13.3
Impact Factor
Theranostics 2023; 13(12):4247-4265. doi:10.7150/thno.86528 This issue Cite
Review
Regulatory mechanisms and therapeutic implications of insulin-like growth factor 2 mRNA-binding proteins, the emerging crucial m6A regulators of tumors
1. Department of Surgical Oncology and General Surgery, Key Laboratory of Precision Diagnosis and Treatment of Gastrointestinal Tumors, Ministry of Education, The First Hospital of China Medical University, Shenyang, Liaoning 110001, People's Republic of China.
2. Department of Anesthesiology, The First Hospital of China Medical University, Shenyang, Liaoning 110001, People's Republic of China.
3. Department of Plastic Surgery, The First Hospital of China Medical University, Shenyang, Liaoning 110001, People's Republic of China.
4. Department of Endoscopy, The First Hospital of China Medical University, Shenyang, Liaoning 110001, People's Republic of China.
5. Department of Gynecology and Obstetrics, Shengjing Hospital of China Medical University, Shenyang, Liaoning 110001, People's Republic of China.
6. Department of Pharmacology, School of Pharmacy, China Medical University, Shenyang, 110122, People's Republic of China.
7. Liaoning Key Laboratory of Molecular Targeted Anti-Tumor Drug Development and Evaluation; Liaoning Cancer Immune Peptide Drug Engineering Technology Research Center; Key Laboratory of Precision Diagnosis and Treatment of Gastrointestinal Tumors, Ministry of Education; China Medical University, Shenyang, 110122, People's Republic of China.
8. Shenyang Kangwei Medical Laboratory Analysis Co. LTD, Liaoning Province, China.
#These authors contributed equally to this work: Heng Zhou, Qiang Sun, Mingliang Feng.
Received 2023-5-26; Accepted 2023-7-17; Published 2023-7-24
Abstract
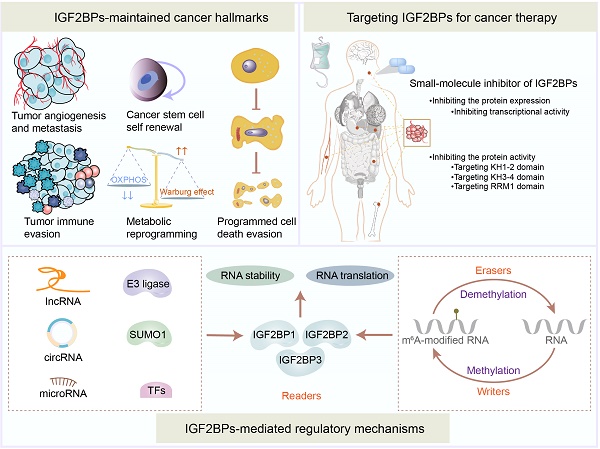
Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) serve essential biological functions as post-transcriptional performers, participating in the acquisition or maintenance of tumor hallmarks due to their distinct protein structures. Emerging evidence indicates that IGF2BPs belong to the class III type of RNA N6-methyladenosine (m6A) modification readers, controlling RNA stability, storage, localization, metabolism, and translation in multiple vital bioprocesses, particularly tumorigenesis and tumor progression. Here, we discuss the underlying regulatory mechanisms and pathological functions of IGF2BPs which act as m6A readers in the context of tumor pathogenesis and multidrug resistance. Furthermore, we highlight the potential of IGF2BPs as drug targets in clinical tumor treatment. Hence, precise and novel tumor therapeutic approaches could be uncovered by targeting epigenetic heterogeneity.
Keywords: Cancer, N6-methyladenosine, IGF2BPs, Reader, Cancer therapy
Introduction
Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs, IMPs), including IGF2BP1/2/3, are localized inside the nucleus as well as the cytoplasm. These proteins comprise two RNA recognition motifs (RRMs) and four K homology (KH) domains. In 1999, IGF2BP1/2/3 were first identified as a group of proteins that could potently bind to IGF2 leader 3 (IGF-II-L3) mRNA [1]. To date, IGF2BPs have been found to be dysregulated in at least 15 types of tumors, such as gastric cancer (GC), colorectal cancer (CRC), and breast cancer, etc. Initially, IGF2BPs were identified by their ability to bind in vitro transcribed (unmodified) RNAs. In recent years, IGF2BPs have been found to also recognize the m6A modification on mRNAs and non-coding RNAs (ncRNAs), promoting their stability or translation and regulating the acquisition and maintenance of tumor hallmarks, such as cell proliferation [2], metabolic reprogramming [3], and immune evasion [4], etc. Diverse regulatory mechanisms, such as ncRNAs, transcription factors (TFs), and post-translational modifications (PTMs), manipulate the dysregulation of IGF2BPs in tumor initiation and development. Moreover, IGF2BPs have been implicated in the development of multi-drug resistance during tumor therapy [5] and can serve as prognostic predictors for the clinical outcomes of patients with tumors [6]. Given the spectrum of oncogenes that these proteins can bind to, developing pharmaceutical inhibitors of IGF2BPs, including small-molecule inhibitors, represents a significant therapeutic avenue.
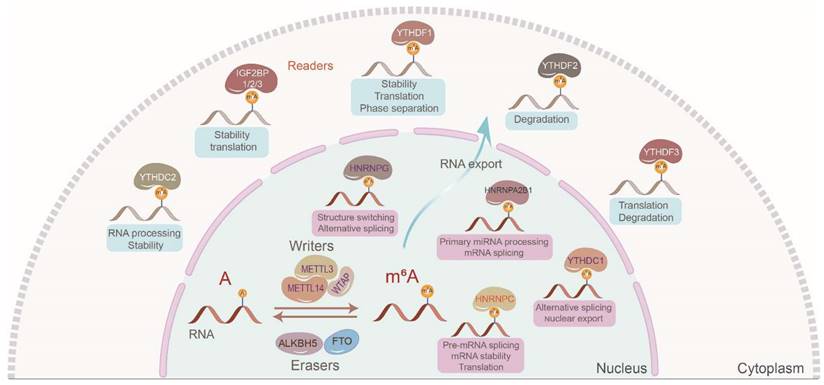
In 2018, IGF2BPs were identified as a novel class III family of m6A readers governing the fate of downstream target RNAs in an m6A-dependent manner [7]. Of the more than 170 types of modifications identified in mRNAs and ncRNAs, m6A is the most abundant in eukaryotes [8]. The m6A modification mostly occurs in the 3′ untranslated region (3′ UTR) and the vicinity of stop codons in target RNAs with a typical consensus sequence of “RRACH” (R = G or A; H = A, C, or U). The canonical m6A regulatory process is dynamic and reversible. Generally, m6A writers including methyltransferase 3 (METTL3), methyltransferase 14 (METTL14), Wilms tumor 1-associated protein (WTAP), etc., m6A readers including IGF2BPs, heterogeneous nuclear ribonucleoproteins (hnRNPs), YT521-B homology (YTH) domain family, etc., and m6A erasers including fat mass and obesity-associated protein (FTO), alkB homolog 5 (ALKBH5), etc., create, recognize, and demolish m6A modifications on target RNAs, respectively [9, 10] (Figure 1). However, we should notice that very few adenosines are likely to be methylated in most RNAs, even in the 3'UTR of RNAs. Additionally, we should also notice that it is difficult to confirm the percent of the RNAs which are actually methylated among those RNAs that can be methylated.
Being novel m6A readers, IGF2BPs can control RNA stability, storage, localization, metabolism, and translation in multiple vital bioprocesses. Herein, we have outlined in detail the structures, functions, mechanisms, and therapeutic potential of IGF2BPs in oncology. This review provides insights into the m6A role of IGF2BPs in cancers and helps identify novel therapeutic strategies.
Subcellular localization and structure of IGF2BPs
In humans, the subcellular localization of IGF2BPs ultimately determines their function and the fate of their target RNAs. IGF2BPs are predominantly observed in the cytoplasm, where they 'cage' target mRNAs and ncRNAs within cytoplasmic ribonucleoprotein complexes (RNPs). In the cytoplasm, IGF2BPs modulate a spectrum of RNA processes, including localization, translation, stability, and metabolism, primarily through granule-like messenger ribonucleoprotein structures located in the perinuclear region [9] (Figure 2A). In contrast, IGF2BPs have only marginally been detected in the nucleus [11].
The canonical regulatory process of m6A modification. The dynamic and reversible m6A modification mostly occurs in the 3′ untranslated region (3′ UTR) and the vicinity of stop codons in target RNAs with a typical consensus sequence of “RRACH” (R = G or A; H = A, C, or U). m6A writers mainly include METTL3, METTL14, and WTAP which install the m6A marks. m6A erasers involving FTO and ALKBH5 demolish m6A marks. m6A readers mainly including IGF2BPs, hnRNPs, and YT521-B homology domain family recognize the m6A marks to manipulate localization, decay, stability, and translation of target RNAs.
Subcellular localization and structure of IGF2BPs. (A) IGF2BPs are predominantly observed in the cytoplasm, where they 'cage' target mRNAs and ncRNAs within cytoplasmic RNPs. And they modulate a spectrum of RNA processes, including translation and stability, primarily through granule-like messenger ribonucleoprotein structures located in the perinuclear region. (B) IGF2BPs are highly conserved RNA-binding proteins that consist of two RRMs in the N-terminal region and four heterogeneous nuclear ribonucleoprotein KH domains in the C-terminal region. (C) The amino acid sequence of the IGF2BPs has been validated with over 56% similarity. Furthermore, IGF2BP1 and IGF2BP3 exhibit the closest similarity with 73% amino acid sequence identity. The sequence similarity of IGF2BPs is highlighted in red.
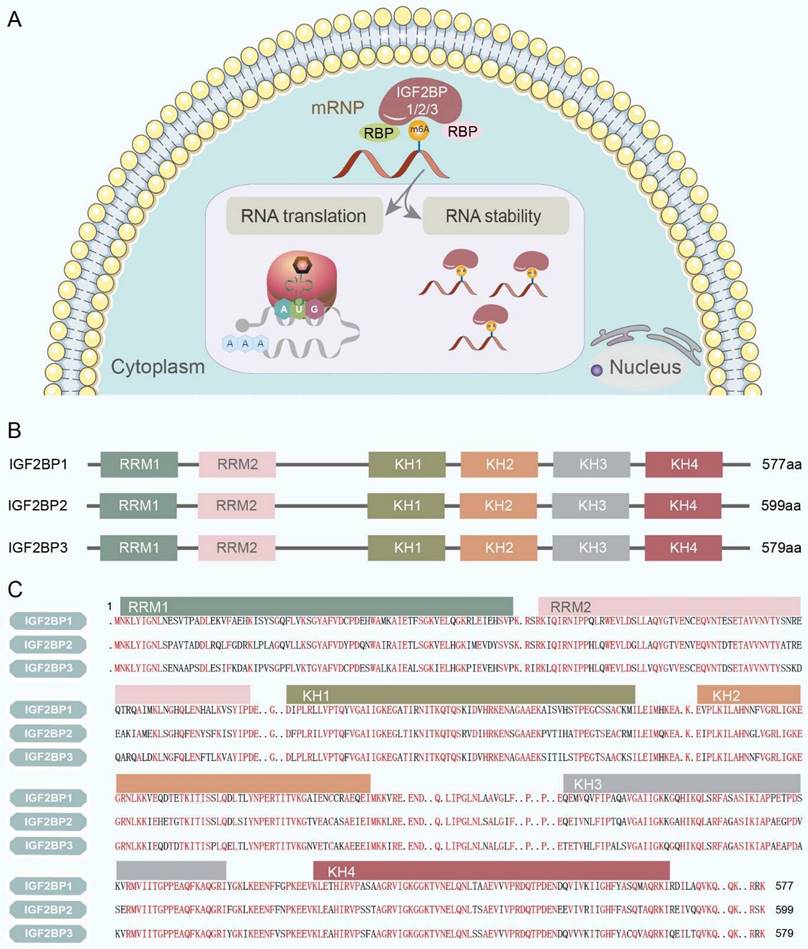
Structurally, IGF2BPs are highly conserved RNA-binding proteins having two RRMs in the N-terminal region and four KH domains in the C-terminal region (Figure 2B). In mammals, their molecular weights range from 58-66 kDa [12]. The amino acid sequences of the IGF2BPs have been validated with a similarity of over 56%. Furthermore, IGF2BP1 and IGF2BP3 exhibit the closest sequence identity (73%; Figure 2C). This extent of sequence similarity among IGF2BPs indicates that they perform analogical biochemical functions, including binding RNA, irrespective of the organism, tissue, or cell type [9].
In vitro studies have demonstrated that the KH domains primarily administrate RNA binding, whereas the RRM domains stabilize the IGF2BP-RNA complexes, leading to a prolonged half-life [13]. Structural analysis has illustrated that the KH3 and KH4 domains of IGF2BP1 assume an anti-parallel pseudo-dimer conformation in which they contact the target RNA and are responsible for the transport and local translation of β-actin mRNA [14] and many other RNAs. Moreover, KH4 is found to identify a non-canonical “GGA” sequence by means of a broadened and dynamic hydrophobic groove, while KH3 binds to a central “CA” sequence with weak nucleotide discrimination [15]. Relying on the representative KH3-4 domain, IGF2BPs can stabilize mRNAs; for example, overexpressed IGF2BP1 is shown to recognize the m6A sites in the 3' UTR of pyruvate kinase M1/2 (PKM2) via the KH3-4 domain to promote cancer progression[16]. Besides mRNAs, IGF2BPs can interact with ncRNAs through the KH3-4 domain; for instance, the KH3-4 domain of IGF2BP2 interacts with the “CAUCAU” m6A sequence motif at the exon 5-exon 4 junction of circNSUN2 to promote colorectal liver metastasis [17]. Moreover, IGF2BP2 activates the Warburg effect in CRC by recognizing the m6A modification on the long ncRNA (lncRNA) ZFAS1 via the KH3-4 domain [18]. The roles of the KH1-2 and RRM1-2 domains in RNA recognition are not clearly understood yet. A single recent study has revealed that IGF2BP1 fortifies the stability of its target RNA by binding to it via its KH1-2 domain. Specifically, IGF2BP1, through its KH1-2 domain, recognizes the METTL3-installed m6A on lncRNA ABL to maintain its stability and further inhibits apoptosome assembly and caspase‐9/3 activation in GC [19]. These findings hint that the functions performed by the KH1-2 and KH3-4 domains may be identical.
Considering the unique biochemical functions of the IGF2BP domains, structure-based screening via docking to the KH domains may be a feasible strategy to identify potential IGF2BP inhibitors.
The biological functions of IGF2BPs in tumors
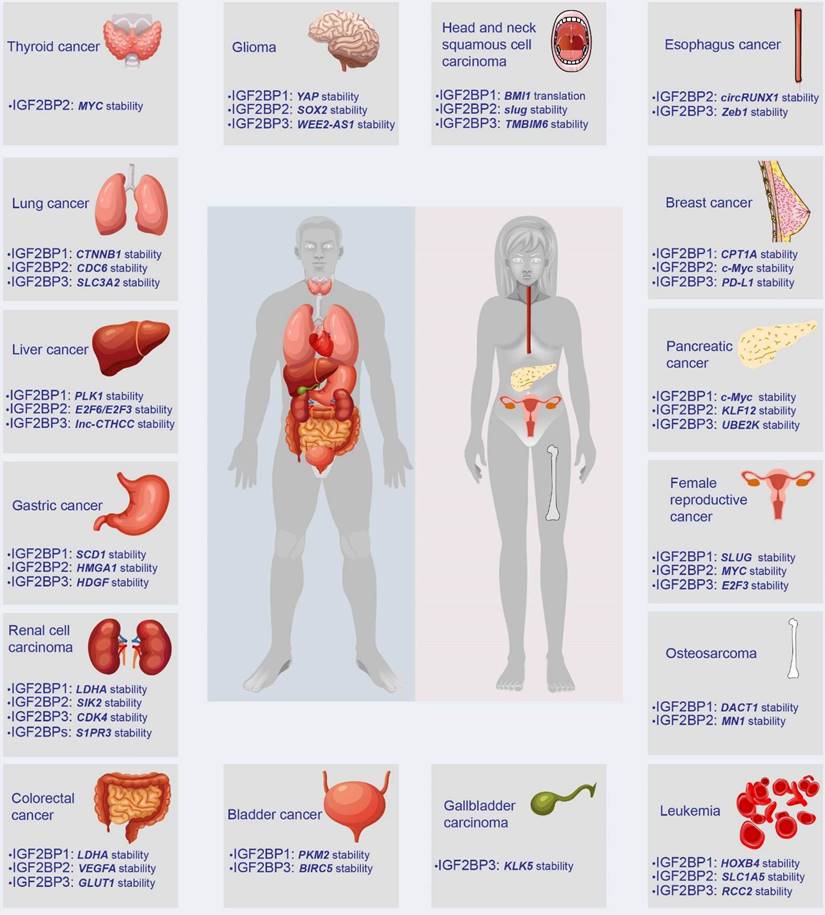
IGF2BPs are observed in most tissues during embryogenesis. Distinguishingly, IGF2BP2 is ubiquitously expressed in normal adult and tumor tissues, whereas IGF2BP1 and IGF2BP3 are de novo synthesized during numerous malignancies, earning themselves the label of bona fide oncofetal proteins. However, IGF2BP1 has also been found to play a tumor-suppressive role in multiple cancers. For instance, decreasing IGF2BP1 enhances the abilities of proliferation and migration of metastatic breast cancer cells [20]. Moreover, loss of IGF2BP1 promotes cell proliferation of leukemia [21]. Additionally, the knockdown of stromal IGF2BP1 facilitates a carcinogenic microenvironment and increases the histologic grade of colitis-associated cancer [22]. This contradiction remains confusing. As oncogenes, IGF2BPs have been reported in various tumors, including breast cancer, GC, and CRC, among others (Figure 3). Accumulating evidence has linked IGF2BPs with tumorigenesis, tumor progression, the establishment of tumor cell hierarchy, and poor prognosis (Table 1). Notably, loss- and gain-of-function models have validated the effect of IGF2BPs on aggressive phenotypes of cancer, robustly evidencing their status as oncogenes influencing cancer cell self-renewal, angiogenesis, apoptosis, metabolic reprogramming, immune evasion, etc. (Figure 4).
Cancer stem cell self-renewal
Self-renewal is a process of division to produce sufficient stem cells that can last throughout life. Remarkably, IGF2BPs play an indispensable role in facilitating the self-renewal of cancer stem cells. Of note, MYC, one of the most frequently activated oncogenes which contribute to the self-renewal of cancer stem cells, has been validated to be a significant target of IGF2BPs [7]. In vivo and in vitro assays have illustrated that IGF2BPs can enhance the recognition of m6A on MYC in CRC, triple-negative breast cancer, GC, acute myeloid leukemia (AML), etc. [23-27]. Specifically, IGF2BP1 maintains the stability of MYC by binding to the coding region determinant of m6A-modified MYC in a hypoxic microenvironment, thereby promoting the self-renewal of breast cancer stem cells [28]. Moreover, the expression of SRY-box transcription factor 2 (SOX2), also essential for the self-renewal of cancer stem cells, is found to be governed by IGF2BPs. IGF2BP1 binds to the m6A sites in the 3′ UTR of SOX2 to suppress its decay in endometrial cancer (EC) [29]. Likewise, IGF2BP2 recognizes the coding sequence (CDS) of SOX2 to prevent its degradation in CRC, thus reinforcing the self-renewal of stem cells [30]. The IGF2BP3-mediated snail family transcriptional repressor 2 (SLUG) activates SOX2 transcription, contributing to the self-renewal of triple-negative breast cancer cells [31]. By maintaining the stability of E2F transcription factor 6 (E2F6)/ E2F transcription factor 3 (E2F3), IGF2BP2 facilitates the stem cell self-renewal of liver cancer [32].
The expression and target genes of IGF2BPs in human tumors. In humans, IGF2BPs are overexpressed in at least 15 types of tumors, such as GC, CRC, liver cancer, and breast cancer. IGF2BPs promote the stability or translation of downstream target RNAs, including mRNA, lncRNA, and circRNA.
Besides managing cancer stemness-related genes, IGF2BPs also participate in cancer stemness-related signaling pathways, such as Wnt/β-catenin, Hedgehog, and Hippo signaling, to promote the self-renewal of cancer stem cells. Activation of the Wnt/β-catenin pathway is shown to sustain the self-renewal of stem cells [33], while Dishevelled-binding antagonist of β-catenin 1 (DACT1) is confirmed to negatively regulate this pathway [34]. Interestingly, IGF2BP1 fails to stabilize DACT1 after the m6A modification is eliminated by FTO, leading to the downregulation of DACT1 and further stimulation of the Wnt/β-catenin pathway in osteosarcoma [35]. Being a target of Wnt/β-catenin pathway, IGF2BP1 stabilizes GLI family zinc finger 1 (GLI1) to activate Hedgehog signaling in CRC and basal cell carcinoma cells [36, 37]. Additionally, overexpression of IGF2BP3 inhibits Hippo signaling in a de-ubiquitination-dependent manner in gallbladder cancer [38].
Thus, IGF2BPs promote cancer stem cell self-renewal which accounts for cancer initiation, progression, recurrence, and metastasis, via related oncogenes and pathways. Targeting these proteins may provide a promising intervention to thwart the self-renewal capacity of cancer stem cells.
Tumor metastasis
Metastasis is the most fatal manifestation of tumors. Of note, IGF2BPs can induce tumor metastasis via regulating tumor angiogenesis, epithelial-mesenchymal transition (EMT), etc. Tumor angiogenesis has been considered an effective therapeutic target for cancers because it critically supports tumor growth and metastasis by supplying urgent requirements of oxygen and nutrients. It is primarily driven by the vascular endothelial growth factor A (VEGFA)-related signal transduction pathway. Strikingly, IGF2BPs have been shown to promote tumor angiogenesis by stabilizing VEGFA. For instance, IGF2BP3 directly enhances the stability of m6A-modified VEGFA to promote angiogenesis in colon cancer [39]. It also indirectly increases VEGFA expression by stabilizing apoptosis inhibitor Survivin (BIRC5), thus contributing to angiogenesis in bladder cancer [40]. Similarly, IGF2BP2/3 manipulate the expression of ephrin type-A receptor 2 (EphA2) and VEGFA to facilitate vasculogenic mimicry in CRC [41]. In addition to VEGFA, IGF2BP3 directly binds to and stabilizes hypoxia inducible factor 1 subunit alpha (HIF1A) to promote angiogenesis in GC [42]. Likewise, IGF2BP2 indirectly regulates HIF1A and matrix metalloproteinase 14 (MMP14) to form vasculogenic mimicry in glioma [43]. Additionally, IGF2BP3 consolidates the stability of hepatoma-derived growth factor (HDGF), and this secreted protein accelerates angiogenesis in GC [44].
IGF2BPs also induce EMT that stimulates metastasis. In intrahepatic cholangiocarcinoma, IGF2BP1 stimulates the AKT/matrix metalloproteinase 2 (MMP2) signaling pathway to facilitate tumor metastasis in an m6A-dependent manner [45]. Furthermore, IGF2BP2 increases the stability of SLUG mRNA which is an EMT-related transcriptional factor via binding to its m6A site in the CDS, leading to lymphatic metastasis in head and neck squamous carcinoma (HNSCC) [46]. Additionally, IGF2BP3-mediated m6A modification stabilizes minichromosome maintenance complex component (MCM5) mRNAs and activates the Notch signaling, leading to EMT and metastasis in lung adenocarcinoma [47].
The expression, function, and prognosis of IGF2BPs in cancers
| IGF2BPs | Cancer type | Expression | Prognosis | Target genes or pathways | Biological function | Ref | |
|---|---|---|---|---|---|---|---|
| IGF2BP1 | Bladder cancer | Upregulated | Unfavorable | MYC and FSCN1 | Promote cell proliferation, migration, and invasion. | [84] | |
| Intrahepatic cholangiocarcinoma | Upregulated | Unfavorable | c-Myc and ZIC2 | Promote tumor growth and metastasis, inhibit senescence. | [45] | ||
| Breast cancer | Upregulated | Unfavorable | CPT1A | Promote tumor metastasis. | [126] | ||
| Gastric cancer | Upregulated | Unfavorable | c-Myc | Accelerate tumor aerobic glycolysis. | [127] | ||
| Colorectal cancer | Upregulated | Unfavorable | LDHA | Promote glucose metabolism. | [64] | ||
| Lung cancer | Upregulated | Unknown | LIN28B | Regulate cell cycle, DNA damage repair, and genome instability. Promote tumor proliferation and metastasis. | [128] | ||
| Liver cancer | Upregulated | Unfavorable | PLK1 | Promote tumor cell proliferation. | [49] | ||
| Glioblastoma | Upregulated | Unfavorable | YAP | Promote stemness, sphere formation, and tumorigenicity. | [102] | ||
| Endometrial cancer | Upregulated | Unfavorable | PEG10 | Accelerate tumor cell proliferation and cell cycle progression. | [129] | ||
| IGF2BP2 | AML | Upregulated | Unfavorable | MYC, GPT2, and SLC1A5 | Promote tumor initiation/progression, stem cell self-renewal, and amino acid metabolism. | [122] | |
| ESCC | Upregulated | Unknown | circRUNX1 | Promote tumor growth and metastasis. | [130] | ||
| HNSCC | Upregulated | Unfavorable | Slug | Promote lymphatic metastasis and epithelial-mesenchymal transition. | [46] | ||
| Colorectal cancer | Upregulated | Unfavorable | ZFAS1 | Activate the Warburg effect. | [18] | ||
| Glioma | Upregulated | Unknown | OIP5-AS1 | Promote vasculogenic mimicry. | [43] | ||
| Cervical cancer | Upregulated | Unknown | MYC | Accelerate tumor aerobic glycolysis. | [62] | ||
| Pancreatic cancer | Upregulated | Unfavorable | PI3K-Akt signaling | Promote tumor cell proliferation. | [107] | ||
| Gastric cancer | Upregulated | Unfavorable | HMGA1 | Promote epithelial to mesenchymal transition and metastasis. | [131] | ||
| Liver cancer | Upregulated | Unknown | FEN1 | Promote tumor growth. | [132] | ||
| IGF2BP3 | Lung cancer | Upregulated | Unfavorable | GPX4, SLC3A2, ACSL3, and FTH1 | Suppress ferroptosis. | [57] | |
| Gastric cancer | Upregulated | Unfavorable | SLC7A5 | Promote tumor proliferation. | [80] | ||
| Nasopharyngeal carcinoma | Upregulated | Unfavorable | KPNA2 | promote proliferation and metastasis. | [104] | ||
| AML | Upregulated | Unfavorable | RCC2 | promote proliferation and tumorigenesis. | [52] | ||
| EC | Upregulated | Unfavorable | E2F3 | Promote tumor growth. | [133] | ||
| Colon cancer | Upregulated | Unfavorable | CCND1 and VEGF | Accelerate cell cycle and angiogenesis. | [39] | ||
| IGF2BPs | Renal cell cancer | Upregulated | Unfavorable | S1PR3 | Drive tumorigenesis and metastasis. | [100] |
The biological functions of IGF2BPs in tumors. IGF2BPs participate in the acquisition or maintenance of tumor hallmarks, including cancer cell self-renewal, tumor angiogenesis, metabolic reprogramming, immune evasion, etc. IGF2BPs are involved in cancer cell self-renewal via managing cancer stemness-related genes and pathways. IGF2BPs induce tumor angiogenesis via stabilizing VEGFA, HIF1A, and HDGF. IGF2BPs stabilize apoptosis related RNAs, including mRNA, lncRNA, and circRNA, to facilitate tumor apoptosis evasion. IGF2BPs maintain the stability of HK2, GLUT1, circQSOX1, and ZFAS1 to activate the Warburg effect. IGF2BP1 and IGF2BP3 elevate the stability and expression of PD-L1, mediating immune evasion. IGF2BP3 diminishes the ability of natural killer cells to recognize transformed cells by downregulating ULBP2.
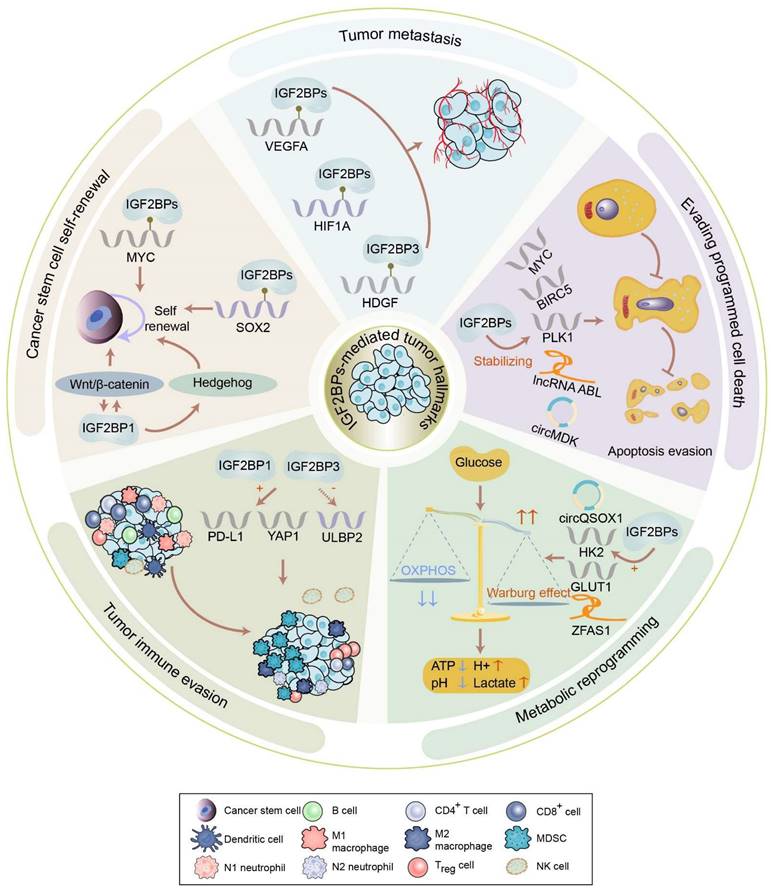
Altogether, these studies demonstrate that IGF2BPs facilitate tumor angiogenesis and EMT, which are crucial for tumor metastasis. Inhibiting IGF2BP expression can simultaneously suppress tumor metastasis, providing a novel strategy for decelerating cancer progression.
Evading programmed cell death
IGF2BPs can dysregulate the programmed cell death, chiefly involving apoptosis, ferroptosis, and autophagy, in cancer. Apoptosis is a programmed response for lysing and obliterating stressed, impaired, malignant, or infected cells in an organism [48]. IGF2BPs have been found to be associated with apoptotic pathways, which dictate cell fate in cancers. For example, the aforementioned MYC and BIRC5, which are upregulated by IGF2BPs, can also inhibit cancer cell apoptosis. Moreover, IGF2BP1 elevates the expression of Polo like kinase 1 (PLK1), thus inhibiting the apoptosis of hepatocellular carcinoma (HCC) cells [49]. Furthermore, IGF2BP1 maintains the stability of the lncRNA ABL, which binds to apoptotic protease-activating factor 1 (APAF1) and prohibits apoptosome assembly as well as caspase-9/3 activation, assisting in the evasion of apoptosis [19]. Moreover, IGF2BP1 facilitates the translation of circMAP3K4 into a novel peptide, circMAP3K4-455aa, which blocks the apoptosis of HCC cells by inhibiting the cleavage and nuclear distribution of the mitochondria-associated apoptosis-inducing factor 1 (AIF) [50]. Additionally, IGF2BP1 stimulates the translation of cellular inhibitor of apoptosis 1 (cIAP1), which regulates caspase-8-mediated cell death in rhabdomyosarcomas [51]. IGF2BP3 elevates the expression of regulator of chromosome condensation 2 (RCC2) in an m6A-dependent manner to suppress apoptosis in AML [52]. Simultaneously, IGF2BP3 stabilizes and upregulates transmembrane BAX inhibitor-1-containing motif 6 (TMBIM6), which negatively regulates the intrinsic apoptotic signaling pathway in laryngeal squamous cell carcinoma [53]. Interestingly, IGF2BP1 promotes PI3K/AKT/mTOR signaling pathway-mediated apoptosis evasion via a circMDK-miR-346/miR-874-3p-ATG16L1 regulatory axis in HCC [54]. In summary, IGF2BPs empower cancer cells to obtain the ability to escape apoptosis.
Ferroptosis is a newly discovered form of iron-dependent cell death. IGF2BPs have been identified to be involved in ferroptosis. For example, IGF2BP1 increases the stability of solute carrier family 7 member 11 (SLC7A11) mRNA which is modified by METTL3 in an m6A-dependent manner to resist ferroptosis in hepatoblastoma [55]. In hypopharyngeal squamous cell carcinoma, IGF2BP2-involved m6A modification stabilizes NFE2L2/NRF2 mRNA, contributing to ferroptosis resistance [56]. Likewise, IGF2BP3 as an m6A reader also participates in anti-ferroptosis. IGF2BP3 could stabilize a spectrum of anti-ferroptosis mRNA, including glutathione peroxidase 4 (GPX4), solute carrier family 3 member 2 (SLC3A2), acyl-CoA synthetase long chain family member 3 (ACSL3), and ferritin heavy chain 1 (FTH1), to inhibit ferroptosis in lung adenocarcinoma cells [57].
Autophagy is the process by which cells meet the cellular metabolism and renew the organelles to maintain homeostasis [58]. However, the role of autophagy in cancer is dichotomous. Recently, accumulating reports have revealed that IGF2BPs participate in the regulation of autophagy during tumor progression. In clear cell renal cell carcinoma (ccRCC), IGF2BP2 stabilizes salt inducible kinase 2 (SIK2) mRNA to enhance autophagic flux via the FTO-mediated m6A modification, diminishing the ccRCC growth and metastasis [59]. On the contrary, IGF2BP2 elevates the stability of a deubiquitinase ubiquitin specific peptidase 13 (USP13) to prolong the half-life of autophagy-related protein 5 (ATG5), inducing pro-survival autophagy and drug resistance in gastrointestinal stromal tumors [60]. Dramatically, whatever the role autophagy plays in tumor development, IGF2BPs have consistently played the role of oncogenes.
Programmed cell death plays a vital role in inducing resistance to radiotherapy and chemotherapy. Therefore, inhibiting IGF2BPs may enhance the sensitivity toward radiotherapy and chemotherapy, improving the overall status quo of anti-tumor therapy resistance.
Metabolic reprogramming
Metabolic reprogramming, mainly that of glucose, fatty acid, and amino acid metabolism, is a mechanism by which cells change their metabolic patterns to satisfy the energy needs, thereby promoting proliferation and growth. This process not only helps cells resist external stress but also endows them with new functions. It is frequently linked with tumorigenesis as well. Remarkably, IGF2BPs can influence metabolic reprogramming.
The Warburg effect or aerobic glycolysis is the most common pathway of metabolic reprogramming in tumors. MYC, which is amplified by IGF2BPs, helps enhance the Warburg effect in cancers [61-63]. IGF2BPs promote glucose metabolism in cancer cells by stabilizing lactate dehydrogenase A (LDHA) [64, 65]. Specifically, IGF2BP1 enhances glycolysis in GC cells by upregulating NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4 (NDUFA4) in an m6A-dependent manner [3]. Likewise, IGF2BP1 also binds to the 3′ UTR of PKM2 to induce the Warburg effect in bladder cancer [16]. Moreover, IGF2BP2 recognizes and upregulates m6A-decorated Apolipoprotein E (APOE) to promote glycolysis and tumor growth in papillary thyroid cancer [66]. Moreover, IGF2BP2-stabilized hexokinase 2 (HK2) and IGF2BP2/3-stabilized solute carrier family 2 member 1 (SLC2A1, GLUT1) activate glycolysis to promote CRC progression [67]. Furthermore, IGF2BP3 directly recognizes HDGF in the nucleus to activate the expression of glucose transporter type 4 (GLUT4) and enolase 2 (ENO2), which enhances glycolysis in GC cells [44]. Additionally, IGF2BP3 could mediate glycolysis in cancer cells by stabilizing pyruvate dehydrogenase kinase 4 (PDK4) [68].
Besides IGF2BPs-mediated mRNAs, ncRNAs can also regulate glucose metabolism in cancer cells. For instance, IGF2BP2 activates the Warburg effect by consolidating the ZFAS1/ Obg like ATPase 1 (OLA1) axis in CRC [18]. circFOXK2 cooperates with IGF2BP3 to accelerate aerobic glycolysis in oral squamous cell carcinoma by stabilizing GLUT1 [69]. circQSOX1, which is stabilized by IGF2BP2, provokes glycolysis via the miR-326/miR-330-5p/ phosphoglycerate mutase 1 (PGAM1) axis in CRC [70].
Acetyl-CoA is an important intermediate metabolite in energy metabolism. Notably, it is a precursor for the biosynthesis of lipid molecules that support cancer cell growth and proliferation. IGF2BP3 sustains the cellular levels of acetyl-CoA by directly binding to lncRNA TINCR and preventing its degradation, which blocks the ubiquitin-mediated degradation of ATP citrate lyase to promote lipid biosynthesis and induces cancer progression and chemoresistance [71]. Glutamine as an anaplerotic substrate replenishes the tricarboxylic acid cycle and enhances tumor proliferation. Remarkably, IGF2BP2 governs glutamine uptake and metabolism by upregulating MYC, glutamic-pyruvic transaminase 2 (GPT2), and solute carrier family 1 member 5 (SLC1A5) in AML, which indicates that IGF2BPs could regulate amino acid metabolism in cancers [26].
These studies highlight that IGF2BPs can influence the metabolic reprogramming of cancer cells in an m6A-dependent manner. Intervention of IGF2BPs may perturb the state of tumor cells and the tumor microenvironment, thereby inhibiting tumor growth.
Tumor immune evasion
Immune evasion has emerged as a crucial phenomenon in cancer progression and immunotherapy resistance. Unsurprisingly, IGF2BPs modulate tumor immune surveillance as well. Of note, the aforementioned pathway of aerobic glycolysis can promote tumor immune evasion [72]. IGF2BPs can also directly or indirectly regulate immune checkpoints to facilitate immune evasion. In HCC, IGF2BP1 not only reduces the infiltration of immune cells, such as CD4+ and CD8+ T cells, CD56+ natural killer cells, and F4/80+ macrophages but also increases the expression of programmed death-ligand 1 (PD-L1), which indicates that IGF2BP1 may act as a new anti-tumor therapeutic target by disrupting the tumor immune microenvironment [73]. Moreover, IGF2BP1 elevates the stability and expression of PD-L1, mediating the immune evasion of bladder cancer [4]. Similarly, IGF2BP3 impairs anti-tumor immunity via PD-L1-mediated T cell activation, exhaustion, and infiltration in breast cancer [74]. Furthermore, IGF2BP3 diminishes the ability of natural killer cells to recognize transformed cells by downregulating the stress-induced ligands UL16 binding protein 2 (ULBP2) and major histocompatibility complex class I polypeptide-related sequence B (MICB), contributing to tumor immune evasion [75]. Additionally, IGF2BP3 could accelerate the polarization of macrophage tumor-promoting phenotypes to mediate immune evasion [76].
Overall, tumor immunotherapy has achieved some success, but many patients remain insensitive to it. IGF2BPs enhance tumor immune evasion, suggesting that a combination of tumor immunotherapy and IGF2BP inhibitors might provide novel insights to overcome resistance to tumor immunotherapy.
Dysregulation of IGF2BPs in tumors
In this section, we will mainly discuss the dysregulation of IGF2BPs during tumorigenesis and progression. These IGF2BPs are involved in multiple regulatory mechanisms, including ncRNA-specific recruitment, RNA-protein complex mechanism, competing endogenous RNA (ceRNA) regulation, transcriptional factor regulation, mutation regulation, protein post-translational modifications (Figure 5A), and manipulation of the fate of RNAs with m6A (Figure 5B).
NcRNA-specific recruitment
NcRNAs expedite the function of IGF2BPs
Increasing evidence has indicated that ectopic lncRNAs and circRNAs exaggerate the function of IGF2BPs via an ncRNA-specific recruitment mechanism [77]. Strikingly, by binding to IGF2BPs, lncRNAs prevent their degradation. Acting as a scaffold, an aberrantly overexpressed lncRNA, KB-1980E6.3, interacts with the KH1-2 domain of IGF2BP1 to constitute a KB-1980E6.3/IGF2BP1/c-Myc signaling axis, which promotes the stability of c-Myc and maintains the stemness of breast cancer stem cells [28]. LINC01021 enhances the stability and expression of IGF2BP2 by recruiting it via the “CCCAC” fragment in the 870-921 nucleotide region. Subsequently, upregulated IGF2BP2 binds to msh homeobox 1 (MSX1) and jumonji and AT-rich interaction domain containing 2 (JARID2) to form a LINC01021-IGF2BP2-MSX1/JARID2 signaling axis, which contributes to CRC tumorigenesis and progression [78]. The lncRNA DMDRMR recruits IGF2BP3 and enhances its stability to drive the progression of ccRCC. Mechanistically, the 51-115 nucleotide region in DMDRMR exon 1 binds to the KH1-2 domain of IGF2BP3 to form a DMDRMR/IGF2BP3/CDK4 axis in ccRCC [2].
(A) Upstream regulatory mechanisms of IGF2BPs. NcRNA-specific recruitment. NcRNAs expedite or attenuate the function of IGF2BPs via ncRNA-specific recruitment. LINC01021 enhances the stability and expression of IGF2BP2 by recruiting it via the “CCCAC” fragment in the 870-921 nucleotide region, forming a LINC01021-IGF2BP2-MSX1/JARID2 signaling axis to promote CRC tumorigenesis and progression. Conversely, LINC01093 occupies the KH3-4 domain of IGF2BP1 via its 1000-1260-nucleotide region to decrease the binding affinity of IGF2BP1 to its target genes in HCC. ceRNA mechanism. IGF2BPs are manipulated by ceRNA machinery regulation at mRNA levels. LncRNA MALAT1 sponges miR-204 to upregulate IGF2BP2, contributing to the overexpression of MYC in an m6A-dependent manner. Similarly, circRNA circEZH2/ miR-133b/IGF2BP2/CREB1 axis orchestrates the CRC progression in an m6A-dependent manner. TF mechanism. TFs enhance the expression of IGF2BPs at transcriptional levels. EGR2 transcriptionally activates IGF2BPs by directly binding to the promoter regions to improve the expression of IGF2BPs, leading to the stabilization of S1PR3. Mutation. The gene fusion involving THADA and LOC389473 genes contributes to strong overexpression of IGF2BP3. PTM mechanism. IGF2BP proteins are governed by PTMs, predominantly including ubiquitination and SUMOylation, to perform multiple cellular processes. E3 ubiquitin ligase FBXO45 interacts with IGF2BP1 and facilitates its activation, leading to the stabilization of PLK1. SUMO1 SUMOylates IGF2BP2 at the lysine residues K497, K505, and K509, promoting cancer progression. (B) Downstream IGF2BPs-mediated m6A modification-dependent RNA metabolism. IGF2BPs recognize the m6A marks to increase the stability and expression of mRNAs, lncRNAs, and circRNAs. Mechanistically, IGF2BPs bind to the typical consensus motifs of RNAs by the unique KH3-4 domain to enhance the stability of RNAs. Additionally, IGF2BPs recognize the m6A marks to facilitate the translation of mRNAs and ncRNAs. IGF2BP2 binds to ERBB2 and promotes its translational efficiency. IGF2BP1 binds to circRNA circMAP3K4 and induces circMAP3K4 to translate into a novel peptide circMAP3K4-455aa.
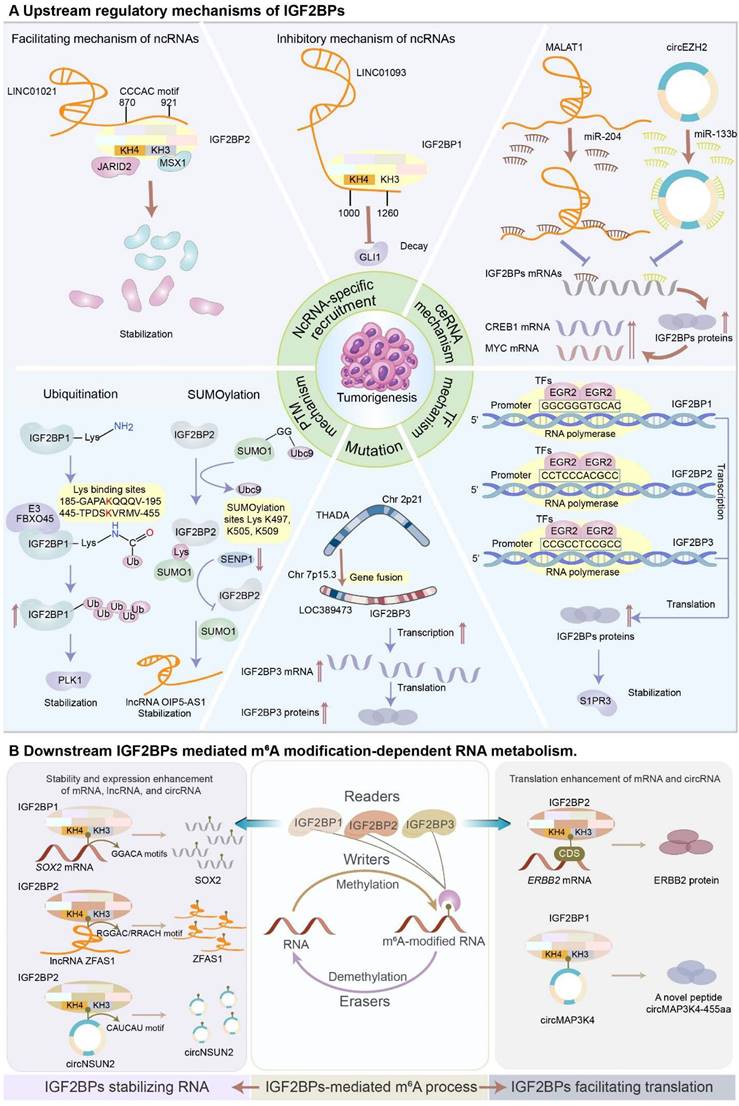
Likewise, circRNAs can also interact with IGF2BPs and enhance the function of RNA-binding proteins. For instance, circXPO1 is upregulated in lung adenocarcinoma and interacts with IGF2BP1 without influencing its expression to promote its function of stabilizing CTNNB1, leading to cancer progression [79]. Upregulated circARHGAP29 interacts with IGF2BP2 to increase the expression of LDHA, which induces aerobic glycolysis in docetaxel-resistant prostate cancer via two different pathways [65]. Firstly, circARHGAP29 interacts with the KH3-4 domain of IGF2BP2 which binds to the “UGGAC” consensus sequence in the 3′ UTR of LDHA, directly forming a circARHGAP29-IGF2BP2-LDHA RNA-protein ternary complex in the cytoplasm to facilitate the stability of LDHA [65]. Secondly, circARHGAP29, IGF2BP2, and c-Myc establish a novel RNA-protein ternary complex, which suppresses the decay of c-Myc protein and transcriptionally activates LDHA [65]. Similarly, circARID1A acts as a scaffold to form a circARID1A-IGF2BP3-SLC7A5 RNA-protein ternary complex in GC, which expedites the proliferation of GC cells [80].
NcRNAs attenuate the function of IGF2BPs
On the contrary, ncRNAs that exert antineoplastic effects can hinder the function of IGF2BPs. For instance, the lncRNA FGF13-AS1 disrupts the association between IGF2BPs and their target mRNA by competitively binding to the “GGAC” m6A core motif to inhibit breast cancer progression [63]. LINC01093 interacting with the KH3-4 domain of IGF2BP1 via its 1000-1260 nucleotide region suppresses the binding affinity of IGF2BP1 to its target genes in HCC [81]. The lncRNA NBAT1 competitively binds to IGF2BP1 in HCC to impede its association with MYC [82].
Similarly, circRNAs can also perturb the oncogenic effect of IGF2BPs. CircNDUFB2 acts as a scaffold to facilitate the binding between IGF2BPs and tripartite motif protein 25 (TRIM25), an E3 ligase, promoting the ubiquitination and degradation of IGF2BPs [83]. Moreover, circPTPRA inhibits the progression of bladder cancer by occupying the KH3-4 domain of IGF2BP1, thus impairing its ability to recognize downstream m6A-modified mRNA [84]. Furthermore, CA-rich sequences in the circRNA CDR1as interact with IGF2BP3 to attenuate the progression of melanoma [85]. Additionally, acting as a protein decoy, circTNPO3 directly binds to IGF2BP2/3 to suppress the metastasis of GC and ccRCC [24, 86].
Taken together, these findings suggest that the dualistic modulations between ncRNAs and IGF2BPs can influence tumorigenesis and cancer progression, offering innovative and precise cancer therapeutic approaches that target regulatory epigenetic characteristic biomarkers.
ceRNA machinery regulation
miRNAs induce the degradation of target mRNAs or inhibit translation through direct reciprocal interactions [87]. Remarkably, miRNAs or ceRNAs can regulate the expression of IGF2BPs to influence tumor progression. Numerous miRNAs have been identified as upstream regulators of IGF2BP1, including miR-372 [88], miR-873 [89], miR-494 [90], and miR-885-5p [91]. Moreover, miR-625, miR-196b, and miR-98-5p are shown to negatively regulate IGF2BP1 by binding to its 3′ UTR, leading to the inhibition of malignant phenotypes in liver cancer [92-94]. Similarly, miRNAs have been demonstrated to serve as tumor suppressors by modulating the expression of IGF2BP2. In HCC, miR-216b directly binds to the 3′ UTR of IGF2BP2 to suppress cell growth [95]. In lung cancer, overexpressed miR-485-5p inhibits growth and invasion by downregulating IGF2BP2 [96]. In CRC, circEZH2, acting as a miR-133b sponge, upregulates IGF2BP2, leading to cancer progression in an m6A-dependent manner [6]. Likewise, the MALAT1/miR-204/IGF2BP2/m6A-MYC axis orchestrates the proliferation, migration, and invasion of thyroid cancer [97]. As expected, miRNAs have also been shown to attenuate the pro-tumor biological functions of IGF2BP3. In GC, miR-34a suppresses cell proliferation and invasion by downregulating IGF2BP3 [98]. Notably, ceRNA signaling involving circIGHG/miR-142-5p/IGF2BP3 plays a critical oncogenic role in oral squamous cell carcinoma [99]. Although developing miRNA-targeting anti-tumor drugs is tough currently, they are promising therapeutic targets.
Overall, these studies reveal that miRNAs also govern the mRNA expression of IGF2BPs. Therefore, manipulating the miRNAs which act as the upstream regulatory molecules of IGF2BPs might inhibit tumor malignancy.
IGF2BPs regulated by Transcriptional factors
TFs control chromatin and transcription by recognizing specific DNA sequences to guide gene expression. TFs can enhance the expression of IGF2BPs. In renal cell cancer, the transcription factor early growth response protein 2 (EGR2) remarkably enhances IGF2BP levels by directly binding to the promoter regions, thereby driving tumorigenesis and metastasis [100]. The transcription of IGF2BPs can be stimulated by different TFs separately. For instance, transcription factor-activating and enhancer-binding protein 4 (TFAP4) binds to the IGF2BP1 promoter to activate its transcription in non-small cell lung cancer [101]. Interestingly, IGF2BP1 stabilizes Yes1 associated transcriptional regulator (YAP) mRNA, leading to the activation of YAP/WW domain containing transcription regulator 1 (TAZ, WWTR1) signaling in an m6A-dependent manner. Reciprocally, TAZ induces the expression of IGF2BP1, creating a feedback loop to facilitate tumorigenesis and the progression of malignancy [102]. Furthermore, Hepatocyte nuclear factor 4 gamma (HNF4G), acting as a TF, enhances the expression of IGF2BP2 [103]. Likewise, MYC effectively activates the expression of IGF2BP3 by binding to its promoter to drive tumor progression and metastasis [104]. Intriguingly, IGF2BP3 could stabilize MYC and increase its expression, establishing a positive feedback mechanism to expedite tumor deterioration.
Collectively, these findings illustrate that TFs influence the expression of IGF2BPs by binding to their promoter regions, adding to the complexity of the regulatory mechanisms governing IGF2BP expression.
IGF2BPs regulated by Protein post-translational modifications
Protein post-translational modifications (PTMs) increase the functional diversity of the proteome and participate in multiple cellular processes. Undoubtedly, IGF2BPs, being RNA-binding proteins, are regulated by PTMs, predominantly involving ubiquitination and SUMOylation. Ubiquitination selectively degrades target proteins through the ubiquitin-proteasome system (UPS). However, an atypical E3 ubiquitin ligase, F-box protein 45 (FBXO45), can prohibit the proteolysis of some proteins. For instance, overexpressed FBXO45 interacts with IGF2BP1 and facilitates its activation, leading to liver tumorigenesis [49]. Contrarily, tripartite motif-containing protein 21 (TRIM21), another E3 ubiquitin ligase, expedites IGF2BP3 decay via the UPS, arresting the growth of CRC [105]. SUMOylation can facilitate numerous pivotal physiological and pathological processes by helping maintain protein stability. SUMOylation improves the expression of IGF2BP2 and prevents its UPS-mediated degradation. Mechanistically, IGF2BP2 is identified to be SUMOylated at the lysine residues K497, K505, and K509 by small ubiquitin-related modifier 1 (SUMO1), promoting the progression of cancer [43].
The use of proteolysis targeting chimera (PROTAC) is a revolutionary, emerging strategy for targeted protein degradation. Technically, PROTAC degrades target proteins through the UPS. PROTAC may tag IGF2BPs for degradation to attenuate cancer progression, offering a new approach for anti-tumor treatment.
Mutation regulation on IGF2BPs
Mutations have been identified as crucial drivers in tumors. As expected, IGF2BPs can be governed by mutations mainly including gene fusion and copy number variation (CNV). In thyroid cancer, a recurrent fusion between the thyroid adenoma-associated (THADA) gene on chromosome 2 and the LOC389473 gene on chromosome 7 located 12 kb upstream of the IGF2BP3 gene contributes to high expression of IGF2BP3, activating the insulin like growth factor 1 receptor (IGF1R) signaling and facilitating the proliferation, invasion, and transformation of thyroid cancer cells [106]. In pancreatic cancer, genomic mutation analysis based on the cBioPortal database reveals that the IGF2BP2 locus is amplified in 15.25% of patients, indicating that the overexpression of IGF2BP2 is highly correlated with CNV [107]. These evidence shows that overexpression of IGF2BPs manipulated by gene fusion and amplification may become novel anti-tumor targets.
RNA m6A modification-dependent metabolism
IGF2BPs enhancing RNA stability
The binding of IGF2BPs to mRNAs can help facilitate mRNA stability, which is predominantly mediated by aberrant writers or erasers in cancer [59, 60, 108]. For example, in bladder cancer, IGF2BP1 recognizes the m6A moiety in the vicinity of the PD-L1 stop codon, enhancing mRNA stability to induce immune escape via a METTL3-mediated m6A mechanism [4]. In CRC, IGF2BP2 mainly binds to the CDS of SOX2 to facilitate tumor progression in a METTL3-dependent manner [30]. Similarly, IGF2BP3 recognizes the m6A signal added by METTL3 in the 3′ UTR of BIRC5 mRNA to stabilize it, resulting in cell growth and metastasis of bladder cancer [40]. Additionally, IGF2BPs also participate in the m6A process mediated by abnormally expressed m6A erasers. For instance, the depletion of FTO increases the binding propensity of IGF2BP2 toward metastasis-associated protein 1 (MTA1) mRNA, leading to CRC metastasis [109]. Notably, IGF2BPs acting as m6A readers have been shown to maintain or enhance the stability of their target transcripts through their KH3-4 domain. Specifically, IGF2BP2 is confirmed to bind MSX1 and JARID2 through its KH3-4 domain in CRC [78]. Moreover, the stabilizing activity of KH3-4 is limited. Once the KH3-4 domain is binding to an RNA, the ability to bind to other RNAs is significantly reduced. Research has shown that circFAM13B competes with PKM2 to bind to the KH3-4 domain of IGF2BP1, leading to the attenuation of PKM2 expression in bladder cancer [16].
NcRNAs such as lncRNAs, circular RNAs (circRNAs), and microRNAs (miRNAs) are critical regulators of tumor initiation and progression [110]. IGF2BPs rely on their characteristic KH domains to recognize the m6A signals on ncRNAs, thereby sustaining their stability [111]. IGF2BP1, with the help of its KH1-2 domain, recognizes and binds to the “GGACCACA” motif in lncRNA ABL to maintain its stability, consequently inhibiting the apoptosis of GC cells [19]. IGF2BP2, depending on its KH3-4 domain, recognizes the m6A modification at adenosine +843 within the “RGGAC/RRACH” element of lncRNA ZFAS1 to activate the Warburg effect in CRC [18]. Furthermore, IGF2BP1 and IGF2BP3 recognize the “GGAC” site within lnc-CTHCC and prevent its decay in an m6A-dependent manner, thereby promoting hepatocellular carcinogenesis [112]. Besides lncRNAs, overexpressed IGF2BP1 could bind to circMDK via its predicted “RRACU” m6A motif at the exon 5 site to augment the stability of this circRNA, subsequently promoting the progression of HCC [54]. The KH3-4 domain of IGF2BP2 specifically interacts with the “CAUCAU” motif at the exon 5-exon 4 junction of circNSUN2 to form an RNA-protein ternary complex, inducing CRC metastasis [17]. Furthermore, IGF2BP3 increases the stability of circCCAR1 via a WTAP-dependent m6A modification to aggravate HCC progression [113].
Taken together, IGF2BPs function as the real executors in the m6A process. KH domains, especially the KH3-4 domain, play a vital role in identifying the m6A site. Disturbing the expression or activity of IGF2BPs may counter aberrant m6A process-induced tumorigenesis.
IGF2BPs facilitating RNA translation
In addition to increasing RNA stability, IGF2BPs also promote the translation of RNAs, including ncRNAs [50, 108]. In oral squamous cell carcinoma, IGF2BP1 cooperates with METTL3 to promote BMI1 translation by recognizing the “AUGGAC” motif in the 3′ UTR of BMI1 [108]. In papillary thyroid cancer, IGF2BP2 enhances the translation efficacy of erb-b2 receptor tyrosine kinase 2 (ERBB2) by binding to m6A motifs in its CDS, acquiring resistance to tyrosine kinase inhibitors [114]. In osteosarcoma, IGF2BP2 interacts with m6A motifs in the CDS of MN1 to promote both mRNA stability and translation [115]. Notably, IGF2BP1 facilitates the translation of circMAP3K4 into a novel peptide of 63 kDa, circMAP3K4-455aa, which inhibits the apoptosis of HCC cells. Mechanistically, m6A mutations at A862C and A787/862C of circMAP3K4 dramatically attenuate the binding affinity of IGF2BP1 and decrease the expression of circMAP3K4-455aa, indicating that IGF2BP1 modulates circMAP3K4 translation via a METTL3-dependent m6A modification [50].
In summary, these findings highlight the role of IGF2BPs in RNA translation and reveal a novel carcinogenic pattern mediated by IGF2BPs, indicating that these proteins could be potential therapeutic targets in tumors.
Targeting IGF2BPs as potential therapeutic biomarkers in tumors
IGF2BPs not only promote cancer progression but also blunt the sensitivity to anti-tumor therapies. In a nutshell, IGF2BPs expedite the development of drug resistance by enhancing tumor stemness [5, 116], blocking apoptosis [19], inducing metabolic reprogramming [16, 64, 70], etc. Logically, it follows that targeting IGF2BPs may be a novel anti-tumor strategy. More researches are required, though, to answer whether IGF2BPs are targetable. Remarkably, a few IGF2BP inhibitors have been screened out from the compound library (Table 2).
So far, merely four IGF2BP1 inhibitors have been discovered. The most representative of them is the small-molecule inhibitor 2-{[(5-bromo-2-thienyl) methylene]amino} benzamide (BTYNB), identified in 2017. It selectively decreases the expression of the mRNA targets of IGF2BP1, including MYC, by limiting the association between IGF2BP1 and the coding region stability determinants of its target mRNAs, eventually suppressing the proliferation of melanomas and ovarian cancer cells [117]. Recently, BTYNB has shown the greatest efficacy in attenuating tumorigenesis and the growth of solid tumors by disrupting the interaction between IGF2BP1 and E2F-driven genes without influencing the abundance of the former [118]. Furthermore, BTYNB is shown to suppress the growth of intrahepatic cholangiocarcinoma by decreasing the expression of MYC in a patient-derived xenograft model [45]. C646, another small-molecule inhibitor, is identified to mitigate the mRNA and protein expression of IGF2BP1 in intrahepatic cholangiocarcinoma by inhibiting the levels of H3K27ac in its promoter [45]. Recently, a high-throughput screen of over 27,000 small molecules leads to the discovery of a new IGF2BP1 inhibitor called '7773'. '7773' is confirmed to directly bind to the KH3-4 domain of IGF2BP1, weakening its binding affinity to Kras and attenuating the oncogenic effect of IGF2BP1 in cancer cells [119]. Additionally, the fourth small-molecule inhibitor, cucurbitacin B, is considered to specifically recognize the Cys253 site in the KH1-2 domain of IGF2BP1, which allosterically impairs the ability of IGF2BP1 to read m6A signals in RNAs, promoting apoptosis and activating immune responses in HCC [73].
Similarly, IGF2BP2 inhibitors are woefully scarce, the first one being discovered as recently as in 2022. Ten compounds belonging to the benzamidobenzoic acid class and ureidothiophene class are screened out using a fluorescence polarization assay. Furthermore, partial compounds are confirmed to interact with the RRM1 and KH3-4 domains of IGF2BP2 and competitively inhibit its RNA-binding abilities without disrupting its expression in CRC. Meanwhile, the three most active compounds are validated to inhibit tumor growth in vivo. All these findings indicate that IGF2BP2 is a druggable anti-tumor target [120]. Thereafter, a molecular docking model is applied to screen small-molecule compounds that could bind to the KH3-4 domain based on the three-dimensional structure of IGF2BP2. Resultantly, JX5, a novel small-molecule inhibitor of IGF2BP2, is identified. JX5 directly binds to IGF2BP2 without influencing its mRNA levels; instead, the inhibitor dramatically attenuates the expression of its downstream target RNA and decelerates the progression of T cell acute lymphoblastic leukemia [121]. A new IGF2BP2 inhibitor named CWI1-2, purified from the compound NSC69557, has been demonstrated to reduce the binding affinity of IGF2BP2 to its RNA targets. Molecular docking illustrates that CWI1-2 docks to the hydrophobic pocket within the KH4 domain and competes with RNAs to bind to IGF2BP2. It could potentially exhibit anti-tumor properties in AML [122].
Unlike IGF2BP1 and IGF2BP2 inhibitors, the mechanism of action of IGF2BP3 inhibitors has not been clarified yet. Nevertheless, a few inhibitors influencing IGF2BP3 expression have been discovered. JQ1 [123] and I-BET151 [27], belonging to the bromodomain and extraterminal domain class of inhibitors, have been validated to downregulate IGF2BP3 expression and impair tumor growth in Ewing sarcoma and mixed-lineage leukemia-rearranged B-acute lymphoblastic leukemia. Berberine, an isoquinoline alkaloid derived from Coptidis Rhizoma, inhibits the proliferation of CRC cells by targeting IGF2BP3 [124]. Similarly, isoliquiritigenin, a flavonoid primarily obtained from licorice root, restrains the malignant phenotype of lung cancer cells by downregulating IGF2BP3 [125].
Small molecule inhibitor of IGF2BPs
| IGF2BPs | Inhibitors | Cancer types | Target site | Effect | Ref |
|---|---|---|---|---|---|
| IGF2BP1 | BTYNB | Melanoma, Ovarian cancer | Unknown | Disrupting the interaction between | [117, 118] |
| IGF2BP1 and target mRNAs | |||||
| 7773 | Lung cancer | A hydrophobic surface at the | Reducing the level of mRNA targets | [119] | |
| boundary of KH3 and KH4 domain | |||||
| Cucurbitacin B | HCC | The Cys253 site in the KH1-2 domain | A pharmacological allosteric effect to inhibit | [73] | |
| recognition of target mRNAs | |||||
| C646 | Intrahepatic | H3K27ac in the promoter of IGF2BP1 | Decreasing the IGF2BP1 mRNA and protein | [45] | |
| cholangiocarcinoma | expression | ||||
| IGF2BP2 | Ten compounds, | CRC, Liver cancer | RRM1 and KH3-4 (Compounds | Reducing tumor cell proliferation | [120] |
| belonging to the | from benzamidobenzoic acid class), | ||||
| benzamidobenzoic acid class | Unknown (Compounds from | ||||
| and ureidothiophene class | ureidothiophene class) | ||||
| JX5 | Leukemia | KH3-4 domain | Suppressing the cancer progression without | [121] | |
| influencing the IGF2BP2 mRNA levels | |||||
| CWI1-2 | Leukemia | The hydrophobic pocket within | Decreasing downstream RNA and protein levels of | [122] | |
| the KH4 domain | IGF2BP2 to exert anti-tumor efficacy | ||||
| IGF2BP3 | JQ1 and I-BET151 | Leukemia, | Unknown | Decreasing levels of IGF2BP3 and its mRNA | [27, 123] |
| Ewing Sarcoma | targets to hinder tumor growth | ||||
| Berberine | CRC | Unknown | Down-regulating IGF2BP3 at the protein level to | [105, 124] | |
| inhibit CRC growth | |||||
| Isoliquiritigenin | Non-small cell lung cancer | Unknown | Reducing IGF2BP3 at the mRNA and protein level | [125] | |
| to inhibit lung cancer progression | |||||
| RIG | Lung adenocarcinoma | Unknown | Reducing IGF2BP3 expression and tumor growth | [57] |
Rigosertib (RIG), another small-molecule IGF2BP3 inhibitor, is identified by screening a library consisting of more than 1800 small molecules approved by the Food and Drug Administration. RIG represses IGF2BP3 expression and sensitizes lung cancer cells to RSL3 and erastin-induced ferroptosis, suggesting that RIG can inhibit tumor growth and promote programmed cell death [57].
In general, the development of virtual high-throughput screening and molecular docking has enabled the discovery of small-molecule inhibitors of IGF2BPs, which play antitumor roles mainly by inhibiting protein expression and activity. However, this class of inhibitors faces many limitations and challenges, such as the development of resistance to small-molecule inhibitors, the unsustainable inhibition of target protein activity, and “untargetable” proteins. The application of new technologies like PROTAC and molecular glue may overcome the limitations faced by small-molecule inhibitors. Thus, promising antitumor drugs targeting IGF2BPs may be screened out via PROTAC and molecular glue technology, providing a new strategy to treat tumors.
Conclusions and perspective
The novel m6A readers IGF2BPs are widely overexpressed in cancers and participate in the regulation of mRNA stabilization and translation. IGF2BPs directly bind to m6A-modified RNAs via their KH domains, which are evolutionarily conserved RNA recognition elements. Dramatically, each IGF2BP can recognize more than 3,000 mRNA transcripts and at least 5,000 mRNAs, with great overlaps among the three IGF2BPs, suggesting that these proteins prominently govern m6A-dependent gene regulation [7]. In addition to mRNAs, IGF2BPs also recognize ncRNAs, including lncRNAs and circRNAs, to exacerbate cancer progression. Intriguingly, the m6A modification, as a type of non-mutational epigenetic reprogramming, participates in the administration of most other cancer hallmarks partially via IGF2BPs. Emerging evidence has consistently demonstrated that IGF2BPs help cancer cells proliferate, invade tissues, metastasize, sustain angiogenesis, evade programmed cell death, dysregulate energy metabolism, and avoid immune destruction. All the evidence indicates that IGF2BPs may be promising druggable targets in tumor therapies. Several IGF2BP inhibitors have been proposed so far. Some directly interact with the KH domains to interfere with the RNA binding capacity without affecting IGF2BP expression, while the mechanism of others is still unclear and needs further research. The expression of IGF2BPs is also regulated by multiple mechanisms, mainly including ncRNAs, TFs, and PTMs, which can further manipulate the fate of oncogenes during tumorigenesis and tumor growth. The expression of IGF2BPs may be suppressed by interfering with their upstream regulatory mechanisms, eventually inhibiting tumor progression, which presents a novel antitumor approach. The discovery of IGF2BP inhibitors and the determination of upstream regulatory mechanisms both demonstrate the potential accessibility of IGF2BPs via exogenous manipulations.
In conclusion, IGF2BPs are crucial factors in tumor progression, chemotherapy resistance, and immunotherapy response. Therefore, incorporating a combination of IGF2BP inhibitors in antitumor therapy may remarkably inhibit cancer invasion, metastasis, and recurrence, or effectively overcome antitumor drug resistance. Unfortunately, fundamental research on IGF2BP inhibitors and their mechanisms is currently lacking, and no such clinical trial has ever been performed. Therefore, multidisciplinary collaboration is necessary to develop and optimize the use of IGF2BP inhibitors in cancer treatment.
Abbreviations
AIF: apoptosis-inducing factor 1;
ALKBH5: alkB homolog 5;
ACSL3: acyl-CoA synthetase long chain family member 3;
AML: acute myeloid leukemia;
APAF1: apoptotic protease-activating factor 1;
APOE: Apolipoprotein E;
ATG5: autophagy-related protein 5;
BIRC5: apoptosis inhibitor Survivin;
BTYNB: 2-{[(5-bromo-2-thienyl) methylene]amino} benzamide;
ccRCC: clear cell renal cell carcinoma;
CDS: coding sequence;
ceRNA: competing endogenous RNA;
cIAP1: cellular inhibitor of apoptosis 1;
CNV: copy number variation;
CRC: colorectal cancer;
DACT1: Dishevelled-binding antagonist of β-catenin 1;
EC: endometrial cancer;
EMT: epithelial-mesenchymal transition;
ENO2: enolase 2;
EphA2: ephrin type-A receptor 2;
FTH1: ferritin heavy chain 1;
FTO: fat mass and obesity-associated protein;
GC: gastric cancer;
GLI1: GLI family zinc finger 1;
GLUT4: glucose transporter type 4;
GPT2: glutamic-pyruvic transaminase 2;
GPX4: glutathione peroxidase 4;
HDGF: hepatoma-derived growth factor;
HIF1A: hypoxia inducible factor 1 subunit alpha;
hnRNPs: heterogeneous nuclear ribonucleoproteins;
HNSCC: head and neck squamous carcinoma;
IGF2BPs: Insulin-like growth factor 2 mRNA-binding proteins;
IGF-II-L3: IGF2 leader 3;
JARID2: jumonji and AT-rich interaction domain containing 2;
KH: K homology;
LDHA: lactate dehydrogenase A;
m6A: N6-methyladenosine;
MCM5: minichromosome maintenance complex component;
METTL14: methyltransferase 14;
METTL3: methyltransferase 3;
MICB: major histocompatibility complex class I polypeptide-related sequence B;
MMP14: matrix metalloproteinase 14;
MMP2: matrix metalloproteinase 2;
MSX1: msh homeobox 1;
ncRNAs: non-coding RNAs;
NDUFA4: NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 4;
OLA1: Obg like ATPase 1;
PDK4: pyruvate dehydrogenase kinase 4;
PD-L1: programmed death-ligand 1;
PGAM1: phosphoglycerate mutase 1;
PKM2: pyruvate kinase M1/2;
PLK1: Polo like kinase 1;
PROTAC: proteolysis targeting chimera;
PTMs: post-translational modifications;
PTMs: post-translational modifications;
RCC2: regulator of chromosome condensation 2;
RNPs: ribonucleoprotein complexes;
RRMs: RNA recognition motifs;
SIK2: salt inducible kinase 2;
SLC1A5: solute carrier family 1 member 5;
SLC2A1: solute carrier family 2 member 1;
SLC3A2: solute carrier family 3 member 2;
SLC7A11: solute carrier family 7 member 11;
SLUG: snail family transcriptional repressor 2;
SOX2: SRY-box transcription factor 2;
TFs: transcription factors;
TMBIM6: transmembrane BAX inhibitor-1-containing motif 6;
ULBP2: stress-induced ligands UL16 binding protein 2;
USP13: ubiquitin specific peptidase 13;
UTR: untranslated region;
VEGFA: vascular endothelial growth factor A;
WTAP: Wilms tumor 1-associated protein;
YTH: YT521-B homology.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (82272915), the project of the fourth batch of science and technology plan of Liaoning province (2021JH210300133), and the project of the applied basic research program of Liaoning province (2022JH2/101300050). We thank Bullet Edits Limited for the linguistic editing and proofreading of the manuscript.
Author contributions
K.L., H.W., and H.Z. conceived the structure of the manuscript. H.Z., Q.S., and M.F. wrote the manuscript. S.G., H.W., K.L., X.Y., and Z.G. provided guidance throughout the revision of this manuscript. K.L., H.W., H.Z., S.J., and L.C. revised the paper. H.Z. and H.W. prepared the figures. All authors read and approved the final manuscript.
Consent for publication
All authors consent to publication.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19:1262-70
2. Gu Y, Niu S, Wang Y, Duan L, Pan Y, Tong Z. et al. DMDRMR-Mediated Regulation of m(6)A-Modified CDK4 by m(6)A Reader IGF2BP3 Drives ccRCC Progression. Cancer Res. 2021;81:923-34
3. Xu W, Lai Y, Pan Y, Tan M, Ma Y, Sheng H. et al. m6A RNA methylation-mediated NDUFA4 promotes cell proliferation and metabolism in gastric cancer. Cell Death Dis. 2022;13:715
4. Ni Z, Sun P, Zheng J, Wu M, Yang C, Cheng M. et al. JNK Signaling Promotes Bladder Cancer Immune Escape by Regulating METTL3-Mediated m6A Modification of PD-L1 mRNA. Cancer Res. 2022;82:1789-802
5. Elcheva IA, Wood T, Chiarolanzio K, Chim B, Wong M, Singh V. et al. RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia. 2020;34:1354-63
6. Yao B, Zhang Q, Yang Z, An F, Nie H, Wang H. et al. CircEZH2/miR-133b/IGF2BP2 aggravates colorectal cancer progression via enhancing the stability of m(6)A-modified CREB1 mRNA. Mol Cancer. 2022;21:140
7. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-95
8. Tan F, Zhao M, Xiong F, Wang Y, Zhang S, Gong Z. et al. N6-methyladenosine-dependent signalling in cancer progression and insights into cancer therapies. J Exp Clin Cancer Res. 2021;40:146
9. Ramesh-Kumar D, Guil S. The IGF2BP family of RNA binding proteins links epitranscriptomics to cancer. Semin Cancer Biol. 2022;86(Pt 3):18-31
10. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635-46
11. Oleynikov Y, Singer RH. Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr Biol. 2003;13:199-207
12. Bell JL, Wachter K, Muhleck B, Pazaitis N, Kohn M, Lederer M. et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70:2657-75
13. Nielsen J, Kristensen MA, Willemoes M, Nielsen FC, Christiansen J. Sequential dimerization of human zipcode-binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability. Nucleic Acids Res. 2004;32:4368-76
14. Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24:148-58
15. Nicastro G, Candel AM, Uhl M, Oregioni A, Hollingworth D, Backofen R. et al. Mechanism of beta-actin mRNA Recognition by ZBP1. Cell Rep. 2017;18:1187-99
16. Lv J, Li K, Yu H, Han J, Zhuang J, Yu R. et al. HNRNPL induced circFAM13B increased bladder cancer immunotherapy sensitivity via inhibiting glycolysis through IGF2BP1/PKM2 pathway. J Exp Clin Cancer Res. 2023;42:41
17. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD. et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695
18. Lu S, Han L, Hu X, Sun T, Xu D, Li Y. et al. N6-methyladenosine reader IMP2 stabilizes the ZFAS1/OLA1 axis and activates the Warburg effect: implication in colorectal cancer. J Hematol Oncol. 2021;14:188
19. Wang Q, Chen C, Xu X, Shu C, Cao C, Wang Z. et al. APAF1-Binding Long Noncoding RNA Promotes Tumor Growth and Multidrug Resistance in Gastric Cancer by Blocking Apoptosome Assembly. Adv Sci (Weinh). 2022;9:e2201889
20. Gu W, Pan F, Singer RH. Blocking beta-catenin binding to the ZBP1 promoter represses ZBP1 expression, leading to increased proliferation and migration of metastatic breast-cancer cells. J Cell Sci. 2009;122:1895-905
21. Liao B, Patel M, Hu Y, Charles S, Herrick DJ, Brewer G. Targeted knockdown of the RNA-binding protein CRD-BP promotes cell proliferation via an insulin-like growth factor II-dependent pathway in human K562 leukemia cells. J Biol Chem. 2004;279:48716-24
22. Hamilton KE, Chatterji P, Lundsmith ET, Andres SF, Giroux V, Hicks PD. et al. Loss of Stromal IMP1 Promotes a Tumorigenic Microenvironment in the Colon. Mol Cancer Res. 2015;13:1478-86
23. Zhu S, Wang JZ, Chen D, He YT, Meng N, Chen M. et al. An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat Commun. 2020;11:1685
24. Yu T, Ran L, Zhao H, Yin P, Li W, Lin J. et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol Ther Nucleic Acids. 2021;26:649-64
25. Li J, Gao X, Zhang Z, Lai Y, Lin X, Lin B. et al. CircCD44 plays oncogenic roles in triple-negative breast cancer by modulating the miR-502-5p/KRAS and IGF2BP2/Myc axes. Mol Cancer. 2021;20:138
26. Weng H, Huang F, Yu Z, Chen Z, Prince E, Kang Y. et al. The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell. 2022;40:1566-82
27. Palanichamy JK, Tran TM, Howard JM, Contreras JR, Fernando TR, Sterne-Weiler T. et al. RNA-binding protein IGF2BP3 targeting of oncogenic transcripts promotes hematopoietic progenitor proliferation. J Clin Invest. 2016;126:1495-511
28. Zhu P, He F, Hou Y, Tu G, Li Q, Jin T. et al. A novel hypoxic long noncoding RNA KB-1980E6.3 maintains breast cancer stem cell stemness via interacting with IGF2BP1 to facilitate c-Myc mRNA stability. Oncogene. 2021;40:1609-27
29. Xue T, Liu X, Zhang M, E Q, Liu S, Zou M. et al. PADI2-Catalyzed MEK1 Citrullination Activates ERK1/2 and Promotes IGF2BP1-Mediated SOX2 mRNA Stability in Endometrial Cancer. Adv Sci (Weinh). 2021;8:2002831
30. Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN. et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112
31. Samanta S, Sun H, Goel HL, Pursell B, Chang C, Khan A. et al. IMP3 promotes stem-like properties in triple-negative breast cancer by regulating SLUG. Oncogene. 2016;35:1111-21
32. Chen Z, Huang L, Wang K, Zhang L, Zhong X, Yan Z. et al. rtcisE2F promotes the self-renewal and metastasis of liver tumor-initiating cells via N(6)-methyladenosine-dependent E2F3/E2F6 mRNA stability. Sci China Life Sci. 2022;65:1840-54
33. Bugter JM, Fenderico N, Maurice MM. Mutations and mechanisms of WNT pathway tumour suppressors in cancer. Nat Rev Cancer. 2021;21:5-21
34. Gao J, Zhao C, Liu Q, Hou X, Li S, Xing X. et al. Cyclin G2 suppresses Wnt/beta-catenin signaling and inhibits gastric cancer cell growth and migration through Dapper1. J Exp Clin Cancer Res. 2018;37:317
35. Lv D, Ding S, Zhong L, Tu J, Li H, Yao H. et al. M(6)A demethylase FTO-mediated downregulation of DACT1 mRNA stability promotes Wnt signaling to facilitate osteosarcoma progression. Oncogene. 2022;41:1727-41
36. Noubissi FK, Kim T, Kawahara TN, Aughenbaugh WD, Berg E, Longley BJ. et al. Role of CRD-BP in the growth of human basal cell carcinoma cells. J Invest Dermatol. 2014;134:1718-24
37. Noubissi FK, Goswami S, Sanek NA, Kawakami K, Minamoto T, Moser A. et al. Wnt signaling stimulates transcriptional outcome of the Hedgehog pathway by stabilizing GLI1 mRNA. Cancer Res. 2009;69:8572-8
38. Liu S, Li H, Zhu Y, Ma X, Shao Z, Yang Z. et al. LncRNA MNX1-AS1 sustains inactivation of Hippo pathway through a positive feedback loop with USP16/IGF2BP3 axis in gallbladder cancer. Cancer Lett. 2022;547:215862
39. Yang Z, Wang T, Wu D, Min Z, Tan J, Yu B. RNA N6-methyladenosine reader IGF2BP3 regulates cell cycle and angiogenesis in colon cancer. J Exp Clin Cancer Res. 2020;39:203
40. Liu H, Gu J, Huang Z, Han Z, Xin J, Yuan L. et al. Fine particulate matter induces METTL3-mediated m(6)A modification of BIRC5 mRNA in bladder cancer. J Hazard Mater. 2022;437:129310
41. Liu X, He H, Zhang F, Hu X, Bi F, Li K. et al. m6A methylated EphA2 and VEGFA through IGF2BP2/3 regulation promotes vasculogenic mimicry in colorectal cancer via PI3K/AKT and ERK1/2 signaling. Cell Death Dis. 2022;13:483
42. Jiang L, Li Y, He Y, Wei D, Yan L, Wen H. Knockdown of m6A Reader IGF2BP3 Inhibited Hypoxia-Induced Cell Migration and Angiogenesis by Regulating Hypoxia Inducible Factor-1alpha in Stomach Cancer. Front Oncol. 2021;11:711207
43. Li H, Wang D, Yi B, Cai H, Wang Y, Lou X. et al. SUMOylation of IGF2BP2 promotes vasculogenic mimicry of glioma via regulating OIP5-AS1/miR-495-3p axis. Int J Biol Sci. 2021;17:2912-30
44. Wang Q, Chen C, Ding Q, Zhao Y, Wang Z, Chen J. et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69:1193-205
45. Xiao P, Meng Q, Liu Q, Lang Q, Yin Z, Li G. et al. IGF2BP1-mediated N6-methyladenosine modification promotes intrahepatic cholangiocarcinoma progression. Cancer Lett. 2023;557:216075
46. Yu D, Pan M, Li Y, Lu T, Wang Z, Liu C. et al. RNA N6-methyladenosine reader IGF2BP2 promotes lymphatic metastasis and epithelial-mesenchymal transition of head and neck squamous carcinoma cells via stabilizing slug mRNA in an m6A-dependent manner. J Exp Clin Cancer Res. 2022;41:6
47. Yang X, Bai Q, Chen W, Liang J, Wang F, Gu W. et al. m(6) A-Dependent Modulation via IGF2BP3/MCM5/Notch Axis Promotes Partial EMT and LUAD Metastasis. Adv Sci (Weinh), in press. doi: 10.1002/advs.202206744
48. Liu L, Li H, Hu D, Wang Y, Shao W, Zhong J. et al. Insights into N6-methyladenosine and programmed cell death in cancer. Mol Cancer. 2022;21:32
49. Lin XT, Yu HQ, Fang L, Tan Y, Liu ZY, Wu D. et al. Elevated FBXO45 promotes liver tumorigenesis through enhancing IGF2BP1 ubiquitination and subsequent PLK1 upregulation. Elife. 2021;10:e70715
50. Duan JL, Chen W, Xie JJ, Zhang ML, Nie RC, Liang H. et al. A novel peptide encoded by N6-methyladenosine modified circMAP3K4 prevents apoptosis in hepatocellular carcinoma. Mol Cancer. 2022;21:93
51. Faye MD, Beug ST, Graber TE, Earl N, Xiang X, Wild B. et al. IGF2BP1 controls cell death and drug resistance in rhabdomyosarcomas by regulating translation of cIAP1. Oncogene. 2015;34:1532-41
52. Zhang N, Shen Y, Li H, Chen Y, Zhang P, Lou S. et al. The m6A reader IGF2BP3 promotes acute myeloid leukemia progression by enhancing RCC2 stability. Exp Mol Med. 2022;54:194-205
53. Wang X, Tian L, Li Y, Wang J, Yan B, Yang L. et al. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J Exp Clin Cancer Res. 2021;40:80
54. Du A, Li S, Zhou Y, Disoma C, Liao Y, Zhang Y. et al. M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma. Mol Cancer. 2022;21:109
55. Liu L, He J, Sun G, Huang N, Bian Z, Xu C. et al. The N6-methyladenosine modification enhances ferroptosis resistance through inhibiting SLC7A11 mRNA deadenylation in hepatoblastoma. Clin Transl Med. 2022;12:e778
56. Ye J, Chen X, Jiang X, Dong Z, Hu S, Xiao M. RNA demethylase ALKBH5 regulates hypopharyngeal squamous cell carcinoma ferroptosis by posttranscriptionally activating NFE2L2/NRF2 in an m(6) A-IGF2BP2-dependent manner. J Clin Lab Anal. 2022;36:e24514
57. Xu X, Cui J, Wang H, Ma L, Zhang X, Guo W. et al. IGF2BP3 is an essential N(6)-methyladenosine biotarget for suppressing ferroptosis in lung adenocarcinoma cells. Mater Today Bio. 2022;17:100503
58. Chen JL, Wu X, Yin D, Jia XH, Chen X, Gu ZY. et al. Autophagy inhibitors for cancer therapy: Small molecules and nanomedicines. Pharmacol Ther, in press. doi: 10.1016/j.pharmthera.2023.108485
59. Xu Y, Zhou J, Li L, Yang W, Zhang Z, Zhang K. et al. FTO-mediated autophagy promotes progression of clear cell renal cell carcinoma via regulating SIK2 mRNA stability. Int J Biol Sci. 2022;18:5943-62
60. Gao Z, Li C, Sun H, Bian Y, Cui Z, Wang N. et al. N(6)-methyladenosine-modified USP13 induces pro-survival autophagy and imatinib resistance via regulating the stabilization of autophagy-related protein 5 in gastrointestinal stromal tumors. Cell Death Differ. 2023;30:544-59
61. Wang Y, Lu JH, Wu QN, Jin Y, Wang DS, Chen YX. et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18:174
62. Hu C, Liu T, Han C, Xuan Y, Jiang D, Sun Y. et al. HPV E6/E7 promotes aerobic glycolysis in cervical cancer by regulating IGF2BP2 to stabilize m(6)A-MYC expression. Int J Biol Sci. 2022;18:507-21
63. Ma F, Liu X, Zhou S, Li W, Liu C, Chadwick M. et al. Long non-coding RNA FGF13-AS1 inhibits glycolysis and stemness properties of breast cancer cells through FGF13-AS1/IGF2BPs/Myc feedback loop. Cancer Lett. 2019;450:63-75
64. Zhang XL, Li KJ, Feng JX, Liu GJ, Feng YL. Blocking the IGF2BP1-promoted glucose metabolism of colon cancer cells via direct de-stabilizing mRNA of the LDHA enhances anticancer effects. Mol Ther Nucleic Acids. 2021;23:835-46
65. Jiang X, Guo S, Wang S, Zhang Y, Chen H, Wang Y. et al. EIF4A3-Induced circARHGAP29 Promotes Aerobic Glycolysis in Docetaxel-Resistant Prostate Cancer through IGF2BP2/c-Myc/LDHA Signaling. Cancer Res. 2022;82:831-45
66. Huang J, Sun W, Wang Z, Lv C, Zhang T, Zhang D. et al. FTO suppresses glycolysis and growth of papillary thyroid cancer via decreasing stability of APOE mRNA in an N6-methyladenosine-dependent manner. J Exp Clin Cancer Res. 2022;41:42
67. Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X. et al. m(6)A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19:72
68. Li Z, Peng Y, Li J, Chen Z, Chen F, Tu J. et al. N(6)-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun. 2020;11:2578
69. Cui Y, Liu J, Liu L, Ma X, Gui Y, Liu H. et al. m(6)A-modified circFOXK2 targets GLUT1 to accelerate oral squamous cell carcinoma aerobic glycolysis. Cancer Gene Ther. 2023;30:163-71
70. Liu Z, Zheng N, Li J, Li C, Zheng D, Jiang X. et al. N6-methyladenosine-modified circular RNA QSOX1 promotes colorectal cancer resistance to anti-CTLA-4 therapy through induction of intratumoral regulatory T cells. Drug Resist Updat. 2022;65:100886
71. Zheng ZQ, Li ZX, Guan JL, Liu X, Li JY, Chen Y. et al. Long Noncoding RNA TINCR-Mediated Regulation of Acetyl-CoA Metabolism Promotes Nasopharyngeal Carcinoma Progression and Chemoresistance. Cancer Res. 2020;80:5174-88
72. Guo D, Tong Y, Jiang X, Meng Y, Jiang H, Du L. et al. Aerobic glycolysis promotes tumor immune evasion by hexokinase2-mediated phosphorylation of IkappaBalpha. Cell Metab. 2022;34:1312-24
73. Liu Y, Guo Q, Yang H, Zhang XW, Feng N, Wang JK. et al. Allosteric Regulation of IGF2BP1 as a Novel Strategy for the Activation of Tumor Immune Microenvironment. ACS Cent Sci. 2022;8:1102-15
74. Wan W, Ao X, Chen Q, Yu Y, Ao L, Xing W. et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N(6)-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol Cancer. 2022;21:60
75. Schmiedel D, Tai J, Yamin R, Berhani O, Bauman Y, Mandelboim O. The RNA binding protein IMP3 facilitates tumor immune escape by downregulating the stress-induced ligands ULPB2 and MICB. Elife. 2016;5:e13426
76. Pan Z, Zhao R, Li B, Qi Y, Qiu W, Guo Q. et al. EWSR1-induced circNEIL3 promotes glioma progression and exosome-mediated macrophage immunosuppressive polarization via stabilizing IGF2BP3. Mol Cancer. 2022;21:16
77. Shen H, Zhu H, Chen Y, Shen Z, Qiu W, Qian C. et al. ZEB1-induced LINC01559 expedites cell proliferation, migration and EMT process in gastric cancer through recruiting IGF2BP2 to stabilize ZEB1 expression. Cell Death Dis. 2021;12:349
78. Wu H, Ding X, Hu X, Zhao Q, Chen Q, Sun T. et al. LINC01021 maintains tumorigenicity by enhancing N6-methyladenosine reader IMP2 dependent stabilization of MSX1 and JARID2: implication in colorectal cancer. Oncogene. 2022;41:1959-73
79. Huang Q, Guo H, Wang S, Ma Y, Chen H, Li H. et al. A novel circular RNA, circXPO1, promotes lung adenocarcinoma progression by interacting with IGF2BP1. Cell Death Dis. 2020;11:1031
80. Ma Q, Yang F, Huang B, Pan X, Li W, Yu T. et al. CircARID1A binds to IGF2BP3 in gastric cancer and promotes cancer proliferation by forming a circARID1A-IGF2BP3-SLC7A5 RNA-protein ternary complex. J Exp Clin Cancer Res. 2022;41:251
81. He J, Zuo Q, Hu B, Jin H, Wang C, Cheng Z. et al. A novel, liver-specific long noncoding RNA LINC01093 suppresses HCC progression by interaction with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Lett. 2019;450:98-109
82. Wei L, Ling M, Yang S, Xie Y, Liu C, Yi W. Long noncoding RNA NBAT1 suppresses hepatocellular carcinoma progression via competitively associating with IGF2BP1 and decreasing c-Myc expression. Hum Cell. 2021;34:539-49
83. Li B, Zhu L, Lu C, Wang C, Wang H, Jin H. et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat Commun. 2021;12:295
84. Xie F, Huang C, Liu F, Zhang H, Xiao X, Sun J. et al. CircPTPRA blocks the recognition of RNA N(6)-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Mol Cancer. 2021;20:68
85. Hanniford D, Ulloa-Morales A, Karz A, Berzoti-Coelho MG, Moubarak RS, Sanchez-Sendra B. et al. Epigenetic Silencing of CDR1as Drives IGF2BP3-Mediated Melanoma Invasion and Metastasis. Cancer Cell. 2020;37:55-70
86. Pan X, Huang B, Ma Q, Ren J, Liu Y, Wang C. et al. Circular RNA circ-TNPO3 inhibits clear cell renal cell carcinoma metastasis by binding to IGF2BP2 and destabilizing SERPINH1 mRNA. Clin Transl Med. 2022;12:e994
87. Doghish AS, Ismail A, El-Mahdy HA, Elkady MA, Elrebehy MA, Sallam AM. A review of the biological role of miRNAs in prostate cancer suppression and progression. Int J Biol Macromol. 2022;197:141-56
88. Huang X, Huang M, Kong L, Li Y. miR-372 suppresses tumour proliferation and invasion by targeting IGF2BP1 in renal cell carcinoma. Cell Prolif. 2015;48:593-9
89. Wang RJ, Li JW, Bao BH, Wu HC, Du ZH, Su JL. et al. MicroRNA-873 (miRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J Biol Chem. 2015;290:8938-48
90. Ohdaira H, Sekiguchi M, Miyata K, Yoshida K. MicroRNA-494 suppresses cell proliferation and induces senescence in A549 lung cancer cells. Cell Prolif. 2012;45:32-8
91. Lixin S, Wei S, Haibin S, Qingfu L, Tiemin P. miR-885-5p inhibits proliferation and metastasis by targeting IGF2BP1 and GALNT3 in human intrahepatic cholangiocarcinoma. Mol Carcinog. 2020;59:1371-81
92. Rebucci M, Sermeus A, Leonard E, Delaive E, Dieu M, Fransolet M. et al. miRNA-196b inhibits cell proliferation and induces apoptosis in HepG2 cells by targeting IGF2BP1. Mol Cancer. 2015;14:79
93. Jiang T, Li M, Li Q, Guo Z, Sun X, Zhang X. et al. MicroRNA-98-5p Inhibits Cell Proliferation and Induces Cell Apoptosis in Hepatocellular Carcinoma via Targeting IGF2BP1. Oncol Res. 2017;25:1117-27
94. Zhou X, Zhang CZ, Lu SX, Chen GG, Li LZ, Liu LL. et al. miR-625 suppresses tumour migration and invasion by targeting IGF2BP1 in hepatocellular carcinoma. Oncogene. 2015;34:965-77
95. Liu FY, Zhou SJ, Deng YL, Zhang ZY, Zhang EL, Wu ZB. et al. MiR-216b is involved in pathogenesis and progression of hepatocellular carcinoma through HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death Dis. 2015;6:e1670
96. Huang RS, Zheng YL, Li C, Ding C, Xu C, Zhao J. MicroRNA-485-5p suppresses growth and metastasis in non-small cell lung cancer cells by targeting IGF2BP2. Life Sci. 2018;199:104-11
97. Ye M, Dong S, Hou H, Zhang T, Shen M. Oncogenic Role of Long Noncoding RNAMALAT1 in Thyroid Cancer Progression through Regulation of the miR-204/IGF2BP2/m6A-MYC Signaling. Mol Ther Nucleic Acids. 2021;23:1-12
98. Zhou Y, Huang T, Siu HL, Wong CC, Dong Y, Wu F. et al. IGF2BP3 functions as a potential oncogene and is a crucial target of miR-34a in gastric carcinogenesis. Mol Cancer. 2017;16:77
99. Liu J, Jiang X, Zou A, Mai Z, Huang Z, Sun L. et al. circIGHG-Induced Epithelial-to-Mesenchymal Transition Promotes Oral Squamous Cell Carcinoma Progression via miR-142-5p/IGF2BP3 Signaling. Cancer Res. 2021;81:344-55
100. Ying Y, Ma X, Fang J, Chen S, Wang W, Li J. et al. EGR2-mediated regulation of m(6)A reader IGF2BP proteins drive RCC tumorigenesis and metastasis via enhancing S1PR3 mRNA stabilization. Cell Death Dis. 2021;12:750
101. Shen Q, Xu Z, Sun G, Wang H, Zhang L. TFAP4 Activates IGF2BP1 and Promotes Progression of Non-Small Cell Lung Cancer by Stabilizing TK1 Expression through m6A Modification. Mol Cancer Res. 2022;20:1763-75
102. Yang J, Wu X, Wang J, Guo X, Chen J, Yang X. et al. Feedforward loop between IMP1 and YAP/TAZ promotes tumorigenesis and malignant progression in glioblastoma. Cancer Sci. 2022;114:2053-62
103. Zhan J, Zhang Q, Tong X, Liu X, Zhao C. HNF4G stimulates the development of pancreatic cancer by promoting IGF2BP2 transcription. Clin Transl Oncol. 2023;25:1472-81
104. Du M, Peng Y, Li Y, Sun W, Zhu H, Wu J. et al. MYC-activated RNA N6-methyladenosine reader IGF2BP3 promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cell Death Discov. 2022;8:53
105. Gui Z, Li J, Li J, Li X, Chen L, Ma Z. et al. Berberine promotes IGF2BP3 ubiquitination by TRIM21 to induce G1/S phase arrest in colorectal cancer cells. Chem Biol Interact. 2023;374:110408
106. Panebianco F, Kelly LM, Liu P, Zhong S, Dacic S, Wang X. et al. THADA fusion is a mechanism of IGF2BP3 activation and IGF1R signaling in thyroid cancer. Proc Natl Acad Sci U S A. 2017;114:2307-12
107. Xu X, Yu Y, Zong K, Lv P, Gu Y. Up-regulation of IGF2BP2 by multiple mechanisms in pancreatic cancer promotes cancer proliferation by activating the PI3K/Akt signaling pathway. J Exp Clin Cancer Res. 2019;38:497
108. Liu L, Wu Y, Li Q, Liang J, He Q, Zhao L. et al. METTL3 Promotes Tumorigenesis and Metastasis through BMI1 m(6)A Methylation in Oral Squamous Cell Carcinoma. Mol Ther. 2020;28:2177-90
109. Ruan DY, Li T, Wang YN, Meng Q, Li Y, Yu K. et al. FTO downregulation mediated by hypoxia facilitates colorectal cancer metastasis. Oncogene. 2021;40:5168-81
110. Dai L, Liang W, Shi Z, Li X, Zhou S, Hu W. et al. Systematic characterization and biological functions of non-coding RNAs in glioblastoma. Cell Prolif. 2023;56:e13375
111. Liu HT, Zou YX, Zhu WJ, Sen L, Zhang GH, Ma RR. et al. lncRNA THAP7-AS1, transcriptionally activated by SP1 and post-transcriptionally stabilized by METTL3-mediated m6A modification, exerts oncogenic properties by improving CUL4B entry into the nucleus. Cell Death Differ. 2022;29:627-41
112. Xia A, Yuan W, Wang Q, Xu J, Gu Y, Zhang L. et al. The cancer-testis lncRNA lnc-CTHCC promotes hepatocellular carcinogenesis by binding hnRNP K and activating YAP1 transcription. Nat Cancer. 2022;3:203-18
113. Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y. et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer. 2023;22:55
114. Sa R, Liang R, Qiu X, He Z, Liu Z, Chen L. IGF2BP2-dependent activation of ERBB2 signaling contributes to acquired resistance to tyrosine kinase inhibitor in differentiation therapy of radioiodine-refractory papillary thyroid cancer. Cancer Lett. 2022;527:10-23
115. Li HB, Huang G, Tu J, Lv DM, Jin QL, Chen JK. et al. METTL14-mediated epitranscriptome modification of MN1 mRNA promote tumorigenicity and all-trans-retinoic acid resistance in osteosarcoma. EBioMedicine. 2022;82:104142
116. Zhang R, Liu L, Wang F, Zhao W, Liu K, Yu H. et al. AKAP8L enhances the stemness and chemoresistance of gastric cancer cells by stabilizing SCD1 mRNA. Cell Death Dis. 2022;13:1041
117. Mahapatra L, Andruska N, Mao C, Le J, Shapiro DJ. A Novel IMP1 Inhibitor, BTYNB, Targets c-Myc and Inhibits Melanoma and Ovarian Cancer Cell Proliferation. Transl Oncol. 2017;10:818-27
118. Muller S, Bley N, Busch B, Glass M, Lederer M, Misiak C. et al. The oncofetal RNA-binding protein IGF2BP1 is a druggable, post-transcriptional super-enhancer of E2F-driven gene expression in cancer. Nucleic Acids Res. 2020;48:8576-90
119. Wallis N, Oberman F, Shurrush K, Germain N, Greenwald G, Gershon T. et al. Small molecule inhibitor of Igf2bp1 represses Kras and a pro-oncogenic phenotype in cancer cells. RNA Biol. 2022;19:26-43
120. Dahlem C, Abuhaliema A, Kessler SM, Krohler T, Zoller BGE, Chanda S. et al. First Small-Molecule Inhibitors Targeting the RNA-Binding Protein IGF2BP2/IMP2 for Cancer Therapy. ACS Chem Biol. 2022;17:361-75
121. Feng P, Chen D, Wang X, Li Y, Li Z, Li B. et al. Inhibition of the m(6)A reader IGF2BP2 as a strategy against T-cell acute lymphoblastic leukemia. Leukemia. 2022;36:2180-8
122. Weng H, Huang F, Yu Z, Chen Z, Prince E, Kang Y. et al. The m(6)A reader IGF2BP2 regulates glutamine metabolism and represents a therapeutic target in acute myeloid leukemia. Cancer Cell. 2022;40:1566-82
123. Mancarella C, Pasello M, Ventura S, Grilli A, Calzolari L, Toracchio L. et al. Insulin-Like Growth Factor 2 mRNA-Binding Protein 3 is a Novel Post-Transcriptional Regulator of Ewing Sarcoma Malignancy. Clin Cancer Res. 2018;24:3704-16
124. Zhang Y, Liu X, Yu M, Xu M, Xiao Y, Ma W. et al. Berberine inhibits proliferation and induces G0/G1 phase arrest in colorectal cancer cells by downregulating IGF2BP3. Life Sci. 2020;260:118413
125. Cui Y, Wu Y, Wang C, Wang Z, Li Y, Jiang Z. et al. Isoliquiritigenin inhibits non-small cell lung cancer progression via m(6)A/IGF2BP3-dependent TWIST1 mRNA stabilization. Phytomedicine. 2022;104:154299
126. Shi J, Zhang Q, Yin X, Ye J, Gao S, Chen C. et al. Stabilization of IGF2BP1 by USP10 promotes breast cancer metastasis via CPT1A in an m6A-dependent manner. Int J Biol Sci. 2023;19:449-64
127. Luo F, Lin K. N(6)-methyladenosine (m(6)A) reader IGF2BP1 accelerates gastric cancer aerobic glycolysis in c-Myc-dependent manner. Exp Cell Res. 2022;417:113176
128. Wang C, Gu Y, Zhang E, Zhang K, Qin N, Dai J. et al. A cancer-testis non-coding RNA LIN28B-AS1 activates driver gene LIN28B by interacting with IGF2BP1 in lung adenocarcinoma. Oncogene. 2019;38:1611-24
129. Zhang L, Wan Y, Zhang Z, Jiang Y, Gu Z, Ma X. et al. IGF2BP1 overexpression stabilizes PEG10 mRNA in an m6A-dependent manner and promotes endometrial cancer progression. Theranostics. 2021;11:1100-14
130. Wang C, Zhou M, Zhu P, Ju C, Sheng J, Du D. et al. IGF2BP2-induced circRUNX1 facilitates the growth and metastasis of esophageal squamous cell carcinoma through miR-449b-5p/FOXP3 axis. J Exp Clin Cancer Res. 2022;41:347
131. Ouyang J, Li J, Li D, Jiang J, Hao T, Xia Y. et al. IGF2BP2 Promotes Epithelial to Mesenchymal Transition and Metastasis through Stabilizing HMGA1 mRNA in Gastric Cancer. Cancers (Basel). 2022;14:5381
132. Pu J, Wang J, Qin Z, Wang A, Zhang Y, Wu X. et al. IGF2BP2 Promotes Liver Cancer Growth Through an m6A-FEN1-Dependent Mechanism. Front Oncol. 2020;10:578816
133. Wang C, Kong F, Ma J, Miao J, Su P, Yang H. et al. IGF2BP3 enhances the mRNA stability of E2F3 by interacting with LINC00958 to promote endometrial carcinoma progression. Cell Death Discov. 2022;8:279
Author contact
![]() Corresponding authors: Kai Li, MD, PhD. Email: cmu_likaicom; Huizhe Wu, Email: wuhzedu.cn; Shan Gao, Email: mount1121com.
Corresponding authors: Kai Li, MD, PhD. Email: cmu_likaicom; Huizhe Wu, Email: wuhzedu.cn; Shan Gao, Email: mount1121com.
 Global reach, higher impact
Global reach, higher impact