13.3
Impact Factor
Theranostics 2022; 12(15):6626-6645. doi:10.7150/thno.77243 This issue Cite
Review
Comprehensive overview of Nrf2-related epigenetic regulations involved in ischemia-reperfusion injury
1. Department of Cardiology, The First Affiliated Hospital of Soochow University, 188 Shizi Street, Suzhou, Jiangsu 215006, China.
2. Department of Orthopedics, The First Affiliated Hospital of Soochow University, Suzhou 215000, China.
3. Department of Cardiology, Kunshan Hospital of Traditional Chinese Medicine, Kunshan Affiliated Hospital of Nanjing University of Chinese Medicine, Suzhou 215000, Jiangsu, China.
*These authors contributed equally to this work.
Received 2022-7-18; Accepted 2022-9-1; Published 2022-9-11
Abstract
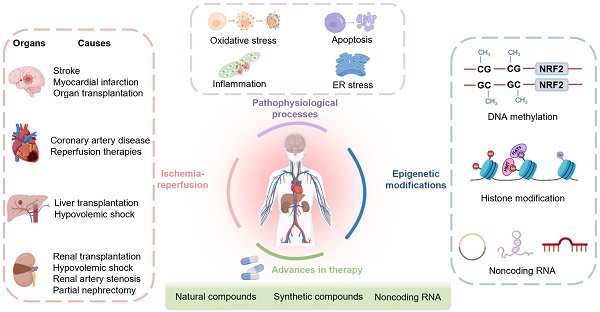
Ischemic disease is a class of diseases in which an organ is ischemic due to vascular occlusion, a major contributor to death and disability worldwide. However, when the blood flow is restored, more severe damage occurs than ischemia alone and is known as ischemic-reperfusion injury (IRI). During reperfusion, the imbalance between the production of reactive oxygen species (ROS) and buffering capacity of the antioxidant defense system results in cell damage and death. Nuclear factor E2-related factor 2 (Nrf2) significantly affects antioxidant stress damage. The function of Nrf2 in the pathological process of IRI has been widely discussed, but the impact of epigenetic modifications associated with Nrf2 remains unclear. This article provides a comprehensive overview of the role and mechanism of Nrf2-related epigenetic modifications in the IRI of various organs, including the brain, heart, liver, and kidney. In addition, we summarize agonists that may target epigenetic regulation of Nrf2, which may be beneficial in seeking more effective strategies to improve IRI.
Keywords: epigenetic modifications, ischemic-reperfusion injury, Nrf2, oxidative stress, therapy
Introduction
Many diseases lead to impaired blood circulation, which can contribute to ischemia in various organs, including the brain, heart, liver, and kidneys [1]. Prolonged ischemia of an organ results in irreversible damage. Blood flow restoration by clinical intervention is the major treatment strategy for ischemic organs, known as reperfusion therapy, which can effectively reduce tissue damage and ameliorate patient mortality. However, reperfusion causes additional damage creating a significant challenge in the current treatment of ischemic disease [2, 3].
The role of oxidative stress, inflammation, and apoptosis in the initiation and development of ischemic-reperfusion injury (IRI) has been extensively studied [4, 5]. It is noteworthy that oxidative stress is perceived as a primary contributor to IRI. The continuous production of oxidative and electrophilic chemicals irreversibly damages cellular lipids, proteins, and nucleic acids [6]. A complex antioxidant defense system has evolved to counteract the stress generated by endogenous and exogenous oxidants and electrophiles that includes nuclear factor erythroid 2-related factor 2 (Nrf2), protein kinase C (PKC), protein kinase B (PKB), and hypoxia-inducible factor-alpha (HIF-α). Among them, Nrf2, encoded by Nfe2l2, is considered to be a master regulator that protects cells from the effect of oxidative stress [7, 8]. Nrf2 activities differ greatly depending on the stage of various pathological processes. Although somatic mutations or single nucleotide polymorphisms (SNPs) could explain this difference, somatic mutations are only present in a small subset of tissues [9-11], indicating the existence of other mechanisms regulating Nrf2 expression.
Epigenetic modifications refer to heritable changes in the phenotype of cells, principally involving DNA methylation, histone modifications, and noncoding RNAs and, are not affected by changes in the DNA sequence. There is substantial evidence that epigenetic mechanisms can modulate Nrf2 expression in cancer and other diseases [7], but the application of epigenetics to IRI is still a relatively young field rapidly gaining attention. With improved research methods, key advances have been made, contributing to the progress of this field. Recently, hypermethylation of CpG islands in the promoter region of the Nrf2 gene was found to reduce Nrf2 expression and increase oxidative stress, inflammation, and apoptosis [12]. Acetylation, methylation, phosphorylation, and ubiquitination are the amino terminus modification of histones, among which acetylation and methylation have a dominant role in IRI by affecting the Nrf2 gene expression [13]. Moreover, modulation of the Nrf2 gene by noncoding RNAs has also been investigated in IRI [14, 15]. Unlike genetic mutations, epigenetic changes are reversible and susceptible to environmental factors. Therefore, developing drugs targeting epigenetic mechanisms represents a potential direction for clinical research.
Currently, preclinical studies and clinical trials are extensively employed to validate effective therapeutic strategies. From this perspective, it is necessary to understand the underlying cellular signaling pathways relevant to the development and treatment of IRI. Herein, we review the structure of Kelch-like ECH-associated protein 1 (Keap1)/Nrf2 and their epigenetic mechanisms, focusing on their functions in oxidative stress, apoptosis, inflammation, and endoplasmic reticulum stress during IRI in various organs, including the brain, heart, liver, and kidney. Furthermore, we summarize recent advances in the pharmacotherapy of Nrf2-related epigenetic modifications that might help prevent IRI.
Structures and interactions of Keap1/Nrf2
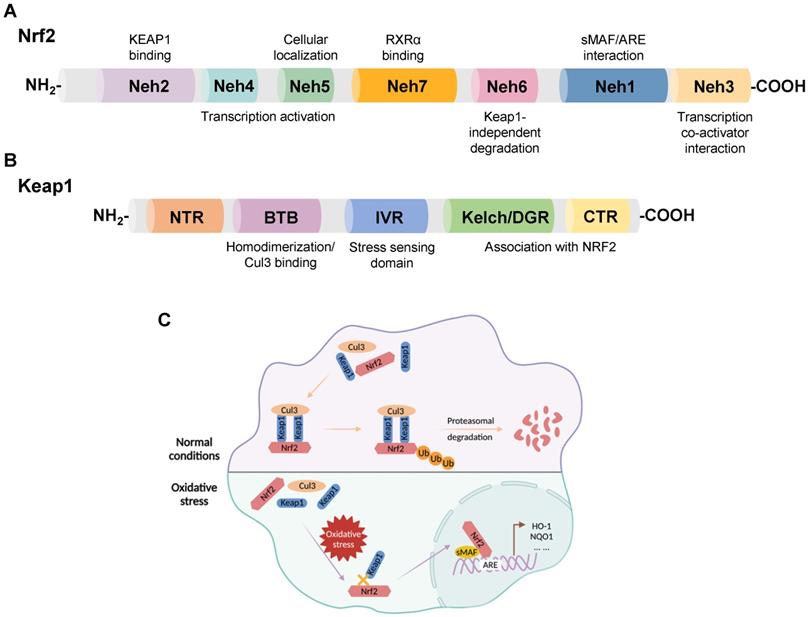
Nrf2, a member of the Cap-n-Collar family of basic leucine zipper proteins, was initially identified as an activator of β-globin gene expression and later shown to be a key sensor of cellular oxidative stress [16, 17]. Nrf2 is widely expressed in oxygen-consuming organs, such as the heart, liver, brain, kidney, muscle, and blood vessels. Nrf2 consists of 605 amino acids with seven functional domains, known as Nrf2-ECH homology (Neh) 1-7, responsible for preserving the stability of Nrf2 and modulating its transcriptional activity [18-20]. Among these seven domains, Neh1, 3, 4, and 5 are associated with transcriptional activation, while Neh2, 6, and 7 are negative regulatory domains [21]. In addition, Nrf2, as a transcription factor, is instrumental in maintaining redox homeostasis by inducing the expression of numerous genes involved in antioxidant defense, such as nicotinamide adenine dinucleotide phosphate (NADPH), glutathione peroxidase (GPx), NQO1, HO-1, and ferritin [22-26] (Figure 1A).
Keap1, a 624 amino acid protein, is an adapter for the Cul3-Rbx E3 ubiquitin ligase complex with five domains: an N-terminal region (NTR); a Broad complex, Tramtrack, and Bric-á-Brac (BTB) domain; an intervening region (IVR); a double glycine repeat (DGR) and a Kelch domain; the C-terminal region (CTR) [27, 28]. DGR and CTR mediate the binding between Keap1 and Nrf2. The BTB domain functions by boosting the homodimerization of Keap1 and facilitating the link between Keap1 and Cul3 [6] (Figure 1B).
Nrf2 is degraded by its negative regulator, Keap1. In physiological states, Keap1 acts as a link between Nrf2 and the ubiquitination ligase Cullin-3 (Cul3), promoting the rapid degradation of Nrf2 in the cytoplasm through the ubiquitin-proteasome pathway. When subjected to oxidative stress, the interaction between Keap1 and Nrf2 is impaired, and Nrf2 translocates from the cytoplasm to the nucleus. Subsequently, Nrf2 forms a heterodimerization complex with the transcriptional regulator musculoaponeurotic fibrosarcoma (sMAF), which recognizes antioxidant response elements (AREs) and promotes the transcription of antioxidant genes, such as heme oxygenase-1 (HO-1), NAD(P)H quinone oxidoreductase 1 (NQO1), and glutathione-s-transferase (GST) [29, 30] (Figure 1C).
The expression and activity of Keap1 and Nrf2 are affected by various factors. Currently, the focus is on regulating their expression and activity at the transcriptional level through epigenetic modifications, providing new ideas for the application of Keap1/Nrf2 in disease treatment.
Epigenetic modifications
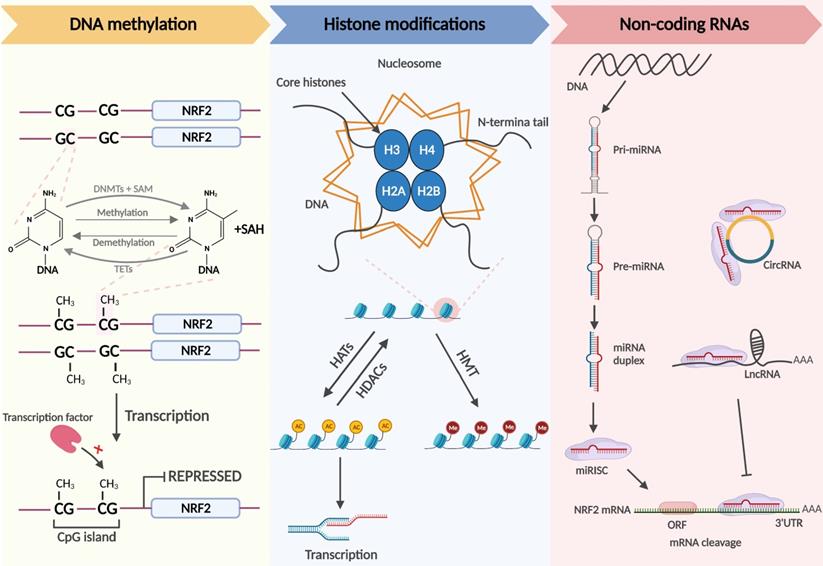
The term epigenetics was originally used to describe the complex mutual effects between the genome and the environment involved in the development and differentiation of higher organisms [31]. Epigenetics is currently used to explain the phenomenon in which gene expression alterations occur without changes to the nucleotide sequence [32]. Similar to genetic factors, it preserves cell differentiation and mammalian development [33]. The most common epigenetic alterations are DNA methylation, histone modifications, and the regulation of noncoding RNAs, which modulate cell differentiation, cell-specific gene expression, and genome stability and structure [34] (Figure 2).
DNA methylation, the transfer of methyl groups onto the C5 position of cytosine, occurs mainly in CpG-rich regions called CpG islands that are present in regulatory regions, such as promoters or enhancers of genes [35]. When CpG islands in the promoter region are heavily methylated, it is difficult for transcription factors to recognize the promoter, and gene expression is shut down. DNA methyltransferases (DNMTs) include DNMT3a, DNMT3b, and DNMT1. DNMT3a and DNMT3b, known as de novo DNA methyltransferases, are methylate double-stranded DNA that have never been methylated. Usually, the nucleosome DNA is the substrate of de novo DNA methyltransferases, and modifications of histones in nucleosomes affect the ability of these enzymes for de novo methylation. DNMT1 maintains DNA methylation and can methylate cytosine to methylcytosine in the double strands of semi-methylated DNA molecules, participating in the methylation of new synthetic chains in DNA replication [35, 36]. Demethylation of DNA boosts subsequent gene expression. Currently, there are two generally accepted methods of DNA demethylation, namely, active and passive demethylation. The former refers to the oxidation of methyl groups to hydroxymethylation that is then repaired in the presence of ten-eleven translocation (TET) methylcytosine dioxygenases; the latter partly occurs in the case of low DNMT activity or a low amount of methyl donor during replication. In addition, passive demethylation may also arise when methylated cytosine is oxidized by TET enzymes to hydroxymethyl-cytosine, which cannot be distinguished by DNMT1, resulting in the unmethylated cytosine on the synthetic chains [34].
Pharmacological targeting of epigenetic modifications of Nrf2
| Drugs | Cell/Animal type | Epigenetic mechanism | Molecular targets | Ref. |
|---|---|---|---|---|
| Quercetin | Transient middle cerebral artery occlusion (tMCAO) rats | Decreased histone acetylation | Increase expression of SIRT1 | [135] |
| Isorhamnetin | HT22 cells | Decreased histone acetylation | Increase expression of AKT and SIRT1 | [136] |
| H9C2 cells | Decreased histone acetylation | Increase expression of SIRT1 | [137] | |
| Honokiol | Type 1 diabetes (T1D) rats | Decreased histone acetylation | Increase expression of SIRT1 | [138] |
| Crocin | Myocardial I/R-induced primary cardiomyocytes and mouse | Reduced expression of miR-34a | Increase expression of SIRT1 and Nrf2 | [139] |
| Total flavonoids from Rosa laevigata Michx fruit | Hepatic I/R-induced rats | Decreased histone acetylation | Increase expression of SIRT1 | [140] |
| Resveratrol | HepG2 cells | Reduce methylation of Nfe2l2 | Suppressed expression of DNMTs | [58] |
| Sulforaphane | N2a/APPswe cells | Reduce methylation of Nfe2l2 | Suppressed expression of DNMTs | [151] |
| JB6 P+ cells | Reduce methylation of Nfe2l2 | Suppressed expression of DNMTs; decreased HDAC1, 2, 3, 4; reduced activities of HDACs | [152] | |
| TRAMP C1 cells | Reduce methylation of Nfe2l2 | Suppressed expression of DNMT1, 3a; decreased HDAC1,4,5,7 | [153, 154] | |
| human Caco-2 cells | Reduce methylation of Nfe2l2 | Suppressed expression and activities of DNMT1 protein | [154] | |
| C57/BL mice | Reduce methylation of Nfe2l2; promoted Ac- H3 accumulation in the Nrf2 promoter region | Suppressed expression and activities of DNMTs; decreased expression and activities of HDACs | [155] | |
| Ursolic acid | JB6 P+ cells | Reduce methylation of Nfe2l2 | Suppressed expression of DNMT1, 3a; decreased expression of HDAC1, 2, 3, 6, 7, 8; inhibit activities of HDACs | [158] |
| LNCaP cells | Increased H3K4me1 enrichment at the Nrf2 promoter | Induced expression of Setd7 | [160] | |
| Luteolin | Renal I/R-induced rats | Reduced expression of miR-320 | Decrease expression of Nrf2 | [164] |
| HCT116 cells | Reduce methylation of Nfe2l2 | Suppressed expression of DNMT1, 3a, 3b; decreased expression and activities of HDACs | [166] | |
| HT-29/SNU-407 cells | Reduce methylation of Nfe2l2 | Suppressed expression of DNMTs; increased TETs | [167] | |
| Curcumin | TRAMP C1/LnCap/HT29 cells | Reduce methylation of Nfe2l2 | Decreased activities of DNMTs | [12, 172, 173] |
| C66 | C57BL/6J mice | Increase miR-200a targeting 3'UTR of Keap1 | Increased expression of miR-200a | [171] |
| Sodium butyrate | Chronic cerebral hypoperfusion rats | Increasing histone H4 acetylation levels | Increasing HDAC1,2 mRNA levels | [177] |
| C57BL/6 mice | Increase occupancy of the AHR and P300 at the Nrf2 gene promoter; inhibit HDACs activity | [179] | ||
| Sprague-Dawley rats | Increased H3K9ac at the Nrf2 promoter | Decreased expression of HDAC1 | [181] |
(A) Domain structures of Nrf2. (B) Domain structures of Keap1. (C) Regulatory mechanism of Keap1 on the cellular abundance of Nrf2. Under physiological conditions, Keap1 serves as a link between Nrf2 and Cul3, promoting the rapid degradation of Nrf2 in the cytoplasm through the ubiquitin-proteasome pathway. When cells suffer oxidative stress damage, the Keap1-Nrf2 interaction is disrupted, and Nrf2 translocates to the nucleus, forming a heterodimerization complex with sMAF. Nrf2-sMAf heterodimers recognize ARE sequences and activate the transcription of antioxidant genes, such as HO-1 and NQO1.
Chromatin, the basic structural units of which are nucleosomes, consists of macromolecular DNA and histone complexes. In each nucleosome, four core histone proteins (H2A, H2B, H3, H4) form an octamer around which DNA is tightly wrapped. The free amino ends of histones in the core region of the nucleosome are called histone tails, which can be modified differently by enzymes and affect the transcriptional activity of genes. Post-translational modifications in histone tails consist of acetylation, methylation, ubiquitination, and phosphorylation. Histone acetylation is modulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). HATs transfer acetyl groups to lysine residues to neutralize the positive charge in histones. Because DNA is negatively charged, increased acetylation results in a weak interaction between histones and DNA, prompting nucleosomes to become loose and enabling transcription. Thus, histone acetylation is usually associated with transcriptional activation, while deacetylation participates in derailing gene expression [37].
Histone methylation refers to the transfer of methyl groups to the histone tail by histone methyltransferases (HMTs) and usually occurs on arginine or lysine residues. The regulatory role of histone methylation on transcription depends on the modification site and the methylation stoichiometry (monomethylation, dimethylation, or trimethylation) [38]. Another epigenetic modification, histone ubiquitination [39], is the process of the sequential actions of E1 activating, E2 conjugating, and E3 ligase enzymes, resulting in the covalent conjugation of ubiquitin, a 76 amino acid protein, to a lysine residue on proteins, which plays a key role in transcription initiation and elongation, silencing, and chromatin structure [40, 41]. Ubiquitination can be reversed by deubiquitinases (DUBs). DUBs hydrolyze ester bonds, peptide bonds, or isopeptide bonds at the carboxyl terminus of ubiquitin, specifically separating ubiquitin from protein substrates and regulating deubiquitination.
Noncoding RNAs, including microRNAs (miRNAs), long noncoding RNAs (lncRNAs), and circular RNAs (circRNAs), are functional RNA molecules that cannot be translated into proteins but have been identified as epigenetic regulators, playing a significant role in controlling Nrf2 gene expression [42, 43]. MiRNAs are composed of 18-25 nucleotides and serve as post-transcriptional regulators of gene expression, can bind to the untranslated regions (UTRs) of mRNA, and this imperfect sequence complementarity contributes to translation inhibition and/or mRNA degradation. Moreover, the expression of miRNAs is also modulated by epigenetic mechanisms [44, 45]. CircRNAs have a stable circular structure that prevents exonuclease-mediated elimination. LncRNAs are large noncoding RNAs > 200 nucleotides in length. Recent studies have shown that circRNAs and lncRNAs hold rich miRNA binding sites that can act as miRNA sponges, indirectly modulating the expression of targeted genes [46].
Nrf2-related epigenetic modifications: DNA methylation. DNMTs methylate the C5 position of cytosine in DNA to generate 5-methylcytosine, which is progressively oxidized to 5-carboxylcytosine by TETs. When the CpG islands in the promoter region are extensively methylated, the transcription factor cannot identify the promoter, and Nrf2 transcription is repressed. Histone modifications. Chromatin consists of DNA and histone complexes. In each nucleosome, four core histone proteins (H2A, H2B, H3, H4) form an octamer around which DNA is tightly wrapped. Histone tails, the free amino ends of histones, can be modified differently under the action of related enzymes, affecting gene expression. Histone acetylation and deacetylation are regulated by HATs and HDACs and hyperacetylated histones are associated with transcriptional activation. Histone tail lysine and arginine residues are methylated by HMTs. The effect of histone methylation on transcription depends on the methylation site and the methylation stoichiometry. Noncoding RNAs. Mature miRNAs identify the complementary bases of the 3'UTR in the Nrf2 mRNA and trigger mRNA cleavage. CircRNAs and lncRNAs contain miRNA binding sites that act as competing endogenous RNAs (ceRNAs) to regulate the expression of miRNA target mRNAs.
The effect of epigenetic modifications on the Keap1/Nrf2 signaling pathway has been demonstrated. In the following section, we discuss the mechanisms of Keap1/Nrf2-related epigenetic modifications and beneficial therapeutic agents in IRI.
Nrf2-related epigenetic modifications in the pathological process
Oxidative stress
Oxidative stress arises when there is an imbalance between the oxidant and antioxidant defense systems of the body [8]. This imbalance results in apoptosis, autophagy, inflammation, and other damage to cells via various signaling pathways, resulting in irreversible cell injury and organ dysfunction [20, 47]. Nrf2 is known as the “master regulator” of the antioxidant process, responsible for modulating the antioxidant system and the expression of cell-protective genes [48-50]. Nrf2 is widely present in oxygen-depleting organs such as the brain, heart, liver, kidneys, and blood vessels. Due to its positive impact on oxidative stress damage, Nrf2 has protective effects on multiple diseases. For example, it has been reported to protect nerve cells from IRI [51], and etomidate, an anesthetic agent, exerts a beneficial effect on myocardial tissue by activating the Nrf2/ARE signaling pathway [52]. Thus, it is necessary to study the mechanism of action of Nrf2 in redox imbalance. In this section, we will summarize the specific mechanisms of Keap1/Nrf2 signaling in oxidative stress from the epigenetic modification perspective (Figure 3).
Nrf2-related epigenetic modifications associated with oxidative stress. Delphinidin and resveratrol play a role in antioxidant reactions by decreasing DNMTs and reducing DNA methylation in the Nfe2l2 promoter region. Colistin reduces the acetylation of histones by inducing HDAC expression and reducing Nrf2. 7-HC and corosolic acid increase histones acetylation to exert antioxidant effects by decreasing HDAC expression. LncRNA HOTAIR increases H4 acetylation on the Nfe2l2 promoter, which, in turn, promotes Nrf2 expression. MiR-28, miR-144, miR-153, miR-27a, and miR-142-5p promote mRNA degradation by binding to the 3'UTR of Nrf2 mRNA, inhibiting Nrf2 expression and increasing oxidative stress. Dexamethasone inhibits the expression of Nrf2 target antioxidant genes by enhancing GR enrichment to AREs and blocking CBP recruitment and histone acetylation at AREs.
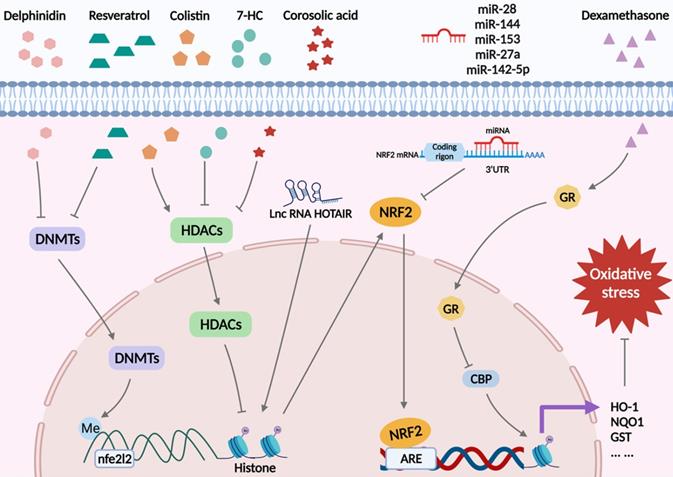
DNA demethylation of the Keap1 promoter CpG islands activates Keap1 protein production in the lens of diabetic cataract patients, increasing Nrf2 proteasomal degradation, altering lens protein redox balance, and blocking the cellular response to stress [53]. In addition, in 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced mouse skin epidermal JB6 P+ cells, decreased DNA methylation in the first 15 CpG sites in the Nfe2l2 promoter region by delphinidin could alleviate oxidative stress injury by restraining DNMT1 and DNMT3a [54]. Also, a recent study showed downregulated Nrf2 expression in a mouse prostate cancer model, and mechanistic studies identified a CpG island in the Nrf2 promoter region in tumorigenic TRAMP C1 cells but not in normal prostate or non-tumorigenic TRAMP C3 cells. Methylation of these CpG islands significantly repressed Nrf2 transcription in TRAMP C1 cells and augmented the recruitment of methyl-CpG-binding protein 2 (MBD2) and trimethyl-histone H3 (Lys9) proteins to the CpG islands in TRAMP C1 cells compared with TRAMP C3 cells. In contrast, the binding of RNA Pol II and acetylated histone H3 to the Nrf2 promoter was reduced. Intervention with 5-aza-29-deoxycytidine (5-aza), which inhibits DNMTs, and trichostatin A (TSA), an HDAC inhibitor, the expression of Nrf2 and NQO1 was restored in TRAMP C1 cells, verifying that hypermethylation of five CpG sites at the Nrf2 promoter was associated with mouse prostate cancer [55]. A similar phenomenon was observed in human prostate cancer tissues, where three specific CpG sites in the Nrf2 promoter were hypermethylated [56]. Interestingly, in mouse skin epidermal JB6 P+ cells, the flavonoid taxifolin showed the same effect [57]. Furthermore, methylation of the Nrf2 promoter region was upregulated in high glucose (HG)-treated HepG2 cells, and resveratrol restricted HG-induced ROS production through demethylation of the Nrf2/ARE signaling pathway [58]. Conversely, the combined therapeutics of trastuzumab and pertuzumab promoted DNA methylation on the Nfe2l2 promoter in human ovarian cancer cells, weakening the expression of Nrf2 and attenuating its antioxidant capacity, thereby achieving an anticancer effect [59].
Recently, Wang et al. showed that in kidney cells, colistin significantly diminished the expression of HDAC1 and deacetylated histone 3 in the Nrf2 promoter region at Lys27, inhibiting Nrf2 signaling, increasing cellular oxidative stress and apoptosis, and leading to renal damage. The coumarin derivative, 7-Hydroxycoumarin (7-HC) could attenuate HDAC1 level and restore histone acetylation in the Nrf2 promoter region to increase Nrf2 expression, thus enhancing the antioxidant capacity of the kidney [60]. A study using TRAMP C1 prostate cells indicated that corosolic acid treatment promoted histone H3 lysine 27 (H3K27ac) acetylation while reducing the trimethylation of histone H3 lysine 27 (H3K27me3) in the promoter region of Nrf2, which might be related to a decrease in HDACs [61]. Furthermore, another study showed that in mouse hepatocellular carcinoma (Hepalclc7) cells, dexamethasone facilitated glucocorticoid receptor (GR) recruitment to AREs, inhibiting the assembly of Nrf2-dependent cAMP-response element-binding protein (CBP) and histone acetylation at AREs and the subsequent transcription of downstream Nrf2, blocking its antioxidant capacity [62].
In reticulocytes of patients with sickle cell disease, miR-144 downregulated Nrf2 expression, thereby impairing tolerance to oxidative stress [63]. Similarly, miR153/miR27a/miR142-5p/miR144 reduced Nrf2 expression by targeting Nfe2l2 and increased the sensitivity of neuronal SH-SY5Y cells to oxidative stress [64]. Yang et al. reported that miR-28 aggravated oxidative stress in breast epithelial cells by targeting the 3′UTR of Nrf2 mRNA [65]. Dioscin, a natural steroid saponin, was shown to activate Nrf2, Sirtuin 2 (SIRT2), and downstream antioxidant stress-related target genes, such as glutamate-cysteine ligase modifier subunit (GCLM), forkhead box O3 (FOXO3a), HO-1, and NQO1 in H9C2 cells by significantly reducing the expression level of miR-140-5p [66]. Moreover, Hox transcript antisense intergenic RNA (HOTAIR) was the first lncRNA documented to modulate Nrf2 expression. In the sperms of asthenozoospermic and oligoasthenozoospermic patients, HOTAIR reduction attenuated the ability to resist oxidative stress by limiting the acetylation of H4 in the Nfe2l2 promoter, which in turn impeded Nrf2 expression [66].
Inflammation
Inflammation underlies various physiological and pathological processes triggered by harmful stimuli, such as infection and tissue damage. In addition, oxidative stress can also promote inflammatory responses. In addition, oxidative stress can also promote inflammatory responses. Cells damaged by ROS release damage-related model molecules (DAMPs), which combine with TLR4 or TLR9 on KCs to activate the NF-kB signaling pathway and thus generate more ROS to amplify the inflammatory response [67, 68]. Epigenetic modifications have been reported to be involved in the inflammatory process, and numerous studies have focused on the effect of epigenetic modifications on Nrf2 during the inflammatory response. One recent study found that environmental enrichment increased DNA methylation and histone H3 and H4 acetylation levels in the SAMP8 mouse hippocampus, likely associated with increased Nrf2 expression and decreased expressions of inflammatory genes, such as IL-6 and Cxcl10 [69]. Significant differences in Nrf2 and histone deacetylase expression were also observed in patients with type 2 diabetes and diabetic foot ulcers compared with healthy controls. Another study found a significantly positive correlation of HDAC4 with IL-6, and sirtuin 1 (SIRT1) was negatively correlated with IL-6, indicating that in the Nrf2 signaling pathway, HDAC4 and SIRT1 had pro-inflammatory anti-inflammatory effects, respectively [70]. Huang et al. reported that HDAC3 inhibition could promote anti-inflammatory gene expression through the Keap1-Nrf2-Nox4 signaling pathway and improve endothelial dysfunction in type 2 diabetes mellitus [71]. Interestingly, Nrf2 could also regulate HDAC expression; in mice exposed to cigarette smoke, deletion of the Nrf2 gene resulted in decreased HDAC2 and increased inflammation in the lung tissue [72].
Apoptosis
Apoptosis, a mode of programmed cell death, plays a significant role in the development of various diseases. Physiologically, apoptosis helps the body remove abnormal and harmful cells. In pathological conditions, injury-induced excessive apoptosis will aggravate tissue damage. Therefore, it is extremely important to study the role of apoptosis and the regulation of apoptosis in diseases. Recently, the regulation of apoptosis by Nrf2-related epigenetic modifications has attracted much attention. In the human lung cancer cell line H292, licochalcone A, a natural flavonoid, could increase endoplasmic reticulum stress and apoptosis by increasing miR-144-3p and downregulating Nrf2 [73]. A commonly used anesthetic agent, sevoflurane inhibited histone methyltransferase G9a and histone H3 lysine 9 (H3K9me2), promoted Nrf2 expression, and reduced the levels of inflammatory factors and neuronal apoptosis, attenuating hypoxic-ischemic brain injury in neonatal rats [74]. In podocytes, overexpression of Sirt6, a histone deacetylase, could increase the expression of Nrf2 and HO-1 genes and inhibit angiotensin (Ang) II-induced ROS generation and DNA double-strand breaks, reducing Ang II-induced podocyte apoptosis [75]. Also, Shao et al. found that in Pc12 cells, overexpression of miR-153 promoted the Nrf2/ARE signaling pathway, which inhibited neuronal apoptosis and promoted cell growth, alleviating isoflurane-induced neurotoxicity [76].
Nrf2-related epigenetic modifications in IRI
IRI is a pathological event that occurs in various diseases, causing severe cellular damage and death. As the name implies, IRI includes two distinct stages. Ischemia, the blockage of the arterial blood supply to an organ by emboli, is the first significant stage, while the second predominant process is reperfusion, the restoration of blood flow and oxygen supply to the affected ischemic area, which aggravates oxidative stress injury and contributes to excessive tissue deterioration. IRI can occur in many organs, such as the heart, brain, kidneys, lungs, and muscle [77, 78]. Besides the ischemic organs, IRI can cause systemic damage and negatively affect other organs, leading to multiple organ failure [79]. There is an imperative need to explore new therapeutic approaches and effective treatments. Nrf2 plays an important role in IRI due to its extensive biological effects, including anti-oxidative stress, anti-inflammation, and improved mitochondrial function. Recently, emerging evidence has indicated that the epigenetic modification of Nrf2 is involved in the pathophysiology of IRI. In this section, we discuss the latest research progress on Nrf2-related epigenetic mechanisms in ischemia-reperfusion (I/R) diseases focusing on the nervous, circulatory, digestive, and urinary systems.
Cerebral IRI
Stroke is the second leading cause of death and one of the major drivers of long-term disability globally [80]. Although various treatment methods restore blood flow in the infarcted area, the prognosis of patients has not been significantly improved, mainly due to IRI. Cerebral IRI often occurs after a clinical stroke, myocardial infarction, and organ transplantation. Ischemia and hypoxia cause neuronal impairment, necrosis, brain tissue damage, and metabolic dysfunction, which are further aggravated after blood flow is restored. The physiological and pathological mechanisms are complex and involve oxidative stress, excessive activation of inflammatory responses, and apoptosis or necrosis [81]. In IRI, regulating the expression of Keap1/Nrf2 through epigenetic modification can affect its downstream targets to alleviate brain tissue injury (Figure 4). Therefore, this section aims to investigate the role of epigenetic modifications of Keap1/Nrf2 in ameliorating cerebral IRI, paving the way for the development of new therapeutic approaches.
Recently, Li et al. observed that the expression of HDAC6, a class IIB HDAC isoenzyme, was increased in the brain tissue of cerebral I/R mice, while Nrf2 content was significantly decreased in the nucleus. Further research found that HDAC6 interference could reduce the infarct size of brain tissue and improve neurological function by considerably increasing the expression levels of Nrf2 and HO-1 [13]. Similarly, another study showed that HDAC6 inhibition could reduce oxidative stress damage and exert neuroprotective effects by activating Nrf2 [82].
SIRT1 is an NAD+-dependent class III histone deacetylase that regulates the acetylation of specific transcription factors and proteins [83]. For example, in cerebral I/R rats subjected to hyperbaric oxygen preconditioning (HBO-PC), increased SIRT1 promoted the expression of Nrf2, HO-1, and superoxide dismutase 1 (SOD1), lowering oxidative stress damage and playing a neuroprotective role in transient focal cerebral ischemia. At the same time, this protective effect was inhibited by SIRT1 or Nrf2 interference [84], suggesting that SIRT1, as a key epigenetic regulator, plays a significant role in ameliorating cerebral IRI by regulating the expression of Nrf2.
In oxygen-glucose-serum deprivation/restoration (OGSD/R)-treated astrocytes, overexpression of the nuclear factor of activated T cells 5 (NFAT5), a member of the Rel family, could significantly decrease histone acetylation and promote Nrf2 nuclear transport to reduce oxidative stress and apoptosis [85]. Additional studies have reported that dexmedetomidine attenuated oxidative stress, inflammation, and apoptosis via the NFAT5/SIRT1/Nrf2 signaling pathway, thus protecting the brain from the effects of IRI aggravated by diabetic hyperglycemia [86].
The protein SET domain-containing lysine methyltransferase 7 (SETD7), a class of lysine methyltransferases, has been identified as a key enzyme in the lysine methylation of histone and non-histone proteins [87-89]. Using an in vitro cell model of cerebral IRI, Pan et al. observed increased SETD7 expression in cells, decreased PC12 cell viability, and increased apoptosis. However, SETD7 knockout could reverse the effects of oxygen-glucose deprivation/reoxygenation (OGD/R) on PC12 cell viability and apoptosis by increasing Nrf2 expression and inactivating NF-κB, simultaneously diminishing the inflammatory response and oxidative stress [90].
TSA increases neuron viability and reduces infarct size in stroke mice after OGD. In vitro studies revealed that TSA reduced Keap1 level and induced Keap1/Nrf2 dissociation and Nrf2 nuclear translocation. Subsequently, Nrf2, combined with the ARE in the HO-1 gene, triggered HO-1 transcription, exerting a protective effect on hypoxic neurons [91].
MiRNAs also regulate neuronal survival during cerebral IRI. For example, in hippocampal neurons, miR-142-5p was involved in OGSD/R-induced cellular damage, and its downregulation relieved oxidative stress and neuronal damage by promoting Nrf2 expression [92]. Ji et al. reported that miR-153 could degrade Nrf2 in neurons, and its downregulation could promote the expression of Nrf2 and HO-1, alleviating OGSD/R-induced neuronal lesions and oxidative stress [93]. MiR-34b was predicted to limit Keap1 expression by binding to the 3'-UTR of Keap1 in rats. After focal I/R of the cerebrum, miR-34b expression was time-dependently downregulated. The overexpression of miR-34b increased Nrf2 and HO-1 protein levels to rectify I/R-induced oxidative stress damage and the volume of cerebral infarction by directly restraining Keap1 expression [94]. Theaflavin, derived from black tea, significantly reduced I/R-induced neuronal damage by eliminating miRNA-128-3p-mediated Nrf2 inhibition and oxidative stress, thereby improving impaired memory and learning ability [95].
Nrf2-related epigenetic modifications in cerebral IRI. TSA increases Nrf2 and its nuclear translocation by decreasing Keap1 levels, and reduces oxidative stress after OGD. MiR-34b increases Nrf2 by decreasing Keap1 to relieve oxidative stress in cerebral IRI. HDAC6 can promote histone deacetylation, reducing the expression of Nrf2. MiR-93, miR-153, and miR-142-5p might restrain the Nrf2/HO-1 pathway to exert an antioxidant effect on OGD/R-induced neurocyte injury. Rg1 may promote Nrf2 expression by restraining miR-144 to protect against I/R-induced oxidative stress damage. Theaflavin activates Nrf2 by restraining miR-128-3p to resist oxidative stress in cerebral IRI. HBO-PC alleviates oxidative stress damage in rats by promoting the SIRT1/Nrf2/HO-1 axis. Activation of the NFAT5/SIRT1/Nrf2 pathway by dexmedetomidine may reduce apoptosis in diabetic cerebral IRI. SETD7 aggravates OGD/R-induced inflammatory responses by inhibiting Nrf2 and activating NF-κB.
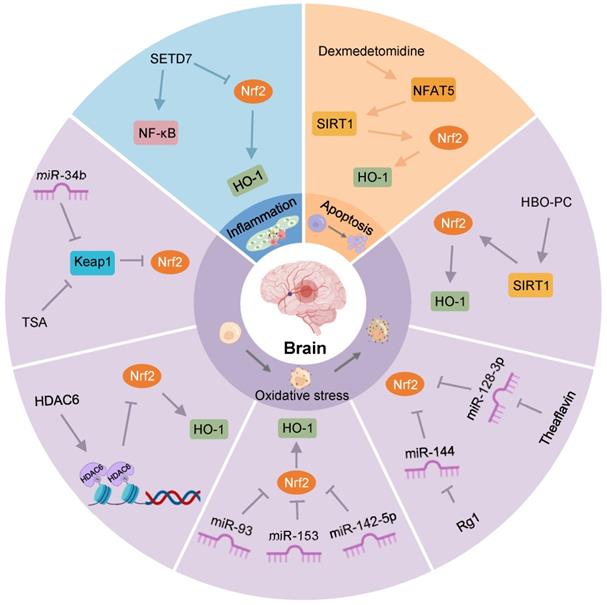
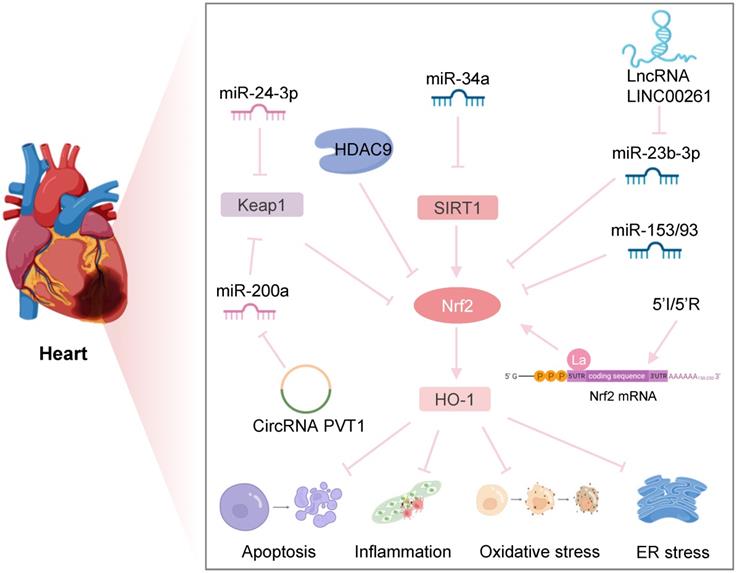
Nrf2-related epigenetic modifications in myocardial IRI. Downregulation of HDAC9 regulates hypoxia-induced cardiomyocyte apoptosis by activating the Nrf2/HO-1 pathway. Inhibition of miR-34a expression increases the SIRT1/Nrf2/HO-1 pathway, protecting against ER stress and apoptosis after myocardial I/R. MiR-24-3p reduces I/R-induced apoptosis of cardiomyocytes by reducing Keap1 expression. CircRNA PVT1 alleviates cardiac I/R damage by inhibiting the miR-200a/Keap1/Nrf2 pathway. lncRNA LINC002261 inhibits I/R-induced myocardial apoptosis by restraining the miR-23b-3p/Nrf2 pathway. Upregulation of miR-153 and miR-93 reduces Nrf2 expression to increase I/R-induced apoptosis of myocardial cells. Transient ischemia and reperfusion (5'I/5'R) preadaptation increase the La protein binding to the 5'UTR of Nrf2, exerting protective effects on myocardial IRI.
Furthermore, ginsenoside Rg1, a saponin extracted from Panax ginseng, has been shown to protect against I/R-induced oxidative stress through the miR-144/Nrf2/ARE pathway. Specifically, in OGSD/R-treated PC12 cells, Rg1 markedly decreased miR-144, downregulating Nrf2 production by targeting its 3′-UTR [15]. Similarly, miR-93 was increased in brain tissue after ischemia and could directly combine with the 3′-UTR of Nrf2 mRNA and reduce the level of Nrf2 and its downstream target genes. Intracerebroventricular injection of miR-93 antagomir decreased cerebral infarct volume, neuronal apoptosis, and oxidative stress after transient middle cerebral artery occlusion (MCAO), providing an attractive prospect for miR-93 as a therapeutic target for ischemic stroke [96].
Myocardial IRI
In an era of increasing reperfusion technology and availability, IRI has become a predominant issue in interventional cardiology [97, 98]. After myocardial I/R, multiple factors, including oxidative stress, mitochondrial dysfunction, and cell death, can lead to myocardial damage, resulting in myocardial shock, arrhythmia, increased infarct size, and heart failure. Therefore, it is necessary to study the underlying IRI mechanisms to improve the prognosis of patients with myocardial ischemia (Figure 5).
HDAC9 is a class II HDAC family member that is abundantly expressed in the brain and cardiac muscle and is involved in the epigenetic regulation of DNA transcription. A recent study found that Nrf2 mRNA and protein levels were significantly reduced in contrast to the marked upregulation of HDAC9 expression in the in vitro and in vivo models of myocardial infarction. Down-regulation of HDAC9 regulated cardiomyocyte proliferation, apoptosis, and myogenesis by activating the Nrf2/Keap1/HO-1 pathway, thus improving hypoxia-induced cardiomyocyte injury and cardiac function [99]. Interestingly, a genome-wide association study indicated that HDAC9 played a critical role in the development of ischemic stroke, and its variants could increase the risk of ischemic stroke by promoting carotid atherosclerosis [100]. These studies suggested that HDAC9 might be a new target for diagnosing and treating ischemic diseases.
A study showed that curcumin treatment exerted an anti-oxidative stress effect by reversing the epigenetic silencing of Nrf2 during prostate tumor development in TRAMP mice. This effect was achieved by curcumin acting as a hypomethylating agent by altering the methylation level of CpG islands in the Nrf2 gene promoter region [12]. Li and colleagues discovered a new antioxidant, novel curcumin analog 14p, which could ameliorate oxidative stress and limit myocardial IRI by activating Nrf2 [101]. However, the specific mechanism by which curcumin analog 14p increases Nrf2 expression and reduces myocardial IRI through demethylation of Nrf2 remains to be elucidated.
In a mouse model of myocardial I/R, Xu et al. observed that overexpression of SIRT1 could significantly reduce Nrf2 acetylation and upregulate the downstream signaling pathway of Nrf2, considerably improving cardiac oxidative stress, cardiac function, and infarct size [102]. Another study showed that miR-34a expression was significantly upregulated after myocardial I/R, and inhibition of miR-34a could reduce myocardial injury by eliminating SIRT1 inhibition, reducing apoptosis and infarct size, and improving left ventricular dysfunction [103].
Dang et al. found that inhibition of SETD7 reduced hypoxia/reoxygenation (H/R)-induced cardiomyocyte damage by downregulating Keap1 and promoting Nrf2-mediated antioxidant signaling [104]. Interestingly, in diabetic retinopathy development, SETD7 knockout inhibited Keap1 expression by preventing lysine 4 methylation on histone H3 (H3K4), stimulating protein-1 (Sp1) binding to the Keap1 promoter, and promoting Nrf2 expression [105]. Furthermore, another study on a human bronchial epithelial cell line (Beas-2B) indicated that SETD7 might directly methylate lysine residues in Nfe2l2 to alter the interaction between Nrf2 and Keap1, reducing overall Nrf2 protein levels. In addition, in SETD7 knockout cells, increased Nrf2 protein might offer a positive feedback mechanism for its own transcription by binding to the ARE region in the Nfe2l2 promoter, resulting in a concurrent increase in Nrf2 mRNA levels [106]. Therefore, Keap1/Nrf2 epigenetic modifications are expected to provide a new approach for exploring the specific mechanism by which SETD7 regulates H/R-induced cardiomyocyte injury. In another study by Xu et al., cycles of brief ischemia and reperfusion (5′I/5′R) could increase Nrf2 expression and protect mice against myocardial IRI. Specifically, cycles of 5′I/5′R or oxidants increased the binding of the La protein to the Nrf2 5′ UTR, promoting de novo Nrf2 protein translation [107].
Noncoding RNAs have been shown to participate in myocardial IRI, and numerous studies have focused on the function of noncoding RNAs on Nrf2 in developing myocardial IRI. Hou et al. found that in a rat model of I/R, inhibition of miR-153 could activate Nrf2 expression and reduce cardiomyocyte apoptosis, attenuating the inflammatory response and oxidative stress [108]. Another study reported that in hypoxic cardiomyocytes, overexpression of miR-200a significantly reduced Keap1, increased nuclear translocation of Nrf2, and the expression of downstream antioxidant enzyme genes, ameliorating cell apoptosis and ROS production and facilitating cardiomyocyte viability. Bioinformatic analysis and a dual-luciferase reporter assay found that miR-200a could bind to the 3'-UTR of Keap1 and inhibited the expression of Keap1 at the post-transcriptional level [109]. Moreover, studies have reported that circular RNA PVT1 limited the expression of miR-200a as a composite endogenous RNA in rat myocardial tissue. Circular RNA PVT1 silencing significantly alleviated apoptosis induced by myocardial IRI by increasing the expression of miR-200a [110]. These findings shed light on the potential of the circular RNA PVT1/miR-200a/Keap1/Nrf2 axis as a treatment strategy for myocardial IRI.
After OGD/R in cardiomyocytes, elevated miR-93 expression increased apoptosis of cardiomyocytes by targeting inhibitory Nrf2 [111]. MiR-24-3p is a tumor suppressor that modulates various tumors. Recent studies have demonstrated that the increased expression of miR-24-3p reduces IRI-induced apoptosis of cardiomyocytes in vitro by targeting inhibition of Keap1, leading to the activation of Nrf2 signaling [112]. Zhang et al. discovered that in I/R myocardial tissue and H/R cardiomyocytes, lncRNA LINC00261 expression was downregulated, while overexpression of LINC00261 reduced the H/R-induced apoptosis of cardiomyocytes. Further evidence suggested that LINC00261 upregulated Nrf2 expression in cardiomyocytes by restraining miR-23b-3p and consequently reducing H/R-induced apoptosis in cardiomyocytes [113].
Liver IRI
Liver IRI is one of the main causes of irreversible injury during liver transplantation and hypovolemic shock. Treatment of hepatic IRI involves multiple aspects, involving genetics, pharmacology, and surgical procedures. This section will focus on the latest treatment methods for hepatic IRI in the context of regulating Nrf2-related epigenetic modifications (Figure 6).
Nrf2-related epigenetic modifications in liver and renal IRI: Sevoflurane alleviates the inhibition of Nrf2 by miR-122, inhibiting I/R-induced oxidative stress and inflammation of cardiomyocytes. Crocin, zinc sulfate, and lncRNA MEG3 could promote the Nrf2/ARE pathway by inhibiting miR-34a to improve oxidative stress in liver IRI. Fucoxanthin and melatonin might alleviate apoptosis induced by renal IRI through the SIRT1/Nrf2/HO-1 pathway. PRMT5 promotes oxidative stress and pyroptosis by inhibiting the Nrf2/HO-1 pathway to aggravate renal IRI. The lncRNA TUG1 inhibits renal I/R-induced oxidative stress and apoptosis by suppressing the miR-144-3p/Nrf2 pathway.
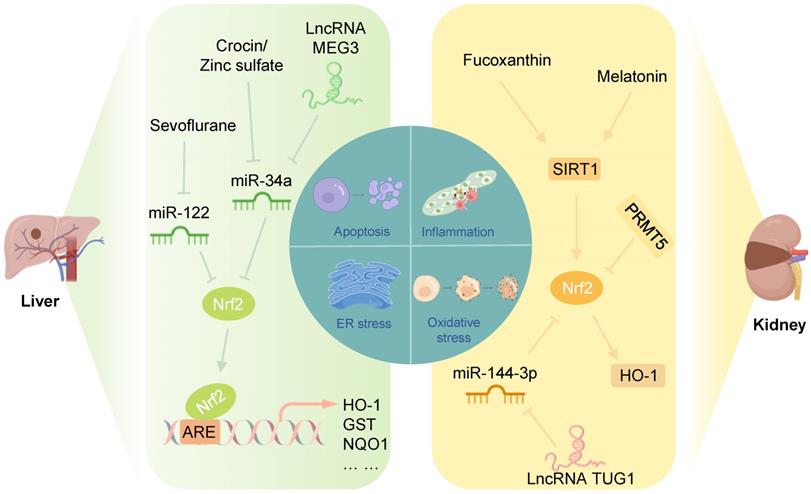
Studies have shown that in a rat model of hepatic I/R, crocin and zinc sulfate could promote Nrf2 expression in the liver and reduce the miR-34a level, improving the antioxidant capacity of the liver. Notably, serum miR-34a levels were negatively correlated with Nrf2 levels in rats in the hepatic I/R group [114, 115]. Furthermore, Huang et al. showed that miR-34a inhibited the activity of Nrf2 and its ARE targets in the liver in a NaHS-preconditioned young rat model of hepatic I/R [116]. These results suggested that inhibition of miR-34a protects against hepatic IRI by increasing the translation of Nrf2 and its downstream antioxidant genes. Another study showed that the lncRNA MEG3, as a ceRNA for miR-34a, protected liver cells from I/R damage by downregulating the expression of miR-34a and upregulating the expression of Nrf2 [117].
MiR-122 is a primary liver-specific miRNA that significantly modulates liver development, liver function, and hepatocyte growth [118]. The level of circulating miR-122 was markedly increased during liver IRI and positively related to the activity of liver function indicators, such as aspartate aminotransferase (AST), alanine aminotransferase (ALT), and lactate dehydrogenase (LDH), suggesting that miR-122 could be a key molecule for liver I/R therapy [119]. A recent study demonstrated that sevoflurane promoted Nrf2 expression in the I/R liver tissue and reduced miR-122 expression, ameliorating I/R-induced oxidative stress, inflammatory responses, and apoptosis in the liver tissue [120]. Interestingly, Nrf2 has been identified as a potential target of miR-122 in acute liver injury [121].
Renal IRI
In the clinical setting, IRI causes acute kidney injury, adverse outcomes after transplantation, and patient morbidity and mortality [122]. After renal I/R, elevated production of ROS in damaged tissues is a crucial cause of tubular epithelial cell death and renal injury [123]. When ROS generation surpasses the antioxidant ability, further tissue damage and apoptosis are induced [124]. Therefore, reducing oxidative stress during renal I/R is crucial for treating renal ischemia. This section will examine the effect of Keap1/Nrf2 on renal IRI and epigenetic modifications of Keap1/Nrf2 to explore new therapeutic targets (Figure 6).
In a mouse model of IRI and a human renal proximal tubular epithelial cell line (HK-2) H/R injury model, the marine carotenoid fucoxanthin inhibited oxidative stress-induced apoptosis through the SIRT1/Nrf2/HO-1 signaling pathway, and its mechanism was likely associated with Nrf2 nuclear translocation [125]. Shi et al. found that melatonin attenuated acute renal IRI in diabetic rats by activating the SIRT1/Nrf2/HO-1 signaling pathway [126]. Also, activation of SIRT1 could stabilize the transcription factor Nrf2 through deacetylation [102, 127]. Future studies should focus on whether SIRT1 can improve renal IRI by regulating promoted by SIRT1 Nrf2 deacetylation. Protein arginine methylation transferase 5 (PRMT5), which regulates arginine methylation and participates in the modulation of epigenetics, aggravated renal IRI by promoting oxidative stress and pyroptosis, inhibiting the Nrf2/HO-1 pathway, and suppressing the proliferation of tubular epithelium [128].
Zhao et al. demonstrated that in mouse renal IRI and mouse renal tubular epithelial cell (TCMK cell) H/R injury models, lncRNA TUG1 expression was increased, while that of miR-144-3p decreased significantly. Mechanistic experiments clarified that miR-144-3p could target and inhibit Nrf2 expression. Furthermore, lncRNA TUG1 could promote Nrf2 expression by sponging miR-144-3p and improving oxidative stress damage and apoptosis of cells and tissues [129]. In summary, the lncRNA TUG1/miR-144-3p/Nrf2 axis provides a new therapeutic approach for renal IRI.
Pharmacological targeting of epigenetic modifications of Nrf2
Currently, natural compounds extracted from fruits, vegetables, tea, and medicinal plants are applied to treat various diseases, such as cancer and cardiovascular, neurological, and kidney diseases. These phytochemicals can modulate Nrf2-related epigenetic modifications and overcome oxidative stress at nontoxic concentrations, providing new approaches for the pharmacotherapy of diseases [130-133]. The protective effect of Nrf2 agonists has been validated in preclinical models and human clinical trials. The regulation of epigenetic modifications of Nrf2 by phytochemicals is an emerging field that will open up avenues for exploring new strategies to target Nrf2 signaling and provide new ideas for treating ischemic diseases and cancer. This section will summarize recent advances in regulating Nrf2 signaling by various phytochemicals targeting DNA methylation, histone modifications, and noncoding RNAs.
SIRT1 agonists
As a deacetylase, SIRT1 can regulate gene expression through histone deacetylation and play a key role in preventing oxidative stress and inflammation. For example, SIRT1 could enhance Nrf2 expression through the deacetylation of Nrf2 [134]. Yang et al. found that quercetin significantly reduced cerebral infarct volume and ROS generation via the SIRT1/Nrf2/HO-1 signaling pathway and exerted a neuroprotective effect on cerebral IRI, which could be reversed by the SIRT1 selective inhibitor EX527 [135]. In addition, studies have demonstrated that HG could aggravate OGD/R-induced apoptosis, the inflammatory response, and oxidative stress in HT22 hippocampal neurons, which could be alleviated by isorhamnetin. Further studies showed that the isorhamnetin effect was exerted by increasing AKT expression, which activated the SIRT1/Nrf2/HO-1 signaling pathway [136]. Isorhamnetin also has positive effects on H/R-stimulated H9C2 cells, ameliorating apoptosis and oxidative stress in H9C2 cells by promoting SIRT1/Nrf2/HO-1-mediated antioxidant signaling [137]. Honokiol has been shown to improve myocardial IRI in diabetic rats by facilitating SIRT1 and increasing Nrf2 expression to reduce oxidative stress and apoptosis [138]. It was also reported that crocin, a water-soluble carotenoid, may protect cardiomyocytes from endoplasmic reticulum stress and IRI-induced apoptosis by regulating the miR-34a/SIRT1/Nrf2 pathway [139]. Furthermore, in rat liver I/R, total flavonoids (TFs) from Rosa laevigata Michx fruit could significantly increase the SIRI1 level in the liver tissue, activating Nrf2 expression and alleviating IRI-induced oxidative stress injury [140].
Resveratrol
Resveratrol (trans-3,5,400-trihydroxystibene, RV) is a naturally occurring polyphenolic compound in daily diets and some herbal medicines and has been proven to have protective effects against cardiovascular diseases, cancer, and aging [141]. In cerebral, myocardial, and renal IRI, RV has been shown to reduce oxidative stress and inflammatory responses by upregulating Nrf2 expression [142-145]. Interestingly, recent research has suggested that epigenetic changes regulated by RV are associated with some pathological processes. For instance, in nonalcoholic fatty liver disease (NAFLD), RV treatment could downregulate Nrf2 promoter methylation by decreasing the expression of DNMTs, facilitating Nrf2 transcription, and attenuating oxidative stress [58]. However, the epigenetic regulation of Nrf2 by RV in IRI injury has not been reported thus far, identifying an area that needs to be addressed.
Sulforaphane
Sulforaphane (SFN), a natural and well-studied isothiocyanate and a plant-derived chemoprotective ingredient, is extracted from cruciferous vegetables such as broccoli, cauliflower, cabbage, and kale [146]. It is a strong activator of Nrf2 that can exert chemotherapeutic effects by regulating oxidative stress, inflammation, cell proliferation, differentiation, and apoptosis. Studies have shown that SFN can protect the brain, liver, heart, and kidney from IRI injury by increasing Nrf2 expression [147-150]. However, the specific mechanism by which SFN regulates Nrf2 expression has not been well explored. Epigenetic modifications provide a new possibility to understand how SFN regulates Nrf2. In studies of Alzheimer's disease, skin cancer, prostate cancer, colon cancer and angiotensin II-induced cardiomyopathy, SFN has been found to exert protective effects through epigenetic modifications. Specifically, SFN reduced methylation of the first 15 CpG islands on the Nrf2 gene promoter, which was associated with reduced DNMT levels, and promoted the enrichment of acetylated histone H3 in the Nrf2 promoter region by inhibiting the expression and activity of HDAC [151-155].
Ursolic acid
Ursolic acid (UA), a representative pentacyclic triterpenoid obtained from berries and the peels of pears, apples, plums, and other fruits, reduces ROS toxicity and enhances antioxidant enzyme activity [156]. Notably, many current studies have revealed that UA can exert a significant effect in treating diseases by modulating the expression of epigenetic modification-related enzymes, such as DNMTs and HDACs [157, 158]. As a classic anti-inflammatory and antioxidant molecule, Nrf2 has been shown to be a key target for UA. For example, UA protected the mouse brain against cerebral ischemic injury through the Nrf2 pathway [159]. Another study showed that UA treatment increased SETD7 expression in LNCaP cells and H3K4me1 enrichment at the Nrf2 promoter, enhancing Nrf2 expression and protecting DNA from oxidative damage [160]. In contrast, SETD7 knockdown ameliorated oxidative stress induced by cerebral IRI by increasing Nrf2 transcriptional activity [90]. Furthermore, SETD7 inhibition could reduce Keap1 expression in retinal endothelial cells by preventing H3K4 methylation, thereby increasing Nrf2 expression and reducing apoptosis and oxidative stress damage [105]. It is possible that UA can regulate Nrf2 by inducing SETD7 expression, which can regulate Nrf2 expression through epigenetic modifications and reduce IRI damage.
Luteolin
Luteolin (3′,4′,5,7-tetrahydroxyflavone, LUT), a natural flavone compound extracted from Chinese herbs and vegetables, has many biological benefits, such as anti-inflammatory, antioxidant, cardioprotective, and neuroprotective effects [161-163]. A recent study found that LUT could reverse the elevated expression of miR-320 and Nrf2, increasing antioxidant enzymes and reducing oxidative stress in renal I/R [164]. In contrast, Zhu et al. found that the miR-320 expression was downregulated in myocardial IRI. Inhibition of miR-320 could enhance the antioxidant capability of the myocardium and reduce apoptosis and IRI by increasing Nrf2 expression [165]. The differences in miR-320 expression in the kidney and heart may be due to the tissue-specific expression of miRNAs. The epigenetic regulation of Nrf2 by LUTs has been extensively studied in cancers. For example, Zuo et al. found that in LUT-treated HCT116 and HT29 colon cancer cells, LUT inhibits the protein level and enzymatic activity of DNMT1, DNMT3a, and DNMT3b in a dose-dependent manner. Then, the demethylation of the Nrf2 promoter leads to the activation of Nrf2 and its downstream genes, reduces ROS levels and DNA damage, and inhibits cancer cell proliferation. In addition, the protein and enzymatic activities of HDACs in HCT116 cells also decreased after LUT treatment. Interestingly, subsequent studies reported that in HT-29 and SNU-407 cells, LUT could enhance the transcription activator TET DNA in addition to repressing the expression of the transcriptional repressor DNMTs. It also promotes the combining of TET1 with the Nrf2 promoter, significantly reduces DNA methylation in the Nrf2 promoter region of colon cancer cells, and upregulates the expression of Nrf2, thereby inducing apoptosis of colon cancer cells. Taken together, these studies demonstrate that LUT plays a series of biological roles by increasing the expression of Nrf2 through epigenetic modifications of Nrf2. Unfortunately, the epigenetic modifications of Nrf2 by LUT are still concentrated in the field of tumors, and its role in other diseases needs to be further explored in the future [166, 167]. These studies demonstrate that LUT plays a series of biological roles by increasing the expression of Nrf2 through epigenetic modifications of Nrf2, which provides a new research direction for treatment of IRI by LUT.
Curcumin
Curcumin, also called diferuloylmethane, is a yellow chemical compound generally isolated from the rhizome of turmeric [168]. Curcumin has antioxidant, antibacterial, anti-inflammatory, and anticancer properties and can also act as an epigenetic regulator for Nrf2 and its downstream genes.
Li et al. reported that curcumin could reduce neurological dysfunction, cerebral infarct volume, cerebral edema, and blood-brain barrier disorder after cerebral IRI. The mechanism might be associated with the upregulation of Nrf2 expression by curcumin [169]. Furthermore, dimethyl fumarate and curcumin combination treatment could significantly reduce serum ALT and AST activities caused by liver I/R and improve liver histopathology. The combined treatment of dimethyl fumarate and curcumin exerted its effect by activating the Nrf2/HO-1 signaling pathway and reducing inflammatory markers (TNF-α, IL-1β, Il-6, and iNOS) [170]. It has been reported that C66, a novel analog of curcumin with extremely high bioavailability, increased miR-200a expression in kidneys of diabetic mice, decreased Keap1 at the transcriptional and translational levels, induced Nrf2 function, and ameliorated diabetes-induced oxidative stress damage [171]. Similarly, Sun et al. demonstrated that the expression of miR-200a was significantly downregulated in ischemic myocardium and hypoxic cardiomyocytes. Overexpression of miR-200a could substantially increase the nuclear translocation of Nrf2 and expression of downstream antioxidant enzyme genes and alleviate hypoxia-induced oxidative stress injury related to the interaction of miR-200a with the 3'-UTR of Keap1 [109]. In addition, studies have shown that the occurrence of prostate cancer is related to the hypermethylation of the Nrf2 promoter region and the decreased expression of Nrf2 and its downstream detoxification and antioxidant genes. In murine prostate cancer TRAMP C1 cells, human prostate cancer LnCap cells and human colon cancer HT29 cells, curcumin exerts a preventive effect on cancer by reducing methylation of CpG islands of Nrf2 and activating its expression [12, 172, 173].
Sodium butyrate
Sodium butyrate (NaB) is a sodium salt metabolite of short-chain fatty acids (SCFAs) produced by microorganisms that degrade dietary fibers in the colonic lumen [174]. SCFAs exert a crucial role in preserving gut health, enter the systemic circulation, and directly affect the metabolism and function of peripheral tissues [175]. As an HDAC inhibitor, first reported in 1978 [176], NaB has shown great potential in oxidative stress, immunomodulation, and cancer prevention and treatment. Recently, NaB has been reported to function as an Nrf2 agonist [177], epigenetic modifications of which play a role in many diseases.
Dong et al. reported that in diabetic mice, NaB treatment ameliorated diabetes-induced renal oxidative damage, apoptosis, inflammation, fibrosis, pathological changes, and renal dysfunction by enhancing Nrf2 and its downstream targets HO-1 and NQO1. Interestingly, the nuclear translocation of Nrf2 or Keap1 expression inhibited HDAC activity, suggesting that NaB promoted the binding of transcription factors to the Nrf2 gene promoter by depressing HDAC activity to activate Nrf2 at the transcriptional level [178]. However, sodium butyrate (SB) treatment could activate Nrf2 and downstream gene transcription by increasing hippocampus HDAC1,2 mRNA and histone H4 acetylation levels in chronic cerebral hypoperfusion rats, thereby protecting cognitive function. This contradictory expression may be due to the following reason. The mechanism of action of HDAC inhibitors mainly depends on the interaction of the enzyme and the inhibitor with zinc in the active site. Therefore, insufficient SB-induced enzymatic activity results in compensatory transcription of mRNA [177]. Similarly, in HG-treated aortic endothelial cells, NaB inhibited HDAC activity. It increased the occupancy of the transcription factor aryl hydrocarbon receptor (AHR) and the co-activator P300, a histone acetylase, at the Nrf2 gene promoter, resulting in increased Nrf2 transcription to exert a protective role against oxidative stress and inflammation [179]. In contrast, in a non-transformed small intestine epithelial IEC-6 cell line, NaB increased the expression of Nrf2 at the gene level and its nuclear translocation [180]. These contradictory results could be explained due to different cell types and processing methods used. Also, in a high-fat diet-induced rat model, NaB activated the transcription of Nrf2 and its downstream antioxidant enzymes, ameliorating high-fat diet-induced weight gain, oxidative stress, and insulin resistance. The upregulation of Nrf2 expression in NaB-treated rat livers was associated with a decrease in HDAC1 protein expression and an increase in histone H3 acetyl K9 (H3K9ac) modification on the Nrf2 promoter [181].
Conclusions and Perspectives
Oxidative stress and inflammation induced by I/R aggravate organ function damage; therefore, preventing and reducing ROS production and inflammatory damage is essential to improve and preserve organ function in IRI. As a major antioxidant factor, Nrf2 protects normal cells from damage by enhancing their antioxidant and detoxification capabilities. Theoretically, during cancer development and progression, Nrf2 could have two functions. Prior to tumorigenesis, Nrf2 could play a crucial role in combating oxidative stress and preventing malignant transformation. After tumor formation, malignantly transformed cells could utilize the Nrf2 defense system to impede cancer cell growth, spread, and resistance to therapy [182, 183]. However, the two-faced nature of Nrf2 in IRI has not yet been reported, emphasizing the need for more exploration in this field.
In recent years, the regulatory effect of epigenetic modifications on Keap1/Nrf2 in various diseases has been elucidated. This review focuses on the specific mechanisms by which methylation/demethylation of CpGs in the promoter region of Keap1/Nrf2, methylation/demethylation and acetylation/deacetylation of histones, or miRNAs regulate Keap1/Nrf2 expression in I/R diseases.
The role of epigenetic modifications associated with Keap1/Nrf2 during I/R remains to be better understood. With the identification of endogenous imprinted genes, genomic imprinting became widely recognized as an epigenetic mechanism in which the expression pattern of a parental allele influences phenotypic expression [184]. Besides the canonical genomic imprinting mediated by allelic DNA methylation, recent studies have shown that maternal H3K27me3 can lead to imprinting unrelated to DNA methylation, termed “non-canonical imprinting”. Abnormal expression of these genes has been found in genetic diseases, cancer, and autoimmune diseases [185]. Future research on genomic imprinting may better explain individual differences in resistance to IRI and provide new ideas for developing therapeutic drugs for IRI. In 2013, Young et al. discovered a new class of gene regulatory elements called "super-enhancers," characterized by clusters of adjacent large enhancers that drive the specific expression of genes [186]. It has been demonstrated that transcriptional programs can be regulated by epigenetic alterations in super-enhancers, which play an important role in maintaining cancer cell properties [187]. Furthermore, a recent study of renal IRI demonstrated that renal injury results in genome-wide alterations in the enhancer and super-enhancer repertoire. Specifically, 216 injury-associated super-enhancers, 164 super-enhancers lost during injury, and 385 SHARED super-enhancers were identified by using the ROSE algorithm, which were present in kidney samples at baseline and IRI day 2 sample. Most of these super-enhancers can play important roles in renal epithelial cells by regulating gene expression [188]. Although the current research on super-enhancers in IRI is limited, a better understanding of the regulation of Nrf2 transcription with the possible involvement of super-enhancers would have far-reaching significance.
We have also reviewed compounds that regulate the Keap1/Nrf2 pathway through epigenetic modifications. However, most of the current research on the epigenetic modification effects of Nrf2 agonists has focused on cancer treatment, and few studies have elaborated on the mechanism of these drugs and their potential in IRI, indicating the need for mechanistic studies in this field.
Owing to the complex pathophysiology of IRI, there is currently no effective approach to counterbalance this injury. Therefore, studying the function of Nrf2 in IRI, especially the role of Nrf2-related epigenetic modifications, will provide a new approach to treating I/R-related diseases.
Abbreviations
AREs: antioxidant response elements; Cul3: Cullin-3; circRNAs: circular RNAs; DNMTs: DNA methyltransferases; GR: glucocorticoid receptor; HATs: histone acetyltransferases; HDACs: histone deacetylases; HMT: histone methyltransferase; H3K27ac: acetylation of histone H3 lysine 27; H3K27me3: trimethylation of histone H3 lysine 27; HOTAIR: Hox transcript antisense intergenic RNA; HO-1: heme oxygenase-1; HG: high glucose; H/R: hypoxia/reoxygenation; H3K4: lysine 4 on histone H3; IRI: ischemic-reperfusion injury; I/R: ischemia‒reperfusion; Keap1: Kelch-like ECH-associated protein 1; lncRNAs: long noncoding RNAs; miRNAs: microRNAs; Nrf2: nuclear factor erythroid 2-related factor 2; NQO1: NAD(P)H quinone oxidoreductase 1; Neh: Nrf2-ECH homology; NFAT5: nuclear factor of activated T cells 5; NaB: Sodium butyrate; OGD/R: oxygen-glucose deprivation/restoration; OGSD/R: oxygen-glucose-serum deprivation/restoration; ROS: reactive oxygen species; RV: resveratrol; SIRT1: silencing information regulator 2 related enzyme 1; SETD7: SET domain-containing lysine methyltransferase 7; sMAF: musculoaponeurotic fibrosarcoma; TET: ten-eleven translocation; TPA: 12-O-tetradecanoylphorbol-13-acetate; TSA: trichostatin A; UTRs: untranslated regions.
Acknowledgements
This work is supported by grants from the National Natural Science Foundation of China (Nos. 81770327 and 81100173).
Author Contributions
All authors have read and approved the final review manuscript. J. Zhang, W. Pan and Y. Zhang conceived and wrote this review. M. Tan, Y. Yin and Y. Li searched and analyzed the literature. L. Zhang and L. Han reviewed and edited this review. H. Li, T. Jiang, J. Bai and W. Pan modified this review.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Tsao CW, Aday AW, Almarzooq ZI, Alonso A, Beaton AZ, Bittencourt MS. et al. Heart Disease and Stroke Statistics-2022 Update: A Report From the American Heart Association. Circulation. 2022;145:e153-e639
2. Doenst T, Haverich A, Serruys P, Bonow RO, Kappetein P, Falk V. et al. PCI and CABG for Treating Stable Coronary Artery Disease: JACC Review Topic of the Week. J Am Coll Cardiol. 2019;73:964-76
3. McCarthy CP, Vaduganathan M, McCarthy KJ, Januzzi JL Jr, Bhatt DL, McEvoy JW. Left Ventricular Thrombus After Acute Myocardial Infarction: Screening, Prevention, and Treatment. JAMA Cardiol. 2018;3:642-9
4. Khan H, Gupta A, Singh TG, Kaur A. Mechanistic insight on the role of leukotriene receptors in ischemic-reperfusion injury. Pharmacol Rep. 2021;73:1240-54
5. Hajjar DP, Gotto AM. Biological relevance of inflammation and oxidative stress in the pathogenesis of arterial diseases. Am J Pathol. 2013;182:1474-81
6. Lu M-C, Ji J-A, Jiang Z-Y, You Q-D. The Keap1-Nrf2-ARE Pathway As a Potential Preventive and Therapeutic Target: An Update. Med Res Rev. 2016;36:924-63
7. Sharma A, Parikh M, Shah H, Gandhi T. Modulation of Nrf2 by quercetin in doxorubicin-treated rats. Heliyon. 2020;6:e03803
8. Thiruvengadam M, Venkidasamy B, Subramanian U, Samynathan R, Ali Shariati M, Rebezov M. et al. Bioactive Compounds in Oxidative Stress-Mediated Diseases: Targeting the NRF2/ARE Signaling Pathway and Epigenetic Regulation. Antioxidants (Basel). 2021 10
9. Cho H-Y, Marzec J, Kleeberger SR. Functional polymorphisms in Nrf2: implications for human disease. Free Radic Biol Med. 2015;88:362-72
10. Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO. et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420
11. Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA. et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16:3743-53
12. Khor TO, Huang Y, Wu T-Y, Shu L, Lee J, Kong A-NT. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochem Pharmacol. 2011;82:1073-8
13. Li J, Zhao Y, Shi J, Ren Z, Chen F, Tang W. Histone deacetylase 6 interference protects mice against experimental stroke-induced brain injury via activating Nrf2/HO-1 pathway. Anim Cells Syst (Seoul). 2019;23:192-9
14. Zhu X, Zhao Y, Hou W, Guo L. MiR-153 regulates cardiomyocyte apoptosis by targeting Nrf2/HO-1 signaling. Chromosome Res. 2019;27:167-78
15. Chu S-F, Zhang Z, Zhou X, He W-B, Chen C, Luo P. et al. Ginsenoside Rg1 protects against ischemic/reperfusion-induced neuronal injury through miR-144/Nrf2/ARE pathway. Acta Pharmacol Sin. 2019;40:13-25
16. Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91:9926-30
17. Martin F, van Deursen JM, Shivdasani RA, Jackson CW, Troutman AG, Ney PA. Erythroid maturation and globin gene expression in mice with combined deficiency of NF-E2 and nrf-2. Blood. 1998;91:3459-66
18. Galicia-Moreno M, Lucano-Landeros S, Monroy-Ramirez HC, Silva-Gomez J, Gutierrez-Cuevas J, Santos A. et al. Roles of Nrf2 in Liver Diseases: Molecular, Pharmacological, and Epigenetic Aspects. Antioxidants (Basel). 2020 9
19. Bellezza I, Giambanco I, Minelli A, Donato R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim Biophys Acta Mol Cell Res. 2018;1865:721-33
20. Tu W, Wang H, Li S, Liu Q, Sha H. The Anti-Inflammatory and Anti-Oxidant Mechanisms of the Keap1/Nrf2/ARE Signaling Pathway in Chronic Diseases. Aging Dis. 2019;10:637-51
21. Zhang B, Ma Z, Tan B, Lin N. Targeting the cell signaling pathway Keap1-Nrf2 as a therapeutic strategy for adenocarcinomas of the lung. Expert Opin Ther Targets. 2019;23:241-50
22. Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401-26
23. Krajka-Kuźniak V, Paluszczak J, Baer-Dubowska W. The Nrf2-ARE signaling pathway: An update on its regulation and possible role in cancer prevention and treatment. Pharmacol Rep. 2017;69:393-402
24. Pandey P, Singh AK, Singh M, Tewari M, Shukla HS, Gambhir IS. The see-saw of Keap1-Nrf2 pathway in cancer. Crit Rev Oncol Hematol. 2017;116:89-98
25. Hu Y-R, Ma H, Zou Z-Y, He K, Xiao Y-B, Wang Y. et al. Activation of Akt and JNK/Nrf2/NQO1 pathway contributes to the protective effect of coptisine against AAPH-induced oxidative stress. Biomed Pharmacother. 2017;85:313-22
26. Ahmed SMU, Luo L, Namani A, Wang XJ, Tang X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim Biophys Acta Mol Basis Dis. 2017;1863:585-97
27. Fuse Y, Kobayashi M. Conservation of the Keap1-Nrf2 System: An Evolutionary Journey through Stressful Space and Time. Molecules. 2017 22
28. Furukawa M, Xiong Y. BTB protein Keap1 targets antioxidant transcription factor Nrf2 for ubiquitination by the Cullin 3-Roc1 ligase. Mol Cell Biol. 2005;25:162-71
29. Guo Y, Yu S, Zhang C, Kong A-NT. Epigenetic regulation of Keap1-Nrf2 signaling. Free Radic Biol Med. 2015;88:337-49
30. Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD. et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76-86
31. Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123:2145-56
32. Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481-6
33. Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662-73
34. Ling C, Rönn T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019;29:1028-44
35. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Reviews Genetics. 2012;13:484-92
36. Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247-57
37. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381-95
38. Hogg SJ, Beavis PA, Dawson MA, Johnstone RW. Targeting the epigenetic regulation of antitumour immunity. Nat Rev Drug Discov. 2020;19:776-800
39. Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653-63
40. Oh E, Akopian D, Rape M. Principles of Ubiquitin-Dependent Signaling. Annu Rev Cell Dev Biol. 2018;34:137-62
41. Mattiroli F, Penengo L. Histone Ubiquitination: An Integrative Signaling Platform in Genome Stability. Trends Genet. 2021;37:566-81
42. Han T-S, Hur K, Cho H-S, Ban HS. Epigenetic Associations between lncRNA/circRNA and miRNA in Hepatocellular Carcinoma. Cancers (Basel). 2020 12
43. Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q. et al. Circular RNAs in immune responses and immune diseases. Theranostics. 2019;9:588-607
44. Kota SK, Kota SB. Noncoding RNA and epigenetic gene regulation in renal diseases. Drug Discov Today. 2017;22:1112-22
45. Yao Q, Chen Y, Zhou X. The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol. 2019;51:11-7
46. Yang Y, Yujiao W, Fang W, Linhui Y, Ziqi G, Zhichen W. et al. The roles of miRNA, lncRNA and circRNA in the development of osteoporosis. Biol Res. 2020;53:40
47. Xiang M, Lu Y, Xin L, Gao J, Shang C, Jiang Z. et al. Role of Oxidative Stress in Reperfusion following Myocardial Ischemia and Its Treatments. Oxid Med Cell Longev. 2021;2021:6614009
48. Hybertson BM, Gao B, Bose SK, McCord JM. Oxidative stress in health and disease: the therapeutic potential of Nrf2 activation. Mol Aspects Med. 2011;32:234-46
49. Shen Y, Liu X, Shi J, Wu X. Involvement of Nrf2 in myocardial ischemia and reperfusion injury. Int J Biol Macromol. 2019;125:496-502
50. Cheng L, Jin Z, Zhao R, Ren K, Deng C, Yu S. Resveratrol attenuates inflammation and oxidative stress induced by myocardial ischemia-reperfusion injury: role of Nrf2/ARE pathway. Int J Clin Exp Med. 2015;8:10420-8
51. Liu H, Wu X, Luo J, Zhao L, Li X, Guo H. et al. Adiponectin peptide alleviates oxidative stress and NLRP3 inflammasome activation after cerebral ischemia-reperfusion injury by regulating AMPK/GSK-3β. Exp Neurol. 2020;329:113302
52. Lv Z, Wang Fe, Zhang X, Zhang X, Zhang J, Liu R. Etomidate Attenuates the Ferroptosis in Myocardial Ischemia/Reperfusion Rat Model via Nrf2/HO-1 Pathway. Shock. 2021;56:440-9
53. Periyasamy P, Shinohara T. Age-related cataracts: Role of unfolded protein response, Ca mobilization, epigenetic DNA modifications, and loss of Nrf2/Keap1 dependent cytoprotection. Prog Retin Eye Res. 2017 60
54. Kuo H-CD, Wu R, Li S, Yang AY, Kong A-N. Anthocyanin Delphinidin Prevents Neoplastic Transformation of Mouse Skin JB6 P+ Cells: Epigenetic Re-activation of Nrf2-ARE Pathway. AAPS J. 2019;21:83
55. Yu S, Khor TO, Cheung K-L, Li W, Wu T-Y, Huang Y. et al. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS One. 2010;5:e8579
56. Khor TO, Fuentes F, Shu L, Paredes-Gonzalez X, Yang AY, Liu Y. et al. Epigenetic DNA methylation of antioxidative stress regulator NRF2 in human prostate cancer. Cancer Prev Res (Phila). 2014;7:1186-97
57. Kuang H, Tang Z, Zhang C, Wang Z, Li W, Yang C. et al. Taxifolin Activates the Nrf2 Anti-Oxidative Stress Pathway in Mouse Skin Epidermal JB6 P+ Cells through Epigenetic Modifications. Int J Mol Sci. 2017 18
58. Hosseini H, Teimouri M, Shabani M, Koushki M, Babaei Khorzoughi R, Namvarjah F. et al. Resveratrol alleviates non-alcoholic fatty liver disease through epigenetic modification of the Nrf2 signaling pathway. Int J Biochem Cell Biol. 2020;119:105667
59. Khalil HS, Langdon SP, Goltsov A, Soininen T, Harrison DJ, Bown J. et al. A novel mechanism of action of HER2 targeted immunotherapy is explained by inhibition of NRF2 function in ovarian cancer cells. Oncotarget. 2016;7:75874-901
60. Wang J, Ishfaq M, Fan Q, Chen C, Li J. 7-Hydroxycoumarin Attenuates Colistin-Induced Kidney Injury in Mice Through the Decreased Level of Histone Deacetylase 1 and the Activation of Nrf2 Signaling Pathway. Front Pharmacol. 2020;11:1146
61. Yang J, Wu R, Li W, Gao L, Yang Y, Li P. et al. The triterpenoid corosolic acid blocks transformation and epigenetically reactivates Nrf2 in TRAMP-C1 prostate cells. Mol Carcinog. 2018;57:512-21
62. Alam MM, Okazaki K, Nguyen LTT, Ota N, Kitamura H, Murakami S. et al. Glucocorticoid receptor signaling represses the antioxidant response by inhibiting histone acetylation mediated by the transcriptional activator NRF2. J Biol Chem. 2017;292:7519-30
63. Sangokoya C, Telen MJ, Chi J-T. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116:4338-48
64. Narasimhan M, Patel D, Vedpathak D, Rathinam M, Henderson G, Mahimainathan L. Identification of novel microRNAs in post-transcriptional control of Nrf2 expression and redox homeostasis in neuronal, SH-SY5Y cells. PLoS One. 2012;7:e51111
65. Yang M, Yao Y, Eades G, Zhang Y, Zhou Q. MiR-28 regulates Nrf2 expression through a Keap1-independent mechanism. Breast Cancer Res Treat. 2011;129:983-91
66. Zhao L, Tao X, Qi Y, Xu L, Yin L, Peng J. Protective effect of dioscin against doxorubicin-induced cardiotoxicity via adjusting microRNA-140-5p-mediated myocardial oxidative stress. Redox Biol. 2018;16:189-98
67. Tsung A, Klune JR, Zhang X, Jeyabalan G, Cao Z, Peng X. et al. HMGB1 release induced by liver ischemia involves Toll-like receptor 4 dependent reactive oxygen species production and calcium-mediated signaling. J Exp Med. 2007;204:2913-23
68. Bamboat ZM, Balachandran VP, Ocuin LM, Obaid H, Plitas G, DeMatteo RP. Toll-like receptor 9 inhibition confers protection from liver ischemia-reperfusion injury. Hepatology. 2010;51:621-32
69. Grinan-Ferre C, Puigoriol-Illamola D, Palomera-Avalos V, Perez-Caceres D, Companys-Alemany J, Camins A. et al. Environmental Enrichment Modified Epigenetic Mechanisms in SAMP8 Mouse Hippocampus by Reducing Oxidative Stress and Inflammaging and Achieving Neuroprotection. Front Aging Neurosci. 2016;8:241
70. Teena R, Dhamodharan U, Ali D, Rajesh K, Ramkumar KM. Gene Expression Profiling of Multiple Histone Deacetylases (HDAC) and Its Correlation with NRF2-Mediated Redox Regulation in the Pathogenesis of Diabetic Foot Ulcers. Biomolecules. 2020 10
71. Huang S, Chen G, Sun J, Chen Y, Wang N, Dong Y. et al. Histone deacetylase 3 inhibition alleviates type 2 diabetes mellitus-induced endothelial dysfunction via Nrf2. Cell Commun Signal. 2021;19:35
72. Adenuga D, Caito S, Yao H, Sundar IK, Hwang JW, Chung S. et al. Nrf2 deficiency influences susceptibility to steroid resistance via HDAC2 reduction. Biochem Biophys Res Commun. 2010;403:452-6
73. Chen G, Ma Y, Jiang Z, Feng Y, Han Y, Tang Y. et al. Lico A Causes ER Stress and Apoptosis via Up-Regulating miR-144-3p in Human Lung Cancer Cell Line H292. Front Pharmacol. 2018;9:837
74. Wang H, Xu Y, Zhu S, Li X, Zhang H. Post-Treatment Sevoflurane Protects Against Hypoxic-Ischemic Brain Injury in Neonatal Rats by Downregulating Histone Methyltransferase G9a and Upregulating Nuclear Factor Erythroid 2-Related Factor 2 (NRF2). Med Sci Monit. 2021;27:e930042
75. Fan Y, Cheng J, Yang Q, Feng J, Hu J, Ren Z. et al. Sirt6-mediated Nrf2/HO-1 activation alleviates angiotensin II-induced DNA DSBs and apoptosis in podocytes. Food Funct. 2021;12:7867-82
76. Shao D, Wu Z, Bai S, Fu G, Zou Z. The function of miRNA-153 against isoflurane-induced neurotoxicity via Nrf2/ARE cytoprotection. Mol Med Rep. 2019
77. Yan H-F, Tuo Q-Z, Yin Q-Z, Lei P. The pathological role of ferroptosis in ischemia/reperfusion-related injury. Zool Res. 2020;41:220-30
78. Lv S, Liu H, Wang H. The Interplay between Autophagy and NLRP3 Inflammasome in Ischemia/Reperfusion Injury. Int J Mol Sci. 2021 22
79. Chazelas P, Steichen C, Favreau F, Trouillas P, Hannaert P, Thuillier R. et al. Oxidative Stress Evaluation in Ischemia Reperfusion Models: Characteristics, Limits and Perspectives. Int J Mol Sci. 2021 22
80. Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S. et al. Heart Disease and Stroke Statistics-2018 Update: A Report From the American Heart Association. Circulation. 2018 137
81. Zheng J-H, Xie L, Li N, Fu Z-Y, Tan X-F, Tao R. et al. PD98059 protects the brain against mitochondrial-mediated apoptosis and autophagy in a cardiac arrest rat model. Life Sci. 2019;232:116618
82. Gaisina IN, Lee SH, Kaidery NA, Ben Aissa M, Ahuja M, Smirnova NN. et al. Activation of Nrf2 and Hypoxic Adaptive Response Contribute to Neuroprotection Elicited by Phenylhydroxamic Acid Selective HDAC6 Inhibitors. ACS Chem Neurosci. 2018;9:894-900
83. Dang R, Wang M, Li X, Wang H, Liu L, Wu Q. et al. Edaravone ameliorates depressive and anxiety-like behaviors via Sirt1/Nrf2/HO-1/Gpx4 pathway. J Neuroinflammation. 2022;19:41
84. Xue F, Huang JW, Ding PY, Zang HG, Kou ZJ, Li T. et al. Nrf2/antioxidant defense pathway is involved in the neuroprotective effects of Sirt1 against focal cerebral ischemia in rats after hyperbaric oxygen preconditioning. Behav Brain Res. 2016;309:1-8
85. Xia X, Qu B, Li Y-M, Yang L-B, Fan K-X, Zheng H. et al. NFAT5 protects astrocytes against oxygen-glucose-serum deprivation/restoration damage via the SIRT1/Nrf2 pathway. J Mol Neurosci. 2017 61
86. Chen L, Cao J, Cao D, Wang M, Xiang H, Yang Y. et al. Protective effect of dexmedetomidine against diabetic hyperglycemia-exacerbated cerebral ischemia/reperfusion injury: An in vivo and in vitro study. Life Sci. 2019;235:116553
87. Pradhan S, Chin HG, Estève P-O, Jacobsen SE. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics. 2009;4:383-7
88. Kwon T, Chang JH, Kwak E, Lee CW, Joachimiak A, Kim YC. et al. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. EMBO J. 2003;22:292-303
89. Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P. et al. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002;16:479-89
90. Pan X, Fan J, Peng F, Xiao L, Yang Z. SET domain containing 7 promotes oxygen-glucose deprivation/reoxygenation-induced PC12 cell inflammation and oxidative stress by regulating Keap1/Nrf2/ARE and NF-kappaB pathways. Bioengineered. 2022;13:7253-61
91. Wang B, Zhu X, Kim Y, Li J, Huang S, Saleem S. et al. Histone deacetylase inhibition activates transcription factor Nrf2 and protects against cerebral ischemic damage. Free Radic Biol Med. 2012;52:928-36
92. Wang N, Zhang L, Lu Y, Zhang M, Zhang Z, Wang K. et al. Down-regulation of microRNA-142-5p attenuates oxygen-glucose deprivation and reoxygenation-induced neuron injury through up-regulating Nrf2/ARE signaling pathway. Biomed Pharmacother. 2017;89:1187-95
93. Ji Q, Gao J, Zheng Y, Liu X, Zhou Q, Shi C. et al. Inhibition of microRNA-153 protects neurons against ischemia/reperfusion injury in an oxygen-glucose deprivation and reoxygenation cellular model by regulating Nrf2/HO-1 signaling. J Biochem Mol Toxicol. 2017 31
94. Huang R, Ma J, Niu B, Li J, Chang J, Zhang Y. et al. MiR-34b Protects Against Focal Cerebral Ischemia-Reperfusion (I/R) Injury in Rat by Targeting Keap1. J Stroke Cerebrovasc Dis. 2019;28:1-9
95. Li R, Li X, Wu H, Yang Z, Fei L, Zhu J. Theaflavin attenuates cerebral ischemia/reperfusion injury by abolishing miRNA-128-3p-mediated Nrf2 inhibition and reducing oxidative stress. Mol Med Rep. 2019;20:4893-904
96. Wang P, Liang X, Lu Y, Zhao X, Liang J. MicroRNA-93 Downregulation Ameliorates Cerebral Ischemic Injury Through the Nrf2/HO-1 Defense Pathway. Neurochem Res. 2016;41:2627-35
97. Neumann FJ, Hochholzer W, Siepe M. [ESC/EACTS guidelines on myocardial revascularization 2018: The most important innovations]. Herz. 2018;43:689-94
98. Valgimigli M, Gragnano F, Branca M, Franzone A, Baber U, Jang Y. et al. P2Y12 inhibitor monotherapy or dual antiplatelet therapy after coronary revascularisation: individual patient level meta-analysis of randomised controlled trials. BMJ. 2021;373:n1332
99. Liu F, Di Y, Ma W, Kang X, Li X, Ji Z. HDAC9 exacerbates myocardial infarction via inactivating Nrf2 pathways. J Pharm Pharmacol. 2022;74:565-72
100. Loci associated with ischaemic stroke and its subtypes (SiGN). a genome-wide association study. Lancet Neurol. 2016;15:174-84
101. Li W, Wu M, Tang L, Pan Y, Liu Z, Zeng C. et al. Novel curcumin analogue 14p protects against myocardial ischemia reperfusion injury through Nrf2-activating anti-oxidative activity. Toxicol Appl Pharmacol. 2015;282:175-83
102. Xu J-J, Cui J, Lin Q, Chen X-Y, Zhang J, Gao E-H. et al. Protection of the enhanced Nrf2 deacetylation and its downstream transcriptional activity by SIRT1 in myocardial ischemia/reperfusion injury. Int J Cardiol. 2021;342:82-93
103. Fu BC, Lang JL, Zhang DY, Sun L, Chen W, Liu W. et al. Suppression of miR-34a Expression in the Myocardium Protects Against Ischemia-Reperfusion Injury Through SIRT1 Protective Pathway. Stem Cells Dev. 2017;26:1270-82
104. Dang Y, Ma X, Li Y, Hao Q, Xie Y, Zhang Q. et al. Inhibition of SETD7 protects cardiomyocytes against hypoxia/reoxygenation-induced injury through regulating Keap1/Nrf2 signaling. Biomed Pharmacother. 2018;106:842-9
105. Mishra M, Zhong Q, Kowluru RA. Epigenetic modifications of Keap1 regulate its interaction with the protective factor Nrf2 in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014;55:7256-65
106. He S, Owen DR, Jelinsky SA, Lin L-L. Lysine Methyltransferase SETD7 (SET7/9) Regulates ROS Signaling through mitochondria and NFE2L2/ARE pathway. Sci Rep. 2015;5:14368
107. Xu B, Zhang J, Strom J, Lee S, Chen QM. Myocardial ischemic reperfusion induces de novo Nrf2 protein translation. Biochim Biophys Acta. 2014;1842:1638-47
108. Hou W, Zhu X, Liu J, Ma J. Inhibition of miR-153 ameliorates ischemia/reperfusion-induced cardiomyocytes apoptosis by regulating Nrf2/HO-1 signaling in rats. Biomed Eng Online. 2020;19:15
109. Sun X, Zuo H, Liu C, Yang Y. Overexpression of miR-200a protects cardiomyocytes against hypoxia-induced apoptosis by modulating the kelch-like ECH-associated protein 1-nuclear factor erythroid 2-related factor 2 signaling axis. Int J Mol Med. 2016;38:1303-11
110. Luo C, Ling G-X, Lei B-F, Feng X, Xie X-Y, Fang C. et al. Circular RNA PVT1 silencing prevents ischemia-reperfusion injury in rat by targeting microRNA-125b and microRNA-200a. J Mol Cell Cardiol. 2021;159:80-90
111. Yan LJ, Fan XW, Yang HT, Wu JT, Wang SL, Qiu CG. MiR-93 inhibition ameliorates OGD/R induced cardiomyocyte apoptosis by targeting Nrf2. Eur Rev Med Pharmacol Sci. 2017;21:5456-61
112. Xiao X, Lu Z, Lin V, May A, Shaw DH, Wang Z. et al. MicroRNA miR-24-3p Reduces Apoptosis and Regulates Keap1-Nrf2 Pathway in Mouse Cardiomyocytes Responding to Ischemia/Reperfusion Injury. Oxid Med Cell Longev. 2018;2018:7042105
113. Zhang R, Li Y, Liu X, Qin S, Guo B, Chang L. et al. FOXO3a-mediated long non-coding RNA LINC00261 resists cardiomyocyte hypoxia/reoxygenation injury via targeting miR23b-3p/NRF2 axis. J Cell Mol Med. 2020;24:8368-78
114. Akbari G, Mard SA, Dianat M, Mansouri E. The Hepatoprotective and MicroRNAs Downregulatory Effects of Crocin Following Hepatic Ischemia-Reperfusion Injury in Rats. Oxid Med Cell Longev. 2017;2017:1702967
115. Mard SA, Akbari G, Dianat M, Mansouri E. Protective effects of crocin and zinc sulfate on hepatic ischemia-reperfusion injury in rats: a comparative experimental model study. Biomed Pharmacother. 2017;96:48-55
116. Huang X, Gao Y, Qin J, Lu S. The role of miR-34a in the hepatoprotective effect of hydrogen sulfide on ischemia/reperfusion injury in young and old rats. PLoS One. 2014;9:e113305
117. Huang X, Gao Y, Qin J, Lu S. The mechanism of long non-coding RNA MEG3 for hepatic ischemia-reperfusion: Mediated by miR-34a/Nrf2 signaling pathway. J Cell Biochem. 2018;119:1163-72
118. Lin H, Ewing LE, Koturbash I, Gurley BJ, Miousse IR. MicroRNAs as biomarkers for liver injury: Current knowledge, challenges and future prospects. Food Chem Toxicol. 2017;110:229-39
119. Van Caster P, Brandenburger T, Strahl T, Metzger S, Bauer I, Pannen B. et al. Circulating microRNA-122, -21 and -223 as potential markers of liver injury following warm ischaemia and reperfusion in rats. Mol Med Rep. 2015;12:3146-50
120. Zhang K, Xu X, Hu L. Sevoflurane attenuates hepatic ischemia reperfusion injury by the miR-122/Nrf2 pathway. Ann Transl Med. 2022;10:350
121. Meng M, Zhang R, Han R, Kong Y, Wang R, Hou L. The polysaccharides from the fruiting body prevent lipopolysaccharide/D-galactosamine-induced acute liver injury the miR-122-Nrf2/ARE pathways. Food Funct. 2021;12:1973-82
122. Pefanis A, Ierino FL, Murphy JM, Cowan PJ. Regulated necrosis in kidney ischemia-reperfusion injury. Kidney Int. 2019;96:291-301
123. Chatterjee PK, Cuzzocrea S, Brown PA, Zacharowski K, Stewart KN, Mota-Filipe H. et al. Tempol, a membrane-permeable radical scavenger, reduces oxidant stress-mediated renal dysfunction and injury in the rat. Kidney Int. 2000;58:658-73
124. Chouchani ET, Pell VR, James AM, Work LM, Saeb-Parsy K, Frezza C. et al. A Unifying Mechanism for Mitochondrial Superoxide Production during Ischemia-Reperfusion Injury. Cell Metab. 2016;23:254-63
125. Mao H, Wang L, Xiong Y, Jiang G, Liu X. Fucoxanthin Attenuates Oxidative Damage by Activating the Sirt1/Nrf2/HO-1 Signaling Pathway to Protect the Kidney from Ischemia-Reperfusion Injury. Oxid Med Cell Longev. 2022;2022:7444430
126. Shi S, Lei S, Tang C, Wang K, Xia Z. Melatonin attenuates acute kidney ischemia/reperfusion injury in diabetic rats by activation of the SIRT1/Nrf2/HO-1 signaling pathway. Biosci Rep. 2019 39
127. Ding Y-W, Zhao G-J, Li X-L, Hong G-L, Li M-F, Qiu Q-M. et al. SIRT1 exerts protective effects against paraquat-induced injury in mouse type II alveolar epithelial cells by deacetylating NRF2 in vitro. Int J Mol Med. 2016;37:1049-58
128. Diao C, Chen Z, Qiu T, Liu H, Yang Y, Liu X. et al. Inhibition of PRMT5 Attenuates Oxidative Stress-Induced Pyroptosis via Activation of the Nrf2/HO-1 Signal Pathway in a Mouse Model of Renal Ischemia-Reperfusion Injury. Oxid Med Cell Longev. 2019;2019:2345658
129. Zhao S, Chen W, Li W, Yu W, Li S, Rao T. et al. LncRNA TUG1 attenuates ischaemia-reperfusion-induced apoptosis of renal tubular epithelial cells by sponging miR-144-3p via targeting Nrf2. J Cell Mol Med. 2021;25:9767-83
130. Vanden Berghe W. Epigenetic impact of dietary polyphenols in cancer chemoprevention: lifelong remodeling of our epigenomes. Pharmacol Res. 2012;65:565-76
131. Lee JH, Khor TO, Shu L, Su Z-Y, Fuentes F, Kong A-NT. Dietary phytochemicals and cancer prevention: Nrf2 signaling, epigenetics, and cell death mechanisms in blocking cancer initiation and progression. Pharmacol Ther. 2013;137:153-71
132. Miceli M, Bontempo P, Nebbioso A, Altucci L. Natural compounds in epigenetics: a current view. Food Chem Toxicol. 2014;73:71-83
133. Schnekenburger M, Dicato M, Diederich M. Plant-derived epigenetic modulators for cancer treatment and prevention. Biotechnol Adv. 2014;32:1123-32
134. Singh V, Ubaid S. Role of Silent Information Regulator 1 (SIRT1) in Regulating Oxidative Stress and Inflammation. Inflammation. 2020;43:1589-98
135. Yang R, Shen Y-J, Chen M, Zhao J-Y, Chen S-H, Zhang W. et al. Quercetin attenuates ischemia reperfusion injury by protecting the blood-brain barrier through Sirt1 in MCAO rats. J Asian Nat Prod Res. 2022;24:278-89
136. Wu Y, Fan L, Wang Y, Ding J, Wang R. Isorhamnetin Alleviates High Glucose-Aggravated Inflammatory Response and Apoptosis in Oxygen-Glucose Deprivation and Reoxygenation-Induced HT22 Hippocampal Neurons Through Akt/SIRT1/Nrf2/HO-1 Signaling Pathway. Inflammation. 2021;44:1993-2005
137. Zhao T-T, Yang T-L, Gong L, Wu P. Isorhamnetin protects against hypoxia/reoxygenation-induced injure by attenuating apoptosis and oxidative stress in H9c2 cardiomyocytes. Gene. 2018;666:92-9
138. Zhang B, Zhai M, Li B, Liu Z, Li K, Jiang L. et al. Honokiol Ameliorates Myocardial Ischemia/Reperfusion Injury in Type 1 Diabetic Rats by Reducing Oxidative Stress and Apoptosis through Activating the SIRT1-Nrf2 Signaling Pathway. Oxid Med Cell Longev. 2018;2018:3159801
139. Wang X, Yuan B, Cheng B, Liu Y, Zhang B, Wang X. et al. Crocin Alleviates Myocardial Ischemia/Reperfusion-Induced Endoplasmic Reticulum Stress via Regulation of miR-34a/Sirt1/Nrf2 Pathway. Shock. 2019;51:123-30
140. Tao X, Sun X, Xu L, Yin L, Han X, Qi Y. et al. Total Flavonoids from Rosa laevigata Michx Fruit Ameliorates Hepatic Ischemia/Reperfusion Injury through Inhibition of Oxidative Stress and Inflammation in Rats. Nutrients. 2016 8
141. Aggarwal BB, Bhardwaj A, Aggarwal RS, Seeram NP, Shishodia S, Takada Y. Role of resveratrol in prevention and therapy of cancer: preclinical and clinical studies. Anticancer Res. 2004;24:2783-840
142. Li J, Li L, Wang S, Zhang C, Zheng L, Jia Y. et al. Resveratrol Alleviates Inflammatory Responses and Oxidative Stress in Rat Kidney Ischemia-Reperfusion Injury and H2O2-Induced NRK-52E Cells via the Nrf2/TLR4/NF-kappaB Pathway. Cell Physiol Biochem. 2018;45:1677-89
143. Xu G, Zhao X, Fu J, Wang X. Resveratrol increase myocardial Nrf2 expression in type 2 diabetic rats and alleviate myocardial ischemia/reperfusion injury (MIRI). Ann Palliat Med. 2019;8:565-75
144. Ren J, Fan C, Chen N, Huang J, Yang Q. Resveratrol pretreatment attenuates cerebral ischemic injury by upregulating expression of transcription factor Nrf2 and HO-1 in rats. Neurochem Res. 2011;36:2352-62
145. Yang J, Huang J, Shen C, Cheng W, Yu P, Wang L. et al. Resveratrol Treatment in Different Time-Attenuated Neuronal Apoptosis After Oxygen and Glucose Deprivation/Reoxygenation via Enhancing the Activation of Nrf-2 Signaling Pathway In vitro. Cell Transplant. 2018;27:1789-97
146. Sivapalan T, Melchini A, Saha S, Needs PW, Traka MH, Tapp H. et al. Bioavailability of Glucoraphanin and Sulforaphane from High-Glucoraphanin Broccoli. Mol Nutr Food Res. 2018;62:e1700911
147. Chen L, Zhang WL, Xie DQ, Jia W. Sulforaphane alleviates hepatic ischemia-reperfusion injury through promoting the activation of Nrf-2/HO-1 signaling. Transpl Immunol. 2021;68:101439
148. Silva-Palacios A, Ostolga-Chavarria M, Sanchez-Garibay C, Rojas-Morales P, Galvan-Arzate S, Buelna-Chontal M. et al. Sulforaphane protects from myocardial ischemia-reperfusion damage through the balanced activation of Nrf2/AhR. Free Radic Biol Med. 2019;143:331-40
149. Yoon HY, Kang NI, Lee HK, Jang KY, Park JW, Park BH. Sulforaphane protects kidneys against ischemia-reperfusion injury through induction of the Nrf2-dependent phase 2 enzyme. Biochem Pharmacol. 2008;75:2214-23
150. Chi X, Zhang R, Shen N, Jin Y, Alina A, Yang S. et al. Sulforaphane reduces apoptosis and oncosis along with protecting liver injury-induced ischemic reperfusion by activating the Nrf2/ARE pathway. Hepatol Int. 2015;9:321-9
151. Zhao F, Zhang J, Chang N. Epigenetic modification of Nrf2 by sulforaphane increases the antioxidative and anti-inflammatory capacity in a cellular model of Alzheimer's disease. Eur J Pharmacol. 2018 824
152. Su Z-Y, Zhang C, Lee JH, Shu L, Wu T-Y, Khor TO. et al. Requirement and epigenetics reprogramming of Nrf2 in suppression of tumor promoter TPA-induced mouse skin cell transformation by sulforaphane. Cancer Prev Res (Phila). 2014;7:319-29
153. Zhang C, Su Z-Y, Khor TO, Shu L, Kong A-NT. Sulforaphane enhances Nrf2 expression in prostate cancer TRAMP C1 cells through epigenetic regulation. Biochem Pharmacol. 2013;85:1398-404
154. Zhou J-W, Wang M, Sun N-X, Qing Y, Yin T-F, Li C. et al. Sulforaphane-induced epigenetic regulation of Nrf2 expression by DNA methyltransferase in human Caco-2 cells. Oncol Lett. 2019;18:2639-47
155. Su X, Wang S, Zhang H, Yang G, Bai Y, Liu P. et al. Sulforaphane prevents angiotensin II-induced cardiomyopathy by activation of Nrf2 through epigenetic modification. J Cell Mol Med. 2021;25:4408-19
156. Ikeda Y, Murakami A, Ohigashi H. Ursolic acid: an anti- and pro-inflammatory triterpenoid. Mol Nutr Food Res. 2008;52:26-42
157. Zhang C, Wang C, Li W, Wu R, Guo Y, Cheng D. et al. Pharmacokinetics and Pharmacodynamics of the Triterpenoid Ursolic Acid in Regulating the Antioxidant, Anti-inflammatory, and Epigenetic Gene Responses in Rat Leukocytes. Mol Pharm. 2017;14:3709-17
158. Kim H, Ramirez CN, Su Z-Y, Kong A-NT. Epigenetic modifications of triterpenoid ursolic acid in activating Nrf2 and blocking cellular transformation of mouse epidermal cells. J Nutr Biochem. 2016;33:54-62
159. Li L, Zhang X, Cui L, Wang L, Liu H, Ji H. et al. Ursolic acid promotes the neuroprotection by activating Nrf2 pathway after cerebral ischemia in mice. Brain Res. 2013;1497:32-9
160. Wang C, Shu L, Zhang C, Li W, Wu R, Guo Y. et al. Histone Methyltransferase Setd7 Regulates Nrf2 Signaling Pathway by Phenethyl Isothiocyanate and Ursolic Acid in Human Prostate Cancer Cells. Mol Nutr Food Res. 2018;62:e1700840
161. Manzoor MF, Ahmad N, Ahmed Z, Siddique R, Zeng X-A, Rahaman A. et al. Novel extraction techniques and pharmaceutical activities of luteolin and its derivatives. J Food Biochem. 2019;43:e12974
162. Juszczak AM, Wöelfle U, Končić MZ, Tomczyk M. Skin cancer, including related pathways and therapy and the role of luteolin derivatives as potential therapeutics. Med Res Rev. 2022;42:1423-62
163. Ali F, Siddique YH. Bioavailability and Pharmaco-therapeutic Potential of Luteolin in Overcoming Alzheimer's Disease. CNS Neurol Disord Drug Targets. 2019;18:352-65
164. Kalbolandi SM, Gorji AV, Babaahmadi-Rezaei H, Mansouri E. Luteolin confers renoprotection against ischemia-reperfusion injury via involving Nrf2 pathway and regulating miR320. Mol Biol Rep. 2019;46:4039-47
165. Zhu XA, Gao LF, Zhang ZG, Xiang DK. Down-regulation of miR-320 exerts protective effects on myocardial I-R injury via facilitating Nrf2 expression. Eur Rev Med Pharmacol Sci. 2019;23:1730-41
166. Zuo Q, Wu R, Xiao X, Yang C, Yang Y, Wang C. et al. The dietary flavone luteolin epigenetically activates the Nrf2 pathway and blocks cell transformation in human colorectal cancer HCT116 cells. J Cell Biochem. 2018;119:9573-82
167. Kang KA, Piao MJ, Hyun YJ, Zhen AX, Cho SJ, Ahn MJ. et al. Luteolin promotes apoptotic cell death via upregulation of Nrf2 expression by DNA demethylase and the interaction of Nrf2 with p53 in human colon cancer cells. Exp Mol Med. 2019 51
168. Aggarwal BB, Sung B. Pharmacological basis for the role of curcumin in chronic diseases: an age-old spice with modern targets. Trends Pharmacol Sci. 2009;30:85-94
169. Li W, Suwanwela NC, Patumraj S. Curcumin by down-regulating NF-kB and elevating Nrf2, reduces brain edema and neurological dysfunction after cerebral I/R. Microvasc Res. 2016;106:117-27
170. Ibrahim SG, El-Emam SZ, Mohamed EA, Abd Ellah MF. Dimethyl fumarate and curcumin attenuate hepatic ischemia/reperfusion injury via Nrf2/HO-1 activation and anti-inflammatory properties. Int Immunopharmacol. 2020;80:106131
171. Wu H, Kong L, Tan Y, Epstein PN, Zeng J, Gu J. et al. C66 ameliorates diabetic nephropathy in mice by both upregulating NRF2 function via increase in miR-200a and inhibiting miR-21. Diabetologia. 2016;59:1558-68
172. Guo Y, Wu R, Gaspar JM, Sargsyan D, Su Z-Y, Zhang C. et al. DNA methylome and transcriptome alterations and cancer prevention by curcumin in colitis-accelerated colon cancer in mice. Carcinogenesis. 2018;39:669-80
173. Shu L, Khor TO, Lee J-H, Boyanapalli SSS, Huang Y, Wu T-Y. et al. Epigenetic CpG demethylation of the promoter and reactivation of the expression of Neurog1 by curcumin in prostate LNCaP cells. AAPS J. 2011;13:606-14
174. Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567-90
175. Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11:577-91
176. Candido EP, Reeves R, Davie JR. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell. 1978;14:105-13
177. Liu H, Zhang J-J, Li X, Yang Y, Xie X-F, Hu K. Post-occlusion administration of sodium butyrate attenuates cognitive impairment in a rat model of chronic cerebral hypoperfusion. Pharmacol Biochem Behav. 2015;135:53-9
178. Dong W, Jia Y, Liu X, Zhang H, Li T, Huang W. et al. Sodium butyrate activates NRF2 to ameliorate diabetic nephropathy possibly via inhibition of HDAC. J Endocrinol. 2017;232:71-83
179. Wu J, Jiang Z, Zhang H, Liang W, Huang W, Zhang H. et al. Sodium butyrate attenuates diabetes-induced aortic endothelial dysfunction via P300-mediated transcriptional activation of Nrf2. Free Radic Biol Med. 2018;124:454-65
180. Yaku K, Enami Y, Kurajyo C, Matsui-Yuasa I, Konishi Y, Kojima-Yuasa A. The enhancement of phase 2 enzyme activities by sodium butyrate in normal intestinal epithelial cells is associated with Nrf2 and p53. Mol Cell Biochem. 2012 370
181. Sun B, Jia Y, Yang S, Zhao N, Hu Y, Hong J. et al. Sodium butyrate protects against high-fat diet-induced oxidative stress in rat liver by promoting expression of nuclear factor E2-related factor 2. Br J Nutr. 2019;122:400-10
182. Sivinski J, Zhang DD, Chapman E. Targeting NRF2 to treat cancer. Seminars In Cancer Biology. 2021;76:61-73
183. Schmidlin CJ, Shakya A, Dodson M, Chapman E, Zhang DD. The intricacies of NRF2 regulation in cancer. Seminars In Cancer Biology. 2021;76:110-9
184. Tucci V, Isles AR, Kelsey G, Ferguson-Smith AC, Erice Imprinting G. Genomic Imprinting and Physiological Processes in Mammals. Cell. 2019;176:952-65
185. Falls JG, Pulford DJ, Wylie AA, Jirtle RL. Genomic imprinting: implications for human disease. Am J Pathol. 1999;154:635-47
186. Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA. et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934-47
187. Thandapani P. Super-enhancers in cancer. Pharmacol Ther. 2019;199:129-38
188. Wilflingseder J, Willi M, Lee HK, Olauson H, Jankowski J, Ichimura T. et al. Enhancer and super-enhancer dynamics in repair after ischemic acute kidney injury. Nat Commun. 2020;11:3383
Author contact
![]() Corresponding authors: Hongxia Li, Department of Cardiology, The First Affiliated Hospital of Soochow University, 188 Shizi Street, Suzhou, Jiangsu 215006, P. R. China. E-mail: shrimpedu.cn; Tingbo Jiang, Department of Cardiology, The First Affiliated Hospital of Soochow University, 188 Shizi Street, Suzhou, Jiangsu 215006, P. R. China. E-mail: 18906201122cn; Jiaxiang Bai, Department of Orthopedics, The First Affiliated Hospital of Soochow University, 188 Shizi Street, Suzhou, Jiangsu 215006, P. R. China. E-mail: jxbai1995com.
Corresponding authors: Hongxia Li, Department of Cardiology, The First Affiliated Hospital of Soochow University, 188 Shizi Street, Suzhou, Jiangsu 215006, P. R. China. E-mail: shrimpedu.cn; Tingbo Jiang, Department of Cardiology, The First Affiliated Hospital of Soochow University, 188 Shizi Street, Suzhou, Jiangsu 215006, P. R. China. E-mail: 18906201122cn; Jiaxiang Bai, Department of Orthopedics, The First Affiliated Hospital of Soochow University, 188 Shizi Street, Suzhou, Jiangsu 215006, P. R. China. E-mail: jxbai1995com.
 Global reach, higher impact
Global reach, higher impact