracell Vesicles. 2015;4:25530
57. Welsh JA, Holloway JA, Wilinson JS, Englyst NA. Extracellular vesicle flow cytometry analysis and standardization. Front Cell Dev Biol. 2017;5:78
58. Inglis HC, Danesh A, Shah A, Lacroix J, Spinella PC, Norris PJ. Techniques to improve detection and analysis of extracellular vesicles using flow cytometry. Cytometry A. 2015;87A:1052-63
59. Grasso L, Wyss R, Weidenauer L, Thampi A, Demurtas D, Prudent M, Lion N, Vogel H. Molecular screening of cancer-derived exosomes by surface plasmon resonance spectroscopy. Anal Bioanal Chem. 2015;407:5425-32
60. Sina AAI, Vaidyanathan R, Dey S, Carrascosa LG, Shiddiky MJA, Trau M. Real time and label free profiling of clinically relevant exosomes. Sci Rep. 2016;6:30460
61. Thakur A, Qiu G, Ng SP, Guan J, Yue J, Lee Y, Wu CL. Direct detection of two different tumor-derived extracellular vesicles by SAM-AuNIs LSPR biosensor. Biosens Bioelectron. 2017;94:400-7
62. Liang K, Liu F, Fan J, Sun D, Liu C, Lyon CJ, Bernard DW, Li Y, Yokoi K, Katz MH, Koay EJ, Zhao Z, Hu Y. Nanoplasmonic quantification of tumour-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat Biomed Engineer. 2017;1:0021
63. Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18:1835-40
64. Doldán X, Fagúndez P, Cayota A, Laíz J, Tosar JP. Electrochemical sandwich immunosensor for determination of exosomes based on surface marker-mediated signal amplification. Anal Chem. 2016;8:10466-73
65. Zhou YG, Mohamadi RM, Poudineh M, Kermanshah L, Ahmed S, Safaei TS, Stojcic J, Nam RK, Sargent EH, Kelley SO. Interrogating circulating microsomes and exosomes using metal nanoparticles. Small. 2016;12:727-32
66. Jeong S, Park J, Pathania D, Castro CM, Weissleder R, Lee H. Integrated magneto-electrochemical sensor for exosome analysis. ACS Nano. 2016;10:1802-9
67. Belov L, Matic KJ, Hallal S, Best OG, Mulligan SP, Christopherson RI. Extensive surface protein profiles of extracellular vesicles from cancer cells may provide diagnostic signatures from blood samples. J Extracell Vesicles. 2016;5:25355
68. Oliveira-Rodríguez M, Serrano-Pertierra E, García AC, Martín SL, Mo MJ, Cernuda-Morollón E, Blanco-López MC. Point-of-care detectionofextracellularvesicles:Sensitivity optimizationandmultiple-targetdetection. Biosens Bioelectron. 2017;87:38-45
69. Li Q, Tofaris GK, Davis JJ. Concentration-normalized electroanalytical assaying of exosomal markers. Anal Chem. 2017;89:3184-90
70. Etayash H, McGee AR, Kaur K, Thundat T. Nanomechanical sandwich assay for multiple cancer biomarkers in breast cancer cell-derived exosomes. Nanoscale. 2016;8:15137-41
71. Kneipp K, Kneipp H, Itzkan H, Dasari RR, Feld MS. Surface-enhanced Raman scattering and biophysics. J Phys Condens Matter. 2002;14:R597-624
72. Nie S, Emory SR. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102-06
73. Kneipp K, Wang Y, Kneipp H, Perelman LT, Itzkan I, Dasari RR, Feld MS. Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS). Phys Rev Lett. 1997;78:1667-70
74. Tirinato L, Gentile F, Di Mascolo D, Coluccio ML, Das G, Liberale C, Pullano SA, Perozziello G, Francardi M, Accardo A, De Angelis F, Candeloro P, Di Fabrizio E. SERS analysis on exosomes using super-hydrophobic surfaces. Microelectron Eng. 2012;97:337-40
75. Lee C, Carney RP, Hazari S, Smith ZJ, Knudson A, Robertson CS, Lam KS, Wachsmann-Hogiu S. 3D plasmonic nanobowl platform for the study of exosomes in solution. Nanoscale. 2015;7:9290-7
76. Stremersch S, Marro M, Pinchasik B, Baatsen P, Hendrix A, De Smedt SC, Loza-Alvarez P, Skirtach AG, Raemdonck K, Braeckmans K. Identifi cation of individual exosome-like vesicles by surface enhanced Raman spectroscopy. Small. 2016;12:3292-301
77. Park J, Hwang M, Choi BH, Jeong H, Jung J, Kim HK, Hong S, Park J, Choi Y. Exosome classification by pattern analysis of surface-enhanced Raman spectroscopy data for lung cancer diagnosis. Anal Chem. 2017;89:6695-701
78. Wang Y, Yan B, Chen L. SERS tags: novel optical nanoprobes for bioanalysis. Chem Rev. 2013;113:1391-428
79. Doering WE, Piotti ME, Natan MJ, Freeman RG. SERS as a foundation for nanoscale, optically detected biological labels. Adv Mater. 2007;19:3100-8
80. Qian X, Peng XH, Ansari DO, Goen QY, Chen GZ, Shin DM, Yang L, Young AN, Wang MD, Nie S. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83-90
81. Keren S, Zavaleta C, Cheng Z, de la Zerda A, Gheysens O, Gambhir SS. Noninvasive molecular imaging of small living subjects using Raman spectroscopy. Proc Natl Acad Sci USA. 2008;105:5844-9
82. Sha MY, Xu HX, Natan MJ, Cromer R. Surface-enhanced Raman scattering tags for rapid and homogeneous detection of circulating tumor cells in the presence of human whole blood. J Am Chem Soc. 2008;130:17214-5
83. Wang X, Qian XM, Beitler JJ, Chen ZG, Khuri FR, Lewis MM, Shin HJC, Nie SM, Shin DM. Detection of circulating tumor cells in human peripheral blood using surface-enhanced Raman scattering nanoparticles. Cancer Res. 2011;71:1526-32
84. Shi W, Paproski RJ, Moore R, Zemp R. Detection of circulating tumor cells using targeted surface-enhanced Raman scattering nanoparticles and magnetic enrichment. J Biomed Opt. 2014;19:056014
85. Nima ZA, Mahmood M, Xu Y, Mustafa T, Watanabe F, Nedosekin DA, Juratli MA, Fahmi T, Galanzha EI, Nolan JP, Basnakian AG, Zharov VP, Biris AS. Circulating tumor cell identification by functionalized silver-gold nanorods with multicolor, super-enhanced SERS and photothermal resonances. Sci Rep. 2014;4:4752
86. Bhana S, Chaffin E, Wang Y, Mishra SR, Huang X. Capture and detection of cancer cells in whole blood with magnetic-optical nanoovals. Nanomedicine(Lond). 2014;9:593-606
87. Huang X, O'Connor R, Kwizera EA. Gold nanoparticle based platforms for circulating cancer marker detection. Nanotheranostics. 2017;1:80-102
88. Bhana S, Huang X. Nanotechnology for enrichment and detection of circulating tumor cells. Nanomedicine (Lond). 2015;10:1973-90
89. Smolsky J, Kaur S, Hayashi C, Batra SK, Krasnoslobodtsev AV. Surface-enhanced Raman scattering-based immunoassay, technologies for detection of disease biomarkers. Biosensors. 2017;7:7
90. Hao G, Schatz GC. Electromagnetic fields around silver nanoparticles and dimers. J Chem Phys. 2004;120:357-66
91. Murphy CJ, Sau TK, Gole AM, Orendorff CJ, Gao J, Gou L, Hunyadi SE, Li T. Anisotropic metal nanoparticles: synthesis, assembly, and optical applications. J Phys Chem. B. 2005;109:13857-70
92. Nikoobakht B, El-Sayed MA. Preparation and growth mechanism of gold nanorods (NRs) using seed-mediated growth method. Chem Mater. 2003;15:1957-61
93. Bhana S, Liu G, Wang L, Starring H, Mishra SR, Liu G, Huang X. Near infrared-absorbing gold nanopopcorns with iron oxide cluster core for magnetically amplified photothermal and photodynamic cancer therapy. ACS Appl Mater Interfaces. 2015;7:11637-47
94. Sivapalan ST, DeVetter BM, Yang TK, van Dijk T, Schulmerich MV, Carney PS, Bhargava R, Murphy CJ. Off-resonance surface-enhanced Raman spectroscopy from gold nanorod suspensions as a function of aspect ratio: not what we thought. ACS Nano. 2013;7:2099-105
95. Nikoobakht B, El-Sayed MA. Evidence for bilayer assembly of cationic surfactants on the surface of gold nanorods. Langmuir. 2001;17:6368-74
96. Bhana S, Rai BK, Mishra SR, Wang Y, Huang X. Synthesis and properties of near infrared-absorbing magnetic-optical nanopins. Nanoscale. 2012;4:4939-42
97. Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg F, Breakefield XO, Lee H, Weissleder R. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun. 2015;6:6999
98. Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH, Goulet Jr R, Badve S, Nakshatri H. CD44+/CD24-breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res. 2006;8:R59
99. Keller PJ, Lin AF, Arendt LM, Klebba I, Jones AD, Rudnick JA, DiMeo TA, Gilmore H, Jefferson DM, Graham RA, Naber SP, Schnitt S, Kuperwasser C. Mapping the cellular and molecular heterogeneity of normal and malignant breast tissues and cultured cell lines. Breast Cancer Res. 2010;12:R87
100. Gostner JM, Fong D, Wrulich OA, Lehne F, Zitt M, Hermann M, Krobitsch S, Martowicz A, Gastl G, Spizzo G. Effects of EpCAM overexpression on human breast cancer cell lines. BMC Cancer. 2011;11:45
101. Martowicz A, Spizzo G, Gastl G, Untergasser G. Phenotype-dependent effects of EpCAM expression on growth and invasion of human breast cancer cell lines. BMC Cancer. 2012;12:501
102. Subik K, Lee JF, Baxter L, Strzepek T, Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG, Tang P. The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast cancer. 2010;4:35-41
103. Soysal SD, Muenst S, Barbie T, Fleming T, Gao F, Spizzo G, Oertli D, Viehl CT, Obermann EC, Gillanders WE. EpCAM expression varies significantly and is differentially associated with prognosis in the luminal B HER2+, basal-like, and HER2 intrinsic subtypes of breast cancer. British J Cancer. 2013;108:1480-7
104. Chen IH, Xue L, Hsu CC, Paez JSP, Pan L, Andaluz H, Wendt MK, Iliuk AB, Zhu JK, Tao WA. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci USA. 2017;114:3175-80
Author contact
![]() Corresponding author: Email: xhuang4edu; Phone: (901) 678 1728; Fax: (901) 678 3744
Corresponding author: Email: xhuang4edu; Phone: (901) 678 1728; Fax: (901) 678 3744
 Global reach, higher impact
Global reach, higher impact