13.3
Impact Factor
Theranostics 2022; 12(7):3150-3177. doi:10.7150/thno.67409 This issue Cite
Research Paper
Glycoproteogenomics characterizes the CD44 splicing code associated with bladder cancer invasion
1. Experimental Pathology and Therapeutics Group, IPO Porto Research Center (CI-IPOP), Portuguese Oncology Institute (IPO Porto), 4200-072 Porto, Portugal.
2. RISE@CI-IPOP (Health Research Network), Portuguese Oncology Institute of Porto (IPO Porto), 4200-072 Porto, Portugal.
3. Porto Comprehensive Cancer Center (P.ccc), 4200-072 Porto, Portugal.
4. Institute of Biomedical Sciences Abel Salazar (ICBAS), University of Porto, 4050-313 Porto, Portugal.
5. Center for Applied Medical Research (Centro de Investigación Médica Aplicada, CIMA), University of Navarra, 31008 Pamplona, Navarra, Spain.
6. REQUIMTE-LAQV, Department of Chemistry, University of Aveiro, 3810-193 Aveiro, Portugal.
7. Institute for Research and Innovation in Health (i3S), University of Porto, 4200-135 Porto, Portugal.
8. Institute for Biomedical Engineering (INEB), University of Porto, 4200-135 Porto, Portugal.
9. REQUIMTE-LAQV, Department of Chemistry and Biochemistry, Faculty of Sciences of the University of Porto, 4169-007 Porto, Portugal.
10. Cancer Genetics Group, IPO Porto Research Center (CI-IPOP), Portuguese Oncology Institute (IPO Porto), 4200-072 Porto, Portugal.
11. FP-I3ID, University Fernando Pessoa, 4249-004 Porto, Portugal.
12. Laboratoire d'Etude du Métabolisme des Médicaments (LEMM), CEA, INRA, Université Paris Saclay, F-91191, Gif-sur-Yvette cedex, France.
13. Immunology Department, Portuguese Oncology Institute (IPO Porto), 4200-072 Porto, Portugal.
14. Faculty of Medicine of the University of Porto, 4200-319 Porto, Portugal.
15. Ipatimup—Institute of Molecular Pathology and Immunology of the University of Porto, University of Porto, 4200-135 Porto, Portugal.
16. GlycoMatters Biotech, 4500-162 Espinho, Portugal.
17. Department of Surgical Oncology, Portuguese Oncology Institute (IPO Porto), 4200-072 Porto, Portugal.
Abstract
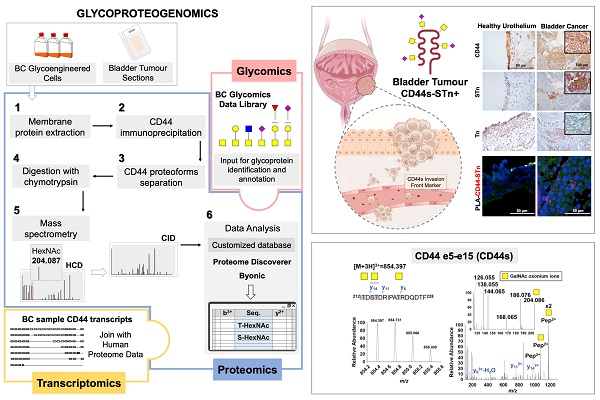
Rationale: Bladder cancer (BC) management demands the introduction of novel molecular targets for precision medicine. Cell surface glycoprotein CD44 has been widely studied as a potential biomarker of BC aggressiveness and cancer stem cells. However, significant alternative splicing and multiple glycosylation generate a myriad of glycoproteoforms with potentially distinct functional roles. The lack of tools for precise molecular characterization has led to conflicting results, delaying clinical applications. Addressing these limitations, we have interrogated the transcriptome and glycoproteome of a large BC patient cohort for splicing signatures.
Methods: CD44 gene and its splicing variants were assessed by Real Time-Polymerase Chain Reaction (RT-PCR) and RNAseq in tumor tissues. The co-localization of CD44 and short O-glycans was evaluated by proximity ligation assay (PLA), immunohistochemistry and double-immunofluorescence. An innovative glycoproteogenomics approach, integrating transcriptomics-customized datasets and glycomics for protein annotation from nanoLC-ESI-MS/MS experiments, was developed and implemented to identify CD44 variants and associated glycosignatures. The impact of CD44 silencing on proliferation and invasion of BC cell lines and glycoengineered cells was determined by BrdU ELISA and Matrigel invasion assays, respectively. Antibody phosphoarrays were used to investigate the role of CD44 and its glycoforms in the activation of relevant oncogenic signaling pathways.
Results: Transcriptomics analysis revealed remarkable CD44 isoforms heterogeneity in bladder cancer tissues, as well as associations between short CD44 standard splicing isoform (CD44s), invasion and poor prognosis. We further demonstrated that targeting short O-glycoforms such as the Tn and sialyl-Tn antigens was key to overcome the lack of cancer specificity presented by CD44. Glycoproteogenomics allowed, for the first time, the comprehensive characterization of CD44 splicing code at the protein level. The concept was applied to invasive human BC cell lines, glycoengineered cells, and tumor tissues, enabling unequivocal CD44s identification as well as associated glycoforms. Finally, we confirmed the link between CD44 and invasion in CD44s-enriched cells in vitro by small interfering RNA (siRNA) knockdown, supporting findings from BC tissues. The key role played by short-chain O-glycans in CD44-mediated invasion was also demonstrated through glycoengineered cell models.
Conclusions: Overall, CD44s emerged as biomarker of poor prognosis and CD44-Tn/ Sialyl-Tn (STn) as promising molecular signatures for targeted interventions. This study materializes the concept of glycoproteogenomics and provides a key vision to address the cancer splicing code at the protein level, which may now be expanded to better understand CD44 functional role in health and disease.
Keywords: glycomics, proteogenomics, glycoproteogenomics, bladder cancer, CD44
 Global reach, higher impact
Global reach, higher impact