13.3
Impact Factor
Theranostics 2013; 3(12):943-952. doi:10.7150/thno.7445 This issue Cite
Review
Micro-RNAs as Potential New Molecular Biomarkers in Oncology: Have They Reached Relevance for the Clinical Imaging Sciences?
Department for Clinical Radiology, Ludwig-Maximilians-University, Campus Innenstadt, Nussbaumstrasse 20, 80336 Munich, Germany
Received 2013-8-18; Accepted 2013-10-21; Published 2013-11-30
Abstract
Minimally invasive biomarkers for early cancer detection and monitoring of personalized therapies are of high importance to further improve prognosis in oncological disease. MicroRNAs (miRNAs) are small regulatory RNAs in humans and play a key role in carcinogenesis. In recent years they have emerged as promising biomarkers in oncology. miRNA profiling has demonstrated its capacity for sub-classifying tumors and monitoring of therapeutic effects. Different expression profiles of miRNAs in cancer and the stability of circulating miRNAs potentially provide a clinically accessible molecular monitoring tool of malignant tissues and its response to therapies. Clinical imaging including the modalities PET/CT and MRI is well established for characterizing tumor tissue and sub-classifying morphological, metabolic or vascular treatment response in cancer. Sophisticated clinical imaging biomarkers for cancer detection and monitoring should now been correlatively applied to further validate the potential of miRNAs as oncologic biomarkers for the clinic.
Keywords: MicroRNA, Treatment Monitoring, Personalized Therapy, Cancer Imaging, Biomarker
Introduction
miRNAs are belonging to the key gene regulators of cells in humans. They play a significant role in directing mRNA and subsequent protein expression both in physiological tissues, and in cancer. It is therefore not surprising that they rise -since discovery of their function- more and more attention with respect to their potential as biomarkers in oncology.
In the last few years, much research has been focussed on addressing miRNA-expression profiling of primary tumors that contribute significantly to the understanding of tumor pathophysiology.
However, both from a basic science and clinical view, repetitive miRNA analysis from peripheral blood would have several advantages and is certainly more broadly clinically transferable. Circulating tumor cells (CTC) in the peripheral blood can provide significant information with respect to treatment effects and prognosis. Analysis of CTC for their possible molecular profile including miRNA expression will further expand the utility of CTC in clinical assessment of tumor stage, metastatic status, and even response to therapy.
Besides these CTC associated miRNAs, free circulating miRNAs have also been discovered in peripheral whole blood, plasma and serum and can be used for tumor characterization.
Analysis of miRNAs in peripheral blood, either cell associated or cell free, holds great potential for longitudinal, repetitive, non-invasive and real-time molecular based monitoring of treatment effects in oncology.
This article mainly reviews results of miRNA expression profiling in primary tumors and in peripheral blood with special emphasis on their potential as biomarkers in oncology. Subsequently we will discuss aspects about how correlative clinical biomedical imaging including the modalities PET/CT and MRI with its well established tools for cancer detection and sub-classification of response to personalized treatment potentially can contribute to further validation and an accelerated clinical transfer of miRNAs as new molecular oncologic biomarkers.
miRNAs
MiRNAs were first discovered in 1993. They are small single stranded RNA (ssRNA) molecules of 21-23 nucleotides in length, and it was long speculated that they are just degraded RNA fragments or non-translated small RNAs [1]. High miRNA expression in a wide range of pathologies and their significance in carcinogenesis has stimulated researchers in oncology to focus on miRNA profiling in different tumor types [1]. One key advantage with respect to their potential role as biomarkers is their stability. Cell free miRNAs in body fluids are stable even after repeated freeze-thaw cycles and prolonged storage [2]. They are also well-preserved in formalin-or paraffin fixed tissue over several years which opens up enormous possibilities for retrospective molecular analysis in large sample size studies [2].
Evaluation of miRNA expression allows assessment of molecular tumor features, which might lay the ground for rationale, on continuous repetitive molecular profiling based optimization of diagnostic and therapeutic strategies in oncology.
MiRNA-biogenesis and function
The biogenesis of miRNAs is a complex multi-step process: In the nucleus, they are first transcribed by RNA polymerase II into large RNA precursors (pri-miRNAs). These pri-miRNAs are subsequently cleaved into precursor miRNAs (pre-miRNAs), which are ~70-90 nucleotides in length, and have an imperfect stem loop hairpin structure. They are then transported into the cytoplasm by exportin-5, where the hairpin precursors are cleaved resulting in a small dsRNA duplex that contains both the mature miRNA strand and its complementary strand [3].
The mature miRNA strand is incorporated into a RNA-induced silencing complex (RISC), which inhibits the function of its target mRNA through degradation or, most commonly, by translational repression after binding of the RISC to the target mRNA, and in very few cases increase the expression by stabilizing the target mRNA. MiRNAs are estimated to regulate with below further detailed processes up to 30% of all human transcripts [3].
MiRNAs regulate post-transcriptional gene expression in a sequence-specific manner, recognizing their mRNA with the 5' end of the mature miRNA strand, which is called the 'seed sequence' [3]. After recognition of the target mRNA, regulation of gene expression can occur through two mechanisms:
- In case of perfect homology between miRNA and the mRNA, cleavage of the mRNA by argonaute, which is present in the RISC complex, is induced.
- In case of imperfect binding to partially complementary sequences in the 3'-untranslated region of the mRNA (which is more frequent than perfect binding), the target mRNA is regulated by repression of protein translation [3].
Proteins are therefore regulated by miRNAs without significantly affecting their respective mRNAs expression levels [3], [4].
MiRNAs and Cancer
MiRNAs are significant players in tumorigenesis and basically every step of carcinogenesis, including invasion, metastasis, apoptosis and drug resistance [5]. In cancer, pathologically altered miRNA regulation can be due to mutations affecting miRNA coding loci, epigenetic alterations, or chromosomal aberrations. All of these pathologic variations of miRNA biogenesis potentially affect miRNA expression. Depending on their downstream target genes, miRNAs can act as tumor suppressors or oncogenes, and therefore miRNA expression in tumors can be either up-or downregulated [6].
Reflecting the central role of miRNAs in tumor biology, there is a wide range of possible applications, from evaluating tumor biology, diagnosis, prognosis, prediction, serving as pharmacodynamic markers, and/or even as potential drug targets. The preserved stability of the miRNAs both in the primary tumor and in circulation, make them attractive possible biomarkers even for repetitive non-invasive longitudinal monitoring for a wide array of cancer types [7].
Table 1 shows a selection of circulating altered miRNAs in various cancer types with possible clinical applications.
The first described miRNAs in pathologically altered tissue were miR15a and miR-16-1, in chronic lymphocytic leukaemia, both target Bcl-2. Their downregulation inhibits apoptosis.
MiR-21 targets PTEN and pro-apoptotic genes, and is one of the most commonly overexpressed miRNAs in cancer [8]. This pathologic molecular alteration in miRNA expression leads to enhance survival signals within the tumor.
Altered cell-free circulating miRNAs in various cancer types (selection)
| MicroRNA | Type of deregulation | Possible application |
|---|---|---|
| let-7a | Decrease in gastric cancer | Discriminate gastric cancer from healthy controls |
| let-7f | Decrease in NSCLC | Associated with overall survival in NSCLC |
| miR-1 | Decrease in NSCLC | Associated with overall survival in NSCLC |
| miR-10b | Increase in breast cancer | Associated with metastases in breast cancer |
| miR-17 | Increase in gastric cancer | Discriminate gastric cancer from healthy controls |
| miRs-17+106a | Increase in gastric cancer | Discriminate gastric cancer from healthy controls |
| miR-17-3p | Increase in CRC | Discriminate CRC from healthy controls |
| miR-17-5p | Increase in gastric cancer | Discriminate gastric cancer from healthy control |
| miR-20b | Decrease in NSCLC | Associated with advanced stages and lymph node metastases in NSCLC |
| miR-21 | Increase in CLL harbouring 17p deletion | Associated with overall survival in CLL |
| miRs-21+126+210+486+5p | Deregulate in NSCLC | Discriminate stage/NSCLC from healthy controls |
| miRs 21+155+196a+210 | Increase in pancreatic adenocarcinoma | Discriminate pancreatic adenocarcinoma from healthy controls |
| miR-29a | Increase in CRC | Discriminate CRC from healthy controls |
| miR-29b, miR-29c | Decrease in CLL harbouring 17p deletion | Associated with progression in CLL |
| miR-30d | Increase in NSCLC | Associated with overall survival inNSCLC |
| miR-30e-3p | Decrease in NSCLC | Associated with short disease free survival in NSCLC |
| miR-34a | Increase in breast cancer | Discriminate advanced stages from early stages in breast cancer |
| miR-92, miR-92a | Increase in CRC | Discriminate CRC from Gastric Cancer, IBD, and healthy controls |
| miR-106a | Increase in gastric cancer | Discriminate gastric cancer from healthy controls |
| miR-106b | Increase in gastric cancer | Discriminate gastric cancer from healthy controls |
| miR-144 | Decrease in CN-AML | Associated with adverse prognostic marker FLT3-ITD in CN-AML |
| miR-181a | Increase in CLL harboring trisomy 12 | Associated with progression in CLL |
| miR-195 | Increase in breast cancer | Discriminate breast cancer from other cancers and from healthy controls |
| miR-200a, miR-200b | Increase in pancreatic cancer | Discriminate pancreatic cancer from healthy controls |
| miR-451 | Increase in breast cancer | Associated with malignancy in breast cancermiR |
| miR-486 | Increase in NSCLC | Associated with overall survival in NSCLC |
| miR-499 | Decrease in NSCLC | Associated with overall survival in NSCLC |
A pilot study in breast cancer patients demonstrated, that the relative concentration of miR-155 in serum significantly discriminated primary breast cancer patients from healthy women [9].
Such results prompted a systematic and comprehensive search for tumor-specific miRNA expression profiles and possible applications of miRNAs in oncology.
In this review, we want to focus on the potential biomarker-application of miRNAs, both in the setting of primary tumor analysis, and miRNA expression profiling in the peripheral blood. The biomarker field might hold the greatest potential for an overlap of miRNA research with biomedical imaging applications.
MiRNAs as biomarkers in solid cancers
What is a biomarker?
Biomarkers define disease states. In cancer, biomarkers can subtype tumors and contribute in monitoring of therapeutic interventions [10]. Useful biomarkers can contribute in dismantling pathways of pathogenesis and are especially in need for development and optimization of new and improved personalized molecular therapies.
Clinically well-established laboratory tumor markers
Clinically well-established laboratory tumor-markers so far include, among others, thyroglobulin, prostate-specific antigen (PSA), carcinoembryonic antigen (CEA), CA125 and alpha-fetoprotein [11].
Potential role of miRNAs
Not yet clinically established, but recently discovered and investigated is the potential role of mRNA and miRNAs as biomarkers in oncology [12].
Alterations of miRNA expression are not exceptional but rather the rule in human cancer. As an attempt to establish whether miRNA could be used for tumor classification, diagnosis and prognosis, different platforms to assess the global expression of miRNA genes in normal and diseased tissue were developed [12]. After an extensive use of custom-made and then commercial miRNA microarrays, the next step was the generation of large-scale profiling methods utilizing high throughput deep sequencing [13]. Genome-wide profiling showed that miRNA expression signatures allowed different types of cancer to be discriminated with high accuracy [14] and the tissue of origin of poorly differentiated tumors to be identified. By contrast, mRNA profiles were highly inaccurate indicators of tissue or cancer type. Indeed MiRNA expression profiling has proven to be helpful in early disease detection, distinguishing different cancer types, such as sub-classification of breast cancer subtypes and prostate cancer subtypes, identifying the tissue of origin in cancer with unknown primary and contributing to the postoperative adjuvant situation. Predictive miRNA signatures have been established for a number of tumors, including lung, prostate, cervical and colon cancers [12]. One study, for example, developed a classifier of 48 miRNAs from a sample of 336 primary and metastatic tumors, and was able to use this classifier to accurately predict the tissue origin in 86% of a blind test set, including 77% of the metastatic tumors [31]. Given that cancers of undefined origin account for approximately 4% of all malignancies and are associated with a poorer prognosis, the continued development of miRNA classifiers has foreseeable benefits for clinical management.
miRNA profiles can distinguish not only between normal and cancerous tissue and identify tissue of origin, but can also discriminate different subtypes of a particular cancer, or even specific histopathological abnormalities. Gene expression profiling has already demonstrated its effectiveness at subtyping various cancers: miRNAs for example, are differently expressed between basal and luminal breast cancer subtypes [15], and can specifically classify estrogen receptor, progesterone receptor and HER2/neu receptor status [16]. Other examples are the differential expression of miRNAs according to specific histotypes of ovarian carcinoma [17] and the ability of miR-205 expression to discriminate squamous from non-squamous non-small cell lung carcinoma [18]. Deep sequencing based screening methods revealed the differential expression of miRNAs in ovarian cancer [19], and in favourable versus unfavourable neuroblastoma [20]. Challenges in this field remain, e.g. time of collection and processing the miRNAs could impact miRNA levels due to the dynamic and immediate regulation of miRNA in stress response and hypoxia or other factors. Although more comprehensive validation needs to be done to address inconsistencies between different studies, which might be based on such challenges, this is unlikely to represent an obstacle to the development of miRNA in diagnostics.
Another major clinical issue is clearly represented by the need of biomarkers for an early diagnosis, extremely important considering that prognosis of patients is closely linked to the stage of the tumor at the time of detection. MiRNAs have great potential as early diagnostic biomarkers in oncology: overexpression of miR-205 and miR-21 in ductal adenocarcinoma as an example are reported to precede morphologic changes of the ducts, thus suggesting the possibility for an early detection of this neoplasm [21]. By analysing plasma samples of lung cancer patients collected 1-2 years before the onset of therapy, miRNA signatures were found with strong predicitive diagnostic and prognostic potentials [22].
Although predicting survival might be important in a more general sense, the prediction of response to a specific therapy is of far greater clinical value. It is very interesting to note that miRNAs have potential not only for longitudinal monitoring during treatment in general, but more specifically also for contributing to rationally select patients for specific subtypes of targeted therapies [12]. For instance, low levels of miR-26 are an independent indicator of poor survival in patients suffering from hepatocellular carcinoma (HCC); however, patients with low miR-26 responded well to interferon-alpha treatment, resulting in improved survival [23]. Therefore miR-26 expression might be a useful biomarker to discriminate patients who could benefit most from an interferon-alpha therapy. Other miRNAs have been correlated with a poor response to specific treatments. In various cancers increased miR-21 expression is an indicator of poor outcome [24] and is also sufficient to predict poor response to adjuvant chemotherapy in adenocarcinomas. High levels of miR-125b in breast cancer predict poor response to taxol-based treatments in vitro [25], and a similar finding has been reported for miR-21 in pancreatic cancer patients treated with gemcitabine [26].
From many more possible applications of miRNA profiling in oncology, we are now summarizing a selection of interesting biomarker-related work - now separated for analysis of miRNAs from primary cancer tissue, and second from miRNA profiling in the peripheral blood.
miRNAs in primary cancer analysis
Stability of miRNA in tissue samples of primary tumors
When considering the analysis of miRNAs from primary tumor samples the important question arises, how fast we need to get access to the samples, what type of processing needs to be done to maintain the miRNA stable, and how feasible the pre-processings in a post operative or in a biopsy based sampling settings are.
FFPE (formalin-fixed paraffin embedded) sampling is a very well established standard histopathology processing procedure for tissue material, usually gained during invasive diagnostic procedures (endoscopic or CT guided biopsies etc.) or surgery. Transcript mRNA profiling is challenging utilizing FFPE samples; main hurdles are degradation due to formalin fixation and deterioration of the mRNA over time from the nucleases [27]. However, FFPE material allows extensive discovery, exploration and validation of potential molecular biomarkers utilizing miRNAs [28]. MiRNAs have the substantial advantage that they are essentially unaffected by the fixation procedure, demonstrate remarkable stability and can be readily extracted from the samples due to their small size. This offers great opportunities to evaluate expression profiles of miRNAs and investigation of their potentially usefulness as biomarkers without much hurdles in getting and handling the samples.
Selected results of miRNA profiling of primary tumors
MiR-21 is a well-established miRNA aberrant in human cancer. It is altered in a wide variety of cancers. Its over-expression commonly induces increased cell proliferation, migration, invasion and survival, its suppression results in induced apoptosis, and a decline of cell proliferation and invasion. MiR-21 overexpression is also reported to be responsible for drug resistance in human cancers [29].
MiR-155 [30] is another commonly and well established dysregulated (in majority of tumors over-expressed) miRNA in a wide variety of tumors.
Besides these two well-established miRNAs, there are numerous alternate dysregulated miRNAs as potential markers under evaluation; challenges in the laborious process to establish markers include limitations in the statistical design of studies, but also individual tumor biologies, population based variation, and still more work is needed to fully establish optimal marker (or marker combinations) of characteristic miRNA profiles within tumor types without much variation between ethnic sub-groups.
Possible applications of miRNA profiling of primary tumors include:
Cancer of unknown origin
miRNA expression profiling can help to clarify the tissue of origin in cases of cancer with unknown primary origin, which is a common clinical problem. This profiling could thus contribute to a more valid selection of the appropriate chemotherapy in such patients. Rosenfeld [31] and Rosenwald [32] reached an overall sensitivity of 72% and specificity of 99% compared to an independent test set in a collective with unknown primaries. They applied a microarray-based classifier utilizing 48 miRNAs, which were selected from analysis of 205 primary tumors and 131 metastases of 22 different tumor origins.
An additional study demonstrated successful classification of poorly differentiated primary tumors utilizing miRNA expression profiling. Interestingly, mRNA profiles analysed in these samples were not helpful for the tumor classification [33].
Sub-classification of cancers
Breast cancer demonstrates a wide variety of histopathologic subtypes. Sempere at al [34] was able to subclassify formalin-fixed breast cancer tissue samples utilizing in situ hybridization to analyse spatial miRNA expression profiles in the tissues.
Prognosis
Prognostic miRNAs would be helpful for optimized individual treatment planning. The previously mentioned MiR-21 is a well-established miRNA aberrant in human cancer and its overexpression reduces prognosis. In the meantime, many prognostic miRNAs were identified. Applications of miRNAs with respect to prognosis are reviewed in detail in reference [35].
Prediction of outcome
Studies with the aim to predict response to a specific therapy are highly interesting, but difficult to design, so the number of convincingly validated specific miRNAs is limited. We are briefly summarizing some key results. In a cell culture study in ovarian cancer, miR-214 was shown to be involved in resistance to cisplatin, through targeting of PTEN [36].
In a very interesting clinical study in 58 patients with non-small cell lung cancer, EGFR mutations and loss of miR-128b in patient samples were associated with improved response to gefitinib, while EGFR expression did not correlate with gefitinib response. miR-128b was chosen based on its regulatory role for EGFR and the fact that loss of chromosome 3p, where miR-128b is located, is one of the most frequent events in lung carcinogenesis [37].
In a study utilizing 246 oestrogen receptor positive primary breast tumors, higher expression of miR-30c was associated with benefit from first line tamoxifen monotherapy and longer progression-free survival [38].
These are very interesting results in various biomarker-settings, however, on the downside all these approaches require prior acquiration of tissue material of the tumor, which usually demands invasive procedures. Moreover, it has to be taken into account that expression profiling in such settings reflect only local tumor situations.
MiRNAs in peripheral blood samples as biomarkers
Over the last couple of years, there is rising interest in the investigation of miRNAs as non-invasive biomarkers in circulating blood. Despite the successes in miRNAs profiling of tumor samples, there are numerous clinical situations in which it is beneficial to gain information about miRNAs from the peripheral blood [39]:
- Measurements require no significant interventional procedure and are reflecting the systemic distribution of the targeted molecular marker, which both is potentially helpful for intended longitudinal monitoring of individualized therapies.
- MiRNA expression of tumor tissue might change during time and after therapy cycles, so continuous follow-up monitoring of blood derived miRNAs (from circulating cancer cells or free miRNAs) might better allow a real time assessment of the molecular situation of the tumor and might be superior for continuous treatment monitoring and outcome evaluation [39].
Another emerging role for repetitively measurable circulating miRNAs might be especially to contribute information for predicting therapeutic response in targeted therapy such as EGFR inhibitors.
The role of KRAS and EGFR expression in this example of targeted therapy has been extensively investigated, but given their central role in tumor progression, it is to be expected that miRNAs might be a future key biomarker for such molecular targeted approaches. Second or third line targeted therapies are available, and it is very well conceivable, that a treatment adaption based on circulating tumor characteristics might be more valid than treatment based on tissue characteristics of an circumscribed and only limited area of the primary tumor [40].
Rapidly adapting a therapy once resistance occurs would be a very attractive future application for molecular profiling of circulating miRNAs. But where do we stand today?
CTCs have already been characterized for the presence of gene amplification, expression of proteins, several mRNA and also miRNA expressions.
MiRNAs have been detected within plasma or serum as free miRNAs or contained within microvesicles such as exosomes, which can also carry functional mRNA.
The observation of elevated levels of miR-155, miR-210 and miR-21 in patient's sera was first described in B-cell lymphoma, with miR-21 associated with relapse-free survival [41].
In solid cancer, Mitchell [39] first identified tumor-related miRNAs in plasma samples, suggesting that amount of miRNAs in the plasma might reflect tumor burden.
Over the past few years, key applications of miRNAs profiling in peripheral blood has emerged and include:
Early detection - discrimination from healthy controls or discrimination from other diseases
Shen et al [42] demonstrated altered expression of miRNAs in plasma in NSCLC at an early stage. He utilized qRT-PCR and a combination of 4 serum miRNAs to distinguish the lung cancer patients from healthy controls (86% sensitivity, 97% specificity). Such approaches might be helpful for early diagnosis of disease or even screening in high-risk collectives.
Plasma investigations in colorectal cancer (CRC) indicated that miR-29a and miR-92a, could significantly discriminate neoplasia from healthy controls, with the highest sensitivity when combining these two markers [43]. This indicates that miR-29a and miR-92a have potential as minimally invasive biomarkers for CRC. In another study [44], plasma miR-92a could significantly discriminate CRC from gastric cancer, normal subjects, and even more interestingly, from inflammatory bowel disease, suggesting to be a potential molecular marker for CRC.
Most pancreatic cancers display hypomethylation, and over-expression of miR-200a and miR200b in their serum, silencing of SIP1 by promoter methylation, and retention of E-cadherin expression. The elevated serum levels of miR-200a and miR-200b in most patients with pancreatic cancer might have diagnostic utility [45]. Profiling miR-21, miR-155, miR-196a and miR-210 demonstrated that miRNA profiling could also differentiate pancreatic adenocarcinoma patients from healthy controls. Plasma miRNA profiling might provide a sensitive and specific blood based biomarker assay for pancreatic cancer which might have potential for further translation into the clinic e.g. for contributing to distinguish inflammatory changes of the pancreas from (recurrent) neoplastic disease [46].
Early stage disease-postoperative follow-up-planning of adjuvant therapy
Elevation of circulating miR-195 was found to be breast cancer specific and, together with let-7a decreased postoperatively, indicating a potential use as a biomarker for early stage breast cancer and in the early follow-up in therapy monitoring [47].
In the peripheral blood samples from post-operative gastric cancer patients, the levels of miR--17 and miR-106a were significantly higher than those in healthy controls [48]. The detection of miRNAs in peripheral blood in this setting indicates a possible role of miRNAs in the postoperative care and planning of adjuvant therapy.
Molecular characterisation of disease
In patients with CN-AML, FLT-3 internal tandem duplication (ITD) was found to be significantly associated with overall survival and shorter disease-free survival. Whitmann et al [49] demonstrated that this is accompanied with miRNA expression signature including overexpression of miR-155 and under-expression of miR-144 and miR-451. These expression signatures may provide new insights for novel therapies in older CN-AML patients with higher molecular risks.
Interestingly, miRNA levels can also relate to hormone receptor status of tumors: oestrogen receptor (ER) negative breast cancer sera samples had higher levels of miR-21 and miR-10b; in contrast to miR-155 which was detected for progesterone receptor (PR) positive patients [50].
Assessment of prognosis
The identification of a patient who is prognostically good or poor is very important for the planning of a therapeutic strategy. Recent studies underlined that there is a great potential of circulating miRNAs signatures as molecular fingerprints to predict survival of cancer patients. Genome-wide serum miRNAs expression analysis found that the levels of four miRNAs (miR-1, miR-30d, miR-486, and miR-499) were significantly associated with the overall survival of non-small cell lung cancer (NSCLC) patients [42.] Many more candidate miRNAs with prognostic relevance have been identified.
Despite of many more interesting insights, it has to be mentioned that the list of potential blood miRNA biomarkers is still diverse and need further refinement and validation.
In summary, there are multitudes of reports describing the potential value of miRNAs as both diagnostic and prognostic biomarkers in oncology. However, to date none has been fully translated to clinical practice with excellent specificity and sensitivity.
CTCs or cell free miRNAs
Most of the studies on circulating miRNAs are carried out utilizing cell free miRNAs, and as detailed above provide interesting and relevant results.
Challenges include that even in healthy donors, there are many and various free circulating miRNAs, so false positive results deem possible. On the other hand circulating miRNAs, which are clearly expressed differently in cancer patients than in healthy donors have been identified. In addition there remain inconsistencies in the level of miRNAs expression from the serum of the same patient collected at different time points.
The fraction of blood in which circulating miRNAs should be measured is still not clearly defined. In most cases miRNAs have been measured in serum plasma or exosome fractions of blood. This helps reducing the leucocyte background, which is mostly present in the whole blood samples. However there are concerns that free miRNAs measured in these samples are not clearly derived from circulating epithelial cells.
So it is still not excluded that CTC associated miRNAs might be superior biomarkers. On the other hand the far majority of miRNAs are present in the cell-free form. Future studies will have to further validate for which application CTC associated miRNAs provide more robust results and in which settings free miRNAs are sufficient.
Challenges of miRNAs as biomarkers
MiRNAs are an innovative and exciting topic in oncology. The application of microarray, microfluidics, qRT-PCR and bioinformatics has identified a number of miRNAs, both in tissues and in circulating tumor cells, as potential biomarkers for cancer early detection, risk assessment, adjuvant therapy monitoring, and prolonged follow-up. There are numerous promising data supporting the potential value of miRNAs as biomarkers, and if these relatively stable small RNAs can be robustly detected by commonly applied PCR methods, they have the potential to become a new class of clinical biomarkers.
Most significant successes so far with respect to the envisioned clinical transfer are the establishment of a prognostic value of certain miRNAs in various cancer types and the initial valid data on their predicative value.
Challenges remain in the process to prepare miRNAs as biomarkers for prognosis or therapy response for the clinical transfer:
- Careful selection for the most promising and stable miRNA candidates for the clinical transfer. Most stable miRNAs have to be selected, and markers with high fluctuation rate have to be excluded. MiRNAs with too subtle upstream effects have to be identified and excluded. This will also require standardized and broadly applied normalization strategies.
- Appropriate statistical analysis must be utilized to identify further candidate miRNAs signatures. Adequately sized independent reference cohorts are key to further confirm the predictive value and for normalization.
- Further analysis of miRNAs in sera and plasma, investigating their exact biological role and correlation to prognostic and therapeutic signatures while using appropriately sized cohorts are warranted to further develop these molecular markers both for prognostic purposes and especially for monitoring and refinement of individualized treatment.
- Further validation on which fraction of the peripheral blood measurements of the miRNAs should be carried is needed; value of free and cell bound miRNAs has to be compared.
MiRNAs as molecular biomarkers in oncology - is there already relevance for the clinical imaging scientist?
MiRNAs have emerged as promising biomarkers in oncology. As summarized above, they demonstrate their capacities in classifying and sub-classify tumors, for prediction and prognosis, as well as for stratifying for and monitoring of personalized therapies.
The most intriguing potential capacity of miRNAs is a continuous molecular monitoring of primary or metastatic tissues utilizing miRNA profiling in circulating tumor cells or free miRNAs from peripheral blood.
Much correlative work in the initial process of validation and clinical translation of miRNAs as biomarkers was carried out by analysing parameters like time to progression or survival data [12]. However, imaging can provide very sophisticated well-established methods to detect and characterize tumor tissue, visualize and quantify extent of disease and sub-classify treatment effects. Tumors can be effectively assessed not only with respect to their size, morphology and spread, but also with respect to functional parameters like tumor metabolism (e.g. FDG-PET, see figure 1) [51], cellular density (MR diffusion) [52], vascularity (CT/MR/PET or US based methods), receptor status (PET), and metabolic tumor pathways (PET/hyperpolarized MRI) [53, 54].
These imaging methods can be very helpful both in targeting the optimal location of biopsy for further primary tumor tissue examinations and sub-classification of treatment response: tumor size, spread, vascularity, metabolism and many other parameters can be assessed non-invasively.
While miRNAs as biomarkers still are in the initial process of preparing for clinical transfer, it's now time that results of miRNA expression profiling is correlated with both morphological and functional clinical imaging data.
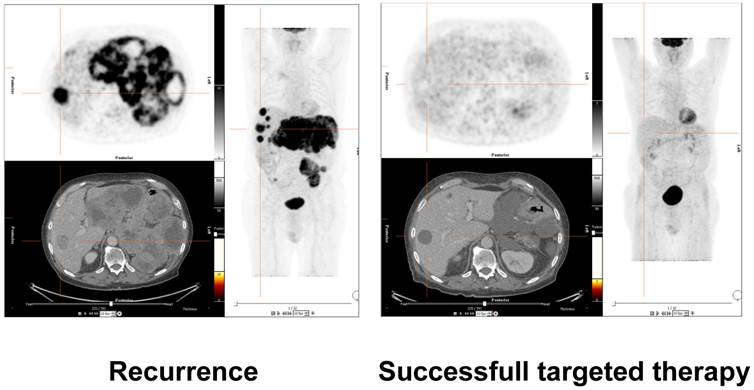
Assessment of treatment response of molecular therapy in a patient with gastrointestinal stromal tumor (GIST). 18-F-FDG-PET/CT images on the left side demonstrate recurrent disease with hypermetabolic liver and mesenteric metastases. The images on the right side validate successful molecular pharmacotherapy with no remaining suspicious hypermetabolism, although morphologic alterations are still visible. Future research efforts will have to correlate clinical results of molecular and morphological imaging with molecular laboratory oncologic biomarkers such as miRNAs. These correlations to in vivo imaging will further validate the capacity of the molecular markers for early disease detection, therapy selection, treatment monitoring and prediction of prognosis.
Such correlative studies will have the potential both to further validate and speed up the process of clinical transfer of miRNAs in oncology. It is now time to reinforce collaborations of molecular biologists and imaging scientists in this field to further push the development of these new promising molecular markers towards the clinic.
Conclusion
While still in the early process of clinical transfer, miRNA profiling has already proven its significant potential as biomarker in oncology.
Imaging scientists should be aware of the new and fascinating possibilities for molecular characterization of tumor tissue and therapeutic response provided by miRNA analysis. Clinical imaging offers sophisticated and well-established techniques for sensitive tumor detection and evaluation of treatment response of tumors to therapy, ranging from assessment of tumor perfusion, metabolism, cell density and many more markers of disease states.
Advanced clinical imaging biomarkers for cancer detection and monitoring should now been correlatively applied to further validate the potential of miRNAs as oncologic biomarkers for the clinic. This will contribute to a more standardized and validated, accelerated and safer transition of these promising new molecular laboratory oncologic biomarkers into clinical reality.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mattick JS, Makunin IV. Non-coding RNA. Human molecular genetics. 2006;15:R17-29
2. Li J, Smyth P, Flavin R. et al. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC biotechnology. 2007;7:36
3. Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer. 2007;43(10):1529-1544
4. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature reviews. 2005;6(5):376-385
5. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281-297
6. Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Developmental biology. 2007;302(1):1-12
7. Chen X, Ba Y, Ma L. et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell research. 2008;18(10):997-1006
8. Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799-2803
9. Roth C, Rack B, Muller V, Janni W, Pantel K, Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12(6):R90
10. Ratain MJ, Glassman RH. Biomarkers in phase I oncology trials: signal, noise, or expensive distraction? Clin Cancer Res. 2007;13(22 Pt 1):6545-6548
11. Glassman RH, Ratain MJ. Biomarkers in early cancer drug development: limited utility. Clinical pharmacology and therapeutics. 2009;85(2):134-135
12. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857-866
13. Farazi TA, Horlings HM, Ten Hoeve JJ. et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011;71:4443-4453
14. Volinia S, Calin GA, Liu CG, Ambs S. et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261
15. Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV. et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612-11620
16. Mattie MD, Benz CC, Bowers J, Sensinger K. et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24
17. Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F. et al. MicroRNA in human ovarian cancer. Cancer Res. 2007;67:8699-8707
18. Lebanony D, Benjamin H, Gilad S, Ezagouri M. et al. Diagnostic assay based on has-miR-205 expression distinguishes squamous from non-squamous non-small cell lung carcinoma. J Clin Oncol. 2009;7:2030-2037
19. Wymann SK, Parkin RK, Mitchell PS, Fritz BR. et al. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLOS ONE. 2009;4:e5311
20. Schulte JH, Marschall T, Martin M, Rosenstiel P. et al. Deep sequencing reveals differential expression of microRNAs in favourable versus unfavourable neuroblastoma. Nucleic Acids Res. 2010;38:5919-5928
21. du Rieu MC, Torrisani J, Selves J, Al Saati T. et al. MicroRNA-21 is induced early in pancreatic ductal adenocarcinoma precursor lesions. Clin Chem. 2010;56:603-612
22. Boeri M, Verri C, Conte D, Roz L. et al. MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108:3713-3718
23. Ji J, Shi J, Budhu H, Yu Z. et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437-1447
24. Schetter AJ, Leung SY, Sohn JJ, Zanetti KA. et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425-436
25. Zhou M, Liu Z, Zhao Y, Ding Y. et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (BAK) expression. J Biol Chem. 2010;285:21496-22507
26. Giovannetti E, Funel N, Peters GJ, Del Chiaro M, Erozenci LA. et al. MicroRNA-21 in pancreatic cancer: correlation with clinical outcome and pharmacological aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 2010;70:4528-4538
27. Cronin M, Pho M, Dutta D. et al. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. The American journal of pathology. 2004;164(1):35-42
28. Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. Journal of cellular and molecular medicine. 2009;13(1):39-53
29. Lu Z, Liu M, Stribinskis V. et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27(31):4373-4379
30. Fan C, Oh DS, Wessels L. et al. Concordance among gene-expression-based predictors for breast cancer. The New England journal of medicine. 2006;355(6):560-569
31. Rosenfeld N, Aharonov R, Meiri E. et al. MicroRNAs accurately identify cancer tissue origin. Nature biotechnology. 2008;26(4):462-469
32. Rosenwald S, Gilad S, Benjamin S. et al. Validation of a microRNA-based qRT-PCR test for accurate identification of tumor tissue origin. Mod Pathol. 2010;23(6):814-823
33. Lu J, Getz G, Miska EA. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834-838
34. Sempere LF, Christensen M, Silahtaroglu A. et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer research. 2007;67(24):11612-11620
35. Ferracin M, Veronese A, Negrini M. Micromarkers: miRNAs in cancer diagnosis and prognosis. Expert review of molecular diagnostics. 2010;10(3):297-308
36. Yang H, Kong W, He L. et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer research. 2008;68(2):425-433
37. Weiss GJ, Bemis LT, Nakajima E. et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008;19(6):1053-1059
38. Rodriguez-Gonzalez FG, Sieuwerts AM, Smid M. et al. MicroRNA-30c expression level is an independent predictor of clinical benefit of endocrine therapy in advanced estrogen receptor positive breast cancer. Breast cancer research and treatment. 2011;127(1):43-51
39. Mitchell PS, Parkin RK, Kroh EM. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(30):10513-10518
40. Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Medicinal research reviews. 2012;32(2):326-348
41. Lawrie CH, Gal S, Dunlop HM. et al. Detection of elevated levels of tumor-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. British journal of haematology. 2008;141(5):672-675
42. Shen J, Todd NW, Zhang H. et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Laboratory investigation, a journal of technical methods and pathology. 2011;91(4):579-587
43. Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. International journal of cancer. 2010;127(1):118-126
44. Ng EK, Chong WW, Jin H. et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58(10):1375-1381
45. Li A, Omura N, Hong SM. et al. Pancreatic cancers epigenetically silence SIP1 and hypomethylate and overexpress miR-200a/200b in association with elevated circulating miR-200a and miR-200b levels. Cancer research. 2010;70(13):5226-5237
46. Wang J, Chen J, Chang P. et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer prevention research. 2009;2(9):807-813
47. Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Annals of surgery. 2010;251(3):499-505
48. Zhou H, Guo JM, Lou YR. et al. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using microRNA as a marker. Journal of molecular medicine. 2010;88(7):709-717
49. Whitman SP, Maharry K, Radmacher MD. et al. FLT3 internal tandem duplication associates with adverse outcome and gene- and microRNA-expression signatures in patients 60 years of age or older with primary cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116(18):3622-3626
50. Yan LX, Huang XF, Shao Q. et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14(11):2348-2360
51. Tomasi G, Rosso L. PET imaging: implications for the future of therapy monitoring with PET/CT in oncology. Current opinion in pharmacology. 2012;12(5):569-575
52. Heijmen L, Verstappen MC, Ter Voert EE. et al. Tumor response prediction by diffusion-weighted MR imaging: ready for clinical use? Critical reviews in oncology/hematology. 2012;83(2):194-207
53. Ehling J, Lammers T, Kiessling F. Non-invasive imaging for studying anti-angiogenic therapy effects. Thrombosis and haemostasis. 2013;109(3):375-390
54. Viale A, Reineri F, Dastru W, Aime S. Hyperpolarized (13)C-pyruvate magnetic resonance imaging in cancer diagnostics. Expert opinion on medical diagnostics. 2012;6(4):335-345
Author contact
![]() Corresponding author: Frank Berger M.D. Department for Clinical Radiology, Ludwig-Maximilians-University, Campus Innenstadt, Nussbaumstrasse 20, 80336 Munich, Germany. Phone: +49-89-5160-9201. Email: Frank.Bergeruni-muenchen.de
Corresponding author: Frank Berger M.D. Department for Clinical Radiology, Ludwig-Maximilians-University, Campus Innenstadt, Nussbaumstrasse 20, 80336 Munich, Germany. Phone: +49-89-5160-9201. Email: Frank.Bergeruni-muenchen.de
 Global reach, higher impact
Global reach, higher impact