13.3
Impact Factor
Theranostics 2025; 15(7):2778-2793. doi:10.7150/thno.107761 This issue Cite
Review
Multiple regulatory mechanisms, functions and therapeutic potential of chaperone-mediated autophagy
1. The College of Basic Medical Science, Health Sciences Institute, China Medical University, Shenyang, Liaoning Province 110122, China.
2. Key Laboratory of Cell Biology of Ministry of Public Health, Key Laboratory of Medical Cell Biology of Ministry of Education, Key Laboratory of Precision Diagnosis and Treatment of Gastrointestinal Tumors of Ministry of Education, Liaoning Province Collaborative Innovation Center of Aging Related Disease Diagnosis and Treatment and Prevention, China Medical University, Shenyang, Liaoning Province 110122, China.
3. Department of Gastroenterology, Shengjing Hospital of China Medical University, Shenyang, China.
4. Department of Ophthalmology, The First Affiliated Hospital of China Medical University, Shenyang, Liaoning, 110001, China.
#These authors contributed equally to this work.
Received 2024-11-27; Accepted 2025-1-25; Published 2025-2-3
Abstract
Autophagy refers to the proteolytic degradation of cytoplasmic components by lysosomes, and includes three defined types: macroautophagy, chaperone-mediated autophagy (CMA), and microautophagy. Although the regulatory pathways of macroautophagy are well defined, how CMA is accurately regulated remains less understood. In recent years, emerging evidence has suggested that chaperone-mediated autophagy is regulated by multiple mechanisms at nucleic acid and protein levels. In this review, we summarized recent progress on multiple regulatory mechanisms and functions concerning CMA, as well as novel treatments targeting specific regulation sites.
Keywords: autophagy, chaperone-mediated autophagy, post-translational modifications, Hsc70, LAMP2A.
Introduction
Autophagy refers to the proteolytic degradation of cytoplasmic components by lysosomes, and includes three defined types: macroautophagy, chaperone-mediated autophagy (CMA) and microautophagy [1]. Autophagy is critical for the maintenance of homeostasis by eliminating degenerated cellular components such as proteins and organelles [2]. Of the three major patterns of autophagy, CMA is the first to be identified as a process that can be selective [3]. The mechanism of CMA includes three main components: the substrate to be degraded, a chaperone molecule, and a receptor on the lysosome membrane [4]. Chaperone-mediated autophagy specifically degrades proteins with a pentapeptide motif, KFERQ. Such a substrate is recognized by heat shock protein family A (HSP70) member 8 (HSPA8; also known as heat shock cognate protein 70 [Hsc70]) and is subsequently delivered to the lysosomal membrane [4-6]. The lysosomal receptor, lysosomal associated membrane protein 2A (LAMP2A), is responsible for transporting the substrate into the lysosomal lumen for degradation [7]. Chaperone-mediated autophagy has been reported to regulate a variety of biological processes [4].
Emerging evidence suggests that chaperone-mediated autophagy is regulated by multiple mechanisms. From the perspective of CMA substrates, the RNA around the substrate [8] or changes in the gene of the substrate itself [9] can affect its degradation. In addition, the occurrence of CMA is closely related to the post-translational modifications (PTMs) of substrates. PTMs of proteins are special chemical groups that attach to amino acid side chains by covalent binding, including phosphorylation, acetylation, ubiquitination, methylation, and so on [10, 11]. PTMs of CMA substrates could change the recognition and binding between the substrates and CMA component molecules (Hsc70 and LAMP2A). Similarly, the chaperone molecules Hsc70 and receptor LAMP2A are also regulated by nucleic acids and PTMs [12-14]
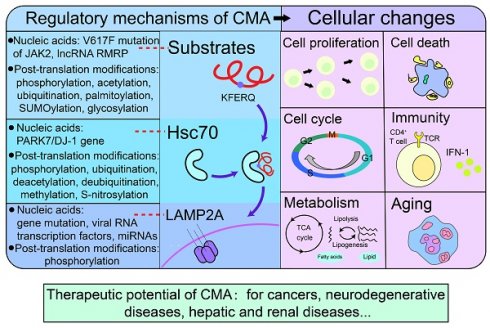
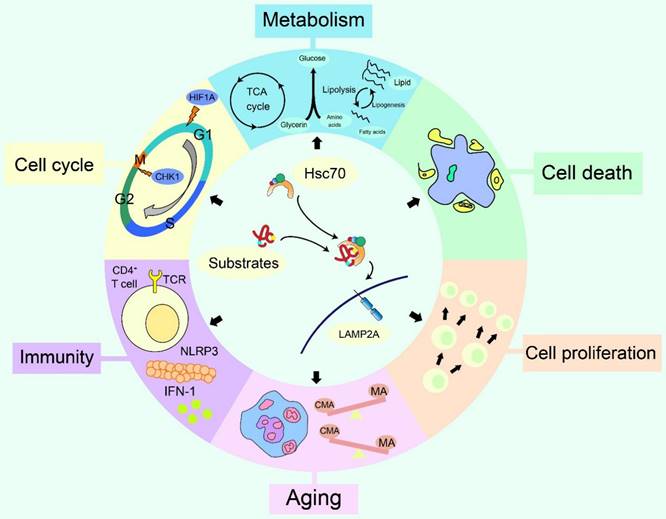
CMA modulates various cellular changes, including cell proliferation [15], cell cycle [16], metabolism [17], immunity [18], aging [19], cell death [20]and so on. Furthermore, targeting CMA could benefit the treatment of cancer [21], neurodegenerative diseases [22, 23], hepatic diseases [24, 25], kidney diseases [26] and diabetic complications [27]. In this review, we systematically summarized recent advances of the regulation, the effects on cellular changes and the therapeutic potential of CMA.
Regulatory mechanisms of chaperone-mediated autophagy
Substrates of chaperone-mediated autophagy
Basic characteristics of CMA substrates
The characteristic of CMA substrates is certain protein with KFERQ motif [28]. Actually, it is the properties of the amino acid residues composing the pentapeptide motif that determines whether a substrate can be degraded by CMA [29]. PTMs of substrates (like phosphorylation, acetylation) could change the motif composition and promote CMA degradation by producing similar targeting motifs [30]. For instance, acetylated K (Lys) can replace Q (Gln) because acetylation enables K to acquire properties similar to Q [30-32].
Regulation of nucleic acids on CMA substrates
DNA mutation of substrates and their regulator, as well as substrate interacting RNAs have been suggested to regulate substrates degradation through CMA. V617F mutation of the Janus kinase 2 (JAK2) gene (JAK2V617F) reduced the degradation of pyruvate kinase M1 (PKM1) through CMA by blocking Hsc70-PKM1 binding, which leads to STAT3 activation and promotes the production of interleukin-6 [33]. The LFSINE insertion sequence in a hematopoietic-specific member of the Rho GTPase family (RhoH) promoted the degradation of RhoH through CMA since deletion of this sequence delayed the degradation of RhoH, suggesting that this sequence may be a signal for lysosomal uptake [9]. In addition, long noncoding RNA (lncRNA) RNA component of mitochondrial RNA-processing endoribonuclease (RMRP) interacts with Small nuclear ribonucleoprotein polypeptide A' (SNRPA1) and isolates it in the nucleus, which attenuates the contact between SNRPA1 and CMA component molecules and inhibits the CMA degradation of SNRPA1 in the cytoplasm [8].
Regulation of phosphorylation on CMA substrates
Phosphorylation is involved in almost every biochemical process, including cell division, regulation of gene expression, and signal transduction [34]. Phosphorylation of CMA substrates can affect the tendency of substrates to be degraded, which can be enhanced or weakened. ULK1 phosphorylation mimic variants (ULK1 S423D) bound more CMA component molecules than those in wild-type cells, which suggests that ULK1 phosphorylation increased its interaction with LAMP2A or Hsc70 [35]. Microrchidia family CW-type zinc finger 2 (MORC2) is phosphorylated at T717 and T733 by cyclin-dependent kinase 1 (CDK1), which is activated by paclitaxel and vincristine. The phosphorylation at T717 and T733 enhances the combination of MORC2 with Hsc70 and LAMP2A [36]. However, in another situation, protein kinase cAMP-activated catalytic subunit alpha (PRKACA), activated by G protein-coupled estrogen receptor 1 (GPER1), phosphorylates MORC2 at T582, thus triggering a reduction in the interaction between MORC2 and CMA component molecules and protecting MORC2 from lysosomal degradation through CMA [37]. Phosphorylation of BCL2 binding component 3 (BBC3) at Ser10, mediated by IKKβ (inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase β), prevents its degradation by CMA [38]. The accumulation of α-synuclein (α-syn) has been implicated in the pathogenesis of Parkinson's disease (PD). S129E mutant α-syn (mimic phosphorylated α-syn) failed to translocate across the lysosomal membrane compared to the wild-type α-syn, indicating that phosphorylated α-syn could not degrade through CMA [39]. Phosphoprotein Enriched in Astrocytes of 15 kDa, a death effector domain family member (PED) is identified as a substrate for CMA. The major phosphorylation of PED at Ser104 and Ser116 attenuates the combination of PED and Hsc70, thus restraining PED degradation through CMA [40]. Proviral Insertion in the murine lymphomas 2 (PIM2) oncogene phosphorylates hexokinase-II (HK2) at T473. Through the knockdown of Hsc70 and blockade of CMA, PIM2-mediated T473 phosphorylation of HK2 was proved to be responsible for the stability of HK2 [41].
Regulation of acetylation on CMA substrates
Acetylation is a biochemical reaction that transfers acetyl groups from acetyl coenzyme A (acetyl-CoA) to specific proteins, thereby regulating the effects and functional diversity of certain proteins [42]. In the acetylation reaction, acetyltransferase and deacetylase are responsible for the modification and removal of acetyl groups to substrate proteins [42]. Acetylation of 17β-hydroxysteroid dehydrogenase type 4 (HSD17B4) at K669 by dihydroxytestosterone enhances its degradation through CMA, thus promoting its degradation in prostate cancer (PCa) cells [43]. Estrone (E1) also enhances K669 acetylation of HSD17B4 and promoted its degradation [44]. M2 isoform of pyruvate kinase (PKM2) acetylation at K305 promoted its binding to Hsc70 and enhanced its degradation through CMA, which ultimately promoted tumor growth [31]. The stress response protein regulated in development and DNA damage response 1 (REDD1) are associated with complications of recognition by Hsc70 [45]. Generally, the above results indicate that substrate acetylation can promote the occurrence of CMA or is a prerequisite for CMA.
The acetylation of the substrate might also exert an inhibitory effect on CMA. Tau is a microtubule-binding protein in cells and its abnormal deposition is associated with the onset of many diseases, collectively referred to as tauopathies [46]. Tau acetylation induced by the loss of TSC1 prevents its clearance through CMA, thereby increasing the risk of tauopathy [47]. However, in another study, an increase in acetylated tau did reduce CMA, but by affecting substrate translocation within the lysosomes, rather than interfering with substrate binding to the surface of the lysosomes [48]. Mammalian hepatitis B X-interacting protein (HBXIP), highly expressed in breast cancer tissues, is involved in cellular proliferation and invasion as an oncoprotein [49]. HBXIP promotes K277 acetylation of homeobox B13 (HOXB13, a transcription factor) and prevents its degradation by CMA, which eventually leads to drug resistance in breast cancer [50].
Not only does acetylation itself affect CMA, but the enzymes (like deacetylase) involved in acetylation also regulate CMA. The expression of silent information regulator 3 (SIRT3) in hepatocytes activates AMP-activated protein kinase (AMPK) and promotes the formation of a LAMP2A-Hsc70-PLN2 complex, which, in turn, promotes the occurrence of CMA [51]. Besides, the activity of CMA is affected by histone deacetylase 6 (HDAC6)-mediated HSP90 deacetylation [52]. In a HeLa cell line with HDAC10 knockdown, the substrate Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was found to be more efficiently degraded by CMA, indicating that knockdown of HDAC10 activated CMA and accelerated substrate degradation [53].
Regulation of other PTMs on CMA substrates
In addition to phosphorylation and acetylation, ubiquitination, palmitoylation, SUMOylation and glycosylation are also involved in the regulation of CMA substrates. Ubiquitination is a type of reversible PTMs using ubiquitin as a substrate, and it is catalyzed by a series of enzymes called E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases [54-56]. K63 ubiquitination of hypoxia inducible factor 1, α subunit (HIF1A) is crucial for its degradation through CMA. The inhibition of this type of ubiquitination hindered the degradation of HIF1A by CMA [57]. Palmitoylation is a reversible special case among many irreversible lipid modification reactions, which modifies a sixteen-carbon saturated fatty acid (palmitate) to the cysteine residue of the protein in the form of thioesterification. Palmitoylation of most proteins is carried out by palmitoyl S-acyltransferases, which are part of the zinc finger DHHC-type containing (ZDHHC) family [58, 59]. ZDHHC12 catalyzes palmitoylation of nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin-domain-containing 3 (NLRP3) at C844. This palmitoylation enhances its recognition by Hsc70, thus promoting its degradation through CMA [60]. SUMOylation is a type of PTMs using small ubiquitin-like modifier (SUMO) as the substrate, which usually modifies lysine residues in protein [61]. Like ubiquitin, a SUMO protein also undergoes a cascade reaction in vivo to modify the target protein, including the action of enzymes E1, E2, and E3 [62, 63]. SUMOylation of stimulator of interferon genes (STING) by the E3 ubiquitin ligase TRIM38 at K337 is located in the attachment of the QEVLR motif (amino acids 326-330) in STING, thus leading to a masking effect of modified SUMO molecules on the motif and inhibition of the interaction of Hsc70 with STING [64]. Glycosylation refers to the selective modification of amino acid residues of substrate proteins by monosaccharides or glycans, including N-glycosylation, O-glycosylation, and C-glycosylation according to the different covalent bonds between proteins and glycogroups [65]. After human papillomavirus (HPV) infection, ULK1 undergoes an O-linked β-N-acetylglucosamine (O-GlcNAc) modification. Since the Ser423 phosphorylation of ULK1 is known to promote the CMA of ULK1 [35], it was demonstrated that the O-GlcNAcylation of ULK1 at Ser409 and Ser410 antagonizes its phosphorylation at Ser423 and thus attenuated its degradation via CMA [66].
Chaperone molecules of chaperone-mediated autophagy
Introduction of chaperone molecules
Hsc70 recognizes proteins with KFERQ-like motif and binds with the protein, thus targeting the substrate for lysosomal degradation [5]. So far, several co-chaperones have been found to help localize substrate proteins to lysosomes in an Hsc70-dependent manner, including HSP40 (also called as Dnajb1), carboxyl terminus of Hsc70-interacting protein (CHIP) and HSP70-HSP90 organizing protein (HOP), but Hsc70 is the only chaperone found to directly recognize the KFERQ-like motif [6, 67]. After Hsc70 recognizes the substrate and binds to it, the HSC70-substrate complex binds to LAMP2A located on the lysosomal membrane [68, 69]. Hsc70 then initiates the substrate unfolding with the assistance of several co-chaperones in preparation for the internalization of the substrate into lysosomes [6]. After the above processes, Hsc70 dissociates from LAMP2A and is ready to recognize and bind new substrates for further CMA cycle [68].
Regulation of chaperone molecules
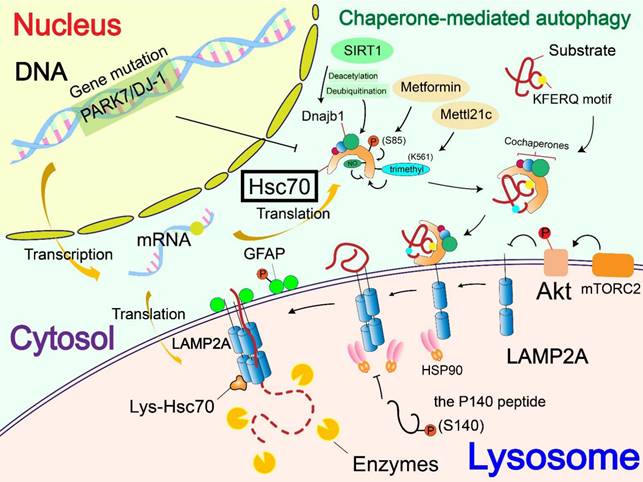
At present, few studies focus on the regulation of nucleic acids on Hsc70. A mutation or deficiency of PARK7/DJ-1 gene regulates CMA through changing the level of Hsc70. The lack of DJ-1 down-regulates the level of Hsc70 and leads to the aggregation of the substrate α-syn, rather than affecting the number of lysosomes in cells [12].
At PTMs levels, Metformin can activate TAK1-IKKα/β signaling, leading to the phosphorylation of Hsc70 at Ser85 and activation of CMA [13]. Generated by the spliceosomal protein, SNRNP70/U1-70K, a P140 peptide is a 21-mer linear peptide (sequence 131-151) that has a phosphorylation site at S140. The P140 peptide inhibits CMA by interfering with the binding of lumenal Hsc70 to substrates [70]. SIRT1 mediated deacetylation and deubiquitination of DnaJ heat shock protein family member B1 (Dnajb1), which promotes its binding to Hsc70 and the activity of CMA[71]. CHIP, as a molecular chaperone, promotes the degradation of mutant p53 through lysosomes. This is carried out through K63 ubiquitination, but the specific form of autophagy was not clear and needs to be investigated [72]. Methylation is a type of PTMs using the methyl group from S-adenoslyl-L-methionine (SAM) [73]. Protein methyltransferases mainly target arginine and lysine residues and are therefore divided into protein arginine methyltransferases (PRMTs) and lysine methyltransferases [74]. An effect of a new type of non-histone methyltransferase called methyltransferase-like 21c (Mettl21c) was found to contribute to the stability of Hsc70. Mettl21c trimethylates Hsc70 at lysine 561, thus enhancing its stability and promoting its function in CMA [75]. S-nitrosylation is a special PTM mediated by nitric oxide (NO), targeting cysteine residues in substrate proteins. In mammals, NO is produced by nitric oxide synthase (NOS), which mediates the linking of the thiol side chain of cysteine (single or multiple) to nitroso, thus forming a structure called S-nitrosothiol [76, 77]. NO-mediated S-nitrosylation of Hsc70 was found to interfere with CMA [78].
Receptor of chaperone-mediated autophagy
Introduction of the receptor
LAMP2A, expressed by the gene LAMP2, is the first identified CMA component molecule located on the lysosome [79, 80]. In addition to LAMP2A, this gene also expresses two other splice variants (LAMP2B and LAMP2C), but LAMP2A is the only protein involved in CMA [80, 81]. LAMP2A participates in several different steps of CMA, including substrate binding, LAMP2A assembly, substrate translocation, and so on [3]. After the formation of the Hsc70-substrate complex, the 12-amino acid tail on the cytoplasmic side of LAMP2A binds to the complex, thus completing the docking of the complex on the lysosome [80, 81]. After that, LAMP2A begins to multimerize, forming a 700kDa protein complex that is required for substrate translocation into the interior of the lysosome [68, 82]. HSP90 is also involved in LAMP2A assembly and is responsible for stabilizing LAMP2A multimers [68].
Regulation of nucleic acids and phosphorylation on LAMP2A
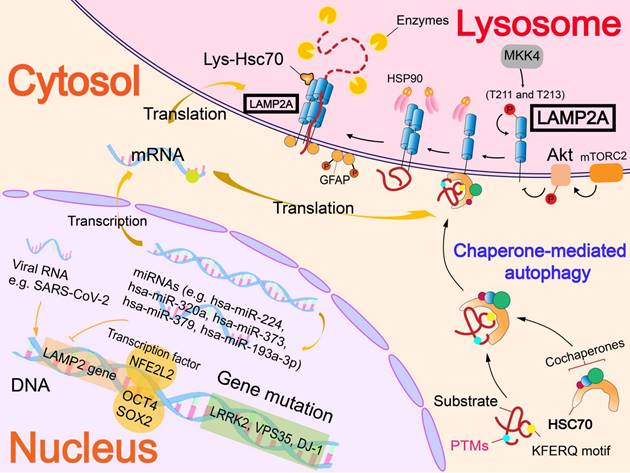
In addition to affecting the level of Hsc70, a mutation or deficiency of PARK7/DJ-1 gene can also regulate CMA by accelerating the degradation of LAMP2A [12]. Deficiency of VPS35 gene also accelerates LAMP2A degradation. In VPS35-deficient dopamine neurons, this effect attenuates the CMA of α-syn, causing the abnormal accumulation of α-syn and the development of PD [83].
Some of the transcription factors can bind to the LAMP2 gene, thereby participating in LAMP2A expression to affect CMA. Nuclear factor, erythroid derived 2, like 2 (NFE2L2/NRF2) was found to interact with two sequences in the LAMP2 gene, which can promote the expression of LAMP2A. Therefore, the level of LAMP2A was significantly decreased in NFE2L2-knockout hepatocytes, as well as the activity of CMA [84]. Besides, in mouse embryonic stem cells, OCT4 and SOX2 bind with the LAMP2 gene and inhibit CMA [85].
At the RNA level, different kinds of RNA can interfere with LAMP2A expression or interact with LAMP2A, thereby affecting CMA. MicroRNAs, including Homo sapiens (hsa)-miR-224, hsa-miR-320a [86], hsa-miR-373, hsa-miR-379[87] and hsa-miR-193a-3p[88] are the main miRNAs that target the LAMP2 gene according to current studies. These miRNAs induced downregulation of CMA and contributed to the accumulation of α-syn in the pathogenesis of PD. In addition, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection induced the upregulation of LAMP2A. Besides, overexpression of LAMP2A significantly decreased the RNA level of SARS-CoV-2, indicating that LAMP2A has a role in reducing the viral RNA level in SARS-CoV-2-infected cells [89].
The effect of PTMs on LAMP2A is poorly understood, and only phosphorylation has been found to regulate LAMP2A. Endoplasmic reticulum stressors result in activation by a double-stranded RNA-activated protein kinase-like ER kinase, which recruits mitogen-activated protein kinase kinase 4 (MKK4) to lysosomes. The MKK4 can activate p38 mitogen activated protein kinase (MAPK) at lysosomes, which phosphorylates LAMP2A at T211 and T213 directly and activates CMA [14]. Beyond that, phosphorylation of upstream molecules also affects LAMP2A. Located on the lysosomal membrane, Akt regulates the phosphorylation of glial fibrillary acidic protein (GFAP) and further affects CMA. Unphosphorylated GFAP promotes the multimerization of LAMP2A by binding the cytosolic tail of LAMP2A, thus accelerating substrate uptake, and facilitates the process of CMA [82, 90]. In addition to affecting Hsc70, the above-mentioned P140 peptide also reduces the expression of LAMP2A. Overall, the P140 peptide has a variety of negative effects on the LAMP2A-Hsc70 lysosomal axis, which ultimately leads to the inhibition of CMA [70].
The effects of chaperone-mediated autophagy on cellular changes
Cell proliferation
In the studies of esophageal squamous cell carcinoma (ESCC) and non-small cell lung cancer (NSCLC), CMA knockdown inhibited cell proliferation and colony formation, and increased the sensitivity of cancer cells to chemotherapeutic drugs, which provides a new potential target for therapeutic research [15, 91]. Using the same approach, it was found that CMA inhibition in colorectal cancer (CRC) cells resulted in a decreased ability of cell migration and invasion, and that overexpression of LAMP2A increased cell viability [92]. Some studies found that CMA changes regulate cell proliferation through the action of other molecules. In studies of glioma, additional activation of CMA caused by upregulation of LAMP2A enhanced the proliferation and invasion of cancer cells by promoting the degradation of SMAD3[93]. Besides, the attenuation of CMA caused by LAMP2A knockdown in gastric cancer cells hindered cell proliferation by promoting Rho Family GTPase 3 (RND3/RhoE) accumulation [94]. LAMP2A was found to be a potential biomarker for early warning of gastric cancer [94]. In addition, co-inhibition of CMA and macroautophagy demonstrated a synergistic effect on the inhibition of proliferation and survival in K-Ras G12V (K-Ras oncogene mutation at G12V) mouse embryonic fibroblasts (MEFs) [95].
Targets and sites of regulatory mechanism on CMA substrates.
| Substrate | Regulatory mechanism | Regulatory target | Site | Effect on CMA | |
|---|---|---|---|---|---|
| PKM1 | Nucleic acids | V617F mutation of JAK2 gene | Hsc70 | inhibited | |
| RhoH | LFSINE insertion sequence | RhoH | enhanced | ||
| SNRPA1 | lncRNA RMRP | SNRPA1 | inhibited | ||
| ULK1 | Post-translational modifications of proteins | Phosphorylation | ULK1 | S423D | enhanced |
| MORC2 | MORC2 | T717 and T733 | enhanced | ||
| MORC2 | MORC2 | T582 | inhibited | ||
| BBC3 | BBC3 | S10 | inhibited | ||
| α-syn | α-syn | S129E | inhibited | ||
| PED | PED | S104 and S116 | inhibited | ||
| HK2 | HK2 | T473 | inhibited | ||
| HSD17B4 | Acetylation | HSD17B4 | K669 | enhanced | |
| PKM2 | PKM2 | K305 | enhanced | ||
| REDD1 | REDD1 | K129 | enhanced | ||
| tau | tau | K174 | inhibited | ||
| HOXB13 | HOXB13 | K277 | inhibited | ||
| HIF1A | Ubiquitination | HIF1A | K63 linked | enhanced | |
| NLRP3 | Palmitoylation | NLRP3 | C844 | inhibited | |
| STING | SUMOylation | STING | K337 | inhibited | |
| ULK1 | Glycosylation | ULK1 | S409 and S410 | inhibited | |
Regulatory mechanisms of CMA substrates. Regulation of CMA substrates is divided into regulation of nucleic acids and regulation of the level of post-translational modifications (PTMs). Regulation of nucleic acids includes gene mutations that affect the binding of substrate to Hsc70 (JAK2 gene) and the interaction with substrate to prevent the substrate from entering the cytosol (RMRP). The effects of PTMs levels include phosphorylation, acetylation, ubiquitination, palmitoylation and SUMOylation which modify the substrates and affect the binding of the substrate to Hsc70. In addition, different PTMs of the substrate can interfere with each other, such as the glycosylation modification of ULK1 inhibits the promotion effect of its own phosphorylation on the CMA process.
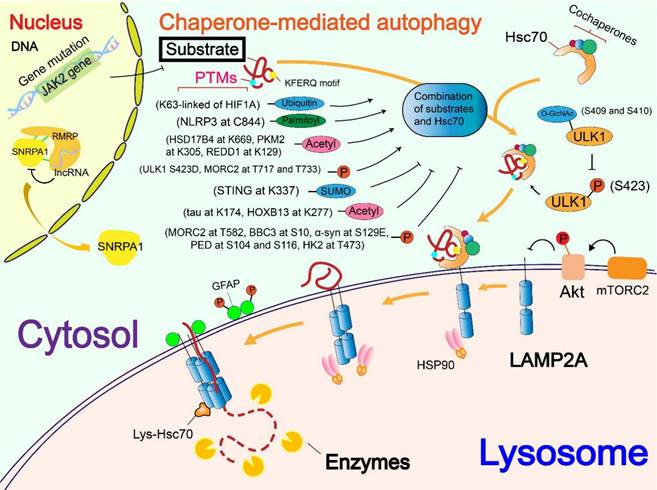
Regulatory mechanisms of chaperone (Hsc70). The regulation of Hsc70 is divided into the level of gene mutations and the regulation of Hsc70 by its own PTMs. The mutation of PARK7/DJ-1 gene down-regulates the level of Hsc70, thereby affecting the occurrence of CMA. Phosphorylation and methylation of Hsc70 activate Hsc70, whereas S-nitrosylation inhibits Hsc70 activity. Besides, deacetylase SIRT1 mediated deacetylation and deubiquitination of the cochaperone Dnajb1, thus enhancing its interaction with Hsc70 and regulating CMA.
Targets and sites of regulatory mechanism on Hsc70.
| Regulatory mechanism | Target | Site | Effect on Hsc70 | Effect on CMA | |
|---|---|---|---|---|---|
| Gene mutation | PARK7/DJ-1 gene | Hsc70 | Down-regulated | inhibited | |
| Phosphorylation | Metformin | Hsc70 | S85 | Activated | enhanced |
| P140 peptide | Hsc70-HSP90AA1 complex | Interfered | inhibited | ||
| Ubiquitination | Dnajb1 | Binding of Dnajb1 to Hsc70 | enhanced | ||
| Deacetylation | SIRT1 | Dnajb1 | enhanced | ||
| Deubiquitination | SIRT1 | Dnajb1 | enhanced | ||
| Methylation | Mettl21c | Hsc70 | K561 (trimethylation) | Activated | enhanced |
| S-nitrosylation | NO | Hsc70 | Interfered | inhibited | |
Targets and effects of regulatory mechanism on LAMP2A.
| Regulatory mechanism | Target | Effect on LAMP2A | Effect on CMA | |
|---|---|---|---|---|
| Gene mutation | PARK7/DJ-1 gene | LAMP2A | Accelerating degradation | inhibited |
| VPS35 gene | LAMP2A | Accelerating degradation | inhibited | |
| GBA1 gene | Lysosome | Causing dysfunction | inhibited | |
| LRRK2 gene | Lysosome | Causing dysfunction | inhibited | |
| Transcription factors | NFE2L2/NRF2 | LAMP2 gene | Up-regulated | enhanced |
| OCT4 and SOX2 | LAMP2 gene | Down-regulated | inhibited | |
| MiRNA | hsa-miR-224 | LAMP2 gene | Down-regulated | inhibited |
| hsa-miR-320a | LAMP2 gene | Down-regulated | inhibited | |
| hsa-miR-373 | LAMP2 gene | Down-regulated | inhibited | |
| hsa-miR-379 | LAMP2 gene | Down-regulated | inhibited | |
| hsa-miR-193a-3p | LAMP2 gene | Down-regulated | inhibited | |
| Viral RNA | SARS-CoV-2 | LAMP2A | Up-regulated | enhanced |
| Phosphorylation | MKK4 | LAMP2A at T211 and T213 | Activated | enhanced |
| Akt | GFAP | Inactivated | inhibited | |
Regulatory mechanisms of receptor (LAMP2A). The regulation of LAMP2A mainly focuses on the level of nucleic acids. Gene mutations can accelerate LAMP2A degradation (PARK7/DJ-1 gene and VPS35 gene) or affect lysosomal function (GBA1 gene and LRRK2 gene). Transcription factors interact with LAMP2 gene to up-regulate or down-regulate the level of LAMP2A. MiRNA and viral RNA also interact with the LAMP2 gene to regulate LAMP2A expression. In addition, phosphorylation of LAMP2A can activate LAMP2A to promote CMA.
It is worth mentioning that downregulation of CMA may also promote cancer cell proliferation due to the lack of degradation of specific molecules. The transcriptional coactivators Yes1 associated transcriptional regulator (YAP1) and interleukin 6 cytokine family signal transducer (IL6ST) were identified as CMA substrates. Knockdown of LAMP2A resulted in increased levels of both the two proteins in hepatocellular carcinoma (HCC) cells, which ultimately promoted the proliferation and migration of HCC cells [96].
Cell death
Cell death refers to the response of cells to internal and external interference factors or biological signals to inactivate themselves, including apoptosis, necrosis, ferroptosis, cuproptosis, pyroptosis, and so on [97]. For example, excessive activation of CMA of hexokinase 2 (HK2, a key enzyme in glucose metabolism) can lead to metabolic disorder of cancer cells and trigger metabolic catastrophe, resulting in the death of cancer cells [98]. Furthermore, CMA activation by LAMP2A overexpression reduces hypoxia-induced apoptosis in cardiomyocytes [99], whereas CMA inhibition results in caspase-induced apoptosis occurring in mouse hepatocytes [100].
In fact, so far, the mode of cell death that has been more frequently studied in relation to CMA is ferroptosis. Among them, glutathione peroxidase 4 (GPX4) is a “bridge” connecting CMA and ferroptosis. GPX4 is considered to be an inhibitor of ferroptosis, and its activity is dependent on glutathione [101]. In acute kidney injury, Legumain promotes CMA in renal tubular cells, leading to increased degradation of GPX4, thereby triggering ferroptosis of tubular cells [102]. CMA inhibition stabilizes the activity of GPX4 and reduces the occurrence of ferroptosis [103]. CMA degradation of GPX4 leads to antimony-induced ferroptosis in neurons [20]. Conjugated fatty acids (CFAs) were found to promote CMA of GPX4 and target mitochondria for lipid peroxidation and GPX4 degradation, which provides a new perspective for ferroptosis-related cancer therapy research [104]. However, in particular, LAMP2A deficiency was found to increase reactive oxygen species (ROS)-induced ferroptosis in retinal pigment epithelial (RPE) cells, and glutathione (GSH) activity was decreased in LAMP2A-deficient RPE cells [105]. In addition to GPX4, CMA of Acyl-CoA synthetase long-chain family member 4 (ACSL4) is also associated with ferroptosis. ACSL4 is an enzyme involved in lipid metabolism and is associated with susceptibility of cells to ferroptosis. Melatonin can promote CMA of ACSL4, thereby reducing the occurrence of ferroptosis [106].
Cell cycle
HIF1A has been identified as a substrate of CMA [16]. As mentioned above, K63 ubiquitination is necessary for HIF1A degradation by CMA [57]. HIF1A is responsible for the adaptive response of cells to anaerobic environment, and its overexpression can arrest cells in G1 phase [107]. Under hypoxic stress, CMA is a negative regulator of HIF1A, and the inhibition of CMA leads to less degradation of HIF1A and cell cycle arrest [108]. Moreover, a pair of cyclin-dependent kinases (CDK1 and CDK2) regulate the degradation of HIF1A through CMA, blocking and promoting HIF1A degradation, respectively [107]. In addition, HIF1A-related cell cycle arrest is also associated with the expression of CDK inhibitors p21 and p27 [109]. The relationship between CMA and the cell cycle was also confirmed by the observation that hypoxia-induced CMA and lysosomal biogenesis in cancer cells are part of a negative feedback regulatory loop [16].
In addition to HIF1A, another CMA substrate implicated in cell cycle regulation is checkpoint kinase 1 (Chk1) [110]. Just like the negative feedback regulatory mechanism related to HIF1A, after the DNA of cells was destroyed by etoposide, CMA could be timely upregulated and degrade Chk1 to help resume the cell cycle. CMA inhibition led to the accumulation of Chk1, which blocked normal cell cycle progression and impaired cell viability upon DNA damage [110, 111].
Immunity
The development and maturation of T cells in the human body is quite essential for maintaining the immune function of the body [112]. Itchy E3 ubiquitin protein ligase (Itch) and regulator of calcineurin 1 (Rcan-1) are negative regulators of the T-cell receptor (TCR) signaling pathway, and both inhibit the activation of CD4+ T cells [113, 114]. CMA maintains CD4+ T cell activation by degrading Itch and Rcan-1 [18]. Itch is a type of E3 ubiquitin ligase responsible for ubiquitinating the Bcl10 Immune Signaling Adaptor (Bcl10), which inhibits the promotion of T-cell activation by the downstream NF-κB signaling pathway [113, 115]. In addition, the deficiency of Itch leads to the excessive activation of T cells and causes autoimmune diseases [116]. Rcan-1 is a type of calcineurin inhibitor that inhibits the Ca2+/ calcineurin/ nuclear factor of activated T-lymphocyte (NFAT) signaling pathway, which promotes T cell activation [114, 117]. Furthermore, T cell activation is abnormally suppressed in the LAMP2A-deficient model, accompanied by increased level of Itch and Rcan-1, as well as immune dysregulation [18].
CMA is not only related to specific immunity, but also to non-specific immunity (innate immunity). CMA degradation of NLRP3 is associated with the palmitoylation of NLRP3 [60]. In fact, NLRP3 is a signaling pathway receptor that regulates the innate immune response by affecting the formation of inflammasomes, which eventually leads to pyroptosis [118]. STING degradation via CMA has also been implicated in the regulation of innate immunity, and its role in the cell is to respond to nucleic acids present in the cytosol [64]. In addition, TANK binding kinase-1(TBK-1), associated with IFN-1 production during cell infection [119], was identified as a substrate of CMA [120]. Inhibition of CMA decreased the degradation of TBK-1 and increased the production of IFN-1, thus improving the antiviral ability of cells [121].
Metabolism
CMA usually affects the metabolic processes by degrading important enzymes in metabolic pathways. As for glucose metabolism, the degradation of HK2 and PKM2, two essential enzymes in glucose metabolism, was mediated by CMA [31, 98]. Besides, the capacity of gluconeogenesis and glycogen storage is weakened by CMA inhibition, while cell glycolysis is also enhanced in LAMP2A-knockout mouse hepatocytes [17]. The inhibition of CMA causes the accumulation of CMA substrates responsible for gluconeogenesis including GADPH and malate dehydrogenase, which promotes the glycolysis process [17].
In addition to glucose metabolism, CMA is also associated with lipid metabolism [17]. Several proteins involved in lipid metabolism, including lipid droplet coat proteins, lipogenic enzymes, lipid carriers, have been identified as CMA substrates [17, 122]. CMA promotes lipolysis by degrading lipid droplet coat proteins such as perilipins 2 and 3(PLIN2 and PLIN3) [122]. In addition, aberrant lipogenesis and lipolysis processes are found to cause hepatosteatosis in LAMP2A-knockout mouse hepatocytes [17]. Furthermore, CMA can also regulate preadipocyte differentiation by degrading MYC and transforming growth factor-β (TGFβ) during adipogenesis [123].
OCT4 and SOX2, as mentioned above, can regulate CMA by affecting LAMP2 expression. In fact, CMA also has a regulatory effect on the differentiation of embryonic stem cells by regulating the glucose metabolism of embryonic stem cells [85, 124].
Aging
Aging-dependent decrease in CMA activity has been found in a wide variety of cells [19, 125, 126]. LAMP2A is a key rate-limiting protein during CMA, which is also a major factor in the aging-dependent reduction of CMA [127]. Alternations in the lipid composition of the lysosomal membrane are found to contribute to the changes in LAMP2A stability during aging [128]. In mouse models of hepatocytes and T cells, macroautophagy was found to compensate for a portion of reduced function of CMA [17, 18]. However, studies in mouse retinal cells found that only CMA could compensate for the loss of macroautophagy, while macroautophagy could not in turn compensate for the deficiency of CMA [126]. Therefore, in general, the loss of CMA function accompanied by aging is irreversible and irreplaceable. Genetic intervention targeting LAMP2A can restore the aging-induced functional deficits. Exogenously induced expression of LAMP2A has been shown to enhance the ability of the aging liver to resist adverse factors, and contribute to the overall improvement of an individual's ability to maintain protein homeostasis [125].
CMA is also involved in the maintenance of bone marrow hematopoietic function during aging. Bone marrow hematopoietic function decreases with aging due to the decline in the number of hematopoietic stem cells and the reduction in the ability of cell renewal. The decrease of CMA also affects the hematopoietic function of bone marrow by causing the glucose metabolism disorder of hematopoietic stem cells. Reactivation of CMA in senescent cells can partially restore the hematopoietic function of bone marrow [129].
Therapeutic potential of chaperone-mediated autophagy
Therapeutic potential of CMA on cancers
Colorectal cancer
It has been suggested that CMA can promote the proliferation, metastasis, and chemotherapy resistance of colorectal cancer (CRC) cells [21]. Targeting and regulation of CMA might contribute to the treatment of CRC. Increased LAMP2A expression was found in colon cancer tissues [92]. CMA promotes the metastasis and proliferation of colorectal cancer cells during oxaliplatin resistance and under oxidative stress conditions, which is associated with the process of cancer cell glycolysis [92]. The Hsc70/caveolin-1 [CAV1]/β-catenin axis was found to participate in the progression of colorectal cancer. Hsc70 promotes the degradation of CAV1 through CMA, which subsequently activates the Wnt/β-catenin pathway and promotes the metastasis of BRAF V600E CRC [130]. Fucosyltransferase 8 (FUT8) is highly expressed in cancer cells, which is responsible for mediating the core fucosylation of the key immune checkpoint molecule, CD276 (B7-H3). FDW028, a specific FUT8 inhibitor, shows potent anticancer activity by promoting degradation of B7-H3 through CMA [131].
Breast cancer
The inhibition of CMA is a potential target for the improvement of drug resistance of breast cancer. LAMP2A expression was found to increase in breast cancer tissues. Inhibition of LAMP2A induces apoptosis and increases the sensitivity of breast cancer cells to the drug [132]. Hepatitis B X-interacting protein (HBXIP) is an oncoprotein that promotes TAM resistance of cancer cells via the inhibition of the degradation of HOXB13 by CMA. Aspirin can overcome tamoxifen (TAM) resistance in estrogen receptor-positive breast cancer through decreasing the overexpression of HBXIP [50]. 17β-estradiol (E2), TAM, and fulvestrant (FUL) stabilized MORC2 through the inhibition of CMA. The knockdown of MORC2 prevented cell proliferation induced by E2 and allowed cancer cells to regain sensitivity to TAM and FUL, thus becoming a potential target of treatment [37].
Lung cancer
Knockdown of LAMP2A inhibited tumor progression and enhanced the sensitivity of NSCLC cells to cisplatin, suggesting that inhibition of CMA is a potential approach for the treatment of NSCLC [91]. Besides, polyphyllin D suppressed the interaction of Hsc70 with LAMP2A and thus disrupted CMA, contributing to the treatment of NSCLC [133]. PI3K/mTOR inhibitors enhanced the interaction of G6PD protein (the rate-limiting enzyme of the pentose phosphate pathway) with Hsc70, thereby enhancing its degradation through CMA, leading to an exacerbation of oxidative stress damage. Therefore, PI3K/mTOR inhibitors may serve as a therapeutic approach to attenuate radioresistance in SCLC [134].
Other cancers
CMA and programmed cell death-ligand 1 (PD-L1) upregulation were found to occur simultaneously in metastatic melanoma. Self-assembling prionoid (SAP) was a prion-like chemical inducer of proximity that was designed artificially to induce PD-L1 close to Hsc70 via decomposing into phytohaemagglitinin after infiltrating tumor cells, thus enhancing PD-L1 degradation through CMA [135]. For papillary thyroid carcinoma (PTC), CMA regulated by the estrogen receptor can increase the expression of stromal cell-derived factor 1 and C-X-C motif chemokine receptor 4 by reducing the expression of peroxisome proliferator-activated receptor γ, thus promoting the proliferation and metastasis of PTC [136]. In the treatment of multiple myeloma, upregulation of CMA contributed to tumor resistance to bortezomib and might serve as a target to overcome resistance [137]. Tumor protein D52 (TPD52) mediated the activation of CMA and promoted the proliferation and metastasis of prostatic cancer (PCa). Romidepsin, an inhibitor of HDAC2, increases TPD52 acetylation, which attenuates TPD52-mediated CMA activation and prevents the growth of PCa [138]. In non-acute myeloid leukemia cells, inhibition of Fms-like tyrosine kinase 3 led to a sensitivity of cancer cells to autophagy inhibition and an excessive activation of CMA. The activation of CMA increased the degradation of HK2, which triggered the metabolic disorder and impaired proliferation of cancer cells [98].
Therapeutic potential of CMA on neurodegenerative diseases
Parkinson's disease
Parkinson's disease is a neurodegenerative disorder with an age-related onset and is characterized by the accumulation of α-syn in the brain. In general, CMA activation can increase the degradation of α-syn, thereby alleviating the symptoms of PD [22]. AR7 [7-chloro-3-(4-methylphenyl)-2H-1,4-benzoxazine], an atypical RARA/RARα (retinoic acid receptor α) antagonist [139], can activate LAMP2A transcription and lysosomal activity, thereby inhibiting the accumulation of α-syn oligomers [23]. Besides, β-asarone has a therapeutic effect in a rat model of PD induced by 6-hydroxydopamine, preventing brain damage by increasing MEF2D and tyrosine hydroxylase (TH), and decreasing α-syn through a Hsc70/MAPK/MEF2D/beclin-1 pathway [140]. Tubastatin A attenuated the toxicity of α-syn by upregulating HSC70 and LAMP2A [141]. In addition to the clearance of α-syn, inhibition of the p38-transcription factor EB pathway can promote the degradation of NLRP3 through CMA, thereby suppressing microglia activation, which is also a potential therapeutic approach of PD [142].
Alzheimer's disease
The treatment strategy for Alzheimer's disease (AD) is also the activation of CMA, which leads to the degradation of abnormally aggregated proteins to alleviate the disease. Metformin was reported to activate CMA and significantly reduce the level of Aβ plaque in the brains of mice, thus alleviating the phenotype and symptoms of AD [13]. Labeling multiple CMA motifs on pathological plaques can promote their recognition by HSC70 so as to protect nerve cells [143]. Lactulose and trehalose were found to reduce neuroinflammation by anti-inflammatory and CMA activation. Moreover, lactulose better promoted synaptic expression in mouse models than trehalose and thus could be used as a potential therapeutic agent for AD [144].
Therapeutic potential of CMA on hepatic and renal diseases
Aiming at the hepatotoxicity caused by lipid accumulation, the combination of N-acetylcysteine (NAC) and baicalin has been reported to stabilize the function of mitochondria in hepatocytes, thereby inhibiting mitochondrial reactive oxygen species-mediated CMA by transcriptional factor A-choline, promoting lipid phagocytosis and alleviating liver toxicity [25]. CMA activators improved proximal tubular function in patients with nephrotic cystinosis. The reason is that CMA activation improved the expression of the endocytic receptor megalin on the cell membrane in proximal tubule cells, thereby restoring its normal function [26].
Therapeutic potential of CMA on diabetic complications
Glucose toxicity caused by hyperglycemia is the main cause of diabetes-related complications. The role of lmp-2 (LAMP2A homolog) was studied in Caenorhabditis elegans. The inhibition of lmp-2 can prevent decreased proteasome activity caused by glucose, thereby preventing glucose toxicity [145]. In diabetic retinopathy (DR), CMA activator QX77 (a derivative of AR7, an RARα antagonist) was found to prevent early DR by promoting the degradation of ACSL4 protein through CMA and preventing the abnormal accumulation that led to the generation of harmful lipid substances [27].
Therapeutic potential of CMA on microbial infectious diseases
Infection with Salmonella enterica serovar Typhimurium (S. Typhimurium) leads to the death of macrophages. This process is resulted by impairing the degradation of tripartite motif 21 (TRIM21) through CMA by this bacterium and elevated TRIM21 levels eventually led to the death of macrophages. This result links CMA to bacterial infections, suggesting that CMA modulation of bacterial infectious diseases may become a novel therapeutic approach [146]. For Group B Coxsackievirus (CVB) infections, anisomycin was reported to inhibit the replication of CVB by promoting the degradation of eukaryotic translation elongation factor 1 alpha 1 (eEF1A1) through CMA, thus becoming a candidate for the treatment of CVB infection [147].
Therapeutic potential of CMA on other diseases
CMA is closely implicated in the immunosuppression of mesenchymal stromal cells induced by inflammatory cytokines, that is, the inhibition of CMA is a key factor in immunosuppression [148]. Besides, pinacidil attenuates myocardial ischemia-reperfusion injury by inhibiting the degradation of calreticulin through CMA [149]. For status epilepticus, CMA activity was reported to be upregulated in a rat model and antioxidants such as vitamin E would partially inhibit CMA, thus potentially relieving status epilepticus [150].
Therapeutic molecules targeting chaperone-mediated autophagy.
| Therapeutic molecule(s) | Targets | Target type | Effect on CMA | Effects on diseases |
|---|---|---|---|---|
| FDW028 | FUT8 | upstream molecule that regulates CMA | enhanced | Inhibition of metastatic CRC |
| Aspirin | HBXIP | upstream molecule that regulates CMA | enhanced | Reducing TAM resistance of breast cancer |
| Polyphyllin D | Hsc70 and LAMP2A | components of CMA | inhibited | Enhancing the sensitivity of NSCLC to cisplatin |
| PI3K/mTOR inhibitors | G6PD and Hsc70 | substrate and component of CMA | enhanced | Attenuating radioresistance of SCLC |
| SAP | PD-L1 and Hsc70 | substrate and component of CMA | enhanced | Restoring the antitumor immune response in metastatic melanoma |
| Romidepsin | HDAC2 and TPD52 | upstream molecules that regulate CMA | inhibited | Preventing the tumor growth of PCa |
| AR7 | LAMP2A | component of CMA | enhanced | Alleviating the symptoms of PD |
| β-asarone | MEF2D and TH | upstream molecules that regulate CMA | enhanced | Preventing brain damage in PD |
| Lactulose | N.A. | N.A. | enhanced | Reducing neuroinflammation and promoting the synaptic expression in AD |
| Trehalose | N.A. | N.A. | enhanced | Reducing neuroinflammation and promoting the synaptic expression in AD |
| NAC | N.A. | N.A. | enhanced | Alleviating NSAID-induced steatosis and hepatotoxicity |
| Baicalin | N.A. | N.A. | enhanced | Alleviating NSAID-induced steatosis and hepatotoxicity |
| QX77 | ACSL4 | substrate | enhanced | Preventing the generation of harmful lipid substances in diabetic retinopathy |
| Anisomycin | eEF1A1 | substrate | enhanced | Inhibiting the replication of CVB in CVB infection |
| Pinacidil | CRT | substrate | inhibited | Attenuating myocardial I-R injury |
| Vitamin E | N.A. | N.A. | inhibited | Relieving status epilepticus |
The effects of CMA on cellular changes. The cellular changes generated by CMA have six aspects, including cell proliferation, cell death, cell cycle, immunity, metabolism and aging. Up-regulation or down-regulation of CMA can accelerate or slow cell proliferation and death. The relationship between CMA and the cell cycle is mainly due to two CMA substrates that regulate the cell cycle, HIF1A and CHK1. CMA regulates the activation of CD4+ T cells and is associated with several processes of innate immunity. In terms of metabolism, CMA interferes with glucose and lipid metabolism and regulates the pluripotency of embryonic stem cells. During aging, a compensatory reaction between CMA and macroautophagy exists to a certain extent, and CMA is also related to the maintenance of bone marrow hematopoietic function during senescence process.
Future perspectives
Recent studies revealed various regulatory mechanisms of CMA as well as the effects of CMA on cellular changes. This review summarized research advances on the regulatory mechanisms of CMA from the perspectives of substrates, chaperone molecule and receptor, and discussed the research and development frontiers of effects on cellular change and therapeutic potential of CMA.
CMA substrates are mainly regulated by PTMs, including phosphorylation, acetylation, ubiquitination and palmitoylation. CMA substrates are characterized by a KFERQ-like motif, which allows the protein to be recognized by Hsc70 for CMA processing. Certain PTMs of the motif can modify the remaining residues to possess properties similar to those in KFERQ, and thus can also be recognized by chaperone molecules. In addition, the modification of residues around the motifs by PTMs can also cause the motifs to be buried or exposed, thereby affecting the recognition of substrates by chaperones. However, little is known on the specific mechanisms of how these PTMs affect the recognition and degradation of CMA component molecules toward substrates. At present, PTMs of Hsc70 phosphorylation, methylation and S-nitrosylation have been studied. Other types of PTMs of Hsc70 might also contribute to the regulation of CMA process. Most researches focused on the regulation of nucleic acids such as miRNA on LAMP2A rather than investigation of PTMs of LAMP2A.
The effects of CMA on cellular changes mainly focused on cell proliferation, cell death, cell cycle, immunity, metabolism, and aging. Other cellular processes such as cell differentiation and cell migration require future investigations. Although most research found that CMA promoted cancer cells proliferation, the relationship of CMA with cell proliferation still need further confirmation as some study came up with contradictory results. Studies on different types of cell death other than ferroptosis might find novel involvement of CMA in cell death regulation. CMA has been reported to maintain normal cell cycle by degradation of relevant proteins. As for immune regulation, CMA could activate CD4+ T cells for specific immune reactions as well as innate immune reactions. The glucose and lipid metabolism processes of cells are related to CMA, but the relationship between other metabolic processes such as nucleic acid or amino acid metabolism and CMA still needs further research. During the aging process, the weakening of CMA activity is the main manifestation, and the activation of CMA enhances the ability of aging cells to maintain protein homeostasis.
Investigations on therapeutic potential of CMA mainly focused on malignant tumors, neurodegenerative diseases, liver and kidney diseases. The relationship between CMA and colorectal cancer, breast cancer, lung cancer has been widely studied. CMA can promote the occurrence of cancer, and inhibiting CMA can benefit cancer treatment. In the research of breast cancer, it is only clear that CMA promotes drug resistance of cancer cells, but whether CMA is related to the occurrence of breast cancer needs further research. In addition, whether CMA is related to gastric cancer, pancreatic cancer, liver cancer and other cancers remains to be studied. The neurodegenerative diseases related to CMA mainly include PD and AD. Activation of CMA promotes the degradation of abnormal proteins in neurons, which benefits the treatment of these diseases. Further research is needed to determine whether CMA activation is beneficial for other neurodegenerative diseases such as Huntington's disease and amyotrophic lateral sclerosis. In the complications of diabetes, CMA activation alleviates early diabetes retinopathy, but the relationship between CMA and other complications such as diabetes nephropathy, atherosclerosis remains largely unknown. The advantage of CMA-targeted drugs mainly includes the accurate degradation/stabilization of certain protein for treatment of diseases. While the limitation of CMA-targeted drugs is the possible adverse effect on other systems caused by the significant change of CMA substrate. The diseases such as neurodegenerative diseases and aging might become potential therapeutic objects of CMA-targeted drugs in the future because of the identified relationship and mechanism.
Overall, CMA is regulated at multiple levels of nucleic acid and post-translational modifications which contribute to various cellular processes. Increasing progresses have been made on therapeutic potential of CMA, which may become promising approaches for the effective treatment of multiple diseases.
Funding
This work was supported by the key project of the National Natural Science Foundation (82030091), the key project of LiaoNing Science Foundation (2022JH6/100100037, 2022JH2/20200034, 2021JH2/10300023), and the National Natural Science Foundation (82102740).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 2010;221:3-12
2. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728-41
3. Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19:365-81
4. Bejarano E, Cuervo AM. Chaperone-mediated autophagy. Proc Am Thorac Soc. 2010;7:29-39
5. Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382-5
6. Agarraberes FA, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci. 2001;114:2491-9
7. Assaye MA, Gizaw ST. Chaperone-Mediated Autophagy and Its Implications for Neurodegeneration and Cancer. Int J Gen Med. 2022;15:5635-49
8. Chen Y, Hao Q, Wang S, Cao M, Huang Y, Weng X. et al. Inactivation of the tumor suppressor p53 by long noncoding RNA RMRP. Proc Natl Acad Sci U S A. 2021;118:e2026813118
9. Troeger A, Chae HD, Senturk M, Wood J, Williams DA. A unique carboxyl-terminal insert domain in the hematopoietic-specific, GTPase-deficient Rho GTPase RhoH regulates post-translational processing. J Biol Chem. 2013;288:36451-62
10. Huang KY, Su MG, Kao HJ, Hsieh YC, Jhong JH, Cheng KH. et al. dbPTM 2016: 10-year anniversary of a resource for post-translational modification of proteins. Nucleic Acids Res. 2016;44:D435-46
11. Lee JM, Hammarén HM, Savitski MM, Baek SH. Control of protein stability by post-translational modifications. Nat Commun. 2023;14:201
12. Xu CY, Kang WY, Chen YM, Jiang TF, Zhang J, Zhang LN. et al. DJ-1 Inhibits α-Synuclein Aggregation by Regulating Chaperone-Mediated Autophagy. Front Aging Neurosci. 2017;9:308
13. Xu X, Sun Y, Cen X, Shan B, Zhao Q, Xie T. et al. Metformin activates chaperone-mediated autophagy and improves disease pathologies in an Alzheimer disease mouse model. Protein Cell. 2021;12:769-87
14. Li W, Zhu J, Dou J, She H, Tao K, Xu H. et al. Phosphorylation of LAMP2A by p38 MAPK couples ER stress to chaperone-mediated autophagy. Nat Commun. 2017;8:1763
15. Cao D, Shan D, Yan W, Zhang Z, Song Q, Jiang Y. et al. Chaperone-mediated autophagy affects tumor cell proliferation and cisplatin resistance in esophageal squamous cell carcinoma. Thorac Cancer. 2021;12:1048-57
16. Hubbi ME, Hu H, Kshitiz, Ahmed I, Levchenko A, Semenza GL. Chaperone-mediated autophagy targets hypoxia-inducible factor-1α (HIF-1α) for lysosomal degradation. J Biol Chem. 2013;288:10703-14
17. Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014;20:417-32
18. Valdor R, Mocholi E, Botbol Y, Guerrero-Ros I, Chandra D, Koga H. et al. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol. 2014;15:1046-54
19. Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505-13
20. Yu S, Li Z, Zhang Q, Wang R, Zhao Z, Ding W. et al. GPX4 degradation via chaperone-mediated autophagy contributes to antimony-triggered neuronal ferroptosis. Ecotoxicol Environ Saf. 2022;234:113413
21. Xuan Y, Zhao S, Xiao X, Xiang L, Zheng HC. Inhibition of chaperone-mediated autophagy reduces tumor growth and metastasis and promotes drug sensitivity in colorectal cancer. Mol Med Rep. 2021;23:360
22. Xilouri M, Brekk OR, Landeck N, Pitychoutis PM, Papasilekas T, Papadopoulou-Daifoti Z. et al. Boosting chaperone-mediated autophagy in vivo mitigates α-synuclein-induced neurodegeneration. Brain. 2013;136:2130-46
23. Ho PW, Leung CT, Liu H, Pang SY, Lam CS, Xian J. et al. Age-dependent accumulation of oligomeric SNCA/α-synuclein from impaired degradation in mutant LRRK2 knockin mouse model of Parkinson disease: role for therapeutic activation of chaperone-mediated autophagy (CMA). Autophagy. 2020;16:347-70
24. Lee W, Kim HY, Choi YJ, Jung SH, Nam YA, Zhang Y. et al. SNX10-mediated degradation of LAMP2A by NSAIDs inhibits chaperone-mediated autophagy and induces hepatic lipid accumulation. Theranostics. 2022;12:2351-69
25. Sun J, Chen Y, Wang T, Ali W, Ma Y, Liu Z. et al. Role of Mitochondrial Reactive Oxygen Species-Mediated Chaperone-Mediated Autophagy and Lipophagy in Baicalin and N-Acetylcysteine Mitigation of Cadmium-Induced Lipid Accumulation in Liver. Antioxidants (Basel). 2024;13:115
26. Zhang J, He J, Johnson JL, Rahman F, Gavathiotis E, Cuervo AM. et al. Chaperone-Mediated Autophagy Upregulation Rescues Megalin Expression and Localization in Cystinotic Proximal Tubule Cells. Front Endocrinol (Lausanne). 2019;10:21
27. Liu C, Sun W, Zhu T, Shi S, Zhang J, Wang J. et al. Glia maturation factor-β induces ferroptosis by impairing chaperone-mediated autophagic degradation of ACSL4 in early diabetic retinopathy. Redox Biol. 2022;52:102292
28. Dice JF. Altered degradation of proteins microinjected into senescent human fibroblasts. J Biol Chem. 1982;257:14624-7
29. Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem Sci. 1990;15:305-9
30. Kirchner P, Bourdenx M, Madrigal-Matute J, Tiano S, Diaz A, Bartholdy BA. et al. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol. 2019;17:e3000301
31. Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H. et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719-30
32. Bonhoure A, Vallentin A, Martin M, Senff-Ribeiro A, Amson R, Telerman A. et al. Acetylation of translationally controlled tumor protein promotes its degradation through chaperone-mediated autophagy. Eur J Cell Biol. 2017;96:83-98
33. Li R, Sun N, Chen X, Li X, Zhao J, Cheng W. et al. JAK2(V617F) Mutation Promoted IL-6 Production and Glycolysis via Mediating PKM1 Stabilization in Macrophages. Front Immunol. 2020;11:589048
34. Liu X, Zhang Y, Wang Y, Yang M, Hong F, Yang S. Protein Phosphorylation in Cancer: Role of Nitric Oxide Signaling Pathway. Biomolecules. 2021;11:1009
35. Wang C, Wang H, Zhang D, Luo W, Liu R, Xu D. et al. Phosphorylation of ULK1 affects autophagosome fusion and links chaperone-mediated autophagy to macroautophagy. Nat Commun. 2018;9:3492
36. Hu SY, Qian JX, Yang SY, Andriani L, Liao L, Deng L. et al. Destabilization of microrchidia family CW-type zinc finger 2 via the cyclin-dependent kinase 1-chaperone-mediated autophagy pathway promotes mitotic arrest and enhances cancer cellular sensitivity to microtubule-targeting agents. Clin Transl Med. 2023;13:e1210
37. Yang F, Xie HY, Yang LF, Zhang L, Zhang FL, Liu HY. et al. Stabilization of MORC2 by estrogen and antiestrogens through GPER1- PRKACA-CMA pathway contributes to estrogen-induced proliferation and endocrine resistance of breast cancer cells. Autophagy. 2020;16:1061-76
38. Xie W, Zhang L, Jiao H, Guan L, Zha J, Li X. et al. Chaperone-mediated autophagy prevents apoptosis by degrading BBC3/PUMA. Autophagy. 2015;11:1623-35
39. Martinez-Vicente M, Talloczy Z, Kaushik S, Massey AC, Mazzulli J, Mosharov EV. et al. Dopamine-modified alpha-synuclein blocks chaperone-mediated autophagy. J Clin Invest. 2008;118:777-88
40. Quintavalle C, Di Costanzo S, Zanca C, Tasset I, Fraldi A, Incoronato M. et al. Phosphorylation-regulated degradation of the tumor-suppressor form of PED by chaperone-mediated autophagy in lung cancer cells. J Cell Physiol. 2014;229:1359-68
41. Yang T, Ren C, Qiao P, Han X, Wang L, Lv S. et al. PIM2-mediated phosphorylation of hexokinase 2 is critical for tumor growth and paclitaxel resistance in breast cancer. Oncogene. 2018;37:5997-6009
42. Dang F, Wei W. Targeting the acetylation signaling pathway in cancer therapy. Semin Cancer Biol. 2022;85:209-18
43. Huang H, Liu R, Huang Y, Feng Y, Fu Y, Chen L. et al. Acetylation-mediated degradation of HSD17B4 regulates the progression of prostate cancer. Aging (Albany NY). 2020;12:14699-717
44. Zhang Y, Xu YY, Yao CB, Li JT, Zhao XN, Yang HB. et al. Acetylation targets HSD17B4 for degradation via the CMA pathway in response to estrone. Autophagy. 2017;13:538-53
45. Miller WP, Sha CM, Sunilkumar S, Toro AL, VanCleave AM, Kimball SR. et al. Activation of Disulfide Redox Switch in REDD1 Promotes Oxidative Stress Under Hyperglycemic Conditions. Diabetes. 2022;71:2764-76
46. Revesz T, Holton JL. Anatamopathological spectrum of tauopathies. Mov Disord. 2003;18(Suppl 6):S13-20
47. Alquezar C, Schoch KM, Geier EG, Ramos EM, Scrivo A, Li KH. et al. TSC1 loss increases risk for tauopathy by inducing tau acetylation and preventing tau clearance via chaperone-mediated autophagy. Sci Adv. 2021;7:eabg3897
48. Caballero B, Bourdenx M, Luengo E, Diaz A, Sohn PD, Chen X. et al. Acetylated tau inhibits chaperone-mediated autophagy and promotes tau pathology propagation in mice. Nat Commun. 2021;12:2238
49. Li Y, Zhang Z, Zhou X, Li L, Liu Q, Wang Z. et al. The oncoprotein HBXIP enhances migration of breast cancer cells through increasing filopodia formation involving MEKK2/ERK1/2/Capn4 signaling. Cancer Lett. 2014;355:288-96
50. Liu B, Wang T, Wang H, Zhang L, Xu F, Fang R. et al. Oncoprotein HBXIP enhances HOXB13 acetylation and co-activates HOXB13 to confer tamoxifen resistance in breast cancer. J Hematol Oncol. 2018;11:26
51. Zhang T, Liu J, Shen S, Tong Q, Ma X, Lin L. SIRT3 promotes lipophagy and chaperon-mediated autophagy to protect hepatocytes against lipotoxicity. Cell Death Differ. 2020;27:329-44
52. Su M, Guan H, Zhang F, Gao Y, Teng X, Yang W. HDAC6 Regulates the Chaperone-Mediated Autophagy to Prevent Oxidative Damage in Injured Neurons after Experimental Spinal Cord Injury. Oxid Med Cell Longev. 2016;2016:7263736
53. Obayashi H, Nagano Y, Takahashi T, Seki T, Tanaka S, Sakai N. et al. Histone deacetylase 10 knockout activates chaperone-mediated autophagy and accelerates the decomposition of its substrate. Biochem Biophys Res Commun. 2020;523:246-52
54. Cruz Walma DA, Chen Z, Bullock AN, Yamada KM. Ubiquitin ligases: guardians of mammalian development. Nat Rev Mol Cell Biol. 2022;23:350-67
55. Ciechanover A. The ubiquitin-proteasome pathway: on protein death and cell life. Embo j. 1998;17:7151-60
56. Zhong T, Lei K, Lin X, Xie Z, Luo S, Zhou Z. et al. Protein ubiquitination in T cell development. Front Immunol. 2022;13:941962
57. Ferreira JV, Soares AR, Ramalho JS, Pereira P, Girao H. K63 linked ubiquitin chain formation is a signal for HIF1A degradation by Chaperone-Mediated Autophagy. Sci Rep. 2015;5:10210
58. Ko PJ, Dixon SJ. Protein palmitoylation and cancer. EMBO Rep. 2018;19:e46666
59. Zhou B, Hao Q, Liang Y, Kong E. Protein palmitoylation in cancer: molecular functions and therapeutic potential. Mol Oncol. 2023;17:3-26
60. Wang L, Cai J, Zhao X, Ma L, Zeng P, Zhou L. et al. Palmitoylation prevents sustained inflammation by limiting NLRP3 inflammasome activation through chaperone-mediated autophagy. Mol Cell. 2023;83:281-97.e10
61. Talamillo A, Barroso-Gomila O, Giordano I, Ajuria L, Grillo M, Mayor U. et al. The role of SUMOylation during development. Biochem Soc Trans. 2020;48:463-78
62. Sheng Z, Zhu J, Deng YN, Gao S, Liang S. SUMOylation modification-mediated cell death. Open Biol. 2021;11:210050
63. Han ZJ, Feng YH, Gu BH, Li YM, Chen H. The post-translational modification, SUMOylation, and cancer (Review). Int J Oncol. 2018;52:1081-94
64. Hu MM, Yang Q, Xie XQ, Liao CY, Lin H, Liu TT. et al. Sumoylation Promotes the Stability of the DNA Sensor cGAS and the Adaptor STING to Regulate the Kinetics of Response to DNA Virus. Immunity. 2016;45:555-69
65. Eichler J. Protein glycosylation. Curr Biol. 2019;29:R229-r31
66. Shi Y, Yan S, Shao GC, Wang J, Jian YP, Liu B. et al. O-GlcNAcylation stabilizes the autophagy-initiating kinase ULK1 by inhibiting chaperone-mediated autophagy upon HPV infection. J Biol Chem. 2022;298:102341
67. Ferreira JV, Fôfo H, Bejarano E, Bento CF, Ramalho JS, Girão H. et al. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy. 2013;9:1349-66
68. Bandyopadhyay U, Kaushik S, Varticovski L, Cuervo AM. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008;28:5747-63
69. Cuervo AM, Dice JF, Knecht E. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997;272:5606-15
70. Macri C, Wang F, Tasset I, Schall N, Page N, Briand JP. et al. Modulation of deregulated chaperone-mediated autophagy by a phosphopeptide. Autophagy. 2015;11:472-86
71. Zhang Z, Zhang X, Wu X, Zhang Y, Lu J, Li D. Sirt1 attenuates astrocyte activation via modulating Dnajb1 and chaperone-mediated autophagy after closed head injury. Cereb Cortex. 2022;32:5191-205
72. Maan M, Pati U. CHIP promotes autophagy-mediated degradation of aggregating mutant p53 in hypoxic conditions. Febs j. 2018;285:3197-214
73. Cheng X, Roberts RJ. AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res. 2001;29:3784-95
74. Malbeteau L, Pham HT, Eve L, Stallcup MR, Poulard C, Le Romancer M. How Protein Methylation Regulates Steroid Receptor Function. Endocr Rev. 2022;43:160-97
75. Wang C, Arrington J, Ratliff AC, Chen J, Horton HE, Nie Y. et al. Methyltransferase-like 21c methylates and stabilizes the heat shock protein Hspa8 in type I myofibers in mice. J Biol Chem. 2019;294:13718-28
76. Sharma V, Fernando V, Letson J, Walia Y, Zheng X, Fackelman D. et al. S-Nitrosylation in Tumor Microenvironment. Int J Mol Sci. 2021;22:4600
77. Guil-Luna S, Sanchez-Montero MT, Rodríguez-Ariza A. S-Nitrosylation at the intersection of metabolism and autophagy: Implications for cancer. Biochim Biophys Acta Rev Cancer. 2023;1878:189012
78. Valek L, Heidler J, Scheving R, Wittig I, Tegeder I. Nitric oxide contributes to protein homeostasis by S-nitrosylations of the chaperone HSPA8 and the ubiquitin ligase UBE2D. Redox Biol. 2019;20:217-35
79. Eskelinen EL, Cuervo AM, Taylor MR, Nishino I, Blum JS, Dice JF. et al. Unifying nomenclature for the isoforms of the lysosomal membrane protein LAMP-2. Traffic. 2005;6:1058-61
80. Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501-3
81. Cuervo AM, Dice JF. Unique properties of lamp2a compared to other lamp2 isoforms. J Cell Sci. 2000 113 Pt 24: 4441-50
82. Bandyopadhyay U, Sridhar S, Kaushik S, Kiffin R, Cuervo AM. Identification of regulators of chaperone-mediated autophagy. Mol Cell. 2010;39:535-47
83. Tang FL, Erion JR, Tian Y, Liu W, Yin DM, Ye J. et al. VPS35 in Dopamine Neurons Is Required for Endosome-to-Golgi Retrieval of Lamp2a, a Receptor of Chaperone-Mediated Autophagy That Is Critical for α-Synuclein Degradation and Prevention of Pathogenesis of Parkinson's Disease. J Neurosci. 2015;35:10613-28
84. Pajares M, Rojo AI, Arias E, Díaz-Carretero A, Cuervo AM, Cuadrado A. Transcription factor NFE2L2/NRF2 modulates chaperone-mediated autophagy through the regulation of LAMP2A. Autophagy. 2018;14:1310-22
85. Xu Y, Zhang Y, García-Cañaveras JC, Guo L, Kan M, Yu S. et al. Chaperone-mediated autophagy regulates the pluripotency of embryonic stem cells. Science. 2020;369:397-403
86. Li G, Yang H, Zhu D, Huang H, Liu G, Lun P. Targeted suppression of chaperone-mediated autophagy by miR-320a promotes α-synuclein aggregation. Int J Mol Sci. 2014;15:15845-57
87. Alvarez-Erviti L, Seow Y, Schapira AH, Rodriguez-Oroz MC, Obeso JA, Cooper JM. Influence of microRNA deregulation on chaperone-mediated autophagy and α-synuclein pathology in Parkinson's disease. Cell Death Dis. 2013;4:e545
88. Izco M, Vettorazzi A, Forcen R, Blesa J, de Toro M, Alvarez-Herrera N. et al. Oral subchronic exposure to the mycotoxin ochratoxin A induces key pathological features of Parkinson's disease in mice six months after the end of the treatment. Food Chem Toxicol. 2021;152:112164
89. Verma R, Saha S, Kumar S, Mani S, Maiti TK, Surjit M. RNA-Protein Interaction Analysis of SARS-CoV-2 5' and 3' Untranslated Regions Reveals a Role of Lysosome-Associated Membrane Protein-2a during Viral Infection. mSystems. 2021;6:e0064321
90. Arias E, Koga H, Diaz A, Mocholi E, Patel B, Cuervo AM. Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Mol Cell. 2015;59:270-84
91. Ichikawa A, Fujita Y, Hosaka Y, Kadota T, Ito A, Yagishita S. et al. Chaperone-mediated autophagy receptor modulates tumor growth and chemoresistance in non-small cell lung cancer. Cancer Sci. 2020;111:4154-65
92. Chen R, Zhang Y, Ge Y, He C, Wu Z, Wang J. et al. LAMP2A overexpression in colorectal cancer promotes cell growth and glycolysis via chaperone-mediated autophagy. Oncol Lett. 2024;27:33
93. Liu H, Yong Y, Li X, Ye P, Tao K, Peng G. et al. Chaperone-mediated Autophagy Regulates Cell Growth by Targeting SMAD3 in Glioma. Neurosci Bull. 2022;38:637-51
94. Zhou J, Yang J, Fan X, Hu S, Zhou F, Dong J. et al. Chaperone-mediated autophagy regulates proliferation by targeting RND3 in gastric cancer. Autophagy. 2016;12:515-28
95. Yadav VK, Awasthi P, Behl R, Kumar A. HSc70 interactome reveal major role of macroautophagy and minor role of chaperone mediated autophagy in K-Ras G12V cell proliferation and survival. J Proteomics. 2022;264:104614
96. Desideri E, Castelli S, Dorard C, Toifl S, Grazi GL, Ciriolo MR. et al. Impaired degradation of YAP1 and IL6ST by chaperone-mediated autophagy promotes proliferation and migration of normal and hepatocellular carcinoma cells. Autophagy. 2023;19:152-62
97. Newton K, Strasser A, Kayagaki N, Dixit VM. Cell death. Cell. 2024;187:235-56
98. Xia HG, Najafov A, Geng J, Galan-Acosta L, Han X, Guo Y. et al. Degradation of HK2 by chaperone-mediated autophagy promotes metabolic catastrophe and cell death. J Cell Biol. 2015;210:705-16
99. Ghosh R, Gillaspie JJ, Campbell KS, Symons JD, Boudina S, Pattison JS. Chaperone-mediated autophagy protects cardiomyocytes against hypoxic-cell death. Am J Physiol Cell Physiol. 2022;323:C1555-c75
100. Wang Y, Singh R, Xiang Y, Czaja MJ. Macroautophagy and chaperone-mediated autophagy are required for hepatocyte resistance to oxidant stress. Hepatology. 2010;52:266-77
101. Chen X, Li J, Kang R, Klionsky DJ, Tang D. Ferroptosis: machinery and regulation. Autophagy. 2021;17:2054-81
102. Chen C, Wang D, Yu Y, Zhao T, Min N, Wu Y. et al. Legumain promotes tubular ferroptosis by facilitating chaperone-mediated autophagy of GPX4 in AKI. Cell Death Dis. 2021;12:65
103. Wu Z, Geng Y, Lu X, Shi Y, Wu G, Zhang M. et al. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc Natl Acad Sci U S A. 2019;116:2996-3005
104. Hirata Y, Yamada Y, Taguchi S, Kojima R, Masumoto H, Kimura S. et al. Conjugated fatty acids drive ferroptosis through chaperone-mediated autophagic degradation of GPX4 by targeting mitochondria. Cell Death Dis. 2024;15:884
105. Lee JJ, Ishihara K, Notomi S, Efstathiou NE, Ueta T, Maidana D. et al. Lysosome-associated membrane protein-2 deficiency increases the risk of reactive oxygen species-induced ferroptosis in retinal pigment epithelial cells. Biochem Biophys Res Commun. 2020;521:414-9
106. Yuan Q, Wang M, Zhang Z, Wang R, Wang D, Sang Z. et al. The ameliorative effects of melatonin against BDE-47-induced hippocampal neuronal ferroptosis and cognitive dysfunction through Nrf2-Chaperone-mediated autophagy of ACSL4 degradation. Ecotoxicol Environ Saf. 2024;290:117542
107. Hubbi ME, Gilkes DM, Hu H, Kshitiz, Ahmed I, Semenza GL. Cyclin-dependent kinases regulate lysosomal degradation of hypoxia-inducible factor 1α to promote cell-cycle progression. Proc Natl Acad Sci U S A. 2014;111:E3325-34
108. Hubbi ME, Semenza GL. An essential role for chaperone-mediated autophagy in cell cycle progression. Autophagy. 2015;11:850-1
109. Goda N, Ryan HE, Khadivi B, McNulty W, Rickert RC, Johnson RS. Hypoxia-inducible factor 1alpha is essential for cell cycle arrest during hypoxia. Mol Cell Biol. 2003;23:359-69
110. Park C, Suh Y, Cuervo AM. Regulated degradation of Chk1 by chaperone-mediated autophagy in response to DNA damage. Nat Commun. 2015;6:6823
111. Lamarche BJ, Orazio NI, Weitzman MD. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010;584:3682-95
112. Kumar BV, Connors TJ, Farber DL. Human T Cell Development, Localization, and Function throughout Life. Immunity. 2018;48:202-13
113. Bonnevier JL, Zhang R, Mueller DL. E3 ubiquitin ligases and their control of T cell autoreactivity. Arthritis Res Ther. 2005;7:233-42
114. Li W, Bell HW, Ahnn J, Lee SK. Regulator of Calcineurin (RCAN-1) Regulates Thermotaxis Behavior in Caenorhabditis elegans. J Mol Biol. 2015;427:3457-68
115. Gerondakis S, Siebenlist U. Roles of the NF-kappaB pathway in lymphocyte development and function. Cold Spring Harb Perspect Biol. 2010;2:a000182
116. Perry WL, Hustad CM, Swing DA, O'Sullivan TN, Jenkins NA, Copeland NG. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat Genet. 1998;18:143-6
117. Park YJ, Yoo SA, Kim M, Kim WU. The Role of Calcium-Calcineurin-NFAT Signaling Pathway in Health and Autoimmune Diseases. Front Immunol. 2020;11:195
118. Mangan MSJ, Olhava EJ, Roush WR, Seidel HM, Glick GD, Latz E. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Discov. 2018;17:588-606
119. Ahmad L, Zhang SY, Casanova JL, Sancho-Shimizu V. Human TBK1: A Gatekeeper of Neuroinflammation. Trends Mol Med. 2016;22:511-27
120. Herhaus L. TBK1 (TANK-binding kinase 1)-mediated regulation of autophagy in health and disease. Matrix Biol. 2021;100-101:84-98
121. Zhao X, Di Q, Yu J, Quan J, Xiao Y, Zhu H. et al. USP19 (ubiquitin specific peptidase 19) promotes TBK1 (TANK-binding kinase 1) degradation via chaperone-mediated autophagy. Autophagy. 2022;18:891-908
122. Kaushik S, Cuervo AM. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015;17:759-70
123. Kaushik S, Juste YR, Lindenau K, Dong S, Macho-González A, Santiago-Fernández O. et al. Chaperone-mediated autophagy regulates adipocyte differentiation. Sci Adv. 2022;8:eabq2733
124. Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413-6
125. Zhang C, Cuervo AM. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med. 2008;14:959-65
126. Rodríguez-Muela N, Koga H, García-Ledo L, de la Villa P, de la Rosa EJ, Cuervo AM. et al. Balance between autophagic pathways preserves retinal homeostasis. Aging Cell. 2013;12:478-88
127. Kiffin R, Kaushik S, Zeng M, Bandyopadhyay U, Zhang C, Massey AC. et al. Altered dynamics of the lysosomal receptor for chaperone-mediated autophagy with age. J Cell Sci. 2007;120:782-91
128. Rodriguez-Navarro JA, Kaushik S, Koga H, Dall'Armi C, Shui G, Wenk MR. et al. Inhibitory effect of dietary lipids on chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2012;109:E705-14
129. Dong S, Wang Q, Kao YR, Diaz A, Tasset I, Kaushik S. et al. Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature. 2021;591:117-23
130. Li B, Ming H, Qin S, Zhou L, Huang Z, Jin P. et al. HSPA8 Activates Wnt/β-Catenin Signaling to Facilitate BRAF V600E Colorectal Cancer Progression by CMA-Mediated CAV1 Degradation. Adv Sci (Weinh). 2024;11:e2306535
131. Wang M, Zhang Z, Chen M, Lv Y, Tian S, Meng F. et al. FDW028, a novel FUT8 inhibitor, impels lysosomal proteolysis of B7-H3 via chaperone-mediated autophagy pathway and exhibits potent efficacy against metastatic colorectal cancer. Cell Death Dis. 2023;14:495
132. Saha T. LAMP2A overexpression in breast tumors promotes cancer cell survival via chaperone-mediated autophagy. Autophagy. 2012;8:1643-56
133. Dong RF, Qin CJ, Yin Y, Han LL, Xiao CM, Wang KD. et al. Discovery of a potent inhibitor of chaperone-mediated autophagy that targets the HSC70-LAMP2A interaction in non-small cell lung cancer cells. Br J Pharmacol. 2023
134. Deng H, Chen Y, Wang L, Zhang Y, Hang Q, Li P. et al. PI3K/mTOR inhibitors promote G6PD autophagic degradation and exacerbate oxidative stress damage to radiosensitize small cell lung cancer. Cell Death Dis. 2023;14:652
135. Yan J, Liu D, Wang J, You W, Yang W, Yan S. et al. Rewiring chaperone-mediated autophagy in cancer by a prion-like chemical inducer of proximity to counteract adaptive immune resistance. Drug Resist Updat. 2024;73:101037
136. Zhou H, Xie X, Chen Y, Lin Y, Cai Z, Ding L. et al. Chaperone-mediated Autophagy Governs Progression of Papillary Thyroid Carcinoma via PPARγ-SDF1/CXCR4 Signaling. J Clin Endocrinol Metab. 2020;105:dgaa366
137. Nikesitch N, Rebeiro P, Ho LL, Pothula S, Wang XM, Khong T. et al. The Role of Chaperone-Mediated Autophagy in Bortezomib Resistant Multiple Myeloma. Cells. 2021;10:3464
138. Fan Y, Hou T, Gao Y, Dan W, Liu T, Liu B. et al. Acetylation-dependent regulation of TPD52 isoform 1 modulates chaperone-mediated autophagy in prostate cancer. Autophagy. 2021;17:4386-400
139. Anguiano J, Garner TP, Mahalingam M, Das BC, Gavathiotis E, Cuervo AM. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol. 2013;9:374-82
140. Huang L, Deng M, He Y, Lu S, Liu S, Fang Y. β-asarone increases MEF2D and TH levels and reduces α-synuclein level in 6-OHDA-induced rats via regulating the HSP70/MAPK/MEF2D/Beclin-1 pathway: Chaperone-mediated autophagy activation, macroautophagy inhibition and HSP70 up-expression. Behav Brain Res. 2016;313:370-9
141. Francelle L, Outeiro TF, Rappold GA. Inhibition of HDAC6 activity protects dopaminergic neurons from alpha-synuclein toxicity. Sci Rep. 2020;10:6064
142. Chen J, Mao K, Yu H, Wen Y, She H, Zhang H. et al. p38-TFEB pathways promote microglia activation through inhibiting CMA-mediated NLRP3 degradation in Parkinson's disease. J Neuroinflammation. 2021;18:295
143. Dou J, Su P, Xu C, Wen Z, Mao Z, Li W. Targeting Hsc70-based autophagy to eliminate amyloid β oligomers. Biochem Biophys Res Commun. 2020;524:923-8
144. Lee YS, Lai DM, Huang HJ, Lee-Chen GJ, Chang CH, Hsieh-Li HM. et al. Prebiotic Lactulose Ameliorates the Cognitive Deficit in Alzheimer's Disease Mouse Model through Macroautophagy and Chaperone-Mediated Autophagy Pathways. J Agric Food Chem. 2021;69:2422-37
145. Eisermann DJ, Wenzel U, Fitzenberger E. Inhibition of chaperone-mediated autophagy prevents glucotoxicity in the Caenorhabditis elegans mev-1 mutant by activation of the proteasome. Biochem Biophys Res Commun. 2017;484:171-5
146. Hos NJ, Fischer J, Hos D, Hejazi Z, Calabrese C, Ganesan R. et al. TRIM21 Is Targeted for Chaperone-Mediated Autophagy during Salmonella Typhimurium Infection. J Immunol. 2020;205:2456-67
147. Shao E, Zhao S, Dong Y, Wang Y, Fei Y, Li S. et al. Anisomycin inhibits Coxsackievirus B replication by promoting the lysosomal degradation of eEF1A1. Antiviral Res. 2023;215:105621
148. Zhang J, Huang J, Gu Y, Xue M, Qian F, Wang B. et al. Inflammation-induced inhibition of chaperone-mediated autophagy maintains the immunosuppressive function of murine mesenchymal stromal cells. Cell Mol Immunol. 2021;18:1476-88
149. Liu M, Li S, Yin M, Li Y, Chen J, Chen Y. et al. Pinacidil ameliorates cardiac microvascular ischemia-reperfusion injury by inhibiting chaperone-mediated autophagy of calreticulin. Basic Res Cardiol. 2024;119:113-31
150. Cao L, Chen R, Xu J, Lin Y, Wang R, Chi Z. Vitamin E inhibits activated chaperone-mediated autophagy in rats with status epilepticus. Neuroscience. 2009;161:73-7
Author contact
![]() Corresponding authors: Xiaoyu Song, xysongedu.cn; Liu Cao, lcaoedu.cn; Hao Feng, haofengedu.cn.
Corresponding authors: Xiaoyu Song, xysongedu.cn; Liu Cao, lcaoedu.cn; Hao Feng, haofengedu.cn.
 Global reach, higher impact
Global reach, higher impact