13.3
Impact Factor
Theranostics 2024; 14(6):2304-2328. doi:10.7150/thno.91700 This issue Cite
Review
Role of the gut microbiota in tumorigenesis and treatment
1. Department of Biotherapy, Cancer Center and State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, 610041, China.
2. Department of Pharmacy, West China Hospital, Sichuan University, Chengdu, 610041, China.
Received 2023-12-1; Accepted 2024-3-1; Published 2024-3-17
Abstract
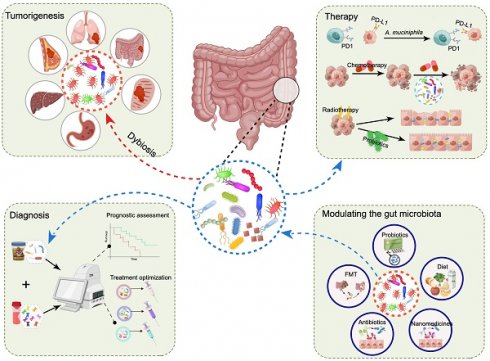
The gut microbiota is a crucial component of the intricate microecosystem within the human body that engages in interactions with the host and influences various physiological processes and pathological conditions. In recent years, the association between dysbiosis of the gut microbiota and tumorigenesis has garnered increasing attention, as it is recognized as a hallmark of cancer within the scientific community. However, only a few microorganisms have been identified as potential drivers of tumorigenesis, and enhancing the molecular understanding of this process has substantial scientific importance and clinical relevance for cancer treatment. In this review, we delineate the impact of the gut microbiota on tumorigenesis and treatment in multiple types of cancer while also analyzing the associated molecular mechanisms. Moreover, we discuss the utility of gut microbiota data in cancer diagnosis and patient stratification. We further outline current research on harnessing microorganisms for cancer treatment while also analyzing the prospects and challenges associated with this approach.
Keywords: gut microbiota, dysbiosis, cancer, probiotics, fecal microbiota transplantation (FMT)
1. Introduction
The human gut microecosystem is a highly intricate system, and the gut microbiota plays a pivotal role in human health through interactions with the host [1,2]. However, cultivating most of the gut microbiota in vitro is challenging, which hinders a comprehensive understanding of these intricate interactions [3,4]. The advent of high-throughput sequencing technology (HTST) has propelled the investigation of uncultured microbial communities. As a result, researchers have shifted their focus from merely characterizing the composition of microbial communities to exploring the intricate interplay between these communities and human health, as well as elucidating the underlying mechanisms [5]. In terms of genetic constitution, the number of bacteria in the gut microbiota is more than 25 times greater than that in the host, and the gut microbiota consists mainly of bacteria [6]. Compared to the human genome, this community can perform a wider range of metabolic functions; however, achieving these metabolic functions through the human genome is difficult [7,8]. For this reason, scientists refer to the gut microbiota as the “second genome”.
A substantial body of research has demonstrated a significant correlation between dysbiosis of the gut microbiota and the prevalence of nervous system disorders [9], cancer [10], gastrointestinal ailments [11], cardiovascular conditions [12], and other diseases [13-15]. Cancer is a major public health issue and remains one of the leading causes of death worldwide [16]. An increasing number of studies indicate that dysbiosis in the gut microbiota is a significant factor in cancer progression; such dysbiosis is now recognized by the scientific community as a hallmark of cancer [17,18]. There is compelling evidence that Fusobacterium nucleatum and Bacteroides fragilis are significantly associated with the pathogenesis of colorectal cancer (CRC) [19,20]. It is plausible that F. nucleatum and B. fragilis may not be the sole potential pathogens. Bullman noted “Every day now there seems to be some new microbe associated with cancer” [21]. Although studies have shown that dysbiosis of the gut microbiota is associated with tumorigenesis, further clinical investigations and preclinical studies are warranted to elucidate the underlying mechanisms involved.
The gut microbiota not only plays a role in tumorigenesis but also actively participates in various physiological processes within the human body, thereby making significant contributions to human health [22,23]. They serve as signaling molecules implicated in regulating host physiological states, including systemic blood pressure control, modulating the inflammatory response, and preserving the functionality of specific cells [24]. Moreover, they facilitate the decomposition of contents within the gastrointestinal tract, where dietary fiber and complex polysaccharides are metabolized into short-chain fatty acids (SCFAs), such as acetate, propionate, and butyrate. Among these SCFAs, butyrate is recognized as a pivotal modulator of the immune response [25]. In addition, the gut microbiota also stimulates the development of the host immune system through other means, subsequently influencing overall immune function within the body [26,27]. The gut microbiota impacts the immune system, thus contributing to its influence on cancer immunotherapy efficacy. The relationship between cancer immunotherapy and the gut microbiota has garnered significant attention since an article published in 2015 by Science reported that the modulation of the immunosuppressive response is influenced by the gut microbiota [28]. In addition to influencing cancer immunotherapy efficacy, the gut microbiota also modulates the efficacy of drugs. Drugs introduced into the gut interact with the resident gut microbiota. These drugs can alter the composition and abundance of the gut microbiota, while the enzymes produced by them can modify drug structure, thereby influencing drug biological activity and toxicity [29,30]. Currently, this topic is being explored as a potential target for optimizing drug therapy.
The gut microbiota has been found to be associated with the physiological and pathological status of the host [2,31]. Can cancer treatment be optimized through the regulation of the gut microbiota? Among the various factors contributing to patients' unresponsiveness to immune checkpoint inhibitors (ICIs), regulation of the gut microbiota appears to be a readily modifiable aspect through human intervention, with the aim of enhancing patient response to ICIs [32]. B. fragilis exhibits potential for enhancing the therapeutic efficacy of CTLA-4 blockade [33]. Supplementation with Akkermansia muciniphila has the potential to augment the efficacy of PD-1 inhibitors [34]. Current regulatory approaches and ongoing research on the gut microbiota include manipulating the dietary composition to selectively promote the growth of specific microorganisms [35], administering probiotics [36], performing FMT [37], and targeting the inhibition of specific microorganisms [38]. These strategies have the potential to enhance cancer treatment efficacy or mitigate the detrimental effects caused by certain pathogenic microorganisms on the host.
In this review, we focus on the influence of dysbiosis of the gut microbiota on tumorigenesis and treatment response across multiple cancer types while also analyzing the associated molecular mechanisms. Additionally, we briefly explored the use of the gut microbiota as a biomarker for cancer diagnosis and treatment. Subsequently, we elucidate the contemporary approaches employed to regulate the gut microbiota and address their respective prospects and challenges.
2. Is the gut microbiota implicated in tumorigenesis?
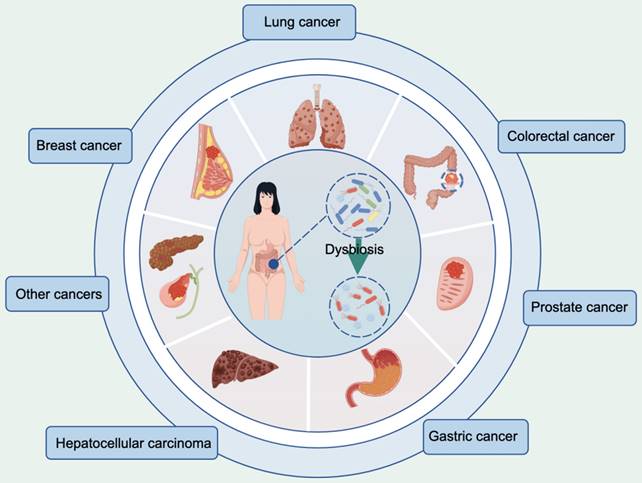
According to the latest global cancer burden figures released by the International Agency for Research on Cancer, breast (11.7%), lung (11.4%), CRC (10.0%), prostate (7.3%), stomach (5.6%) and liver (4.7%) cancers account for the top six new cases of cancer worldwide [39]. Substantial experimental and epidemiological evidence strongly supports the influence of the gut microbiota on these types of cancer, as shown in Figure 1. Alterations in the composition of the gut microbiota and associated metabolites have the potential to modulate cellular metabolism and human immune function, thereby establishing a plausible link to cancer [40,41]. Despite the increasing interest in this field, there is currently no standardized framework for investigating the correlation between the gut microbiota and cancer incidence, particularly regarding the interpretation of research findings [42,43]. Here, we explored the influence of the gut microbiota on these six cancers while analyzing the underlying mechanisms involved.
2.1 Breast cancer
The incidence of breast cancer (BC) has surpassed that of lung cancer (LC), making it the leading type of cancer worldwide [39]. Recent research has suggested that the gut microbiota may significantly influence BC pathogenesis [17,44]. Hou et al. conducted an analysis of the gut microbiota of 267 BC patients and observed that premenopausal individuals with BC exhibited reduced diversity in their gut microbiota and a decreased abundance of probiotics containing tumor suppressor factors and that postmenopausal patients displayed increased levels of pathogenic bacteria. Additionally, they identified 14 microbial markers associated with different menopausal statuses in patients with BC [45]. Goedert et al. conducted a comparative analysis of the fecal microbiota of 48 postmenopausal BC patients before treatment and 48 women with normal mammography results. Consistent with the findings of Hou et al., their study also revealed diminished diversity in the gut microbiota among BC patients. Furthermore, they identified higher levels of Clostridiaceae, Faecalibacterium, and Ruminococcaceae as well as lower levels of Dorea and Lachnospiraceae in BC patients [46].
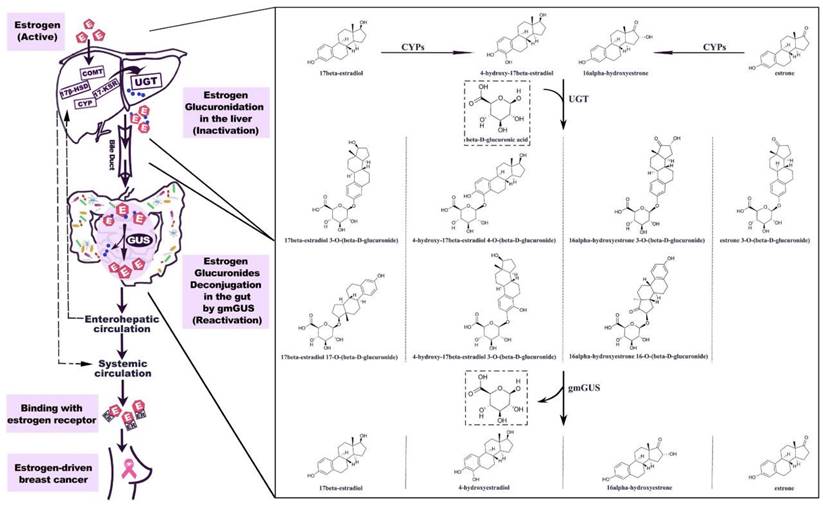
Currently, the impact of the gut microbiota on BC can be attributed to its influence on estrogen metabolism [47,48]. The incidence of BC is predominantly observed in postmenopausal women; thus, investigations into the relationship between the gut microbiota and BC have primarily concentrated on postmenopausal patients. Postmenopausal women with elevated endogenous estrogen levels are at increased risk of developing BC [49]. In 1976, a pioneering study revealed increased excretion of estrogen in the feces of individuals administered ampicillin, suggesting that the gut microbiota enhances estrogen metabolism [47]. Gut microbial metabolites, such as β-glucuronidase, influence the levels of nonovarian estrogens through the enterohepatic circulation [48]. The human gut microbiota harbors a diverse range of microorganisms, including species such as Bacteroidetes, Firmicutes, and Escherichia, which can encode and produce β-glucuronidases [50,51]. Estrogen glucuronides are the primary metabolites of estrogen in liver phase II metabolism. They are excreted into the gut via bile, where β-glucuronidase catalyzes their conversion to free estrogen, which can be absorbed by the gut mucosa and enter the enterohepatic circulation before being distributed to various organs, such as the mammary gland (Figure 2) [51]. Among postmenopausal women, an increase in circulating estrogen levels is associated with increased susceptibility to BC. We anticipate that β-glucuronidase-producing microorganisms could be used clinically as biomarkers for predicting BC; alternatively, such microorganisms could be eliminated or β-glucosidase inhibitors could be employed to mitigate the risk of BC development and facilitate treatment [52,53].
Cancer types associated with dysbiosis of the gut microbiota. This figure was created using Figdraw.
Estrogen metabolism is mediated by gut microbial β-glucuronidase (gmGUS: gut microbial β-glucuronidase). The hepatic metabolism of estrogen is facilitated by a cascade of enzymes. The conjugation of parent estrogens and their phase I metabolites with glucuronic acid can be catalyzed by uridine 5′-diphospho-glucuronosyltransferase (UGT). Estrogen glucuronides are biologically inactive; upon bile excretion, they undergo gastrointestinal transit, during which gmGUS enzymatically hydrolyzes the conjugates to release active estrogens. The reactivated estrogens enter the hepatic circulation and are subsequently reabsorbed into the body. CYP, cytochrome P-450 enzyme. Adapted with permission from [51], Copyright 2021, Sui, Wu and Chen.
Additionally, the modulation of immune responses by the human gut microbiota also influences BC pathology [54]. The gut tract serves as the primary immune organ in the human body and exerts a profound influence on human immunity through its pivotal role in promoting the development and maturation of the host immune system, as well as actively participating in regulating overall immune responses [8,55]. Neutrophils, a crucial component of the innate immune system, have been implicated in BC progression [56]. Clarke et al. demonstrated that the administration of broad-spectrum antibiotics results in the dysregulation of the gut microbiota, thereby impacting neutrophil function in both the serum and bone marrow, consequently compromising innate immune defense [57]. The impact of antibiotics on innate immunity suggests that patients receiving immunotherapy in clinical practice should use antibiotics with caution [58,59]. Neutrophils are abundant in malignant tumor lesions and play an important role in tumor initiation and progression by generating angiogenic factors, promoting metastasis, and suppressing the immune response to tumors [56,60]. A study by Rutkowski et al. provides further evidence of the impact of the gut microbiota on neutrophils. These authors found that Allobaculum and Lactobacillus were enriched in Toll-like receptor (TLR) 5-deficient mice and that Bacteroides was enriched in WT mice. These authors demonstrated that the TLR5-dependent commensal gut microbiota in BC patients can stimulate the systemic upregulation of IL-6, thereby promoting the mobilization of granulocytic myeloid-derived suppressor cells and suppressing tumor immunity, ultimately accelerating the malignant progression of tumors [54]. Clinical testing is necessary to ascertain the potential of these microorganisms in BC detection and treatment outcome prediction.
Research findings indicate that breast tissue is not entirely free of bacteria [61]. A study by Parida et al. suggested that the promotion of cancer metastasis by microorganisms in breast tissue is associated with specific genera of the gut microbiota [62]. They observed that B. fragilis was consistently detected in all breast tissue samples, including those from benign and malignant breast cancer patients, as well as in nipple aspirate fluid. The authors fed a cohort of mice harboring B. fragilis, which colonizes the gut. As a result, the mice exhibited a significant increase in thickening of the breast duct lining and hyperproliferation of the breast epithelium. The virulence of B. fragilis is attributed to the presence of B. fragilis toxin [62,63]. BC cells exposed to B. fragilis toxin for 72 hours retained memory and were capable of initiating cancer development and forming metastatic lesions in mouse models. They also observed the presence of B. fragilis in the mammary ducts of mice harboring gut B. fragilis infection; however, it remains unclear whether B. fragilis translocated internally from the gut to the breast or whether gut-infected mice acquired mammary gland infection through environmental exposure [62].
2.2 Lung cancer
The incidence of LC ranks second globally among all types of cancer, yet it remains the primary cause of cancer-related mortality [39]. The histological subtypes of LC are categorized as non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). Globally, NSCLC accounts for approximately 85% of all lung cancers, with SCLC accounting for the remaining 15% of lung cancers [64]. The morphological, etiological, and molecular characteristics of LC have been extensively investigated. In addition to genetic and environmental factors, the gut microbiota plays a pivotal role in the development of LC [65]. Qin et al. discovered that, compared with healthy individuals, LC patients exhibit reduced bacterial diversity and that as LC progresses, the levels of SCFAs and anti-inflammatory bacteria decrease; additionally, certain pathogenic bacteria associated with inflammation or tumor promotion were found to be more prevalent among LC patients [66]. Zheng et al. recruited 42 early-stage LC patients and 65 healthy individuals to analyze the gut microbiota using 16S ribosomal RNA (rRNA) gene sequencing analysis. They found that Ruminococcus, Enterobacteriaceae, and Lachnospiraceae were highly enriched in the cancer group and that Faecalibacterium, Streptococcus, Bifidobacterium, and Veillonella were significantly enriched in the healthy group [67].
The results of the aforementioned clinical studies suggest a potential association between the gut microbiota and the progression of LC. It is widely acknowledged that there are inseparable associations between chronic inflammation and the onset and progression of LC [68]. Dysbiosis of the gut microbiota and its metabolites can induce systemic chronic inflammation, thereby contributing to the initiation and progression of LC [69,70]. Research by He et al. indicated that antibiotics modulate the gut microbiota to suppress lung inflammation in Treg-deficient mice [71]. To investigate the impact of antibiotic-modulated microbiota on suppressing lung inflammation in Treg-deficient SF mice, a treatment regimen involving three different antibiotics was administered to these mice. The results demonstrated that antibiotics reversed the decreases in the relative abundances of the genus Sutterella and the family Mycoplasmataceae associated with Treg deficiency, thus altering cytokine expression through microbiota-associated metabolites; furthermore, both ampicillin and vancomycin reduced IL-6 levels [71]. Sandri's examination of lung tissue revealed that interstitial fibroblasts express IL-6 and contribute to the promotion of cancer [72]. The suppression of inflammation in the lungs is achieved through IL-6 blockade. To further validate these findings, IL-6 knockout mice were used to confirm that the deletion of IL-6 confers protection against Treg-induced inflammation [71].
Numerous studies have demonstrated that dysbiosis of the gut microbiota can lead to impaired immune surveillance in lung tissue and create a microenvironment that facilitates the formation of LC cells [73]. The impact of the gut microbiota on pulmonary immune function may lie in the activation of gut immunity by the gut microbiota, which leads to the migration of these activated immune cells to the lungs and their involvement in pulmonary immunity. Chemokine-induced homing of lymphocytes plays an important role in this process [74]. Congenital lymphocytes in the intestine are closely related to lung homeostasis. The gut microbiota enhances resistance against lung infection by facilitating the recruitment of interleukin-22 (IL-22)-producing group 3 innate lymphoid cells (IL-22+ILC3) into the lungs of neonatal mice [75]. The interaction between the gut microbiota and intestinal dendritic cells (DCs) (CD103+CD11b+DCs) induces the upregulation of CCR4-related homing receptors in gut IL-22+ILC3s, facilitating the selective migration of gut IL-22+ILC3s to the lungs. CCR4 is a chemokine receptor that is commonly identified as a key mediator in the trafficking of T cells and Treg cells to the lungs [76]. The chemokine CCL17, which is expressed in lung epithelial cells, activates the CCR4 receptor, thereby facilitating the recruitment of IL-22+ILC3s into the lungs of neonatal mice. Elevated levels of IL-22 within the lung environment can impede pathogen proliferation [75]. Disruption of commensal bacteria interrupts the migratory program of IL-22+ILC3s, impairing their ability to traffic to the lungs and rendering newborn mice more susceptible to pneumonia.
2.3 Colorectal cancer
CRC is the fourth most common cancer diagnosed worldwide, while it is the third most common cancer [39]. A distinctive characteristic of CRC is its close association with the gut microbiota, which constitutes an integral component of the tumor microenvironment [20,77]. The gut microbiota in patients with CRC has been extensively investigated, making it arguably the most exemplary illustration of the role played by the gut microbiota in cancer [78]. The initial evidence supporting the involvement of the gut microbiota in CRC emerged in 1975 when both germ-free and conventional mice were administered 1,2-dimethylhydrazine. A significantly greater percentage (93%) of conventional mice developed CRC than did germ-free mice (21%) [79]. Subsequent studies have demonstrated that specific strains of the gut microbiota, such as Enterococcus, Bacteroidetes, and Clostridium, may contribute to the development of CRC by enhancing crypt lesions induced by 1,2-dimethylhydrazine [80]. However, FMT from CRC patients into germ-free mice promoted gut cell proliferation and facilitated the progression of azoxymethane-induced crypt lesions to CRC [81]. Genomic sequencing of fecal samples from CRC patients across different regions revealed several core pathogenic species. Notably, these strains, which include F. nucleatum, Parvimonas micra, Peptostreptococcus stomatis, Peptostreptococcus anaerobius, Porphyromonas asaccharolytica, Solobacterium moorei and Prevotella intermedia, are also enriched in the oral cavity [82,83]. The relationship between the gut microbiota and CRC has been elucidated, encompassing the factors depicted in Figure 3.
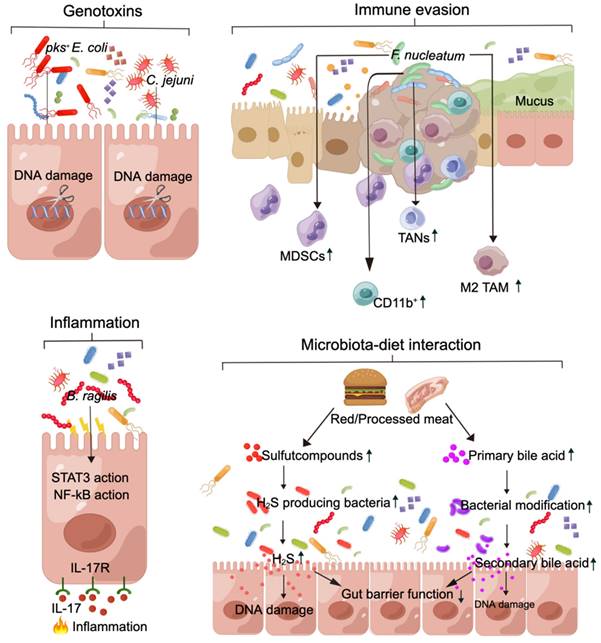
Genotoxins. Genetic alterations in the activation of oncogenes and/or inactivation of tumor suppressor genes, which are mediated by toxins produced by the gut microbiota, contribute to tumorigenesis [84,85]. For example, colibactin is produced by pks+ Escherichia coli strains [86], and the cytolethal distending toxin is produced by Campylobacter jejuni [85]. These toxins induce double-stranded DNA breaks in host cells, triggering a signaling cascade of DNA damage that results in persistent mitosis, chromosomal aberrations, and an increased frequency of gene mutations [86-88]. Cao et al. showed that gene toxins within the gut microbiota continuously induce DNA damage in host epithelial cells synergistically with chronic inflammation and other environmental factors within the gut microenvironment, ultimately facilitating the initiation and progression of CRC [87].
Immune evasion. Gut pathogenic bacteria have been shown to promote an immunosuppressive tumor microenvironment that facilitates the growth of CRC, with a particular emphasis on the role of F. nucleatum in promoting immune evasion in CRC [89,90]. Research findings indicate that an F. nucleatum inhibitor protein effectively suppresses human T-cell activity by impeding the G1 phase of the cell cycle, thereby fostering an immunosuppressive microenvironment conducive to tumor cell proliferation [90]. Additionally, F. nucleatum modulates the tumor immune microenvironment and leads to the expansion of myeloid-derived suppressor cells (MDSCs), CD11b+ cells, M2-like tumor-associated macrophages (M2 TAMs), and tumor-associated neutrophils (TANs). These cells play crucial roles in suppressing antitumor immunity and promoting tumor progression [91]. Jiang et al. demonstrated that succinic acid derived from F. nucleatum inhibits the cGAS-interferon-β pathway, thereby attenuating the antitumor response by restricting CD8+ T-cell trafficking to the tumor microenvironment in vivo [92].
Inflammation. Gut microbiota dysbiosis is strongly linked to inflammation of the gastrointestinal tract and plays a crucial role in the initiation of colitis-associated CRC [87,93]. The inflammation induced by gut pathogenic bacteria often involves the activation of the IL-17, NF-κB, and pattern recognition receptor (PRR) signaling pathways, as well as disruption of gut barrier function [94,95]. These interconnected cascades collectively contribute to a proinflammatory phenotype. Chung et al. demonstrated that the induction of inflammation by enterotoxigenic B. fragilis begins with the disruption of gut barrier function and the subsequent activation of STAT3 and NF-κB signaling in IL17Rexpressing colonic epithelial cells in a cascade of inflammation-related responses [95]. Thus, myeloid cell-dependent distal colon tumorigenesis is triggered by myeloid cells.
Diet. Studies indicate that the initiation of CRC is associated with dietary constituents, such as the consumption of red meat and processed meat [96,97]. Hydrogen sulfide (H2S) is produced in the gut by sulfur-reducing bacteria from inorganic sulfur, which is commonly used as a preservative in processed meat, or by fermentative bacteria that metabolize organic sulfur compounds found in animal products such as red meat [96]. The microbiota metabolizes these meats to generate nitroso compounds, H2S, and other procarcinogens, thereby contributing to the initiation of CRC [98,99]. In a cohort of elderly men, there was an association between increased dietary intake of organic sulfur and an increase in the fecal abundance of H2S-producing Clostridium clostridioforme [100]. In addition, fiber and resistant starch are decomposed by the gut microbiota to produce SCFAs. A reduction in the intake of these substances leads to a decrease in SCFA levels, which has been extensively demonstrated in numerous studies to inhibit the development of CRC [101,102]. Elevated levels of secondary bile acids, such as deoxycholic acid, in individuals adhering to a high-fat diet have been linked to increased susceptibility to CRC [96,103].
2.4 Prostate cancer
The incidence of prostate cancer (PCa), a prevalent malignancy that poses a significant threat to men's health, ranks fourth globally among newly diagnosed cancers [39], and age, race, and family history are the main risk factors [104]. Diet and physical activity also play important roles in the development and progression of PCa, particularly in relation to ethnicity, at different incidence rates [105]. Additionally, an increasing body of research over the past decade has demonstrated the significant role that the gut microbiota plays in the occurrence and development of PCa [106,107]. Liss et al. conducted an analysis of the gut microbiota in 133 U.S. men who underwent prostate biopsy; they performed 16S rRNA amplicon sequencing on 105 samples (64 with cancer and 41 without cancer) [108]. These findings revealed enrichment of Bacteroides and Streptococcus species among PCa patients. Furthermore, a subsequent metagenomic analysis demonstrated significant alterations in the arginine and folate pathways within the gut microbiota. Consequently, the authors propose that the gut microbiota may influence the risk of developing PCa. Matsushita et al. conducted an analysis of 152 Japanese men who underwent prostate biopsies; 96 had cancer, and 56 did not [109]. The results showed that the relative abundances of Rikenellaceae, Alistipes, and Lachnospira significantly increased in the high-risk group (men with grade 2 or above PCa) and the negative + low-risk group (men with biopsy negative or grade 1 PCa).
Gut microbiota dysbiosis contributes to the development of CRC through a diverse range of molecular mechanisms. (A) pks+ E. coli and C. jejuni produce genotoxins, which induce DNA damage and increase the frequency of gene mutations, thus contributing to CRC. (B) F. nucleatum leads to the expansion of MDSCs, CD11b+ cells, M2 TAMs, and TANs. These cells play a crucial role in suppressing antitumor immunity. (C) B. fragilis triggers a procarcinogenic, multistep inflammatory cascade involving the IL-17R, NF-kB, and STAT3 signaling pathways in colonic epithelial cells. (D) Red/processed meat can potentially modify the structure and function of the microbiota, leading to increased production of H2S and secondary bile acids by microorganisms. These alterations can result in damage to gut barrier function and DNA, thereby elevating the risk of CRC. MDSCs: myeloid-derived suppressor cells; TANs: tumor-associated neutrophils; M2 TAMs: M2-like tumor-associated macrophages; H2S: hydrogen sulfide. This figure was created using Figdraw.
The results of the aforementioned studies provide preliminary evidence suggesting a correlation between the gut microbiota and PCa incidence, but the impact of the gut microbiota on PCa incidence is still under investigation. The etiology of PCa primarily involves excessive androgen production, and androgen deprivation therapy (ADT) is commonly used for treatment [110]. Although initially effective, this treatment can lead to a transition in patients' condition from hormone-sensitive prostate cancer to castration-resistant prostate cancer (CRPCa) as therapy progresses [111]. Pernigoni et al. reported that the gut microbiota contributes to the development of CRPCa by providing an alternative source of androgens [112]. Surgical castration (CT) was performed on mice, which subsequently progressed to the castration-sensitive phase (CS), characterized by a rapid decrease in prostate cancer tumor volume. Subsequently, the mice progressed to the castration-resistant phase (CR), which was characterized by a gradual increase in tumor volume. The subsequent depletion of the gut microbiota in CT mice through antibiotic treatment resulted in a significant reduction in tumor volume. 16S rDNA sequencing analysis revealed a significant increase in the abundance of Ruminococcus gnavus and Bacteroides acidifaciens in CR mice. Furthermore, it has been confirmed that Ruminococcus sp. DSM_100440 is capable of metabolizing pregnenolone and hydroxypregnenolone into androgens such as dehydroepiandrosterone and testosterone in a mouse model.
2.5 Gastric cancer
Gastric cancer (GC) is the fifth most prevalent and fourth most deadly malignancy worldwide, making it one of the leading causes of death globally [39]. The gastrointestinal microbiota plays a crucial role in the occurrence and progression of GC [113]. Helicobacter pylori infection significantly increases the risk of GC [114]; however, it cannot solely account for all cases of GC [115]. The advancement of high-throughput sequencing technology has facilitated an increasing number of studies investigating the association between the gut microbiota and GC [116,117]. Li et al. analyzed the gut microbiota of 130 patients with gastrointestinal tumors and 147 healthy controls and found significant differences in the composition and abundance of the gut microbiota between the two groups [118]. Zhou et al. sequenced 16S rRNA target genes from tumor tissue and fecal samples of 1043 participants from 10 hospitals. Streptococcus anginosus and Streptococcus constellatus were enriched in both the tumor tissue and feces of GC patients, with a stronger enrichment signal observed in the feces than in the tissue samples [119].
With the advancement of related research, the underlying mechanism by which H. pylori contributes to GC pathogenesis has been progressively elucidated. Studies have shown that H. pylori can induce the production of ROS and that excessive ROS can lead to oxidative stress, resulting in DNA damage and thus the formation of tumor precursors [120]. The proliferation and apoptosis of gastric epithelial cells are normal physiological phenomena. H. pylori infection leads to increased apoptosis and proliferation of gastric epithelial cells, but proliferation still dominates, which may be one of the causes of GC [121]. H. pylori induces a strong inflammatory response, which may play an important role in the progression from chronic inflammation to gastric malignancy [122]. An important characteristic of H. pylori infection is the rapid recruitment of regulatory T cells and myeloid cells (including dendritic cells, neutrophils, and M1 macrophages) to the stomach for the secretion of a series of cytokines (such as IFN-γ, IL-17, and IL-21), which collectively establish an immunosuppressive microenvironment prior to the development of gastric epithelial cell malignancy [123].
A study conducted by Zhou et al. demonstrated the enrichment of S. anginosus in both tumor tissue and the intestinal microbiota among patients with GC [119]. It is an emerging pathogen with previously unrecognized pathogenic potential that has recently garnered increased amounts of attention in the scientific community. Asam et al. demonstrated that streptolysin, produced by S. anginosus, functions as a broad-spectrum hemolysin and cytolysin capable of facilitating bacterial translocation across the epithelial barrier, inducing tissue damage, and destroying neutrophils and macrophages to evade host immune escape [124]. Sasaki et al. isolated and purified an antigen from the bacterial supernatant of S. anginosus [125]. This antigen can stimulate macrophages to synthesize nitric oxide (NO), leading to intracellular oxidative stress and lipid peroxidation, ultimately resulting in DNA damage and subsequent tumorigenesis [126].
2.6 Hepatocellular carcinoma
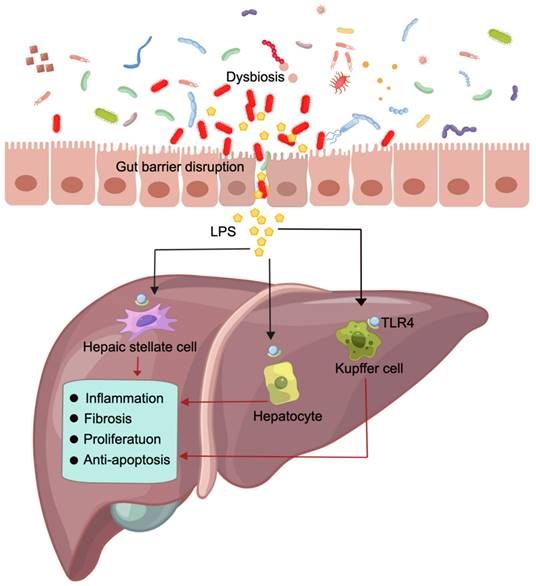
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer mortality worldwide, accounting for approximately 90% of liver cancer cases and posing a significant global healthcare challenge [18,127]. HCC predominantly arises in patients with underlying chronic liver disease and is propelled by an intricate interplay of hepatic injury, inflammation, and regeneration that typically spans several decades [127,128]. Emerging evidence strongly supports the pivotal role of alterations in the gut barrier and the composition of the gut microbiota in driving the progression of chronic liver disease and facilitating HCC development [129]. Research findings indicate that a significant increase in lipopolysaccharide (LPS) levels in HCC patients is accompanied by gut barrier disruption [129,130]. Impairment of the gut barrier may contribute to the excessive production of LPS by the gut microbiota into the portal vein and liver, further triggering HCC [131]. Additionally, Ren et al. conducted a comprehensive analysis by collecting 419 samples, revealing the enrichment of Actinobacteria, Gemmiger, and Parabacteroides in early HCC [132]. Ni et al. demonstrated that patients with primary HCC exhibited an increase in proinflammatory bacteria within their fecal microbiota, and the degree of dysbiosis in the gut microbiota was significantly greater than that in healthy controls [133]. These research findings provide evidence supporting a potential association between HCC and the gut barrier, as well as the composition of the gut microbiota, thereby contributing to the elucidation of this relationship through scientific investigation.
The hepatic artery in the abdominal cavity and the portal vein delivered by the gut and spleen constitute the dual blood supply of the liver, with 75% of its blood being supplied by the portal vein [129]. The blood from the gut portal vein not only contains nutrients but also carries substances such as LPS and peptidoglycan from the gut microbiota [134,135]. Usually, these substances are present in minimal quantities and can be efficiently cleared by Kupffer cells in the liver without eliciting any detrimental effects on the host [135]. The maintenance of these physiological conditions relies on the integrity of the gut barrier.
Dysbiosis of the gut microbiota can reduce the diversity and abundance of probiotics while promoting the growth of pathogenic bacteria [136,137]. Consequently, this disrupts the integrity of the gut barrier, facilitates bacterial translocation, and allows for substantial entry of LPS into the portal vein and liver. The impairment of the gut barrier promotes hepatic inflammation, fibrosis, proliferation and the activation of antiapoptotic signals by activating LPS and its receptor TLR. This process thereby facilitates the development of HCC (Figure 4) [131,138]. Administering dextran sulfate sodium disrupts the gut barrier, which not only results in increased systemic LPS levels and liver fibrosis but also promotes HCC formation in mice [139]. However, inhibiting TLR4 signaling suppressed liver inflammation, fibrosis, and HCC formation in both mice and rats [131]. Experiments conducted on TLR4 chimeric mice have demonstrated that the expression of TLR4 on liverresident cells, including hepatocytes and Kupffer cells, is accountable for promoting fibrogenesis and HCC [140]. Activation of the LPS-TLR4 signaling pathway in Kupffer cells has been shown to induce TNF-dependent and IL-6-dependent compensatory hepatocyte proliferation while also reducing oxidative stress and apoptosis [141]. Additionally, the activation of TLR4 in HCC cell lines induced by LPS enhances the invasive potential of these cells and induces epithelial-mesenchymal transition [142,143].
The involvement of the gut microbiota in the pathogenesis of various types of cancer, including BC [46], LC [65] and CRC [77], has been confirmed in clinical and preclinical studies. However, the clinical lineups in some studies had a limited sample size [46,67]. Moreover, the incidence rates of the same cancer also vary across different regions worldwide. For instance, in Southern Europe, the incidence rate of CRC is 25.3 per 100,000 males and 16.4 per 100,000 females, whereas in Middle Africa, it is only 2.9 per 100,000 males and 2.3 per 100,000 females [39]. It is necessary to include larger and more regional populations to further refine and validate the association between the gut microbiota and corresponding cancers. The ultimate goal is to provide a new method based on the gut microbiota for early cancer screening, enabling its prompt diagnosis and treatment [132].
3. Questioning the impact of the gut microbiota on cancer treatment
While some gut microbiota may promote the initiation and progression of cancer, not all gut microbiota are harmful; in fact, certain types of gut microbiota can be beneficial for cancer treatment [144,145]. Research on the use of the gut microbiota for treating cancer has focused primarily on enhancing human immunity [145,146], but the benefits of the gut microbiota are not limited to immune enhancement; it can also improve chemotherapeutic efficacy and mitigate adverse effects [147,148], as shown in Figure 5.
3.1 Immunotherapy
The host acquires microbiota from the environment at birth; this microbiota interacts with the immune system during the first 2-3 years of life and subsequently establishes a stable microbiota [149]. This community promotes both innate and adaptive immunity at multiple levels [150]. The evolution of the innate and adaptive immune systems leads not only to the elimination of specific pathogens but also to the shaping of the composition of the commensal gut microbiota [149]. The immune system also undergoes its own progression [26].
Contribution of the gut microbiota to HCC and the underlying mechanisms involved Dysbiosis of the gut microbiota and impairment of the gut barrier result in the translocation of LPS from the gut lumen to the bloodstream, leading to increased hepatic exposure to LPS. This promotes hepatic inflammation, fibrosis, proliferation and the activation of antiapoptotic signals. This figure was created using Figdraw.
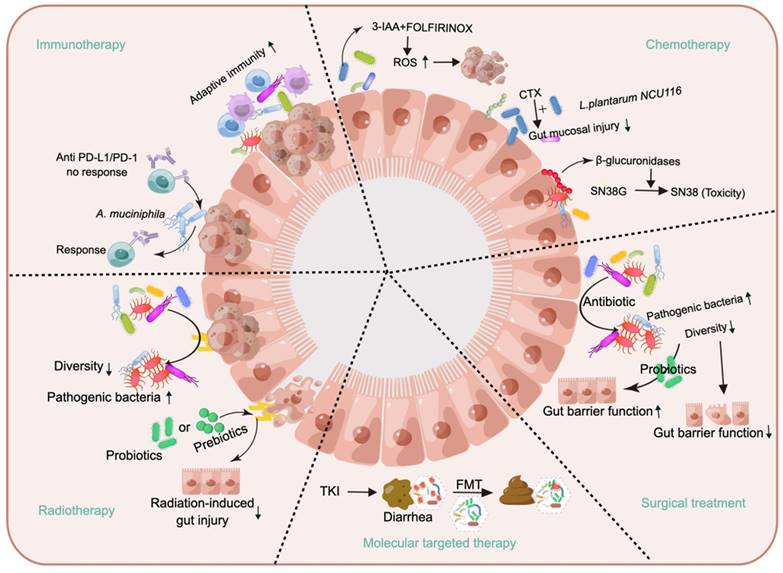
Considering the history of microorganisms as anticancer tools, they were first recognized for having such effects in the 19th century. Busch observed tumor regression in cancer patients after infection with erysipelas, and Fehleisen identified Streptococcus pyogenes as the causative agent of erysipelas infection. Afterward, Coley developed the first cancer immunotherapy drug (“Coley toxin”) based on heat-killed bacteria [151]. Over 1000 patients, many or most of whom had sarcomas, exhibited degenerative changes and cures. However, this “medicine” gradually failed because it was not administered following a scientific protocol and could not consistently achieve reproducible results [152]. The advent and advancement of radiotherapy, chemotherapy, and targeted therapy have long overshadowed immunotherapy as effective methods for cancer treatment [153,154]. After nearly a century of effort, with the emergence of immunosuppressants, initial results have been achieved in the application of immunotherapy in cancer treatment. The Food and Drug Administration (FDA) has approved several immune drugs, either alone or in combination with other drugs, for the treatment of various malignancies, e.g., ipilimumab [155], nivolumab [156], and imiquimod [157]. However, clinical studies have shown that not all ICIs are effective for every patient [28,158]. Since the publication of an article in Science in 2015 demonstrating that the gut microbiota can reverse nonresponse to immunosuppression, there has been renewed interest in exploring the influence of the gut microbiota on tumor immunotherapy [28].
The adaptive immune response induced by the gut microbiota primarily occurs through interactions between pathogen-associated molecular patterns (PAMPs) and PRRs [159,160]. Local immune responses are initiated when PRRs (e.g., TLRs) recognize PAMPs from the gut microbiota [161]. Microorganisms or their metabolites (e.g., SCFAs) can serve as inducers of local immunity. During this process, SCFAs activate the plasma cell production of IgA, which hinders bacterial adhesion, aggregation, and invasion while also directly affecting bacterial virulence [26]. In addition, PAMPs induce DC maturation. Mature DCs then migrate to mesenteric lymph nodes where they interact with naive T cells, facilitating their development into CD4+ T cells. The stimulation of CD8+ T cells is also directly induced by DCs, and activated T cells play a crucial role in maintaining the stability of the gut environment and preventing gut infections [26].
The survival rate of patients with epithelial tumors who did not receive antibiotic treatment during anti-PD1/PD-L1 therapy was significantly greater than that of patients who received antibiotics [162,163]. A comparison of the gut microbiota between PD-1 inhibitor responders and nonresponders revealed significant differences, particularly in terms of the greater diversity observed among responders [164]. Additionally, the abundance of A. muciniphila in fecal samples from patients who exhibited a positive response to PD-1 inhibitors was significantly greater than that in nonresponders [162]. Oral administration of A. muciniphila can ameliorate patient responsiveness to PD-1 inhibitors through the promotion of immune cell infiltration into tumors through supplementation with A. muciniphila. Specifically, CR9+, CXCR3+, and CD4+ T cells are recruited to the tumor microenvironment, where they restore the efficacy of PD-L1 inhibitors [165].
Chimeric antigen receptor (CAR) T-cell therapy, a state-of-the-art immunotherapy, represents a novel therapeutic avenue for patients with refractory and recurrent B-cell leukemia or lymphoma. However, the efficacy of this treatment remains heterogeneous, with only 40% of patients achieving complete and durable remission at best, thereby impeding its widespread clinical application [166,167]. Stein-Thoeringer et al. demonstrated that exposure to broad-spectrum, high-risk antibiotics (such as meropenem, piperacillin-tazobactam or cefepime) within 3 weeks prior to CAR-T-cell therapy is associated with worse progression-free survival and overall survival [59]. Next, they analyzed the gut microbiota and found that Bacteroides, Ruminococcus, Eubacterium and Akkermansia are the most important genera for determining CAR-T-cell responsiveness. Additionally, Akkermansia was shown to be associated with preinfusion peripheral T-cell counts in these patients. Luu et al. reported that the frequency of IFN-γ+TNF-α+CD8+ T cells within cytotoxic T lymphocytes significantly increased after treatment with supernatants derived from Megasphaera massiliensis, and they demonstrated that this increase was caused by SCFAs in the supernatant [168]. Further studies have shown that treatment with butyrate or pentanoate can enhance the expression of CD25 and the production of TNF-α and IFN-γ in receptor tyrosine kinase-like orphan receptor 1 CAR-T cells upon stimulation, thereby enhancing the antitumor efficacy of CAR-T cells.
CpG-oligodeoxynucleotide (CpG-ODN) is a TLR9 agonist with an immunoactivating effect that can directly induce the activation and maturation of plasmacytoid dendritic cells as an adjuvant for tumor immunotherapy [169]. Studies have shown that CpG-ODNs activate the immune response to tumor cells by inducing the secretion of proinflammatory cytokines such as TNF-α and IL-12 from myeloid cells [170]. Iida et al. reported that in this proinflammatory microenvironment, antigen-specific T-cell activation occurs, resulting in the effective clearance of most conventional mouse tumors. However, tumor-infiltrating myeloid cells in germ-free mice fail to produce proinflammatory agents that respond to CpG-ODNs, leading to diminished therapeutic efficacy of CpG-ODN therapy [171]. Additionally, they investigated the association between the gut microbiota and CpG-ODN efficacy and revealed a positive correlation between Ruminococcus obeum and Alistipes and TNF-α secretion, with Lactobacillus sp. exhibiting a negative correlation. Subsequent administration of Alistipes following antibiotic treatment restored CpG-ODN-induced TNF-α secretion, whereas oral administration of Lactobacillus sp. reduced TNF-α secretion.
3.2 Chemotherapy
Currently, chemotherapy is the conventional therapeutic approach for treating pancreatic ductal adenocarcinoma; however, approximately 50% of patients do not respond to this therapeutic approach [172,173]. Genetic alterations in patients cannot explain the difference between responsive and nonresponsive patients to chemotherapy [174,175]. Emerging evidence highlights the pivotal role of the gut microbiota in determining the response to chemotherapy [173]. Tintelnot et al. reported that the microbiota-derived tryptophan metabolite indole-3-acetic acid (3-IAA) is enriched in patients who respond to chemotherapy [173]. Furthermore, they demonstrated that the efficacy of 3-IAA and chemotherapy is mediated by neutrophil-derived myeloperoxidase. In conjunction with chemical treatment, myeloperoxidase oxidizes 3-IAA, which leads to the downregulation of the enzymes glutathione peroxidase 3 and glutathione peroxidase 7, which are responsible for degrading reactive oxygen species (ROS). The accumulation of ROS and the downregulation of autophagy compromise the metabolic fitness of cancer cells, ultimately impeding their proliferation. The gut microbiota not only enhances the effectiveness of chemical drugs but also alleviates their adverse effects [148,170]. When chemotherapy drugs act on rapidly proliferating tumor cells, gut mucosal cells are also affected by their high proliferation rate, resulting in disruption of the gut barrier [176]. Cyclophosphamide (CTX) is a commonly used chemical drug that is widely employed in the treatment of patients with solid tumors and hematological malignancies [177]. However, it is known to induce acute gut mucosal injury [178]. Oral administration of Lactobacillus plantarum NCU116 has significant efficacy in ameliorating CTX-mediated gut mucosal injury and improving gut metabolism and the gut microbiota [179].
The gut microbiota exerts both beneficial and detrimental effects on chemotherapy outcomes [179,180]. Relevant studies have demonstrated that irinotecan, a chemical drug frequently used for treating CRC, can induce severe diarrhea due to the presence of bacterial β-glucuronidase in the gut [180]. Carboxylesterases, which are present in various tissues, catalyze the conversion of CPT-11 into SN-38, thereby killing cancer cells. Moreover, SN-38 can be inactivated in the liver by the uridine diphosphoglucuronosyltransferase 1A1, resulting in the formation of SN-38G. Subsequently, SN-38G is excreted into the gut via bile. Although SN-38G does not exhibit toxicity toward the gut mucosa, the enzymatic activity of β-glucuronidases produced by the gut microbiota leads to the metabolic conversion of SN-38G into SN-38, which damages the gut mucosa [180,181].
3.3 Radiotherapy
Radiotherapy is an important method for treating tumors because it induces DNA damage in both tumor cells and normal cells through indirect energy transfer, which involves the production of reactive oxygen and nitrogen [182]. Gastrointestinal cells exhibit rapid turnover and are highly susceptible to radiation, making them the primary target of injury during radiotherapy and significantly impacting patient quality of life [183]. Numerous studies have demonstrated the crucial role of the gut microbiota in regulating the physiological and pathological states of the host [1,2], suggesting its potential involvement in radiation-induced damage [184]. Ferreira et al. conducted a cohort study to investigate the relationship between the gut microbiota and radiation-induced enteropathy. They found that patients with radiation enteropathy had a reduced diversity of gut microbiota, as well as a significantly greater abundance of Clostridium IV, Roseburia, and Phascolarctobacterium [185].
Guo et al. reported that only a small percentage of mice were able to survive a high dose of radiation. Subsequent research revealed enrichment of Lachnospiraceae and Enterococcaceae, which can mitigate radiation-induced gastrointestinal damage, in these elite survivors. Through nontargeted metabolomics research, they discovered that downstream metabolites of the gut microbiota, such as propionate and tryptophan, contribute substantially to radioprotection [184]. Considering the correlation between the gut microbiota and radiation-induced gut damage, probiotics and prebiotics have been used in clinical interventions to prevent or treat radiation-induced gut injury [186,187]. The results indicate the beneficial effects of the gut microbiota and its metabolites on radiation-induced gut damage; however, these findings are not yet sufficient to influence clinical practice [26]. However, additional research is needed to confirm the protective effect of the gut microbiota and its metabolites on radiation-induced gut injury.
3.4 Molecular targeted therapy
Molecular targeted therapy has increasingly been utilized in the treatment of malignant tumors, establishing itself as a novel paradigm for tumor drug therapy. Compared to conventional therapies such as radiotherapy and chemotherapy, targeted therapy has superior efficacy and reduced toxicity [188]. However, the adverse reactions induced by molecular targeted drugs cannot be disregarded. Diarrhea represents a prevalent clinical manifestation that not only compromises patient quality of life but also imposes limitations on the safe utilization of these drugs [189]. Increasing evidence suggests that the gut microbiota could influence the development of tyrosine kinase inhibitor (TKI)-induced diarrhea [190]. Pal et al. collected stool samples from patients with metastatic renal cell carcinoma who were receiving vascular endothelial growth factor (VEGF)-TKIs and evaluated the relationship between VEGF-TKI-related diarrhea and the gut microbiota. They discovered higher levels of Bacteroides spp. and lower levels of Prevotella spp. in patients with diarrhea [191]. Alterations in the gut microbiota can be observed in patients who experience TKI-induced diarrhea, and regulating these changes may reduce the occurrence of these side effects. Ianiro et al. reported findings from a randomized clinical trial (ClinicalTrials.gov number: NCT04040712) of FMT for the treatment of diarrhea induced by TKIs in patients with metastatic renal cell carcinoma [192]. In this study, twenty patients were randomly assigned to receive FMT from either healthy donors or placebo-treated FMT. These findings demonstrate that donor FMT exhibits superior efficacy to placebo FMT in the treatment of TKI-induced diarrhea; additionally, successful engraftment is observed in recipients receiving donor feces.
Histone deacetylase (HDAC) inhibitors not only induce cellular differentiation, apoptosis, autophagy, and cell cycle arrest but also modulate immune responses and inhibit angiogenesis in various hematologic malignancies and some solid tumors [193]. Butyric acid, an SCFA, accelerates histone acetylation and participates in the apoptosis and proliferation of various cancer cells. It has been extensively investigated as an HDAC inhibitor in the field of antitumor research [194,195]. He et al. revealed that butyrate, a metabolite of the gut microbiota, can enhance the immune response of CD8+ T cells in an ID2-dependent manner, thereby improving the effectiveness of oxaliplatin in antitumor therapy [25]. Luu et al. also demonstrated that pentanoate and butyrate modulate CD8+ T-cell responses, enhancing the antitumor activity of cytotoxic T lymphocytes and CAR-T cells [168]. Moreover, their research suggested that M. massiliansis may be a potential probiotic for the production of pentanoate and butyrate.
3.5 Surgical treatment
In the early stages of cancer, surgery is commonly used as a treatment method and significantly impacts patient microbiota, especially the gut microbiota [196]. Research has shown that in patients undergoing tumor surgery, the use of preoperative antibiotics may lead to a reduction in gut microbial diversity and the growth of pathogenic bacteria, potentially resulting in complications such as increased gut permeability [197,198]. Modulating the gut microbiota can be considered a potential strategy to alleviate this issue. Relevant studies have shown that certain microorganisms, such as Lactobacillus spp. and A. muciniphila, can regulate gut barrier healing through mechanisms dependent on ROS or formyl peptide receptors [199,200]. The Bacteroides thetaiotaomicron is an important component of the gut microbiota in normal mice and humans. It has been demonstrated that these commensal bacteria can successfully colonize germ-free mice and significantly regulate the expression of genes associated with various gut functions, such as nutrient absorption, mucosal barrier reinforcement, and angiogenesis [201]. In contrast to probiotics that preserve the integrity of the gut barrier, certain pathogenic bacteria, such as Serratia marcescens and Pseudomonas aeruginosa, can exacerbate damage to the gut barrier [202]. Therefore, selectively eliminating pathogenic bacteria and preserving probiotics before surgery can effectively mitigate surgical complications. Currently, several ongoing studies are investigating the impact of perioperative probiotics and commensal bacteria on surgical complications in patients undergoing tumor resection [203].
Mounting evidence suggests that the gut microbiota plays a crucial role in modulating both the efficacy and toxicity of cancer therapy [30,148]. However, the field is in its infancy, and tremendous opportunities exist to further elucidate the mechanisms through which these microorganisms impact cancer therapy. Therefore, it is crucial to explore a comprehensive approach that integrates the regulation of the gut microbiota with cancer immunotherapy, intensive chemotherapy, radiotherapy, targeted therapy, and surgery to achieve enhanced therapeutic outcomes while minimizing adverse effects. Currently, numerous ongoing clinical trials are underway to translate research findings from laboratory experiments to practical applications [147]. Moreover, considering the substantial variation in bacterial strains among different healthy individuals and the limited functional understanding of the gut microbiota, coupled with a lack of knowledge regarding the composition of an “optimal” bacterial consortium, caution should be exercised when regulating the gut microbiota in cancer patients.
4. The gut microbiota: A new force in cancer diagnosis?
An increasing number of animal experiments and clinical studies have demonstrated that the diversity and abundance of the gut microbiota are associated with cancer pathogenesis and treatment [10,17]. These data illustrate the potential use of the gut microbiota as a biomarker for understanding cancer pathogenesis and guiding cancer treatment [18,132,204].
4.1 Screening for cancer
The utilization of the gut microbiota as a biomarker for cancer diagnosis is being extensively studied in both preclinical and clinical research [205]. Timely treatment for early-stage cancer can lead to effective therapeutic outcomes. For example, the 5-year survival rate for patients with localized CRC is 90%, while that for patients with distal metastatic CRC is only 14% [16]. Many HCC patients are already in the advanced stage at the time of diagnosis, and the gut microbiota could serve as a reliable biomarker for the early screening of HCC [132,133]. Researchers collected fecal samples from individuals in East China, Central China, and Northwest China and analyzed the fecal microbiota using HTST. These feces were obtained from healthy individuals, patients with liver cirrhosis, and patients in the early stage of liver cancer. Screening with a random forest model revealed that 30 gut microbial markers can most accurately reflect the progression of liver cancer; the area under the curve (AUC) was 0.8. The model was validated in liver cancer patients from Northwest and Central China, and the AUC for differentiating between healthy individuals and those with early-stage liver cancer was 0.768, while it was 0.804 for differentiating between healthy individuals and those with advanced-stage liver cancer [132]. This model establishes a connection between changes in the gut microbiota and liver cancer screening, emphasizing the potential of the gut microbiota as a diagnostic tool for liver cancer.
Given that gut dysbiosis is typically an early event in the development of CRC, numerous studies have been conducted to explore the fecal microbiome to identify potential diagnostic markers [205,206]. Kong et al. conducted metagenomic and metabolomic analyses of the interactions among the gut microbiota, metabolites and microbial enzymes in 130 individuals with late-onset CRC (LO-CRC), 114 individuals with early-onset colorectal cancer (EO-CRC), and age-matched healthy controls to assess the potential of those factors to serve as noninvasive biomarkers for EO-CRC [97]. Compared to that in the control group, the alpha diversity in both the LO-CRC and EO-CRC groups was lower. The enrichment of F. nucleatum and depletion of SCFAs are characteristic features observed in LO-CRC. In comparison, the multiomics signatures of EO-CRC exhibited a tendency toward an increased presence of Flavonifractor plauti and elevated levels of tryptophan, bile acid and choline metabolism. Yu et al performed HTST on a cohort of CRC patients (n=74) and healthy individuals (n=54) and reported that 20 types of gut microbiota were associated with CRC; the AUC was 0.84 [204].
This article provides an overview of gut microbiota-cancer therapy interactions. The gut microbiota induces adaptive immunity, and A. muciniphila reverses the nonresponse to PD-1/PD-L inhibitors. The metabolite 3-IAA, produced by the gut microbiota, enhances the efficacy of chemotherapy. Supplementation with Lactobacillus plantarum NCU116 can reduce the damage caused by CTX to the gut mucosa. However, β-glucuronidases produced by the gut microbiota can convert SN-38G into SN-38, which is toxic to the gut. Radiation therapy can result in a reduction in the diversity of the gut microbiota and an increase in the abundance of pathogenic bacteria, whereas supplementation with probiotics and prebiotics exerts a protective effect against radiation-induced damage. The use of FMT is indicated for the treatment of diarrhea resulting from TKI therapy. The administration of preoperative antibiotics may result in a reduction in gut microbial diversity and the proliferation of pathogenic bacteria, which can compromise gut barrier function. Conversely, probiotic supplementation has been shown to enhance gut barrier function. This figure was created using Figdraw.
4.2 Predictive biomarkers
The available data suggest a correlation between specific gut bacteria and cancer prognosis, indicating that microbial markers have the potential to predict the treatment response of cancer patients. However, there is a paucity of data from studies that specifically investigate the longitudinal changes in the gut microbiota during chemotherapy, radiotherapy, and molecular targeted therapy. Given the pivotal role of the gut microbiota in facilitating an efficacious response to ICIs, numerous studies have endeavored to establish correlations between specific gut microbial signatures and ICI responses and survival outcomes. Associations between specific bacterial species, such as A. muciniphila [162,165] and Bifidobacterium spp. [28], and the response to ICIs have been extensively documented. Martini et al. compared responders and nonresponders in a cohort of 14 patients with CRC who received cetuximab plus the antiPDL1 antibody avelumab [207]. They found that, compared to nine patients with shorter progression-free survival (PFS) (2-6 months), five long-term responding patients (those with PFS 9-24 months) had significantly greater abundances of two butyrate-producing bacteria, Agathobacter M104/1 and Blautia SR1/5. These findings were validated in the CAVE-Lung validation cohort. In addition, improved predictors of ICI response are essential for optimizing the efficacy of this therapeutic approach.
The administration of antibiotics compromises the therapeutic efficacy of PD-1 blockade in cancer patients [163]; antibiotic treatment leads to a decrease in gut microbiota diversity and an increase in the abundance of Enterocloster clostridioformis, which subsequently downregulates the serum mucosal addressin cell adhesion molecule-1 (MAdCAM-1) level. A low serum concentration of MAdCAM-1 has a negative impact on prognosis, which has been verified in a cohort of NSCLC patients [163,208]. Therefore, it is imperative to investigate the impact of antibiotic exposure on the outcome of ICI treatment.
Treatment with combined immune checkpoint blockade (CICB) agents targeting both CTLA-4 and PD-1 has demonstrated remarkable clinical efficacy across various tumor types; however, this efficacy comes at the cost of frequent, severe immune-related adverse events [209]. Andrews et al. demonstrated that there is a correlation between gut microbiota signatures and the toxicity associated with CICB [210]. Gut microbiota profiling revealed a significantly greater abundance of Bacteroides intestinalis in patients experiencing toxicity than in patients without toxicity, and the gut microbiota mediated CICB-induced intestinal toxicity through IL-1β; however, the underlying mechanism requires further elucidation.
4.3 Future directions
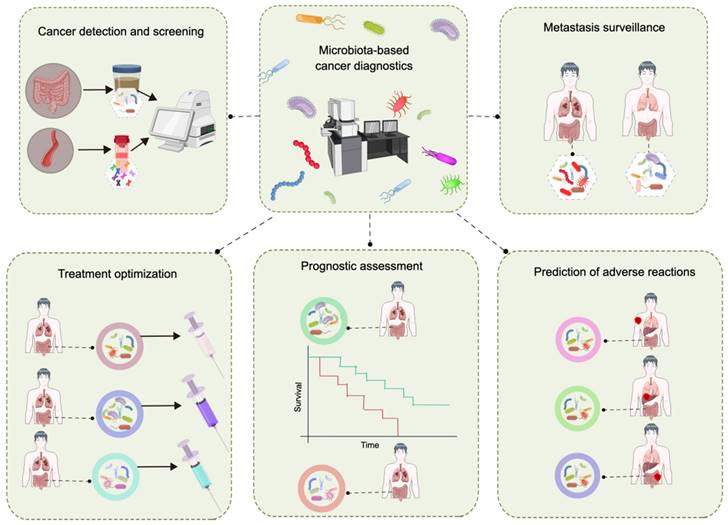
Extensive research has confirmed the relationship between the gut microbiota and cancer. As research continues to elucidate the corresponding mechanisms, unprecedented opportunities arise for exploring the use of the gut microbiota in cancer diagnosis and management; however, challenges remain. The primary limitation lies in the accuracy of utilizing fecal microbial markers for cancer screening [77,211]. To address this issue, the integration of the gut microbiota with other biomarkers has been employed to increase the precision of detection. Numerous studies have demonstrated the close relationship between F. nucleatum and the initiation of CRC. The combination of fecal immunochemical detection with the abundance of F. nucleatum significantly enhances the efficacy and sensitivity of fecal immunochemical detection [212,213]. The fecal immunochemical test has an AUC value of 0.86 for CRC detection; however, incorporating the abundance of F. nucleatum into the model further increases the AUC value to 0.95 [214]. In addition, the integration of the gut microbiota and diagnostic biomarkers in serum enhances the precision of cancer detection via the gut microbiota. The fecal metagenomic classifier had an AUC of 0.84 for accurately identifying pancreatic ductal adenocarcinoma (PDAC) within a Spanish cohort, and the accuracy improved (AUC of 0.94) when combined with the less specific carbohydrate antigen (CA) 19-9 serum marker [215]. CA19-9 is currently the only FDA-approved noninvasive diagnostic biomarker for PDAC and has a low specificity for diagnosing PDAC [215,216]. The incorporation of the gut microbiota and other biomarkers enhances the precision of cancer detection. In future investigations, evaluating the combined utilization of the gut microbiota and additional biomarkers will be particularly crucial. The ultimate goal is to develop a method based on the gut microbiota for early cancer detection, metastasis surveillance, treatment optimization, etc. (Figure 6).
5. Modulating the gut microbiota
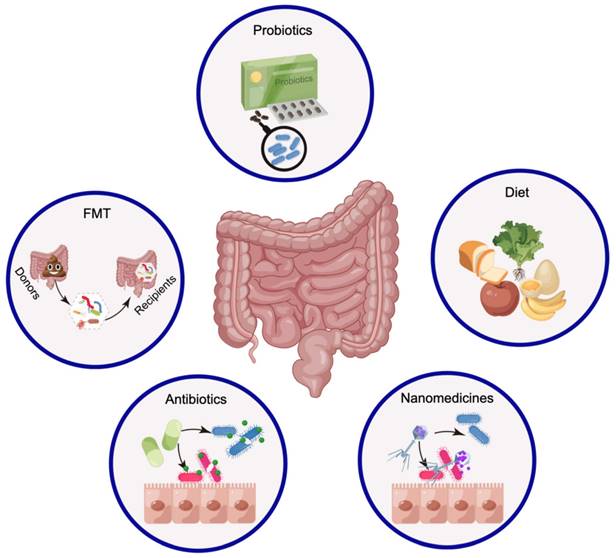
Numerous studies have shown that cancer- and host-related factors combine in different ways, revealing the heterogeneity of cancer pathogenesis and clinical treatment outcomes. How can the gut microbiota be utilized for cancer treatment? Modulating the gut microbiota may be a manipulable and beneficial approach to cancer treatment when considering all factors [52,217]. Although people hope to improve the efficacy of tumor treatment by modulating the gut microbiota, there is currently a lack of consensus on how to regulate this process. Currently, FMT [217], probiotics [26], dietary adjustments [218], and antibiotics [219] are utilized primarily for modifying the composition of the gut microbiota (Figure 7). Notably, nanomedicine is prepared in an interdisciplinary manner with the aim of targeting and eliminating specific pathogenic bacteria [38].
Utilization of gut microbiota data in cancer diagnosis and patient stratification. This figure was created using Figdraw.
5.1 Fecal microbiota transplantation
FMT has been studied in the context of cancer treatment, and restoring the recipient's gut microbiota to an optimal health status is the most direct method; this approach poses both a major challenge and an urgent opportunity [217]. FMT preparations can be administered through oral delivery via freeze-dried or frozen pellets, as well as invasive procedures such as colonoscopy or gastroscopy [26]. FMT transplants a complete gut microbiota from the donor to the recipient, and the introduced microbiota is more stable in the recipient's environment and less competitive against the recipient's own microbiota, thereby facilitating microbial interdependence and collaboration [203]. FMT has achieved excellent results in the treatment of Clostridium difficile infections, demonstrating a greater cure rate than standard therapy [203]. Moreover, FMT is considered a viable treatment option for various diseases, including diabetes, metabolic syndrome, and inflammatory bowel disease [220,221]. Currently, clinical trials of ICI therapy involving FMT are underway. A study by Routy et al. demonstrated that FMT enhances the effectiveness of anti-PD-1 therapy [37].
While research on FMT is flourishing, a patient in the U.S. died after receiving FMT in 2019, and as a result, the U.S. FDA suspended some clinical trials involving FMT until safety was fully confirmed. The poor efficacy of FMT raises concerns regarding the potential risk of infection [222]. The common adverse effects of FMT often pertain to gastrointestinal discomfort, which can include abdominal cramps, constipation, bloating, hiccups, nausea, vomiting, diarrhea, hematochezia and so on. However, these symptoms usually resolve quickly and do not pose a significant threat to patient health [223]. The improper screening of donors and inadequate analysis of fecal donor material may also result in serious adverse reactions. For example, donor feces were not screened, and as a result, two patients contracted multidrug-resistant bacteria after FMT, leading to one death. Therefore, the FDA has warned researchers to expand fecal screening in FMT studies to include specific antibiotic-resistant bacteria. However, this measure alone is insufficient for predicting adverse events caused by specific pathogen infections. These infections may not contain antibiotic-resistant bacteria, but pathogenic bacteria derived from the donor could still possess inherent virulence and pose a threat to recipients' health [222]. Conducting thorough screening tests on FMT donors is essential for reducing and preventing the incidence of adverse events [221,222].
Strategies to modify the gut microbiota for cancer treatment The modulation of the gut microbiota through FMT, probiotics, and dietary regulation primarily contributes to the enrichment of probiotics. The current practice primarily involves the use of antibiotics for eradicating pathogenic bacteria, which may have detrimental effects on treatment outcomes. However, the application of nanomedicines offers opportunities for targeted elimination of pathogenic bacteria. This figure was created using Figdraw.
Additionally, successful FMT requires not only the transplantation of microbiota into the recipient's gut but also long-term colonization to maintain therapeutic efficacy [224,225]. After FMT, the gut microbiota of the recipient and donor exhibit the highest similarity on the first day, but the composition changes over time [226]. The average duration for maintaining a clinical response in patients with Crohn's disease is 125 days after the initial FMT and 176.5 days after the second transplantation [227]. These findings suggest that FMT can be regularly performed to maintain clinical efficacy. Currently, there are limited clinical studies on the application of FMT as an adjuvant cancer treatment; however, ensuring its safety and long-term efficacy are important concerns.
5.2 Probiotics
Although FMT is the most direct method for altering the gut microbiota, the complex microbial community increases the risk of infection in patients [222,223]. Compared to FMT, probiotic transplantation provides a more practical approach for regulating the gut microbiota in clinical treatment [26]. The term “probiotics” refers to live microorganisms that, when administered in appropriate quantities, provide a safe beneficial effect on the host's health [228]. The earliest commercial probiotic supplements were derived from easily cultivable single strains of food sources, such as Bifidobacterium and Lactobacillus, which have well-established uses in the treatment of numerous gastrointestinal disorders [229,230]. Given such observations, is it feasible to utilize probiotics in cancer treatment?
Research on the use of probiotics for cancer treatment has focused mainly on their ability to enhance immune function, potentially helping combat cancer [203]. In CRC patients treated with Lactobacillus johnsonii during the perioperative period, bacteria adhere to the colonic mucosa, reducing the concentration of pathogens in feces and regulating local immune function [231,232]. Additionally, A. muciniphila is capable of restoring mouse responsiveness to PD-1 inhibitors [165]. Bifidobacterium has shown potential in enhancing antitumor immunity and improving the effectiveness of anti-PD-L1 treatments [28]. The administration of probiotics enhances the immune response and mitigates the adverse effects of radiation therapy. Treatment with Lactobacillus acidophilus LAC-361 and Bifidobacterium longum BB-536 can reduce radiation-induced diarrhea [233]. The aforementioned cases exemplify the promising potential of probiotics in the field of cancer therapy.
Despite the demonstrated benefits of probiotics in cancer treatment, there are still numerous challenges that need to be addressed. Probiotics vary in their ability to survive gastric acid treatment and colonize the gut, just as their species, dosage, preparation method, and host microbiota also differ [228]. The vast majority of clinical trials on probiotics reported in the literature have not raised significant safety concerns; however, there are still some serious adverse reactions caused by probiotics that draw our attention to their potential risks. These reported cases involve incidents of bacterial sepsis associated with Lactobacilli-containing probiotic supplements, as well as the death of a preterm infant from gastrointestinal mucormycosis associated with mold contamination in a probiotic supplement [234]. In addition, probiotics are used to regulate the gut microbiota but are largely unregulated in both the EU and the US, potentially resulting in significant variations in quality [234]. Currently, there are no universally applicable probiotics available for modulating the gut microbiota. Prior to administering probiotics to cancer patients, individual analysis and cautious usage are warranted, with tailored strategies developed for specific populations [26]. Strategies for selecting probiotics should be considered. The safety assessment of probiotics is of paramount importance. In what combinations can probiotics be used (such as joint synbiotics), what is the timing of use, and what are the mechanisms of action [235]? How can successful microbial treatments be efficiently packaged, deployed, and dosed over time to achieve effective treatment or reduce treatment side effects?
5.3 Diet
Considering the crucial role of the gut microbiota in preventing cancer and the limitations associated with FMT and probiotics, most researchers have incorporated diet into their studies on regulating the gut microbiota. Diet plays a crucial role in determining the structure and function of the gut microbiota, and the interaction between diet and microorganisms determines whether they are beneficial or detrimental to host health [218,236]. Considering a series of parameters, the Mediterranean diet is associated with a lower risk of cancer initiation and death, primarily by enhancing immune function mediated by cytotoxic cells and helper T cells [237]. Is it possible to modulate the gut microbiota in a way that is beneficial to human health through specific dietary components?
Analyses of dietary components have revealed that certain ingredients can influence the composition and abundance of specific gut microbiota [203,236]. The supplementation of dietary fibers, such as fructan and galactooligosaccharide, alters the composition of the gut microbiota, increases the abundance of Bifidobacterium and Lactobacillus spp., subsequently increases the concentration of butyrate in feces, and inhibits CRC [238-240]. Resistant starch is a substance that benefits gut health by serving as a valuable substrate for numerous beneficial gut microorganisms, including the genera Bifidobacterium, Akkermansia, and Megasphaera [241,242]. The combination of resistant starch with arabinoxylan increases the abundance of Bifidobacterium while decreasing the abundance of other undesirable genera in the gut microbiota, thus modulating the concentration of SCFAs in the gut and exerting beneficial effects on colon health [243]. The results of that study suggest, to some extent, that the use of resistant starch and arabinoxylan for modulating the gut microbiota may enhance the therapeutic effect of CRC treatment. Preclinical and clinical studies have demonstrated that both the type and quantity of protein in the diet impact the composition of the gut microbiota [218]. Studies have shown that casein acts as a growth factor for Lactobacilli and Bifidobacteria; additionally, these bacteria have been shown to be beneficial in cancer treatment [218].
5.4 Elimination of pathogenic microorganisms
Research indicates that the gut microbiota plays a significant role in the initiation, progression and treatment of cancers such as BC [44], CRC [78], and HCC [129,131]. The main way to regulate the gut microbiota mentioned above is by increasing the abundance of beneficial bacteria, aiming to prevent cancer initiation and aid in cancer treatment. However, eradicating pathogenic microorganisms is equally crucial for both cancer prevention and treatment. Antibiotic treatment is the most common method for eliminating pathogenic microorganisms, but this indiscriminate elimination can harm probiotics [219,244]. The targeted elimination of pathogenic microorganisms through nanomedicine offers greater possibilities for effectively regulating the gut microbiota [38].
5.4.1 Antibiotics
The use of antibiotics in the treatment of diseases is becoming increasingly prevalent among humans. The scavenging effects of antibiotics on the microbiota are well known; they can eliminate pathogenic microorganisms but may also disrupt the structure of the microbial community in the human body, leading to the dysregulation of host-microbiota interactions [219,244]. Even at sublethal concentrations, antibiotics can cause significant and nonselective changes in the gut microbiota. Furthermore, Parthasarathy et al. reported that the impact of antibiotics on slow-growing and aggregating microorganisms is more pronounced than that on rapidly growing microorganisms [245]. A study conducted by Hagan et al. demonstrated that antibiotic usage led to a 10,000-fold decrease in the gut microbiota load [58]. Additionally, both the diversity and abundance of the gut microbiota decreased, and the bacteria failed to fully recover within six months. These findings suggested that certain specific microbial species may be absent for a prolonged period following antibiotic use. They also found that antibiotics not only affect the composition of the gut microbiota but also disrupt blood metabolism, such as bile acid and tryptophan metabolism. The nonselective eradication of the gut microbiota by antibiotics compromises the therapeutic efficacy of PD-1 blockade in cancer patients [162]. Fidelle et al. reported that antibiotic treatment leads to a decrease in gut microbiota diversity and an increase in the abundance of Enterocloster clostridioformis, resulting in a low serum soluble MAdCAM-1 level and thus a negative impact on prognosis [163]. Therefore, it is particularly important to selectively eliminate pathogenic microorganisms to minimize this impact.
5.4.2 Nanomedicines
Nanomaterials can serve as carriers for delivering various therapeutic drugs to target sites, thereby prolonging their circulation time, protecting drugs from degradation, and reducing drug accumulation at nontarget sites to minimize side effects [246-248]. Utilizing these advantages of nanomaterials to prepare nanomedicines makes it possible to selectively eliminate pathogenic microorganisms. The targeting effect of phages is noteworthy for the specific eradication of pathogenic microorganisms [249,250]. Inspired by this, Zhang et al. designed a targeted nanomedicine to eradicate F. nucleatum, as multiple studies have shown its association with the initiation and progression of CRC [38]. They isolated a phage that specifically lysed F. nucleatum, which was subsequently modified with azide. Dextran nanoparticles were used to coat anti-CRC drugs, and these drugs were covalently linked to the azide-modified phages. These phage-mediated nanoparticles can target F. nucleatum without binding to Bacillus thuringiensis, E. coli, or Clostridium butyricum. This bacteriophage-mediated nanomedicine specifically targets and modulates the composition of the gut microbiota, thereby enhancing the therapeutic efficacy of chemotherapy for CRC. In another study, the capsid protein of this phage was electrostatically assembled with silver nanoparticles to achieve specific clearance of F. nucleatum in the gut and reshape the tumor immune microenvironment, significantly extending overall survival in CRC mice [89]. These studies demonstrate the potential of utilizing nanomaterials to selectively eliminate pathogenic microorganisms, which could be an effective therapeutic strategy for modulating the gut microbiota in the future.
The era of the gut microbiota is ongoing, and the role of the gut microbiota in cancer therapy has been extensively reported in preclinical and clinical studies, suggesting that the gut microbiota may become a potential factor in cancer treatment [26]. However, investigations of the impact of modulating the gut microbiota on cancer treatment have relied primarily on murine models, with few clinical trials being carried out [203]. The human gut microbiota differs significantly from that of mice; therefore, safety assessments should be conducted before extrapolating results from mouse trials to humans [251].
6. Summary and perspectives
The gut microbiota, referred to as the “second genome” of the human body, plays an undeniable role in human health; however, it is also closely associated with various diseases. There is substantial evidence suggesting that the gut microbiota is associated with the initiation of CRC, HCC, BC, and other types of cancer. Recent research on the role of pathogenic microorganisms in cancer has focused primarily on determining the correlation between the abundance of specific strains and cancer using HTST and elucidating the underlying mechanisms that contribute to tumorigenesis. Eliminating carcinogenic microorganisms can prevent cancer or benefit patients during cancer treatment. However, there are still many unresolved issues, such as how to remove pathogenic microorganisms without affecting other probiotics. The scavenging effects of antibiotics on the microbiota are widely acknowledged; however, their administration for eradicating the gut microbiota may inadvertently compromise probiotic populations, potentially leading to unintended consequences. The targeted elimination of detrimental gut microbiota constituents through nanomedicine represents a highly promising method for future exploration and necessitates further comprehensive investigation.
While certain gut microorganisms may be associated with the initiation of cancer, importantly, there are numerous beneficial gut microorganisms that play crucial roles in the body's defense against cancer. This includes enhancing the effectiveness of ICIs and chemotherapy, as well as reducing gut damage caused by chemotherapy and radiation therapy. Considering the advantageous characteristics of the gut microbiota, modulating the gut microbiota is expected to enhance the effectiveness of anticancer therapies. The current primary approaches employed include FMT, probiotic administration, and dietary interventions. The application of these methods can increase the abundance of probiotics, thereby strengthening the effectiveness of cancer treatment; however, they may also induce certain adverse reactions. The future holds promise for enhancing the efficacy of cancer treatment through personalized modulation of the gut microbiota through the use of appropriate interventions while minimizing intolerable adverse reactions.
Several researchers have proposed listing the gut microbiota as a biomarker for the diagnosis and management of cancer based on its impact on cancer pathogenesis and treatment. The sensitivity of the gut microbiota alone as a marker for cancer diagnosis and management may not be high; however, when combined with other markers, the detection sensitivity of the gut microbiota is significantly enhanced. The future holds promise for utilizing the gut microbiota as a noninvasive approach for cancer detection and assessment of treatment efficacy.
Abbreviations
FMT: fecal microbiota transplantation; HTST: high-throughput sequencing technology; CRC: colorectal cancer; SCFAs: short-chain fatty acids; ICIs: immune-checkpoint inhibitors; BC: breast cancer; LC: lung cancer; UGT: uridine 5′-diphospho-glucuronosyltransferase; gmGUS: gut microbial β-glucuronidase; TLR: toll-like receptors; NSCLC: non-small cell lung cancer; SCLC: small cell lung cancer; rRNA: ribosomal RNA; MDSCs: myeloid-derived suppressor cells; TAM: tumor-associated macrophages; TANs: tumor-associated neutrophils; PRR: pattern recognition receptor; H2S: hydrogen sulfide; PCa: prostate cancer; ADT: androgen deprivation therapy; CRPCa: castration-resistant prostate cancer; CT: surgical castration; CS: castration-sensitive phase; CR: castration-resistant phase; GC: gastric cancer; NO: nitric oxide; HCC: hepatocellular carcinoma; LPS: lipopolysaccharide; FDA: Food and Drug Administration; PAMPs: pathogen-associated molecular patterns; DCs: dendritic cells; CAR: chimeric antigen receptor; CpG-ODN: CpG-oligodeoxynucleotide; 3-IAA: indole-3-acetic acid; ROS: reactive oxygen species; CTX: Cyclophosphamide; AUC: area under the curve; LO-CRC: late-onset CRC; EO-CRC: early-onset colorectal cancer; PFS: progression-free survival; MAdCAM-1: mucosal addressin cell adhesion molecule-1; CICB: combined immune checkpoint blockade; PDAC: pancreatic ductal adenocarcinoma; CA: carbohydrate antigen.
Acknowledgements
This work was funded by the National Natural Science Foundation of China Regional Innovation and Development Joint Fund (NSFCU21A20417), the National Natural Science Foundation of China (NSFC31930067, NSFC32001003), the Natural Science Foundation of Sichuan Province (2022NSFSC1282), and the 135 Project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYGD18002). We also thank the support by Post-Doctor Research Project, West China Hospital, Sichuan University (2023HXBH011), Postdoctoral Interdisciplinary Innovation Foundation Project of Sichuan University (1082204112G23), and China Postdoctoral Science Foundation (2023M742476).
Competing Interests
The authors have declared that no competing interest exists.
References
1. O'Toole PW, Paoli M. The human microbiome, global health and the Sustainable Development Goals: opportunities and challenges. Nat Rev Microbiol. 2023;21(10):624-625
2. Caballero-Flores G, Pickard JM, Núñez G. Microbiota-mediated colonization resistance: mechanisms and regulation. Nat Rev Microbiol. 2023;21(6):347-360
3. Elzinga J, van der Oost J, de Vos WM, Smidt H. The Use of Defined Microbial Communities To Model Host-Microbe Interactions in the Human Gut. Microbiol Mol Biol Rev. 2019;83(2):e00054
4. He ZY, Wu H, Yan XH, Liu W. Recent advances in droplet microfluidics for microbiology. Chin Chem Lett. 2022;33(4):1729-1742
5. Wang M, Osborn LJ, Jain S, Meng X, Weakley A, Yan J. et al. Strain dropouts reveal interactions that govern the metabolic output of the gut microbiome. Cell. 2023;186(13):2839-2852.e21
6. Skelly AN, Sato Y, Kearney S, Honda K. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat Rev Immunol. 2019;19(5):305-323
7. Pascal Andreu V, Augustijn HE, Chen L, Zhernakova A, Fu J, Fischbach MA. et al. gutSMASH predicts specialized primary metabolic pathways from the human gut microbiota. Nat Biotechnol. 2023;41(10):1416-1423
8. Chen F, Stappenbeck TS. Microbiome control of innate reactivity. Curr Opin Immunol. 2019;56:107-113
9. Sun SQ, Mao JW, Wang YD. The Role of Gut Microbiota in the Pathogenesis of Alzheimer's Disease. J Biomater Tissue Eng. 2022;12(12):2483-2488
10. Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat Med. 2019;25(3):377-388
11. Braun T, Di Segni A, BenShoshan M, Neuman S, Levhar N, Bubis M. et al. Individualized Dynamics in the Gut Microbiota Precede Crohn's Disease Flares. Am J Gastroenterol. 2019;114(7):1142-1151
12. Nemet I, Saha PP, Gupta N, Zhu W, Romano KA, Skye SM. et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell. 2020;180(5):862-877.e22
13. Miyauchi E, Shimokawa C, Steimle A, Desai MS, Ohno H. The impact of the gut microbiome on extra-intestinal autoimmune diseases. Nat Rev Immunol. 2023;23(1):9-23
14. Diaz Heijtz R, Gressens P, Swann JR. Targeting microbial metabolites to treat autism. Nat Med. 2022;28(3):448-450
15. Li LJ, Sun N, Hao ZL, Sun PP, Fan KH, Yin W. et al. The bio-derived material acacetin ameliorated hyperlipidemia and intestinal barrier damage in mice by modulating gut microbiota. Mater Express. 2023;13(5):753-769
16. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17-48
17. El Tekle G, Garrett WS. Bacteria in cancer initiation, promotion and progression. Nat Rev Cancer. 2023;23(9):600-618
18. Greten TF, Villanueva A, Korangy F, Ruf B, Yarchoan M, Ma L. et al. Biomarkers for immunotherapy of hepatocellular carcinoma. Nat Rev Clin Oncol. 2023;20(11):780-798
19. Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM. et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292-298
20. Janney A, Powrie F, Mann EH. Host-microbiota maladaptation in colorectal cancer. Nature. 2020;585(7826):509-517
21. Dolgin E. Fighting cancer with microbes. Nature. 2020;577(7792):S16-S18
22. Scepanovic P, Hodel F, Mondot S, Partula V, Byrd A, Hammer C. et al. A comprehensive assessment of demographic, environmental, and host genetic associations with gut microbiome diversity in healthy individuals. Microbiome. 2019;7(1):130
23. Shalon D, Culver RN, Grembi JA, Folz J, Treit PV, Shi H. et al. Profiling the human intestinal environment under physiological conditions. Nature. 2023;617(7961):581-591
24. Tang WHW, Bäckhed F, Landmesser U, Hazen SL. Intestinal Microbiota in Cardiovascular Health and Disease: JACC State-of-the-Art Review. J Am Coll Cardiol. 2019;73(16):2089-2105
25. He Y, Fu L, Li Y, Wang W, Gong M, Zhang J. et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8+ T cell immunity. Cell Metab. 2021;33(5):988-1000.e7
26. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell. 2018;33(4):570-580
27. McCoy KD, Geuking MB. Microbiota regulates intratumoral monocytes to promote anti-tumor immune responses. Cell. 2021;184(21):5301-5303
28. Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084-1089
29. Weersma RK, Zhernakova A, Fu J. Interaction between drugs and the gut microbiome. Gut. 2020;69(8):1510-1519
30. Feng W, Liu J, Ao H, Yue S, Peng C. Targeting gut microbiota for precision medicine: Focusing on the efficacy and toxicity of drugs. Theranostics. 2020;10(24):11278-11301
31. Hou K, Wu ZX, Chen XY, Wang JQ, Zhang D, Xiao C. et al. Microbiota in health and diseases. Signal Transduct Target Ther. 2022;7(1):135
32. Simpson RC, Shanahan ER, Scolyer RA, Long GV. Towards modulating the gut microbiota to enhance the efficacy of immune-checkpoint inhibitors. Nat Rev Clin Oncol. 2023;20(10):697-715
33. Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C. et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079-1084
34. Ansaldo E, Slayden LC, Ching KL, Koch MA, Wolf NK, Plichta DR. et al. Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science. 2019;364(6446):1179-1184
35. Zeng X, Xing X, Gupta M, Keber FC, Lopez JG, Lee YJ. et al. Gut bacterial nutrient preferences quantified in vivo. Cell. 2022;185(18):3441-3456.e19
36. Bird L. Probiotics promote local antitumour immunity. Nat Rev Immunol. 2023;23(6):343
37. Routy B, Lenehan JG, Miller WH Jr, Jamal R, Messaoudene M, Daisley BA. et al. Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: a phase I trial. Nat Med. 2023;29(8):2121-2132
38. Zheng DW, Dong X, Pan P, Chen KW, Fan JX, Cheng SX. et al. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat Biomed Eng. 2019;3(9):717-728
39. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209-249
40. Peek RM Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2(1):28-37
41. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195-206
42. Gilbert JA, Lynch SV. Community ecology as a framework for human microbiome research. Nat Med. 2019;25(6):884-889
43. Sepich-Poore GD, Zitvogel L, Straussman R, Hasty J, Wargo JA, Knight R. The microbiome and human cancer. Science. 2021;371(6536):eabc4552
44. Terrisse S, Derosa L, Iebba V, Ghiringhelli F, Vaz-Luis I, Kroemer G. et al. Intestinal microbiota influences clinical outcome and side effects of early breast cancer treatment. Cell Death Differ. 2021;28(9):2778-2796
45. Hou MF, Ou-Yang F, Li CL, Chen FM, Chuang CH, Kan JY. et al. Comprehensive profiles and diagnostic value of menopausal-specific gut microbiota in premenopausal breast cancer. Exp Mol Med. 2021;53(10):1636-1646
46. Yang J, Tan Q, Fu Q, Zhou Y, Hu Y, Tang S. et al. Gastrointestinal microbiome and breast cancer: correlations, mechanisms and potential clinical implications. Breast Cancer. 2017;24(2):220-228
47. Adlercreutz H, Martin F, Pulkkinen M, Dencker H, Rimer U, Sjoberg NO. et al. Intestinal metabolism of estrogens. J Clin Endocrinol Metab. 1976;43(3):497-505
48. Fernández-Murga ML, Gil-Ortiz F, Serrano-García L, Llombart-Cussac A. A New Paradigm in the Relationship between Gut Microbiota and Breast Cancer: β-glucuronidase Enzyme Identified as Potential Therapeutic Target. Pathogens. 2023;12(9):1086
49. Cleary MP, Grossmann ME. Minireview: Obesity and breast cancer: the estrogen connection. Endocrinology. 2009;150(6):2537-2542
50. Hu S, Ding Q, Zhang W, Kang M, Ma J, Zhao L. Gut microbial beta-glucuronidase: a vital regulator in female estrogen metabolism. Gut Microbes. 2023;15(1):2236749
51. Sui Y, Wu J, Chen J. The Role of Gut Microbial β-Glucuronidase in Estrogen Reactivation and Breast Cancer. Front Cell Dev Biol. 2021;9:631552
52. Fernandes MR, Aggarwal P, Costa RGF, Cole AM, Trinchieri G. Targeting the gut microbiota for cancer therapy. Nat Rev Cancer. 2022;22(12):703-722
53. Arnone AA, Cook KL. Gut and Breast Microbiota as Endocrine Regulators of Hormone Receptor-positive Breast Cancer Risk and Therapy Response. Endocrinology. 2022;164(1):bqac177
54. Rutkowski MR, Stephen TL, Svoronos N, Allegrezza MJ, Tesone AJ, Perales-Puchalt A. et al. Microbially driven TLR5-dependent signaling governs distal malignant progression through tumor-promoting inflammation. Cancer Cell. 2015;27(1):27-40
55. Allaire JM, Crowley SM, Law HT, Chang SY, Ko HJ, Vallance BA. The Intestinal Epithelium: Central Coordinator of Mucosal Immunity. Trends Immunol. 2018;39(9):677-696
56. Wellenstein MD, Coffelt SB, Duits DEM, van Miltenburg MH, Slagter M, de Rink I. et al. Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis. Nature. 2019;572(7770):538-542
57. Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16(2):228-231
58. Hagan T, Cortese M, Rouphael N, Boudreau C, Linde C, Maddur MS. et al. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell. 2019;178(6):1313-1328.e13
59. Stein-Thoeringer CK, Saini NY, Zamir E, Blumenberg V, Schubert ML, Mor U. et al. A non-antibiotic-disrupted gut microbiome is associated with clinical responses to CD19-CAR-T cell cancer immunotherapy. Nat Med. 2023;29(4):906-916
60. Zhang D, Frenette PS. Cross talk between neutrophils and the microbiota. Blood. 2019;133(20):2168-2177
61. Fu A, Yao B, Dong T, Chen Y, Yao J, Liu Y. et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. 2022;185(8):1356-1372.e26
62. Parida S, Wu S, Siddharth S, Wang G, Muniraj N, Nagalingam A. et al. A Procarcinogenic Colon Microbe Promotes Breast Tumorigenesis and Metastatic Progression and Concomitantly Activates Notch and β-Catenin Axes. Cancer Discov. 2021;11(5):1138-1157
63. Ross BD. Bacteroides fragilis uses toxins for gut success. Nat Microbiol. 2024;9(1):11-12
64. Khan FH, Bhat BA, Sheikh BA, Tariq L, Padmanabhan R, Verma JP. et al. Microbiome dysbiosis and epigenetic modulations in lung cancer: From pathogenesis to therapy. Semin Cancer Biol. 2022;86(Pt 3):732-742
65. Chen H, Lai Y, Ye C, Wu C, Zhang J, Zhang Z. et al. Global research trends between gut microbiota and lung cancer from 2011 to 2022: A bibliometric and visualization analysis. Front Oncol. 2023;13:1137576
66. Qin X, Bi L, Yang W, He Y, Gu Y, Yang Y. et al. Dysbiosis of the Gut Microbiome Is Associated With Histopathology of Lung Cancer. Front Microbiol. 2022;13:918823
67. Zheng Y, Fang Z, Xue Y, Zhang J, Zhu J, Gao R. et al. Specific gut microbiome signature predicts the early-stage lung cancer. Gut Microbes. 2020;11(4):1030-1042
68. Jia L, Gao F, Hu G, Fang Y, Tang L, Wen Q. et al. A Novel Cytochrome P450 2E1 Inhibitor Q11 Is Effective on Lung Cancer via Regulation of the Inflammatory Microenvironment. Adv Sci. 2023;10(35):e2303975
69. Ubachs J, Ziemons J, Soons Z, Aarnoutse R, van Dijk DPJ, Penders J. et al. Gut microbiota and short-chain fatty acid alterations in cachectic cancer patients. J Cachexia Sarcopenia Muscle. 2021;12(6):2007-2021
70. Chakradhar S. A curious connection: Teasing apart the link between gut microbes and lung disease. Nat Med. 2017;23(4):402-404
71. He B, Liu Y, Hoang TK, Tian X, Taylor CM, Luo M. et al. Antibiotic-modulated microbiome suppresses lethal inflammation and prolongs lifespan in Treg-deficient mice. Microbiome. 2019;7(1):145
72. Sandri BJ, Masvidal L, Murie C, Bartish M, Avdulov S, Higgins L. et al. Distinct Cancer-Promoting Stromal Gene Expression Depending on Lung Function. Am J Respir Crit Care Med. 2019;200(3):348-358
73. Dessein R, Bauduin M, Grandjean T, Le Guern R, Figeac M, Beury D. et al. Antibiotic-related gut dysbiosis induces lung immunodepression and worsens lung infection in mice. Crit Care. 2020;24(1):611
74. Liu X, Cheng Y, Zang D, Zhang M, Li X, Liu D. et al. The Role of Gut Microbiota in Lung Cancer: From Carcinogenesis to Immunotherapy. Front Oncol. 2021;11:720842
75. Gray J, Oehrle K, Worthen G, Alenghat T, Whitsett J, Deshmukh H. Intestinal commensal bacteria mediate lung mucosal immunity and promote resistance of newborn mice to infection. Sci Transl Med. 2017;9(376):eaaf9412
76. Mikhak Z, Strassner JP, Luster AD. Lung dendritic cells imprint T cell lung homing and promote lung immunity through the chemokine receptor CCR4. J Exp Med. 2013;210(9):1855-1869
77. Wong CC, Yu J. Gut microbiota in colorectal cancer development and therapy. Nat Rev Clin Oncol. 2023;20(7):429-452
78. Thomas AM, Fidelle M, Routy B, Kroemer G, Wargo JA, Segata N. et al. Gut OncoMicrobiome Signatures (GOMS) as next-generation biomarkers for cancer immunotherapy. Nat Rev Clin Oncol. 2023;20(9):583-603
79. Reddy BS, Weisburger JH, Narisawa T, Wynder EL. Colon carcinogenesis in germ-free rats with 1,2-dimethylhydrazine and N-methyl-n'-nitro-N-nitrosoguanidine. Cancer Res. 1974;34(9):2368-2372
80. Onoue M, Kado S, Sakaitani Y, Uchida K, Morotomi M. Specific species of intestinal bacteria influence the induction of aberrant crypt foci by 1,2-dimethylhydrazine in rats. Cancer Lett. 1997;113(1-2):179-186
81. Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W. et al. Gavage of Fecal Samples From Patients With Colorectal Cancer Promotes Intestinal Carcinogenesis in Germ-Free and Conventional Mice. Gastroenterology. 2017;153(6):1621-1633.e6
82. Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M. et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 2019;25(4):667-678
83. Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25(4):679-689
84. Nesić D, Hsu Y, Stebbins CE. Assembly and function of a bacterial genotoxin. Nature. 2004;429(6990):429-433
85. He Z, Gharaibeh RZ, Newsome RC, Pope JL, Dougherty MW, Tomkovich S. et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut. 2019;68(2):289-300
86. Pleguezuelos-Manzano C, Puschhof J, Rosendahl Huber A, van Hoeck A, Wood HM, Nomburg J. et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature. 2020;580(7802):269-273
87. Cao Y, Oh J, Xue M, Huh WJ, Wang J, Gonzalez-Hernandez JA. et al. Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science. 2022;378(6618):eabm3233
88. Dougherty MW, Jobin C. Intestinal bacteria and colorectal cancer: etiology and treatment. Gut Microbes. 2023;15(1):2185028
89. Dong X, Pan P, Zheng DW, Bao P, Zeng X, Zhang XZ. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci Adv. 2020;6(20):eaba1590
90. Shenker BJ, Datar S. Fusobacterium nucleatum inhibits human T-cell activation by arresting cells in the mid-G1 phase of the cell cycle. Infect Immun. 1995;63(12):4830-4836
91. Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M. et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207-215
92. Jiang SS, Xie YL, Xiao XY, Kang ZR, Lin XL, Zhang L. et al. Fusobacterium nucleatum-derived succinic acid induces tumor resistance to immunotherapy in colorectal cancer. Cell Host Microbe. 2023;31(5):781-797.e9
93. Sharma BR, Kanneganti TD. Inflammasome signaling in colorectal cancer. Transl Res. 2023;252:45-52
94. Thiele Orberg E, Fan H, Tam AJ, Dejea CM, Destefano Shields CE, Wu S. et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2017;10(2):421-433
95. Chung L, Thiele Orberg E, Geis AL, Chan JL, Fu K, DeStefano Shields CE. et al. Bacteroides fragilis Toxin Coordinates a Pro-carcinogenic Inflammatory Cascade via Targeting of Colonic Epithelial Cells. Cell Host Microbe. 2018;23(2):203-214.e5
96. Song M, Chan AT, Sun J. Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology. 2020;158(2):322-340
97. Kong C, Liang L, Liu G, Du L, Yang Y, Liu J. et al. Integrated metagenomic and metabolomic analysis reveals distinct gut-microbiome-derived phenotypes in early-onset colorectal cancer. Gut. 2023;72(6):1129-1142
98. Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T. et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968-976
99. Yazici C, Wolf PG, Kim H, Cross TL, Vermillion K, Carroll T. et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut. 2017;66(11):1983-1994
100. Nguyen LH, Abu-Ali G, Mehta RS, Lloyd-Price J, Song MY, Yan Y. et al. Dietary patterns, sulfur intake, and the abundance of sulfate-reducing bacteria. Gastroenterology. 2018;154(6):S640
101. Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K. et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. 2013;98(1):111-120
102. Lang T, Zhu R, Zhu X, Yan W, Li Y, Zhai Y. et al. Combining gut microbiota modulation and chemotherapy by capecitabine-loaded prebiotic nanoparticle improves colorectal cancer therapy. Nat Commun. 2023;14(1):4746
103. Zeng H, Umar S, Rust B, Lazarova D, Bordonaro M. Secondary Bile Acids and Short Chain Fatty Acids in the Colon: A Focus on Colonic Microbiome, Cell Proliferation, Inflammation, and Cancer. Int J Mol Sci. 2019;20(5):1214
104. Patel AR, Klein EA. Risk factors for prostate cancer. Nat Clin Pract Urol. 2009;6(2):87-95
105. Chan JM, Gann PH, Giovannucci EL. Role of diet in prostate cancer development and progression. J Clin Oncol. 2005;23(32):8152-8160
106. Miya TV, Marima R, Damane BP, Ledet EM, Dlamini Z. Dissecting Microbiome-Derived SCFAs in Prostate Cancer: Analyzing Gut Microbiota, Racial Disparities, and Epigenetic Mechanisms. Cancers (Basel). 2023;15(16):4086
107. Liu Y, Zhou Q, Ye F, Yang C, Jiang H. Gut microbiota-derived short-chain fatty acids promote prostate cancer progression via inducing cancer cell autophagy and M2 macrophage polarization. Neoplasia. 2023;43:100928
108. Liss MA, White JR, Goros M, Gelfond J, Leach R, Johnson-Pais T. et al. Metabolic Biosynthesis Pathways Identified from Fecal Microbiome Associated with Prostate Cancer. Eur Urol. 2018;74(5):575-582
109. Matsushita M, Fujita K, Motooka D, Hatano K, Fukae S, Kawamura N. et al. The gut microbiota associated with high-Gleason prostate cancer. Cancer Sci. 2021;112(8):3125-3135
110. Lara PN Jr, Mayerson E, Gertz E, Tangen C, Goldkorn A, van Loan M. et al. Bone Biomarkers and Subsequent Survival in Men with Hormone-sensitive Prostate Cancer: Results from the SWOG S1216 Phase 3 Trial of Androgen Deprivation Therapy with or Without Orteronel. Eur Urol. 2024;85(2):171-176
111. Terrisse S, Goubet AG, Ueda K, Thomas AM, Quiniou V, Thelemaque C. et al. Immune system and intestinal microbiota determine efficacy of androgen deprivation therapy against prostate cancer. J Immunother Cancer. 2022;10(3):e004191
112. Pernigoni N, Zagato E, Calcinotto A, Troiani M, Mestre RP, Calì B. et al. Commensal bacteria promote endocrine resistance in prostate cancer through androgen biosynthesis. Science. 2021;374(6564):216-224
113. Alsina M, Arrazubi V, Diez M, Tabernero J. Current developments in gastric cancer: from molecular profiling to treatment strategy. Nat Rev Gastroenterol Hepatol. 2023;20(3):155-170
114. Zhou XM, Ding SZ, Hu RB. The Related Study on the Pathogenesis of Gastrointestinal Diseases in Gastrointestinal Flora and the Risk of Gastric Ulcer Carcinogenesis. J Biomater Tissue Eng. 2021;11(7):1418-1428
115. Chen HN, Wang Z, Li X, Zhou ZG. Helicobacter pylori eradication cannot reduce the risk of gastric cancer in patients with intestinal metaplasia and dysplasia: evidence from a meta-analysis. Gastric Cancer. 2016;19(1):166-175
116. Nabavi-Rad A, Yadegar A, Sadeghi A, Aghdaei HA, Zali MR, Klionsky DJ. et al. The interaction between autophagy, Helicobacter pylori, and gut microbiota in gastric carcinogenesis. Trends Microbiol. 2023;31(10):1024-1043
117. Kashyap S, Pal S, Chandan G, Saini V, Chakrabarti S, Saini NK. et al. Understanding the cross-talk between human microbiota and gastrointestinal cancer for developing potential diagnostic and prognostic biomarkers. Semin Cancer Biol. 2022;86(Pt 3):643-651
118. Li N, Bai C, Zhao L, Ge Y, Li X. Characterization of the fecal microbiota in gastrointestinal cancer patients and healthy people. Clin Transl Oncol. 2022;24(6):1134-1147
119. Zhou CB, Pan SY, Jin P, Deng JW, Xue JH, Ma XY. et al. Fecal Signatures of Streptococcus anginosus and Streptococcus constellatus for Noninvasive Screening and Early Warning of Gastric Cancer. Gastroenterology. 2022;162(7):1933-1947.e18
120. Wu S, Chen Y, Chen Z, Wei F, Zhou Q, Li P. et al. Reactive oxygen species and gastric carcinogenesis: The complex interaction between Helicobacter pylori and host. Helicobacter. 2023;28(6):e13024
121. Tong T, Zhou Y, Huang Q, Xiao C, Bai Q, Deng B. et al. The regulation roles of miRNAs in Helicobacter pylori infection. Clin Transl Oncol. 2023;25(7):1929-1939
122. Chonwerawong M, Ferrand J, Chaudhry HM, Higgins C, Tran LS, Lim SS. et al. Innate Immune Molecule NLRC5 Protects Mice From Helicobacter-induced Formation of Gastric Lymphoid Tissue. Gastroenterology. 2020;159(1):169-182.e8
123. Zavros Y, Merchant JL. The immune microenvironment in gastric adenocarcinoma. Nat Rev Gastroenterol Hepatol. 2022;19(7):451-467
124. Asam D, Mauerer S, Spellerberg B. Streptolysin S of Streptococcus anginosus exhibits broad-range hemolytic activity. Med Microbiol Immunol. 2015;204(2):227-237
125. Sasaki M, Ohara-Nemoto Y, Tajika S, Kobayashi M, Yamaura C, Kimura S. Antigenic characterisation of a novel Streptococcus anginosus antigen that induces nitric oxide synthesis by murine peritoneal exudate cells. J Med Microbiol. 2001;50(11):952-958
126. Lala PK, Chakraborty C. Role of nitric oxide in carcinogenesis and tumour progression. Lancet Oncol. 2001;2(3):149-156
127. Filliol A, Saito Y, Nair A, Dapito DH, Yu LX, Ravichandra A. et al. Opposing roles of hepatic stellate cell subpopulations in hepatocarcinogenesis. Nature. 2022;610(7931):356-365
128. Brunner SF, Roberts ND, Wylie LA, Moore L, Aitken SJ, Davies SE. et al. Somatic mutations and clonal dynamics in healthy and cirrhotic human liver. Nature. 2019;574(7779):538-542
129. Zhou A, Tang L, Zeng S, Lei Y, Yang S, Tang B. Gut microbiota: A new piece in understanding hepatocarcinogenesis. Cancer Lett. 2020;474:15-22
130. Huo R, Chen Y, Li J, Xu Q, Guo J, Xu H. et al. Altered Gut Microbiota Composition and Its Potential Association in Patients with Advanced Hepatocellular Carcinoma. Curr Oncol. 2023;30(2):1818-1830
131. Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat Rev Gastroenterol Hepatol. 2017;14(9):527-539
132. Ren Z, Li A, Jiang J, Zhou L, Yu Z, Lu H. et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut. 2019;68(6):1014-1023
133. Ni J, Huang R, Zhou H, Xu X, Li Y, Cao P. et al. Analysis of the Relationship Between the Degree of Dysbiosis in Gut Microbiota and Prognosis at Different Stages of Primary Hepatocellular Carcinoma. Front Microbiol. 2019;10:1458
134. Seki E, Schnabl B. Role of innate immunity and the microbiota in liver fibrosis: crosstalk between the liver and gut. J Physiol. 2012;590(3):447-458
135. Remetic J, Ghallab A, Hobloss Z, Brackhagen L, Hassan R, Myllys M. et al. Loss of bile salt export pump aggravates lipopolysaccharide-induced liver injury in mice due to impaired hepatic endotoxin clearance. Hepatology. 2022;75(5):1095-1109
136. Liu Q, Lu Y, Xiao Y, Yuan L, Hu D, Hao Y. et al. Effects of Docetaxel Injection and Docetaxel Micelles on the Intestinal Barrier and Intestinal Microbiota. Adv Sci (Weinh). 2021;8(24):e2102952
137. Schwabe RF, Greten TF. Gut microbiome in HCC - Mechanisms, diagnosis and therapy. J Hepatol. 2020;72(2):230-238
138. Manilla V, Di Tommaso N, Santopaolo F, Gasbarrini A, Ponziani FR. Endotoxemia and Gastrointestinal Cancers: Insight into the Mechanisms Underlying a Dangerous Relationship. Microorganisms. 2023;11(2):267
139. Achiwa K, Ishigami M, Ishizu Y, Kuzuya T, Honda T, Hayashi K. et al. DSS colitis promotes tumorigenesis and fibrogenesis in a choline-deficient high-fat diet-induced NASH mouse model. Biochem Biophys Res Commun. 2016;470(1):15-21
140. Dapito DH, Mencin A, Gwak GY, Pradere JP, Jang MK, Mederacke I. et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell. 2012;21(4):504-516
141. Yu LX, Yan HX, Liu Q, Yang W, Wu HP, Dong W. et al. Endotoxin accumulation prevents carcinogen-induced apoptosis and promotes liver tumorigenesis in rodents. Hepatology. 2010;52(4):1322-1333
142. Jing YY, Han ZP, Sun K, Zhang SS, Hou J, Liu Y. et al. Toll-like receptor 4 signaling promotes epithelial-mesenchymal transition in human hepatocellular carcinoma induced by lipopolysaccharide. BMC Med. 2012;10:98
143. Tang S, Jiang X, Wu L, Chen S, Chen L, Jiang J. et al. Toll-like receptor 4 shRNA attenuates lipopolysaccharide-induced epithelial-mesenchymal transition of intrahepatic biliary epithelial cells in rats. Biomed Pharmacother. 2018;107:1210-1217
144. Seo YD, Ajami N, Wargo JA. Using gut microorganisms to treat cancer. Nat Med. 2023;29(8):1910-1911
145. Di Luccia B, Colonna M. Precision Probiotic Medicine to Improve ICB Immunotherapy. Cancer Discov. 2022;12(5):1189-1190
146. Kawanabe-Matsuda H, Takeda K, Nakamura M, Makino S, Karasaki T, Kakimi K. et al. Dietary Lactobacillus-Derived Exopolysaccharide Enhances Immune-Checkpoint Blockade Therapy. Cancer Discov. 2022;12(5):1336-1355
147. Liu L, Shah K. The Potential of the Gut Microbiome to Reshape the Cancer Therapy Paradigm: A Review. JAMA Oncol. 2022;8(7):1059-1067
148. Chrysostomou D, Roberts LA, Marchesi JR, Kinross JM. Gut Microbiota Modulation of Efficacy and Toxicity of Cancer Chemotherapy and Immunotherapy. Gastroenterology. 2023;164(2):198-213
149. DeWeerdt S. How baby's first microbes could be crucial to future health. Nature. 2018;555(7695):S18-S19
150. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492-506
151. Guo L, Ding JS, Zhou WH. Harnessing bacteria for tumor therapy: Current advances and challenges. Chin Chem Lett. 2023;35(2):108557
152. Littman DR. Releasing the Brakes on Cancer Immunotherapy. Cell. 2015;162(6):1186-1190
153. Sun Q, Barz M, De Geest BG, Diken M, Hennink WE, Kiessling F. et al. Nanomedicine and macroscale materials in immuno-oncology. Chem Soc Rev. 2019;48(1):351-381
154. Waldman AD, Fritz JM, Lenardo MJ. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol. 2020;20(11):651-668
155. Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ. et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19(3):151-172
156. Wrangle JM, Velcheti V, Patel MR, Garrett-Mayer E, Hill EG, Ravenel JG. et al. ALT-803, an IL-15 superagonist, in combination with nivolumab in patients with metastatic non-small cell lung cancer: a non-randomised, open-label, phase 1b trial. Lancet Oncol. 2018;19(5):694-704
157. Lei H, Kim JH, Son S, Chen L, Pei Z, Yang Y. et al. Immunosonodynamic Therapy Designed with Activatable Sonosensitizer and Immune Stimulant Imiquimod. ACS Nano. 2022;16(7):10979-10993
158. Su J, Zhou L, Zhang Z, Xiao X, Qin Y, Zhou X. et al. The components of tumor microenvironment as biomarker for immunotherapy in metastatic renal cell carcinoma. Front Immunol. 2023;14:1146738
159. Potrykus M, Czaja-Stolc S, Stankiewicz M, Kaska Ł, Małgorzewicz S. Intestinal Microbiota as a Contributor to Chronic Inflammation and Its Potential Modifications. Nutrients. 2021;13(11):3839
160. Negi S, Das DK, Pahari S, Nadeem S, Agrewala JN. Potential Role of Gut Microbiota in Induction and Regulation of Innate Immune Memory. Front Immunol. 2019;10:2441
161. Burgueño JF, Abreu MT. Epithelial Toll-like receptors and their role in gut homeostasis and disease. Nat Rev Gastroenterol Hepatol. 2020;17(5):263-278
162. Pinato DJ, Howlett S, Ottaviani D, Urus H, Patel A, Mineo T. et al. Association of Prior Antibiotic Treatment With Survival and Response to Immune Checkpoint Inhibitor Therapy in Patients With Cancer. JAMA Oncol. 2019;5(12):1774-1778
163. Fidelle M, Rauber C, Alves Costa Silva C, Tian AL, Lahmar I, de La Varende AM. et al. A microbiota-modulated checkpoint directs immunosuppressive intestinal T cells into cancers. Science. 2023;380(6649):eabo2296
164. Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97-103
165. Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91-97
166. Locke FL, Ghobadi A, Jacobson CA, Miklos DB, Lekakis LJ, Oluwole OO. et al. Long-term safety and activity of axicabtagene ciloleucel in refractory large B-cell lymphoma (ZUMA-1): a single-arm, multicentre, phase 1-2 trial. Lancet Oncol. 2019;20(1):31-42
167. Schuster SJ, Bishop MR, Tam CS, Waller EK, Borchmann P, McGuirk JP. et al. Tisagenlecleucel in Adult Relapsed or Refractory Diffuse Large B-Cell Lymphoma. N Engl J Med. 2019;380(1):45-56
168. Luu M, Riester Z, Baldrich A, Reichardt N, Yuille S, Busetti A. et al. Microbial short-chain fatty acids modulate CD8+ T cell responses and improve adoptive immunotherapy for cancer. Nat Commun. 2021;12(1):4077
169. Hartimath SV, Ramasamy B, Xuan TY, Rong TJ, Khanapur S, Cheng P. et al. Granzyme B PET Imaging in Response to In Situ Vaccine Therapy Combined with αPD1 in a Murine Colon Cancer Model. Pharmaceutics. 2022;14(1):150
170. Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017;17(5):271-285
171. Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA. et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967-970
172. Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M. et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369(18):1691-1703
173. Tintelnot J, Xu Y, Lesker TR, Schönlein M, Konczalla L, Giannou AD. et al. Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature. 2023;615(7950):168-174
174. Raghavan S, Winter PS, Navia AW, Williams HL, DenAdel A, Lowder KE. et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell. 2021;184(25):6119-6137.e26
175. Aung KL, Fischer SE, Denroche RE, Jang GH, Dodd A, Creighton S. et al. Genomics-Driven Precision Medicine for Advanced Pancreatic Cancer: Early Results from the COMPASS Trial. Clin Cancer Res. 2018;24(6):1344-1354
176. Wardill HR, Bowen JM. Chemotherapy-induced mucosal barrier dysfunction: an updated review on the role of intestinal tight junctions. Curr Opin Support Palliat Care. 2013;7(2):155-161
177. Buske C, Dimopoulos MA, Grunenberg A, Kastritis E, Tomowiak C, Mahé B. et al. Bortezomib-Dexamethasone, Rituximab, and Cyclophosphamide as First-Line Treatment for Waldenström's Macroglobulinemia: A Prospectively Randomized Trial of the European Consortium for Waldenström's Macroglobulinemia. J Clin Oncol. 2023;41(14):2607-2616
178. Zuo T, Cao L, Xue C, Tang QJ. Dietary squid ink polysaccharide induces goblet cells to protect small intestine from chemotherapy induced injury. Food Funct. 2015;6(3):981-986
179. Xie JH, Fan ST, Nie SP, Yu Q, Xiong T, Gong D. et al. Lactobacillus plantarum NCU116 attenuates cyclophosphamide-induced intestinal mucosal injury, metabolism and intestinal microbiota disorders in mice. Food Funct. 2016;7(3):1584-1592
180. Patel AG, Kaufmann SH. Cancer. Targeting bacteria to improve cancer therapy. Science. 2010;330(6005):766-767
181. Chamseddine AN, Ducreux M, Armand JP, Paoletti X, Satar T, Paci A. et al. Intestinal bacterial β-glucuronidase as a possible predictive biomarker of irinotecan-induced diarrhea severity. Pharmacol Ther. 2019;199:1-15
182. Lee SY, Jeong EK, Ju MK, Jeon HM, Kim MY, Kim CH. et al. Induction of metastasis, cancer stem cell phenotype, and oncogenic metabolism in cancer cells by ionizing radiation. Mol Cancer. 2017;16(1):10
183. Cui M, Xiao H, Li Y, Zhou L, Zhao S, Luo D. et al. Faecal microbiota transplantation protects against radiation-induced toxicity. EMBO Mol Med. 2017;9(4):448-461
184. Guo H, Chou WC, Lai Y, Liang K, Tam JW, Brickey WJ. et al. Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science. 2020;370(6516):eaay9097
185. Reis Ferreira M, Andreyev HJN, Mohammed K, Truelove L, Gowan SM, Li J. et al. Microbiota- and Radiotherapy-Induced Gastrointestinal Side-Effects (MARS) Study: A Large Pilot Study of the Microbiome in Acute and Late-Radiation Enteropathy. Clin Cancer Res. 2019;25(21):6487-6500
186. Liu D, Zhuang B, Wei M, Yuan T, Li J, Deng P. et al. Oral konjac glucomannan for prevention of ionizing radiation-induced injury by regulating gut microbiota and increasing short chain fatty acids. Int J Biol Macromol. 2023;240:124402
187. Jian YP, Yang G, Zhang LH, Liang JY, Zhou HL, Wang YS. et al. Lactobacillus plantarum alleviates irradiation-induced intestinal injury by activation of FXR-FGF15 signaling in intestinal epithelia. J Cell Physiol. 2022;237(3):1845-1856
188. Huang R, Zhou PK. DNA damage repair: historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct Target Ther. 2021;6(1):254
189. Ou SI, Nishio M, Ahn MJ, Mok T, Barlesi F, Zhou C. et al. Efficacy of Brigatinib in Patients With Advanced ALK-Positive NSCLC Who Progressed on Alectinib or Ceritinib: ALK in Lung Cancer Trial of brigAtinib-2 (ALTA-2). J Thorac Oncol. 2022;17(12):1404-1414
190. Secombe KR, Van Sebille YZA, Mayo BJ, Coller JK, Gibson RJ, Bowen JM. Diarrhea Induced by Small Molecule Tyrosine Kinase Inhibitors Compared With Chemotherapy: Potential Role of the Microbiome. Integr Cancer Ther. 2020;19:1534735420928493
191. Pal SK, Li SM, Wu X, Qin H, Kortylewski M, Hsu J. et al. Stool Bacteriomic Profiling in Patients with Metastatic Renal Cell Carcinoma Receiving Vascular Endothelial Growth Factor-Tyrosine Kinase Inhibitors. Clin Cancer Res. 2015;21(23):5286-5293
192. Ianiro G, Rossi E, Thomas AM, Schinzari G, Masucci L, Quaranta G. et al. Faecal microbiota transplantation for the treatment of diarrhoea induced by tyrosine-kinase inhibitors in patients with metastatic renal cell carcinoma. Nat Commun. 2020;11(1):4333
193. Shanmugam G, Rakshit S, Sarkar K. HDAC inhibitors: Targets for tumor therapy, immune modulation and lung diseases. Transl Oncol. 2022;16:101312
194. Zhao A, Zhou H, Yang J, Li M, Niu T. Epigenetic regulation in hematopoiesis and its implications in the targeted therapy of hematologic malignancies. Signal Transduct Target Ther. 2023;8(1):71
195. Qu R, Zhang Y, Ma Y, Zhou X, Sun L, Jiang C. et al. Role of the Gut Microbiota and Its Metabolites in Tumorigenesis or Development of Colorectal Cancer. Adv Sci (Weinh). 2023;10(23):e2205563
196. Koliarakis I, Athanasakis E, Sgantzos M, Mariolis-Sapsakos T, Xynos E, Chrysos E. et al. Intestinal Microbiota in Colorectal Cancer Surgery. Cancers (Basel). 2020;12(10):3011
197. Bachmann R, Leonard D, Delzenne N, Kartheuser A, Cani PD. Novel insight into the role of microbiota in colorectal surgery. Gut. 2017;66(4):738-749
198. Zheng Z, Hu Y, Tang J, Xu W, Zhu W, Zhang W. The implication of gut microbiota in recovery from gastrointestinal surgery. Front Cell Infect Microbiol. 2023;13:1110787
199. Jones RM, Luo L, Ardita CS, Richardson AN, Kwon YM, Mercante JW. et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013;32(23):3017-3028
200. Wade H, Pan K, Duan Q, Kaluzny S, Pandey E, Fatumoju L. et al. Akkermansia muciniphila and its membrane protein ameliorates intestinal inflammatory stress and promotes epithelial wound healing via CREBH and miR-143/145. J Biomed Sci. 2023;30(1):38
201. Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291(5505):881-884
202. Hyoju SK, Klabbers RE, Aaron M, Krezalek MA, Zaborin A, Wiegerinck M. et al. Oral Polyphosphate Suppresses Bacterial Collagenase Production and Prevents Anastomotic Leak Due to Serratia marcescens and Pseudomonas aeruginosa. Ann Surg. 2018;267(6):1112-1118
203. McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 2019;20(2):e77-e91
204. Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y. et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70-78
205. Wong SH, Yu J. Gut microbiota in colorectal cancer: mechanisms of action and clinical applications. Nat Rev Gastroenterol Hepatol. 2019;16(11):690-704
206. Liang JQ, Wong SH, Szeto CH, Chu ES, Lau HC, Chen Y. et al. Fecal microbial DNA markers serve for screening colorectal neoplasm in asymptomatic subjects. J Gastroenterol Hepatol. 2021;36(4):1035-1043
207. Martini G, Ciardiello D, Dallio M, Famiglietti V, Esposito L, Corte CMD. et al. Gut microbiota correlates with antitumor activity in patients with mCRC and NSCLC treated with cetuximab plus avelumab. Int J Cancer. 2022;151(3):473-480
208. Fidelle M, Tian AL, Zitvogel L, Kroemer G. Bile acids regulate MAdCAM-1 expression to link the gut microbiota to cancer immunosurveillance. Oncoimmunology. 2023;12(1):2224672
209. Perez-Ruiz E, Minute L, Otano I, Alvarez M, Ochoa MC, Belsue V. et al. Prophylactic TNF blockade uncouples efficacy and toxicity in dual CTLA-4 and PD-1 immunotherapy. Nature. 2019;569(7756):428-432
210. Andrews MC, Duong CPM, Gopalakrishnan V, Iebba V, Chen WS, Derosa L. et al. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat Med. 2021;27(8):1432-1441
211. Huang X, Hu M, Sun T, Li J, Zhou Y, Yan Y. et al. Multi-kingdom gut microbiota analyses define bacterial-fungal interplay and microbial markers of pan-cancer immunotherapy across cohorts. Cell Host Microbe. 2023;31(11):1930-1943.e4
212. Liang QY, Chiu J, Chen YX, Huang YQ, Higashimori A, Fang JY. et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin Cancer Res. 2017;23(8):2061-2070
213. Zhao R, Xia D, Chen Y, Kai Z, Ruan F, Xia C. et al. Improved diagnosis of colorectal cancer using combined biomarkers including Fusobacterium nucleatum, fecal occult blood, transferrin, CEA, CA19-9, gender, and age. Cancer Med. 2023;12(13):14636-14645
214. Wong SH, Kwong TNY, Chow TC, Luk AKC, Dai RZW, Nakatsu G. et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66(8):1441-1448
215. Kartal E, Schmidt TSB, Molina-Montes E, Rodríguez-Perales S, Wirbel J, Maistrenko OM. et al. A faecal microbiota signature with high specificity for pancreatic cancer. Gut. 2022;71(7):1359-1372
216. Qader G, Aali M, Smail SW, Mahmood K, Hasan B, M-Amen K. et al. Cardiac, Hepatic and Renal Dysfunction and IL-18 Polymorphism in Breast, Colorectal, and Prostate Cancer Patients. Asian Pac J Cancer Prev. 2021;22(1):131-137
217. Pitt JM, Vétizou M, Daillère R, Roberti MP, Yamazaki T, Routy B. et al. Resistance Mechanisms to Immune-Checkpoint Blockade in Cancer: Tumor-Intrinsic and -Extrinsic Factors. Immunity. 2016;44(6):1255-1269
218. Yang Q, Liang Q, Balakrishnan B, Belobrajdic DP, Feng QJ, Zhang W. Role of Dietary Nutrients in the Modulation of Gut Microbiota: A Narrative Review. Nutrients. 2020;12(2):381
219. Nguyen LH, Örtqvist AK, Cao Y, Simon TG, Roelstraete B, Song M. et al. Antibiotic use and the development of inflammatory bowel disease: a national case-control study in Sweden. Lancet Gastroenterol Hepatol. 2020;5(11):986-995
220. Ng SC, Kamm MA, Yeoh YK, Chan PKS, Zuo T, Tang W. et al. Scientific frontiers in faecal microbiota transplantation: joint document of Asia-Pacific Association of Gastroenterology (APAGE) and Asia-Pacific Society for Digestive Endoscopy (APSDE). Gut. 2020;69(1):83-91
221. Matsuoka K. Fecal microbiota transplantation for ulcerative colitis. Immunol Med. 2021;44(1):30-34
222. Papanicolas LE, Gordon DL, Wesselingh SL, Rogers GB. Improving Risk-Benefit in Faecal Transplantation through Microbiome Screening. Trends Microbiol. 2020;28(5):331-339
223. Marcella C, Cui B, Kelly CR, Ianiro G, Cammarota G, Zhang F. Systematic review: the global incidence of faecal microbiota transplantation-related adverse events from 2000 to 2020. Aliment Pharmacol Ther. 2021;53(1):33-42
224. Wilson BC, Vatanen T, Cutfield WS, O'Sullivan JM. The Super-Donor Phenomenon in Fecal Microbiota Transplantation. Front Cell Infect Microbiol. 2019;9:2
225. Danne C, Rolhion N, Sokol H. Recipient factors in faecal microbiota transplantation: one stool does not fit all. Nat Rev Gastroenterol Hepatol. 2021;18(7):503-513
226. Weingarden A, González A, Vázquez-Baeza Y, Weiss S, Humphry G, Berg-Lyons D. et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome. 2015;3:10
227. Li P, Zhang T, Xiao Y, Tian L, Cui B, Ji G. et al. Timing for the second fecal microbiota transplantation to maintain the long-term benefit from the first treatment for Crohn's disease. Appl Microbiol Biotechnol. 2019;103(1):349-360
228. Kristensen NB, Bryrup T, Allin KH, Nielsen T, Hansen TH, Pedersen O. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med. 2016;8(1):52
229. Hempel S, Newberry SJ, Maher AR, Wang Z, Miles JN, Shanman R. et al. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: a systematic review and meta-analysis. JAMA. 2012;307(18):1959-1969
230. Lukasik J, Dierikx T, Besseling-van der Vaart I, de Meij T, Szajewska H. Multispecies Probiotic in AAD Study Group. Multispecies Probiotic for the Prevention of Antibiotic-Associated Diarrhea in Children: A Randomized Clinical Trial. JAMA Pediatr. 2022;176(9):860-866
231. Gianotti L, Morelli L, Galbiati F, Rocchetti S, Coppola S, Beneduce A. et al. A randomized double-blind trial on perioperative administration of probiotics in colorectal cancer patients. World J Gastroenterol. 2010;16(2):167-175
232. Bereswill S, Ekmekciu I, Escher U, Fiebiger U, Stingl K, Heimesaat MM. Lactobacillus johnsonii ameliorates intestinal, extra-intestinal and systemic pro-inflammatory immune responses following murine Campylobacter jejuni infection. Sci Rep. 2017;7(1):2138
233. Demers M, Dagnault A, Desjardins J. A randomized double-blind controlled trial: impact of probiotics on diarrhea in patients treated with pelvic radiation. Clin Nutr. 2014;33(5):761-767
234. de Simone C. The Unregulated Probiotic Market. Clin Gastroenterol Hepatol. 2019;17(5):809-817
235. Wilkinson JE, Franzosa EA, Everett C, Li C, Hu FB, Wirth DF. et al. A framework for microbiome science in public health. Nat Med. 2021;27(5):766-774
236. Matson V, Gajewski TF. Dietary modulation of the gut microbiome as an immunoregulatory intervention. Cancer Cell. 2022;40(3):246-248
237. Küçük AN, Çiftçi S. The role of intermittent fasting and the ketogenic diet in cancer disease: can they replace the Mediterranean diet? Eur J Cancer Prev. 2023;32(6):533-543
238. So D, Whelan K, Rossi M, Morrison M, Holtmann G, Kelly JT. et al. Dietary fiber intervention on gut microbiota composition in healthy adults: a systematic review and meta-analysis. Am J Clin Nutr. 2018;107(6):965-983
239. Donohoe DR, Holley D, Collins LB, Montgomery SA, Whitmore AC, Hillhouse A. et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014;4(12):1387-1397
240. Song M, Wu K, Meyerhardt JA, Ogino S, Wang M, Fuchs CS. et al. Fiber Intake and Survival After Colorectal Cancer Diagnosis. JAMA Oncol. 2018;4(1):71-79
241. Reuter MA, Tucker M, Marfori Z, Shishani R, Bustamante JM, Moreno R. et al. Dietary resistant starch supplementation increases gut luminal deoxycholic acid abundance in mice. Gut Microbes. 2024;16(1):2315632
242. Li S, Meng Y, Wang C, Suonan Z, Zhang X, Wu T. et al. Effect of structural characteristics of resistant starch prepared by various methods on microbial community and fermentative products. Int J Biol Macromol. 2024;254(Pt 1):127725
243. Hald S, Schioldan AG, Moore ME, Dige A, Lærke HN, Agnholt J. et al. Effects of Arabinoxylan and Resistant Starch on Intestinal Microbiota and Short-Chain Fatty Acids in Subjects with Metabolic Syndrome: A Randomised Crossover Study. PLoS One. 2016;11(7):e0159223
244. Schwartz DJ, Shalon N, Wardenburg K, DeVeaux A, Wallace MA, Hall-Moore C. et al. Gut pathogen colonization precedes bloodstream infection in the neonatal intensive care unit. Sci Transl Med. 2023;15(694):eadg5562
245. Schlomann BH, Wiles TJ, Wall ES, Guillemin K, Parthasarathy R. Sublethal antibiotics collapse gut bacterial populations by enhancing aggregation and expulsion. Proc Natl Acad Sci U S A. 2019;116(43):21392-21400
246. Su M, Dai Q, Chen C, Zeng Y, Chu C, Liu G. Nano-Medicine for Thrombosis: A Precise Diagnosis and Treatment Strategy. Nanomicro Lett. 2020;12(1):96
247. Shi Y, Lammers T. Combining Nanomedicine and Immunotherapy. Acc Chem Res. 2019;52(6):1543-1554
248. Liu ZH, Liu H, Cheng JL, Wang HL, Yang YF, Le J. et al. Strategies and opportunities of micro/nano delivery systems for targeted therapy of ulcerative colitis: Focus on underlying mechanisms and future perspectives. Chin Chem Lett. 2023;35(2):109074
249. Gencay YE, Jasinskytė D, Robert C, Semsey S, Martínez V, Petersen AØ. et al. Engineered phage with antibacterial CRISPR-Cas selectively reduce E. coli burden in mice. Nat Biotechnol. 2024;42(2):265-274
250. Federici S, Kredo-Russo S, Valdés-Mas R, Kviatcovsky D, Weinstock E, Matiuhin Y. et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell. 2022;185(16):2879-2898.e24
251. Hugenholtz F, de Vos WM. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci. 2018;75(1):149-160
Author contact
![]() Corresponding author: Prof. Zhiyong Qian, E-mail addresses: anderson-qiancom.
Corresponding author: Prof. Zhiyong Qian, E-mail addresses: anderson-qiancom.
 Global reach, higher impact
Global reach, higher impact