13.3
Impact Factor
Theranostics 2013; 3(12):930-942. doi:10.7150/thno.7026 This issue Cite
Review
MicroRNAs -the Next Generation Therapeutic Targets in Human Diseases
1. Lee Kong Chian School of Medicine, Nanyang Technological University, 50 Nanyang Drive, Research Techno Plaza, Level 4, X-Frontier Block, Singapore-637553
2. Institute of Materials Research and Engineering, 3 Research Link, Singapore-117602.
3. Centre for Pheromone Technology (CPT), Department of Animal Science, Bharathidasan University, Tiruchirappalli- 620 024, India.
Received 2013-6-27; Accepted 2013-10-8; Published 2013-11-29
Abstract
MicroRNAs (miRNAs), an abundant class of ~22-nucleotide non-coding RNAs, regulate the expression of genes at post transcriptional level. MiRNAs are important regulators of eukaryotic gene expression and therefore implicated in a wide range of biological processes. The miRNA-related genetic alterations are possibly more implicated human diseases than currently appreciated. Genetic variants in miRNA target sites, called miRNA genes are identified to be associated with human diseases. This review discusses about the role of micro-RNA genes in various human diseases such as neurodegenerative disorders, cardio-vascular diseases, and metabolic disorders, and how they can be targeted as a new therapeutic tool in future with reference to drug discoveries/ development.
Keywords: miRNA therapy, novel therapeutics, Neurodegenerative disorders, Cardio-vascular diseases and metabolic disorders
INTRODUCTION
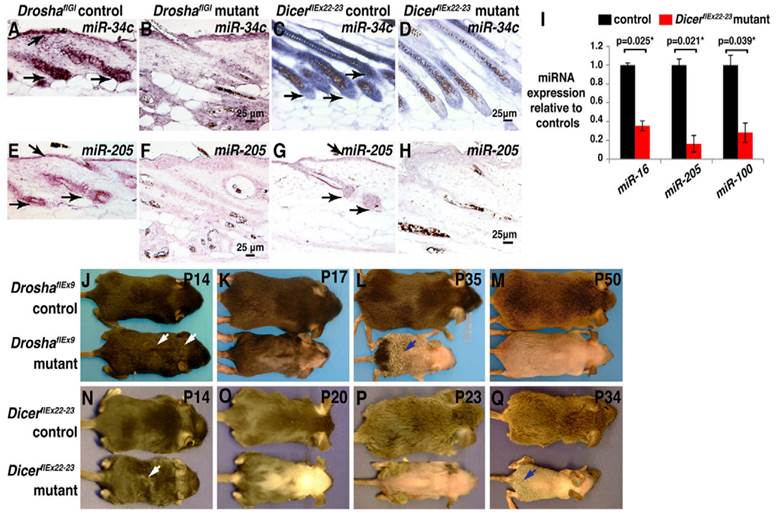
MicroRNAs (miRNAs or miRs), the post transcriptional regulators constitute a class of small RNAs are found in plants, animals, and in a number of DNA viruses. Mature microRNAs are a class of about 21-25 nucleotides in length and small non-coding RNA molecules, and occur naturally. They were first described in 1993[1], and the term microRNA was created in 2001[2]. The main functions of miRNAs are to down-regulate the gene expression in translational repression, cleavage of messenger RNA (mRNA) and in a variety of other processes. Each miRNA is partially or completely accompanied by one or more mRNAs. MiRNAs target around 60% of all genes and are profusely present in all human cells, and are able to repress hundreds of each target. The processed mature miRNA molecules are shorter than the final mature functional miRNAs. The regulatory elements, primary transcripts of miRNAs are shared with their respective pre-miRNA, since the residence of them in the introns of its specific Pre-miRNA host genes, so that they will have similar expression profiles. Few primary transcripts have been fully identified for the residue of miRNA genes that are transcribed from their specific promoters. The transcription processes of microRNAs are performed by Pri-miRNAs, and RNA precursors that comprise of cap and poly-A tail[3]. Microprocessor complexes processes pri-miRNAs which comprises of enzyme RNase III Drosha[4] and the RNA-binding double-stranded protein called DGCR8in nucleus[5].An imperfect stem-loop structure is formed from the resulted pre-miRNAs that are 70-nucleotides in length. Ran-GTP and Karyopherin exportin complexes export the pre-miRNAs into cytoplasm[6]. The translocation of RNA with protein needs ras-related nuclear protein (RAN), and a small GTP binding protein that occurs through the nuclear pore complex[7]. The nuclear heterotrimer with the pre-miRNAs are formed by the binding of Ran GTPase and Exp5 [8]. The double stranded miRNA, of 22-nucleotides in length generated by the processing of pre-miRNA undergoes an extra step by the RNAse III enzyme Dicer[9]. The formation of the RNA-induced silencing complex (RISC) occurs by the Dicer[10]. Due to the miRNA expression and RNA interference, the gene silencing comes under the responsibility of RISC. Fig. 1 illustrates the defects caused by the deletion of 'Drosha' or 'Dicer' in anagen, the foremost phase of hair cycle during the synthesis of hair in which the active product of hair growth occurs in the hair follicle. MiRNAs have all types of functions in regulating the cell physiology, proliferation, cell differentiation, and apoptosis to the endocrine system, hematopoiesis, limb morphogenesis and fat metabolism. By reflecting the diversity of cellular phenotypes, different expression profiles are displayed by them from tissue to tissue.
Nomenclature of miRNA
Since hundreds and thousands of miRNA occur in each species, it is important that each new miRNA is assigned a specific name, as per the established system of miRNA nomenclature. In this system, miRNAs that are confirmed by the experiments are given a number following the prefix of 'mir-'. The capitalized miR- denotes the mature form of miRNA and the un-capitalized mir- refers to the pre-miRNA. MiRNAs with similar structures are annotated to show their nucleotide structures in number with the added lower case letter as miR-1a and miR-1b. As miRNAs has the ability to produce the same miRNA at different loci, such cases are mentioned with the additional numbers and shown as miR-1-1 and miR-1-2 [11]. The main annotation in the system of nomenclature of miRNA is regarding the Species annotation in which the miRNA are observed; for example if it is observed in 'Homosapiens', it should be denoted as, 'hsa-miR-xxx'. Other miRNAs that are commonly seen in the species of viral and Drosophila are denoted as v-miRNA and d-miRNA. MiRNAs that arises for the 3' or 5' end are specified with a suffix of -3p or -5p (miR-116-5p, miR- 116-3p). The nomenclature of miRNA can be registered on miRBase.
(A-H) miRNA depletion, hair shaft defects and permanent hair loss are caused by the deletion of Drosha or Dicer in anagen. (A-D) In-situ hybridization for miR-34c in skin. (E-H) In-situ hybridization for miR-205 in skin. (I) After treatment of deoxycycline from P1, this illustrates the qPCR for indicated miRNAs using Dicer deleted epidermis. (J-Q) Deoxycycline treatment of Drosha (J-M) and Dicer (N-Q) control and mutant mice. Reproduced with permission from Ref. [86].
Cellular functions of miRNAs
The function of miRNAs is a complementary part of one or more mRNAs in gene regulation. By causing DNA methylation in promoter sites, the main expression of target genes gets affected by miRNAs infrequently. MicroRNAs that function as counterparts of proteins are collectively called miRNPs. Human miRNPs contains eIF2C2 (Argonaute 2), DDX20, GEMIN4 and MiRNA[12, 13].
On the other hand, in as much as functional genes are common to all cells, there are only limited miRNA target sites, therefore, there are possibilities that functional genes may not always be targeted by miRNAs [14]. The microarray dubbed MM-chips contains all known miRNA for human, mouse, rat, dog and 'Drosophila'. The class of miRNAs located in the introns of the mRNA programming host genes is called as Mirtron which plays a bypass role and enter as pre-miRNA in miRNA maturation pathway.
MicroRNAs ROLE IN VARIOUS HUMAN DISEASES
Though ten years got over after the identification of first miRNA, only now the scientists have begun to comprehend the possibility and diversity of these regulatory molecules. Decisive regulatory functions exhibited by the miRNA are associated with a wide variety of human diseases such as cancer[15, 16] and heart disease[17]. MicroRNAs are liberalized in lung, breast, and colon cancer, and up-regulated in human B-cell lymphomas. MiRNAs are likely to be highly applied in future cancer diagnostics, and are attractive targets for disease intrusion. In addition to the link with cancer, microRNAs play a vital role in the control of cardiac - related diseases. The integration of the ventricular wall, gene expression, contractility, and maintenance of cardiac rhythm including myocyte growth, can be achieved. For multiple forms of heart disease, the mis-expressions of miRNAs were shown to be necessary[17].
This review focuses on the research of miRNA in human disease that includes the methodologies for gene identification and miRNA targets, and the role of miRNA in human diseases. Other than atherosclerosis, it mainly focuses on three captions namely,
- Neurodegenerative disorders
- Cardiovascular diseases
- Metabolic disorders
These details explain on the future role of miRNA gene and its therapeutic potential for various human diseases through the currently available applications.
NEURODEGENERATIVE DISORDERS
In the neurodegenerative disease, the substantial role of miRNA gene in their pathogenesis had been documented [18]. The pathogenic mechanisms in psychiatric deficits and neuro-degeneration development are not always different, but they are preferably suited for therapeutic targeting by pleiotropic miRNAs. Dicer like miRNA processing machinery, as well as the mature miRNA and the precursor subjects to the commotion with neurological disease progression.
Altered miRNA profiles and the summary of derestricted miRNAs have been identified in the psychiatric diagnoses like Schizophrenia, autism, Fragile X syndrome, depression, addiction, and anxiety[19]. MiRNA derestriction has also been observed in peripheral blood, plasma, serum and cerebral spinal fluid (CSF), and furthermore number of studies regarding miRNA profiling has been performed for neurological diseases[20]. Remarkably, the population analyses have been interrupted in some miRNA families that include miR-15 and miR-30 families[21].MiRNA disruption has been observed in the patients with psychiatric symptoms that co-exist with neurodegenerative pathologies such as Parkinson's disease (PD) Alzheimer's disease (AD) and Huntington's disease (HD) [22].In some cases, miRNA interruption can be seen in both the psychiatric and neurodegenerative diseases. The best example is miR-132 that has been linked with schizophrenia[23]. Regular disruption of miR-132 is seen in wide range of neurological diseases with the principal role of miRNA in neuronal plasticity[24].
Alzheimer's disease
MicroRNAs have recently been involved mostly in neurodegenerative disorders including Alzheimer's disease[25]. Alzheimer's disease brains comprise rod-like structures called Hirano bodies[26], which consists of actin binding proteins with large actions. These proteins are called as Cofilin, which in abnormal condition affects the function of cytoskeleton[27, 28]. However, the formation mechanism of Cofilin-actin is still unknown and this annexation plays a dynamic role in pathogenesis of Alzheimer's disease. As the expressions of genes are regulated by the large number of miRNA expression in brain, miR-107 is decreased in postmortem in Alzheimer's disease human brain. It gets allied with the increased expression of Beta-site amyloid cleavage enzyme 1 (BACE1), where miRNAs plays a major role in the pathogenesis of Alzheimer's disease.
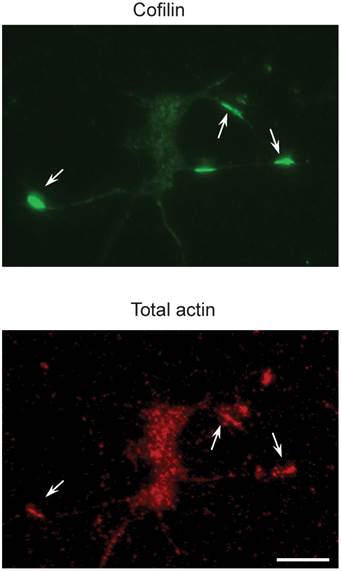
Also, miR-103 and miR-107 suppress Cofilin translation with the increased level of active Cofilin protein, which leads in the Cofilin rods formation. The reduction of miR-103 or miR-107 results in an increase in Cofilin protein levels in Alzheimer's disease brains[29]. Fig. 2 represents the recognition of Cofilin rods in the primary neurons .Most of the studies focus on the derivatives of soluble-A and tau in Alzheimer's disease[30, 31], but a few focus on blood and CSF and have reconnoitered the role of miRNA[32, 33]. It will be really interesting to cartel with circulating miRNAs in the future researches and even most of the studies discussed about the miRNA in Alzheimer's disease; there is no conclusive evidence for the miRNA target gene regulation in-vivo. These are addressed to gain an advanced knowledge of the contribution of miRNAs in Alzheimer's disease. As mice are genetically homogenous comparing to humans, it's quite easy to study miRNA related gene pathways in the Alzheimer's disease development. Due to the indications of profiling studies in human, that Alzheimer's disease brain gets altered by the miRNA gene expression, and it is to be patterned whether the changes are specific to this particular disease or it tends to overall reduction of miRNA function remains to be detected. MiRNA gene knockout is required to address these and other enthralling suggestions.
Cofilin rods in the primary neurons (top) are recognized by an antibody to total actins (bottom). The scale bar representation is 100 µm. Reproduced with from Ref. [29].
Parkinson's disease
MiRNAs are accompanying with the pathogenesis of Parkinson's disease, as found in many other diseases[34, 35]. Nowadays, many unfathomable perceptions into neuro-pathogenesis in Parkinson's disease have been studied in more number; however this review describes about a few increasingly studied miRNA that deals with the Parkinson's disease, which is generally associated with Aging and hence has a high incidence in elderly individuals. The obliteration of dopaminergic neurons in the mid-brain or the insertions of proteins and Lewy bodies results in Parkinson's disease. The inspection of molecular processes in normal control and human diseases can be obtained by a novel approach of the study of miRNA.
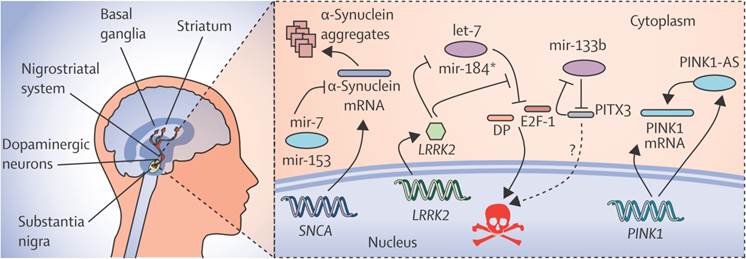
N-Myc down-regulated gene 1 (NDRG1) and junction plakoglobin (JUP) are the two new transcriptional factors noted in eleven genes, which are identified by miRNA and the transcription factors associated with Parkinson's disease, in which the regulation of NDRG1 is achieved by miRNA-133[36]. MiR-7, miR-153 and miR-433 modify the expression of α-synuclein in Parkinson's disease as shown in Fig. 3. The decreased expression of miR-34b and miR-34c was indicated by the miRNA profile analysis and the changes were found in the frontal cortex, cerebellum, and amygdala in Area 17 of brain. The depletion of miR-34b or miR34c results in the decreased expression of Parkinson protein 2 (Park2) and Parkinson protein 7 (DJ1) and Cell death. Mitochondrial dysfunction is finally resulted by these kinds of cellular changes in Parkinson's disease[35].As the studies of miRNA microarrays results about 4% of miRNAs were from healthy brain tissues, it should be checked also in the plasma of healthy controls. For the process of Validation and Evaluation of the biomarkers from the new set of replication in Parkinson's disease and controls, the quantitative real-time Polymerase Chain Reaction technique (qRT-PCR) can be done. To lower the biomarker performance, the biomarkers at the high predictive performance can be combined and applied to a new panel of independent set of validation.
For the future research, most of the detailed participation of miRNA should be studied in the field of Neuro-degeneration. Here are some suggestions that may virtue some reflection in the future research of miRNAs in Neurodegeneration:
- With convoluted functions and requirement of protein translation long, neurons are differentiated in a geometrical manner by their respective asymmetric shape. It takes many centimeters in some places from the cell nucleus. It should be noted for the perturbation of the translational control contributed to Neurodegeneration, if miRNA plays a vital role in localized control of messenger RNA (mRNA).
- Due to the alterations of miRNA related to Neurodegeneration, alteration of biochemical function should be checked, whether the miRNA alteration occurs because of the alteration in RNAs in Neurodegeneration.
- The miRNAs related pathways like Apoptosis, cell division, etc., relevant to earlier development may be pathogenic in the adult brain.
- It should be checked whether the Risk factors of Neurodegeneration affects the miRNA brain expression and miRNA-related biochemical predictions for therapeutics.
Schizophrenia
Schizophrenia is a devastating complex neuropsychiatric disorder categorized by cognitive impairments that involves interruptions in neural circuitry and synaptic function. The neural network necessitates coordination by an intricate intracellular network of molecular signal transduction systems. The potential of combinational gene variants cause system dysfunction as related to neurobehavioral syndromes that are meant by the neural networks termination. MicroRNAs are mainly used for the topography shaping of these networks, which have a potential for the regulation of target genes by the function of miRNA in Intracellular gene-silencing machinery as their specificity factor. Later in the mammalian brain, miRNA displays composite temporospatial expression patterns. During development ubiquitous changes occurs in the network due to the dysregualtion of miRNA which also happens in the matured brain. This is highly significant in the pathophysiology of Schizophrenia and can be the regulatory factor in most of the human genes in the tissue-specific stages of development [37, 38].
A surfeit of postmortem studies using cases spotted with Schizophrenia delivered a solid support for deviant expressions of miRNA in psychiatric disorders [39-41]. miR-132 and miR-212 are involved in several abnormalities like synaptic plasticity and connectivity in Schizophrenia and in neuronal cultures, the mature miR-132 stability is affected by NMDA inhibition[42]. Similarly, the pharmacological blockage in NMDA receptors leads to the down-regulated expression of miR-219[43]. Other important miRNAs that play a major role in Schizophrenia are miR-195, which regulate more number of Schizophrenia-related genes[44] and miR-137 the expression of which gets inhibited by Mecp2 that targets other chromatin modifying genes.
Epilepsy
Epilepsy is a chronic neurological disorder that forms unprovoked seizures; in some cases, it forms a single seizure combined with brain alterations that later forms more number of seizures in brain. MiRNAs takes their roles in the pathogenesis of seizure-induced epilepsy and the change of multiple miRNAs expression are detected by miRNA profiling, which in late follows miRNA regulation in human epilepsy [45, 46]. The novel targets for neuro-protection and anti-epileptogenesis can be attained by the identification of miRNAs regulating seizure-induced neuronal death [47, 48]. MiR-34a explored its role as up-regulation due to its presence in p-53 dependent control of apoptosis[49]. Several other miRNAs like miR-21, miR-29a and miR-132, were identified as p53 regulated and also they gets regulated after seizures formation. Future studies in miR-34a can help to achieve the cure for Epilepsy, as it only has the ability to control by medication.
The regulatory network of miRNAs in parkinson's disease showing the expression of α-synuclein and its modification by specific miRNAs like miR-7, miR-153 and miR-433. Reproduced with permission from Ref.[87].
Down syndrome
Down's syndrome disease, also called trisomy 21, is a chromosomal genetic disorder caused by the presence of the third copy of Chromosome 21 and it is more common in human that leads to intellectual disability. Down's syndrome is associated with mental retardation and delay in physical growth. The rapid proliferation of astrocytes called Astrogliosis, is associated with the Down's syndrome. In the brain tissues of Down's syndrome and Astrogliosis induced human astrocytes, the levels of miR-125b gets up-regulated in its enrichment. This kind of miRNA can be studied in future for their contribution in Neurodegenerative disorders, as miR-125b has multiple targets in the brain affected with Neurodegenerative disorders [50, 51].
CARDIOVASCULAR DISEASES
The growth, survival, function and gene expression of Cardiomyocyte gets regulated by the disruption in normal mechanisms, intricate in the pathogenesis of heart failure. The heat map diagram for the illustration of circulating miRNA profile in Coronary Artery Disease (CAD) patients and healthy volunteers has been compared [50]. Cardiac Fibrosis and Myocardial Infarction results with the participation of Cardio-vascular cells and Cardiac-interstitial cells, but some novel therapeutic targets by miRNA have proven to be clinically applicable and that is described in the following, as disease segmented.
Myocardial Infarction
MiR-21 is an Oncomir, since it exerts oncogenic activity and it is the only miRNA that got gradual up-regulation in both cancer and cardio-vascular diseases like Myocardial Infarction. This particular miRNA contributes to the progression of fibrosis and heart failure or block and highly seen in the fibroblasts, in the cardiovascular system. Against heart diseases like infarction, miR-21 serves as a major part in the development of therapeutic strategies. The inhibition of this specific miRNA (miR-21) by the synthetic miRNA antagonist results in the improvement of heart function in cardiac disease model. The same constructive effects were observed in miR-21 knockout mice exposed to pressure-overload of the left ventricle emphasizing the key role of miR-21 as a therapeutic target. Comparing to the healthy controls, coronary artery disease patients undergo a reduction of miR-145, miR-17, inflammation-accompanying miR155 which reduces the circulating levels of angiogenesis-related miR-126 and miR-92a;Coronary artery disease patients shown an elevation in the miR-133a and miR-208a levels of cardiac muscle enrichment.
Of the genome, 50% is allotted for the miRNA to function as regulators and that's the main reason for probable use of miRNA as novel therapeutic targets to modulate cardiovascular signal transduction. After the cardiac stress, the signal transduction may be helpful for the cardiac regeneration. Mammalian miRNAs show a partial complement to respective mRNAs due to the small interference RNA mediated gene silencing. Also the mammalian miRNAs possess stronger biological effects which facilitate their use as multiple targets. Cardiac remodeling and the development of heart failure are attained by Myocardial infarction (MI); the perilous event is, after the Myocardial Infarction, it is noted that the myocardial capillary density is not sufficient. However, the Cardiac mechanisms are not well understood. To regenerate Endothelial cells after Myocardial Infarction, a new miRNA has been identified which increases cardiac function and decreases the infarct size [37]. To regenerate cardiac tissue after the Myocardial Infarction, Stem cell based transplantation ideas are currently hindered within the affected area and inflated fibrosis by the ischemic environment. These processes can be targeted now by miRNAs and as the miRNA approaches got into clinical phase studies in a short time, thus indicating the high translational impact of miRNA therapeutics in future, but it remains as a great challenge to interpret such tactics into a therapeutic cardiovascular locale.
Cardiac fibrosis
Another important cardio-vascular disease with high incidence is myocardial fibrosis, which is a vital complication of most forms of human heart disease. By regulating cardiomyocyte apoptosis and discharge of pro-fibrotic factors, myocardial fibrosis gets influenced by myocyte-specific miRNAs. It also gets regulated by miRNAs that are expressed in Cardiac fibroblasts[38]. Some of the major examples that play a high role in this process is two miRNAs, miR-21 and miR-29, which are extensively expressed as miRNAs that are highly augmented in apparently cardiac fibroblasts compared to myocytes [39, 40]. In an experimental heart failure model, up-regulation of miR-21 was narrowed to cardiac interstitial cells and it shown to promote cardiac fibrosis[39]. Down-regulation of miR-29 in limit zone myocardium has been associated with the contribution in myocardial fibrosis[40]. Predicted miR-29 targets encompassed fibrosis-related mRNAs that include collagens with increased expression in heart of miR-29b antagomir knockdown mice. This overexpression of miR-29b in cultured cardiac fibroblasts contracted collagen expression[38].
Cardiomyocyte are reliant on an opulent vascular complex, maintenance of which is dependent upon complex signaling between cardiac interstitial, cardiomyocyte, and endothelial cells. Vascular homeostasis is determined by the proangiogenic and antiangiogenic signaling balance. miR-126 is the only endothelial-specific miRNA that regulates angiogenesis[41, 42]. Isolated non-myocytes from the hypertrophied myocardium of calcineurinin transgenic mice, miR-126 was also down-regulated[43]. MiR-23a was up-regulated by the activation of calcium-dependent pathway which acts as a vital facilitator of cardiac hypertrophy, called Calcineurin-NFAT pathway[44]. Calcineurin -NFAT pathway block was attained by antagomir-mediated loss of miR-23a activity[44] and the overexpression of miR-23a drives cardiomyocyte hypertrophy[45]. MiR-24, miR-195 and miR-214 are other significant miRNAs that regulates cardiac hypertrophy[45]. In Smooth muscle cells, miR-143 and miR-145 are co-transcribed miRNAs that get expressed, which were down-regulated in atherosclerotic vessels for the promotion of differentiation and for the process of inhibiting proliferation[46].
Atherosclerosis
Hemodynamic forces like hydrostatic and fluid shear stress, plays a major role in the development of physiology and vessel maturation. They are subjected with the blood vessels and the monolayer that is in straight contact with flowing blood, vascular endothelial cells are continuously exposed to blood flow-induced shear stress. Atherosclerosis occurs normally in arterial curvatures and branches that have very low and dynamic shear stress[47]. The dysfunction of endothelial cells is the preliminary step of Atherosclerosis and an inflammatory response through peroxisome proliferators-activated receptor-α by 3'-UTR targeting achieved as a result of induced expression of miR-21 in cultured endothelial cells at the transcriptional level, due to the Oscillatory Shear Stress (OSS)[48]. In endothelial cells, the expression of miR-92a is down-regulated by the Pulsatile Shear Stress (PSS), whereas the same gets up-regulated by Oscillatory Shear Stress down-regulated. KLF-2 gene serves as a target for miR-92a, which get up-regulated by athero-protective shear flow like Pulsatile Shear Stress and Laminar Shear stress, whereas it gets down-regulated by Oscillatory shear stress stimulated endothelial cells and it is confirmed by the demonstration of bioinformatics analysis. Thrombomodulin (TM) and eNOS are the regulated genes of KLF-2, which were repressed in the endothelial cells by the overexpression of miR92a. So, the regulatory responses of KLF-2 and miRNAs towards athero-protective shear flow can be predicted by a new concept. Another miRNA is also shown to be regulated by shear stress;, MiR-663[49] plays a vital role in the modulation of inflammation and proliferation of endothelial cells.
By various cultured cells and animal models, the involvement of several miRNAs in the biological progressions have been identified and that aid to study the expression profiles of circulating miRNAs[50] in Cardiovascular disease patients. The Peripheral blood mononuclear cells (PBMCs)[51] that underlies with the involvement of miRNAs; unfortunately, the involvement of miRNAs in atherosclerotic plaques has not received fine consideration. In the human atherosclerotic plaques from peripheral arteries, the expression profiles of miRNA were investigated in comparison to the non-atherosclerotic Left internal thoracic arteries (LITA)[52]. Also the relationship between the miRNA expression profiles and biological processes in atherosclerosis was elucidated. It is found that miR-21, -34a, -146a, -146b-5p, and -210 were expressed at substantial levels and in human atherosclerotic plaques and these mentioned miRNA's frequent prophesied targets were down-regulated. MiR-34a was recognized as a novel therapeutic target for the pathogenesis of atherosclerosis, mainly because of its function in apoptosis.
MiR-146a family (miR-1461/b) is highly expressed in atherosclerotic plaques and gets regulated downstream through a negative-feedback regulation loop. By utilizing the Lipopolysaccharide - stimulated human monocytic cell line that an elevated miR-146 expression has been noted[53]. The contribution of the activity of endothelial cells to the lower levels of circulating miRNAs may be required, since the perfect mechanisms of reduction of circulating miRNAs is uncertain. In blood, by leading to the reduction of circulating miRNAs, as another insinuation, circulating miRNAs are taken up into atherosclerotic lesions.
METABOLIC DISORDERS
Biological functions of microRNA is much less, though hundreds of miRNA were identified. In greater organisms, miRNAs affect the fundamental pathways for the metabolic control such as adipocyte and skeletal muscle differentiation. It is a completely requisite for all living cells to make changes in the internal or external environment in response for the ability to control the rates of metabolic processes. Transcriptional, translational, posttranslational and allosteric regulations are the Complex mechanisms that are highly involved in the metabolic control and the maintenance of homeostasis. To add an innovative level of regulation and fine-tuning for gene expressions, miRNA aids a foremost part by comprising the novel class of genes that is significant for a wide sort of metabolism. The possibilities of miRNAs to contribute themselves on metabolic disorders highly points on more common human metabolic control like diabetes and obesity, which are detailed below to list-out the novel therapeutic chances by targeting miRNAs
Diabetes mellitus
Pancreatic β-cells play a vital role in diabetes mellitus and in the complex mechanism of glucose homeostasis by involving in the Insulin production. In the development and maintenance of the pancreatic islets, miRNAs commences to involve in the insulin pathway. The flaw in the production of insulin and in β-cells with an imperfection of the endocrine pancreatic lineage development was attained by the ablation of Dicer 1 which causes miRNA processing block[54]. In diabetic controls, both the mRNA and protein levels were decreased by the Dicer-1-deificient β-cells and the levels of transcriptional repressors, Bhlhe22 and Sox6, were increased by silencing miR-24, miR-26, miR-148 or miR-182 in autonomous[55]. This leads to the down-regulation of mRNA levels and specific insulin promoter activity, by affecting the insulin transcription levels which assures that miRNAs therapeutic targets in Diabetes mellitus whereby modulation in their expression regulates insulin production.
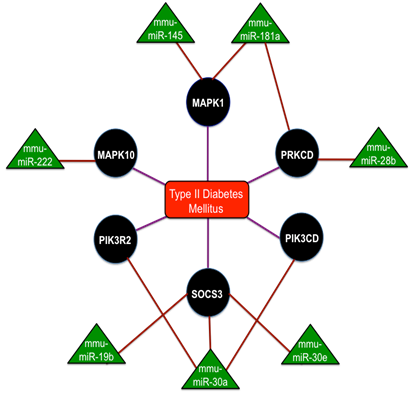
MiR-375 is the most studied miRNA that are expressed in the pancreas and are overexpressed in the islet cells. These miR-375 functions on the control over glucose-stimulated insulin release and the overexpression of miR-375 inhibits insulin secretion[56]. Reduced β-cell mass and increased a-cell number was seen in miR-375 knockout mice that exhibited the diabetic phenotypes like hyperglycemia and glucose intolerance [57]. A section of ten miRNAs such as miR-27a, miR-130a, miR-192, miR-200a, miR-320, miR-337, miR-369-5p, miR-379, miR-410 and miR-532, were down-regulated in the concluding group[58]. Reduced glucose-stimulated insulin secretion was obtained by the knockdown of miR-410, miR-200a and miR-130a separately and a significant reduction in the expression with glucose treatment was confirmed by miR-690, miR-484 and miR-296 and the up-regulation of miR-124a, miR-107 and miR-30d. Thus the complete interaction network of miRNAs in Type-2 Diabetes mellitus is shown in Fig. 4 and collectively these studies discovered the network of miRNAs for the maintenance of insulin synthesis and secretion by controlling the range of mRNAs and miRNAs potentially contribute to the manifestation of diabetes.
Obesity
Obesity is categorized with diminished adipocyte function and aberrations in adipokine secretion that are associated with insulin resistance. The lean individuals with more peripheral distribution of fat are more insulin sensitive than the obese individuals who have their fat distributed in the central abdomen. miRNA profiling of human fat cells recognized an increase in the expression of miR-100, miR-125b, miR-221 and miR-34a and the down-regulation of miR-130b, miR-185 and miR-210 in obese subjects[59]. During adipocyte differentiation and maturation, miR-100, miR-125b, miR-221 gets down-regulated, whereas miR-130b and miR-210 decreased in their expression[60], but miR-143 showed a positive association with bodyweight and mesenteric fat weight[60]. Important correlation between the morphology of adipose tissue and the expression levels of miR-17-5p, miR-132, miR-134, miR-145, miR-181a, miR-197 and miR-99a were reported with the fasting plasma glucose and circulating leptin, adiponectin and IL-6[61]. Significant differences between obese and non-obese fat were reported by the expression levels of miR-17-5p and miR-132[62]. Thus these studies provide the insinuations of the miRNA in obesity.
As the wide medical vocation is very alarmed by the growing incidence of overweight and obesity and its main association with the increased health risks, miRNA researches will be more helpful in its specific clinical applications and they can be pointed-out by focusing the future perspective miRNA researches in them. Also, this can be attained by revealing studies regarding the expression criteria.
The interaction network of miRNAs in Type - 2 Diabetes mellitus with the triangular regions representing the miRNAs that negatively regulate the refined genes and the round regions representing the genes of the refined gene set. Adopted from Ref. [88].
Methodologies used in Gene Identification
Various successful methods and techniques to identify the computational part of the gene, is a major thrust research area. Due to many imperative functions of these tiny gene regulators, like patterning of nervous system[63], miRNA genes are planned to be identified, which develops miRscan and conserved in more than one genome. The conserved intronic and intragenic regions in D. melanogaster and D. pseudoobscuraare followed by additional algorithm[64]. The development of that algorithm can be achieved by the conversation of those short sequences. Based on the sequence conservation between Arabidopsis and Oryzagenome, a wide computational approach got developed[65], to detect miRNA genes present in the Arabidopsis genome. An additional approach is based on stiff complementarities between miRNAs and their targets, for the detection of miRNAs in A. thaliana.
The factors for the development of algorithm other than the Conservation of sequence were also described. Bentwitch combined Sequence directed cloning and microarray analysis with bioinformatics prediction in the year 2005, which was an integrative approach[66]. The distant/close homolog identification can be attained by the probabilistic co-learning method that got developed by the information at sequence and structural level[67]. Wang and his group were able to develop a novel computational approach with miRAlign for the detection of new miRNA based on structure and sequence alignment[68]. For the identification of candidate miRNA homolog in a set of sequences, the developed Micro-Harvester by similar sequence and structure information can be used and additional concept, that miRNAs are repeatedly found in groups, was the basis of mirAbela[69] that took into consideration only those genomic regions that are present around known miRNAs from mouse, rat and human.
As new theories were formed by the Novel concepts, Berezikov[70] reported a characteristic conservation profile used for the detection of known miRNAs efficiently, which was revealed by the phylogenetic shadowing of miRNAs in primate species. It predicts a wide-ranging set of miRNAs based on human- mouse-rat genome wide comparisons.
Most of the predictor algorithms depend on miRNA sequence conversations among diverse species. Such approaches are obviously limited to detecting only the conserved microRNAs. For the identification of non-conserving miRNAs, the machine learning concept entered, which has a difficult pattern-recognition challenging. For unique true pre-miRNAs from other pre-miRNA like hairpin structures taking into account a novel local connecting structure sequence feature, and developed a first method[71] called as Triplet-SVM, and used SVM with these structures to classify real and pseudo pre-miRNAs. To improve the calculations by Triplet-SVM, additional tool MiPred was developed in a combined way with other features with the MFE of the secondary structure[72].
Potential miRNA targets
The product of the genes that miRNA regulates can be alleged by the specific miRNA's function. The information regarding the genomic targets of miRNA is also included in the miRBase with the miRNA sequence data, from the miRNA gene identification shifted up to the functional characterization. For the extrapolation of miRNA targets in mRNA sequences, there are number of computational techniques existed, even though it is very difficult to detect the targets for the large-scale experiments [73-78]. Both for the selection of targets for experimental validation and for the prediction of potential targets, the existed methods can be used but for the most part, with a large degree of miRNA complement, the potential binding sites of miRNA are detected as the first step by the computational techniques. This is then followed by filtering out of the sites that do not appear to be unspoiled in various numbers of species. However, the species that not possess closely related genome sequences has deprived protection measure, regarding the species having orthologs clearly defined and closely related species like human, mouse and rat, this approach appears to work well for them specifically.
MicroRNAs APPLICATIONS
The upcoming paragraphs will describe about the applications of MicroRNAs and its complete role as Composite biological links, unintentional cancer genes at Genomic divisions, tumor suppressing potential and in tumor invasion metastasis. The overall expressions of various miRNAs that are discussed in this review are figured out in Fig. 5. The future role of MicroRNAs details about the suggestions and measures that can be taken in the research of various human diseases like Alzheimer's disease (AD), Parkinson's disease (PD) and Cardio-vascular diseases like Myocardial Infarction.
A cartoon showing the major miRNAs described in this review with their target specific functions and related human diseases.
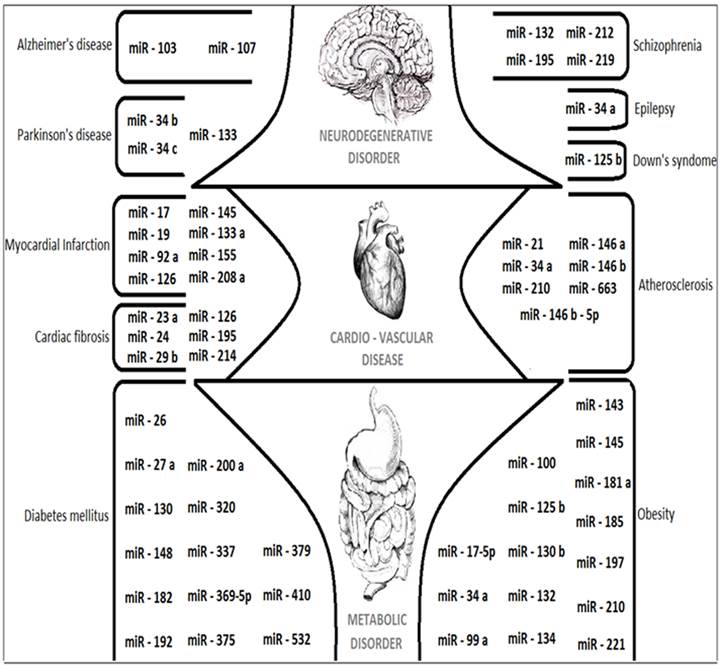
Composite biological links
The expressions of miRNA may differ on comparing the metastatic lesions with the malignancies harbor distinct patterns, which have been studied in more number and consisted of keen profiling for the investigation of global patterns of miRNA expression in malignancies. But, to have an essential role in Cancer biology, the appearance of selected miRNAs occurs [79, 80]. By understanding the roles of miRNAs in transcriptome and proteomic levels, helps to shape the modulation of the multiple biological links, as all the miRNAs don't undergo an isolated function. More than 4000 human cancers were evaluated for the miRNA expression with an example of hiring miRNA in an example of placing miRNAs in the perspective of pathways. For example, multiple numbers of MiRNAs inclines to be up-regulated namely, miR-21, miR-17, and miR-92a, whereas miR-205 and miR-145 like miRNAs tends to be down-regulated [81] in several kinds of cancers. In addition, bunches of select miRNAs inclined to differentiate subtypes of cancers. This assures that the multiple miRNAs are more active than the single miRNA function in Cancer. The subtypes of cancers are differentiated by the cluster of miRNAs and by assimilating the data of genome helps for the identification of the factual signal, so that that can be even more perplexing. Using mathematical calculations, Zhang group[82] analyzed the expression of mRNA and miRNA by multiple number of ovarian cancer samples which underwent DNA methylation and identified about the multi-dimensional modules.
Genomic divisions as unintentional Cancer genes
An involvement of miRNAs in cancer was explained as the first indication in the field of miRNA's applications in Cancer[83] and the deletion of abnormal chromosome 13 in Chronic lymphocytic leukemia (CLL) contribute to leukemogenesis. Calin group identified miR-15 and miR-16 in the deletion part of the chromosome as an unintentional gene and their expression in CLL patient's blood samples were analyzed with the result of absence of both the miRNAs when compared to normal lymphocytes. This confirms that miR-15 and miR-16 are involved in the CLL pathogenesis. Three major breakthroughs have been pointed out in the year 2005, firstly, the contribution of miRNAs to carcinogenesis and secondly, about the relationship between miR-17-92 and the Myc oncogenic pathway[84].
Metastasis and tumor incursion
In cancer, miRNA expression may be driven by the transcriptional links as reported by Ma and group[85] with the suppression of the direct target and activation of pro-metastatic gene by the induction of specific miRNA expression due to the pleiotropic transcription factor twist and leads to tumor cell incursion and metastasis. This pleiotropic transcriptional factor twist induces the expression of miR-10b which promotes cell migration and invades in breast cancer cells, which gets correlated with clinical progression. These findings of Ma group suggest that specific miRNAs have a role on the event of tumor initiation by contributing directly in tumor incursion and metastasis.
List of MicroRNAs and their corresponding functions in various human diseases
| MicroRNAs | Disease | Function | Reference |
|---|---|---|---|
| miR-132 | Neurodegenerative and Psychiatric disorder | Helps in Neuronal plasticity for both of the disorders | [21] |
| miR-103 miR-107 | Neurodegenerative disorder - Alzheimer's disease | Repress Cofilin translation | [28] |
| miR-133 | Neurodegenerative disorder - Parkinson's disease | Regulation of NDRG1 | [35] |
| miR-34b miR-34c | Neurodegenerative disorder - Parkinson's disease | Decrease Park2 and DJ1 expression with Cell Death | [34] |
| miR-132 miR-212 | Neurodegenerative disorder - Schizophrenia | Causes abnormalities in synaptic plasticity | [41] |
| miR-219 | Neurodegenerative disorder - Schizophrenia | Blocks NMDA receptors | [42] |
| miR-195 | Neurodegenerative disorder - Schizophrenia | Regulates more no. of Schizophrenia related genes | [43] |
| miR-34a | Neurodegenerative disorder - Epilepsy | Apoptosis control | [48] |
| miR-125b | Neurodegenerative disorder - Down's syndrome | Has multiple targets in the diseased brain | [49, 50] |
| miR-17, 21, 92a, 126, 133a, 145, 155, 208a | Cardio-vascular disease - Myocardial Infarction | Develops therapeutic strategies | [51, 52] |
| miR-29b | Cardio-vascular disease - Cardiac fibrosis | Contracts Collagen expression | [52] |
| miR-126 | Cardio-vascular disease - Cardiac fibrosis | Regulates Angiogenesis | [55, 56] |
| miR-23a | Cardio-vascular disease - Cardiac fibrosis | Blocks calcineurin-NFAT pathway | [58] |
| miR-24, 195, 214 | Cardio-vascular disease - Cardiac fibrosis | Regulates Cardiac hypertrophy | [59] |
| miR-663 | Cardio-vascular disease - Atherosclerosis | Modulates inflammation and proliferation of Endothelial cells | [63] |
| miR-21, 34a, 146a, 146b-5p, 210 | Cardio-vascular disease - Atherosclerosis | Expressed at substantial levels and in Atherosclerosis plaques | [64] |
| miR-146a, 146b | Cardio-vascular disease - Atherosclerosis | Expressed in Atherosclerosis plaques | [67] |
| miR-24, 26, 148, 182 | Metabolic disorder - Diabetes mellitus | Increases the levels of transcriptional repressors | [69] |
| miR-375 | Metabolic disorder - Diabetes mellitus | Controls the release of over glucose stimulated Insulin | [70] |
| miR-27a, 130a, 192, 200a, 320, 337, 369-5p, 379, 410, 532 | Metabolic disorder - Diabetes mellitus | Gets down-regulated in the concluding group | [72] |
| miR-100, 125b, 221, 34a, 130b, 185, 210 | Metabolic disorder - Obesity | Increases miRNA profiling of human fat cells | [73] |
| miR-143 | Metabolic disorder - Obesity | Positively associated with body weight and mesenteric fat weight | [74] |
| miR-17-5p, 99a, 132, 134, 145, 181a, 197 | Metabolic disorder - Obesity | Alters the morphology of adipose tissue | [75] |
| miR-17-5p, 132 | Metabolic disorder - Obesity | Differentiates obese and non-obese omental fat | [76] |
CONCLUSION
In this review, we have addressed in detail the contribution of miRNA genes in earlier studies with the future perceptions, the methodologies for gene identification and the applications of miRNA genes and their therapeutic potential for the human diseases like neuropsychiatric and neurodegenerative disorders, cardio-vascular diseases and metabolic disorders. In conclusion miRNAs are critical regulators in human diseases, holding the promise of being a new class of therapeutic targets for current drug discovery and developmental biotechnology applications.
Acknowledgements
The authors (PP and BG) acknowledge the support from the LKC School of Medicine, NTU Singapore. STS acknowledges the support from the A*STAR (Agency for Science, Technology and Research), Singapore, Science Engineering Research Council (SERC) Biomedical Engineering Program (BEP) Grant No. 103 149 0011.
COMPETING INTERESTS
The authors have declared that no competing interest exists.
References
1. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-54
2. Ruvkun G. Molecular biology. Glimpses of a tiny RNA world. Science. 2001;294:797-9
3. Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH. et al. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051-60
4. Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016-27
5. Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231-5
6. Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011-6
7. Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661-3
8. Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95-8
9. Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363-6
10. Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822-9
11. Victor Ambros, Bonnie Bartel, David P. Bartel, Christopher B. Burge, et al. A uniform system for microRNA annotation. RNA. 2003;9(3):277-279
12. Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209-16
13. Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C. et al. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557-60
14. Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201-14
15. Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358:502-11
16. Meltzer PS. Cancer genomics - Small RNAs with big impacts. Nature. 2005;435:745-6 doi:Doi 10.1038/435745a
17. van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J Clin Invest. 2007;117:2369-76
18. Bilen J, Liu N, Burnett BG, Pittman RN, Bonini NM. MicroRNA pathways modulate polyglutamine-induced neurodegeneration. Mol Cell. 2006;24:157-63
19. Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA. et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113-7
20. Schipper HM, Maes OC, Chertkow HM, Wang E. MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul Syst Bio. 2007;1:263-74
21. Beveridge NJ, Cairns MJ. MicroRNA dysregulation in schizophrenia. Neurobiol Dis. 2012;46:263-71
22. Wang H, Liu J, Zong Y, Xu Y, Deng W, Zhu H. et al. miR-106b aberrantly expressed in a double transgenic mouse model for Alzheimer's disease targets TGF-beta type II receptor. Brain Res. 2010;21:166-74
23. Perkins T, Stokes M, McGillivray J, Bittar R. Mirror neuron dysfunction in autism spectrum disorders. J Clin Neurosci. 2010;17:1239-43
24. Magill ST, Cambronne XA, Luikart BW, Lioy DT, Leighton BH, Westbrook GL. et al. microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci U S A. 2010;107:20382-7
25. Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E. et al. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220-4
26. Hirano A. Hirano bodies and related neuronal inclusions. Neuropathol Appl Neurobiol. 1994;20:3-11
27. Minamide LS, Striegl AM, Boyle JA, Meberg PJ, Bamburg JR. Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat Cell Biol. 2000;2:628-36
28. Maloney MT, Bamburg JR. Cofilin-mediated neurodegeneration in Alzheimer's disease and other amyloidopathies. Mol Neurobiol. 2007;35:21-44
29. Yao J, Hennessey T, Flynt A, Lai E, Beal MF, Lin MT. MicroRNA-related cofilin abnormality in Alzheimer's disease. PLoS One. 2010;5:0015546
30. Marksteiner J, Hinterhuber H, Humpel C. Cerebrospinal fluid biomarkers for diagnosis of Alzheimer's disease: beta-amyloid(1-42), tau, phospho-tau-181 and total protein. Drugs Today. 2007;43:423-31
31. Reitz C, Brayne C, Mayeux R. Epidemiology of Alzheimer disease. Nat Rev Neurol. 2011;7:137-52
32. Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:13
33. Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B. et al. Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis. 2008;14:27-41
34. Tanzer A, Stadler PF. Molecular evolution of a microRNA cluster. J Mol Biol. 2004;339:327-35
35. Lal A, Kim HH, Abdelmohsen K, Kuwano Y, Pullmann R Jr, Srikantan S. et al. p16(INK4a) translation suppressed by miR-24. PLoS One. 2008;3:0001864
36. Zhou X, Duan X, Qian J, Li F. Abundant conserved microRNA target sites in the 5'-untranslated region and coding sequence. Genetica. 2009;137:159-64
37. Fiedler J, Jazbutyte V, Kirchmaier BC, Gupta SK, Lorenzen J, Hartmann D. et al. MicroRNA-24 regulates vascularity after myocardial infarction. Circulation. 2011;124:720-30
38. Ikeda S, Pu WT. Expression and function of microRNAs in heart disease. Curr Drug Targets. 2010;11:913-25
39. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M. et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980-4
40. van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS. et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027-32
41. Wang S, Olson EN. AngiomiRs--key regulators of angiogenesis. Curr Opin Genet Dev. 2009;19:205-11
42. Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA. et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15:261-71
43. Ikeda S, He A, Kong SW, Lu J, Bejar R, Bodyak N. et al. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol. 2009;29:2193-204
44. Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li PF. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci U S A. 2009;106:12103-8
45. van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD. et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255-60
46. Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN. et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705-10
47. Chiu JJ, Chen LJ, Lee PL, Lee CI, Lo LW, Usami S. et al. Shear stress inhibits adhesion molecule expression in vascular endothelial cells induced by coculture with smooth muscle cells. Blood. 2003;101:2667-74
48. Zhou J, Wang KC, Wu W, Subramaniam S, Shyy JY, Chiu JJ. et al. MicroRNA-21 targets peroxisome proliferators-activated receptor-alpha in an autoregulatory loop to modulate flow-induced endothelial inflammation. Proc Natl Acad Sci U S A. 2011;108:10355-60
49. Ni CW, Qiu H, Jo H. MicroRNA-663 upregulated by oscillatory shear stress plays a role in inflammatory response of endothelial cells. Am J Physiol Heart Circ Physiol. 2011;300:4
50. Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C. et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677-84
51. Guo M, Mao X, Ji Q, Lang M, Li S, Peng Y. et al. miR-146a in PBMCs modulates Th1 function in patients with acute coronary syndrome. Immunol Cell Biol. 2010;88:555-64
52. Raitoharju E, Lyytikainen LP, Levula M, Oksala N, Mennander A, Tarkka M. et al. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis. 2011;219:211-7
53. Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481-6
54. Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938-45
55. Melkman-Zehavi T, Oren R, Kredo-Russo S, Shapira T, Mandelbaum AD, Rivkin N. et al. miRNAs control insulin content in pancreatic beta-cells via downregulation of transcriptional repressors. Embo J. 2011;30:835-45
56. Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE. et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226-30
57. Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P. et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci U S A. 2009;106:5813-8
58. Hennessy E, Clynes M, Jeppesen PB, O'Driscoll L. Identification of microRNAs with a role in glucose stimulated insulin secretion by expression profiling of MIN6 cells. Biochem Biophys Res Commun. 2010;396:457-62
59. Ortega FJ, Moreno-Navarrete JM, Pardo G, Sabater M, Hummel M, Ferrer A. et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS One. 2010;5:0009022
60. Takanabe R, Ono K, Abe Y, Takaya T, Horie T, Wada H. et al. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem Biophys Res Commun. 2008;376:728-32
61. Kloting N, Berthold S, Kovacs P, Schon MR, Fasshauer M, Ruschke K. et al. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS One. 2009;4:4
62. Heneghan HM, Miller N, McAnena OJ, O'Brien T, Kerin MJ. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J Clin Endocrinol Metab. 2011;96:2010-701
63. Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW. et al. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991-1008
64. Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:30
65. Bonnet E, Wuyts J, Rouze P, Van de Peer Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci U S A. 2004;101:11511-6
66. Bentwich I. Prediction and validation of microRNAs and their targets. FEBS Lett. 2005;579:5904-10
67. Nam JW, Shin KR, Han J, Lee Y, Kim VN, Zhang BT. Human microRNA prediction through a probabilistic co-learning model of sequence and structure. Nucleic Acids Res. 2005;33:3570-81
68. Wang X, Zhang J, Li F, Gu J, He T, Zhang X. et al. MicroRNA identification based on sequence and structure alignment. Bioinformatics. 2005;21:3610-4
69. Dezulian T, Remmert M, Palatnik JF, Weigel D, Huson DH. Identification of plant microRNA homologs. Bioinformatics. 2006;22:359-60
70. Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21-4 doi:10.1016/j.cell.2004.12.031
71. Xue C, Li F, He T, Liu GP, Li Y, Zhang X. Classification of real and pseudo microRNA precursors using local structure-sequence features and support vector machine. BMC Bioinformatics. 2005;6:310
72. Jiang P, Wu H, Wang W, Ma W, Sun X, Lu Z. MiPred: classification of real and pseudo microRNA precursors using random forest prediction model with combined features. Nucleic Acids Res. 2007;35:6
73. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20 doi:DOI 10.1016/j.cell.2004.12.035
74. Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. Rna. 2004;10:1507-17
75. Rajewsky N, Socci ND. Computational identification of microRNA targets. Dev Biol. 2004;267:529-35
76. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787-98
77. Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005 3
78. Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ. et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495-500
79. Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-8
80. Ferracin M, Pedriali M, Veronese A, Zagatti B, Gafa R, Magri E. et al. MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J Pathol. 2011;225:43-53
81. Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A. et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589-99
82. Zhang S, Liu CC, Li W, Shen H, Laird PW, Zhou XJ. Discovery of multi-dimensional modules by integrative analysis of cancer genomic data. Nucleic Acids Res. 2012;40:9379-91
83. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E. et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524-9
84. He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S. et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828-33
85. Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682-8
86. Teta M, Choi YS, Okegbe T, Wong G, Tam OH, Chong MM. et al. Inducible deletion of epidermal Dicer and Drosha reveals multiple functions for miRNAs in postnatal skin. Development. 2012;139:1405-16
87. Salta E, De Strooper B. Non-coding RNAs with essential roles in neurodegenerative disorders. Lancet Neurol. 2012;11:189-200
88. Han H, Wang YH, Qu GJ, Sun TT, Li FQ, Jiang W. et al. Differentiated miRNA expression and validation of signaling pathways in apoE gene knockout mice by cross-verification microarray platform. Exp Mol Med. 2013;8:31
Author contact
![]() Corresponding author: Parasuraman Padmanabhan. Tel: +65-6592 2583; E-mail: ppadmanabhanedu.sg or ppadmanabhan1com
Corresponding author: Parasuraman Padmanabhan. Tel: +65-6592 2583; E-mail: ppadmanabhanedu.sg or ppadmanabhan1com
 Global reach, higher impact
Global reach, higher impact