13.3
Impact Factor
Theranostics 2021; 11(15):7175-7187. doi:10.7150/thno.59056 This issue Cite
Research Paper
CXCL10 potentiates immune checkpoint blockade therapy in homologous recombination-deficient tumors
1. Obstetrics and Gynecology Hospital, NHC Key Laboratory of Reproduction Regualtion, Shanghai Institute of Planned Parenthood Research, State Key Laboratory of Genetic Engineering at School of Life Sciences, Institute of Reproduction and Development, Fudan University, Shanghai 200032, China.
2. Shanghai Key Laboratory of Female Reproductive Endocrine Related Diseases, Institute of Metabolism and Integrative Biology, Institutes of Biomedical Sciences, Fudan University, Shanghai 200433, China.
3. Department of Ophthalmology, Ninth People's Hospital, Shanghai JiaoTong University School of Medicine, Shanghai, 200025, China.
4. Shanghai Key Laboratory of Orbital Diseases and Ocular Oncology, Shanghai, 200025, China.
5. Children's Hospital of Fudan University, Shanghai 201100, China.
Received 2021-2-4; Accepted 2021-5-12; Published 2021-5-24
Abstract
Background: Homologous recombination deficiency (HRD) is a common molecular characteristic of genomic instability, and has been proven to be a biomarker for target therapy. However, until now, no research has explored the changes in the transcriptomics landscape of HRD tumors.
Methods: The HRD score was established from SNP array data of breast cancer patients from the cancer genome atlas (TCGA) database. The transcriptome data of patients with different HRD scores were analyzed to identify biomarkers associated with HRD. The candidate biomarkers were validated in the gene expression omnibus (GEO) database and immunotherapy cohorts.
Results: Based on data from the gene expression profile and clinical characteristics from 1310 breast cancer patients, including TCGA database and GEO database, we found that downstream targets of the cGAS-STING pathway, such as CXCL10, were upregulated in HRD tumors and could be used as a predictor of survival outcome in triple-negative breast cancer (TNBC) patients. Further comprehensive analysis of the tumor immune microenvironment (TIME) revealed that the expression of CXCL10 was positively correlated with neoantigen load and infiltrating immune cells. Finally, in vivo experimental data and clinical trial data confirmed that the expression of CXCL10 could be used as a biomarker for anti-PD-1/PD-L1 therapy.
Conclusions: Together, our study not only revealed that CXCL10 is associated with HRD but also introduced a potential new perspective for identifying prognostic biomarkers of immunotherapy.
Keywords: HRD, cGAS-STING, TIME, CXCL10, immunotherapy
Introduction
Immune checkpoint blockade (ICB) therapy produces an average objective response rate of up to 50% in various types of solid tumors with DNA deficient mismatch repair [1,2]. This is an important finding, indicating that ICB inhibitors can be used as “broad-spectrum drugs” for tumor treatment. Generally, dMMR tumors are characterized by an increased number of mutations and enhanced T-cell infiltration; however, a considerable number of dMMR tumors have a high mutation burden but lack sufficient T-cell infiltration and respond poorly to ICB therapy [3]. Usually, the response to ICB therapy depends on the following biomarkers: 1) the number of neoantigens derived from genome instability, such as the tumor mutation burden (TMB) and dMMR, and 2) tumor-infiltrating lymphocytes (TILs), mainly CD4+ T cells, CD8+ T cells and gene signals that interfere with the activity of T cells, such as those affecting PD-1/PD-L1/CTLA4 function [4,5]. The effective activation of T cells not only depends on neoantigens but also requires costimulatory molecular signals recognized by the innate immune system. As an important part of innate immune recognition, cytoplasmic DNA recognition mediated by the cGAS-STING pathway was reported in 2013 as a breakthrough finding [6].
In the recent issue of Cancer Cell, Lu and Guan et al. demonstrated that the activation of the cGAS-STING pathway in tumor tissues was significantly and positively correlated with the prognosis of patients bearing dMMR tumors but not that of patients with pMMR (proficient MMR) tumors. The results of their in vitro experiments showed that antigen-presenting cells nurtured by dMMR but not pMMR tumors could strongly promote T-cell proliferation, whereas this phenomenon was abolished when STING was ablated in dMMR tumor cells [7].
In addition to dMMR, homologous recombination deficiency (HRD) also induces genomic instability and serves as an effective therapeutic biomarker for breast cancer and ovarian cancer [8-11].
To explore whether there is a possible connection between the ICB therapeutic biomarkers in genomically unstable tumors elicited by HRD, we extracted the HRD score and transcriptome data from the Cancer Genome Atlas breast cancer cohort (TCGA-BRCA). By analyzing the transcriptome data of patients with HRD tumors, we found that downstream targets of the cGAS-STING pathway, such as CXCL10, were positively associated with HRD. Furthermore, we discovered a positive relationship between the CXCL10 expression and tumor immune microenvironment (TIME), including infiltrating immune cells, neoantigen load and immune checkpoint blockade (ICB). Moreover, high CXCL10 expression was able to be used as biomarkers for anti-PD-1/PD-L1 therapy, and the predictive effect of CXCL10 was better than that of PD-1/PD-L1. Our research perspectives and methods provide a possible direction for immunotherapy. The results of this study may be valuable for understanding the relationship between the genomic instability and TIME and improving the clinical outcome of patients receiving anti-PD-1/PD-L1 therapy.
Materials and Methods
Data collection and processing
Patients' RNA sequencing data, SNP array data and corresponding clinical follow-up information were downloaded from the publicly available the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov) and the NCBI Gene Expression Omnibus (GEO) database [12]. RNA sequencing data were normalized as transcripts per million (TPM) by using the R. SNP array data were processed using Affymetrix Power Tools and PennCNV. The somatic mutation counts, copy number variation (CNV), fraction genome altered scores (FGA: percentage of copy number altered chromosome regions out of measured regions) and MSIsensor score (microsatellite instability detection using paired tumor-normal sequence data) were obtained from the cBioPortal database (http://www.cbioportal.org/study?id = brca_tcga_pan_can_atlas_2018). In total, 1055 TCGA-BRCA samples data were extracted; 255 GEO samples data were extracted (E-MTAB-365, GSE19615, GSE21653, GSE2603 and GSE31519) (http://kmplot.com/analysis/index.php?p = service&cancer = breast ). Please refer to Table S1 for the clinical information of patients included in this study. Single-cell RNA-seq of Triple-negative breast cancer (TNBC) patients were obtained from GSE118389 [13]. The transcriptome profile and clinical information from immunotherapy cohorts were obtained from Imvigor210 [14,15]. The RNA-seq data of immune checkpoint treated tumors from TNBC murine models were downloaded from GSE124821 [16].
HRD score analysis
Loss of heterozygosity (LOH) was defined as the number of counts of chromosomal LOH regions shorter than whole chromosome and longer than 15 Mb [17]. Large-scale State Transitions (LST) were defined as chromosome breakpoint (change in copy number or allelic content ) between adjacent regions each of at least 10 megabases obtained after smoothing and filtering shorter than 3 Mb small-scale copy number variation [18]. Telomeric Allelic Imbalance (TAI) was defined as the number of regions with allelic imbalance which extend to the sub-telomere but do not cross the centromere [19]. The HRD score was defined as the sum of TAI, LST, and LOH scores [20-22]. The HRD score of each patient was shown in Table S2.
Neoantigen load
The 4-digit HLA type for each sample was inferred using POLYSOLVER [23]. Neo-epitopes were predicted for each patient by defining all novel amino acid 9mers and 10mers resulting from mutation in expressed genes (median >10 TPM in the tumor type) and determining whether the predicted binding affinity to the patient's germline HLA alleles was < 500 nM using NetMHCpan [24-26]. The Neoantigen load of each patient was shown in Table S3.
KEGG and Gene Ontology enrichment analysis
RNA-seq data (raw counts) analysis was conducted using the “edgeR” package of R [27] . Fold change > 1.5, adj. p < 0.05, TPM > 1 and genes with the first 75% of median absolute deviation (MAD) were set as the cutoffs to screen for differentially expressed genes (DEGs). Functional enrichment analysis of DEGs was performed by DAVID [28] to identify GO categories by their biological processes (BP), molecular functions (MF), or KEGG pathways.
Identification of prognostic DEGs positively associated with the HRD score
Kaplan-Meier plots were generated to illustrate the relationship between patients' overall survival and gene expression levels of DEGs. The relationship was tested by log-rank test.
Immune cells infiltration in bulk tumor gene expression data
In order to study the enrichment of immune cells, we used TIMER [29], an efficient algorithm for predicting immune cell infiltration of bulk tumor gene expression data (https://cistrome.shinyapps.io/timer/). For each sample, TIMER quantified the relative abundance of six types of infiltrating immune cells, including T cells, B cells, macrophages, neutrophiles and dendritic cells.
Results
The HRD Score reflects patients' genomic instability and can be used as a prognostic marker in patients with TNBC
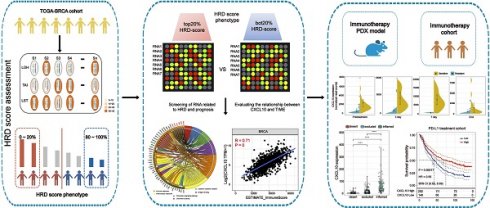
According to the HRD-algorithm, LOH, TAI and LST were used as the basis for calculating the HRD score (Table S2). To explore the correlation between the HRD score and other hallmarks of genomic instability, including somatic mutation counts, fraction genome altered and microsatellite instability (MSI), the breast cancer patients were sorted in ascending order of HRD scores; and the bottom 20% and the top 20% of the patients were selected. As shown in Figure 2, the hallmarks of genomic instability were significantly higher in the top 20% HRD-score group than the bottom 20% HRD-score group (Wilcoxon signed-rank test, P < 0.0001, Figure 2A-C). However, the HRD score was not a good prognostic marker in the whole breast cancer cohort (Figure S1A). This result might be due to the endocrine therapy of breast cancer. Therefore, we analyzed the prognosis of triple-negative breast cancer (TNBC) patients by the HRD score. In the TNBC cohort, the Kaplan-Meier survival curve (Figure 2D) showed that overall survival of patients in the HRD-positive group (HRD scores > 26) was much longer than the cases in the HRD-negative group (HRD scores were ≤ 26) (log-rank test, P < 0.0001). Receiver operating characteristic (ROC) analysis showed that the HRD score had a good predictive effect on the prognosis of TNBC (Figure 2E).
Downstream targets of the cGAS-STING pathway are associated with HRD
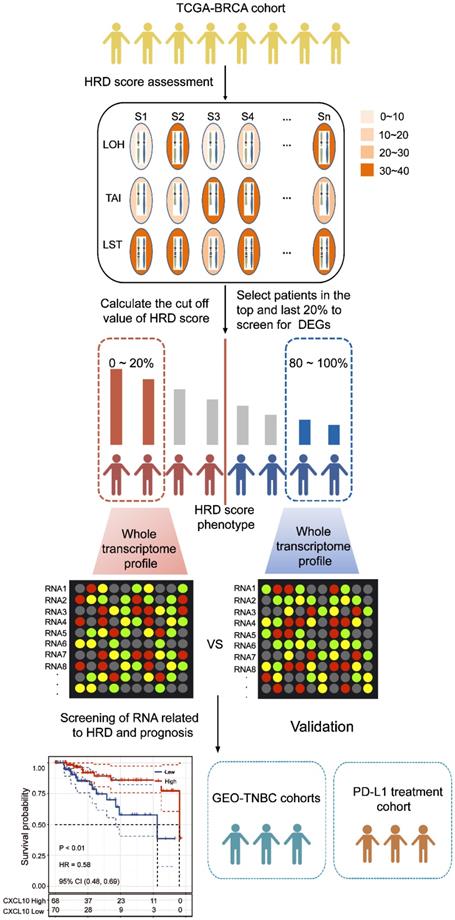
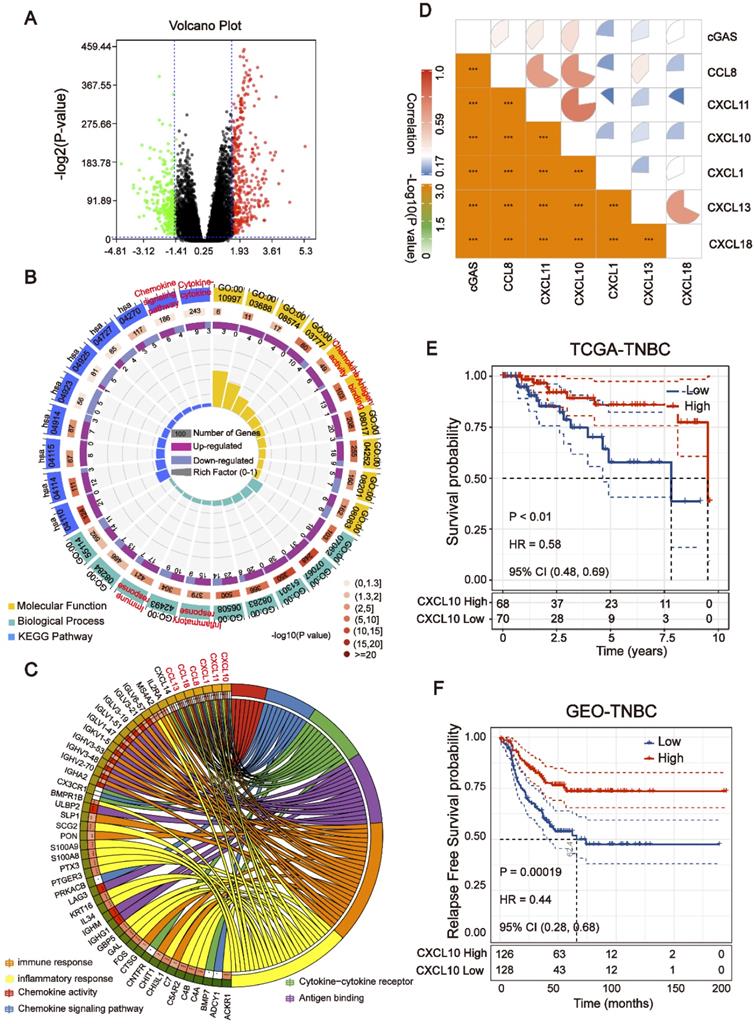
To explore the transcriptomic signatures associated with HRD, the breast cancer patients were sorted in ascending order of HRD scores; and the bottom 20% and the top 20% of the patients were selected. We compared the difference of whole transcriptome between the top 20% HRD-score group and the bottom 20% HRD-score group (Figure 1). Utilizing the egdeR method, a total of 632 differentially expressed genes (DEGs) were screened out in breast cancer (Figure 3A). In breast cancer, the KEGG and GO cluster plots revealed that the up-regulated DEGs were enriched in immune-related signaling pathways such as immune response, chemokine signaling pathway and cytokine-cytokine receptor interaction in the top 20% HRD-score group (Figure 3B). Intriguingly, six upregulated genes (CXCL1, CXCL10, CXCL11, CCL8, CCL13 and CCL18) among the immune-related signaling pathways appear repeatedly (Figure 3C). Notably, the six upregulated genes were closely correlated with the transcriptional data, indicating cGAS-STING signaling activation in the TCGA-BRCA cohort, as CXCL10/11 had been reported to be downstream targets of the cGAS-STING pathway (Figure 3D, Pearson correlation coefficient, R > 0.4, P ≈ 0). The results from the Kaplan-Meier analysis showed that TCGA-TNBC patients with high expression of these six genes had prolonged survival, and CXCL10 expression had the strongest predictive power for survival (Figure 3E and Figure S1B-F). The prognostic value of CXCL10 was further verified in the GEO-TNBC cohort (Figure 3F). To confirm that up-regulated CXCL10 expression is derived from HRD tumor cells, we re-analyzed single-cell RNA-seq data of TNBC patients [13]. The box plot showed that the expression of CXCL10 in the epithelial cells of the patient with BRCA1 loss-of-function mutations was significantly higher than the expression of CXCL10 in the epithelial cells of the HR proficient patient (Wilcoxon signed-rank test, **** P < 0.0001, Figure S2C).
The CXCL10 expression signature is positively associated with the tumor immune microenvironment
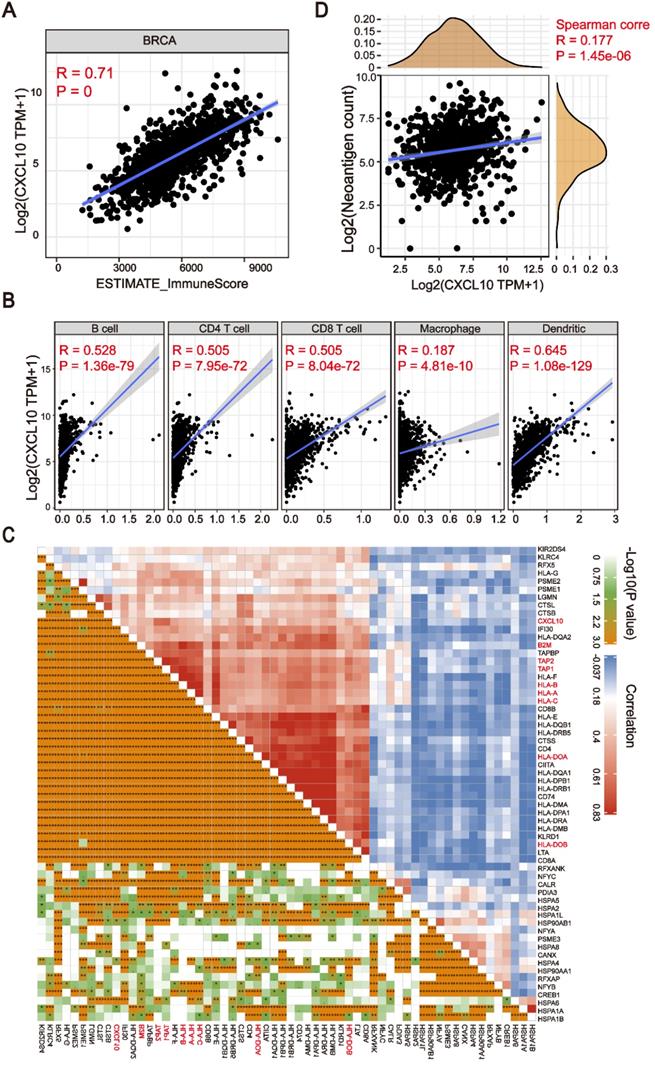
The expression of Cytokines/Chemokines is essential for attracting immune cells [30,31]. In order to explore the relationship between the chemokine signatures and tumor infiltrating immune cells, we first used the ESTIMATE algorithm to calculate the correlation between downstream targets of the cGAS-STING pathway and immune scores [32]. As shown in Figure 4A, the expression of CXCL10 was positively correlated with immune scores in breast cancer patients (Spearman's rank correlation coefficient, R > 0.6, P ≈ 0). More importantly, we found that this correlation existed not only in the TCGA-BRCA cohort, but also in other cancer types (Figure S3). To further clarify the subtype of tumor infiltrating cells, the TIMER algorithm [33] was applied to estimate the association of various immune cell types with the CXCL10 expression signature. As depicted in the scatter plot, the CXCL10 upregulation correlated to the infiltration of dendritic cells and anti-tumor lymphocyte subpopulations (Spearman's rank correlation coefficient, P < 0.0001) (Figure 4B). Dendritic cells play a crucial role in antigen processing and presentation pathways, hinting that the expression of antigen-related genes might be associated with the expression of CXCL10. To prove this assumption, we explored the correlation of antigen-related genes with the CXCL10 expression signature by using the Pearson correlation coefficient. We found that the expression of MHC class I/II (I: HLA-A, HLA-B, and HLA-C; II: HLA-DP, HLA-DM, HLA-DOA, HLA-DOB, HLA-DQ, and HLA-DR) and the key antigen binding (B2M, TAP1/2 and so on) molecules were positively correlated with the CXCL10 expression signature (Spearman's rank correlation coefficient, P < 0.0001) (Figure 4C). We further identified that the predicted neoantigen load was positively correlated with the CXCL10 expression signature (Spearman's rank correlation coefficient, P < 0.0001) (Figure 4D).
Computational overview of HRD-related RNAs detection. The columns reflected TCGA-BRCA samples, and the rows reflected three biomarkers of the HRD score. The color reflects the scores for each biomarker on each sample. HRD-related RNAs were detected by comparing the RNA expression profile between the top 20% patients with high HRD scores and the bottom 20% patients with low HRD scores.
The CXCL10 expression signature is positively associated with ICB-related genes
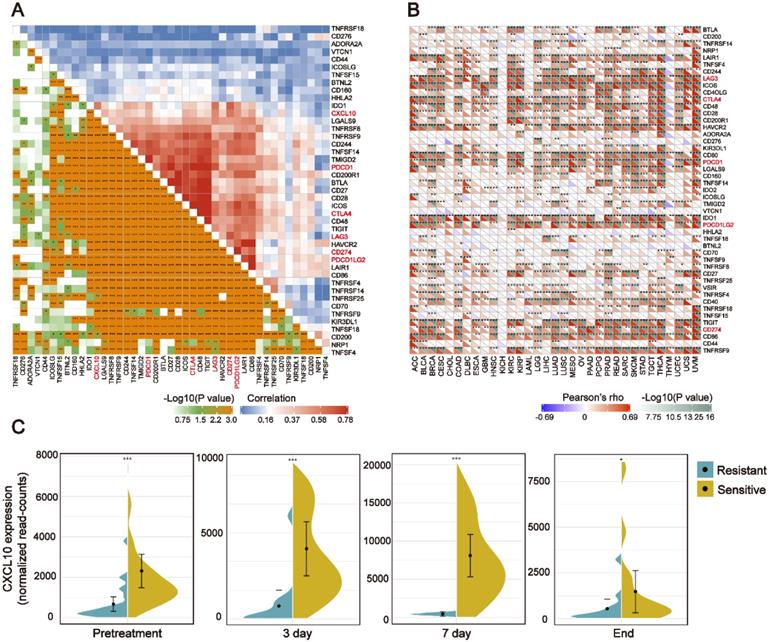
In recent years, ICB therapy, represented by anti-PD-1/L1, has played an increasingly important role in anti-tumor treatment [34]. The characteristics of TIME and ICB-related genes have a profound impact on ICB therapy. Therefore, we collected more than 40 common ICB-related genes and analyzed the relationship between the CXCL10 expression signature and ICB-related genes [35]. As displayed by heatmap, the CXCL10 expression was positively correlated with the expression of multiple ICB-related genes in the TCGA-BRCA cohort (Figure 5A). Ten of the most relevant ICB-related genes were: LAG3, ICOS, CTLA4, CD48, HAVCR2, PDCD1(PD-1), PDCDILG2(PD-L2), TIGIT, CD274(PD-L1) and CD86. Generally, the key regulatory factors involved in immunity perform similar functions in different tissues. We thus explored the CXCL10 expression signature and ICB-related genes across the cancer types. We found that the positive relationship between CXCL10 and the ICB-related genes was not only present in breast cancer, but also in 32 other cancer types (Figure 5B).
The CXCL10 expression signature could be used as a potential biomarker for ICB therapy
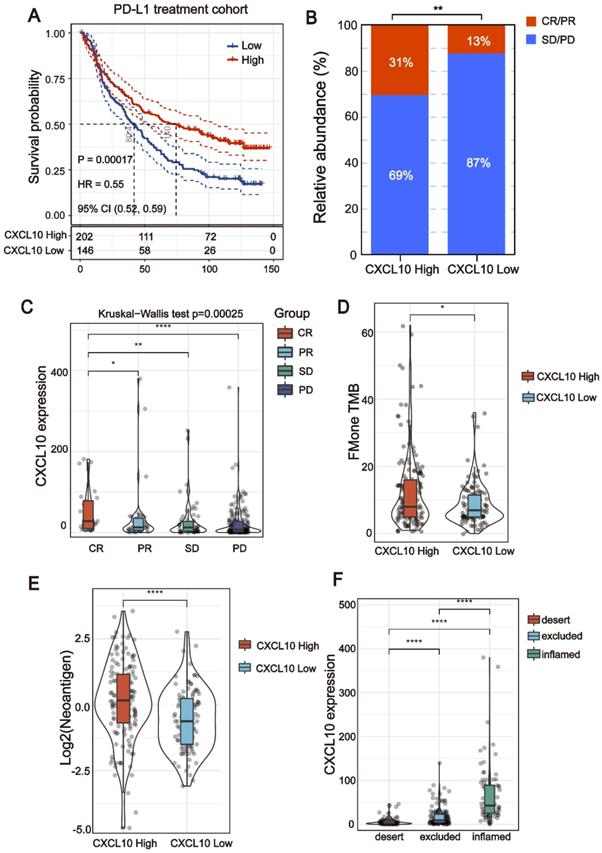
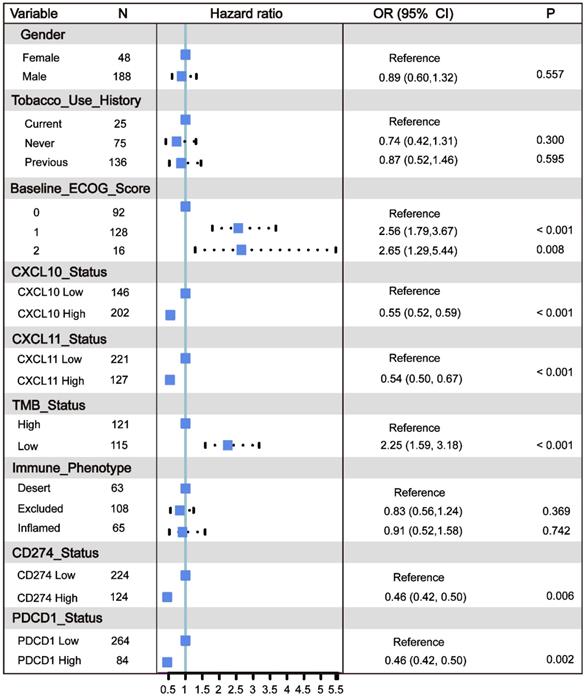
All of the above results indicate that the CXCL10 expression signature is closely related to the biomarkers for ICB therapy. Therefore, the RNA-seq data of immune checkpoint treated tumors from TNBC murine models [16] were used to investigate the role of CXCL10 in ICB Therapy. As shown in Figure 5C, the expression of CXCL10 in sensitive tumor tissues is significantly up-regulated at different time points of treatment compared with resistant tumor tissues (Wilcoxon signed-rank test, *** P < 0.001). To further confirm the predictive effect of CXCL10 on ICB treatment, we collected the transcriptome profile and clinical information from an immunotherapy cohort (Imvigor210) of urothelial cancer (UC) treated with atezolizumab [14]. In this cohort, tumor patients with high CXCL10 expression exhibited markedly improved clinical benefits and significantly prolonged survival (Figure 6A). Significant therapeutic advantages and immune responses to PD-L1 blockades were observed in samples with high expression of CXCL10 compared to those with low expression (Fisher extract test, P < 0.01, Figure 6B; Kruskal-Wallis H test, P < 0.001, Figure 6C). Further analysis revealed that TMB, neoantigen load and tumor infiltrating immune phenotype were significantly elevated in tumors with high expression of CXCL10, which was closely linked to immunotherapeutic efficacy (Figures 6D-F). Besides, the association between the expression of CXCL10 and immunotherapy survival remained statistically significant after taking into account gender, smoking, ECOG score, immunophenotype and, PD-1/PD-L1 status (Figure 7).
The HRD Score reflects patients' genomic instability and can be used as a prognostic marker in patients with TNBC. (A) Violin plot of somatic mutations in the top 20% HRD-score group and the bottom 20% HRD-score group. Somatic mutation counts in the top 20% HRD-score group were significantly higher than those in the bottom 20% HRD-score group (Wilcoxon signed-rank test, **** P < 0.0001). (B) Violin plot of MSI in the top 20% HRD-score group and the bottom 20% HRD-score group (Wilcoxon signed-rank test, **** P < 0.0001). (C) Violin plot of fraction genome altered in the top 20% HRD-score group and the bottom 20% HRD-score group (Wilcoxon signed-rank test, **** P < 0.0001). (D) Kaplan-Meier estimates of overall survival of patients with the HRD-positive or HRD-negative tumors calculated by the HRD score in the TCGA-TNBC cohort. (E) ROC curves analysis of the HRD score in the TCGA-TNBC cohort.
Downstream targets of the cGAS-STING pathway are associated with HRD. (A) Volcano plot of DEGs in the top 20% HRD-score group and the bottom 20% HRD-score group. The horizontal line at false discovery rate (FDR) < 0.05; vertical line at |log2FC| = 1.5; TPM > 1; MAD > 75%. (B) KEGG and Gene Ontology enrichment analysis of DEGs. The outermost ring represented the name of signaling pathways. The second outer ring represented the number of genes in signaling pathways, the heights of the columns in the inner ring indicate the proportion of DEGs in the total number of genes in the signaling pathway and the color depth represented the number of differential genes. (C) Chord plot depicting the relationship between genes and immune-related signaling pathways. The genes marked in red fonts refer to the most frequently repeated genes in immune-related signaling pathways. (D) Correlation between the expression of cGAS and the most frequently repeated DEGs in TCGA-BRCA cohort. The size and color of the pie chart represent the correlation coefficient. (E and F) Curve for overall survival is shown for high and low CXCL10 expression in the TCGA-TNBC cohort and GEO-TNBC cohort.
The CXCL10 expression signature is associated with TIME. (A) Positive correlation between the CXCL10 expression signature and ImmuneScore in the TCGA-BRCA cohort (Spearman's rank correlation coefficient, R = 0.71, P ≈ 0). (B) Positive correlation between the CXCL10 expression signature and immune cell subpopulations in the TCGA-BRCA cohort (Spearman's rank correlation coefficient, P < 0.0001). (C) Positive correlation between the CXCL10 expression signature and antigen-related genes in the TCGA-BRCA cohort. (D) Positive correlation between the CXCL10 expression signature and neoantigen load in the TCGA-BRCA cohort (Spearman's rank correlation coefficient, P < 0.0001).
The CXCL10 expression signature is positively associated with ICB-related genes (A) Correlation between the CXCL10 expression signature and ICB-related genes in the TCGA-BRCA cohort. (B) Correlation between the CXCL10 expression signature and ICB-related genes in the TCGA-pan cancer cohorts. (C) Violin plot depicting the expression of CXCL10 in sensitive or resistant tumor tissues. Immunotherapy: anti-PD1 and anti-CTLA-4 combination therapy. From left to right: Before receiving immunotherapy, on the 3rd day of receiving immunotherapy, on the 7th day of receiving immunotherapy, and at the time point of the end of immunotherapy (Wilcoxon signed-rank test, ** P < 0.01, *** P < 0.001).
Discussion
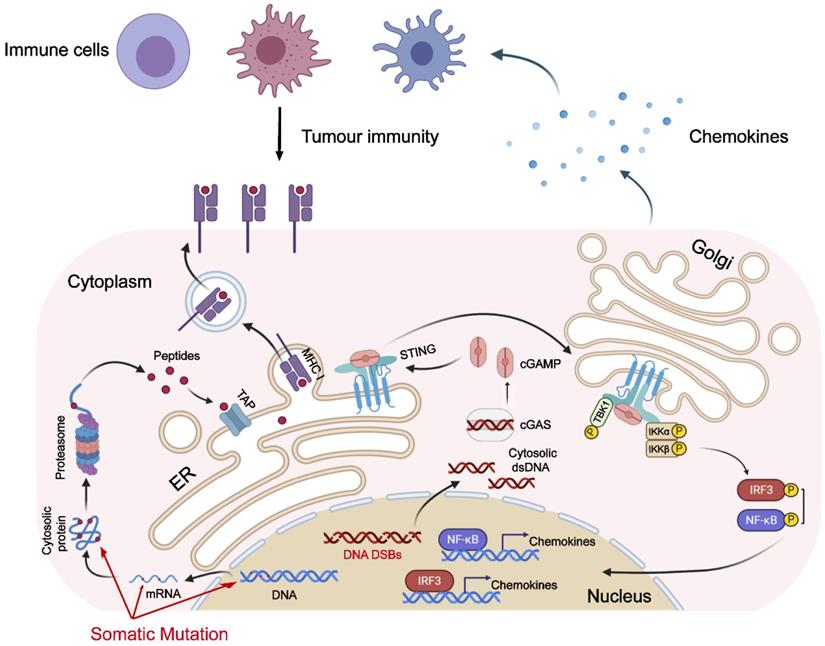
Emerging evidence has shown the importance of the cGAS-STING pathway in tumor immunotherapy [36,37]. Recent studies have found that the cGAS-STING pathway being activated in the setting of genome instability can be attributed to dMMR [38], PARP inhibitor treatment, or functional loss of BRCA1/2 genes [39], and it predicts a better prognosis of tumor patients. While these previous studies, combined with our research, fully prove that genomic instability is associated with the activation of the cGAS-STING pathway. As one of the hallmarks of malignant tumors, genomic instability plays an important role in the occurrence and development of tumors [40,41]. Genomic instability could facilitate the evolution of tumors. However, as the old saying goes, “every coin has two sides”: genomic instability also makes tumor cells bear a higher neoantigen load, which is more easily recognized by the immune system [42,43]. The activation of the anti-tumor immune response requires the participation of the innate immune response. The instability of the tumor genome also causes the up-regulation of the cGAS-STING pathway [44,45], which in turn activates the innate immune response (Figure 8).
By analyzing the expression profile of patients with different HRD scores, we identified that CXCL1, CXCL10, CXCL11, CCL8, CCL13 and CCL18 expression were enriched in patients with high HRD scores, and that the expression of CXCL10 had the strongest ability to predict the prognosis of the patients. Although recent studies have found that in different types of cancer, patients with high expression of CXCL10 have a better clinical prognosis [46-48], but no one has explained the molecular mechanism of CXCL10 high expression. Our cohort analysis combined with the single-cell RNA-seq results fully demonstrated the correlation between HRD and CXCL10. As a downstream target gene of the cGAS-STING pathway [49,50], CXCL10 is a small cytokine belonging to the CXC chemokine family that is also known as Interferon-inducible T-cell alpha chemoattractant and Interferon-gamma-inducible proteins [51,52]. CXCL10 has been attributed to several roles, such as chemoattraction for monocytes/macrophages, T cells, NK cells, dendritic cells, and promotion of T cell adhesion to endothelial cells [53-56]. Our results demonstrated that the correlation between genomic instability and activated the cGAS-STING signaling in dMMR tumors [7] can be extended to HRD tumors. Most importantly, we introduced CXCL10 as a potentially reliable biomarker for the efficacy of ICB therapy. The CXCL10 can be used as a barometer of the HRD score with strong predictive power.
The CXCL10 expression signature could be used as potential biomarkers for ICB therapy. (A) Curve for overall survival is shown for high and low CXCL10 expression in the PD-L1 treatment cohort. (B and C) The proportion of immune response to anti-PD-L1 treatment in high versus low CXCL10 expression subgroups. CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease. (D and E) TMB and neoantigen load in the immunotherapy cohort were compared among distinct CXCL10 expression signature subgroups (Wilcoxon signed-rank test, * P < 0.05, **** P < 0.0001). (F) The CXCL10 expression signature in different immune phenotype subgroups. The tumor immunophenotype was defined according to immunohistochemistry results of the CD8 antibody (Wilcoxon signed-rank test, **** P < 0.0001) [14].
Multivariate Cox regression analysis of the CXCL10 expression signature with gender, smoking, ECOG score and immunophenotype were taken into account. ECOG: Eastern Cooperative Oncology Group. TMB: Tumor Mutation Burden. Immune phenotype: Desert, CD8+ T cells are absent from the tumor and its periphery; Excluded, CD8+ T cells accumulated but do not efficiently infiltrate; Inflamed, CD8+ T cells infiltrate but their effects are inhibited.
Under this situation, the determination of whether upregulated CXCL10 unavoidably results from cGAS-STING activation needs further experimental testing. As a downstream target of the cCAS-STING pathway, upregulated CXCL10 showed superior predictive power compared with the HRD score. Importantly, the clinical examination of CXCL10 in tumor tissues or serum is more feasible than applying the steps necessary for calculating the HRD score, and the prediction accuracy of upregulated CXCL10 is even better than the HRD score itself. We introduced, for the first time, a prospective biomarker associated with the efficacy of immunotherapy in HRD tumors, which merits further investigation in multiple cohorts.
Abbreviations
HR: homologous recombination; HRD: homologous recombination deficiency; PAPRi: PAPR inhibitor; TIME: tumor immune microenvironment; ICB: immune checkpoint blockade; OSC: ovarian serous cystadenocarcinoma; TNBC: triple-negative breast cancer; HR: homologous recombination repair; TCGA: the Cancer Genome Atlas; GEO: Gene Expression Omnibus; CXCL10: C-X-C motif chemokine 10; TPM: transcripts per million; CNV: copy number variation; LOH: loss of heterozygosity; LST: Large-scale State Transitions; TAI: Telomeric Allelic Imbalance; GSEA: Gene set enrichment analysis; DEGs: differentially expressed genes; MSI: microsatellite instability; MAD: median absolute deviation.
Model for genomic instability-induced cGAS activation and immune response. Genomic instability, caused by dMMR or HRD, leads to tumor cells bearing a higher neoantigen load. Additionally, the instability of the tumor genome also causes the activation of the cGAS-STING pathway, which in turn up-regulates the expression of chemokines and attracts immune cells to migrate to tumor tissue.
Supplementary Material
Supplementary table S1.
Supplementary table S2.
Supplementary table S3.
Acknowledgements
Authors' contributions
Designing research studies: Zhiwen Shi, Keqin Hua and Hongyan Wang. Analyzing data: Zhiwen Shi and Jianfeng Shen. Preparing the manuscript: Zhiwen Shi and Jianfeng Shen. Grammar Check: Junjun Qiu and Qingguo Zhao. Supervision: Keqin Hua and Hongyan Wang. Funding Acquisition: Keqin Hua and Hongyan Wang. The authors read and approved the final manuscript.
Funding
This work was jointly supported by grants from the Shanghai Shenkang Hospital Development Center Funding (SHDC2020CR1045B), the Artificial Intelligence Innovation Project of the Shanghai Municipal Commission of Economy and Informatization (No. 2018-RGZN-02041) to K.H and the National Natural Science Foundation of China (81930036, 31521003 and 31771669) and National key research and development program (2016YFC1000502) to H.W.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409-13
2. Lee V, Murphy A, Le DT, Diaz Jr LA. Mismatch repair deficiency and response to immune checkpoint blockade. Oncologist. 2016;21:1200
3. Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J. et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018 362
4. Darvin P, Toor SM, Nair VS, Elkord E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp Mol Med. 2018;50:1-11
5. van de Donk PP, de Ruijter LK, Lub-de Hooge MN, Brouwers AH, van der Wekken AJ, Oosting SF. et al. Molecular imaging biomarkers for immune checkpoint inhibitor therapy. Theranostics. 2020;10:1708
6. Li X-D, Wu J, Gao D, Wang H, Sun L, Chen ZJ. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390-4
7. Lu C, Guan J, Lu S, Jin Q, Rousseau B, Lu T. et al. DNA sensing in mismatch repair-deficient tumor cells is essential for anti-tumor immunity. Cancer Cell. 2020
8. Vanderstichele A, Busschaert P, Olbrecht S, Lambrechts D, Vergote I. Genomic signatures as predictive biomarkers of homologous recombination deficiency in ovarian cancer. Eur J Cancer. 2017;86:5-14
9. Chopra N, Tovey H, Pearson A, Cutts R, Toms C, Proszek P. et al. Homologous recombination DNA repair deficiency and PARP inhibition activity in primary triple negative breast cancer. Nat Commun. 2020;11:1-12
10. den Brok WD, Schrader KA, Sun S, Tinker AV, Zhao EY, Aparicio S. et al. Homologous recombination deficiency in breast cancer: a clinical review. JCO Precis Oncol. 2017;1:1-13
11. da Cunha Colombo Bonadio RR, Fogace RN, Miranda VC, Diz MdPE. Homologous recombination deficiency in ovarian cancer: a review of its epidemiology and management. Clinics. 2018 73
12. Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol. 2015;19:A68
13. Mihriban K, Simona C, Gillespie SM, Patel AP, Ravindra M, Luo CC. et al. Unravelling subclonal heterogeneity and aggressive disease states in TNBC through single-cell RNA-seq. Nat Commun. 2018;9:3588
14. Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y. et al. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature. 2018;554:544-8
15. Hwang S, Kwon AY, Jeong JY, Kim S, Kang H, Park J. et al. Immune gene signatures for predicting durable clinical benefit of anti-PD-1 immunotherapy in patients with non-small cell lung cancer. Sci Rep;10.
16. Hollern DP, Xu N, Thennavan A, Glodowski C, Garcia-Recio S, Mott KR. et al. B Cells and T Follicular Helper Cells Mediate Response to Checkpoint Inhibitors in High Mutation Burden Mouse Models of Breast Cancer. Cell. 2019;179:1191-206.e21
17. Abkevich V, Timms K, Hennessy B, Potter J, Carey M, Meyer LA. et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107:1776-82
18. Manié E, Popova T, Battistella A, Tarabeux J, Caux-Moncoutier V, Golmard L. et al. Genomic hallmarks of homologous recombination deficiency in invasive breast carcinomas. Int J Cancer. 2016;138:891-900
19. Birkbak NJ, Wang ZC, Kim J-Y, Eklund AC, Li Q, Tian R. et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2:366-75
20. Takaya H, Nakai H, Takamatsu S, Mandai M, Matsumura N. Homologous recombination deficiency status-based classification of high-grade serous ovarian carcinoma. Sci Rep. 2020;10:1-8
21. Sztupinszki Z, Diossy M, Krzystanek M, Reiniger L, Csabai I, Favero F. et al. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer. 2018;4:1-4
22. Von Minckwitz G, Timms K, Untch M, Elkin EP, Fasching PA, Schneeweiss A. et al. Prediction of pathological complete response (pCR) by Homologous Recombination Deficiency (HRD) after carboplatin-containing neoadjuvant chemotherapy in patients with TNBC: Results from GeparSixto. Am Soc Clin Oncol. 2015
23. Matey-Hernandez ML, Brunak S, Izarzugaza JM. Benchmarking the HLA typing performance of Polysolver and Optitype in 50 Danish parental trios. BMC Bioinformatics. 2018;19:1-12
24. Li X, Wen D, Li X, Yao C, Chong W, Chen H. Identification of an Immune Signature Predicting Prognosis Risk and Lymphocyte Infiltration in Colon Cancer. Front Immunol. 2020;11:1678
25. Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449-W54
26. Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48-61
27. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139-40
28. Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC. et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:1-11
29. Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS. et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108-e10
30. Surmi BK, Hasty AH. The role of chemokines in recruitment of immune cells to the artery wall and adipose tissue. Vascul Pharmacol. 2010;52:27-36
31. Lippitz BE. Cytokine patterns in patients with cancer: a systematic review. Lancet Oncol. 2013;14:e218-e28
32. Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H, Torres-Garcia W. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:1-11
33. Li B, Severson E, Pignon J-C, Zhao H, Li T, Novak J. et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:1-16
34. Adams S, Gatti-Mays ME, Kalinsky K, Korde LA, Sharon E, Amiri-Kordestani L. et al. Current landscape of immunotherapy in breast cancer: a review. JAMA Oncol. 2019;5:1205-14
35. Petitprez F, Meylan M, de Reyniès A, Sautès-Fridman C, Fridman WH. The tumor microenvironment in the response to immune checkpoint blockade therapies. Front Immunol. 2020 11
36. Li A, Yi M, Qin S, Song Y, Chu Q, Wu K. Activating cGAS-STING pathway for the optimal effect of cancer immunotherapy. J Hematol Oncol. 2019;12:35
37. Pellegrino B, Musolino A, Llop-Guevara A, Serra V, De Silva P, Hlavata Z. et al. Homologous recombination repair deficiency and the immune response in breast cancer: a literature review. Transl Oncol. 2020;13:410-22
38. Guan J, Lu C, Jin Q, Lu H, Chen X, Tian L. et al. MLH1 deficiency-triggered DNA hyperexcision by exonuclease 1 activates the cGAS-STING pathway. Cancer Cell. 2020
39. Reisländer T, Groelly FJ, Tarsounas M. DNA Damage and Cancer Immunotherapy: a STING in the Tale. Mol Cell. 2020
40. Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability - an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220-8
41. Andor N, Maley CC, Ji HP. Genomic instability in cancer: teetering on the limit of tolerance. Cancer Res. 2017;77:2179-85
42. Mardis ER. Neoantigens and genome instability: impact on immunogenomic phenotypes and immunotherapy response. Genome Med. 2019;11:1-12
43. Germano G, Lamba S, Rospo G, Barault L, Magrì A, Maione F. et al. Inactivation of DNA repair triggers neoantigen generation and impairs tumour growth. Nature. 2017;552:116-20
44. Chen H, Zhang J, Wang Y, Simoneau A, Yang H, Levine AS. et al. cGAS suppresses genomic instability as a decelerator of replication forks. Sci Adv. 2020;6:eabb8941
45. Kwon J, Bakhoum SF. The cytosolic DNA-sensing cGAS-STING pathway in cancer. Cancer Discov. 2020;10:26-39
46. Qin X-J, Lin X, Xue G, Fan H-L, Wang H-Y, Wu J-F. et al. CXCL10 is a potential biomarker and associated with immune infiltration in human papillary thyroid cancer. Biosci Rep. 2021 41
47. Chuan T, Li T, Yi C. Identification of CXCR4 and CXCL10 as Potential predictive biomarkers in triple negative breast cancer (TNBC). Med Sci Monit. 2020;26:e918281-1
48. Kaesler S, Wölbing F, Kempf WE, Skabytska Y, Köberle M, Volz T. et al. Targeting tumor-resident mast cells for effective anti-melanoma immune responses. JCI Insight. 2019 4
49. Chen Y-A, Shen Y-L, Hsia H-Y, Tiang Y-P, Sung T-L, Chen L-Y. Extrachromosomal telomere repeat DNA is linked to ALT development via cGAS-STING DNA sensing pathway. Nat Struct Mol Biol. 2017;24:1124
50. Carroll EC, Jin L, Mori A, Muñoz-Wolf N, Oleszycka E, Moran HB. et al. The vaccine adjuvant chitosan promotes cellular immunity via DNA sensor cGAS-STING-dependent induction of type I interferons. Immunity. 2016;44:597-608
51. Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S. et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation-a target for novel cancer therapy. Cancer Treat Rev. 2018;63:40-7
52. Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. IFN-γ-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol. 2002;168:3195-204
53. Dufour JH, Dziejman M, Liu MT, Leung JH, Lane TE, Luster AD. IFN-γ-Inducible Protein 10 (IP-10; CXCL10)-Deficient Mice Reveal a Role for IP-10 in Effector T Cell Generation and Trafficking. J Immunol. 2002;168:3195-204
54. Bronger H, Singer J, Windmüller C, Reuning U, Zech D, Delbridge C. et al. CXCL9 and CXCL10 predict survival and are regulated by cyclooxygenase inhibition in advanced serous ovarian cancer. Br J Cancer. 2016;115:553-63
55. House IG, Savas P, Lai J, Chen AX, Oliver AJ, Teo ZL. et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res. 2020;26:487-504
56. Fujita M, Zhu X, Ueda R, Sasaki K, Kohanbash G, Kastenhuber ER. et al. Effective immunotherapy against murine gliomas using type 1 polarizing dendritic cells—significant roles of CXCL10. Cancer Res. 2009;69:1587-95
Author contact
![]() Corresponding authors: Hongyan Wang, E-mail: wanghylabedu.cn; Keqin Hua, E-mail: huakeqinedu.cn.
Corresponding authors: Hongyan Wang, E-mail: wanghylabedu.cn; Keqin Hua, E-mail: huakeqinedu.cn.
 Global reach, higher impact
Global reach, higher impact