13.3
Impact Factor
Theranostics 2021; 11(10):5061-5076. doi:10.7150/thno.56202 This issue Cite
Research Paper
Identification and validation of hypoxia-derived gene signatures to predict clinical outcomes and therapeutic responses in stage I lung adenocarcinoma patients
1. Department of Radiation Oncology, University Hospital, LMU Munich, Munich D-81377, Germany.
2. Department of Medical Oncology, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou 310003, China.
3. Research Unit Radiation Cytogenetics, Helmholtz Center Munich, German Research Center for Environmental Health GmbH, Neuherberg D-85764, Germany.
4. Department of Radiotherapy, Sichuan Cancer Hospital, School of Medicine, University of Electronic Science and Technology of China, Chengdu 610041, China.
5. Laboratory of Chinese Herbal Pharmacology, Oncology Center, Renmin Hospital, Hubei Key Laboratory of Wudang Local Chinese Medicine Research, Hubei University of Medicine, Shiyan 442000, China.
* These authors contributed equally to this work.
Abstract
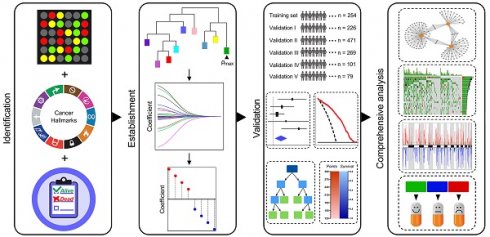
Rationale: The current tumour-node-metastasis (TNM) staging system is insufficient for precise treatment decision-making and accurate survival prediction for patients with stage I lung adenocarcinoma (LUAD). Therefore, more reliable biomarkers are urgently needed to identify the high-risk subset in stage I patients to guide adjuvant therapy.
Methods: This study retrospectively analysed the transcriptome profiles and clinical parameters of 1,400 stage I LUAD patients from 14 public datasets, including 13 microarray datasets from different platforms and 1 RNA-Seq dataset from The Cancer Genome Atlas (TCGA). A series of bioinformatic and machine learning approaches were combined to establish hypoxia-derived signatures to predict overall survival (OS) and immune checkpoint blockade (ICB) therapy response in stage I patients. In addition, enriched pathways, genomic and copy number alterations were analysed in different risk subgroups and compared to each other.
Results: Among various hallmarks of cancer, hypoxia was identified as a dominant risk factor for overall survival in stage I LUAD patients. The hypoxia-related prognostic risk score (HPRS) exhibited more powerful capacity of survival prediction compared to traditional clinicopathological features, and the hypoxia-related immunotherapeutic response score (HIRS) outperformed conventional biomarkers for ICB therapy. An integrated decision tree and nomogram were generated to optimize risk stratification and quantify risk assessment.
Conclusions: In summary, the proposed hypoxia-derived signatures are promising biomarkers to predict clinical outcomes and therapeutic responses in stage I LUAD patients.
Keywords: Stage I lung adenocarcinoma, Hypoxia, Clinical outcomes, Genomic alterations, Machine learning.
 Global reach, higher impact
Global reach, higher impact