13.3
Impact Factor
Theranostics 2023; 13(13):4526-4558. doi:10.7150/thno.87266 This issue Cite
Review
Organ-on-a-chip meets artificial intelligence in drug evaluation
1. Beijing Key Laboratory of Traditional Chinese Medicine Basic Research on Prevention and Treatment for Major Diseases, Experimental Research Center, China Academy of Chinese Medical Sciences, Beijing 100700, China.
2. Institute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Beijing 100700, China.
3. Yunnan Biovalley Pharmaceutical Co., Ltd, Kunming 650503, China.
4. Robot Intelligent Laboratory of Traditional Chinese Medicine, Experimental Research Center, China Academy of Chinese Medical Sciences & MEGAROBO, Beijing 100700, China.
Received 2023-6-17; Accepted 2023-8-2; Published 2023-8-15
Abstract
Drug evaluation has always been an important area of research in the pharmaceutical industry. However, animal welfare protection and other shortcomings of traditional drug development models pose obstacles and challenges to drug evaluation. Organ-on-a-chip (OoC) technology, which simulates human organs on a chip of the physiological environment and functionality, and with high fidelity reproduction organ-level of physiology or pathophysiology, exhibits great promise for innovating the drug development pipeline. Meanwhile, the advancement in artificial intelligence (AI) provides more improvements for the design and data processing of OoCs. Here, we review the current progress that has been made to generate OoC platforms, and how human single and multi-OoCs have been used in applications, including drug testing, disease modeling, and personalized medicine. Moreover, we discuss issues facing the field, such as large data processing and reproducibility, and point to the integration of OoCs and AI in data analysis and automation, which is of great benefit in future drug evaluation. Finally, we look forward to the opportunities and challenges faced by the coupling of OoCs and AI. In summary, advancements in OoCs development, and future combinations with AI, will eventually break the current state of drug evaluation.
Keywords: Organ-on-a-chip, Microfluidics, Drug evaluation, Artificial intelligence, In vitro model.
Introduction
Drug discovery and development is one of the most significant translational science activities contributing to human health and well-being. Nevertheless, the discovery and development pipelines are time-consuming and incur massive costs, primarily because of the preclinical validation as well as clinical trials involved [1, 2]. It is estimated that over 10 years are needed to evaluate a new drug before it enters the market, and the average cost will be $2.5-5 billion [3, 4]. Generally, a standard drug discovery process can be conceptually divided into three parts: target selection, lead identification, and preclinical studies [5]. In the early preclinical stage of drug development, drug evaluation is crucial for confidently advancing a new drug candidate. Drug evaluation mainly focuses on physicochemical properties, biological activity, toxicity, safety, metabolism, pharmacological efficacy, and medicinal value of newly developed drugs, which in order to preliminarily verify their safety and effectiveness for further clinical trials, and to protect people from drugs which are unsafe, ineffective, or both [6-8]. Traditional drug evaluation has mainly relied on cellular monolayer planar culture models and animal experiments. However, traditional methods face several challenges, in part due to the intrinsic limitations of two-dimensional (2D) cell culture models that may not be able to mimic the microenvironment in an organ, and animal models may not accurately represent what occurs in humans [9-11]. In addition, animal models are often not suitable for high-throughput bioassays as well as large-scale drug screening [12], and are also often cost-prohibitive. A bill signed in December 2022 allows the United States Food and Drug Administration (FDA) to approve new drugs without being tested on animals. This marks a major change in people's use of animals after more than 80 years of drug safety supervision. Thus, it necessitates quick and robust methods with the goal of discovering, analyzing, and optimizing a reliable drug candidate [1, 13].
Microfluidics is the science and technology of manipulating and detecting fluids on a micro-scale [14]. With its obvious advantages, including fast processing speed, high spatial resolution, sensitivity, and integration, easy control, and low cost of reagents, microfluidics has become an increasingly attractive tool for both fundamental and practical research [15]. Furthermore, microfluidics has already been utilized to create more in vivo-like models of cell culture because of the dimensional comparisons with biological cells [16, 17]. Notably, microfluidics has the ability to capture, align, and manipulate single cells in drug discovery. Furthermore, microfluidic system has the capability for higher-throughput screening, and it could be used for screening drugs at different species and concentrations. As a valuable tool for developing more in vitro models which capture cellular and organ-level responses, microfluidic technology is widely used for fast and animal-free risk evaluation of new drugs [18].
As a product of microfluidic technology gradually developed, OoCs could faithfully mimic the pathophysiological microenvironment of target organs in vivo, offering exciting potential to bridge the gap between in vitro evaluation models and in vivo pathophysiological complexity [19, 20]. In 2004, adapting microfluidic technology for modeling organs and systemic-level functions of human physiology or disease research was first published [21]. Then, the most famous and landmark OoC device, known as the 'breathing lung' (lung-on-a-chip) was designed in 2010 [22], which initiated the advancement of the biologically inspired OoCs today. Since then, examples of single OoCs include brain/blood-brain barrier [23-25], lung [22, 26, 27], heart [28-30], liver [31-33], kidney [34-36], gut [37-39], vasculature [40-42], skin [43, 44], bone/bone marrow [45, 46], retina [47, 48], muscle [49, 50], fat [51, 52], and tumor/cancer [53-56] have been successfully developed, all of these can be used for drug research. Furthermore, it is possible to investigate organ-organ interactions and systemic diseases like drug off-target toxicity, cancer metastasis, and inflammation by coupling multiple OoC platforms together through vascular perfusion of supernatant exchange or a shared blood substitute [57].
The mechanism of action of drugs is diverse, with various phenotypic effects on cells and organs. Simply recognizing and categorizing these features from the perspective of molecular indicator detection is time-consuming and laborious, which has become a challenge for large-scale molecular library (estimated to be more than 1060 molecules [58]) drug screening, and it is even more difficult to display real-time changes in cellular mechanisms. Nowadays, the functional disclosure of drug targets tends to reveal their functions in the dynamic process of life. During this process, drug evaluation with the OoC platform will generate many images and datasets, and the feature extraction of these dynamic data cannot be completed manually. In recent years, the application of AI in microfluidics has achieved significant results, with new deep learning methods and deep neural network models constantly emerging. OoCs are now starting to attract AI, especially the machine learning (ML) and deep learning (DL) approaches to experimental design and data interpretation [57]. Deep learning was introduced into the field of machine learning by Rina Dechter as early as 1986, and in 2000, Aizenberg introduced Artificial Neural Networks in the field of machine learning [59]. Visual recognition and data processing based on AI will bring possibilities to solve the above problems, including culture conditions optimization, image detection and tracking, and processing such a large volume of data.
At present, OoCs and AI are hot topics in research, and researchers hope to generate more possibilities through the combination of the two. Drug discovery and pharmacological researchers also hope to see this type of review article to obtain relevant knowledge simply and directly. However, most of the current reviews are still focused on discussing the combination of microfluidics and AI (machine learning and deep learning) [60-64]. Although a recent review focused on the combination of OoCs and deep learning, the core of this review was not specifically on the application of integration in drug evaluation [19]. In fact, drug evaluation is one of the most important areas of OoCs application. Thus, a summary of the application of OoCs in drug evaluation, as well as a timely and comprehensive review of the driving role of AI in this field, will facilitate the combination of both for drug evaluation in the future. In this review, we first give a brief overview of basic information on microfluidics-based organ-on-a-chip. Then, we introduce the most recent advances in the field of OoCs, which exhibit clinical mimicry by simulating human patient responses or have utilized this technology to further drug development and personalized medicine. Moreover, we reviewed typical cases of AI application in drug evaluation using OoCs, which will pave the way for future drug development (Figure 1). Finally, we discuss opportunities and challenges for the future of the field. In addition to cells and tissues, the “organ” here also includes organoids, and organoid-on-a-chip has been included in here OoCs.
The background of OoCs invention
In traditional, 2D cultures cells grow in a culture flask or dish as an adherent monolayer, attached to a plastic surface [65]. Although 2D monolayer-based assays have proven to be a valuable method for cell-based studies of low cost, ease to use, and high throughput, adherent culture also has numerous disadvantages, and its limitations have been increasingly recognized [66, 67]. One such key limitation is that 2D cultured cells fail to accurately reproduce the natural human physiology, which prevents this culture method from replicating the cell-cell and cell-environment interactions present in native tissue. As a result, drugs respond differently between cells cultured in 2D and corresponding tissues [68]. In addition, a drawback is that the cells in the monolayer have unrestricted access to the components of the medium, such as nutrients, metabolites, oxygen, and signal molecules [69]. Meanwhile, adherent culture usually allows the study of only a single cell type, which results in cells lacking the microenvironment, or niches, in which they reside in vivo. Thus, the predictive value of 2D monoculture models is quite limited.
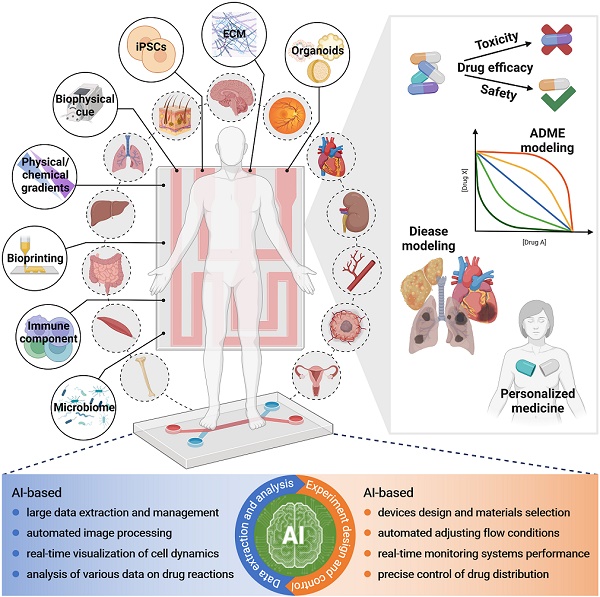
Schematic of organ-on-a-chip meets artificial intelligence in drug evaluation. OoCs have been utilized to model almost all organs in humans for drug testing, disease modeling, personalized medicine, and others. To improve the physiological relevance of OoCs, various factors including cell types, stimulations, and materials are considered and incorporated. Finally, OoCs combine with AI will be of great benefit in experiment design and control as well as data extraction and analysis, which holds exciting promise for drug evaluation with OoCs. Abbreviations: iPSCs: induced pluripotent stem cells; ECM: extracellular matrix. Created with BioRender (www.biorender.com).
Efforts to address some limitations of 2D culture models, 3D culture models have been developed, which provide in vivo-like microenvironments and have received much attention. These models use synthetic or natural cell scaffolds (decellularized) to support cell attachment, growth, and morphogenesis in a 3D environment [70]. Synthetic cell scaffolds typically contain biocompatible polymer materials, such as a variety of fiber and hydrogel scaffolds [71]. Natural cell scaffolds are made from extracellular matrix gels, which contain such as collagen and glycoproteins, and minerals including hydroxyapatite [72]. While the 3D conditions more closely resemble the in vivo state, these models remain lack the multiscale structures and tissue interfaces that are meaningful for organ function. In addition, the lack of controlled and precise application of nutrient supply gradients and chemical cues, results in the poor modeling of an in vivo physiological microenvironment. Importantly, cells are typically not exposed to physical stimuli that are essential for organ development and functioning [6]. Of note, one of the most significant paradigm changes in medicine recently has been the recognition of the central role displayed by the microbiome, which is made up of host-specific communities of commensal microbes, in human health and disease [39]. However, it is not yet possible for human cells to co-culture with complex microbial that come into direct contact, as this frequently leads to culture contamination and cell death within hours [73]. These all limit their use in drug screening.
Preclinical animal models are an essential component of the drug discovery and development process. Although animal models have offered a living system to assess the efficacy of drugs on target site and non-target organ toxicity, it captures the physiological complexity with a high degree of fidelity. However, it is not really representative of human physiology, pathological, and genetic characteristics, thus failing to accurately anticipate drug response in humans [74], as the pharmaceutical industry is gradually discovering. Of note, recent systematic studies on the correlation between animal data and human outcomes have shown a weak predictive ability of animal models [75], and the clinical translatability of drug efficacy tests conducted on animal models is highly controversial [76]. Furthermore, animal models have been associated with ethical concerns, high costs, and low yields, as well as difficulty in performing high-throughput evaluations of drugs. Thus, preclinical drug testing models with better physiologically relevant are needed to simulate complex human-relevant conditions, enable high-throughput assessment of drug candidates, improve the success of clinical trials, and ultimately deliver safe and effective drugs to the market.
Organ-on-a-chip is an in vitro microphysiological system (MPS) used for mimicking the human body environment, representing a simplified but realistic model of its organ-level and even organism-level functional counterpart with functionality read-outs matching the intended application [77]. Microfluidics-based OoCs take advantage of control strategies and multiparametric approaches designed for microfluidics, compared to static culture models, which allows better oxygen perfusion, continuous nutrient exchange, physiological microenvironments, and tissue mechanical forces to provide sufficient nutrients and necessary chemical/mechanical stimuli to better emulation of conditions within the organisms [78, 79]. Notably, OoCs have realized co-culture with microorganisms [39, 80-82]. Animal models often lack the ability to predict results in human drug response. Humans and animals differ substantially in physiological structure, complexity, tissue/organ function, and other parameters, resulting in reduced accuracy and reproducibility of experimental results [83]. For instance, drug metabolism can lead to the production of metabolites with physicochemical and pharmacological properties significantly different from the parent drug, thereby enhancing biological activity or producing adverse biological consequences [84, 85]. Thus, species differences in metabolism may result in an inability to predict the efficacy/toxicity of a drug in humans. For the same drug, it may have different or even opposite pharmacological effects between humans and animals due to differences in the species' target expression, binding capacity, and drug pharmacokinetics and pharmacodynamics (PK/PD). Furthermore, other problems such as ethical concerns, which have also greatly limited progress in drug development. To that end, as an emerging in vitro model, OoCs have been envisioned to replace animal studies. Meanwhile, OoCs may improve the current lack of female individuals in human clinical trials [73].
As a type of microfluidic device, OoCs are created with microchip-manufacturing methods with a miniaturization feature. Owing to the intrinsic characteristics of microfluidics (e.g., compact microchannels), OoCs can provide accurate control of biophysical, biochemical, and cellular parameters [86], and reduces the sample sizes and materials consumption required for drug testing [65]. Importantly, OoCs can simulate chemical concentration gradients, which are essential for the regulation of various biological processes and drug studies. Furthermore, OoCs with a physiological barrier function can better mimic the delivery and absorption of drug compounds in vivo [87]. Polydimethylsiloxane (PDMS) is the preferred choice for manufacturing OoCs, with advantages such as ease of fabrication and handling, gas permeability, low cost, and optical transparency for real-time culture monitoring [88]. Finally, membranes can be integrated into the chips to create multiple channels and separate cells [65].
To date, researchers have developed single-organ-on-a-chip for almost organs in the human body, all of which can be used for drug research. Nevertheless, they lack both a systemic dimension and cross-organ communication [89]. As the human body is a physiologically complicated system, thus necessary to evaluate drug disposition throughout the whole body as well as to quantify PK/PD parameters that contribute to direct clinical trial design, and try to gain more understanding of diseases which is caused by multiorgan interactions [90, 91]. Multi-organ-on-a-chip, coupled single organs by flow, have been created to recapitulate organ-organ interactions and potentially whole-body responses to drugs and to serve as models for diseases [92].
After rapid developments in recent years, OoCs that replicate human organ functions are a promising technology for drug evaluation (e.g., drug transport, metabolism, toxicity, and therapeutic effects), disease modeling, and personalized medicine, which indicates its potential role in all phases of the drug development (Figure 2). The rise of OoCs has brought a new dawn to drug evaluation. Therefore, in the following, we will outline instances of various single and multi-OoCs examples to discuss recent advances in OoCs development, with a focus on their application in drug evaluation in a human-relevant manner.
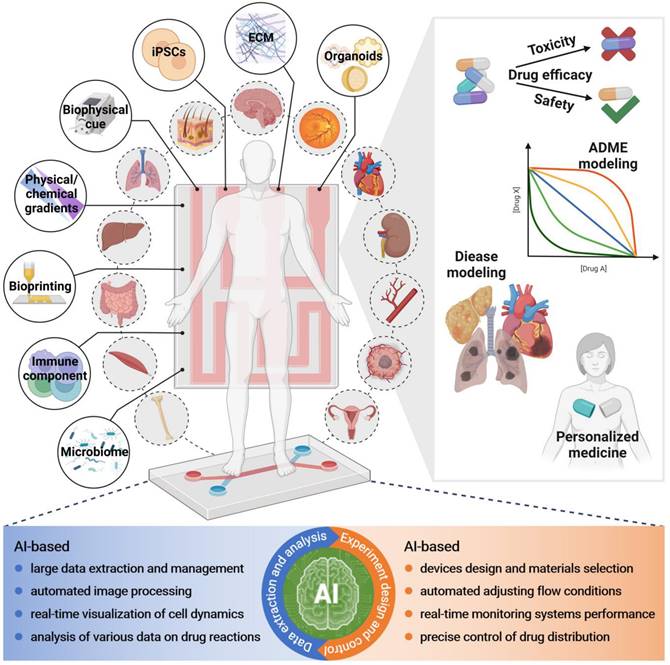
At various phases of drug development, the comparison of throughput and reproducibility with physiological relevance and complexity of different in vitro drug evaluation models. Created with BioRender (www.biorender.com).
The application of single-organ-on-a-chip in drug evaluation
The design guidelines for OoCs are founded on the objective of recapitulating the physiology of the organ system under study. Ideally, the OoCs environment should be created using a minimally functional (simplest feasible) unit of each organ system [78]. Since 2010, almost all organ systems have been modeled using OoCs to gain a new understanding of the molecular and cellular underpinnings of various physiological and pathophysiological processes, and to recapitulate clinical responses to therapeutics seen in human patients [73]. In this section, we review the key human single-organ-on-a-chip studies, especially under the background of drug development (Figure 3 and Table 1).
Brain/BBB-on-a-chip
The structural and functional complexity of the human brain presents unique challenges for neurological drug development. A major obstacle is the blood-brain barrier (BBB), which selectively controls the passage of drugs into the central nervous system (CNS) and prevents it from blood-borne neurotoxic substances as well as maintains homeostasis for optimal brain function [93]. In addition, the complexity also makes it challenging to research in non-human models. In this context, OoCs emulating the function of BBB is of particular interest as they enable testing of whether drugs used for the treatment of neuro-related diseases could act across the BBB to their designated targets [78, 94]. The very first BBB model design consisted of an upper and a lower PDMS channel divided by a porous membrane, similar to a sandwich structure [95]. Usually, astrocytes, pericytes, or other types of brain cells are cultured in the lower channel, while endothelial cells are seeded in the upper channel. Moreover, the neurovascular unit OoC systems were created to develop a more faithful model of the BBB [96], as the BBB is a significant obstacle to the delivery of a lot of neuroactive therapeutics. Although sometimes used interchangeably, the BBB is described as the neurovascular unit free of microglial and neuronal components [97]. These models employ transendothelial resistance (TEER) as a functional readout, which is a gold standard method for measuring the 'tightness' of the constructed BBB [94]. Previously, it has been demonstrated that using microfluidic perfusion improves physiological barrier function and offers a more predictive drug reaction [98]. For instance, hypoxia-enhanced BBB OoC platform outlines the shuttling of CNS-targeting drugs and antibodies in vivo, which may contribute to the development of drugs or delivery vehicles (Figure 3B) [23]. More recently, a BBB OoC device was employed to investigate stem cell-based therapies' therapeutic potential for ischemic stroke. This model demonstrated clinically relevant responses to an ischemic injury, and recapitulated the interactions between therapeutic stem cells and host cells [24]. Therefore, a human-specific model of the BBB would enhance the comprehension of human neurodegenerative diseases and the discovery of neurological drugs.
Lung-on-a-chip
As the lung fills with air, the respiratory regions cyclically expand and contract to increase the surface area accessible for gas exchange. When the alveoli were considered the smallest functional unit of the lung, cyclic expansion can be simulated by applying mechanical stretch to the gas exchange surface [78]. The most well-known organ-on-a-chip, known as the 'breathing lung' (lung-on-a-chip) was designed in 2010 (Figure 3A) [22]. This device has a microporous membrane between two layers of a channel construction which in human alveolar epithelial cells lined the upper layer of the membrane and human pulmonary endothelial cells lined the bottom layer. Once the alveolar cells were confluent, the medium inhaled from the upper channel formed an air-liquid interface with the alveolar cells. The lung structure is replicated on a platform using flowing air and culture medium, respectively, and the extension and contraction of the porous membrane are achieved by varying the internal pressure of the channels on either side of the channel during particular cycles to mimic physiological respiration [70, 99, 100]. The subsequent model used a similar chip design and cell seeding with modifications and additional improvements for various applications, including replicating the drug toxicity seen in cancer patients receiving IL-2 [26], and investigating the pulmonary toxicity of nanoparticles [101]. In addition, the model of lung airway OoCs was designed to reproduce the lung airway microenvironment [102]. Taking the presently well-known COVID-19 as an example, the lung airway OoC system was rapidly being used to repurpose FDA-approved drugs as possible treatments against SARS-CoV-2 [103], and amodiaquine was discovered through this platform to be a potential entry inhibitor for SARS-CoV-2 [27]. More recently, a model that simulates alveoli in vivo using collagen and elastin has been developed, which was called the second-generation lung OoCs [104].
Heart-on-a-chip
The heart is one of the least regenerative organs in the body [105], which is also a significant target organ for toxicity. Cardiotoxicity as one of the most common causes of drug failures [106], drives the development of heart OoCs. Cardiac muscle is a highly ordered dense tissue that is susceptible to interference from drugs, drug-drug interactions, or off-target side effects [79]. So far, a variety of heart OoC platforms have been developed, including co-culture of multiple cell types such as cardiomyocytes, endothelial cells, and cardiac fibroblasts, focused on establishing biomimetic and functional aspects of the heart [107]. Interestingly, most cardiac OoCs are primarily used in cardiotoxicity research. In order to improve the assembly of functional tissue models, anchoring pillars, posts, and wires were utilized to stretch cardiac tissues [108]. A platform that used a 'Biowire' model showed it enabled the generation of highly aligned heart tissues and matured these microtissues by electrical stimulation to achieve functional characteristics resembling those of native human cardiac muscle [109]. In addition, a novel based on 3D bioprinting was used to construct endothelialized human myocardium for cardiovascular toxicity evaluation, reproducing the cancer drug doxorubicin-related myocardial toxicity that has been clinically observed [28]. However, a challenge is the limited ability of mature cardiomyocytes to self-renew [110]. In this framework, induced pluripotent stem cells-derived cardiomyocytes (iPSC-CMs) hold great promise; yet, the limitation of its immaturity still remains, which ultimately affects the pharmacological response [28, 107]. To obtain a cardiac model with adult-like features, methods such as mechanical, electrical, and hydrodynamic stimulation were used to improve tissue maturation [108]. Despite this, it still further expands the potential application of heart OoCs in the field of cardiotoxicity. Heart OoCs are also used to evaluate potential treatments for COVID-19. A study found that azithromycin and hydroxychloroquine, two drugs considered to have therapeutic promise for SARS-CoV-2, when used separately or together as a therapy both have a proarrhythmic potential, which is in accordance with clinical literature [29]. Another comparable study came to a similar conclusion [111].
Representative examples of drug evaluation in single-organ-on-a-chip.
| Single-organ | Materials | Channel | Cell sources | Applications | Ref. |
|---|---|---|---|---|---|
| Brain/BBB | PDMS | Two | Human brain microvascular endothelial cells (HBMVECs) (iPSCs), pericytes, astrocytes | Drug and antibody transport | [23] |
| PDMS | Three | Microglia cells (HMC3), HBMVECs, astrocytes, pericytes | Stem cell therapy efficacy | [24] | |
| PDMS | Two | HBMVECs (iPSCs), brain pericytes, astrocytes | Drug transport | [25] | |
| Lung | PDMS | Two | Human pulmonary microvascular endothelial cells (HPMECs), alveolar epithelial cells, neutrophils | Nanoparticulate toxicity | [22] |
| PDMS | Two | HPMECs, alveolar epithelial cells | Drug toxicity | [26] | |
| PDMS | Two | Human lung bronchial-airway epithelial basal stem cells, HPMECs, neutrophils | Drug efficacy | [27] | |
| Heart | PDMS and PMMA | - | Human umbilical vein endothelial cells (HUVECs) (3D printed), cardiomyocytes (iPSCs) | Drug toxicity | [28] |
| PDMS | Two | Cardiomyocytes (iPSCs) | Drug toxicity | [29] | |
| Gelatin | Two | Cardiomyocytes (iPSCs) | Drug toxicity | [30] | |
| Liver | Glass and plastic | Two | Hepatocytes (iPSCs), HMEC-1 endothelial cells, THP-1 | Drug toxicity | [31] |
| PDMS | Three | Hepatocytes, Kupffer cells, liver sinusoidal endothelial cells (HLSECs), hepatic stellate cells | Drug efficacy | [32] | |
| PDMS | Two | HLSECs, hepatocytes, stellate cells, Kupffer cells | Human and cross-species (rat, dog) drug toxicities | [33] | |
| Kidney | PDMS | Two | Human proximal tubular epithelial cells | Drug transport and toxicity | [34] |
| Plastic | Three | Podocytes, glomerular endothelial cells | Drug efficacy and toxicity | [35] | |
| PDMS | Two | Podocytes, vascular endothelial cells | Drug toxicity | [36] | |
| Gut | PDMS | Two | Caco2 | Drug permeability | [37] |
| PDMS | Two | HUVECs, intestinal epithelial cells, Caco2 | Drug efficacy | [38] | |
| PDMS | Two | Human intestinal microvascular endothelial cells, Caco2 | Microbiome-host interactions | [39] | |
| Vasculature | PDMS | Four | HUVECs, lung fibroblasts | Nanomedicine efficacy | [40] |
| PDMS | Two | HUVECs | mAb therapy toxicity | [41] | |
| PDMS | Three | HUVECs | Drug efficacy | [42] | |
| Skin | PDMS | One | Fibroblasts, keratinocytes | Drug efficacy | [43] |
| PMMA | Two | Keratinocytes | Drug toxicity | [44] | |
| Bone/Bone barrow | PDMS | Two | Bone marrow stromal cells, HUVECs, CD34+ cells | Drug toxicity | [45] |
| PDMS | Five | Bone marrow mesenchymal stem cells, HUVECs, CD34+ cells | Radiation toxicity | [46] | |
| Retina | PDMS | Two | Retinal pigmented epithelial cells, seven essential retinal cells (iPSCs) | Drug toxicity | [47] |
| PDMS | Four | Retinal pigment epithelium cells (ARPE-19), HUVECs, lung fibroblasts | mAb therapy efficacy | [48] | |
| Muscle | PDMS | Three | Human aortic smooth muscle cells (HAoSMCs) | Drug efficacy | [49] |
| PDMS | - | HAoSMCs | Drug efficacy | [50] | |
| Fat | PDMS | - | Adipocytes, peripheral blood mononuclear cells (PBMCs) | Drug efficacy, cell-cell interaction | [51] |
| PDMS | Two | Adipocytes | Drug efficacy | [52] | |
| Tumor/Cancer | PDMS | Two | Human lung microvascular endothelial cells, lung alveolar epithelial cells, non-small-cell lung cancer cell line (H1975) | Drug efficacy | [53] |
| PDMS | Three | HBMVECs, microglia cells (HMC3 or patients), PBMCs, macrophages | Immunotherapy efficacy | [54] | |
| PDMS | Two | Human colonic microvascular endothelial cells (HCoMECs), colorectal cancer cell line (HCT-116) | Nanomedicine delivery | [55] | |
| PDMS | Two | Human gastric epithelial cells (NCI-N87) | Drug efficacy | [56] | |
| Spinal | Plastic | - | Human embryonic stem cells (WA09) | Drug efficacy | [275] |
| Cartilage | PDMS | Two | HUVECs, synovial fibroblasts, articular chondrocytes, monocytes, synovial fluid | Drug efficacy | [276] |
| Placenta | PDMS | Two | Human placental villous endothelial cells (HPVECs), trophoblast cells (BeWo b30) | Drug transport | [277] |
| Pancreas | PDMS and glass | Two | Pancreatic ductal epithelial cells (PDECs), pancreatic islets (all patient) | Disease modeling | [278] |
| Teeth | PDMS | Three | Stem cells from the apical papilla (SCAPs), dentinal tubules | Biomaterials toxicity | [279] |
| Uterus | PDMS | Five | HUVECs, endometrial epithelial cells, endometrial stromal fibroblasts | Drug efficacy | [280] |
| Vagina | PDMS | Two | Human vaginal epithelial cells, uterine fibroblasts | Microbiome-host interactions | [82] |
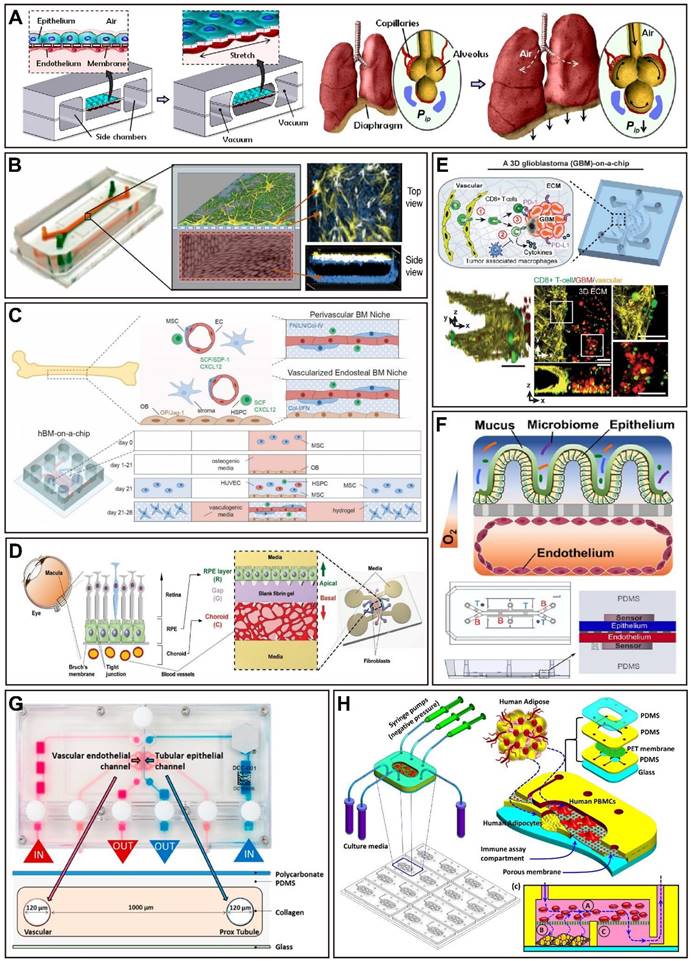
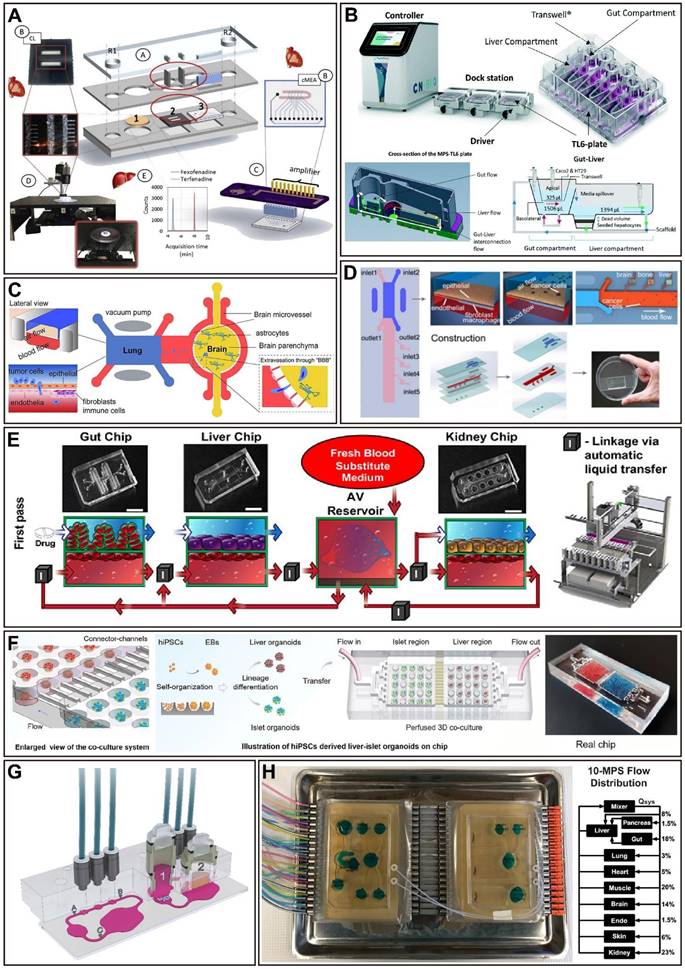
Representative examples of single OoCs. (A) An OoC platform simulates physiological breathing movements by applying a vacuum to the lateral chambers to produce mechanical stretching of the PDMS membrane that forms the alveolar-capillary barrier. Adapted with permission from [22], copyright 2010 American Association for the Advancement of Science. (B) Reconstitution of BBB in a microfluidic device showed that hypoxia-enhanced BBB OoC platform reproduces the barrier function and outlines the shuttling of drugs and antibodies. Adapted with permission from [23], copyright 2019 Nature Publishing Group. (C) A bone marrow (BM) OoC platform could summarize both the central perivascular BM niche (without OBs) and the vascularized endosteal BM niche (with OBs) that is discovered in the cavities of long bones. Adapted with permission from [46], copyright 2021 Elsevier. (D) A retina microfluidic platform including an RPE monolayer and adjacent perfusable blood vessel network with barrier function of oBRB successfully mimics the pathogenesis of CNV, especially in terms of morphogenesis. Adapted with permission from [48], copyright 2018 Wiley. (E) A patient-specific glioblastoma OoC platform with immunosuppressive tumor microenvironments was used to dissect the heterogeneity of immunosuppressive tumor microenvironments to optimize PD-1 immunotherapy. Adapted with permission from [54], copyright 2020 eLife Sciences Publications. (F) A gut OoC device contains a complex human microbiome, which makes the study of host-microbiome interactions possible. Adapted with permission from [39], copyright 2019 Springer Nature. (G) A vascularized dual-channel microphysiological system provides a platform to evaluate the renal secretion of novel drug candidates. Adapted with permission from [128], copyright 2020 American Chemical Society. (H) A fat OoC platform combines adipocyte and immune cells to model the inflamed adipose tissue for the analysis of immune-metabolic in type II diabetes. Adapted with permission from [51], copyright 2019 Nature Publishing Group.
Liver-on-a-chip
The liver has a complex microarchitecture with various functions and displays a central role in the synthesis and metabolism of various substances [112]. Drug-induced liver injury (DILI) is the most frequent reason for drug candidate failure in preclinical and clinical trials, as well as a common reason for withdrawal from the market after drug approval [113]. Thus, during the drug discovery process, accurate prediction of its metabolic capacity and toxicity is extremely important [99]. Today, a variety of in vitro models have already been developed to accurately mimic the complex liver architecture and physiology, and to generalize the human liver's response to drugs [114]. Notably, these liver organoids successfully reproduce express cytochrome P450 and secrete serum albumin of hepatocytes, recapitulating the function of the native liver [115]. The liver is constituted of approximately 1 million lobules which are its constitutional unit, and contain the hepatocytes responsible for drug metabolism [116]. However, liver OoC systems usually use primary human hepatocytes or cell lines that decline in function with increasing culture time, which challenge could be overcome by co-cultures with like Kupffer cells, fibroblasts, stellate cells, and endothelial cells, as well as perfusion [88]. Recently, a high-throughput hepatotoxicity screening OoC device, OrganoPlate LiverTox™, which contains iPSC-derived hepatocytes, endothelial cells, and Kupffer cells, was used to evaluate 159 compounds known to cause hepatotoxicity, and the toxicological prioritization scores were computed (Figure 5A) [31]. Another collagen-based liver OoC platform showed better predictive sensitivity than all previously reported in vitro models after screening 122 clinical drugs for liver toxicity [117]. Furthermore, a study that investigated the impacts of human population variability on liver drug metabolism with the use of hepatocytes from different donors, and an analysis of six drugs confirmed significant inter-donor variability in hepatocyte function. The predicted clearance values and those observed in vivo had excellent correlations [118]. Of note, one of the beneficial applications of liver OoCs is to mimic human-specific hepatotoxicities, which is frequently overlooked in preclinical animal models [73]. A study comparing human, dog, and rat liver OoC platform highlighting demonstrated species-specific differences in drug metabolism and toxicity (including hepatocellular injury, steatosis, cholestasis, and fibrosis) [33], showing the significance of employing human-specific cells in some experiments, while confirming the relevance of using non-human models. Meanwhile, the largest OoCs study to date, in which 780 liver OoC devices were used to evaluate the toxicity risk of a blinded group of 27 known hepatotoxic and nontoxic drugs, showed a sensitivity and specificity of 87% and 100% for liver OoCs, respectively [119]. These results are superior to animal and microsphere models, and support the application of OoCs in preclinical toxicology evaluation. With further development, liver OoCs will contribute to predicting drug toxicity early and reduce the occurrence of adverse drug events.
Kidney-on-a-chip
The kidney is a significant organ responsible for metabolism, excretion, and reabsorption, which is a frequent site of toxicity during drug discovery [120]. Drug-induced kidney injury (DIKI) is frequently observed in drug therapy and may as a dose-limiting factor [121]. Accurately identifying nephrotoxic compounds during the preclinical testing stage would enable effectively avoiding nephrotoxic drugs during development. The minimal functional unit of the kidney is the nephron, which contains the glomerulus, proximal convoluted tubule, loop of Henle, distal convoluted tubule, and collecting duct [122, 123]. In 2008, a nephron-on-a-chip containing the glomerulus, the proximal tubule, and the loop of Henle was designed to replicate the function of a single nephron [124]. Because of their physiological functions and high energy requirements, proximal tubule cells are particularly susceptible to drug toxicity [125]. The first nephrotoxicity study was performed using proximal tubule OoC device consisting of Human Renal Proximal Tubular Epithelial Cells (HRPTEpiC), exposed to fluid flow [34, 126]. After that, the proximal tubular model is the main type of OoCs to predict drug-induced nephrotoxicity. Recently, a study showed that proximal tubular OoC platform successfully predicted the nephrotoxicity of a drug (SPC5001). Of note, the drug exhibited nephrotoxicity in phase I clinical trials but not in preclinical animal testing on mice and non-human primates [127]. A vascularized human proximal tubule model was developed in a dual-channel OoC system, which is an advancement of previous studies (Figure 3G) [128]. In addition, glomerulus OoCs have been developed in the OoCs field in recent years. In a model, human glomerular endothelial cells and podocytes were seeded to reproduce the glomerular filtration barrier [35]. However, other kidney structures, including the distal tubules and collecting duct, have not yet been replicated by human cells and used for toxicological applications [129].
Gut-on-a-chip
For drug administration, the oral route is the most common. As the first step of ADME (absorption, distribution, metabolism, and excretion), absorption is the vital precondition to play the therapeutic effects of oral drugs [130]. The gut is the main digestive organ, responsible for the digestion and absorption of drugs. Thus, understanding the absorption and metabolism of drugs in the gut is critical to drug discovery and development [68]. The development of gut OoCs has made it feasible to study the absorption, metabolism, and transport of oral drugs. Early gut OoCs consisted of two overlapping cell culture chambers divided by a membrane lined with Caco-2 cells. To reproduce the dynamic mechanical microenvironment of the gut, this system included symbiotic microbial flora and utilized negative pressure-driven membrane stretching to simulate peristaltic movements. Under these physiological conditions, the cultured cells were reprogrammed to undergo spontaneous 3D villus morphogenesis and small intestinal cell differentiation [131, 132]. Importantly, except for the barrier function of the human intestine, the model also has absorption properties that can be used for drug absorption studies [87], for example, to analyze the intestinal permeability of the model drug curcumin in real-time and generate data that are consistent with prior research on the function of the human intestinal barrier [37]. Furthermore, exposure to associated biomechanical forces, like flow and peristalsis, can mimic some aspects of the drug's bioavailability and activity [79]. What's more, gut OoCs are stable to create the physiologically relevant oxygen gradient support co-culture of epithelium cells with stable communities of aerobic and anaerobic gut microbiota (Figure 3F) [39, 80], which is critical for true human relevance.
Tumor/Cancer-on-a-chip
It is still a challenge to predict clinical responses to anticancer drugs in cancer treatment [133]. Tumors possess a complicated microenvironment, which contains a dense extracellular matrix (ECM), various stromal/stem and immune cells, irregular blood vessels, and limited perfusion of nutrients, all of which have a significant effect on the efficacy of administered therapies [134, 135]. The advancement of cancer OoCs has significantly contributed to the capacity of in vitro models to reproduce the tumor microenvironment in vivo, as multiple factors in the tumor microenvironment (TME) can be controlled separately and precisely in microfluidic platforms, which is essential to improve anti-cancer drug selection strategies [65]. A breast OoC platform mimicking cancer mammary ducts showed that tumor cells grown in channels have distinct morphologies and exhibit various sensitivities to two anticancer drugs (bleomycin and doxorubicin) compared to traditional flat surface culture [136], which provides novel insight into the development and testing of cancer therapies. Human orthotopic models of non-small-cell lung cancer OoCs can be able to simulate growth patterns observed in patients, and is consistent with the published results of human clinical trials, indicating that under physiological breathing motions, the growth and invasion of cancer cell were suppressed, and almost completely resistant to the inhibitory effects of the rociletinib [53]. In addition, a pancreatic ductal adenocarcinoma OoC platform was developed to further comprehend pancreatic ductal adenocarcinoma-vascular interactions. The authors identified the activin-ALK7 pathway as a mediator of endothelial ablation by pancreatic ductal adenocarcinoma, which results in the limitation of the delivery of chemotherapeutic drugs to the tumors at later stages, and they replicated their findings in mice [137]. Researchers utilized patient-specific glioblastoma OoCs to anatomize the heterogeneity of immunosuppressive tumor microenvironments and personalize anti-PD-1 immunotherapy for various glioblastoma subtypes (Figure 3E) [54]. Hypoxia (Oxygen content below 3%) is a key feature of tumors, which can influence the cancer response to therapies and facilitate immune escape [138]. Another bioprinted patient-specific glioblastoma OoC device reproduces clinically reported patient-specific resistances to concurrent chemoradiation and temozolomide treatment by selectively using materials with different gas-permeable properties to generate an oxygen gradient, and exhibits patient-specific sensitivity to possible drug combinations [139].
At present, tumor OoCs also include colorectal [55, 140], ovarian [141, 142], prostate [143, 144], bladder [145, 146], cervical [147, 148], gastric [56, 149], and skin [150, 151] cancer. As described, tumor OoCs have the capacity to reconstruct major tumor microenvironment characteristics and have great potential to study the mechanisms of tumor development, screen anticancer drugs, and evaluate cancer therapeutics, as well as toward precision medicine.
Other single-organ-on-a-chip
The vasculature is important for providing adequate gas, transporting nutrients, removing waste, and offering a selective barrier for drugs introduced through the circulatory system [78, 152]. As a 3D metabolically active matrix in vitro, which contains a capillary network for the first time that allows operating within physiological pressure gradients and interstitial flow ranges, showed an application in drug discovery [153]. Meanwhile, perfusable 3D microvascular networks were successfully designed, promoting the development of vasculature OoCs [154]. At present, vasculature OoCs are formed by providing endothelial cells with various chemical, cellular, or biophysical substances to induce self-assembly of the microvascular network; seeding endothelial cells onto preformed support structures (e.g., by injection molding, 3D printing, and the use of sacrificial network) or embedding cells into hydrogels, and then inducing germination by flow and chemical factors (e.g., hypoxia, VEGF, and nutrient deprivation). Recently, a vasculature OoC platform with innate immunity identified angiopoietin-1 derived peptide that can be used to therapeutic SARS-CoV-2 induced inflammation [155].
The skin acts as the largest organ in the human body and severs as the main barrier to the environment, which is critical for evaluating the cutaneous effects of drugs and modeling transdermal drug absorption [78]. In recent years, drug delivery through the skin has also been a hot topic of research [112]. Thus, when designing skin OoCs, the reproduction of multiple layers of skin is crucial (i.e., epidermis, dermis, and hypodermis). However, because of its complexity, it is challenging to develop a suitable substitute that can simulate all the skin's properties [156]. In this context, the most common skin OoCs has been those generated by introducing directly the tissue inside the model, which continues to be regarded as the gold standard method for simulating physiological situations in a realistic setting [157, 158]. Nonetheless, the variability of donor skin could affect the analysis and present challenges in evaluating compounds over time, as is typical in the drug development process. In addition, the availability is another limitation [159]. Given this, both reconstructed human epidermis (including EpiDermTM, EpiSkinTM, and SkinEthicTM) and full-thickness skin models have been used for many applications, such as pharmacological [160]. Despite this, challenges still remain in these models to evaluate the absorption or permeability of drugs and systemic exposure with the use of topically applied drugs.
The bone is one of the active organs which is undergoing a carefully choreographed remodeling process throughout the life course [161]. In recent, a microfluidic device fabricated from hydroxyapatite and PDMS provided a highly bionic bone environment. This model successfully produced a concentration gradient of the model drug, demonstrating the tremendous potential for bone-related drug screening in high-throughput [162]. In addition, a vascularized human bone marrow OoC platform containing bone marrow-derived stromal cells and CD34+ cells, which can generalize myeloerythroid toxicity following exposure to chemotherapeutic agents [45]. Another bone marrow OoC device consists of the human endosteal, central marrow, and perivascular niches, which can be utilized to obtain a better understanding of normal and impaired hematopoiesis, and a variety of bone marrow pathologies (Figure 3C) [46].
Human donor retinal explants offer a fully functional model, but due to inter-donor variability, limited availability, and poor cultivability, it is not suitable for drug development and testing [47]. Recently, a study demonstrated that the interaction of mature photoreceptor segments with the retinal pigment epithelium (RPE) can be reproduced in vitro by a retina OoC platform, which was integrated with over seven different hiPSC-derived essential retinal cell types. Importantly, the model recapitulated the retinopathic side effects of the antibiotic gentamicin and the anti-malaria drug chloroquine, exhibiting the potential of facilitating drug development [47]. As another example, a model supporting the outer blood-retinal barrier (oBRB) barrier function successfully mimicked the pathogenesis of choroidal neovascularization (CNV, a key pathological step in a variety of ophthalmic diseases), and proved that bevacizumab alleviated pathological angiogenesis (Figure 3D) [48].
Nowadays, muscle OoCs have been employed in mechanistic research to better comprehend the human skeletal muscles and assess the effects and toxicity of drugs [163]. The development of safer and more effective drugs could be helped with the obtained accurate contractility data. Given this, a muscle thin-film technology-based muscle OoC platform demonstrated its ability to simultaneously analyze the contractility of both striated and smooth muscle on the same chip [164]. Recently, a high-throughput aorta smooth muscle OoC device replicated the abnormal activation of HIF-1α observed in aortas from thoracic aortic aneurysm patients, and finally identified the two most effective drugs (2-methoxyestradiol and digoxin) from the seven specific HIF-1α inhibitors [49].
Fat tissue, as a major energy reserve, will contribute to obesity due to the imbalance between energy intake and expenditure, and its associated comorbidities present a looming challenge to healthcare delivery throughout the world [165, 166]. The interaction of immune cells and adipocytes may lead to chronic low-grade inflammation, which will then result in insulin resistance. An OoC system for characterizing the interaction of adipocytes with immune cells displayed increased pro-inflammatory cytokine secretion and insulin resistance, relative to adipocytes alone. Compared to the previously reported data, the known diabetic drug metformin and the nutraceutical compound omega-3 showed satisfactory results (Figure 3H) [51]. Another OoC platform allows monitoring of the intake of fatty acids and quantification of metabolite released into the effluent media in real-time, and its applicability for pharmaceutical research has been assessed by using isoproterenol, which is known to induce lipolysis [52].
Representative examples of multi-organ-on-a-chip for applications in drug evaluation.
| Number | Multi-organ | Cell types | Medium | Duration | Applications | Ref. |
|---|---|---|---|---|---|---|
| Two | Liver-lung | Primary cell, cell line | PneumaCult™-ALI | 28d | Drug toxicity | [169] |
| Liver-heart | Primary cell, iPSCs | Serum-free medium (HSL2 and HLS3) | 28d | Drug toxicity | [170] | |
| Liver-skin | EpiDermTM, primary cell, cell line | “Co-culture Medium”: EPI-100-NMM-WE | 6d | Drug PK/PD analysis | [179] | |
| Live-heart | Primary cell, iPSCs | RPMI 1640 and DMEM (1:1 ratio) | 5d | Drug toxicity | [197] | |
| Lung-skin | Primary cell, cell line | E3 medium supplemented with glucose | 5d | mAb therapy efficacy and toxicity | [198] | |
| Liver-gut | Primary cell, cell line | Serum-free common medium contained Williams E medium, Gibco Cocktail B, and hydrocortisone | 3d | Drug PK modeling | [281] | |
| Liver-testis | Primary cell, cell line | William's medium E supplemented with CTSTM KnockOutTM SR XenoFree medium | 7d | Drug toxicity | [282] | |
| Liver-gut/skin | Primary cell, cell line, tissue | N.A | 14d | Oral or transdermal drug absorption | [283] | |
| Liver-pancreas | iPSCs | Co‐culture medium: RPMI 1640 with glucose, N-acetylcysteine, B27 supplement, N2 supplement, GlutaMAX, and non‐essential amino acids | 30d | Glucose‐stimulated insulin secretion, drug efficacy | [183] | |
| Liver-kidney | Cell line | DMEM (high glucose) | 1d | Drug metabolism | [284] | |
| Three | Liver-kidney-gut/bone marrow | Primary cell, cell line | “Blood substitute”: DMEM/F12 with EGM-2 supplements, growth factors, and FBS | 10d | Drug PK/PD analysis and toxicity | [177] |
| Liver-kidney-gut | Cell line | DMEM (high glucose) | 3d | Drug PK analysis, and metabolism | [178] | |
| Liver-lung-heart | Primary cell, iPSCs | α-MEM with FBS and L-glutamine | 9d | Drug efficacy, toxicity, and metabolism | [285] | |
| Liver-heart-skeletal muscle | Primary cell, iPSCs | Serum-free medium (blood surrogate) | 7d | Drug PK/PD analysis, immune response | [286] | |
| Liver-lung-colon cancer | Cell line | DMEM-10 and EGM-2 with FBS (3:1 ratio) | 15d | Cancer metastasis | [287] | |
| Liver-lung-breast cancer | Cell line | “Device medium”: EMEM supplement with FBS | 2d | Inhalation and intravenous therapy, drug efficacy and toxicity | [288] | |
| Liver-lung-gut | Cell line | DMEM supplement with FBS and MEM non-essential amino acids | 3d | Oral administration, drug efficacy | [289] | |
| Four | Liver-heart-neuronal-muscle | Primary cell, iPSCs, stem cell, cell line | Serum-free medium supplemented with growth factors | 14d | Drug toxicity | [172] |
| Liver-gut-colon cancer-connective tissue | Cell line | Medium 670 | 3d | Drug metabolism and efficacy | [290] | |
| Liver-kidney-gut-brain | iPSCs | HepaRG medium | 14d | Personalized medicine | [199] | |
| Liver-kidney-BBB-gut | Primary cell, cell line, iPSCs | Functional coupling medium | N.A | Drug metabolism and PK analysis | [291] | |
| Liver-heart-breast-vulva cancer | Primary cell, cell line, iPSCs | Custom serum-free medium formulation | 14d | Drug metabolism, efficacy, and toxicity | [292] | |
| Five | Liver-fallopian tube-uterine-cervix-ovary (mouse) | Primary cell | Maturation medium (with prolactin, day 0 to day 14) | 28d | Human menstrual cycle | [173] |
| Six | Liver-heart-lung- vasculature-testis-brain/colon (rabbit) | Primary cell, iPSCs, cell line, stromal mesenchymal cell, stem cell | Testis organoid media and EGM media (with supplements, without FBS) (1:1 ratio) | 28d | Drug toxicity | [293] |
| Liver-heart-lung- vasculature-brain-testis | Primary cell, iPSCs, stem cell | Testis organoid media and EGM media (with supplements, without FBS) (1:1 ratio) | 21d | Drug metabolism and toxicity | [294] | |
| Seven | Liver-brain-pancreas-lung-heart-gut-endometrium | Primary cell, cell line | N.A | 14d | Drug toxicity | [295] |
| Eight | Liver-intestine-lung-brain-heart-skin-kidney-BBB | Primary cell, iPSCs, cell line | DMEM/F12 with EGM-2 supplements, FBS, and growth factors | 21d | Drug PK analysis | [181] |
| Ten | Liver-intestine-lung-endometriumbrain-heart-pancreas (rat)-skin-kidney-muscle | Primary cell, iPSCs, cell line, tissue construct | Mixed medium | 28d | Drug PK analysis | [182] |
Representative examples of multi-OoCs. (A) A Liver-heart platform for studying the effect of liver metabolism on off-target cardiotoxicity. Adapted with permission from [170], copyright 2018 Elsevier. (B) PhysioMimix gut-liver MPS consists of a controller machine with a pump system and a touchscreen that interacts with the user. The system was used for the quantitative pharmacokinetic study of mycophenolate mofetil. Adapted with permission from [180], copyright 2022 Royal Society of Chemistry. (C) A multi-OoC platform consisted of two bionic organ modules, an upstream 'lung' and a downstream 'brain', allowing to study of lung cancer brain metastasis. Adapted with permission from [187], copyright 2019 Elsevier. (D) A four-organ system for mimicking lung cancer cell metastasis to the liver, bone, and brain. Adapted with permission from [188], copyright 2016 American Chemical Society. (E) A multiple vascularized OoC platform utilizing fluid transfer coupling enables quantitative prediction of human PK responses. Adapted with permission from [177], copyright 2020 Springer Nature. (F) The differentiation and generation of hiPSCs-derived liver and islet organoids in a microfluidic device to simulate human-relevant liver-islet axis under both physiological and pathological conditions for future T2DM study and drug development. Adapted with permission from [183], copyright 2022 Wiley. (G) A 3D co-culture microfluidic model for simultaneous assessment of anti-EGFR-induced tumor and adverse skin impacts. Adapted with permission from [198], copyright 2018 Nature Publishing Group. (H) A multi-OoC system containing up to 10 different organs with different flow configurations, which include epithelial barrier tissues and non-barrier organs, for PK analysis of diclofenac metabolism. Adapted with permission from [182], copyright 2018 Nature Publishing Group.
The application of multi-organ-on-a-chip in drug evaluation
As the development of single OoCs matures, when these single organs are fully functionally characterized (i.e., when they show the key characteristics of the desired simulated organ), they can be combined to create the proposed multi-organ-on-a-chip (often referred to as body-on-a-chip or human-on-a-chip). Connecting a single OoCs to another by microfluidics simulates the in vivo role of vascular perfusion and allows control of the culture environment to reproduce some aspects of homeostasis [78]. The main advantage of multi-OoCs is obvious, that is, these connections enable the complex and dynamic crosstalk between interested organs and promote a more physiological method for drug delivery, distribution, and absorption [167]. As reported, there are three main strategies for connecting single OoCs: 1) connecting the single organ modules with the use of capillary tubing; 2) attaching single organ modules to a microfluidic motherboard that contains all fluidic connections; 3) employing a user-friendly plate with all organ models connected to a channel that controls fluid flow in a manner similar to the vasculature [89]. So far, multi-OoCs may have from 2 to 10 different organs, generally between 2 to 4 organs, which have been capable of simulating complex physiological and pathophysiological responses in an impressive manner, and also offer new in vitro tools for assessing drug toxicity and PK/PD [73], finally towards personalized medicine. In this section, we focus on reviewing the main application scenarios of multi-OoCs (Figure 4 and Table 2).
Drug safety evaluation
In most cases, many drugs fail in phase III clinical trials or have serious side effects after marketing [78], leading to failure in the development of new medicine. Thus, the evaluation of toxicity is critical in late-stage preclinical and clinical research. Toxicity is closely related to liver metabolism, so multi-OoCs designed for toxicity purposes typically include a liver (as the primary site of drug metabolism) and at least one other (target) organ. For example, a biomimetic human liver OoC platform with lobule-like microarchitectures successfully analyzed unfavorable reactions caused by drug-drug interactions of clinical pharmaceuticals during hepatic metabolism, providing an evaluation device to assess drug-induced hepatotoxicity in vitro, especially during combinational therapies [168]. In addition, the use of the lung-liver OoC system in acute and chronic toxicity studies of drugs provides new opportunities for demonstrating the security and effectiveness of new drug candidates that target the lung [169]. A model with primary hepatocytes and iPSC-CMs allows non-invasive readouts of the cardiotoxicity of drugs and their metabolites while also exploring the impact of liver metabolism on off-target cardiotoxicity, which demonstrates the heart-liver crosstalk (Figure 4A) [170]. In a subsequent study, a heart-liver platform containing a skin mimic showed the differential effects of acute and chronic drug exposure, which can be utilized to assess potential drug toxicity from dermal absorption [171]. Moreover, multi-organ toxicity was exhibited in a four-organ system made up of neuronal, muscle, cardiac, and liver modules, and all drug treatments generally agreed with published toxicity results based on human and animal data [172]. Additionally, a system that integrates liver, lung, cardiac, colon, testis, vascular, and brain derived from human primary cells and stem cells, which can stay viable for at least 28 days, responding appropriately to a series of drugs, including those because of toxicity in humans that the FDA has removed from the market [173]. The promise of OoCs to promote drug development lies in their ability to provide humanized drug toxicity information, which can be used as a useful tool to assess drug toxicity effectively and accurately prior to the drug being approved for use in clinical trials.
Drug PK/PD modeling
After identifying candidate molecules and targets, PK and PD studies are conducted. On the one hand, PK researches describe drug concentrations at various organ sites during metabolism, which is referred to as the absorption, distribution, metabolism, and elimination (ADME) of drug candidates. On the other hand, PD researches investigate the effects of the drug on target organs or tissues, such as a correlation between drug dose and pharmacological or toxicological response [174]. The combination of PK/PD parameters is critical for new drug development because it can predict the drug response that will occur, thus minimizing the production of toxic metabolites and the side effects of drugs [175, 176]. For instance, a multi-OoC platform allowed recapitulation of physiological PK modeling of drug absorption, metabolism, and excretion which drugs first-pass. The model was verified using orally administered nicotine (using gut, liver, and kidney chips) and intravenously injected cisplatin (using coupled bone marrow, liver, and kidney chips). Also, the cisplatin PD predictions are consistent with previously published patient data [177]. Determination of drug-administration schemes for phase I clinical trials may be improved by the quantitative in-vitro-to-in-vivo translation of PK and PD parameters via fluidically coupled OoCs (Figure 4E) [177]. In a recent study, a platform that adopted a multi-layer structure was used to systematic analysis the absorption, metabolism, and toxicity of ginsenoside compound K, and the PK results were consistent with previous reports [178]. Another multi-OoC platform called 'HUMIMIC Chip2' was used to integrate liver spheroids and a skin model for PK-PD studies with local exposure to chemicals of hyperforin and permethrin [179]. Moreover, integrating gut-liver OoC platform data with in silico modeling allows to investigate complex combinations of intestinal and hepatic processes for quantitative in vitro PK studies (Figure 4B) [180]. Recently, OoCs that combine more organs are designed to study PK/PD. A robotic interrogator maintained the viability and organ-specific functions of eight vascularized, two-channel OoC devices (liver, heart, kidney, intestine, skin, lung, brain, and BBB) for 3 weeks in culture, and predicted the distribution of an inulin tracer throughout the entire system [181]. Furthermore, a three-layer microbioreactor-based platform containing up to ten different organs, including epithelial barrier tissues and non-barrier organs, which can sustain cell cultures for more than four weeks. The functionality of the platform has been verified by modeling the PK of a nonsteroidal anti-inflammatory drug diclofenac, which revealed that both diclofenac and 4-OH-diclofenac were distributed throughout all the representative organs (Figure 4H) [182]. In summary, the outcomes from multi-OoCs provided insightful data that eventually be applied to evaluate the PK/PD of potential new drugs, leading to a more dependable preclinical stage in drug development.
Disease modeling
The dearth of clinically applicable models is a challenge for many human diseases, especially those complex diseases that involve multiorgan interactions. As a systematic multi-organ metabolic disease, Type 2 diabetes mellitus (T2DM) is characterized by the dynamic interplay of various organs [183], and a clinical cure is not yet available. Recently, a multi-OoC platform was used to model the liver-pancreatic islet axis under both normal and type 2 diabetes conditions, which successfully mimicked the functional coupling of the liver and islet organs' response to external hyperglycemic stimulus and drugs is relevant to humans (Figure 4F) [183]. An ulcerative colitis multi-organ system was created by connecting the liver, gut, and circulating immune cells, showing that short-chain fatty acids (SCFAs) derived from the microbiome could either improve or worsen the severity of ulcerative colitis, and these converse results resting with the participation of effector CD4+ T cells [184]. This study brought new insights into the immune and metabolic regulation of pathophysiology. Moreover, multi-OoCs connected to the vasculature and circulatory system are critical for understanding local and distant disease development, like cancer initiation and metastasis [185], the latter contributes to up to 90% of cancer-related mortality [186]. For instance, a methodological platform that was used to study brain metastasis demonstrated that the protein Aldo-keto reductase family 1 B10 (AKR1B10) contributes to brain metastasis of lung cancer cells (Figure 4C) [187]. Furthermore, a four-organ platform that reproduced lung cancer metastasis to the liver, bone, and brain, revealed tumor-induced tissue damage in the targeted bone and liver compartments (Figure 4D) [188]. These suggest that multi-OoCs are a practical alternative for predicting cancer metastasis and evaluating antimeatstatic therapies. In addition, the advancement of degenerative brain diseases including Parkinson's or Alzheimer's disease has been linked with gut microbiota, this functional relation is often referred to as the microbiota-gut-brain axis (MGBA) [189]. However, a comprehensive in vitro model was lacking for researchers to elucidate potential microbiota-neurodegeneration mechanisms. The European Research Council has funded a project called 'MINERVA' (ID 724734), which seeks to build the first multi-OoC device for microbiome-gut-brain engineering to assess the effect of intestinal microflora on neurodegeneration [190]. More importantly, multi-OoCs are particularly valuable for clarifying mechanisms and developing treatments for rare diseases affecting multiple organ systems, where drug development is incredibly difficult because of the available human subjects being scarce, like Churg-Strauss syndrome and POEMS syndrome [191]. Thus, the application of multi-OoCs to model diseases improves disease comprehension, diagnosis, prevention, and treatment.
Personalized medicine
Although various in vitro platforms have been developed for drug development screening, there are few that exist for clinical deployment to benefit unique patients. This is an unmet clinical need because patient responses to drugs are frequently unpredictable due to genetic and microenvironmental heterogeneities [192]. Remarkable strides in the hiPSCs field allow for the development of patient-specific personalized therapies, making it possible to identify more efficient drugs for a particular individual or patient group [175]. Recently, the integration of a heart chip and a liver chip, both created with the same hiPSC line, was reported to investigate the drug-drug interaction (DDI) of the fungicide ketoconazole and the arrhythmogenic gastroprokinetic cisapride, which facilitates the screening of DDI [193]. In the treatment of COVID-19, more attention should be paid to comorbidities. A lung OoC platform comprising infected cells from COVID-19 patients has the promise to overcome the potential effects, such as liver, cardiovascular, and kidney disease, or malignant tumors, as which have occurred in patients reported previously, and may assist in providing effective treatment for individual patients [175, 194]. Oncology diseases, which are characterized by rapid mutations that lead to morphological changes and various phenotypes of multidrug-resistant that affect the patient's response to treatment, are another area where multi-OoCs have gained great attention in personalized treatment [195, 196]. A multisensor-integrated multi-OoC system was developed, and by linking iPSC-CMs and primary hepatocytes together to achieve automated sensing of APAP-induced organoid toxicity. Using this model, hepatocytes were replaced by hepatocarcinoma cells to assess the chemotherapeutic drug doxorubicin treatment-induced pronounced cardiotoxicity [197]. Thus, this platform can be used in predicting the cardiotoxicity of drugs by using patient-specific iPSC-CMs. Moreover, a commercially available multi-OoC platform coupling two organs (lung cancer and skin) culture compartments fluidically for evaluating the efficacy of therapeutic anti-EGFR monoclonal antibodies while analyzing a side effect of dermatological toxicities. The results showed that it is possible to detect several key side effects on the cetuximab-exposed skin microtissues at a very early stage, as well as reproduce the inhibition of keratinocyte growth and altered expression of CXCL8 and CXCL10 observed in patients [198]. We believe it will be achievable to personalize the screening of drugs using patient-specific preclinical models prior to treatment, while monitoring the adverse effects of all organ systems in the platform, and improving treatment outcomes. Of note, a four organ model integrated predifferentiated organs from the same human iPSCs and successful coculture over 14 days (although the renal model did not further differentiate) [199], which demonstrates the promise of taking advantage of OoCs to optimize the selection of therapeutics in a personalized manner.
Integration OoC technology with artificial intelligence
OoC technology and deep learning are frontier fields in biomedical engineering and AI, respectively, and represent an ideal combination of experimental and analytical throughput [60]. Here, we introduce various applications of AI to OoCs, trying to illustrate the power and versatility of integrating OoCs with AI. Although the integration of these two disciplines has not been extensively explored so far, especially in the field of drug evaluation, we can still get a glimpse of the great potential of OoCs combined with AI in future drug evaluation from the existing research.
The challenges faced by OoCs in higher-throughput
High-throughput platforms for preclinical drug screening are crucial to reducing the cost of drug discovery [200]. Nowadays, the relatively low throughput of the majority of OoC platforms has hampered the widespread adoption of organ-on-a-chip for drug screening. More reliable statistical data requires a large number of tests and results, hence the need for higher-throughput studies on OoCs. Recently, a few studies have been proposed to address this need (Figure 5). For instance, a microfluidic device for modeling the human microcirculation was demonstrated, as a protocol extension. This device can self-organize human microvascular networks, and then perfuse the tumor to summarize discrete steps of early metastatic seeding. Combined with high-resolution imaging, reliable and quick scoring of extravascular cells can be easily achieved. In addition, the ability to manufacture and seed up to 36 devices at once while not affecting cell viability was reported, which further allows for highly parametric studies, and generating a significant amount of data [201]. A high-throughput OoC platform with 96 devices integrated programmable fluid flow and real-time sensing for physiologically relevant tissue generation and measurement, enabling accelerated optimization of in vitro models (Figure 5C) [202]. Another 96-device platform (PREDICT96-ALI) is compatible with high-resolution in situ imaging and real-time sensing for rapid assessment of drug efficacy against viruses including coronaviruses (Figure 5D) [203]. In perfused microfluidic devices, extracellular matrix-supported intestinal tubules were introduced. The OrganoPlate platform is a standard 384-well microtiter plate format with 40 microfluidic channel structures. On this platform, a study containing 357 gut tubes was conducted to test against drug compounds at various concentrations to evaluate the impact on epithelial barrier integrity. Notably, the study produced more than 20,000 data points, which makes it the largest reported OoC platform data set to date (Figure 5B) [204]. Another microfluidic platform named IFlowPlate was also built on a 384-well plate, which can be used to culture up to 128 organoids, achieved in vitro perfusable culture and vascularization of patient-derived colon organoids, and successfully developed a colon inflammation model with an innate immune function [205]. Thus, the higher-throughput nature of these studies suggests the potential of OoCs as novel, effective, and dependable preclinical models with applications in drug evaluation.
However, it must be recognized that growing throughput typically causes large data generation, leading to labor-intensive and time-consuming processes. Thus, in order to streamline the experimental procedures, it is crucial to develop protocols that facilitate efficient device operation, data collection, and data analysis. For instance, robotics can be used to automate tasks (e.g., operating chips and gathering data), while machine learning can be used to speed up data analysis [93].
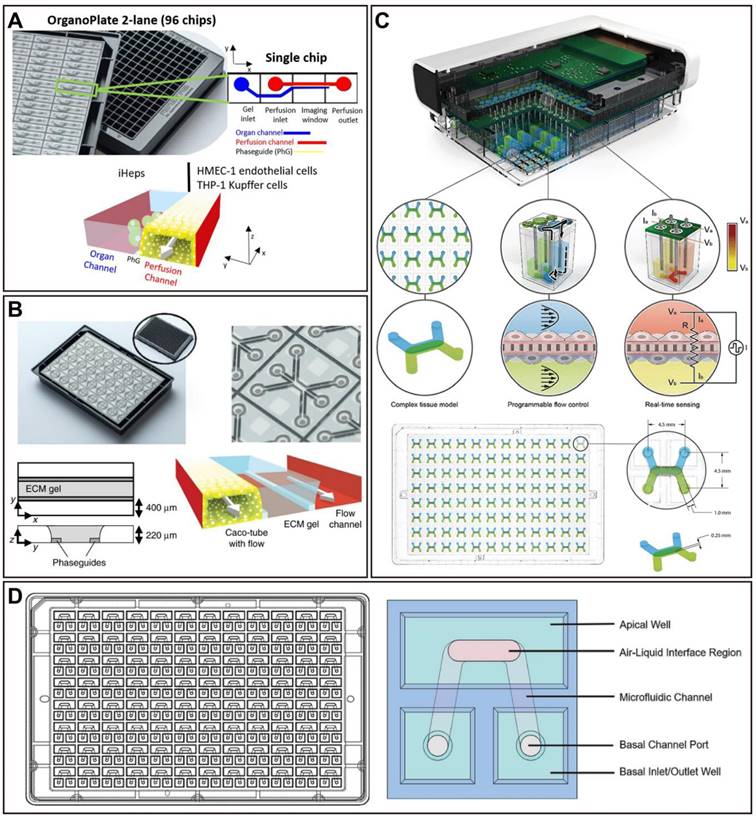
Representative examples of high-throughput implemented in OoCs. (A) An OrganoPlate 2-lane has 96 chip units, with the perfusion and organ channels divided by a Phaseguide. Adapted with permission from [31], copyright 2021 Elsevier. (B) An OrganoPlate contains 40 microfluidic channel networks, each of which consists of three channels (including a tubule with flow, an extracellular matrix gel, and a flow channel) that join in the center. Adapted with permission from [204], copyright 2017 Nature Publishing Group. (C) A platform incorporates 96 independent microfluidics-based organ models, each with two channels separated by a permeable membrane, and the micropumps integrated with the trans-epithelial electrical resistance electrodes and electronics of the micro-pump sensor array. Adapted with permission from [202], copyright 2021 Royal Society of Chemistry. (D) A PREDICT96-ALI platform is a standard 384-well plate layout, which is an individual airway model with an oval-shaped upper chamber and an inverse U-shaped bottom chamber with inlet and outlet ports. Adapted with permission from [203], copyright 2021 Nature Publishing Group.
The increasingly prominent advantages of AI
In the past few years, AI has supplied significant advantages in many areas of healthcare in research and clinical settings, such as disease diagnosis, precision medicine, and drug discovery and development. Notably, opportunities for applying AI arise at all stages of drug discovery and development, including clinical trials [206]. Applications include identification and validation of drug targets, designing of new drugs, quantifying structure-activity relationship, drug repurposing, improving the research and development (R&D) efficiency, as well as evaluation of absorption, distribution, metabolism, excretion, and toxicity, and even aggregating and analyzing biomedicine information and refining the decision-making process to recruit patients for clinical trials and so on [207-210]. Furthermore, the identification of new disease genes, pathways, and targets using omics analysis with AI becomes possible [211, 212], thereby providing new mechanisms for future drug discovery and development, as well as precision medicine. Facing massive volumes of accumulated data (e.g., medical images and gene expression data), AI-based approaches can further transform these enormous amounts of data into usable knowledge, thus facilitating systematical discovery, understanding, and learning [213]. Importantly, the application of AI offers the opportunity to overcome the inefficiencies and uncertainties in traditional drug discovery and development approaches, while also reducing human intervention and personal bias in the process [208]. Today, advances in areas OoCs and AI are increasingly providing the basis for more efficient and successful drug evaluation. Notably, multi-OoCs, and future coupling with AI, will provide a powerful tool for the pharmacological research of drugs, especially complex chemical drugs, botanical medicines, and Traditional Chinese Medicines.
ML is a common technical means to achieve AI, and DL is a type of ML algorithm. Of note, DL is the most representative research field in AI [19]. ML could be categorized into supervised, unsupervised, semi-supervised, and reinforcement learning based on labels [214]. ML provides automated analytical statistical/model-building approaches for machines to make decisions by extracting information from data or identifying patterns (i.e., learning), without explicit human programming [215]. ML has been growing utilized to analyze data (e.g., to clarify processes and predict outcomes), which may reduce inter-operator variability during data analysis. Deep learning is a machine learning technique encompassing a variety of learning models known as deep neural networks (DNNs), which are referred to as 'deep' since containing multiple processing layers [213, 216]. The blooming of algorithms, including Deep Belief Networks (DBNs), Autoencoder Networks (AEs), Convolutional Neural Networks (CNNs), Recurrent Neural Networks (RNNs), and Generative Adversarial Networks (GANs) [217, 218], leading to various studies with the use of DL-based AI in drug evaluation. In comparison to traditional ML, which has a limited ability in processing natural data in its raw form, while DL directly performs the feature extraction of the data [216], and is easier to have high accuracy by minimizing errors. Moreover, DL models previously trained on one task commonly could be retrained to execute similar tasks, named transfer learning, which typically needs raw data and fewer computational resources, making DL applicable to a variety of tasks [219].
With the development of OoCs, especially utilizing higher-throughput, highly parallelized microfluidic systems, generating unprecedented quantities of data; however, the large amount of data generated has far exceeded researchers' capacity to process it efficiently, creating a bottleneck in the analysis [60]. Typically, manually analyzing data is inefficient and is likely to miss trends that are nonobvious or of interest [220], hence the need for appropriate systems to manage and analyze the data. Thus, AI has been applied to address the challenge of analyzing large and multidimensional datasets, and assists researchers to derive meaningful insights. In brief, new data processing systems should contain four main components: 1) the suitable measuring hardware and microchips, with precise sensors and microsystems to effectively monitor the required parameters; 2) the provided appropriate forms of data collection, transmission, and storage; 3) the improved machine learning algorithms which enable to extract desired information from the obtained massive amounts of data sets; 4) the proper explanation of the collected data, and applied to the discovery of new results [65, 213].
AI-based visual recognition in data analysis of OoCs
To date, the most common data output structure for OoCs is fluorescence microscopic images [60], due to the PDMS's transparency and great compatibility with fluorescence microscopy. This is traditionally handled by manual methods, which are frequently inefficient, time-consuming, and error-prone [221]. However, it is important to note that DNN has been trained with organ autofluorescence images to achieve virtual histological staining for relevant assays, including hematoxylin-eosin (H&E), Masson's trichrome, and Jones silver stain. The outcomes showed that these computational stains are almost indistinguishable from the corresponding histologically stained tissue [222]. Thus, virtual staining of label-free samples on OoCs could be achieved by utilizing deep learning in the near future.
Representative examples of OoCs integrated with AI for data analysis. (A) A CNN model was developed to identify important tumor cell behavior parameters from fluorescence images in a glioblastoma OoC platform, and combined with an in vitro-in silico approach to achieve real-time prediction of tumor evolution. Adapted with permission from [223], copyright 2021 Elsevier. (B) A deep prior algorithm, called Recursive Deep Prior Video, was developed for addressing the challenge of the low resolution in time-lapse microscope in OoC applications, and the approach was successfully validated. Adapted with permission from [225], copyright 2021 Elsevier. (C) A tumor OoC platform combining cancer, immune, endothelial, and fibroblasts recapitulated an anti-tumoral antibody-dependent cell-mediated cytotoxicity, and CellHunter method was used to track cancer-immune cell interactions. Adapted with permission from [233], copyright 2018 Cell Press. (D) The cells on the OoC are located and tracked through the video sequence obtained by time-lapse microscopy, and then extract relevant features from the visual atlas are for classification tasks with the use of a pre-trained DL-based algorithm. Adapted with permission from [228], copyright 2020 Nature Publishing Group.
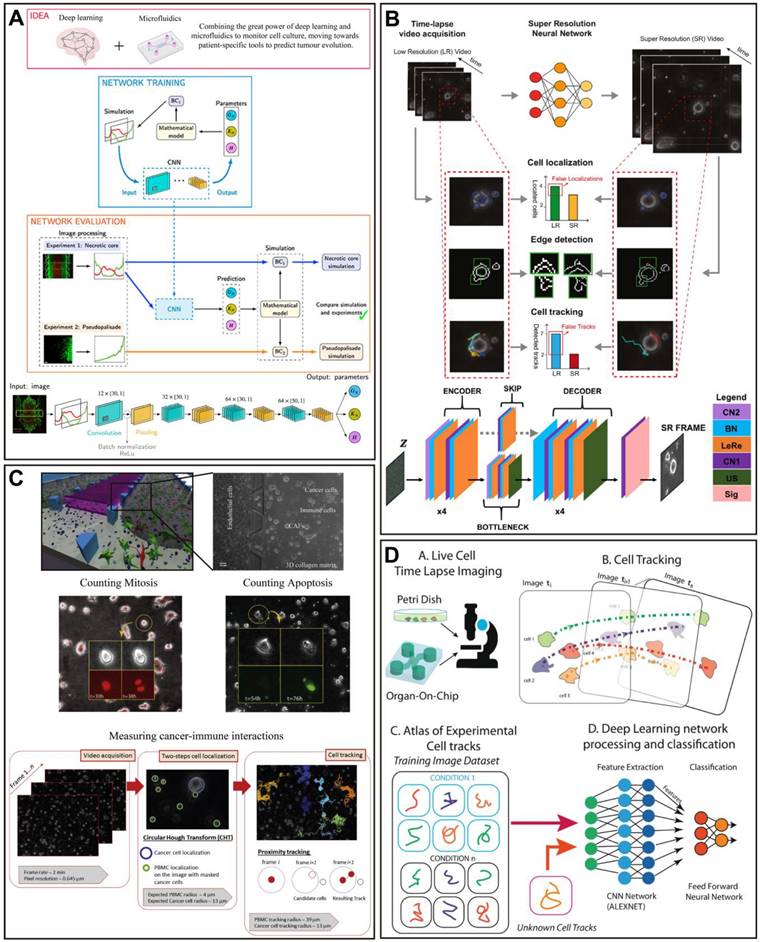
In a recent study, a CNN model was developed to identify important tumor cell behavior parameters from fluorescence images in a glioblastoma OoC platform, and combined with an in vitro-in silico approach to achieve real-time prediction of tumor evolution (Figure 6A) [223]. A microfluidic multicellular coculture array (MCA) was developed and combined with ML to assess skin sensitivity to drugs. The performance prediction of MCA and support vector machine (SVM) classification algorithm demonstrated that the model has 87.5% accuracy, 75% specificity, and 100% sensitivity in predicting skin adverse drug reactions, with the potential as a platform for high-throughput drug screening (Figure 8A) [224]. For some OoC platforms, the segment of the special parts of the image with great significance is important for the analysis of experimental results. In this case, applying DL models to accomplish pixel-level segmentation of images acquired from OoCs, will help in the analysis of drug therapy [19]. Given this, researchers have developed a new DL-based algorithm without requiring any training, called Recursive Deep Prior Video, to address the challenge of the low resolution in time-lapse microscope in OoC applications, and the approach was successfully validated on real videos of OoC experiments associated with tumor-immune interactions (Figure 6B) [225]. Furthermore, a biomimetic bone OoC device has been developed to achieve simultaneous and high-throughput drug testing for osteoporosis. This device was integrated with CNN-based image segmentation algorithms for fluorescence image analysis, and successfully validated its feasibility for drug evaluation (Figure 7A) [226], although mouse-derived cells were used, human-derived cells can be used for future studies.
At the same time, the possibility to visualize cell morphology and trajectory in real-time is essential for the study of drug therapy in OoCs, especially in cancer OoCs. For example, in a muscle OoC device, the authors studied the temporal prediction of muscle cell morphology with the use of an RNN model with long short-term memory blocks. Next, a CNN model was trained with the temporal images of the RNN to judge the cell function (Figure 7B) [227]. Besides, a cancer OoC system with the use of a DL-based CNN architecture was used to evaluate the effectiveness of cancer drug therapies by discovering the hidden information within cell trajectories. The Deep Tracking was capable of accurately classifying the cells (91.5% on average), indicating that the DL approach is very proficient at identifying how drug treatments affect cell motility behaviors (Figure 6D) [228]. Notably, the use of a CNN algorithm for recognition achieved to perform accurate tumor boundary detection and analysis of tumor invasion [229], which could be further applied in OoCs. Another platform supported by automated image acquisition and cropping analysis has successfully implemented a label-free approach to evaluate the viability of tumor spheroids on a microfluidic platform with up to 1920 tumor spheroids. The authors trained a CNN model to estimate sphere viability based on bright-field images, and accurately evaluated the efficacy of three chemotherapeutic drugs, adriamycin, oxaliplatin, and irinotecan. It is important to note that the training networks of doxorubicin and oxaliplatin have been cross validated, indicating the possibility of using representative drugs to train a universal network and applying it to numerous different drugs in large-scale screening (Figure 7C) [230].
A key challenge in cancer studies is the increased complexity of the tumor microenvironment. Recently, a novel 3D microfluidic blood brain niche (µBBN) platform quantified the microenvironment of brain metastatic tumors (breast cancer) by using confocal tomography and machine learning (neural networks and random forest learning algorithms) to identify intrinsic phenotypic differences in tumor cells capable of metastasizing through the model [231]. The same group performed a similar study prior to this one, in which breast cancer cell extravasation was analyzed within a μBBN device using an advanced live cell imaging algorithm to detect small differences between cells with and without brain metastasis potential (Figure 7D) [232]. The device enables further utilization to assess the molecular determinants of metastatic cancer cell migration and survival, and to evaluate the effectiveness of drug therapy. In addition, a more complex and better replicating breast tumor microenvironment was built and CellHunter was employed to track intercellular interactions between immune cells and tumor cells in OoCs (Figure 6C). Deep learning enables visualization and quantification of the complex dynamics of tumor OoCs, for characterizing drug responses at the ecosystem level, and for dissecting the roles of stromal components [233].
CellHunter is a DL-based cell tracking analysis algorithm. As reported, CellHunter has been successfully applied to reveal the interactions between human peripheral blood mononuclear cells (PBMCs) and tumor cells in OoCs and showed that only cells from 'wild type' donors (FPR1 normal expression) establish sustained interactions with chemotherapy-treated cancer cells [234]. Another similar study investigated the effect of spatial and temporal resolutions of cell-cell interaction analysis in OoCs based on the same platform [235]. Furthermore, a microfluidic platform combining advanced microscopy and revised CellHunter was developed to assess the effective migration of interferon-α-conditioned dendritic cells (IFN-DCs) toward drug-treated cancer cells, while discovering the involvement of major underlying factors, such as CXCR4 [236]. The oncolytic vaccinia virus is an emerging agent in cancer immunotherapy. Recently, a tumor OoC device revealed cooperative antitumoral activity of immune cells and oncolytic vaccinia virus by direct imaging and automatic analysis. In this work, the CellHunter algorithm was applied to high-resolution video analysis to localize and track cancer cells, which high-resolution video is aimed at measuring immune kinematics and cancer-immune interactions (Figure 8B) [237].
Representative examples of OoCs integrated with AI for data analysis. (A) A biomimetic bone OoC device for high-throughput osteoporosis drug testing with AI-assisted image analysis. Adapted with permission from [226], copyright 2022 Wiley. (B) In a muscle OoC device, temporal prediction of muscle cell morphology was studied with the RNN model with long short-term memory blocks and trained a CNN model with the temporal images of the RNN to judge the cell function. Adapted with permission from [227], copyright 2019 Elsevier. (C) A microfluidic platform with up to 1920 tumor spheres integrated with a CNN model to assess the efficacy of chemotherapy drugs. Adapted with permission from [230], copyright 2019 American Chemical Society. (D) A µBBN platform for identifying the extravasation potential of cancer cells to brain metastasis niches with advanced live cell imaging algorithm and artificial intelligence. Adapted with permission from [232], copyright 2019 Royal Society of Chemistry.
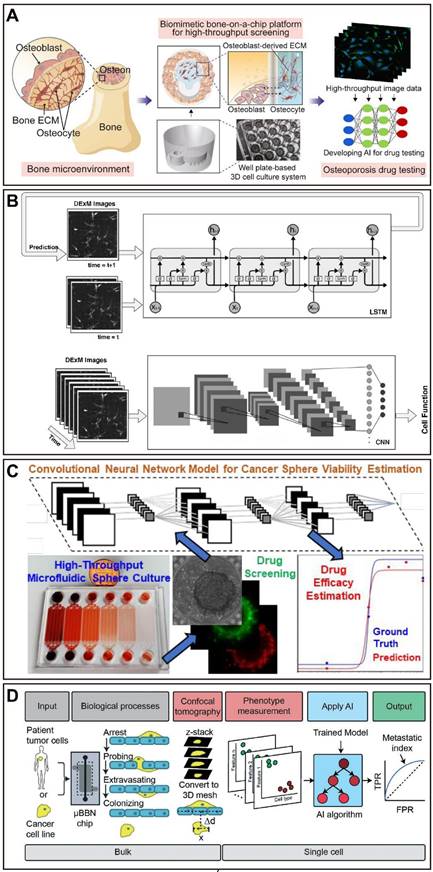
Representative examples of OoCs integrated with AI for data analysis. (A) A microfluidic multicellular coculture array (MCA) was developed and combined with ML to assess adverse skin drug responses. Adapted with permission from [224], copyright 2022 Royal Society of Chemistry. (B) The combined anti-tumor activity of immune cells and oncolytic vaccinia virus in tumor OoC device was revealed by direct imaging and automated analysis. Adapted with permission from [237], copyright 2022 Elsevier.
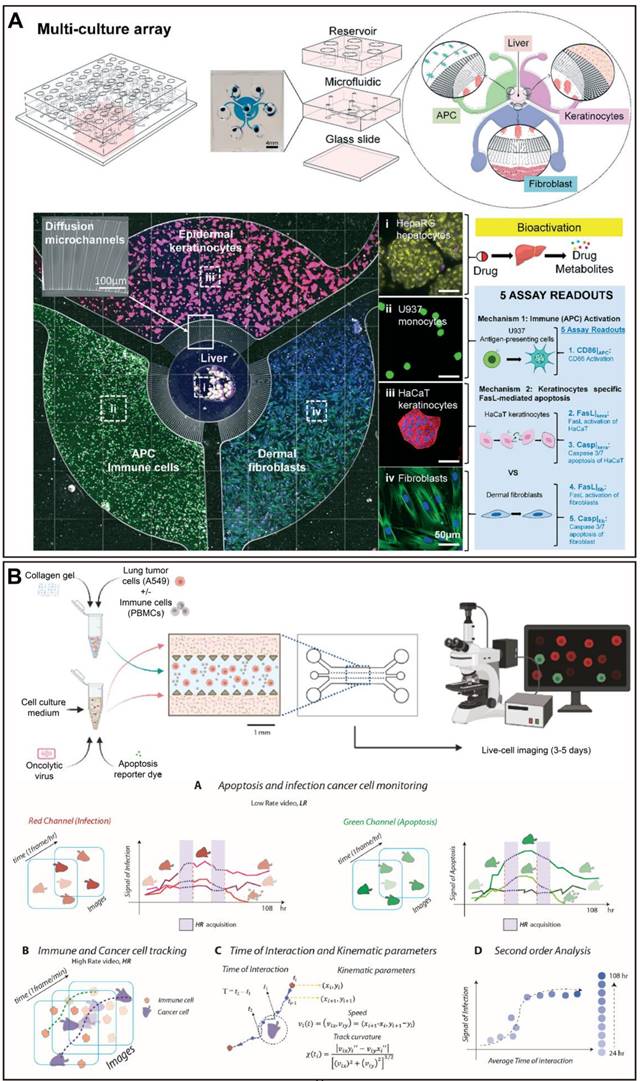
Angiogenesis has been reported to be associated with more than 70 diseases. Lately, a study has utilized DL-based image processing algorithms to analyze and quantify angiogenesis on a chip. This method allows the assessment of angiogenesis using up to 16 angiogenesis-related metrics and the extraction of 3D indicators from 2D images. In this work, the model successfully showed changes in response to biochemical gradients and can be applied to drug development [238]. Moreover, a new OoC platform for growing, vascularized, and perfused microtissues suitable for large-scale drug screening has been applied to assess the effectiveness of antiangiogenic compounds. This platform is based on a CNN method that enables fast and accurate flagging of compounds that effectively disrupt vascular networks from images of before and following drug applications with near-perfect accuracy. The CNN model significantly outperformed well-trained human raters, representing a substantive step in the automated analysis of data toward high-throughput drug screening [239].
AI-based in electrochemical detection and analysis of OoCs
Except for microscopic images, various data types, including electrochemical detection data, can serve as inputs for DL model development for training, enabling the detection of organ functions and drug treatment endpoints on OoCs. Electrochemical monitoring technology is automated and noninvasive, making it suitable for long-term operation in the monitoring of OoCs. By integrating a variety of online physical and biological electrochemical sensors to accomplish continual, automated, and in situ sensing of microenvironment biophysical and biochemical parameters (Figure 9A) [197], noninvasive monitoring OoCs and performing real-time data analysis (e.g., measurement of nutrients, metabolites, proteins, growth factors, exosomes, shear forces, current/electrical resistance, pH, oxygen levels, and drug uptake) could be achieved for autonomous decision-making [240]. Importantly, all these real-time and continuously collected data could be combined with AI-based data processing for studying and optimizing closed-loop feedback-based experimental parameters. Eventually, the system will be able to automatically regulate and control various functional parameters of OoCs, achieving the development of intelligent OoCs [62, 241].
AI-based experimental design and control of OoCs
Although the most of current applications focus on post-experimental data analysis, DL has increasing potential for designing microfluidic systems and controlling systems during experiments [60]. Microfluidics represents an excellent platform for supporting automation and intelligent control of reaction conditions [242]. In the preparation phase of a project, DL can be employed for devices design and materials selection to make OoCs more suitable for specific applications [19]. Traditionally, soft lithography, photolithography, and etching techniques have been widely utilized in the manufacture of OoC devices. However, these methods presented serious limitations that hinder the pace of development and innovation of microfluidic applications [243]. Given this, 3D printing has the potential to be a promising solution. This technique provides high-throughput and scalability, and allows for industrial means of mass production. To date, a number of studies have applied 3D printing technology to build molds for manufacturing OoC devices. In a study, 3D printing was utilized in the manufacture of almost all chip components [244]. To realize high precision microscale structures, material properties, and printing parameters require delicate control [245]. During this process, AI could be applied to provide support to improve accuracy and efficiency, such as 1) the optimization of processes; 2) the detection of manufacturing defects; 3) the evaluation of dimensional accuracy; and 4) the prediction of material properties [246]. For instance, a computer vision-based (CNN-aided calibration) approach was used to rapidly and precisely design microfluidic devices and minimize absolute errors in device manufacturing, which offers a convenient, effective, and efficient solution for 3D printing of OoC platforms [247].
During the experiment, reinforcement learning (RL)-based deep Q network can utilize image feedback to assist in retaining stable flow conditions for prolonged periods by automatically adjusting flow conditions to mitigate the inconsistent system performance exhibited in microfluidic platforms during extended experiments (Figure 9B) [248], which could be applied to the control of culture media in OoCs. An emerging direction in the development of OoCs is vascularization, which is becoming a significant and necessary physiological level feature of most OoCs [249]. Recently, a study assessed the oxygen transport capacity of vascular network association with the most common morphological indicators through ML algorithms such as multiple linear regression and random forest. This approach will assist in measuring the performance or biological function of vascularized networks in OoCs (Figure 9C) [249]. For lung OoCs, mechanical stretch is used to mimic the cyclical expansion, resulting in tissue mechanical force control being critical. Therefore, automatic control of the applied tissue mechanical forces can be achieved by DL of cell morphology and microenvironment. Importantly, DL allows for the real-time monitoring of the entire system performance while continuously detecting cell processes and biomarkers without harming cell viability [19]. Researchers have proposed to regulate microenvironmental parameters through spectroscopy, automated multisensor, and microscopy monitoring systems, as well as through machine-intelligent data-driven optimization, as the cell microenvironment is crucial for maintaining physiologically relevant organ functions and responses to drugs [62]. A multi-OoC system achieved exact control of flow distribution and drug distribution to different organs using an on-board pneumatically-driven pump with independently programmable flow rates [182]. Furthermore, an automated microfluidic platform was developed to accomplish dynamic and combinatorial drug screening, which allows for highly dynamic, reproducible, and reliable analyses of patient-derived organoids (Figure 9E) [250]. What's exciting is that liquid-handling robotics control systems allowed the automated culture, perfusion, medium addition, fluidic linking, sample collection, and in situ microscopy imaging of up to ten organ models (Figure 9D) [181], which will be easier to integrate with pharmaceutical robotic pipelines [73]. Therefore, automation allows real-time data collection and analysis for feedback on target results. In the near future, it is conceivable that AI-guided organ-on-a-chip may in fact be more fundamentally natural than human control.
In conclusion, even though only a few OoC studies have incorporated AI, we believe that these existing studies are sufficient to indicate exciting prospects for synergy between OoCs and AI in future drug evaluation (Figure 6-9). Nevertheless, much follow-up work and collaboration are still needed to drive the development of the combination of OoCs and AI, and ultimately to contribute to the advancement in the field of drug evaluation.
The future prospect of OoCs and AI in drug evaluation
OoCs will become an indispensable part of the future drug evaluation system
OoCs using human cell sources (e.g., primary cells, cell lines, iPSCs, or organoids) could possibly eliminate the effects of cross-species differences introduced by utilizing animal models for clinical drug studies. A major obstacle of OoCs is the limited lifespan of cells in the device, and such limitation is exacerbated when not using immortalized cell lines [251]. Despite the widespread view that cell lines typically lack the ability to simulate tissue-specific functions with high fidelity; however, cell lines are able to provide a practically limitless supply of similar cells that could be utilized for studies in higher-throughput. This could potentially improve the reproducibility of results, making it highly valuable for drug development in the early stages [73]. A study demonstrated that the reproducibility was greatly dependent on the cell source [252]. It is noteworthy to mention that iPSCs, while iPSCs provide the same advantages, they are frequently limited the wide applicability by their failure to display a fully mature differentiated phenotype, and the necessary purity for many tissues [73].
OoCs have been extensively used to build a variety of in vitro disease models, an important aspect of which is rare disease models, and OoCs could fill the gaps where animal models do not work or even do not exist. To date, only about 400 of over 7,000 identified rare diseases have active research programs due to the absence of animal models for others, resulting in significant hindrances to the development of new drugs in the field [253]. In the past, OoCs have been used to successfully model many rare diseases. Recently, TNT005, a drug received clinical approval from the FDA based on preclinical efficacy data obtained from rare disease-on-a-chip, showing that OoCs have great potential in the field of rare diseases leading to the generation of IND [254]. OoCs-based models for rare diseases have the potential to produce significant data that are typically not observable in in vitro and in vivo models or clinical samples, as OoCs enable long-term and real-time monitoring of changes in physiological processes. Through further analysis of these data by ML/DL in real-time, it is possible to analyze the progression at the molecular level of such diseases, and eventually discover the specific mechanisms by which the diseases occur [19]. Thus, OoCs have a promising opportunity in rare diseases. Besides, similar approaches are applicable to other diseases as well. Furthermore, OoCs could be seeded with iPSCs and patient-derived organoids to develop patient-specific models, and deliver on the promise of advanced personalized medicine.
Perhaps more importantly, OoCs can be used first time in emergencies for disease mechanism research and drug repurposing, discovery, and toxicity evaluation. For instance, during the COVID-19 global pandemic, several OoC platforms have been successfully used [27, 29, 111, 155, 203, 255-258]. A vascularized lung OoC platform was utilized for SARS-CoV-2 infection and to identify the virus-induced vascular damage, including inflammatory response and loss of barrier integrity, the latter can be alleviated by tocilizumab treatment [255]. The same team also revealed the SARS-CoV-2-induced intestinal injury and immune responses with the use of a gut OoC platform [256]. Moreover, lung airway OoCs have also been used for the reconstruction of clinically relevant influenza virus evolution [257]. Meanwhile, OoCs have also been successfully used in the study of other viruses [259, 260]. Thus, the development of antiviral drugs will continue to be a major focus of drug discovery in the post-COVID-19 era [261].
Representative examples of OoCs combined with AI or automation for experimental design and control. (A) A multi-OoC system includes a physical, biochemical, and optical sensing platform that operates organ units continuously, dynamically, and automatically, achieving in situ monitoring of organoid behaviors. Adapted with permission from [197], copyright 2017 National Academy of Sciences. (B) Automatically adjusts flow conditions with reinforcement learning to assist microfluidic platforms in maintaining stable flow conditions over time. Adapted with permission from [248], copyright 2018 American Chemical Society. (C) The utilization of supervised ML to evaluate the morphological and biological functions of vascularized OoCs. Adapted with permission from [249], copyright 2023 Springer International Publishing. (D) A robotic system consists of a 3-axis motion system, automated liquid handler, peristaltic pump, and custom microscope stage that enables the continuous perfusion, linking, and image analysis of up to ten organ models. Adapted with permission from [181], copyright 2020 Springer Nature. (E) A microfluidic platform based on a programmable membrane-valve allows to provision of combinatorial and dynamic drug therapies and enables analysis of organoids in real-time for personalized drug screening. Adapted with permission from [250], copyright 2020 Nature Publishing Group.
New AI for new OoCs
AI itself is a rapidly developing discipline, and deep learning networks and automated machine learning have promoted the development of generative AI. AI has shown unprecedented creativity, based on OoCs' own needs and new creative needs, the development of OoCs will be greatly accelerated.
Firstly, AI can address the bottleneck issue in the development of OoCs hardware. PDMS is one of the most popular materials for fabricating microfluidic devices. However, there is a common concern that OoCs made out of PDMS are unable to be utilized efficiently for drug research because of drug absorption [262]. But this issue has not proven to be as serious a concern as first thought, because only hydrophobic drugs are absorbed by PDMS, which is only a small portion of the drug development pipeline [73]. In fact, as previously mentioned, those showing in vivo simulations in response to clinically relevant drug exposures, as well as OoC platforms capable of quantitatively predicting human pharmacokinetic parameters, were manufactured almost by PDMS (Table 1). Despite this, the absorption of PDMS remains a significant challenge for drug screening of hydrophobic compounds like small molecule drugs, which could result in biased experimental results. Now, researchers could avoid the risk of small molecule drugs being absorbed by using alternative materials such as inorganic (e.g., glass and silicon), elastomeric (e.g., polyesters and polyurethane), and thermoplastic (e.g., poly(methyl methacrylate) and polycarbonate) materials, or coating PDMS with non-absorbent coatings [31, 44, 263, 264]. Nevertheless, a careful characterization of the adsorption/absorption curves is required for OoC platforms, regardless of the material of manufacture chosen [79]. Furthermore, PDMS-based devices frequently need to be manually cast, punched, and assembled, thus significantly reducing the reproducibility and throughput of the fabrication process [265], and that could be addressed by 3D printing/bioprinting [266, 267]. 3D bioprinting technology has been widely utilized in the construction of in vitro tissue/organ models and testing devices for drug screening [6], including OoCs. It makes it possible to precisely distribute cells or biomaterial in a target region, which allows the creation of more complex structures and microenvironments that more accurately mimic the function of living organisms [6, 268]. In fact, in other fields, AI and 3D bioprinting have been widely combined. AI-assisted artificial design of structures has a much faster optimization iteration speed than engineers, and the identification and discovery of material defects based on AI feature recognition are also faster and more accurate. In the future, generative AI will bring deeper technological changes to the development of OoCs in terms of drawing design, structural optimization, and even full process production.
Despite this, OoCs remain facing various challenges that must get past in order to promote their physiological relevance and facilitate their translation into the clinic. For example, introducing more cell subtypes, metabolites, and microbiomes, as well as biochemical and biophysical gradients [175], or vascularization and innervation of organs [269], to increase the complexity of the models. More importantly, the body-on-a-chip system needs to be scaled according to the sizes of actual organs in vivo, relying on appropriate scaling rules and methods, and the fluid volume and dynamics in chips should be adjusted in accordance with specific human organs [175], which are key to simulate physiological responses. Of note, in the experiment design, a balance between the system's feasibility and complexity should be considered carefully [174]. In addition, the use of robust culture media, especially for multi-OoCs, which can help to promote cell survival in different organ types and keep the various organs functional [175], is equally significant, as a key challenge. In microfluidic devices, organ function often declines in long-term culture, and tissue necrosis caused by a lack of oxygen diffusion continues to be a barrier to the use of larger or more complex organs, although vascularization of most organs has been achieved. In view of this, vascularized constructs with a tissue-specific vasculature and perfusable vascular network will offer the foundation for reliable OoCs with sustained functionality [269]. Notably, AI can use reinforcement learning to identify optimal medium compositions and dynamic culture conditions for particular cell types, possibly extending the lifespan and functionality of present in vitro organ models [62]. More importantly, for integrated multiorgan models, identifying optimal medium compositions and dynamic culture conditions for co-culture of various cell types, would make more sense.
Secondly, AI will increase the detection throughput of OoCs. Typically, OoC devices are low-throughput, which limits their applicability in the early phases of drug discovery [270]. However, considering that there are multiple stages in drug discovery and development where OoCs could be implemented, more complex low-throughput to medium-throughput OoC devices may be more useful at later stages, such as drug efficacy studies [79]. For pharmaceutical companies, the model can be selected according to the stage of drug development, after considering time, cost, and benefits. Although physiological relevance may be compromised in exchange for high-throughput [93], in fact, the recently developed high-throughput OoC devices still retain the key features required for drug developers [31, 203, 205]. Yet, the increase of larger data due to the use of high-throughput platforms is a new challenge, this could probably be solved with AI, as previously discussed. In addition, automation is another key requirement for developing reliable and high-throughput platforms [271], while such devices have been developed, they are extremely rare [181]. Importantly, OoCs successfully integrated with AI will be invaluable for its applications in drug development, and investigating how better to integrate is worthwhile.
Finally, AI further promotes the convenience of OoCs data processing. At present, the iterative development speed of AI is astonishing. A simpler, more open, and more user-friendly AI-based algorithm platform will be developed, making image and big data analysis for OoCs dynamic processes simpler. Recently, especially with the emergence of open AI, it will also be applied in the scientific field. The design of OoCs and the extraction of more biological-related features will be completed by AI. Imagine the future, where people will become emitters of commands, AI-controlled automated OoCs production and analysis platforms will present the data we need, and drug evaluation will become an easy task at that time.
We can foresee that AI will further help us invent new OoCs products in the design, production, and control of OoCs, as well as data processing, to expand the application of OoCs. While OoCs' potential is exciting, the technology is still in its early stage [79]. Also, despite the significant benefits of coupling OoCs with AI, there are still some challenges ahead, especially as this fall within the healthcare field. For instance, as mentioned in section 5.2, how to achieve the proper explanation of the collected data. A typical issue of machine learning, particularly deep learning models is known as “black box approach”; that is, the lack of interpretability, which limits the obtaining of a suitable explanation from such models on how they arrive at their results [206]. This lack of interpretability may significantly hinder their application in the short term. Thus, explainable models are necessary to be developed to improve trust, which will promote the application of models in OoCs. Meanwhile, repeatability is another important issue. It may produce different results by using different algorithms, thereby increasing uncertainty [206]. To date, some solutions have been proposed to move AI towards reproducible [272]. The quality of the model depends on the quality and characteristics of the data, so the datasets collection and selection are very significant for building a model, which directly impacts the accuracy and predictability of the model. Of note, training, validation, and testing datasets are crucial for model development, but the amounts of datasets required depends on the complexity of the data type and the task [93, 273]. Despite such challenges, as both field progresses, the application of AI will undoubtedly bring greater vitality and impetus to the development of OoCs, especially in the area of drug evaluation.
Last but not the least, while OoCs have advanced significantly in the academic environment, and a few OoC platforms have even been successfully converted into commercial products, numerous challenges prevent its complete deployment in an industrial setting, and OoCs are still marginalized in the pharmaceutical industry [14, 174]. To bridge the academia-to-industry gap and facilitate the adoption and implementation of OoCs in the drug development process, ongoing engagement, and discussions with OoCs developers, end users, and regulatory bodies are critical [79, 274]. From a long-term viewpoint, as technology improves and cost decrease, OoCs will finally be better accepted and adopted by the pharmaceutical industry.
Conclusion
In summary, organ-on-a-chip successfully replicates the critical physiological functions and environment of the human organs, as a state-of-the-art in vitro model, showing encouraging performances in a variety of drug evaluation platforms. Nevertheless, the current OoCs still must face many challenges that take it from academia to industry. Continuing development with AI, it is undeniable that OoCs will likely dramatically change drug development, disease modeling, and personalized medicine.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82274224), China Academy of Chinese Medical Sciences Innovation Fund (CI2021A00610) and the Fundamental Research Funds for the Central Public Welfare Research Institutes of China (JBGS2021001, CI2021B017-2).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Wang Y, Chen Z, Bian F, Shang L, Zhu K, Zhao Y. Advances of droplet-based microfluidics in drug discovery. Expert Opin Drug Discov. 2020;15:969-79
2. Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4:35-44
3. DiMasi JA, Hansen RW, Grabowski HG. The price of innovation: new estimates of drug development costs. J Health Econ. 2003;22:151-85
4. Stewart WC, Kruft B, Nelson LA, Stewart JA. Success of ophthalmic pharmaceutical start-up companies. Acta Ophthalmol. 2018;96:e266-e8
5. Kang L, Chung BG, Langer R, Khademhosseini A. Microfluidics for drug discovery and development: from target selection to product lifecycle management. Drug Discov Today. 2008;13:1-13
6. Nie J, Gao Q, Fu J, He Y. Grafting of 3D Bioprinting to In Vitro Drug Screening: A Review. Adv Healthc Mater. 2020;9:e1901773
7. Liu Y, Sun L, Zhang H, Shang L, Zhao Y. Microfluidics for Drug Development: From Synthesis to Evaluation. Chem Rev. 2021;121:7468-529
8. Rome BN, Avorn J. Drug Evaluation during the Covid-19 Pandemic. N Engl J Med. 2020;382:2282-4
9. Liu X, Zheng W, Jiang X. Cell-Based Assays on Microfluidics for Drug Screening. ACS Sens. 2019;4:1465-75
10. Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483:531-3
11. Wang Y, Kankala RK, Ou C, Chen A, Yang Z. Advances in hydrogel-based vascularized tissues for tissue repair and drug screening. Bioact Mater. 2022;9:198-220
12. Zhao Y, Demirci U, Chen Y, Chen P. Multiscale brain research on a microfluidic chip. Lab Chip. 2020;20:1531-43
13. Dittrich PS, Manz A. Lab-on-a-chip: microfluidics in drug discovery. Nat Rev Drug Discov. 2006;5:210-8
14. Zou Z, Luo X, Chen Z, Zhang YS, Wen C. Emerging microfluidics-enabled platforms for osteoarthritis management: from benchtop to bedside. Theranostics. 2022;12:891-909
15. Du Z, Mi S, Yi X, Xu Y, Sun W. Microfluidic system for modelling 3D tumour invasion into surrounding stroma and drug screening. Biofabrication. 2018;10:034102
16. Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368-73
17. Meyvantsson I, Beebe DJ. Cell culture models in microfluidic systems. Annu Rev Anal Chem (Palo Alto Calif). 2008;1:423-49
18. Kohl Y, Biehl M, Spring S, Hesler M, Ogourtsov V, Todorovic M. et al. Microfluidic In Vitro Platform for (Nano)Safety and (Nano)Drug Efficiency Screening. Small. 2021;17:e2006012
19. Li J, Chen J, Bai H, Wang H, Hao S, Ding Y. et al. An Overview of Organs-on-Chips Based on Deep Learning. Research (Wash D C). 2022;2022:9869518
20. Tian C, Zheng S, Liu X, Kamei KI. Tumor-on-a-chip model for advancement of anti-cancer nano drug delivery system. J Nanobiotechnology. 2022;20:338
21. Sin A, Chin KC, Jamil MF, Kostov Y, Rao G, Shuler ML. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol Prog. 2004;20:338-45
22. Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662-8
23. Park TE, Mustafaoglu N, Herland A, Hasselkus R, Mannix R, FitzGerald EA. et al. Hypoxia-enhanced Blood-Brain Barrier Chip recapitulates human barrier function and shuttling of drugs and antibodies. Nat Commun. 2019;10:2621
24. Lyu Z, Park J, Kim KM, Jin HJ, Wu H, Rajadas J. et al. A neurovascular-unit-on-a-chip for the evaluation of the restorative potential of stem cell therapies for ischaemic stroke. Nat Biomed Eng. 2021;5:847-63
25. Sahtoe DD, Coscia A, Mustafaoglu N, Miller LM, Olal D, Vulovic I. et al. Transferrin receptor targeting by de novo sheet extension. Proc Natl Acad Sci U S A. 2021;118:e2021569118
26. Huh D, Leslie DC, Matthews BD, Fraser JP, Jurek S, Hamilton GA. et al. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med. 2012;4:159ra47
27. Si L, Bai H, Rodas M, Cao W, Oh CY, Jiang A. et al. A human-airway-on-a-chip for the rapid identification of candidate antiviral therapeutics and prophylactics. Nat Biomed Eng. 2021;5:815-29
28. Zhang YS, Arneri A, Bersini S, Shin SR, Zhu K, Goli-Malekabadi Z. et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials. 2016;110:45-59
29. Charrez B, Charwat V, Siemons B, Finsberg H, Miller EW, Edwards AG. et al. In vitro safety "clinical trial" of the cardiac liability of drug polytherapy. Clin Transl Sci. 2021;14:1155-65
30. Kujala VJ, Pasqualini FS, Goss JA, Nawroth JC, Parker KK. Laminar ventricular myocardium on a microelectrode array-based chip. J Mater Chem B. 2016;4:3534-43
31. Bircsak KM, DeBiasio R, Miedel M, Alsebahi A, Reddinger R, Saleh A. et al. A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate(R). Toxicology. 2021;450:152667
32. Freag MS, Namgung B, Reyna Fernandez ME, Gherardi E, Sengupta S, Jang HL. Human Nonalcoholic Steatohepatitis on a Chip. Hepatol Commun. 2021;5:217-33
33. Jang KJ, Otieno MA, Ronxhi J, Lim HK, Ewart L, Kodella KR. et al. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci Transl Med. 2019;11:eaax5516
34. Jang KJ, Mehr AP, Hamilton GA, McPartlin LA, Chung S, Suh KY. et al. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr Biol (Camb). 2013;5:1119-29
35. Petrosyan A, Cravedi P, Villani V, Angeletti A, Manrique J, Renieri A. et al. A glomerulus-on-a-chip to recapitulate the human glomerular filtration barrier. Nat Commun. 2019;10:3656
36. Roye Y, Bhattacharya R, Mou X, Zhou Y, Burt MA, Musah S. A Personalized Glomerulus Chip Engineered from Stem Cell-Derived Epithelium and Vascular Endothelium. Micromachines (Basel). 2021;12:967
37. Gao D, Liu H, Lin JM, Wang Y, Jiang Y. Characterization of drug permeability in Caco-2 monolayers by mass spectrometry on a membrane-based microfluidic device. Lab Chip. 2013;13:978-85
38. Jalili-Firoozinezhad S, Prantil-Baun R, Jiang A, Potla R, Mammoto T, Weaver JC. et al. Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-Chip. Cell Death Dis. 2018;9:223
39. Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A. et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng. 2019;3:520-31
40. Lee S, Kim S, Koo DJ, Yu J, Cho H, Lee H. et al. 3D Microfluidic Platform and Tumor Vascular Mapping for Evaluating Anti-Angiogenic RNAi-Based Nanomedicine. ACS Nano. 2021;15:338-50
41. Barrile R, van der Meer AD, Park H, Fraser JP, Simic D, Teng F. et al. Organ-on-Chip Recapitulates Thrombosis Induced by an anti-CD154 Monoclonal Antibody: Translational Potential of Advanced Microengineered Systems. Clin Pharmacol Ther. 2018;104:1240-8
42. Kim C, Kasuya J, Jeon J, Chung S, Kamm RD. A quantitative microfluidic angiogenesis screen for studying anti-angiogenic therapeutic drugs. Lab Chip. 2015;15:301-10
43. Kim K, Jeon HM, Choi KC, Sung GY. Testing the Effectiveness of Curcuma longa Leaf Extract on a Skin Equivalent Using a Pumpless Skin-on-a-Chip Model. Int J Mol Sci. 2020;21:3898
44. Zhang J, Chen Z, Zhang Y, Wang X, Ouyang J, Zhu J. et al. Construction of a high fidelity epidermis-on-a-chip for scalable in vitro irritation evaluation. Lab Chip. 2021;21:3804-18
45. Chou DB, Frismantas V, Milton Y, David R, Pop-Damkov P, Ferguson D. et al. On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology. Nat Biomed Eng. 2020;4:394-406
46. Nelson MR, Ghoshal D, Mejias JC, Rubio DF, Keith E, Roy K. A multi-niche microvascularized human bone marrow (hBM) on-a-chip elucidates key roles of the endosteal niche in hBM physiology. Biomaterials. 2021;270:120683
47. Achberger K, Probst C, Haderspeck J, Bolz S, Rogal J, Chuchuy J. et al. Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform. Elife. 2019;8:e46188
48. Chung M, Lee S, Lee BJ, Son K, Jeon NL, Kim JH. Wet-AMD on a Chip: Modeling Outer Blood-Retinal Barrier In Vitro. Adv Healthc Mater. 2018;7:1700028
49. Zhu S, Abudupataer M, Yan S, Wang C, Wang L, Zhu K. Construction of a high-throughput aorta smooth muscle-on-a-chip for thoracic aortic aneurysm drug screening. Biosens Bioelectron. 2022;218:114747
50. Ma W, Zhang J, Liu S, Yan S, Xu K, Zhang YS. et al. Patient-derived microphysiological model identifies the therapeutic potential of metformin for thoracic aortic aneurysm. EBioMedicine. 2022;81:104080
51. Kongsuphol P, Gupta S, Liu Y, Bhuvanendran Nair Gourikutty S, Biswas SK, Ramadan Q. In vitro micro-physiological model of the inflamed human adipose tissue for immune-metabolic analysis in type II diabetes. Sci Rep. 2019;9:4887
52. Rogal J, Binder C, Kromidas E, Roosz J, Probst C, Schneider S. et al. WAT-on-a-chip integrating human mature white adipocytes for mechanistic research and pharmaceutical applications. Sci Rep. 2020;10:6666
53. Hassell BA, Goyal G, Lee E, Sontheimer-Phelps A, Levy O, Chen CS. et al. Human Organ Chip Models Recapitulate Orthotopic Lung Cancer Growth, Therapeutic Responses, and Tumor Dormancy In Vitro. Cell Rep. 2017;21:508-16
54. Cui X, Ma C, Vasudevaraja V, Serrano J, Tong J, Peng Y. et al. Dissecting the immunosuppressive tumor microenvironments in Glioblastoma-on-a-Chip for optimized PD-1 immunotherapy. Elife. 2020;9:e52253
55. Carvalho MR, Barata D, Teixeira LM, Giselbrecht S, Reis RL, Oliveira JM. et al. Colorectal tumor-on-a-chip system: A 3D tool for precision onco-nanomedicine. Sci Adv. 2019;5:eaaw1317
56. Bakhchova L, Jantaree P, Gupta A, Isermann B, Steinmann U, Naumann M. On-a-Chip-Based Sensitive Detection of Drug-Induced Apoptosis in Polarized Gastric Epithelial Cells. ACS Biomater Sci Eng. 2021;7:5474-83
57. Vunjak-Novakovic G, Ronaldson-Bouchard K, Radisic M. Organs-on-a-chip models for biological research. Cell. 2021;184:4597-611
58. Mullard A. The drug-maker's guide to the galaxy. Nature. 2017;549:445-7
59. Lu H, Luo M. Survey on new progresses of deep learning based computer vision. J Data Acquis Process. 2022;37:247-78
60. Riordon J, Sovilj D, Sanner S, Sinton D, Young EWK. Deep Learning with Microfluidics for Biotechnology. Trends Biotechnol. 2019;37:310-24
61. McIntyre D, Lashkaripour A, Fordyce P, Densmore D. Machine learning for microfluidic design and control. Lab Chip. 2022;22:2925-37
62. Galan EA, Zhao H, Wang X, Dai Q, Huck WTS, Ma S. Intelligent Microfluidics: The Convergence of Machine Learning and Microfluidics in Materials Science and Biomedicine. Matter. 2020;3:1893-922
63. Liu H, Nan L, Chen F, Zhao Y, Zhao Y. Functions and applications of artificial intelligence in droplet microfluidics. Lab Chip. 2023;23:2497-2513
64. Zheng J, Cole T, Zhang Y, Kim J, Tang SY. Exploiting machine learning for bestowing intelligence to microfluidics. Biosens Bioelectron. 2021;194:113666
65. Fetah KL, DiPardo BJ, Kongadzem EM, Tomlinson JS, Elzagheid A, Elmusrati M. et al. Cancer Modeling-on-a-Chip with Future Artificial Intelligence Integration. Small. 2019;15:e1901985
66. Breslin S, O'Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today. 2013;18:240-9
67. Hachey SJ, Hughes CCW. Applications of tumor chip technology. Lab Chip. 2018;18:2893-912
68. Parasrampuria DA, Benet LZ, Sharma A. Why Drugs Fail in Late Stages of Development: Case Study Analyses from the Last Decade and Recommendations. AAPS J. 2018;20:46
69. Kapalczynska M, Kolenda T, Przybyla W, Zajaczkowska M, Teresiak A, Filas V. et al. 2D and 3D cell cultures - a comparison of different types of cancer cell cultures. Arch Med Sci. 2018;14:910-9
70. Francis I, Shrestha J, Paudel KR, Hansbro PM, Warkiani ME, Saha SC. Recent advances in lung-on-a-chip models. Drug Discov Today. 2022;27:2593-602
71. Yang X, Li K, Zhang X, Liu C, Guo B, Wen W. et al. Nanofiber membrane supported lung-on-a-chip microdevice for anti-cancer drug testing. Lab Chip. 2018;18:486-95
72. Wang Y, Jeon H. 3D cell cultures toward quantitative high-throughput drug screening. Trends Pharmacol Sci. 2022;43:569-81
73. Ingber DE. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat Rev Genet. 2022;23:467-91
74. Ahadian S, Civitarese R, Bannerman D, Mohammadi MH, Lu R, Wang E. et al. Organ-On-A-Chip Platforms: A Convergence of Advanced Materials, Cells, and Microscale Technologies. Adv Healthc Mater. 2018;7:1700506
75. Namdari R, Jones K, Chuang SS, Van Cruchten S, Dincer Z, Downes N. et al. Species selection for nonclinical safety assessment of drug candidates: Examples of current industry practice. Regul Toxicol Pharmacol. 2021;126:105029
76. Urbanczyk M, Zbinden A, Schenke-Layland K. Organ-specific endothelial cell heterogenicity and its impact on regenerative medicine and biomedical engineering applications. Adv Drug Deliv Rev. 2022;186:114323
77. Donnelly H, Salmeron-Sanchez M, Dalby MJ. Designing stem cell niches for differentiation and self-renewal. J R Soc Interface. 2018;15:20180388
78. Ronaldson-Bouchard K, Vunjak-Novakovic G. Organs-on-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development. Cell Stem Cell. 2018;22:310-24
79. Low LA, Mummery C, Berridge BR, Austin CP, Tagle DA. Organs-on-chips: into the next decade. Nat Rev Drug Discov. 2021;20:345-61
80. Kim R, Attayek PJ, Wang Y, Furtado KL, Tamayo R, Sims CE. et al. An in vitro intestinal platform with a self-sustaining oxygen gradient to study the human gut/microbiome interface. Biofabrication. 2019;12:015006
81. Jing B, Wang ZA, Zhang C, Deng Q, Wei J, Luo Y. et al. Establishment and Application of Peristaltic Human Gut-Vessel Microsystem for Studying Host-Microbial Interaction. Front Bioeng Biotechnol. 2020;8:272
82. Mahajan G, Doherty E, To T, Sutherland A, Grant J, Junaid A. et al. Vaginal microbiome-host interactions modeled in a human vagina-on-a-chip. Microbiome. 2022;10:201
83. Tan J, Guo Q, Tian L, Pei Z, Li D, Wu M. et al. Biomimetic lung-on-a-chip to model virus infection and drug evaluation. Eur J Pharm Sci. 2023;180:106329
84. Kirchmair J, Goller AH, Lang D, Kunze J, Testa B, Wilson ID. et al. Predicting drug metabolism: experiment and/or computation? Nat Rev Drug Discov. 2015;14:387-404
85. Ai X, Zhao L, Lu Y, Hou Y, Lv T, Jiang Y. et al. Integrated Array Chip for High-Throughput Screening of Species Differences in Metabolism. Anal Chem. 2020;92:11696-704
86. Shroff T, Aina K, Maass C, Cipriano M, Lambrecht J, Tacke F. et al. Studying metabolism with multi-organ chips: new tools for disease modelling, pharmacokinetics and pharmacodynamics. Open Biol. 2022;12:210333
87. Esch EW, Bahinski A, Huh D. Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov. 2015;14:248-60
88. Polini A, Prodanov L, Bhise NS, Manoharan V, Dokmeci MR, Khademhosseini A. Organs-on-a-chip: a new tool for drug discovery. Expert Opin Drug Discov. 2014;9:335-52
89. Picollet-D'hahan N, Zuchowska A, Lemeunier I, Le Gac S. Multiorgan-on-a-Chip: A Systemic Approach To Model and Decipher Inter-Organ Communication. Trends Biotechnol. 2021;39:788-810
90. Joseph X, Akhil V, Arathi A, Mohanan PV. Comprehensive Development in Organ-On-A-Chip Technology. J Pharm Sci. 2022;111:18-31
91. Sung JH. Multi-organ-on-a-chip for pharmacokinetics and toxicokinetic study of drugs. Expert Opin Drug Metab Toxicol. 2021;17:969-86
92. Sung JH, Wang YI, Kim JH, Lee JM, Shuler ML. Application of chemical reaction engineering principles to 'body-on-a-chip' systems. AIChE J. 2018;64:4351-60
93. Kawakita S, Mandal K, Mou L, Mecwan MM, Zhu Y, Li S. et al. Organ-On-A-Chip Models of the Blood-Brain Barrier: Recent Advances and Future Prospects. Small. 2022;18:e2201401
94. Oddo A, Peng B, Tong Z, Wei Y, Tong WY, Thissen H. et al. Advances in Microfluidic Blood-Brain Barrier (BBB) Models. Trends Biotechnol. 2019;37:1295-314
95. Booth R, Kim H. Characterization of a microfluidic in vitro model of the blood-brain barrier (muBBB). Lab Chip. 2012;12:1784-92
96. Brown JA, Pensabene V, Markov DA, Allwardt V, Neely MD, Shi M. et al. Recreating blood-brain barrier physiology and structure on chip: A novel neurovascular microfluidic bioreactor. Biomicrofluidics. 2015;9:054124
97. Cameron T, Bennet T, Rowe EM, Anwer M, Wellington CL, Cheung KC. Review of Design Considerations for Brain-on-a-Chip Models. Micromachines (Basel). 2021;12:441
98. Griep LM, Wolbers F, de Wagenaar B, ter Braak PM, Weksler BB, Romero IA. et al. BBB on chip: microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed Microdevices. 2013;15:145-50
99. Kimura H, Sakai Y, Fujii T. Organ/body-on-a-chip based on microfluidic technology for drug discovery. Drug Metab Pharmacokinet. 2018;33:43-8
100. Shrestha J, Razavi Bazaz S, Aboulkheyr Es H, Yaghobian Azari D, Thierry B, Ebrahimi Warkiani M. et al. Lung-on-a-chip: the future of respiratory disease models and pharmacological studies. Crit Rev Biotechnol. 2020;40:213-30
101. Zhang M, Xu C, Jiang L, Qin J. A 3D human lung-on-a-chip model for nanotoxicity testing. Toxicol Res (Camb). 2018;7:1048-60
102. Humayun M, Chow CW, Young EWK. Microfluidic lung airway-on-a-chip with arrayable suspended gels for studying epithelial and smooth muscle cell interactions. Lab Chip. 2018;18:1298-309
103. Tang H, Abouleila Y, Si L, Ortega-Prieto AM, Mummery CL, Ingber DE. et al. Human Organs-on-Chips for Virology. Trends Microbiol. 2020;28:934-46
104. Zamprogno P, Wuthrich S, Achenbach S, Thoma G, Stucki JD, Hobi N. et al. Second-generation lung-on-a-chip with an array of stretchable alveoli made with a biological membrane. Commun Biol. 2021;4:168
105. Naumova AV, Yarnykh VL. Assessment of heart microstructure: from mouse to man. Circulation. 2014;129:1720-2
106. Vargas R, Egurbide-Sifre A, Medina L. Organ-on-a-Chip systems for new drugs development. ADMET DMPK. 2021;9:111-41
107. Paloschi V, Sabater-Lleal M, Middelkamp H, Vivas A, Johansson S, van der Meer A. et al. Organ-on-a-chip technology: a novel approach to investigate cardiovascular diseases. Cardiovasc Res. 2021;117:2742-54
108. Tavakol DN, Fleischer S, Vunjak-Novakovic G. Harnessing organs-on-a-chip to model tissue regeneration. Cell Stem Cell. 2021;28:993-1015
109. Nunes SS, Miklas JW, Liu J, Aschar-Sobbi R, Xiao Y, Zhang B. et al. Biowire: a platform for maturation of human pluripotent stem cell-derived cardiomyocytes. Nat Methods. 2013;10:781-7
110. Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326-35
111. Charrez B, Charwat V, Siemons BA, Goswami I, Sakolish C, Luo YS. et al. Heart Muscle Microphysiological System for Cardiac Liability Prediction of Repurposed COVID-19 Therapeutics. Front Pharmacol. 2021;12:684252
112. Li Z, Hui J, Yang P, Mao H. Microfluidic Organ-on-a-Chip System for Disease Modeling and Drug Development. Biosensors (Basel). 2022;12:370
113. Mancio-Silva L, Fleming HE, Miller AB, Milstein S, Liebow A, Haslett P. et al. Improving Drug Discovery by Nucleic Acid Delivery in Engineered Human Microlivers. Cell Metab. 2019;29:727-35 e3
114. Moradi E, Jalili-Firoozinezhad S, Solati-Hashjin M. Microfluidic organ-on-a-chip models of human liver tissue. Acta Biomater. 2020;116:67-83
115. Ramamurthy RM, Atala A, Porada CD, Almeida-Porada G. Organoids and microphysiological systems: Promising models for accelerating AAV gene therapy studies. Front Immunol. 2022;13:1011143
116. Messelmani T, Morisseau L, Sakai Y, Legallais C, Le Goff A, Leclerc E. et al. Liver organ-on-chip models for toxicity studies and risk assessment. Lab Chip. 2022;22:2423-50
117. Xiao RR, Lv T, Tu X, Li P, Wang T, Dong H. et al. An integrated biomimetic array chip for establishment of collagen-based 3D primary human hepatocyte model for prediction of clinical drug-induced liver injury. Biotechnol Bioeng. 2021;118:4687-98
118. Tsamandouras N, Kostrzewski T, Stokes CL, Griffith LG, Hughes DJ, Cirit M. Quantitative Assessment of Population Variability in Hepatic Drug Metabolism Using a Perfused Three-Dimensional Human Liver Microphysiological System. J Pharmacol Exp Ther. 2017;360:95-105
119. Ewart L, Apostolou A, Briggs SA, Carman CV, Chaff JT, Heng AR. et al. Performance assessment and economic analysis of a human Liver-Chip for predictive toxicology. Commun Med (Lond). 2022;2:154
120. Schetz M, Dasta J, Goldstein S, Golper T. Drug-induced acute kidney injury. Curr Opin Crit Care. 2005;11:555-65
121. Wilmer MJ, Ng CP, Lanz HL, Vulto P, Suter-Dick L, Masereeuw R. Kidney-on-a-Chip Technology for Drug-Induced Nephrotoxicity Screening. Trends Biotechnol. 2016;34:156-70
122. Tian Z, Liang M. Renal metabolism and hypertension. Nat Commun. 2021;12:963
123. Cerqueira DM, Tayeb M, Ho J. MicroRNAs in kidney development and disease. JCI Insight. 2022;7:e158277
124. Weinberg E, Kaazempur-Mofrad M, Borenstein J. Concept and computational design for a bioartificial nephron-on-a-chip. Int J Artif Organs. 2008;31:508-14
125. Ashammakhi N, Wesseling-Perry K, Hasan A, Elkhammas E, Zhang YS. Kidney-on-a-chip: untapped opportunities. Kidney Int. 2018;94:1073-86
126. Yan J, Li Z, Guo J, Liu S, Guo J. Organ-on-a-chip: A new tool for in vitro research. Biosens Bioelectron. 2022;216:114626
127. Nieskens TTG, Magnusson O, Andersson P, Soderberg M, Persson M, Sjogren AK. Nephrotoxic antisense oligonucleotide SPC5001 induces kidney injury biomarkers in a proximal tubule-on-a-chip. Arch Toxicol. 2021;95:2123-36
128. Chapron A, Chapron BD, Hailey DW, Chang SY, Imaoka T, Thummel KE. et al. An Improved Vascularized, Dual-Channel Microphysiological System Facilitates Modeling of Proximal Tubular Solute Secretion. ACS Pharmacol Transl Sci. 2020;3:496-508
129. Doi K, Kimura H, Matsunaga YT, Fujii T, Nangaku M. Glomerulus-on-a-Chip: Current Insights and Future Potential Towards Recapitulating Selectively Permeable Filtration Systems. Int J Nephrol Renovasc Dis. 2022;15:85-101
130. Huang Y, Deng S, Luo X, Liu Y, Xu W, Pan J. et al. Evaluation of Intestinal Absorption Mechanism and Pharmacokinetics of Curcumin-Loaded Galactosylated Albumin Nanoparticles. Int J Nanomedicine. 2019;14:9721-30
131. Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165-74
132. Kim HJ, Ingber DE. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr Biol (Camb). 2013;5:1130-40
133. Majumder B, Baraneedharan U, Thiyagarajan S, Radhakrishnan P, Narasimhan H, Dhandapani M. et al. Predicting clinical response to anticancer drugs using an ex vivo platform that captures tumour heterogeneity. Nat Commun. 2015;6:6169
134. Liu X, Fang J, Huang S, Wu X, Xie X, Wang J. et al. Tumor-on-a-chip: from bioinspired design to biomedical application. Microsyst Nanoeng. 2021;7:50
135. Sun W, Luo Z, Lee J, Kim HJ, Lee K, Tebon P. et al. Organ-on-a-Chip for Cancer and Immune Organs Modeling. Adv Healthc Mater. 2019;8:e1801363
136. Vidi PA, Maleki T, Ochoa M, Wang L, Clark SM, Leary JF. et al. Disease-on-a-chip: mimicry of tumor growth in mammary ducts. Lab Chip. 2014;14:172-7
137. Nguyen DT, Lee E, Alimperti S, Norgard RJ, Wong A, Lee JJ. et al. A biomimetic pancreatic cancer on-chip reveals endothelial ablation via ALK7 signaling. Sci Adv. 2019;5:eaav6789
138. Zhang J, Tavakoli H, Ma L, Li X, Han L, Li X. Immunotherapy discovery on tumor organoid-on-a-chip platforms that recapitulate the tumor microenvironment. Adv Drug Deliv Rev. 2022;187:114365
139. Yi HG, Jeong YH, Kim Y, Choi YJ, Moon HE, Park SH. et al. A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy. Nat Biomed Eng. 2019;3:509-19
140. Jeong SY, Lee JH, Shin Y, Chung S, Kuh HJ. Co-Culture of Tumor Spheroids and Fibroblasts in a Collagen Matrix-Incorporated Microfluidic Chip Mimics Reciprocal Activation in Solid Tumor Microenvironment. PLoS One. 2016;11:e0159013
141. Rizvi I, Gurkan UA, Tasoglu S, Alagic N, Celli JP, Mensah LB. et al. Flow induces epithelial-mesenchymal transition, cellular heterogeneity and biomarker modulation in 3D ovarian cancer nodules. Proc Natl Acad Sci U S A. 2013;110:E1974-83
142. Wang HF, Ran R, Liu Y, Hui Y, Zeng B, Chen D. et al. Tumor-Vasculature-on-a-Chip for Investigating Nanoparticle Extravasation and Tumor Accumulation. ACS Nano. 2018;12:11600-9
143. Kerr SC, Morgan MM, Gillette AA, Livingston MK, Lugo-Cintron KM, Favreau PF. et al. A bioengineered organotypic prostate model for the study of tumor microenvironment-induced immune cell activation. Integr Biol (Camb). 2020;12:250-62
144. Chakrabarty S, Quiros-Solano WF, Kuijten MMP, Haspels B, Mallya S, Lo CSY. et al. A Microfluidic Cancer-on-Chip Platform Predicts Drug Response Using Organotypic Tumor Slice Culture. Cancer Res. 2022;82:510-20
145. Kim JH, Lee S, Kang SJ, Choi YW, Choi SY, Park JY. et al. Establishment of Three-Dimensional Bioprinted Bladder Cancer-on-a-Chip with a Microfluidic System Using Bacillus Calmette-Guerin. Int J Mol Sci. 2021;22:8887
146. Liu PF, Cao YW, Zhang SD, Zhao Y, Liu XG, Shi HQ. et al. A bladder cancer microenvironment simulation system based on a microfluidic co-culture model. Oncotarget. 2015;6:37695-705
147. Wang N, Wang J, Meng X, Bao Y, Wang S, Li T. 3D microfluidic in vitro model and bioinformatics integration to study the effects of Spatholobi Caulis tannin in cervical cancer. Sci Rep. 2018;8:12285
148. Bachal K, Yadav S, Gandhi P, Majumder A. Design and validation of a flowless gradient generating microfluidic device for high-throughput drug testing. Lab Chip. 2023;23:261-271
149. Chung M, Ahn J, Son K, Kim S, Jeon NL. Biomimetic Model of Tumor Microenvironment on Microfluidic Platform. Adv Healthc Mater. 2017;6:1700196
150. Ayuso JM, Sadangi S, Lares M, Rehman S, Humayun M, Denecke KM. et al. Microfluidic model with air-walls reveals fibroblasts and keratinocytes modulate melanoma cell phenotype, migration, and metabolism. Lab Chip. 2021;21:1139-49
151. Patel D, Gao Y, Son K, Siltanen C, Neve RM, Ferrara K. et al. Microfluidic co-cultures with hydrogel-based ligand trap to study paracrine signals giving rise to cancer drug resistance. Lab Chip. 2015;15:4614-24
152. Haase K, Kamm RD. Advances in on-chip vascularization. Regen Med. 2017;12:285-302
153. Moya ML, Hsu YH, Lee AP, Hughes CC, George SC. In vitro perfused human capillary networks. Tissue Eng Part C Methods. 2013;19:730-7
154. Kim S, Lee H, Chung M, Jeon NL. Engineering of functional, perfusable 3D microvascular networks on a chip. Lab Chip. 2013;13:1489-500
155. Lu RXZ, Lai BFL, Rafatian N, Gustafson D, Campbell SB, Banerjee A. et al. Vasculature-on-a-chip platform with innate immunity enables identification of angiopoietin-1 derived peptide as a therapeutic for SARS-CoV-2 induced inflammation. Lab Chip. 2022;22:1171-86
156. Varga-Medveczky Z, Kocsis D, Naszlady MB, Fonagy K, Erdo F. Skin-on-a-Chip Technology for Testing Transdermal Drug Delivery-Starting Points and Recent Developments. Pharmaceutics. 2021;13:1852
157. Risueno I, Valencia L, Jorcano JL, Velasco D. Skin-on-a-chip models: General overview and future perspectives. APL Bioeng. 2021;5:030901
158. Planz V, Lehr CM, Windbergs M. In vitro models for evaluating safety and efficacy of novel technologies for skin drug delivery. J Control Release. 2016;242:89-104
159. Hardwick RN, Betts CJ, Whritenour J, Sura R, Thamsen M, Kaufman EH. et al. Drug-induced skin toxicity: gaps in preclinical testing cascade as opportunities for complex in vitro models and assays. Lab Chip. 2020;20:199-214
160. Zoio P, Oliva A. Skin-on-a-Chip Technology: Microengineering Physiologically Relevant In Vitro Skin Models. Pharmaceutics. 2022;14:682
161. Mansoorifar A, Gordon R, Bergan R, Bertassoni LE. Bone-on-a-chip: microfluidic technologies and microphysiologic models of bone tissue. Adv Funct Mater. 2021;31:2006796
162. Tang Q, Li X, Lai C, Li L, Wu H, Wang Y. et al. Fabrication of a hydroxyapatite-PDMS microfluidic chip for bone-related cell culture and drug screening. Bioact Mater. 2021;6:169-78
163. Jodat YA, Kang MG, Kiaee K, Kim GJ, Martinez AFH, Rosenkranz A. et al. Human-Derived Organ-on-a-Chip for Personalized Drug Development. Curr Pharm Des. 2018;24:5471-86
164. Grosberg A, Nesmith AP, Goss JA, Brigham MD, McCain ML, Parker KK. Muscle on a chip: in vitro contractility assays for smooth and striated muscle. J Pharmacol Toxicol Methods. 2012;65:126-35
165. Compera N, Atwell S, Wirth J, von Torne C, Hauck SM, Meier M. Adipose microtissue-on-chip: a 3D cell culture platform for differentiation, stimulation, and proteomic analysis of human adipocytes. Lab Chip. 2022;22:3172-86
166. McCarthy M, Brown T, Alarcon A, Williams C, Wu X, Abbott RD. et al. Fat-On-A-Chip Models for Research and Discovery in Obesity and Its Metabolic Comorbidities. Tissue Eng Part B Rev. 2020;26:586-95
167. Chramiec A, Teles D, Yeager K, Marturano-Kruik A, Pak J, Chen T. et al. Integrated human organ-on-a-chip model for predictive studies of anti-tumor drug efficacy and cardiac safety. Lab Chip. 2020;20:4357-72
168. Ma C, Zhao L, Zhou EM, Xu J, Shen S, Wang J. On-Chip Construction of Liver Lobule-like Microtissue and Its Application for Adverse Drug Reaction Assay. Anal Chem. 2016;88:1719-27
169. Bovard D, Sandoz A, Luettich K, Frentzel S, Iskandar A, Marescotti D. et al. A lung/liver-on-a-chip platform for acute and chronic toxicity studies. Lab Chip. 2018;18:3814-29
170. Oleaga C, Riu A, Rothemund S, Lavado A, McAleer CW, Long CJ. et al. Investigation of the effect of hepatic metabolism on off-target cardiotoxicity in a multi-organ human-on-a-chip system. Biomaterials. 2018;182:176-90
171. Pires de Mello CP, Carmona-Moran C, McAleer CW, Perez J, Coln EA, Long CJ. et al. Microphysiological heart-liver body-on-a-chip system with a skin mimic for evaluating topical drug delivery. Lab Chip. 2020;20:749-59
172. Oleaga C, Bernabini C, Smith AS, Srinivasan B, Jackson M, McLamb W. et al. Multi-Organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep. 2016;6:20030
173. Xiao S, Coppeta JR, Rogers HB, Isenberg BC, Zhu J, Olalekan SA. et al. A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nat Commun. 2017;8:14584
174. Ma C, Peng Y, Li H, Chen W. Organ-on-a-Chip: A New Paradigm for Drug Development. Trends Pharmacol Sci. 2021;42:119-33
175. Jalili-Firoozinezhad S, Miranda CC, Cabral JMS. Modeling the Human Body on Microfluidic Chips. Trends Biotechnol. 2021;39:838-52
176. Lave T, Parrott N, Grimm HP, Fleury A, Reddy M. Challenges and opportunities with modelling and simulation in drug discovery and drug development. Xenobiotica. 2007;37:1295-310
177. Herland A, Maoz BM, Das D, Somayaji MR, Prantil-Baun R, Novak R. et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat Biomed Eng. 2020;4:421-36
178. Liu D, Jiao S, Wei J, Zhang X, Pei Y, Pei Z. et al. Investigation of absorption, metabolism and toxicity of ginsenosides compound K based on human organ chips. Int J Pharm. 2020;587:119669
179. Kuhnl J, Tao TP, Brandmair K, Gerlach S, Rings T, Muller-Vieira U. et al. Characterization of application scenario-dependent pharmacokinetics and pharmacodynamic properties of permethrin and hyperforin in a dynamic skin and liver multi-organ-chip model. Toxicology. 2021;448:152637
180. Milani N, Parrott N, Ortiz Franyuti D, Godoy P, Galetin A, Gertz M. et al. Application of a gut-liver-on-a-chip device and mechanistic modelling to the quantitative in vitro pharmacokinetic study of mycophenolate mofetil. Lab Chip. 2022;22:2853-68
181. Novak R, Ingram M, Marquez S, Das D, Delahanty A, Herland A. et al. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat Biomed Eng. 2020;4:407-20
182. Edington CD, Chen WLK, Geishecker E, Kassis T, Soenksen LR, Bhushan BM. et al. Interconnected Microphysiological Systems for Quantitative Biology and Pharmacology Studies. Sci Rep. 2018;8:4530
183. Tao T, Deng P, Wang Y, Zhang X, Guo Y, Chen W. et al. Microengineered Multi-Organoid System from hiPSCs to Recapitulate Human Liver-Islet Axis in Normal and Type 2 Diabetes. Adv Sci (Weinh). 2022;9:e2103495
184. Trapecar M, Communal C, Velazquez J, Maass CA, Huang YJ, Schneider K. et al. Gut-Liver Physiomimetics Reveal Paradoxical Modulation of IBD-Related Inflammation by Short-Chain Fatty Acids. Cell Syst. 2020;10:223-39 e9
185. Sontheimer-Phelps A, Hassell BA, Ingber DE. Modelling cancer in microfluidic human organs-on-chips. Nat Rev Cancer. 2019;19:65-81
186. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559-64
187. Liu W, Song J, Du X, Zhou Y, Li Y, Li R. et al. AKR1B10 (Aldo-keto reductase family 1 B10) promotes brain metastasis of lung cancer cells in a multi-organ microfluidic chip model. Acta Biomater. 2019;91:195-208
188. Xu Z, Li E, Guo Z, Yu R, Hao H, Xu Y. et al. Design and Construction of a Multi-Organ Microfluidic Chip Mimicking the in vivo Microenvironment of Lung Cancer Metastasis. ACS Appl Mater Interfaces. 2016;8:25840-7
189. Raimondi I, Izzo L, Tunesi M, Comar M, Albani D, Giordano C. Organ-On-A-Chip in vitro Models of the Brain and the Blood-Brain Barrier and Their Value to Study the Microbiota-Gut-Brain Axis in Neurodegeneration. Front Bioeng Biotechnol. 2019;7:435
190. Raimondi MT, Albani D, Giordano C. An Organ-On-A-Chip Engineered Platform to Study the Microbiota-Gut-Brain Axis in Neurodegeneration. Trends Mol Med. 2019;25:737-40
191. Wang YI, Carmona C, Hickman JJ, Shuler ML. Multiorgan Microphysiological Systems for Drug Development: Strategies, Advances, and Challenges. Adv Healthc Mater. 2018;7:1701000
192. Skardal A, Shupe T, Atala A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov Today. 2016;21:1399-411
193. Lee-Montiel FT, Laemmle A, Charwat V, Dumont L, Lee CS, Huebsch N. et al. Integrated Isogenic Human Induced Pluripotent Stem Cell-Based Liver and Heart Microphysiological Systems Predict Unsafe Drug-Drug Interaction. Front Pharmacol. 2021;12:667010
194. Wang T, Du Z, Zhu F, Cao Z, An Y, Gao Y. et al. Comorbidities and multi-organ injuries in the treatment of COVID-19. Lancet. 2020;395:e52
195. Banda M, McKim KL, Myers MB, Inoue M, Parsons BL. Outgrowth of erlotinib-resistant subpopulations recapitulated in patient-derived lung tumor spheroids and organoids. PLoS One. 2020;15:e0238862
196. Kankala RK, Wang SB, Chen AZ. Microengineered Organ-on-a-chip Platforms towards Personalized Medicine. Curr Pharm Des. 2018;24:5354-66
197. Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Mousavi Shaegh SA. et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc Natl Acad Sci U S A. 2017;114:E2293-E302
198. Hubner J, Raschke M, Rutschle I, Grassle S, Hasenberg T, Schirrmann K. et al. Simultaneous evaluation of anti-EGFR-induced tumour and adverse skin effects in a microfluidic human 3D co-culture model. Sci Rep. 2018;8:15010
199. Ramme AP, Koenig L, Hasenberg T, Schwenk C, Magauer C, Faust D. et al. Autologous induced pluripotent stem cell-derived four-organ-chip. Future Sci OA. 2019;5:FSO413
200. Criscione J, Rezaei Z, Hernandez Cantu CM, Murphy S, Shin SR, Kim DH. Heart-on-a-chip platforms and biosensor integration for disease modeling and phenotypic drug screening. Biosens Bioelectron. 2023;220:114840
201. Chen MB, Whisler JA, Frose J, Yu C, Shin Y, Kamm RD. On-chip human microvasculature assay for visualization and quantification of tumor cell extravasation dynamics. Nat Protoc. 2017;12:865-80
202. Azizgolshani H, Coppeta JR, Vedula EM, Marr EE, Cain BP, Luu RJ. et al. High-throughput organ-on-chip platform with integrated programmable fluid flow and real-time sensing for complex tissue models in drug development workflows. Lab Chip. 2021;21:1454-74
203. Gard AL, Luu RJ, Miller CR, Maloney R, Cain BP, Marr EE. et al. High-throughput human primary cell-based airway model for evaluating influenza, coronavirus, or other respiratory viruses in vitro. Sci Rep. 2021;11:14961
204. Trietsch SJ, Naumovska E, Kurek D, Setyawati MC, Vormann MK, Wilschut KJ. et al. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat Commun. 2017;8:262
205. Rajasekar S, Lin DSY, Abdul L, Liu A, Sotra A, Zhang F. et al. IFlowPlate-A Customized 384-Well Plate for the Culture of Perfusable Vascularized Colon Organoids. Adv Mater. 2020;32:e2002974
206. Vamathevan J, Clark D, Czodrowski P, Dunham I, Ferran E, Lee G. et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18:463-77
207. Paul D, Sanap G, Shenoy S, Kalyane D, Kalia K, Tekade RK. Artificial intelligence in drug discovery and development. Drug Discov Today. 2021;26:80-93
208. Mak KK, Pichika MR. Artificial intelligence in drug development: present status and future prospects. Drug Discov Today. 2019;24:773-80
209. Tripathi A, Misra K, Dhanuka R, Singh JP. Artificial Intelligence in Accelerating Drug Discovery and Development. Recent Pat Biotechnol. 2023;17:9-23
210. Chan HCS, Shan H, Dahoun T, Vogel H, Yuan S. Advancing Drug Discovery via Artificial Intelligence. Trends Pharmacol Sci. 2019;40:592-604
211. Pun FW, Ozerov IV, Zhavoronkov A. AI-powered therapeutic target discovery. Trends Pharmacol Sci. 2023 [Epub ahead of print]
212. Singh AV, Chandrasekar V, Paudel N, Laux P, Luch A, Gemmati D. et al. Integrative toxicogenomics: Advancing precision medicine and toxicology through artificial intelligence and OMICs technology. Biomed Pharmacother. 2023;163:114784
213. Zare Harofte S, Soltani M, Siavashy S, Raahemifar K. Recent Advances of Utilizing Artificial Intelligence in Lab on a Chip for Diagnosis and Treatment. Small. 2022;18:e2203169
214. Huang B, Huang H, Zhang S, Zhang D, Shi Q, Liu J. et al. Artificial intelligence in pancreatic cancer. Theranostics. 2022;12:6931-54
215. Zhang P, Fonnesbeck C, Schmidt DC, White J, Kleinberg S, Mulvaney SA. Using Momentary Assessment and Machine Learning to Identify Barriers to Self-management in Type 1 Diabetes: Observational Study. JMIR Mhealth Uhealth. 2022;10:e21959
216. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-44
217. Lin Q, Li T, Cao C, Cao Y, Man Z, Wang H. Deep learning based automated diagnosis of bone metastases with SPECT thoracic bone images. Sci Rep. 2021;11:4223
218. Ho TKK, Gwak J. Feature-level ensemble approach for COVID-19 detection using chest X-ray images. PLoS One. 2022;17:e0268430
219. Fink O, Wang Q, Svensén M, Dersin P, Lee W-J, Ducoffe M. Potential, challenges and future directions for deep learning in prognostics and health management applications. Engineering Applications of Artificial Intelligence. 2020;92:103678
220. Zhou X, Qu M, Tebon P, Jiang X, Wang C, Xue Y. et al. Screening Cancer Immunotherapy: When Engineering Approaches Meet Artificial Intelligence. Adv Sci (Weinh). 2020;7:2001447
221. Du X, Dua S. Segmentation of fluorescence microscopy cell images using unsupervised mining. Open Med Inform J. 2010;4:41-9
222. Rivenson Y, Wang H, Wei Z, de Haan K, Zhang Y, Wu Y. et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat Biomed Eng. 2019;3:466-77
223. Perez-Aliacar M, Doweidar MH, Doblare M, Ayensa-Jimenez J. Predicting cell behaviour parameters from glioblastoma on a chip images. A deep learning approach. Comput Biol Med. 2021;135:104547
224. Chong LH, Ching T, Farm HJ, Grenci G, Chiam KH, Toh YC. Integration of a microfluidic multicellular coculture array with machine learning analysis to predict adverse cutaneous drug reactions. Lab Chip. 2022;22:1890-904
225. Cascarano P, Comes MC, Mencattini A, Parrini MC, Piccolomini EL, Martinelli E. Recursive Deep Prior Video: A super resolution algorithm for time-lapse microscopy of organ-on-chip experiments. Med Image Anal. 2021;72:102124
226. Paek K, Kim S, Tak S, Kim MK, Park J, Chung S. et al. A high-throughput biomimetic bone-on-a-chip platform with artificial intelligence-assisted image analysis for osteoporosis drug testing. Bioengineering & Translational Medicine. 2022;8:e10313
227. Jena BP, Gatti DL, Arslanturk S, Pernal S, Taatjes DJ. Human skeletal muscle cell atlas: Unraveling cellular secrets utilizing 'muscle-on-a-chip', differential expansion microscopy, mass spectrometry, nanothermometry and machine learning. Micron. 2019;117:55-9
228. Mencattini A, Di Giuseppe D, Comes MC, Casti P, Corsi F, Bertani FR. et al. Discovering the hidden messages within cell trajectories using a deep learning approach for in vitro evaluation of cancer drug treatments. Sci Rep. 2020;10:7653
229. Chen Z, Ma N, Sun X, Li Q, Zeng Y, Chen F. et al. Automated evaluation of tumor spheroid behavior in 3D culture using deep learning-based recognition. Biomaterials. 2021;272:120770
230. Zhang Z, Chen L, Wang Y, Zhang T, Chen YC, Yoon E. Label-Free Estimation of Therapeutic Efficacy on 3D Cancer Spheres Using Convolutional Neural Network Image Analysis. Anal Chem. 2019;91:14093-100
231. Oliver CR, Westerhof TM, Castro MG, Merajver SD. Quantifying the Brain Metastatic Tumor Micro-Environment using an Organ-On-A Chip 3D Model, Machine Learning, and Confocal Tomography. J Vis Exp. 2020 162, e61654
232. Oliver CR, Altemus MA, Westerhof TM, Cheriyan H, Cheng X, Dziubinski M. et al. A platform for artificial intelligence based identification of the extravasation potential of cancer cells into the brain metastatic niche. Lab Chip. 2019;19:1162-73
233. Nguyen M, De Ninno A, Mencattini A, Mermet-Meillon F, Fornabaio G, Evans SS. et al. Dissecting Effects of Anti-cancer Drugs and Cancer-Associated Fibroblasts by On-Chip Reconstitution of Immunocompetent Tumor Microenvironments. Cell Rep. 2018;25:3884-93 e3
234. Biselli E, Agliari E, Barra A, Bertani FR, Gerardino A, De Ninno A. et al. Organs on chip approach: a tool to evaluate cancer -immune cells interactions. Sci Rep. 2017;7:12737
235. Comes MC, Casti P, Mencattini A, Di Giuseppe D, Mermet-Meillon F, De Ninno A. et al. The influence of spatial and temporal resolutions on the analysis of cell-cell interaction: a systematic study for time-lapse microscopy applications. Sci Rep. 2019;9:6789
236. Parlato S, De Ninno A, Molfetta R, Toschi E, Salerno D, Mencattini A. et al. 3D Microfluidic model for evaluating immunotherapy efficacy by tracking dendritic cell behaviour toward tumor cells. Sci Rep. 2017;7:1093
237. Mencattini A, Lansche C, Veith I, Erbs P, Balloul JM, Quemeneur E. et al. Direct imaging and automatic analysis in tumor-on-chip reveal cooperative antitumoral activity of immune cells and oncolytic vaccinia virus. Biosens Bioelectron. 2022;215:114571
238. Choi DH, Liu HW, Jung YH, Ahn J, Kim JA, Oh D. et al. Analyzing angiogenesis on a chip using deep learning-based image processing. Lab Chip. 2023;23:475-84
239. Urban G, Bache KM, Phan D, Sobrino A, Shmakov AK, Hachey SJ. et al. Deep Learning for Drug Discovery and Cancer Research: Automated Analysis of Vascularization Images. IEEE/ACM Trans Comput Biol Bioinform. 2019;16:1029-35
240. Young AT, Rivera KR, Erb PD, Daniele MA. Monitoring of Microphysiological Systems: Integrating Sensors and Real-Time Data Analysis toward Autonomous Decision-Making. ACS Sens. 2019;4:1454-64
241. Sabate Del Rio J, Ro J, Yoon H, Park TE, Cho YK. Integrated technologies for continuous monitoring of organs-on-chips: Current challenges and potential solutions. Biosens Bioelectron. 2023;224:115057
242. Ma S, Zhao H, Galan EA. Integrating Engineering, Automation, and Intelligence to Catalyze the Biomedical Translation of Organoids. Adv Biol (Weinh). 2021;5:e2100535
243. Tabatabaei Rezaei N, Kumar H, Liu H, Lee SS, Park SS, Kim K. Recent Advances in Organ-on-Chips Integrated with Bioprinting Technologies for Drug Screening. Adv Healthc Mater. 2023: e2203172.
244. Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan H, Yadid M. et al. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat Mater. 2017;16:303-8
245. Lee RY, Wu Y, Goh D, Tan V, Ng CW, Lim JCT. et al. Application of Artificial Intelligence to In Vitro Tumor Modeling and Characterization of the Tumor Microenvironment. Adv Healthc Mater. 2023: e2202457.
246. Yu C, Jiang J. A Perspective on Using Machine Learning in 3D Bioprinting. Int J Bioprint. 2020;6:253
247. Wang J, Liang K, Zhang N, Yao H, Ho TY, Sun L. Automated calibration of 3D-printed microfluidic devices based on computer vision. Biomicrofluidics. 2021;15:024102
248. Dressler OJ, Howes PD, Choo J, deMello AJ. Reinforcement Learning for Dynamic Microfluidic Control. ACS Omega. 2018;3:10084-91
249. Tronolone JJ, Mathur T, Chaftari CP, Jain A. Evaluation of the Morphological and Biological Functions of Vascularized Microphysiological Systems with Supervised Machine Learning. Ann Biomed Eng. 2023;51:1723-1737
250. Schuster B, Junkin M, Kashaf SS, Romero-Calvo I, Kirby K, Matthews J. et al. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat Commun. 2020;11:5271
251. Ashammakhi N, Nasiri R, Barros NR, Tebon P, Thakor J, Goudie M. et al. Gut-on-a-chip: Current progress and future opportunities. Biomaterials. 2020;255:120196
252. Sakolish C, Weber EJ, Kelly EJ, Himmelfarb J, Mouneimne R, Grimm FA. et al. Technology Transfer of the Microphysiological Systems: A Case Study of the Human Proximal Tubule Tissue Chip. Sci Rep. 2018;8:14882
253. Moutinho S. Researchers and regulators plan for a future without lab animals. Nat Med. 2023 [Epub ahead of print]
254. Rumsey JW, Lorance C, Jackson M, Sasserath T, McAleer CW, Long CJ. et al. Classical Complement Pathway Inhibition in a "Human-On-A-Chip" Model of Autoimmune Demyelinating Neuropathies. Adv Ther (Weinh). 2022;5:2200030
255. Thacker VV, Sharma K, Dhar N, Mancini GF, Sordet-Dessimoz J, McKinney JD. Rapid endotheliitis and vascular damage characterize SARS-CoV-2 infection in a human lung-on-chip model. EMBO Rep. 2021;22:e52744
256. Guo Y, Luo R, Wang Y, Deng P, Song T, Zhang M. et al. SARS-CoV-2 induced intestinal responses with a biomimetic human gut-on-chip. Sci Bull (Beijing). 2021;66:783-93
257. Si L, Bai H, Oh CY, Jin L, Prantil-Baun R, Ingber DE. Clinically Relevant Influenza Virus Evolution Reconstituted in a Human Lung Airway-on-a-Chip. Microbiol Spectr. 2021;9:e0025721
258. Zhang M, Wang P, Luo R, Wang Y, Li Z, Guo Y. et al. Biomimetic Human Disease Model of SARS-CoV-2-Induced Lung Injury and Immune Responses on Organ Chip System. Adv Sci (Weinh). 2021;8:2002928
259. Si L, Bai H, Oh CY, Jiang A, Hong F, Zhang T. et al. Self-assembling short immunostimulatory duplex RNAs with broad-spectrum antiviral activity. Mol Ther Nucleic Acids. 2022;29:923-40
260. Bai H, Si L, Jiang A, Belgur C, Zhai Y, Plebani R. et al. Mechanical control of innate immune responses against viral infection revealed in a human lung alveolus chip. Nat Commun. 2022;13:1928
261. Li H, Wei W, Xu H. Drug discovery is an eternal challenge for the biomedical sciences. Acta Materia Medica. 2022;1:1-3
262. Toepke MW, Beebe DJ. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip. 2006;6:1484-6
263. van Meer BJ, de Vries H, Firth KSA, van Weerd J, Tertoolen LGJ, Karperien HBJ. et al. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem Biophys Res Commun. 2017;482:323-8
264. Nahak BK, Mishra A, Preetam S, Tiwari A. Advances in Organ-on-a-Chip Materials and Devices. ACS Appl Bio Mater. 2022;5:3576-607
265. Li ZA, Sant S, Cho SK, Goodman SB, Bunnell BA, Tuan RS. et al. Synovial joint-on-a-chip for modeling arthritis: progress, pitfalls, and potential. Trends Biotechnol. 2022;41:511-527
266. Weisgrab G, Ovsianikov A, Costa PF. Functional 3D Printing for Microfluidic Chips. Advanced Materials Technologies. 2019;4:1900275
267. Ho CM, Ng SH, Li KH, Yoon YJ. 3D printed microfluidics for biological applications. Lab Chip. 2015;15:3627-37
268. Rothbauer M, Eilenberger C, Spitz S, Bachmann BEM, Kratz SRA, Reihs EI. et al. Recent Advances in Additive Manufacturing and 3D Bioprinting for Organs-On-A-Chip and Microphysiological Systems. Front Bioeng Biotechnol. 2022;10:837087
269. Park D, Lee J, Chung JJ, Jung Y, Kim SH. Integrating Organs-on-Chips: Multiplexing, Scaling, Vascularization, and Innervation. Trends Biotechnol. 2020;38:99-112
270. Escriba R, Ferrer-Lorente R, Raya A. Inborn errors of metabolism: Lessons from iPSC models. Rev Endocr Metab Disord. 2021;22:1189-200
271. Dellaquila A, Le Bao C, Letourneur D, Simon-Yarza T. In Vitro Strategies to Vascularize 3D Physiologically Relevant Models. Adv Sci (Weinh). 2021;8:e2100798
272. Carter RE, Attia ZI, Lopez-Jimenez F, Friedman PA. Pragmatic considerations for fostering reproducible research in artificial intelligence. NPJ Digit Med. 2019;2:42
273. Moving towards reproducible machine learning. Nat Comput Sci. 2021;1:629-30
274. Ching T, Toh YC, Hashimoto M, Zhang YS. Bridging the academia-to-industry gap: organ-on-a-chip platforms for safety and toxicology assessment. Trends Pharmacol Sci. 2021;42:715-28
275. Cai H, Ao Z, Tian C, Wu Z, Kaurich C, Chen Z. et al. Engineering human spinal microphysiological systems to model opioid-induced tolerance. Bioact Mater. 2023;22:482-90
276. Mondadori C, Palombella S, Salehi S, Talo G, Visone R, Rasponi M. et al. Recapitulating monocyte extravasation to the synovium in an organotypic microfluidic model of the articular joint. Biofabrication. 2021;13:045001
277. Blundell C, Yi YS, Ma L, Tess ER, Farrell MJ, Georgescu A. et al. Placental Drug Transport-on-a-Chip: A Microengineered In Vitro Model of Transporter-Mediated Drug Efflux in the Human Placental Barrier. Adv Healthc Mater. 2018;7:1700786
278. Shik Mun K, Arora K, Huang Y, Yang F, Yarlagadda S, Ramananda Y. et al. Patient-derived pancreas-on-a-chip to model cystic fibrosis-related disorders. Nat Commun. 2019;10:3124
279. Franca CM, Tahayeri A, Rodrigues NS, Ferdosian S, Puppin Rontani RM, Sereda G. et al. The tooth on-a-chip: a microphysiologic model system mimicking the biologic interface of the tooth with biomaterials. Lab Chip. 2020;20:405-13
280. Ahn J, Yoon MJ, Hong SH, Cha H, Lee D, Koo HS. et al. Three-dimensional microengineered vascularised endometrium-on-a-chip. Hum Reprod. 2021;36:2720-31
281. Tsamandouras N, Chen WLK, Edington CD, Stokes CL, Griffith LG, Cirit M. Integrated Gut and Liver Microphysiological Systems for Quantitative In Vitro Pharmacokinetic Studies. AAPS J. 2017;19:1499-512
282. Baert Y, Ruetschle I, Cools W, Oehme A, Lorenz A, Marx U. et al. A multi-organ-chip co-culture of liver and testis equivalents: a first step toward a systemic male reprotoxicity model. Hum Reprod. 2020;35:1029-44
283. Maschmeyer I, Hasenberg T, Jaenicke A, Lindner M, Lorenz AK, Zech J. et al. Chip-based human liver-intestine and liver-skin co-cultures-A first step toward systemic repeated dose substance testing in vitro. Eur J Pharm Biopharm. 2015;95:77-87
284. Theobald J, Abu El Maaty MA, Kusterer N, Wetterauer B, Wink M, Cheng X. et al. In vitro metabolic activation of vitamin D3 by using a multi-compartment microfluidic liver-kidney organ on chip platform. Sci Rep. 2019;9:4616
285. Skardal A, Murphy SV, Devarasetty M, Mead I, Kang HW, Seol YJ. et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci Rep. 2017;7:8837
286. Sasserath T, Rumsey JW, McAleer CW, Bridges LR, Long CJ, Elbrecht D. et al. Differential Monocyte Actuation in a Three-Organ Functional Innate Immune System-on-a-Chip. Adv Sci (Weinh). 2020;7:2000323
287. Aleman J, Skardal A. A multi-site metastasis-on-a-chip microphysiological system for assessing metastatic preference of cancer cells. Biotechnol Bioeng. 2019;116:936-44
288. Miller PG, Chen CY, Wang YI, Gao E, Shuler ML. Multiorgan microfluidic platform with breathable lung chamber for inhalation or intravenous drug screening and development. Biotechnol Bioeng. 2020;117:486-97
289. Kimura H, Ikeda T, Nakayama H, Sakai Y, Fujii T. An on-chip small intestine-liver model for pharmacokinetic studies. J Lab Autom. 2015;20:265-73
290. Satoh T, Sugiura S, Shin K, Onuki-Nagasaki R, Ishida S, Kikuchi K. et al. A multi-throughput multi-organ-on-a-chip system on a plate formatted pneumatic pressure-driven medium circulation platform. Lab Chip. 2017;18:115-25
291. Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA. et al. Functional Coupling of Human Microphysiology Systems: Intestine, Liver, Kidney Proximal Tubule, Blood-Brain Barrier and Skeletal Muscle. Sci Rep. 2017;7:42296
292. McAleer CW, Long CJ, Elbrecht D, Sasserath T, Bridges LR, Rumsey JW. et al. Multi-organ system for the evaluation of efficacy and off-target toxicity of anticancer therapeutics. Sci Transl Med. 2019;11:eaav1386
293. Skardal A, Aleman J, Forsythe S, Rajan S, Murphy S, Devarasetty M. et al. Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication. 2020;12:025017
294. Rajan SAP, Aleman J, Wan M, Pourhabibi Zarandi N, Nzou G, Murphy S. et al. Probing prodrug metabolism and reciprocal toxicity with an integrated and humanized multi-tissue organ-on-a-chip platform. Acta Biomater. 2020;106:124-35
295. Wang X, Cirit M, Wishnok JS, Griffith LG, Tannenbaum SR. Analysis of an Integrated Human Multiorgan Microphysiological System for Combined Tolcapone Metabolism and Brain Metabolomics. Anal Chem. 2019;91:8667-75
Author contact
![]() Corresponding author: Peng Chen (chenpengac.cn); Hongjun Yang (hongjun0420sina.com); Zheng Fu (fuzhengtech).
Corresponding author: Peng Chen (chenpengac.cn); Hongjun Yang (hongjun0420sina.com); Zheng Fu (fuzhengtech).
 Global reach, higher impact
Global reach, higher impact