13.3
Impact Factor
Theranostics 2023; 13(8):2384-2407. doi:10.7150/thno.79493 This issue Cite
Research Paper
Non-canonical integrin signaling activates EGFR and RAS-MAPK-ERK signaling in small cell lung cancer
1. Université de Lorraine, CNRS, Laboratoire IMoPA, UMR 7365; F-54000 Nancy, France.
2. Lung Cancer Epigenetics, Max-Planck-Institute for Heart and Lung Research; 61231 Bad Nauheim, Germany.
3. Department of Pathology, Massachusetts General Hospital and Harvard Medical School; Charlestown, MA, 02129, USA.
4. International Laboratory EPIGEN, Consejo de Ciencia y Tecnología del Estado de Puebla (CONCYTEP), Instituto de Ciencias, EcoCampus, Benemérita Universidad Autónoma de Puebla; Puebla 72570, Mexico.
5. Institute of Medical Microbiology and Hospital Hygiene, Department of Medicine, Philipps-University Marburg; Marburg, Germany.
6. Independent Researcher, collaborator of International Laboratory EPIGEN-CONCYTEP.
7. Emmy Noether Research Group Epigenetic Machineries and Cancer, Division of Chronic Inflammation and Cancer, German Cancer Research Center (DKFZ), 69120 Heidelberg, Germany.
8. Department of Cardiovascular Genomics and Epigenomics, European Center for Angioscience (ECAS), Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
9. German Centre for Cardiovascular Research (DZHK).
10. ECCPS Bioinformatics and Deep Sequencing Platform, Max-Planck-Institute for Heart and Lung Research; 61231 Bad Nauheim, Germany.
11. Department of Cardiac Development, Max-Planck-Institute for Heart and Lung Research; 61231 Bad Nauheim, Germany.
12. Pharmaceutical Technology and Biopharmaceutics, Department of Pharmacy, Ludwig-Maximilians-University of Munich; Munich, Germany.
13. Institute for Pathology, Justus Liebig University; 35392 Gießen, Germany.
14. Institute for Pathology, Hannover Medical School; Hanover, Germany.
15. Biomedical Research in Endstage and Obstructive Lung Disease Hannover (BREATH) Research Network; Hanover, Germany.
16. Center for Infection and Genomics of the Lung (CIGL), Universities of Giessen and Marburg Lung Center; Giessen, Germany.
17. Institute of Lung Health, German Center for Lung Research (DZL); Giessen, Germany.
18. CECAD, University of Cologne; Cologne, Germany.
19. Faculty of Medicine and University Hospital, University of Cologne; Cologne, Germany.
20. Center for Data and Simulation Science, University of Cologne; Cologne, Germany.
21. Center for Cancer Research, Massachusetts General Hospital and Harvard Medical School; Charlestown, MA, 02129, USA.
22. Biomolecular Mass Spectrometry, Max-Planck-Institute for Heart and Lung Research; 61231 Bad Nauheim, Germany.
23. Institute of Translational Proteomics, Department of Medicine, Philipps-University Marburg; 35043 Marburg, Germany.
†Lead contact +33 7 55 34 78 11.
Abstract
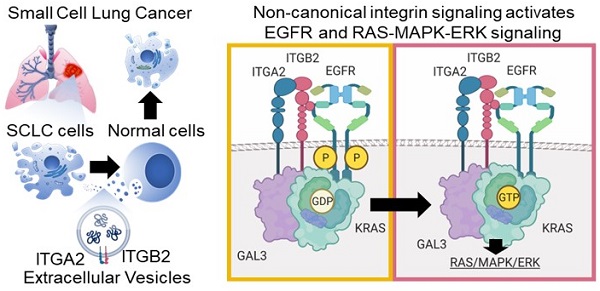
Background: Small cell lung cancer (SCLC) is an extremely aggressive cancer type with a patient median survival of 6-12 months. Epidermal growth factor (EGF) signaling plays an important role in triggering SCLC. In addition, growth factor-dependent signals and alpha-, beta-integrin (ITGA, ITGB) heterodimer receptors functionally cooperate and integrate their signaling pathways. However, the precise role of integrins in EGF receptor (EGFR) activation in SCLC remains elusive.
Methods: We analyzed human precision-cut lung slices (hPCLS), retrospectively collected human lung tissue samples and cell lines by classical methods of molecular biology and biochemistry. In addition, we performed RNA-sequencing-based transcriptomic analysis in human lung cancer cells and human lung tissue samples, as well as high-resolution mass spectrometric analysis of the protein cargo from extracellular vesicles (EVs) that were isolated from human lung cancer cells.
Results: Our results demonstrate that non-canonical ITGB2 signaling activates EGFR and RAS/MAPK/ERK signaling in SCLC. Further, we identified a novel SCLC gene expression signature consisting of 93 transcripts that were induced by ITGB2, which may be used for stratification of SCLC patients and prognosis prediction of LC patients. We also found a cell-cell communication mechanism based on EVs containing ITGB2, which were secreted by SCLC cells and induced in control human lung tissue RAS/MAPK/ERK signaling and SCLC markers.
Conclusions: We uncovered a mechanism of ITGB2-mediated EGFR activation in SCLC that explains EGFR-inhibitor resistance independently of EGFR mutations, suggesting the development of therapies targeting ITGB2 for patients with this extremely aggressive lung cancer type.
Keywords: small cell lung cancer, integrin, EGFR, KRAS, extracellular vesicles
 Global reach, higher impact
Global reach, higher impact