13.3
Impact Factor
Theranostics 2024; 14(19):7309-7332. doi:10.7150/thno.102437 This issue Cite
Review
Emerging advanced approaches for liquid biopsy: in situ nucleic acid assays of extracellular vesicles
1. Aoyang Institute of Cancer, Affiliated Aoyang Hospital of Jiangsu University, 279 Jingang Road, Suzhou Jiangsu 215600, China.
2. Zhenjiang Key Laboratory of High Technology Research on Exosomes Foundation and Transformation Application, Jiangsu Key Laboratory of Medical Science and Laboratory Medicine, School of Medicine, Jiangsu University, Zhenjiang Jiangsu 212013, China.
† Equal contributors.
Received 2024-8-16; Accepted 2024-10-20; Published 2024-10-28
Abstract
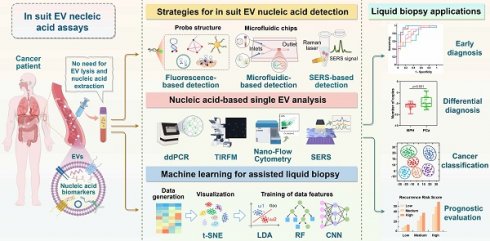
Extracellular vesicles (EVs) have emerged as valuable biomarkers in liquid biopsies owing to their stability, accessibility, and ability to encapsulate nucleic acids. The majority of existing methodologies for detecting EV nucleic acid biomarkers require the lysis of EVs to extract DNA or RNA. This process is labor-intensive and may lead to the loss and degradation of nucleic acids. However, the emerging field of in situ EV assays offers innovative tools for liquid biopsy, facilitating direct profiling of nucleic acids within intact EVs and reducing sample handling procedures. This review focuses on the promising and innovative field of in situ EV nucleic acid analysis. It examines the translational potential of in situ EV nucleic acid analysis in liquid biopsies from detection strategies, diagnostic applications, and diagnostic aids for single EV analysis and machine learning techniques. We highlight the innovative approach of in situ EV nucleic acid assays and provide novel insights into advancing liquid biopsy technology. This approach shows a promising avenue for improving EV-based cancer diagnosis and guiding personalized treatment with minimal invasiveness.
Keywords: extracellular vesicles, cancer, liquid biopsy, detection, diagnosis
Introduction
Cancer is one of the leading causes of death in humans [1]. Traditional tumor diagnosis methods, such as histopathological biopsy, are invasive and can pose risks and financial burdens to patients. As a result, clinics are turning to liquid biopsy, which is a non-invasive or minimally invasive, convenient, rapid, and cost-effective alternative [2]. Extracellular vesicles (EVs) are particles released by all cell types, characterized by their lipid bilayer membranes. These vesicles carry a diverse range of biologically active substances, including proteins, lipids, nucleic acids, and metabolites, which provide crucial insights into the physiological status of diseases and play a significant role in mediating cellular communication [3-6]. Research has shown that EVs contribute to tumor development by promoting cancer cell proliferation, metastasis, angiogenesis, drug resistance, and immune evasion [7, 8]. Furthermore, EVs are abundant and stable in body fluids, making them promising biomarkers for cancer diagnosis, treatment, and monitoring [9].
EV-based liquid biopsy presents a promising approach for diagnosing tumors in clinical patients. However, accurately and rapidly detecting these biomarkers in bodily fluids is a major challenge in clinical practice. The quantitative analysis of EV nucleic acids is usually conducted using traditional methods such as quantitative real-time PCR (qRT-PCR), which is cumbersome and low sensitive [10]. Recently, a variety of innovative techniques for detecting EV nucleic acids have started to emerge. These biosensors eliminate the need for conventional cDNA synthesis and enzymatic amplification, offering advantages in sensitivity, specificity, and ease of operation over qRT-PCR [11]. For instance, Lei Zheng's team have developed an electrochemical biosensor that employs localized DNA tetrahedron-assisted catalytic hairpin assembly for efficient and specific detection of plasma-derived EV miRNAs in gastric cancer [12]. Kang et al. have designed a SERS-based sensor using a gold octahedra array as a substrate for sensitive detection of breast cancer-derived exosomal let-7a [13]. A peptide nucleic acid-functionalized nanochannel biosensor has shown good agreement with qRT-PCR in detecting exosomal miR-10b for early diagnosis of pancreatic cancer [14]. An SPR biosensor using Au-on-Ag heterostructure and DNA tetrahedral framework enables ultra-sensitive detection of multiple miRNAs of exosomal origin from NSCLC [15]. Nevertheless, due to the protective nature of nucleic acids in EVs by lipid bilayers, the above EV nucleic acid assays still require labor-intensive processes of EV lysis and nucleic acid extraction, and they are susceptible to nucleic acid loss and degradation [16]. Consequently, direct analysis of nucleic acids from intact EVs is crucial for advancing liquid biopsy techniques. In situ EV nucleic acid detection is a technique that directly analyzes nucleic acids within EVs while preserving EV integrity. It eliminates the need for EV lysis and nucleic acid extraction, thereby streamlining sample handling procedures.
Over the past few years, many reviews have been published on EV liquid biopsy techniques, including fluorescence, electrical or electrochemical, surface plasmon resonance (SPR), surface-enhanced Raman scattering (SERS), colorimetry, microfluidics, and various nanomaterial biosensors [17-22]. Nevertheless, these reviews have mainly focused on detection methods and protein analyses, with only a few specifically addressing the detection of EV nucleic acid biomarkers. Furthermore, the development of in situ detection techniques for directly analyzing nucleic acid from intact EVs is still in its early stages, and a comprehensive investigation of their potential for application in liquid biopsies has not been documented yet. Notably, in situ EV nucleic acid analysis emphasizes the importance of probes being able to directly penetrate EVs for effective nucleic acid detection, which is not essential for accurately identifying EV subtypes. Single EV analysis has been developed to elucidate EV heterogeneity, with methods such as fluorescent labelling for identifying nucleic acids in single intact EVs playing a significant role in reflecting disease progression and improving the timeliness of disease diagnosis [23, 24]. This serves as a vital complement to in situ EV nucleic acid assays. Nonetheless, there is currently a lack of comprehensive summaries on single EV analysis techniques from the perspective of in situ EV nucleic acid detection. As a result, we have intentionally included a discussion on this topic in the manuscript to advance the development of in situ EV nucleic acid assays.
This review begins with an introduction to the biological role of EVs in tumors and their potential as biomarkers for tumor diagnosis. Subsequently, it offers a comprehensive overview of current strategies for in situ analysis of EV nucleic acids. These strategies encompass fluorescence detection employing various probe structures, microfluidics chips, and SERS detection. Furthermore, the review explores recent advancements in single EV analysis and machine learning-assisted in situ EV nucleic acid detection. Additionally, it summarizes the application of in situ EV nucleic acid analysis in liquid biopsies. Finally, the review delves into the prospects and existing challenges related to the implementation of in situ EV nucleic acid assays in liquid biopsies, and provides potential recommendations for enhancing their transition to clinical practice. In conclusion, this review aims to promote the progress of EV-based liquid biopsy techniques, offering valuable guidance and insights for the diagnosis and treatment of cancer.
Extracellular vesicles
Characteristics and biological roles of EVs
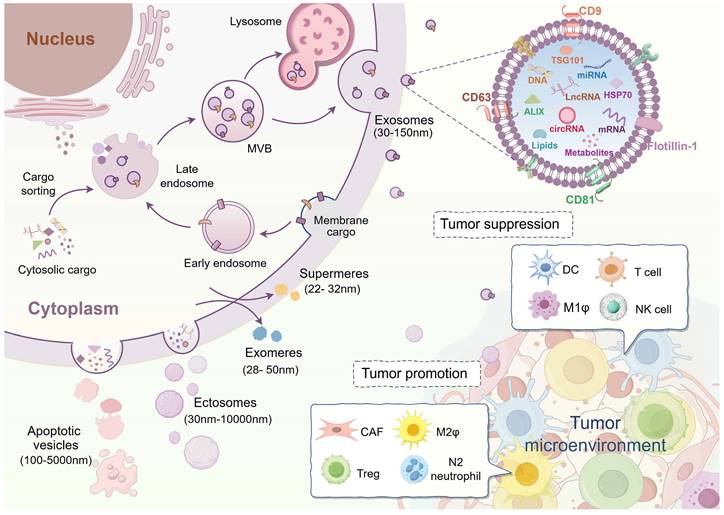
EVs are categorized into distinct subtypes based on variations in size and origin. Specifically, vesicles with a diameter of smaller than 200 nm are often referred to as small extracellular vesicles (sEV). Depending on how they are formed, EVs can be classified as exosomes (30-150 nm) originating from the endosomal system and ectosomes (30-10,000 nm) originating from cytoplasmic membrane. Exosomes, a subset of small extracellular vesicles (sEVs), form through fusion with the cytoplasmic membrane following the inward budding of late endosomes to create multivesicular bodies. Ectosomes exhibit a diverse range of sizes, including various EVs found in cell-specific conditions, such as apoptotic bodies or vesicles (100-5000 nm). They are produced through the direct secretion from apoptotic cell membranes or the cellular debris following cell death [3, 5, 25]. Notably, in addition to the membrane vesicles mentioned above, recent research has identified smaller-sized (<50 nm) non-membrane nanoparticles, like exomeres and supermeres, although the mechanism underlying their formation remains unknown (Figure 1) [26, 27]. EVs exhibit heterogeneity in terms of size, quantity, composition, cellular sources, and physiological roles [28]. Factors contributing to variations in EV sizes include differences in cytoplasmic membrane invagination, membrane budding, and isolation methodologies. The diverse cellular sources and differences in size directly impact the content and composition of EVs. Since EV-carried cargoes are derived from parental cells, EVs from different cellular sources exert different biological functions on target cells. For example, EVs from stem cells are involved in disease repair mechanisms, while EVs from tumor cells contribute to cancer progression [29-35].
In recent years, EVs have emerged as crucial players in intercellular communication within the tumor microenvironment, exhibiting both promoting and inhibitory effects on cancer progression (Figure 1) [36]. EVs released by cells in the cancer microenvironment have been shown to enhance tumor growth, angiogenesis, metastasis, drug resistance, and immune evasion, thereby contributing to tumor progression. For example, EVs from M2-like macrophages (M2ф) stimulate angiogenesis and facilitate the growth of pancreatic ductal adenocarcinoma (PDAC) by targeting E2F2 [37]. Similarly, nicotine-activated N2-neutrophils secrete EV miR-4466 to promote stemness and metabolism of brain metastatic tumor cells in lung cancer [38]. Cancer-associated fibroblast (CAF)-derived EVs inhibit ferroptosis via the miR-432-5p/CHAC1 axis, thereby increasing resistance to docetaxel in prostate cancer [39]. Additionally, breast cancer-derived EV lncRNA SNHG16 induces amplification of CD73+γδ1 regulatory T cells (Tregs) and consequently exerts an immunosuppressive effect on tumors [40].
Currently, tumor EVs with anti-cancer properties predominantly originate from immune cells, and researchers are focusing on cancer immunotherapy by modifying or enhancing the functionality of these immune cells. Dendritic cells (DCs), which are highly efficient antigen-presenting cells, release a significant quantity of EVs that trigger strong anti-cancer responses. For example, dendritic cell-derived EVs enriched with RAE-1γ can stimulate NK cells and T cells to eliminate chronic myeloid leukemia (CML) cells, regardless of the presence of the T315I mutation, resulting in effective anti-cancer outcomes [41]. Other sources of EVs capable of exerting anti-cancer effects include T cells [42], NK cells [43], and M1-like macrophages (M1ф) [44].
Biogenesis of extracellular vesicles and their roles in the tumor microenvironment. Extracellular vesicles (EVs) with lipid bilayers consist of various isoforms. Among these subtypes, exosomes, originating from the endosomal system, are the most extensively researched category of small extracellular vesicles (sEV). Ectosomes, derived from the cytoplasmic membrane, exhibit a range of sizes. Apoptotic vesicles are generated by cells undergoing apoptosis. Exomeres and supermeres are the latest discovery of non-membranous nanoparticles that can be secreted into the extracellular compartment. Although they are much smaller in size, the process of their formation remains unclear. EVs represented by exosomes contain a variety of cargoes, including nucleic acids, proteins, lipids, and metabolites. EVs serve as critical mediators of intercellular communication and assume a biphasic role within the tumor microenvironment. Tumor-associated fibroblasts (CAF), M2-like macrophages (M2ф), regulatory T cells (Tregs), and N2 neutrophils-derived EVs predominantly contribute to the promotion of tumor progression. By contrast, EVs from dendritic cells (DCs), T cells, M1-like macrophages (M1ф), and natural killer (NK) cells function as tumor-suppressive effects. (Created with Figdraw.com).
EVs, with their double-layer membrane structure, serve as optimal cargo transportation carriers, protecting cargo from degradation. Moreover, EVs exhibit excellent biocompatibility and prolonged circulation, making them suitable for delivering therapeutic drugs or molecules to specific cells for disease treatment [45]. For instance, in vivo administration of EV-mediated si-ciRS-122 reversed oxaliplatin resistance and suppressed the proliferation of colorectal cancer (CRC) tumors [34]. Small extracellular vesicles (sEVs) derived from human umbilical cord mesenchymal stem cells enhanced retinopathy recovery in diabetic mice by loading miR-5068 and miR-10228 through electroporation [29]. Engineered EVs, such as those utilizing the tumor-targeting peptide RGD, have emerged as a promising platform for targeted drug delivery in tumor therapies. By encapsulating circDIDO1, these engineered EVs effectively suppressed the growth of gastric tumors in nude mice [46]. Neutrophil-derived EVs engineered with superparamagnetic iron oxide nanoparticles (SPIONs) improved tumor localization accuracy. Similarly, nano-sized vesicles derived from neutrophils (NNVs), functionalized with SPIONs and encapsulated with DOX, showed targeted accumulation at the tumor site under an external magnetic field. This resulted in robust suppression of gastric cancer (GC) progression [47]. In a comprehensive review, Zhang et al. provided detailed insights into the strategies and methodologies involved in engineering EVs [48].
EV-based liquid biopsy of cancer
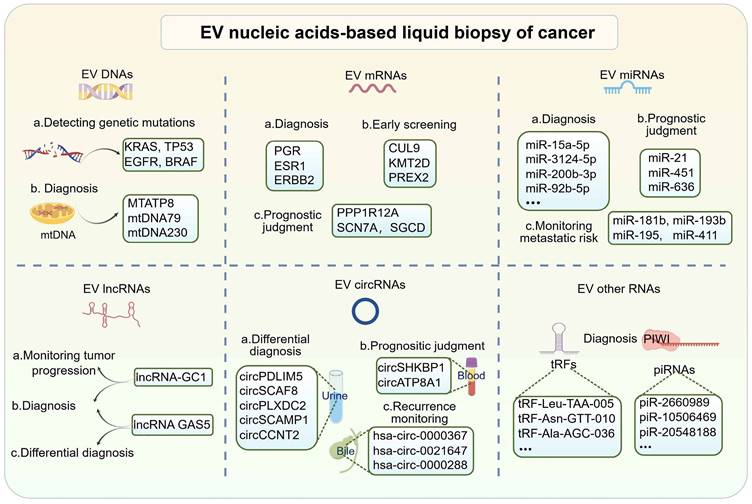
In recent years, EV-based liquid biopsies have gained traction in clinical diagnostic research. Compared to circulating tumor cells (CTCs), EVs offer advantages such as smaller size, wider distribution, and more accessible enrichment in bodily fluids. Additionally, the unique lipid bilayer structure of EVs protects their cargo from degradation, distinguishing them from circulating tumor DNA (ctDNA) or cell-free RNA (cfRNA) and making them ideal components for liquid biopsies [49]. EVs contain abundant nucleic acid components, including DNA, mRNA, miRNA, lncRNA, circRNA, and others. This section provides an overview of prevalent nucleic acid biomarkers in EVs (Figure 2).
EV-derived DNAs
Currently, DNA liquid biopsies primarily focus on ctDNA. The study of EV DNAs, despite its data, biological stability, and clinical relevance, has been limited due to its minimal content of large DNA fragments [50]. DNAs from EVs primarily serve as a diagnostic biomarker for identifying tumor gene mutations in clinical settings. Studies have demonstrated that identifying mutations in plasma EV-derived KRAS can predict colorectal cancer (CRC) and pancreatic ductal adenocarcinoma (PDAC) [51, 52]. Similarly, mutations in EV TP53 have been linked to hepatocellular carcinoma (HCC) prognosis [53]. Due to the low concentration of EV DNAs, digital PCR is commonly used to enhance the sensitivity of detecting these rare DNA mutations. For instance, Choi et al. achieved a higher detection rate of KRAS mutations in EVs from CRC patients compared to cfDNA using digital PCR, with a sensitivity of 76% and specificity of 100% [52]. Additionally, EGFR and BRAF mutations in EVs are implicated in cancer diagnosis [54, 55]. Mitochondrial DNA (mtDNA) is another non-invasive molecule with diagnostic significance in cancer, and it can be detected in circulating bodily fluids due to its high copy number [56]. Recent studies have demonstrated the biomarker potential of mtDNA in EVs. Lou et al. identified elevated levels of specific mtDNA fragments (mtDNA79, mtDNA230, and MTATP8) in plasma EVs from non-small cell lung cancer (NSCLC). These mtDNA fragments exhibited a strong association with more aggressive NSCLC traits, including larger tumors, advanced stage, lymph node metastasis, and distant metastasis, indicating significant potential for NSCLC diagnosis [57].
EV-derived mRNAs
mRNAs can be transferred between cells through EVs. mRNAs from EVs have shown potential as a liquid biopsy method. Studies have demonstrated that EV mRNAs in blood and urine can be used to diagnose urinary tract diseases. For example, a recent study on breast cancer found that a combination of specific mRNA signatures (PGR, ESR1, ERBB2, and GAPDH) derived from EVs significantly enhanced the accuracy of breast cancer diagnosis when used in conjunction with multiplexed detection using ddPCR and a machine-learning algorithm (AUC=0.95). This approach has the potential to expedite the discovery of low-abundance nucleic acid biomarkers discovery in EVs, thereby advancing the prospect of early cancer screening based on EV mRNA profile [58]. He et al. identified five EV mRNAs (CUL9, KMT2D, PBRM1, PREX2, and SETD2) as novel potential biomarkers for clear cell renal cell carcinoma (ccRCC). Among these, KMT2D and PREX2 showed an early diagnosis of ccRCC (AUC=0.83), while CUL9, KMT2D, and PREX2 could differentiate between patients with ccRCC and those with benign renal masses (AUC=0.81) [59]. Moreover, EV mRNAs excel in prognostic assessment. A study identified a prognostic EV mRNA signature (PPP1R12A, SCN7A, and SGCD) through transcriptomic analysis of plasma EV from pancreatic cancer patients. This signature correlated with a decreased overall survival rate (p = 0.014) in high-risk PDAC patients, establishing it as an independent prognostic biomarker for PDAC [60].
EV-derived miRNAs
MicroRNAs (miRNAs) are short non-coding RNAs (~22 bases) that regulate protein-coding genes at the post-transcriptional level [61]. They play a crucial role in tumor development and liquid biopsy by destabilizing target mRNAs and suppressing their translation [62]. Many studies have investigated the potential of EV miRNAs as tumor biomarkers. For instance, elevated levels of plasma-derived EV miR-15a-5p, miR-106b-5p, and miR-107 have been observed in endometrial cancer (EC). Notably, miR-15a-5p exhibited an AUC value of 0.813 in distinguishing between stage I EC patients and healthy donors. Combining miR-15a-5p with CEA and CA125 improved the AUC value of 0.899 [63]. In another study, up-regulation of serum-derived EV miRNAs (miR-3565, miR-3124-5p, miR-200b-3p, miR-6515, miR-3126-3p, and miR-9-5p) were found in patients with small-cell lung cancer (SCLC), while miR-92b-5p showed down-regulation. The combined measurement of three miRNAs (miR-200b-3p, miR-3124-5p, and miR-92b-5p) significantly improved early diagnostic efficiency (AUC = 0.93) [64]. Additionally, Sun et al. established a metastatic risk score model using miR-21, miR-451, and miR-636 extracted from urinary EVs in prostate cancer. This model accurately predicted metastasis with an AUC value of 0.925, surpassing the predictive ability of the preoperative PSA levels or clinical Gleason scores. It shows promise as a noninvasive biomarker for predicting prognosis in PCa patients [65]. Moreover, four EV miRNAs (miR-181b, miR-193b, miR-195, and miR-411) have shown robustness in detecting lymph node metastasis (LNM) in CRC patients. A risk stratification model was established by incorporating key pathologic features, which reduced the false-positive rate of LNM to 76% without missing any actual LNM-positive patients [66]. Thus, EV miRNA can be used to monitor tumor progression and predict risk stratification for metastasis.
EV-derived lncRNAs
Long non-coding RNAs (LncRNAs) are nucleotide sequences longer than 200 nucleotides that do not encode proteins. They play crucial roles in regulating gene expression at the transcriptional and post-transcriptional levels, contributing to cancer pathogenesis [67]. EV-derived lncRNAs hold promise as biomarkers for tumors [68]. For example, circulating EV lncRNA-GC1 has been shown to diagnose early GC and monitor disease progression. Compared to CEA, CA72-4, and CA19-9, lncRNA-GC1 showed higher diagnostic accuracy for GC with an AUC of 0.89, along with sufficient specificity (84.97%) and sensitivity (84.77%). Furthermore, the level of EV lncRNA-GC1 was significantly correlated with GC progression [69]. Additionally, lncRNA-GC1 levels in circulating EVs showed strong agreement (>50% reductions) with imaging response (Cohen's κ, 0.704), serving as a biomarker for evaluating the efficacy of neoadjuvant chemotherapy (neoCT) and predicting survival outcomes in GC patients receiving neoCT [70]. Moreover, the tumor-derived EV lncRNA GAS5 can serve as an early diagnostic biomarker for NSCLC. EV-GAS5 was down-regulated in NSCLC patients compared to healthy controls and exhibited an AUC of 0.822 in identifying stage I tumors. When combined with CEA, the AUC value of Exo-GAS5 reached 0.929 [71].
EV-derived circRNAs
Circular RNAs (circRNAs) are stable and abundant single-stranded, non-coding RNA molecules with covalent closed-loop structures. They play a crucial role in regulating various cellular processes in mammals, acting as miRNA sponges, protein decoys, molecular scaffolds, transcriptional regulators, and translational polypeptides [72]. Additionally, circRNAs are enriched in EVs and can be transported to target cells, contributing to tumor development. The exceptional stability and tissue-specific expression patterns of EV circRNAs make them highly potential and advantageous biomarkers for liquid biopsy in cancer [73, 74]. Initial studies identified over 1000 circRNAs in serum EVs that distinguish CRC patients from individuals [75]. Subsequently, additional circRNA biomarkers in EVs have been discovered in different biological samples. For instance, the elevated levels of circSHKBP1 and circATP8A1 in blood-derived EVs from gastric cancer (GC) patients are associated with advanced TNM staging and unfavorable prognosis, suggesting their promise as a diagnostic and prognostic biomarker [76, 77]. In urine, an extracellular vesicular circRNA classifier (Ccirc) containing circPDLIM5, circSCAF8, circPLXDC2, circSCAMP1, and circCCNT2 demonstrated the ability to detect high-grade prostate cancer (PCa) at initial biopsy, achieving an NPV of 87.50% and a sensitivity of 66.39% in the validation cohort, reducing the need for puncture biopsies by 56.34% [78]. Furthermore, researchers discovered a panel of circRNAs (has-circ-0000367, has-circ-0021647, and has-circ-0000288) in serum and bile EVs for early recurrence monitoring. Both bile-derived ERS and serum-derived ERS showed significant correlations with relapse-free survival (RFS) based on the computation of the early recurrence score (ERS), achieving AUCs of 0.851 and 0.759 and accuracies of 0.720 and 0.762 for bile-ERS and serum-ERS, respectively, in the recurrence monitoring model [79].
Significance of EV nucleic acid biomarkers in liquid biopsy. EV nucleic acid-based liquid biopsies for cancer have demonstrated substantial potential. DNA analysis in EVs has primarily targeted mutation detection in tumors, highlighting mitochondrial DNA (mtDNA) as a promising innovation for liquid biopsy biomarkers. A variety of RNA types within EVs have been identified as biomarker potential. These RNA biomarkers, such as mRNAs, miRNAs, lncRNAs, circRNAs, and others, have exhibited promising utility in tumor diagnosis, identification, screening, prognosis, and recurrence monitoring. (Created with Figdraw.com).
EV-derived other RNAs
In addition to the extensively investigated EV nucleic acid biomarkers mentioned above, EVs from tumor liquid biopsies have also been found to contain other nucleic acids such as piRNA and tsRNA. tsRNA are small non-coding RNAs derived from tRNA molecules, specifically tRNA-derived fragments (tRFs) and tRNA-derived stress-induced RNAs (tiRNAs), which are produced through enzymatic cleavage of mature tRNAs or tRNA precursors [80]. piRNAs, on the other hand, are a type of RNA associated with proteins from the PIWI branch of the argonaute family and play a crucial role in germ cell development [81]. Both tiRNAs and piRNAs, as small RNA molecules, have been detected in EVs and show promise as diagnostic biomarkers. For example, a study found that certain EV-derived tRFs, tRF-Leu-TAA-005, tRF-Asn-GTT-010, tRF-Ala-AGC-036, tRF-Lys-CTT-049, and tRF-Trp-CCA-057, were significantly down-regulated in early-stage NSCLC patients compared to healthy controls. When these five tRFs were combined, their diagnostic accuracy increased, suggesting their potential as diagnostic biomarkers for NSCLC [82]. Furthermore, analysis of plasma EVs from cholangiocarcinoma and gallbladder cancer patients revealed upregulation of piRNAs, including piR-2660989, piR-10506469, piR-20548188, piR-10822895, piR-has-23209, and piR-18044111, as identified through RNA sequencing. Interestingly, the postoperative analysis revealed a significant reduction in the expression of piR-10506469 and piR-20548188, suggesting their potential utility as diagnostic biomarkers for cholangiocarcinoma and gallbladder cancer [83]. Overall, these findings highlight the potential of tiRNAs and piRNAs in EVs as valuable diagnostic biomarkers for various types of cancer.
Apart from those above nucleic acid-based liquid biopsies, EV nucleic acid-based multi-omics characterization has shown promise in precise tumor diagnosis. For instance, a 6-EV-RNA panel (let-7i-5p, miR-1307-3p, LZIC, SRSF6, lncFTH1-211, and lncPTMA-209) identified through EV multi-omics RNA sequencing was able to robustly identified advanced GC patients treated with fluorouracil-based neoadjuvant chemotherapy. The panel achieved 100% sensitivity, specificity, and area under the ROC curve in the training cohort after analysis using the COX regression algorithm [84]. Furthermore, in-depth molecular characterization of tumor heterogeneity has proven effective in risk stratification of patients [85]. The application of a robust corroborative analysis for biomarker discovery (RCABD) strategy has been proposed for identifying EV molecules, validating differential expression, modeling risk prediction, and conducting heterogenous dissection with multi-omics. A panel of 10 EV signatures (RTCA, SLC1A5, miR-324-5p, AURKA, HLA-DQB2, P2RX1, COL17A1, miR-99a-5p, LINC01055, and C3) enables effective risk stratification in breast cancer, serving as a novel and precise prognostic marker with significant CoxP value, hazard ratio, and confidence interval. This provides valuable insights into exploring the relationships between biological heterogeneity, risk stratification, and prognosis prediction in cancer [86].
Despite the proven importance of EV biomarkers in tumor progression, their potential for cancer diagnosis still faces challenges. Firstly, the complexity of biological fluids impacts the purity and yield of EV isolation, thereby influencing the efficiency of extracting nucleic acid biomarkers. Secondly, more clinical samples are needed to validate the feasibility of these biomarkers. Additionally, sensitive, stable, and user-friendly assay kits are needed to facilitate the clinical application of EV nucleic acid biomarkers.
Emerging techniques for in situ nucleic acid assays of EVs
Presently, there have been significant advancements in EV-based nucleic acid analysis techniques. Along with traditional PCR amplification, next-generation sequencing, microarray chip, and northern blotting, newer detection techniques such as fluorescence, electrochemical, colorimetric, SERS, SPR, and microfluidic biosensors have shown promising performance [11, 87, 88]. However, these methodologies are constrained by several limitations, such as the necessity for a substantial sample size to ensure effective EV isolation, as well as labor-intensive and time-consuming procedures like EV lysis, RNA extraction, and cDNA synthesis. Moreover, RNA degradation and loss due to the lack of protection from the EV membrane are also concerns [16]. Conversely, in situ detection strategies for EV nucleic acids have emerged as a solution, avoiding these limitations and improving diagnostic reliability. This section will discuss various in situ detection strategies reported thus far, including fluorescence assays based on different probe structures, microfluidic chips integrating fluorescence, and SERS assays (Figure 3).
Fluorescence-based detection
Fluorescence-based analysis offers rapid speed, high sensitivity, excellent selectivity, and convenient operation, making it widely used in biology, medicine, environment, and food-related applications in recent years [89-91]. The essential components for fluorescence detection include probes that hybridize with target molecules and active fluorescent groups attached to the probes. Researchers have optimized the probe's structure and enhanced fluorescence signal output efficiency to enable effective target analysis. Here, we summarize fluorescence detection strategies based on the probes' structural characteristics (Table 1).
Molecular beacon-based fluorescence detection
Molecular beacons (MBs) are labeled stem-loop oligonucleotide chains with fluorescent and quenching groups at the end. In their unbound state, MBs form a hairpin conformation where the fluorescent and quenching groups are close, leading to fluorescence quenching. When they hybridize with the target, the stem region of the MBs unfolds, increasing the distance between the fluorescent and quenching group, resulting in the restoration of fluorescence [92]. MB-based fluorescence detection has been demonstrated for in situ detection of EV miRNAs. For example, Wu et al. developed a modularized DNAzyme-amplified two-stage cascaded hybridization chain reaction (CHCR-DNAzyme) circuit using multiple MBs. The output of the first hybridization chain reaction (HCR1) triggers the subsequent hybridization chain reaction (HCR2), producing numerous Mg2+-dependent DNAzyme branched nanowires for enhanced fluorescence detection. Through electroporation, their setup successfully detected miR-21 in oral squamous cell carcinoma cells, indicating potential for cancer diagnosis [93].
To overcome the heterogeneity of EVs and the limitation of a single biomarker and accurately reflect disease symptoms and stages, methods that simultaneously detect surface protein and nucleic acid biomarkers are needed. Cho et al. reported a multiplexed in situ EV assay platform using a CD63 antibody for EV capture combined with fluorescent-labeled MBs to detect miR-21 and miR-574-3p within EVs. This enables quantitative analysis of PCa cell-derived EV miRNAs and proteins at the single EV level [94]. Additionally, He et al. constructed a Mg2+-dependent split DNAzyme probe (SDP) containing two divided DNAzyme fragments (D1 and D2) and an MB. Upon entering EVs, the SDP activates and cleaves the MB's stem-loop region in the presence of miR-21, generating fluorescence signals [95].
However, the efficiency of MB penetration into EVs alone is limited. Though electroporation or treatment with streptolysin O (SLO) can facilitate MB transport into EVs, it may disrupt the EV membrane and cause internal cargo leakage.
Schematic illustration of various strategies for in situ EV nucleic acid assays. Presently, there are three predominant approaches for the in situ detection of EV nucleic acids: fluorescence-based detection, microfluidic chips integrating fluorescence strategies, and SERS-based detection. Fluorescence detection, being the most common approach, relies on unique structural properties of various probes to enable sensitive in situ EV nucleic acid detection. The array of functional materials utilized in fluorescence detection includes molecular beacons (MBs) (A) for their sequence-specific recognition, DNA nanostructures (B) for their strong membrane permeability and customizable design, gold nanoparticles (AuNPs) (C) known for their conductivity and surface easy modification properties, liposomes or virus-mimicking fusogenic vesicle (Vir-FVs) (D) for their biocompatibility and ability to encapsulate cargo, and innovative nanomaterials such as BP (black phosphorus) (E) for its unique surface-to-volume ratio, excellent biocompatibility and degradability properties. The microfluidic chip triggers fluorescence detection by loading liposomes or Vir-FVs fused with EVs to promote the binding of molecular beacons to the target (F). Surface-enhanced Raman scattering (SERS) has emerged as a potent technique for in situ EV nucleic acid detection, achieved by modifying Au@DTNB nanoparticles on the surface of EVs to act as Raman tags, thereby enabling SERS detection with high specificity and signal sensitivity (G). (Created with Figdraw.com).
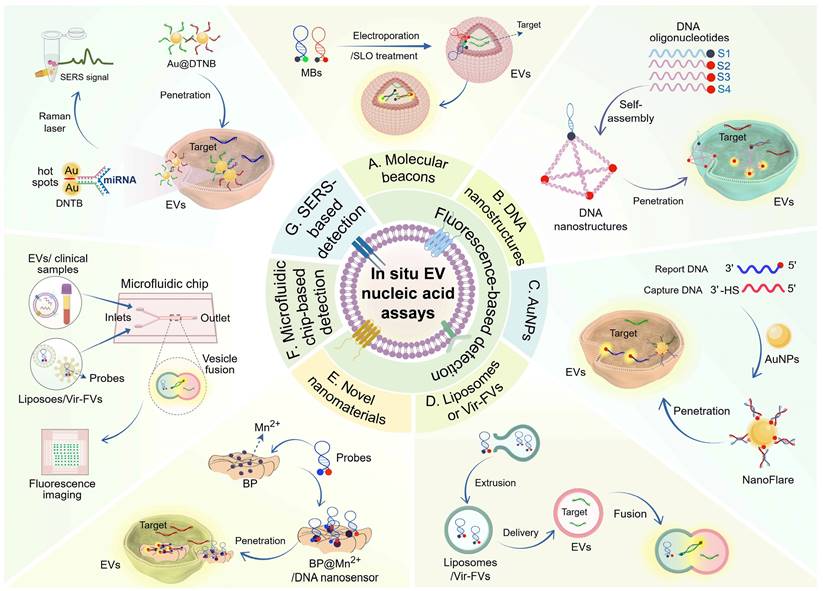
DNA nanostructure-based fluorescence detection
DNA nanostructures, such as nanowires, tetrahedrons, and cubes, have shown promise in cellular drug delivery and biosensors. These structures are programmable and stable, allowing them to penetrate biological membranes and protect substrates from interference effectively. They also confine molecular probes, reducing the risk of quenching and degradation [96-98]. For example, DNA nanowires composed of complementary sequences are resistant to nuclease degradation and can be used for in situ detection of EV nucleic acids [99]. Zhang et al. have developed a novel NgCHA nanoprobe using DNA nanowires assembled from two hairpins and a single-stranded DNA in combination with catalytic hairpin assembly (CHA), which enables direct detection of EV-miRNAs with high stability and penetration, showing great potential in EV-based disease management. Within the CHA process, two hairpins (H1, H2) are immobilized on the nanowire, and the miRNA target triggers the toehold strand displacement assembly of the two hairpins, thus recycling the miRNA and CHA products [100]. DNA tetrahedrons, known for their endocytosis capabilities, have also been utilized in nanoprobe design [101]. One study designed a fLIGHT nanoprobe with an integrated DNA tetrahedron and hairpin probe. The fLIGHT nanoprobe consists of three vertices with fluorescence donors, while the fourth vertex is located near the quencher of the hairpin probe. In the absence of the target, the three fluorescent groups can bind to the quenching group, resulting in the fluorescence turning off. When the target miRNA is present, it disrupts the stem-loop structure, leading to the quencher moving away from the vertices and activating the fluorescence. The fLIGHT nanoprobes can directly visualize miRNAs in EVs without damaging the EV membrane or extracting the EV cargo, creating a novel approach for in situ tracking of EV cargoes [102]. In addition, DNA cubes have been utilized for in situ detection of EV nucleic acids. Chen et al. developed a DNA cube-based DDCA nano platform for the rapid, reliable, and sensitive detection of EV miRNA-21, showing potential in cancer screening and cellular communication studies. This approach utilizes a DNA nanocube and two hairpin DNAs (H1 and H2) within the EV. The target miRNA binds to H1, forming an intermediate structure, which then hybridizes with H2 to create an H1-H2, releasing the target miRNA for further cycles. This process brings the fluorescent groups of the two hairpins close together, enabling fluorescence resonance energy transfer (FRET) [103]. Another study also showed that three-dimensional MBs based on DNA nanocages can effectively and non-destructively detect EV miRNAs in situ [104].
While DNA nanostructure-based probes enhance EV uptake efficiency and prevent cargo leakage, nanostructures that can assemble multiple probes for signal amplification to improve detection sensitivity are needed. Further understanding of the self-assembly mechanism and penetration process of nanostructures into the EV interior is also necessary.
Au nanoparticle-based fluorescence detection
Gold nanoparticles (AuNPs) have great potential in biomedicine due to their electrical conductivity, surface plasmon resonance, and accessible surface modification [105, 106]. AuNPs-based nanoprobes with high loading capacity and efficient membrane penetration enable sensitive analysis of EVs through different amplification strategies. For example, Liu et al. developed a dual miRNA-activated, entropy-driven catalysis (EDC)-enhanced system for accurate detection of HCC cell-derived EV miR-21 and miR-122. This system consists of miRNA detection modules (SN), a reporter module (TA), and a signal amplification module (FA). Target EVs trigger a toehold-mediated strand displacement (TMSD) reaction, releasing the initiator (N) and initiating a cyclic reaction to generate an amplified fluorescent signal [107]. Moreover, the combination of Au nanoflare and CHA amplification allows in situ and sensitive detection of EV miRNAs. Three different fluorescently labeled H1 hairpins (specifically binding miR-21, miR-122, and miR-375) ligated to AuNPs allow entry into the EVs directly. The fluorescence is activated upon recognition and binding of the target miRNAs by H1, and hairpin H2 subsequently initiates the hybridization chain reaction (CHA) to amplify the fluorescent signal response [108]. Thermophoresis, induced by localized laser irradiation, derives the directional migration of fluorescently labeled EVs toward the center of the laser spot, leading to amplified fluorescence signals [109]. Professor Sun's team implemented a thermophoretic sensor in nanoflare for in situ sensitive detection of breast cancer-derived EV miR-375. The miRNA reporter probe-modified AuNPs on nanoflares bind to the target miRNA in the EVs, generating a fluorescent signal. Subsequent thermophoretic enrichment of nanoflare-loaded EVs through localized laser heating enhances the fluorescence signal [110]. Although AuNPs show promise for in situ EV detection, their penetration efficiency into EVs needs improvement. The size of particles directly influences the toxicity and biocompatibility of AuNPs. Ultrasmall AuNPs with diameters less than 10 nm have been found to exhibit superior cell penetration capabilities [111].
Liposome or Vir-FV -based fluorescence detection
Recent advancements in targeted drug delivery have been facilitated by employing liposomes and membrane vesicles, which exhibit outstanding biocompatibility [112, 113]. Leveraging the membrane characteristics of EVs, researchers have employed membrane fusion strategies to deliver molecular probes enclosed in liposomes and virus-mimicking fusogenic vesicles (Vir-FVs) to enable in situ detection of nucleic acids. For instance, Liu et al. developed a sensitive and tethered cationic lipoplex nanoparticles (tCLN) biochip for identifying EV miRNAs in NSCLC patient serum. Within the tCLN biochip, cationic lipoplex nanoparticles with molecular fiducials are anchored on a gold-coated glass surface. Through electrostatic interactions, positively charged liposome nanoparticles and negatively charged EVs undergo fusion, followed by the hybridization of MBs in the liposomes with targets and the generation of fluorescent signals. This enables quantitative detection of EV miRNAs using TIRF microscopy [114]. Another study utilized the innate RNase activity of bacterial Cas13a to directly measure EV miRNA in plasma by combining liposomes packaging CRISPR/Cas13a and reporter probes with EVs [115]. However, the efficiency of membrane fusion mediated by these strategies requires improvement. Directing specific membrane fusions through DNA zipper hybridization enables rapid fusion kinetics. Researchers used liposomes and MBs to prepare probe-containing lipid vesicles and cholesterol-modified A/B′ double-stranded DNA (ZDC) through co-extrusion. At the same time, EV ZDC complementary to the liposomal ZDC was also prepared. Liposomes and EVs emit fluorescence upon hybridization of the probe to the target molecule through ZDC-mediated membrane fusion, enabling rapid detection of EV miRNAs for cancer diagnosis [116]. Aside from liposome-mediated in situ EV detection, virus-mimicking fusogenic vesicles (Vir-FVs) can also facilitate efficient fusion with EVs. Gao et al. constructed Vir-FVs expressing hemagglutinin-neuraminidase (HN) protein and fusion (F) protein, allowing Vir-FVs directly target the salivary acid receptor on EVs and induce effective fusion. MBs enclosed in Vir-FVs can specifically target EV miRNAs, providing a promising strategy for cancer diagnosis and therapeutic monitoring [117]. The studies mentioned above have demonstrated the possibility of using liposome- or Vir-FVs-based assays for in situ EV detection. However, it is essential to note that their instability could potentially lead to the premature release of the delivered MBs.
Novel nanomaterial-based fluorescence detection
In addition to AuNPs, various new materials have emerged in recent years for the development of biosensors, including quantum dots, graphene, hydrogels, metal oxide nanoparticles, carbon nanotubes, and upconversion nanoparticles. Black phosphorus (BP), a novel two-dimensional nanomaterial, possesses a honeycomb folded structure formed by strong intralayer P-P covalent bonds and weak interlayer van der Waals forces. It has a large surface-to-volume ratio, as well as good biocompatibility and degradability [118]. The stability of BP can be enhanced by modifying it with Mn2+, which also promotes the adsorption of single-stranded DNA on the BP surface [119]. Xia et al. recently developed a BP@Mn2+/DNA nanosensor by adsorbing miRNA fluorescent probes and EpCAM aptamers onto Mn2+-modified BP. This nanosensor enables direct detection of cancer-specific EV miRNAs and can easily penetrate the lipid bilayer membrane. Upon membrane penetration, the adsorbed DNA probe hybridizes with EV miR-21 and is released from the BP@Mn2+ surface, leading to the recovery of fluorescence signals [120]. The development of the BP@Mn2+/DNA nanosensor provides a new approach for the rapid and efficient detection of in situ EV nucleic acids. Future advancements are expected to yield more sensitive and innovative 2D nanosensor platforms for cancer diagnosis.
Microfluidic chips integrating fluorescence strategies
Microfluidics enables precise manipulation of fluids in small channels, offering high sensitivity, integration, and minimal sample and reagent consumption. It allows for the consolidation of multiple functions on a single chip, streamlining analysis and simplifying operational procedures [125]. Currently, most of the microfluidic-based assay platforms incorporate fluorescence principles, with many designed for both EV isolation and nucleic acid detection [126, 127]. However, only a limited number of these platforms have been documented to be utilized for in situ analysis of EV nucleic acids. Qian et al. introduced the isExoCD, a microfluidic chip based on agarose for EV concentration and in situ detection of EV miRNA. The chip utilizes capillary effect and agarose gel's permeability to concentrate the loaded mixture at one end of the microchannel, and any excess probes exit through the agarose gel nanopore. The isExoCD incorporates a CHA-based amplification strategy, enabling highly sensitive detection of EV miRNAs without complex elution steps or sample destruction [128].
Performance comparison of four popular fluorescence strategies.
| Methods | Probe Stability | Detection flux | Penetration efficiency | EV integrity | Ref. |
|---|---|---|---|---|---|
| Molecular beacon-based fluorescence detection | Stable at room temperature | Multiple molecular beacons can be transported to EVs | Requires membrane treatment to facilitate penetration | Electroporation and SLO treatment destroy membrane structure | [93, 94, 121] |
| DNA nanostructure-based fluorescence detection | Inefficient stabilization of self-assembly | Confined space restricts the carrying of probes | Rigidity and hardness of nanostructures provide better permeation efficiency | Have better integrity | [101, 103, 122] |
| Au nanoparticle-based fluorescence detection | Related to the way of probe modification onto the surface of AuNPs | Surface of AuNP can be modified with multiple probes | Related to the size of AuNPs | Have better integrity | [108, 123, 124] |
| Liposome or Vir-FV -based fluorescence detection | Poor stability, needs to be ready to use | Generally, only one type of probe is loaded | Instability of liposomes may affect their fusion efficiency with EV membranes | Membrane is fused | [112, 114, 115] |
EV: extracellular vesicle; SLO: streptolysin O; Vir-FV: virus-mimicking fusogenic vesicle.
Additionally, incorporating liposomes or Vir-FVs fusion with EVs has improved probe detection efficiency. For example, the mCLN-based assay with a micro-mixer biochip enables fast and sensitive detection of EV TTF-1 mRNA and miR-21 [129]. Another study developed a hydrogel microfluidic device that encapsulates liposomes containing probes, facilitating the release of the target gene as the hydrogel degrades and activating the CHA reaction for fluorescence signal amplification [130]. Furthermore, Zhou et al. designed a 3D microfluidic chip with dual channels for the simultaneous detection of EV proteins and miRNAs. The chip recognizes tumor EV proteins using quantum dot-labeled antibodies (CD81, EphA2, and CA19-9) and detects EV miRNAs in situ by preparing Vir-FVs containing MBs fused with the EV [131].
Microfluidic-based in situ EV nucleic acid sensing platforms offer significant advantages in throughput, speed, and sample consumption. However, current detection methods heavily rely on liposomes or Vir-FVs for in situ analysis, highlighting the need to develop analytical strategies with improved permeability efficiency and non-destructive EV detection.
SERS-based detection
Surface-enhanced Raman scattering (SERS) is a powerful technique that amplifies Raman signals near plasma nanostructures by attaching the analyte to its surface. It has been widely applied in food safety, environmental monitoring, and disease diagnosis due to its high sensitivity, signal specificity, and resistance to photobleaching [132]. Although researchers have developed various SERS-based EV detection strategies for cancer diagnosis, only one report has been published on in situ EV analysis. Jiang et al. developed a Fe3O4@TiO2-based SERS sensing platform using LNA probe-modified Au@DTNB nanoparticles as tags. These tags were transferred to EVs through incubation and hybridization with target miRNAs, generating strong SERS signals. Additionally, Fe3O4@TiO2 nanoparticles were incorporated to bind the phosphate group of EVs, enabling Raman laser-based SERS detection. This SERS sensor demonstrated high sensitivity (LOD= 0.21 fM) and simplicity in directly quantifying EV miR-10b in PDAC-derived serum, providing a rapid and accurate method for EV-based cancer diagnosis [133]. Further exploration of SERS strategies with high throughput and strong Raman signals is needed.
In summary, we present a comprehensive overview of the different strategies currently employed for in situ EV analysis, with fluorescence-based methods showing promising outcomes. However, probes enter EVs randomly, which may introduce interference from impurities in the sample, resulting in inaccurate target identification and potential false positive signals. Therefore, researchers must address background interference, develop sensitive and efficient probes, and facilitate EV translocation and target-specific recognition.
Nucleic acid-based single EV assays
The heterogeneity of EVs brings significant challenges in achieving precise tumor diagnosis [134]. Traditionally, EV analyses have often treated them as a homogeneous group, potentially masking the intricate biomarker details present in tumor-specific EVs and thus compromising the sensitivity and specificity of assays such as nanoparticle tracking analysis (NTA) and microfluidics. In contrast, single EV analysis, which focuses on the phenotype and characteristics of individual EV particles, offers a significant advantage in addressing this vesicular heterogeneity. This approach allows for a more nuanced understanding of the EV population, enhancing the potential for accurate biomarker detection and analysis.
Various methodologies are utilized for single EV analysis, including label-free analysis based on the physical characteristics of EVs and fluorescent labeling approaches rooted in their biological properties. Among these approaches, fluorescent labelling-based analyses can quantify protein and nucleic acid expression in single EVs, such as microdroplet digital PCR [135], digital ELISA [136], SERS [137], flow cytometry [138], and microscopic imaging [139]. While there is some overlap between fluorescent labelling-based single EV analysis and in situ EV analysis, the current literature predominantly focuses on surface protein identification in single EV analysis. Furthermore, the integration of single EV analysis with in situ EV nucleic acid assays has not been extensively explored in existing studies. This section synthesizes recent advances in single EV analysis techniques through the lens of in situ EV nucleic acid assays (Figure 4).
Droplet digital PCR
Droplet digital PCR (ddPCR) is a highly sensitive, specific, and quantitative polymerase chain reaction method that has become crucial for detecting rare targets. It involves dividing a sample into numerous microdroplets and distributing them evenly among different reaction units for amplification. The fluorescence signals produced by these reaction units are absolutely quantified by the statistical method of Poisson distribution [140]. In RNA analysis within EVs, ddPCR typically requires extraction of EV RNA. Excitingly, ddPCR can be used to analyze RNA in individual EVs recently. Pasini et al. utilized the TaqMan-based ddPCR approach to analyze RNAs at the single EV level efficiently. By co-packing EV and PCR reaction solutions into microdroplets using the QX200™ Microdroplet Digital™ PCR System, they demonstrated a sensitivity exceeding 90% for EV-RNA-based EGFR mutation detection, with methods accuracy confirmed through Sanger sequencing [141]. However, the mechanism by which the probe and PCR reaction solution enter the individual EV for target amplification remains unclear.
In addition to RNA, ddPCR can also detect DNA in single EVs. Recently, Jiao et al. developed a hydrogel-based droplet digital multiple displacement amplification (ddMDA) assay for precise analysis of DNA cargo in individual EVs [142]. MDA is a highly effective whole genome amplification technique that uses Phi 29 DNA polymerase and randomly clustered primers to amplify even a single copy of DNA. In the ddMDA assay, hydrogel droplets containing individual EVs are generated using a vibrating sharp-tip capillary system and immobilized on a microfluidic chip. The cross-linked hydrogel traps the EVs while allowing the diffusion of small molecules and enzymes. By cleaving the EVs, the hydrogel enables the amplification of EV-DNA using ddMDA within a single droplet. This hydrogel-based ddMDA strategy not only enables the absolute quantification of DNA-containing EVs but also allows for the extraction of droplets containing fluorescent clusters for DNA sequencing. This provides a simple, sensitive, and powerful tool for early cancer detection and monitoring of treatment response.
Schematic representation of single EV analysis strategy for in situ nucleic acid detection. Presently, nucleic acid-based single extracellular vesicle assays are classified into four categories: droplet digital PCR (ddPCR), total internal reflection fluorescence (TIRF) microscopy (TIRFM), Nano-flow cytometry, and surface-enhanced Raman scattering (SERS). In ddPCR systems, the EVs and the PCR reaction mixture are co-encapsulated within droplets, which are then evenly distributed across various reaction chambers to facilitate signal amplification (top left). TIRF microscopy elicits evanescent waves, which in turn excite fluorescent moieties within the EVs to generate fluorescent signals for subsequent imaging and quantitative assessment of the EVs (top right). Nano-flow cytometry can detect nanoparticles smaller than 200 nm, facilitating the capture and subsequent analysis of fluorescently labeled extracellular vesicles (EVs) to determine their concentration with the nano-flow cytometer (nFCM) (bottom left). The MoSERS microchip transforms the cargo information present on and within the EVs into a unique spectral fingerprint through the application of Raman laser illumination (bottom right). (Created with Figdraw.com).
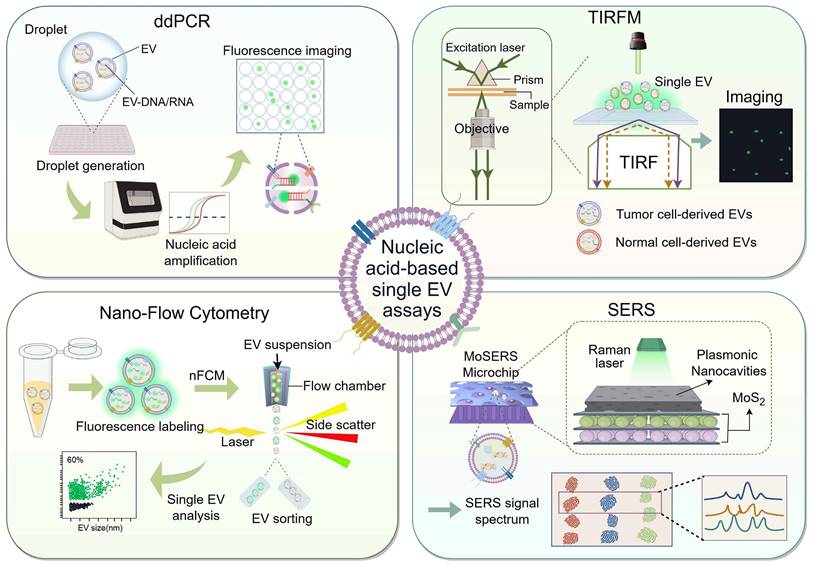
Total internal reflection fluorescence microscopy
Microscopic imaging methods, although capable of directly visualizing EVs through fluorescent labeling, are limited in their ability to differentiate between different EV subtypes and perform quantitative analysis. In recent years, total internal reflection fluorescence microscopy (TIRFM) has emerged as a promising technique for single EV analysis. By utilizing an evanescent wave produced by total internal reflection to excite fluorophores near the sample surface at a hundred-nanometer scale, TIRFM enables quantitative analysis of the emitted fluorescence signals [143]. To detect EV RNA in situ, He et al. developed a TIRFM-based single vesicle imaging platform for quantitative and chemometric analysis of EV miRNA. This platform utilizes split DNAzyme probes that selectively attach to melanoma cell-derived EV miR-21, resulting in the emission of fluorescent signals captured by TIRFM. The TIRFM imaging assay allows for precise quantification of target miRNA at the single vesicle level and determination of the miRNA and EV stoichiometry. This platform is poised to become a universal and valuable tool for in situ quantification and stoichiometric analysis of disease-related EV miRNA biomarkers [144]. Similarly, another study utilized TIRFM to simultaneously detect protein and mRNA/miRNA in a single EV, validated in a cohort of lung adenocarcinoma patients [145].
In addition, Zhang's lab proposed a single-EV and particle (siEVP) protein and RNA assay (siEVPPRA) assay to simultaneously detect protein and RNA biomarkers in multiple EV and lipoproteins subpopulations, revealing heterogeneity between vesicles and particles. The siEVPPRA assay is manufactured from PRIMO optical modules containing circular micropattern arrays, and the signals of siEVP are visualized using TIRFM. The micropatterns include immobilized antibodies targeting the surface protein of EVP, enabling selective sorting of siEVP, while mRNAs and miRNAs are labeled with MBs. The siEVPPRA assay has superior sensitivity compared to qRT-PCR (linear range of 106-1011 particles/mL), and transcriptomic analysis reveals the heterogeneity of miRNAs in single EVs. Additionally, siEVPPRA facilitates better differentiation between patients with glioblastoma (GBM) and healthy donors by detecting miRNA biomarkers in serum, presenting a novel approach for liquid biopsy and biomarker discovery [146].
Nano-flow cytometry
Flow cytometry holds promise in EV analysis due to its capacity to assess data from individual cells. However, conventional flow cytometers have a detection limit for scattered light in the ranges from 300 to 500 nm, making it challenging to quantify and characterize nanoscale vesicles accurately. Although protein analysis of single EVs has been achieved by optimizing flow cytometers and incorporating fluorescence imaging, background interference and limited sensitivity remain issues [147, 148].
In recent years, nano-flow detectors have emerged as a potential solution for analyzing single EVs. These detectors overcome the limitations of conventional flow cytometry for particles smaller than 200 nm, enabling the characterization of individual particle sizes, distributions, concentrations, and biochemical properties with high sensitivity and throughput. This advancement herald new prospects in nanoscale flow detection technology [149]. For example, Oliveira et al. employed a cell-penetrating peptide to deliver MBs into the EV and utilized a nano-flow assay to rapidly analyze the fluorescence signal of miRNA-451a, creating a new tool for MB-based RNA detection [150]. However, the fast flow rate of the instrument and the need to detect faint fluorescent signals within a limited timeframe (average of 22 μs) pose challenges.
In terms of DNA detection, another study developed a nano-flow cytometer (nFCM) that can detect single EVs as small as 40 nm and single DNA fragments of 200 bp. This nFCM assay utilized SYTO 16 staining to analyze EV-DNA at the single vesicle level. The study identified DNA heterogeneity, including differences in DNA localization, quantity, and isoforms. Additionally, it revealed that cancer cells release a higher proportion of external DNA EVs, highlighting the potential of EV-DNA as a liquid biopsy strategy [151]. This investigation provides direct experimental evidence for a comprehensive understanding of the relationship between DNA and EVs, in-depth offering a fresh insight into liquid biopsy strategies based on EV-DNA.
Surface-enhanced Raman scattering
The rapid, non-destructive, and non-invasive nature of surface-enhanced Raman scattering (SERS) highlights its advantages in biosensing applications. Label-free SERS-based methods have the potential to convert the complete biochemical profile of EV into a unified spectral pattern or "fingerprint," making it an attractive tool for liquid biopsy applications [152, 153]. However, the detection of biomarkers at the level of single EVs using SERS has not yet been achieved.
Recent studies have introduced a SERS-based microfluidic device called the MoSERS microchip, which incorporates embedded nanocavity arrays. These nanocavity arrays are equipped with MoS2 monolayers and layered plasma cavities, resulting in a significant enhancement of the electromagnetic field within the cavities. This allows for the acquisition of SERS spectra with single EV resolution without the need for additional biometric components. Additionally, the MoSERS microchip has been successfully used to characterize and analyze the cargo of single EVs, including the identification of mutations in glioma cells and circulating blood samples. These findings demonstrate the heterogeneity of EVs derived from glioblastoma cells and the potential of non-invasive liquid biopsies [154].
Single EV analysis is an emerging and fascinating research field that has the potential to revolutionize early cancer diagnosis. However, existing technologies often come with high costs and complicated procedures, and the purity of EV isolation is crucial for assay accuracy. It is essential to highlight that while our focus is on single EV-based nucleic acid analysis, the detection of multiple biomarkers can provide a more comprehensive understanding of the diversity of tumor EVs, particularly by examining proteins and nucleic acids.
Machine learning for assisted in situ EV nucleic acid detection
In recent years, machine learning (ML) has emerged as a promising tool for enhancing disease diagnosis, cancer classification, survival prediction, and treatment decision-making, leading to more reliable and accurate disease-specific diagnostic systems [155, 156]. Several ML algorithms have been extensively employed in biomedicine, including principal component analysis (PCA), t-distributed stochastic neighbor embedding (t-SNE), linear discriminant analysis (LDA), support vector machine (SVM), random forest (RF), K-nearest neighbors (KNN), convolutional neural network (CNN), decision tree, logistic regression, and various others. In this context, we provide an overview of some widely adopted machine learning algorithms currently utilized for in situ EV detection (Figure 5).
Linear discriminant analysis
LDA is a classical statistical machine learning algorithm used for classifying or distinguishing between different event features by identifying linear combinations of these features [157]. It has been applied successfully in cancer classification, as demonstrated in a study involving 64 cases of five cancer types, including breast cancer, lung cancer (LC), hepatocellular carcinoma (HCC), colorectal cancer (CRC), and cervical cancer (CC). A nanoflare & CHA-based sensing platform integrated with the LDA algorithm can quickly and accurately differentiate five types of cancers in a non-invasive manner. Patients diagnosed with the five types of cancers exhibited minimal overlap in the LDA graph, achieving an overall accuracy of 99% [108]. Moreover, Zhou et al. developed a 3D microfluidic chip that integrates two channels for simultaneous detection of EV proteins and miRNAs. By applying LDA analysis to evaluate the expression levels of EV biomarkers, the chip successfully differentiated patients with advanced pancreatic cancer (PC) from those with early-stage PC and benign controls. The classification results demonstrated a remarkable overall accuracy of 100% for cancer diagnosis and clinical staging, with individual EV biomarkers showing error rates ranging from 35% to 44% [131]. In another exciting development, Ray et al. developed the SORTER assay, which detects a specific 6-miRNA signature in EVs derived from PCa. Validated using LDA, the assay exhibited 100% sensitivity, specificity, and accuracy in both training and validation cohorts, enabling the discrimination of PCa from benign prostatic hyperplasia patients. Interestingly, the identified PCa signature by SORTER was found to be unrelated to serum PSA levels [158].
Random forest
RF algorithm combines multiple weak classifiers to achieve high accuracy and robust generalization performance through voting or averaging [159]. It has been shown to predict tumor recurrence risk and contribute to cancer classification effectively. For instance, Zhang et al. used RF in conjunction with EV-miRNA signatures (miR-1246, miR-375, miR-221, miR-21) detected by the NgCHA nanoprobe assay to evaluate the recurrence risk in breast cancer patients and guide personalized treatment strategies. The RF-based risk assessment achieved an overall accuracy of 87% in the training cohort and 82% in the validation cohort, with EV-miR-1246 exhibiting significant influence. The model also accurately differentiated between different tumor types, with optimal accuracy observed in identifying breast and lung cancers. In comparison to other machine learning algorithms such as NN, SVM, and LDA, RF outperformed with a classification diagnosis accuracy of 58% in the validation cohort [100].
Convolutional neural network
CNN is an advanced deep-learning algorithm that specializes in image analysis, a subfield of machine learning known for its robust auto-learning capabilities, making it particularly advantageous for analyzing intricate samples [160, 161]. Jalali et al. used a CNN model to train the spectral fingerprint output from their MoSERS microarrays for distinguishing glioblastoma mutations. By collecting 946 SERS fingerprints of glioma cell EVs and dividing them into a training dataset (70% of the total data) and a test dataset (30% of the total data), the findings showed that CNN accurately classified individual EV spectra of different mutation types, achieving an overall classification accuracy of 89.3%. Moreover, CNN achieved an accuracy of 0.85 in the classification of circulating blood EVs containing various glioblastoma variants (EGFR amplification, EGFRvIII, and MGMT methylation) while also achieving an AUC value of 0.91 in discriminating between patients with GBM with genetic variants and healthy donors. Additionally, the MoSERS validation using binary CNN algorithms for EV segmentation showcased the predictive efficacy in identifying GBM variants (AUC=0.89), enhancing diagnostic precision of GBM mutations and presenting a promising analytical approach [154].
Machine learning algorithms for assisted in situ EV nucleic acid analysis employed in liquid biopsies. The incorporation of four machine learning algorithms, including linear discriminant analysis (LDA), random forest (RF), convolutional neural network (CNN), and t-distributed stochastic neighbor embedding (t-SNE), enhances the procedures involved in tumor diagnosis, differentiation, prognostic evaluation, and mutation identification utilizing the in situ EV nucleic acid analysis platform. Specifically, t-SNE enables visualization by reducing the dimensionality of raw data, and subsequently integrates with other algorithms to aid in distinguishing data characteristics, thereby enhancing the diagnostic efficacy of the model.
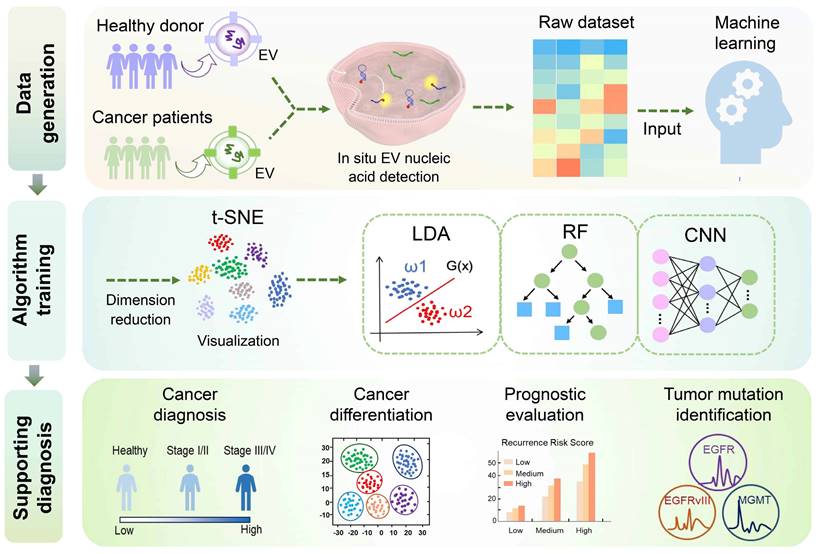
T-distributed stochastic neighbor embedding
t-SNE stands out as a cutting-edge machine learning algorithm designed for dimensionality reduction. This method effectively transforms high-dimensional datasets into two-dimensional or three-dimensional representations for visualization, exhibiting superior dimensionality reduction compared to PCA [162]. While the t-SNE algorithm excels at preserving the local structures with complex data sets but lacks predictive capabilities, it is often combined with complementary algorithms to enhance model performance. For example, the collaborative use of t-SNE and RF algorithms has demonstrated notable efficacy in distinguishing between various types of cancer. Zhang et al. prioritized the use of the t-SNE algorithm to map the properties of EV-miRNA panels in the training cohort to a two-dimensional plane for visualization in verifying the application potential of their constructed NgCHA in situ detection platform. Subsequently, the RF algorithm was utilized to differentiate the EV-miRNA features for identifying breast, lung, liver, gastric, and colorectal cancers [100]. In addition, the t-SNE enables data classification post-dimensionality reduction. In the context of in situ detection of EV biomarkers utilizing the SORTER platform, researchers observed no significant correlation among multiple miRNAs associated with distinct EV subgroups. Therefore, they applied the t-SNE to distinguish between prostate cancer and benign prostatic hyperplasia across different EV subtypes and employed LDA to identify EV-miRNA profiles to improve the diagnostic performance of the platform [158].
The application of machine learning in EV liquid biopsy has opened up a new chapter in cancer diagnosis assistance, particularly in addressing the heterogeneity of EVs for cancer classification. In addition to the four ML algorithms described above, SVM, KNN, and other deep learning algorithms are expected to be utilized in the future for assisted diagnosis relying on in situ EV nucleic acid analysis. It is important to note that machine learning models require sufficient data for effective training. Limited clinical samples in current studies may reduce the model's capacity and distort the predicted outcomes. The choice of interpretable algorithms to help physicians understand the decision-making process also needs to be considered. Therefore, further improvements are needed to enhance the accuracy of these models.
Application of in situ EV nucleic acid detection in liquid biopsy
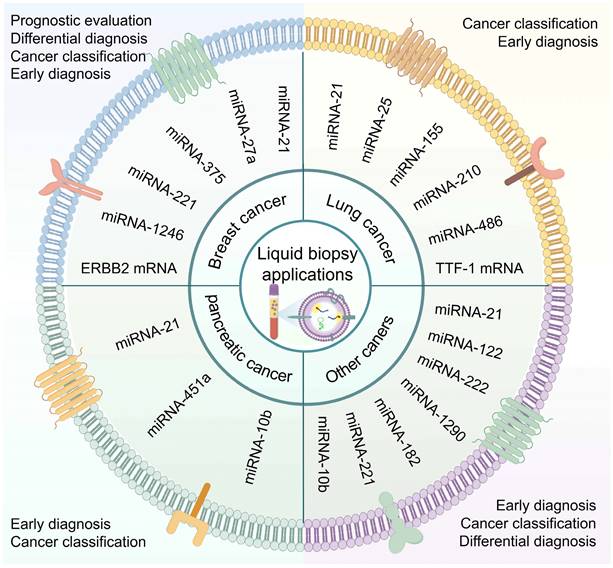
EV-based liquid biopsies have made significant progress, with certain biomarkers advancing to clinical trials [163-165]. However, detecting these biomarkers is often time-consuming, labor-intensive, and expensive. The development of in situ EV nucleic acid analysis technology has gained attention for its simplicity, speed, and affordability. Here, we illustrate the application of in situ EV nucleic acid analysis methods in liquid biopsies (Figure 6) and summarize their diagnostic performance in different types of cancer (Table 2).
Application of in situ EV nucleic acid detection in liquid biopsies. In situ detection of EV nucleic acids shows promising prospects in early diagnosis, differential diagnosis, cancer classification, and prognostic evaluation in cancer. These assays have been validated across various types of cancer, including breast cancer, lung cancer, pancreatic cancer, and other cancers (hepatocellular carcinoma, colorectal cancer, prostate cancer), with EV miRNAs derived from EVs in circulating blood serving as the primary biomarkers. (Created with Figdraw.com).
Breast cancer
Globally, early detection and treatment of breast cancer (BC) are crucial in reducing morbidity and mortality rates among women [1]. Research has identified miR-21 and miR-375 as up-regulation biomarkers in various cancers, including BC, with implications for tumor progression and prognosis [167-171]. In situ EV biomarker identification in BC has primarily focused on miR-21, followed by miR-375. For instance, Liu et al. developed a DNA cube-based DDCA nano platform capable of sensitive in situ detection of EV miR-21 in BC within 30 min. Clinical validation demonstrated higher FRET signals in BC patients compared to healthy individuals, as well as in advanced-stage patients compared to early-stage patients. This suggests the potential application of DDCA for early diagnosis of BC [103]. In estrogen receptor (ER)-positive BC, the most prevalent subtype with a better prognosis, miR-375 plays a regulatory role. Zhao et al. reported the use of a thermophoretic sensor (TSN) utilizing nanoflares detecting breast EV miRNA. TSN achieved high sensitivity, with the ability to analyze miRNAs from small serum samples. The clinical application showed that EV miR-375 exhibited 90% accuracy in distinguishing between ER-positive BC patients and healthy donors, with an impressive AUC value of 0.96. Moreover, EV miR-375 demonstrated good sensitivity, specificity, accuracy, and AUC value of 0.94 for early-stage (stage I and II) detection of ER-positive BC, providing a potential tool for improving BC diagnosis [110]. Additionally, liposome-based MFS-CRISPR platform, DNA-functionalized AuNPs probes, and multi-branched localized catalytic hairpin assembly (MLCHA) probes have shown promising diagnostic capabilities in discriminating BC patients from healthy donors [115, 123, 166].
Individual biomarker assays often lack accuracy and sensitivity. However, combining multiple biomarkers has significant advantages in precise cancer classification. Wang et al. developed a Y-scaffold probe that can detect multiple EV miRNAs (miR-21, miR-375, miR-27a) simultaneously in situ using competitive strand replacement. This approach showed higher miRNA fluorescence intensity in BC serum EVs compared to healthy controls, effectively distinguishing BC patients from healthy donors [121]. Recently, NgCHA nanoprobes based on DNA nanowire-guided catalyzed hairpin assembly have also been used for BC diagnosis and recurrence risk assessment. Zhang et al. employed a panel of 4-EV-miRNAs (miR-221, miR-375, miR-1246, and miR-21) as potential biomarkers for NgCHA detection, and analysis of 36 clinical samples revealed significant variations in the expression levels of these four EV-miRNAs among patients. Discriminating BC from healthy donors, the combination of these four EV-miRNAs demonstrated the highest accuracy with an AUC of 0.945, 95% sensitivity, and 95% specificity in the early stages of BC. Moreover, EV-miRNA-1246 played a crucial role in recurrence risk assessment modeling [100].
Lung cancer
Liquid biopsy plays a crucial role in the diagnosis and management of lung cancer (LC), which is a prevalent malignancy [172]. One effective approach is utilizing DNA tetrahedrons as nanocarriers to transport hairpin probes into EVs. Chen et al. employed this method, known as fLIGHT, to detect EV miR-21. The nanoprobe demonstrated comparable performance to RT-qPCR in differentiating non-small cell lung cancer (NSCLC) patients from healthy individuals. The fLIGHT assay exhibited a strong correlation with RT-qPCR, suggesting its suitability for EV miRNA quantification [102]. In addition, microfluidic-based biochips offer fast and sensitive detection of EV RNA. In one study, a microfluidic cationic lipid complex nanoparticle (mCLN)-based assay successfully detected miR-21 and TTF-1 mRNA in serum EVs, effectively distinguishing NSCLC patients from normal controls. Compared to the conventional qRT-PCR, the mCLN assay required a smaller sample volume (30ul vs. 100ul) and significantly reduced the processing time (10 minutes vs. 4 hours), demonstrating superior diagnostic accuracy in NSCLC [129]. Moreover, a study utilizing a tCLN biochip validated the diagnostic value of five miRNAs (miR-21, miR-25, miR-155, miR-210, miR-486) in serum EVs from both early-and late-stage NSCLC patients. The tCLN assay exhibited excellent diagnostic performance when utilizing the combined panel of five miRNA in differentiating normal controls from all NSCLC patients. Notably, the tCLN assay showed absolute sensitivity and specificity in differentiating early-stage NSCLC patients from normal controls, highlighting its potential as a liquid biopsy assay for early NSCLC detection [114].
Pancreatic cancer
MiR-10b is a commonly utilized diagnostic and prognostic biomarker for pancreatic ductal adenocarcinoma (PDAC) detected in EVs [173]. Researchers focused on validating miR-10b as a target for in situ EV assays. Jiang et al. developed a SERS-based sensing strategy, incorporating an LNA-Au@DTNB label into serum and enriching EVs using Fe3O4@TiO2. The Raman assay demonstrated significantly higher miRNA-10b signals in the sera of PDAC patients, with an AUC of 0.996 for distinguishing PDAC from healthy controls. This was highly correlated (0.996) with qRT-PCR results [133]. In addition, a comprehensive cancer diagnostic platform was designed to analyze multiple biomarkers at the single EV level by using a 3D microfluidic device, six EV markers (CD81, EphA2, CA199, miR-451a, miR-21, miR-10b) were simultaneously identified in 40 plasma specimens. The combined expression of CD81 (EphA2, miRNA-451a, miRNA-21, miRNA-10b) showed the best diagnostic value (AUC=1). Distinctly, EphA2, miRNA-451a, and miRNA-21 profiles could differentiate between healthy controls and early (stage I/II) pancreatic cancer patients. Moreover, miR-451a and miR-10b signatures were effective in discriminating between early-stage (I/II) and advanced-stage (III/IV) pancreatic cancer, indicating the strong potential of this platform for early cancer diagnosis [131].
Other cancer (Hepatocellular carcinoma, Colorectal cancer, Prostate cancer)
In addition to the mentioned cancer types, in situ detection techniques have been used to diagnose other malignancies. For example, in hepatocellular carcinoma (HCC), an entropy-driven catalysis (EDC)-enhanced DNA Logical device based on AuNPs was employed to detect serum EV miR-21 and miR-122. Simultaneous detection of these two EV miRNAs effectively distinguished HCC patients from healthy individuals with an accuracy rate of 93.3% [107]. In colorectal cancer (CRC), Xia et al. employed a BP@Mn2+/DNA nanosensor capable of differentiating EV miR-21 in plasma samples of CRC patients from healthy individuals. The sensor integrated an EpCAM aptamer to recognize CRC-specific EVs [120]. Additionally, Lei et al. designed a SORTER assay for identifying tumor-derived EVs and analyzing miRNAs to improve diagnostic accuracy in prostate cancer (PCa). By detecting six specific miRNAs (miR-222, miR-1290, miR-182, miR-21, miR-221, and miR-10b) in EVs, SORTER achieved a flawless discrimination between PCa and benign prostatic hyperplasia (BPH) with an impressive accuracy rate of 100%. Moreover, it achieved a diagnostic accuracy of 90.6% in distinguishing metastatic from non-metastatic PCa, demonstrating the potential of miRNA-based liquid biopsy in clinical settings [158].
In conclusion, in situ EV analysis shows promise for early cancer diagnosis, cancer classification, differential diagnosis, and predicting recurrence risks. However, most detection methodologies primarily focus on established miRNAs as target markers, often using breast cancer models and overlooking the exploration of novel biomarkers in various circulating body fluids. Larger clinical sample sizes are needed to ensure the reliability of the results. Looking ahead, these detection platforms have the potential to expand their applications, including disease screening, prognosis determination, and monitoring treatment responses in the future.
The significance of in situ extracellular vesicle nucleic acid analysis in liquid biopsy of cancer.
| Biomarker | Sample | Test Methods | Cases of tumors | Linear range | LOD | Time | AUC | Sample volume | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| miR-21, miR-122, miR-375 | BC, LC, HCC, CRC, CC derived plasma | Fluorescence | 64 | 0.0195 fM-19.5 pM | 5 aM | 2h | 0.429, 0.561, 0.556 (combined= 0.944) | 500ul plasma | [108] |
| miRNA-21 | BC derived serum | Fluorescence | 22 | 2.5×105-1.5×107 particles/ul | 9.8×104 particles/ul | 30min | / | / | [103] |
| miR-21, miR-122 | HCC derived serum | Fluorescence | 15 | 2.4×105-1.7×106 particles/ul | 1.2×105 particles/ul | 3.5h | 0.92 | / | [107] |
| miR-375 | BC derived serum | Fluorescence | 15 | / | 6.53×103 particles/ul | 3h | 0.92 | / | [123] |
| miR-375, miR-1246, miR-221, miR-21 | BC derived plasma | Fluorescence | 21 | 1 pM-50 nM | 0.8 pM | 30 min | 0.756, 0.894, 0.810, 0.875 (combined=0.965) | 1ml plasma | [100] |
| miRNA-27a | MCF-7-tumor-bearing mice derived plasma | Fluorescence | 10 | 0.76×106-15.24×106 particles/ul | 1.9×104 particles/ul | 3.5h | 0.95 | 5ul plasma | [166] |
| miR-21, miR-25, miR-155, miR-210, miR-486 | NSCLC derived serum | Fluorescence | 64 | / | / | 2.5h | 0.97 | 60ul resum | [114] |
| miR-21, miR-27a, miR-375 | BC derived serum | Fluorescence | 3 | / | 0.116 μg/mL, 125 μg/mL, 0.287 μg/mL | 30min | / | / | [121] |
| miR-21 | BC derived serum | Fluorescence | 4 | 1.25-200 nM | 0.5 nM | 30min | / | 5ml serum | [116] |
| miR-21 | BC derived plasma | Fluorescence | 10 | 101-105 particles/ul | 1.2 particles/ul | 3.75h | 0.84 | 500ul plasma | [115] |
| miR-21 | Melanoma, CC, BC derived serum | Fluorescence | 3 | 3×104-107 particles/ μL | 378 copies/μL | 1h | / | / | [144] |
| miR-21- 5p | NSCLC derived serum | Fluorescence | 10 | 1×10-12-10×10-9 M | 45.4×10-15 M | 20min | / | / | [102] |
| miR-21 | BC derived serum | Fluorescence | 5 | / | / | 2h | / | / | [117] |
| miR-21 | CRC derived plasma | Fluorescence | 10 | 5×102-2×104 particles/μl | 1.86×102 particles/ul | 30min | / | 200ul plasma | [120] |
| miR-375 | BC derived serum | Fluorescence | 17 | 0.17-170 fM | 0.36 fM | 2h | 0.96 | 0.5ul serum | [110] |
| miR-451a, miR-21, miR-10b | PC derived plasma | Microfluidic chip and fluorescence | 30 | / | 0.71×10-9 M, 1.74×10-9 M, 1.28×10-9 M | 1h | 0.930, 0.939, 0.875 | 2ul plasma | [131] |
| miR-222, miR-1290, miR-182, miR-21, miR-221, miR-10b | PCa derived plasma | Microfluidic chip and fluorescence | 47 | / | 0.8×105 particles/ul | 2h | combined=1 | 0.2ul plasma | [158] |
| miRNA-10b | PDAC derived serum | SERS | 15 | 0.33 fM-1.65 pM | 0.21 fM | 4h | 0.996 | 4ul serum | [133] |
| miR-21, TTF-1 mRNA | NSCLC derived serum | Microfluidic chip and fluorescence | 10 | 8×104-8×107 particles/ul | 3.71×106 particles/ul, 2.06×106 particles/ul | 10min | / | 30ul serum | [129] |
| ERBB2 | MCF-7-tumor-bearing mice derived plasma | Microfluidic chip and fluorescence | 8 | 100 fM-1 μM | 58.3 fM | 2h | / | 400ul plasma | [130] |
BC: breast cancer; LC: lung cancer; HCC: hepatocellular carcinoma; CRC: colorectal cancer; CC: cervical carcinoma; NSCLC: non-small cell lung cancer; PC: pancreatic cancer; PCa: prostate cancer; PDAC: pancreatic ductal adenocarcinoma.
Challenges and perspectives
EVs have gained significant attention in liquid biopsy due to their abundance, stability, and diverse information reflecting disease progression in bodily fluids. These vesicles carry various biomarkers, with RNA-based molecules, particularly miRNAs, playing a crucial role. miRNAs, enriched in EVs and characterized by short sequences, show promise in early cancer diagnosis, prognostic assessment, and progress monitoring. Although other biomarkers have shown improved diagnostic capabilities, the utilization of DNA, mRNA, and lncRNA is less common due to EV volume limitation. CircRNAs, known for their superior stability and tissue specificity, are highly abundant in EVs, making them a potential focus for liquid biopsy strategies [73]. Despite these advancements, clinical implementation of EV biomarkers still faces challenges, such as the lack of validated clinical samples, suboptimal diagnostic sensitivity and specificity, impurities in EV isolation, and limited sensitivity and reproducibility of detection methodologies. Most of the EV biomarkers studied so far have been identified in sEVs or exosomes present in different circulating body fluids. However, nucleic acid biomarkers derived from microvesicles, apoptotic vesicles, and platelet vesicles have not yet been detected in situ.
Numerous methodologies for EV nucleic acid detection have been extensively studied. However, these methods require demanding procedures involving EV lysis and nucleic acid extraction. Therefore, our attention is directed towards approaches for in situ EV nucleic acid analysis and adjunctive diagnostic strategies that combine single EV analysis and machine learning, particularly focusing on their prospective utility in liquid biopsy. These innovative tools have significant potential in contemporary liquid biopsies for rapid and direct acquisition of information on EV biomarkers for tumor diagnosis, classification, and prognostic evaluation. The goal of in situ EV nucleic acid analysis is to develop sensitive, precise, stable, and high-throughput assay platforms integrated with single EV analysis to identify tumor-specific EVs and implement them effectively in liquid biopsies. Nevertheless, several challenges need to be addressed in the future. The following insights emphasize the unaddressed needs of these strategies in the context of diagnostic applications for in situ nucleic acid analysis of EVs.
1. The advancement of fluorescence-based in situ EV nucleic acid detection has made rapid progress. DNA nanostructured and AuNPs probes can directly penetrate EVs without requiring membrane treatment. However, further refinement is needed to enhance EV penetration efficiency and increase the stability of the assembled probes. Moreover, fluorescence detection is prone to interference from impurities. So, it is advisable to minimize self-induced fluorescence by using ratiometric fluorescence detection. Furthermore, the effectiveness of this approach should be confirmed by employing conventional detection techniques.
2. In situ EV nucleic acid detection requires high sensitivity. Current signal amplification strategies, such as CHA and CHR, involve the use of multiple probes. Therefore, it is imperative to explore methods that enable efficient integration of more probes into EVs while facilitating EV translocation. Additionally, future advancements are expected to introduce novel nanomaterials for application in in situ detection.
3. Microfluidic chips integrating fluorescence strategies demonstrate excellent performance in terms of high throughput, rapid detection, and minimal sample consumption. However, there is a need to enhance their sensitivity and stability. In addition to the promising SERS assays, it is expected that in situ analysis strategies for EV nucleic acids employing electrochemical, colorimetric, and surface plasmon resonance (SPR) methods will be developed and applied in liquid biopsies.
4. Single EV analysis is valuable for comprehending cancer heterogeneity and pinpointing diagnostic biomarkers. However, current platforms for directly analyzing nucleic acids at the single EV level, such as digital PCR, nano-flow cytometry, and TIRFM, are expensive and face challenges in clinical adoption. Microfluidic devices based on SERS exhibit promising potential for application. It is crucial to characterize multiple biomarkers at the single EV level to improve diagnostic accuracy.
5. The machine learning methods (LDA, RF, CNN, t-SNE) employed for assisting in situ EV nucleic acid detection demonstrate excellent analytical capabilities in discriminating between cancer and healthy donors and categorizing different cancer types, thereby enhancing diagnostic accuracy. However, the small sample size of the proposed detection method may pose a challenge and require algorithm optimization or integration with other algorithms for more realistic outcomes. Training before validation is advisable to enhance the algorithm's learning capacity and assess accuracy effectively. Additionally, simultaneous detection of multiple biomarkers combined with machine learning enables fast and accurate cancer diagnosis. Integrating microfluidic-based high-throughput methods with machine learning presents a promising strategy.
6. In situ EV nucleic acid assays promise to advance liquid biopsy technology, but factors such as sample volume and assay time hinder clinical implementation. Techniques to identify various biomarkers and the integration of multiple biomarkers can facilitate the precise characterization of tumor-derived EVs for accurate diagnosis. Additionally, the in situ detection of DNA mutation and novel biomarkers (e.g., circRNA, lncRNA) will offer potential for comprehensive integration into future liquid biopsy applications.
In conclusion, we emphasize the importance of in situ EV nucleic acid-based detection techniques in tumor liquid biopsy. Integration of solutions for single EV analysis and machine learning is expected to enhance in situ detection methods, enabling swift and effective precision medicine practices.
Abbreviations
EV: extracellular vesicle; SERS: surface-enhanced Raman scattering; SPR: surface plasmon resonance; M2ф: M2-like macrophages; PDAC: pancreatic ductal adenocarcinoma; CAF: Cancer-associated fibroblast; Treg: regulatory T cell; DC: Dendritic cell; M1ф: M1-like macrophages; CRC: colorectal cancer; GC: gastric cancer; HCC: hepatocellular carcinoma; mtDNA: mitochondrial DNA; NSCLC: non-small cell lung cancer; EC: endometrial cancer; SCLC: small-cell lung cancer; PCa: prostate cancer; MB: molecular beacon; SLO: streptolysin O; AuNP: gold nanoparticle; Vir-FV: virus-mimicking fusogenic vesicle; BP: black phosphorus; ddPCR: droplet digital PCR; TIRFM: total internal reflection fluorescence microscopy; GBM: glioblastoma; LDA: linear discriminant analysis; RF: random forest; CNN: convolutional neural network; t-SNE: t-distributed stochastic neighbor embedding; BC: breast cancer; LC: lung cancer.
Acknowledgements
This work was supported by the Jiangsu Province Science and Technology Plan Key Projects (BE2020680), the Technology Development Project of Suzhou (SKY2021018), the Innovation Foundation for Medicine and Education Synergy of Jiangsu University (JDY2022020), the Technology Project of Zhangjiagang (ZKHBZ2216).
Author contributions
Dongli Wang: Writing - original draft, Conceptualization, Methodology, Investigation, Visualization. Ye Shen: Writing - original draft, Methodology, Investigation. Hui Qian: Methodology, Visualization. Jiajia Jiang: Conceptualization, Funding acquisition, Project administration, Supervision. Wenrong Xu: Writing - review & editing, Funding acquisition, Project administration, Supervision.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I. et al. Global cancer statistics 2022: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229-263
2. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18:297-312
3. Jeppesen DK, Zhang Q, Franklin JL, Coffey RJ. Extracellular vesicles and nanoparticles: emerging complexities. Trends Cell Biol. 2023;33:667-681
4. Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ. et al. Reassessment of exosome composition. Cell. 2019;177:428-445.e418
5. Arya SB, Collie SP, Parent CA. The ins-and-outs of exosome biogenesis, secretion, and internalization. Trends Cell Biol. 2024;34:90-108
6. Isaac R, Reis FCG, Ying W, Olefsky JM. Exosomes as mediators of intercellular crosstalk in metabolism. Cell Metab. 2021;33:1744-1762
7. Kalluri R, Lebleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977
8. Han QF, Li WJ, Hu KS, Gao J, Zhai WL, Yang JH. et al. Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer. Mol Cancer. 2022;21:207
9. Yu D, Li Y, Wang M, Gu J, Xu W, Cai H. et al. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer. 2022;21:56
10. Lee H, He X, Le T, Carnino JM, Jin Y. Single-step RT-qPCR for detection of extracellular vesicle microRNAs in vivo: a time- and cost-effective method. Am J Physiol Lung Cell Mol Physiol. 2020;318:L742-L749
11. Deng Y, Sun Z, Wang L, Wang M, Yang J, Li G. Biosensor-based assay of exosome biomarker for early diagnosis of cancer. Front Med. 2022;16:157-175
12. Zhang Y, Li W, Ji T, Luo S, Qiu J, Situ B. et al. Localized DNA tetrahedrons assisted catalytic hairpin assembly for the rapid and sensitive profiling of small extracellular vesicle-associated microRNAs. J Nanobiotechnology. 2022;20:503
13. Kang T, Zhu J, Luo X, Jia W, Wu P, Cai C. Controlled self-assembly of a close-packed gold octahedra array for SERS sensing exosomal microRNAs. Anal Chem. 2021;93:2519-2526
14. Xiao PP, Wan QQ, Liao T, Tu JY, Zhang GJ, Sun ZY. Peptide nucleic acid-functionalized nanochannel biosensor for the highly sensitive detection of tumor exosomal microRNA. Anal Chem. 2021;93:10966-10973
15. Wu W, Yu X, Wu J, Wu T, Fan Y, Chen W. et al. Surface plasmon resonance imaging-based biosensor for multiplex and ultrasensitive detection of NSCLC-associated exosomal miRNAs using DNA programmed heterostructure of Au-on-Ag. Biosens Bioelectron. 2021;175:112835
16. Sun Z, Wu Y, Gao F, Li H, Wang C, Du L. et al. In situ detection of exosomal RNAs for cancer diagnosis. Acta Biomater. 2023;155:80-98
17. Xu L, Shoaie N, Jahanpeyma F, Zhao J, Azimzadeh M, Al Jamal KT. Optical, electrochemical and electrical (nano)biosensors for detection of exosomes: a comprehensive overview. Biosens Bioelectron. 2020;161:112222
18. Amrollahi P, Zheng W, Monk C, Li CZ, Hu TY. Nanoplasmonic sensor approaches for sensitive detection of disease-associated exosomes. ACS Appl Bio Mater. 2021;4:6589-6603
19. Khaksari S, Abnous K, Hadizadeh F, Ramezani M, Taghdisi SM, Mousavi Shaegh SA. Signal amplification strategies in biosensing of extracellular vesicles (EVs). Talanta. 2023;256:124244
20. Shen J, Ma Z, Xu J, Xue T, Lv X, Zhu G. et al. Exosome isolation and detection: from microfluidic chips to nanoplasmonic biosensor. ACS Appl Mater. Interfaces. 2024;16:22776-22793
21. Ma X, Hao Y, Liu L. Progress in nanomaterials-based optical and electrochemical methods for the assays of exosomes. Int J Nanomedicine. 2021;16:7575-7608
22. Sun Z, Zhang B, Tu H, Pan C, Chai Y, Chen W. Advances in colorimetric biosensors of exosomes: novel approaches based on natural enzymes and nanozymes. Nanoscale. 2024;16:1005-1024
23. Zhang J, Wu J, Wang G, He L, Zheng Z, Wu M. et al. Extracellular vesicles: techniques and biomedical applications related to single vesicle analysis. ACS Nano. 2023;17:17668-17698
24. Morales RT, Ko J. Future of digital assays to resolve clinical heterogeneity of single extracellular vesicles. ACS Nano. 2022;16:11619-11645
25. Welsh JA, Goberdhan DCI, O'driscoll L, Buzas EI, Blenkiron C, Bussolati B. et al. Minimal information for studies of extracellular vesicles (MISEV2023): from basic to advanced approaches. J Extracell Vesicles. 2024;13:e12404
26. Clancy JW, Boomgarden AC, D'souza-Schorey C. Profiling and promise of supermeres. Nat Cell Biol. 2021;23:1217-1219
27. Zhang Q, Jeppesen DK, Higginbotham JN, Graves-Deal R, Trinh VQ, Ramirez MA. et al. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat Cell Biol. 2021;23:1240-1254
28. Van De Wakker SI, Meijers FM, Sluijter JPG, Vader P. Extracellular vesicle heterogeneity and its impact for regenerative medicine applications. Pharmacol Rev. 2023;75:1043-1061
29. Sun F, Sun Y, Wang X, Zhu J, Chen S, Yu Y. et al. Engineered mesenchymal stem cell-derived small extracellular vesicles for diabetic retinopathy therapy through HIF-1α/EZH2/PGC-1α pathway. Bioact Mater. 2024;33:444-459
30. Wang M, Yang D, Li L, Wu P, Sun Y, Zhang X. et al. A dual role of mesenchymal stem cell derived small extracellular vesicles on TRPC6 protein and mitochondria to promote diabetic wound healing. ACS Nano. 2024;18:4871-4885
31. Ji C, Zhang J, Zhu Y, Shi H, Yin S, Sun F. et al. Exosomes derived from hucMSC attenuate renal fibrosis through CK1δ/β-TRCP-mediated YAP degradation. Cell Death Dis. 2020;11:327
32. Chen SW, Zhu SQ, Pei X, Qiu BQ, Xiong D, Long X. et al. Cancer cell-derived exosomal circUSP7 induces CD8(+) T cell dysfunction and anti-PD1 resistance by regulating the miR-934/SHP2 axis in NSCLC. Mol Cancer. 2021;20:144
33. Sun H, Wang C, Hu B, Gao X, Zou T, Luo Q. et al. Exosomal S100A4 derived from highly metastatic hepatocellular carcinoma cells promotes metastasis by activating STAT3. Signal Transduct Target Ther. 2021;6:187
34. Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T. et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539-555
35. Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y. et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol. 2020;13:156
36. Paskeh MDA, Entezari M, Mirzaei S, Zabolian A, Saleki H, Naghdi MJ. et al. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J Hematol Oncol. 2022;15:83
37. Yang Y, Guo Z, Chen W, Wang X, Cao M, Han X. et al. M2 macrophage-derived exosomes promote angiogenesis and growth of pancreatic ductal adenocarcinoma by targeting E2F2. Mol Ther. 2021;29:1226-1238
38. Tyagi A, Wu SY, Sharma S, Wu K, Zhao D, Deshpande R. et al. Exosomal miR-4466 from nicotine-activated neutrophils promotes tumor cell stemness and metabolism in lung cancer metastasis. Oncogene. 2022;41:3079-3092
39. Zhao J, Shen J, Mao L, Yang T, Liu J, Hongbin S. Cancer associated fibroblast secreted miR-432-5p targets CHAC1 to inhibit ferroptosis and promote acquired chemoresistance in prostate cancer. Oncogene. 2024;43:2104-2114
40. Ni C, Fang QQ, Chen WZ, Jiang JX, Jiang Z, Ye J. et al. Breast cancer-derived exosomes transmit lncRNA SNHG16 to induce CD73+gammadelta1 Treg cells. Signal Transduct Target Ther. 2020;5:41
41. Du Z, Huang Z, Chen X, Jiang G, Peng Y, Feng W. et al. Modified dendritic cell-derived exosomes activate both NK cells and T cells through the NKG2D/NKG2D-L pathway to kill CML cells with or without T315I mutation. Exp Hematol Oncol. 2022;11:36
42. Shin S, Jung I, Jung D, Kim CS, Kang SM, Ryu S. et al. Novel antitumor therapeutic strategy using CD4(+) T cell-derived extracellular vesicles. Biomaterials. 2022;289:121765
43. Hashemi ZS, Ghavami M, Kiaie SH, Mohammadi F, Barough MS, Khalili S. et al. Novel delivery of sorafenib by natural killer cell-derived exosomes-enhanced apoptosis in triple-negative breast cancer. Nanomedicine. 2023;18:437-453
44. Choo YW, Kang M, Kim HY, Han J, Kang S, Lee JR. et al. M1 macrophage-derived nanovesicles potentiate the anticancer efficacy of immune checkpoint inhibitors. ACS Nano. 2018;12:8977-8993
45. Wu P, Zhang B, Ocansey DKW, Xu W, Qian H. Extracellular vesicles: a bright star of nanomedicine. Biomaterials. 2021;269:120467
46. Guo Z, Zhang Y, Xu W, Zhang X, Jiang J. Engineered exosome-mediated delivery of circDIDO1 inhibits gastric cancer progression via regulation of miR-1307-3p/SOCS2 axis. J Transl Med. 2022;20:326
47. Zhang J, Ji C, Zhang H, Shi H, Mao F, Qian H. et al. Engineered neutrophil-derived exosome-like vesicles for targeted cancer therapy. Sci Adv. 2022;8:eabj8207
48. Zhang X, Zhang H, Gu J, Zhang J, Shi H, Qian H. et al. Engineered extracellular vesicles for cancer therapy. Adv Mater. 2021;33:e2005709
49. Yu W, Hurley J, Roberts D, Chakrabortty SK, Enderle D, Noerholm M. et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann Oncol. 2021;32:466-477
50. Sharma A, Johnson A. Exosome DNA: critical regulator of tumor immunity and a diagnostic biomarker. J Cell Physiol. 2020;235:1921-1932
51. Lucchetti D, Zurlo IV, Colella F, Ricciardi-Tenore C, Di Salvatore M, Tortora G. et al. Mutational status of plasma exosomal KRAS predicts outcome in patients with metastatic colorectal cancer. Sci Rep. 2021;11:22686
52. Choi J, Cho HY, Jeon J, Kim KA, Han YD, Ahn JB. et al. Detection of circulating KRAS mutant DNA in extracellular vesicles using droplet digital PCR in patients with colon cancer. Front Oncol. 2022;12:1067210
53. Li Y, Wu J, Li E, Xiao Z, Lei J, Zhou F. et al. TP53 mutation detected in circulating exosomal DNA is associated with prognosis of patients with hepatocellular carcinoma. Cancer Biol Ther. 2022;23:439-445
54. Batool SM, Muralidharan K, Hsia T, Falotico S, Gamblin AS, Rosenfeld YB. et al. Highly sensitive EGFRvIII detection in circulating extracellular vesicle RNA of glioma patients. Clin Cancer Res. 2022;28:4070-4082
55. Zocco D, Bernardi S, Novelli M, Astrua C, Fava P, Zarovni N. et al. Isolation of extracellular vesicles improves the detection of mutant DNA from plasma of metastatic melanoma patients. Sci Rep. 2020;10:15745
56. Afrifa J, Zhao T, Yu J. Circulating mitochondria DNA, a non-invasive cancer diagnostic biomarker candidate. Mitochondrion. 2019;47:238-243
57. Lou C, Ma X, Chen Z, Zhao Y, Yao Q, Zhou C. et al. The mtDNA fragments within exosomes might be novel diagnostic biomarkers of non-small cell lung cancer. Pathol Res Pract. 2023;249:154718
58. Liu C, Li B, Lin H, Yang C, Guo J, Cui B. et al. Multiplexed analysis of small extracellular vesicle-derived mRNAs by droplet digital PCR and machine learning improves breast cancer diagnosis. Biosens Bioelectron. 2021;194:113615
59. He X, Tian F, Guo F, Zhang F, Zhang H, Ji J. et al. Circulating exosomal mRNA signatures for the early diagnosis of clear cell renal cell carcinoma. BMC Med. 2022;20:270
60. Han Y, Drobisch P, Kruger A, William D, Grutzmann K, Bothig L. et al. Plasma extracellular vesicle messenger RNA profiling identifies prognostic EV signature for non-invasive risk stratification for survival prediction of patients with pancreatic ductal adenocarcinoma. J Hematol Oncol. 2023;16:7
61. Diener C, Keller A, Meese E. Emerging concepts of miRNA therapeutics: from cells to clinic. Trends Genet. 2022;38:613-626
62. Pontecorvi G, Bellenghi M, Puglisi R, Care A, Mattia G. Tumor-derived extracellular vesicles and microRNAs: functional roles, diagnostic, prognostic and therapeutic options. Cytokine Growth Factor Rev. 2020;51:75-83
63. Zhou L, Wang W, Wang F, Yang S, Hu J, Lu B. et al. Plasma-derived exosomal miR-15a-5p as a promising diagnostic biomarker for early detection of endometrial carcinoma. Mol Cancer. 2021;20:57
64. Kim DH, Park H, Choi YJ, Im K, Lee CW, Kim DS. et al. Identification of exosomal microRNA panel as diagnostic and prognostic biomarker for small cell lung cancer. Biomark Res. 2023;11:80
65. Shin S, Park YH, Jung SH, Jang SH, Kim MY, Lee JY. et al. Urinary exosome microRNA signatures as a noninvasive prognostic biomarker for prostate cancer. NPJ Genom Med. 2021;6:45
66. Miyazaki K, Wada Y, Okuno K, Murano T, Morine Y, Ikemoto T. et al. An exosome-based liquid biopsy signature for pre-operative identification of lymph node metastasis in patients with pathological high-risk T1 colorectal cancer. Mol Cancer. 2023;22:2
67. Ahmad M, Weiswald LB, Poulain L, Denoyelle C, Meryet-Figuiere M. Involvement of lncRNAs in cancer cells migration, invasion and metastasis: cytoskeleton and ECM crosstalk. J Exp Clin. Cancer Res. 2023;42:173
68. Nie H, Liao Z, Wang Y, Zhou J, He X, Ou C. Exosomal long non-coding RNAs: emerging players in cancer metastasis and potential diagnostic biomarkers for personalized oncology. Genes Dis. 2021;8:769-780
69. Guo X, Lv X, Ru Y, Zhou F, Wang N, Xi H. et al. Circulating exosomal gastric cancer-associated long noncoding RNA1 as a biomarker for early detection and monitoring progression of gastric cancer: a multiphase study. JAMA Surg. 2020;155:572-579
70. Guo X, Gao Y, Song Q, Wei J, Wu J, Dong J. et al. Early assessment of circulating exosomal lncRNA-GC1 for monitoring neoadjuvant chemotherapy response in gastric cancer. Int J Surg. 2023;109:1094-1104
71. Li C, Lv Y, Shao C, Chen C, Zhang T, Wei Y. et al. Tumor-derived exosomal lncRNA GAS5 as a biomarker for early-stage non-small-cell lung cancer diagnosis. J Cell Physiol. 2019;234:20721-20727
72. Misir S, Wu N, Yang BB. Specific expression and functions of circular RNAs. Cell Death Differ. 2022;29:481-491
73. Zhang F, Jiang J, Qian H, Yan Y, Xu W. Exosomal circRNA: emerging insights into cancer progression and clinical application potential. J Hematol Oncol. 2023;16:67
74. Wang D, Li R, Jiang J, Qian H, Xu W. Exosomal circRNAs: novel biomarkers and therapeutic targets for gastrointestinal tumors. Biomed Pharmacother. 2023;157:114053
75. Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981-984
76. Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F. et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19:112
77. Deng C, Huo M, Chu H, Zhuang X, Deng G, Li W. et al. Exosome circATP8A1 induces macrophage M2 polarization by regulating the miR-1-3p/STAT6 axis to promote gastric cancer progression. Mol Cancer. 2024;23:49
78. He YD, Tao W, He T, Wang BY, Tang XM, Zhang LM. et al. A urine extracellular vesicle circRNA classifier for detection of high-grade prostate cancer in patients with prostate-specific antigen 2-10 ng/mL at initial biopsy. Mol Cancer. 2021;20:96
79. Wen N, Peng D, Xiong X, Liu G, Nie G, Wang Y. et al. Cholangiocarcinoma combined with biliary obstruction: an exosomal circRNA signature for diagnosis and early recurrence monitoring. Signal Transduct Target Ther. 2024;9:107
80. Fu M, Gu J, Wang M, Zhang J, Chen Y, Jiang P. et al. Emerging roles of tRNA-derived fragments in cancer. Mol Cancer. 2023;22:30
81. Wang X, Ramat A, Simonelig M, Liu MF. Emerging roles and functional mechanisms of PIWI-interacting RNAs. Nat Rev Mol Cell Biol. 2023;24:123-141
82. Zheng B, Song X, Wang L, Zhang Y, Tang Y, Wang S. et al. Plasma exosomal tRNA-derived fragments as diagnostic biomarkers in non-small cell lung cancer. Front Oncol. 2022;12:1037523
83. Gu X, Wang C, Deng H, Qing C, Liu R, Liu S. et al. Exosomal piRNA profiling revealed unique circulating piRNA signatures of cholangiocarcinoma and gallbladder carcinoma. Acta Biochim Biophys Sin. 2020;52:475-484
84. Guo T, Tang XH, Gao XY, Zhou Y, Jin B, Deng ZQ. et al. A liquid biopsy signature of circulating exosome-derived mRNAs, miRNAs and lncRNAs predict therapeutic efficacy to neoadjuvant chemotherapy in patients with advanced gastric cancer. Mol Cancer. 2022;21:216
85. Russano M, Napolitano A, Ribelli G, Iuliani M, Simonetti S, Citarella F. et al. Liquid biopsy and tumor heterogeneity in metastatic solid tumors: the potentiality of blood samples. J Exp Clin Cancer Res. 2020;39:95
86. Long F, Ma H, Hao Y, Tian L, Li Y, Li B. et al. A novel exosome-derived prognostic signature and risk stratification for breast cancer based on multi-omics and systematic biological heterogeneity. Comput Struct Biotechnol J. 2023;21:3010-3023
87. Xiong H, Huang Z, Yang Z, Lin Q, Yang B, Fang X. et al. Recent progress in detection and profiling of cancer cell-derived exosomes. Small. 2021;17:e2007971
88. Shao B, Xiao Z. Recent achievements in exosomal biomarkers detection by nanomaterials-based optical biosensors - a review. Anal Chim Acta. 2020;1114:74-84
89. Dou WT, Han HH, Sedgwick AC, Zhu GB, Zang Y, Yang XR. et al. Fluorescent probes for the detection of disease-associated biomarkers. Sci Bull. 2022;67:853-878
90. Qu B, Sun J, Li P, Jing L. Current advances on g-C3N4-based fluorescence detection for environmental contaminants. J Hazard Mater. 2022;425:127990
91. Chen Z, Ma J, Sun DW. Aggregates-based fluorescence sensing technology for food hazard detection: Principles, improvement strategies, and applications. Compr Rev Food Sci Food Saf. 2023;22:2977-3010
92. Zheng J, Yang R, Shi M, Wu C, Fang X, Li Y. et al. Rationally designed molecular beacons for bioanalytical and biomedical applications. Chem Soc Rev. 2015;44:3036-3055
93. Wu Q, Wang H, Gong K, Shang J, Liu X, Wang F. Construction of an autonomous nonlinear hybridization chain reaction for extracellular vesicles-associated microRNAs discrimination. Anal Chem. 2019;91:10172-10179
94. Cho S, Yang HC, Rhee WJ. Simultaneous multiplexed detection of exosomal microRNAs and surface proteins for prostate cancer diagnosis. Biosens Bioelectron. 2019;146:111749
95. Ma Y, Wang X, Liu H, Wei L, Xiao L. Recent advances in optical microscopic methods for single-particle tracking in biological samples. Anal Bioanal Chem. 2019;411:4445-4463
96. Hu Q, Li H, Wang L, Gu H, Fan C. DNA nanotechnology-enabled drug delivery systems. Chem Rev. 2019;119:6459-6506
97. Zhang Y, Pan V, Li X, Yang X, Li H, Wang P. et al. Dynamic DNA structures. Small. 2019;15:e1900228
98. Shen L, Wang P, Ke Y. DNA nanotechnology-based biosensors and therapeutics. Adv Healthc Mater. 2021;10:e2002205
99. Zhang C, Belwal T, Luo Z, Su B, Lin X. Application of nanomaterials in isothermal nucleic acid amplification. Small. 2022;18:e2102711
100. Zhang Y, Wu Y, Luo S, Yang C, Zhong G, Huang G. et al. DNA nanowire guided-catalyzed hairpin assembly nanoprobe for in situ profiling of circulating extracellular vesicle-associated microRNAs. ACS Sens. 2022;7:1075-1085
101. Zhang T, Tian T, Lin Y. Functionalizing framework nucleic-acid-based nanostructures for biomedical application. Adv Mater. 2022;34:e2107820
102. Chen X, Jia M, Liu L, Qiu X, Zhang H, Yu X. et al. High-fidelity determination and tracing of small extracellular vesicle cargoes. Small. 2020;16:e2002800
103. Chen J, Xie M, Shi M, Yuan K, Wu Y, Meng HM. et al. Spatial confinement-derived double-accelerated DNA cascade reaction for ultrafast and highly sensitive in situ monitoring of exosomal miRNA and exosome tracing. Anal Chem. 2022;94:2227-2235
104. Mao D, Zheng M, Li W, Xu Y, Wang C, Qian Q. et al. Cubic DNA nanocage-based three-dimensional molecular beacon for accurate detection of exosomal miRNAs in confined spaces. Biosens Bioelectron. 2022;204:114077
105. Ramalingam V. Multifunctionality of gold nanoparticles: plausible and convincing properties. Adv Colloid Interface Sci. 2019;271:101989
106. Lee KX, Shameli K, Yew YP, Teow SY, Jahangirian H, Rafiee-Moghaddam R. et al. Recent developments in the facile bio-synthesis of gold nanoparticles (AuNPs) and their biomedical applications. Int J Nanomedicine. 2020;15:275-300
107. Liu B, Zhao D, Chen J, Shi M, Yuan K, Sun H. et al. DNA logical device combining an entropy-driven catalytic amplification strategy for the simultaneous detection of exosomal multiplex miRNAs in situ. Anal Chem. 2024;96:1733-1741
108. Zhang XW, Du L, Liu MX, Wang JH, Chen S, Yu YL. All-in-one nanoflare biosensor combined with catalyzed hairpin assembly amplification for in situ and sensitive exosomal miRNA detection and cancer classification. Talanta. 2024;266:125145
109. Deng J, Zhao S, Li J, Cheng Y, Liu C, Liu Z. et al. One-step thermophoretic AND gate operation on extracellular vesicles improves diagnosis of prostate cancer. Angew Chem Int Ed Engl. 2022;61:e202207037
110. Zhao J, Liu C, Li Y, Ma Y, Deng J, Li L. et al. Thermophoretic detection of exosomal microRNAs by nanoflares. J Am Chem Soc. 2020;142:4996-5001
111. Fan M, Han Y, Gao S, Yan H, Cao L, Li Z. et al. Ultrasmall gold nanoparticles in cancer diagnosis and therapy. Theranostics. 2020;10:4944-4957
112. Guimarães D, Cavaco-Paulo A, Nogueira E. Design of liposomes as drug delivery system for therapeutic applications. Int J Pharm. 2021;601:120571
113. Herrmann IK, Wood MJA, Fuhrmann G. Extracellular vesicles as a next-generation drug delivery platform. Nat Nanotechnol. 2021;16:748-759
114. Liu C, Kannisto E, Yu G, Yang Y, Reid ME, Patnaik SK. et al. Non-invasive detection of exosomal microRNAs via tethered cationic lipoplex nanoparticles (tCLN) biochip for lung cancer early detection. Front Genet. 2020;11:258
115. Zhang J, Guan M, Ma C, Liu Y, Lv M, Zhang Z. et al. Highly effective detection of exosomal miRNAs in plasma using liposome-mediated transfection CRISPR/Cas13a. ACS Sens. 2023;8:565-575
116. Xie M, Wang F, Yang J, Guo Y, Ding F, Lu X. et al. DNA zipper mediated membrane fusion for rapid exosomal miRNA detection. Anal Chem. 2022;94:13043-13051
117. Gao X, Li S, Ding F, Fan H, Shi L, Zhu L. et al. Rapid detection of exosomal microRNAs using virus-mimicking fusogenic vesicles. Angew Chem Int Ed Engl. 2019;58:8719-8723
118. Zeng G, Chen Y. Surface modification of black phosphorus-based nanomaterials in biomedical applications: strategies and recent advances. Acta Biomater. 2020;118:1-17
119. Jiang H, Xia Q, Zheng J, Bu J, Li R, Cai Z. et al. Mn2+ modified black phosphorus nanosheets with enhanced DNA adsorption and affinity for robust sensing. Biosens Bioelectron. 2022;216:114622
120. Xia Q, Zheng J, Bu J, Li R, Li X, Fan S. et al. Mn2+-modified black phosphorus nanosensor for detection of exosomal microRNAs and exosomes. Mikrochim Acta. 2023;190:295
121. Wang H, He D, Wan K, Sheng X, Cheng H, Huang J. et al. In situ multiplex detection of serum exosomal microRNAs using an all-in-one biosensor for breast cancer diagnosis. Analyst. 2020;145:3289-3296
122. Zuo H, Mao C. A minimalist's approach for DNA nanoconstructions. Adv Drug Deliv Rev. 2019;147:22-28
123. Shi MQ, Wang X, Wu YA, Yuan K, Li ZJ, Meng HM. et al. Exploring the size of DNA functionalized gold nanoparticles for high efficiency exosome uptake and sensitive biosensing. Sens Actuators B Chem. 2022;355:131315
124. Ma X, Li X, Luo G, Jiao J. DNA-functionalized gold nanoparticles: modification, characterization, and biomedical applications. Front Chem. 2022;10:1095488
125. Ayuso JM, Virumbrales-Muñoz M, Lang JM, Beebe DJ. A role for microfluidic systems in precision medicine. Nat Commun. 2022;13:3086
126. Jia Y, Yu L, Ma T, Xu W, Qian H, Sun Y. et al. Small extracellular vesicles isolation and separation: current techniques, pending questions and clinical applications. Theranostics. 2022;12:6548-6575
127. Lin B, Lei Y, Wang J, Zhu L, Wu Y, Zhang H. et al. Microfluidic-based exosome analysis for liquid biopsy. Small Methods. 2021;5:e2001131
128. Qian CG, Xiao YJ, Wang J, Li YW, Li SJ, Wei B. et al. Rapid exosomes concentration and detection of exosomal microRNA on agarose-based microfluidic chip. Sens Actuators B Chem. 2021;333:129559
129. Yang Y, Kannisto E, Patnaik SK, Reid ME, Li L, Wu Y. Ultrafast detection of exosomal RNAs via cationic lipoplex nanoparticles in a micromixer biochip for cancer diagnosis. ACS Appl Nano Mater. 2021;4:2806-2819
130. Lim J, Kang B, Son HY, Mun B, Huh YM, Rho HW. et al. Microfluidic device for one-step detection of breast cancer-derived exosomal mRNA in blood using signal-amplifiable 3D nanostructure. Biosens Bioelectron. 2022;197:113753
131. Zhou S, Hu T, Han G, Wu Y, Hua X, Su J. et al. Accurate cancer diagnosis and stage monitoring enabled by comprehensive profiling of different types of exosomal biomarkers: surface proteins and miRNAs. Small. 2020;16:e2004492
132. Liu H, Gao X, Xu C, Liu D. SERS tags for biomedical detection and bioimaging. Theranostics. 2022;12:1870-1903
133. Jiang S, Li Q, Wang C, Pang Y, Sun Z, Xiao R. In situ exosomal microRNA determination by target-triggered SERS and Fe3O4@TiO2-based exosome accumulation. ACS Sens. 2021;6:852-862
134. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487-514
135. Ko J, Wang Y, Carlson JCT, Marquard A, Gungabeesoon J, Charest A. et al. Single extracellular vesicle protein analysis using immuno-droplet digital polymerase chain reaction amplification. Adv Biosyst. 2020;4:e1900307
136. Yang Z, Atiyas Y, Shen H, Siedlik MJ, Wu J, Beard K. et al. Ultrasensitive single extracellular vesicle detection using high throughput droplet digital enzyme-linked immunosorbent assay. Nano Lett. 2022;22:4315-4324
137. Xia Z, Zhang X, Yao J, Liu Z, Jin Y, Yin H. et al. Giant enhancement of raman scattering by a hollow-core microstructured optical fiber allows single exosome probing. ACS Sens. 2023;8:1799-1809
138. Görgens A, Bremer M, Ferrer-Tur R, Murke F, Tertel T, Horn PA. et al. Optimisation of imaging flow cytometry for the analysis of single extracellular vesicles by using fluorescence-tagged vesicles as biological reference material. J Extracell Vesicles. 2019;8:1587567
139. Amrhein K, Taylor ML, Wilson R, Gallops CE, Annamer A, Vinduska V. et al. Dual imaging single vesicle surface protein profiling and early cancer detection. ACS Appl Mater Interfaces. 2023;15:2679-2692
140. Olmedillas-López S, Olivera-Salazar R, García-Arranz M, García-Olmo D. Current and emerging applications of droplet digital PCR in oncology: an updated review. Mol Diagn Ther. 2022;26:61-87
141. Pasini L, Notarangelo M, Vagheggini A, Burgio MA, Crino L, Chiadini E. et al. Unveiling mutational dynamics in non-small cell lung cancer patients by quantitative EGFR profiling in vesicular RNA. Mol Oncol. 2021;15:2423-2438
142. Jiao Y, Gao L, Zhang T, He Z, Zheng SY, Liu W. Profiling DNA cargos in single extracellular vesicles via hydrogel-based droplet digital multiple displacement amplification. Anal Chem. 2024;96:1293-1300
143. Colson L, Kwon Y, Nam S, Bhandari A, Maya NM, Lu Y. et al. Trends in single-molecule total internal reflection fluorescence imaging and their biological applications with lab-on-a-chip technology. Sensors (Basel). 2023;23:7691
144. He D, Wang H, Ho SL, Chan HN, Hai L, He X. et al. Total internal reflection-based single-vesicle in situ quantitative and stoichiometric analysis of tumor-derived exosomal microRNAs for diagnosis and treatment monitoring. Theranostics. 2019;9:4494-4507
145. Zhou J, Wu Z, Hu J, Yang D, Chen X, Wang Q. et al. High-throughput single-EV liquid biopsy: rapid, simultaneous, and multiplexed detection of nucleic acids, proteins, and their combinations. Sci Adv. 2020;6:eabc1204
146. Zhang J, Rima XY, Wang X, Nguyen LTH, Huntoon K, Ma Y. et al. Engineering a tunable micropattern-array assay to sort single extracellular vesicles and particles to detect RNA and protein in situ. J Extracell Vesicles. 2023;12:e12369
147. Ricklefs FL, Maire CL, Reimer R, Dührsen L, Kolbe K, Holz M. et al. Imaging flow cytometry facilitates multiparametric characterization of extracellular vesicles in malignant brain tumours. J Extracell Vesicles. 2019;8:1588555
148. De Rond L, Van Der Pol E, Bloemen PR, Van Den Broeck T, Monheim L, Nieuwland R. et al. A systematic approach to improve scatter sensitivity of a flow cytometer for detection of extracellular vesicles. Cytometry A. 2020;97:582-591
149. Choi D, Montermini L, Jeong H, Sharma S, Meehan B, Rak J. Mapping subpopulations of cancer cell-derived extracellular vesicles and particles by nano-flow cytometry. ACS Nano. 2019;13:10499-10511
150. Oliveira GP Jr, Zigon E, Rogers G, Davodian D, Lu S, Jovanovic-Talisman T. et al. Detection of extracellular vesicle RNA using molecular beacons. iScience. 2020;23:100782
151. Liu H, Tian Y, Xue C, Niu Q, Chen C, Yan X. Analysis of extracellular vesicle DNA at the single-vesicle level by nano-flow cytometry. J Extracell Vesicles. 2022;11:e12206
152. Li J, Li Y, Li P, Zhang Y, Du L, Wang Y. et al. Exosome detection via surface-enhanced Raman spectroscopy for cancer diagnosis. Acta Biomater. 2022;144:1-14
153. Carmicheal J, Hayashi C, Huang X, Liu L, Lu Y, Krasnoslobodtsev A. et al. Label-free characterization of exosome via surface enhanced Raman spectroscopy for the early detection of pancreatic cancer. Nanomedicine. 2019;16:88-96
154. Jalali M, Del Real Mata C, Montermini L, Jeanne O, I IH, Gu Z. et al. MoS2-plasmonic nanocavities for Raman spectra of single extracellular vesicles reveal molecular progression in glioblastoma. ACS Nano. 2023;17:12052-12071
155. Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019;20:e262-e273
156. Swanson K, Wu E, Zhang A, Alizadeh AA, Zou J. From patterns to patients: advances in clinical machine learning for cancer diagnosis, prognosis, and treatment. Cell. 2023;186:1772-1791
157. Xu L, Raitoharju J, Iosifidis A, Gabbouj M. Saliency-based multilabel linear discriminant analysis. IEEE Trans Cybern. 2022;52:10200-10213
158. Lei Y, Fei X, Ding Y, Zhang J, Zhang G, Dong L. et al. Simultaneous subset tracing and miRNA profiling of tumor-derived exosomes via dual-surface-protein orthogonal barcoding. Sci Adv. 2023;9:eadi1556
159. Hu J, Szymczak S. A review on longitudinal data analysis with random forest. Brief Bioinform. 2023;24:bbad002
160. Li Z, Liu F, Yang W, Peng S, Zhou J. A survey of convolutional neural networks: analysis, applications, and prospects. IEEE Trans Neural Netw Learn Syst. 2022;33:6999-7019
161. Cui F, Yue Y, Zhang Y, Zhang Z, Zhou HS. Advancing biosensors with machine learning. ACS Sens. 2020;5:3346-3364
162. Wang Z, Xie J, Nie F, Wang R, Jia Y, Liu S. T-distributed stochastic neighbor network for unsupervised representation learning. Neural Netw. 2024;179:106520
163. Tutrone R, Donovan MJ, Torkler P, Tadigotla V, Mclain T, Noerholm M. et al. Clinical utility of the exosome based ExoDx prostate(intelliscore) EPI test in men presenting for initial biopsy with a PSA 2-10 ng/mL. Prostate Cancer Prostatic Dis. 2020;23:607-614
164. Tutrone R, Lowentritt B, Neuman B, Donovan MJ, Hallmark E, Cole TJ. et al. ExoDx prostate test as a predictor of outcomes of high-grade prostate cancer - an interim analysis. Prostate Cancer Prostatic Dis. 2023;26:596-601
165. Liu C, Hu C, Chen T, Jiang Y, Zhang X, Liu H. et al. The role of plasma exosomal lnc-SNAPC5-3:4 in monitoring the efficacy of anlotinib in the treatment of advanced non-small cell lung cancer. J Cancer Res Clin Oncol. 2022;148:2867-2879
166. Wu T, Liu X, Chen H, Liu Y, Cao Y. An in situ exosomal miRNA sensing biochip based on multi-branched localized catalytic hairpin assembly and photonic crystals. Biosens Bioelectron. 2023;222:115013
167. Bautista-Sánchez D, Arriaga-Canon C, Pedroza-Torres A, De La Rosa-Velázquez IA, González-Barrios R, Contreras-Espinosa L. et al. The promising role of miR-21 as a cancer biomarker and its importance in RNA-based therapeutics. Mol Ther Nucleic Acids. 2020;20:409-420
168. Yuan X, Qian N, Ling S, Li Y, Sun W, Li J. et al. Breast cancer exosomes contribute to pre-metastatic niche formation and promote bone metastasis of tumor cells. Theranostics. 2021;11:1429-1445
169. Wang H, Tan Z, Hu H, Liu H, Wu T, Zheng C. et al. microRNA-21 promotes breast cancer proliferation and metastasis by targeting LZTFL1. BMC Cancer. 2019;19:738
170. Zhao Q, Liu Y, Wang T, Yang Y, Ni H, Liu H. et al. MiR-375 inhibits the stemness of breast cancer cells by blocking the JAK2/STAT3 signaling. Eur J Pharmacol. 2020;884:173359
171. Liu Y, Wang Q, Wen J, Wu Y, Man C. MiR-375: a novel multifunctional regulator. Life Sci. 2021;275:119323
172. Li W, Liu JB, Hou LK, Yu F, Zhang J, Wu W. et al. Liquid biopsy in lung cancer: significance in diagnostics, prediction, and treatment monitoring. Mol Cancer. 2022;21:25
173. Jia E, Ren N, Shi X, Zhang R, Yu H, Yu F. et al. Extracellular vesicle biomarkers for pancreatic cancer diagnosis: a systematic review and meta-analysis. BMC Cancer. 2022;22:573
Author biographies
 Dongli Wang is currently a Ph. D. student under the supervision of Prof. Wenrong Xu. Her research interests include digging for tumor-derived extracellular vesicle biomarkers and exploring the mechanisms of their involvement in tumor progression.
Dongli Wang is currently a Ph. D. student under the supervision of Prof. Wenrong Xu. Her research interests include digging for tumor-derived extracellular vesicle biomarkers and exploring the mechanisms of their involvement in tumor progression.
 Ye Shen is a general surgeon at Aoyang Hospital, affiliated with Jiangsu University, and graduated from the Medical School of Nantong University with an M.S. degree. His research interest focuses on the diagnostic potential and role of extracellular vesicles in gastrointestinal tumors.
Ye Shen is a general surgeon at Aoyang Hospital, affiliated with Jiangsu University, and graduated from the Medical School of Nantong University with an M.S. degree. His research interest focuses on the diagnostic potential and role of extracellular vesicles in gastrointestinal tumors.
 Dr. Hui Qian is currently the dean of the School of Medicine at Jiangsu University and a full professor. Her current research interests focus on the role of stem cell-derived exosomes in the treatment of diabetic kidney disease and the construction of engineered exosomes. She was honored as an Elsevier China Highly Cited Researcher from 2020 to 2023.
Dr. Hui Qian is currently the dean of the School of Medicine at Jiangsu University and a full professor. Her current research interests focus on the role of stem cell-derived exosomes in the treatment of diabetic kidney disease and the construction of engineered exosomes. She was honored as an Elsevier China Highly Cited Researcher from 2020 to 2023.
 Dr. Jiajia Jiang received her Ph.D. degree from Jiangsu University. She is currently the director of the Aoyang Institute of Cancer, Affiliated Aoyang Hospital of Jiangsu University. Her primary research interests are extracellular vesicle-based tumor molecular biology and cancer therapeutics.
Dr. Jiajia Jiang received her Ph.D. degree from Jiangsu University. She is currently the director of the Aoyang Institute of Cancer, Affiliated Aoyang Hospital of Jiangsu University. Her primary research interests are extracellular vesicle-based tumor molecular biology and cancer therapeutics.
 Dr. Wenrong Xu is a second-grade professor at Jiangsu University, as well as a member of the Society of Medical Laboratory Science of the Chinese Medical Association. His research revolves around stem cells and tumor-associated extracellular vesicles in various complications of diabetes, spinal cord injuries, and treatment of tumors. He was honored as an Elsevier China Highly Cited Researcher from 2020 to 2023. Besides, he has now published more than 200 scientific papers with a total of more than 9,000 citations.
Dr. Wenrong Xu is a second-grade professor at Jiangsu University, as well as a member of the Society of Medical Laboratory Science of the Chinese Medical Association. His research revolves around stem cells and tumor-associated extracellular vesicles in various complications of diabetes, spinal cord injuries, and treatment of tumors. He was honored as an Elsevier China Highly Cited Researcher from 2020 to 2023. Besides, he has now published more than 200 scientific papers with a total of more than 9,000 citations.
![]() Corresponding authors: Jiajia Jiang, jiangjiajia_2001com; Wenrong Xu, iclsedu.cn.
Corresponding authors: Jiajia Jiang, jiangjiajia_2001com; Wenrong Xu, iclsedu.cn.
 Global reach, higher impact
Global reach, higher impact