13.3
Impact Factor
Theranostics 2024; 14(8):3127-3149. doi:10.7150/thno.97162 This issue Cite
Review
Distinctive tumorigenic significance and innovative oncology targets of SUMOylation
1. Department of Surgical Oncology and General Surgery, The First Hospital of China Medical University, Shenyang, Liaoning 110001, China; Key Laboratory of Molecular Pathology and Epidemiology of Gastric Cancer in the Universities of Liaoning Province, Shenyang, Liaoning 110001, China.
2. Department of Anesthesiology, The First Hospital of China Medical University, Shenyang, Liaoning 110001, China.
3. Department of Hematology, The Fourth Affiliated Hospital of China Medical University, Shenyang, Liaoning 110001, China.
4. Department of Cell Biology, Key Laboratory of Cell Biology, National Health Commission of the PRC and Key Laboratory of Medical Cell Biology, Ministry of Education of the PRC, China Medical University, Shenyang, Liaoning 110122, China.
5. Scientific Experimental Center, School of Pharmacy, China Medical University, Shenyang, Liaoning 110122, China.
6. Department of Gynecology and Obstetrics, Shengjing Hospital of China Medical University, Shenyang, Liaoning 110001, China.
7. Department of Pharmacology, School of Pharmacy, China Medical University, Shenyang, Liaoning 110122, China.
8. Liaoning Key Laboratory of Molecular Targeted Anti-Tumor Drug Development and Evaluation; Liaoning Cancer Immune Peptide Drug Engineering Technology Research Center; Key Laboratory of Precision Diagnosis and Treatment of Gastrointestinal Tumors, Ministry of Education; China Medical University, Shenyang, Liaoning 110122, China.
9. Shenyang Kangwei Medical Laboratory Analysis Co. LTD, Liaoning Province, China.
#These authors contributed equally to this work: Heng Zhou, Na Deng, Yanshu Li, Xiaoyun Hu.
Received 2024-4-10; Accepted 2024-5-13; Published 2024-5-19
Abstract
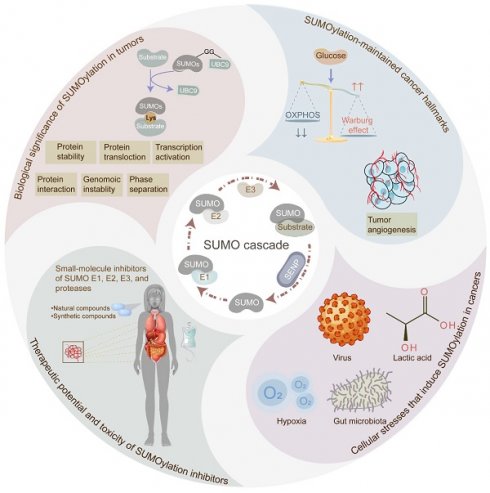
Protein SUMOylation, a post-translational modification, intricately regulates diverse biological processes including gene expression, cell cycle progression, signaling pathway transduction, DNA damage response, and RNA metabolism. This modification contributes to the acquisition of tumorigenicity and the maintenance of cancer hallmarks. In malignancies, protein SUMOylation is triggered by various cellular stresses, promoting tumor initiation and progression. This augmentation is orchestrated through its specific regulatory mechanisms and characteristic biological functions. This review focuses on elucidating the fundamental regulatory mechanisms and pathological functions of the SUMO pathway in tumor pathogenesis and malignant evolution, with particular emphasis on the tumorigenic potential of SUMOylation. Furthermore, we underscore the potential therapeutic benefits of targeting the SUMO pathway, paving the way for innovative anti-tumor strategies by perturbing this dynamic and reversible modifying process.
Keywords: Cancer, SUMOylation, Post-translational modification, Cancer hallmarks, Cancer therapy
Introduction
SUMOylation, identified in the 1990s, is a dynamic and reversible protein post-translational modification [1]. In a nutshell, this process is a multi-step enzymatic cascade catalyzing the covalent attachment of small ubiquitin-like modifier (SUMO) proteins to specific lysine (K) residues of substrate proteins [2]. Typically, SUMOylation and ubiquitination compete for substrate proteins; however, unlike ubiquitination, which often targets proteins for degradation, SUMOylation primarily maintains protein stability. Dysregulation of SUMOylation has been validated across more than 10 types of tumors, including colorectal cancer (CRC) [3], lung cancer [4], and hepatic cancer [5]. In most cases, SUMOylation exerts an oncogenic effect in cancers by modulating critical biological processes such as transcriptional regulation, protein-protein interaction, protein translocation, phase separation, and protein stability. Consequently, SUMOylation influences significant biological processes, involving gene expression, cell cycle progression, signaling pathway transduction, DNA damage response, and RNA metabolism, thus contributing to tumorigenicity and sustaining cancer hallmarks such as tumor invasion, metastasis, programmed cell death escape, metabolic reprogramming, tumor immune evasion, and epigenetic reprogramming. Additionally, diverse cellular stressors, such as hypoxia, viral infection, gut microbiota alterations, and lactic acid levels, contribute to the dysregulation of SUMOylation in tumor initiation and development. Furthermore, SUMOylation has been implicated in the development of multidrug resistance during tumor therapy [6]. Thus, targeting the enzymes involved in SUMOylation, through the development of pharmaceutical inhibitors comprising natural and synthetic compounds, emerges as a crucial avenue for anti-tumor strategies. This review provides a comprehensive overview of the process and biological significance of SUMOylation in cancer, elucidates the cancer hallmarks maintained by SUMOylation, explores the cellular stressors triggering SUMOylation, and discusses the potential of targeting SUMOylation for anti-tumor therapy. By shedding light on the oncogenic role of SUMOylation, this review contributes to the exploration of new therapeutic strategies in cancer treatment.
SUMOylation cascade and biological significance in tumors
Overview of the SUMOylation process
SUMOylation involves multiple proteins, including SUMOs, SUMO E1 activating enzymes, SUMO E2 conjugating enzyme, SUMO E3 ligases, and SUMO-specific proteases. Among mammals, the SUMO family comprises five members: SUMO1, SUMO2, SUMO3, SUMO4, and SUMO5. Among these, SUMO2 exhibits the highest abundance, compensating for the reduced levels of SUMO1 and SUMO3 [7, 8]. Notably, SUMO2 and SUMO3 share a striking similarity in amino acid sequences, exceeding 95%, and are collectively referred to as SUMO2/3. Currently, the functionality of SUMO4 in substrate protein conjugation remains debated. While one study suggests that the unique proline-90 residue of SUMO4 may hinder its maturation, potentially impeding substrate conjugation [9], subsequent research has failed to provide conclusive evidence for the functional significance of the proline-90 residue of SUMO4 [10]. Recently, SUMO5 has emerged as a novel poly-SUMO isoform implicated in SUMOylation, enhancing the conjugation capacity of SUMO2/3 in human cells [11]. However, its classification as a human SUMO pseudogene or a functional isoform necessitates further investigation.
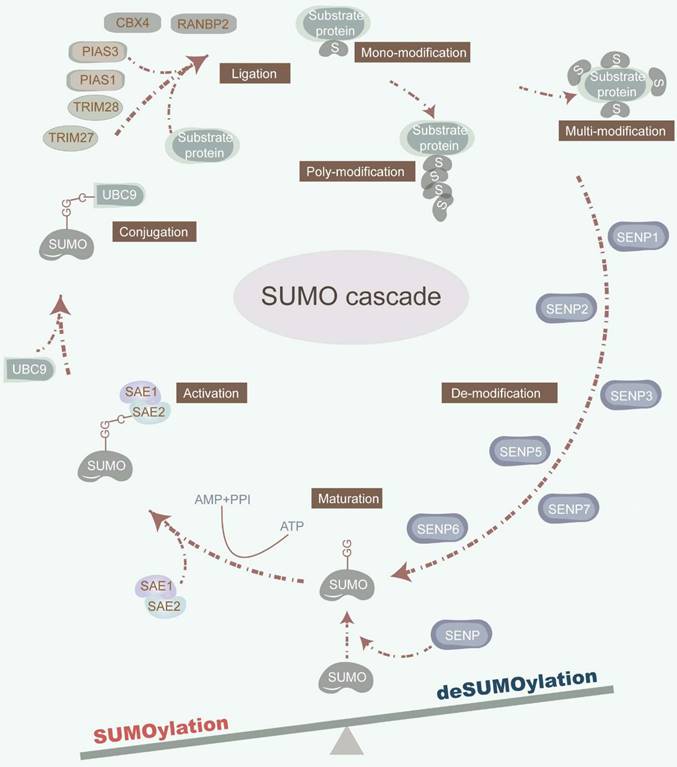
The dynamic and reversible nature of SUMOylation involves a series of enzymatic steps, including maturation, activation, conjugation, ligation, and deconjugation [2] (Figure 1). Mechanistically, sentrin-specific proteases (SENPs) catalyze the proteolytic cleavage of the C-terminal amino acids of inactive SUMO precursor proteins, exposing their diglycine (-GG) motif and promoting maturation. The dimeric SUMO-activating enzyme E1 (SAE1/2) interacts with the C-terminal of SUMO, facilitated by ATP hydrolysis. Activated SUMOs are then transferred to a cysteine residue of ubiquitin carrier protein 9 (UBC9), a SUMO E2 conjugating enzyme. Finally, with the assistance of UBC9 and SUMO E3 ligase, SUMOs are covalently attached to a lysine (K) within the SUMOylation consensus motif ψKx[E/D] (where ψ represents a hydrophobic residue; x denotes any amino acid; K signifies lysine; E refers to glutamic acid; D refers to aspartic acid) or in the reversed motif [E/D]xK of target proteins. Of note, SUMO E3 ligases are primarily composed of the protein inhibitors of activated signal transducer and activator of transcription (PIAS) with SP-RING and SP-CTD domains, bind to UBC9 or SUMO, respectively [12]. Furthermore, SUMO can non-covalently bind to proteins harboring SUMO-interacting motifs (SIM). The deconjugation of SUMOs is manipulated by SENPs, which also facilitate the maturation of SUMO precursors. Mammals possess six SENP family members, including SENP1-3 and SENP5-7, each exhibiting distinct activities toward individual SUMOs, which orchestrate the deSUMOylation by removing SUMOs from substrate proteins [13]. For instance, SENP1 displays heightened activity on SUMO1, while SENP2 prefers SUMO2 [14]. Furthermore, SENP3 plays a critical role in SUMO2/3 precursor maturation and deconjugation [15]. Additionally, SENP5 predominantly cleaves SUMO2, while SENP6 and SENP7 preferentially disassemble SUMO polymers [13] (Table 1).
Biologically significant manipulation by SUMOylation in tumors
In tumor cells, protein SUMOylation orchestrates essential molecular processes, predominantly influencing protein stability, protein-protein interaction, protein translocation, phase separation, and transcriptional activity. It also participates in cell homeostasis disruption via aberrant gene expression induction, cell cycle progression, signaling pathway transduction, DNA damage response, and RNA metabolism (Figure 2).
The SUMOylation cascade. SUMOylation is a multi-step enzymatic cascade catalyzing SUMOs covalently conjugating to substrate proteins. This dynamic and reversible SUMOylation process consists of multi-step actions, involving maturation, activation, conjugation, ligation, and deconjugation. SENPs cleave the amino acids in the C-terminal of inactive precursors of SUMO proteins to expose their diglycine (-GG) motif. SAE1/2 combines its cysteine site with the C-terminal of SUMO under the premise that ATP hydrolysis provides energy. Under the synergy of UBC9 and SUMO E3 ligase, the SUMOs are covalently conjugated to a lysine (K) in the SUMOylation consensus motif ψKx[E/D]. SENPs also orchestrate the deSUMOylation by removing SUMOs from substrate proteins.
Protein stability
SUMO modifiers, which are analogous to ubiquitin, administer protein stability within tumors. Characteristically, SUMOylation blocks the ubiquitin-proteasome system (UPS)-induced protein degradation. Specifically, insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2), which is SUMOylated at K497, K505, and K509 by SUMO1, avoids degradation by UPS and enhances its protein stability, thereby facilitating vasculogenic mimicry in glioma [16]. Similarly, the SUMOylation of myeloid cell leukemia 1 (MCL1) at K234 and K238 increases its protein stability through the prevention of UPS-mediated MCL1 protein degradation, leading to the proliferation of cancer cells [17]. Moreover, SUMO1-mediated SUMOylation of RNA-binding protein Raly (RALY) at K175 improves its protein stability, thereby facilitating vasculogenic mimicry in glioma cells [18]. Additionally, the noncovalent attachment of SUMO-2/3 to the SIM (236-240aa) of 5-methylcytosine (m5C) RNA methyltransferase NOL1/NOP2/Sun domain family member 2 (NSUN2) enhances its protein stability and induces m5C modification, exacerbating gastric cancer [19]. Of note, SUMOylation can also facilitate protein degradation via the proteasome pathway in rare cases. For example, SUMO2-mediated SUMOylation of GLUT1 promotes the ubiquitination of GLUT1 in nasopharyngeal carcinoma [20]. Additionally, transcription factor CP2c, which is involved in human malignancies, is SUMOylated at its K50 and SIM158 in a SUMO1-dependent manner. The SUMOylated CP2c is subsequently decayed by the ubiquitin-independent PSME3/20S proteasome system [21]. These studies confirm the dual role of SUMOylation in maintaining protein stability.
Molecules and molecular characteristics in the SUMO pathway
| Subsets | Enzymatic activity | Molecules | Molecular weights | Cellular location |
|---|---|---|---|---|
| SUMO | None | SUMO1 | 11.56 kDa | Nucleus, cytoplasm |
| SUMO2 | 10.87 kDa | Nucleus | ||
| SUMO3 | 11.64 kDa | Cytoplasm, nucleus | ||
| SUMO4 | 10.65 kDa | Nucleus | ||
| SUMO5 | 11.53 kDa | Nucleus | ||
| E1 | Activating SUMO | SAE1 | 38.45 kDa | Nucleus |
| SAE2 | 71.22 kDa | Cytoplasm, nucleus | ||
| E2 | Conjugating SUMO | UBC9 | 18.00 kDa | Nucleus, cytoplasm |
| E3 | Ligating SUMO | SP-RING Family | ||
| PIAS1 | 71.84 kDa | Nucleus | ||
| PIAS2 | 68.24 kDa | Nucleus | ||
| PIAS3 | 68.02 kDa | Cytoplasm, nucleus | ||
| PIAS4 | 56.50 kDa | Nucleus | ||
| MMS21 | 27.93 kDa | Nucleus | ||
| SIM Family | ||||
| RANBP2 | 358.20 kDa | Nucleus | ||
| KIAA1586 | 89.67 kDa | Cytoplasm, nucleus | ||
| CBX4 | 61.37 kDa | Nucleus | ||
| ZNF451 | 121.48 kDa | Nucleus | ||
| SLX4 | 200.01 kDa | Nucleus | ||
| TRIM Family | ||||
| TRIM1 | 83.21 kDa | Cytoplasm | ||
| TRIM27 | 58.49 kDa | Nucleus, cytoplasm, mitochondrion | ||
| TRIM28 | 88.55 kDa | Nucleus | ||
| TRIM38 | 53.42 kDa | Cytoplasm | ||
| PML | 97.55 kDa | Nucleus, cytoplasm | ||
| SUMO protease | SUMO precursor | SENP1 | 73.48 kDa | Nucleus, cytoplasm |
| maturation, | SENP2 | 67.86 kDa | Nucleus, cytoplasm | |
| deconjugating | SENP3 | 65.01 kDa | Nucleus, cytoplasm | |
| SUMO | SENP5 | 86.69 kDa | Nucleus | |
| SENP6 | 126.15 kDa | Nucleus | ||
| SENP7 | 119.67 kDa | Cytoplasm | ||
Protein-protein interaction
SUMOylation enhances protein-protein interactions which are crucial for tumor initiation and progression. For instance, SUMO3-modified SUMOylation of E3 ubiquitin ligase RING finger protein 146 (RNF146) at K19, K61, K174, and K175 facilitates the association of RNF146 with axis inhibition protein 1 (Axin) to expedite the ubiquitination and degradation of Axin, resulting in the progression of hepatocellular carcinoma (HCC) [22]. Similarly, SUMO1-mediated SUMOylation of promyelocytic leukemia (PML) protein at K65, K160, and K490 increases its interaction with proto-oncogene c-Myc (c-Myc) and further stabilizes c-Myc protein, contributing to the deterioration of glioblastoma (GBM) malignancy [23]. Furthermore, SUMO-2/3 modification of Flotillin-1 (Flot-1) at K195 boosts its interaction with nuclear zinc finger protein SNAI1 (Snail) and inhibits Snail proteasomal degradation, thereby promoting epithelial-to-mesenchymal transition (EMT) of metastatic prostate cancer [24]. Additionally, SUMOylation of OTU domain-containing ubiquitin aldehyde-binding protein 2 (OTUB2) at K233 anchors it to yes-associated protein (YAP)/PDZ-binding motif (TAZ) via a “V/I-X-V/I-V/I” SIM in YAP and TAZ, contributing to tumor metastasis in a Hippo-independent manner [25].
Protein translocation
Proteins perform their specific biological functions in different cell sub-regions to help cells accurately respond to different physiological, pathological, or environmental stimuli. Strikingly, SUMOylation regulates the occurrence and development of tumors via altering the location of proteins. Concretely, in HCC, SUMO1-mediated SUMOylation via the IKII265-268 SIM site of pyruvate kinase M2 (PKM2) relocates PKM2 from the cytoplasm to the nucleus, leading to glycolytic reprogramming and cancer progression via EMT induction and signal transducer and activator of the transcription 3 (STAT3) signaling pathway [26]. Likewise, Ran-binding protein 2 (RanBP2)-mediated SUMOylation of interleukin-33 (IL-33) at K54 facilitates its nuclear shuttling, therefore resulting in immune evasion in HCC [27]. Furthermore, in prostate cancer, SUMOylation of tumor suppressor gene p53, which is mediated by the RanBP2/SUMO1/Ubc9 complex, facilitates its translocation from the nucleus to the cytoplasm, promoting malignancy progression [28]. Also in prostate cancer, SUMO-2 modification of extracellular-signal-related kinase 5 (ERK5) at K6 and K22 improves the ability of ERK5 to translocate to the nucleus, ultimately facilitating tumor cell proliferation [29]. Additionally, SUMOylation influences nuclear translocation by impacting protein phosphorylation. For instance, SUMO1-mediated p65 SUMOylation at K37, K122, K123, and K221 sites increases p65 phosphorylation at the S276 site, and further stimulates the nuclear import of p65 and NF-κB transcriptional activity, ultimately contributing to the malignant phenotype of liver cancer cells [30].
Biological significance manipulated by SUMOylation in tumors. Protein SUMOylation governs a spectrum of biological effects in tumors, involving protein stability, protein-protein interaction, protein translocation, phase separation, transcriptional activation, and genomic instability, by which it participates in the disruption of cell homeostasis via inducing aberrance of gene expression, cell cycle progression, signaling pathway transduction, DNA damage response, RNA metabolism, etc.
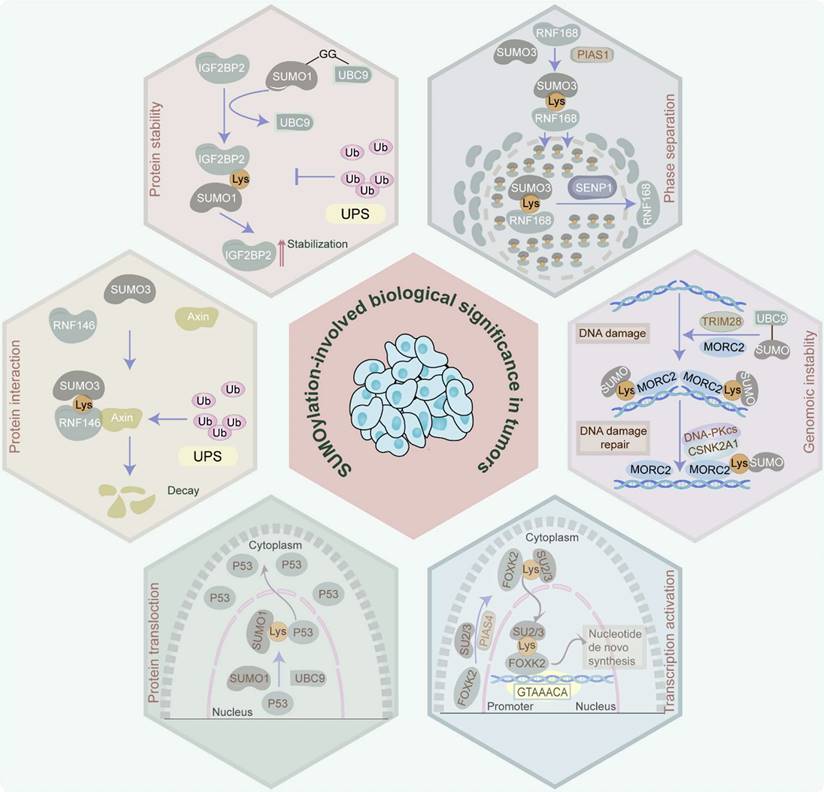
Phase separation
Liquid-liquid phase separation (LLPS) is essentially a physical and chemical phenomenon. In living cells, LLPS drives the inhomogeneous spatiotemporal coordination of individual molecules into membrane-free, droplet-like biomolecular condensates (BMCs) [31]. Notably, the establishment of LLPS plays a significant role in tumor initiation, progression, metastasis, and drug resistance. In fact, in vitro assay demonstrates that the rate of SUMOylation is tremendously improved in the LLPS-driven condensate model that is artificially engineered [32]. Furthermore, in silico analysis indicates that the strong and weak SUMO-SIM interactions can create conditions for the formation of condensates [33]. Additionally, intrinsically disordered region (IDR) is a general hallmark of proteins that participate in LLPS. Meanwhile, the proteomics strategy reveals that lysine residues residing in the disordered regions of proteins are prone to be SUMOylated [34]. All these results indirectly prove that SUMOylation may be a driving factor of LLPS. However, whether SUMOylation is the authentic cause of the formation of LLPS needs further proof. Recently, SUMOylation was identified to facilitate the LLPS via specifically targeting lysine residues within the IDRs of proteins in tumors [35]. Concretely, SUMOylation of RING finger protein 168 (RNF168) which is triggered by SUMO3 and PIAS family at K210 in a long IDR sequence isolates RNF168 from DNA damage sites, attenuates RNF168-mediated ubiquitination, and sequesters the repair protein TP53-binding protein 1 (53BP1) in nuclear condensates via evoking LLPS, eventually attenuating DNA damage repair efficiency. However, overexpressed SENP1-induced deSUMOylation reverses these biological processes, contributing to drug resistance in colon cancer [6].
Transcriptional activation
Transcription factors are indispensable for cancer-related signaling pathways. Of note, SUMOylation represents a dual influence on the transcriptional activation of transcription factors. Typically, SUMO1-mediated SUMOylation of transcription factor forkhead box protein M1 B (FOXM1B) at K463 represses p21 transcription but facilitates JNK1 transcription in breast cancer cells, indicating that its key involvement in the transcriptional activity of FOXM1B [36]. Moreover, transcription factor (forkhead box K2) FOXK2 which is SUMOylated by SUMO2/3 at K527 and K633 contributes to its nuclear translocation and expedites the transcription of nucleotide synthetic genes, leading to tumorigenesis and chemoresistance in HCC [37]. Conversely, SUMO1-mediated SUMOylation of Eyes absent homolog 1 (Eya1) at K43 and K146 hampers its transcription activity in triple-negative breast cancer cells [38].
Genomic instability
Genomic instability induced by ectopic DNA damage repair mechanisms is a vital hallmark of tumor occurrence, deterioration, and anti-tumor drug resistance. Remarkably, SUMOylation has also been demonstrated to play a role in DNA damage repair and genome maintenance. For instance, SUMOylation of microrchidia CW-type zinc finger 2 (MORC2) by SUMO1/2/3 and tripartite motif containing 28 (TRIM28) at K767 elevates its interplay with casein kinase II subunit alpha (CSNK2A1), triggering DNA-dependent protein kinase catalytic subunit, therefore stimulating DNA repair and chemoresistance in breast cancer [39]. Furthermore, the SUMO2/3-mediated SUMOylation of minichromosome maintenance protein 10 (MCM10) at K669 contributes to the proliferation and metastasis of esophageal squamous cell carcinoma through the induction of aberrant DNA replication licensing and genomic instability [40]. Of note, deSUMOylation can also dysregulate the DNA damage repair process. Specifically, SENP5 can deSUMOylate histone H2A.Z (H2AZ) via interacting with K121, K122, and K126 within the DNA binding domains in the C-terminal of H2AZ to facilitate DNA damage repair in a homologous recombination-dependent manner and mediate CRC resistance [41].
In summary, the SUMOylation cascade exerts multifaceted effects in cancer cells, influencing critical processes such as gene expression, cell metabolism, and tumor cell proliferation. Targeting SUMOylation holds promise for developing novel cancer therapeutics, and a comprehensive understanding of SUMOylation provides insights into identifying predictive biomarkers and personalized therapeutic strategies.
The role of SUMOylation in sustaining malignancy
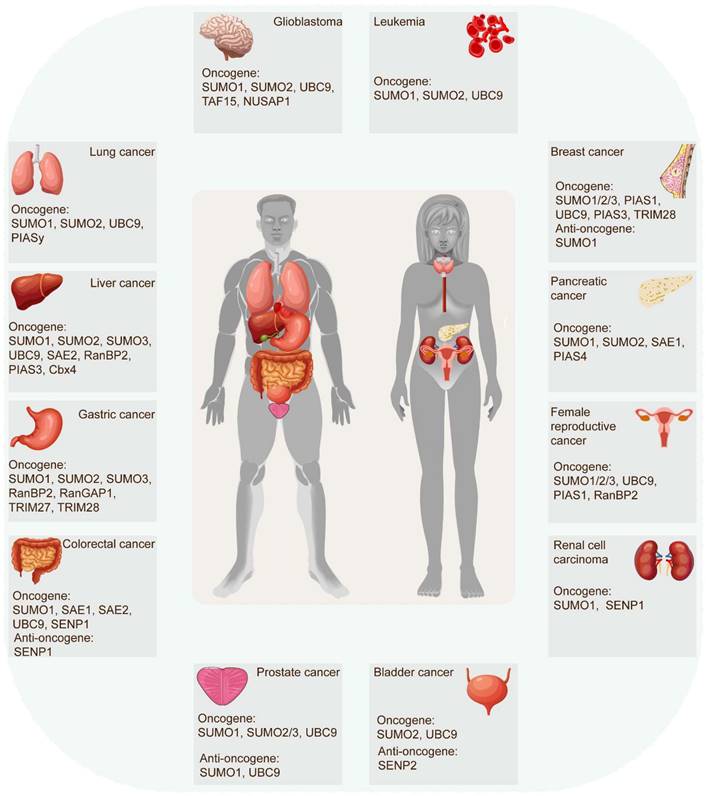
The SUMOylation cascade frequently emerges as a key player in tumor initiation, progression, and response to therapies. In most cases, SUMOylation exerts an indispensable role in driving cancer progression. However, certain SUMOylation-related enzymes and proteins demonstrated tumor-suppressive roles in specific contexts, sometimes even within the same tumor (Figure 3) (Table 2). For example, SUMO1-mediated SUMOylation of p65 recruits mesencephalic astrocyte‐derived neurotrophic factor (MANF) and increases their interaction, thus restraining the NF‐κB/Snail pathway, EMT, and HCC progression [42]. Conversely, the SUMO1-mediated SUMOylation of methyltransferase-like 3 (Mettl3) stabilizes Snail mRNA in an N6-methyladenosine (m6A)-dependent manner, promoting EMT and HCC progression [43]. These findings underscore the complexity of SUMOylation's effect on tumorigenicity. Recent evidence increasingly links SUMOylation with tumorigenesis, tumor progression, anti-tumor therapy response, and poor survival. Accumulating research has robustly demonstrated SUMOylation's oncogenic role in tumor invasion and metastasis, cancer stem cell self-renewal, angiogenesis, evasion of programmed cell death, metabolic reprogramming, and more. (Figure 4).
The role of SUMOylation in tumor invasion and metastasis
The characteristic phenotype of tumor cell dissemination is the invasive-metastatic cascade which is the most lethal factor of tumors. Strikingly, the SUMOylation cascade facilitates tumor metastasis by stimulating tumor angiogenesis, EMT, etc. Tumor angiogenesis is the formation of new blood vessels to provide tumors with sufficient oxygen and nutrients for growth and metastasis.
The function and mechanism of SUMO Molecules in cancers
| Cancer type | SUMO enzyme | Substrate | SUMOylation sites | Function and mechanism | Refs |
|---|---|---|---|---|---|
| Bladder cancer | SUMO2, UBC9 | hnRNPA2B1 | K108 | promote lymphatic metastasis via driving TBK1 mRNA circularization | [142] |
| SUMO2 | DDX39B | K53 | promote lymph node metastasis via driving nuclear export of circNCOR1 | [143] | |
| SUMO2, UBC9 | hnRNPA1 | K113 | promote lymph node metastasis via packaging ELNAT1 into EVs | [144] | |
| SENP2 | TGF-βRI | K389 | inhibit EMT and metastasis via deSUMOylatiing TGF-βRI | [145] | |
| BRCA | SUMO1/2/3, TRIM28 | MORC2 | K767 | promote chromatin remodelling and DNA repair | [39] |
| UBC9 | SYNJ2BP-COX16 | K107 | promote tumor progression via DRP1-mediated mitochondrial fission | [146] | |
| SENP1 | HIF-1α | K391, K477 | promote metastasis via activating HIF-1 signalling | [147] | |
| UBC9, PIAS3 | Rac1 | K183, K184, K186, K188 | promote metastasis via activating Rac1 | [148] | |
| PIAS1 | DDX5 | K53 | promote tumor progression via forming DDX5/Drosha/DGCR8 complex | [149] | |
| SUMO1/2/3, PIAS1 | TFAP2A | K10 | promote tumor outgrowth via inhibiting transcriptional activity of TFAP2A | [150] | |
| SUMO1 | Eya1 | K43, K146 | inhibit tumorigenesis via repressing Eya1 transcription activity | [38] | |
| SUMO1 | FOXM1 | K201, K218, K460, K478, K495 | delay mitotic transition via enhancing APC/Cdh1-mediated ubiquitination | [151] | |
| Cervical cancer | SUMO1, RanBP2 | TCF4 | Unknown | promote metastasis via activating Wnt/β-catenin signaling | [48] |
| SUMO1/2/3, UBC9, PIAS1 | FoxM1b | Unknown | inhibit tumorigenesis by inducing destabilization and cytoplasmic localization of FoxM1b | [152] | |
| CRC | SAE1, SAE2, UBC9 | IRF1 | K78 | promote maintenance and self-renewal of cancer stem cells via stabilizing Oct-1 | [50] |
| SUMO1 | IQGAP1 | K1445 | enhance tumorigenesis and tumor progression via activating AKT-ERK signaling | [153] | |
| SENP1 | RNF168 | K210 | promote DNA damage repair via decreasing RNF168 phase separation | [6] | |
| SENP1 | ELOC | K32 | promote stemness via facilitating the deubiquitination and stabilization of HIF1A | [154] | |
| SENP1 | p16, p19, p21, p27 | Unknown | facilitate tumor growth via downregulating CDK inhibitors | [155] | |
| ESCC | SUMO2/3 | MCM10 | K669 | promote metastasis via inducing genomic instability | [40] |
| SUMO2/3 | HSP27 | Unknown | promote tumor progression via upregulating PKM2 | [156] | |
| GC | TRIM28 | TBK1 | K63 | promote immune escape via increasing PD-L1 abundance | [74] |
| SUMO2/3 | NSUN2 | SIM (236-240aa) | promote tumor progression via regulating mRNA m5C methylation | [19] | |
| SUMO2/3, RanBP2, RanGAP1 | DAXX | Unknown | promote tumor progression via modulating the subcellular localization of DAXX | [157] | |
| SUMO1 | p38α | K152 | promote metastasis via activating MK2 and accelerating ROS accumulation | [158] | |
| SUMO1 | hnRNP K | K422 | promote tumorigenicity and metastasis via stabilizing β-catenin | [159] | |
| SUMO2/3 | hnRNPM | Unknown | promote tumor progression via regulating alternative splicing | [160] | |
| SUMO1, TRIM27 | TUFT1 | K79 | promote tumor progression via activating AKT/mTOR signaling | [161] | |
| GBM | SUMO1 | PML | K65, K160, K490 | facilitate malignancy via stabilizing c-Myc protein | [23] |
| NUSAP1 | ATR | Unknown | promote tumor progression via suppressing the ubiquitin-dependent proteolysis of ATR | [162] | |
| SUMO1, UBC9 | CYLD | K40 | promote proneural-to-mesenchymal transition via unleashing NF-κB signaling | [163] | |
| SUMO2, TAF15 | NOP58 | Unknown | promote stem cell maintenance and tumorigenicity via regulating 2'-O-methylation | [164] | |
| UBC9 | hnRNP A2/B1 | K108 | promote angiogenesis via eliminating miR-204-3p | [47] | |
| SUMO1 | CDK6 | K216 | promote tumor progression via stabilizing CDK6 protein | [165] | |
| HCC | Cbx4 | HIF-1α | K391, K477 | promote angiogenesis via enhancing VEGF expression | [45] |
| SUMO1, UBC9 | PEPCK1 | K124 | promote tumor progression via mediating glucose metabolism | [166] | |
| SUMO1 | Lats1 | K751 | promote tumor progression via inhibiting Hippo signaling | [55] | |
| SUMO2 | LKB1 | K178 | promote tumor progression via altering LKB1 localization | [83] | |
| SUMO1 | NRF2 | K110 | maintaining tumorigenesis via promoting de novo serine synthesis | [76] | |
| SUMO1, UBC9 | Mettl3 | K177, K211, K212, K215 | promote tumor progression via increasing Snail expression in an m6A-dependent manner | [43] | |
| RANBP2 | FTO | K216 | promote tumorigenesis via regulating m6A modification | [5] | |
| SUMO1, SAE2 | PKM2 | SIM (IKII265-268) | promote tumor progression and metastasis via inducing the Warburg effect | [26] | |
| RanBP2 | IL-33 | K54 | promote immune escape via stabilizing IRF1 | [27] | |
| SUMO3, UBC9, PIAS3 | RNF146 | K19, K175 | promote tumor progression via degrading Axin and activating Wnt/β-catenin signaling | [22] | |
| SENP1 | EIF3I | K298 | promote metastasis via inducing EMT | [167] | |
| HNSCC | SENP1 | ACSL4 | Unknown | promote tumor progression via inhibiting ferroptosis | [64] |
| SENP1 | STAT1 | K110, K703 | promote tumor progression | [168] | |
| ICC | SUMO1, SAE1 | AKT | Unknown | promote tumor progression | [99] |
| UBC9 | p27kip1 | Unknown | promote tumorigenesis via governing p27kip1 nuclear export | [169] | |
| Leukemia | SUMO1 | PKM2 | K270 | inhibit myeloid differentiation via degrading RUNX1 | [170] |
| SUMO1, UBC9 | IGF-1R | K1025, K1100 | promote proliferation of leukemia cells | [171] | |
| SUMO1 | sPRDM16 | K568 | promote tumor progression | [172] | |
| SUMO2 | ERG | K37, K74, K289 | promote tumor progression via enhancing ERG stability | [173] | |
| Lung cancer | SUMO1, UBC9 | YTHDF2 | K571 | promote tumor progression by increasing binding affinity of YTHDF2 with m6A-modified mRNAs | [78] |
| SUMO2, UBC9 | hnRNPA2B1 | K108 | promote lymphatic metastasis via modulating extracellular vesicles | [174] | |
| SUMO1, UBC9, PIASy | Slug | K239, K240, K244, K248, K258 | promote metastasis via enhancing the transcriptional repression activity of Slug | [4] | |
| SUMO1 | KEAP1 | K39 | promote tumor growth via increasing NRF2-target gene expression | [175] | |
| PDAC | SUMO2, SAE1 | hnRNPA1 | K113 | promote lymphangiogenesis via packaging hnRNPA1 into the extracellular vesicle | [176] |
| SUMO1, PIAS4 | VHL | Unknown | promote tumor growth via activating hypoxia signaling | [177] | |
| Prostate cancer | UBC9 | STAT4 | K350 | inhibit immunostimulatory macrophage activation and antitumor T cell response via hindering transcriptional activity of STAT4 | [72] |
| SUMO1, UBC9 | HK2 | K315, K492 | inhibit tumorigenesis via decreasing glycolysis | [68] | |
| SENP1 | HIF-1α | Unknown | promote progression and metastasis via HIF1α signaling | [178] | |
| SUMO2/3, UBC9 | Flot-1 | K51, K195 | promote EMT via inhibiting Snail degradation | [24] | |
| RCC | SUMO1 | HAF | K94, K141 | promote growth and metastasis via activating HIF-2 | [80] |
| SENP1 | HIF2α | Unknown | promote metastasis via inducing EMT | [179] |
Role of SUMO molecules in tumors. SUMOylation cascade is frequently identified in tumor initiation, progression, and response to therapies. In most instances, SUMOylation plays an indispensable role in exacerbating cancer progression. However, some SUMOylation molecules have been proven to act as tumor-suppressive roles in certain human tumors, even playing distinct roles in the same tumor.
The biological functions of SUMOylation cascade in tumors. SUMOylation participates in the acquisition or maintenance of cancer hallmarks, such as tumor invasion, metastasis, cancer stem cell self-renewal, epigenetic reprogramming, immune evasion, metabolic reprogramming, and programmed cell death escape. SUMOylation is involved in tumor invasion and metastasis via administrating tumor angiogenesis, EMT, etc. SUMOylation induces cancer stem cell maintenance and self-renewal via governing cancer stemness-related genes and pathways. SUMOylation manages epigenetic reprogramming via perturbing transcriptional activation, protein interaction, protein localization, protein stability, and RNA metabolism. SUMOylation facilitates tumor immune evasion via altering the tumor microenvironment and abolishing immune surveillance. SUMOylation promotes metabolic reprogramming via stimulating the Warburg effect. SUMOylation enhances the escape of programmed cell death via blocking apoptosis and autophagy. EMT: epithelial-to-mesenchymal transition.
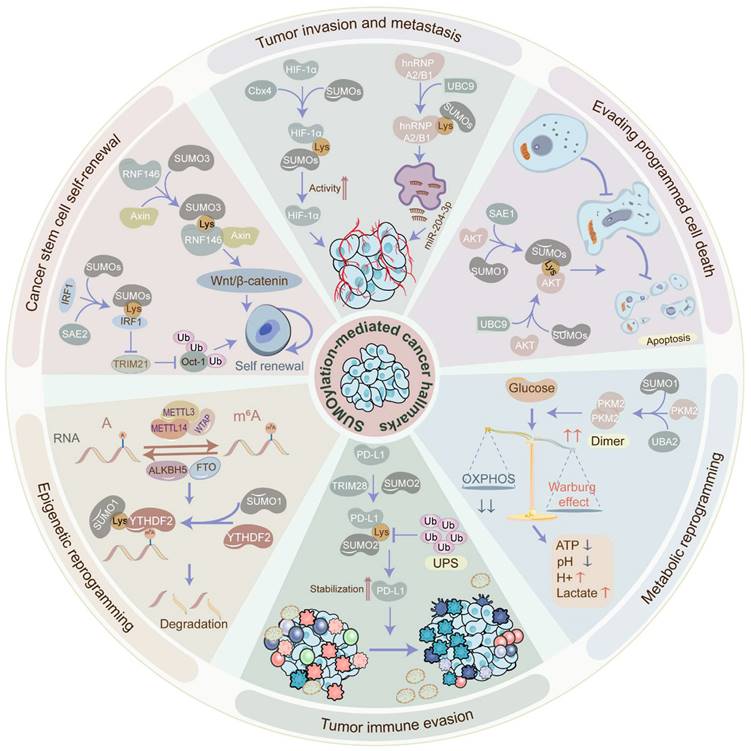
Thus, tumor angiogenesis inhibition has been speculated as a promising anti-tumor therapeutic strategy. While the role of SUMOylation in tumor angiogenesis is not fully elucidated, some evidence suggests its involvement. For instance, the most representative driven pathway is the vascular endothelial growth factor (VEGF) signal transduction pathway. However, the SUMOylation of VEGF is yet to be verified. Merely one study reports that SUMO1-induced SUMOylation of vascular endothelial growth factor receptor 2 (VEGFR2) at K1270 hampers the activity of VEGFR2 and the angiogenesis signaling pathway [44], indicating an inhibitory effect of the SUMOylation cascade on tumor invasion and metastasis. Conversely, the SUMOylation of hypoxia-inducible factor-1alpha (HIF-1α) can upregulate VEGF in the hypoxic environment to stimulate angiogenesis. For instance, SUMO E3 ligase chromobox protein homolog 4 (Cbx4) boosts the transcriptional activity of HIF-1α via SUMOylating HIF-1α at K391 and K477 under hypoxic conditions, following which HIF-1α further transcriptionally activates VEGF, potentiating angiogenesis in HCC [45]. However, SUMOylating of HIF-1α also shows a converse role in VEGF expression. Specifically, hypoxia-mediated SUMOylation of HIF-1α facilitates its binding to E3 ubiquitin ligase and promotes its ubiquitination and degradation in a proline hydroxylation-independent manner, contributing to the downregulation of VEGF [46]. Additionally, SUMOylation of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP A2/B1) in hypoxic conditions facilitates its translocation from the nucleus to the cytoplasm and further enhances the exosome-sorting process of miR-204-3p via binding to its RRM1 motif segment, expediting angiogenesis in GBM [47].
EMT, characterized by increased invasiveness and metastatic potential, is regulated by SUMOylation in various cancers. For instance, overexpressed nucleolar and spindle-associated protein 1 (NUSAP1), associated with SUMO E3 ligase RanBP2, robustly triggers the SUMOylation of transcription factor 4 (TCF4) and the Wnt/β-catenin signaling pathway, contributing to the advancement of EMT and cervical cancer cell metastasis [48]. On the contrary, SUMOylation can hinder the EMT and tumor metastasis. Concretely, SUMO E3 ligase PIAS3-mediated SUMOylation of E3 ubiquitin ligase SMAD specific E3 ubiquitin protein ligase 2 (Smurf2) at K26 and K369 enhances Smurf2 protein stability and further degrades transforming growth factor-β (TGFβ) receptor, thus attenuating EMT, cancer cell invasiveness, and metastasis [49].
Collectively, these findings highlight the dual role of the SUMOylation cascade in tumor angiogenesis and EMT, adding complexity to the regulatory mechanisms governing the invasive-metastatic cascade through SUMOylation.
The role of SUMOylation in cancer stem cell maintenance and self-renewal
Cancer stem cells (CSCs) represent a small subset of undifferentiated cells within tumor tissues, distinguished by their robust self-renewal and tumorigenic potential. The maintenance of CSCs relies heavily on self-renewal mechanisms, crucial for tumor cell survival and proliferation. Intriguingly, the global SUMOylation level exhibits distinct patterns in CSCs, exerting a notable influence on CSC maintenance and self-renewal. Indeed, the global SUMOylation level in CSCs has been observed to be higher compared to non-CSCs. Mechanistically, SUMOylation of interferon regulatory factor 1 (IRF1), a transcriptional activator of ubiquitin E3 ligase tripartite motif containing 21 (TRIM21), at K78 hinders its transcription activity, leading to reduced expression of TRIM21, thereby attenuating the ubiquitination of Oct-1, which is a transcriptional activator of aldehyde dehydrogenases, and eventually facilitating CSC maintenance and self-renewal in CRC cells [50]. Moreover, SUMO1-induced SUMOylation of PML protein augments its interplay with c-Myc, stabilizing c-Myc oncoprotein, and contributes to the CSC self-renewal in glioma [23]. Additionally, the SUMOylation cascade has been implicated in expanding the CSC pools in breast cancer and CRC [51].
In addition to manipulating cancer stemness-related proteins, the SUMOylation cascade also engages in the well-known signaling pathways that favor the maintenance and self-renewal of CSCs. Notably, the activation of the Wnt/β-catenin signaling and inhibition of the Hippo signaling are depicted to sustain the self-renewal of stem cells [52, 53]. Specifically, E3 ubiquitin ligase RNF146 SUMOylated at K19, K61, K174, and K175 enhances its nuclear localization and the interaction with Axin, facilitating Axin degradation and Wnt/β-catenin signaling activation in HCC [22]. Moreover, E3 ubiquitin-protein ligase UHRF2-induced SUMOylation of TCF4 stabilizes TCF4 and activates Wnt signaling in CRC [54]. Furthermore, SUMOylation of Large tumor suppressor 1 (Lats1) at K751 undermines the kinase activity, therefore impeding Hippo signaling in tumors [55].
Taken together, the SUMOylation cascade plays a crucial role in maintaining and self-renewal of CSCs, which are essential for tumor initiation, progression, recurrence, and metastasis. This is achieved through the modification of oncoproteins and the regulation of associated pathways. Inhibiting the SUMOylation cascade may hold promise in impairing the self-renewal ability of CSCs.
The role of SUMOylation in evading programmed cell death
Programmed cell death (PCD) such as apoptosis, autophagy, and ferroptosis is essential for maintaining cellular homeostasis. Remarkably, the SUMOylation cascade contributes to the dysregulation of PCD in cancer. Apoptosis, a highly regulated form of cell death, is committed to removing non-functional, harmful, abnormal, and misplaced cells promptly. SUMOylation cascade has been involved in apoptotic pathways, administrating the destiny of tumor cells. Of note, the AKT signaling pathway, known to inhibit apoptosis in tumors, is directly influenced by SUMOylation. For instance, the SUMOylation of AKT at K276 in the SUMOylation consensus motif dramatically motivates its kinase activity in cancers [56]. Similarly, the SUMOylation of AKT at K276 improves AKT kinase activity without influencing its phosphorylation level but directly phosphorylating Ubc9 at Thr35 and SUMO1 at Thr76 to govern the global SUMOylation status, ultimately contributing to tumorigenesis [57]. Additionally, SUMO E1 activating enzyme SAE1 facilitates AKT SUMOylation and phosphorylation, aiding in apoptosis evasion in glioma [58]. Autophagy is a process of self-degradation to recycle cellular components [59]. Dramatically, the SUMOylation cascade also modulates the autophagy of tumor cells. In breast cancer cells, disruption of SUMO1 complexes contributes to autophagy-mediated tumor cell death, potentially through the upregulation of Tribbles pseudokinase-3 (TRIB3) [60]. Conversely, the deSUMOylation of m6A demethylase AlkB Homolog 5 (ALKBH5) enhances its activity and further increases the stability of DDIT4 mRNA, inducing autophagy and tumorigenesis in head-neck squamous cell carcinoma (HNSCC) [61]. Ferroptosis, an iron-dependent PCD driven by excessive lipid peroxidation, has participated in tumorigenesis and tumor development [62]. Increasing evidence indicates that SUMOylation is involved in ferroptosis in cancer. For instance, SENP1-mediated deSUMOylation of A20 disturbs its interplay with Acyl-CoA synthetase long-chain family member 4 (ACSL4) and solute carrier family 7 member 11 (SLC7A11), contributing to the inhibition of ferroptosis in lung cancer [63]. Similarly, SENP1-induced deSUMOylation of ACSL4 hinders ferroptosis via decreasing ACSL4 protein stability in HNSCC [64]. Additionally, PIAS4-mediated SUMOylation of SLC7A11 at K500 inhibits ferroptosis in breast cancer [65].
Altogether, PCD exerts an essential effect on tumorigenesis, tumor progression, recurrence, and chemoradiotherapy resistance. Therefore, targeting the SUMOylation cascade may offer a promising approach to sensitize tumors to therapy and suppress tumor progression, thereby ameliorating the overall status quo of tumor treatment.
The role of SUMOylation in metabolic reprogramming
Metabolic reprogramming, the process by which cells adapt their metabolism to support survival and growth, plays a crucial role in malignant transformation and tumor progression. Strikingly, the SUMOylation cascade can determine metabolic reprogramming, such as the Warburg effect and pentose phosphate pathway [66]. Warburg effect also known as aerobic glycolysis, a typical abnormality of glucose metabolism in tumors, is manipulated by SUMOylation. Concretely, SUMO1-induced SUMOylation of PKM2 via binding to the SUMO-interacting motif site IKII265-268 promotes PKM2 dimerization and nuclear translocation, leading to glycolysis in HCC [26]. Similarly, IKII265-268-mediated SUMOylation of PKM2 conduces to its distribution into ectosomes, inducing the metabolic reprogramming in monocytes and reshaping the tumor microenvironment [67]. Furthermore, a novel circRNA circRNF13 which is downregulated in nasopharyngeal carcinoma diminishes the expression of SUMO2 and the SUMOylation level of glucose transporter type 1 (GLUT1), finally inhibiting GLUT1 degradation and promoting glycolysis [20]. However, the SUMOylation cascade can restrain glycolysis in tumors. Hexokinase 2 (HK2), the first rate-limiting enzyme of glycolysis, is SUMOylated at K315 and K492 in prostate cancer cells, restraining its binding to the mitochondria, thereby reducing the tumor cell glycolysis [68]. In addition to aerobic glycolysis, SUMOylation may also dysregulate the pentose phosphate pathway in tumors. Specifically, glucose-6-phosphate dehydrogenase (G6PD), a rate-limiting enzyme in the pentose phosphate pathway, is SUMOylated in a SUMO1-dependent manner, therefore enhancing G6PD protein stability and promoting the progression of clear-cell renal cell cancer (ccRCC) [69].
Overall, these findings highlight the crucial role of the SUMOylation cascade in regulating metabolic reprogramming in tumors, exerting both promoting and inhibitory effects. Hence, targeting the SUMOylation cascade may therefore offer a promising strategy to suppress metabolic reprogramming, improve the tumor microenvironment, inhibit tumor growth, and enhance the sensitivity of anti-tumor drugs.
The role of SUMOylation in tumor immune evasion
Tumor immune evasion, characterized by the ability of tumor cells to evade immune surveillance, is a major factor in tumor progression and immunotherapy resistance. Functionally, the SUMOylation cascade expedites tumor immune evasion via altering the tumor microenvironment and abolishing immune surveillance. Aerobic glycolysis, a central factor in regulating the tumor microenvironment, is conducive to tumor immune evasion as well [70]. Of note, the SUMOylation cascade regulates antigen presentation deficiency and lymphocyte inactivation. For example, SUMO2/3-mediated SUMOylation of scaffold attachment factor B (SAFB) at K294 declines the abundance and activity of tumor-infiltrating T cells and blocks MHC class I antigen presentation, inducing immune evasion in cancer [71]. Moreover, UBC9-mediated SUMOylation of signal transducer and activator of transcription 4 (STAT4) at K350 attenuates its nuclear translocation and stability, further blocking the pro-inflammatory activation of macrophages and impeding the anti-tumor T cell response in prostate cancer [72]. Furthermore, cholesterol-induced SUMOylation of liver X receptors (LXRs) hinders IL-9 expression via inhibiting p65-IL-9 binding, contributing to the reduction of IL-9-producing CD8+ T cell differentiation and anti-tumor response [73]. In addition to immune cells, SUMOylation occurring in tumor cells can also facilitate tumor immune evasion. Specifically, SUMOylation of programmed cell death protein-1 ligand (PD-L1) by TRIM28, an E3 ubiquitin ligase and E3 SUMO ligase, stabilizes PD-L1 via hampering PD-L1 ubiquitination and enhancing PD-L1 SUMOylation, leading to the T cell inactivation and immune evasion in gastric cancer [74]. Additionally, E3 ligase RanBP2-induced SUMOylation of nuclear factor IL-33 at K54 prevents the degradation of transcription factor IRF1 which elevates the expression of PD-L1, a T cell immune checkpoint ligand, in HCC cells, ultimately leading to the inactivation of T cells and immune surveillance [27].
In conclusion, immunotherapy represents a novel paradigm in cancer therapy. However, a significant number of patients with tumors remain unresponsive to this new anti-tumor strategy. Notably, the SUMOylation cascade contributes to tumor immune evasion, indicating that targeting SUMO modification could enhance anti-tumor immunity. Combining tumor immunotherapy with SUMOylation inhibitors may offer a promising approach to overcoming resistance to immunotherapy.
The role of SUMOylation in epigenetic reprogramming
Epigenetic reprogramming, encompassing transcriptional factor regulation, RNA epigenetic modification, protein translational modification, etc., orchestrates changes in cell fate through epigenetic modifications without altering the DNA sequence [75]. Of interest, SUMOylation plays a pivotal role in manipulating epigenetic reprogramming, impacting transcriptional activation, protein interaction, protein localization, protein stability, and RNA metabolism as mentioned above. Moreover, SUMOylation, as a form of epigenetic modification itself, participates in the dysregulation of epigenetic reprogramming in cancers, fostering tumor heterogeneity. At the transcriptional level, SUMOylation governs gene expression via SUMOylating transcription factors. For example, SUMO1-induced SUMOylation of the transcription factor nuclear factor erythroid-2 related factor 2 (NRF2) at K110 stimulates the transcription of glutathione peroxidase 2 (Gpx2) and motivates the intracellular reactive oxygen species (ROS)-phosphoglycerate dehydrogenase (PHGDH) signaling, promoting the de novo serine synthesis and tumorigenesis in HCC [76]. Notably, histone SUMOylation is also significant for gene expression. For instance, SUMO1-induced SUMOylation of histone H4 upregulates the expression of the progesterone receptor (PR) gene, promoting the proliferation of Ishikawa Cells and inhibiting the apoptosis of tumor cells in endometrial cancer [77]. Moreover, SUMOylation can manipulate gene expression post-transcriptionally influencing the m6A and m5C modifications. Specifically, SUMO1-mediated SUMOylation of the m6A reader YT521-B homology domain-containing family protein 2 (YTHDF2) at K571 improves its binding affinity for m6A-modified mRNAs, facilitating cancer progression [78]. Strikingly, the SUMOylation of the E3 ubiquitin ligases can boost their interaction with substrate proteins. For instance, SUMO3-mediated SUMOylation of RING-type E3 ubiquitin ligase RNF146 at K19, K61, K174, and K175 enhances the association between RNF146 and its substrate Axin, expediting the ubiquitination and degradation of Axin and consequently deteriorating the progression of HCC [22]. Likewise, deubiquitinating enzymes can be SUMOylated to promote tumor progression. For instance, SUMOylation of the deubiquitinating enzyme OTUB2 at K233 heightens its interaction with YAP/TAZ and stabilizes YAP/TAZ via deubiquitination, contributing to cancer metastasis [25].
Collectively, elucidating the molecular mechanisms of SUMOylation-mediated epigenetic reprogramming is crucial for uncovering new interventions to conquer tumor heterogeneity.
Cellular stresses that induce SUMOylation in cancers
Protein SUMOylation, critical for maintaining cellular homeostasis, is evoked by diverse cellular stresses. Nevertheless, what cellular stresses can provoke SUMOylation and how SUMOylation responds to them remain enigmas. Herein, we delve into the stimuli and mechanisms underlying SUMOylation induction in cancers (Figure 5).
Hypoxia and SUMOylation
Hypoxia, a prevalent feature in solid tumors, is closely related to tumor proliferation, differentiation, angiogenesis, energy metabolism, anti-tumor drug resistance, and poor prognosis. Remarkably, hypoxia has been identified as a trigger for SUMOylation [79]. Yet, how hypoxia sustains the malignant phenotypes via activating SUMOylation requires further elaboration. Of note, hypoxia-inducible factors (HIFs), indispensable regulators of the cellular response to hypoxia, link hypoxia to SUMOylation-mediated tumorigenesis. Specifically, the SUMOylation of E3 ligase HAF induced by hypoxia specifically facilitates HIF-2 binding to the DNA promoters and driving transcription, thus leading to the growth and metastasis of ccRCC [80]. In addition to HIFs, hypoxia-mediated SUMOylation is responsible for cancer progression via supervising other transcription activators or transcriptional repressors. In lung cancer, hypoxia facilitates SUMOylation-dependent competition between transcriptional repressor HIC1 and transcription activator Sp1, leading to the transcriptional repression of SIRT1 and subsequently promoting tumor metastasis [81]. Analogously, hypoxia promotes the SUMOylation of transcriptional repressor Slug via diminishing its interaction with SUMO proteases, contributing to lung cancer metastasis [4]. Moreover, hypoxia-induced SUMOylation impairs the biogenesis pathway of tumor suppressor microRNAs, promoting tumorigenesis and cancer progression. For instance, SUMO1-mediated SUMOylation of KH-type splicing regulatory protein (KHSRP) modified at the site K87 in the hypoxia microenvironment segregates KHSRP from the pri-miRNA/Drosha-DGCR8 complex and disturbs the transformation of pre-miRNAs from pri-miRNAs, contributing to the inhibition of TL-G-Rich microRNA biogenesis and the advancement of tumorigenesis [82]. Furthermore, hypoxia-induced SUMOylation could manipulate the RNA epigenetic modification such as m6A.
Cellular stressors that induce SUMOylation in cancers. SUMOylation which plays a critical role in maintaining cellular homeostasis is evoked by diverse cellular stresses, including viruses, hypoxia, gut microbiota, and lactic acid. Viruses such as HBV, EBV, and HPV have been confirmed to contribute to cancers via manipulating the SUMOylation process. Hypoxia has been identified as the inducement the SUMOylation in tumors. Gut microbiota and its metabolites such as pks+ E. coli and SCFAs participate in the tumorigenesis via modulating the SUMOylation in p53 and NF-κB signaling. Lactate may promote tumor progression via stabilizing the anaphase-promoting complex (APC/C) in a SUMOylation-dependent manner. SCFA: short-chain fatty acid.
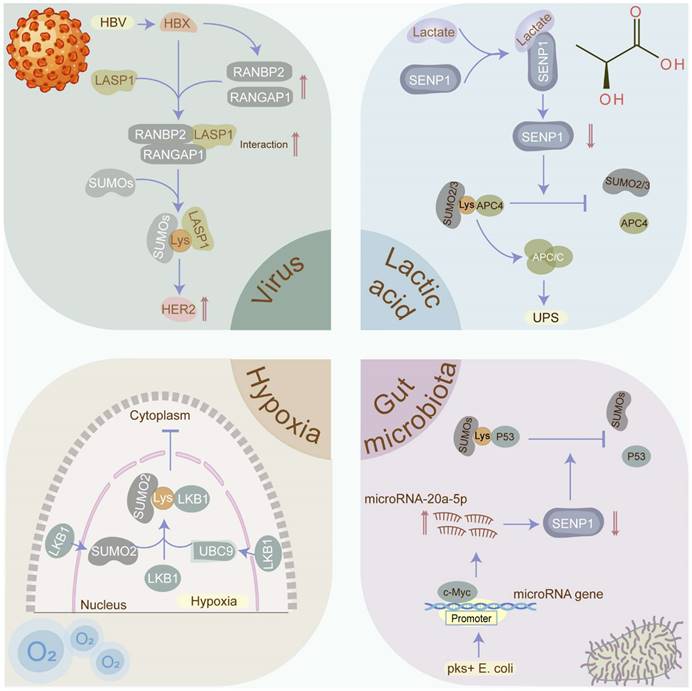
Concretely, hypoxia facilitates the cancer progression via inducing the SUMOylation of YTHDF2 [78]. Additionally, SUMOylation in response to hypoxic stress can regulate tumorigenesis and progression via altering protein localization. In liver cancer, liver kinase B1 (LKB1) which is SUMOylated by SUMO-2 at Lys178 under a hypoxic microenvironment expedites tumor growth via impeding LKB1 nucleocytoplasmic shuttling [83]. Intriguingly, in vivo research has been utilized to intervene in intratumoral hypoxia, with hyperoxic breathing at 60 % O2 demonstrated to reverse hypoxia in tumor microenvironments and hamper the expression of HIF-1α and its downstream target proteins [84]. However, it is yet to be determined whether hyperoxic breathing manipulates the SUMOylation process.
In summary, SUMOylation is a hypoxia-related post-translational modification. Targeting intratumoral hypoxia, such as by improving tumor oxygenation, may reduce the overall SUMOylation levels, thereby hindering cancer progression.
Viruses and SUMOylation
Approximately 10% of cancers are associated with virus infections, principally including Epstein-Barr virus (EBV), human papilloma virus (HPV), hepatitis B virus (HBV), etc. [85]. Intriguingly, these viruses contribute to carcinogenesis in part via manipulating the SUMOylation process. EBV, a known human oncogenic virus, consistently infects individuals globally, opportunistically leading to tumorigenesis. Strikingly, EBV promotes tumorigenesis partially through EBV-mediated SUMOylation. In EBV-positive lymphomas, the EBV oncoprotein latent membrane protein-1 (LMP1) triggers NF-κB signaling to elevate the levels of SUMOs [86], simultaneously hijacking SUMO-conjugating enzyme UBC9 [87] and diminishing SUMO-protease SENP2 activity and turnover [88], collectively contributing to the enhancement of overall SUMOylated protein levels and tumorigenesis. Similarly, HPV facilitates the malignant transformation of the host's infected cells via administrating SUMOylation. For instance, HPV E6/E7 oncoproteins up-regulate SUMO-conjugating enzyme UBC9 via inhibiting the autophagy-lysosome pathway to govern SUMOylation during HPV-mediated tumorigenesis and induce apoptosis evasion [89]. Likewise, HBV infection plays a significant role in hepatocarcinogenesis via controlling SUMOylation. Mechanistically, HBV X (HBX) protein facilitates the SUMOylation of LASP1 through the RANBP2-RANGAP1 complex and enhances the interaction between LASP1 and human epidermal growth factor receptor 2 (HER2) to upregulate HER2 by hampering ubiquitination-mediated proteasomal degradation, resulting in hepatocarcinogenesis [90].
Given the distinctive capability of viruses to manipulate the SUMO pathway and enhance tumorigenesis, the eradication of oncoviruses through vaccination may impede their capacity to increase global cellular SUMOylation and induce malignant transformation.
Gut microbiota and SUMOylation
Emerging evidence suggests a connection between the gut microbiota and its metabolites with multiple malignancies. Escherichia coli harboring the pks island (pks+ E. coli) has been closely implicated in the tumorigenesis and progression of CRC. Mechanistically, pks+ E. coli transcriptionally activates miR20a-5p which hijacks SUMO-protease SENP1 and further declines SENP1 expression, ultimately reducing the p53 SUMOylation and colon tumor growth [91]. Multiple research indicates that NF-κB signaling is a canonical mechanism for inflammation-associated colorectal tumorigenesis. A recent study has validated that the SUMOylation in intestinal cells, which prevents IκBα degradation and hinders the NF-κB signaling, is triggered by the short-chain fatty acids (SCFAs) produced by gut microbiota in a pH-dependent manner, thus attenuating the inflammatory responses and maintaining the intestinal epithelial integrity [92]. However, the abundance of SCFAs and SCFA-producing bacteria is tremendously shrunk in CRC [93], with low fecal SCFA concentration associated with a higher CRC incidence [94].
These findings suggest that gut microbiota and its metabolites may promote colorectal tumorigenesis and progression through the modulation of SUMOylation, leading to the activation of the NF-κB signaling pathway or disruption of the p53 pathway in intestinal cells. Interventions targeting the gut microbiota and its metabolites, such as SCFA-producing probiotics and dietary fiber supplementation, hold promise for inhibiting tumor initiation and progression.
Tumor metabolite and SUMOylation
Lactic acid, considered as the most representative tumor metabolite and traditionally viewed as a metabolic waste product of aerobic glycolysis or Warburg effect in cancers, has emerged as a significant oncometabolite. Lactic acid is exported to the tumor microenvironment (TME) to shape an environment with accumulated lactate and decreased pH value, promoting invasion, metastasis, angiogenesis, immune evasion, etc. [95]. Strikingly, a recent study highlighted that cumulative lactate can stabilize the anaphase-promoting complex (APC/C) via mediating SUMOylation to expedite cell proliferation. Mechanistically, increasing lactate directly interacts with zinc in the SUMO protease SENP1 active site to induce SENP1 inhibition and manipulate the activity of E3 ubiquitin ligase APC/C, resulting in vigorous cell proliferation [96]. Meanwhile, APC/C has been validated to play the role of oncoprotein in cancers principally via degrading substrate proteins such as Axin [97] and SMAR1 [98]. Comprehensively, we speculate that lactate produced via the Warburg effect may facilitate the SUMOylation of APC/C, contributing to tumorigenesis and progression by targeting multiple tumor suppressors. Moreover, lactate-driven SUMOylation may extend beyond APC/C, necessitating further exploration of potential substrate proteins. Thus, decreasing lactate levels or inversing the Warburg effect may reduce the global SUMOylation level in cancers, providing a novel anti-tumor strategy.
Therapeutic potential and toxicity of SUMOylation inhibitors
The proteins involved in the SUMOylation enzymatic cascade not only sustain cancer hallmarks but also diminish the sensitivity of anti-tumor therapies. Of note, dysregulation of SUMOylation-related proteins such as SAE1/2, UBC9, SUMO E3-ligases, and SENPs has been observed in multiple cancers [24, 74, 99], leading to anti-tumor drug resistance via fortifying tumor stemness [50], promoting DNA damage repair [6], and provoking nucleotide de novo synthesis [37], etc. Consequently, targeting these enzymes in the SUMOylation cascade presents an attractive anti-tumor strategy [100, 101]. Over the past two decades, a plethora of natural and synthetic SUMOylation inhibitors have been identified (Table 3). However, except for TAK-981, all the SUMOylation inhibitors are in the preclinical research and development stage.
SUMO E1 inhibitors
SUMO E1, which functions as activating enzymes in the form of SAE1/2 dimer, has been linked to cancer exacerbation. For instance, upregulation of SAE1 or SAE2 facilitates tumor proliferation, metastasis, or apoptosis evasion in intrahepatic cholangiocarcinoma (ICC) [99], HCC [102], glioma [58], and lung cancer [103], etc. So far, several SUMO E1 inhibitors have been discovered or synthesized. The foremost discovered SAE1/2 inhibitors include ginkgolic acid extracted from Ginkgo biloba leaves, anacardic acid, and kerriamycin B from microbial metabolites [104, 105]. These natural compounds impede protein SUMOylation via blocking the formation of the SAE1/2-SUMO intermediate and the conjugation of SUMOs to substrates. Remarkably, in vivo and in vitro studies have corroborated that exposure to ginkgolic acid or anacardic acid inhibits the proliferation, migration, and apoptosis escape of GC, breast cancer, CRC, etc. [51, 106]. Subsequently, two novel natural compounds were identified, namely Davidiin (extracted from the plant Davidia involucrate) [107] and tannic acid (purified from Gallotannin) [108], as SUMO E1 inhibitors, sharing analogous mechanisms of action with previously discovered inhibitors. Experimentally, both inhibitors restrain the growth of multiple cancer cells, involving GC MKN-45 cells, prostate cancer DU-145 cells, and lung cancer NCI-H460 cells [107], highlighting their anti-tumor ability. However, these natural compounds lack efficiency and specificity due to the half maximal inhibitory concentration (IC50) ranging in micromole and wide spectrum of targets.
There is therefore an immediate demand for inhibitors targeting the SUMO E1 enzymes that are both highly efficient and highly specific. To date, synthetic E1 inhibitors, which predominantly include compound-21 [109], COH000 [110], ML-792 [111], ML-93 [112], and TAK-981 [113], have garnered attention. Mechanistically, compound-21 occupies the ATP binding site of SUMO E1 and further hampers the formation of the E1-biotinylated SUMO-1 thioester intermediate. COH000 covalently binds to the Cys30 of Uba2 and concomitantly induces structural changes of SUMO E1, shaping an inactive conformation. In 2017, ML-792 was authenticated to selectively decline SAE1/2 enzyme activity and global SUMOylation level via establishing an adduct with SUMO in an ATP-dependent manner catalyzed by the enzyme itself [111]. In acute myeloid leukemia, ML-792 treatment induces the deconjugation of all the SUMO-2/3 targets and blunts Daunorubicin-mediated transcriptional reprogramming [114]. ML-93 as the derivative of ML-792 manifests a robust selectivity to hinder the SUMOylation via an identical mechanism of action in pancreatic cancer, contributing to G2/M phase arrest and apoptosis [112].
Notably, a great breakthrough in targeting the SUMO pathway is TAK-981, a derivative of ML-792. Mechanistically, TAK-981 impedes the activity of SUMO-activating enzyme via shaping a SUMO-TAK-981 adduct in the enzyme catalytic site [113].
Inhibitors of SUMO molecules
| Compound | Target | Cancer type | Compound source | Compound type | Refs |
|---|---|---|---|---|---|
| Ginkgolic acid | E1 | Gastric cancer, Breast cancer, Uveal melanoma | Ginkgo biloba leaves | Alkylphenol | [104] |
| Anacardic acid | E1 | Thyroid cancer, Nonpromyelocytic acute myeloid leukemia, Breast cancer, Colon cancer, B-cell lymphoma | Microbial metabolites | Analog of ginkgolic acid | [104] |
| Kerriamycin B | E1 | Unknown | Microbial metabolites | Antibiotic | [105] |
| Davidiin | E1 | Gastric cancer, Prostate cancer, Lung cancer | Davidia involucrate | Ellagitannin | [107] |
| Tannic acid | E1 | Unknown | Gallotannin | Gallotannin | [108] |
| Compound-21 | E1 | Unknown | Synthetic | Phenyl urea | [109] |
| COH000 | E1 | Unknown | Synthetic | Dimethyl 1-((R)-1-(phenylamino)-2-(p-tolyl)ethyl)-7-oxabicyclo [2.2.1]hepta-2,5-diene-2,3-dicarboxylate | [110] |
| ML-792 | E1 | Hepatocellular carcinoma, Pancreatic cancer | Synthetic | Pyrazole- carbonylpyrimidine | [111] |
| ML-93 | E1 | Pancreatic cancer | Synthetic | Derivative of ML-792 | [112] |
| TAK-981 | E1 | Acute myeloid | Synthetic | Derivative of ML-792, Pyrazole- carbonylpyrimidine | [113] |
| Leukemia, Hepatocellular carcinoma, Chronic lymphocytic leukemia, Glioblastoma, Pancreatic cancer, Multiple myeloma | |||||
| Spectomycin B1 | E2 | Nasopharyngeal carcinoma | Streptomyces spectabilis | Antibiotic | [124] |
| 2-D08 | E2 | Prostate cancer, Acute myeloid leukemia | Synthetic | Oxygenated flavonoid | [125] |
| GSK145A | E2 | Unknown | Synthetic | Unknown | [126] |
| Compound 2 | E2 | Unknown | Synthetic | Pyridine | [127] |
| WNN0605-F008 | E2 | Unknown | Synthetic | Heterocycle | [128] |
| UNC3866 | E3, CBX4 | Hepatocellular carcinoma | Synthetic | Unknown | [129] |
| Streptonigrin | SENP1 | Unknown | Streptomyces flocculus | Antibiotic | [131] |
| Triptolide | SENP1 | Prostate cancer | root of Tripterygium wilfordii | Tripterygium wilfordii Hook F | [132] |
| Momordin Ic | SENP1 | Acute myeloid leukemia, Colon cancer, Prostate cancer | Kochia scoparia | Triterpenoid glycoside | [133] |
| GN6958 | SENP1 | Unknown | Synthetic | Phenyl urea | [134] |
| Compound 3 | SENP1 | Unknown | Synthetic | Phenyl | [135] |
| Compound 13m | SENP1 | Unknown | Synthetic | Phenyl | [136] |
| Ebselen | SENP2 | Unknown | Synthetic | Organo-selenium | [137] |
| Compound 69 | SENP2 | Unknown | Synthetic | Oxadiazoles | [138] |
| Compound 117 | SENP2 | Unknown | Synthetic | Oxadiazoles | [138] |
Of note, In vivo and ex vivo assays have demonstrated that TAK-981 directly evokes anti-tumor immune responses via the pharmacological reactivation of IFN1 signaling [115-118]. Specifically, TAK-981 facilitates T cell sensitivity, macrophage phagocytosis, and NK cell cytotoxicity. A recent study revealed that TAK981 disrupts immune suppressive activities of regulatory T (Treg) cells in an IFN-alpha receptor 1 (IFNAR1)-dependent manner [119]. Moreover, TAK981 can restrain trogocytosis and increase the viability of endogenous and immunotherapeutic cytotoxic T cells via upregulating cholesterol 25-hydroxylase (CH25H) [120]. Furthermore, TAK-981 effectively motivates the antitumor immune responses via driving T cell activation and augmenting the percentage of activated CD8 T cells and natural killer (NK) cells in preclinical models [115, 118]. Additionally, an in vivo study reported that TAK-981 in combination with the anti-CD20 antibody rituximab can potentiate macrophage phagocytosis and NK cell cytotoxicity in lymphoma models [116]. These studies demonstrate that the SUMOylation inhibitor TAK981 plays a crucial role in the recuperation of immune-killing competence. In addition to type 1 interferon and immune-dependent mechanism, TAK-981 also provokes apoptosis and cell-cycle arrest in acute myeloid leukemia [121]. Additionally, TAK-981 dampens the expression of SUMOylated hnRNP A2/B1 and total hnRNP A2/B1, further thwarting exosome sorting of miR-204-3p as well as resulting in the prohibition of tumor growth and angiogenesis in GBM [47].
Strikingly, multiple phase 0/1/2 clinical trials of TAK-981 have been implemented. For instance, a phase 0 clinical trial (NCT04065555) was utilized to evaluate the biological effects on the TME of intratumoral TAK-981 injection and TAK-981 injection combined with cetuximab or avelumab in 12 patients with head and neck carcinoma, revealing that both can switch the TME from immune-suppressive to immune-permissive in an IFN pathway-dependent manner [122]. Moreover, a phase 1/2 clinical trial (NCT03648372) was performed to assess the safety, tolerability, efficacy, and pharmacokinetics of TAK-981 in patients with hematologic malignancies or solid tumors. Furthermore, another three TAK-981 combination medication studies (namely NCT04381650, NCT04074330, and NCT04776018) are already undergoing phase 1 clinical trials. NCT04381650 (TAK-981 in combination with pembrolizumab) focuses on the safety, tolerability, and anti-tumor activity of combination medications in patients with solid tumors, while NCT04074330 (TAK-981 in combination with rituximab) evaluates the safety of the drug and the efficacy of combination medications in patients with CD20-positive non-Hodgkin lymphoma. Additionally, NCT04776018 (TAK-981 in combination with anti-CD38 monoclonal antibodies) is centered on the safety and efficacy of combination medications in patients with multiple myeloma. However, these clinical trials either have limitations due to the small number of recruited patients or failure to produce any substantial results.
Collectively, these in vitro, in vivo, and clinical studies reveal that natural and synthetic compounds targeting SUMO E1, especially TAK-981, perturb the state of tumor cells and the TME and impede tumorigenesis and cancer progression, thereby offering a new approach for anti-tumor treatment.
SUMO E2 inhibitors
UBC9, as the sole SUMO E2 conjugating enzyme, receives SUMO proteins delivered by E1 and cooperates with E3 to identify the substrates. In most cases, UBC9 acts as an oncogene. Therefore, silencing UBC9 has the potential to restrain proliferation and migration and facilitate apoptosis of tumor cells, such as osteosarcoma U-2OS cells [123]. Of note, several SUMO E2 inhibitors have been screened out, involving Spectomycin B1, 2-D08, GSK145A, etc. Spectomycin B1, a natural compound derived from Streptomyces spectabilis initially recognized as an antibiotic against gram-positive bacteria, straightly attaches to UBC9, specifically impeding the establishment of the E2-SUMO intermediate and leading to the proliferative inhibition of MCF7 human breast-cancer cells [124]. Similarly, 2-D08, a synthetic oxygenated flavonoid, blocks the conjugation of SUMOs from the UBC9-SUMO thioester to substrates, thereby hindering SUMOylation in tumor cells [125], such as MDA-MB-231 breast-cancer cells [60]. Although GSK145A is not a specific noncovalent inhibitor of UBC9-dependent SUMOylation, it acts as a competitive substrate of SUMOylation, blocking the substrate recognition for Tricho-rhino-phalangeal syndrome type I protein (TRPS1) [126]. Compound 2, a synthesized UBC9-binding compound identified by small-molecule microarray (SMM)-based screening using fluorescently tagged UBC9, prevents the SUMOylation of RanGAP1 [127]. Additionally, WNN0605-F008 disturbs the catalytic activity of UBC9 via occupying its catalytic pocket, leading to the inhibition of RanGAP1 SUMOylation [128].
Altogether, the advancement of high-throughput screening assays has led to the discovery of inhibitors for the “undruggable” E2 enzyme UBC9. However, their clinical value requires further evaluation.
SUMO E3 inhibitors
SUMO E3-ligases ensure the specificity of target substrates and facilitate the transfer of SUMOs from the E2 conjugating enzyme to the substrates. Unfortunately, small-molecule inhibitors for SUMO E3-ligases are scarce. UNC3866 is one such inhibitor that binds preferentially to the CBX chromodomains of SUMO E3-ligase CBX4, inhibiting PC3 prostate cancer cells [129]. Additionally, UNC3866 exerts a powerful anti-tumor effect via hindering CBX4, contributing to the suppression of tumor cell growth and cancer stem cell properties [130]. Further development of SUMO E3 inhibitors with enhanced specificity is needed to disrupt the SUMOylation cascade effectively.
SUMO protease inhibitors
SUMO proteases act as both oncogenes and anti-oncogenes. In some tumors, the high expression of partial SUMO proteases exacerbates the malignant phenotypes, making them anti-tumor targets. Therefore, several natural and synthetic small-molecule inhibitors targeting SENP1 and SENP2 have been identified. For instance, streptonigrin, a natural product, directly binds to SENP1, thwarting its interaction with SUMO1 and leading to the diminution of hypoxia-inducible factor alpha (HIF1α) [131]. Triptolide, another natural small-molecule inhibitor, augments the cellular SUMOylation in prostate cancer cells via mitigating the mRNA and protein levels of SENP1, contributing to the inhibition of tumor cell growth in vivo and in vitro [132]. Similarly, a natural product Momordin Ic reduces prostate cancer cell proliferation and induces cell apoptosis via decreasing SENP1 expression and disrupting its SUMO2-RanGAP1-induced cleavage [133]. Correspondingly, several synthetic small-molecule compounds, such as GN6958 [134], compound 3 [135], and compound 13m [136], have also been developed to inhibit SENP1 activity; however, further verification is warranted using in vivo studies. Furthermore, three SENP2 inhibitors have been developed, namely ebselen [137], Compound 69 and 117 [138]. However, there is no evidence that these SENP2 inhibitors exert anti-tumor properties.
Taken together, SUMO proteases play dual roles in the SUMOylation cascade, involving promoting the SUMO maturation and expediting the SUMO deconjugation. Thus, inhibiting SUMO proteases may induce the antithetical outcomes of the SUMOylation cascade. Therefore, the anti-tumor effect of SUMO protease inhibitors requires not only in vivo and in vitro verification but also further clinical validation.
An in vivo study reported that loss of Ubc9 in adult mice can lead to severe diarrhea or death [139], indicating that SUMOylation inhibitors may possess potential toxicity and induce adverse effects. Moreover, the unanticipated tumor-suppressive role of SUMOylation may have significant implications for the appropriate use of SUMOylation inhibitors in clinical practice [140].
In summary, while most SUMOylation inhibitors hold promise in anti-tumor treatment, their potential toxicity and unexpected side effects necessitate further investigation. Considering the dual role of SUMOylation in tumors and physiological homeostasis, it is crucial to assess the safety and efficacy of these inhibitors comprehensively before their clinical use.
Conclusions and perspective
SUMOylation, as one of the critical PTMs, modulates protein function and localization, exerting tremendous influence on tumor initiation and progression. SUMOs are covalently conjugated to the lysine (K) residues of proteins with a SUMOylation consensus motif. To date, more than 3,600 SUMOylated proteins, with at least 7,300 SUMOylation sites, have been authenticated utilizing mass spectrometry and bioinformatics technology [141], underscoring the pivotal impact of this dynamic and reversible SUMOylation cascade on protein fate. To date, the SUMOylation process has been depicted adequately after about 30 years of in-depth research. However, there are still some enigmas in this multi-step enzymatic cascade. First, why do such a small series of SUMO enzymes catalyze thousands of protein substrates? Second, what exactly is SUMO5? a human SUMO pseudogene or a functional isoform? Third, what other cellular stress can induce SUMOylation in tumors? Fourth, what specific functions does each SUMO E3 ligase perform? Will there be new SUMO E3 ligases identified? Fifth, why does SUMOylation degrade substrate protein in some cases? Is this protein degradation induced by specific SUMO E3 ligase-mediated SUMOylation? With the development of multi-omics sequencing and deep learning, a better understanding of protein SUMOylation will be provided in future years.
In tumors, the SUMOylation cascade possesses the characteristic of exacerbating the cancer progression relying on its multifunctional biological effects, such as regulating transcriptional activity, increasing protein stability, etc. Intriguingly, SUMOylation can be triggered by various cellular stresses, involving hypoxia, viral infections, gut microbiota, and lactic acid accumulation, indicating that interfering with these cellular stresses holds promise for inhibiting tumor progression. However, the specific mechanisms of SUMOylation induced by cellular stresses remain unclear. Additionally, SUMOylation, as a form of non-mutational epigenetic reprogramming, engages in the management of various cancer hallmarks. Increasing evidence has successively illustrated that SUMOylation assists tumor cells in proliferation, invasion, metastasis, angiogenesis, PCD escape, metabolic reprogramming, and immune evasion. Thus, targeting enzymes involved in the SUMOylation cascade emerges as a promising anti-tumor strategy, with multiple small-molecule inhibitors targeting SUMOylation enzymes identified. However, merely one small-molecule inhibitor has progressed to clinical trials, highlighting the need for further research to address potential adverse effects using advanced techniques such as virtual high-throughput screening, molecular docking, proteolysis targeting chimera (PROTAC), and molecular glue. By leveraging multidisciplinary collaboration, the concurrent application of SUMOylation inhibitors in anti-tumour interventions holds the promise to impede tumor invasion, metastasis, and recurrence, and overcome drug resistance and immune evasion, ultimately advancing tumor therapeutics.
Abbreviations
SUMO: small ubiquitin-like modifier
CRC: colorectal cancer
SENP: sentrin-specific protease
UBC9: ubiquitin carrier protein 9
SIM: SUMO-interacting motif
UPS: ubiquitin-proteasome system
MCL1: myeloid cell leukemia 1
IGF2BP2: insulin-like growth factor 2 mRNA-binding protein 2
RALY: RNA-binding protein Raly
m5C: 5-methylcytosine
NSUN2: NOL1/NOP2/Sun domain family member 2
RNF146: RING finger protein 146
Axin: axis inhibition protein 1
HCC: hepatocellular carcinoma
PML: promyelocytic leukemia
c-Myc: proto-oncogene c-Myc
Flot-1: flotillin-1
Snail: zinc finger protein SNAI1
EMT: epithelial-to-mesenchymal transition
OTUB2: OTU domain-containing ubiquitin aldehyde-binding protein 2
YAP: yes-associated protein
TAZ: PDZ-binding motif
PKM2: pyruvate kinase M2
STAT3: signal transducer and activator of the transcription 3
RanBP2: Ran-binding protein 2
IL-33: interleukin-33
ERK5: extracellular-signal-related kinase 5
LLPS: liquid-liquid phase separation
BMC: biomolecular condensate
RNF168: RING finger protein 168
PIAS: protein inhibitors of activated signal transducer and activator of transcription
53BP1: TP53-binding protein 1
FOXM1B: forkhead box protein M1 B
JNK1: Jun amino-terminal kinases 1
FOXK2: forkhead box K2
Eya1: eyes absent homolog 1
MORC2: microrchidia CW-type zinc finger 2
TRIM28: tripartite motif containing 28
CSNK2A1: casein kinase II subunit alpha
MCM10: minichromosome maintenance protein 10
H2AZ: histone H2A.Z
MANF: mesencephalic astrocyte‐derived neurotrophic factor
Mettl3: methyltransferase-like 3
m6A: N6-methyladenosine
VEGF: vascular endothelial growth factor
VEGFR2: vascular endothelial growth factor receptor 2
HIF-1α: hypoxia-inducible factor-1alpha
Cbx4: chromobox protein homolog 4
hnRNP: heterogeneous nuclear ribonucleoprotein
NUSAP1: nucleolar and spindle-associated protein 1
TCF4: transcription factor 4
Smurf2: SMAD specific E3 ubiquitin protein ligase 2
TGFβ: transforming growth factor-β
CSC: cancer stem cell
IRF1: interferon regulatory factor 1
TRIM21: tripartite motif containing 21
Lats1: large tumor suppressor 1
PCD: programmed cell death
HNSCC: head-neck squamous cell carcinoma
TRIB3: tribbles pseudokinase-3
ALKBH5: AlkB Homolog 5
GLUT1: glucose transporter type 1
HK2: hexokinase 2
SAFB: scaffold attachment factor B
LXRs: liver X receptors
PD-L1: programmed cell death protein-1 ligand
IFNAR1: IFN-alpha receptor 1
NRF2: nuclear factor erythroid-2 related factor 2
Gpx2: glutathione peroxidase 2
ROS: reactive oxygen species
PHGDH: phosphoglycerate dehydrogenase
YTHDF2: YT521-B homology domain-containing family protein 2
KHSRP: KH-type splicing regulatory protein
LKB1: liver kinase B1
EBV: Epstein-Barr virus
HPV: human papilloma virus
HBV: hepatitis B virus
LMP1: latent membrane protein-1
HER2: human epidermal growth factor receptor 2
SCFA: short-chain fatty acid
TME: tumor microenvironment
APC/C: anaphase-promoting complex
ICC: intrahepatic cholangiocarcinoma
TRPS1: tricho-rhino-phalangeal syndrome type I protein
BRCA: breast cancer
GC: gastric cancer
ESCC: esophageal squamous-cell carcinoma
PDAC: pancreatic ductal adenocarcinoma
RCC: renal cell cancer
Acknowledgements
We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82272915, 82073884), the project of the fourth batch of the science and technology plan of Liaoning province (2021JH210300133), the project of the applied basic research program of Liaoning province (2022JH2/101300050), and China Postdoctoral Science Foundation (2023MD734246).
Author contributions
K.L., H.W., S.G., and H.Z. conceived the structure of the manuscript. H.Z., N.D., and Y.L. drafted the manuscript. S.G., H.W., K.L., and X.Y. supervised the revision of the manuscript. X.H. and H.Z. revised the manuscript. H.Z., N.D., Y.L., S.J., C.Z., and H.W. prepared the figures. All authors read and approved the final manuscript.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97-107
2. Gu Y, Fang Y, Wu X, Xu T, Hu T, Xu Y. et al. The emerging roles of SUMOylation in the tumor microenvironment and therapeutic implications. Exp Hematol Oncol. 2023;12:58
3. Li B, Kang H, Xiao Y, Du Y, Xiao Y, Song G. et al. LncRNA GAL promotes colorectal cancer liver metastasis through stabilizing GLUT1. Oncogene. 2022;41:1882-94
4. Hung PF, Hong TM, Chang CC, Hung CL, Hsu YL, Chang YL. et al. Hypoxia-induced Slug SUMOylation enhances lung cancer metastasis. J Exp Clin Cancer Res. 2019;38:5
5. Liu X, Liu J, Xiao W, Zeng Q, Bo H, Zhu Y. et al. SIRT1 Regulates N(6) -Methyladenosine RNA Modification in Hepatocarcinogenesis by Inducing RANBP2-Dependent FTO SUMOylation. Hepatology. 2020;72:2029-50
6. Wei M, Huang X, Liao L, Tian Y, Zheng X. SENP1 Decreases RNF168 Phase Separation to Promote DNA Damage Repair and Drug Resistance in Colon Cancer. Cancer Res. 2023;83:2908-23
7. Wang L, Wansleeben C, Zhao S, Miao P, Paschen W, Yang W. SUMO2 is essential while SUMO3 is dispensable for mouse embryonic development. EMBO Rep. 2014;15:878-85
8. Evdokimov E, Sharma P, Lockett SJ, Lualdi M, Kuehn MR. Loss of SUMO1 in mice affects RanGAP1 localization and formation of PML nuclear bodies, but is not lethal as it can be compensated by SUMO2 or SUMO3. J Cell Sci. 2008;121:4106-13
9. Owerbach D, McKay EM, Yeh ET, Gabbay KH, Bohren KM. A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation. Biochem Biophys Res Commun. 2005;337:517-20
10. Wei W, Yang P, Pang J, Zhang S, Wang Y, Wang MH. et al. A stress-dependent SUMO4 sumoylation of its substrate proteins. Biochem Biophys Res Commun. 2008;375:454-9
11. Liang YC, Lee CC, Yao YL, Lai CC, Schmitz ML, Yang WM. SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies. Sci Rep. 2016;6:26509
12. Chang HM, Yeh ETH. SUMO: From Bench to Bedside. Physiol Rev. 2020;100:1599-619
13. Vertegaal ACO. Signalling mechanisms and cellular functions of SUMO. Nat Rev Mol Cell Biol. 2022;23:715-31
14. Kunz K, Piller T, Muller S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J Cell Sci. 2018;131:jcs211904
15. Long X, Zhao B, Lu W, Chen X, Yang X, Huang J. et al. The Critical Roles of the SUMO-Specific Protease SENP3 in Human Diseases and Clinical Implications. Front Physiol. 2020;11:558220
16. Li H, Wang D, Yi B, Cai H, Wang Y, Lou X. et al. SUMOylation of IGF2BP2 promotes vasculogenic mimicry of glioma via regulating OIP5-AS1/miR-495-3p axis. Int J Biol Sci. 2021;17:2912-30
17. Li S, Wang J, Hu G, Aman S, Li B, Li Y. et al. SUMOylation of MCL1 protein enhances its stability by regulating the ubiquitin-proteasome pathway. Cell Signal. 2020;73:109686
18. Cao S, Wang D, Wang P, Liu Y, Dong W, Ruan X. et al. SUMOylation of RALY promotes vasculogenic mimicry in glioma cells via the FOXD1/DKK1 pathway. Cell Biol Toxicol. 2023;39:3323-40
19. Hu Y, Chen C, Tong X, Chen S, Hu X, Pan B. et al. NSUN2 modified by SUMO-2/3 promotes gastric cancer progression and regulates mRNA m5C methylation. Cell Death Dis. 2021;12:842
20. Mo Y, Wang Y, Zhang S, Xiong F, Yan Q, Jiang X. et al. Circular RNA circRNF13 inhibits proliferation and metastasis of nasopharyngeal carcinoma via SUMO2. Mol Cancer. 2021;20:112
21. Son SH, Kim MY, Lim YS, Jin HC, Shin JH, Yi JK. et al. SUMOylation-mediated PSME3-20S proteasomal degradation of transcription factor CP2c is crucial for cell cycle progression. Sci Adv. 2023;9:eadd4969
22. Li W, Han Q, Zhu Y, Zhou Y, Zhang J, Wu W. et al. SUMOylation of RNF146 results in Axin degradation and activation of Wnt/beta-catenin signaling to promote the progression of hepatocellular carcinoma. Oncogene. 2023;42:1728-40
23. Zhang A, Tao W, Zhai K, Fang X, Huang Z, Yu JS. et al. Protein sumoylation with SUMO1 promoted by Pin1 in glioma stem cells augments glioblastoma malignancy. Neuro Oncol. 2020;22:1809-21
24. Jang D, Kwon H, Choi M, Lee J, Pak Y. Sumoylation of Flotillin-1 promotes EMT in metastatic prostate cancer by suppressing Snail degradation. Oncogene. 2019;38:3248-60
25. Zhang Z, Du J, Wang S, Shao L, Jin K, Li F. et al. OTUB2 Promotes Cancer Metastasis via Hippo-Independent Activation of YAP and TAZ. Mol Cell. 2019;73:7-21 e7
26. Zhou Q, Yin Y, Yu M, Gao D, Sun J, Yang Z. et al. GTPBP4 promotes hepatocellular carcinoma progression and metastasis via the PKM2 dependent glucose metabolism. Redox Biol. 2022;56:102458
27. Wang Z, Pan B, Qiu J, Zhang X, Ke X, Shen S. et al. SUMOylated IL-33 in the nucleus stabilizes the transcription factor IRF1 in hepatocellular carcinoma cells to promote immune escape. Sci Signal. 2023;16:eabq3362
28. Ashikari D, Takayama K, Tanaka T, Suzuki Y, Obinata D, Fujimura T. et al. Androgen induces G3BP2 and SUMO-mediated p53 nuclear export in prostate cancer. Oncogene. 2017;36:6272-81
29. Erazo T, Espinosa-Gil S, Dieguez-Martinez N, Gomez N, Lizcano JM. SUMOylation Is Required for ERK5 Nuclear Translocation and ERK5-Mediated Cancer Cell Proliferation. Int J Mol Sci. 2020;21:2203
30. Jiang C, Zhang C, Dai M, Wang F, Xu S, Han D. et al. Interplay between SUMO1-related SUMOylation and phosphorylation of p65 promotes hepatocellular carcinoma progression. Biochim Biophys Acta Mol Cell Res. 2024;1871:119595
31. Zheng LW, Liu CC, Yu KD. Phase separations in oncogenesis, tumor progressions and metastasis: a glance from hallmarks of cancer. J Hematol Oncol. 2023;16:123
32. Cheng X, Yang W, Lin W, Mei F. Paradoxes of Cellular SUMOylation Regulation: A Role of Biomolecular Condensates? Pharmacol Rev. 2023;75:979-1006
33. Bhandari K, Cotten MA, Kim J, Rosen MK, Schmit JD. Structure-Function Properties in Disordered Condensates. J Phys Chem B. 2021;125:467-76
34. Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal AC, Nielsen ML. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol. 2017;24:325-36
35. Yan X, Zhang M, Wang D. Interplay between posttranslational modifications and liquid-liquid phase separation in tumors. Cancer Lett. 2024;584:216614
36. Wang CM, Liu R, Wang L, Nascimento L, Brennan VC, Yang WH. SUMOylation of FOXM1B alters its transcriptional activity on regulation of MiR-200 family and JNK1 in MCF7 human breast cancer cells. Int J Mol Sci. 2014;15:10233-51
37. Li Y, Chen J, Wang B, Xu Z, Wu C, Ma J. et al. FOXK2 affects cancer cell response to chemotherapy by promoting nucleotide de novo synthesis. Drug Resist Updat. 2023;67:100926
38. Sun Y, Kaneko S, Li XK, Li X. The PI3K/Akt signal hyperactivates Eya1 via the SUMOylation pathway. Oncogene. 2015;34:2527-37
39. Zhang FL, Yang SY, Liao L, Zhang TM, Zhang YL, Hu SY. et al. Dynamic SUMOylation of MORC2 orchestrates chromatin remodelling and DNA repair in response to DNA damage and drives chemoresistance in breast cancer. Theranostics. 2023;13:973-90
40. Tian J, Lu Z, Niu S, Zhang S, Ying P, Wang L. et al. Aberrant MCM10 SUMOylation induces genomic instability mediated by a genetic variant associated with survival of esophageal squamous cell carcinoma. Clin Transl Med. 2021;11:e485
41. Liu T, Wang H, Chen Y, Wan Z, Du Z, Shen H. et al. SENP5 promotes homologous recombination-mediated DNA damage repair in colorectal cancer cells through H2AZ deSUMOylation. J Exp Clin Cancer Res. 2023;42:234
42. Liu J, Wu Z, Han D, Wei C, Liang Y, Jiang T. et al. Mesencephalic Astrocyte-Derived Neurotrophic Factor Inhibits Liver Cancer Through Small Ubiquitin-Related Modifier (SUMO)ylation-Related Suppression of NF-kappaB/Snail Signaling Pathway and Epithelial-Mesenchymal Transition. Hepatology. 2020;71:1262-78
43. Xu H, Wang H, Zhao W, Fu S, Li Y, Ni W. et al. SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating Snail mRNA homeostasis in hepatocellular carcinoma. Theranostics. 2020;10:5671-86
44. Wang M, Jiang X. SUMOylation of vascular endothelial growth factor receptor 2 inhibits the proliferation, migration, and angiogenesis signaling pathway in non-small cell lung cancer. Anticancer Drugs. 2020;31:492-9
45. Li J, Xu Y, Long XD, Wang W, Jiao HK, Mei Z. et al. Cbx4 governs HIF-1alpha to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell. 2014;25:118-31
46. Cheng J, Kang X, Zhang S, Yeh ET. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 2007;131:584-95
47. Guo Q, Fan Y, Wang Q, Li B, Qiu W, Qi Y. et al. Glioblastoma upregulates SUMOylation of hnRNP A2/B1 to eliminate the tumor suppressor miR-204-3p, accelerating angiogenesis under hypoxia. Cell Death Dis. 2023;14:147
48. Li H, Zhang W, Yan M, Qiu J, Chen J, Sun X. et al. Nucleolar and spindle associated protein 1 promotes metastasis of cervical carcinoma cells by activating Wnt/beta-catenin signaling. J Exp Clin Cancer Res. 2019;38:33
49. Chandhoke AS, Karve K, Dadakhujaev S, Netherton S, Deng L, Bonni S. The ubiquitin ligase Smurf2 suppresses TGFbeta-induced epithelial-mesenchymal transition in a sumoylation-regulated manner. Cell Death Differ. 2016;23:876-88
50. Du L, Li YJ, Fakih M, Wiatrek RL, Duldulao M, Chen Z. et al. Role of SUMO activating enzyme in cancer stem cell maintenance and self-renewal. Nat Commun. 2016;7:12326
51. Bogachek MV, Park JM, De Andrade JP, Lorenzen AW, Kulak MV, White JR. et al. Inhibiting the SUMO Pathway Represses the Cancer Stem Cell Population in Breast and Colorectal Carcinomas. Stem Cell Reports. 2016;7:1140-51
52. Bugter JM, Fenderico N, Maurice MM. Mutations and mechanisms of WNT pathway tumour suppressors in cancer. Nat Rev Cancer. 2021;21:5-21
53. Hao J, Zhang Y, Jing D, Li Y, Li J, Zhao Z. Role of Hippo signaling in cancer stem cells. J Cell Physiol. 2014;229:266-70
54. Li L, Duan Q, Zeng Z, Zhao J, Lu J, Sun J. et al. UHRF2 promotes intestinal tumorigenesis through stabilization of TCF4 mediated Wnt/beta-catenin signaling. Int J Cancer. 2020;147:2239-52
55. Mei L, Yuan L, Shi W, Fan S, Tang C, Fan X. et al. SUMOylation of large tumor suppressor 1 at Lys751 attenuates its kinase activity and tumor-suppressor functions. Cancer Lett. 2017;386:1-11
56. Li R, Wei J, Jiang C, Liu D, Deng L, Zhang K. et al. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res. 2013;73:5742-53
57. Lin CH, Liu SY, Lee EH. SUMO modification of Akt regulates global SUMOylation and substrate SUMOylation specificity through Akt phosphorylation of Ubc9 and SUMO1. Oncogene. 2016;35:595-607
58. Yang Y, Liang Z, Xia Z, Wang X, Ma Y, Sheng Z. et al. SAE1 promotes human glioma progression through activating AKT SUMOylation-mediated signaling pathways. Cell Commun Signal. 2019;17:82
59. Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495-504
60. Lorente M, Garcia-Casas A, Salvador N, Martinez-Lopez A, Gabicagogeascoa E, Velasco G. et al. Inhibiting SUMO1-mediated SUMOylation induces autophagy-mediated cancer cell death and reduces tumour cell invasion via RAC1. J Cell Sci. 2019 132
61. Yu F, Zhu AC, Liu S, Gao B, Wang Y, Khudaverdyan N. et al. RBM33 is a unique m(6)A RNA-binding protein that regulates ALKBH5 demethylase activity and substrate selectivity. Mol Cell. 2023;83:2003-19 e6
62. Wang Y, Hu J, Wu S, Fleishman JS, Li Y, Xu Y. et al. Targeting epigenetic and posttranslational modifications regulating ferroptosis for the treatment of diseases. Signal Transduct Target Ther. 2023;8:449
63. Gao C, Xiao F, Zhang L, Sun Y, Wang L, Liu X. et al. SENP1 inhibition suppresses the growth of lung cancer cells through activation of A20-mediated ferroptosis. Ann Transl Med. 2022;10:224
64. Xu X, Mao Y, Feng Z, Dai F, Gu T, Zheng J. SENP1 inhibits ferroptosis and promotes head and neck squamous cell carcinoma by regulating ACSL4 protein stability via SUMO1. Oncol Rep. 2024;51:34
65. Luo N, Zhang K, Li X, Hu Y, Guo L. Tanshinone IIA destabilizes SLC7A11 by regulating PIAS4-mediated SUMOylation of SLC7A11 through KDM1A, and promotes ferroptosis in breast cancer. J Adv Res. 2024 [Epub ahead of print]
66. Zhu S, Gu H, Peng C, Xia F, Cao H, Cui H. Regulation of Glucose, Fatty Acid and Amino Acid Metabolism by Ubiquitination and SUMOylation for Cancer Progression. Front Cell Dev Biol. 2022;10:849625
67. Hou PP, Luo LJ, Chen HZ, Chen QT, Bian XL, Wu SF. et al. Ectosomal PKM2 Promotes HCC by Inducing Macrophage Differentiation and Remodeling the Tumor Microenvironment. Mol Cell. 2020;78:1192-206 e10
68. Shangguan X, He J, Ma Z, Zhang W, Ji Y, Shen K. et al. SUMOylation controls the binding of hexokinase 2 to mitochondria and protects against prostate cancer tumorigenesis. Nat Commun. 2021;12:1812
69. Ni Y, Yang Z, Agbana YL, Bai H, Wang L, Yang L. et al. Silent information regulator 2 promotes clear cell renal cell carcinoma progression through deacetylation and small ubiquitin-related modifier 1 modification of glucose 6-phosphate dehydrogenase. Cancer Sci. 2021;112:4075-86
70. Guo D, Tong Y, Jiang X, Meng Y, Jiang H, Du L. et al. Aerobic glycolysis promotes tumor immune evasion by hexokinase2-mediated phosphorylation of IkappaBalpha. Cell Metab. 2022;34:1312-24 e6
71. Demel UM, Boger M, Yousefian S, Grunert C, Zhang L, Hotz PW. et al. Activated SUMOylation restricts MHC class I antigen presentation to confer immune evasion in cancer. J Clin Invest. 2022;132:e152383
72. Xiao J, Sun F, Wang YN, Liu B, Zhou P, Wang FX. et al. UBC9 deficiency enhances immunostimulatory macrophage activation and subsequent antitumor T cell response in prostate cancer. J Clin Invest. 2023;133:e158352
73. Ma X, Bi E, Huang C, Lu Y, Xue G, Guo X. et al. Cholesterol negatively regulates IL-9-producing CD8(+) T cell differentiation and antitumor activity. J Exp Med. 2018;215:1555-69
74. Ma X, Jia S, Wang G, Liang M, Guo T, Du H. et al. TRIM28 promotes the escape of gastric cancer cells from immune surveillance by increasing PD-L1 abundance. Signal Transduct Target Ther. 2023;8:246
75. Zhao Y, Chen Y, Jin M, Wang J. The crosstalk between m(6)A RNA methylation and other epigenetic regulators: a novel perspective in epigenetic remodeling. Theranostics. 2021;11:4549-66
76. Guo H, Xu J, Zheng Q, He J, Zhou W, Wang K. et al. NRF2 SUMOylation promotes de novo serine synthesis and maintains HCC tumorigenesis. Cancer Lett. 2019;466:39-48
77. Zheng J, Liu L, Wang S, Huang X. SUMO-1 Promotes Ishikawa Cell Proliferation and Apoptosis in Endometrial Cancer by Increasing Sumoylation of Histone H4. Int J Gynecol Cancer. 2015;25:1364-8
78. Hou G, Zhao X, Li L, Yang Q, Liu X, Huang C. et al. SUMOylation of YTHDF2 promotes mRNA degradation and cancer progression by increasing its binding affinity with m6A-modified mRNAs. Nucleic Acids Res. 2021;49:2859-77
79. Comerford KM, Leonard MO, Karhausen J, Carey R, Colgan SP, Taylor CT. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc Natl Acad Sci U S A. 2003;100:986-91
80. Koh MY, Nguyen V, Lemos R Jr, Darnay BG, Kiriakova G, Abdelmelek M. et al. Hypoxia-induced SUMOylation of E3 ligase HAF determines specific activation of HIF2 in clear-cell renal cell carcinoma. Cancer Res. 2015;75:316-29
81. Sun L, Li H, Chen J, Dehennaut V, Zhao Y, Yang Y. et al. A SUMOylation-dependent pathway regulates SIRT1 transcription and lung cancer metastasis. J Natl Cancer Inst. 2013;105:887-98
82. Yuan H, Deng R, Zhao X, Chen R, Hou G, Zhang H. et al. SUMO1 modification of KHSRP regulates tumorigenesis by preventing the TL-G-Rich miRNA biogenesis. Mol Cancer. 2017;16:157
83. Zubiete-Franco I, Garcia-Rodriguez JL, Lopitz-Otsoa F, Serrano-Macia M, Simon J, Fernandez-Tussy P. et al. SUMOylation regulates LKB1 localization and its oncogenic activity in liver cancer. EBioMedicine. 2019;40:406-21
84. Hatfield SM, Kjaergaard J, Lukashev D, Belikoff B, Schreiber TH, Sethumadhavan S. et al. Systemic oxygenation weakens the hypoxia and hypoxia inducible factor 1alpha-dependent and extracellular adenosine-mediated tumor protection. J Mol Med (Berl). 2014;92:1283-92
85. Zapatka M, Borozan I, Brewer DS, Iskar M, Grundhoff A, Alawi M. et al. The landscape of viral associations in human cancers. Nat Genet. 2020;52:320-30
86. Salahuddin S, Fath EK, Biel N, Ray A, Moss CR, Patel A. et al. Epstein-Barr Virus Latent Membrane Protein-1 Induces the Expression of SUMO-1 and SUMO-2/3 in LMP1-positive Lymphomas and Cells. Sci Rep. 2019;9:208
87. Bentz GL, Whitehurst CB, Pagano JS. Epstein-Barr virus latent membrane protein 1 (LMP1) C-terminal-activating region 3 contributes to LMP1-mediated cellular migration via its interaction with Ubc9. J Virol. 2011;85:10144-53
88. Selby TL, Biel N, Varn M, Patel S, Patel A, Hilding L. et al. The Epstein-Barr Virus Oncoprotein, LMP1, Regulates the Function of SENP2, a SUMO-protease. Sci Rep. 2019;9:9523
89. Mattoscio D, Casadio C, Miccolo C, Maffini F, Raimondi A, Tacchetti C. et al. Autophagy regulates UBC9 levels during viral-mediated tumorigenesis. PLoS Pathog. 2017;13:e1006262
90. You H, Yuan D, Li Q, Zhang N, Kong D, Yu T. et al. Hepatitis B virus X protein increases LASP1 SUMOylation to stabilize HER2 and facilitate hepatocarcinogenesis. Int J Biol Macromol. 2023;226:996-1009
91. Cougnoux A, Dalmasso G, Martinez R, Buc E, Delmas J, Gibold L. et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut. 2014;63:1932-42
92. Ezzine C, Loison L, Montbrion N, Bole-Feysot C, Dechelotte P, Coeffier M. et al. Fatty acids produced by the gut microbiota dampen host inflammatory responses by modulating intestinal SUMOylation. Gut Microbes. 2022;14:2108280
93. Hou H, Chen D, Zhang K, Zhang W, Liu T, Wang S. et al. Gut microbiota-derived short-chain fatty acids and colorectal cancer: Ready for clinical translation? Cancer Lett. 2022;526:225-35
94. Alvandi E, Wong WKM, Joglekar MV, Spring KJ, Hardikar AA. Short-chain fatty acid concentrations in the incidence and risk-stratification of colorectal cancer: a systematic review and meta-analysis. BMC Med. 2022;20:323
95. Ippolito L, Morandi A, Giannoni E, Chiarugi P. Lactate: A Metabolic Driver in the Tumour Landscape. Trends Biochem Sci. 2019;44:153-66
96. Liu W, Wang Y, Bozi LHM, Fischer PD, Jedrychowski MP, Xiao H. et al. Lactate regulates cell cycle by remodelling the anaphase promoting complex. Nature. 2023;616:790-7
97. Zhang Q, Huang H, Liu A, Li J, Liu C, Sun B. et al. Cell division cycle 20 (CDC20) drives prostate cancer progression via stabilization of beta-catenin in cancer stem-like cells. EBioMedicine. 2019;42:397-407
98. Paul D, Ghorai S, Dinesh US, Shetty P, Chattopadhyay S, Santra MK. Cdc20 directs proteasome-mediated degradation of the tumor suppressor SMAR1 in higher grades of cancer through the anaphase promoting complex. Cell Death Dis. 2017;8:e2882
99. Zheng J, Wang Y, Tao L, Cai J, Shen Z, Liu Y. et al. Circ-RAPGEF5 promotes intrahepatic cholangiocarcinoma progression by stabilizing SAE1 to facilitate SUMOylation. J Exp Clin Cancer Res. 2023;42:239
100. Kukkula A, Ojala VK, Mendez LM, Sistonen L, Elenius K, Sundvall M. Therapeutic Potential of Targeting the SUMO Pathway in Cancer. Cancers (Basel). 2021;13:4402
101. Kroonen JS, Vertegaal ACO. Targeting SUMO Signaling to Wrestle Cancer. Trends Cancer. 2021;7:496-510
102. Chen Y, Peng W, Tao Q, Li S, Wu Z, Zhou Y. et al. Increased Small Ubiquitin-like Modifier-Activating Enzyme SAE1 Promotes Hepatocellular Carcinoma by Enhancing mTOR SUMOylation. Lab Invest. 2023;103:100011
103. Liu X, Xu Y, Pang Z, Guo F, Qin Q, Yin T. et al. Knockdown of SUMO-activating enzyme subunit 2 (SAE2) suppresses cancer malignancy and enhances chemotherapy sensitivity in small cell lung cancer. J Hematol Oncol. 2015;8:67
104. Fukuda I, Ito A, Hirai G, Nishimura S, Kawasaki H, Saitoh H. et al. Ginkgolic acid inhibits protein SUMOylation by blocking formation of the E1-SUMO intermediate. Chem Biol. 2009;16:133-40
105. Fukuda I, Ito A, Uramoto M, Saitoh H, Kawasaki H, Osada H. et al. Kerriamycin B inhibits protein SUMOylation. J Antibiot (Tokyo). 2009;62:221-4
106. Liu D, Li Z, Yang Z, Ma J, Mai S. Ginkgoic acid impedes gastric cancer cell proliferation, migration and EMT through inhibiting the SUMOylation of IGF-1R. Chem Biol Interact. 2021;337:109394
107. Takemoto M, Kawamura Y, Hirohama M, Yamaguchi Y, Handa H, Saitoh H. et al. Inhibition of protein SUMOylation by davidiin, an ellagitannin from Davidia involucrata. J Antibiot (Tokyo). 2014;67:335-8
108. Suzawa M, Miranda DA, Ramos KA, Ang KK, Faivre EJ, Wilson CG. et al. A gene-expression screen identifies a non-toxic sumoylation inhibitor that mimics SUMO-less human LRH-1 in liver. Elife. 2015;4:e09003
109. Kumar A, Ito A, Hirohama M, Yoshida M, Zhang KY. Identification of sumoylation activating enzyme 1 inhibitors by structure-based virtual screening. J Chem Inf Model. 2013;53:809-20
110. Lv Z, Yuan L, Atkison JH, Williams KM, Vega R, Sessions EH. et al. Molecular mechanism of a covalent allosteric inhibitor of SUMO E1 activating enzyme. Nat Commun. 2018;9:5145
111. He X, Riceberg J, Soucy T, Koenig E, Minissale J, Gallery M. et al. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat Chem Biol. 2017;13:1164-71
112. Biederstadt A, Hassan Z, Schneeweis C, Schick M, Schneider L, Muckenhuber A. et al. SUMO pathway inhibition targets an aggressive pancreatic cancer subtype. Gut. 2020;69:1472-82
113. Langston SP, Grossman S, England D, Afroze R, Bence N, Bowman D. et al. Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer. J Med Chem. 2021;64:2501-20
114. Boulanger M, Aqrouq M, Tempe D, Kifagi C, Ristic M, Akl D. et al. DeSUMOylation of chromatin-bound proteins limits the rapid transcriptional reprogramming induced by daunorubicin in acute myeloid leukemias. Nucleic Acids Res. 2023;51:8413-33
115. Lightcap ES, Yu P, Grossman S, Song K, Khattar M, Xega K. et al. A small-molecule SUMOylation inhibitor activates antitumor immune responses and potentiates immune therapies in preclinical models. Sci Transl Med. 2021;13:eaba7791
116. Nakamura A, Grossman S, Song K, Xega K, Zhang Y, Cvet D. et al. The SUMOylation inhibitor subasumstat potentiates rituximab activity by IFN1-dependent macrophage and NK cell stimulation. Blood. 2022;139:2770-81
117. Gabellier L, De Toledo M, Chakraborty M, Akl D, Hallal R, Aqrouq M. et al. SUMOylation inhibitor TAK-981 (subasumstat) synergizes with 5-azacitidine in preclinical models of acute myeloid leukemia. Haematologica. 2023;109:98-114
118. Kumar S, Schoonderwoerd MJA, Kroonen JS, de Graaf IJ, Sluijter M, Ruano D. et al. Targeting pancreatic cancer by TAK-981: a SUMOylation inhibitor that activates the immune system and blocks cancer cell cycle progression in a preclinical model. Gut. 2022;71:2266-83
119. Zhang H, Tomar VS, Li J, Basavaraja R, Yan F, Gui J. et al. Protection of Regulatory T Cells from Fragility and Inactivation in the Tumor Microenvironment. Cancer Immunol Res. 2022;10:1490-505
120. Lu Z, McBrearty N, Chen J, Tomar VS, Zhang H, De Rosa G. et al. ATF3 and CH25H regulate effector trogocytosis and anti-tumor activities of endogenous and immunotherapeutic cytotoxic T lymphocytes. Cell Metab. 2022;34:1342-58 e7
121. Kim HS, Kim BR, Dao TTP, Kim JM, Kim YJ, Son H. et al. TAK-981, a SUMOylation inhibitor, suppresses AML growth immune-independently. Blood Adv. 2023;7:3155-68
122. Derry JMJ, Burns C, Frazier JP, Beirne E, Grenley M, DuFort CC. et al. Trackable Intratumor Microdosing and Spatial Profiling Provide Early Insights into Activity of Investigational Agents in the Intact Tumor Microenvironment. Clin Cancer Res. 2023;29:3813-25
123. Zhang D, Yu K, Yang Z, Li Y, Ma X, Bian X. et al. Silencing Ubc9 expression suppresses osteosarcoma tumorigenesis and enhances chemosensitivity to HSV-TK/GCV by regulating connexin 43 SUMOylation. Int J Oncol. 2018;53:1323-31
124. Hirohama M, Kumar A, Fukuda I, Matsuoka S, Igarashi Y, Saitoh H. et al. Spectomycin B1 as a novel SUMOylation inhibitor that directly binds to SUMO E2. ACS Chem Biol. 2013;8:2635-42
125. Kim YS, Keyser SG, Schneekloth JS Jr. Synthesis of 2',3',4'-trihydroxyflavone (2-D08), an inhibitor of protein sumoylation. Bioorg Med Chem Lett. 2014;24:1094-7
126. Brandt M, Szewczuk LM, Zhang H, Hong X, McCormick PM, Lewis TS. et al. Development of a high-throughput screen to detect inhibitors of TRPS1 sumoylation. Assay Drug Dev Technol. 2013;11:308-25
127. Zlotkowski K, Hewitt WM, Sinniah RS, Tropea JE, Needle D, Lountos GT. et al. A Small-Molecule Microarray Approach for the Identification of E2 Enzyme Inhibitors in Ubiquitin-Like Conjugation Pathways. SLAS Discov. 2017;22:760-6
128. Wang YZ, Liu X, Way G, Madarha V, Zhou QT, Yang DH. et al. An in vitro Forster resonance energy transfer-based high-throughput screening assay identifies inhibitors of SUMOylation E2 Ubc9. Acta Pharmacol Sin. 2020;41:1497-506
129. Stuckey JI, Dickson BM, Cheng N, Liu Y, Norris JL, Cholensky SH. et al. A cellular chemical probe targeting the chromodomains of Polycomb repressive complex 1. Nat Chem Biol. 2016;12:180-7
130. Zhao W, Ma B, Tian Z, Han H, Tang J, Dong B. et al. Inhibiting CBX4 efficiently protects hepatocellular carcinoma cells against sorafenib resistance. Br J Cancer. 2021;124:1237-48
131. Ambaye N, Chen CH, Khanna S, Li YJ, Chen Y. Streptonigrin Inhibits SENP1 and Reduces the Protein Level of Hypoxia-Inducible Factor 1alpha (HIF1alpha) in Cells. Biochemistry. 2018;57:1807-13
132. Huang W, He T, Chai C, Yang Y, Zheng Y, Zhou P. et al. Triptolide inhibits the proliferation of prostate cancer cells and down-regulates SUMO-specific protease 1 expression. PLoS One. 2012;7:e37693
133. Wu J, Lei H, Zhang J, Chen X, Tang C, Wang W. et al. Momordin Ic, a new natural SENP1 inhibitor, inhibits prostate cancer cell proliferation. Oncotarget. 2016;7:58995-9005
134. Uno M, Koma Y, Ban HS, Nakamura H. Discovery of 1-[4-(N-benzylamino)phenyl]-3-phenylurea derivatives as non-peptidic selective SUMO-sentrin specific protease (SENP)1 inhibitors. Bioorg Med Chem Lett. 2012;22:5169-73
135. Xie W, Wang Z, Zhang J, Wang L, Zhao Y, Zhou H. Development and evaluation of a highly reliable assay for SUMO-specific protease inhibitors. Bioorg Med Chem Lett. 2016;26:2124-8
136. Zhao Y, Wang Z, Zhang J, Zhou H. Identification of SENP1 inhibitors through in silico screening and rational drug design. Eur J Med Chem. 2016;122:178-84
137. Bernstock JD, Ye D, Smith JA, Lee YJ, Gessler FA, Yasgar A. et al. Quantitative high-throughput screening identifies cytoprotective molecules that enhance SUMO conjugation via the inhibition of SUMO-specific protease (SENP)2. FASEB J. 2018;32:1677-91
138. Kumar A, Ito A, Takemoto M, Yoshida M, Zhang KY. Identification of 1,2,5-oxadiazoles as a new class of SENP2 inhibitors using structure based virtual screening. J Chem Inf Model. 2014;54:870-80
139. Demarque MD, Nacerddine K, Neyret-Kahn H, Andrieux A, Danenberg E, Jouvion G. et al. Sumoylation by Ubc9 regulates the stem cell compartment and structure and function of the intestinal epithelium in mice. Gastroenterology. 2011;140:286-96
140. Lopez I, Chalatsi E, Ellenbroek SIJ, Andrieux A, Roux PF, Cerapio JP. et al. An unanticipated tumor-suppressive role of the SUMO pathway in the intestine unveiled by Ubc9 haploinsufficiency. Oncogene. 2020;39:6692-703
141. Hendriks IA, Vertegaal AC. A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol. 2016;17:581-95
142. Zhao Y, Chen J, Zheng H, Luo Y, An M, Lin Y. et al. SUMOylation-Driven mRNA Circularization Enhances Translation and Promotes Lymphatic Metastasis of Bladder Cancer. Cancer Res. 2024;84:434-48
143. An M, Zheng H, Huang J, Lin Y, Luo Y, Kong Y. et al. Aberrant Nuclear Export of circNCOR1 Underlies SMAD7-Mediated Lymph Node Metastasis of Bladder Cancer. Cancer Res. 2022;82:2239-53
144. Chen C, Zheng H, Luo Y, Kong Y, An M, Li Y. et al. SUMOylation promotes extracellular vesicle-mediated transmission of lncRNA ELNAT1 and lymph node metastasis in bladder cancer. J Clin Invest. 2021;131:e146431
145. Tan M, Zhang D, Zhang E, Xu D, Liu Z, Qiu J. et al. SENP2 suppresses epithelial-mesenchymal transition of bladder cancer cells through deSUMOylation of TGF-betaRI. Mol Carcinog. 2017;56:2332-41
146. Wang M, Wei R, Li G, Bi HL, Jia Z, Zhang M. et al. SUMOylation of SYNJ2BP-COX16 promotes breast cancer progression through DRP1-mediated mitochondrial fission. Cancer Lett. 2022;547:215871
147. Jia Y, Guo Y, Jin Q, Qu H, Qi D, Song P. et al. A SUMOylation-dependent HIF-1alpha/CLDN6 negative feedback mitigates hypoxia-induced breast cancer metastasis. J Exp Clin Cancer Res. 2020;39:42
148. Wei L, Wang W, Yao J, Cui Z, Xu Z, Ding H. et al. PACT promotes the metastasis of basal-like breast cancer through Rac1 SUMOylation and activation. Oncogene. 2022;41:4282-94
149. Li Y, Xing Y, Wang X, Hu B, Zhao X, Zhang H. et al. PAK5 promotes RNA helicase DDX5 sumoylation and miRNA-10b processing in a kinase-dependent manner in breast cancer. Cell Rep. 2021;37:110127
150. Bogachek MV, Chen Y, Kulak MV, Woodfield GW, Cyr AR, Park JM. et al. Sumoylation pathway is required to maintain the basal breast cancer subtype. Cancer Cell. 2014;25:748-61
151. Myatt SS, Kongsema M, Man CW, Kelly DJ, Gomes AR, Khongkow P. et al. SUMOylation inhibits FOXM1 activity and delays mitotic transition. Oncogene. 2014;33:4316-29
152. Jaiswal N, John R, Chand V, Nag A. Oncogenic Human Papillomavirus 16E7 modulates SUMOylation of FoxM1b. Int J Biochem Cell Biol. 2015;58:28-36
153. Liang Z, Yang Y, He Y, Yang P, Wang X, He G. et al. SUMOylation of IQGAP1 promotes the development of colorectal cancer. Cancer Lett. 2017;411:90-9
154. Mu M, Zhang Q, Li J, Zhao C, Li X, Chen Z. et al. USP51 facilitates colorectal cancer stemness and chemoresistance by forming a positive feed-forward loop with HIF1A. Cell Death Differ. 2023;30:2393-407
155. Xu Y, Li J, Zuo Y, Deng J, Wang LS, Chen GQ. SUMO-specific protease 1 regulates the in vitro and in vivo growth of colon cancer cells with the upregulated expression of CDK inhibitors. Cancer Lett. 2011;309:78-84
156. Zhang X, Liu T, Zheng S, Liu Q, Shen T, Han X. et al. SUMOylation of HSP27 regulates PKM2 to promote esophageal squamous cell carcinoma progression. Oncol Rep. 2020;44:1355-64
157. Chen C, Sun X, Xie W, Chen S, Hu Y, Xing D. et al. Opposing biological functions of the cytoplasm and nucleus DAXX modified by SUMO-2/3 in gastric cancer. Cell Death Dis. 2020;11:514
158. Wang Q, Xu C, Fan Q, Yuan H, Zhang X, Chen B. et al. Positive feedback between ROS and cis-axis of PIASxalpha/p38alpha-SUMOylation/MK2 facilitates gastric cancer metastasis. Cell Death Dis. 2021;12:986
159. Han J, Nie M, Chen C, Cheng X, Guo T, Huangfu L. et al. SDCBP-AS1 destabilizes beta-catenin by regulating ubiquitination and SUMOylation of hnRNP K to suppress gastric tumorigenicity and metastasis. Cancer Commun (Lond). 2022;42:1141-61
160. Birladeanu AM, Rogalska M, Potiri M, Papadaki V, Andreadou M, Kontoyiannis DL. et al. The scaffold protein IQGAP1 links heat-induced stress signals to alternative splicing regulation in gastric cancer cells. Oncogene. 2021;40:5518-32
161. Wang T, Min L, Gao Y, Zhao M, Feng S, Wang H. et al. SUMOylation of TUFT1 is essential for gastric cancer progression through AKT/mTOR signaling pathway activation. Cancer Sci. 2023;114:533-45
162. Zhao Y, He J, Li Y, Lv S, Cui H. NUSAP1 potentiates chemoresistance in glioblastoma through its SAP domain to stabilize ATR. Signal Transduct Target Ther. 2020;5:44
163. Chen Z, Wang S, Li HL, Luo H, Wu X, Lu J. et al. FOSL1 promotes proneural-to-mesenchymal transition of glioblastoma stem cells via UBC9/CYLD/NF-kappaB axis. Mol Ther. 2022;30:2568-83
164. Liu L, Liu Z, Liu Q, Wu W, Lin P, Liu X. et al. LncRNA INHEG promotes glioma stem cell maintenance and tumorigenicity through regulating rRNA 2'-O-methylation. Nat Commun. 2023;14:7526
165. Bellail AC, Olson JJ, Hao C. SUMO1 modification stabilizes CDK6 protein and drives the cell cycle and glioblastoma progression. Nat Commun. 2014;5:4234
166. Bian XL, Chen HZ, Yang PB, Li YP, Zhang FN, Zhang JY. et al. Nur77 suppresses hepatocellular carcinoma via switching glucose metabolism toward gluconeogenesis through attenuating phosphoenolpyruvate carboxykinase sumoylation. Nat Commun. 2017;8:14420
167. Li B, Xiong X, Xu J, Peng D, Nie G, Wen N. et al. METTL3-mediated m(6)A modification of lncRNA TSPAN12 promotes metastasis of hepatocellular carcinoma through SENP1-depentent deSUMOylation of EIF3I. Oncogene. 2024;43:1050-1062
168. Zhang J, Tan GL, Jiang M, Wang TS, Liu GH, Xiong SS. et al. Effects of SENP1-induced deSUMOylation of STAT1 on proliferation and invasion in nasopharyngeal carcinoma. Cell Signal. 2023;101:110530
169. Huang J, Tan X, Liu Y, Jiang K, Luo J. Knockdown of UBE2I inhibits tumorigenesis and enhances chemosensitivity of cholangiocarcinoma via modulating p27kip1 nuclear export. Mol Carcinog. 2023;62:700-15
170. Xia L, Jiang Y, Zhang XH, Wang XR, Wei R, Qin K. et al. SUMOylation disassembles the tetrameric pyruvate kinase M2 to block myeloid differentiation of leukemia cells. Cell Death Dis. 2021;12:101
171. Zhang J, Huang FF, Wu DS, Li WJ, Zhan HE, Peng MY. et al. SUMOylation of insulin-like growth factor 1 receptor, promotes proliferation in acute myeloid leukemia. Cancer Lett. 2015;357:297-306
172. Dong S, Chen J. SUMOylation of sPRDM16 promotes the progression of acute myeloid leukemia. BMC Cancer. 2015;15:893
173. Chen X, Qin Y, Zhang Z, Xing Z, Wang Q, Lu W. et al. Hyper-SUMOylation of ERG Is Essential for the Progression of Acute Myeloid Leukemia. Front Mol Biosci. 2021;8:652284
174. Diao X, Guo C, Zheng H, Zhao K, Luo Y, An M. et al. SUMOylation-triggered ALIX activation modulates extracellular vesicles circTLCD4-RWDD3 to promote lymphatic metastasis of non-small cell lung cancer. Signal Transduct Target Ther. 2023;8:426
175. Yang H, Du Y, Fei X, Huang S, Yimiti M, Yang X. et al. SUMOylation of the ubiquitin ligase component KEAP1 at K39 upregulates NRF2 and its target function in lung cancer cell proliferation. J Biol Chem. 2023;299:105215
176. Luo Y, Li Z, Kong Y, He W, Zheng H, An M. et al. KRAS mutant-driven SUMOylation controls extracellular vesicle transmission to trigger lymphangiogenesis in pancreatic cancer. J Clin Invest. 2022;132:e157644
177. Chien W, Lee KL, Ding LW, Wuensche P, Kato H, Doan NB. et al. PIAS4 is an activator of hypoxia signalling via VHL suppression during growth of pancreatic cancer cells. Br J Cancer. 2013;109:1795-804
178. Wang Q, Xia N, Li T, Xu Y, Zou Y, Zuo Y. et al. SUMO-specific protease 1 promotes prostate cancer progression and metastasis. Oncogene. 2013;32:2493-8
179. Lee MH, Sung K, Beebe D, Huang W, Shapiro D, Miyamoto S. et al. The SUMO protease SENP1 promotes aggressive behaviors of high HIF2alpha expressing renal cell carcinoma cells. Oncogenesis. 2022;11:65
Author contact
![]() Corresponding author: Kai Li, MD, PhD. Email: kliedu.cn; Huizhe Wu, Email: wuhzedu.cn; Shan Gao, Email: mount1121com.
Corresponding author: Kai Li, MD, PhD. Email: kliedu.cn; Huizhe Wu, Email: wuhzedu.cn; Shan Gao, Email: mount1121com.
 Global reach, higher impact
Global reach, higher impact