13.3
Impact Factor
Theranostics 2019; 9(7):2036-2055. doi:10.7150/thno.32738 This issue Cite
Research Paper
UBE2C, Directly Targeted by miR-548e-5p, Increases the Cellular Growth and Invasive Abilities of Cancer Cells Interacting with the EMT Marker Protein Zinc Finger E-box Binding Homeobox 1/2 in NSCLC
1. Clinical Medical Laboratory, Binzhou Medical University Hospital, Binzhou, 256603, P.R. China
2. Cancer research institute, Binzhou Medical University Hospital, Binzhou, 256603, P.R. China
3. Department of Thyroid and Breast Surgery, Binzhou Medical University Hospital, Binzhou, 256603, P.R. China
4. Department of Clinical Laboratory, Binzhou Medical University Hospital, Binzhou, 256603, P.R. China
5. Department of Gastrointestinal Surgery, Binzhou Medical University Hospital, Binzhou, 256603, P.R. China
6. Department of Hepatobiliary Surgery, Binzhou Medical University Hospital, Binzhou, 256603, P.R. China
*These authors contributed equally to this work
Received 2019-1-2; Accepted 2019-2-3; Published 2019-3-17
Abstract
Background: Recent evidence indicates that UBE2C participates in carcinogenesis by regulating the cell cycle, apoptosis, metastasis, and transcriptional processes. Additionally, miR-548e-5p dysregulation plays a vital role in tumor progression. However, the molecular mechanism via which UBE2C is directly targeted by miR-548-5p, resulting in increase in cellular growth and invasiveness of cancer cells, and its interactions with the epithelial-mesenchymal transition (EMT) marker protein ZEB1/2 in non-small cell lung cancer (NSCLC) is not understood.
Methods: Expression of UBE2C and miR-548e-5p was analyzed using reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The protein level of UBE2C and ZEB1/2 was analyzed using western blotting and immunofluorescence staining. Cellular proliferation was detected using the cell counting kit 8 (CCK8) and 3-(4,5-dimethylthiazol-2-Yl)-2,5-diphenyltetrazolium bromide (MTT) assays. Cell migration, invasion, and growth were analyzed using the wound healing and transwell assay. Promoter activity and transcription was analyzed using the luciferase reporter assay. Chromatin immunoprecipitation was used to detect binding of UBE2C to 5′UTR-ZEB1/2.
Results: We observed that 4,5-ubiquitin-conjugating enzyme E2C (UBE2C) expression was higher in NSCLC tissue than in the adjacent normal tissue and was associated with increased cell proliferation and invasion. UBE2C enhanced NSCLC progression and metastasis by affecting the cell cycle and inhibiting apoptosis. We also observed that miR-548e-5p was significantly downregulated in lung cancer tissue specimens, which decreased the expression of its direct substrate, UBE2C. Moreover, miR-548e-5p overexpression and UBE2C under-expression significantly suppressed lung cancer cell proliferation, migration, and invasion. Luciferase reporter and chromatin immunoprecipitation assays indicated that miR-548e-5p directly binds to the 3′-UTR of UBE2C and decreases UBE2C mRNA expression. Furthermore, UBE2C knockdown downregulated the mesenchymal marker vimentin and upregulated the epithelial marker E-cadherin. Bioinformatics assays, coupled with western blotting and luciferase assays, revealed that UBE2C directly binds to the 5′-untranslated region (UTR) of the transcript of the E-cadherin repressor ZEB1/2 and promotes EMT in lung cancer cells.
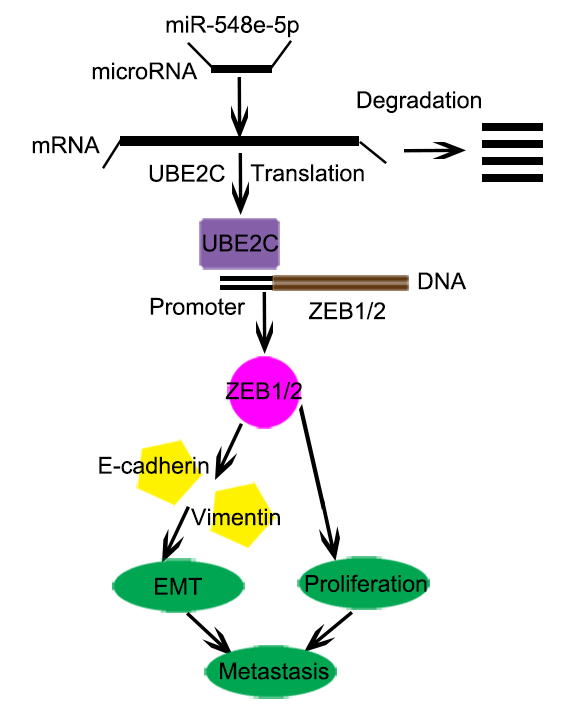
Conclusion: miR-548e-5p directly binds to the 3′-UTR of UBE2C and decreases UBE2C mRNA expression. UBE2C is an oncogene that promotes EMT in lung cancer cells by directly targeting the 5′-UTR of the transcript encoding the E-cadherin repressor ZEB1/2. miR-548e-5p, UBE2C, and ZEB1/2 constitute the miR-548e-5p-UBE2C-ZEB1/2 signal axis, which enhances cancer cell invasiveness by directly interacting with e EMT marker proteins. We believe that the miR-548e-5p-UBE2C-ZEB1/2 signal axis may be a suitable diagnostic marker and a potential target for lung cancer therapy.
Keywords: miR-548e-5p, UBE2C, ZEB1/2, cellular growth, EMT, lung cancer metastasis
Introduction
Non-small-cell lung cancer (NSCLC) is a major histological type of lung cancer. It is characterized by elusive carcinogenesis and resistance to the therapeutic interventions currently in use [1, 2]. Improvements in diagnosis and therapeutic options have substantially extended the survival of patients with lung cancer. However, most patients experience recurrences within 5 years without more effective targeted therapy [3]. Precise molecular characterization of the abnormally deregulated signal cascades involved in lung cancer development and progression is necessary to identify novel molecular targets of NSCLC, which may assist in innovating and revolutionizing lung cancer therapeutic options.
miRNA deregulation and dysfunction play a significant role in human cancer pathogenesis [4-6]. Mature miRNAs, a class of ~22 nucleotide-long RNA molecules, are cleaved from long hairpin miRNA precursors by the cytoplasmic RNase III, Dicer. Aberrant miRNA expression may be linked to tumorigenesis as miRNAs play important roles in various cellular processes [7-9]. Recent studies have considerably unraveled the molecular mechanism of lung cancer [10]. Reports show that various miRNAs are significantly deregulated in human lung cancer tissues [11-13]. Some miRNAs act as novel biomarkers and therapeutic targets for lung cancer [11]. Certain studies indicated that miR-548e-5p is associated with human tumorigenesis and cancer development [14, 15]. Downregulation of miR-548e-5p may promote cell proliferation and inhibit apoptosis, subsequently accelerating metastatic lung cancer [14]. However, the underlying mechanisms via which miR-548e-5p inhibits lung cancer progression and metastasis remain unknown. UBE2C is a ubiquitin-conjugating enzyme that acts with the ubiquitin activating enzyme E1 and ubiquitin protein ligase E3 to catalyze the degradation of proteins into smaller polypeptides, amino acids, and ubiquitin in the 26S proteasome. UBE2C participates in carcinogenesis by regulating the cell cycle, apoptosis, and transcriptional processes. UBE2C upregulation has been correlated with poor overall survival (OS) and progression-free survival (PFS) in patients with NSCLC [16-18]. Previous studies have shown that UBE2C overexpression promotes cell proliferation. In various cell lines, short interfering (si)RNA-mediated UEB2C knockdown decreased cell proliferation [19-21]. Therefore, UBE2C expression is associated with the degree of malignancy of breast, lung, ovary, and bladder cancers and lymphoma. UBE2C downregulation inhibited proliferation, clone formation, and malignant transformation and promoted senescence in tumor cells [22], although the underlying mechanisms are not clear.
Epithelial-mesenchymal transition (EMT) is a crucial event in the progression toward cancer metastasis. It triggers cellular mobility and induces the invasion of tumor cells [23, 24]. EMT is mediated by the EMT-inducing transcriptional factors ZEB1/2. During this process, epithelial cells lose E-cadherin expression and cell-cell contact, change their apical-basal polarity, and transdifferentiate into mesenchymal cells [25-27]. The most prominent characteristics of an EMT event are loss in the expression of E-cadherin and epithelial markers and increase in the expression of the mesenchymal markers, N-cadherin and vimentin [25]. Reports show that the EMT-activator ZEB1/2 promotes metastasis by interacting with some transcription factors [27-30]. Furthermore, some reports indicated that EMT is regulated at multiple levels, including transcriptional control of gene expression, regulation of RNA splicing, and translational/post-translational control [31, 32]. ZEB1 plays an important role in this process as it is a central element in the network of transcription factors that control EMT. Therefore, the etiology of fatal tumors such as lung cancers can be elucidated by targeting ZEB1/2 and certain molecular networks. Here we report that the downregulation of miR-548e-5p expression correlates with UBE2C upregulation in lung cancer tissues and cell lines. UBE2C increases ZEB1/2 transcription and protein levels. Therefore, miR-548e-5p, UBE2C, and ZEB1/2 constitute a signal transduction pathway known as the miR548e-UBE2C-ZEB1/2 signal axis, which regulates EMT in lung cells and lung cancer cell migration and invasion. Our study demonstrated that the miR548e-UBE2C-ZEB1/2 signal axis enhances lung cancer cell invasiveness by directly interacting with the EMT maker proteins, E-cadherin and vimentin.
Materials and Methods
Molecular biology
Flag-tagged UBE2C, Flag-tagged ZEB1 and Flag-tagged ZEB2 constructs were made using the pcDNA 3.1 vector (Invitrogen, Carlsbad, CA, USA). Sequences encoding the Flag epitope (DYKDDDDK) were added by PCR through replacement of the first Met-encoding codon in the respective cDNA clones.
Cell lines and culture
Human lung normal cell line HBEC and NSCLC cell lines A549, H1299, Calu6 and H520 were purchased from American Type Culture Collections (Manassas, VA). 95-D cells were purchased from the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Science (Shanghai, China). Cell lines were cultivated in RPMI-1640 medium supplemented with 10% FBS (Hyclone, USA), penicillin /streptomycin (100 mg/mL). Culture flasks were kept at 37 ˚C in a humid incubator with 5% CO2.
microRNAs and the process of its processing and maturation
The miRNAs are noncoding single‐stranded small molecules of 19-22 nucleotides. The biogenesis of miRNAs begins in the nucleus and plays a role in cytoplasm. Its mature process contains of catalyzing, cleaving, transporting, which includes three stages: pri‐miRNA (1-3 kb), pre‐miRNAs (60-70 bp), and mature miRNAs (19-22 bp). The microRNAs and the process of its processing and maturation were described in Atena Soleimani. et al. [33].
Over-expression and knockdown of genes
Overexpressing plasmid (2 μg) or siRNA (1.5 μg) of indicated genes were transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) for over-expression and knockdown of indicated genes, followed by analysis 48-72 h later. The selected sequences for knockdown as follow: siUBE2C1 were: 5′-CCUGCAAGAAACCUACUCA-3′, siUBE2C2 were 5′-CUUCUAGGAGAACCCAACA-3′, siZEB1-1 were: 5′-GGAUCAACCACCAAUGGUU-3′, siZEB1-2 were: 5′-AGAUGAUGAAUGCGAGUCG-3′, siZEB2-1 were: 5′-CGAGAUAUGUAAACUAAGGA-3′,siZEB2-2 were: 5′- GTTAATATTCATAGCTTCATC-3′.
Western blot analysis
Human lung cancer cells were transfected with the relevant plasmids and cultured for 36 h. For western blot analysis, cells were lysed in NP-40 buffer (10 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA pH 8.0, 1 mM EGTA pH 8.0, 1 mM PMSF, and 0.5% NP-40) at 25 ˚C for 40 min. The lysates were added to 5× loading dye and then separated by electrophoresis. The primary antibodies used in this study were 1:1000 rabbit anti-Flag (sc-166384, Santa Cruz, Dallas, TX, USA) and 1:1000 Abcam (Cambridge, UK) antibody of UBE2C (ab12290), ZEB1 (ab203829), ZEB2 (ab138222), Vimentin (ab45939), E-cadherin (ab1416), cleaved Capase-3 (ab32042) and Tubulin (ab6046).
Immunofluorescent staining
To examine the protein expression by immunofluorescent staining, lung cancer cells were seeded onto coverslips in a 24-well plate and left overnight. Cells were then fixed using 4% formaldehyde for 30 min at 25 ˚C and treated with 2% bovine serum albumin (BSA) in phosphate buffered saline (PBS) for 30 min. The coverslips were incubated with rabbit anti-UBE2C, Ki67, Annexin V, ZEB1, ZEB2, Vimentin and mouse anti-E-cadherin monoclonal antibody (Abcam) at 1:200 dilution in 3% BSA. Alexa-Fluor 467 (green, 1:500, A‑11029; Invitrogen, USA) and 594 (red, 1:500, A‑11032; Invitrogen, USA) tagged anti-rabbit or -mouse monoclonal secondary antibody at 1:1000 dilution in 3% BSA. Hoechst (3 µg/mL, cat. no. E607328; Sangon Biotech Co., Ltd.) was added for nuclear counterstaining. Images were obtained with a Zeiss Axio Imager Z1 Fluorescent Microscope (Zeiss, Oberkochen, Germany).
Cell flow cytometry assays
A549 cells were transfected with the relevant plasmids culturing for 36 h, harvested and fixed with 70% ethanol. These cells were then stained using propidium iodide (PI) and the cell cycle stage assessed by flow cytometry. Data were collected by BD FACSC Flow Cytometer using FACSD software (BD Biosciences, San Jose, CA, USA) and analyzed by a software FlowJo (http://www.flowjo.com).
RNA isolation and reverse transcription (RT)-PCR assay
We used TRIzol reagent (TransGen Biotech, Beijing, China) to isolate total RNA from the samples. RNA was reverse transcribed into first-strand cDNA using a TransScript All-in-One First-Strand cDNA Synthesis Kit (TransGen Biotech). cDNAs were used in RT-PCR and RT-qPCR assay with the human GAPDH gene as an internal control. The final RT-qPCR reaction mix contained 10 μL Bestar® SYBR Green qPCR Master Mix, Amplification was performed as follows: a denaturation step at 94 ˚C for 5 min, followed by 40 cycles of amplification at 94 ˚C for 30 sec, 58 ˚C for 30 sec and 72 ˚C for 30 sec. The reaction was stopped at 25 ˚C for 5 min. The relative expression levels were detected and analyzed by ABI Prism 7900HT / FAST (Applied Biosystems, USA) based on the formula of 2-ΔΔct. We got the images of RT-PCR by Image LabTM Software (ChemiDocTM XRS+, BiO-RAD) and these images were TIF with reversal color format. The RT-PCR primers were: UBE2C forward primer: 5′-GGATTTCTGCCTTCCCTGAA-3′, UBE2C reverse primer: 5′-GATAGCAGGGCGTGAGGAAC-3′, E-cadherin forward primer: 5′-ACCATTAACAGGAACACAGG -3′, E-cadherin reverse primer: 5′-CAGTCACTTTCAGTGTGGTG-3′, Vimentin forward primer: 5′- CGCCAACTACATCGACAAGGTGC-3′, Vimentin reverse primer: 5′-CTGGTCCACCTGCCGGCGCAG-3′, GAPDH forward primer: 5′-CTCCTCCTGTTCGACAGTCAGC-3′, GAPDH reverse primer: 5′-CCCAATACGACCAAATCCGTT-3′, ZEB1 forward primer: 5′-GATGATGAATGCGAGTCAGATGC-3′, ZEB1 reverse primer: 5′-CTGGTCCTCTTCAGGTGCC-3′, ZEB2 forward primer: 5′-AACAACGAGATTCTACAAGCCTC-3′, ZEB2 reverse primer: 5′-TCGCGTTCCTCCAGTTT TCTT-3′.
Subcellular Fraction
Transfected A549 cells were harvest in PBS and resuspended for 10min on ice in 500 μL CLB Buffer (10 mM Hepes, 10 mM NaCl, 1 mM KH2PO4, 5 mM NaHCO3, 5 mM EDTA, 1 mM CaCl2, 0.5mM MgCl2). Thereafter, 50 μL of 2.5 M sucrose was added to restore isotonic conditions. The first round of centrifugation was performed at 6300 g for 5 min at 4 ˚C. The pellet washed with TSE buffer (10 mM Tris, 300 mM sucrose, 1 mM EDTA, 0.1% NP40, PH 7.5) at 4000 g for 5 min at 4 ˚C until the supernatant was clear. The resulting supernatant was discarded, and the pellets were nucleus. The resulting supernatant from the first round of differential centrifugation was sedimented for 30 min at 14000 rpm. The resulting pellets were membranes and the supernatant were cytoplasm.
SA-β-gal staining
SA‑β‑gal was detected using the Senescence β‑Galactosidase Staining kit (C0602; Beyotime) following the manufacturer's instructions: In brief, the cells were washed twice with PBS and then fixed with PBS containing 2% formaldehyde and 0.2% glutaraldehyde for 10 min. The cells were then incubated at 37 ˚C for 12 h with staining solution. After being washed twice with PBS, the SA‑β‑gal‑positive cells were observed under an optical microscope (IX53, Olympus, Tokyo, Japan) and assessed using the ImageJ software (http://imagej.nih.gov/ij/).
MTT and CCK8 assays
Cell viability was determined using 3-(4,5-dimethylthiazol-2yl)-2,5-diphenyltetrazolium bromide (MTT) and CCK8 assays in 96-well plates in a manner. Cells were transfected with the relevant plasmids culturing for 36 h, followed by incubation with MTT and CCK8 for 4 h. Next 100 μL isopropanol (in 0.04 N-hydrochloric acid) was added to dissolve the formazan crystals for the MTT assay. Absorbance was read at 450 nm for CCK8 and 570 nm for MTT assay using a spectrophotometer (Tecan, Männedorf, Switzerland).
Luciferase reporter assay
To construct the core region of UBE2C, ZEB1 and ZEB2 promoters, the regions of UBE2C, ZEB1 or ZEB2 were amplified by PCR from the human genomic DNA of A549 cells and were inserted into the upstream of the pGL3-Basic vector (Promega, Madison, WI, USA) via KpnI and XhoI sites to generate UBE2C luc, ZEB1 luc and ZEB2 luc. Thereafter, we use the Firefly Luciferase Reporter Gene Assay Kit (Beyotime, RG005) to detect the promoter activities using a spectrophotometer (Tecan, Männedorf, Switzerland). The PCR primers were: UBE2C forward primer: 5′-GATATGAACCTGTGTTGT-3′, UBE2C reverse primer: 5′-GGCTCGGCTCAGCTCCTTTACGG-3′, ZEB1 forward primer: 5′-GAAACCAGGCGTCCCTGG-3′, ZEB1 reverse primer: 5′-CAACCGTGGGCACTGCTGAA-3′, ZEB2 forward primer: 5′-TTGGTGTACCAAGAGGC-3′, ZEB2 reverse primer: 5′-CAACCCTGAAACAGAGG-3′.
CHIP assay
ChIP experiments were performed according to the laboratory manual. Immunoprecipitation was performed for 6 h or overnight at 48 °C with specific antibodies. After immunoprecipitation, 45 μL protein A-Sepharose and 2 μg of salmon sperm DNA were added and the incubation was continued for another 1 h. Precipitates were washed sequentially for 10 min each in TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 150 mM NaCl), TSE II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1, 500 mM NaCl), and buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl, pH 8.1). Precipitates were then washed three times with TE buffer and extracted three times with 1% SDS, 0.1 M NaHCO3. Eluates were pooled and heated at 65 ˚C for at least 6 h to reverse the formaldehyde cross-linking. DNA fragments were purified with a QIAquick Spin Kit (Qiagen, CA). For PCR, 2 μL from a 5 mL extraction and 21-25 cycles of amplification were used. The sequences of the primers used are provided as follows: ZEB1 forward primer: 5′-GAAGTCACTTCCCATCCCGG-3′, ZEB1 reverse primer: 5′-CGCGGCTGCCCGGGGCAGG-3′, ZEB2 forward primer: 5′-AAGTATGTACTGACATAACC-3′, ZEB2 reverse primer: 5′-GCTCTAAAGGAAGCAATCAT-3′.
Wound healing assays
To assess the cellular migration, 104 cells were seeded onto 6-well plates with transfection of the relevant plasmids. These were then incubated in 5% CO2 at 37 ˚C for 48 h. A wound was scraped into the cells using a plastic 200 μL tip and then washed by PBS. The cells were then incubated in RPMI-1640 medium containing 2% FBS. Images were captured at the time points of 0 and 36 h after wounding. The relative distance of the scratches was observed under an optical microscope (IX53, Olympus, Tokyo, Japan) and assessed using the ImageJ software.
Transwell migration assays
Transwell migration assays were performed using a 24-well chamber (Costar 3422; Corning Inc., Corning, NY, USA). The lower and upper chambers were partitioned by a polycarbonate membrane (8-μm pore size). Lung cancer cells (5 × 103) were seeded into RPMI-1640 without FBS in the upper chamber. RPMI-1640 containing 10% FBS was added to the lower chamber. The cells were allowed to migrate for 36 h at 37 ˚C in a humidified atmosphere containing 5% CO2. Cells remaining on the upper side of the membrane were removed using PBS-soaked cotton swabs. The membrane was then fixed in 4% paraformaldehyde for 20 min at 37 ˚C and then stained with crystal violet. Cells on the lower side of the membrane were counted under an Olympus light microscope (Olympus, Tokyo, Japan).
Analysis of publicly available datasets
To analyze correlation between UBE2C, ZEB1, ZEB2 or miR-548e-5p expression level and prognostic outcome of patients, Kaplan-Meier survival curves of NSCLC patients with low and high expression of UBE2C, ZEB1, ZEB2 or miR-548e-5p were generated using Kaplan-Meier Plotter (www.kmplot.com/analysis and www.oncolnc.org) [34].
Human lung cancer specimen collection
All the human lung cancer and normal lung specimens were collected in Affiliated Hospital of Binzhou Medical College with written consents of patients and the approval from the Institute Research Ethics Committee.
In vivo experiments
To assess the in vivo effects of miR-548e-5p, 3 to 5-week old female BALB/c athymic (NU/NU) nude mice were housed in a level 2 biosafety laboratory and raised according to the institutional animal guidelines of Binzhou Medical University. All animal experiments were carried out with the prior approval of the Binzhou Medical University Committee on Animal Care. For the experiments, mice were injected with 5 × 106 lung cancer cells with stably expression of relevant plasmids and randomly divided into two groups (five mice per group) after the diameter of the xenografted tumors had reached approximately 5 mm in diameter. Xenografted mice were then intraperitoneally administered for three times a week and tumor volume and body weight were measured every second day. Tumor volume was estimated as 0.5 × a2 × b (where a and b represent a tumors short and long diameter, respectively). Mice were euthanized after six weeks and the tumors were measured a final time. Tumor and organ tissue were then collected from xenograft mice and analyzed by immunohistochemistry.
Immunohistochemical analysis
Tumor tissues were fixed in 4% paraformaldehyde overnight and then embedded in paraffin wax. Four-micrometer thick sections were and stained using hematoxylin and eosin (H&E) for histological analysis.
Statistical analysis
Each experiment was repeated at least three times. The statistical analyses of the experiment data were performed by using a two-tailed Student's paired T-test and one-way ANOVA. Statistical significance was assessed at least three independent experiments and significance was considered at either P-value < 0.05 was considered statistically significant and highlighted an asterisk in the figures, while P-values < 0.01 were highlighted using two asterisks and P-values < 0.001 highlighted using three asterisks in the figures.
Ethics approval and consent to participate
The experimental protocol was approved by the Research Ethics Committee of Binzhou Medical University, China (No. 2017-016-01 for human lung cancer specimen and No. 2017-009-09 for mouse experiments in vivo) and the written informed consent was obtained from all subjects. Informed consent was obtained from all individual participants included in the study. All patients were staged based on the criteria of the 7th Edition of the AJCC Cancer Staging Manual (2010).
Results
Aberrant UBE2C activation in lung tumors and UBE2C dysfunction affected cell proliferation and invasion
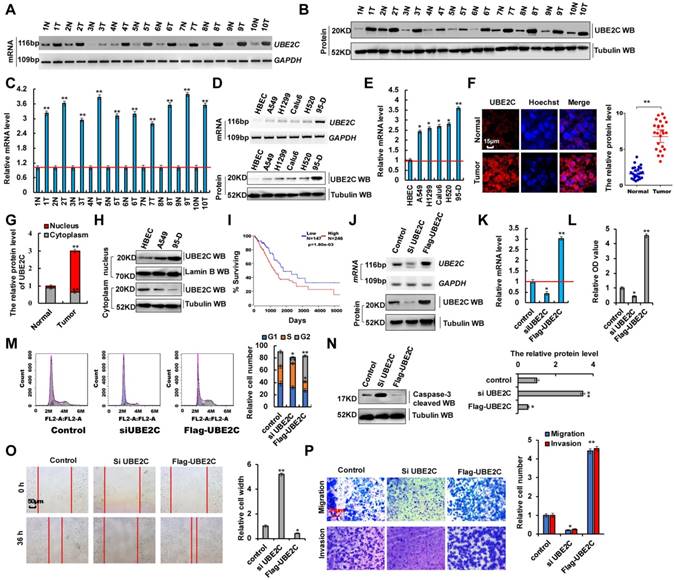
In total, 50 samples were obtained from patients who underwent lung resection surgery at the Affiliated Hospital of Binzhou Medical College (Binzhou, China) between January 2014 and January 2016. Each sample was examined, and the clinicopathological findings of UBE2C are summarized in Table 1. To investigate the endogenous UBE2C mRNA and protein levels in human lung cancer tissues, we performed reverse transcription-polymerase chain reaction (RT-PCR), reverse transcription-quantitative PCR (RT-qPCR) and western blotting analysis, respectively. We observed that UBE2C mRNA and protein levels were higher in human lung cancer tissues than in their normal adjacent lung tissues (Figure 1A-C). We also observed that UBE2C mRNA and protein levels were higher in lung cancer cells than in normal human bronchial epithelial cell (HBEC) controls (Figure 1D, E). UBE2C expression was highest in 95-D cells. This subline showed stronger cellular proliferation and metastasis than other lung cancer cells (Figure S1A-D). Immunohistochemical assay using frozen and paraffin embedded sections showed that UBE2C protein levels were significantly higher in human lung tumor tissues than in adjacent normal lung tissues (Figure 1F and 2E). The relative level of cytoplasmic UBE2C was lower in these lung tumor samples than in adjacent normal lung tissues. Immunohistochemical and immunoblotting assay indicated that UBE2C accumulated to higher concentrations in NSCLC cell nuclei than in control cell (HBEC) nuclei (Figure 1G, H). Immunofluorescence staining indicated that the nuclear UBE2C concentrations were higher in 95-D cells than in A549- or HBEC cells (Figure S2A). Publicly available datasets were screened and used to determine the prognostic correlation between UBE2C expression and survival of patients with lung cancer. Kaplan-Meier analyses indicated that higher UBE2C expression level correlated highly with shorter overall survival (OS) (n = 491, P = 1.08 × 10-3) [34, 35] (Figure 1I). These results showed that UBE2C, as a probable growth factor, promoted lung tumorigenesis and lung cancer development. UBE2C silencing using siRNA (siUBE2C-1 and siUBE2C-2) and UBE2C overexpression using pcDNA-Flag UBE2C were used to determine whether UBE2C oncogene activation underlies lung cancer initiation, progression, and metastasis (Figure 1J, K and S1E-G). We observed that UBE2C knockdown was better using siUBE2C-2 (Figure S1E-G), and therefore, siUBE2C-2 was used for knocking down UBE2C in subsequent experiments. These approaches were also used in experiments regarding ZEB1/2 or miR-548e-5p. Cell counting kit 8 (CCK8) assay (Figure 1L) and Ki67 immunoblotting (Figure S2B) showed that UBE2C knockdown and overexpression decreased and increased A549 cell proliferation, respectively. The 3-(4,5-dimethylthiazol-2-Yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Figure S2C) indicated that UBE2C overexpression promoted cell growth. UBE2C depletion and overexpression significantly decreased and increased clone formation, respectively (Figure S2D). Furthermore, siUBE2C induced significant G2 arrest, whereas UBE2C increased the number of A549 cells in the S phase, as shown in histograms representing cell cycle distribution and quantitation analysis (Figure 1M). Results of the immunoblotting assay indicated that the level of the pro-apoptotic protein caspase-3 increased with siUBE2C treatment and decreased with UBE2C overexpression in A549 cells (Figure 1N). Immunofluorescence staining revealed that annexin V level increased with siUBE2C treatment and decreased with UBE2C overexpression in A549 cells (Figure S2E). Furthermore, results of the scratch assay (Figure 1O and S1H) and transwell assay (Figure 1P and S1I) showed that UBE2C knockdown in 95-D cells or overexpression in A549 cells decreased or increased cellular migration and invasion, respectively. To facilitate subsequent research, the scratch and transwell assays were performed only in A549 cells with overexpression or knockdown of UBE2C. These results demonstrated that aberrant UBE2C activation occurred in lung tumors and UBE2C dysfunction affected cell proliferation and invasion.
Patient's demographics and tumor characteristics and association of miR-548e-5p and UBE2C level with clinicopathological features in lung tumor population.
| Characteristics | No. of patients, N=50 (%) | P value |
|---|---|---|
| Patients Parameter | ||
| Age (years) | 0.142 | |
| Average [range] | 55 [30-81] | |
| <55 | 20 (40.0) | |
| ≥55 | 30 (60.0) | |
| Gender | 0.0681 | |
| Male | 35 (70.0) | |
| Female | 15 (30.0) | |
| Tumor Characteristics | ||
| Tumor size (cm) | 0.005** | |
| <4 | 12 (24.0) | |
| ≥4 | 38 (76.0) | |
| Differentiation | 0.056 | |
| Poor | 35 (70.0) | |
| Well-moderate | 15 (30.0) | |
| Lymph node metastasis | 0.034* | |
| N- | 10 (20.0) | |
| N+ | 40 (80.0) | |
| Distant metastasis | 0.014* | |
| M- | 18 (36.0) | |
| M+ | 32 (64.0) | |
| Level of UBE2C | ||
| Protein level | (Figure 1B) | |
| High | 39 (78.0) | 0.003** |
| median | 7 (14.0) | 0.051 |
| low | 4 (8.0) | 0.132 |
| mRNA level | (Figure 1A) | |
| High | 42 (84.0) | 0.001** |
| median | 5 (10.0) | 0.053 |
| low | 4 (6.0) | 0.132 |
| Level of miR-548e-5p | ||
| mRNA level | (Figure 6A) | |
| High | 2 (4.0) | 0.167 |
| median | 4 (8.0) | 0.021 |
| low | 44 (88.0) | 0.003** |
Abnormal ZEB1 or ZEB2 expression in lung tumors, and ZEB1/2 overexpression promoted cellular proliferation and invasion via EMT
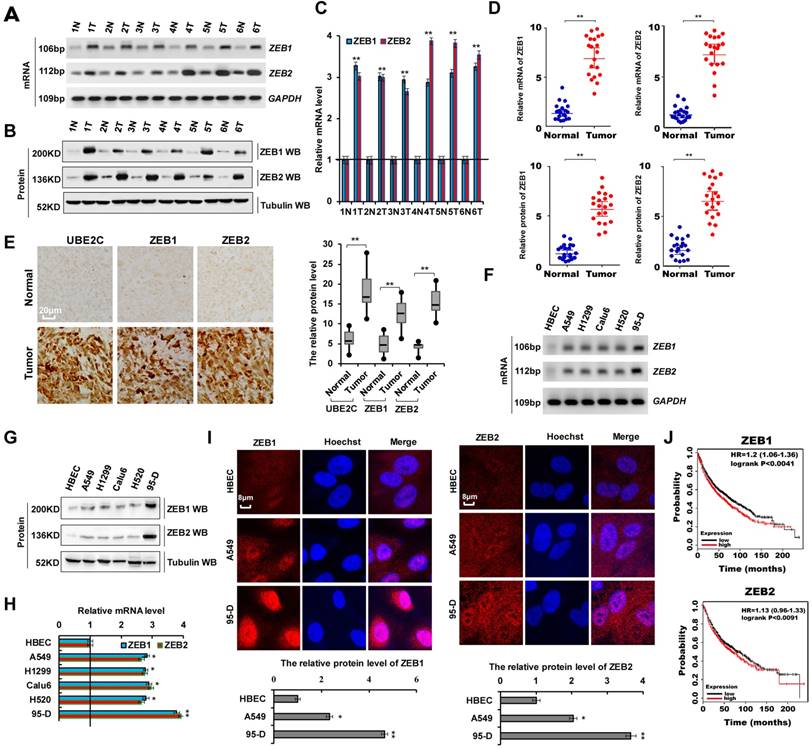
As shown in Supplementary Table 1, we analyzed the patient demographics and tumor characteristics and the association of ZEB1/2 level with clinicopathological features in lung tumor population from patients (n = 45) who underwent a lung resection surgery between January 2016 and January 2018. These data indicated that ZEB1/2 expression was higher in lung cancers and ZEB1/2 overexpression promoted cell invasion. To assess ZEB1 and ZEB2 expression in human lung cancer tissues, we performed RT-PCR, immunoblotting, and RT-qPCR. We observed that ZEB1 and ZEB2 mRNA (Figure 2A, C, D) and protein (Figure 2B, D) levels were higher in human lung cancer tissues than in normal adjacent lung tissues. A semi-quantitative immunohistochemical analysis of ZEB1 and ZEB2 expression revealed that ZEB1 and ZEB2 protein levels were higher in human lung cancer tissues than in normal adjacent tissues (Figure 2E). Furthermore, RT-PCR, immunoblotting, and RT-qPCR assays indicated that the ZEB1 and ZEB2 mRNA and protein levels were higher in lung cancer cells than in normal HBEC controls (Figure 2F-H). Immunofluorescence assay demonstrated higher ZEB1 and ZEB2 expression in A549 and 95-D cells than in normal HBEC controls (Figure 2I). Publicly available datasets were screened and used to analyze the prognostic correlation between ZEB1 or ZEB2 and lung cancer patient survival. Kaplan-Meier analyses indicated that higher ZEB1 or ZEB2 expression levels correlated highly with shorter overall survival (OS) (n = 1926, P = 1.0 × 10-6) (31) (Figure 2J). Furthermore, according to the scratch assay (Figure S1 A) and transwell assay (Figure S1B), cellular migration and invasion were higher in 95-D cells with elevated ZEB1 and ZEB2 expression levels than in A549 cells (Figure 2F). These results showed that ZEB1 or ZEB2 expression was abnormal in lung tumors and cell lines and that ZEB1/2 overexpression promoted cell invasion.
Aberrant activation of UBE2C in lung tumors from patients and dysfunction of UBE2C affected cell proliferation and invasion. (A-C) RT-PCR (A), western blot (B) and RT-qPCR (C) indicated that the mRNA and protein levels of UBE2C were higher in human lung cancer tissues compared with their normal adjacent lung tissues. (D, E) Gel-based RT-PCR and immunoblotting with densitometric quantitation (D) and RT-qPCR (E) demonstrating elevated mRNA and protein expressions of UBE2C in human lung cancer cells compared with their normal control cell HBEC. (F, G) Immunohistochemistry with frozen sections indicated that increased the protein level of UBE2C in lung cancer samples compared with their normal adjacent lung tissues (F) and UBE2C was accumulated in nuclear of lung cancer samples while more UBE2C was localized in cytoplasm of those normal adjacent lung tissues (G). (H) Immunoblotting with densitometric quantitation demonstrating increased nuclear and decreased cytoplasmic UBE2C in A549 and 95-D cells than normal cell line HBEC. (*p<0.05, **p<0.01 vs control group correspond to two-tailed Student's tests). (I) Kaplan Meier overall survival (OS) curves of UBE2C (p=1.80e-03 by log-rank test for significance) for human lung cancers. (J, K) RT-PCR, western blot (J) and RT-qPCR (K) assays indicated that the mRNA and protein levels of UBE2C were decreased by using siRNA but over expressing UBE2C increased the mRNA and protein levels of UBE2C in A549 cells compared with the control cells. (L) In vitro proliferation assay by CCK8 demonstrating that knockdown of UBE2C using the siRNA or over expressing UBE2C significantly arrested or promoted cellular proliferation in A549 cells. (M) Knockdown of UBE2C using the siRNA or over expressing UBE2C significantly influenced cellular S and G2 phase as showing in both representative histograms of cell cycle distribution and their quantitation analysis. (N) Western blot indicated that pro-apoptosis protein caspase-3 was more increased by using siRNA of UBE2C or significantly decreased by over expressing UBE2C in A549 cells compared with their control cells, respectively. (O) Scratch assay showing that over expressing UBE2C or knockdown of UBE2C using the siRNA dramatically increased or decreased cell migration in A549 cells for 36h. (P) Trans-well assay identified that over expressing UBE2C or knockdown of UBE2C using the siRNA significantly increased or decreased cell invasive growth and migration compared with the A549 control cells. (*p<0.05, **p<0.01, ANOVA with Bonferroni correction). Results were presented as mean ± SD, and the error bars represent the SD of three independent experiments.
Abnormal ZEB1 or ZEB2 expression in lung tumors and ZEB1/2 overexpression promoted cellular proliferation. (A-C) RT-PCR (A), western blot (B) and RT-qPCR (C) indicated that the mRNA and protein levels of ZEB1 and ZEB2 were higher in human lung cancer tissues compared with their normal adjacent lung tissues. (D) the mRNA and protein expressions of ZEB1 and ZEB2 in 20 lung cancer tissues and adjacent normal tissues. (E) Immunohistochemical assay of human lung tissues and their normal adjacent lung tissues shows that UBE2C, ZEB1 and ZEB2 expression was more increased in human lung tissues, (n=15). (F-I) Gel-based RT-PCR (F), immunoblotting with densitometric quantitation (G), RT-qPCR (H) and immunofluorescent staining (I) demonstrating elevated mRNA and protein expressions of ZEB1 and ZEB2 in human lung cancer cells compared with their normal control cell HBEC. (J) Kaplan Meier overall survival (OS) curves of ZEB1 and ZEB2 (n=1926, p=0.0041 for ZEB1 and p=0.0091 for ZEB2 by log-rank test for significance) for lung cancers. Results were presented as mean ± SD, and the error bars represent the SD of three independent experiments. (*p<0.05, **p<0.01 vs control group correspond to two-tailed Student's tests).
ZEB1 and ZEB2 promote cellular proliferation and invasion via EMT
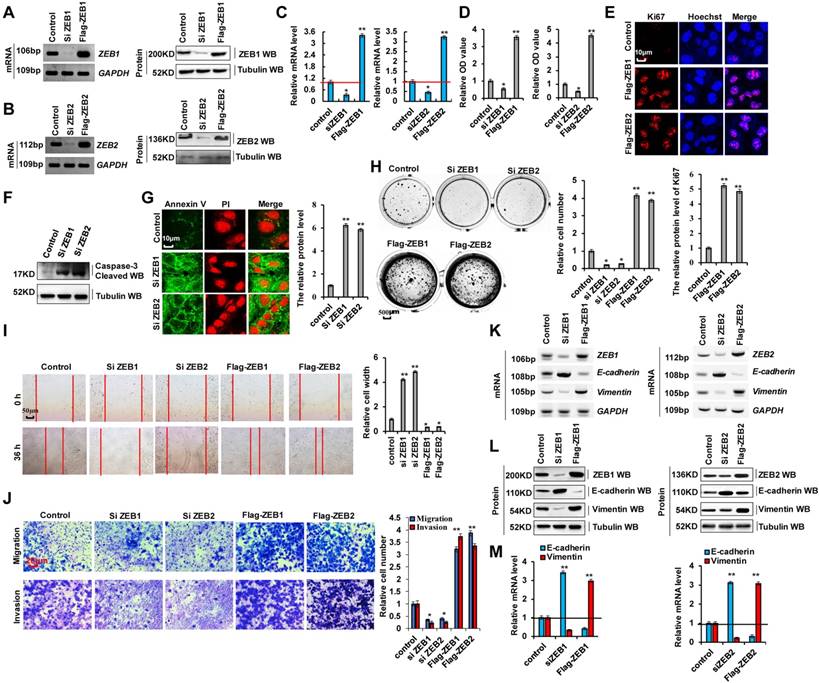
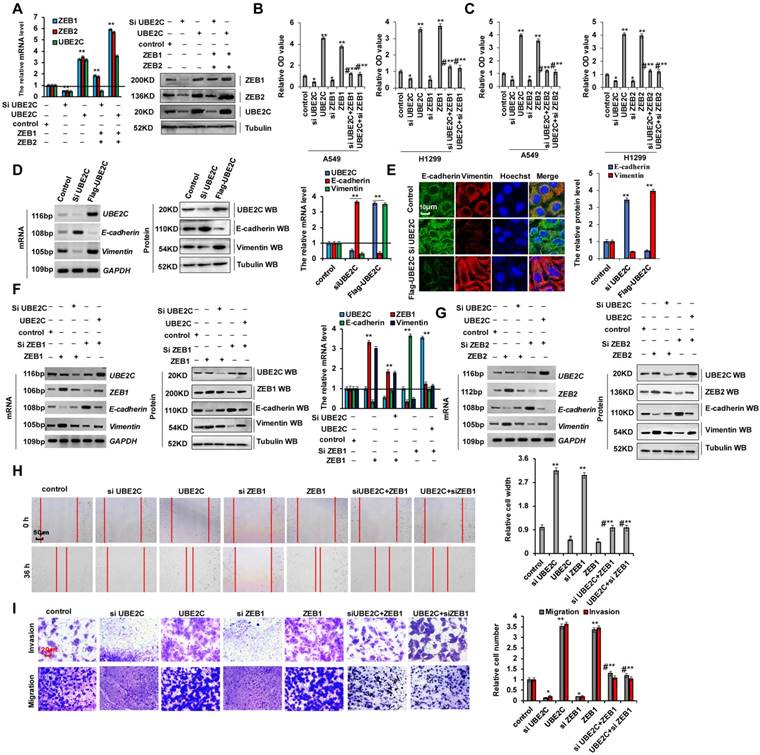
As cellular invasion and migration were higher in 95-D cells than in A549 cells, we determined whether EMT marker protein expression was relatively higher in 95-D cells. RT-PCR, western blotting, and RT-qPCR assays indicated that vimentin mRNA and protein levels were higher in 95-D cells than in A549 cells. In contrast, those of E-cadherin were lower in 95-D cells than in A549 cells (Figure S3A-C). To determine the molecular mechanism via which ZEB1/2 regulate cellular invasion and migration, we silenced ZEB1(siZEB1-1 and siZEB1-2) or ZEB2 (siZEB2-1 and siZEB2-2) with siRNAs and induced transient ectopic ZEB1 (pcDNA Flag-ZEB1) or ZEB2 (pcDNA Flag-ZEB2) overexpression (Figure 3A-C and S3D-I). Our objective was to determine whether ZEB1 or ZEB2 oncogene activation affected lung cancer initiation, progression, and metastasis. Furthermore, we observed that ZEB1 or ZEB2 knockdown using siZEB1-1 or siZEB2-1 was better, and therefore siZEB1-1 or siZEB2-1 were used for knocking down ZEB1 or ZEB2 in further experiments (Figure S3D-I). We observed that A549 and 95-D cell migration and invasion were significantly reduced after transfection with siZEB1 or siZEB2 (Figure S3J, K). These data suggested that higher ZEB1/2 expression levels promote cellular invasion and migration in 95-D cells than in A549 cells by regulating the expression of EMT marker proteins vimentin and E-cadherin. In addition, ZEB1 or ZEB2 knockdown decreased cell growth (Figure 3D), Ki67 protein levels (Figure 3E), clone formation (Figure 3H), cell migration (Figure 3I), and cell invasiveness (Figure 3J) but increased cleaved caspase-3 (Figure 3F) and annexin V (Figure 3G) protein levels in A549 cells. The opposite effects were observed for all these factors in A549 cells overexpressing ectopic ZEB1 or ZEB2 (Figure 3D-J). Furthermore, ZEB1 or ZEB2 depletion decreased vimentin mRNA and protein levels but increased those of E-cadherin, and opposite effects were observed for all these factors in A549 cells overexpressing ectopic ZEB1 or ZEB2 (Figure 3K-M). These observations demonstrate that ZEB1 and ZEB2 play important roles in lung cancer cell growth, EMT, and invasion.
ZEB1 and ZEB2 promotes cellular proliferation and invasion via EMT. (A-C) RT-PCR (A, B), western blot (A, B) and RT-qPCR (C) assays indicated that decreased or increased the protein levels of ZEB1 (A, C) and ZEB2 (B, C) by using siRNA or over expressing ZEB1 and ZEB2 in A549 cells compared with the control cells. (D) In vitro proliferation assay by CCK8 demonstrating that knockdown of ZEB1 or ZEB2 using the siRNA or over expressing ZEB1 or ZEB2 significantly arrested or promoted cellular proliferation in A549 cells, respectively. (E) Immunofluorescence assay showed that cellular proliferative maker protein of Ki67 was increased in A549 cells by over expressing ZEB1 or ZEB2 compared with control cells. (F) Immunoblotting assay indicated that the protein of active Caspase-3 was increased in A549 cells with knockdown of ZEB1 or ZEB2 using siRNA compared with control cells. (G) Immunofluorescent staining indicated that the protein of Annexin V was increased in A549 cells with knockdown of ZEB1 or ZEB2 using siRNA compared with control cells. (H-M) A549 cells were transfected with Flag-ZEB1 and Flag-ZEB2 or knockdown of ZEB1 and ZEB2 using the siRNA, separately. (H) Colony formation was analyzed by colony formation assay. (I) Cell migration growth was analyzed by scratch assay. (J) the cell invasion and migration were analyzed by transwell assay. (K-M) the mRNA and protein levels of E-cadherin and Vimentin were analyzed by RT-PCR (K), western blot (L) and RT-qPCR (M) assay. Results were presented as mean ± SD, and the error bars represent the SD of three independent experiments. (*p<0.05, **p<0.01, ANOVA with Bonferroni correction).
UBE2C directly binds the 5′-UTR of ZEB1 and ZEB2 to upregulates their transcription
siUBE2C inhibited lung cancer cell proliferation, migration, invasion, and EMT, which contributed to lung cancer progression and metastasis (Figure 1). siZEB1/2 played a similar important role in tumorigenesis and development of NSCLC (Figure 2 and 3). Therefore, we hypothesized that UBE2C plays an important role in lung cancer progression and metastasis via regulation of ZEB1/2 expression. To investigate the relationship between UBE2C and ZEB1/2 in lung cancer cell growth, EMT, and invasion, PROMO analysis (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promo.cgi?dirDB=TF_8.3) [36, 37] was used to predict the putative binding site of UBE2C (most predicted factors bind to these sites) in the 5′ untranslated region (5′-UTR) of ZEB1 and ZEB2. UBE2C may directly target ZEB1and ZEB2 (Figure S4A, B). To further confirm the interaction between UBE2C and ZEB1/2, we identified the ZEB1 and ZEB2 promoters' core regions responsive to UBE2C. Various lengths of the ZEB1 5′-flanking region, including-1156/+180 (pGL3-1336), -1156/-250 (pGL3-950), -250/-108 (pGL3-142), and -108/+180 (pGL3-288) were cloned and transiently transfected into A549 cells expressing UBE2C to determine promoter activity. The luciferase reporter gene assays indicated that pGL3-142 exhibited maximum luciferase activity (Figure 4A), indicating that the region encompassing -250/-108 is the promoter core region of ZEB1 interacted with UBE2C. We further observed that UBE2C was able to increase the activities of pGL3-142 in A549 cells in a dose-dependent manner (Figure 4B), suggesting that the -250/-108 region (pGL3-142) of the ZEB1 promoter might be responsible for UBE2C binding-mediated ZEB1 promoter activity. Similarly, the -80/+95 region of the ZEB2 promoter might be responsible for the UBE2C-mediated ZEB2 promoter activity (Figure 4A, B). We further performed a deletion scan and mutational analysis of the region -250/-108 to determine the ZEB1 promoter region regulated by UBE2C. Deletion of nucleotides -155 to -128 (PGL3-142Δ) and mutation of nucleotides -145 to -138 (PGL3-142*) abolished UBE2C-mediated activation (Figure 4A, C and S4C, D). Within this region, we identified a putative UBE2C-response element spanning positions -145 to -138 (TTTCCTTG) (Figure 4C). Mutation of this region dramatically reduced the activation of the ZEB1 promoter by UBE2C (Figure 4D). Similarly, we identified a putative UBE2C-response element of ZEB2 spanning positions -48 to -41 (AATTGTTA) (Figure 4A, C). Deletion and mutation of this region dramatically reduced the activation of the ZEB2 promoter by UBE2C (Figure 4A, C and S4E, F). Next, we constructed luciferase reporter plasmids containing either wild type 5′-UTR of ZEB1/2 (ZEB1WT and ZEB2WT) or the UBE2C response element mutant (ZEB1Mut and ZEB2Mut) sequences (Figure 4C). Co-transfection of ZEB1W-5′-UTR and UBE2C into A549 or H1299 cells significantly increased luciferase activity compared to co-transfection with the control plasmid, whereas luciferase activity was reduced in A549 or H1299 cells co-transfected with ZEB1W-5′-UTR and siUBE2C. Relative to the control group, luciferase activity changed in neither A549 nor H1299 cells after co-transfection of ZEB1M-5′-UTR and UBE2C or siUBE2C (Figure 4D). Therefore, UBE2C directly targets the indicated positions of the ZEB1 promoter. Similar luciferase activity was observed after co-transfection of the 5′-UTR of ZEB2 and UBE2C in A549 and H1299 cells. Therefore, UBE2C directly targets the indicated positions of ZEB2 promoter (Figure 4E). To determine whether UBE2C endogenously regulates ZEB1/2, A549 cells were collected and their ZEB1 and ZEB2 mRNA and protein levels were analyzed 48 h after UBE2C or siUBE2C transfection. Results showed that the ZEB1 or ZEB2 mRNA and protein levels in A549 cells were significantly elevated after UBE2C overexpression as detected using RT-PCR, western blotting (Figure 4F-H), and immunofluorescence (Figure 4I, J) assay. Their simulative effects were suppressed when UBE2C was downregulated (Figure 4F-J). UBE2C increased ZEB1 and ZEB2 mRNA and protein levels in a dose-dependent manner (Figure 4K, L). Spearman's rank correlation analysis of the results of western blotting showed significant positive correlation between UBE2C and ZEB1 or ZEB2 protein levels (Figure 4M). Furthermore, quantitative chromatin immunoprecipitation (ChIP) assays were performed to determine whether UBE2C directly binds to ZEB1/2-5′-UTR. Consistent with the results of the luciferase activity assay, results of the ChIP assay indicated that ZEB1W-5′-UTR and UBE2C co-transfection into A549 or H1299 cells significantly promoted UBE2C binding to ZEB1W-5′-UTR, whereas co-transfection with siUBE2C did not. Therefore, UBE2C directly targets the ZEB1 promoter (Figure 4N). Similar results were obtained for ZEB2 using A549 and H1299 cell lines (Figure 4N). Overall, we demonstrated that UBE2C upregulates ZEB1 and ZEB2 by directly targeting the 5′-UTR region.
UBE2C promotes EMT and increases cell proliferation and invasion by regulating ZEB1 and ZEB2
ZEB1 and ZEB2 are recognized as important EMT regulators in lung cancer and were confirmed to be the targets of UBE2C (Figure 4). We separately transfected siUBE2C or UBE2C into A549 cells and used western blotting and RT-qPCR to determine ZEB1 and ZEB2 expression. We observed that ZEB1 or ZEB2 levels were reduced by siUBE2C transfection and increased by UBE2C overexpression. However, opposite effects were observed for each of these factors in A549 cells ectopically transfected with ZEB1 or ZEB2, which indicated that ZEB1/2 were the downstream targets of UBE2C (Figure 5A). Lung cancer cell proliferation substantially increased when UBE2C was overexpressed; however, CCK8 assays in A549 and H1299 cells showed that this effect was reversed after the cells were co-transfected with siZEB1or siZEB2. Cell proliferation significantly decreased after siUBE2C transfection, which, however, was reversed by ZEB1 or ZEB2 transfection (Figure 5B, C). Immunofluorescence staining and clone formation assay showed similar results for Ki67 protein levels (Figure S5A) and clone formation (Figure S5B). In contrast, opposite effects were observed for each of these treatments in A549 cells for cleaved caspase-3 (Figure S5C), annexin V (Figure S5D), and cellular senescence (Figure S5E). As UBE2C promotes cell invasion and migration, we determined whether it also regulates E-cadherin and vimentin expression. UBE2C depletion decreased vimentin mRNA and protein levels but increased those of E-cadherin. However, the effects were opposite in A549 cells ectopically overexpressing UBE2C as determined using RT-PCR, western blotting, RT-qPCR, and immunofluorescence assay (Figure 5D, E). To determine whether UBE2C regulates the expression of EMT marker proteins via ZEB1/2, we co-transfected siUBE2C and ZEB1 or UBE2C and siZEB1 and assessed E-cadherin and vimentin mRNA and protein levels. RT-PCR, western blotting, RT-qPCR, and immunofluorescence staining indicated that E-cadherin level was increased by siUBE2C transfection and decreased by UBE2C overexpression; opposite effects were observed for each of these factors in A549 cells ectopically transfected with ZEB1 or siZEB1. In contrast, siUBE2C transfection reduced vimentin expression, which was however increased by UBE2C overexpression; opposite effects were observed for each of these factors in A549 cells ectopically transfected with ZEB1 or siZEB1 (Figure 5F and S5G). Similar results were obtained for RT-PCR, western blotting, RT-qPCR, and immunofluorescence assay for A549 cell lines transfected with ZEB2 (Figure 5G and S5F, H). These data indicated that ZEB1/2 is located downstream of UBE2C, and that UBE2C plays an important function in EMT via regulation of ZEB1/2. Furthermore, scratch assays (Figure 5H) and transwell assays (Figure 5I) indicated that siUBE2C transfection reduced cellular migration and invasion, which were increased by UBE2C overexpression; however, the opposite effects were observed for each of these factors in A549 cells ectopically transfected with ZEB1 or siZEB1 (data for ZEB2 not shown) (Figure 5H, I). These results demonstrated that UBE2C promoted EMT and increased cell invasion by regulating ZEB1 and ZEB2.
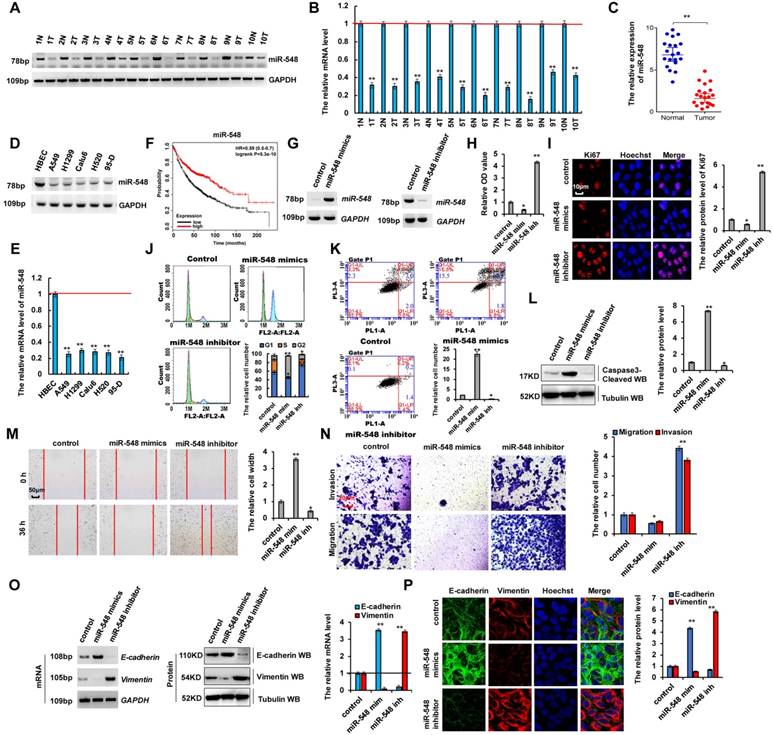
miR-548e-5p expression was lower in lung cancer and it inhibited proliferation, migration, and invasion of lung cancer cells
Recent studies showed that microRNAs play critical roles in human tumorigenesis and cancer development [8-13, 38]. To determine whether miR-548e-5p (miR-548) was related to the occurrence of lung cancers, we performed RT-PCR and RT-qPCR assays and observed that the mRNA levels of miR-548e-5p were lower in human lung cancer tissues than in their normal adjacent lung tissues, indicating that miR-548e-5p played an important role in lung cancer as a tumor suppressor (Figure 6A-C). Moreover, the correlate the expression of miR-548e-5p, UBE2C, ZEB1 and ZEB2 in same individual patient were shown in the Figure S6A. Furthermore, RT-PCR and RT-qPCR assays indicated that miR-548e-5p expression was lower in lung cancer cells than in normal HBEC controls (Figure 6D, E). We analyzed the patient's demographics and tumor characteristics and association of miR-548e-5p level with clinicopathological features in lung tumor population from patients (n = 50) who underwent lung resection surgery between January 2014 and January 2016 (Table 1). Publicly available datasets (http://www.kmplot.com/analysis/ and http://www.oncolnc.org/) (31) were filtered and used to analyze the prognostic correlation between survival of patients with lung cancer and miR-548e-5p expression. Kaplan-Meier analyses revealed that miR-548e-5p expression levels correlated positively with survival. High expression levels were associated with longer overall survival (OS) (n = 1926, P = 5.3 × 10-10) (Figure 6F). miR-548e-5p-mimics (miR-548 mim) and miR-548-5p-inhibitor (miR-548 inh) were used to determine whether miR-548e-5p tumor suppressor activity underlies lung cancer cell initiation, progression, and metastasis (Figure 6G and S6B, C). In A549 cells, the miR-548e-5p-mimics decreased cell growth (Figure 6H), Ki67 protein level (Figure 6I), clone formation (Figure S6D), cell migration (Figure 6M), and cell invasion (Figure 6N), but increased apoptosis (Figure 6K), cleaved caspase-3 level (Figure 6L), annexin V level (Figure S6E), and cellular senesce (Figure S6F). The opposite effects were observed for all these factors in A549 cells with downregulated ectopic miR-548e-5p expression (Figure 6G-N and S6C-E). Furthermore, our analysis of cell cycle profile in lung cancer cells treated with miR-548e-5p mimics revealed that the treatment induced significant G2 arrest in A549 cells; however, treatment with the miR-548e-5p inhibitor did not change the pattern of the cell cycle but increased the cell number in the G1 phase compared to that in the control group (Figure 6J). Furthermore, we determined whether miR-548e-5p regulated E-cadherin and vimentin expression. miR-548e-5p mimics decreased the mRNA and protein levels of vimentin but increased those of E-cadherin; opposite effects were observed for each of these factors in A549 cells treated with the miR-548e-5p inhibitor as observed using RT-PCR, western blotting, RT-qPCR, and immunofluorescent staining assay (Figure 6O, P). These results indicated that miR-548e-5p expression was lower in lung cancer and that it inhibited the proliferation, migration, and invasion of lung cancer cells.
UBE2C directly binds the 5′-UTR of ZEB1 and ZEB2 to up-regulate their transcription. (A) the activities of different fragments of ZEB1 promoter (pGL3-1336, pGL3-950, pGL3-288, pGL3-142 and pGL3-142Δ) and ZEB2 (pGL3-1155, pGL3-830, pGL3-150, GL3-175 and GL3-175Δ) were measured by luciferase reporter gene assays in A549 cells. (B) the activities of pGL3-142 (-250~-108) for ZEB1 and pGL3-175 (-80~+95) for ZEB2 were measured by luciferase reporter gene assays in A549 cells with transfection of UBE2C. (C) Putative UBE2C binding sites in the 5′UTR sequence of ZEB1 and ZEB2. (D, E) Luciferase activity of A549 or H1299 cells were transfected with plasmids carrying a wild-type or mutant 5′UTR of ZEB1 (D) or ZEB2 (E) in response to overexpressing UBE2C or knockdown of UBE2C using the siRNA. (F-J) the mRNA and protein levels of ZEB1 and ZEB2 were analyzed by RT-PCR, western blotting (F, G), RT-qPCR (H) and immunofluorescent staining (I, J) assay in A549 cells with overexpressing UBE2C or knockdown of UBE2C using the siRNA. (K, L) RT-PCR, western blot (K) and RT-qPCR (L) result shows that UBE2C dose-dependently increased the mRNA and protein levels of ZEB1 and ZEB2. (M) the relationship between the UBE2C and ZEB1 or ZEB2 was analyzed based on immunoblotting assay, respectively. (N) Quantitative ChIP analysis demonstrating that knockdown of UBE2C using the siRNA decreases but overexpressing UBE2C increases UBE2C levels within the promoter region of ZEB1 or ZEB2 in A549 or H1299 cells. Results were presented as mean ± SD, and the error bars represent the SD of three independent experiments. (*p<0.05, **p<0.01, ***p<0.001, ANOVA with Bonferroni correction).
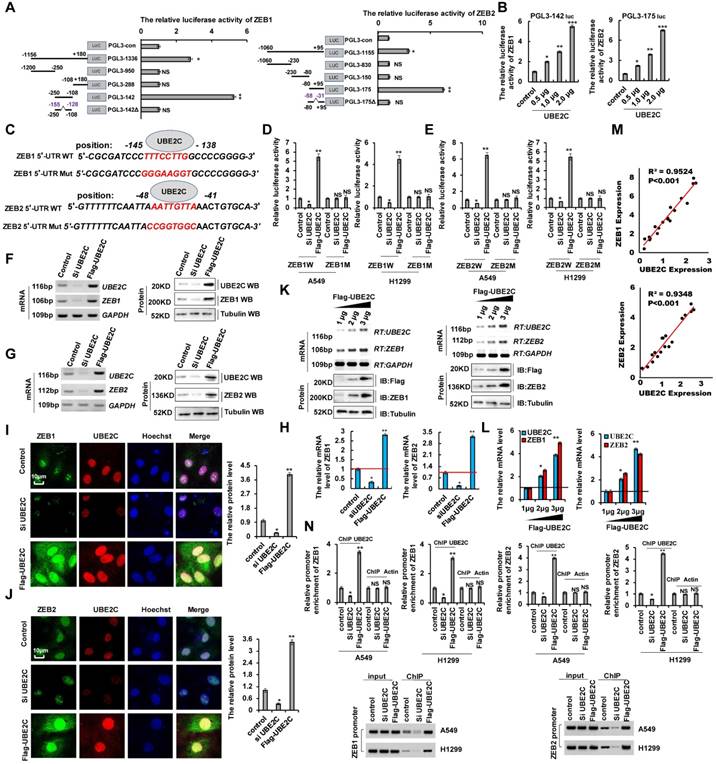
UBE2C promotes EMT and increases cell proliferation and invasion via regulating ZEB1 and ZEB2. A549 or H1299 cells were transfected with UBE2C or siUBE2C. ZEB1 or ZEB2 were used for upregulated the protein level of UBE2C target genes, respectively. (A) the mRNA and protein levels of ZEB1, ZEB2 and UBE2C were analyzed by RT-qPCR and immunoblotting assay. (B, C) the cellular growth was analyzed by CCK8 assay. (D, E) A549 cells were transfected with UBE2C or siUBE2C, respectively. The mRNA and protein levels of EMT-related molecules, E-cadherin and Vimentin were analyzed by RT-PCR, immunoblotting, RT-qPCR (D) and immunofluorescent staining (E) assays. (F-I) A549 cells were transfected with UBE2C or siUBE2C. ZEB1 or ZEB2 were used for upregulated the protein level of UBE2C target genes, respectively. (F, G) the mRNA and protein expression levels of UBE2C, ZEB1, ZEB2, E-cadherin and Vimentin were analyzed by RT-PCR, immunoblotting and RT-qPCR assays. (H, I) Cellular migration and invasion ability was analyzed by scratch assay (H) and transwell assay (I). Results were presented as mean ± SD, and the error bars represent the SD of three independent experiments. (*p<0.05, **p<0.01, ANOVA with Bonferroni correction).
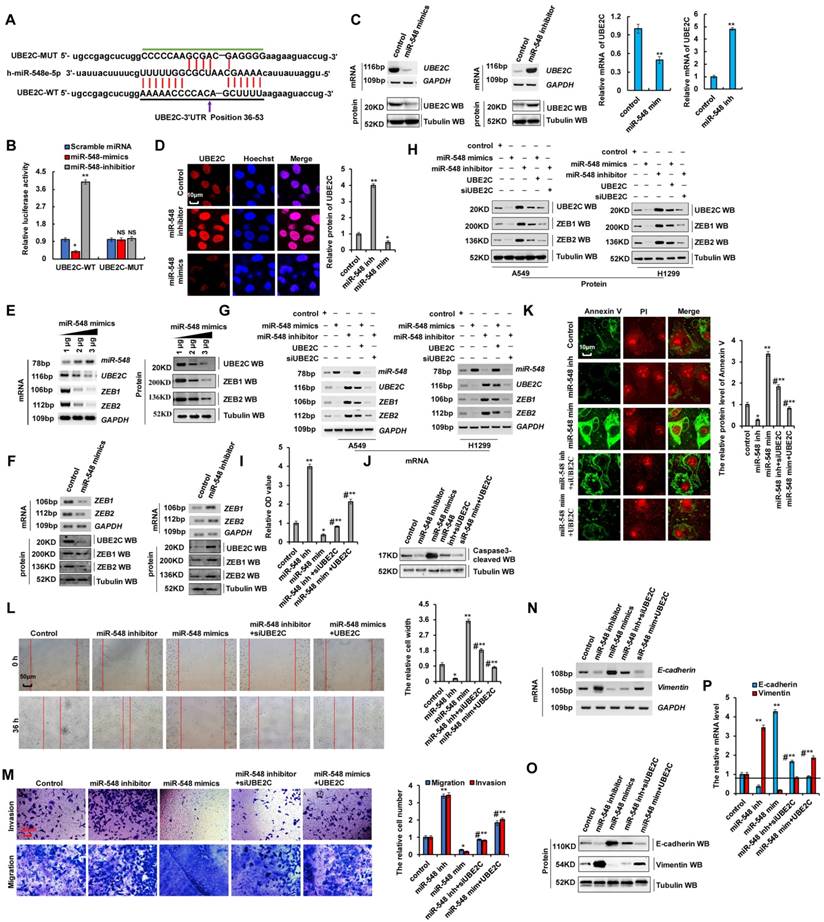
miR-548e-5p affected cell proliferation and invasion by regulating UBE2C mRNA stabilization
Our results showed that UBE2C binds to the 5′-UTR of ZEB1 and ZEB2 promoters to regulate their transcription, and promoted EMT and increased cell proliferation and invasion by regulating ZEB1 and ZEB2 expression (Figure 4 and 5). The miR-548e-5p inhibitor and UBE2C exhibit similar biological functions in terms of lung cancer cell proliferation, apoptosis, migration, invasion, and EMT. Therefore, we hypothesized that miR-548e-5p suppresses the invasive abilities of lung cancer cells via regulation of ZEB1 and ZEB2 by directly targeting UBE2C, and that the miR-548e-5p-UBE2C-ZEB1/2 axis decreased cell proliferation and invasion. To test this hypothesis, the candidate miRNAs targeting UBE2C were predicted using a combination of three databases: miRbase, miRanda, and TargetScan. Results showed that miR-548e-5p was consistently predicted by all three servers (Figure S7A and Supplementary Table 2) to bind to the 3′-UTR of UBE2C. Therefore, miR-548e-5p may directly target UBE2C (Figure 7A). To confirm the interaction between miR-548e-5p and UBE2C, we constructed luciferase reporter plasmids containing either the sequences of the 3′-UTR of UBE2C (UBE2C-WT) or the miR-548 response element mutant (UBE2C-MUT) (Figure 7A). Co-transfection of UBE2C-3′-UTR-WT and miR-548e-5p mimics in A549 cells resulted in significantly lower luciferase activity than co-transfection with scrambled miRNA. This reduction was rescued in cells transfected with UBE2C-3′-UTR-MUT or the miR-548e-5p inhibitor. Therefore, miR-548e-5p directly targets UBE2C (Figure 7B). To determine whether miR-548e-5p endogenously regulates UBE2C, the UBE2C mRNA and protein levels in A549 cells were determined 48 h after the transfection of miR-548e-5p mimics- or miR-548e-5p inhibitor. Results showed that the UBE2C mRNA and protein levels were significantly reduced in the A549 cells after miR-548e-5p mimic overexpression. These inhibitory effects were suppressed when miR-548e-5p expression was downregulated (Figure 7C, D). miR-548e-5p reduced UBE2C mRNA and protein levels in a dose-dependent manner (Figure 7E and Figure S7B). Furthermore, results of RT-PCR, western blotting, and RT-qPCR indicated that the ZEB1/2 mRNA and protein levels were significantly reduced in A549 cells after miR-548e-5p overexpression. These inhibitory effects were suppressed when miR-548e-5p was downregulated (Figure 7F and S7C, D). miR-548e-5p dose-dependently reduced the mRNA and protein level of ZEB1/2 (Figure 7E and S7B), indicating that miR-548e-5p regulated ZEB1/2 by binding to the 3′-UTR of UBE2C. To confirm whether miR-548e-5p inhibits EMT, cell migration, and invasion by targeting complementary sites in the 3′-UTR of UBE2C, we co-transfected miR-548e-5p mimics and UBE2C or miR-548e-5p inhibitor and siUBE2C into A549 and H1299 cells and used RT-PCR, western blotting, and RT-qPCR to detect UBE2C, ZEB1 and ZEB2. We observed that miR-548e-5p transfection reduced UBE2C, ZEB1, or ZEB2 levels, which were however elevated by miR-548e-5p inhibitor expression. However, opposite effects were observed for each of these factors in A549 cells ectopically transfected with UBE2C or siUBE2C (Figure 7G, H and S7E, F). These results indicated that UBE2C is located downstream of miR-548e-5p in the miR-548e-5p-UBE2C-ZEB1/2 signal axis and that miR-548e-5p plays an important function in EMT by binding to the 3′-UTR of UBE2C. Furthermore, the CCK8 assay in A549 cells indicated that lung cancer cell proliferation significantly increased after miR-548e-5p inhibitor transfection, although this inhibitory effect was suppressed after siUBE2C co-transfection. Similarly, lung cancer cell proliferation significantly decreased after miR-548e-5p mimic transfection, although this inhibitory effect was suppressed after UBE2C co-transfection (Figure 7I). Similar results were obtained from Edu staining (Figure S8A) and cell cycle profile assay in A549 cells (Figure S8B). In addition, western blotting and immunofluorescence staining demonstrated opposite effects on the protein levels of active caspase-3 (Figure 7J and S8C) and annexin V (Figure 7K) in A549 cells; similar results were also obtained for apoptotic cells using flow cytometry (Figure S8D). However, the similar results for cell migration (Figure 7L) and cell invasion (Figure 7M) likewise of cell growth were obtained in in A549 cells by scratch and transwell assays. As the miR-548e-5p inhibitor promotes cell invasion and migration, we used RT-PCR and western blotting to confirm whether it regulates E-cadherin and vimentin via UBE2C. miR-548e-5p overexpression upregulated E-cadherin but downregulated vimentin. In contrast, the expression levels of E-cadherin and vimentin were significantly reversed in A549 cells co-transfected with miR-548e-5p mimics and UBE2C (Figure 7N-P and S8E). Opposite effects were obtained for cells co-transfected with the miR-548e-5p inhibitor and siUBE2C (Figure 7N-P). Therefore, miR-548e-5p functions as a tumor suppressor in lung cancer cells by targeting the 3′-UTR of UBE2C, and the miR-548e-5p-UBE2C-ZEB1/2 axis decreases proliferation and invasion of cancer cells.
miR-548e-5p inhibits lung cancer metastasis in vivo by regulating ZEB1/2 via UBE2C
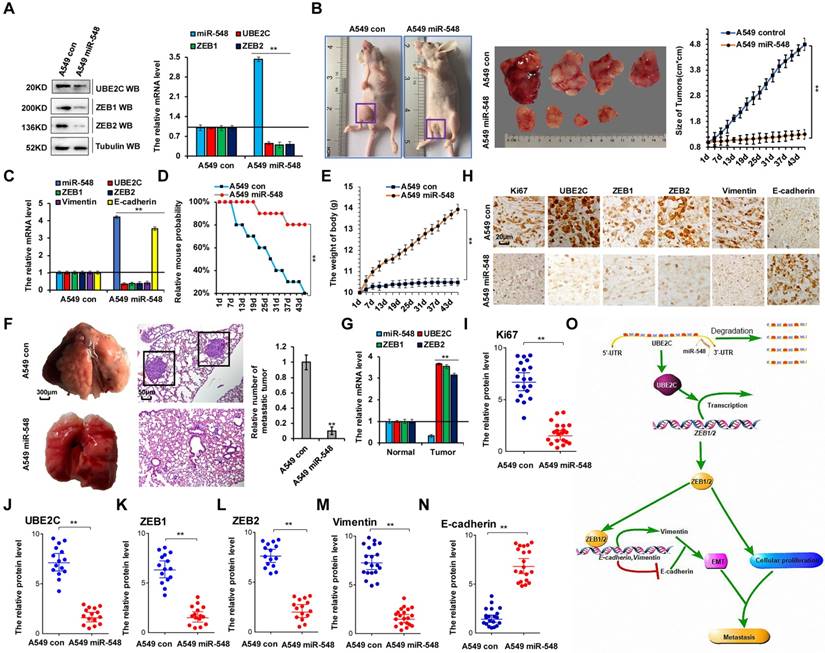
To confirm the correlation between miR-548e-5p, UBE2C, and ZEB1/2 in human lung cancer metastasis, we established A549 cell lines stably overexpressing miR-548e-5p mimics (A549 miR-548) and control cells (A549 con). We used these cells to generate a mouse xenograft cell metastasis model. First, UBE2C and ZEB1/2 protein levels were analyzed using western blotting and RT-qPCR to validate the constructed cell lines (Figure 8A). Approximately 2 weeks after subcutaneous implantation of A549 con and A549 miR-548e cells into concave mouse cecum niches, the tumor volume of the A549 miR-548e group was significantly smaller than that of the control group (Figure 8B) and the mRNA levels of UBE2C, ZEB1, ZEB2, and vimentin were lower in the A549 miR-548e tumor tissues than in A549 con tumor tissues; however, RT-qPCR assay revealed opposite effects on miR-548e and E-cadherin expression in these tumor tissues (Figure 8C). Furthermore, the A549 miR-548e mice had longer survival times (Figure 8D) and relatively higher body weight (Figure 8E) than the control mice, suggesting that miR-548e improved the quality of life in mice. In addition, significantly fewer lung cancers originated from the xenografted tumors in the A549 miR-548e nude mice than in the A549 control nude mice (Figure 8F). Results of the RT-qPCR assay showed that the mRNA levels of UBE2C, ZEB1, and ZEB2 were higher in lung cancer tissues than in their normal adjacent lung tissues in A549 control nude mice (Figure 8G). Furthermore, semi-quantitative immunohistochemical analysis of Ki67 (Figure 8I), UBE2C (Figure 8J), ZEB1 (Figure 8K), ZEB2 (Figure 8L), and vimentin (Figure 8M) expression in the xenografts revealed that the levels of these proteins were lower in the A549 miR-548e group than in the control mice (n = 15, Figure 8H-M). In contrast, E-cadherin expression was higher in the A549 miR-548e group than in the A549 con mice (n = 15, Figure 8H, N). Therefore, our results indicated miR-548e inhibits lung cancer metastasis in vivo by regulating ZEB1/2 via UBE2C.
miR-548e-5p was lower expression in lung cancer tissues and miR-548e-5p inhibitor promoted cancer cell proliferation, migration, invasion in lung cancer cells. (A, B) RT-PCR (A) and RT-qPCR (B) assays showed that the mRNA level of miR-548e-5p (miR-548) was lower in human lung cancer tissues compared with their normal adjacent lung tissues. (C) the mRNA level of miR-548e-5p was showed in 20 lung cancer tumor tissues and adjacent normal tissues. (D, E) Gel-based RT-PCR with densitometric quantitation (D) and RT-qPCR (E) demonstrating reduced the expression of miR-548e-5p in human lung cancer cells compared with their normal control cell HBEC. (*p<0.05, **p<0.01 vs control group correspond to two-tailed Student's tests). (F) Kaplan Meier overall survival (OS) curves of miR-548e-5p (p=5.3 x 10-10 by log-rank test for significance) for human lung cancers. (G-P) A549 cells were transfected with miR-548e-5p-mimics (miR-548 mim) and miR-548e-5p inhibitor (miR-548 inh). (G) the mRNA level of miR-548e-5p was analyzed by RT-PCR assay. (H) the cellular proliferation was analyzed by CCK8 assay. (I) the protein of Ki67 was analyzed by immunofluorescent staining. (J, K) Cell cycle profile (J) and apoptosis (K) were analyzed by cell flow cytometry. (L) the protein of cleaved Caspase3 was analyzed by immunoblotting assay. (M, N) Cellular migration and invasion ability was analyzed by cell scratch assay (M) and transwell assay (N). (O, P) the mRNA and protein levels of E-cadherin and Vimentin were analyzed by RT-PCR, immunoblotting, RT-qPCR (O) and immunofluorescent staining (P). (*p<0.05, **p<0.01, ANOVA with Bonferroni correction). Results were presented as mean ± SD, and the error bars represent the SD of three independent experiments. *p<0.05; **p<0.01 vs control group.
miR-548e-5p affected cell proliferation and invasion via regulating the stabilization of mRNA of UBE2C. (A) Putative miR-548e-5p binding sites in the 3′-UTR sequences of UBE2C. (B) Luciferase activity of A549 cells transfected with plasmids carrying a wild-type or mutant 3'UTR of UBE2C in response to miR-548e-5p mimics or inhibitor. (C, D) A549 cells were transfected with miR-548e-5p mimics or miR-548e-5p inhibitor. The mRNA and protein levels of UBE2C were analyzed by RT-PCR, immunoblotting, RT-qPCR (C) and immunofluorescent staining (D). (E) RT-PCR and western blot result shows that miR-548e-5p dose-dependently decreased the mRNA and protein levels of UBE2C, ZEB1 and ZEB2. (F) A549 cells were transfected with miR-548e-5p mimics or miR-548e-5p inhibitor. The mRNA and protein expression levels of UBE2C, ZEB1 and ZEB2 were analyzed by RT-PCR and immunoblotting. (G-P) A549 or H1299 cells were transfected with miR-548e-5p mimics or miR-548e-5p inhibitor. UBE2C or siUBE2C were used for upregulating or downregulating the protein level of miR-548e-5p target genes, respectively. (G, H) the mRNA and protein expression levels of miR-548e-5p, UBE2C, ZEB1 and ZEB2 were analyzed by RT-PCR (G) and immunoblotting (H) assays. (I) the cellular proliferation was analyzed by CCK8 assay in the A549 cells. (J) the protein of cleaved Caspase3 was analyzed by immunoblotting assay in the A549 cells. (K) the protein of Annexin V was analyzed by immunofluorescent staining assay in the A549 cells. (L, M) Cellular migration and invasion ability was analyzed by cell scratch assay (L) and transwell assay (M) in the A549 cells. (N-P) the mRNA and protein levels of E-cadherin and Vimentin were analyzed by RT-PCR (N), immunoblotting (O) and RT-qPCR (P) assay in the A549 cells. Results were presented as mean ± SD, and the error bars represent the SD of three independent experiments. (*p<0.05, **p<0.01, ANOVA with Bonferroni correction).
miR-548e-5p inhibits lung cancer metastasis in vivo by regulation of ZEB1/2 via UBE2C. (A) Western blot and RT-qPCR analysis indicated the protein and mRNA levels of UBE2C, ZEB1 and ZEB2 in the A549 cells with stable overexpression of miR-548e-5p mimics (A549 miR-548) and control mimics (A549 con). (B) Xenografted A549 cell tumors with stably expressing control mimics or miR-548e-5p mimics in mice and the dimensions and weights measured at regular intervals. (C) the mRNA level of miR-548, UBE2C, ZEB1, ZEB2, E-cadherin and Vimentin was analyzed by RT-qPCR in the tumor tissues of mice injected A549 cells with stably expressing control mimics or miR-548e-5p mimics. (D) Kaplan-Meier overall survival (OS) curves of mice injected with stably expressing control mimics or miR-548e-5p mimics. (E) the mice body weights of nude mice injected with A549 miR-548 cells compared to A549 control cells demonstrating that miR-548e-5p significantly alter the body weight of these xenografted animals. (F) Representative phase and H&E stained microscopic images of the mice lung tumors originated from xenografted A549 miR-548e-5p cells or A549 control cells by subcutaneous injection. (G) the mRNA level of miR-548e-5p, UBE2C, ZEB1 and ZEB2 was analyzed by RT-qPCR in the mice lung tumors and normal adjacent lung tissues originated from xenografted A549 control cells by subcutaneous injection. (H-N) Immunohistochemical microscopy of tumor nodules in nude mice injected A549 cells with stably expressing miR-548e-5p mimics or control mimics (H). The protein expression levels of Ki67 (I), UBE2C (J), ZEB1 (K), ZEB2 (L), Vimentin (M) and E-cadherin (N) were analyzed by immunohistochemical staining (n=15). (**p<0.01 vs control group correspond to two-tailed Student's tests). (O) the diagram of miR548e-5p-UBE2C-ZEB1/2 axis regulated cellular growth and metastasis in lung cancer. Results were presented as mean ± SD, and the error bars represent the SD of three independent experiments.
Discussion
In recent decades, substantial improvements have been made in the early diagnosis and therapeutic intervention of lung cancer, which have increased the survival rates and quality of life of patients with lung cancer. Nevertheless, NSCLC is highly aggressive and malignant and is associated with one of the lowest survival rates [3, 39]. Till date, almost all patients with advanced and unresectable NSCLC have few treatment options except chemotherapy and radiotherapy, both of which are associated with severe side effects [39, 40]. Hence, the demand for novel tumor-selective drugs with low or no toxicity is high. In this study, we evaluated the specificity and efficacy of the miR-548e-5p-UBE2C-ZEB1/2 signal axis in regulating NSCLC progression and metastasis (Fig. 8o). Based on our findings, we defined it as a novel candidate NSCLC therapy.
UBE2C belongs to the E2 gene family and encodes a 19 kDa protein associated with ubiquitin-dependent proteolysis. Overexpression of UBE2C leads to chromosome mis-segregation and alteration of the cell cycle profile, which facilitates cell proliferation and inhibits cellular apoptosis. UBE2C has been reported to be highly expressed in various types of cancers, including metastatic prostate carcinoma, ovarian carcinoma, thyroid anaplastic carcinoma, breast cancer, hepatocellular carcinoma, and lung cancer [16-21]. Furthermore, some studies have suggested a close relationship between drug resistance and protein ubiquitination and degradation; for instance, C-X-C chemokine receptor type 4 was shown to promote the ubiquitin-mediated degradation of cyclooxygenase 1, leading to aspirin resistance [41]. UBE2C induced the transcription of ubiquitin-specific peptidase 1 to reduce DDP-associated cytotoxicity in response to DNA damage in NSCLC [42]. In addition, UBE2C inhibition blocked caspase-3/8 signaling to regulate c-Jun N-terminal kinase activity, increasing sensitivity of chemotherapeutic drugs and improving treatment outcome in NSCLC [43]. Ubiquitination is an important cellular mechanism for targeted degradation of proteins that are instrumental for a variety of cellular processes including, but not limited to, transcription, cell cycle progression, programmed cell death, and antigen presentation. Additionally, results our previous studies have shown that abnormal regulation of UBE2C results in cisplatin resistance in NSCLC via ABCG2 and ERCC1 [42]. However, till date, reports on the direct involvement of ubiquitination with invasiveness of lung cancer are lacking. Furthermore, our data indicated that UBE2C, as a novel transcription factor, promoted human tumorigenesis and cancer development, although the specific molecular mechanisms remain to be further investigated.
Growing evidence show that miRNAs regulate the expression of target genes in lung cancer cells. Therefore, elucidation of the molecular mechanisms of lung cancer is an interesting area of research [44-46]. miR-548e-5p participates in lung cancer progression. We used RT-PCR to determine miR-548e-5p expression in lung cancer tissues and in matched normal tissues adjacent to the tumors. We showed that miR-548e-5p expression in tumor-adjacent tissues was significantly higher than in lung cancer tissues. In addition, miR-548e-5p was expressed at significantly lower levels in lung cancer cell lines than in normal cells. Cell migration and invasion assays in lung cancer cells overexpressing miR-548e-5p indicated that overexpressed miR-548e-5p significantly suppressed cell proliferation, migration, and invasion in lung cancer cells. Our data also revealed that UBE2C mRNA and protein levels were elevated in human lung cancer cells compared to in normal cells and that UBE2C overexpression promoted cell growth and invasion. Therefore, miR-548e-5p and UBE2C play important roles in lung cancer tumorigenesis and metastasis.
Our data showed negative correlation between miR-548e-5p expression and UBE2C protein level. We deduced that low miR-548e-5p expression was associated with higher UBE2C levels. We also performed proliferation, migration, and invasion assays on lung cancer cells co-transfected with miR-548e-5p and UBE2C. Results indicated that, via UBE2C, miR-548e-5p significantly affected lung cancer cell proliferation, migration, and invasion. Therefore, the correlation between miR-548e-5p and UBE2C is significant in lung cancer tumorigenesis and metastasis.
In this study, we observed a positive correlation between UBE2C expression and ZEB1/2 protein levels. EMT plays a crucial role in tumor metastasis. Hence, we also determined the effects of changes in UBE2C expression on EMT by analyzing E-cadherin and vimentin levels. The significant reduction in E-cadherin expression and simultaneous increase in vimentin protein level in response to UBE2C downregulation indicated that UBE2C is a positive EMT regulator in lung cancer cells. In contrast, UBE2C upregulation increased lung cancer cell migration and invasion. In summary, siUBE2C inhibited lung cancer cell migration and invasion by suppressing EMT. In addition, ZEB1 is a master regulator of EMT, and its deregulation has been observed in various cancer types and other pathological processes, such as reversible embryonic transdifferentiation that allows partial or complete transition from an epithelial to a mesenchymal state [47, 48]. Aberrant ZEB1 expression has been reported in several human cancers, where it is generally believed to foster migration, invasion, and metastasis [49, 50]. Over the past few years, in vitro and in vivo observations have highlighted unsuspected intrinsic oncogenic functions of ZEB1 that impact tumorigenesis from its earliest stages [49, 50]. ZEB1 is also a key determinant of cell plasticity, endowing cells with the capacity to withstand aberrant mitogenic activity, with a profound impact on the genetic history of tumorigenesis, and to the ability to adapt to the multiple constraints encountered over the course of tumor development [31, 32]. Furthermore, ZEB1/2 repress several master epithelial polarity regulators and, therefore, control E-cadherin expression and EMT progression. Luciferase reporter assays confirmed that UBE2C directly binds to the 5′-UTR of ZEB1/2 transcripts. Furthermore, ZEB1/2 overexpression rescues the migration of a lung cancer cell line when UBE2C is ectopically knocked down. Therefore, UBE2C disruption may be a critical event in lung cancer metastasis. However, whether UBE2C contains a domain that directly interacts with the promoter of ZEB1/2 is not clearly understood and warrants further investigation.
In conclusion, miR-548e-5p downregulation and UBE2C and ZEB1/2 upregulation occur in lung cancer tissues and cells in vitro. miR-548e-5p directly binds to the 3′-UTR of UBE2C and decreases UBE2C mRNA expression. UBE2C mRNA directly binds to the 5′-UTR of ZEB1/2. Reduced miR-548e-5p expression is physiologically significant because miR-548e-5p delays lung cancer cell proliferation, migration, and invasion via UBE2C by suppressing ZEB1/2-induced EMT. Therefore, UBE2C is an oncogene that promotes EMT in lung cancer cells by directly targeting the E-cadherin repressor ZEB1/2. miR-548e-5p, UBE2C, and ZEB1/2 constitute the miR-548e-5p-UBE2C-ZEB1/2 signal axis, which enhances cancer cell invasiveness by directly interacting with the EMT maker protein (Figure 8O). However, whether other microRNAs reduced the stability of UBE2C mRNA and UBE2C directly regulated other molecules affecting the EMT (e.g. twist, snail, slug et. al) need to be further explored. But yet we believe that the miR-548e-5p-UBE2C-ZEB1/2 signal axis may be a suitable diagnostic marker and a potential target for lung cancer therapy.
Conclusions
This study was to elucidate the functions, direct target genes, and molecular mechanisms of miR-548e- 5p in non-small-cell lung cancer. It was found that miR-548e-5p downregulation naturally occurs in human lung cancer tissues and that miR-548e-5p plays significant roles in suppressing or delaying lung cancer cell proliferation, migration, and invasion. It does so by suppression of the E-cadherin repressor, ZEB1/2, via the downregulating the oncogene UBE2C. Moreover, miR-548e-5p directly binds to the 3'-UTR of UBE2C and decreases UBE2C mRNA expression. UBE2C directly binds to the 5'-UTR of ZEB1/2 to promote EMT in lung cancer cells by directly targeting the E-cadherin repressor ZEB1/2. Thus, the miR-548e-5p, UBE2C, and ZEB1/2 constitute the miR-548e-5p-UBE2C-ZEB1/2 signal axis, which enhances cancer cell invasiveness by directly interacting with the EMT maker protein. We believe that the miR-548e-5p-UBE2C-ZEB1/2 signal axis may be a suitable diagnostic marker and a potential target in lung cancer therapy.
Abbreviations
miR-548e-5p, microRNA-548e-5p; UBE2C, ubiquitin-conjugating enzyme E2 C; EMT, epithelial-to-mesenchymal transition; 3′-UTR, 3′-untranslated regions; 5′-UTR, 5′-untranslated regions; ZEB1, zinc finger E-box binding homeobox 1; ZEB2, zinc finger E-box binding homeobox 2; HBEC, human bronchial epithelial cells; NSCLC, non-small cell lung cancer; siRNA, short interfering RNA.
Acknowledgements
We appreciate Professor Sichuan. Xi (National Institutes of Health, USA) for critical reading of the manuscript. This work was supported by National Natural Science Foundation of China (No.31801085), the Science and Technology Development Foundation of Yantai (2015ZH082), Natural Science Foundation of Shandong Province (ZR2018QH004, ZR2016HB55, ZR2017PH067, ZR2017MH125 and ZR2015PH004), and Research Foundation of Binzhou Medical University (BY2015KYQD25 and BY2015KJ14).
Contributions
Jiwei, Guo and Dan, Jin designed the experiments. Jiwei Guo, Dan Jin, Yan Wu, Jing Du, Xiaohong Wang, Jiajia An, Baoguang Hu, Lingqun Kong, Cuijie Shao, Weihua Di and Wansheng Wang performed the work. Jiwei, Guo and Dan, Jin analyzed the data and competed the figures. Jiwei, Guo wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Supplementary figures and tables.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Barton MK. Human immunodeficiency virus status has no effect on survival in patients with non-small cell lung cancer. CA Cancer J Clin. 2013;63:145-146
2. Barton MK. Encouraging long-term outcomes reported in patients with stage I non-small cell lung cancer treated with stereotactic ablative radiotherapy. CA Cancer J Clin. 2017;67:349-350
3. Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: New biological insights and recent herapeutic advances. CA Cancer J Clin. 2011;61:91-112
4. Xu L, Li Y, Yin L, Qi Y, Sun H, Sun P. et al. miR-125a-5p ameliorates hepatic glycolipid metabolism disorder in type 2 diabetes mellitus through targeting of STAT3. Theranostics. 2018;8:5593-5609
5. Xie Y, Wang Y, Li J, Hang Y, Jaramillo L, Wehrkamp CJ. et al. Cholangiocarcinoma therapy with nanoparticles that combine downregulation of MicroRNA-210 with inhibition of cancer cell invasiveness. Theranostics. 2018;8:4305-4320
6. Kim MK, Moon YA, Song CK, Baskaran R, Bae S, Yang SG. Tumor-suppressing miR-141 gene complex-loaded tissue-adhesive glue for the locoregional treatment of hepatocellular carcinoma. Theranostics. 2018;8:3891-3901
7. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK. et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384-388
8. Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC. et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527:100-104
9. Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C. et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107-110
10. Garofalo M, Croce CM. Role of microRNAs in maintaining cancer stem cells. Adv Drug Deliv Rev. 2015;81:53-61
11. Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543-550
12. Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2011;481:190-194
13. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333-338
14. Shi Y, Qiu M, Wu Y, Hai L. MiR-548-3p functions as an anti-oncogenic regulator in breast cancer. Biomed Pharmacother. 2015;75:111-116
15. Liang T, Guo L, Liu C. Genome-wide analysis of mir-548 gene family reveals evolutionary and functional implications. J Biomed Biotechnol. 2012;2012:679563
16. Guo L, Ding Z, Huang N, Huang Z, Zhang N, Xia Z. Forkhead Box M1 positively regulates UBE2C and protects glioma cells from autophagic death. Cell Cycle. 2017;16:1705-1718
17. Psyrri A, Kalogeras KT, Kronenwett R, Wirtz RM, Batistatou A, Bournakis E. et al. Prognostic significance of UBE2C mRNA expression in high-risk early breast cancer. A Hellenic Cooperative Oncology Group (HeCOG) Study. Ann Oncol. 2012;23:1422-1427
18. Chen Z, Zhang CP, Wu DY, Chen HY, Rorick A, Zhang XT. et al. Phospho-MED1-enhanced UBE2C locus looping drives castration-resistant prostate cancer growth. EMBO J. 2011;30:2405-2419
19. Sivakumar S, Lucas FAS, McDowell TL, Lang W, Xu L, Fujimoto J. et al. Genomic Landscape of Atypical Adenomatous Hyperplasia Reveals Divergent Modes to Lung Adenocarcinoma. Cancer Res. 2017;77:6119-6130
20. Esmerats JF, Villa-Roel N, Kumar S, Gu L, Salim MT, Ohh M. et al. Disturbed Flow Increases UBE2C (Ubiquitin E2 Ligase C) via Loss of miR-483-3p, Inducing Aortic Valve Calcification by the HIF-1α (Hypoxia-Inducible Factor-1α) Pathway in Endothelial Cells. Arterioscler Thromb Vasc Biol. 2019 doi: 10.1161/ATVBAHA.118.312233
21. van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol. 2010;188:83-100
22. Xie C, Powell C, Yao M, Wu J, Dong Q. Ubiquitin-conjugating enzyme E2C: a potential cancer biomarker. Int J Biochem Cell Biol. 2014;47:113-117
23. Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611-629
24. Guo J, Wu Y, Yang L, Du J, Gong K, Chen W. et al. Repression of YAP by NCTD disrupts NSCLC progression. Oncotarget. 2017;8:2307-2319
25. Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F. et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61-74
26. Krebs AM, Mitschke J, Lasierra LM, Schmalhofer O, Boerries M, Busch H. et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol. 2017;19:518-529
27. Su W, Xu M, Chen X, Chen N, Gong J, Nie L. et al. Long noncoding RNA ZEB1-AS1 epigenetically regulates the expressions of ZEB1 and downstream molecules in prostate cancer. Mol Cancer. 2017;16:142
28. Zheng G, Peng C, Jia X, Gu Y, Zhang Z, Deng Y. et al. ZEB1 transcriptionally regulated carbonic anhydrase 9 mediates the chemoresistance of tongue cancer via maintaining intracellular pH. Mol Cancer. 2015;14:84
29. Liu Y, Yin JJ, Abou-Kheir W, Hynes PG, Casey OM, Fang L. et al. MiR-1 and miR-200 inhibit EMT via Slug-dependent and tumorigenesis via Slug-independent mechanisms. Oncogene. 2013;32:296-306
30. Li Y, Wang L, Pappan L, Galliher-Beckley A, Shi J. IL-1beta promotes stemness and invasiveness of colon cancer cells through Zeb1 activation. Mol Cancer. 2012;11:87
31. Simeone P, Trerotola M, Franck J, Cardon T, Marchisio M, Fournier I. et al. The multiverse nature of epithelial to mesenchymal transition. Semin Cancer Biol. 2018 doi: 10.1016/j.semcancer.2018.11.004
32. Caramel J, Ligier M, Puisieux A. Pleiotropic Roles for ZEB1 in Cancer. Cancer Res. 2018;78:30-35
33. Soleimani A, Khazaei M, Ferns GA, Ryzhikov M, Avan A, Hassanian SM. Role of TGF-beta signaling regulatory microRNAs in the pathogenesis of colorectal cancer. J Cell Physiol. 2019 doi: 10.1002/jcp.28169
34. Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q. et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725-731
35. Guo J, Wu Y, Du J, Yang L, Chen W, Gong K. et al. Deregulation of UBE2C-mediated autophagy repression aggravates NSCLC progression, Oncogenesis. 2018; 7: 49.
36. Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333-334
37. Farré D, Roset R, Huerta M, Adsuara JE, Roselló L, Albà MM. et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic acids Res. 2003;31:3651-3653
38. Zhang Y, Tian S, Li X, Ji Y, Wang Z, Liu C. UBE2C promotes rectal carcinoma via miR-381. Cancer Biol Ther. 2018;19:230-238
39. Barton MK. Adjuvant chemotherapy benefits older and younger non-small cell lung cancer patients alike. CA Cancer J Clin. 2012;62:279-280
40. Hurria A, Kris MG. Management of lung cancer in older adults. CA Cancer J Clin. 2003;53:325-341
41. Garvanska DH, Larsen MS, Nilsson J. Synergistic inhibition of the APC/C by the removal of APC15 in HCT116 cells lacking UBE2C. Biol Open. 2016;5:1441-1448
42. Guo J, Jin D, Wu Y, Yang L, Du J, Gong K. et al. The miR 495-UBE2C-ABCG2/ERCC1 axis reverses cisplatin resistance by downregulating drug resistance genes in cisplatin-resistant non-small cell lung cancer cells. EBioMedicine. 2018;35:204-221
43. Shen Z, Jiang X, Zeng C, Zheng S, Luo B, Zeng Y. et al. High expression of ubiquitin-conjugating enzyme 2C (UBE2C) correlates with nasopharyngeal carcinoma progression. BMC Cancer. 2013;13:192
44. Wiemer EAC. Prognostic Circulating MicroRNA Biomarkers in Early-Stage Non-Small Cell Lung Cancer: A Role for miR-150. Clin Pharmacol Ther. 2018;103:968-970
45. Hsu YL, Hung JY, Chang WA, Jian SF, Lin YS, Pan YC. et al. Hypoxic Lung-Cancer-Derived Extracellular Vesicle MicroRNA-103a Increases the Oncogenic Effects of Macrophages by Targeting PTEN. Mol Ther. 2018;26:568-581
46. Liu X, Huang YG, Jin CG, Zhou YC, Chen XQ, Li J. et al. MicroRNA-370 inhibits the growth and metastasis of lung cancer by down-regulating epidermal growth factor receptor expression. Oncotarget. 2017;8:88139-88151
47. Qi XK, Han HQ, Zhang HJ, Xu M, Li L, Chen L. et al. OVOL2 links stemness and metastasis via fine-tuning epithelial-mesenchymal transition in nasopharyngeal carcinoma. Theranostics. 2018;8:2202-2216
48. Wang C, Jin H, Wang N, Fan S, Wang Y, Zhang Y. et al. Gas6/Axl Axis Contributes to Chemoresistance and Metastasis in Breast Cancer through Akt/GSK-3beta/beta-catenin Signaling. Theranostics. 2016;6:1205-1219
49. Lee SY, Jeong EK, Ju MK, Jeon HM, Kim MY, Kim CH. et al. Induction of metastasis, cancer stem cell phenotype, and oncogenic metabolism in cancer cells by ionizing radiation. Mol Cancer. 2017;16:10
50. Voutsadakis IA. Epithelial-Mesenchymal Transition (EMT) and Regulation of EMT Factors by Steroid Nuclear Receptors in Breast Cancer: A Review and in Silico Investigation. J Clin Med. 2016;5:11
Author contact
![]() Corresponding author: Jiwei Guo, Cancer research institute, Binzhou Medical University Hospital, Binzhou 256603, P.R. China; E-mail: gjw0510nankai.edu.cn, Tel and Fax: +86 05433258276.
Corresponding author: Jiwei Guo, Cancer research institute, Binzhou Medical University Hospital, Binzhou 256603, P.R. China; E-mail: gjw0510nankai.edu.cn, Tel and Fax: +86 05433258276.
 Global reach, higher impact
Global reach, higher impact