13.3
Impact Factor
Theranostics 2021; 11(10):4992-5009. doi:10.7150/thno.55006 This issue Cite
Research Paper
Single-cell transcriptome dissection of the toxic impact of Di (2-ethylhexyl) phthalate on primordial follicle assembly
College of Life Sciences, Key Laboratory of Animal Reproduction and Germplasm Enhancement in Universities of Shandong, Qingdao Agricultural University, Qingdao 266109, China
Received 2020-10-25; Accepted 2021-1-22; Published 2021-3-5
Abstract
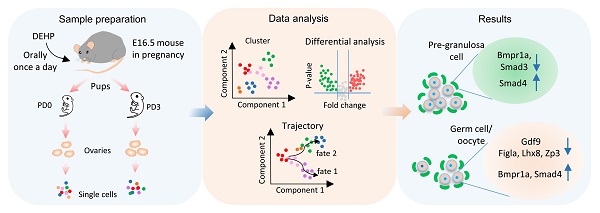
Rationale: Accumulated evidence indicates that environmental plasticizers are a threat to human and animal fertility. Di (2-ethylhexyl) phthalate (DEHP), a plasticizer to which humans are exposed daily, can trigger reproductive toxicity by acting as an endocrine-disrupting chemical. In mammals, the female primordial follicle pool forms the lifetime available ovarian reserve, which does not undergo regeneration once it is established during the fetal and neonatal period. It is therefore critical to examine the toxicity of DEHP regarding the establishment of the ovarian reserve as it has not been well investigated.
Methods: The ovarian cells of postnatal pups, following maternal DEHP exposure, were prepared for single cell-RNA sequencing, and the effects of DEHP on primordial follicle formation were revealed using gene differential expression analysis and single-cell developmental trajectory. In addition, further biochemical experiments, including immunohistochemical staining, apoptosis detection, and Western blotting, were performed to verify the dataset results.
Results: Using single-cell RNA sequencing, we revealed the gene expression dynamics of female germ cells and granulosa cells following exposure to DEHP in mice. Regarding germ cells: DEHP impeded the progression of follicle assembly and interfered with their developmental status, while key genes such as Lhx8, Figla, and others, strongly evidenced the reduction. As for granulosa cells: DEHP likely inhibited their proliferative activity, and activated the regulation of cell death. Furthermore, the interaction between ovarian cells mediated by transforming growth factor-beta signaling, was disrupted by DEHP exposure, since the expression of GDF9, BMPR1A, and SMAD3 was affected. In addition, DNA damage and apoptosis were elevated in germ cells and/or somatic cells.
Conclusion: These findings offer substantial novel insights into the reproductive toxicity of DEHP exposure during murine germ cell cyst breakdown and primordial follicle formation. These results may enhance the understanding of DEHP exposure on reproductive health.
Keywords: DEHP, primordial follicle assembly, female germ cell, granulosa cell, single-cell transcriptome
Introduction
Environmental pollution is evident worldwide and is obviously threatening human health, including increasing the risk of cancer. Among various pollutants, ubiquitous plastics are annually utilized in staggeringly large volumes; this has raised a public health concern. Phthalates are widely used as a plasticizer for polyvinyl chloride resin (PVC) in the production of plastics [1]. As phthalates are not covalently bond to polymers [2, 3], they can easily migrate and penetrate human and animal bodies through various routes, such as oral ingestion, inhalation, and dermal exposure [4, 5]. Worryingly, human exposure to phthalate occurs on a daily basis [1, 6]. Moreover, robust evidence has heightened a link between plasticizer exposure and some reproductive disorders [7-10]. Thus, an investigation into the reproductive toxicity of phthalates is deemed necessary to heighten awareness of the need for protection against detrimental phthalates, and to propose potential preventive and therapeutic strategies for toxicity mitigation.
In females, primordial follicle (PF) formation (or assembly) occurs perinatally in mice and around mid-gestation in humans; this results in a finite follicle reserve termed the ovarian reserve or PF pool [11-13]. After primordial germ cells arrive at the murine gonad, they undergo mitosis to produce germ cell cysts (or nests), from which oogonia mature after sexual differentiation. Subsequently, oogonia enter meiosis around 13.5 days post coitum (dpc) to become oocytes. Shortly afterwards, germ cell cysts breakdown following the invasive activity of proliferative pre-granulosa cells from 17.5 dpc to a few days postnatally. A substantial loss of germ cells, and the surviving oocytes become enclosed within a layer of flattened granulosa cells, which signifies PF formation [14, 15]. It is known that the PF pool is non-renewable once established and that it progressively diminishes following the first-wave of follicle recruitment during puberty, and is almost exhausted at menopause in humans [16-18]. Thus, the size of the PF pool is the primary determinant of reproductive lifespan in females [19]. Notably, the available oocytes within PFs naturally undergo irreversible exhaustion or atresia during development; however, they are prone to damage from adverse external environmental chemicals (such as plasticizers) as well as cellular aging [14, 20]. Meanwhile, the ovarian dysgenesis syndrome hypothesis proposes that an early disruption to ovarian development can potentially lead to ovarian disease and disorders in adulthood; this is particularly pertinent at times that are sensitive to disruption by exogenous chemicals, such as PF assembly [21-23]. Consequently, attention needs to be focused on the protection of female fertility from toxic materials.
Di (2-ethylhexyl) phthalate (DEHP), the most commonly used plasticizer, is known to be an endocrine-disrupting chemical (EDC) that can trigger reproductive and developmental toxicity in females [1, 21]. Importantly, DEHP exposure impairs establishment of the ovarian reserve by delaying or reducing PF assembly [24, 25]. Similar observations in other studies suggest the deregulation of estrogen receptor (ER) and progesterone receptor expression, and attenuation of NOTCH signaling [26]. Furthermore, the number of apoptotic germ cells increases during in vitro culture of newborn ovaries exposed to DEHP for three days [25]. Specifically, autophagy can be induced by DEHP exposure via activating AMPK-SKP2-CARM1 signaling, thus further reducing the PF pool and damaging female fertility [27]; these results indicate that DEHP affects oocyte survival during PF formation. From another aspect, in vivo and in vitro studies both suggest that DEHP damages steroidogenesis and exerts toxicity during different stages of folliculogenesis [9, 28]. For example, plasticizer exposure could accelerate the development of PFs and therefore result in premature ovarian failure (POF) [29], and antral follicle depletion is increased as a result of estrogen deficiency, estrous cycle disorder, and impaired aromatase activity [4, 7, 30, 31]. Meanwhile, the transgenerational inheritance of DEHP has attracted considerable attention [32-34]. Through maternal exposure, DEHP has been shown to exert adverse reproductive effects across generations: it reduces oocyte quality and follicle reserve in F1 female mice, and alters the same ovarian phenotype in F2 and F3 offspring; moreover, it accelerates follicular recruitment [24, 35].
The development of single-cell transcriptomics has revolutionized the dissection of cellular heterogeneity and cell fate transition [36-38]. From such studies, many novel insights have been proposed regarding reproductive system development, derived from single cell datasets in both males [39-42] and females [43-46]. Recently, the credo of a limited ovarian reserve has been reinforced by single-cell analysis of human ovarian cortical tissue, where no oogonial stem cells were detected [47]. In addition, the transcriptomic landscapes of germ cells during early murine oogenesis have been delineated, which concentrates on two vital germline events, meiotic initiation [43, 48] and PF assembly [49]. Furthermore, the respective waves of PF formation in the ovarian cortex and medulla disclose two distinct pathways for pre-granulosa cell differentiation [50]. To date, the effect of DEHP on PF assembly has not been thoroughly examined and deciphered.
The current study set out to characterize the molecular mechanism of the toxic effects of DEHP exposure on PF assembly, at a daily dose of 40 µg/kg body weight DEHP through oral administration to pregnant mice at 16.5 dpc. Subsequently, the DEHP-treated ovaries at PD0 (PD0-DEHP) and PD3 (PD3-DEHP) were subjected to single-cell RNA sequencing (scRNA-seq) using a 10× Genomics platform. At a single-cell level, the toxicity of DEHP exposure to PF assembly was further investigated, and compared to previously reported normal murine developmental ovarian datasets at PD0 and PD3 [49].
Results
scRNA-seq identified six main cell types in post-natal ovaries
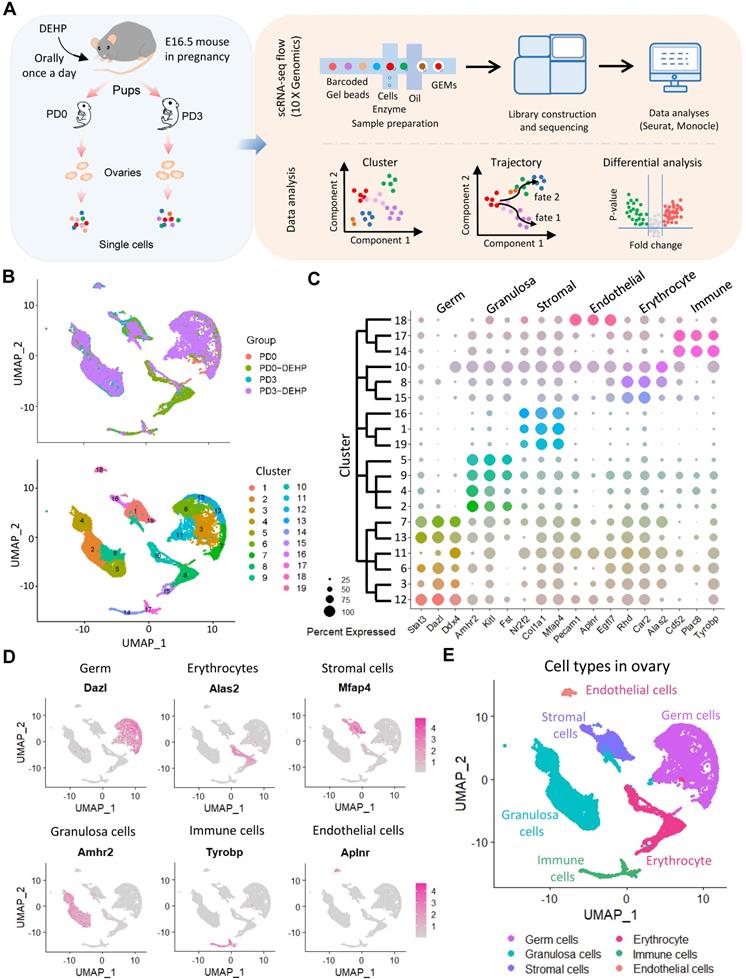
To characterize the transcriptome dynamics of ovarian cells affected by DEHP during PF formation, pregnant mice were given DEHP daily from 16.5 dpc, and the ovaries were collected at postnatal day 0 (PD0-DEHP) and 3 (PD3-DEHP), from which single-cell suspensions were prepared for scRNA-seq (Figure 1A, left panel). Following the combination of barcoded gel beads, enzyme mixtures, and oil, the nanoliter-scale droplets (termed “gel bead-in emulsions”, GEMs) were generated for library construction and sequencing (upper part of right panel). Then, single-cell datasets were conducted for cell cluster, pseudotime trajectory, and differential analysis of gene expression using Seurat and Monocle (Figure 1A, bottom part of right panel). In addition, after data pre-progressing and potential doublets filtration (Figure S1A), 4,558 high-quality ovarian cells were obtained at PD0-DEHP and 5,252 at PD3-DEHP, and the number of high-quality cells from collected datasets of normal ovarian tissues at PD0 was 4,270, with 5,375 at PD3 (Figure S1B). The harvested high-quality cells were used for subsequent analysis.
Based on uniform manifold approximation and projection (UMAP) technology, 19 cell clusters were generated (Figure 1B), and several cell clusters were at least partially displayed in a dependent manner for the developmental stage or DEHP treatment. To determine the ovarian cell composition at an investigated period, marker genes were identified in cell clusters (Table S1); the top five marker genes in each cluster are shown in Figure S1C. According to their transcription characteristics, cell clusters were divided into six cell types, and the representative genes in each cell type are shown in Figure 1C. In addition, the cell-type-specific genes are presented in Figure 1D and Figure S1D; six main cell types were identified (Figure 1E). Moreover, germ cells and granulosa cells were examined as the major participants of PF assembly using immunohistochemistry; STAT3 for germ cells (Figure S2A) and AMHR2 for granulosa cells (Figure S2B), respectively.
DEHP impacted the progress of PF assembly and germ cell development
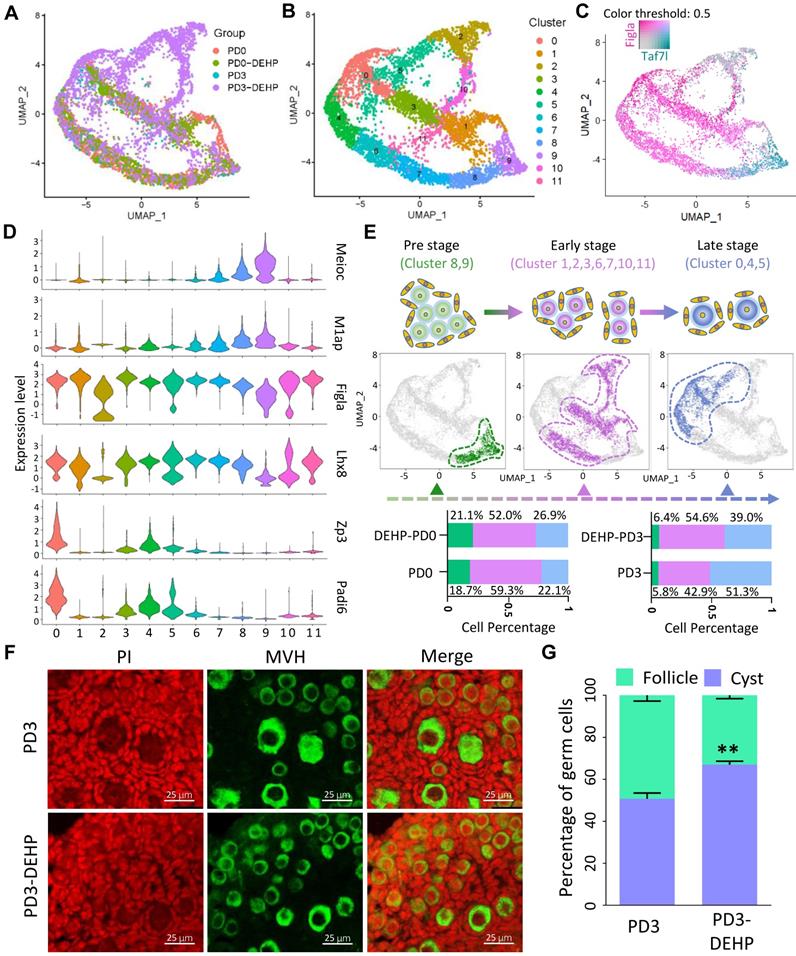
Following the cell type classification above, the germ cell population was extracted for in-depth analysis; according to the results, cell heterogeneity was not dependent on cell cycle genes (Figure S3A) and cells in the four groups were classified as twelve cell clusters (Figure 2A-B). In addition, marker genes in each germ cell cluster were identified (Table S2), and the top five genes are displayed in a heatmap in Figure S3B. Among them, several key genes involved in biological events were highlighted, including meiosis (Meioc, M1ap), PF assembly (Figla, Lhx8), and oocyte growth (Zp3, Padi6; Figure S3C). Specifically, the representative genes, Taf7l and Figla, were capable of separating cell clusters into two developmental stages (Figure 2C). Meanwhile, based on the expression pattern of genes associated with meiosis, PF assembly, and oocyte growth in clusters (Figure 2D and Figure S3C), the germ cells were divided into three developmental stages: pre-, early-, and late-stage of PF assembly (Figure 2E). The pre-stage consisted of cells in clusters 8 and 9; the early-stage contained cells in clusters 1, 2, 3, 6, 7, 10, and 11; and the late-stage comprised cells in clusters 0, 4, and 5 (Figure 2E upper). Moreover, the percentage of cells at the three stages indicated a decline in cell percentage at the early-stage for PD0 and the late-stage for PD3 after DEHP treatment (Figure 2E bottom); furthermore, the results suggested that DEHP impacted the progress of PF formation. In particular, the immunostaining of ovarian sections at PD3 (Figure 2F) with or without DEHP suggested that DEHP exposure indeed decreased the percentage of germ cells within follicles (Figure 2G).
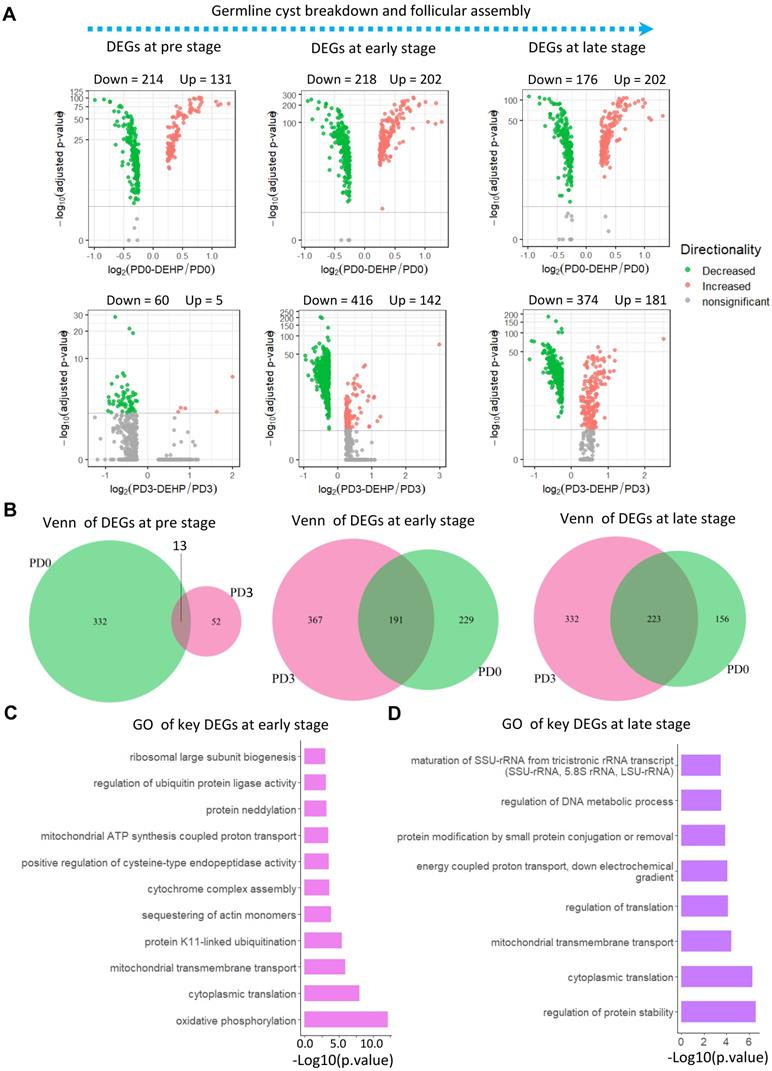
Furthermore, gene differential expression was carried out at the three identified stages (Figure 3A). At the pre-stage of PF formation (Table S3), there were 214 down- and 131 up-regulated genes for PD0-DEHP vs. PD0, and 65 differentially expressed genes (DEGs) between the PD3-DEHP and PD3 groups (Figure 3A left). For the early-stage (Table S4) with DEHP treatment, there were 202 up- and 218 down-regulated DEGs at PD0, and 142 up- and 416 down-regulated DEGs at PD3, respectively (Figure 3A middle). At the late-stage (Table S5), DEHP significantly increased 202 and 181 DEGs, and decreased 176 and 374 genes at PD0 and PD3, respectively (Figure 3A right). Furthermore, the representative genes involved in PF assembly and oocyte growth dramatically declined (Figure S3D). These results implied that DEHP tended to influence the transcription of germ cells during PF assembly. In addition, 13, 191, and 223 key DEGs (this refers to the common genes in Venn diagrams that were both affected by DEHP at PD0 and PD3) were produced at the pre-, early-, and late-stages of PF formation, respectively (Figure 3B). Enrichment analysis demonstrated that key genes at the pre-stage were related to “Ribosome”, “DNA repair”, and “Regulation of cellular response to stress” (Figure S3E); 191 key DEGs at the early-stage were mostly enriched in the biological processes of “oxidative phosphorylation”, and “mitochondrial ATP synthesis coupled proton transport” (Figure 3C). Meanwhile, at the late-stage, DEHP largely affected “regulation of protein stability” (Figure 3D), as well as “energy coupled proton transport”. Moreover, both were enriched in “cytoplasmic translation” and “mitochondrial transmembrane transport”.
Molecular characterization of post-natal ovaries. (A) A schematic pipeline of sample collection, 10× Genomics platform sequencing, and data analysis. (B) UMAP plots of ovarian cells based on sample group (upper) and Seurat cluster (below). (C) Dot plot of the three marker genes identified from ovarian cell types in each cluster. Dot size represents the percentage of gene expression in each cell cluster; color intensity refers to the intensity of expression, with strong color indicating high gene expression. The dendrogram of cell clusters is presented in the left panel. (D) Feature plots of specific marker genes from six main ovarian cell types. (E) The six main ovarian cell types are identified on a UMAP plot.
DEHP impacted the progress of germline cyst breakdown and follicular assembly. (A) Clustering of germ cell population with UMAP, colored based on sample groups. (B) Clustering of germ cell population with UMAP, colored based on 12 cell clusters. (C) Expression of Taf7l and Figla separating the germ cell clusters into different developmental stages based on UMAP. The color intensity of cyan or maroon represents gene expression level; grey refers to the double-negative cells and the color threshold of gene expression is 0.5. (D) Expression of representative genes in cell clusters for germ cell-specific developmental stages, the pre-stage of follicle formation with highly expressed meiosis related genes: Meioc, M1ap; the early-stage marked by high expression of key genes for follicle formation: Figla, Lhx8; the late-stage contained genes required for oocyte growth: Zp3, Padi6. (E) The three separated developmental stages in germ cell clusters. Cell cluster classification, feature plots and cell percentages of sample groups for the three stages are shown in the upper, middle, and bottom panels. (F) Representative images of ovarian sections with normal and DEHP-treated groups at PD3. Germ cells were marked by MVH (green) and cell nuclei were counterstained with propidium iodide (PI, red). Scale bar = 25 μm. (G) The percentages of germ cells within cysts and follicles at PD3 with and without DEHP exposure. Data are presented as mean ± SD (N = 3 for independent repeats, the total germ cell counting at PD3 in number was 1,710, 1,355, and 1,401, and 2,004, 1,828, and 2,341 at PD3-DEHP). **P < 0.01.
Differential expression of stage-specific germ cells with or without DEHP treatment. (A) Volcano plots of germ cells with three developmental stages. The left panel illustrates differentially expressed genes (DEGs) of germ cells at the pre-stage of PF formation with or without DEHP treatment for PD0 and PD3, respectively. The middle and right panels refer to pre- and late-stage. (B) Venn plots of germ cells with three developmental stages. The light green and violet red circles represent the number of DEGs at PD0 and PD3. (C-D) GO terms of key DEGs at early- (C) and late-stages (D) of PF formation.
In a separate analysis, the DEG lists at PD0 and PD3, that were associated with the DEHP effect, were examined. Results showed that at the pre-stage, the biological process of DEG lists at PD0 and PD3 varied massively (Figure S4A), and the difference at the early- and late-stages of PF formation were not distinct (Figure S4B-C). Furthermore, DEHP always impacted the processes of “cellular responses to stress”, “chromatin organization”, “oxidative phosphorylation”, “cytoplasmic translation”, “cellular response to DNA damage stimulus”, and “transcriptional regulation by TP53” throughout both PD0 and PD3 stages. At the same time, the transforming growth factor-beta (TGF-beta) signal, cell cycle, and mRNA processing were all likely associated with DEHP effect on germ cell development.
DEHP disordered germ cell fate revealed by single cell trajectory
To further dissect the fate transition of germ cells during PF formation, the pseudotime trajectory of germ cells was established to reveal five developmental States and two branching cell fates (Figure 4A). According to the distribution of germ cells in States, the differentiated status of DEHP-treated germ cells at PD3 underwent a remarkable change, with an apparent increase of cells at State 5 being observed (Figure 4B-C). Gene expression along with cell trajectory showed at State 1 that the Taf7l gene was actively expressed at the pre-stage of PF formation, while the Figla gene was expressed at all investigated stages, while at State 3 the Zp3 and Padi6 genes were specifically and actively expressed in follicular oocytes (late-stage; Figure S5A). This evidence implied that PF assembly was impeded by DEHP.
To investigate the inherent mechanism, gene expression patterns at two branches was performed (Figure 4D). Four gene sets regarding two cell fates were generated: set 1 consisted of 282 genes, mostly and highly expressed in cell fate 1, partly expressed at cell fate 2, and slightly expressed at pre-branch (State 1); furthermore, the GO terms of the top 100 genes in the set were mostly enriched in biological processes such as “Translation”, “Ribosomal small subunit biogenesis”, “Regulation of translation”, “Negative regulation of ubiquitin protein ligase activity” and “rRNA processing”. Set 2 contained 643 genes that were mainly expressed in pre-branch and fate 1 cells, and hugely decreased in fate 2, which acted on “Translation”, “Ribosome assembly”, “Oxidative phosphorylation”, “Ribosome biogenesis”, and “Positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator”. For the 396 genes in set 3, they were mainly expressed at pre-branch, showing a slight decline at fate 2 and were dramatically down-regulated at fate 1, but largely enriched in “Synapsis”, “Protein-DNA complex subunit organization”, “Mitochondrial genome maintenance”, “Protein refolding”, and “Negative regulation of mRNA metabolic process”. The 1,371 genes in set 4 seemed to be elevated by DEHP at fate 2 and were related to the processes of “ATP synthesis coupled electron transport”, “Histone H3-K4 monomethylation”, “Cellular response to steroid hormone stimulus” and “Supramolecular fiber organization” (Figure 4E). Therefore, following DEHP exposure during PF formation, these results suggest that some biological consequences (such as “organization cell cycle” and “sexual reproduction”) persisted from pre-branch (Figure 4F, set 3), the “process metabolic biosynthetic” decreased (set 2), and “process regulation metabolic” increased (set 4). Moreover, the representative genes in each gene set are shown in Figure S5B; for set 4, with enrichment analysis of protein-protein interaction, the genes seemed to be involved in “chromatin organization”, “covalent chromatin modification”, and “histone modification” (Figure S5C).
Another noteworthy branch point existed in germ cell trajectory. In the comparative analysis, the heatmap of gene expression dynamics from State 4 to State 3 or 1 comprised two large gene sets (Figure S5D): one of known meiosis-related genes that were highly marked and expressed in State 1, such as Taf7l, Sycp1, Sycp3, and Smc1b; while the other included more key genes (Dppa3, Zp3, and Padi6) critical to PF assembly. Importantly, the gene expression level along with pseudotime indicated that State 4 was almost equidistant between States 1 and 3 (Figure S5E), therefore, the cells at State 4 appeared to be a transitional stage of female germ cells between cyst and follicle.
DEHP impacted the development of pre-granulosa cells at the prenatal stage
It is already known that pre-granulosa (PG) cells are required for PF formation. PG cells were clustered based on UMAP (Figure 5A). Given that the occurrence of two waves of ovarian follicles is supported by two distinct pathways of PG cells [44, 50], the PG cells in this study were divided into bipotential (BPG) and epithelial (EPG) types (Figure 5B). There were seven clusters of PG cells that showed a high expression of Amhr2 in the study, which were distinguished by Foxl2 and Lgr5, the marker genes of BPG and EPG cells, respectively (Figure 5B, right panel). Further, the clustering of BPG and EPG cells were separated, and six cell clusters for BPG and five for EPG were produced (Figure 5C-D); their expression patterns were consistent with previous research (Figure S6A-B) [50].
DEHP influenced the developmental status of germ cells during PF formation. (A) The pseudotime trajectory of germ cell populations colored by five States. Three branches were identified with two developmental fates. (B) The pseudotime trajectory of germ cell population colored by four sample groups. (C) Cell percentages of five States among four sample groups. (D) Pseudotime ordered heatmap of four differentially expressed gene sets between two obvious fates at branch point one. Several representative genes of each gene set are listed in the right panel. (E) The enrichment of GO terms of the top 100 genes in each gene set. (F) Enrichmentmap illustrated that DEHP exposure disturbed multiple processes in germ cells.
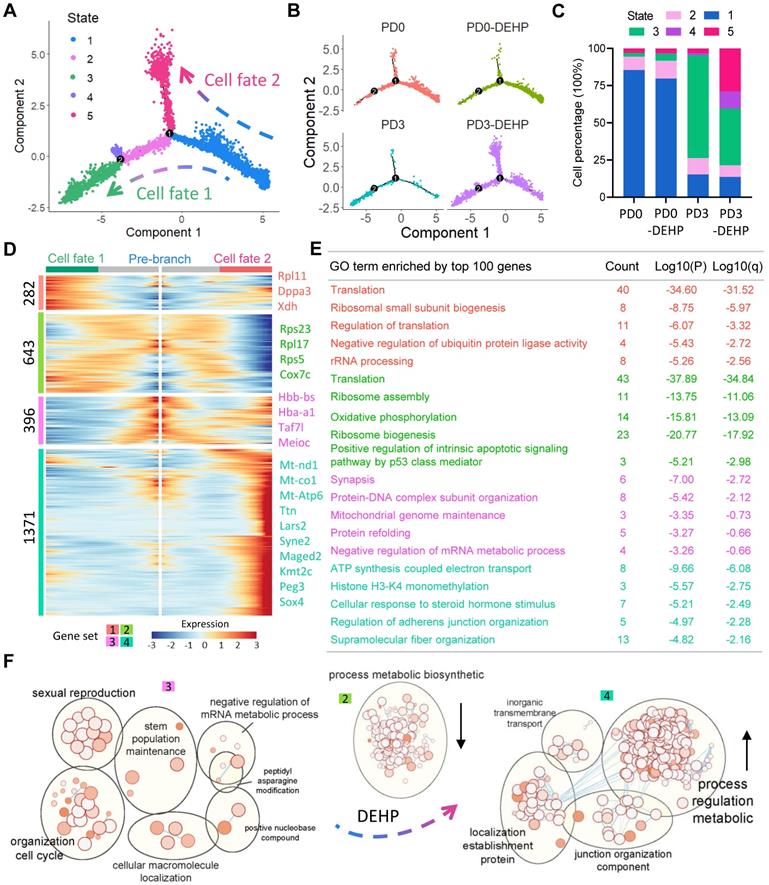
Gene differential expression analysis was applied to uncover the effects of DEHP exposure to PG cells. In BPG cells, exposure to DEHP significantly increased 186 genes and decreased 159 genes at PD0, while the number of DEGs up-regulated by DEHP at PD3 reached 279 and with 571 down-regulated (Figure 5E, Table S6). Similarly, for EPG cells, 149 and 474 genes were up-regulated by DEHP at PD0 and PD3, respectively, and 236 and 313 genes were down-regulated (Figure 5F, Table S7). In addition, 192 key DEGs in BPG and 248 key DEGs in EPG were affected by DEHP both at PD0 and PD3, respectively (Figure S6C-D). GO enrichment analysis indicated that the key DEGs of BPG cells were involved in the processes of “ribosome assembly”, “catabolic process”, “positive regulation of cell death”, and “reproductive system development” (Figure 5G). Meanwhile, genes of EPG cells were enriched in “cytoplasmic translation” and “regulation of transition”, as well as “positive regulation of cell death” and “reproductive system development” (Figure 5H). Furthermore, Metascape enrichment analysis of protein-protein interaction revealed that the key DEGs of BPG were enriched in terms of “regulation of cyclin-dependent protein kinase activity” and “positive regulation of cell cycle” (Figure S6E); while those of EPG were associated with “regulation of binding”, “protein localization to cell surface”, and “cellular response to cadmium ion” (Figure S6F). Notably, TRRUST analysis in Metascape indicated both DEGs of BPG and EPG were under the regulation of transcriptional factor Stat3 (Figure S6E-F, bottom panel).
DEHP impaired the interaction of female germ cells and pre-granulosa cells during PF formation
The two datasets of female germ cells and pre-granulosa cells during PF formation, were integrated to investigate the interaction (Figure 6A, and Figure S7A-B). Using the gene differential expression analysis above, the key DEGs affected by DEHP produced 194 genes in germ cells (DEGs detected by at least two strategies) and 138 genes in granulosa cells (Figure S7C-D). Enrichment of germ cells in the processes of “oxidative phosphorylation”, “cytoplasmic translation” and “cellular responses to stress”, and mTORC1 signaling were also included (Figure S7E). For granulosa cells, the terms of “ribosome assembly”, regulation of catabolic process and cell death, as well as WNT signaling and reactive oxygen species were also enriched (Figure S7F). It is noteworthy that the “Downregulation of SMAD2/3: SMAD4 transcriptional activity” term was enriched in both cell types. Thus, components of the TGF-beta pathway were highlighted in subsequent analysis (Figure 6B). In the results, most known elements regarding TGF-beta signaling were expressed by both or either of germ cells or granulosa cells. Results suggested that Gdf9 and Bmp2 were slightly expressed by germ cells and granulosa cells, respectively, while the receptors were mostly expressed by granulosa cells, and the target genes and effectors were highly expressed by germ cells. Moreover, the effect of DEHP on germ cells was not apparent, except for a reduction of Gdf9 at PD3 and a rise in the levels of Bmpr1a and Smad4 (Figure 6C, left). However, it caused upregulation of Smad4 in granulosa cells, as well as a reduction of other genes such as the receptors of Bmpr1a and Bmpr2, targets of Id2 and Id3, and effectors of Smad2 and Smad3 (Figure 6C, right). Furthermore, the ovarian section staining of GDF9 protein revealed its location in germ cells (Figure 6D), Western blot in the protein level exhibited a decline of GDF9 and LHX8 after DEHP treatment (Figure 6E), and BMPR1A and SMAD3 expression was also affected by DEHP exposure (Figure S7G-H).
DEHP leveraged apoptosis in both germ cells and somatic cells
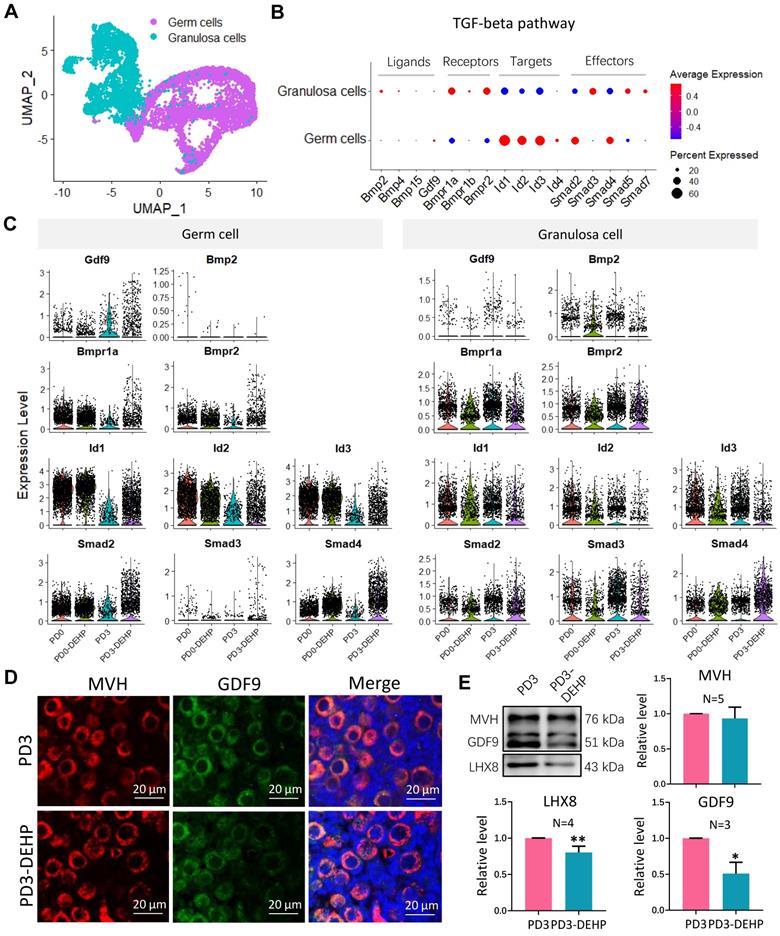
To uncover the destiny of germ cells in lagging cysts affected by DEHP, the DNA double strand break (DSB) and apoptotic processes were examined. The staining of ovarian sections showed that the double-positive of MVH and γH2AX (phosphorylated histone H2AX, served as a marker of DSB) in germ cells was raised by DEHP at PD3, as well as PD5 and PD7 (Figure 7A). In addition, the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay detected that germ cells and ovarian somatic cells both underwent apoptosis (Figure 7B), and the numbers of TUNEL positive cells in these cell types increased (Figure 7C). These results suggested that DEHP exposure of pregnant mothers would trigger the apoptosis of pup ovarian cells.
DEHP effects on immune cells and stromal cells during PF assembly
Further, the effects of DEHP on ovarian somatic cells were investigated. For immune cells, eight clusters were produced, and DEHP-treated cells at PD3 seemed to be more distinct from others (Figure S8A). Gene differential expression revealed that DEHP up- and down-regulated 10 genes respectively at PD0, and 26 genes were reduced but 88 genes were increased at PD3 (Figure S8B). GO terms of DEGs at PD0 enriched the biological processes of “multi-organism”, “immune system” and “localization”, while the top terms at PD3 were “immune system process”, “cell proliferation”, and “response to stimulus”. In terms of stromal cells, six clusters were generated, and the expression pattern of cells in the PD3-DEHP group was largely different from others (Figure S8D). Particularly, DEHP hardly affected stromal cells at PD0, with only one DEG (Birc5) being detected; at PD3, there were 12 down- and 75 up-regulated genes as a result of DEHP exposure (Figure S8E). Functional enrichment of DEGs at PD3 suggested the greatest enrichment of “cell proliferation”, “reproductive process”, and “cellular component organization or biogenesis” (Figure S8F).
DEHP impacted transcriptome characteristics of two groups of PGs during follicular assembly. (A) Clustering of pre-granulosa cells based on UMAP colored by the sample groups. (B) Two groups of PGs. Seven clusters with high Amhr2 expression (common marker gene of granulosa cell) are identified as PGs in the left panel; two groups of PGs are separated by the expression of Foxl2 (cyan) and Lgr5 (maroon). The color intensity represents gene expression level; grey refers to the double-negative cells with color threshold of gene expression at 0.5. (C) UMAP plot of bipotential PGs (BPGs) colored by sample groups (left) and cell clusters (right). (D) UMAP plot of epithelial PGs (EPGs) colored by sample groups (left) and cell clusters (right). (E) Volcano plots of gene differential expression at BPGs between pairs of PD0-DEHP vs. PD0 and PD3-DEHP vs. PD3. (F) Volcano plots of gene differential expression of EPGs between PD0-DEHP vs. PD0 and PD3-DEHP vs. PD3. (G) The GO term enrichment of key DEGs in BPGs. (H) The GO term enrichment of key DEGs in EPGs.
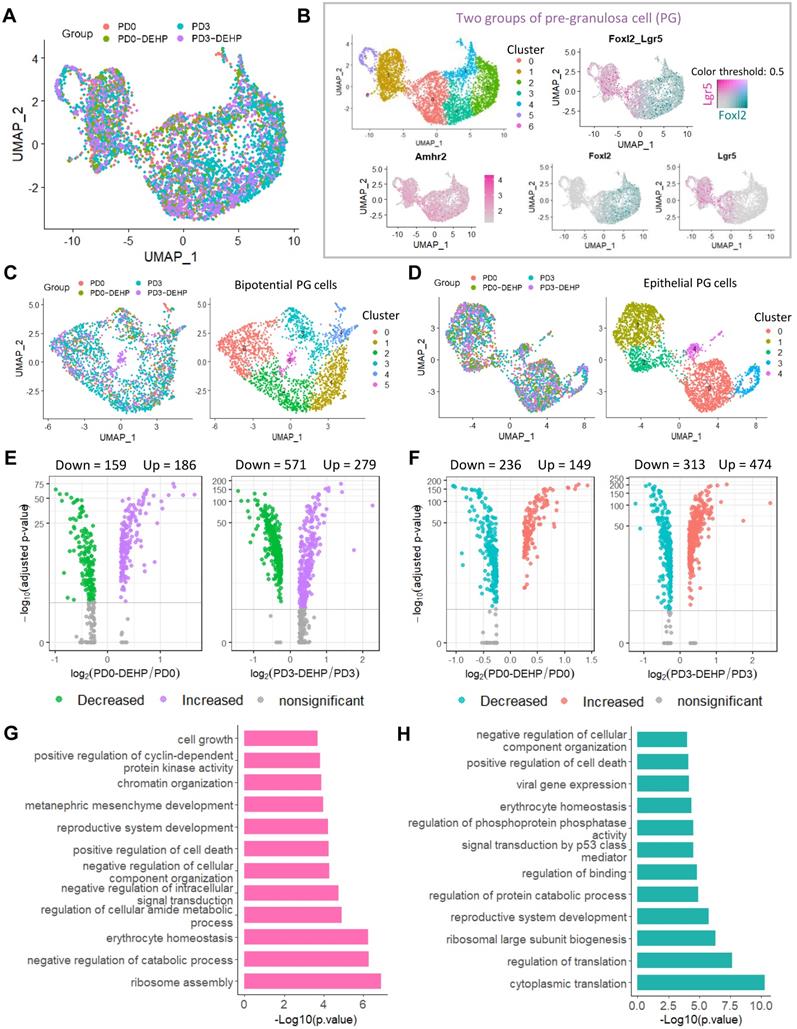
DEHP devastated TGF-beta signal between germ cells and granulosa cells and further impaired follicular assembly. (A) UMAP plot of separated germ cell and granulosa cell subpopulations. (B) Dot plot of components of TGF-beta signal (including signal factors, receptors, target genes, and effectors) between germ cells and granulosa cells. (C) Components of TGF-beta signal between sample groups in germ cells (left) and granulosa cells (right). (D) Histological section staining of MVH (red) and GDF9 (green) with and without DEHP treatment at PD3. Cell nuclei were stained with Hoechst 33342 (blue). Scale bar = 20 μm. (E) Detection of MVH, GDF9, and LHX8 protein levels at PD3 with and without DEHP exposure. MVH was used as a loading control to calculate relative protein levels of GDF9 and LHX8. Data are shown as mean ± SD (N represents independent repeats). * P < 0.05; **P < 0.01.
Discussion
DEHP is widely used as a plasticizer and is ubiquitous in every-day products; its toxicity has been well demonstrated by numerous studies [28, 51], including multiple human body systems, such as neural, endocrine, cardiotoxic, and reproductive [31, 52, 53]. Meanwhile, during PF assembly in mammals, the multicellular regulatory networks make the parsing of its mechanism tough and many biological enigmas remain unknown. In the current study, using a maternal perinatal exposure of DEHP from 16.5 dpc to postnatal day 3, scRNA-seq was used to investigate the alteration of transcriptome programs in pup ovaries to explore how the gene regulation networks and cell interaction were affected by DEHP during PF formation in mice.
Great progress has been made since single-cell transcriptomics has been applied to studies of the female reproductive system [43, 45, 49, 50]. Specifically, the transcriptome landscape of the whole process of folliculogenesis in humans has been described, and single-cell transcriptomic analyses of monkey ovaries have identified ovarian aging associated dysregulation of antioxidative pathways [54]. The current study identified six types of ovarian cells, which was similar to previous scRNA-seq reports using the same timeline in murine ovaries [49, 50]. Subsequently, the biological toxicity of DEHP on germ cells was highlighted. Firstly, from a single-cell perspective, DEHP decreased the expression levels of genes essential for PF assembly (Figure 8, right panel for germ cell), especially at PD3, such as Figla and Lhx8. Similarly, it is reported that DEHP exposure also decreased the levels of oocyte-specific genes, such as Lhx8, Figla, Sohlh1, and Nobox [25]. Furthermore, DEHP inhibited germline cyst breakdown and impaired PF assembly [26, 55, 56]. Nevertheless, the current study displayed an impeded progression of PF assembly through a decline of the cell percentages of early-stage follicles in PD0-DEHP and late-stage in the PD3-DEHP group. Further, the ovarian sections revealed that DEHP treatment significantly reduced the cell percentage of germ cells within follicles.
DEHP triggered apoptosis in germ cells and ovarian somatic cells. (A) Immunohistochemical staining of MVH (red) and γH2AX (green) in ovaries with and without DEHP treatment at PD3, PD5, and PD7. Cell nuclei were stained with Hoechst 33342 (blue). Scale bar = 50 μm. (B) TUNEL (green) and MVH (red) staining of ovarian sections at PD3. Cell nuclei were stained with Hoechst 33342 (blue). White arrows and pentagram mark the germ cells and granulosa cells with TUNEL, respectively. Scale bar = 50 μm. (C) The number of TUNEL positive germ cells (upper) and granulosa cells (lower) for each section at PD3. Data are shown as mean ± SD. **P < 0.01.
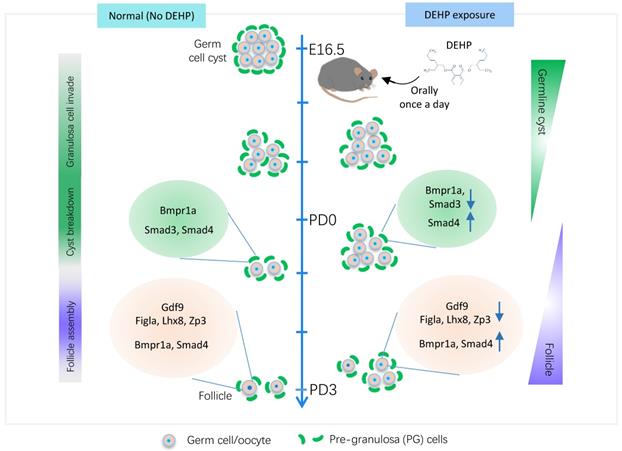
The transcriptional dynamics of DEHP decaying primordial follicle formation. The middle line marks the time window of primordial follicle formation, which represents germ cell fate transition from germline cysts to follicles (right annotation text), however, it consists of three main biological events, granulosa cell invasion, cyst breakdown, and follicle assembly (left annotation text). The left panel indicates normal follicle formation without DEHP, consisting of normal expression levels of Figla, Lhx8, and Zp3, and TGF-beta members (Gdf9, Bmpr1a, and Smad4) in germ cells, as well as TGF-beta receptors (Bmpr1a) and effectors (Smad3 and Smad4) in pre-granulosa cells. The right panel displays follicle assembly impaired by DEHP, which shows the decreasing transcripts of key genes for follicle formation (Figla, Lhx8, and Zp3) and of TGF-beta components in germ cells (Gdf9) and pre-granulosa cells (Bmpr1a and Smad3), but the increasing levels of Smad4 in both cell types and Bmpr1a only in germ cells.
Moreover, the process of cellular response to stress and DNA damage stimulus, as well as oxidative phosphorylation and transcriptional regulation by TP53, were all enriched after DEHP exposure. All these changes seemed to be associated with cellular stress, because TP53 (also termed tumor protein p53) is a DNA sequence-specific transcriptional regulator responsive to various forms of cellular stress and is involved in cell cycle arrest and apoptosis [57, 58]. Moreover, oxidative phosphorylation is the main source of reactive oxygen species (ROS) in mitochondria [59]. Oxidative phosphorylation and mitochondrial activity were also observed in pseudotime analysis, which were largely affected by DEHP. The in vitro study demonstrated that the oxidative stress-related gene Xdh (xanthine dehydrogenase) may serve as the downstream target of DEHP, whose reduction (through Xdh RNAi or melatonin supplement) can inhibit the increasing levels of ROS and mitigate the damage to PF assembly caused by DEHP [55]. Thus, DEHP, as one EDC member, might impair PF assembly through oxidative stress. In terms of molecular mechanisms, previous research suggests that ERs, particularly ER-beta, participate in the DEHP-mediated effect on ovarian tissue [26, 55]. In addition, the alteration of DNA methylation and transgenerational actions by EDCs in germ cells has been well documented [8, 24, 60-62]. In fact, H3K4 mono-methylation was likely affected by DEHP in the present study (Figure 4E). A recent study of Caenorhabditis elegans reports that DEHP exposure alters the expression of the reproduction-related gene spr-5, which encodes H3K4me2 demethylase [63]; however, a similar effect of DEHP on H3K4 mono-methylation in the mouse still needs to be studied.
The effect of DEHP on PG cells and its interaction with germ cells during PF formation was also explored. It is known that DEHP impairs the interaction of germ cells and PG cells via decreasing NOTCH2 signaling and impeding the proliferation of PG cells during PF assembly [26], which seemed to be evidenced, at least partially, by the enriched process of the regulation of cyclin-dependent protein kinase activity in the current study, as well as a positive regulation of the cell cycle in BPG. In addition, the death of both EPG and BPG cells was potentially affected by DEHP (Figure 5G-H). Previous research shows that DEHP exposure increases apoptosis of granulosa cells in adults, which leads to an increase in the number of atretic follicles [64, 65]. Similarly, during the perinatal period, TUNEL tests previously suggested that DEHP exposure in vitro increases apoptosis in oocytes of neonatal ovaries [25], which is consistent with this study (Figure 7B). Meanwhile, the components of the TGF-beta pathway in germ cells and PG cells were disturbed by DEHP (Figure 8, right panel for PG cells). For instance, Gdf9 on germ cells, and BMP receptors (Bmpr1a and Bmpr2) on PG cells, as well as targets (Id2 and Id3 on germ cells) and effectors (Smd2, Smd3 and Smad4 both on germ cells and PG cells), were all influenced by DEHP exposure (Figure 6B-C). Furthermore, GDF9 protein level was more lowly expressed, whilst BMPR1A and SMAD3 were up more highly expressed following DEHP exposure. Growing evidence indicates that TGF-beta signaling is important in the regulation of cyst breakdown and PF assembly [14, 15, 66]. Despite some available connections between DEHP and TGF-beta signaling [67, 68], DEHP toxicity mediated by TGF-beta signaling is still poorly understood, and needs to be further explored.
In conclusion, using scRNA-seq, the current study investigated the transcriptional dynamics of germ cells and PG cells following prenatal DEHP exposure during PF assembly. The results highlighted that DEHP impaired germline cyst breakdown and PF formation might be caused by oxidative stress in germ cells. Moreover, DNA damage and apoptosis were increased, as well as TGF-beta signaling being disturbed between both cell types. The current research presents a systematic understanding of DEHP damage on murine PF formation, and offers a preventive and therapeutic schedule for its biotoxicity in humans.
Materials and methods
Animals, reagents, and preparation for cell suspension
C57BL/6J strain mice, used only for single-cell sequencing, were purchased from Vital River Laboratory Animal Technology Co. Ltd (Beijing, China). For biochemical experiments, CD-1 mice were purchased from Jinan Pengyue Experimental Animal Breeding Co. Ltd (Jinan, China). The mice were raised in cages with ad-libitum food and water, a 12-h light:12-h dark cycle at a constant temperature (24 ℃) and humidity. Females were mated with reproductive males around 17:00 h. The following morning, a vaginal plug indicated a successful mating, and the time was recorded as 0.5 dpc. At 16.5 dpc, mice were treated with 40 μg/kg body weight DEHP via oral gavage once a day. Postnatal female pups (birth on 19.5 dpc) at 0 and 3 days (PD0 and PD3) were sacrificed for subsequent isolation of ovarian tissue. In accordance with national guidelines, all the murine experimental procedures were approved by the Animal Care and Ethical Committee of Qingdao Agricultural University.
For humans, the estimated DEHP dose of daily exposure ranges from 3 to 30 μg/kg body weight [7]. In the current study, DEHP was purchased from Sigma (Sigma-Aldrich, 36735-1G, Shanghai, China), the stock solution of which was prepared in DMSO. Before oral gavage, the working solution was diluted with sterile water. Regarding DEHP dose, 40 μg/kg body weight daily was adopted in this study, following reference to many in vivo publications [21, 51, 69] and previous papers [30, 34, 35].
For the preparation of single cells, the ovarian tissues (4-6 ovaries per sample) were cut into pieces within the digestive solution, which comprised 0.25% trypsin (Hyclone, Beijing, China) and collagenase (2 mg/ml, Sigma-Aldrich, C5138). Subsequently, the mixture was transferred into centrifuge tubes and placed in a 37 ℃ incubator for 6-8 min, and then agitated with a pipette once every 3 min to produce single cells. After the termination of digestion with serum, the mixture was then filtered using 40 μm cell strainers (BD Falcon, 352340, CA, USA), and flushes were performed three times with PBS containing 0.04% BSA. Finally, cell viability and counts were assessed to meet the requirements of sequencing.
Single-cell libraries and sequencing
The single cells were then loaded onto a 10× Chromium chip B (10× Genomics, Pleasanton, CA, USA) to generate single-cell gel beads in emulsion (GEMs), which was followed by cDNA amplification. A 3ʹ gene expression library was then constructed using a 10× Genomics Chromium Single Cell system with v3 chemistry (10× Genomics) according to the manufacturer's instructions. Sequencing was performed on a NovaSeq 6000 (Illumina) by Berry Genomics (Chengdu, China). Output reads (150 bp pair-ended) were yielded through Cell Ranger v3.1.5 using default parameters, including “mkfastq” and “count” pipelines. The detailed information of captured cells for each sample after “count” flow is shown in Figure S1B.
Data analysis using the Seurat and Monocle package
R package “Seurat” v3.1.2 was used for data pre-processing, and parameters were set and adjusted according to the features of captured transcripts in each sample. Doublets were examined by DoubletFinder v2.0.3 with its default threshold value of 0.075 [70]. Meanwhile, a double-check for every type was executed to discard cells with abnormal gene expression (for example, cells that were over-high in a number of unique detected genes, or cells within a single-cell type but with highly expressed specific marker genes in other cell types). The four objects were integrated via “Anchors” (an integration strategy between single-cell datasets for identification of cell pairwise correspondences and constructing atlases into their shared space) [71]; through normalization, datasets scaling, and test for optimal parameter combinations of “dim.use” and “resolution”, clustering was generated with a series commands of “FindNeighbors”, “FindClusters”, and “RunUMAP”. In addition, the “FindAllMarkers” function was applied to identify marker genes in clusters, while gene differential expression analysis was performed with default parameters; the significantly expressed genes (DEGs) were defined with |avg_logFC| > 0.25 and “p_val_adj” < 0.01.
The Monocle package (v2.10) [72] was used to construct a single-cell pseudotime trajectory using the functions of “reduceDimension” and “orderCells”. Ordering genes were selected based on highly variable genes identified by Seurat. In particular, “State” is Monocle's jargon for the segment of the trajectory tree in pesudotime analysis. Moreover, the “BEAM” function was used to calculate the dynamic genes at the branch point, and the genes with “qval < 1e-4” were displayed using heatmaps.
Functional enrichment and mapping
The online tool, Metascape [73] was used to perform the enrichment analyses of gene ontology (GO), and Kyoto encyclopedia and genomes (KEGG), including protein-protein interaction and transcriptional factor of TRRUST. EnrichmentMap was established using the g.profiler (https://biit.cs.ut.ee/gprofiler/gost) with the gene list as input and results were visualized in Cytoscape (v3.7.2) with cutoff at 0.01 of “FDR q-value” [74].
Histological section staining
Ovaries were collected and fixed in 4% paraformaldehyde (Solarbio, P1110, Beijing, China) overnight. Ovaries were embedded in paraffin and 5 µm sections were prepared. Following a standard procedure of gradient rehydration and antigen retrieval, sections were blocked at a room temperature around 20 ℃ for 45 min, then primary antibody incubation was performed overnight at 4 ℃. The primary antibodies included rabbit polyclonal to MVH (Abcam, ab13840, Shanghai, China), AMHR2 (OriGene, TA323994, Rockville, USA), GDF9 (Abcam, ab93892), and mouse monoclonal to γH2AX (Abcam, ab26350), STAT3 (Cell Signaling Technology, #91395, Danvers, USA), and MVH (Abcam, ab27591). The next day, the secondary antibodies (Abcam, Goat Anti-Rabbit IgG H&L Alexa Fluor 488, ab150077, and Anti-Mouse, ab150113; Donkey Anti-Rabbit IgG H&L Alexa Fluor 555, ab150074, and Anti-Mouse, ab150106) were incubated at 37 ℃ for 30 min. After nuclei staining with propidium iodide (PI, Sigma-Aldrich, P4170) or Hoechst 33342 (Beyotime, C1022, Shanghai, China), each slide was sealed with anti-fluorescence attenuation solution (Boster, AR1109, Wuhan, China). Photographs were then taken with a LEICA TCS SP5 II confocal microscope (Leica, Wetzlar, Germany). In addition, a single-germ cell without connection to other germ cells was considered as a germ cell in a follicle, while inter-connected germ cells including two or more in number were regarded as germ cells in cysts. Meanwhile, every one of five ovarian sections was counted, and a total of eight to ten sections for each ovary was viewed as one independent biological duplicate.
TUNEL assay
The stained ovarian sections were examined using a TUNEL BrightGreen Apoptosis Detection Kit (Vazyme, A112-03, Nanjing, China). According to the instructions for usage, after incubation of the secondary antibody for MVH in histological staining workflow, the slides were washed twice with PBS and treated with proteinase K for 10 min. Following two further washes with PBS, the slides were incubated with 1 × Equilibration Buffer for 25 min at room temperature, and the BrightGreen Labeling Mix was applied at 37 ℃ for 80 min. Then, the samples were counterstained with Hoechst 33342 for cell nuclei and sealed for observation under a fluorescence microscope (Olympus, BX51, Japan).
Western blot
Reference was made to previously developed protocols [55, 75]; proteins from six ovaries were extracted with RIPA lysis solution (Beyotime, P0013C). After denaturation treatment, the protein of each sample was separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene fluoride (PVDF, Millipore, ISEQ00010, USA) membrane. Subsequently, protein bands were blocked at a temperature of approximately 20 ℃ for 2 h, and the primary antibody was incubated at 4 ℃ overnight. The following day, after three washes, the secondary antibodies were incubated. Finally, a BeyoECL Plus kit (Beyotime, P0018) was used for chemiluminescence. The primary antibodies used for Western blot consisted of MVH (Abcam, ab13840), GDF9 (Abcam, ab93892), LHX8 (Abcam, ab137036), BMPR1A (Affinity, DF6634, Shanghai, China), SMAD3 (Abcam, ab40854), and GAPDH (Affinity, AF7021).
Statistical analysis
Results are shown as mean ± SD from at least three independent experiments. Statistical analyses were performed with GraphPad Prism software (version 8.0). Significant differences were determined using an unpaired t-test; significant and highly significant differences were defined at * P < 0.05 and ** P < 0.01, respectively.
Supplementary Material
Supplementary figures.
Supplementary table 1: Top-50 marker genes of cell clusters in ovaries.
Supplementary table 2: Top-50 marker genes of germ cells.
Supplementary table 3: Result of differential gene expression analysis at pre-stage of follicle formation in germ cells.
Supplementary table 4: Result of differential gene expression analysis at early-stage of follicle formation in germ cells.
Supplementary table 5: Result of differential gene expression analysis at late-stage of follicle formation in germ cells.
Supplementary table 6: Result of differential gene expression analysis in BPG cells.
Supplementary table 7: Result of differential gene expression analysis in EPG cells.
Acknowledgements
This work was funded by National Nature Science Foundation of China (31970788) and Taishan Scholar Construction Foundation of Shandong Province of China (ts20190946).
Author Contributions
JW, YT, ML, YF, LK, and FZ conducted the animal experiments; JW and YT analyzed the single-cell data; JW and WS proposed the manuscript; WS designed the experiments. All authors reviewed the manuscript and reached a common agreement.
Data availability
The raw data of single-cell sequencing at PD0 and PD3 for ovarian tissues following DEHP exposure have been deposited in the Genome Sequence Archive of National Genomics Data Center (https://bigd.big.ac.cn), under the accession number: CRA003354. The R scripts used for data processing are publicly available on GitHub at https://github.com/WangLab401/DEHP-scRNA.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Lovekamp Swan T, Davis BJ. Mechanisms of phthalate ester toxicity in the female reproductive system. Environ Health Perspect. 2003;111:139-45
2. Chiang Hc, Kuo YT, Shen CC, Lin YH, Wang SL, Tsou TC. Mono (2-ethylhexyl) phthalate accumulation disturbs energy metabolism of fat cells. Arch Toxicol. 2016;90:589-601
3. Peijnenburg W. Phthalates. Encyclopedia of Ecology. 2008:2733-8
4. Li N, Zhou L, Zhu J, Liu T, Ye L. Role of the 17β-hydroxysteroid dehydrogenase signalling pathway in di-(2-ethylhexyl) phthalate-induced ovarian dysfunction: An in vivo study. Sci Total Environ. 2020;712:134406
5. Liu JC, Lai FN, Li L, Sun XF, Cheng SF, Ge W. et al. Di (2-ethylhexyl) phthalate exposure impairs meiotic progression and DNA damage repair in fetal mouse oocytes in vitro. Cell Death Dis. 2017;8:e2966
6. Heudorf U, Mersch Sundermann V, Angerer J. Phthalates: toxicology and exposure. Int J Hyg Environ Health. 2007;210:623-34
7. Hannon PR, Flaws JA. The effects of phthalates on the ovary. Front Endocrinol. 2015;6:8
8. Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466-9
9. Trubo R. Endocrine-disrupting chemicals probed as potential pathways to illness. JAMA. 2005;294:291-3
10. Yilmaz B, Terekeci H, Sandal S, Kelestimur F. Endocrine disrupting chemicals: exposure, effects on human health, mechanism of action, models for testing and strategies for prevention. Rev Endocr Metab Disord. 2020;21:127-47
11. Monniaux D, Clément F, Dalbiès-Tran R, Estienne A, Fabre S, Mansanet C. et al. The ovarian reserve of primordial follicles and the dynamic reserve of antral growing follicles: what is the link? Biol Reprod. 2014;90:85 1-11
12. Ford EA, Beckett EL, Roman SD, McLaughlin EA, Sutherland JM. Advances in human primordial follicle activation and premature ovarian insufficiency. Reproduction. 2020;159:R15-R29
13. Grive KJ, Freiman RN. The developmental origins of the mammalian ovarian reserve. Development. 2015;142:2554-63
14. Wang C, Zhou B, Xia G. Mechanisms controlling germline cyst breakdown and primordial follicle formation. Cell Mol Life Sci. 2017;74:2547-66
15. Pepling ME. Follicular assembly: mechanisms of action. Reproduction. 2012;143:139-49
16. Yan H, Wen J, Zhang T, Zheng W, He M, Huang K. et al. Oocyte-derived E-cadherin acts as a multiple functional factor maintaining the primordial follicle pool in mice. Cell Death Dis. 2019;10:1-14
17. Kerr JB, Myers M, Anderson RA. The dynamics of the primordial follicle reserve. Reproduction. 2013;146:R205-15
18. Zhang H, Panula S, Petropoulos S, Edsgärd D, Busayavalasa K, Liu L. et al. Adult human and mouse ovaries lack DDX4-expressing functional oogonial stem cells. Nat Med. 2015;21:1116-8
19. Findlay JK, Hutt KJ, Hickey M, Anderson RA. How is the number of primordial follicles in the ovarian reserve established? Biol Reprod. 2015;93:111 1-7
20. Hsueh AJ. Fertility: the role of mTOR signaling and KIT ligand. Curr Biol. 2014;24:R1040-2
21. Johansson HKL, Svingen T, Fowler PA, Vinggaard AM, Boberg J. Environmental influences on ovarian dysgenesis—developmental windows sensitive to chemical exposures. Nat Rev Endocrinol. 2017;13:400-14
22. Skakkebæk NE, Meyts RD, Main K. Testicular dysgenesis syndrome: an increasingly common developmental disorder with environmental aspects: Opinion. Hum Reprod. 2001;16:972-8
23. Fowler PA, Bellingham M, Sinclair KD, Evans NP, Pocar P, Fischer B. et al. Impact of endocrine-disrupting compounds (EDCs) on female reproductive health. Mol Cell Endocrinol. 2012;355:231-9
24. Pocar P, Fiandanese N, Berrini A, Secchi C, Borromeo V. Maternal exposure to di (2-ethylhexyl) phthalate (DEHP) promotes the transgenerational inheritance of adult-onset reproductive dysfunctions through the female germline in mice. Toxicol Appl Pharmacol. 2017;322:113-21
25. Zhang T, Li L, Qin XS, Zhou Y, Zhang XF, Wang LQ. et al. Di-(2-ethylhexyl) phthalate and bisphenol A exposure impairs mouse primordial follicle assembly in vitro. Environ Mol Mutagen. 2014;55:343-53
26. Mu X, Liao X, Chen X, Li Y, Wang M, Shen C. et al. DEHP exposure impairs mouse oocyte cyst breakdown and primordial follicle assembly through estrogen receptor-dependent and independent mechanisms. J Hazard Mater. 2015;298:232-40
27. Zhang Y, Mu X, Gao R, Geng Y, Liu X, Chen X. et al. Foetal-neonatal exposure of Di (2-ethylhexyl) phthalate disrupts ovarian development in mice by inducing autophagy. J Hazard Mater. 2018;358:101-12
28. Ge W, Li L, Dyce PW, De Felici M, Shen W. Establishment and depletion of the ovarian reserve: physiology and impact of environmental chemicals. Cell Mol Life Sci. 2019;76:1729-46
29. Rodríguez HA, Santambrosio N, Santamaría CG, Muñoz-de-Toro M, Luque EH. Neonatal exposure to bisphenol A reduces the pool of primordial follicles in the rat ovary. Reprod Toxicol. 2010;30:550-7
30. Lai FN, Liu JC, Li L, Ma JY, Liu XL, Liu YP. et al. Di (2-ethylhexyl) phthalate impairs steroidogenesis in ovarian follicular cells of prepuberal mice. Arch Toxicol. 2017;91:1279-92
31. Li XN, Li HX, Yang TN, Li XW, Huang YQ, Zhu SY. et al. Di-(2-ethylhexyl) phthalate induced developmental abnormalities of the ovary in quail (Coturnix japonica) via disruption of the hypothalamic-pituitary-ovarian axis. Sci Total Environ. 2020;741:140293
32. Doyle TJ, Bowman JL, Windell VL, McLean DJ, Kim KH. Transgenerational effects of di-(2-ethylhexyl) phthalate on testicular germ cell associations and spermatogonial stem cells in mice. Biol Reprod. 2013;88:112 1-15
33. Zhou C, Gao L, Flaws JA. Exposure to an environmentally relevant phthalate mixture causes transgenerational effects on female reproduction in mice. Endocrinology. 2017;158:1739-54
34. Li L, Zhang T, Qin XS, Ge W, Ma HG, Sun LL. et al. Exposure to diethylhexyl phthalate (DEHP) results in a heritable modification of imprint genes DNA methylation in mouse oocytes. Mol Biol Rep. 2014;41:1227-35
35. Zhang XF, Zhang T, Han Z, Liu JC, Liu YP, Ma JY. et al. Transgenerational inheritance of ovarian development deficiency induced by maternal diethylhexyl phthalate exposure. Reprod Fertil Dev. 2015;27:1213-21
36. Stévant I, Kühne F, Greenfield A, Chaboissier MC, Dermitzakis ET, Nef S. Dissecting Cell Lineage Specification and Sex Fate Determination in Gonadal Somatic Cells Using Single-Cell Transcriptomics. Cell Rep. 2019;26:3272-83 e3
37. Luecken MD, Theis FJ. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol Syst Biol. 2019;15:e8746
38. Choi YH, Kim JK. Dissecting cellular heterogeneity using single-cell RNA sequencing. Mol Cells. 2019;42:189-99
39. Wang M, Liu X, Chang G, Chen Y, An G, Yan L. et al. Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell. 2018;23:599-614 e4
40. Hermann BP, Cheng K, Singh A, Roa De La Cruz L, Mutoji KN, Chen IC. et al. The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 2018;25:1650-67 e8
41. Sohni A, Tan K, Song HW, Burow D, de Rooij DG, Laurent L. et al. The neonatal and adult human testis defined at the single-cell level. Cell Rep. 2019;26:1501-17 e4
42. Law NC, Oatley MJ, Oatley JM. Developmental kinetics and transcriptome dynamics of stem cell specification in the spermatogenic lineage. Nat Commun. 2019;10:1-14
43. Ge W, Wang JJ, Zhang RQ, Tan SJ, Zhang FL, Liu WX. et al. Dissecting the initiation of female meiosis in the mouse at single-cell resolution. Cell Mol Life Sci. 2020:1-19
44. Zheng W, Zhang H, Gorre N, Risal S, Shen Y, Liu K. Two classes of ovarian primordial follicles exhibit distinct developmental dynamics and physiological functions. Hum Mol Genet. 2013;23:920-8
45. Fan X, Bialecka M, Moustakas I, Lam E, Torrens Juaneda V, Borggreven N. et al. Single-cell reconstruction of follicular remodeling in the human adult ovary. Nat Commun. 2019;10:3164
46. Zhang Y, Yan Z, Qin Q, Nisenblat V, Chang HM, Yu Y. et al. Transcriptome Landscape of Human Folliculogenesis Reveals Oocyte and Granulosa Cell Interactions. Mol Cell. 2018;72:1021-34 e4
47. Wagner M, Yoshihara M, Douagi I, Damdimopoulos A, Panula S, Petropoulos S. et al. Single-cell analysis of human ovarian cortex identifies distinct cell populations but no oogonial stem cells. Nat Commun. 2020;11:1-15
48. Zhao ZH, Ma JY, Meng TG, Wang ZB, Yue W, Zhou Q. et al. Single-cell RNA sequencing reveals the landscape of early female germ cell development. The FASEB Journal. 2020;34:12634-45
49. Wang JJ, Ge W, Zhai QY, Liu JC, Sun XW, Liu WX. et al. Single-cell transcriptome landscape of ovarian cells during primordial follicle assembly in mice. PLoS Biol. 2020;18:e3001025
50. Niu W, Spradling AC. Two distinct pathways of pregranulosa cell differentiation support follicle formation in the mouse ovary. Proc Natl Acad Sci U S A. 2020;117:20015-26
51. Chiang C, Flaws JA. Subchronic exposure to Di (2-ethylhexyl) phthalate and Diisononyl phthalate during adulthood has immediate and long-term reproductive consequences in female mice. Toxicol Sci. 2019;168:620-31
52. Xu X, Yang Y, Wang R, Wang Y, Ruan Q, Lu Y. Perinatal exposure to di-(2-ethylhexyl) phthalate affects anxiety-and depression-like behaviors in mice. Chemosphere. 2015;124:22-31
53. Posnack NG. The adverse cardiac effects of di (2-ethylhexyl) phthalate and bisphenol A. Cardiovasc Toxicol. 2014;14:339-57
54. Wang S, Zheng Y, Li J, Yu Y, Zhang W, Song M. et al. Single-cell transcriptomic atlas of primate ovarian aging. Cell. 2020;180:585-600 e19
55. Liu JC, Li L, Yan HC, Zhang T, Zhang P, Sun ZY. et al. Identification of oxidative stress-related Xdh gene as a di (2-ethylhexyl) phthalate (DEHP) target and the use of melatonin to alleviate the DEHP-induced impairments in newborn mouse ovaries. J Pineal Res. 2019;67:e12577
56. Zhang T, Shen W, De Felici M, Zhang XF. Di-(2-ethylhexyl) phthalate: Adverse effects on folliculogenesis that cannot be neglected. Environ Mol Mutagen. 2016;57:579-88
57. Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951-61
58. Hollstein M, Hainaut P. Massively regulated genes: the example of TP53. J Pathol. 2010;220:164-73
59. Scialò F, Fernández Ayala DJ, Sanz A. Role of mitochondrial reverse electron transport in ROS signaling: potential roles in health and disease. Front Physiol. 2017;8:428
60. Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253-62
61. Iqbal K, Tran DA, Li AX, Warden C, Bai AY, Singh P. et al. Deleterious effects of endocrine disruptors are corrected in the mammalian germline by epigenome reprogramming. Genome Biol. 2015;16:59
62. Chao HH, Zhang XF, Chen B, Pan B, Zhang LJ, Li L. et al. Bisphenol A exposure modifies methylation of imprinted genes in mouse oocytes via the estrogen receptor signaling pathway. Histochem Cell Biol. 2012;137:249-59
63. Li SW, How CM, Liao VHC. Prolonged exposure of di (2-ethylhexyl) phthalate induces multigenerational toxic effects in Caenorhabditis elegans. Sci Total Environ. 2018;634:260-6
64. Xu C, Chen JA, Qiu Z, Zhao Q, Luo J, Yang L. et al. Ovotoxicity and PPAR-mediated aromatase downregulation in female Sprague-Dawley rats following combined oral exposure to benzo [a] pyrene and di-(2-ethylhexyl) phthalate. Toxicol Lett. 2010;199:323-32
65. Inada H, Chihara K, Yamashita A, Miyawaki I, Fukuda C, Tateishi Y. et al. Evaluation of ovarian toxicity of mono-(2-ethylhexyl) phthalate (MEHP) using cultured rat ovarian follicles. J Toxicol Sci. 2012;37:483-90
66. Trombly DJ, Woodruff TK, Mayo KE. Roles for transforming growth factor beta superfamily proteins in early folliculogenesis. Semin Reprod Med. 2009 27. 14-23
67. Ma H, Li J, Yang JJ, Tian WC, Liu H, Zhou YZ. et al. Di-2-ethylhexyl phthalate and its metabolite single-ethylhexyl phthalate affect TGF-beta 1 expression and telomerase activity in the testis of young male rats. Zhonghua Nan Ke Xue. 2012;18:783-8
68. Zhou Y, Huang F, Liu Y, Li D, Zhou Y, Shen L. et al. TGF-β1 relieves epithelial-mesenchymal transition reduction in hypospadias induced by DEHP in rats. Pediatr Res. 2020;87:639-46
69. Hannon PR, Peretz J, Flaws JA. Daily exposure to Di (2-ethylhexyl) phthalate alters estrous cyclicity and accelerates primordial follicle recruitment potentially via dysregulation of the phosphatidylinositol 3-kinase signaling pathway in adult mice. Biol Reprod. 2014;90:136 1-11
70. McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019;8:329-37 e4
71. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck III WM. et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888-902 e21
72. Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381-6
73. Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1-10
74. Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A. et al. Pathway enrichment analysis and visualization of omics data using g: Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. 2019;14:482-517
75. Sun ZY, Zhang P, Wang JJ, Liu JC, Li L, Shen W. et al. Melatonin alleviates meiotic defects in fetal mouse oocytes induced by Di (2-ethylhexyl) phthalate in vitro. Aging. 2018;10:4175-87
76. Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189-96
Author contact
![]() Corresponding author: Prof. Wei Shen, E-mail: wshenedu.cn; shenwei427com
Corresponding author: Prof. Wei Shen, E-mail: wshenedu.cn; shenwei427com
 Global reach, higher impact
Global reach, higher impact