13.3
Impact Factor
Theranostics 2016; 6(8):1119-1130. doi:10.7150/thno.13944 This issue Cite
Research Paper
Recapitulating the Size and Cargo of Tumor Exosomes in a Tissue-Engineered Model
1. Department of Biomedical Engineering, Columbia University, New York, New York 10032, USA;
2. Department of Pediatrics, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10065, USA;
3. Department of Cell Biology, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10065, USA;
4. Department of Chemistry, Materials and Chemical Engineering "G Natta", Politecnico di Milano, Milano, Italy.
Received 2015-9-22; Accepted 2015-12-20; Published 2016-5-21
Abstract

There is a growing interest in the pivotal role of exosomes in cancer and in their use as biomarkers. However, despite the importance of the microenvironment for cancer initiation and progression, monolayer cultures of tumor cells still represent the main in vitro source of exosomes. As a result, their environmental regulation remains largely unknown. Here, we report a three-dimensional tumor model for studying exosomes, using Ewing's sarcoma type 1 as a clinically relevant example. The bioengineered model was designed based on the hypothesis that the 3-dimensionality, composition and stiffness of the tumor matrix are the critical determinants of the size and cargo of exosomes released by the cancer cells. We analyzed the effects of the tumor microenvironment on exosomes, and the effects of exosomes on the non-cancer cells from the bone niche. Exosomes from the tissue-engineered tumor had similar size distribution as those in the patients' plasma, and were markedly smaller than those in monolayer cultures. Bioengineered tumors and the patients' plasma contained high levels of the Polycomb histone methyltransferase EZH2 mRNA relatively to their monolayer counterparts. Notably, EZH2 mRNA, a potential tumor biomarker detectable in blood plasma, could be transferred to the surrounding mesenchymal stem cells. This study provides the first evidence that an in vitro culture environment can recapitulate some properties of tumor exosomes.
Keywords: Exosomes, tumor models, microenvironment, tissue engineering, EZH2.
Introduction
Exosomes are small membrane vesicles of endocytic origin that are released into the extracellular environment and circulate in the blood stream [1, 2]. They contain cell-specific cargo molecules (i.e. proteins, mRNA, miRNA, DNA), membrane proteins, and lipids. Consequently, exosomes are finding application as diagnostic biomarkers in a number of cancers [3-6]. Also, tumor-derived exosomes were shown to transfer a variety of bioactive molecules to other cells, inducing modifications of their environment and facilitating tumor growth and invasion [7]. There is an increasing interest in understanding how exosomes modulate their environment, after the discovery of their role in the pre-metastatic niche formation in lung [8] and liver [9].
Our knowledge about the putative roles of the microenvironment on tumor exosomes is limited, due to a lack of experimental models that efficiently mimic the human in vivo situation. Animal models used to study the effects of exosomes on cancer development often fail in representing the context of human disease [10]. In vitro, cancer cells are typically cultured under conditions not recapitulating the 3D tumor environment [11]. The absence of physiological cell-cell and cell-matrix-interactions and the currently used non-physiological substrates cause disparity from the in vivo situation and lead to changes in cell morphology, proliferation and cellular processes, such as endo- and exocytosis [11, 12]. Despite the growing notion of the importance of cell microenvironment for cancer signaling [13], supernatants from monolayer cultures still represent the main source of tumor-derived exosomes, such that their microenvironmental regulation remains largely unknown. Bioengineering methods are just about starting to bridge the gap between studies in cell monolayers and experimental animals, providing the models of human tumors that enable studies of how the microenvironment modulates cancer biology [14, 15].
Here we describe a controllable 3D tissue-engineered model for studying tumor exosomes, designed to mimic the native tumor microenvironment. As a clinically relevant example, we selected Ewing's sarcoma (ES), a solid tumor with aggressive biologic behavior [16], that affects children and young adults, and is associated with frequent metastases and poor prognosis [17]. ES is characterized by chromosomal rearrangements of the EWSR1 (22q12) gene with one of the members of the ETS family of transcription factors: the FLI1 gene (11q24) in 85% of cases [18]. Expression of EWSR1-FLI1 fusion protein has been the main approach to study the development of ES [19-22]. Recent studies also demonstrated the presence of EWSR1-FLI1 mRNA in ES-derived exosomes [23, 24].
Human mesenchymal stem cells (hMSC) were the only cell type found to provide an appropriate cellular context for EWSR1-FLI1 expression, supporting the notion that Ewing's sarcoma is derived from hMSCs [20, 21]. Surprisingly, hMSCs were unable to form tumors in immunocompromised mice [21]. Taken together, the published studies show that EWSR1-FLI1 is necessary to activate the oncogenic program, but not sufficient for oncogenic transformation of hMSCs [17]. Therefore, recent research has focused on downstream transcriptional targets [25-27] such as EZH2. EWSR1-FLI1 was shown to bind to the EZH2 promoter and to induce EZH2 expression in Ewing's sarcoma in vivo and hMSCs in vitro [28]. The EZH2 methyltransferase is a major component of the Polycomb repressive complex 2 (PRC2) that is related to transcriptional repression of tumor suppressors such as p14ARF and p16INK4a [29]. EZH2 is involved in the maintenance of cell pluripotency [30-32] and oncogenic transformation of Ewing's sarcoma cells [28]. Additionally, expression of EZH2 correlates with poor prognosis in several tumor types [33-35] including ES [28]. Thus far, the presence of EZH2 in ES-derived exosomes has not been documented.
Our goal was to capture both the effects of the microenvironment on tumor-derived exosomes, and the effects of exosomes on cell populations in the bone niche. To this end, we cultured ES cells in 3-dimensional biomaterial scaffolds designed to mimic the biological and mechanical properties of ES. We analyzed and compared the size distributions and EZH2 mRNA cargo in exosomes from the plasma of patients and culture medium from 2D monolayers (in culture dishes with different matrix coatings), 3D cell aggregates (in polypropylene), and 3D tissue-engineered tumors (in scaffolds resembling native tumor matrix) (Fig.1A). We then investigated the transfer of EZH2 mRNA from tumor-secreted exosomes to the mesenchymal stem cells, osteoblasts and osteoclasts of the ES bone niche.
Materials and Methods
Collection of the tissue samples from patients. Fully de-identified Ewing's sarcoma tumors were obtained from the Columbia University Tissue Bank, on an IRB-approved protocol. Frozen tissue samples from three different patients were cut into sets of contiguous sections for mechanical, histological, and immunohistochemical studies.
Fully de-identified blood plasma samples from Ewing's sarcoma patients for exosome isolation and characterization were collected in Dr. Moore's laboratory on an IRB-approved protocol at Memorial Sloan-Kettering Cancer Centre (New York, USA).
Scaffold preparation. Highly porous scaffolds were produced from Col1-HA solutions by freeze-drying. A 1% (wt/v) solution was prepared from low molecular weight (10-20 kDa) or high molecular weight (500 kDa) Sodium Hyaluronate (HA, Lifecore, US) in distilled water. Four parts of Collagen 1 solution (8-11 mg/ml in 0.02 N Acetic acid Corning, US) were mixed with one part of HA solution (4:1). After mixing, 200 μl of the solution was spread over a 8 mm x 5.5 mm mold, frozen at -40 °C for 4 hours, and sublimed at -40°C overnight under a vacuum of < 100 mTorr.
Lyophilized collagen-HA scaffolds were cross-linked with a water-soluble carbodiimide using a previously described method [36]. Scaffolds were immersed in 95% ethanol solution containing 33 mM EDC (Sigma-Aldrich Co. Ltd., UK) and 6 mM NHS (Sigma-Aldrich Co. Ltd., UK) for 4 h at 25 °C. After crosslinking, the scaffolds were washed thoroughly in distilled water (5 min × 5 times), refrozen and re-lyophilized at the same freeze-drying cycle as specified above.
Preparation of matrix-coated plates. Three different types of solutions were prepared for coating culture plates. For collagen-coated plates, a solution of collagen 1 (8-10 mg/mL, BDTM) was diluted in distilled water (4:1 dilution ratio). For HA coated plates, a suspension of HA (1% weight) was prepared from the low molecular weight sodium hyaluronate (10-20 kDa, Lifecore biomedical) in distilled water. For Col 1/HA coated plates, the above solutions of collagen 1 and HA were mixed in the 4:1 ratio of Col1: HA. 2mL of each of the three above solutions were added into each well of a 6-well plate, and left for 1h at room temperature in a sterile hood. The remaining unattached solutions were carefully aspirated. Each well was plated with 0.3 x106 SK-N-MC cells.
Culture of cells in aggregates and in 3D scaffolds. Ewing's sarcoma cell line SK-N-MC (HTB-10) was purchased from the American Type Culture Collection (ATCC) and cultured according to the manufacturer's specifications, in ATCC-formulated Eagle's Minimum Essential Medium (EMEM) supplemented with 10% (v/v) Hyclone FBS and 1% penicillin/streptomycin.
To form tumor cell aggregates, 0.3 x106 SK-N-MC cells were centrifuged in 15 mL Falcon tubes, 5 minutes at 1200 rpm, with 4 mL of medium and cultured for 7 days at 37°C in a humidified incubator at 5% CO2.
To seed 3D Collagen 1-HA scaffolds, single-cell suspension of SK-N-MC cells was adjusted to the cell concentration of 1 x106 cells/mL in a 50 ml Falcon tube. A total of 15 scaffolds were added to 30 mL of cell suspension, and the Falcon were set onto a rotary platform for 3h at 37°C/5% CO2. Cell seeded scaffolds were then transferred to non-treated wells in 12-multiwell plates (Nunc) and cultured in 2 mL of medium at 37°C / 5% CO2. Cell numbers and were determined by Quant-iT PicoGreen dsDNA Assay Kit (Life technologies) according to the manufacturer's instructions.
Mechanical testing. The mechanical properties of native Ewing's sarcoma tumors collected from the patients at the Memorial Sloan-Kettering Cancer Centre (New York, USA) were measured using a previously established protocol [37]. Briefly, the Young's modulus was determined under unconfined compression in phosphate-buffered saline (PBS) at room temperature. An initial tare load of 0.2 N was applied, and followed by a series of stress-relaxation steps, where specimens were compressed at a ramp velocity of 1% per second up to the 10% strain, and maintained at each position for 1,800 s. The Young's modulus was calculated from the equilibrium force measured at the 10% strain.
Scanning Electron Microscopy (SEM). The morphology of the bioengineered tumors was examined by SEM. Samples were washed twice in PBS and fixed in 4% paraformaldheyde in PBS (Santa Cruz, US) for 1 hour. Fixed specimens underwent a graded dehydration series of ethanol (70, 85, 95, 100% for 5 min each) and hexamethyldisilazane drying for 15 min (HMDS, Sigma). Samples were dried over night in the fume hood, sputter-coated with gold and palladium, and imaged using SEM (Hitachi S-4700).
Fluid Uptake by the Scaffolds. Dried samples were weighed (Wd) and immersed in distilled water at 37°C for different periods of time (2 hours, 3, 7 and 10 days). At each time point, specimens were removed from distilled water and the ability of the scaffold structure to absorb water was measured using a previously described method [36]. At each time point, the samples were removed from water and weighed (Ww). The water uptake was calculated as: Fluid uptake (%) = (Ww-Wd)/Wd x 100. Each sample was measured in triplicate.
Scaffold degradation. Dried samples were weighed (Wd) and immersed in distilled water at 37°C in a humid atmosphere for timed intervals (2 hours, 3, 7 and 10 days). At each time point, specimens were removed from distilled water, air-dried for 24 h and weighed (Wa). The weight loss was calculated as: Weight loss (%) = (Wd-Wa)/Wd x 100. Each sample was measured in triplicate.
Histology and immunohistochemistry (IHC). Frozen sections of the native Ewing's sarcoma tumors and bioengineered tumors were fixed in pre-cooled acetone (-20 oC) for 10 min. Sections were washed with PBS and treated with 0.3% H2O2 solution in PBS at room temperature for 10 min to block endogenous peroxidase activity, and incubated with a blocking buffer from Vectastain Elite ABC Kit (Vector Labs), according to the manufacturer's instructions. Then, sections were stained for CD99 (dilution 1:500; Signet antibodies, SIG-3620) and Collagen 1 (dilution 1:500; Abcam, ab34710). Slides were counterstained with Hematoxylin QS (Vector Labs).
For the hyaluronan acid binding protein (HABP) staining, the sections were blocked using 1% BSA in HBSS at room temperature for 30 min, and incubated with a biotinylated HABP antibody (dilution 1:100; Millipore #385911). A Streptavidin Alexa fluor 488 conjugate (dilution 1:500; Molecular Probes) was used as the secondary antibody.
Live-Dead assay. At timed intervals (day 3 and day 7), Bioengineered tumor models were incubated in EMEM medium containing 2μM Calcein and 4μM of ethidium homodimer-1 for 30 min at 37°C, 5% CO2, as indicated by the manufacturer's protocol (LIVE/DEAD® Viability/Cytotoxicity Kit, Molecular Probes). Samples were imaged with a fluorescence microscope (Olympus IX81 light microscope, Center Valley PA).
Exosome isolation and size analysis. Cells cultured in monolayers, aggregates and 3D scaffolds were washed with PBS twice and cultured in EMEM supplemented with 10% (v/v) Exosome-depleted FBS (SBI) and 1% penicillin/streptomycin for 12h. The supernatants were collected and exosomes were isolated from cell culture media using the total exosome isolation kit (Invitrogen), according to the manufacturer's protocol. Exosomes from plasma samples were also isolated using the total exosome isolation kit (Invitrogen). The size distributions of exosomes were determined by Nanoparticle Tracking Analysis (NTA) using the Nanosight machine.
Genomics Analysis. Overexpression of EZH2 in Ewing's sarcoma tumors at mRNA levels were compared using the R2 Genomics Analysis and Visualization Platform (http://r2.amc.nl.) The R2 platform is an online genomics analysis tool that can analyze a large collection of public data. We selected EZH2 as gene of interest to generate a MegaSampler using the following dataset:
Tumor Ewing Sarcoma-Francesconi (37 samples). Source: GEO ID: gse34620 Dataset Date: 2000-01-01. Pubmed link: 22327514. A genome-wide association study of at least 401 French ES patients compared to either 684 French or 3668 US self-described Caucasian controls consistently revealed candidate loci at chromosomes 1 and 10 (p<10-6).
Tumor Ewing Sarcoma-Delattre (117 samples). Source: GEO ID: gse12102 Dataset Date: 2008-06-15. Pubmed link: 22327514. Available tracks in R2: group (CAT) [ ews metastasis tumor (metastasis) | ews primary tumor (no evidence of disease) | ews primary tumor (relapse)]
Healthy: Normal Various -Roth- (353 samples). Source: GEO ID: GSE3526 Dataset Date: 2006-03-30. Pubmed link: 16572319. Normal human tissue samples from ten post-mortem donors were processed to generate total RNA, which was subsequently analyzed for gene expression using Affymetrix U133 plus 2.0 arrays. Donor information: Donor 1 - 25 year old male; donor 2 - 38 year old male; donor 3 - 39 year old female; donor 4 - 30 year old male; donor 5 - 35 year old male; donor 6 - 52 year old male; donor 7 - 50 year old female; donor 8 - 48 year old female; donor 9 - 53 year old female; donor 10 - 23 year old female.
Quantitative real-time PCR (qRT-PCR). Total RNA from cells was obtained using Trizol (Life Technologies) and total RNA from exosomes was obtained using the Total Exosome RNA & Protein Isolation Kit (ThermoFisher scientific) following the manufacturer's instructions. RNA preparations were treated with “Ready-to-go you-prime first-strand beads” (GE Healthcare) to generate cDNA. Quantitative real-time PCR was performed using DNA Master SYBR Green I mix (Applied Biosystems). mRNA expression levels were quantified applying the ΔCt method, ΔCt = (Ct of gene of interest - Ct of Actin). EZH2 primers were obtained from the PrimerBank database (http://pga.mgh.harvard.edu/primerbank/).
RNA quality. Total RNA quality and size distribution from cells and exosomes were determined by electropherograms from the Bioanalyzer 2100 using the RNA Pico Chip kit (Agilent Technologies).
Western blot. Cells were lysed in RIPA buffer containing protease inhibitors (Sigma-Aldrich, P8340) and exosomes extracts were obtained using the Total Exosome RNA & Protein Isolation Kit (ThermoFisher scientific) following the manufacturer's instructions. Cell preparations were centrifugated at 12,000 g for 10 min and supernatants containing soluble proteins were collected for analysis. 20μg of cells and exosomes extracts were loaded on 4-12% gradient Bis-Tris gels (BioRad), transferred to a nitrocellulose membrane and incubated with antibodies against EZH2 (1:500; Millipore 07-689), Calnexin (1:500; Santa Cruz, sc-11397, CD81 (1:500; Santa Cruz, sc-7637) at 4 degrees over night and GAPDH (1:5000; Invitrogen 437000) at room temperature for one hour. For detection, membranes were incubated with a secondary antibody anti-rabbit or anti-mouse conjugated with Alexa Fluor 680 dye (1:5000; ThermoFisher Scientific) at room temperature for one hour and imaged on Licor Odyssey scanner.
Exosome-mediated transfer of RNA. SKNMC cells were cultured on Col1-HA scaffolds for 7 days in ATCC-formulated Eagle's Minimum Essential Medium (EMEM) supplemented with 10% (v/v) Hyclone FBS and 1% penicillin/streptomycin. For exosome isolation, cells were cultured with 10% Exosome-depleted FBS (SBI) and 1% penicillin/streptomycin for 12h. Supernatants were harvested and exosomes were isolated. To measure protein concentration (by Bradford assay), the concentration of protein was adjusted to ~ 0.1 µg/µL in PBS, and the samples were diluted 1:50 (20μl in 1ml of PBS) for NTA analysis. The same volumes, dilutions and the same camera shutter were used to obtain similar concentrations of particles for measuring size distributions in cell monolayer and TE-Tumors.10µg of exosomes protein were labeled with SYTO RNA Select green fluorescent (Invitrogen) during 30 min at 37°C/5% CO2 at a final dye concentration of 10 μM. Exosome Spin Columns (MW 3000) were used to remove unincorporated dye from exosome labeling. The same volume of PBS without exosomes was also treated with SYTO RNA and Exosome spin columns to serve as a control. Cells (5,000 cells/well) were seeded in a 8-wells chamber slide the day before the exosome-mediated transferring assay. 10 µg of labeled exosomes in PBS, or same volume of PBS control, were incubated with hMSC passage 3, human osteoblasts or human osteoclasts during 2h at 37°C/5% CO2. Cells were fixed for 20 min with 4% PFA in PBS and mounting with Vectashield-DAPI.
Results
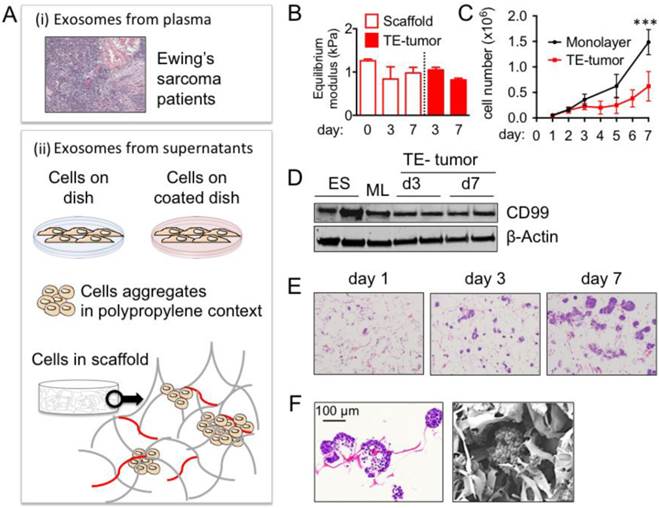
Bioengineered tumor model. Native Ewing's sarcoma (ES) is a pediatric tumor rich in collagen 1 (Col1) and hyaluronic acid (HA) proteins (Fig. S1A), and soft tissue matrix characterized by an equilibrium modulus of ~2 kPa (Fig. S1B). In order to mimic the composition and mechanical properties of the ES extracellular matrix, we used purified preparations of natural Col1 and HA (low molecular weight, LMW; high molecular weight, HMW) with a stiffness matching that of the native tumor (Fig. S1B). Two types of 3D porous scaffolds (Col1-HA LMW; Col1-HA HMW) were made by freeze-drying of Col1/HA solutions, and cross-linking with 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride, EDC, in the presence of N-hydroxysuccinimide, NHS (Fig. S1C).
The swelling behavior, measured by the liquid uptake, was similar for the two porous scaffolds, and in agreement with the previous studies [36]. The rate of degradation was much slower for Col1-HA LMW than Col1-HA HMW scaffolds, presumably due to the higher density of chemical cross-links (Fig. S1D). These results demonstrated that the Col1-HA LMW scaffold was suitable for supporting the in vitro culture of tumor cells. In previous studies, LMW HA was shown to play a role in tumor progression in a number of cancers [38-42]. Therefore, we selected the Col1-HA LMW scaffold as an appropriate biomimetic environment for culturing ES cells.
To bioengineer the most common ES tumor type, we cultured the SK-N-MC cell line (type 1 rearrangement) in Col1-HA LMW scaffolds. Mechanical properties of the TE-tumor did not change over time (Fig. 1B), and the model was stable over one week of culture. The proliferation of ES cells cultured within the TE-tumor model was slower than when the same cells were cultured in monolayer (Fig. 1C), consistent with the known lower rates of cell proliferation in native tumors compared to cancer cells cultured in monolayers [43]. Live/Dead analysis demonstrated uniform distribution of cells throughout the scaffolds at day 3 and day 7, and showed that most of the cells were viable after 7 days of culture (Fig. S2).
Notably, the levels of expression of CD99 in the TE tumor model were comparable to those measured in the samples of patients' tumors (Fig. 1D). These data show that cell culture on Col1/HA scaffolds does not modify the levels of this important membrane protein that is highly expressed in most cases of Ewing's sarcoma and maintains them at levels similar to those in tumors from patients. The cells cultured in the TE-tumor model formed small avascular aggregates that increased in size over time, mimicking the initiation of native tumor formation (Fig. 1 E, F).
Evaluation of the purity of exosomes preparations. In order to check the purity of the exosome preparations, we performed two sets of analysis consisting in protein composition and total RNA profiles [44, 45]. Toward this end, first we analyzed the levels of the CD81 (exosomal marker) and calnexin (only detectable in cellular and apoptotic bodies extracts), in monolayer and the TE tumor model at day 3 and day 7 (Fig. S3A). We also determined GAPDH levels to address the possibility of using GAPDH as a loading control of the technique. We confirmed the absence of calnexin in the extracellular preparations. This suggests that there is not cellular or apoptotic bodies contamination in the exosomes preparations. CD81 was detectable in exosomes preparations from cells in monolayer but not from TE-tumors preparations. GAPDH levels were similar between samples that points GAPDH as a good loading control.
Then, we further analyzed the quality of the exosomes isolation by analyzing RNA profiles from cells and exosomes preparations from cells in monolayer and TE-tumor at day 7, using the Bioanalyzer 2100 (Fig. S3B). As expected, electropherograms showed different RNA size distributions between samples. The RNA profile from cells revealed two dominant peaks, corresponding to the ribosomal RNA (rRNA) subunits 18S and 28S. Both peaks are also observed in RNA profiles from preparations of apoptotic bodies [46]. The RNA profile from extracellular vesicles lacked of both rRNA peaks and showed and enrichment in small RNAs, accordingly with the literature [46].
Exosome size. Using the Nanoparticle Tracking Analysis (NTA), we determined the size distributions of exosomes released into the culture media from the bioengineered tumor and from cell monolayers, and compared these to the size distributions of exosomes secreted into the blood plasma of ES patients. The sizes of exosomes isolated from human plasma (average mean ± SD: 88.7 ± 22 nm; average mode ± SD: 70.0 ± 20 nm, n=7 patients, Fig. 2A) were consistent with the previously reported data [2], and significantly smaller than the exosomes from monolayer cultures of ES cells (average mean ± SD: 149.2 ± 19 nm; average mode ± SD = 103.3 ± 23 nm, n=3, ∗∗p < 0.01; Fig. 2A). In addition, the numbers of particles per unit protein were not statistically different for cell monolayers and tissue engineered tumors (Fig. S4). Notably, the sizes of exosomes released from tumor models (average mean ± SD: 113.4 ± 10 nm, average mode ± SD: 76.7, ± 10.3 n = 6; Fig. 2A) were indistinguishable from those in the patients' plasma. These data suggest that the 3-dimensionality or composition of the scaffold (or both of these factors) regulate the exosomes to reach their native size. To distinguish the relative contributions of the matrix 3-dimensionality and composition, we investigated the sizes of exosomes in multiple model systems.
To evaluate the role of 3-dimensionality, we generated ES cell aggregates in a generic polypropylene context, in the range of sizes that we have observed for bioengineered tumors at day 7 (Fig. 2B). Neither the average mean nor the mode size of exosomes isolated from these aggregates recapitulated the values found in the patients' plasma (Fig. 2B). Mimicking the tumor size and morphology using 3D models without a biomimetic context was thus not sufficient to recapitulate the native exosome size.
To evaluate the role of matrix composition, we cultured ES cells in monolayers formed on polystyrene dishes coated with different extracellular matrix proteins (HA LMW, Col1, Col1-HA LMW, Fig. 2C). We could not observe any difference in the mean size or mode of exosomes secreted by the ES cells cultured on uncoated polystyrene dishes and on dishes coated with the proteins used for fabricating the scaffolds (Fig. 2D). These results indicate that mimicking the native matrix composition without providing the native stiffness and 3D context was also not sufficient for reproducing the native size of exosomes. Providing both the 3-dimensionality of cell culture and the composition of extracellular matrix found in ES was necessary for recapitulating the exosome size.
Ewing's sarcoma type 1 model in a 3-dimensional Collagen 1- Hyaluronic acid scaffold (A) Model systems used to isolate and analyze exosome size and cargo: plasma from patients and supernatants from cells cultured in vitro. (i) Hematoxylin and Eosin staining of Ewing's sarcoma (ii) cartoons representing the different sources of exosomes isolation from supernatants (cells on dish, cells on coated dish, aggregates and cells in scaffold) (B) Equilibrium modulus of Col1-HA LMW initial scaffolds (n=3) and cell-seeded scaffolds (n=3) at indicated time points. (C) Cell proliferation evaluated by changes in numbers of cells over time for SK-N-MC cells cultured in monolayers and in scaffolds. Statistical significance was determined by the two-tailed Student's t test at day 7; ***p < 0.001. (D) CD99 protein levels in SK-N-MC cells cultured in scaffold at day 3 (TE-tumor d3) and day 7 (TE-tumor d7) compared to cell monolayers (ML) and Ewing's sarcoma tumors (ES). (E) Hematoxylin and Eosin staining of bioengineered tumors (SK-N-MC cultured in Col1-HA scaffolds) at day 3 and day 7. (F) Formation of cell aggregates and cell-matrix integration in bioengineered tumors at day 7, by Hematoxylin and Eosin staining (left) and Scanning Electron Microscopy (SEM) (right).
Recapitulation of exosomes' size in the bioengineered tumors. (A) Exosome size analysis for SK-N-MC cells cultured in monolayers (n=3) and into Col1-HA scaffolds (n=6) at day 3 (ML d3 and SF d3, respectively), compared to the plasma from Ewing's sarcoma patients (n=7), by Nanoparticle Tracking Analysis (NTA, Nanosight instruments). (B) Distribution of exosome sizes in plasma of Ewing's sarcoma patients (n=7) and cell aggregates generated in polypropylene environment at day 7 (n=7), by Nanoparticle Tracking Analysis. Inset: Hematoxylin and Eosin staining of a representative SK-N-MC cell aggregate in polypropylene context, with a necrotic core, at day 7. (C) Bright field images of SK-N-MC cells cultured for 3 days in monolayers, on plates coated with Hyaluronic acid (+HA), Collagen 1 (+ Col-1), and Collagen 1-Hyaluronic acid (+Col1/HA). Representative images are shown (n=3 per condition). (D) Exosome size distribution for SK-N-MC cells cultured in monolayers HA-coated (n=3), Col-1-coated (n=3) and Col1-HA-coated (n=3) compared to SK-M-C cells cultured on uncoated culture plates (control; n=3) (E) Analysis of the effect of blebbistatin on cell aggregates in Col1-HA scaffolds (Hematoxylin and Eosin staining), and (F) on exosome's size, by NTA; (n=3 per condition).
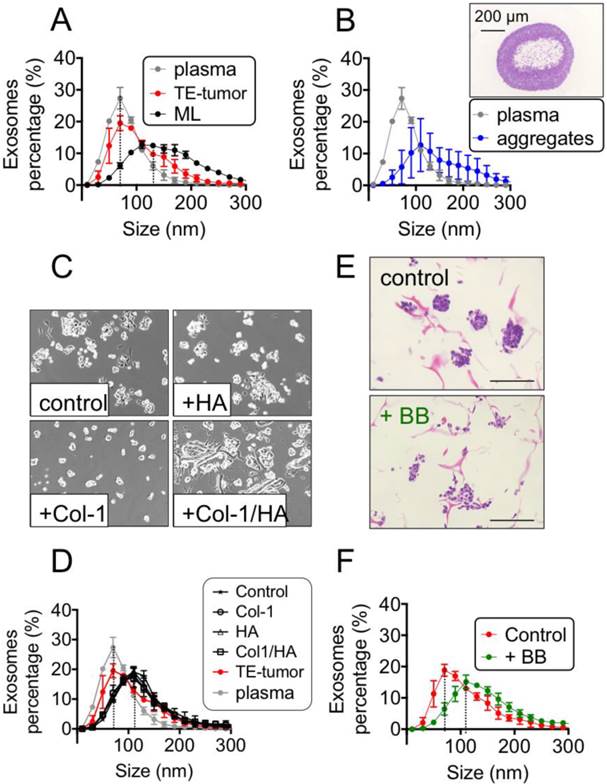
To probe a possible mechanism underlying the observed effects of the tumor environment on exosome size, we modified the tension forces within the cells. To this end, we maintained the 3-dimensionality, composition and stiffness of the microenvironment at levels comparable to the native tumor matrix, while eliminating tension-dependent changes in cell shape by using blebbistatin, a well-known selective inhibitor of non-muscle myosin II [47]. Cell morphology in blebbistatin-treated samples was different from untreated controls (Fig. 2E), with a partial disassembly of cell aggregates (Fig. 2E) and a shift of the exosome size distribution curve to higher values (Fig. 2F) when tensional forces within the cells were modified in a 3D setting.
Exosome cargo. Based on these findings, one could hypothesize that the exosome size is not the only property controlled by the microenvironment, and that their cargo is also a subject to regulation. To test this hypothesis, we analyzed the exosomal mRNA cargo and focused on EZH2, one of the most important mediators of Ewing's sarcoma tumor growth and progression.
First, we confirmed the overexpression of EZH2 in ES tumors at mRNA levels using the R2 Genomics Analysis and Visualization Platform (http://r2.amc.nl), by comparing the gene profiles for ES tumors (arrays from Francesconi n=37, and Delattre; n=117) and healthy tissues (array from Roth n=353) (Fig.3A). We also checked EZH2 overexpression in ES tumors by Immunohistochemistry (data not shown). Interestingly, the EZH2 protein was almost undetectable by Western blot in ES cells cultured in monolayers (Fig. 3B), which also expressed low levels of EZH2 mRNA by qRT-PCR (Fig. 3C). However, EZH2 mRNA and EZH2 protein increased in TE-tumors, both at the protein level (Fig. 3B) and at the mRNA level (Fig. 3C). These data supported the notion that a native-like environment can modulate cancer biology and mimic, at least in part, the properties of real tumors.
Effects of engineered microenvironment on exosome cargo. (A) EZH2 mRNA expression in Ewing's Sarcoma tumors and healthy tissues using publicly available expression array data; BoxBlot presentation using a comparative study of the amc onco-genomics software tool (www.amc.com). Ewing's sarcoma samples (ES) used in the analysis belong to Francesconi's array and to Delattre's array and Healthy set to the Roth's array. The numbers of samples in each cohort are indicated. (B) Protein levels of EZH2 in SK-N-MC cell monolayers (day 3) and bioengineered tumors (day 3 and day 7). (C) qRT-PCR of EZH2 expression indicates 2-fold increase of expression in bioengineered tumors over 7 days of cultivation (SF d7) as compared to cell monolayers (ML). Relative endogenous expression of EZH2 was normalized to actin; error bars represent standard deviation of relative expression. Statistical significance was determined by the two-tailed Student's t test. *p < 0.05; ns, not significant. (D) EZH2 mRNA in exosomes released from SK-N-MC cell monolayers and bioengineered tumors at days 3 and 7 (by qRT-PCR). The measured amounts of EZH2 mRNA were normalized to U6SnRNA; error bars represent standard deviation of relative expression. Statistical significance was determined by the two- tailed Student's t test. ∗∗p < 0.01 (E) Levels of EZH2 mRNA in exosomes isolated from blood plasma from Ewing's sarcoma type 1 patients, healthy donors, non-type 1 Ewing sarcoma patients, and an osteosarcoma patient.
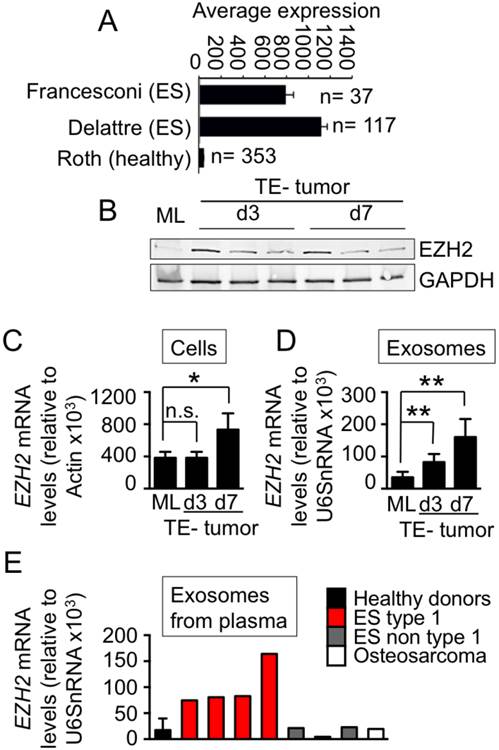
Then, we isolated exosomes released from the ES cells cultured in monolayers and bioengineered tumors. We found high levels of EZH2 mRNA in exosomes from TE-tumors, both at day 3 and day 7, when compared to monolayers (Fig. 3D). Importantly, the measured levels of EZH2 in bioengineered tumors corresponded to those in the blood plasma of ES patients. EZH2 mRNA was detected in exosomes from Ewing's sarcoma type-1 plasma (n=4), but not in plasma of healthy donors (n=4), non-type 1 patients (n=3) or an osteosarcoma patient (n=1) (Fig. 3E).
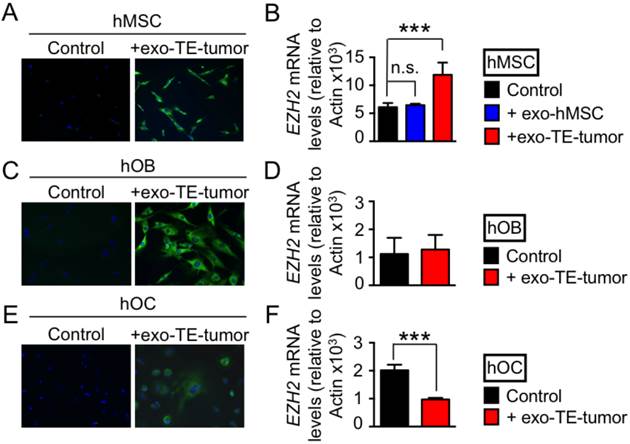
Transfer of exosome cargo. Because EZH2 induces an aberrant phenotype of Ewing's sarcoma in vivo and also affects the hMSCs cultured in vitro [28], we investigated whether the exosomes containing EZH2 mRNA can transfer their cargo to the cells hMSCs normally present in the bone niche. To this end, we labeled exosomes derived from the TE-tumor (Exo-TE-tumor) with the green RNA-selective nucleic acid stain SYTO RNASelect at day 7, the time point at which we observed high levels of EZH2 mRNA in these exosomes. The exosomes from the TE-tumors were taken up by bone marrow derived hMSCs, after 12 hours of incubation compared to the technical control (PBS treated with SYTO RNASelect) (Fig. 4A). Significant increases in EZH2 mRNA levels were detected in hMSC treated with exosomes from TE-tumors, when compared with untreated hMSCs or hMSCs treated with hMSC-derived exosomes (Fig. 4B).
Finally, we analyzed the effects of exosomes secreted by bioengineered tumors on human osteoblasts (hOB) and human osteoclasts (hOC). Labeled exosomes from TE-tumors were taken up by both hOB (4C) and hOC (4E). However, this uptake had no effect on EZH2 mRNA levels in hOB (4D), and resulted in down-regulation of EZH2 in hOC (4F). These data confirm that EZH2 mRNA-loaded exosomes can be transferred in vitro from cancer cells to cell populations from the bone niche, with different effects on hMSC (upregulation of EZH2), hOC (downregulation of EZH2) and hOB (no effect).
Discussion
We studied tumor-secreted exosomes, from two complementary views: effects of the tumor microenvironment on exosome size and cargo, and the ability of exosomes to transfer key regulators from their cargo to the surrounding non-cancer cells in the tumor niche. To this end, we report the first bioengineering study of tumor-secreted exosomes and show that these exosomes more closely match in vitro the size distribution and mRNA cargo found in the exosomes from blood plasma of tumor patients. This result was achieved using a tumor model, formed by culturing tumor cells in scaffolds designed to mimic the native tumor niche (Fig. 1A), and could not be achieved in tumor cell monolayers or tumor cell aggregates. The studies were done using Ewing's Sarcoma as a clinically relevant example of aggressive bone tumor characterized by expression of EWSR1-FLI1 fusion protein [48], which induces expression of EZH2 [28]. We therefore focused the studies of exosome cargo on EZH2 mRNA.
Exosome-mediated transfer of EZH2 mRNA. Confocal images of (A) human mesenchymal stem cells (hMSC), (B) human osteoblasts (hoB), and (C) human osteoclasts (hOC) after 2h of incubation with exosomes released from bioengineered tumors after 7 days of cultivation (+exo TE-tumor) labeled with SYTO RNA Select green fluorescent. PBS treated with SYTO RNA Select was used as technical control of the experiment. Data show representative images from 3 independent studies. EZH2 mRNA levels in (D) hMSC, (E) hOB and (F) hOC treated for 12 hours with exosomes released from bioengineered tumors after 7 days of cultivation (+exo TE-tumor). Statistical significance was determined by the two-tailed Student's t test. ∗∗∗p < 0.001; ns, not significant.
In recent years, major progress has been made with developing advanced 3D models that capture in vitro selected factors of the in vivo tumor microenvironment, with the aid of tissue engineering methods [14]. In line with the view that a bioengineered tumor should be a simplified version of reality tailored towards addressing a specific biological question, we aimed to engineer a “minimally functional unit” with capability to recapitulate the size and cargo of tumor exosomes.
First, we evaluated the individual and collective effects of the bioengineered tumor niche on the size of exosomes. In a multicellular organism, tissue cells are highly organized in a 3-dimensional fashion, and are surrounded by the extracellular matrix [13]. Tumors are also organ-like structures made up of many individual cells, where the 3-dimensionality and the extracellular matrix composition and stiffness play critical roles in tumor development [13, 49-51]. It is not surprising that multiple microenvironmental signals (including the 3-dimensionality of the extracellular matrix, matrix composition and stiffness, and the acting tensile forces) are important for recapitulating the size of exosomes found in native tumors. Neither 3-dimensionality alone (tumor aggregates) nor the composition alone (cell monolayers on polystyrene dishes coated with extracellular matrix components) recapitulated the size values found in plasma of ES patients. Interestingly, we also observed different expression levels in the conventional exosomal marker CD81 between cells in monolayer and in TE-tumor models. Interestingly, CD81 levels were decreased and were almost undetectable in the TE-tumor-derived exosomes. This phenomenon is also observed in Peinado et. al. [8] but with the exosomal HSC70 marker that fluctuates in levels between patients and stages. This suggests that patients conditions and in our case, culture conditions (3-dimensionality, hypoxia, microenvironment composition, stiffness…) may regulate the conventional or established markers for exosomes characterization.
In summary, these findings reinforce the idea that a simplified but realistic version of the tumor complexity, designed to address a specific question of interest, could in fact provide a platform for cancer research.
We found that EZH2 mRNA was present in exosomes secreted from TE-tumors at much higher levels than in exosomes secreted from tumor cells cultured in monolayers. Thus, our data suggest that ES tumors in vivo may also produce exosomes enriched in EZH2 mRNA. Importantly, we confirmed that EZH2 mRNA was only detectable in exosomes from plasma in Ewing's sarcoma type 1, and not in plasma of healthy donors or patients with other types of sarcoma tumors. Again, these findings support the utility of the TE tumor model for cancer research and suggest that exosomes should be studied in native-like experimental settings, rather than in cell monolayers or cell aggregates.
Our data are consistent with the known roles of exosomes as mediators of cancer progression. Transfer of paracrine signals to the healthy surrounding cells was shown to modulate the behavior of these cells and to create paths of low resistance for tumor invasion. Uptake of exosomes secreted by the TE tumor by the cells present in the bone niche resulted in different effects for different cell types with respect to the expression of EZH2 mRNA: up-regulation in hMSC, down-regulation in osteoclasts and no effect in osteoblasts.
MSC can be recruited in large numbers to the tumor stroma, to enhance tumor growth and metastasis [52]. A very interesting recent result is that the melanoma-derived exosomes favor the metastasis of primary tumors by modulating bone marrow progenitors [8]. On the other hand, MSC can be transformed by expression of EWSR1-FLI1 fusion protein [53], to create a permissive cellular environment for mutations and oncogene up-regulation, with significant molecular plasticity.[17, 53, 54]. A number of reports suggested that hMSCs are the cells of origin that give rise to Ewing's sarcoma [20]. In our system, hMSCs displayed increased levels of EZH2 mRNA after treatment with exosomes secreted from TE-tumors. Based on these results, we postulate that tumor cells may induce epigenetic changes in hMSCs from the stromal compartment, by releasing EZH2 mRNA from their exosomes. Epigenetic changes such as gene promoter methylation in stromal cells have been associated with malignancy. However, histone modifications (i.e., H3K27me3 established by EZH2) and chromatin remodeling in the cells comprising tumor microenvironment remain largely unknown and will require further studies.
Interestingly, tumor-secreted exosomes down-regulated EZH2 mRNA in the bone osteoclasts, an effect not previously documented. However, a correlation was recently established between EZH2 levels and the expression of the microRNA-34a[55], a microRNA involved in suppressing osteoclastogenesis, bone resorption and bone metastasis [56]. The mechanism for the observed downregulation of EZH2 mRNA in healthy osteoclasts could thus involve miR34a, with direct effects on bone resorption and tumor progression. However, systematic studies will be needed to determine if there is a link between EZH2 and osteoclast homeostasis.
This study also has limitations. For example, we report that there were no changes in EZH2 levels in hOB after these cells were treated with exosomes secreted from TE-tumors. However, the studies of EZH2 mRNA transfer were performed in monolayer cultures, which do not provide a proper context of the bone niche. At this point, one cannot exclude additional or alternate effects of EZH2 mRNA on the cells from the bone niche within the Ewing's sarcoma tumor context. Further studies using models of the ES tumor and the surrounding bone may be necessary to unravel the direct role of EZH2 in bone homeostasis.
In summary, we developed a bioengineered tumor model for broad use in cancer research, using Ewing sarcoma as a clinically relevant example. Unlike cell monolayers and aggregates, this model recapitulates the size and cargo of exosomes found in patients' blood plasma. We show that the microenvironmental signals (including the 3-dimensionality, composition, and stiffness of the tumor matrix) are necessary for recapitulating the properties of exosomes found in native tumors. Using this model, we investigated the effects of the tumor microenvironment on exosome size and cargo, the role of physical signals, and the ability of exosomes to transfer key regulators to the surrounding non-cancer cells of the bone niche (bone marrow mesenchymal stem cells, osteoblasts, osteoclasts). We suggest that the proposed bioengineered model could serve as an effective tool in studies of human tumor exosomes.
Supplementary Material
Supplementary figures.
Acknowledgements
The authors gratefully acknowledge NIH funding of this work (EB002520, EB 17103), and expert help of Dr. A. Saxena (NTA, exosome isolation, RNA extraction), Molecular Pathology Tumor Bank (tumor samples) Histology Facility at Columbia University (histological services) and Dr. Tao Sue at Columbia University (MPSR Molecular Biology service) for performing Agilent Bioanalyzer analyses.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Thery C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nature reviews Immunology. 2002;2:569-79
2. Caby M-P, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879-87
3. Nawaz M, Camussi G, Valadi H, Nazarenko I, Ekstrom K, Wang X. et al. The emerging role of extracellular vesicles as biomarkers for urogenital cancers. Nat Rev Urol. 2014;11:688-701
4. Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa-Silva B. et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766-9
5. Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J. et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177-82
6. Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G. et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113:275-81
7. Braicu C, Tomuleasa C, Monroig P, Cucuianu A, Berindan-Neagoe I, Calin GA. Exosomes as divine messengers: are they the Hermes of modern molecular oncology? Cell Death Differ. 2015;22:34-45
8. Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G. et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883-91
9. Costa-Silva B, Aiello NM, Ocean AJ, Singh S, Zhang H, Thakur BK. et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17:816-26
10. Kovar H. Context matters: the hen or egg problem in Ewing's sarcoma. Semin Cancer Biol. 2005;15:189-96
11. Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A. et al. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241-54
12. Diz-Munoz A, Fletcher DA, Weiner OD. Use the force: membrane tension as an organizer of cell shape and motility. Trends Cell Biol. 2013;23:47-53
13. Bissell MJ, Hines WC. Why don't we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat Med. 2011;17:320-9
14. Burdett E, Kasper FK, Mikos AG, Ludwig JA. Engineering tumors: a tissue engineering perspective in cancer biology. Tissue Eng Part B Rev. 2010;16:351-9
15. Schuessler TK, Chan XY, Chen HJ, Ji K, Park KM, Roshan-Ghias A. et al. Biomimetic Tissue Engineered Systems for Advancing Cancer Research: NCI Strategic Workshop Report. Cancer Res. 2014
16. Arndt CA, Crist WM. Common musculoskeletal tumors of childhood and adolescence. N Engl J Med. 1999;341:342-52
17. Toomey EC, Schiffman JD, Lessnick SL. Recent advances in the molecular pathogenesis of Ewing's sarcoma. Oncogene. 2010;29:4504-16
18. Lewis TB, Coffin CM, Bernard PS. Differentiating Ewing's sarcoma from other round blue cell tumors using a RT-PCR translocation panel on formalin-fixed paraffin-embedded tissues. Mod Pathol. 2007;20:397-404
19. Braunreiter CL, Hancock JD, Coffin CM, Boucher KM, Lessnick SL. Expression of EWS-ETS fusions in NIH3T3 cells reveals significant differences to Ewing's sarcoma. Cell Cycle. 2006;5:2753-9
20. Potikyan G, France KA, Carlson MRJ, Dong J, Nelson SF, Denny CT. Genetically defined EWS/FLI1 model system suggests mesenchymal origin of Ewing's family tumors. Lab Invest. 2008;88:1291-302
21. Riggi N, Suva ML, Suva D, Cironi L, Provero P, Tercier S. et al. EWS-FLI-1 expression triggers a Ewing's sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res. 2008;68:2176-85
22. von Levetzow C, Jiang X, Gwye Y, von Levetzow G, Hung L, Cooper A. et al. Modeling initiation of Ewing sarcoma in human neural crest cells. PLoS One. 2011;6:e19305
23. Miller IV, Raposo G, Welsch U, Prazeres da Costa O, Thiel U, Lebar M. et al. First identification of Ewing's sarcoma-derived extracellular vesicles and exploration of their biological and potential diagnostic implications. Biol Cell. 2013;105:289-303
24. Tsugita M, Yamada N, Noguchi S, Yamada K, Moritake H, Shimizu K. et al. Ewing sarcoma cells secrete EWS/Fli-1 fusion mRNA via microvesicles. PLoS One. 2013;8:e77416
25. Hu-Lieskovan S, Zhang J, Wu L, Shimada H, Schofield DE, Triche TJ. EWS-FLI1 fusion protein up-regulates critical genes in neural crest development and is responsible for the observed phenotype of Ewing's family of tumors. Cancer Res. 2005;65:4633-44
26. Zwerner JP, Joo J, Warner KL, Christensen L, Hu-Lieskovan S, Triche TJ. et al. The EWS/FLI1 oncogenic transcription factor deregulates GLI1. Oncogene. 2008;27:3282-91
27. Smith R, Owen LA, Trem DJ, Wong JS, Whangbo JS, Golub TR. et al. Expression profiling of EWS/FLI identifies NKX2.2 as a critical target gene in Ewing's sarcoma. Cancer Cell. 2006;9:405-16
28. Richter GHS, Plehm S, Fasan A, Rossler S, Unland R, Bennani-Baiti IM. et al. EZH2 is a mediator of EWS/FLI1 driven tumor growth and metastasis blocking endothelial and neuro-ectodermal differentiation. Proc Natl Acad Sci U S A. 2009;106:5324-9
29. Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C. et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525-30
30. Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM. et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301-13
31. Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006;16:455-62
32. Villasante A, Piazzolla D, Li H, Gomez-Lopez G, Djabali M, Serrano M. Epigenetic regulation of Nanog expression by Ezh2 in pluripotent stem cells. Cell Cycle. 2011;10:1488-98
33. Chase A, Cross NC. Aberrations of EZH2 in cancer. Clin Cancer Res. 2011;17:2613-8
34. McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108-12
35. Sellers WR, Loda M. The EZH2 polycomb transcriptional repressor-a marker or mover of metastatic prostate cancer? Cancer Cell. 2002;2:349-50
36. Davidenko N, Campbell JJ, Thian ES, Watson CJ, Cameron RE. Collagen-hyaluronic acid scaffolds for adipose tissue engineering. Acta Biomater. 2010;6:3957-68
37. Bhumiratana S, Grayson WL, Castaneda A, Rockwood DN, Gil ES, Kaplan DL. et al. Nucleation and growth of mineralized bone matrix on silk-hydroxyapatite composite scaffolds. Biomaterials. 2011;32:2812-20
38. Kumar S, West DC, Ponting JM, Gattamaneni HR. Sera of children with renal tumours contain low-molecular-mass hyaluronic acid. Int J Cancer. 1989;44:445-8
39. Voelcker V, Gebhardt C, Averbeck M, Saalbach A, Wolf V, Weih F. et al. Hyaluronan fragments induce cytokine and metalloprotease upregulation in human melanoma cells in part by signalling via TLR4. Exp Dermatol. 2008;17:100-7
40. Kouvidi K, Berdiaki A, Nikitovic D, Katonis P, Afratis N, Hascall VC. et al. Role of receptor for hyaluronic acid-mediated motility (RHAMM) in low molecular weight hyaluronan (LMWHA)-mediated fibrosarcoma cell adhesion. The Journal of biological chemistry. 2011;286:38509-20
41. Wu M, Cao M, He Y, Liu Y, Yang C, Du Y. et al. A novel role of low molecular weight hyaluronan in breast cancer metastasis. Faseb J. 2015;29:1290-8
42. Wu M, Du Y, Liu Y, He Y, Yang C, Wang W. et al. Low molecular weight hyaluronan induces lymphangiogenesis through LYVE-1-mediated signaling pathways. PLoS One. 2014;9:e92857
43. Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, Polverini PJ. et al. Engineering tumors with 3D scaffolds. Nat Methods. 2007;4:855-60
44. Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C. et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3:26913
45. Lasser C, Eldh M, Lotvall J. Isolation and characterization of RNA-containing exosomes. J Vis Exp. 2012:e3037
46. Crescitelli R, Lasser C, Szabo TG, Kittel A, Eldh M, Dianzani I. et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. 2013:2
47. Mammoto T, Ingber DE. Mechanical control of tissue and organ development. Development. 2010;137:1407-20
48. Riggi N, Stamenkovic I. The Biology of Ewing sarcoma. Cancer Lett. 2007;254:1-10
49. Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell. 2010;18:884-901
50. Vidi PA, Bissell MJ, Lelievre SA. Three-dimensional culture of human breast epithelial cells: the how and the why. Methods Mol Biol. 2013;945:193-219
51. Baker EL, Bonnecaze RT, Zaman MH. Extracellular matrix stiffness and architecture govern intracellular rheology in cancer. Biophys J. 2009;97:1013-21
52. Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW. et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557-63
53. Riggi N, Suva M-L, Suva D, Cironi L, Provero P, Tercier S. et al. EWS-FLI-1 expression triggers a Ewing's sarcoma initiation program in primary human mesenchymal stem cells. Cancer Res. 2008;68:2176-85
54. Galie M, Konstantinidou G, Peroni D, Scambi I, Marchini C, Lisi V. et al. Mesenchymal stem cells share molecular signature with mesenchymal tumor cells and favor early tumor growth in syngeneic mice. Oncogene. 2008;27:2542-51
55. Chen SLaY. Enhancer of zeste homolog 2 couples with HOTAIR to inhibit tumor suppressor miR-34a in human pancreatic ductal adenocarcinoma. Proceedings of the 106th Annual Meeting of the American Association for Cancer Research; Philadelphia, PA. 2015:18-22
56. Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang T-C. et al. miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature. 2014;512:431-5
Author contact
![]() Corresponding author: Gordana Vunjak-Novakovic, Mikati Foundation Professor of Biomedical Engineering, Professor of Medical Sciences, Columbia University, 622 west 168th Street, VC12-234 New York NY 10032. Tel: 1-212-305-2304, fax: 1-212-305-4692 gv2131edu.
Corresponding author: Gordana Vunjak-Novakovic, Mikati Foundation Professor of Biomedical Engineering, Professor of Medical Sciences, Columbia University, 622 west 168th Street, VC12-234 New York NY 10032. Tel: 1-212-305-2304, fax: 1-212-305-4692 gv2131edu.
 Global reach, higher impact
Global reach, higher impact