13.3
Impact Factor
Theranostics 2018; 8(8):2147-2160. doi:10.7150/thno.22726 This issue Cite
Research Paper
Release of MicroRNAs into Body Fluids from Ten Organs of Mice Exposed to Cigarette Smoke
1. Department of Health Sciences, University of Genoa, 16132 Genoa, Italy
2. IRCCS Ospedale Policlinico San Martino, 16132 Genoa, Italy
3. National Center of Oncology, Sofia-1756, Bulgaria
4. Division of Cancer Prevention, National Cancer Institute, Rockville, MD 20850, USA
Received 2017-9-7; Accepted 2018-2-6; Published 2018-3-8
Abstract
Purpose: MicroRNAs are small non-coding RNAs that regulate gene expression, thereby playing a role in a variety of physiological and pathophysiological states. Exposure to cigarette smoke extensively downregulates microRNA expression in pulmonary cells of mice, rats, and humans. Cellular microRNAs are released into body fluids, but a poor parallelism was previously observed between lung microRNAs and circulating microRNAs. The purpose of the present study was to validate the application of this epigenetic biomarker by using less invasive collection procedures.
Experimental design: Using microarray analyses, we measured 1135 microRNAs in 10 organs and 3 body fluids of mice that were either unexposed or exposed to mainstream cigarette smoke for up to 8 weeks. The results obtained with selected miRNAs were validated by qPCR.
Results: The lung was the main target affected by smoke (190 dysregulated miRNAs), followed by skeletal muscle (180), liver (138), blood serum (109), kidney (96), spleen (89), stomach (36), heart (33), bronchoalveolar lavage fluid (32), urine (27), urinary bladder (12), colon (5), and brain (0). Skeletal muscle, kidney, and lung were the most important sources of smoke-altered microRNAs in blood serum, urine, and bronchoalveolar lavage fluid, respectively.
Conclusions: microRNA expression analysis was able to identify target organs after just 8 weeks of exposure to smoke, well before the occurrence of any detectable histopathological alteration. The present translational study validates the use of body fluid microRNAs as biomarkers applicable to human biomonitoring for mechanistic studies, diagnostic purposes, preventive medicine, and therapeutic strategies.
Keywords: microRNAs, cigarette smoke, interorgan distribution, body fluids
Introduction
microRNAs (miRNAs) are a class of endogenous, small (18-25 nucleotides), non-coding, single-stranded RNAs that modulate messenger RNA (mRNA) levels either by translational repression or post-transcriptionally induced mRNA decay. Each miRNA regulates up to several hundred mRNAs simultaneously and affects a number of target transcripts. These molecular biomarkers have been implicated in almost every biological process, including development, cell cycle regulation, cell growth and differentiation, stress response, and apoptosis. As such, miRNAs regulate a variety of physiological processes and play a role in the pathogenesis of cancer and of cardiac, metabolic, neurologic, immune-related, and other diseases [1]. In particular, miRNA profiles are dysregulated in cancers [2] by a variety of mechanisms including amplification, deletion, mutation, and epigenetic silencing [3]. Several genes encoding miRNAs have been characterized as novel proto-oncogenes [4].
The study of cellular miRNAs requires the availability of target tissues, which are easily accessible in preclinical studies but require the use of biopsy material involving invasive collection procedures in humans. In addition, miRNAs have been found in a variety of body fluids, where they are remarkably stable [5-7]. Extracellular miRNAs could serve as molecular biomarkers for diagnostic purposes applicable to both prevention and therapy of human diseases. However, extensive research is necessary for identifying the characteristics of extracellular miRNAs to delineate their roles in cancer pathobiology and cancer prevention [5]. The main problem in pursuing this approach is that extracellular miRNAs are not very specific, being released from a variety of tissues either under physiological conditions or the effect of exogenous agents (diet, drugs, toxicants, protective factors, etc.) or the influence of pathological conditions. Accordingly, there may be a poor correlation between cellular and extracellular miRNAs and between miRNAs detectable in different biological fluids [8, 9].
Therefore, there is need for translational studies aimed at assessing the suitability of extracellular miRNAs as a signature of tissue damage and of dysregulation of cellular miRNA profiles. In addition, application of miRNA monitoring to humans as a diagnostic tool for the prevention of cancer and other diseases implies that no pathological condition is clinically apparent. The goal of the present study was to evaluate miRNA expression in multiple mouse organs, including lungs, liver, heart, kidney, spleen, urinary bladder, skeletal muscle, colon, stomach, and brain, and to establish the correlation with the levels of extracellular miRNAs in body fluids. In humans, the examined fluids can be collected by means either of non-invasive procedures (urine), minimally invasive procedures (blood serum), or semi-invasive procedures (bronchoalveolar lavage fluid or BALF).
Besides being the most important threat to human health, smoking typically targets multiple systems, organs, and tissues. In the oncological domain, according to the International Association for Research on Cancer (IARC), there is evidence for a causal association of smoking with cancers affecting (a) the respiratory system (nasal cavity and paranasal sinuses, nasopharynx, oropharynx and hypopharynx, larynx, and lung), (b) the urinary tract (kidney pelvis, ureter, and bladder), (c) the digestive system (oral cavity, oesophagus, stomach, colon-rectum, liver, and pancreas), (d) the reproductive tract (ovary and uterine cervix), and (e) the hematopoietic system (myeloid leukemia) [10]. In addition, either alone or in synergy with other risk factors, smoking plays important roles in the pathogenesis of other chronic degenerative diseases, including chronic obstructive pulmonary diseases (COPD), such as emphysema and chronic bronchitis, cardiovascular and cerebrovascular diseases, neurodegenerative disorders, and reproductive effects [11]. In the present study, the mice were either untreated or exposed, for varying periods of time, to cigarette smoke (CS), in the form of mainstream CS (MCS). It is noteworthy that the mice were not exposed to selected MCS components but to MCS as a complex mixture of more than 5000 identified chemical compounds [12] in order to reproduce the systemic spread, the multiple mechanisms, and the interplay between its components.
Materials and Methods
Study design
Young mice were either untreated or exposed to MCS for up to 8 weeks. RNA was purified from 10 organs and 3 biological fluids, and the expression of 1135 miRNAs was evaluated by microarrays. For selected miRNAs, the results were validated by qPCR.
Mice
Two-month old Swiss ICR (CD-1) mice were purchased from Harlan Laboratories (San Pietro al Natisone, Udine, Italy). The mice were housed in MakrolonTM cages on sawdust bedding and maintained on standard rodent chow (Teklad 9607, Harlan Laboratories) and tap water ad libitum. The animal room temperature was 23 ± 2°C, with a relative humidity of 55% and a 12 h day-night cycle. Housing and treatments of mice were in accordance with NIH, European (2010/63 UE Directive), and institutional guidelines. The issuance of the NIH Office of Laboratory Animal Welfare (OLAW) with the University of Genoa bears the identification number A5899-01 and is effective until February 28, 2021. The IACUC protocol was approved by the Fox Chase Cancer Center Committee on April 13, 2015.
Experimental groups
The following 8 groups of mice were used: Groups A, B, C, and D, mice kept in filtered air for 2, 4, 6, and 8 weeks, respectively (sham-exposed mice); Groups E, F, G, and H, mice exposed to MCS for 2, 4, 6, and 8 weeks, respectively (MCS-exposed mice). Groups A-C and E-G were composed of 10 females each, whereas Groups D and H were composed of 10 males and 10 females each.
Exposure to MCS
A whole-body exposure of mice to MCS was achieved by using Kentucky 2R4F reference cigarettes (College of Agriculture, The Reference Cigarette Program, University of Kentucky, Lexington, KY), having a declared content of 9.4 mg tar, 0.73 mg nicotine, and delivering 12 mg of CO each when burned. MCS was delivered to the exposure chambers by drawing 15 consecutive puffs, each of 60 mL and lasting 6 s. Each daily session involved 6 consecutive exposures, lasting 10 min each, with 1 min intervals, during which a total air change was made. The average concentration of total particulate matter measured in the exposure chambers was 784 mg/m3.
Collection of organs and body fluids
At the indicated times, the mice were euthanized. Each mouse was transferred into a separate chamber and isolated from the other mice. The 2013 AVMA guidelines on euthanasia were followed using slow introduction of CO2 asphyxiation. Death was confirmed by absence of respiration and/or heartbeat. Immediately after sacrifice, the BAL was performed by intubating the trachea and lavaging with 5 mL of PBS per mouse. The BAL samples were centrifuged and the supernatants (BALF) were pooled within each experimental group. The blood was collected by heart puncture and used for preparing serum. The urine was collected for 8 h and pooled from the mice belonging to each experimental group by using metabolic cages during the day preceding euthanasia of mice. The urine was centrifuged in order to remove the sediment. Ten organs, including lungs, liver, heart, kidney, spleen, urinary bladder, skeletal muscle (left gastrocnemius), colon, stomach, and brain, were collected. Immediately after collection, each organ or fluid was immersed in RNAlater and kept at -80°C.
RNA extraction
RNA was extracted individually from the 10 organs of mice belonging to all experimental groups, whereas the body fluids were pooled within each group, irrespective of gender due to the low amounts of samples available. RNA was extracted from mouse organs (miRNeasy, Qiagen, Hilden, Germany) and body fluids (Exiqon's miRCURY™ RNA Isolation Kit - Biofluids (Exiqon, Vedbaek, Denmark) according to the manufacturers' instructions. The Qubit quantitation assay was used to standardize miRNA quantities to be processed by microarray and qPCR (Qubit™ 3.0 Fluorometer, Life Technologies, Gent, Belgium).
Evaluation of miRNA expression analysis by microarray
For evaluating the expression of miRNAs, we used the 7th generation miRCURY LNATM microRNA Array (Exiqon), which contains 3100 capture probes covering human, mouse, and rat miRNAs. In particular, this microarray analyzes the expression of 1135 mouse miRNAs. The total RNA from each sample was labeled with Label IT® miRNA Labeling Kits, Version 2 (Mirus Bio, WI) following the standard protocol. Total RNA (500 ng) was mixed with 10 µL 10x labeling buffer, 4 µL Label IT reagent (containing Cy 3 or Cy 5 fluorescent tracers), and water to 86 µL. The samples were incubated at 36°C for 1 h and the reaction was stopped by adding 10 µL Stop Reagent. Two differently labeled samples (with Cy3 and Cy5, respectively) were collected together, purified onto a chromatographic column, and then eluted in 25 µL elution buffer. Then, 25 µL of 2x Hybridization Solution (Exiqon) was added and mixed thoroughly, and the resulting mixture was denatured at 65°C for 3 min. The labeled mix was transferred to the microarray and covered with coverslips. The hybridization was performed in GlassArray Hybridization Cassettes (Invitrogen Ltd, Paisley, UK) in a water bath at 37°C for 16 h and then a wash sequence was performed. The array was dried by centrifugation and scanned by a laser scanner (ScanArray, PerkinElmer, Waltham, MA) to record fluorescent signals produced by each spotted probe effectively hybridized with the corresponding miRNA.
Microarray data have been recorded in the GEO database (GEO accession No. GSE107962).
Evaluation of miRNA expression analysis by qPCR
Validation of microarray data was performed by real time-qPCR for let-7e, miR-125b, and miR-322-3p. SYBR GREEN fluorescent tracers were used to identify amplicons whose identity was checked by melting curve analysis. Primer sequences (TIB Molbiol, Genoa, Italy) were identified according to http://www.ncbi.nlm.nih.gov/tools/primer-blast/database. cDNAs were prepared by using Superscript II Reverse Transcription kit (Invitrogen, Carlsbard, CA). PCR was performed in a Rotor-Gene 3000 (Corbett Research, Mortlake, Australia). Each reaction was carried out using 10x PCR buffer, 50 mM MgCl2, dNTM mix, primerA 10 µM, primerS 10 µM, Platinum® Taq DNA polymerase (Invitrogen), cDNA (diluted 1:10), and SYBR GREEN® (Invitrogen) in a 50 μL reaction volume. The thermal profile consisted of hot-start enzyme activation at 95°C for 2 min, 45 cycles of PCR at 94°C for 45 s (denaturation), gene-specific temperature annealing for 30 s, and 72°C for 30 s (elongation). Gene expression was normalized to the RNU6 housekeeping gene. Each sample was tested in triplicate and the results were expressed as relative gene expression intensities as obtained from the first positive amplification cycle.
Statistical analysis
Differences in body weight gain between MCS-exposed and sham-exposed mice were evaluated for statistical significance by ANOVA followed by Student's t test for unpaired data. miRNA microarray data, after local background subtraction, log transformation, and normalization were analyzed by GeneSpring software (Agilent, Santa Clara, CA). Expression data were median centered by using the GeneSpring normalization option. Comparisons between experimental groups were done by evaluating the fold variations of duplicate data generated for each miRNA. In addition, the statistical significance of the differences was evaluated by means of the GeneSpring ANOVA applied by using Bonferroni multiple testing correction. As inferred from volcano-plot analysis, differences between sets of data were taken as significant when they were statistically significant (P < 0.05) and showed >2-fold variation. The microarray data were processed by GeneSpring software and their overall variability, as related to treatments, was examined by scatter-plot analysis (SPA), hierarchical cluster analysis (HCA), and principal component analysis (PCA). MA-plots were calculated by using the R.Net Inferno RDN software (Pacific NorthWest National Lab, Richland, WA, USA). Due to the huge number of datasets to be analyzed, pooled analysis of microarray data was performed without taking into account intergender differences, a goal that was out of the scope of the present study and that had been evaluated in previous studies [9]. However, intergender differences were evaluated for selected miRNAs by using qPCR analysis. The significance of the differences in qPCR data between MCS-exposed and sham-exposed mice was evaluated by Student's t test for unpaired data.
Results
Body weights of mice as related to exposure to MCS
All mice were weighed on the 1st day of treatment (time 0) and thereafter at weekly intervals. The body weights at time 0 (mean ± SE) of sham-exposed mice were 38.7 ± 1.15 g for males and 28.9 ± 0.79 g for females. Thereafter, exposure to MCS tended to slightly decrease the body weights, and from the second week onwards the differences were statistically significant at most time points. Thus, after 8 weeks the body weights of sham-exposed mice were 42.3 ± 1.09 g for males and 37.5 ± 1.16 g for females, and those measured of MCS-exposed mice were 39.4 ± 0.70 g for males (P < 0.05 as compared to sham-exposed males) and 31.7 ± 1.22 g for females (P < 0.01 as compared to sham-exposed females).
Evaluation of RNA purity
Evaluation by nanospectrophotometry showed that the RNA purified from all organs was of good quality, as inferred from the calculation of the 260/280 and 260/230 absorbance ratios, which were consistently higher than 1.9 and 1.3, respectively (data not shown). The RNA recovery efficiency from each one of the 10 organ specimens was satisfactory, ranging between 15.9 ± 0.6 µg for skeletal muscle to 156.6 ± 5.5 µg for kidney. As for the body fluids specimens, the overall 260/280 ratio was 1.3 ± 0.04 for blood serum, 1.7 ± 0.3 for BALF, and 1.3 ± 0.1 for urine, and the amount of purified RNA was 178.0 ± 12.0 ng for blood serum, 4.3 ± 12.0 ng for BALF, and 95.9 ± 70.2 ng for urine. The presence of miRNAs in sufficient amounts to perform microarray analyses was further confirmed by Qubit quantitative fluorescence analysis, which indicated that the collected samples were adequate to perform microarrays.
Comparison of global miRNA profiles in organs and body fluids of sham-exposed mice and MCS-exposed mice
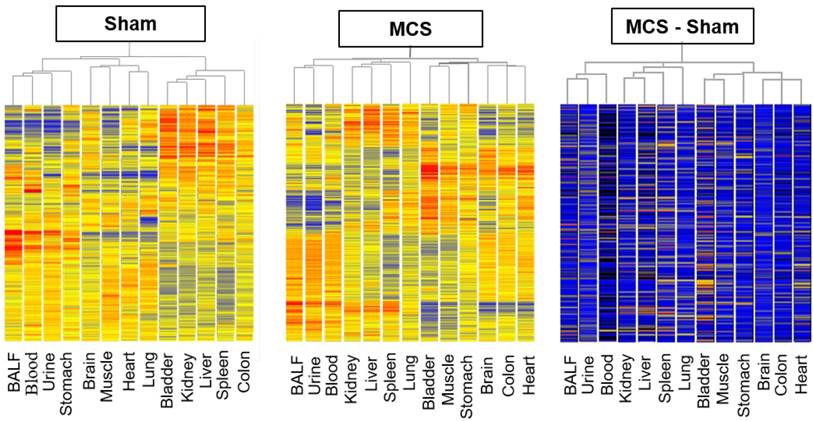
The overall profiles of miRNA expression, as evaluated by microarray analyses, were compared by HCA and PCA. Due to the huge amount of data, these analyses were carried out by pooling the results obtained in males and females, whereas intergender differences were evaluated for the miRNAs analyzed in parallel by microarray and qPCR (see below). HCA indicated that, under the basal conditions of sham-exposed mice (Fig. 1, left panel), body fluids have different miRNA expression profiles as compared to body organs. In fact, BALF, blood serum, and urine clustered together in the hierarchical tree on the left part of the dendrogram, separately from body organs. Some organs belonging to the same system, such as the urinary bladder and kidney, or having a similar histological structure, such as skeletal muscle and heart, clustered together or in nearby branches in the central part of the dendrogram. Likewise, in MCS-exposed mice (Fig. 1, middle panel) BALF, blood serum, and urine clustered together in the hierarchical tree on the left part of the dendrogram, separately from body organs. Positioning of the 10 organs in the dendrogram was considerably altered as compared with the pattern observed in sham-exposed mice. These patterns did not change when evaluating the difference between MCS and Sham values in order to detect the net effects of MCS (Fig. 1, right panel).
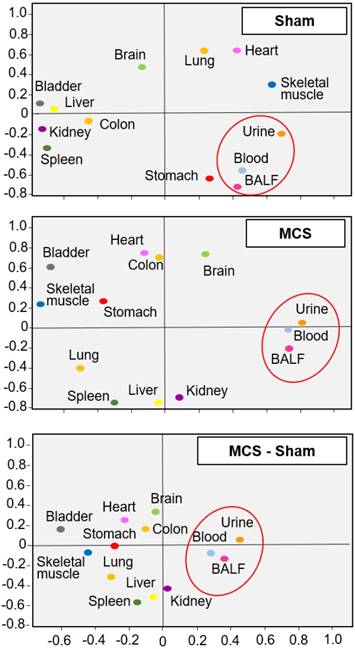
Similar findings were obtained by PCA, which indicated that the miRNA profiles of body fluids were located close to each other but far away from most organs in sham-exposed mice (Fig. 2, upper panel). The only exception was the stomach of sham-exposed mice, which may contain some residual amounts of gastric mucus. Organs having similar tissue composition, i.e., skeletal muscle and heart, were located nearby each other in the same quadrant, irrespective of exposure to MCS. These results indicate that miRNA profiles are tissue-specific. Exposure of mice to MCS (Fig. 2, middle panel) completely modified the position of the lung miRNA profile, which moved from the right-upper quadrant to the left-bottom quadrant. Other organs changing PCA quadrant were the skeletal muscle, stomach, heart, liver, and kidney. The influence of MCS on body fluids was less remarkable. In fact, BALF, blood serum and urine merely moved their position from the lower limit to the upper limit of the right bottom quadrant. A similar picture was observed by calculating the differences between MCS and Sham values (bottom panel).
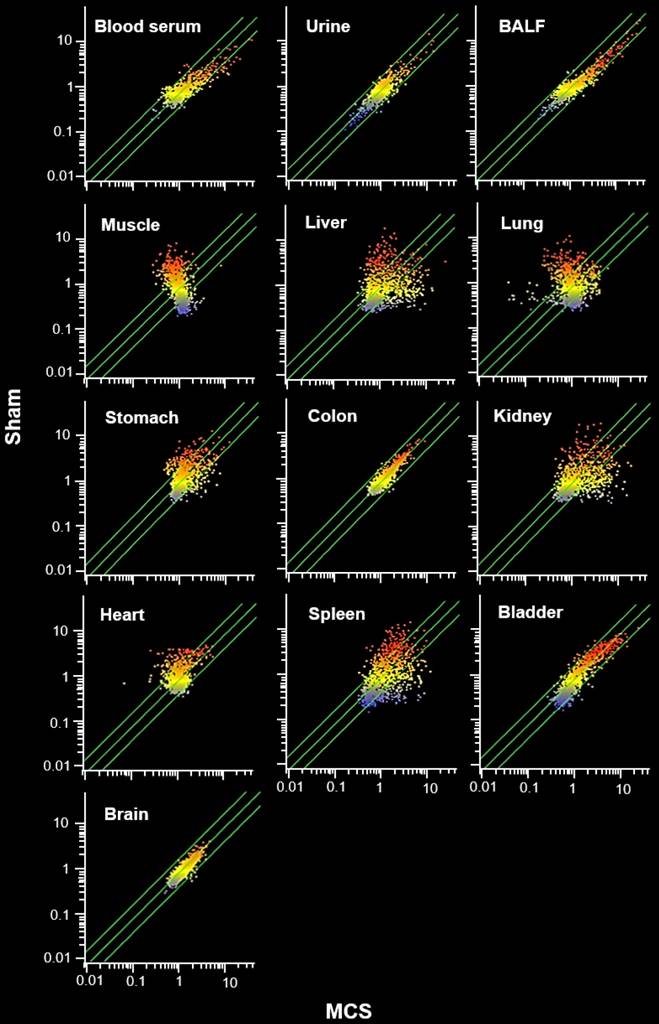
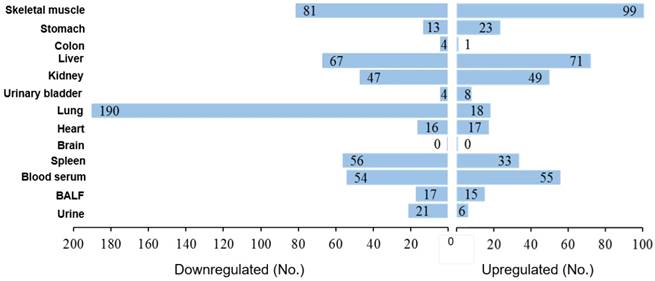
The MCS organotropism was evaluated in detail by comparative scatter-plot analysis (SPA, Fig. 3). According to the number of miRNAs varying their expression more than 2-fold as a consequence of exposure to MCS (i.e., being located outside the diagonal area limited by green lines in Fig. 3), the preferentially hit organ by MCS was the lung, followed by skeletal muscle, liver, kidney, and spleen. An intermediate effect was detected in heart, stomach, and urinary bladder, and only minimal alterations of miRNA profiles occurred in colon and especially in brain. Blood serum, BALF, and urine were altered in their miRNA profiles in decreasing order of magnitude. The remarkable effect of MCS on lung, heart, and skeletal muscle as compared to other organs was also detected by MA-plots (Fig. 1S).
The "translatability" of the results obtained to humans was evaluated comparing by SPA the MCS effect on mouse miRNAs (No. = 1135) and human (No. = 1928) miRNAs spotted on the microarray used. A remarkable overlap was observed (Fig. 2S).
Comparison of individual miRNAs in organs and body fluids of sham-exposed mice and MCS-exposed mice
As to individual miRNAs, we evaluated by volcano-plot analysis (Fig. 3S) the miRNAs that were significantly dysregulated in mice exposed to MCS compared to sham-exposed mice. Table S1 shows the results, in terms of MCS/Sham fold ratio, of all miRNAs out of the 1135 miRNAs tested that were either upregulated or downregulated by MCS in the 10 organs and 3 body fluids examined.
Hierarchical cluster analysis (HCA) of miRNA profiles. HCA compared the profiles of 1135 miRNAs in 10 organs and 3 body fluids of mice aged 4 months, either sham-exposed (left panel) or exposed whole-body to MCS for 8 weeks (middle panel). The right panel was generated by calculating the difference between MCS and Sham values.
Bidimensional principal component analysis (PCA) of miRNA profiles. PCA compared the profiles of 1135 miRNAs in 10 organs and 3 body fluids from mice aged 4 months, either sham-exposed (upper panel) or exposed whole-body to MCS for 8 weeks (middle panel). The bottom panel was generated by calculating the difference between MCS and Sham. The contributions to variance in Sham were 40.7% for PCA1 (X axis), 26.1% for PCA2 (Y axis), and 4.9% for PCA3 (not shown), whereas the contributions to variance in MCS were 39.2% for PCA1, 27.9% for PCA2, and 5.3% for PCA3 (not shown).
Based on the data reported in Table S1, we drew a figure (Fig. 4) showing the number of miRNAs that were significantly upregulated or downregulated by MCS in each organ and body fluid. The rank was as follows: lung 208 dysregulated miRNAs (18.3% of the 1135 analyzed miRNAs), skeletal muscle 180 (15.9%), liver 138 (12.2%), blood serum 109 (9.6%), kidney 96 (8.5%), spleen 89 (7.8%), stomach 36 (3.2%), heart 33 (2.9%), BALF 32 (2.8%), urine 27 (2.4%), urinary bladder 12 (1.1%), colon 5 (0.4%), and brain 0. As shown in Fig. 4, there was a fair balance between upregulated and downregulated miRNAs in liver (51.4% upregulated and 48.6% downregulated), skeletal muscle (55.0% and 45.0%), blood serum (50.5% and 49.5%), kidney (51.0% and 49.0%), heart (51.5% and 48.5%), and BALF (46.9% and 53.1%). There was a prevalence of upregulated over downregulated miRNAs in stomach (63.9% vs. 36.1%), whereas downregulation was prevalent in spleen (62.9% downregulated and 37.1% upregulated), urine (77.8% and 22.2%) and, with sharp differences, in lung (91.3% and 8.7%).
Common miRNA profiles in organs and body fluids
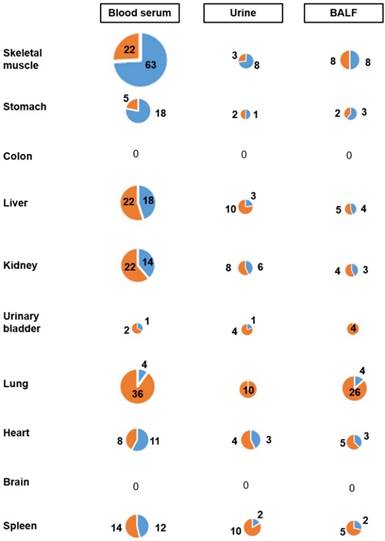
A major problem in evaluating body fluid miRNAs as biomarkers in preventive medicine is to assess whether or not they reflect alterations occurring in target organs of carcinogens. To assess this, we determined by Venn Diagram analysis the contribution of the 10 body organs to the miRNA profiles of the 3 body fluids examined. Such an approach compares the list of miRNAs altered by MCS more than 2-fold in organs or fluids and indicates the number of those miRNAs that are common in both lists. The results of this analysis are shown in the pie charts reported in Fig. 5. The contribution of skeletal muscle to MCS-altered miRNAs detectable in blood serum was by far the highest as compared with other organs. In fact, 85 miRNAs were dysregulated both in skeletal muscle and blood serum, followed by liver and lung (40 each), kidney (36), spleen (26), heart (19), urinary bladder (3), colon and brain (both 0). In urine, the contribution of all examined organs was poor and no striking difference was detectable among different organs, although the contribution of kidney was the highest one. The situation was different in the BALF of MCS-exposed mice, in which the contribution of lung miRNAs was the most remarkable. Except for colon and brain, other organs gave some contribution as well.
Another issue was the extent of miRNA overlap among the body fluids tested. As detected by comparative Venn Diagram analyses, there was limited overlap of miRNAs modulated by MCS in the 3 body fluids. In particular, 11 miRNAs were modulated by MCS in both blood serum and BALF, and 13 miRNAs were modulated in both blood serum and urine. Four miRNAs were modulated in both BALF and urine (see Table S1).
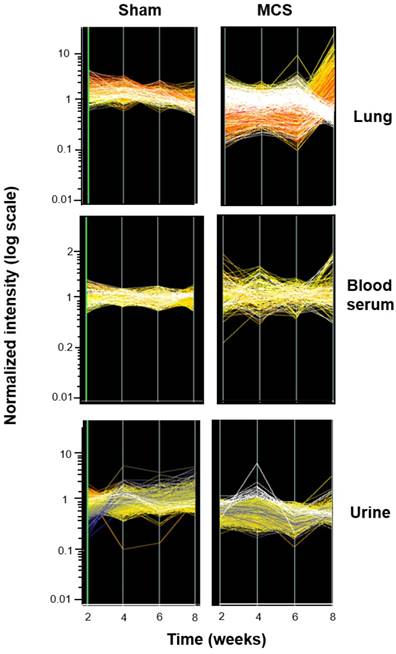
Fig. 6 shows the time course of miRNA profiles in the lung, blood serum, and urine of both sham-exposed and MCS-exposed mice after 2, 4, 6, and 8 weeks. In the lung, the effects of MCS on miRNA expression were mainly detectable after 8 weeks of treatment. A similar situation was observed in blood serum, although the differences among different time points were less evident. All differences were attenuated in urine, but the effects of MCS were clearly detectable after 4 weeks.
Comparative scatter plot analyses (SPA) of miRNA profiles. SPA evaluated the organotropism of MCS in modulating the profiles of 1135 miRNAs in 10 organs and 3 body fluids from mice aged 4 months, either sham-exposed (Y axis) or exposed whole-body to MCS for 8 weeks (X axis). Each dot represents a miRNA, whose expression intensity can be inferred from the position in the X and Y axes, according to a color scale (blue, low; yellow, medium; orange to red, high). Dots falling outside the diagonal two-fold variation interval (diagonal green lines) correspond to miRNAs that were dysregulated more than two-fold in their expression by MCS. Upregulated miRNAs are located in the lower-right area and downregulated miRNAs are in the upper-left area.
Organotropism of miRNAs in MCS-exposed mice. The values indicate the numbers of miRNAs that were either downregulated or upregulated by MCS in each one of the examined mouse organs and body fluids.
Contribution of 10 organs to the miRNA profiles of 3 body fluids from mice exposed whole-body to MCS for 8 weeks. The size of circles refers to the numbers of miRNAs that were significantly altered by MCS in their expression both in the indicated organ (Y axis) and body fluid (X axis). The reported values refer to the numbers of miRNAs that were either upregulated (blue portions) or downregulated (orange portions) both in the indicated organ and body fluid.
Validation of microarray data by qPCR analysis
Three miRNAs (let-7e, miR-125b, and miR-322-3p), all of them significantly dysregulated in selected organs of MCS-exposed mice as assessed by microarray, were further evaluated by qPCR. A total of 22 samples collected from both male and female mice were analyzed in parallel by means of the two techniques. The results, expressed in terms of MCS/ Sham ratios, are summarized in Table 1. In general, the results of qPCR analyses confirmed the trends (either downregulation or upregulation) generated by microarray analyses. Five samples only (let-7e in the heart of both males and females and in the spleen of males; miR-125b in the heart of males; and miR-322-3p in the lung and skeletal muscle of females) did not attain the statistical significance threshold of the MCS/Sham ratio when analyzed by qPCR. Microarray data were well correlated with qPCR data (r = 0.864, P < 0.01), with a regression equation of y = 0.52 + 0.832x, where x is microarray data and y is qPCR data.
Comparative evaluation by microarray and qPCR analyses of 3 miRNAs in organs of mice of both genders exposed to MCS. The results are expressed as MCS/Sham ratios.
| let -7e | miR-125b | miR-322-3p | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Organ | Gender | microarray | qPCR | microarray | qPCR | microarray | qPCR | ||
| Lung | Males | 0.45* | 0.11* | 0.41* | 0.32* | 0.44* | 0.22* | ||
| Females | 0.27* | 0.69* | 0.38* | 0.29* | 0.52* | 0.81 | |||
| Heart | Males | 0.67* | 0.93 | 2.24* | 1.51 | ||||
| Females | 0.31* | 1.07 | 2.01* | 2.27* | |||||
| Spleen | Males | 0.31* | 0.73 | 7.08* | 7.64* | ||||
| Females | 0.38* | 0.42* | 4.42* | 8.32* | |||||
| Muscle | Males | 3.94* | 3.46* | 2.63* | 5.33* | ||||
| Females | 6.41* | 6.61* | 2.41* | 1.69 | |||||
| Liver | Males | 5.50* | 2.09* | ||||||
| Females | 9.84* | 7.14* | |||||||
| Kidney | Males | 9.36* | 7.06* | ||||||
| Females | 5.40* | 7.11* | |||||||
* P < 0.05, MCS vs. Sham
Time-course modulation of miRNA profiles. The lines show the profiles of 1135 miRNAs in the lung, blood serum, and urines from mice aged 4 months, either sham-exposed (panels on the left) or exposed whole-body to MCS (panels on the right) for either 2, 4, 6, or 8 weeks. The cluster of the miRNAs that were most strikingly modulated by MCS are highlighted in white.
The differences between males and females for the analyzed samples were not statistically significant and consistently showed the same dysregulation trends. In particular, the means ± SE of MCS/Sham ratios evaluated by microarray for the 6 samples showing upregulation were 5.1 ± 1.12 in males and 5.1 ± 1.18 in females; for the 6 samples showing downregulation they were 0.5 ± 0.06 in males and 0.4 ± 0.04 in females. Those evaluated by qPCR for the 6 samples showing upregulation were 4.5 ± 1.05 in males and 5.5 ± 1.15 in females; for the 5 samples showing downregulation they were 0.4 ± 0.11 in males and 0.7 ± 0.14 in females.
Discussion
Goals and limitations of the study
The present study evaluated the expression profiles of 1135 miRNAs in 10 organs and 3 body fluids of mice, either under physiological conditions or following exposure to MCS for up to 8 weeks. The herein described results highlighted the multiorgan miRNA distribution as assessed in a single comparative experiment and, above all, at an exposure time when no MCS-related histopathological alterations are apparent. However, at this stage, early molecular changes can be detected as MCS-related increases of both bulky DNA adducts and oxidative DNA damage in the lung [13]. In addition, as shown in this study, miRNA expression analysis was able to identify the target organs of MCS-related molecular damage. An obvious limitation of the study is that all organs are composed of multiple tissues and cell types that may provide differential contributions to the overall miRNA profile of each of them. For instance, within the respiratory tract, nucleotide alterations have been shown to be higher in both tracheal epithelium and BAL cells of rats exposed to environmental CS (ECS) as compared to whole lung [14]. In any case, since the lung tissue used in the present study was obtained from lavaged mice, BAL cells were absent in the lung samples examined. This whole-organ approach does not exclude any tissue compartment, thus providing a comprehensive evaluation of the overall contribution of each organ to miRNA body fluids. A further goal of the present study was to ascertain the extent of miRNA release from the analyzed organs into 3 body fluids in order to predict the translatability of miRNA analysis as a molecular tool for human biomonitoring.
Predictivity of early miRNA alterations induced by MCS and pathological consequences
Evaluation of miRNAs under basal conditions showed that organs belonging to the same system or sharing a similar histological structure had matching overall expression profiles. In MCS-exposed mice, there were variable miRNA response patterns depending on the organ. For instance, consistent with previous studies in both rats [15, 16] and mice [9, 17-19] exposed either to MCS or to ECS, downregulation of miRNA expression was by far most prevalent in the lung. Downregulation of miRNAs in the lung correlates with an upregulation of proteins targeted by the altered miRNAs, which reflects an attempt to defend the respiratory tract by triggering a variety of protective mechanisms, such as antioxidant pathways, detoxification of carcinogens, DNA repair, anti-inflammatory pathways, apoptosis, etc. However, a long-lasting exposure to CS causes irreversible miRNA alterations that activate toxic and carcinogenic mechanisms, such as modulation of oncogenes and oncosuppressor genes, cell proliferation, recruitment of undifferentiated stem cells, inflammation, inhibition of intercellular communication, angiogenesis, invasion, and metastasis [20]. In addition, at least when exposure starts soon after birth, in the medium term (7-9 months) MCS induces a significant increase in several histopathological alterations, malignant tumors among them [reviewed in ref. 21].
In addition, exposure of mice to MCS caused, after 7-9 months, kidney tubular epithelial hyperplasia and adenomas as well as epithelial hyperplasia and papillomas in the urinary bladder, whose increase was statistically significant in some studies [21]. In the present study, kidney and urinary bladder miRNAs exhibited similar global expression profiles in sham-exposed mice. However, the kidney reacted to MCS more intensely than the bladder and exhibited both upregulation and downregulation of miRNAs. It is noteworthy that the kidney was the main source of MCS-dysregulated miRNAs in urine and gave an appreciable contribution to blood serum miRNAs as well. This conclusion is consistent with the notion that several miRNAs are detectable in kidneys, where they are crucial for renal development and normal physiological functions and contribute significantly to the pathogenesis of renal disorders [22]. On the other hand, the alterations of miRNAs in the urinary bladder of MCS-exposed mice were poorly detectable in all examined biological fluids. It is noteworthy that the urine of smoking humans invariably contains mutagenic substances [23], but in the present study no important early miRNA alterations were detected in the bladder of MCS-exposed mice. This apparent discrepancy is presumably due to the fact that miRNAs were analyzed in the bladder as an organ in toto, rather than in the isolated bladder epithelium, and to the circumstance that mutagenic metabolites eliminated with the urine are formed in upstream organs, such as liver and lungs.
Mixed miRNA alterations also occurred in other organs of the mice exposed to MCS. The differential patterns of response of lung and liver in ECS-exposed mice had already been pointed out in a previous study in which the liver reacted to ECS exposure by activating an array of miRNA-related defensive and detoxifying activities [18]. These data correlate with the absence of DNA adducts in the liver of rodents exposed to either MCS [14] or ECS [24]. It is noteworthy that no tumors are detectable in the liver even after a medium-term exposure of mice to MCS, which on the other hand often causes hepatotoxicity [21]. miRNA upregulation prevailed in the stomach of MCS-exposed mice, and an appreciable proportion of dysregulated miRNAs were released into the bloodstream. Smoking is one of the risk factors for gastric cancer, and nicotine promotes gastric tumor growth. Treatment of human gastric cancer cells with nicotine altered the expression of some miRNAs, in particular by upregulating miR-16 and miR-21, which are targeted by the transcription factor NF-κB [25]. Other miRNAs, such as miR-1, miR-133 and miR-206, have been reported to play a key role in tumorigenesis, progression, invasion and metastasis of gastric cancer [26].
Once more, we emphasize that our study evaluated miRNA profiles at an early stage of the carcinogenesis process. Accordingly, the observed effects are likely to be ascribed to dysregulation of miRNA expression by MCS rather than to pathological conditions triggered by this agent. In contrast with the stomach, MCS induced only slight variations in the miRNA expression profile in the colon, and no MCS-altered miRNA detected in this organ was found in any of the examined body fluids. Important alterations of miRNA profiles, especially in the sense of downregulation, occurred in the spleen of MCS-exposed mice and were clearly detectable by analyzing blood serum and urine. These alterations are likely to reflect the induction of inflammatory alterations in white pulp, accompanied by the entrapment of red blood cells altered by MCS exposure in red pulp as a consequence of hemoglobin monocarboxylation and lipid membrane peroxidation. In addition, spleen contains a remarkable leukocyte population, which may play a role in the systemic inflammation produced by MCS.
Multiorgan dysregulation of miRNA expression induced by MCS
As detailed in Table S1, dysregulation by MCS of some miRNAs selectively occurred in certain organs, whereas other miRNAs were dysregulated in multiple organs and body fluids, sometimes with opposite trends in organs and body fluids. To give a few examples, let-7e was downregulated in lung, heart, and spleen. Downregulation of members of the let-7 family in the lung has previously been reported in both mice exposed to MCS [17] and rats exposed to ECS [15]. It is noteworthy that members of this family, acting as tumor suppressor miRNAs, are typically downregulated in human lung cancer, whereas the oncomiRNA miR-21, which was downregulated in the lung of MCS-exposed mice, is upregulated in lung cancer [27]. miR-1a-1 was downregulated in lung, liver, kidney, spleen, and stomach, and was increased in the blood serum of MCS-exposed mice. Note that miR-1a was downregulated in urethane-induced pulmonary carcinogenesis in mice, but at the same time its levels were decreased in the blood serum [28]. The tumor suppressor miR-126, which was upregulated in the liver and downregulated in the urinary bladder of MCS-exposed mice, is downregulated in gastric cancer [27]. Likewise, miR-22, a possible tumor suppressor whose downregulation was proposed as a diagnostic biomarker for gastric cancer [29], was downregulated by MCS in the stomach and additionally in liver, spleen, and blood serum. miR-24, whose upregulation was shown to inhibit proliferation, metastasis, and to induce apoptosis in bladder cancer cells [30], was downregulated by MCS in liver, skeletal muscle, kidney, and urinary bladder, and was significantly decreased in urine. Within the miR-125 family, which has diverse functions in different cell contexts [31], miR-125b-1 was upregulated by MCS in multiple organs, including liver, kidney, heart, skeletal muscle, and spleen, but its levels were decreased in both lung and blood serum. Some miRNAs were regulated by MCS with similar trends in various organs. For instance, miR-293 was upregulated in liver, kidney, spleen, urinary bladder, and skeletal muscle, and miR-335-5p was strongly upregulated in liver, kidney, and spleen. miR-466o was downregulated in liver, kidney, and spleen. miR-494 was strongly upregulated in liver, kidney, spleen, and skeletal muscle, and was increased in blood serum, whereas miR-331-5p, miR-344d-3, and miR-370 were downregulated in liver, kidney, and spleen. miR-154, a potential tumor suppressor in liver carcinogenesis [32], was upregulated in both liver and kidney. On the other hand, miR-155, an important regulator of inflammation promoting alcohol-induced steatohepatitis and fibrosis in vivo [33], was strongly downregulated in the liver, kidney, and spleen of MCS-exposed mice, while its levels were increased in blood serum. The opposite trend of MCS-induced dysregulation observed in blood serum and lung is in line with our previous finding obtained in similar experimental models [9]. These patterns are amenable to the alteration induced by MCS in the miRNA maturation machinery resulting in lack of production of mature miRNAs accompanied by upstream accumulation of miRNA precursors.
Besides dysregulation of miRNAs in the lung of MCS-exposed mice, which may be involved in the pathogenesis of lung cancer and COPD, of special interest is dysregulation in striated muscles, with the heart and skeletal muscles displaying similar trends at a global analysis as well as at the level of some individual miRNAs. miR-206 was the most typical miRNA, among the canonical miRNAs detectable in striated muscles, called myomiRs, which was upregulated by MCS in the skeletal muscle of MCS-exposed mice. This miRNA and other miRNAs that were upregulated in the skeletal muscle of MCS-exposed mice, such as miR-182, are targeted to myocytes and modulate the physiology and pathology of these cells by altering gene expression [34]. miR-206, whose physiological expression significantly decreases in mouse quadriceps during the first 12 weeks of life [35], plays a role in promoting muscle differentiation and regeneration [36] and in repairing exercise-induced muscle injury [37]. Interestingly, miR-206 was upregulated also in the lung, liver, and stomach of MCS-exposed mice, whereas it was decreased in the kidney, blood serum, and BALF. Dysregulation of this miRNA plays a key role in tumorigenesis, progression, invasion, and metastasis of gastric cancer, a knowledge that might be exploited for the development of future miRNA-based interventions for preventing or treating this type of cancer [26]. miR-501 is a novel muscle-specific miRNA, which works as a regulator of myosin heavy chain during muscle regeneration and might serve as a novel serum biomarker for the activation of adult muscle stem cells [38]. This miRNA was significantly decreased in both skeletal muscle and blood serum of MCS-exposed mice. Another miRNA, miR-183, was upregulated in both heart and skeletal muscle of MCS-exposed mice, while it was downregulated in lung, liver, and spleen, and was decreased in blood serum, BALF, and urine. This miRNA is part of the regulatory network revealing the mechanism of inflammation in atrial fibrillation [39].
It has been argued that non-physiological expression of miRNAs may contribute to both development and progression of cardiovascular diseases [40]. As demonstrated in rats, heart and lungs are the organs where the highest levels of bulky DNA adducts are detectable following exposure to ECS, and these nucleotide alterations are more persistent in the heart, presumably because of the less efficient DNA repair capacities in this organ [14]. The clinical outcome of these molecular lesions depends on the rate of cell proliferation. Thus, alterations in the lung are correlated with the development of lung cancers and BPCO, while alterations in both heart and skeletal muscle hardly evolve into cancer, at least in adults, because these organs are composed of perennial cells, and proliferation is a condicio sine qua non for the growth of a neoplastic mass. In fact, both heart tumors and skeletal muscle tumors, such as rhabdomyosarcomas, mainly occur at a young age, when muscle cells are affected at prenatal or perinatal stages and are still replicating. On the other hand, it is likely that molecular alterations in heart and skeletal muscle may be involved in the pathogenesis of degenerative diseases, such as cardiomyopathies [41] and skeletal muscle atrophy [42, 43].
Together with the colon, the brain was the only organ in which exposure of adult mice to MCS resulted in minimal alterations of miRNA profiles, which were mainly oriented in the sense of upregulation. This conclusion contrasts with our previous finding that exposure to MCS of neonatal mice of the same strain used in the present study induced DNA damage and altered base-excision repair and tau levels in the brain [44]. Presumably, this discrepancy is amenable to the fact that there are differences in the blood-brain barrier (BBB) between newborns and adults. In fact, studies in rabbits have demonstrated that, although a selective BBB is operative at birth, considerably higher uptake rates for some compounds occur in newborns [45]. Brain endothelial miRNAs play critical roles in the regulation of BBB function under normal and neuroinflammatory conditions [46], and certain miRNAs, such as miR-143 [47] and miR-150 [48], increase the permeability of BBB. The fact that no MCS-dysregulated miRNA was detectable in any of the three examined biological fluids, besides the poor dysregulation of miRNAs in the brain of MCS-exposed mice, demonstrates that the BBB was not damaged by MCS in adult mice after 8 weeks of exposure.
miRNA signatures in body fluids
The mechanisms underlying the release of miRNAs into body fluids are still a matter of discussion, having been ascribed either to cell death or to other cellular biological mechanisms [7], and some extracellular miRNA species might carry cell-cell signaling functions during various physiological and pathological processes [49]. The body fluids examined in the present study had been deprived of their cellular components by centrifugation in order to assess the release of miRNAs by the organs into fluids without the possible confounding contribution of cells. miRNAs may be either free in body fluids or associated with fluid components, such as lipoproteins and ribonucleoprotein complexes, or packaged within extracellular vesicles such as exosomes, apoptotic bodies, and microvesicles or exosomes, which provide RNase protection [7, 50]. In the present study, RNA was purified from body fluids in such a way to include all forms of extracellular miRNAs. It should be taken into account that the miRNAs found in urine are eliminated from the body, whereas those found in the bloodstream are either eliminated through emunctory organs or transmitted to distant organs. Those found in BALF are removed from the lower respiratory spaces via the mucociliary escalator and either eliminated from the body by expectoration or swallowed. Swallowed miRNAs are expected to contact the mucosae of the GI tract, thus being either detoxified by the liver or ultimately eliminated with feces.
The herein reported results clearly show that the numbers of MCS-dysregulated miRNAs released into each one of the examined body fluids are affected by pharmacokinetic mechanisms. Thus, the miRNAs detectable in the BALF are mainly of pulmonary origin, and the kidney is the main source of miRNAs detectable in urine. The skeletal muscle gave a striking contribution to the presence of MCS-dysregulated miRNAs in the blood serum, which was much higher as compared to the contribution given by the lungs and other organs. The comparison of MCS-dysregulated miRNAs in the three examined body fluids reflected the direct physiological relationship between blood and urine and the lack of a direct relationship between urine and BALF. Time-course experiments showed that the effects of MCS on miRNA expression in lung and blood serum were mainly detectable after 8 weeks of exposure to MCS, whereas the effects in urine, albeit less striking, were clearly detectable after 4 weeks. The rapidity of excretion of miRNAs with urine correlates with the finding that the appearance of mutagenicity in smokers' urine is very fast [23].
The massive release of circulating miRNAs from the skeletal muscle of MCS-exposed mice is attributable to the fact that the skeletal muscle mass is by far the greatest as compared to the mass of other organs, accounting for about 40% of the body mass in humans. Accordingly, the number of cells that release altered miRNAs is higher than that of other organs. miRNAs have been reported to control mitochondrial function and insulin signaling in skeletal muscle cells [51], and mitochondria are known to be particularly susceptible to MCS [52, 53]. Circulating miRNAs exhibit a potential role as mediators of the cross-talk between the skeletal muscle and other tissues/organs [51]. myomiRs regulate myoblast proliferation and differentiation [54]. In humans, circulating heart and muscle myomiRs increased to a great extent in advanced heart failure, in parallel with an increase of cardiac troponin I, which emphasizes the usefulness of measuring circulating miRNAs as biomarkers for heart injury [55]. Our finding that exposure to MCS results in an early and considerable dysregulation of miRNAs in the skeletal muscle, from where they are massively released into the bloodstream, is not surprising, because patients suffering from COPD, of which smoking is the most important risk factor, commonly suffer from skeletal muscle dysfunction. Interestingly, consistent with our experimental data, CS exposure may contribute to the development of skeletal muscle dysfunction even before overt pulmonary pathology [42]. Exposure to MCS caused skeletal muscle atrophy also in a mouse model of COPD [43] and upregulated several pathways of metabolic processes and tissue degradation in muscles [56]. Evaluation of circulating miRNAs in myotonic dystrophy patients suggested that miRNAs might be useful as humoral biomarkers of that muscle disease [57]. The poor correlation between early alterations of miRNA profiles in the lung and in the blood serum of MCS-exposed mice is in agreement with the results of a previous study in mice exposed to the same agent during the first 4 months of life and thereafter kept in filtered air for an additional 3.5 months, when smoking-related lung tumors were detectable [9]. Likewise, it has been reported that miRNA signatures in plasma do not correspond with miRNA signatures in BAL samples of lung cancer patients [8] and that extracellular miRNAs in pooled BAL samples from cancer patients are differentially expressed as compared with samples from noncancer patients [6]. The stability of miRNAs in BAL samples after bronchoscopy is noteworthy [58]. Therefore, in spite of the fact that the collection of BALF is semi-invasive, the analysis of this fluid appears to be more appropriate to detect smoking-related miRNA alterations of pulmonary origin than analysis of blood serum and urine, where the contribution of lung is confounded by the release of miRNAs from other organs. These data are applicable for diagnostic purposes and mechanistic studies in humans, also because the miRNA specificity of human tissues is remarkably correlated with that of rodents [59]. In addition, the translatability of our results to humans is also warranted by the overlap of mouse and human miRNAs in the microarrays used.
The comparison of microarray data and qPCR data did not show any significant differences between males and females, at least for the selected miRNAs and organs. In any case, evaluation of intergender differences was beyond the scope of the present study, and the distinctive miRNA response to MCS of mouse lung and blood serum had been investigated in our previous studies [9, 60].
Conclusions
The results obtained show that after just 8 weeks of exposure of mice to MCS, when no histopathological damage was apparent, miRNAs profiles were altered in many organs, where they are predictive of possible pathological consequences. The lungs, skeletal muscle, and kidneys were the most important sources of MCS-dysregulated miRNAs in the BALF, blood serum, and urine, respectively. These data should be taken into account in studies evaluating miRNA profiles in body fluids for diagnostic purposes and preventive medicine strategies.
Abbreviations
BAL: bronchoalveolar lavage; BALF: bronchoalveolar lavage fluid; BBB: blood-brain barrier; COPD: chronic obstructive pulmonary diseases; CS: cigarette smoke; ECS: environmental cigarette smoke; HCA: hierarchical cluster analysis; IARC: International Agency for Research on Cancer; MCS: mainstream cigarette smoke; miRNA: microRNA; PCA: principal component analysis; SPA: scatter-plot analysis.
Acknowledgements
This work was supported by the U.S. National Cancer Institute (Contract #HHSN261201200015I).
Supplementary Material
Supplementary figures and tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D. et al. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review). Int J Oncol. 2012;41:1897-1912
2. Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A. et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589-599
3. Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167-179
4. Irmak-Yazicioglu MB. Mechanisms of microRNA deregulation and microRNA targets in gastric cancer. Oncol Res Treat. 2016;39:136-139
5. Cheng G. Circulating miRNAs: roles in cancer diagnosis, prognosis and therapy. Adv Drug Deliv Rev. 2015;81:75-93
6. Rehbein G, Schmidt B, Fleischhacker M. Extracellular microRNAs in bronchoalveolar lavage samples from patients with lung diseases as predictors for lung cancer. Clin Chim Acta. 2015;450:78-82
7. Izzotti A, Carozzo S, Pulliero A, Zhabayeva D, Ravetti JL, Bersimbaev R. Extracellular microRNA in liquid biopsy: applicability in cancer diagnosis and prevention. Am J Cancer Res. 2016;6:1461-1493
8. Molina-Pinelo S, Suárez R, Pastor MD, Nogal A, Márquez-Martín E, Martín-Juan J. et al. Association between the miRNA signatures in plasma and bronchoalveolar fluid in respiratory pathologies. Dis Markers. 2012;32:221-230
9. Izzotti A, Balansky R, Ganchev G, Iltcheva M, Longobardi M, Pulliero A. et al. Blood and lung microRNAs as biomarkers of pulmonary tumorigenesis in cigarette smoke-exposed mice. Oncotarget. 2016;7:84758-84774
10. International Agency for Research on Cancer. A review of human carcinogens: personal habits and indoor combustions IARC. Monographs on the Evaluation of the Carcinogenic Risks to Humans 100, part E. Lyon: IARC. 2012
11. De Flora S, Izzotti A, D'Agostini F, La Maestra S, Micale RT, Ceccaroli C. et al. Rationale and approaches to the prevention of smoking-related diseases: overview of recent studies on chemoprevention of smoking-induced tumors in rodent models. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2014;32:105-120
12. Hecht SS. Lung carcinogenesis by tobacco smoke. Int J Cancer. 2012;131:2724-2732
13. Izzotti A, Balansky R, D'Agostini F, Longobardi M, Cartiglia C, Micale RT. et al. Modulation by metformin of molecular and histopathological alterations in the lung of cigarette smoke-exposed mice. Cancer Med. 2014;3:719-730
14. Izzotti A, Bagnasco M, D'Agostini F, Cartiglia C, Lubet RA, Kelloff GJ. et al. Formation and persistence of nucleotide alterations in rats exposed whole-body to environmental cigarette smoke. Carcinogenesis. 1999;20:1499-1505
15. Izzotti A, Calin GA, Arrigo P, Steele VE, Croce CM, De Flora S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke. FASEB J. 2009;23:806-812
16. Izzotti A, Calin GA, Steele VE, Cartiglia C, Longobardi M, Croce CM. et al. Chemoprevention of cigarette smoke-induced alterations of microRNA expression in rat lung. Cancer Prev Res. 2010;3:62-72
17. Izzotti A, Calin GA, Steele VE, Croce C, De Flora S. MicroRNA expression in mouse lung, as related to age and exposure to cigarette smoke and light. FASEB J. 2009;23:3243-3250
18. Izzotti A, Larghero P, Cartiglia C, Longobardi M, Pfeffer U, Steele VE. et al. Modulation of microRNA expression by budesonide, phenethyl isothiocyanate, and cigarette smoke in mouse liver and lung. Carcinogenesis. 2010;31:894-901
19. Izzotti A, Larghero P, Longobardi M, Cartiglia C, Camoirano A, Steele VE. et al. Dose-responsiveness and persistence of microRNA expression alterations induced by cigarette smoke in mouse lung. Mutat Res. 2011;717:9-16
20. De Flora S, Balansky R, D'Agostini F, Cartiglia C, Longobardi M, Steele VE. et al. Smoke-induced microRNA and related proteome alterations and their modulation by chemopreventive agents. Int Cancer. 2012;131:2763-2773
21. De Flora S, Ganchev G, Iltcheva M, La Maestra S, Micale RT, Steele VE. et al. Pharmacological modulation of lung carcinogenesis in smokers: preclinical and clinical evidence. Trends Pharmacol Sci. 2016;37:120-142
22. Bhatt K, Kato M, Natarajan R. Mini-review: emerging roles of microRNAs in the pathophysiology of renal diseases. Am J Physiol Renal Physiol. 2016;310:F109-F118
23. De Flora S, Camoirano A, Bagnasco M, Bennicelli C, van Zandwijk N, Wigbout G. et al. Smokers and urinary genotoxins: implications for selection of cohorts and modulation of endpoints in chemoprevention trials. J Cel Biochem Suppl. 1996;25:92-98
24. De Flora S, Izzotti A, Randerath K, Randerath E, Bartsch H, Nair J. et al. DNA adducts and chronic degenerative disease. Pathogenetic relevance and implications in preventive medicine. Mutat Res. 1996;366:197-238
25. Shin VY, Jin H, Ng EK, Cheng AS, Chong WW, Wong CY. et al. NF-κB targets miR-16 and miR-21 in gastric cancer: involvement of prostaglandin E receptors. Carcinogenesis. 2011;32:240-245
26. Xie M, Dart DA, Owen S, Wen X, Ji J, Jiang W. Insights into roles of the miR-1, -133 and -206 family in gastric cancer (Review). Oncol Rep. 2016;36:1191-1198
27. Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer-a brief overview. Adv Biol Regu. 2015;57:1-9
28. Li X, Wu J, Zheng J, Li Y, Yang T, Hu G. et al. Altered miRNA expression profiles and miR-1a associated with urethane-induced pulmonary carcinogenesis. Toxicol Sci. 2013;135:63-71
29. Jafarzadeh-Samani Z, Sohrabi S, Shirmohammadi K, Effatpanah H, Yadegarazari R, Saidijam M. Evaluation of miR-22 and miR-20a as diagnostic biomarkers for gastric cancer. Chin Clin Oncol. 2017;6:16
30. Zhang S, Zhang C, Liu W, Zheng W, Zhang Y, Wang S. et al. MicroRNA-24 upregulation inhibits proliferation, metastasis and induces apoptosis in bladder cancer cells by targeting CARMA3. Int J Oncol. 2015;47:1351-1360
31. Sun M, Lin KY, Chen YQ. Diverse functions of miR-125 family in different cell contexts. J Hematol Oncol. 2013;6:6
32. Pang X, Huang K, Zhang QW, Zhang Y, Niu J. miR-154 targeting ZEB2 in hepatocellular carcinoma functions as a potential tumor suppressor. Oncol Rep. 2015;34:3272-3279
33. Bala S, Csak T, Saha B, Zatsiorsky J, Kodys K, Catalano D. et al. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J Hepatol. 2016;64:1378-1387
34. Wang H, Wang B. Extracellular vesicle microRNAs mediate skeletal muscle myogenesis and disease. Biomed Rep. 2016;5:296-300
35. Lamon S, Zacharewicz E, Butchart LC, Orellana L, Mikovic J, Grounds MD. et al. MicroRNA expression patterns in post-natal mouse skeletal muscle development. BMC Genomics. 2017;8:52
36. Amirouche A, Jahnke VE, Lunde JA, Koulmann N, Freyssenet DG, Jasmin BJ. Muscle-specific microRNA-206 targets multiple components in dystrophic skeletal muscle representing beneficial adaptations. Am J Physiol Cell Physiol. 2017;312:C209-C221
37. Wu W. Changes in expression of specific miRNAs and their target genes in repair of exercise-induced muscle injury in rats. Genet Mol Res. 2016;16:15
38. Mizbani A, Luca E, Rushing EJ, Krützfeldt J. MicroRNA deep sequencing in two adult stem cell populations identifies miR-501 as a novel regulator of myosin heavy chain during muscle regeneration. Development. 2016;143:4137-4148
39. Zhang H, Liu L, Hu J, Song L. MicroRNA regulatory network revealing the mechanism of inflammation in atrial fibrillation. Med Sci Monit. 2015;14:3505-3513
40. Lee S, Choi E, Kim SM, Hwang KC. MicroRNAs as mediators of cardiovascular disease: Targets to be manipulated. World J Biol Chem. 2015;6:34-38
41. De Flora S, Izzotti A. Modulation of genomic and postgenomic alterations in noncancer diseases and critical periods of life. Mutat Res. 2009;667:15-26
42. Degens H, Gayan-Ramirez G, van Hees HV. Smoking-induced skeletal muscle dysfunction: from evidence to mechanisms. Am J Respir Crit Care Med. 2015;191:620-625
43. Ma R, Gong X, Jiang H, Lin C, Chen Y, Xu X. et al. Reduced nuclear translocation of serum response factor is associated with skeletal muscle atrophy in a cigarette smoke-induced mouse model of COPD. Int J Chron Obstruct Pulmon Dis. 2017;12:581-587
44. La Maestra S, Kisby GE, Micale RT, Johnson J, Kow YW, Bao G. et al. Cigarette smoke induces DNA damage and alters base-excision repair and tau levels in the brain of neonatal mice. Toxicol Sci. 2011;123:471-479
45. Braun LD, Cornford EM, Oldendorf WH. Newborn rabbit blood-brain barrier is selectively permeable and differs substantially from the adult. J Neurochem. 1980;34:147-152
46. Lopez-Ramirez MA, Reijerkerk A. et al. Regulation of brain endothelial barrier function by microRNAs in health and neuroinflammation. FASEB J. 2016;30:2662-2672
47. Bai Y, Zhang Y, Hua J, Yang X, Zhang X, Duan M. et al. Silencing microRNA-143 protects the integrity of the blood-brain barrier: implications for methamphetamine abuse. Sci Rep. 2016;6:35642
48. Fang Z, He QW, Li Q, Chen XL, Baral S, Jin HJ. et al. MicroRNA-150 regulates blood-brain barrier permeability via Tie-2 after permanent middle cerebral artery occlusion in rats. FASEB J. 2016;30:2097-2107
49. Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell-cell communication function? Front Genet. 2013;4:119
50. Natarajan SK, Pachunka JM, Mott JL. Role of microRNAs in alcohol-induced multi-organ injury. Biomolecules. 2015;5:3309-3338
51. Lima TI, Araujo HN, Menezes ES, Sponton CH, Araújo MB, Bomfim LH. et al. Role of microRNAs on the regulation of mitochondrial biogenesis and insulin signaling in skeletal muscle. J Cell Physiol. 2017;232:958-966
52. Balansky R, Izzotti A, Scatolini L, D'Agostini F, De Flora S. Induction by carcinogens and chemoprevention by N-acetylcysteine of adducts to mitochondrial DNA in rat organs. Cancer Res. 1996;56:1642-1647
53. Izzotti A. Gene-environment interactions in non-cancer degenerative diseases. Mutat Res. 2009;667:1-3
54. Coenen-Stass AM, Betts CA, Lee YF, Mäger I, Turunen MP, El Andaloussi S. et al. Selective release of muscle-specific, extracellular microRNAs during myogenic differentiation. Hum Mol Genet. 2016;25:3960-3974
55. Akat KM, Moore-McGriff D, Morozov P, Brown M, Gogakos T, Correa Da Rosa J. et al. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc Natl Acad Sci USA. 2014;111:11151-11156
56. Krüger K, Dischereit G, Seimetz M, Wilhelm J, Weissmann N, Mooren FC. Time course of cigarette smoke-induced changes of systemic inflammation and muscle structure. Am J Physiol Lung Cell Mol Physiol. 2015;309:L119-L128
57. Perfetti A, Greco S, Cardani R, Fossati B, Cuomo G, Valaperta R. et al. Validation of plasma microRNAs as biomarkers for myotonic dystrophy type 1. Sci Rep. 2016;6:38174
58. Brock M1, Rechsteiner T, Kohler M, Franzen D, Huber LC. Kinetics of microRNA expression in bronchoalveolar lavage fluid samples. Lung. 2015;193:381-385
59. Ludwig N, Leidinger P, Becker K, Backes C, Fehlmann T, Pallasch C. et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016;44:3865-3877
60. Izzotti A, Balansky R, Ganchev G, Iltcheva M, Longobardi M, Pulliero A. et al. Early and late effects of aspirin and naproxen on microRNA profiles in the lung and blood of mice, either unexposed or exposed to cigarette smoke. Oncotarget. 2017;8:85716-85748
Author contact
![]() Corresponding author: PD Dr. Silvio De Flora, Department of Health Sciences, University of Genoa, Via A. Pastore 1, 16132 Genoa, Italy sdfit Phone 0039 010 3538500 Fax 0039 010 3538482
Corresponding author: PD Dr. Silvio De Flora, Department of Health Sciences, University of Genoa, Via A. Pastore 1, 16132 Genoa, Italy sdfit Phone 0039 010 3538500 Fax 0039 010 3538482
 Global reach, higher impact
Global reach, higher impact