13.3
Impact Factor
Theranostics 2023; 13(7):2088-2113. doi:10.7150/thno.81488 This issue Cite
Review
Diagnosis and biomarkers for ocular tuberculosis: From the present into the future
1. Lee Kong Chian School of Medicine, Nanyang Technological University of Singapore, Singapore.
2. Yong Loo Lin School of Medicine, National University of Singapore, Singapore, Singapore.
3. National Healthcare Group Eye Institute, Tan Tock Seng Hospital, Singapore, Singapore.
4. Department of Ophthalmology, Faculty of Medicine Universitas Indonesia - CiptoMangunkusmoKirana Eye Hospital, Jakarta, Indonesia.
5. Laboratory Medical Immunology, Department of Immunology, ErasmusMC, UniversityMedical Centre, Rotterdam, the Netherlands.
6. Department of Internal Medicine, Division of Clinical Immunology, Erasmus MC, University Medical Center, Rotterdam, The Netherlands.
7. Department of Ophthalmology, Erasmus MC, University Medical Center, Rotterdam, The Netherlands.
8. Immunology Programme, Life Sciences Institute, National University of Singapore, Singapore, Singapore.
9. Institute of Molecular and Cell Biology (IMCB), Agency for Science, Technology and Research (A*STAR), Singapore, Singapore.
10. Department of Biochemistry, Post Graduate Institute of Medical Education and Research, Chandigarh, India.
11. A*SATR Infectious Diseases Labs (A*STAR ID Labs), Agency for Science, Technology and Research (A*STAR), Singapore, Singapore.
12. Singapore Immunology Network (SIgN), Agency for Science, Technology and Research (A*STAR), Singapore, Singapore.
13. University of Indonesia Hospital (RSUI), Depok, West Java, Indonesia.
14. Advanced Eye Centre, Post-Graduate Institute of Medical Education and Research (PGIMER), Chandigarh, India.
15. Duke NUS Medical School, Singapore, Singapore.
16. Singapore Eye Research Institute, Singapore, Singapore.
17. National Institute for Health Research Biomedical Research Centre, Moorfields Eye Hospital, London, UK.
18. School of Pharmacy, Nantong University, Nantong, P. R. China.
19. Department of Mechanical Engineering, University College London, London, United Kingdom.
Received 2022-12-2; Accepted 2023-3-19; Published 2023-4-1
Abstract
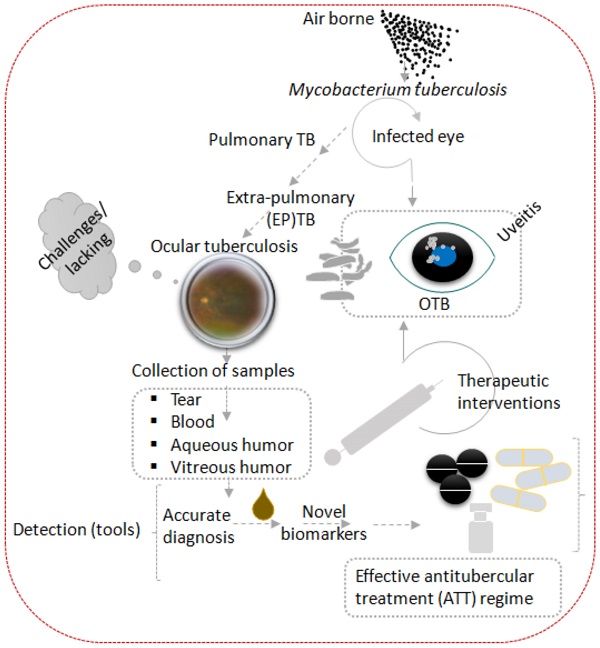
Tuberculosis is an airborne disease caused by Mycobacterium tuberculosis (Mtb) and can manifest both pulmonary and extrapulmonary disease, including ocular tuberculosis (OTB). Accurate diagnosis and swift optimal treatment initiation for OTB is faced by many challenges combined with the lack of standardized treatment regimens this results in uncertain OTB outcomes. The purpose of this study is to summarize existing diagnostic approaches and recently discovered biomarkers that may contribute to establishing OTB diagnosis, choice of anti-tubercular therapy (ATT) regimen, and treatment monitoring. The keywords ocular tuberculosis, tuberculosis, Mycobacterium, biomarkers, molecular diagnosis, multi-omics, proteomics, genomics, transcriptomics, metabolomics, T-lymphocytes profiling were searched on PubMed and MEDLINE databases. Articles and books published with at least one of the keywords were included and screened for relevance. There was no time limit for study inclusion. More emphasis was placed on recent publications that contributed new information about the pathogenesis, diagnosis, or treatment of OTB. We excluded abstracts and articles that were not written in the English language. References cited within the identified articles were used to further supplement the search.
We found 10 studies evaluating the sensitivity and specificity of interferon-gamma release assay (IGRA), and 6 studies evaluating that of tuberculin skin test (TST) in OTB patients. IGRA (Sp = 71-100%, Se = 36-100%) achieves overall better sensitivity and specificity than TST (Sp = 51.1-85.7%; Se = 70.9-98.5%). For nuclear acid amplification tests (NAAT), we found 7 studies on uniplex polymerase chain reaction (PCR) with different Mtb targets, 7 studies on DNA-based multiplex PCR, 1 study on mRNA-based multiplex PCR, 4 studies on loop-mediated isothermal amplification (LAMP) assay with different Mtb targets, 3 studies on GeneXpert assay, 1 study on GeneXpert Ultra assay and 1 study for MTBDRplus assay for OTB. Specificity is overall improved but sensitivity is highly variable for NAATs (excluding uniplex PCR, Sp = 50-100%; Se = 10.5-98%) as compared to IGRA. We also found 3 transcriptomic studies, 6 proteomic studies, 2 studies on stimulation assays, 1 study on intraocular protein analysis and 1 study on T-lymphocyte profiling in OTB patients. All except 1 study evaluated novel, previously undiscovered biomarkers. Only 1 study has been externally validated by a large independent cohort.
Future theranostic marker discovery by a multi-omics approach is essential to deepen pathophysiological understanding of OTB. Combined these might result in swift, optimal and personalized treatment regimens to modulate the heterogeneous mechanisms of OTB. Eventually, these studies could improve the current cumbersome diagnosis and management of OTB.
Keywords: Ocular tuberculosis, molecular diagnostic techniques, biomarkers, multi-omic, precision medicine
1. Background of the clinical problems 1.1. Epidemiology of Tuberculosis and Ocular Tuberculosis (OTB)
Tuberculosis (TB), caused by Mycobacterium tuberculosis (Mtb), is a major public health problem, especially in developing countries. Currently the World Health Organization (WHO) estimates that one-quarter of the world's population is latently infected with Mtb, of these 10% will eventually progress to active tuberculosis [1-3]. Furthermore, the WHO has estimated that 10 million people suffered from active TB in 2021, resulting in 1.6 million deaths [4]. Despite the advancement of GeneXpert MTB/RIF, a nucleic acid assay for rapid TB detection that simultaneously tests for rifampicin sensitivity in pulmonary samples, extrapulmonary TB (EPTB) detection remains challenging due to pauci-bacillary specimens [5]. In addition, suboptimal diagnostic performance of chest x-ray (sensitivity 87%, specificity 89%), sputum smear (sensitivity 32-94%, specificity 50-99%), culture (sensitivity 73-89%, specificity >99%), MTB/RIF (sensitivity 67-98%, specificity 98-99%) further complicates the diagnosis of pulmonary TB. Sputum smear, the classic method of detecting pulmonary TB, requires 5000 to 10,000 bacteria per ml for reliable detection. On top of mentioned assay limitation for the detection of TB only 5-10% of TB-infected individuals display clinical signs and symptoms associated with pulmonary TB [6]. Currently available data on ocular tuberculosis (OTB) varies widely due to the lack of specific diagnostic criteria [7]. Prevalence of ocular tuberculosis (OTB) on a global scale is estimated to account for 4.0% (95% CI 3.0-5.0%) of all uveitis cases and isreported to be one of the leading causes of infectious uveitis, ranging from 22.9-48.0% in Indonesiaand India [8-10]. OTB can affect almost all parts of the eye and may cause sight-threatening complications, such as glaucoma, cataract, and cystoid macular edema in the absence of swift and appropriate treatment [11].
1.2. Clinical features and diagnostic challenges for OTB
OTB is an extrapulmonary manifestation of Mtb infection, which generally is transmitted through inhaled bioaerosols containing Mtb bacilli. Following inhalation, a primary infectious foci is formed in the lungs, dissemination can subsequently occur through the lymphatic and hematologic compartment resulting in EPTB, which can also include the eye's uveal tract (iris, ciliary body, and choroid) [12]. It is hypothesized that Mtb-laden macrophages deposit in the first available capillary beds upon entering the eye, which most likely is the highly vascularized choroid, creating an oxygen rich environment analogous to the apex of the lung [13]. These characteristics likely account for the fact that posterior uveitis is the most common presentation of OTB [14].
Apart from a secondary infection caused by haematogenous spread from a distant infectious site (i.e. lung), OTB can also occur as a primary infection in conjunctiva, cornea, sclera, adnexa, lids, and lacrimal apparatus [13]. Due to the rarity of microbiological evidence of Mtb in ocular fluids two additional pathophysiological mechanisms of OTB have been suggested [15]. The first one indicates that OTB could be a manifestation of a hypersensitivity response to Mtb antigens in the setting of local ocular infection [16]. The second advocate that OTB is an autoimmune reaction to ocular antigens that results from antigenic mimicry between Mtb and ocular antigens (e.g. interphotoreceptor retinoid-binding protein-specific autoantigen), resulting in ocular inflammation even in the absence of mycobacterial products in the eye [17]. How the various pathophysiological mechanism of OTB contribute to the disease and if typical clinical phenotypes can be attributed to a specific mechanism has not been clearly established yet. Despite Mtb's paucibacillary nature, a strong chronic and recurrent intraocular inflammatory response, often in need of adjunctive corticosteroid therapy, has been described. These observations heightened interest in immunopathogenic studies to identify novel biomarkers, such as proteins and metabolites, involved in disease activity and treatment efficacy [18].
OTB is considered as an imitator of various non-inflammatory and infectious or non-infectious inflammatory ocular pathologies, requiring a very high index of suspicion to be diagnosed [16, 19]. Moreover, OTB can occur without clinically apparent pulmonary TB or other signs suggestive of active TB disease [20]. Occult TB elsewhere in the body is generally restricted to paucibacillary and clinically silent sites such as the intrathoracic lymph nodes [18]. Altogether, these difficulties contribute to diagnostic delays, which increase morbidity and vision loss.[19]
Recently, the current view of dichotomized “latent” and active “TB” has been challenged by the hypothesis that there are more than two different stages of TB: eliminated, incipient, subclinical form, and active TB [21]. In ocular TB, many diagnostic confirmation relies on the positivity of immunoreactivity to TB antigens. A previous study found that even in histopathologically-proven ocular TB, active systemic TB may not be present in all cases [22].
As defined by WHO, a bacteriologically confirmed case of TB is whose biological specimen is positive by either smear microscopy, or culture or WHO-approved rapid diagnostic test. These tests fulfills the target product profile (TPP) criteria of goal sensitivity of ≥ 98% % in smear‐positive, culture‐positive pulmonary TB, and ≥ 68% in smear‐negative, culture‐positive adults [23]. However, no optimal TPP diagnostic sensitivity for ocular fluid specimens have been recommended, although the revised recommended TPP for EPTB is 80% sensitivity for all forms of microbiologically-confirmed EPTB [24]. Apart from this, even if a patient does not fulfil the criteria for bacteriological confirmed TB but has been diagnosed with active TB by a clinician, who then decided to initiate anti-tubercular therapy (ATT), is also considered as a clinically diagnosed case of TB. In this scenario the clinical diagnosis could be on the basis of Chest X-ray abnormalities or suggestive histology without laboratory confirmation [23].
On the other hand, global uveitis experts of the Collaborative Ocular Tuberculosis Study (COTS) group have agreed that there is currently no single gold standard diagnostic test for OTB. Clinical diagnosis of OTB is challenging due to highly variable clinical phenotypes, local prevalence of TB, immigration from a highly-endemic country and variable interferon-gamma release assay (IGRA), or tuberculin skin test (TST) or chest x-ray (CXR) findings [25]. The key diagnostic criteria for OTB as defined by the Standardization of Uveitis Nomenclature (SUN) Working Group is a compatible uveitic syndrome, including: 1) anterior uveitis with iris nodules, 2) serpiginous-like tubercular choroiditis, 3) choroidal nodule (tuberculoma), 4) occlusive retinal vasculitis, and 5) in hosts with evidence of active systemic TB, multifocal choroiditis; including: 1) histologically- or microbiologically-confirmed infection, 2) positive IGRA test, or 3) positive TST [26]. Putting all these together, the COTS group published an online, cost-effective, web-based clinical scoring system known as the COTS Calculator (https://www.oculartb.net/cots-calc) to guide the crucial decision of initiating ATT in clinically suspected OTB cases [27].
1.3. Corroborative tests for presumptive diagnosis of OTB
Confirmed OTB requires detection of Mtb from ocular samples. However, OTB is often paucibacillary and difficult to confirm by conventional tests such as nucleic acid amplification tests (NAAT), smear microscopy, or culture [18]. Collection of ocular fluid (tears, aqueous humour, vitreous fluid) or retinal biopsy specimens to confirm the presence of Mtb in ocular tissue samples is not routinely performed due to the invasive nature of the procedures needed to acquire these ocular specimens which in addition form a potential risk for loss of visual acuity among patients with existing ocular inflammation [28]. Therefore, clinical diagnosis of OTB is often presumptive, relying on indirect evidence of TB infection and exclusion of other possible uveitis causes. Corroborative tests like IGRA and TST and chest imaging, assessing the presence of TB suggestive lesions, are usually performed in the presence of a clinical OTB presentation and have a supportive role in the diagnostic workup [18].
Moreover, for most patients with TB-associated immune-induced retinal vasculopathy, actual detection of Mtb is uncommon because the yield of organisms from intraocular specimens is too low [19]. The clinical presentation of tubercular retinal vasculitis can be variable, (1) as an exudative, segmental, hemorrhagic retinal vasculitis, usually associated with peri- or sub-vascular choroiditis, and vitritis, or (2) as a peripheral, minimally exudative, non-hemorrhagic, isolated retinal vasculitis with extensive (not segmental) swathes of pipestem-like sheathing with minimal or no vitritis. The latter is also referred to as “Eales disease” and is proposed to be more related to a hypersensitivity response to tubercular protein, hence proving the presence of Mtb may be difficult in this condition and the diagnosis is mainly based on immunoreactivity to TB antigen [14].
1.3.1. Immunological skin and blood tests
TST also known Mantoux test is a century-old test to assess the presence of immunological memory against Mtb antigens (purified protein derivative (PPD)) during latent and active infection. It is low-cost and readily available in clinics worldwide. However, TST specificity is limited and may produce a false positive result in persons vaccinated with Bacillus Calmette-Guérin (BCG) or those infected with non-tuberculous Mycobacterium (NTM) [29]. Of note, limitations of TST include false negative results in immunocompromised patients or populations with impaired cellular immunity, such as young children and the elderly [30]. TST results can also be confounded by concurrent dermatological diseases such as psoriasis [18, 31]. Moreover, significant variation in the administration and interpretation of TST affects the uniformity and objective reliability of the test. While the recommended cut-off diagnosis for latent TB infection (LTBI) is 10mm, this cut-off value is adjusted to more than 15mm in endemic countries to reduce the rate of false positives and over-treatment with anti-tubercular therapy [32]. Overall, TST has a limited reported specificity of 51.1-85.7% and sensitivity of 70.9- 98.5% for diagnosing OTB (see Table 1).
IGRAs are full blood tests that asses IFN-γ production by T-lymphocytes in response to Mtb antigens, early secreted antigenic target 6 (ESAT-6) and culture filtrate protein 10 (CFP10). These antigens are encoded within the region of difference 1 (RD1) which is absent in BCG and most NTM. Hence, IGRA shows a better specificity for diagnosing OTB than TST (Table 1). The sensitivity of IGRA when compared to TST is more contentious with varying results in available reports. In five head-to-head comparisons of IGRA and TST in TB-endemic countries, IGRA sensitivity underperformed as compared to TST [33-36], while a non-comparative study by Ahn et al. reported a sensitivity of 100% [37]. Amongst two head-to-head comparisons of IGRA and TST in non-endemic countries, only Llorenc et al. reported an improved IGRA sensitivity as compared to TST [38, 39]. Overall, studies more often report a reduced IGRA sensitivity when compared to TST. In addition, the limited number of OTB studies, consistently reported underperforming IGRA sensitivities in TB-endemic countries. This is concordant with the WHO observation that IGRA sensitivity for TB is lower in TB-endemic countries [40]. In practice, low-resources in most TB endemic regions, prevent the use of IGRA due to high cost, technical challenges and IGRA's inability to distinguish active from latent TB [33, 41]. While current literature generally recommends IGRA as a routine screening tool for OTB, due to the low sensitivity of IGRA, negative or indeterminate IGRA results in patients with clinical characteristics strongly suggestive of OTB should be interpreted with caution as the presence of anti-IFN-γ autoantibodies might interfere with detectable IFN-γ levels [28]. TST and IGRA tests demonstrate biomarker use in the form of indirectly and directly quantifying IFN-γ responses to aid OTB diagnosis.
Studies evaluating the specificity and sensitivity of tuberculin skin test (TST), and three different brands of interferon-gamma release assays (IGRA) - QFT-GIT, QFT-PLUS, T-SPOT.TB.
| Test Name | n | Country | Endemic | Inclusion criteria for Controls | Specificity | Sensitivity | |
|---|---|---|---|---|---|---|---|
| TST | |||||||
| Fernández-Zamora et al. (2022) [33] | TST | 191 | Brazil | Yes | Diagnosed with uveitis secondary to other non-infectious or non-TB infectious cause; Non-responsive to ATT. | 71.8 | 98.5 |
| Llorenc et al. (2013) [38] | TST | 103 | Spain | No | Diagnosed with uveitis secondary to other non-infectious or non-TB infectious cause; Non-responsive to ATT. | 85.7 | 87.8 |
| Ang et al. (2009) [34] | TST | 157 | Singapore | Yes | Non-responsive to ATT. | 72.7 | 95.5 |
| Cordero-Coma et al. (2010) [42] | TST | 83 | Spain | No | Non-inflammatory eyes. | 84 | 81 |
| Ang et al. (2012) [35] | TST | 138 | Singapore | Yes | Non-responsive to ATT. | 51.1 | 72.0 |
| Ang et al. (2014) [36] | TST | 106 | Singapore | Yes | Bayesian analysis in the absence of gold standard diagnostic. | 68.3 | 70.9 |
| IGRA | |||||||
| Fernández-Zamora et al. (2022) [33] | QFT-GIT, QFT-PLUS | 191 | Brazil | Yes | Diagnosed with uveitis secondary to other non-infectious or non-TB infectious cause; Non-responsive to ATT. | 99.2 | 90.8 |
| Llorenc et al. (2013) [38] | QFT-PLUS | 103 | Spain | No | Diagnosed with uveitis secondary to other non-infectious or non-TB infectious cause; Non-responsive to ATT. | 82.8 | 90.9 |
| Ang et al. (2009) [34] | QFT-GIT | 157 | Singapore | Yes | Non-responsive to ATT. | 81.8 | 90.9 |
| Cordero-Coma et al. (2010) [42] | QFT-GIT | 83 | Spain | No | Non-inflammatory eyes. | 100 | 81 |
| Babu et al. (2009) [43] | QFT-GIT | 60 | India | Yes | Diagnosed with uveitis secondary to other non-infectious or non-TB infectious cause; Not a close contact of TB patient. | 76 | 82 |
| Ang et al. (2012) [35] | T-SPOT.TB | 138 | Singapore | Yes | Non-responsive to ATT. | 75.0 | 36.0 |
| Ang et al. (2014) [36] | QFT-GIT | 106 | Singapore | Yes | Bayesian analysis in the absence of gold standard diagnostic. | 99.6 | 64.2 |
| Ang et al. (2014) [36] | T-SPOT.TB | 106 | Singapore | Yes | Bayesian analysis in the absence of gold standard diagnostic. | 90.6 | 50.0 |
| Ahn et al. (2014) [37] | QFT-GIT | 181 | Korea | Yes | Non-responsive to ATT. | 72.0 | 100 |
| Gineys et al. (2011) [39] | QFT-GIT | 96 | France | No | Diagnosed with uveitis secondary to other non-infectious or non-TB infectious cause; Non-responsive to ATT. | 87.0 | 84.0 |
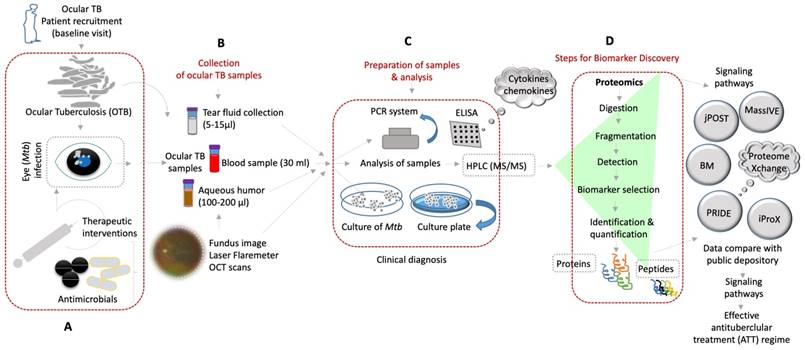
Shows the molecular diagnosis by various approaches of the ocular samples obtained from OTBpatients and identification of novel biomarkers for better treatment of ocular tuberculosis. (A-D) Diverse clinical approaches and diagnostic tools that are used to detect OTB from the patient's samples by using mass spectrometry based-proteomics/peptidomics and others for discovery of novel biomarkers (BM) of potential therapeutic targets and better treatment options of OTB.
1.3.2. Radiographic tests
Other investigative tests include chest X-ray, fluorescein angiography, and ultrasonography. Chest X-ray is used to evaluate patients with suspected intraocular TB, since the lungs are most often the primary site of TB infection. Chest computed tomography and positron emission tomography scans are not routinely performed due to high costs, even though superior delineation between concomitant parenchymal, hilar, or pleural lesions in normal or inconclusive chest X-rays [44]. However, chest radiologic evidence of post-inflammatory lesions such as parenchymal scarring or hilar lymphadenopathy is not specific to TB [45]. Fluorescein angiography can assist in the evaluation of retinal vascular leakage and active choroidal lesions while ultrasonography can support differentiation between uveal tumors and tuberculomas [46].
As corroborative tests, IGRA/TST and chest radiography are insufficient to diagnose OTB. A previous study reported the presence of 1-2 Mtb bacilli at the level of the retinal pigment epithelium layer [22]. This paucibacillary infection in RPE was sufficient to cause ocular inflammation, with some cases having a negative IGRA and TST underlining their ancillary nature [22]. Novel molecular approaches in the diagnosis of OTB can be complementary to (rather than a substitute for) these existing and older tools [22, 47]. Therefore, this review summarizes the published literature for existing confirmatory diagnostic tools in Section 2 and covers the technological development of molecular diagnostics using novel biomarkers for OTB in Section 3 (Figure 1).
1.4. Role of HIV co-infection on OTB diagnosis
An observational study conducted in a tertiary hospital in India reported elevated ocular morbidity (23.8%) in patients co-infected with TB and human immunodeficiency virus (HIV) as compared to those only infected with TB [48]. Nevertheless, it is important to remember that studies on OTB diagnostic tests have largely been carried out in the HIV-uninfected populations, or the study designs do not distinguish patients with HIV from those without as the sample sizes are already small. Hence, most of the adjusted cut-offs in HIV patients come from studies observing HIV patients co-infected with other forms of EPTB, not restricted to OTB.
A significant proportion of HIV patients display CD4 counts <100 cells/μl. Therefore, a Mantoux test can underestimate the prevalence of TB when HIV patients have lower immune reactivity. The cut-off value for positive Mantoux therefore needs to be adjusted to 5mm for HIV patients unlike 10mm for immunocompetent patients [49]. In a systemic review of 14 studies not limited to OTB patients, IGRA testing has a pooled sensitivity of 81% in the general population, but co-infection with HIV reduces sensitivity up to 63% [50]. As HIV induces immunosuppression, extrapulmonary TB (EPTB) disease, mediastinal lymphadenopathy, and miliary tuberculosis become more common, whereas chest x-ray is less likely to show signs of upper lobe cavitation. PET/CT scans and unenhanced CT scans were found to both have similar sensitivity (33.3%) and specificity (100%) for EPTB in 29 OTB patients (5 were HIV-positive) [51]. These findings clearly demonstrate that HIV infection affects interpretation of TB diagnostic tests. It would be prudent of ophthalmologists to check the HIV status of patients suspicious for OTB infection.
2. Existing diagnostic tools for OTB
2.1. Detecting Mtb using traditional bacteriologic tests
Traditional bacteriologic diagnosis used Ziehl-Neelsen or auramine-rhodamine staining's of ocular fluids or tissue sections to detect acid-fast bacilli (AFB) to diagnose OTB.[52] Regarding pulmonary TB, acid-fast bacilli (AFB) smears in sputum samples display a sensitivity of 50% [53]. Hence, the likelihood of detecting AFB in aqueous or vitreous fluid is even lower due to the paucibacillary nature of EPTB and intrinsic antimicrobial properties of conjunctiva and tears that further make the ocular surface paucibacterial with few culturable bacteria [19, 54, 55]. Direct identification of Mtb by culture is considered the gold standard in diagnosis, however, only very few if any bacilli can be obtained from aqueous or vitreous humor and culture results may take 6-8 weeks, significantly delaying diagnosis and treatment initiation [52]. Even though, costly semi-automated or fully-automated systems with liquid media can provide earlier results; diagnosis of OTB based on AFB smear and culture lacks accuracy and speed [53].
2.2. Significance of Molecular Signatures and Novel Biomarkers
A biomarker is defined as a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention [56]. Before diving deep into ongoing research on novel biomarkers, the following subsections aim to emphasize molecular and serological biomarker-based tools already used in clinical practice to directly identify Mtb. (Figure 2). These tests are not routinely performed as triage, unlike the corroborative tests described in the previous sections; however, they have been validated for clinical practice.
2.2.1. Cellular and serological immuno-assays
It has been shown that antibodies against purified cord factor antigen, trehalose-6,6'-dimycolate (TDM), the most abundant cell wall component of Mtb bacilli, can be used for rapid serodiagnosis of pulmonary TB [57]. A study by Sakai et al reported a sensitivity and specificity of 100% for the detection OTB by measuring anti-TDM IgG antibodies in serum samples of nine presumed OTB, three sarcoidosis and three Behcet's disease patients [58]. However, validation studies with larger sample sizes are needed. A decline in anti-TDM antibody levels has been reported during pulmonary TB after ATT initiation, suggesting applicability to monitor treatment efficacy [59]. Detection of anti-TDM antibodies suggests the presence of Mtb bacilli even in the absence of active systemic disease [58]. However, as with IGRA and TST, detection of anti-TDM- antibodies does not differentiate between active, latent, or past TB infection (Figure 2).
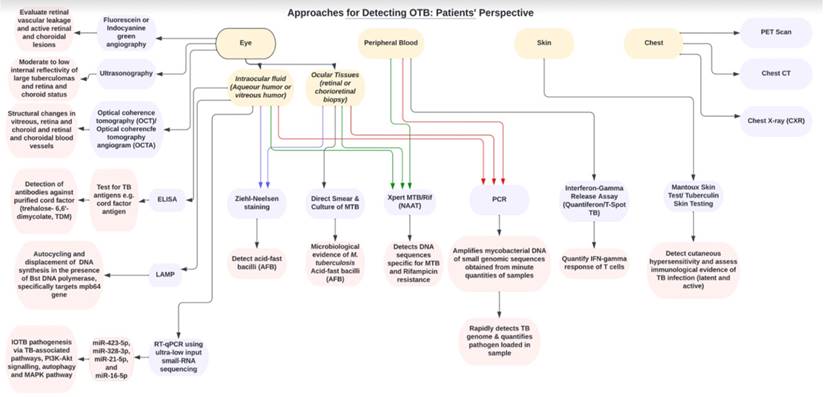
Flow chart showing the clinical approaches and diagnostic tools for detecting OTB from patient's perspective.
2.2.2. Detecting Mtb using nucleic acid amplification tests
Nucleic Acid Amplification Tests (NAAT) enables the amplification of pathogen-specific genomic DNA sequences from minute quantities of ocular fluid/tissue or blood samples. NAAT displays an improved sensitivity over traditional bacteriologic tests, smear microscopy and culture techniques described above (See Table 2). PCR, a form of NAAT, is increasingly commonly applied to rapid detect Mtb-specific DNA sequences such as IS6110 and MPB64 yielding an excellent specificity range of 80-100% (see table 2). However, Mtb-specific PCR performed on ocular fluid is relatively time-consuming and associated with a low to moderate sensitivity of 37.7-73.3%, resulting in false-negatives when either one of the IS6110 or MPB64 gene targets were used [60-64].
More advanced NAAT with improved diagnostic accuracy for detecting Mtb is also available. Multiplex PCR that is based on the amplification of three Mtb target genes (i.e. IS6110, MPB64, and rpoB or protein b) showed improved sensitivity (71.8-77.8%) compared to single-gene target PCR analysis due to the variable presence of IS6110 gene copy in North Indian populations, and false-negative detection of MPB64 gene in simultaneous M. bovis and M. fortuitum infection (see Table 2) [63-67]. Another advantage of multiplex PCR is the possibility to test for multiple pathogens at once in very limited volumes of aqueous or vitreous fluid, which contributes to differential diagnoses.
Another type of NAAT, the loop-mediated isothermal amplification (LAMP) assay, targets specific Mtb genomic sequences to produce reliable results within 1 hour. The LAMP assay does not require expensive PCR instrumentation but rather uses Bacilluss tearothermophilus DNA polymerase under isothermal conditions (60-65ºC) in an ordinary laboratory water bath or heating block [64]. This makes LAMP assay attractive to detect Mtb, especially in resource-limited settings. Based on DNA synthesis of the Mtb MPB64 gene, a 100% specificity and 67.1-85.7% sensitivity were reached in intraocular samples, while this was 100% and 64.3-70% respectively in the case of targeting the IS6110 gene [64, 68, 69]. Sharma et al reported that IS1081, a multi-copy gene in the Mtb genome, has higher sensitivity (71.4%) than other single gene target LAMP (IS6110 and MPB64), while the multiplex LAMP consisting of all three gene targets yielded the highest sensitivity of 77.1% [64].
Although NAAT for Mtb detection in intraocular fluids displays a high specificity, the sensitivity is variable and limited. The latter may be explained by the following: (1) the limited (low) volume of ocular fluids and consequent minute quantity of genomic material obtained, (2) the non-uniform distribution of Mtb genomic material within specimens, (3) the presence of inhibitory factors within the specimens that interfere with NAAT efficacy, and (4) the lack of proper gold standard such that the diagnostic potential of NAAT is often evaluated against a presumptive OTB diagnosis [70]. Data from the COTS-1 illustrated the clinical utility of PCR results. A positive PCR result may support the decision for ATT initiation to prevent the recurrence of OTB flare [47]. Of note, a negative PCR result is insufficient to exclude OTB, and a decision to initiate ATT should be considered in conjunction with other corroborative test results and clinical presentation. In a recent meta-analysis, when the diagnostic value of the PCR result was calculated in relation to ATT treatment response in patients, the overall sensitivity and specificity were 88% (95% CI 83-92) and 71% (95% CI 60-80), respectively [71]. This stresses the limited ability of the current PCR technique to diagnose ocular TB, even though it is still considered valuable in the diagnostic algorithm for OTB where it is available.
2.2.3. Detecting drug resistance gene mutations using NAAT
Multidrug-resistant (MDR) OTB can be a reason for a worsening clinical presentation upon ATT initiation [72]. Emerging molecular genetic tools are useful to rapidly detect drug resistance to ATT as opposed to drug sensitivity tests from bacterial cultures [73]. Swift identification of drug resistance is critical to guide the proper choice of anti-tubercular treatment and to stop misuse of drugs that might perpetuate the evolution of additional drug resistance, which carries serious risks of MDR-TB transmission that is even more difficult to treat [74]. While drug susceptibility testing using GeneXpert and Line Probe Assays (LPA), such as MTBDR plus, are well-documented in published literature for pulmonary TB, there is scanty evidence of ocular sample usage in LPAs.
The GeneXpert MTB/RIF assay is a WHO-endorsed, cartridge-based, automated assay based on real-time PCR or quantitative RT-PCR technology with a rapid turn-around time of 2-3 hours [63]. It employs molecular beacons to detect Mtb-specific DNA and evaluate rpoB gene mutations responsible for rifampicin resistance. Even though, GeneXpert demonstrated a lower sensitivity of 10.5-17.2% in detecting Mtb from vitrectomy samples, it potentially provides rapid information on drug resistance in patients with OTB and is a recommended first-line screening test when available [63, 66, 67]. In a large systematic review evaluating rifampicin resistance in adult sputum samples, GeneXpert pooled sensitivity for TB was 88% while in the HIV-positive sub-group, the sensitivity was only 80% [75]. As this study did not evaluate ocular fluids in HIV patients with ocular inflammation, the sensitivity of GeneXpert in sputum samples of HIV patients seems to be much higher than the sensitivity in vitreous samples of OTB patients with unknown HIV status (see Table 2).
Studies evaluating the specificity and sensitivity of uniplex PCR with only one gene target, loop mediated amplification assays (LAMP), multiplex PCR with multiple gene (DNA or mRNA-based) targets, GeneXpert Assay, GeneXpert Ultra Assay, MTBDRplus Assay.
| Country (endemicity) | N | Samples | Presumed ocular TB criteria in each study | Targets used | Specificity | Sensitivity | Advantages | Limitations | |
|---|---|---|---|---|---|---|---|---|---|
| Uniplex PCR | |||||||||
| Gupta et al. (1998) [79] | India (Yes) | 10 | Aqueous | (a) vasculitis, (b) anterior vitreous cells, (c) snowball, (d) snowbanking, or (e) retinochoroiditis | H37RA DNA (150 bp fragment) | 33 | 95 | Faster turn-around-time than smear | All NAATs are limited by the inability to differentiate live and dead bacilli, the latter causing false-positivity hence decreasing specificity. Sophisticated and costly electricity-driven equipment required for amplification with limited throughput for PCR, thus it is unsuitable for resource-limited high-endemic regions [64]. Variable sensitivity and specificity with different gene targets. IS1081 is a multi-copy gene in the Mtb genome, hence increases the yield of detection than other genes that are present as single copy. Up to 40% of North Indian strains of Mtb lack IS6110 gene copy, hence explaining variable sensitivity of PCR in different populations. MPB64 is known to give false-negative results in the concurrent presence of other member(s) mycobacterium family other than Mtb (e.g. M bovis or M. fortuitum) [64]. |
| Arora et al. (1999) [60] | India (Yes) | 53 | Aqueous | Anterior chamber inflammation, with at least one of the following: (a) vasculitis, (b) anterior vitreous cells, (c) snowball, (d) snowbanking, or (e) retinochoroiditis | H37RA DNA (150 bp fragment) | 95.3 | 37.7 | ||
| Gupta et al. (2003) [80] | India (Yes) | 5 | Aqueous or vitreous | Serpiginous choroiditis | IS6110 | 33 | 89 | ||
| Singh et al. (2012) [81] | India (Yes) | 11 | Vitreous | Eales disease | MPB64 | 33 | 95 | ||
| Murugan et al. (2016) [61] | India (Yes) | 22 | Aqueous or vitreous | Clinical history comprehensive ophthalmic examination, systemic and ocular investigations | MPB64 | 80 | 42 | ||
| Sudheer et al. 2018 [62] | India (Yes) | 56 | Aqueous and/or vitreous | Clinical features: hypopyon, granulomatous keraticpercipitate, iris, choroid, or disc granulomas, active vasculitis, choroiditis, and healed chorioretinal scars along blood vessels; minimum 6 months of follow-up; no response to oral steroids | MPB64 | 92.3 | 73.3 | ||
| Sharma et al. (2019) [63] | India (Yes) | 200 | India (Yes) | (2) all known causes of infectious uveitis except TB and known noninfectious uveitic syndromes ruled out; (3) positive tuberculin skin test (4) received antitubercular therapy for a minimum of 12 months (6) no recurrence of uveitis. Out of these 70 cases, 3 were culture-positive and 67 were culture negative for M. tuberculosis. | MPB64 | 100 | 68.2 | ||
| Sharma et al. (2019) [63] | India (Yes) | 200 | India (Yes) | IS6110 | 100 | 66.4 | |||
| Sharma et al. (2020) [64] | India (Yes) | 120 | Vitreous | IS1081 | 100 | 61.42 | |||
| LAMP | |||||||||
| Balne et al. (2013) [68] | India (Yes) | 14 | Aqueous or vitreous | Criteria as per described in [19] | MPB64 | 100 | 85.7 | Easily administered in the rural setting through battery-operated water baths, simple instructions requiring minimal staff training, and rapid results within one hour. [64] Cost less than 1 USD per sample. [64] High specificity. Higher sensitivity than PCR using the same target gene(s) as LAMP uses three primer pairs to recognize more regions of the target gene(s). [64] | Specificity is still limited, although more than four-fold that of GeneXpert. Insufficient external validation with different geographical populations harboring different genotypes of the Mtb complex (MTBC) to be WHO-endorsed [64]. Unable to detect rifampicin or isoniazid resistance. |
| Sharma et al. (2020) [64] | India (Yes) | 120 | Vitreous | (2) all known causes of infectious uveitis except TB and known noninfectious uveitic syndromes ruled out; (3) positive tuberculin skin test (4) received antitubercular therapy for a minimum of 12 months (6) no recurrence of uveitis. Out of these 70 cases, 3 were culture-positive and 67 were culture negative for M. tuberculosis. | MPB64 | 100 | 67.14 | ||
| Sharma et al. (2015) [82] | India (Yes) | 30 | Vitreous and 1 iris biopsy | Confirmed by positive multitargeted PCR for M tuberculosis from intraocular samples | IS6110 | 100 | 70 | ||
| Sharma et al. (2020) [64] | India (Yes) | 120 | Vitreous | (2) all known causes of infectious uveitis except TB and known noninfectious uveitic syndromes ruled out; (3) positive tuberculin skin test (4) received antitubercular therapy for a minimum of 12 months (6) no recurrence of uveitis. Out of these 70 cases, 3 were culture-positive and 67 were culture negative for M. tuberculosis. | IS6110 | 100 | 64.28 | ||
| Sharma et al. (2020) [64] | IS6110, MPB64, IS1081 | 100 | 77.14 | ||||||
| Sharma et al. (2020) [64] | IS1081 | 100 | 71.42 | ||||||
| DNA-based Multiplex PCR | |||||||||
| Biswas et al. (2016) [83] | India (Yes) | 21 | Aqueous | MSC or choroiditis suspected for TB | IS6110, MPB64 | 50 | 98 | Combination of multiple gene targets has better yield of detection (sensitivity) than compared to a single gene target used. Rapid results within2-3 hours. [64] | Same limitations as Uniplex PCR. Costs nearly 10 times that of LAMP [64]. In settings endemic for MDR-TB, MPCR is unable to detect rifampicin or isoniazid resistance, hence an additional step of gene sequencing to search for resistance genes will cost approximately 20 USD with a turn-around time of 2-3 days [63]. Specificity is still limited, although more than four-fold that of GeneXpert, and more than two-fold that of MTBDR Assay. |
| Mohan et al. (2014) [84] | India (Yes) | 13 | Aqueous | MSC or choroiditis suspected for TB | IS6110, MPB64, and protein b | 60 | 62 | ||
| Agarwal et al. (2019) [47] | Multiple countries (most samples from India | 49 | Aqueous and/or vitreous | Clinical signs suggestive of uveitis TB and others where the specific cause had been excluded; corroborative evidence suggestive of uveitis TB | IS6110, MPB64, and protein b | 80 | 93 | ||
| Sharma et al. (2013) [65] | India (Yes) | 9 | Aqueous or vitreous | Clinical signs suggestive of uveitis TB with other specific causes excluded; corroborative evidence suggestive of uveitis TB | IS6110, MPB64, and protein b | 100 | 77.77 | ||
| Sharma et al. (2019) [63] | India (Yes) | 200 | vitreous | (2) all known causes of infectious uveitis except TB and known noninfectious uveitic syndromes ruled out; (3) positive tuberculin skin test (4) received antitubercular therapy for a minimum of 12 months (6) no recurrence of uveitis. Out of these 70 cases, 3 were culture-positive and 67 were culture negative for M. tuberculosis. | IS6110, MPB64 and protein b | 100 | 71.8 | ||
| Sharma et al. (2022) [66] | India (Yes) | 75 | vitreous | IS6110, MPB64 and protein b | 100 | 72 | |||
| Sharma et al. (2022b) [67] | India (Yes) | 39 | vitreous | Signs of active uveitis or A positive tuberculin skin test as per the Center of disease control (CDC) guidelines, or chest X-ray suggestive of TB was present. All other known causes of infectious or non-infectious uveitis were excluded. | IS6110, MPB64 and protein b | 100 | 73.7 | ||
| mRNA-based Multiplex PCR | |||||||||
| Sharma et al. (2022b) [67] | India (Yes) | 39 | vitreous | Signs of active uveitis or A positive tuberculin skin test as per the Center of disease control (CDC) guidelines, or chest X-ray suggestive of TB was present. All other known causes of infectious or non-infectious uveitis were excluded. | IS6110, MPB64 and protein b | 100 | 42.1 | Able to detect “viable” Mtb bacilli from dead bacilli as the half-life of bacterial mRNA is extremely short (∼9.5 minutes in vitro), hence better reflecting mycobacterial viability and potentially useful for monitoring susceptibility to ATT [67] | Degradation of mRNA during storage and extraction procedures renders poorer sensitivity than DNA-based MPCR, thus may not be sufficient to rule out OTB. |
| GeneXpert Assay | |||||||||
| Sharma et al. (2019) [63] | India (Yes) | 200 | vitreous | (2) all known causes of infectious uveitis except TB and known noninfectious uveitic syndromes ruled out; (3) positive tuberculin skin test (4) received antitubercular therapy for a minimum of 12 months (6) no recurrence of uveitis. Out of these 70 cases, 3 were culture-positive and 67 were culture negative for M. tuberculosis. | IS6110, rpoB | 100 | 17.2 | Rapid results within2-3 hours. [63] Simple cartridge-based real-time PCR eliminated the problem of cross-contamination because of self-contained cartridges. [64] This confers low biosafety risk and staff require minimal training. [63] Detects rifampicin resistance. | Costs 15 USD per sample [63]. Poor sensitivity due to lower analytical sensitivity of Xpert (131 CFU/ml in spiked sputum) in comparison to 2-3 CFU/ml for MPCR [63]. Specimen may be diluted when reagent is added for DNA extraction. Sensitivity is too low to serve as a reliable test for ruling out OTB. False-positive rifampicin resistance due to silent mutations that probe-based molecular tests like Xpert cannot differentiate from true-positive resistance [63]. Does not detect isoniazid resistance. |
| Sharma et al. (2022) [66] | India (Yes) | 75 | vitreous | IS6110, rpoB | 100 | 16 | |||
| Sharma et al. (2022b) [67] | India (Yes) | 39 | vitreous | Signs of active uveitis or A positive tuberculin skin test as per the Center of disease control (CDC) guidelines, or chest X-ray suggestive of TB was present. All other known causes of infectious or non-infectious uveitis were excluded. | IS6110, rpoB | 100 | 10.5 | ||
| GeneXpert Ultra Assay | |||||||||
| Sharma et al. (2022) [66] | India (Yes) | 75 | vitreous | (2) all known causes of infectious uveitis except TB and known noninfectious uveitic syndromes ruled out; (3) positive tuberculin skin test (4) received antitubercular therapy for a minimum of 12 months (6) no recurrence of uveitis. Out of these 70 cases, 3 were culture-positive and 67 were culture negative for M. tuberculosis. | IS108, IS6110, rpoB | 100 | 50 | Circumvents the identification of silent mutations as rpoB gene mutations by using high resolution melt (HRM) curve analysis rather than probe-based chemistry. [66] | Same limitations as GeneXpert. |
| MTBDRplus Assay | |||||||||
| Sharma et al. (2019) [63] | India (Yes) | 200 | vitreous | (2) all known causes of infectious uveitis except TB and known noninfectious uveitic syndromes ruled out; (3) positive tuberculin skin test (4) received antitubercular therapy for a minimum of 12 months (6) no recurrence of uveitis. Out of these 70 cases, 3 were culture-positive and 67 were culture negative for M. tuberculosis. | rpoB, katG and inhA | 100 | 34.5 | Able to establish diagnosis of MDR-TB by detecting rifampicin and isoniazid resistance, especially in areas endemic for isolated rifampicin resistance. Separate working stations required, hence reducing risk of cross- contamination. [63] | Poor sensitivity due to lower analytical sensitivity of MTBDRplus assay (160 CFU/ml in spiked sputum) in comparison to 2-3 CFU/ml for MPCR [63]. Sensitivity is too low to serve as a reliable test for ruling out OTB. False-positive rifampicin resistance due to silent mutations that probe-based molecular tests like MTBDR Assay cannot differentiate from true-positive resistance [63]. Space-consuming requiring designated areas for a proper setup [63]. Time-consuming as turn-around time is 24 hours [63]. More expensive than LAMP, MPCR and GeneXpert costs 22 USD per isolate [63]. Limited accessibility and throughput in endemic resource-limited regions [66]. |
MTBDR plus assay performed on vitreous fluid samples could detect rifampicin resistance based on mutations in the rpoB gene and isoniazid resistance based on mutations in the katG and inhA genes, with a reported sensitivity of 34.5% and specificity of 100% [63]. Probes detecting wild-type and mutant variations of these genes are added as part of the quantitative RT-PCR technique, which has a turnaround time of 24 hours, thus making the MTBDR plus assay an effective alternative to gene sequencing for the proper diagnosis of MDR-TB especially in isolated-rifampicin resistance regions.
With the rapidly expanding field of systems biology, where computational and mathematical analysis of large genomic and/or proteomic datasets are utilized to model complex biological systems, new mechanistic insights into the pathogenesis of TB as well as the discovery of novel biomarkers have been recently obtained. These discoveries can help to optimize diagnosis and development of personalized TB treatment tailored to specific host and/or pathogen factors [76]. Next-generation sequencing (NGS) technologies that generate comprehensive TB drug-resistance profiles will likely be the first-line diagnostic tool to guide early initiation of ATT [77]. In coming years, genomic technologies may detect particular mutations in the mycobacterial genome linked to resistance or unresponsiveness to ATT resulting in disease relapse or treatment failure [78]
3. Novel biomarkers for diagnosis, treatment effectiveness, and prognostication of OTB
As described above none of the available tests, especially in light of OTB, displays optimal diagnostic accuracy, therefore, novel biomarkers are still needed to optimize diagnosis and predict or monitor treatment response and outcome. The following section will discuss the role of novel discovered biomarkers in the pathogenesis, disease activity, and modulation of OTB. In addition, molecular mechanisms within the ocular immune system in response to OTB will be discussed (Figure 3). These novel biomarkers have not yet found their way into a clinical application, as they need to be validated to show accuracy in distinguishing individuals with OTB from patients with recurrent ocular inflammation. Point-of-care (POC) diagnostic tests to incorporate novel biomarkers in the real-life settings are also still being developed. Nonetheless, novel biomarkers discovery might identify new therapeutic ocular targets that contribute to improved diagnosis and personalized OTB treatment regimens, thus avoiding the use of unnecessary drugs and reducing systemic side effects [85]. In this section, the advantages and disadvantages of each omic approach has been analyzed (see Table 3), and a further breakdown of the insights and limitations of each OTB novel biomarker study has been presented (see Supplementary Table 1).
3.1. Mycobacterial genomics: Future improvements
In a previous section on NAAT, different existing techniques for Mtb detection and drug-resistance screening have been discussed. Validation of resistance prediction is a crucial step to bring NAAT into clinical practice for pulmonary TB, as is the case for OTB in certain endemic or resource-sufficient regions [86, 87]. However, even in the treatment of pulmonary TB, routine and full-scale implementation of NAAT-based drug-resistance profiling into clinical and public health practices is hampered by low bacterial loads and possible contaminations in sputum specimens [88, 89]. Additional limitations include the lack of standardized genetic markers indicative of drug resistance that warranting induction of a personalized ATT and high laboratory costs associated with NAAT implementation [90]. In this section, we discuss the possible solutions to overcome these limitations with the aim of NAAT Mtb genome analysis to assess treatment response, risk of relapse and/or treatment failure of OTB.
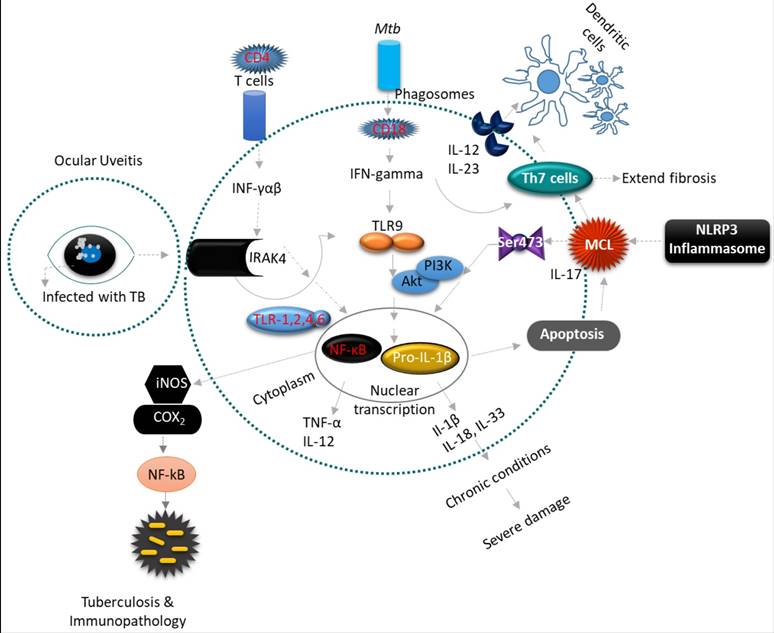
Uveitis is a form of eye inflammation that affects the middle layer of tissue in the eye wall (uvea). It presents with eye redness, pain, blurred vision and deterioration of visual acuity occurs quickly. (A-B) Inflammatory cytokine Interleukin 1 (IL-1) is an essential mediator of innate immunity and promotes inflammatory tissue damage in ocular uveitis (OU). The inflammatory constituents include Th1 and Th17 lymphocytes, which produce pro-inflammatory cytokines such as IL-1β, IL-2, IL-17, IL-18, IL-23, iNOS, COX2, TNF-α and INF-gamma that recruit leukocytes from circulation to result in tissue damage of the eye. The IFN-gamma-driven antimicrobial properties of phagocytes are augmented by IL-18 and IL-1β, and inflammatory cytokines processed by caspase-1, which are recruited to the inflammasomes. Inflammasomes are multimeric protein complexes that serve as a platform for caspase-1, the enzyme responsible for proteolytic cleavage of IL-1β and IL-18 precursors. However, this inflammasome activation triggers the multifaceted action of pro-inflammatory cytokines, a prerequisite for developing an effective inflammatory response against Mtb. The NLRP3 and AIM2 inflammasomes play an important role in innate immunity against Mtb.
Selection criteria of sample and control groups in each novel OTB biomarker study, advantages and disadvantages of each -omics method.
| Author (Year) | Biomarkers discovered | Specimen | Subject (n) | Control (n) | Advantages | Disadvantages | |
|---|---|---|---|---|---|---|---|
| Transcriptomic | Chandalawada et al. (2022) [103] | Four miRNAs, miR-423-5p, miR-328-3p, miR-21-5p, and miR-16-5p, were significantly dysregulated in aqueous humor of OTB patients | Aqueous humor | Confirmed OTB as defined with positive TB PCR test (n = 5). Presumed OTB is with another positive result of follow-up examinations or positive ATT response (n = 2). | Patients with only cataract (n = 2). | Generally speaking, the nucleic acid-based omics approaches for data generation rely on five major steps: appropriate sample collection, high-quality nucleic acid extraction, library preparation, clonal amplification, and sequencing. For the last step, sequencing-based technologies, the most advanced of the omics technologies in terms of availability of laboratory reagents for standardized protocols, analytical tools and public databases for data sharing, provide unique opportunities to obtain high quality from small amounts of tissues or individual cells to address a wide range of biological questions [194]. Peripheral blood samples are easy to obtain. | Micro-RNA to date lack external validation and their findings cannot be compared due to relevant methodological differences in processes such as RNA extraction or data analysis [102]. Heterogeneous datasets pose challenges because quality assurance, quality control, data normalization and data reduction methods differ among the various types of individual datasets [195]. Small sample size for OTB. Larger validation studies are needed to externally validify discovery dataset. |
| La Distia Nora et al. (2018) [120] | IFN signature based on 10 interferon-stimulated genes (UBE2L6, FCGR1B, GBP1, IL1B, MYD88, TLR8, IRF7, STAT1, SERPING1, and IFIT2) could discriminate between active pulmonary TB and healthy controls with a sensitivity of 100% and a specificity of 91%. | Peripheral blood | Active Pulmonary TB Without uveitis, Uveitis with clinically diagnosed active pulmonary TB, QFT (+) uveitis of unknown cause. (n = 80) Mtb sputum-positive active pulmonary TB patients (HIV negative) without uveitis or a history of ATT as a positive control group. (n = 10) | QFT-negative, had no history of uveitis and did not use any medication at the time of the study. (n = 23) | |||
| Schrijver et al. [121] | 10-gene type 1 IFN signature (UBE2L6, FCGR1B, GBP1, IL1B, MYD88, TLR8, IRF7, STAT1, SERPING1, and IFIT2) displayed an inverse correlation with serum complement component C1q. | Peripheral blood | APTB uveitis unknown (n = 50) QFT- Uveitis (n = 51) QFT+ Uveitis of unknown aetiology (n = 58) APTB‐assoc. uveitis (n = 12) APTB w/o uveitis (n = 10) | Healthy controls (n = 73) Primary Sjögren's syndrome (n = 86) Systemic lupus erythematosus (n = 30) Systemic sclerosis (n = 23) | |||
| Proteomic | Ang et al. (2012) [134] | Inflammatory cytokines such as IL-6 and CXCL8/IL-8 and Th1 associated chemokines CXCL9, and CXCL10 were significantly increased in the TB-associated uveitis group compared to the non- inflammatory controls, and it is also distinct from the cytokine profiles of idiopathic uveitis. | Aqueous humour | TB-associated uveitis by presenting with acute, active uveitis, with clinical signs of granulomatous inflammation, broad-based posterior synechiae, or retinal vasculitis, with or without choroiditis, and positive QFT or T-SPOT.TB, and TST indurations ≥15 mm, and respond to ATT. (n = 10) Idiopathic uveitis with no evidence of TB (negative TST (TST<10 mm) and negative IGRA) or other diseases. (n = 13) | Patients with no ocular pathology other than cataract were enrolled as non-inflammatory controls. (n =23) | The development of improved methods in quantitative proteomics (Mass Spectrometry‐based techniques) has increased the relevance of proteins that complements both genomics and traditional biochemical techniques to aid better understanding of the complex interaction between Mtb and host [5]. Recent advances in instrument sensitivity while decreasing the amount of sample required for high-throughput analyses and now allow for the detection of minimal differences in protein abundances [194]. The development of effective isotopic labeling tools for tissue samples have significantly improved the accuracy and reproducibility of peptide and protein quantification using MS [194]. | There is no consensus in terms of data formatting, cleaning and normalization [194]. Proteomics approaches still require significant amounts of sample due to the lack of protein amplification methods, and face difficulties in isolation of membrane proteins, detection of low abundance proteins and insoluble proteins. The reliance on separation of complex chemistries (i.e., different charged states and post-translational modifications) using chromatography adds to variability in protein quantification in top-down and bottom-up proteomics [194]. There is variability in peptide identification due to variation in peptide structure, charge and hydrophobicity, and these biochemical properties of peptides and proteins affect their ability to be detected and identified by NMR or MS. Analysis pipelines for proteomic data must deal with absent data (i.e., is the peptide not detected because it is not ionized efficiently, or is it truly not present in the sample), normalization and absolute versus relative quantification [194]. |
| De Simone et al. (2022).[135] | Lower concentrations CXCL13, CXCL-8, CXCL-10 in AH samples for TBU and Q +OS groups (with no significant difference between groups) than definite OS group. The three chemokines were elevated in AH samples than in peripheral blood, suggesting an intraocular production and supporting their possible role as therapeutic targets. | Aqueous humour, Peripheral blood | Presumptive TBU was based on the positivity of the Q-Gold test and compatible clinical ocular features accompanied by a negative workup for other causes of uveitis. (n = 12) | Controls underwent phacoemulsification intervention for cataract and cornea surgery and who were not affected by any other concomitant inflammatory and/or infectious disease nor had a prior history of uveitis. (n = 9) Definite OS based on the histopathological identification of non-necrotizing epithelioid cell granulomas, negative AFB and clinically compatible ocular signs. (n = 15) Definite OS associated with Q-Gold positivity (Q + OS). (n = 5) | |||
| Abu El-Asrar et al. (2012) [138] | Elevated CXCL8 and CXCL10 levels in aqueous humour samples of presumed OTB patients. | Aqueous humour | Presumed TBU if consistent ocular findings, no other cause of their uveitis, positive TST (⩾15mm), response to ATT. (n = 14) | Cataract extraction with no prior history of uveitis. (n = 30) | |||
| Schrijver et al. (2022) [139] | Vitreous CCL17 and CXCL13 levels were found to distinguish sarcoid uveitis from TB‐associated uveitis (significantly lower), with a sensitivity of 67% and a specificity of 78%. | Vitreous humour | Cohort 1: TB‐associated uveitis (n = 6). Cohort 2 (blinded): TB‐associated uveitis (n = 0). | Cohort 1: sarcoid uveitis (n = 15), (P)VRL (n = 7), TB‐associated uveitis (n = 6), non‐TB infectious uveitis (n = 6), uveitis associated with systemic disease (n = 2), idiopathic uveitis (n = 8), masquerade syndrome other than (P)VRL (n = 3). | |||
| Bansal et al. (2021) [141] | Vitreous protein analysis found that OTB patients showed 11 upregulated differentially expressed proteins (DEPs) and 21 downregulated DEPs compared to a non-TB uveitis or non-uveitis patients. | Vitreous humour | Presumed TBU, which was diagnosed by the presence of active uveitis with characteristic clinical ocular signs, and supported by corroborative evidence of TST ≥ 10 mm of induration at 48-72 h and/or a positive X ray/CT scan of chest. (n = 3) Confirmed TBU with triplex PCR. (n = 10) | Positive control groups consisted of with active uveitis with clinical signs suggestive of causes other than TBU. All these samples also had a negative triplex PCR for MTB. (n = 7) Negative control group had no evidence of any intraocular inflammation, and underwent vitreous surgery for various vitreoretinal disorders such as macular hole, epiretinal membrane, dropped nucleus, etc. (n = 9) | |||
| Van der Colff et al. (2023) [142] | 29 biomarkers were tested on both serum and urine samples. Most biomarker concentrations were significantly higher in serum than in urine (p < 0.01). Only 2 (IL-1RA and IL-2) showed higher concentrations in urine than serum (p < 0.01). Three biomarkers (sIL-2Ra, sTNFRI and IFNγ) showed no difference in concentration between urine and serum (p > 0.05). | Peripheral blood sample and urine sample. | Most patients were diagnosed with possible OTB and only one with confirmed OTB (n = 14) | No control (n = 0) | |||
| Stimulation Assay | Makhoba et al. (2021) [167] | Four-marker biosignature comprising of CD40 ligand, IL-33, IFN-γ, and SAP, which showed potential in diagnosing OTB. | Peripheral blood | OTB diagnosed when other causes of ocular inflammation excluded, at least one suggestive clinical sign as well as evidence of TB on CXR or elsewhere in the body. (n = 32) | Other intra-ocular diseases (n = 60) | Peripheral blood samples are easy to obtain. In vitro stimulation is safe for the patient; no risk of triggering infectious or allergic reaction. Potentially able to differentiate active and latent OTB by detecting the difference in cytokine levels in both states. | Small sample size for OTB. Larger validation studies are needed to externally validify discovery dataset. High TB burden setting may misdiagnose latently infected OTB with active OTB. As latently infected individuals will habour T cells which will recognize the antigens used in QFT tubes, and also secrete host markers into QFT supernatants irrespective of the primary ocular diagnosis. T cell responses may not be limited to the use of single peptides for eliciting both the antimycobacterial and retinal antigen-specific responses. T cell responses to other immunodominant peptides (non-mycobacterial and non-retinal) may confound biomarker discovery specific to immune response triggered by OTB. Does not provide the phenotype of immune cells producing biomarkers. Obtaining vitreous humour via pars plana vitrectomy is invasive. |
| Alam et al. (2022) [168] | TST-positive undifferentiated uveitis (UNK) generates a stronger monofunctional and polyfunctional (dual-cytokine) intraocular cytokine response than active OTB. | Vitreous humour | Patients who fulfilled the SUN classification criteria for OTB (retinal vaculitis, SLC, MFC, intermediate or panuveitis with positive TST and negative tests for sarcoidosis and syphilis) and/or tested positive for TB-PCR. (n = 23) | Patients with a positive TST who did not fulfill the SUN criteria and who had a negative TB-PCR study, classified as uveitis of unknown origin (UNK). (n = 24) TST-negative patients, with or without a well-defined non-TB uveitis entity were classified as non-TB control subjects. (n = 24) | |||
| Serum and intraocular protein analysis | Singh et al. (2021) [177] | Raised VEGF and decreased FGF levels in RPE cells and vitreous humour, but not in tears. | Vitreous humour and tears. | IS6110 PCR-positive vitreous samples were considered confirmed OTB. (n = 15) Tears collected from clinically suspected OTB patients. (n = 15) | Clinically non-OTB uveitis and IS6110 PCR-negative vitreous samples. (n = 27) Tears collected from clinically non- OTB patients. (n = 20) | Potential to improve clarity on pathogenesis of OTB presentation (e.g. retinal vasculitis) and may spur research for other therapeutic agents (e.g. anti-VEGF). | Small sample size. Serial dilution of vitreous samples may cause variability in bacteria density. Tears, an easily obtainable sample, unfortunately did not show similar changes in VEGF and FGF levels as in RPE cells and vitreous humour. Obtaining vitreous humour via pars plana vitrectomy is invasive. Unable to confirm diagnosis of OTB; still relies on presumable diagnosis of OTB. |
| T-lymphocyte profiling | Hutchinson et al. (2021) [192] | Only increased CD38 and HLA-DR expression on Mtb-specific CD4 T cells were significant to discriminate different OTB phenotypes and predict treatment response. | Peripheral blood | At least one of the three tests (chest radiography, Mantoux test, IGRA) positive. Two more patients were diagnosed with OTB, 1 likely had diagnosis of TB uveitis due to presence of clinical finding with panuveitis and occlusive vasculitis in a patient from area endemic with TB (Bangladesh) while the other 1 had previously known history of positive Mantoux test and T-SPOT.TB. (n = 36) | Not Applicable (n = 0) | Provides the phenotype of immune cells as well as their change in cytokine expression (upregulation or downregulation). Potential to discriminate treatment responders from non-responders. In other words, this can differentiate those with active OTB who will benefit from ATT versus those with latent ATT that can experience harmful side effects from overly-aggressive treatment. Peripheral blood samples are easy to obtain. | Small sample size. Unable to confirm diagnosis of OTB; still relies on presumable diagnosis of OTB. |
Targeted sequencing of Mtb genes associated with drug-resistance, as opposed to whole genome sequencing might improve diagnostic limitations due to limited recovery of Mtb DNA from obtained clinical samples. This is especially relevant in the case of paucibacillary infection usually observed in OTB [91]. In recent years, the abundant cleavage activities of activated viz. Cas (CRISPR-associated proteins) has been harnessed for in vitro diagnosis of viral pathogens (e.g. zika, dengue) [92]. As part of the adaptive immune systems of bacteria and archaea, CRISPR-associated proteins can detect target DNA molecules at concentrations as low as 5aM10which makes them excellent diagnostic tools for detection of (paucibacillary) infections [93-97]. A rapid CRISPR-based assay for Mtb detection from various clinical samples was developed and its diagnostic performance was compared to Mtb cultures and GeneXpert MTB/RIF assay [98]. The study found that CRISPR-MTB had a higher sensitivity of 79% as compared to 33% and 66% of culture and GeneXpert, respectively [98]. CRISPR-MTB allowed for near single-copy sensitivity without compromising specificity (98%). The test requires less sample input and a shorter turnaround time for TB diagnosis and drug resistance compared to current clinical used test [98]. The applicability of CRISPR-MTB technology on ocular fluid samples for OTB remains undetermined, warranting clinical evaluation.
However, partial characterized Mtb mutations still require culture-based drug-susceptibility phenotyping to determine the minimum inhibitory concentrations (MIC) of ATT drugs needed for optimal personalized therapy. Hence, there is increasing interest in “big data” analysis of genotypic and phenotypic datasets to improve the accuracy of resistance predictions in MDR TB [76]. Machine learning algorithms and genome-wide association studies are often employed to determine the phenotypic impact of nonstandard mutations to new and repurposed second-line drugs such as bedaquiline, clofazimine or linezolid to which resistance is not yet widespread [99, 100]. These developments can guide future direction of personalized ATT regimens for OTB patients who may also contract these increasingly common and problematic MDR Mtb strains.
3.2. Human and mycobacterial transcriptomics
Among the host-based biomarkers, microRNAs (miRNAs) have emerged as an important candidate to diagnose infectious diseases, including TB [101, 102]. Chadalawada et al. showed that four miRNAs, miR-423-5p, miR-328-3p, miR-21-5p, and miR-16-5p, were significantly dysregulated in aqueous humor of OTB patients [103]. These four miRNAs could contribute to the pathogenesis of OTB via tuberculosis-associated pathways like phosphatidylinositol 3-kinase protein kinase B (PI3K-Akt) signaling, autophagy, and the mitogen-activated protein kinase (MAPK) pathway [103].
Human and Mtb transcriptomic insights obtained from pulmonary TB studies, might be of value in the context of OTB after validation. Firstly, changes in Mtb's transcriptome in response to 75 different anti-TB agents was evaluated by RNA sequencing and related to the mechanism of action of each agent [104, 105]. Secondly, dual RNA sequencing allows simultaneous and unbiased profiling of human and Mtb transcription by capturing the transcriptome in its entirety, hence allowing a deeper understanding of the molecular host-pathogen interaction during Mtb infection [106, 107]. Thirdly, highly sensitive (10 CFU/mL-1) detection of 16S ribosomal RNA (rRNA) might be applicable to monitor treatment response, even after weeks of ATT treatment [108]. Since 16S rRNA is more stable and abundant than mRNA, 16S rRNA might be a superior biomarker to detect the presence and quantity of Mtb bacilli in ocular fluid specimens [109]. Lastly, there is an increasing number of combination host RNA gene signatures from whole blood published in the literature for pulmonary TB [110-114]. None of these transcriptomic signatures have been implemented into routine clinical practice so far [115].
Importantly, many of the pulmonary TB biomarker genes have been shown to relate to interferon (IFN) signaling [116, 117]. Type I IFN-signaling has been shown to be involved in the pathogenesis of TB based on the observation of neutrophil-driven IFN-inducible gene profiling [118]. Type 1 IFN contributes to the death of Mtb-infected macrophages, which might potentiate subsequent Mtb spread to the other cells [119]. In depth understanding of IFN signatures might provide additional options to distinguish between active and latent TB. Profiling of IFN-inducible genes has been done in the context of uveitis of unknown cause with a positive QFT-Gold In-Tube test. La Distia Nora et al. proposed a whole blood IFN signature based on 10 interferon-stimulated genes as an applicable tool to stratify QFT-positive patients with uveitis of unknown cause into groups of high and low risk of having active TB-associated uveitis [120]. Schrijver et al. further evaluated that the peripheral blood type 1 IFN gene signature score displayed an inverse correlation with serum complement component C1q. Combined measurement resulted in improved identification of ocular TB from uveitis with coincidental QFT positivity yet without other signs of active TB infection especially in high TB-endemic regions [121]. In addition to multiple studies showing an important role of IFN signaling in the pathogenesis of pulmonary TB [122, 123] an in vitro study also displayed strong IFN signaling in RPE cells after Mtb infection [124].
Overall, miRNA targets, rRNA targets and Type 1 IFN gene signatures are emerging as potential biomarkers involved in OTB pathogenesis pathways to confirm the presence and activity of Mtb infection in the eye. However, these markers remain in the nascent trial phase and have yet been externally validated with a large independent cohort.
3.3. Proteomics: Human inflammatory cytokines and chemokines
Proteins, peptides, and protein post-translational modifications are robust biomarkers for early disease detection, disease classification, patient stratification, diagnosis, prognosis, and even monitoring disease activity and treatment efficacy [125]. Advanced mass spectrometry (MS)-based clinical proteomics has emerged as a powerful and influential technological platform for the confident identification of such markers in complex biological samples [125]. Proteomic analysis presents the opportunity to identify novel theranostic biomarkers with the potential for targeted therapy in the ocular field [126].
Vitreous proteomics is a rapidly emerging and promising field aimed at advancing the diagnostics and therapeutics of various debilitating ocular diseases [127-130]. For example, a recent study showed discovery of novel therapeutics targets for diabetic retinopathy through systematic analysis of vitreous proteomics [131]. Another study based on vitreous proteomics reported that elevated levels of extracellular carbonic anhydrase-I (CA-I) in vitreous from individuals with diabetic retinopathy that led to develop plasma kallikrein (PKal) inhibitors as a potential treatment for DME [132, 133]. Proteomics analysis on various intraocular structures other than vitreous humour, such as aqueous humour,tears, cornea, lens, ciliary body, retina and retinal pigment epithelium can also be utilized to gain valuable insights into complex molecular signaling pathways of OTB. In addition, a combination of several biomarkers, a so-called disease-defining biosignature, can render a more accurate diagnosis of OTB. The following subsections compile the published literature wherein proteomics was performed on aqueous and vitreous humour and serum. We also explore future possibilities of tear proteomics in OTB.
3.3.1. Ocular fluid proteomics for OTB
Several studies reported protein analysis from aqueous and vitreous humor in OTB patients. Ang et al. performed an aqueous humour cytokine and chemokine analysis in tubercular anterior uveitis patients and reported a significant increase in inflammatory cytokines such as IL-6 and CXCL8/IL-8 and Th1 associated chemokines CXCL9, and CXCL10 [134], which is more consistent with an autoimmune-related ocular inflammation triggered by Mtb rather than an active ocular tuberculous infection [134]. De Simone et al. recently studied the cytokine profile of the aqueous humour and discovered a potential biosignature that can guide more accurate diagnosis and treatment of difficult overlapping cases such as ocular sarcoidosis (OS) and OTB which share remarkably similar epidemiology, pathogenesis, and ocular presentations [135-137]. Thirty-two patients, 15 with OS, five with QFT-positive OS, and 12 with presumed OTB, had blood and aqueous humour samples collected pre-treatment for the analysis of selected cytokines [135]. Results confirmed that CXCL8, CXCL10, and IL-6 levels were higher in aqueous humour samples than in peripheral blood of all three patient groups, suggesting an intraocular source of cytokine/chemokine production which could serve as a localized therapeutic target for both OS and OTB. Aqueous humour CXCL8, CXCL10, and CXCL13 levels were significantly higher in definite OS than in presumptive OTB. However, there were no statistically significant differences in terms of cytokine levels among QFT-positive OS as well as presumptive OTB sample groups. These two sample groups showed similar aqueous humour levels of CXCL9, CXCL10, IFN-γ, IL-2, and IL-15. On the other hand, definitive OS samples showed prevalent aqueous humour expression of CCL20/ MIP-3α, CXCL13, and IL-10. These observed differences in aqueous humour and serum chemokine profiles could contribute to differentiate patients with OS from patients with OTB or concurrent OS and OTB as these display overlapping clinical phenotypes of granulomatous uveitis [135]. These findings are in line with a previous study that reported elevated CXCL8/IL-8 and CXCL10 levels in aqueous humour samples of presumed OTB patients [138].
These recent findings highlighted the use of human vitreous proteomic analysis to differentiate many forms of intraocular inflammation, including OTB. Several differentially-expressed proteins could help differentiate granulomatous uveitis from other entities, along with the usefulness of vitreous humour CCL17 and CXCL13 to distinguish OTB from sarcoidosis [139]. A recent study showed significantly reduced vitreous humour levels of CXCL13 in OTB as compared to OS and QuantiFERON-TB Gold-positive OS [135]. Future extensive analysis of the vitreous phospho-proteome may represent another level of protein analysis worth exploring to gain additional pathophysiological insight into OTB and reveal potential novel biomarkers [140].
Bansal et al. analyzed disease-specific protein biosignatures in vitreous samples of 13 OTB patients. They found that OTB patients showed 11 upregulated proteins and 21 downregulated proteins compared to the combined non-TB uveitis and non-uveitis groups [141]. The upregulated proteins included insulin-like growth factor 2 messenger RNA binding protein 3 (IGF2BP3), Complement component C8 beta chain (C8B), and other proteins involved in complement activation and the coagulation cascade. The downregulated proteins included Glucose-6-phosphate isomerases (GPI) and other proteins involved in carbohydrate-metabolism, gluconeogenesis, and glycolysis. A sub-group analysis showed that a signature consisting of 21 upregulated proteins (related to apoptosis and KRAS signaling), along with 37 downregulated DEPs (related to mTORC1 signaling, gluconeogenesis, and glycolysis) differentiated OTB from non-OTB [141]. Future in depth validation and interpretation of the identified DEPs and associated pathways are needed to confirm the signatures robustness and accuracy and evaluate the potential implementation as a POC test [141].
3.3.2. Serum proteomics for OTB
To our knowledge, there is only one published study on serum proteomics in OTB. Van de Colff et al. conducted a small pilot study comparing the levels of 29 known potential candidate biomarkers for pulmonary TB in serum and urine samples [142]. Most biomarker concentrations were significantly higher in serum than in urine, hence these 2 biofluids cannot be used interchangeably when studying biomarker profiles in future [142]. Although the study found that 2 biomarkers (IL-1RA and IL-2) showed higher concentrations in urine than serum and that three biomarkers (sIL-2Ra, sTNFRI and IFNγ) showed no difference in concentration between urine and serum, this study is not conclusive for candidate biomarkers specific for OTB as there is no negative control group [142].
Three studies described the potential of serum proteomics analysis in pulmonary TB. Mateos et al. showed that C-reactive protein (CRP), haptoglobin (HPT) alpha-1-acid glycoprotein 1 (A1AGP1), complement component C9 (C9), neutrophil defensin 1 (DEF1), and serum amyloid P component (SAA2-4) were elevated where apolipoprotein A (APOA1 and 2), serotransferrin (TRFE) and plasma kallikrein (KLK1B) were significantly decreased in active TB patients as compared to LTBI and healthy controls. Each of the markers: CRP, A1AGP1, KLKB1, TRFE, or APOA1 had an area under the curve value >70% [143]. Another study by Peng et al. determined the presence of serum antibodies against 100 different Mtb antigens. Antibody levels against 15 Mtb antigens were significantly elevated and could distinguish between active and LTBI: MT1560.1-IgM, Rv0049-IgM, Rv0270-IgM, Rv0350-IgG, Rv0350-IgM, Rv0494-IgM, Rv1597-IgM, Rv1860-IgG, Rv1876-IgM, Rv2031c-IgG, Rv2352c-IgM, Rv2450c-IgM, Rv2511-IgG, Rv2688c-IgM, and Rv3480c-IgM. Combining the 15 markers to differentiate active TB from LTBI resulted in a good sensitivity of 85.4% and specificity of 90.3% [144]. Future studies on OTB patients are needed to reveal whether serological proteomes could identify OTB biomarkers and provide pathophysiological insights. Garay-Baquero et al. reports a comprehensive TB plasma proteome by profiling 5022 proteins with diverse biochemical and molecular properties [145]. Novel candidate biomarkers (CFHR5, ILF2) were verified in two independent cohorts, leading to the development of a 5-protein biosignature (CFHR5, LRG1, CRP, LBP, and SAA1) capable of discriminating TB from other respiratory diseases (AUC = 0.81) [145].
3.3.3. Possibility of tear proteomics for OTB
Tears are attractive biofluids to study uveitis with two major advantages: tears are localized to the source organ, and tears are easily obtainable due to the non-invasive nature of sample collection. Hence, tear-proteomic profiling provides an attractive and non-invasive opportunity to discover novel biomarkers associated with pathologies of the ocular surface (e.g. dry eye diseases), posterior eye (e.g. diabetic retinopathy, age-related macular degeneration) and systemic diseases (e.g. Alzheimer's dementia, Parkinson's disease, multiple sclerosis, breast cancer) [126, 146]. Tear proteome profiling in keratoconus patients identified 58 proteins related to process of cell death, oxidative damage and inflammation specific to tears of patients with keratoconus; 41 proteins involved in iron pathways was found to be underexpressed in keratoconus patients and identified in control samples only [147]. Besides improving the knowledge of the disease's pathophysiology, Goni et al. also analysed the tear proteome profile before and after treatments and identified biomarkers, mainly A-kinase anchor protein 13 (AKAP-13), altered by surgical treatment [148]. O'Leary et al. discovered a panel of 13 discriminant tear protein markers to distinguish mild ocular graft versus host disease to moderate-to-severe disease [149]. Also, the use of novel targeted proteomic approaches are of interest to unravel disease-specific tear proteomes, such as in non-infectious uveitis (our data, unpublished) [150]. Although extensive tear-proteome profiling data in OTB is still not provided in the current literature, we believe that such studies are warranted as these would contribute to improved diagnosis in material that can be non-invasively obtained, thereby contributing to patient well-being [126].
3.3.4. Future of proteomics for OTB
Plasma and sputum proteomics allowed differentiation between active and latent pulmonary TB [151, 152]. Protein signatures, including those in stimulated whole blood cultures and sputum samples, could also help distinguish MDR from drug-susceptible pulmonary TB [153-155]. Increased levels of serum matrix metalloproteinases were also shown to be associated with disease severity [156]. In addition, distinct proteomic signatures found in bronchoalveolar lavage fluid can also stratify treatment response by distinguishing clinically cured pulmonary TB patients from patients with active pulmonary TB [157]. These obtained results in the light of pulmonary TB pave the way for future directions of proteomic studies investigating aqueous-and vitreous humour and tears from OTB.
3.4. Possibility of metabolomics for OTB
Only a few studies explored the potential of metabolites in blood, urine, or cerebrospinal fluid as biomarkers for pulmonary TB progression and treatment response [158-161]. The level of seryl-leucine core 1 O-glycosylated peptide (SLC1G) in urine was significantly higher in TB patients than household contacts, and decreased in ATT responders [161]. Hence, SLC1G could be a potential POC biomarker to monitor treatment response which needs further validation. Another study associated low tryptophan cerebrospinal fluid levels with improved survival outcome in TB meningitis [160].
Due to the blood-aqueous and blood-retinal barriers, it is assumed that eye tissue has its own distinct metabolic regulation [162]. Hence, the metabolome of aqueous and vitreous humour in OTB may represent the local metabolic environment and may vary greatly from metabolomes of other bodily fluids present in systemic TB. Metabolomics has been used to analyze healthy ocular fluids, study tissue metabolism, and assess the effect of radiation on the eye [162, 163]. Metabolomics from various sources, including aqueous humor, tears, and retina, have also been studied in other ophthalmic diseases such as keratoconus, glaucoma, and diabetic retinopathy using nuclear magnetic resonance (NMR) spectrometry and liquid and gas chromatography MS techniques [164-166]. Metabolome studies in intraocular fluids of patients with OTB will be an exciting next step to understand Mtb pathogenesis and identify novel OTB-associated biomarkers.
3.5. Stimulation assay
Makhoba et al. recently assessed the diagnostic potential of previously identified biomarkers in the context of OTB for the first time by measuring 47 host biomarkers in a blood sample of 92 patients after stimulation with Mtb antigens (i.e. ESAT6, CFP10, and TB7.7) which are used in the QuantiFERON (QFT) In Tube test [167]. Fourteen of these host derived biomarkers showed a significant difference between patients with probable and possible OTB versus those patients with other ocular diseases. The most promising individual biomarkers for diagnosing OTB and excluding other diseases are CCL4/MIP-1β, CCL8/MCP2, IFN-γ, IL-2, IL-22, and CD258 (LIGHT) which were shown to have sensitivities of less than 50% and specificity approaching 98%. The most accurate four-analyte biosignature included CD40L, IFN-γ, IL-33, and serum amyloid P (SAP), which combined had a sensitivity of 56.3% and specificity of 90%. Biomarker signatures can be further optimized to increase the sensitivity and specificity and ensure identification of patients misclassified by an individual marker [167]. We noticed that this study was performed in South Africa, a TB-endemic country where a lot of patients may be latently infected with Mtb and thus will harbor T-lymphocytes that recognize Mtb antigens. Hence, cells from these latent-infected subjects would secrete host biomarkers after QFT-antigen stimulation even in the absence of active ocular or systemic infection [167].
A study by Alam et al. demonstrated evaluated intracellular cytokine responses by CD4+ T-lymphocytes isolated from vitreous, when stimulated with ESAT-6 and IRBP (interphotoreceptor retinoid-binding protein) peptides. They used the recently published SUN classification criteria for OTB, which divided patients into three groups: OTB, undifferentiated with IGRA/TST-positive (unknown), and controls. An increased percentage of IL-17 positive vitreous infiltrating CD4+ T cells was detected in the OTB group upon stimulation with ESAT-6 while double positive (i.e. IFNγ+IL-17+, TNFα+IL-17+, and TNFα+IFNγ+) vitreous infiltrating T cells were significantly more abundant in the unknown group. However, in all groups, TNF-α, IL-17A, and IFN-γ production upon stimulation with IRBP peptides were comparable [168]. This suggested the applicability of utilizing vitreous T cell responses as a good biomarker for confirming OTB, but could be challenging in a routine clinical lab.
3.6. Serum and intraocular protein analysis
Recently, we described serum CCL17 as a biomarker with the potential to support the stratification of OTB from OS and QFT-negative uveitis. Consistent findings were also reported from TB and sarcoidosis patients in general and thus warrant further exploration to prove clinical applicability [169]. Some reports suggest combining serum measurement of IFN-α, IL-10, and IL-2 with other cytokines like TNF-α, thereby improving the discriminating potential between active and LTBI [170, 171]. TNF-α is also possibly a key marker in the reactivation of OTB when immunosuppressive anti-TNF-α agents are used for other confounding or concurrent ophthalmic inflammatory conditions [172]. The phenomenon of paradoxical worsening in both OTB and pulmonary TB presents as a continued progression of TB lesions despite ATT initiation with unclear pathophysiology. A plausible explanation for this phenomenon includes the release of TNF-α and IL-1 by activated macrophages and monocytes exacerbating inflammation, which indicates increased immunosuppression with steroids and continuation of ATT [173]. Another study tried to measure serum type-1 IFN levels but did not identify a difference in IFNα or IFNβ levels comparing LTBI to active TB, suggesting a discordance between type-1 IFN-induced RNA expression in peripheral blood leukocytes and circulation IFN levels in TB [174].
Several studies explored the potential of vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF) measurements from intraocular fluid samples. VEGF is crucial in angiogenesis and lymphangiogenesis as it is induced in response to tissue inflammation and hypoxia [175]. Thayil et al. showed in guinea pig ocular models that hypoxia within TB granulomas was directly related to the extent of inflammation and associated with vascular occlusion resulting in reduced lesional oxygen tension [176]. This drives excessive VEGF production within the inflamed granulomatous ocular tissues. Singh et al. demonstrated raised VEGF and decreased FGF vitreous humour levels in samples obtained from TB uveitis patients, and in supernatant of human RPE cell cultures infected by Mtb [177]. Clinically, intra-vitreal anti-VEGF therapy was found beneficial in TB granuloma treatment in some case reports. This treatment response was associated with a reduction in granuloma vascularity, causing caseation and tissue necrosis of the granulomatous lesions [178, 179]. Mtb induced suppression of the FGF pathway may represent a host immune evasive mechanism and enable Mtb proliferation [180]. In this regard the role of FGF in the pathogenesis of Mtb-induced ocular inflammation and its application as a biomarker or therapeutic target in OTB remains to be explored.
3.7. T-lymphocyte profiling
TB-related immunity is primarily cell-mediated, involving IFN-γ production by CD4+ and CD8+ T-lymphocytes resulting in macrophage activation and improved intracellular Mtb killing [49]. The role of humoral immunity in TB pathogenesis remains obscure. In pulmonary TB, T-lymphocyte-related biomarkers have been identified to distinguish active from LTBI [181]. At present, increased membrane expression of CD38 and HLA-DR [182] by Mtb-specific CD4+ T-lymphocytes (HLA-DR+IFNγ+, CD38+IFNγ+) [183], or other CD4+ T-lymphocytes such as TNFα+IFNγ-IL2-[184] and IFNγ+IL2+[185] has been reported to distinguish active from latent pulmonary TB infection. A previous study in Singapore also showed an increased frequency of TNFα+CD154+IFNγ+CD27- and TNFα+CD154+GM-CSF+CD27-Mtb-specific CD4+ T-lymphocytes in active TB as compared to LTBI [184]. IL-27, a cytokine that stimulates Th1 differentiation, tend to be higher in tuberculous pleural effusion; whether IL-27 play a significant role in other forms of TB is currently unclear and required further investigation [186]. The frequency of CD38+HLA-DR+Ki67+ CD4 T-lymphocytes was found to decrease with ATT, and their detection may thus provide a tool to monitor treatment response and predict treatment success [187].
Previous studies have shown a correlation between ocular inflammation and different immunophenotypes of T-lymphocytes [188-191]. Hutchinson et al. recently investigated the utility of blood T-lymphocytes specific for Mtb antigen (i.e. PPD or ESAT-6 + CFP-10) as an immune surveillance tool to diagnose and manage OTB [192]. An increased percentage of PPD-specific CD4+ T-lymphocytes from OTB patients (who responded to ATT) expressed CD38 and HLA-DR, which decreased during treatment [192]. These CD38 and HLA-DR Mtb-specific CD4+ T-lymphocytes were additionally identified in peripheral blood, pleural fluid, bronchoalveolar lavage fluid and lung tissue obtained from pulmonary TB patients suggesting a more generalized, non-tissue specific, pathophysiological role of these cells during an Mtb infection [193]. These studies suggest a potential role for T-lymphocyte profiling in assessing treatment guidance and response and warrants future clinical trials to validate these findings.
4. Conclusion
OTB is a serious intraocular disease worldwide with unpredictable treatment outcomes due to an extremely broad clinical spectrum and the lack of reliable, sensitive and specific, yet convenient confirmatory diagnostic tests to help guide the prompt initiation of ATT. Many years of research has introduced many biomarker-based quantitative tools such as IGRA, multiplex PCR and LAMP since the use of the century-old TST. However, distinguishing the underlying pathogenesis of a specific ocular TB phenotype, whether it is due to true Mtb infection in ocular structures, a triggered autoimmune response targeting ocular antigens, or a combination of both remains a future objective. Furthermore, more studies are needed to specifically address this potential autoimmune reaction in OTB and its effect on clinical phenotypes and diagnostics. In clinical practice, existing biomarker-based diagnostic approaches continue to be challenged by the issue of cost in low resource settings, lack of immune reaction in concomitant HIV patients, difficulty differentiating latent and active infection and multi-drug resistant Mtb infection.
Novel biomarkers ignite hope for a new era of precision-guided molecular diagnosis and individualized treatment approaches. However, precision and personalized medicine are currently limited to genetic approaches without effective integration of individual profiling and disease subtyping at the transcriptome and proteome level [110]. The multi-“omics” approach - genomics, transcriptomics, proteomics, and metabolomics, along with cytokines and immunophenotyping through T-lymphocyte profiling has been useful to offer insight into previously unexplainable clinical observations such as strong inflammatory responses despite the paucibacillary nature of OTB and recurrent intraocular inflammation despite prolonged ATT. The immune-mediated inflammatory response suggests more complexity to OTB than simply a precise Mtb directed response. Recent studies looking into the complex immunogenesis of OTB postulate that beyond a pathogen-specific immune response triggered by Mtb infection, there is a role for autoreactive and mycobacterial specific T-lymphocytes, monocytes/macrophages, natural killer, and dendritic cells as evidenced by the expression of cell-specific chemokines, pro-inflammatory cytokines, and activation markers [135]. A broader search for empiric biosignatures is thus needed, which are part of this immunopathogenesis. These markers serves not only to identify the presence of Mtb bacilli but also to guide the choice of therapy regimen, monitor progression and treatment response, and predict disease outcome.
While novel multi-omics insights in the Mtb directed immune response can provide biomarker signatures beneficial for diagnosis and discrimination between different Mtb induced ocular inflammatory processes, these should first be extensively validated in prospective clinical trials and various cohorts. Currently multi-omics approaches remain in the trial phase and the clinical utility of omics-discovered biomarkers in OTB diagnosis and treatment is still to be determined.
There are a number of limitations preventing the widespread use of omics-derived biomarkers. Firstly, most uveitis secondary to autoimmune diseases are also primarily mediated by cellular immunity involving T-lymphocytes. For example, Campisi et al. found that apoptosis of Mtb-infected host cells enabled generation of self-reactive TH17 cells, a subset of helper T-lymphocytes associated with autoimmune diseases [196]. Zhong et al. established that Mtb exposure or genetic susceptibility to TB is a risk factor for uveitis secondary to Behçet disease [197]. Moreover, Previously identified pulmonary TB transcriptomic biomarker signatures were upregulated in HIV positive patients and correlated with viral loads in the absence of TB infection [198]. Therefore the same molecular profiles or biomarkers of OTB can be presented in the autoimmune and infective conditions and thus may be misleading.
Secondly, as aqueous or vitreous sampling to confirm the presence of Mtb are not routinely performed in clinical practice, the lack of a diagnostic gold standard will render the evaluation of predictive parameters (sensitivity, specificity, positive predictive value, negative predictive value) of biomarkers unproductive given the different “gold standards” each independent cohort study compares the same biomarker against. In addition, large heterogeneity in cohort design without comparable output and variation in endemicity may affect the diagnostic potential of identified biomarkers in different populations. Moreover, there are only a limited number of molecular studies available that specifically focus on OTB, each performed on a limited number of clinically presumed OTB patients. The OTB-specific studies mostly comprise HIV-uninfected populations or only minimal numbers of HIV-positive patients. This lowers the power to determine differences and utility of the same biomarker(s) in this sub-group that is prominent in many low-resource, TB-endemic nations. The extent of immunodeficiency from HIV infection also varies widely, hence a large and diverse population of co-infected patients need to be recruited to study the relationship between the accuracy of biomarkers and the extent of immunodeficiency [199]. Therefore, multinational centre-based collaborations are essential to increase patient groups, introduce geographic diversity and introduce an international consortium for standardized and feasible “gold standard” diagnostic comparison. The accuracy and utility of biomarkers as diagnostic tools for OTB may improve when integrated with clinical features to generate a composite scoring system informing personalised therapy.
Lastly, though the multi-omics approach is promising, it requires expensive technological and analytic infrastructures which may be unavailable in most settings [200]. Some considerations to mitigate the issue of cost include using easily accessible specimens such as tears, and investing in integrated and minimalist platforms with internationally standardized operating procedures.
Moving forward, some current “multi-omics” approaches in pulmonary TB research can potentially be refined and re-aligned for ocular applications. The focus should be on integrating “omics” data from different levels of the biological process and take system-biology approach to find markers that may boost diagnostic accuracy and render a more biologically meaningful understanding of ocular diseases with large heterogeneity. More extensive prospective clinical studies exploring metabolomics in OTB and externally validifying current research in genomics, transcriptomics, proteomics, and T-lymphocyte profiling in OTB beyond the discovery dataset would aid the ultimate development of POC diagnostic tests with high specificity and sensitivity. Eventually, only innovations that are able to translate the empiric findings into large-scale clinical interventions can make a real difference in the burden and outcomes of OTB in high-incidence countries. Therefore, focusing on identifying disease-specific marker panels rather than single marker may help speed up progress in delivering personalized care for patients with OTB.
Abbreviations
IS6110: Insertion sequence 6110
rpoB: Ribonucleic Acid Polymerase Beta Subunit
katG: Tuberculosis catalase-peroxidase enzyme
inhA: Inhibin Subunit Alpha
C1q: complement component 1q
CD4+: Cluster of differentiation 4
CD27: Cluster of differentiation negative 27
CD38+: Cluster of differentiation 38
CD40L: Cluster of differentiation 40 Ligand
CD154+: Cluster of differentiation 154
IL-1β: Interleukin 1 beta
IL-2: Interleukin 2
IL-6: Interleukin 6
IL-8: Interleukin 8
IL-10: Interleukin 10
IL-15: Interleukin 15
IL-17: Interleukin 17
IL-22: Interleukin 22
IL-33: Interleukin 33
CXCL8: chemokine (C-X-C motif) ligand 8
CXCL9: chemokine (C-X-C motif) ligand 9
CXCL10: chemokine (C-X-C motif) ligand 10
CXCL13: chemokine (C-X-C motif) ligand 13
TNF-α: Tumour necrosis factor-alpha
TGF-β: Transforming growth factor beta
TB7.7: Tuberculosis 7.7 antigen
CCL4: Chemokine (C-C motif) ligand 4
CCL8: Chemokine (C-C motif) ligand 8
CCL17: Chemokine (C-C motif) ligand 17
MIP-1β: Macrophage inflammatory protein-1 beta
MIP-3α: Macrophage inflammatory protein-3 alpha
KRAS: Kirsten Rat Sarcoma viral oncogene homolog
MTORC1: Mechanistic target of rapamycin complex 1
MCP2: monocyte chemoattractant protein 2
SAP: Serum Amyloid P-Component
HLA-DR+: Human Leukocyte Antigen - DR isotype
GM-CSF+: Granulocyte-macrophage colony-stimulating factor
Supplementary Material
Supplementary table.
Acknowledgements
RA is supported by a grant from the National Medical Research Council (NMRC) by Ministry of Health, Singapore, for the Clinician Scientist Award (CSA) from 2020 to 2023. He has not received funding for his work in this publication. AS is supported by A*STAR ID Labs. RLDN is supported by a grant from RisetInovatifProduktif - Lembaga Pengelola Dana Pendidikan (RISPRO LPDP).
The open access publication of this study is supported by Riset Inovatif Produktif - Lembaga Pengelola Dana Pendidikan (RISPRO LPDP) grant number: RISPRO/KI/B1/KOM/5/15219/2020.
Author contributions
All authors contributed to the intellectual development of this paper. ZL, AAS, RPS, IP, RA conceived and planned the review. RPS wrote the first draft of the paper. All authors contributed to the literature review and provided critical feedback to the paper. The final version of the paper has been seen and approved by all authors.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Cunningham ET Jr, Rathinam SR, Albini TA, Chee SP, Zierhut M. Tuberculous uveitis. Ocul Immunol Inflamm. 2015;23:2-6
2. Goletti D, Lee MR, Wang JY, Walter N, Ottenhoff THM. Update on tuberculosis biomarkers: From correlates of risk, to correlates of active disease and of cure from disease. Respirology. 2018;23:455-466
3. Organization WH. 10 facts on tuberculosis. 12 October 2021. https://www.who.int/news-room/facts-in-pictures/detail/tuberculosis
4. Organization WH. Global tuberculosis report 2022. 27 October 2022. https://apps.who.int/iris/rest/bitstreams/1474924/retrieve
5. Banaei-Esfahani A, Nicod C, Aebersold R, Collins BC. Systems proteomics approaches to study bacterial pathogens: Application to mycobacterium tuberculosis. Curr Opin Microbiol. 2017;39:64-72
6. Acharya B, Acharya A, Gautam S, Ghimire SP, Mishra G, Parajuli N. et al. Advances in diagnosis of tuberculosis: An update into molecular diagnosis of Mycobacterium tuberculosis. Mol Biol Rep. 2020;47:4065-4075
7. Teixeira-Lopes F, Alfarroba S, Dinis A, Gomes MC, Tavares A. Ocular tuberculosis - a closer look to an increasing reality. Pulmonology. 2018;24:289-293
8. Alli HD, Ally N, Mayet I, Dangor Z, Madhi SA. Global prevalence and clinical outcomes of tubercular uveitis: A systematic review and meta-analysis. Surv Ophthalmol. 2022;67:770-792
9. Putera I, La Distia Nora R, Utami N, Karuniawati A, Yasmon A, Wulandari D. et al. The impact of aqueous humor polymerase chain reaction and serological test results for establishing infectious uveitis diagnosis: An indonesian experience. Heliyon. 2022;8:e10988
10. Dogra M, Singh R, Agarwal A, Sharma A, Singh SR, Gautam N. et al. Epidemiology of uveitis in a tertiary-care referral institute in North India. Ocul Immunol Inflamm. 2017;25:S46-S53
11. Gunasekeran DV, Gupta B, Cardoso J, Pavesio CE, Agrawal R. Visual morbidity and ocular complications in presumed intraocular tuberculosis: An analysis of 354 cases from a non-endemic population. Ocul Immunol Inflamm. 2018;26:865-869
12. Moule MG, Cirillo JD. Mycobacterium tuberculosis dissemination plays a critical role in pathogenesis. Front Cell Infect Microbiol. 2020;10:65
13. Heiden D, Saranchuk P, Keenan JD, Ford N, Lowinger A, Yen M. et al. Eye examination for early diagnosis of disseminated tuberculosis in patients with aids. Lancet Infect Dis. 2016;16:493-9
14. Shukla D, Kalliath J, Dhawan A. Tubercular retinal vasculitis: Diagnostic dilemma and management strategies. Clin Ophthalmol. 2021;15:4681-4688
15. Basu S, Monira S, Modi RR, Choudhury N, Mohan N, Padhi TR. et al. Degree, duration, and causes of visual impairment in eyes affected with ocular tuberculosis. J Ophthalmic Inflamm Infect. 2014;4:3
16. Garip A, Diedrichs-Mohring M, Thurau SR, Deeg CA, Wildner G. Uveitis in a patient treated with bacille-calmette-guerin: Possible antigenic mimicry of mycobacterial and retinal antigens. Ophthalmology. 2009;116:2457-62 e1-2
17. Forrester JV, Kuffova L, Dick AD. Autoimmunity, autoinflammation, and infection in uveitis. Am J Ophthalmol. 2018;189:77-85
18. Agrawal R, Testi I, Rousselot A, Chen EJ, Lakshminarayanan R, Singhal A. et al. Insights into the molecular pathogenesis of ocular tuberculosis. Tuberculosis (Edinb). 2021;126:102018
19. Gupta V, Gupta A, Rao NA. Intraocular tuberculosis-an update. Surv Ophthalmol. 2007;52:561-87
20. N Singh N AB, R Wajahat. Ocular tuberculosis without a lung primary. Cureus. 2020;12:e7920
21. Khabibullina NF, Kutuzova DM, Burmistrova IA, Lyadova IV. The biological and clinical aspects of a latent tuberculosis infection. Trop Med Infect Dis. 2022 7
22. Wroblewski KJ, Hidayat AA, Neafie RC, Rao NA, Zapor M. Ocular tuberculosis: A clinicopathologic and molecular study. Ophthalmology. 2011;118:772-7
23. Organization WH. Global tuberculosis report 2015. https://apps.who.int/iris/handle/10665/191102
24. Organization WH. High-priority target product profiles for new tuberculosis diagnostics: Report of a consensus meeting. 29 April 2019. https://apps.who.int/iris/handle/10665/135617
25. Agrawal R, Agarwal A, Jabs DA, Kee A, Testi I, Mahajan S. et al. Standardization of nomenclature for ocular tuberculosis - results of collaborative ocular tuberculosis study (COTS) workshop. Ocul Immunol Inflamm. 2019:1-11
26. Standardization of Uveitis Nomenclature Working G. Classification criteria for tubercular uveitis. Am J Ophthalmol. 2021;228:142-151
27. Agrawal R, Ludi Z, Betzler BK, Testi I, Mahajan S, Rousellot A. et al. The collaborative ocular tuberculosis study (COTS) calculator-a consensus-based decision tool for initiating antitubercular therapy in ocular tuberculosis. Eye (Lond). 2022
28. Wu UI, Chuang YC, Sheng WH, Sun HY, Jhong YT, Wang JY. et al. Use of QuantiFERON-TB Gold In-tube assay in screening for neutralizing anti-interferon-γ autoantibodies in patients with disseminated nontuberculous mycobacterial infection. Clin Microbiol Infect. 2018;24:159-165
29. Morimura Y, Okada AA, Kawahara S, Miyamoto Y, Kawai S, Hirakata A. et al. Tuberculin skin testing in uveitis patients and treatment of presumed intraocular tuberculosis in Japan. Ophthalmology. 2002;109:851-7
30. Lalvani A, Pareek M. A 100 year update on diagnosis of tuberculosis infection. Br Med Bull. 2010;93:69-84
31. Tsiouri G, Gaitanis G, Kiorpelidou D, Dionysiou A, Efthymiou A, Daskalopoulos G. et al. Tuberculin skin test overestimates tuberculosis hypersensitivity in adult patients with psoriasis. Dermatology. 2009;219:119-25
32. Chee CB, Soh CH, Boudville IC, Chor SS, Wang YT. Interpretation of the tuberculin skin test in Mycobacterium bovis BCG-vaccinated Singaporean schoolchildren. Am J Respir Crit Care Med. 2001;164:958-61
33. Fernandez-Zamora Y, Finamor LP, Silva LMP, D SR, Casaroli-Marano RP, Muccioli C. Role of interferon-gamma release assay for the diagnosis and clinical follow up in ocular tuberculosis. Ocul Immunol Inflamm. 2022:1-8
34. Ang M, Htoon HM, Chee SP. Diagnosis of tuberculous uveitis: Clinical application of an interferon-gamma release assay. Ophthalmology. 2009;116:1391-6
35. Ang M, Wong W, Ngan CC, Chee SP. Interferon-gamma release assay as a diagnostic test for tuberculosis-associated uveitis. Eye (Lond). 2012;26:658-65
36. Ang M, Wong WL, Kiew SY, Li X, Chee SP. Prospective head-to-head study comparing 2 commercial interferon gamma release assays for the diagnosis of tuberculous uveitis. Am J Ophthalmol. 2014;157:1306-14 1314 e1-4
37. Ahn SJ, Kim KE, Woo SJ, Park KH. The usefulness of interferon-gamma release assay for diagnosis of tuberculosis-related uveitis in Korea. Korean J Ophthalmol. 2014;28:226-33
38. Llorenc V, Gonzalez-Martin J, Keller J, Rey A, Pelegrin L, Mesquida M. et al. Indirect supportive evidence for diagnosis of tuberculosis-related uveitis: From the tuberculin skin test to the new interferon gamma release assays. Acta Ophthalmol. 2013;91:e99-e107
39. Gineys R, Bodaghi B, Carcelain G, Cassoux N, Boutin LTH, Amoura Z. et al. QuantiFERON-TB Gold cut-off value: Implications for the management of tuberculosis-related ocular inflammation. Am J Ophthalmol. 2011;152:433-440 e1
40. Organization WH. Use of tuberculosis interferon-gamma release assays (IGRAs) in low- and middle-income countries: Policy statement. https://apps.who.int/iris/handle/10665/44759
41. Agrawal R, Testi I, Mahajan S, Yuen YS, Agarwal A, Kon OM. et al. Collaborative ocular tuberculosis study consensus guidelines on the management of tubercular uveitis-report 1: Guidelines for initiating antitubercular therapy in tubercular choroiditis. Ophthalmology. 2021;128:266-276
42. Cordero-Coma M, Calleja S, Torres HE, del Barrio I, Franco M, Yilmaz T. et al. The value of an immune response to Mycobacterium tuberculosis in patients with chronic posterior uveitis revisited: Utility of the new IGRAs. Eye (Lond). 2010;24:36-43
43. Babu K, Satish V, Satish S, Subbakrishna DK, Abraham MP, Murthy KR. Utility of QuantiFERON TB gold test in a south indian patient population of ocular inflammation. Indian J Ophthalmol. 2009;57:427-30
44. Ganesh SK, Roopleen, Biswas J, Veena N. Role of high-resolution computerized tomography (HRCT) of the chest in granulomatous uveitis: A tertiary uveitis clinic experience from India. Ocul Immunol Inflamm. 2011;19:51-7
45. Joshi R, Patil S, Kalantri S, Schwartzman K, Menzies D, Pai M. Prevalence of abnormal radiological findings in health care workers with latent tuberculosis infection and correlations with T cell immune response. PLoS One. 2007;2:e805
46. Agarwal A, Mahajan S, Khairallah M, Mahendradas P, Gupta A, Gupta V. Multimodal imaging in ocular tuberculosis. Ocul Immunol Inflamm. 2017;25:134-145
47. Agarwal A, Agrawal R, Gunasekaran DV, Raje D, Gupta B, Aggarwal K. et al. The collaborative ocular tuberculosis study (COTS)-1 report 3: Polymerase chain reaction in the diagnosis and management of tubercular uveitis: Global trends. Ocul Immunol Inflamm. 2019;27:465-473
48. Bisht D, Pande R. Study of ocular manifestations in tuberculosis and its association with HIV AIDS in a tertiary care hospital. Indian J Tuberc. 2020;67:320-326
49. Mehta S, Peters RP, Smit DP, Gupta V. Ocular tuberculosis in HIV-infected individuals. Ocul Immunol Inflamm. 2020;28:1251-1258
50. Moon HW, Hur M. Interferon-gamma release assays for the diagnosis of latent tuberculosis infection: An updated review. Ann Clin Lab Sci. 2013;43:221-9
51. Burger C, Holness JL, Smit DP, Griffith-Richards S, Koegelenberg CFN, Ellmann A. The role of (18)F-FDG PET/CT in suspected intraocular sarcoidosis and tuberculosis. Ocul Immunol Inflamm. 2021;29:530-536
52. Yan WJ, Zhou HY, Yan H. Characterization of and advanced diagnostic methods for ocular tuberculosis and tuberculosis. Int J Ophthalmol. 2020;13:1820-1826
53. Dinnes J, Deeks J, Kunst H, Gibson A, Cummins E, Waugh N. et al. A systematic review of rapid diagnostic tests for the detection of tuberculosis infection. Health Technol Assess. 2007;11:1-196
54. Ung L, Bispo PJM, Doan T, Van Gelder RN, Gilmore MS, Lietman T. et al. Clinical metagenomics for infectious corneal ulcers: Rags to riches? Ocul Surf. 2020;18:1-12
55. Zegans ME, Van Gelder RN. Considerations in understanding the ocular surface microbiome. Am J Ophthalmol. 2014;158:420-2
56. Califf RM. Biomarker definitions and their applications. Exp Biol Med (Maywood). 2018;243:213-221
57. He H, Oka S, Han YK, Yamamura Y, Kusunose E, Kusunose M. et al. Rapid serodiagnosis of human mycobacteriosis by elisa using cord factor (trehalose-6,6'-dimycolate) purified from Mycobacterium tuberculosis as antigen. FEMS Microbiol Immunol. 1991;3:201-4
58. Sakai J, Matsuzawa S, Usui M, Yano I. New diagnostic approach for ocular tuberculosis by ELISA using the cord factor as antigen. Br J Ophthalmol. 2001;85:130-3
59. Maekura R, Nakagawa M, Nakamura Y, Hiraga T, Yamamura Y, Ito M. et al. Clinical evaluation of rapid serodiagnosis of pulmonary tuberculosis by ELISA with cord factor (trehalose-6,6'-dimycolate) as antigen purified from Mycobacterium tuberculosis. Am Rev Respir Dis. 1993;148:997-1001
60. Arora SK, Gupta V, Gupta A, Bambery P, Kapoor GS, Sehgal S. Diagnostic efficacy of polymerase chain reaction in granulomatous uveitis. Tuber Lung Dis. 1999;79:229-33
61. Murugan BBSP, U; Arya, LK; Gulbert JI. Comparison of polymerase chain reaction results with treatment response in the diagnosis of infectious uveitis. Int J Contemp Med Res. 2016;3:3335-8
62. Sudheer B, Lalitha P, Kumar AL, Rathinam S. Polymerase chain reaction and its correlation with clinical features and treatment response in tubercular uveitis. Ocul Immunol Inflamm. 2018;26:845-852
63. Sharma K, Gupta A, Sharma M, Sharma A, Bansal R, Sharma SP. et al. The emerging challenge of diagnosing drug-resistant tubercular uveitis: Experience of 110 eyes from north india. Ocul Immunol Inflamm. 2019;29:107-114
64. Sharma M, Singh R, Sharma A, Gupta V, Sharma K. IS1081-based multi-targeted LAMP: An opportunity to detect tubercular uveitis. Ocul Immunol Inflamm. 2020;30:168-173
65. Sharma K, Gupta V, Bansal R, Sharma A, Sharma M, Gupta A. Novel multi-targeted polymerase chain reaction for diagnosis of presumed tubercular uveitis. J Ophthalmic Inflamm Infect. 2013;3:25
66. Sharma K, Sharma M, Ayyadurai N, Dogra M, Sharma A, Gupta V. et al. Comparative evaluation of geneXpert MTB/RIF ultra and geneXpert MTB/RIF for detecting tuberculosis and identifying rifampicin resistance in pars plana vitrectomy samples of patients with ocular tuberculosis. Ocul Immunol Inflamm. 2022:1-7
67. Sharma K, Gupta A, Sharma M, Singh S, Sharma A, Singh R. et al. Detection of viable Mycobacterium tuberculosis in ocular fluids using mRNA-based multiplex polymerase chain reaction. Indian J Med Microbiol. 2022;40:254-257
68. Balne PK, Barik MR, Sharma S, Basu S. Development of a loop-mediated isothermal amplification assay targeting the mpb64 gene for diagnosis of intraocular tuberculosis. J Clin Microbiol. 2013;51:3839-40
69. Balne PK, Basu S, Sharma S. Loop-mediated isothermal amplification for rapid diagnosis of tubercular uveitis. JAMA Ophthalmol. 2015;133:225-6
70. Figueira L, Fonseca S, Ladeira I, Duarte R. Ocular tuberculosis: Position paper on diagnosis and treatment management. Rev Port Pneumol (2006). 2017;23:31-38
71. La Distia Nora R, Putera I, Khalisha DF, Septiana I, Sitompul R. The diagnostic value of polymerase chain reaction for ocular tuberculosis diagnosis in relation to antitubercular therapy response: A meta-analysis. Int J Infect Dis. 2021;110:394-402
72. Sharma K, Bansal R, Sharma A, Gupta A, Fiorella PD. Successful treatment of rifampicin-resistant intraocular tuberculosis. Ocul Immunol Inflamm. 2015;23:93-6
73. Abdelaal A, El-Ghaffar HA, Zaghloul MH, El Mashad N, Badran E, Fathy A. Genotypic detection of rifampicin and isoniazid resistant Mycobacterium tuberculosis strains by DNA sequencing: A randomized trial. Ann Clin Microbiol Antimicrob. 2009;8:4
74. Hirama T, Sabur N, Derkach P, McNamee J, Song H, Marras T. et al. Risk factors for drug-resistant tuberculosis at a referral centre in Toronto, Ontario, Canada: 2010-2016. Can Commun Dis Rep. 2020;46:84-92
75. Steingart KR, Schiller I, Horne DJ, Pai M, Boehme CC, Dendukuri N. Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2014;2014:CD009593
76. Kontsevaya I, Lange C, Comella-Del-Barrio P, Coarfa C, DiNardo AR, Gillespie SH. et al. Perspectives for systems biology in the management of tuberculosis. Eur Respir Rev. 2021 30
77. Safi H, Gopal P, Lingaraju S, Ma S, Levine C, Dartois V. et al. Phase variation in mycobacterium tuberculosis glpK produces transiently heritable drug tolerance. Proc Natl Acad Sci U S A. 2019;116:19665-19674
78. Colangeli R, Jedrey H, Kim S, Connell R, Ma S, Chippada Venkata UD. et al. Bacterial factors that predict relapse after tuberculosis therapy. N Engl J Med. 2018;379:823-833
79. Gupta V, Arora S, Gupta A, Ram J, Bambery P, Sehgal S. Management of presumed intraocular tuberculosis: Possible role of the polymerase chain reaction. Acta Ophthalmol Scand. 1998;76:679-82
80. Gupta V, Gupta A, Arora S, Bambery P, Dogra MR, Agarwal A. Presumed tubercular serpiginouslike choroiditis: Clinical presentations and management. Ophthalmology. 2003;110:1744-9
81. Singh R, Toor P, Parchand S, Sharma K, Gupta V, Gupta A. Quantitative polymerase chain reaction for Mycobacterium tuberculosis in so-called eales' disease. Ocul Immunol Inflamm. 2012;20:153-7
82. Sharma K, Sharma A, Gupta A. Loop-mediated isothermal amplification for rapid diagnosis of tubercular uveitis - reply. JAMA Ophthalmol. 2015;133:226
83. Biswas J, Kazi MS, Agarwal VA, Alam MS, Therese KL. Polymerase chain reaction for Mycobacterium tuberculosis DNA detection from ocular fluids in patients with various types of choroiditis in a referral eye center in India. Indian J Ophthalmol. 2016;64:904-907
84. Mohan N, Balne PK, Panda KG, Sharma S, Basu S. Polymerase chain reaction evaluation of infectious multifocal serpiginoid choroiditis. Ocul Immunol Inflamm. 2014;22:384-90
85. Balamurugan S, Das D, Hasanreisoglu M, Toy BC, Akhter M, Anuradha VK. et al. Interleukins and cytokine biomarkers in uveitis. Indian J Ophthalmol. 2020;68:1750-1763
86. Consortium CR, the GP, Allix-Beguec C, Arandjelovic I, Bi L, Beckert P. et al. Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N Engl J Med. 2018;379:1403-1415
87. Papaventsis D, Casali N, Kontsevaya I, Drobniewski F, Cirillo DM, Nikolayevskyy V. Whole genome sequencing of Mycobacterium tuberculosis for detection of drug resistance: A systematic review. Clin Microbiol Infect. 2017;23:61-68
88. Votintseva AA, Bradley P, Pankhurst L, Del Ojo Elias C, Loose M, Nilgiriwala K. et al. Same-day diagnostic and surveillance data for tuberculosis via whole-genome sequencing of direct respiratory samples. J Clin Microbiol. 2017;55:1285-1298
89. Jouet A, Gaudin C, Badalato N, Allix-Beguec C, Duthoy S, Ferre A. et al. Deep amplicon sequencing for culture-free prediction of susceptibility or resistance to 13 anti-tuberculous drugs. Eur Respir J. 2021 57
90. Meehan CJ, Goig GA, Kohl TA, Verboven L, Dippenaar A, Ezewudo M. et al. Whole genome sequencing of Mycobacterium tuberculosis: Current standards and open issues. Nat Rev Microbiol. 2019;17:533-545
91. Ng KCS, Supply P, Cobelens FGJ, Gaudin C, Gonzalez-Martin J, de Jong BC. et al. How well do routine molecular diagnostics detect rifampin heteroresistance in Mycobacterium tuberculosis? J Clin Microbiol. 2019;57:e00717-19
92. Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM. et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded dnase activity. Science. 2018;360:436-439
93. Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234-44
94. Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181-90
95. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816-21
96. Liu Y, Xu H, Liu C, Peng L, Khan H, Cui L. et al. CRISPR-Cas13a nanomachine based simple technology for avian influenza a (H7N9) virus on-site detection. J Biomed Nanotechnol. 2019;15:790-798
97. Qin P, Park M, Alfson KJ, Tamhankar M, Carrion R, Patterson JL. et al. Rapid and fully microfluidic ebola virus detection with CRISPR-Cas13a. ACS Sens. 2019;4:1048-1054
98. Ai JW, Zhou X, Xu T, Yang M, Chen Y, He GQ. et al. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg Microbes Infect. 2019;8:1361-1369
99. Chen ML, Doddi A, Royer J, Freschi L, Schito M, Ezewudo M. et al. Beyond multidrug resistance: Leveraging rare variants with machine and statistical learning models in Mycobacterium tuberculosis resistance prediction. EBioMedicine. 2019;43:356-369
100. Farhat MR, Freschi L, Calderon R, Ioerger T, Snyder M, Meehan CJ. et al. GWAS for quantitative resistance phenotypes in Mycobacterium tuberculosis reveals resistance genes and regulatory regions. Nat Commun. 2019;10:2128
101. Tribolet L, Kerr E, Cowled C, Bean AGD, Stewart CR, Dearnley M. et al. Microrna biomarkers for infectious diseases: From basic research to biosensing. Front Microbiol. 2020;11:1197
102. Sabir N, Hussain T, Shah SZA, Peramo A, Zhao D, Zhou X. MiRNAs in tuberculosis: New avenues for diagnosis and host-directed therapy. Front Microbiol. 2018;9:602
103. Chadalawada S, Kathirvel K, Lalitha P, Rathinam SR, Devarajan B. Dysregulated expression of microRNAs in aqueous humor from intraocular tuberculosis patients. Mol Biol Rep. 2022;49:97-107
104. Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE 3rd. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: Novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174-84
105. Briffotaux J, Liu S, Gicquel B. Genome-wide transcriptional responses of mycobacterium to antibiotics. Front Microbiol. 2019;10:249
106. Rienksma RA, Suarez-Diez M, Mollenkopf HJ, Dolganov GM, Dorhoi A, Schoolnik GK. et al. Comprehensive insights into transcriptional adaptation of intracellular mycobacteria by microbe-enriched dual RNA sequencing. BMC Genomics. 2015;16:34
107. Pisu D, Huang L, Grenier JK, Russell DG. Dual rna-seq of mtb-infected macrophages in vivo reveals ontologically distinct host-pathogen interactions. Cell Rep. 2020;30:335-350 e4
108. Honeyborne I, McHugh TD, Phillips PP, Bannoo S, Bateson A, Carroll N. et al. Molecular bacterial load assay, a culture-free biomarker for rapid and accurate quantification of sputum Mycobacterium tuberculosis bacillary load during treatment. J Clin Microbiol. 2011;49:3905-11
109. Sabiiti W, Azam K, Farmer ECW, Kuchaka D, Mtafya B, Bowness R. et al. Tuberculosis bacillary load, an early marker of disease severity: The utility of tuberculosis molecular bacterial load assay. Thorax. 2020;75:606-608
110. Warsinske H, Vashisht R, Khatri P. Host-response-based gene signatures for tuberculosis diagnosis: A systematic comparison of 16 signatures. PLoS Med. 2019;16:e1002786
111. Penn-Nicholson A, Mbandi SK, Thompson E, Mendelsohn SC, Suliman S, Chegou NN. et al. RISK6, a 6-gene transcriptomic signature of tb disease risk, diagnosis and treatment response. Sci Rep. 2020;10:8629
112. Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM. et al. A blood RNA signature for tuberculosis disease risk: A prospective cohort study. Lancet. 2016;387:2312-2322
113. Turner CT, Gupta RK, Tsaliki E, Roe JK, Mondal P, Nyawo GR. et al. Blood transcriptional biomarkers for active pulmonary tuberculosis in a high-burden setting: A prospective, observational, diagnostic accuracy study. Lancet Respir Med. 2020;8:407-419
114. Thompson EG, Du Y, Malherbe ST, Shankar S, Braun J, Valvo J. et al. Host blood rna signatures predict the outcome of tuberculosis treatment. Tuberculosis (Edinb). 2017;107:48-58
115. Warsinske HC, Rao AM, Moreira FMF, Santos PCP, Liu AB, Scott M. et al. Assessment of validity of a blood-based 3-gene signature score for progression and diagnosis of tuberculosis, disease severity, and treatment response. JAMA Netw Open. 2018;1:e183779
116. Moreira-Teixeira L, Mayer-Barber K, Sher A, O'Garra A. Type 1 interferons in tuberculosis: Foe and occasionally friend. J Exp Med. 2018;215:1273-1285
117. Donovan ML, Schultz TE, Duke TJ, Blumenthal A. Type 1 interferons in the pathogenesis of tuberculosis: Molecular drivers and immunological consequences. Front Immunol. 2017;8:1633
118. Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T. et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973-7
119. Zhang L, Jiang X, Pfau D, Ling Y, Nathan CF. Type 1 interferon signaling mediates Mycobacterium tuberculosis-induced macrophage death. J Exp Med. 2021 218
120. La Distia Nora R, Sitompul R, Bakker M, Versnel MA, Swagemakers SMA, van der Spek PJ. et al. Type 1 interferon-inducible gene expression in QuantiFERON Gold TB-positive uveitis: A tool to stratify a high versus low risk of active tuberculosis? PLoS One. 2018;13:e0206073
121. Schrijver B, Dijkstra DJ, Borggreven NV, La Distia Nora R, Huijser E, Versnel MA. et al. Inverse correlation between serum complement component c1q levels and whole blood type-1 interferon signature in active tuberculosis and QuantiFERON-positive uveitis: Implications for diagnosis. Clin Transl Immunology. 2020;9:e1196
122. Dorhoi A, Yeremeev V, Nouailles G, Weiner J 3rd, Jörg S, Heinemann E. et al. Type 1 IFN signaling triggers immunopathology in tuberculosis-susceptible mice by modulating lung phagocyte dynamics. Eur J Immunol. 2014;44:2380-93
123. Moreira-Teixeira L, Stimpson PJ, Stavropoulos E, Hadebe S, Chakravarty P, Ioannou M. et al. Type 1 IFN exacerbates disease in tuberculosis-susceptible mice by inducing neutrophil-mediated lung inflammation and netosis. Nat Commun. 2020;11:5566
124. La Distia Nora R, Walburg KV, van Hagen PM, Swagemakers SMA, van der Spek PJ, Quinten E. et al. Retinal pigment epithelial cells control early Mycobacterium tuberculosis infection via interferon signaling. Invest Ophthalmol Vis Sci. 2018;59:1384-1395
125. Bansal R, Gupta A. Protein biomarkers in uveitis. Front Immunol. 2020;11:610428
126. Jackson CJ, Gundersen KG, Tong L, Utheim TP. Dry eye disease and proteomics. Ocul Surf. 2022;24:119-128
127. Baumann S, Ceglarek U, Fiedler GM, Lembcke J, Leichtle A, Thiery J. Standardized approach to proteome profiling of human serum based on magnetic bead separation and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chem. 2005;51:973-80
128. Steely HT Jr. and Clark AF. The use of proteomics in ophthalmic research. Pharmacogenomics. 2000;1:267-80
129. Lam TC, Chun RK, Li KK, To CH. Application of proteomic technology in eye research: A mini review. Clin Exp Optom. 2008;91:23-33
130. Agrawal R, Iyer J, Connolly J, Iwata D, Teoh S. Cytokines and biologics in non-infectious autoimmune uveitis: Bench to bedside. Indian J Ophthalmol. 2014;62:74-81
131. Alli-Shaik A, Qiu B, Lai SL, Cheung N, Tan G, Neo SP. et al. System-wide vitreous proteome dissection reveals impaired sheddase activity in diabetic retinopathy. Theranostics. 2022;12:6682-6704
132. Gao BB, Clermont A, Rook S, Fonda SJ, Srinivasan VJ, Wojtkowski M. et al. Extracellular carbonic anhydrase mediates hemorrhagic retinal and cerebral vascular permeability through prekallikrein activation. Nat Med. 2007;13:181-8
133. Van Bergen T, Hu TT, Little K, De Groef L, Moons L, Stitt AW. et al. Targeting plasma kallikrein with a novel bicyclic peptide inhibitor (THR-149) reduces retinal thickening in a diabetic rat model. Invest Ophthalmol Vis Sci. 2021;62:18
134. Ang M, Cheung G, Vania M, Chen J, Yang H, Li J. et al. Aqueous cytokine and chemokine analysis in uveitis associated with tuberculosis. Mol Vis. 2012;18:565-73
135. De Simone L, Bonacini M, Aldigeri R, Alessandrello F, Mastrofilippo V, Gozzi F. et al. Could different aqueous humor and plasma cytokine profiles help differentiate between ocular sarcoidosis and ocular tuberculosis? Inflamm Res. 2022;71:949-961
136. Agrawal R, Kee AR, Ang L, Tun Hang Y, Gupta V, Kon OM. et al. Tuberculosis or sarcoidosis: Opposite ends of the same disease spectrum? Tuberculosis (Edinb). 2016;98:21-6
137. Gupta D, Agarwal R, Aggarwal AN, Jindal SK. Sarcoidosis and tuberculosis: The same disease with different manifestations or similar manifestations of different disorders. Curr Opin Pulm Med. 2012;18:506-16
138. Abu El-Asrar AM, Struyf S, Kangave D, Al-Obeidan SA, Opdenakker G, Geboes K. et al. Cytokine and cxc chemokine expression patterns in aqueous humor of patients with presumed tuberculous uveitis. Cytokine. 2012;59:377-81
139. Schrijver B, Kolijn PM, Ten Berge J, Nagtzaam NMA, van Rijswijk A, Swagemakers SMA. et al. Vitreous proteomics, a gateway to improved understanding and stratification of diverse uveitis aetiologies. Acta Ophthalmol. 2022;100:403-413
140. Sun C, Zou H, Yang Z, Yang M, Chen X, Huang Y. et al. Proteomics and phosphoproteomics analysis of vitreous in idiopathic epiretinal membrane patients. Proteomics Clin Appl. 2022: e2100128.
141. Bansal R, Khan MM, Dasari S, Verma I, Goodlett DR, Manes NP. et al. Proteomic profile of vitreous in patients with tubercular uveitis. Tuberculosis (Edinb). 2021;126:102036
142. van der Colff FJ, Snyders C, Walzl G, Chegou N, Smit D. Differences in biomarker concentrations in serum and urine of patients with ocular tuberculosis - a prospective descriptive study. Tuberculosis (Edinb). 2023;138:102290
143. Mateos J, Estévez O, González-Fernández Á, Anibarro L, Pallarés Á, Reljic R. et al. Serum proteomics of active tuberculosis patients and contacts reveals unique processes activated during Mycobacterium tuberculosis infection. Sci Rep. 2020;10:3844
144. Peng Z, Chen L, Zhang H. Serum proteomic analysis of Mycobacterium tuberculosis antigens for discriminating active tuberculosis from latent infection. J Int Med Res. 2020;48:300060520910042
145. Garay-Baquero DJ, White CH, Walker NF, Tebruegge M, Schiff HF, Ugarte-Gil C. et al. Comprehensive plasma proteomic profiling reveals biomarkers for active tuberculosis. JCI Insight. 2020;5:e137427
146. Ponzini E, Santambrogio C, De Palma A, Mauri P, Tavazzi S, Grandori R. Mass spectrometry-based tear proteomics for noninvasive biomarker discovery. Mass Spectrom Rev. 2021
147. Lopez-Lopez M, Regueiro U, Bravo SB, Chantada-Vazquez MDP, Pena C, Diez-Feijoo E. et al. Shotgun proteomics for the identification and profiling of the tear proteome of keratoconus patients. Invest Ophthalmol Vis Sci. 2022;63:12
148. Goni N, Martinez-Soroa I, Ibarrondo O, Azkargorta M, Elortza F, Galarreta DJ. et al. Tear proteome profile in eyes with keratoconus after intracorneal ring segment implantation or corneal crosslinking. Front Med (Lausanne). 2022;9:944504
149. O'Leary OE, Schoetzau A, Amruthalingam L, Geber-Hollbach N, Plattner K, Jenoe P. et al. Tear proteomic predictive biomarker model for ocular graft versus host disease classification. Transl Vis Sci Technol. 2020;9:3
150. D.P.C. Vergouwen JCTB, A. Rothova, M.W.J. Schreurs, Tear fluid and serum proteomics providing potential novel biomarkers in non-infectious scleritis. Manuscript in preparation. 2022
151. Kumar NP, Moideen K, Nancy A, Viswanathan V, Shruthi BS, Sivakumar S. et al. Plasma chemokines are biomarkers of disease severity, higher bacterial burden and delayed sputum culture conversion in pulmonary tuberculosis. Sci Rep. 2019;9:18217
152. Mateos J, Estevez O, Gonzalez-Fernandez A, Anibarro L, Pallares A, Reljic R. et al. Serum proteomics of active tuberculosis patients and contacts reveals unique processes activated during mycobacterium tuberculosis infection. Sci Rep. 2020;10:3844
153. Chen J, Han YS, Yi WJ, Huang H, Li ZB, Shi LY. et al. Serum sCD14, PGLYRP2 and FGA as potential biomarkers for multidrug-resistant tuberculosis based on data-independent acquisition and targeted proteomics. J Cell Mol Med. 2020;24:12537-12549
154. Yang Q, Chen Q, Zhang M, Cai Y, Yang F, Zhang J. et al. Identification of eight-protein biosignature for diagnosis of tuberculosis. Thorax. 2020;75:576-583
155. Ota MO, Mendy JF, Donkor S, Togun T, Daramy M, Gomez MP. et al. Rapid diagnosis of tuberculosis using ex vivo host biomarkers in sputum. Eur Respir J. 2014;44:254-7
156. Ugarte-Gil CA, Elkington P, Gilman RH, Coronel J, Tezera LB, Bernabe-Ortiz A. et al. Induced sputum MMP-1, -3 & -8 concentrations during treatment of tuberculosis. PLoS One. 2013;8:e61333
157. Beltran CGG, Heunis T, Gallant J, Venter R, du Plessis N, Loxton AG. et al. Investigating non-sterilizing cure in TB patients at the end of successful anti-TB therapy. Front Cell Infect Microbiol. 2020;10:443
158. Weiner J 3rd, Maertzdorf J, Sutherland JS, Duffy FJ, Thompson E, Suliman S. et al. Metabolite changes in blood predict the onset of tuberculosis. Nat Commun. 2018;9:5208
159. Weiner J 3rd, Parida SK, Maertzdorf J, Black GF, Repsilber D, Telaar A. et al. Biomarkers of inflammation, immunosuppression and stress with active disease are revealed by metabolomic profiling of tuberculosis patients. PLoS One. 2012;7:e40221
160. van Laarhoven A, Dian S, Aguirre-Gamboa R, Avila-Pacheco J, Ricano-Ponce I, Ruesen C. et al. Cerebral tryptophan metabolism and outcome of tuberculous meningitis: An observational cohort study. Lancet Infect Dis. 2018;18:526-535
161. Fitzgerald BL, Islam MN, Graham B, Mahapatra S, Webb K, Boom WH. et al. Elucidation of a human urine metabolite as a seryl-leucine glycopeptide and as a biomarker of effective anti-tuberculosis therapy. ACS Infect Dis. 2019;5:353-364
162. Tan SZ, Begley P, Mullard G, Hollywood KA, Bishop PN. Introduction to metabolomics and its applications in ophthalmology. Eye (Lond). 2016;30:773-83
163. Tessem MB, Bathen TF, Cejkova J, Midelfart A. Effect of UV-a and UV-b irradiation on the metabolic profile of aqueous humor in rabbits analyzed by 1H NMR spectroscopy. Invest Ophthalmol Vis Sci. 2005;46:776-81
164. Karamichos D, Zieske JD, Sejersen H, Sarker-Nag A, Asara JM, Hjortdal J. Tear metabolite changes in keratoconus. Exp Eye Res. 2015;132:1-8
165. Mayordomo-Febrer A, Lopez-Murcia M, Morales-Tatay JM, Monleon-Salvado D, Pinazo-Duran MD. Metabolomics of the aqueous humor in the rat glaucoma model induced by a series of intracamerular sodium hyaluronate injection. Exp Eye Res. 2015;131:84-92
166. Santiago AR, Garrido MJ, Cristovao AJ, Duarte JM, Carvalho RA, Ambrosio AF. Evaluation of the impact of diabetes on retinal metabolites by NMR spectroscopy. Curr Eye Res. 2010;35:992-1001
167. Makhoba NS, Smit DP, Walzl G, Chegou NN. Evaluation of potential antigen-specific host biomarkers in quantiferon supernatants as candidates for the diagnosis of ocular tuberculosis. Ocul Immunol Inflamm. 2021;29:1480-1488
168. Alam K, Sharma G, Forrester JV, Basu S. Antigen-specific intraocular cytokine responses distinguish ocular TB from undifferentiated uveitis in TB-immunoreactive patients. Am J Ophthalmol. 2023;246:31-41
169. Schrijver B, La Distia Nora R, Nagtzaam NMA, van Laar JAM, van Hagen PM, Dik WA. Serum CCL17 distinguishes sarcoid uveitis from TB-uveitis and QFT-negative uveitis. Acta Ophthalmol. 2022;100:e1533-e1534
170. Biselli R, Mariotti S, Sargentini V, Sauzullo I, Lastilla M, Mengoni F. et al. Detection of interleukin-2 in addition to interferon-gamma discriminates active tuberculosis patients, latently infected individuals, and controls. Clin Microbiol Infect. 2010;16:1282-4
171. Suzukawa M, Akashi S, Nagai H, Nagase H, Nakamura H, Matsui H. et al. Combined analysis of IGN-gamma, il-2, IL-5, IL-10, IL-1RA and MCP-1 in QFT supernatant is useful for distinguishing active tuberculosis from latent infection. PLoS One. 2016;11:e0152483
172. Gunasekeran DV, Agrawal R, Agarwal A, Carreno E, Raje D, Aggarwal K. et al. The collaborative ocular tuberculosis study (COTS)-1: A multinational review of 251 patients with tubercular retinal vasculitis. Retina. 2019;39:1623-1630
173. Moreno C, Taverne J, Mehlert A, Bate CA, Brealey RJ, Meager A. et al. Lipoarabinomannan from Mycobacterium tuberculosis induces the production of tumour necrosis factor from human and murine macrophages. Clin Exp Immunol. 1989;76:240-5
174. Llibre A, Bilek N, Bondet V, Darboe F, Mbandi SK, Penn-Nicholson A. et al. Plasma type 1 IFN protein concentrations in human tuberculosis. Front Cell Infect Microbiol. 2019;9:296
175. Koch S, Tugues S, Li X, Gualandi L, Claesson-Welsh L. Signal transduction by vascular endothelial growth factor receptors. Biochem J. 2011;437:169-83
176. Thayil SM, Albini TA, Nazari H, Moshfeghi AA, Parel JM, Rao NA. et al. Local ischemia and increased expression of vascular endothelial growth factor following ocular dissemination of Mycobacterium tuberculosis. PLoS One. 2011;6:e28383
177. Singh N, Singh R, Sharma RK, Kumar A, Sharma SP, Agarwal A. et al. Mycobacterium tuberculosis modulates fibroblast growth factor and vascular endothelial growth factor in ocular tuberculosis. Ocul Immunol Inflamm. 2021;29:1445-1451
178. Invernizzi A, Franzetti F, Viola F, Meroni L, Staurenghi G. Optic nerve head tubercular granuloma successfully treated with anti-VEGF intravitreal injections in addition to systemic therapy. Eur J Ophthalmol. 2015;25:270-2
179. Jain S, Agarwal A, Gupta V. Resolution of large choroidal tuberculoma following monotherapy with intravitreal ranibizumab. Ocul Immunol Inflamm. 2020;28:494-497
180. Heslop R, Bojang AL, Jarju S, Mendy J, Mulwa S, Secka O. et al. Changes in host cytokine patterns of TB patients with different bacterial loads detected using 16s rRNA analysis. PLoS One. 2016;11:e0168272
181. de Martino M, Lodi L, Galli L, Chiappini E. Immune response to Mycobacterium tuberculosis: A narrative review. Front Pediatr. 2019;7:350
182. Adekambi T, Ibegbu CC, Cagle S, Kalokhe AS, Wang YF, Hu Y. et al. Biomarkers on patient T cells diagnose active tuberculosis and monitor treatment response. J Clin Invest. 2015;125:1827-38
183. Riou C, Berkowitz N, Goliath R, Burgers WA, Wilkinson RJ. Analysis of the phenotype of Mycobacterium tuberculosis-specific CD4+ T cells to discriminate latent from active tuberculosis in HIV-uninfected and HIV-infected individuals. Front Immunol. 2017;8:968
184. Harari A, Rozot V, Bellutti Enders F, Perreau M, Stalder JM, Nicod LP. et al. Dominant TNF-alpha+ Mycobacterium tuberculosis-specific CD4+ T cell responses discriminate between latent infection and active disease. Nat Med. 2011;17:372-6
185. Sester U, Fousse M, Dirks J, Mack U, Prasse A, Singh M. et al. Whole-blood flow-cytometric analysis of antigen-specific CD4 T-cell cytokine profiles distinguishes active tuberculosis from non-active states. PLoS One. 2011;6:e17813
186. Porcel JM. Biomarkers in the diagnosis of pleural diseases: A 2018 update. Ther Adv Respir Dis. 2018;12:1753466618808660
187. Ahmed MIM, Ntinginya NE, Kibiki G, Mtafya BA, Semvua H, Mpagama S. et al. Phenotypic changes on Mycobacterium tuberculosis-specific CD4 T cells as surrogate markers for tuberculosis treatment efficacy. Front Immunol. 2018;9:2247
188. Tagirasa R, Parmar S, Barik MR, Devadas S, Basu S. Autoreactive T cells in immunopathogenesis of TB-associated uveitis. Invest Ophthalmol Vis Sci. 2017;58:5682-5691
189. Sharma RK, Gupta A, Kamal S, Bansal R, Singh N, Sharma K. et al. Role of regulatory T cells in tubercular uveitis. Ocul Immunol Inflamm. 2018;26:27-36
190. Sharma RK, Sharma J, Khan ZK, Pattekar A, Gupta V, Bansal R. et al. Diminished TLR2-TLR9 mediated CD4+ T cell responses are associated with increased inflammation in intraocular tuberculosis. Sci Rep. 2018;8:13812
191. Schrijver B, Hardjosantoso H, Ten Berge J, Schreurs MWJ, Van Hagen PM, Brooimans RA. et al. No evidence for circulating retina specific autoreactive T-cells in latent tuberculosis-associated uveitis and sarcoid uveitis. Ocul Immunol Inflamm. 2021;29:883-889
192. Hutchinson PE, Kee AR, Agrawal R, Yawata N, Tumulak MJ, Connolly JE. et al. Singapore ocular tuberculosis immunity study (SPOTIS): Role of T-lymphocyte profiling in patients with presumed ocular tuberculosis. Ocul Immunol Inflamm. 2021;29:1489-1495
193. Yang Q, Zhang M, Chen Q, Chen W, Wei C, Qiao K. et al. Cutting edge: Characterization of human tissue-resident memory T cells at different infection sites in patients with tuberculosis. J Immunol. 2020;204:2331-2336
194. Biswapriya B Misra CL, Michael Olivier, Laura A Cox. Integrated omics: Tools, advances and future approaches. J Mol Endocrinol. 2019;62:21-45
195. Ruiz-Tagle C, Naves R, Balcells ME. Unraveling the role of micrornas in Mycobacterium tuberculosis infection and disease: Advances and pitfalls. Infect Immun. 2020;88:e00649-19
196. Campisi L, Barbet G, Ding Y, Esplugues E, Flavell RA, Blander JM. Apoptosis in response to microbial infection induces autoreactive Th17 cells. Nat Immunol. 2016;17:1084-92
197. Zhong Z, Su G, Zhou Q, Meguro A, Takeuchi M, Mizuki N. et al. Tuberculosis exposure with risk of behcet disease among patients with uveitis. JAMA Ophthalmol. 2021;139:415-422
198. Mendelsohn SC, Mbandi SK, Fiore-Gartland A, Penn-Nicholson A, Musvosvi M, Mulenga H. et al. Prospective multicentre head-to-head validation of host blood transcriptomic biomarkers for pulmonary tuberculosis by real-time PCR. Commun Med (Lond). 2022;2:26
199. Nogueira BMF, Krishnan S, Barreto-Duarte B, Araujo-Pereira M, Queiroz ATL, Ellner JJ. et al. Diagnostic biomarkers for active tuberculosis: Progress and challenges. EMBO Mol Med. 2022;14:e14088
200. Yong YK, Tan HY, Saeidi A, Wong WF, Vignesh R, Velu V. et al. Immune biomarkers for diagnosis and treatment monitoring of tuberculosis: Current developments and future prospects. Front Microbiol. 2019;10:2789
Author contact
![]() Corresponding author: A/Prof (Dr) Rupesh Agrawal, Senior Consultant, National Healthcare Group Eye Institute, Tan Tock Seng Hospital, Singapore 308433, Email: Rupesh_agrawalcom.sg.
Corresponding author: A/Prof (Dr) Rupesh Agrawal, Senior Consultant, National Healthcare Group Eye Institute, Tan Tock Seng Hospital, Singapore 308433, Email: Rupesh_agrawalcom.sg.
 Global reach, higher impact
Global reach, higher impact