13.3
Impact Factor
Theranostics 2021; 11(10):4872-4893. doi:10.7150/thno.56409 This issue Cite
Review
Recent progress of surface-enhanced Raman spectroscopy for subcellular compartment analysis
1. State Key Laboratory of Supramolecular Structure and Materials, Institute of Theoretical Chemistry, College of Chemistry, Jilin University, Changchun 130012, People's Republic of China.
2. School of Pharmaceutical Sciences, Key Laboratory of Innovative Drug Development and Evaluation, Hebei Medical University, Shijiazhuang, 050017, China.
3. Department of Molecular Sciences, ARC Centre of Excellence for Nanoscale BioPhotonics (CNBP), Macquarie University, Sydney, New South Wales 2109, Australia.
Received 2020-11-25; Accepted 2021-1-25; Published 2021-3-4
Abstract
Organelles are involved in many cell life activities, and their metabolic or functional disorders are closely related to apoptosis, neurodegenerative diseases, cardiovascular diseases, and the development and metastasis of cancers. The explorations of subcellular structures, microenvironments, and their abnormal conditions are conducive to a deeper understanding of many pathological mechanisms, which are expected to achieve the early diagnosis and the effective therapy of diseases. Organelles are also the targeted locations of drugs, and they play significant roles in many targeting therapeutic strategies. Surface-enhanced Raman spectroscopy (SERS) is a powerful analytical tool that can provide the molecular fingerprint information of subcellular compartments and the real-time cellular dynamics in a non-invasive and non-destructive way. This review aims to summarize the recent advances of SERS studies on subcellular compartments, including five parts. The introductions of SERS and subcellular compartments are given. SERS is promising in subcellular compartment studies due to its molecular specificity and high sensitivity, and both of which highly match the high demands of cellular/subcellular investigations. Intracellular SERS is mainly cataloged as the labeling and label-free methods. For subcellular targeted detections and therapies, how to internalize plasmonic nanoparticles or nanostructure in the target locations is a key point. The subcellular compartment SERS detections, SERS measurements of isolated organelles, investigations of therapeutic mechanisms from subcellular compartments and microenvironments, and integration of SERS diagnosis and treatment are sequentially presented. A perspective view of the subcellular SERS studies is discussed from six aspects. This review provides a comprehensive overview of SERS applications in subcellular compartment researches, which will be a useful reference for designing the SERS-involved therapeutic systems.
Keywords: organelles, SERS, label, label-free, subcellular analysis
Introduction
Subcellular compartments
Subcellular compartments, including cell membrane, nucleus, mitochondrion, lysosome, and endoplasmic reticulum (ER), etc., are the essential functional subunits within eukaryote cells [1, 2]. Each of them plays a specific and indispensable role in supporting the normal functions of cells. As the control center of the cell, the nucleus plays prominent roles in cell proliferation, growth, differentiation, metabolism, and apoptosis [3]. Mitochondrion, the “powerhouse” of cells, its primary task is to generate adenosine triphosphate (ATP) and the necessary metabolites to fulfill the survival demands of the cell [4]. The lysosome is an acidic subcellular compartment containing numerous hydrolytic enzymes, such as proteases and nucleases. Moreover, lysosome is also involved in various physiological processes, such as cholesterol homeostasis, plasma membrane repair, secretion, cell death, and cell signaling [5, 6].
Given the essential roles of these cellular compartments in cellular activities, it can be speculated that any dysfunction or disruption of the structures and functions of subcellular compartments can result in abnormal intracellular homeostasis and various cellular disorders. For example, disturbances to the nucleus may lead to the abnormal regulation of cell activity and even cause cell death. Various diseases, such as cancers, heart disorders, and brain disorders, are also related to nuclear dysfunction [7]. The deficiency of mitochondrion directly affects the energy supply and leads cells to die. Moreover, cell survival and aging are also related to the function of the mitochondrion. Its abnormal function would accompany neurodegenerative diseases such as Alzheimer's disease and multiple sclerosis [8, 9]. Many cell functions are related to the lysosome. It is unsurprising that lysosomal dysfunction causes many diseases [10, 11], especially the lysosomal storage disease, one of the most severe diseases that can cause secondary changes, such as autophagy dysfunction, mitochondrial dysfunction, and inflammation. Therefore, the precise and accurate detections of the structures, functions, and microenvironments of organelles are of great significance for deeply understanding their roles in cell life activities and the diagnosis and treatment of diseases.
Besides, targeting therapeutic agents at specific subcellular organelles is the key to maximize the therapeutic index, completely eradicate tumors, and prevent invasion, metastasis, and recurrence. In the context of cancer therapy, the biggest bottleneck is how to deliver the drugs to the lesion locations, since the simple transportations of therapeutic agents into the tumors or cancer cells are not efficient and enough to yield the expected therapeutic outcome [12]. During chemotherapy [13, 14], reactive oxygen species (ROS) generated in the therapeutic processes are suggested to be close to these bioactive substances, so that the therapeutic effect and the weak side effect can be ensured [15].
At present, the organelle-related analyses mainly focus on two aspects: (1) the detections of structures and compositions of organelles, and (2) the explorations of microenvironments of organelles (e.g., pH, ROS, reactive nitrogen, and ATP, etc.) and the dynamic changes of their structures and microenvironments during therapeutic processes. Traditional subcellular compartment analytical methods are protein immunoblotting [16, 17] and gel electrophoresis [18, 19], in which the cellular components need to be extracted for further analyses. Mass spectroscopy and deoxyribonucleic acid (DNA) sequencing-based genomics spectrometry [20] are widely applied for the quantitative determinations of specific elements. Besides these methods, many in situ characterization techniques have been involved due to the merits of monitoring the dynamic changes of a living cell. Most of them are microscopic techniques since the cells and bacteria are in a micrometer size range. Established microscopic technologies, e.g., electron microscope [21], atomic force microscope [22], and optical technologies, e.g., super-resolution fluorescence microscopy [23] and surface-enhanced Raman spectroscopy (SERS) [24], have been widely applied.
SERS
Raman spectroscopy is one kind of vibrational spectroscopies, which can provide native and rich chemical information of cellular compositions, including nucleic acids, proteins, and lipids. Compared to broad fluorescence bands, Raman bands are narrow and easy to identify. Besides, water has fewer impacts on the Raman spectra. Therefore, compared to infrared spectroscopy, Raman spectroscopy is more suitable for the detections of biological systems.
However, Raman signals are too weak to be detected because only one Raman photon can be excited out of 107 incident photons, limiting its application in biofields. One of the remarkable achievements in the developing history of Raman scattering is the discovery of SERS, first reported by Fleishmann et al. in 1974 [25]. This technique can amplify the Raman signal of molecules by 4 to 11 orders [26]. It gains ultrahigh sensitivity even down to the single-molecule level under the strong electromagnetic enhancement of plasmonic nanomaterials [27], which endows SERS a high detection sensitivity comparable to fluorescence no matter in sensing or imaging. Besides many improvements in the SERS detection sensitivity, SERS substrates make significant progress as well. They start from the silver electrodes to diverse metal nanostructures in the recent forty years. With the continuous efforts on metal nanostructures/nanoparticles (NPs) in preparation, surface modification, assembly, and grafting technologies within near twenty years, the SERS-active nanomaterials have become flexible, adjustable, and designable, which promote SERS advances in bio and medical applications [28-31].
In this review, SERS as a useful analytical tool in subcellular components based detection and therapy were retrospected and summarized (Scheme 1). Two commonly used SERS strategies in biological applications, labeling and label-free methods, were briefly introduced. Some experimental notes were given, and SERS applications for the subcellular compartment detections, including cell membrane, nucleus, mitochondrion, and lysosome, were reviewed. The organelle-based therapy mechanism and the molecular dynamics during the therapeutic processes were presented as a frontier topic. We also described our concerns and perspectives on the future of subcellular SERS applications.
A scheme of the concepts/strategies of the SERS-based subcellular detections.
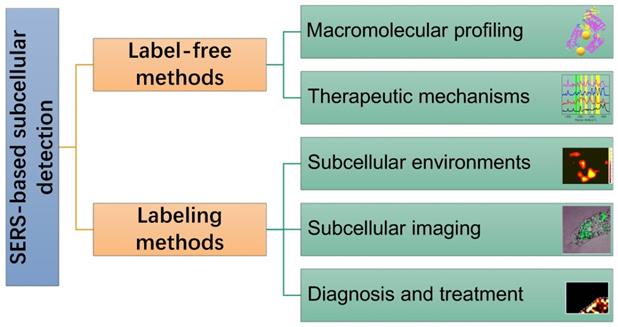
Strategies for subcellular SERS studies
Uniqueness and advantages of SERS for subcellular compartment analyses
SERS has gained ever-increasing attention, recently focusing on cellular studies. This high interest arises mainly from the uniqueness of SERS (molecular specificity and great sensitivity two-in-one) and highly matching between the SERS specialties and the demands of cellular/subcellular investigations. SERS exhibits applicability in subcellular detections from the following aspects.
(1) SERS has an acceptable spatial resolution level for main cell compartments. The cells and prominent organelles, such as cell nuclei (1-20 μm), mitochondria (0.5-1 μm), and lysosome (less than 1 μm), are all at the micrometer level [32], while the plasmonic nanometals are at the nanometer level. Thus, SERS can satisfy the spatial resolution requirements in subcellular detections and give good spatial discrimination on the subcellular domains via the commercial confocal Raman microspectrometers. SERS micro-imaging not only can provide two-dimensional information, but also can reveal surface, interface, or even layer-by-layer information of cells and organelles, enabling three-dimensional chemical structure reconstruction [33].
(2) The localized field enhancement effect allows for selective measurements of aimed locations. SERS is a highly surface-dependent effect [34]. The physical mechanism discloses that the localized surface plasmon resonance effect provides an amplified electromagnetic field that only localizes around the metal surface. Moreover, the chemical mechanism of SERS that exists in typical charge transfer processes can't stand without the surface. Thus, only the probed molecules extremely close to the nanometal surfaces can be detected. It becomes possible to selectively analyze the interested analytes with the help of the organelle-targeted metal nanoparticles (MNPs) previously internalized in a specific organelle.
(3) SERS can explore living cells in an in-situ way. SERS features the common points of optical approaches that are non or less invasive and show minimum biological sample damage [33, 35]. It can be applied for the living cell analysis and the dynamic tracking of physiological and metabolism processes.
Label-free/labeling methods
Subcellular SERS studies follow the conventional SERS detection strategies, which can be sorted as direct/indirect measurements. In some descriptions, they are mentioned as label-free/labeling methods, as shown in Scheme 2.
The label-free method discloses the molecular vibration information of analytes by placing them near the SERS substrates, or the analytes directly contacting the SERS substrates [26]. The most significant advantage of the label-free SERS is that it can provide the intrinsic information of the molecules and avoid false-positive. The direct SERS can analyze molecular dynamic changes in some life activities and the interactions between the analytes and other substances [36]. Besides, the label-free method can provide the conformation and orientation information of adsorbed biomolecules. People have successfully applied the label-free SERS for the differentiation and identification of cells or bacteria [37, 38], cell activities such as cell mitosis [24], apoptosis [39], metabolism [40], intracellular biological macromolecules responding to external stimuli, and other metabolites in living cells [41].
Illustration of the label-free and labeling detection methods.
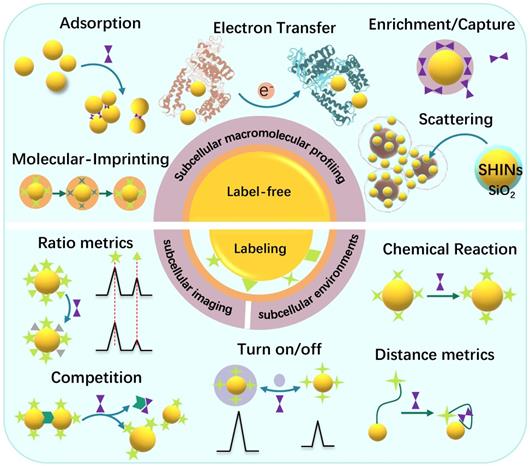
Besides the label-free SERS, another SERS analytical strategy is the SERS labeling method. The SERS reporters and the recognition ligands are modified on the surface of plasmonic NPs, and these integrated complexes are called SERS tags or SERS labels [42, 43]. In a stimuli-responsive process, when the target appears, the ligands can respond, causing the SERS signal changes of reporters [26]. Alternatively, both SERS activity and recognition function are integrated into one, simplifying the labeling strategy [44]. Either the target molecules or the physical or chemical properties of the samples can trigger the SERS signals of the reporter molecules to change. SERS labeling methods are suitable for the detection systems that the analyte without identifiable Raman signal or the target can't be detected by the label-free methods, especially some small molecules or subcellular microenvironments, e.g., local pH, and intracellular temperature changes and some molecules with low Raman activity.
The quantitative sensing mechanisms of SERS nanosensors are mainly cataloged in two types: (1) intensity (ratio)-dependent SERS nanosensors. By constructing or destroying the hotspot structures of the targets, or the signal changes relative to the internal standards, the “SERS on” or “SERS off” is entirely dependent on the concentration of analytes [45-47]. (2) Raman shift changes of the SERS reporters. According to the interactions between the reporters with the analytes or the environments, e.g., charge transfer [48], chemical reaction [49], and host-guest interaction [50], the Raman band shifts of the SERS reporter will be observed, and the correlation between the peak shift and analyte concentration will be built.
SERS labeling methods exhibit broad usage in multi-component detection and imaging [51-53]. The labeling methods lose the advantage of providing material intrinsic information relative to the label-free detections; however, they possess ultra-high detection sensitivity for quantitative determinations. In SERS imaging and mapping experiments, traceable SERS tags can provide high sensitivity comparable to many fluorescence dyes. Still, its narrow fingerprint bands allow the greater possibility for multiplex detections than the fluorescent dyes that are limited to only three channels (blue, green, and red) in the visible light range.
Fundamentals of subcellular targeted detection and therapy
For subcellular targeting researches, delivering the SERS-active nanoplatforms into specific subcellular compartments is one of the most critical steps. For nanomaterials internalization, phagocytosis and pinocytosis are two major cellular internalization mechanisms for exogenous NPs. Most particles will arrive in the lysosome or cytosol eventually. To help the SERS-active nanoplatforms arriving in our interested organelles, so far, two main strategies have been employed and developed to deliver nanocarriers to specific subcellular organelles, noted as passive targeting and active targeting mechanisms.
Passive targeting
Passive targeting refers to the selective accumulation of NPs or nanosystems in specific organelles due to the physical, chemical, and pharmacological factors of the organelles (membrane permeability, potential, and internal environment, etc.) and the inherent properties of NPs, including size, shape, and surface charge. For example, small molecules such as water and some ions can freely enter and exit the nucleus since the nuclear pores on the surface of the nuclear membrane are about 9 nm in size [54]. Accordingly, the nuclear-targeted delivery of nanomaterials can be successfully achieved by controlling their molecular weight or size [54]. With the rapid development of nanotechnology, various ultra-small nanomaterials were developed as potential nuclear pore penetration materials [55]. The internalization of nanomaterials in mitochondria is through a different way. Mitochondrion has a membrane potential of nearly -180 mV, and such a negative potential allows positively charged nanomaterials in the mitochondria autonomously [56]. Accordingly, many mitochondrion-targeting synthetic materials were designed by enriching them with positive charges, e.g., a positively ratiometric Raman probe was prepared to achieve the selective mitochondria targeting and live-cell imaging of hydrogen sulfide in cancer cells [57]. Although passive targeting shows apparent organelle-targeting to a certain degree, it is still limited in its applicability for all kinds of NPs. For example, only ultra-small nanomaterials can be delivered into the nucleus; however, for the NPs with larger sizes, it will be challenging to deliver them to the nucleus if no targeting ligand modification. Therefore, passive targeting is not a universal method to achieve nucleus targeting, and the applications are limited.
Active targeting
Developing universal targeting strategies that are independent of the inherent natures of NPs and organelles is in great demand. Active targeting by modifying targeting ligands such as aptamers and targeting peptide sequences on the surface of the nanomaterials has been extensively exploited, in which the specific targeting peptide sequences are the most commonly used targeting ligands. The surface decoration of the cell-penetrating peptide (RGD, RGDRGDRGDRGDPGC) is helpful for the internalization of nanomaterials. RGD is known to target αvβ6, an RGD receptor, as well as other αv integrins on the cell surface and assists the receptor-mediated endocytosis in cancer cells [58-60].
Shi's group reported a variety of nuclear targeting nano-delivery systems by conjugating the nucleus targeting peptide on the surface of the nanomaterials for cancer targeting therapy [61-63]. They used nuclear targeting sequence is the TAT peptide (YGRKKRRQRRR). This sequence can translocate NPs into cell nuclei by binding the import receptors importin α and β (karyopherin) and subsequently targeting the nuclear pore complex of cancer cells through entering their nuclei [64-66].
In order to obtain the mitochondrion-targeting, the mitochondrion-targeting peptide (MLALLGWWWFFSRKKC) [67, 68] and the membrane potential targeting based on the mitochondriotropics molecules are commonly used to facilitate the selective accumulation in mitochondria, such as the TPP cation conjugated NPs, [69, 70].
At present, the modification of nanocarriers with intelligent response-type lipophilic amines is the most effective strategy for realizing the long-stay of nanocarriers in the lysosome and prohibiting the nanocarriers from escaping from lysosomes. Lysosomes give an acidic microenvironment, which makes amino groups protonated. Thus, NPs with the protonated amino groups easily penetrate the lysosomal membrane, resulting in abundant accumulation of NPs in lysosomes. For other organelles except lysosomes, the microenvironment is in a neutral condition, which can't trigger the protonation reactions, and thus the lysosome-selective targeting is realized. 4-(2-hydroxyethyl) morpholine and N, N-dimethylethylenediamine are two commonly used lysosomal targeting groups, and both of them have been used for the surface decoration of nanomaterials [71, 72].
Less-invasive interventional operation
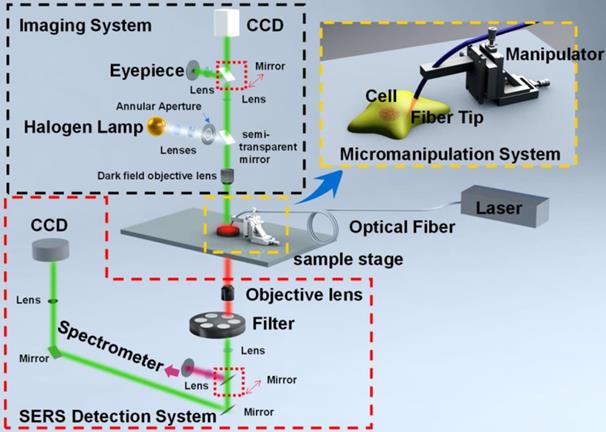
Optical fiber tip [73, 74], carbon nanotube [75], needle tip [76, 77], can be applied for micro-operation-based in vivo cell SERS measurements [79]. These methods break through the traditional SERS substrates in detecting micro-regions by the initiative or passive internalization of MNPs. Differently, they can accurately reach the interesting positions of cells. The tips are mainly composed of single-core quartz fiber, hollow borosilicate glass tubes, or hollow quartz glass tubes, which can be easily fabricated into sharp tips and have good toughness during cell insertion. The ends of microprobes are functionalized with plasmonic NPs, which are highly sensitive to the intracellular microenvironments, enabling the SERS detections of characteristic spectral signatures of cell nucleus and cytoplasm. These nanotips were fixed on a microscope, while a micro-manipulation system is needed (Figure 1). The microscope with 10× objective was used to observe and guide the insertion of the nanotip under the white light. After the tip approaching the surface of cells, a high power objective was switched to record the SERS spectra of the cellular compartments with the laser irradiation and high-resolution spectrometer. By dynamic spectral processing and analysis, these microprobes provide an effective method for monitoring the dynamics and physiology of a single cell [40, 79, 80].
The schematic diagram of the micromanipulation-Raman detection system. Adapted with permission from [80] Copyright 2019 Elsevier B.V.
Notes for experimental aspects
Impacts of SERS-active MNPs on cell physiological activity
Since these internalized MNPs are exogenous matters for cells, they will bring certain interference to the cells. Commonly, for the intracellular SERS measurements, the AuNPs are more acceptable than AgNPs for the consideration of better biocompatibility of Au, especially for the nanotherapeutic experiments in living bodies. So far, Au nanorods (AuNRs) [81], stars [82], spheres [36], and core/shell NPs [83], gap-enhanced Raman tags (GERTs) [84, 85], have been reported for intracellular compartment studies.
The concentration of the SERS-active nanoprobes should be assessed before intracellular SERS measurements, by testing the cellular viability after the cells co-culture with nanoprobes for different time (experimental time). The cell viability can be simply determined by using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) or WST-1 (2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2, 4-disulfophenyl)-2H, tetrazolium monosodium salt) etc. assays, in which the cells were properly metabolized by a dye. They undergo visible color changes that are monitored by spectrophotometer, while the cells incapable of metabolizing the dye remain colorless. Also, flow cytometry is another powerful tool for cell viability tests. Taking spherical AuNPs as an example, over 85% of cellular viability based on the MTT assay is considered low/no toxicity and widely acceptable for intracellular experiments. Above 90% of cell viability is preferred. Otherwise, they will cause severe cell apoptosis. However, in order to achieve high SERS sensitivity, more SERS nanoprobes will be helpful. Thus, a trade-off concentration of nanoprobe is expected.
Surface engineering of MNPs
SERS tags are usually functionalized by both Raman reporters with distinguished signatures and the functional coating layers that endow the SERS tags with biocompatibility/stability and/or targeting bioconjugation. In many reported SERS nanoprobes, the surface coating of polyethylene glycol (PEG) is preferred, which offers protection from the chemical and physical environments, minimizing non-specific binding and prolonging blood circulation lifetimes in living body experiments [86]. PEG is also a feasible linker for the conjugation of biomolecules (e.g., antibodies and peptides), and it can anchor biomolecules to noble metal surfaces through the carboxyl or amino groups. However, some ligands, e.g., cetyltrimethylammonium bromide (CTAB), may give identifiable peaks in the SERS profiles. These functional ligands may compete for the limited surface of nanoprobes with the probed components, which goes against the SERS detections of cellular compartments. So, the Raman signals of the used ligands should be confirmed in advance to make sure their contributions are distinguishable. In some cases, the bands of ligands can also be the internal standards for SERS intensity calibration in quantitative determinations.
The addition of functional coating molecules onto the NP surface tends to affect colloidal stability and brings irreversible NP aggregation, especially in the centrifugation step. This issue also limits the large-scale SERS tag synthesis. How to avoid the colloid aggregation during the SERS nanoprobe preparation is very skillful and bothered each beginner. Thus, the ligand amount must be strictly controlled, and the surface decoration of colloids needs well designed.
The surfaces of MNPs are active for the unspecific bindings of uninterested molecules. To avoid these SERS background signals, commonly used biocompatible protective layers - silica or bovine serum albumin (BSA) coatings [87] were utilized to block the nanoprobe surface and both of which can minimize the particle aggregations. Ren et al. proposed an iodide-modified AgNPs, and the iodide can avoid the nonspecific binding of molecules [88], remarkably improving the reliability and reproducibility of the label-free SERS detections.
Photostability and chemical stability of nanoprobes
Similar to the photobleaching of the fluorescent dyes used for imaging and bio-labeling, the SERS nanoprobes also face the fading problem. The irregular variation or fluctuation of SERS signals may cause the SERS blinking effect, which is the typical behavior of a single molecule caused by the diffusion effects of analytes (Brownian diffusion) [89] or the surface charge fluctuation-caused vibronic coupling [90]. During long-term imaging, the intensity decay was also commonly observed, which is explained by the situations that the reporter molecules diffuse away from the hot spots or they are gradually destroyed by the local photothermal effect around hot spots. To maintain the SERS photostability of nanoprobes upon continuous laser irradiation, people synthesized the core-shell NPs with the embedded Raman reporters, which exhibit excellent photostability performance during the SERS imaging due to the metal protection shell layer and the off-resonance based low photothermal effect [91, 92].
A silica shell can also be a protective layer to maintain colloidal stability and prevent interaction with the external environments. The primary strategy for silica shell decoration is achieved by the mercaptotrimethoxysilane or aminopropyltrimethoxysilane (APTMS) and then reacted with sodium silicate or tetraethylorthosilicate. A remarkable example is the shell-isolated NPs (SHINs, Scheme 2). The chemical stability of the MNPs was significantly improved, and long-term storage was achieved [93].
Photodamage
The local photothermal energy not only burns the Raman reporters but also directly damages the cells and tissues during the SERS measurements. Though the photodamage effect of MNPs can be utilized for killing cancer cells according to their large absorption cross-sectional area of near-infrared light, in most SERS measurements of living cells or bodies, photodamage is undesirable. Laser power should be optimized for safety. Lim et al. [94] set different laser powers for the Raman imaging of human oral cancer cells (HSC-3), and the significant change of cell morphology and the cell death (red staining) was observed when 4.0 and 2.0 mW laser were set (785 nm wavelength Ti: Sapphire laser, 3900S, Spectra-Physics), while for 0.2 mW of laser power, no significant changes of cell morphology were observed. Xia et al. [95] quantified the photothermal treatment effect of 65 nm immune gold nanocages on SK-BR-3 breast cancer cells with a pulsed near-infrared laser (805 nm with a bandwidth of 54 nm, 4.77 W/cm2). In the absence of Au cages, the cells could alive under the laser power of 6.0 W/cm2 for 5 min, but they started to die at a laser power of 1.5 W/cm2 if the Au cages exist. In order to avoid the photodamage on the biosamples, a defocusing way can be applied to minimize the effects of laser damage if the vertical spatial resolution can be ignored [96].
Subcellular compartment SERS detections
In this section, more detailed examples applying SERS label-free and labeling technologies for subcellular compartment detections will be provided, including subcellular macromolecular profiling, subcellular imaging, subcellular environments, cell membrane detections, and isolated organelles.
Label-free SERS for subcellular macromolecular profiling
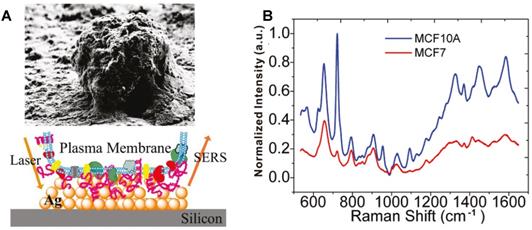
Researchers designed and synthesized various subcellular targeting nanoprobes to obtain the SERS spectra of different subcellular organelles by delivering noble metal nanomaterials (Au and Ag NPs) to the target locations. The surface of the cell membrane is rich in receptors and targets. In 2010, Yan et al. obtained the SERS spectra of the cellular surface of the tumor and nontumor cell lines under the assistance of the drop-coating AgNPs on a silicon wafer (Figure 2A). The quantitative analysis revealed that the characteristic differences between tumor and nontumor cells, especially in the spectral range from 600 to 900 cm-1 (Figure 2B), indicating the possibility of improving the precision of current cancer detection and staging approaches through the spectroscopic profiling of the cell surface cancer biomarkers [97].
The SEM image of an individual cell and the field enhancement generated in the vicinity of AgNPs on the silicon. B SERS spectra of the MCF7 (red) and MCF10A (blue) breast cell lines obtained from the Ag decorated silicon. Adapted with permission from [97]. Copyright 2010 American Chemical Society.
Oyelere et al. prepared the AuNR-based nuclear targeting probes by the grafting of a simian virus (SV40) NLS peptide (CGGPKKKRKVGG) on the AuNRs through a thioalkyl-triazole linker [98]. Based on these nanoprobes, the SERS spectra of malignant and normal epithelial cells were collected to reveal their differences in molecular structure information, which is expected to provide a new way for cancer cell detections. Xu et al. [99] developed a modified strategy in which 5-ethynyl-2'-deoxyuridine (Edu) was adopted as an internal label to locate the nuclear region accurately. In the case of the nuclear targeting AuNRs (decorated with NLS peptide), the accompanying signal of Edu in the Raman silent range can verify whether the SERS spectrum was recorded from the nucleus or not. Thus, the nanoprobe locating and in situ SERS detection of the cellular nucleus can be synchronously completed.
To obtain the SERS information of the entire mitochondria, Karatas et al. isolated mitochondria from A549 lung cancer cells and mixed them with AuNPs to obtain the SERS spectra of mitochondria [100]. Since the redox state and conformation of cytochrome c in mitochondria can affect the overall state of the mitochondrial electron transport chain, as well as the lipid composition and the apoptosis-related processes, Brazhe et al. prepared a silver layered ring substrate to record the conformation and the redox state of the cytochrome c under various conditions [101].
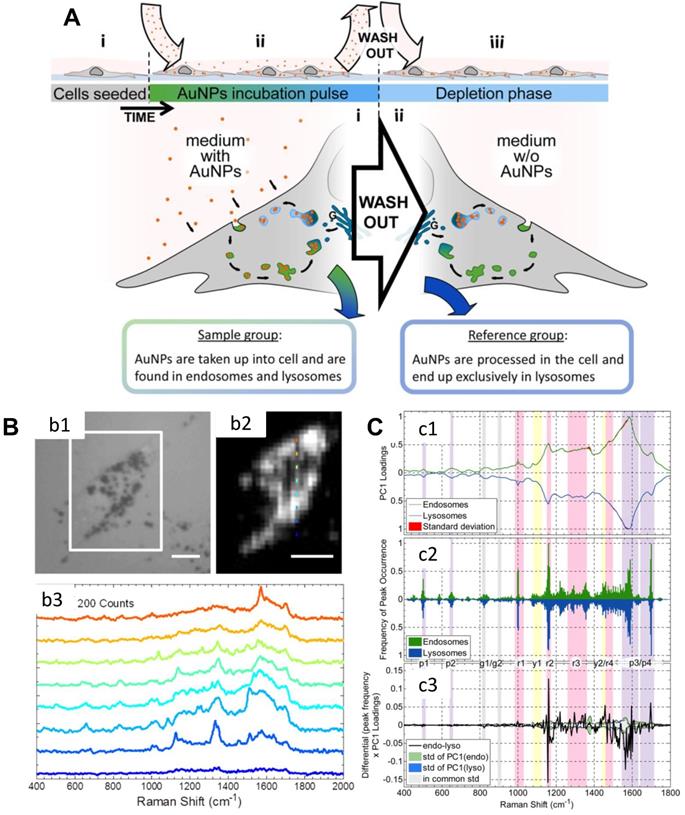
Nanomaterials enter cells through the clathrin-mediated endocytosis pathway, and they will accumulate in the lysosome or cytoplasm at the first stage [102]. Mahajan et al. used label-free SERS to identify and visualize the stages of endocytosis of AuNPs in cells (Figure 3) [103]. They cultured AuNPs with the cells by a pulse method to allow AuNPs to accumulate inside endosomes and lysosomes. Thus, they obtained the SERS signals of endosomes and lysosomes during the AuNPs' endocytosis. With the principal component analysis-linear discriminant analysis (PCA-LDA), several important life processes can be revealed by SERS [103].
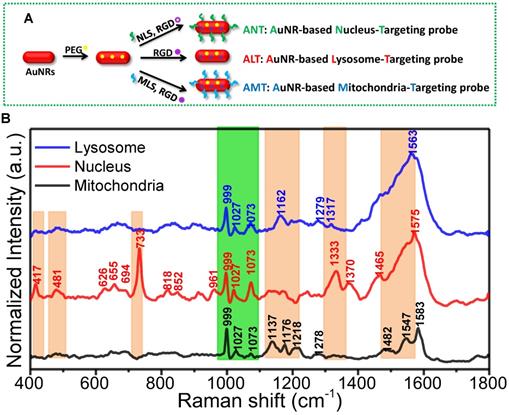
To simultaneously observe and compare the molecular information of different organelles, Xu and Liang et al. designed and prepared the nuclear, mitochondrial and lysosomal targeting AuNR nanoprobes for recording the SERS spectra of three organelles (as shown in Figure 4A). First, the targeting ability of these nanoprobes was identified by super-resolution imaging and transmission electron microscopy (TEM). Under the assistance of these AuNRs-based nanoprobes, the macromolecular profiles of the three subcellular organelles were partly profiled and compared for the first time [104] (Figure 4B).
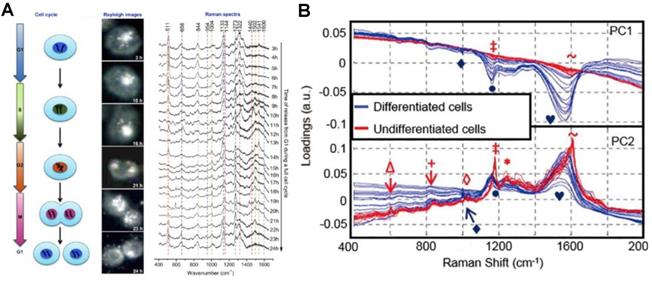
In addition to the subcellular molecular profiling, the SERS strategy can also be used to record the subcellular spectra during dynamic cellular processes, including cell division, differentiation, and apoptosis. El-Sayed group demonstrated the real-time SERS monitoring of the molecular dynamics of mitosis in healthy and cancer cells at the molecular level (Figure 5A) [24]. The nuclear targeting AuNPs were used to track the molecular changes of cancer and normal cells during the cell mitotic process. The results showed that the conformation rate of mitotic proteins from their α-helix structure to the β-sheet is higher in cancer cells during the mitosis process. However, in healthy cells, the existence of proteins is mainly in the α-helix conformation. The roles of protein conformation dynamics during mitosis in cancer development and many other diseases were revealed by this study. In 2013, Mahajan's group used the nuclear targeting probes to detect SERS spectra of different neuronal cells to obtain the SERS spectra of the cytoplasm and nucleus of neuronal cells (Figure 5B). SERS spectra were analyzed with the PCA to distinguish the differentiated and undifferentiated neuronal cells [105].
Although modifying the targeting peptide on the MNP surface can achieve effective organelle targeting, inevitably, some of them will randomly distribute in the cytoplasm, not in the targeted organelles, which limits the accuracy of detection in organelle based studies. The development of nano-tip technology effectively resolves this problem. In 2009, Vitol et al. developed the SERS-active nanoneedles for in situ molecular observation in cells for the first time [106]. The prepared SERS-active nanoneedles can accurately locate in the nucleus and cytoplasm to accurately record the SERS spectra of the cytoplasm and nucleus. This approach provides a new path for cell injection, single-cell surgery, or in situ studies of organelle based cell processes.
The applications of label-free SERS in subcellular studies are mainly about the molecular information profiling of organelles such as proteins, DNA, lipids, and their dynamic changes in critical cellular processes [26, 35]. In order to improve detection sensitivity, nonlinear spectroscopic techniques have been employed, e.g., stimulated Raman scattering (SRS) and coherent anti-Stokes Raman scattering (CARS) [107, 108], which are promising not only in cellular imaging but also in tissue imaging.
A) Scheme of the experimental design. B) (b1) The bright-field image of a cell; (b2) The corresponding SERS map of (b1) (integrated over 400-2000 cm-1); Raman spectra of the different positions of (b2) are shown in (b3). Scale bars: 10 µm. C) Analysis of spectra obtained from the cellular pathway. (c1) Normalized PC1 loadings of all extracted spectra originating from endosomes (green) and lysosomes (blue). (c2) The summary of the (normalized) occurrence probability of the SERS peaks of endosomes and lysosomes. (c3) Difference between the PC1 loadings (c1) according to the occurrence frequency of the peak (c2). The corresponding standard deviations of endosomes, lysosomes and both groups are shown in green, blue and gray. Adapted with permission from [103] Copyright 2015 American Chemical Society.
A) The preparation of the different organelle targeting nanoprobes. B) The normalized SERS spectra of the nucleus, mitochondria, and lysosome under the assistance of the subcellular targeted nanoprobes. The common and different spectra were shown in orange and green bands. Adapted with permission from [104]. Copyright 2018 American Chemical Society.
A) The process of a cell cycle, real-time dark-field images and the SERS spectra of the nucleus in a single HSC-3 cell during the complete cycle. B) PCA analysis of nuclear SERS spectra of undifferentiated and differentiated cells, shown as a scatter plot and 1D intensity plots for PC1 against PC2 scores. Adapted with permission from [24] and [105]. Copyright 2012 and 2013 American Chemical Society.
SERS labels for subcellular imaging
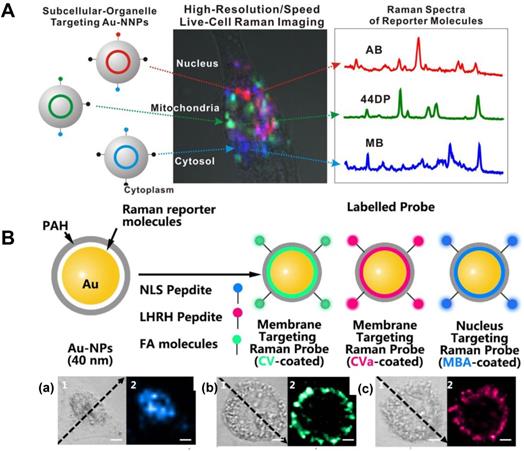
SERS tags with high sensitivity can be one type of location tracers at a subcellular level. In 2015, Lim et al. inserted three different Raman-active molecules into the gap of the Au@Au core-shell structure and further modified them with different organelle targeting peptides for organelle location and imaging [94]. The hot spots generated by the nanogap allow them to create high-speed, high-resolution, multiplexed subcellular organelle imaging (Figure 6A).
The expression of the cell surface species, such as glycoproteins and glycolipids, can be the indexes of various diseases. In situ visualization of them on cell membranes may provide the correlations of proteins, lipids, and glycosylation with the developing states and the metastasis of diseases. Shen et al. prepared the nucleus and membrane targeting nanoprobes by modifying AuNPs with different targeting groups and reporters [53]. The high-resolution 3D image of a single cell, especially the cell membrane and nucleus, were respectively obtained. The chemical properties of the nucleus and the distribution of folic acid (FA) and luteinizing hormone-releasing hormone (LHRH) on the surface of the cell membrane were accurately located and detected (Figure 6B). A zone-controllable SERS approach was developed by Ju et al. for SERS imaging the protein-specific glycosylation on the cell surface using two typed nanoprobes. The AuNPs (10 nm)-based nanoprobe was designed by the surface functionalization of Raman reporters and dibenzocyclooctyne-amine. This nanoprobe not only displays negligible SERS signals, but also can recognize and link the azide-tagged glycan on the cell membrane surface via the click reaction. The aptamer modified AuNPs (30 or 40 nm) that can specifically identify the target proteins were then used for amplifying SERS signal by forming nanoaggregates. This method was used for in situ imaging of sialic acid (SA) on the target protein epithelial cell adhesion molecule (EpCAM), as well as their dynamic changes during drug treatment [109]. Subsequently, Ju's group developed another nanoprobe by decorating the Au nanoflowers with the benzoic group functionalization, which were used as a bridge for both recognition of the target SA and assembly of poly(nacetylneuraminic acid) modified AuNPs. Via the plasmonic coupling of these two kinds of nanoprobes in a single core-multi satellite nanostructure, they realized the sensitive SERS imaging of SA on the surface of living cells. Moreover, they also designed a micro-competition system for the simultaneous quantification of multiple glycans on cell surfaces with SERS [110].
A Scheme of the Au@Au core-shell organelle targeting nanoprobes and the SERS spectra of the nucleus, mitochondria, and cytosol after the cell cultured with different organelle targeting nanoprobes. Adapted with permission from [94] Copyright 2015 American Chemical Society. B Scheme for the AuNPs coated with CV and CVa for targeting the membrane, and the MBA-coated AuNPs for targeting the nucleus (top). The corresponding bright-field and SERS imaging of a HeLa cell treated with the targeted nanoprobes of MBA-coated AuNPs (a), CV-coated AuNPs (b), and CVa-coated AuNPs (c) (bottom). Adapted with permission from [53] Copyright 2016 Macmillan Publishers Limited.
Based on the recognition of the benzoic group with SA, Xu et al. developed a 4-mercaptophenylboronic acid (MPBA)-functionalized AgNP-based SERS nanosensor to in situ study sialoglycan levels in different cell types and the dynamic expression processes of them under the physiological conditions [44]. This nanosensor is unique and multifunctional due to its three-in-one role involving the Raman signal enhancer (AgNPs), the sensing reporter of MPBA, and the target receptor. When it binds to sialoglycans, the vibrational modes of MPBA will change, which can be used to track the sialoglycan expression on cell membranes. Recently, in combination with the microfluidic droplet system, they established a plasmonic microdroplet platform for the SA expression in a single cell via the MPBA@AgNPs SERS nanosensor [111], which avoids the interference of the neighbor cells. The single-cell SA expressions on different cell lines (HepG2, SGC-7901, MCF-7, and BNL.CL2) were compared [111]. The single-cell analysis of the sialoglycan expression reveals the cellular diversity and heterogeneity.
SERS nanosensors for subcellular environments
Subcellular microenvironments such as pH, the content of ROS, biomolecules, and redox potential are important parameters for ensuring normal cell growth, proliferation, and others [112, 113]. However, any fluctuation of subcellular microenvironments can result in various disorders. Therefore, the in situ detections of the subcellular microenvironments will facilitate the understanding of the mechanisms of subcellular related diseases during the occurrence and development processes. In this section, we will introduce some recent progress in SERS tag-based subcellular microenvironment sensing, especially intracellular pH detections.
In 2020, Choi et al. reported an efficient SERS-based immunoassay technique for protein quantification and imaging. Their method can reliably estimate the protein level at a sub-picomolar detection limit [114]. Besides, Ye et al. designed the SERS core-shell NPs with an embedded standard for the non-destructive detection of cholesterol [115]. Wang et al. developed a simple SERS-based nanotip system to perform subcellular localization and detect nucleolin (NCL) in a single living cell based on the aptamer recognition [116]. Their detection platform was available for multiplex targets by decorating the tips with different aptamers. Similarly, in combination with SERS technology and nanotip technology, Xu et al. designed an optical waveguide excited pH SERS-active system by modifying a pH-responsive molecule, 4-mercaptopyridine (Mpy), on the optical fiber tip [80]. The system can detect the pH of the nucleus and cytoplasm. The results showed that the nucleus had a lower pH value than the cytoplasm. Moreover, the probe was used to evaluate the pH environment of normal cells and cancer cells inside/outside the cell. In short, these developed platforms realize the real-time monitoring of the distribution and the content of biomolecules at the subcellular level.
The preparation of organelle targeting pH nanosensors (A) and the procedure for subcellular pH determinations by SERS spectroscopy with the organelle targeting pH nanosensors (B). Adapted with permission from [117]. Copyright 2018 Royal Society of Chemistry.
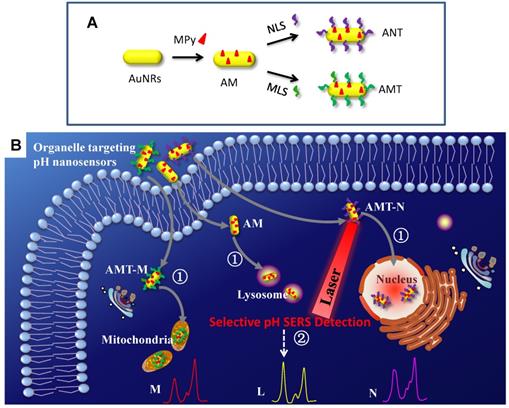
A series of SERS nanosensors with high sensitivity and organelle-targeting ability have been developed for monitoring the subcellular pHs [117]. As shown in Figure 7, three nanosensors were obtained (Figure 7A) by modifying a pH-responsive molecule Mpy and different organelle targeting peptides on the surface of AuNRs. By monitoring the vibrational spectral changes of Mpy, the nuclear, mitochondrial, and lysosomal pHs were obtained and compared (Figure 7B). The developed SERS sensors can also explore more crucial physiological and biological processes related to subcellular pH.
In addition to those nanoprobes that should be internalized in cells for subsequent subcellular environment analyses, many needle-like optrodes and nanopipettes were utilized for tracing the single-cell low-copy proteins and components. Wang et al. decorated the outer surface of glass nanotubes with Au and Raman-active molecules to detect cancer biomarker, free iron ions and hemoglobin in a single cell via the aptamer hybridization [118]. Liu et al. [119] modified monoclonal antibody or molecularly imprinted polymer on the surface of Au-coated nanotubes to achieve specific minimally invasive extraction and in vitro labeling detections. Gogotsi et al. [75] developed a carbon nanotube-tipped endoscope for intracellular biomolecular detection and realized the in-situ determination of the molecular composition of the organelles. Compared with the intracellular SERS applied by nanoprobes, the nanotip-based method can arrive at interesting locations. However, since the micromanipulation under the microscope has limited imaging resolution and low contrast for other organelles except for the cell nucleus, these optrodes are only applicable for detecting cell nucleus, cytoplasm, and extracellular regions.
SERS measurement of isolated organelles
Although the rich and dynamic biomacromolecular information can be obtained by in situ SERS, there is still a problem unsolved on the difficulty to assign these bands because the intracellular biological environment is incredibly complicated, and there are some interference signals from other biomolecules. Therefore, some ex-situ strategy has emerged for structural analysis and quantitative analysis of the isolated organelles.
Culha et al. investigated the feasibility of SERS to understand the mode of interaction of AuNPs with the extracted mitochondria. They found that the AuNPs did not cause significant damage to mitochondria [100]. Besides, Xu's group investigated the molecular information of mitochondria from cancer cells in situ with the assistant of mitochondrion-targeting nanoprobes, compared with the isolation-based ex situ method [120]. In the following study, Xu et al. investigated the dynamic molecular changes of the extracted living mitochondria upon phototherapy by SERS, to elucidate the underlying molecular mechanisms associated with cell death induced by subcellular dysfunction. The result demonstrated that during the cell death process caused by phototherapy, the content of Phe in mitochondria started to increase. The cell apoptosis in this process was mainly due to the changes in DNA structures [121]. Brazhe et al. probed cytochrome c and monitored the redox state and conformation of cytochrome c in the electron transport chain in living isolated mitochondria by SERS [101]. They further studied living cells and the isolated mitochondria by SERS, and they achieved the redox state and the conformation of cytochromes in a natural cell environment [122]. The accurate molecular information of the isolated mitochondria is essential for profoundly understanding the structure and function of mitochondria in cellular biological processes.
Nucleus is a place of genetic material storage and possesses a high-density place in one cell. Cytobiological methods for cell nucleus related studies start with the extraction processes of intranuclear components with many cell lysis buffers, following by the structural characterizations and quantitative analysis of the extracted components. The in situ SERS strategy to selectively measure intracellular biomolecules was compared with the ex-situ SERS strategy [123]. This study described the accurate SERS profiles of the pure cell nuclei, the whole nuclear proteins/DNAs, and the DNA/protein extraction reagents as well [123]. This is the first step in establishing a Ramanomics database of nuclear molecules.
Therapeutic mechanisms and effects at subcellular compartment level revealed by SERS
At present, with a deeper understanding of the structure and function of organelles, the designs of the organelle targeting platforms based on chemotherapy, photothermal therapy (PTT), photodynamic therapy (PDT), and other multi-mode therapeutic platforms have attracted much attention. The most significant advantage of SERS technology in this field is that it can realize the real-time monitor of the biomolecular changes and the quantitative assay of small molecules in living cells.
Subcellular compartment information revealed by SERS during therapies
It is unique to look at the process and mechanism of disease treatments from the organelles' Ramanomics. In 2013, the El-Sayed group developed a nuclear targeting plasmon-enhanced single-cell imaging technology to study the molecular changes of the nucleus, before and after the cells were treated with anticancer drugs [39]. They further used the nuclear-targeted AuNPs as the PTT reagents to study the biomolecular changes in the nucleus during PTT [124]. The results proved that during the PTT process, the molecular structures of proteins and lipids in the nuclei would change, causing cell death. The molecular structure changes have nothing to do with the size of nanomaterials and the intensity of light. This study provides us useful information to understand the mechanism of PTT induced cell death at the molecular level, which is vital for understanding the therapeutic mechanism of nuclear-targeted PTT and efficacy optimization. A further study based on mass spectrometry combined metabolomics and proteomics with SERS spectroscopy to discuss the PTT process [125]. They also studied the changes of nuclear molecules under the action of ultraviolet light [41], high oxygen content [126], and AgNPs [127]. These advances paved a new way for the SERS analyses of therapeutic mechanisms from the subcellular level.
Moreover, the therapeutic plasmonic SERS nanoprobes with organelle-targeting features were designed to study the different responses of cancer cells and normal cells upon to PTT. The nanoprobe composed of porous Ag/Au nanoshells and carbon dots, was prepared for the PTT of cells (Hela, L929, and H8 cells) at organelle level (nucleus and mitochondrion) to assess the heat tolerance mechanism by in situ SERS [128]. The contents of tryptophan, phenylalanine (Phe), and tyrosine in HeLa cells were found more evidently increased than those of L929 and H8 cells during the PPT-induced cell death process. From the mitochondrial point of view, the PPT-induced cell apoptosis for HeLa cells mainly stems from (or is regulated through) cellular thermal stress-responsive proteins, while the damages of L929 and H8 cells seem more related to DNA. Different molecular stress responses at the single-cell level will help us understand the underlying mechanisms of the cell death induced by PTT.
In addition to PTT, changes in the SERS spectrum of the nucleus during PDT were also investigated. The B16 cell line (a murine melanoma cell line) was chosen. The results evidenced the protein degradation and the DNA fragmentation in the therapeutic process, which is meaningful not only for exploring the PDT molecular mechanisms but also for the assessment of the dosage of photosensitive reagents [129].
Besides phototherapies, electrical stimulation is another clinically permitted therapeutic mode. Its curative effect was also revealed by subcellular SERS, reported by Jin et al. [130]. The different responses of the nuclei and mitochondria between normal and cancer cells under electrical stimulation were investigated by the nucleus- and mitochondrion-targeting AuNPs. The results disclosed a common electrosensory and self-repair biofunction of cells and its related Phe metabolism response.
A The schematic illustration of the exploration of the therapeutic effect of aptamer/drug conjugates acting on HepG2 cells by SERS and dark-field imaging with the of nuclear targeting nanoprobes. B SERS spectra of HepG2 cells treated with nuclear-targeted nanoprobes and (a) and further with 4 µM TLS11a-GC-Dox conjugates (b) or 15 µM free Dox (c) for 24 h, respectively. Adapted with permission from [131] Copyright 2017 American Chemical Society.
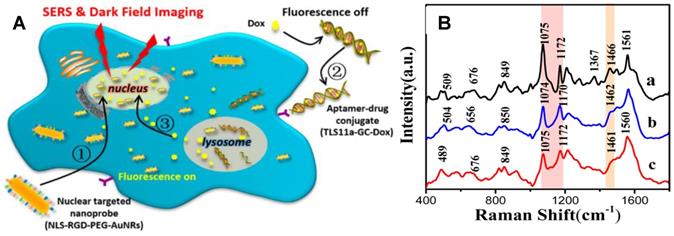
Chemotherapy is an important way for tumor treatments. Xu et al. used AuNRs-based nuclear targeting probes to study the effects of DNA targeting molecules (Hoechst33342) and anticancer drugs (doxorubicin, Dox) on the nucleus [81]. Spectral analysis showed that both molecules acting on the nucleus could change the spectral information of the nucleus. The DNA and protein structures in the nucleus were both influenced; however, the molecular mechanisms of the two molecules acting on the nucleus were different. Hoechst33342 executes its role mainly through acting on the DNA bases to affect vibrational modes of DNA and proteins. However, Dox principally induces DNA damage to achieve anticancer effects. Subsequently, the treatment process of a targeting aptamer/Dox (TLS11a-GC-Dox) drug delivery system on the nucleus was explored by SERS and dark-field technology (Figure 8) [131]. The cell morphology shrunk severely during the treatment, and the DNA and protein structures in the nuclei also changed accordingly. This is the first report to gain insight into the treatment mechanism of a targeted drug by SERS.
Subcellular microenvironments revealed by SERS during therapy
In addition to directly detecting the changes of biomolecules in organelles, detecting the intracellular microenvironment during treatments can help us better understand the therapeutic mechanism and the important role of organelles in the therapeutic processes. In 2016, Yang's group used the AuNRs with the decoration of pH-responsive molecules and a layer of the peptide as a protective layer to carry out the SERS detection for lysosomal pH during PTT [132]. The intramitochondrial pH evolutions during the PDT process were also revealed by Xu's study, in which the Mpy-coated AuNRs with the mitochondrion-targeting peptide decoration were cultured with three kinds of cells before the PDT [133]. When cells were irradiated with a LED for 1 min to trigger the photosensitive reagents releasing ROS, the mitochondrial pH significantly decreased, causing the early stage of apoptosis. With the continuous extension of the illumination time, the pH value in mitochondria increased, suggesting that the cells had self-defense and self-regulation function. The ROS SERS assay suggested that the LO2 cells have better resistance to ROS than two kinds of cancer cells (HepG2 and MCF-7).
Besides the pH environment, ROS changes are also crucial for the understanding of the mechanism of PTT and PDT. Recently, an interference-free SERS-active nanoprobe for intracellular ROS detection was developed by Xu et al [134]. This nanoprobe was composed of an Au core-Ag shell (Au@Ag NPs) with a SERS reporter (4-mercaptobenzonitrile) resided in the gap of the core-shell. Intracellular ROS can etch the Ag shell and dramatically decreases the SERS intensity of the SERS reporters. The Raman band of the reporter used in this study is located in the cellular Raman-silent region (1800-2800 cm-1), which eliminates interference possibility from cellular molecules. By using these Au@Ag NPs, the ROS dynamic changes at mitochondria during the PTT process could be revealed by the Raman mapping. Results show that after irradiation by an 808 nm laser, the SERS intensities of the peak at 2226 cm-1 gradually decreased due to the ROS production during the PTT process. This study also demonstrated that during the PTT process, the ROS-induced cell death is a major reason in comparison to the heat-induced cell death.
Integration of SERS diagnosis and treatment
As a powerful spectroscopic tool, SERS has been employed to detect the organelles and components at the subcellular level, and reveal the action mechanisms of some therapeutic means through the molecular fingerprint information. By integrating the diagnosis and therapy, SERS nanotags could track the release of the drugs in cells and realize tumor treatment at the same time.
In 2016, Cui et al. fabricated a multifunctional nanocarrier for tracking the intracellular Dox release process based on the acidic environment in lysosomes, achieving the killing effect on cancer cells. Furthermore, the detailed release process and the carrier distribution were recorded by SERS via the environmental pH changes during cell endocytosis [135]. In the same way, Chen et al. investigated the intracellular SERS performance of the Dox loaded Ag-mesoporous silica nanocarriers, and the results demonstrated that the nanocarriers could preserve the SERS signal to realize the targeted detection and traceable drug delivery, which was favorable for easy endocytosis, pH-responsive drug release and simultaneous SERS imaging. This work highlighted the potential of Janus Ag-MSN nanocarriers as a practical tool for efficient cancer therapy [136].
In addition to the implementation of tracking drug delivery by SERS, some cancer cell treatment strategies with SERS tags were also developed to support therapy or feedback therapeutic efficacy. For example, Wang et al. prepared a novel traceable and targeted drug delivery nanosystem based on Au nanoflowers (AuNFs). This nanosystem has a high drug encapsulation capability and an intra-lysosome pH-controlled release feature. The AuNF-based nanocarriers were applied for efficient intracellular SERS imaging-guided chemo-phototherapy, showing great potential in facilitating the accurate in vivo treatment of tumors [137]. In a recent study, Jin and Xu et al. developed an intelligent biodegradable nanoreactor based on the tumor microenvironmental response for the chemo-starvation synergistic treatment of tumors. The nanoreactor was composed of an assembly of the 4-mercaptobenzonitrile-decorated AgNPs (AgNPs@MBN) of the glucose oxidase (GOx)-loaded metal-organic-framework (ZIF-8@GOx). The ZIF-8@GOx part has an intelligent response to the acidic microenvironment of the cancerous lysosomes, enabling the release of drugs (GOx) to trigger the catalytic cascade reaction in the tumor. The surface element (AgNPs@MBN) of the nanoreactor has the function of in-situ SERS reporting glucose level in cells based on a ''turn-off'' mechanism (intensity decrease) of the Raman reporter (MBN) induced by the H2O2 etching effect on the AgNPs. Thus, the enzymatic activity of GOx and the degradability of the nanoreactor in an acidic environment can be monitored; that is, this nanosensor can immediately feedback the curative effect by SERS [138].
Conclusions and future directions
This review summarized recent progress in subcellular compartment studies using SERS as a powerful analytical tool. The applications of the SERS-based detection schemes in subcellular structures studies were reviewed (Table 1), including the direct subcellular analysis, investigation of biological processes, as well as SERS-based indirect analysis including subcellular imaging and the SERS sensing of the cellular environment, including pH, ROS, and other vital components. Moreover, the organelle-targeting therapeutic mechanisms including the anticancer drugs, PTT, PDT, and electric stimulation. The integration of SERS diagnosis and treatment were also discussed. These might be a reference for researchers interested in the intracellular SERS studies.
Looking back on the above publications, people have made progress in many aspects; however, many efforts should be exerted no matter on the bio-SERS experimental aspects or on the extensive SERS applications in the subcellular studies, because this subject is full of challenges and many technical problems still exist. Many new technologies are involved in Raman analytical methods, and their integration will bring a brighter future for intracellular SERS studies. In the following, we listed six unsolved problems in the intracellular SERS experiments. According to these problems, a perspective for future studies is given. And some of them are already on the way to reality.
Automation of synthesis of SERS nanoprobes
SERS tags and nanoprobes are all plasmonic MNP-based. So, to prepare SERS nanoprobes, we firstly synthesize metallic Au or Ag NPs. The shape and size are different in trials, which is one of the trouble issues leading to the poor repeatability and low accuracy of the SERS results. A robust method to synthesize monodisperse MNPs with high reproducibility and stability is in great demand. Except for many parameters, such as reagents, temperature, pH, concentration, charging sequence and mixing time, etc., the synthesis method is also essential. The seed-growth method [139] is an applicable and controllable method for the NP preparation due to its better size control. We can envision that automatic production lines and more rigorous quality control management are expected, which will make precise manufacturing of high-quality MNPs possible in the near future.
A summary of the experimental methods and results adopted in publications
| Category | Methods & Nanosensors | Results | Ref. |
|---|---|---|---|
| Label-free method | The drop-coating Ag NPs on a silicon wafer | The SERS spectra of the cellular surface of the tumor and nontumor cell lines were obtained. | [97] |
| The nuclear targeting AuNRs-based probes | The SERS spectra of the nucleus, malignant and normal epithelial cells were obtained. | [98] | |
| The pure AuNPs | The SERS information of the entire mitochondria was obtained. | [100] | |
| The mitochondrial targeting AuNRs-based probes and AuNRs | The molecular information of mitochondria from cancer cells in situ and ex situ method. | [120] | |
| The pure AuNRs | The dynamic molecular changes of the living mitochondria upon phototherapy by SERS was investigated. | [121] | |
| The silver layered ring substrate | Record the conformation and redox state of the cytochrome c under various conditions. | [101] | |
| The Ag@SiO2 colloidosomes | [122] | ||
| The pure AuNPs | Identified and visualized the stages of endocytosis of AuNPs in cells. | [103] | |
| The organelles targeting AuNRs-based nanoprobes | The molecular information of different organelles such as nucleus, mitochondria, and lysosome were observed and compared. | [104] | |
| The nuclear targeting AuNPs | The real-time SERS monitoring of the molecular dynamics of mitosis in healthy and cancer cells at the molecular level was demonstrated. | [24] | |
| The nuclear targeting probes | The SERS spectra of different neuronal cells, and the cytoplasm and nucleus of neuronal cells were obtained. | [105] | |
| The SERS-active nanoneedles | Accurately recorded the SERS spectra of the cytoplasm and nucleus. | [106] | |
| The nuclear targeting AgNPs | The SERS spectral changes of the nucleus during chemotherapy, PTT, and under the action of ultraviolet light, high oxygen content, and AgNP was studied. | [39, 41, 124-127] | |
| The nucleus and mitochondrion-targeting porous Ag/Au with surface decoration of carbon nanodots | The different responses of cancer cells and normal cells upon to PTT | [128] | |
| The nucleus and mitochondrion-targeting AuNPs-based probes | Two electrical stimulation modes at organelle level (nucleus and mitochondria) were studied. | [130] | |
| The nuclear targeting AuNRs-based probes | The treatment mechanism of Dox and PDT by SERS spectroscopy was reported. | [81, 129, 131] | |
| SERS label method | The nuclear targeting preloaded Edu in nucleus AuNRs-based probes | The accompanying signal of Edu in the Raman silent range can verify the SERS spectrum of the nucleus. | [99] |
| The organelle-targeting Au@Au core-shell nanoprobes with three different Raman-active molecules into the gap | High-speed, high-resolution, multiplexed subcellular organelle imaging were obtained. | [94] | |
| The nuclear and membranous targeting AuNPs with different reporters | FA and LHRH on the surface of the cell membrane were accurately located and detected. | [53] | |
| The Raman reporters functionalized nanoprobes | SERS imaging the protein-specific glycosylation on the cell surface | [109, 110] | |
| The AgNPs@MPBA nanosensors | SA levels in different cell types and the dynamic expression processes of them were studied. | [44, 111] | |
| The nano-tip technology based on SERS | The pH of the nucleus and cytoplasm were detected based on 4-mpy | [80] | |
| The organelle targeting AuNRs-based nanoprobes decorated of Mpy | Quantified and monitored of the subcellular ph environment in mitochondria, nucleus, and lysosome | [117] | |
| The AuNRs with the decoration of pH-responsive molecules | Changes of lysosomal ph during PTT were revealed. | [132] | |
| The intramitochondrial ph evolution, ROS and mitochondrial membrane potential during PDT were revealed. | [133] | ||
| The Au core-Ag shell nanoparticle (Au@Ag NPs) with 4-MBN in the gap | The intracellular ROS was detected during PTT | [134] | |
| The multifunctional nanocarrier based on SERS tags | The intracellular Dox release process was tracked and the killing effect on cancer cells was achieved. | [135, 136] | |
| The AuNFs@MBA@RGD with high DOX encapsulation | Efficient intracellular SERS imaging-guided chemo-phototherapy was realized. | [137] | |
| The AgNPs@MBN on the ZIF-8@glucose oxidase NPs | The chemo-starvation synergistic treatment of tumor was achieved and the curative effect was got feedback. | [138] |
The targeting ability of nanoplatforms in living cells
The SERS-based platform has been successfully used in the diagnosis and treatment of cancer in vivo due to SERS tags showing excellent signal intensity. Some organelle targeted-based drug release methods are also designed based on this point [140]. However, the living cell has a complicated and crowded intracellular environment, and its physiological metabolism keeps going all the time. All of these bring uncertainty for the targeting ability of MNPs. Thus, the location of the SERS-based nanoplatforms should be carefully confirmed before SERS measurements and organelle-specific therapies. The optical fiber-based SERS probes and nanotip with the micromanipulators can accurately arrive in the interesting locations while tiny damage on the living cell membrane will occur, which might be a desirable solution for targeting the sites relative to the targeting MNPs for subcellular analysis. However, even a fine tip has a large apex angle, which will be an obstacle when it is applied to many indistinguishable organelles. Atomic force microscopy-based tip-enhanced Raman spectroscopy probe permits cell penetration [141, 142]. Though the tip is very smaller, the shortcoming of this method is that the tip only can probe the location from top to down randomly. Thus, this topic is still open to welcome the integration of new techniques.
Because the targeting ability of the nanoprobes is one of the key issues for ensuring the reproducibility and accuracy of intracellular SERS, more specific targeting ligands and techniques are required to reduce the off-target effect of the nanoprobes. This progress often depends on the discoveries in bionics and the imitation of physiological processes such as viruses and bacteria. Recently, many precise diagnosis and medical methods, for example, gene-editing technique, click chemistry approach and fluorescence in situ hybridization methods, have been successfully applied for illuminating genome-scale and transcriptional activity in a single cell by fluorescent imaging [143]. We believe many examples might be presented in combining these new techniques with subcellular SERS studies in the near future.
Needs for more Raman reporters responding to small intracellular molecules
There are still too few molecules available for intracellular detection. Compared with fluorescent sensing molecules, the choice of responsive Raman reporting molecules is very limited, especially in the detection of small molecules in cells. Also, it is difficult for small molecules to trigger an effective signal amplification mechanism. So, it is still challenging to sense small intracellular molecules, as well as cellular metabolites. More efforts are required to involve in in the synthesis of sensing probe molecules and Raman reporters.
Since the limited surface area of a SERS-active NP, except giving the signal reporter with a larger Raman scattering cross-section and preferably having a band in bio-window range, the Raman reporters are expected to endow multi roles, for example, the bridging linker for identifying elements, the hydrophilic/hydrophobic regulator, the surface blocker and protector, and the modular for the plasmon feature of NPs, etc. The binding ways to metal are not limited to thoil, amino, and carboxyl groups anymore. In a recent study, Li and Tang et al. [144] developed the selenol-based reporter molecules, and found that the Au-Se interface is more stable than the biosensors constructed by the Au-S bond, displaying good resistance to abundant thiol under the biological condition and show better stability. The increasingly demanding molecular designs based on full consideration of all these issues is an irresistible trend, and many organic and pharmaceutical chemists might engage in developing high-quality and multifunctional Raman reporters.
High-speed and ultra-high-resolution SERS imaging
Due to the Raman scattering cross-section of the molecules is about 10-30 cm2/sr, Raman-based subcellular imaging requires a relatively long exposure or integration time, making it difficult to image a subcellular compartment whose cellular changes can occur on the scale of milliseconds. The current SERS imaging speed seriously limits the detection in a high temporal resolution. Some promising improvements were achieved by using highly bright SERS nanotags. In 2019, Ye et al. [145, 146] reported GERTs with strong electromagnetic hot spots from interior subnanometer gaps, and these GERTs reached a Raman enhancement factor beyond 5×109, the detection sensitivity down to a single-NP level, high-resolution cell imaging within 6 s and high-contrast. These nanoprobes may open new opportunities for rapid ultrasensitive biosensing and bioimaging. In addition to the highly bright SERS nanotags, Brolo's group [147] adopted the Airyscan photocell detector—a system composed of an array of 32 photomultiplier elements organized in a hexagonal tiling arrangement, and they demonstrated a single-molecule SERS imaging technique that could probe hotspot dynamics in an individual NP at 800,000 frames per second, gathering tens of millions of SERS data points, with ~7 nm spatial resolution [147]. These results provide the SERS community with a high-speed experimental approach, indicating that rapid SERS imaging is achievable if neglecting ultra-high spectral resolution. Therefore, each advance of high-speed imaging technology will illuminate the SERS imaging field, which requires unremitting efforts on more delicate optical designs and devices.
Besides the imaging speed, spatial resolution is always pursued by researchers. Supra-resolution fluorescence imaging technology has become influential in visualizing the organelles and their interactions. Inspiringly, supra-resolution SERS imaging is already on the way. Using a high-density, uniform hot spot substrate (hot spot density 1011/cm2, 20 to 35 nm gap), Dana et al. [148] developed a SERS-stochastic optical reconstruction (STORM) technique that exhibits a spatially uniform distribution of spot locations with no identifiable sharp features to obtain the 20 nm resolution super-resolved SERS images, and they applied this technique for several complex biological architectures, e.g., the supramolecular self-assembled peptide networks, microalgae membranes, and eukaryotic cells. Lindquist's group [149] combined SERS direct detection with STORM by a 700 nm hexagonal periodic metal nanopore SERS hotspot array with a unit diameter of 150 nm to match a 660 nm laser, which causes the hotspot scintillation locating at 10 nm regions of the substrate to achieve the SERS-STORM imaging. They further employed an ultrathin silver island film to identify the molecular structures of the cell walls of two types of bacteria (Gram-negative bacteria and Gram-positive bacteria) [150]. So far, there are no reports on SERS supra-resolution imaging to visualize the distributions of intracellular compartments with the help of SERS tags. SERS and Raman micro-imaging have been widely reported [151]. Therefore, we believe that the rapid development of the supra-resolution imaging technology will one day be combined with SERS imaging technology, which will become a powerful tool for studying cell structure, morphology, mass transfer and so on.
Establishment of an extensive and accurate subcellular biomolecule database for spectral assignments
Establishing a consolidated SERS database is imperative toward the practical application of SERS. Most subcellular SERS studies are currently based on the workgroup-specific database obtained under different technical and biological parameters or the experience summed up by the predecessors for cellular analysis. This hinders validation in large-scale studies and implementation in clinics. It can't work for the information of unknown samples, or selectively identifying molecular changes resulting from cellular processes in large and multidimensional data sets. Following SERS methodology standardization for subcellular studies, the most critical step is to establish a consolidated subcellular biomolecules SERS database. This database will provide the comprehensive spectral profiles of organelles, including the data of subcellular structures in different cells and the same subcellular compartment at different physiological states, for instance, in response to various stresses. It will be a reliable tool for the identification, discrimination of subcellular compartments, in-depth deciphering of mechanism, and even prediction.
Combination of SERS and chemometric/intelligence algorithms
Although the sensitivity of Raman spectroscopy is sufficiently high to identify significant cellular macromolecular information and their variations existing between different cellular samples and different cellular processes via label-free SERS strategy, reasonable analysis and processing of the obtained data is still a challenge, no matter in the cellular level or in the sub-cellular level. Therefore, combined with advanced algorithms for demultiplexing or decoding the spectra of components will be promising in the near future, e.g., PCA-LDA [103]. The application of chemometric methods in SERS spectroscopy will help us determine the changes between spectra and biomarkers, and even extract representative spectral features used to characterize and/or classify samples [152]. In addition, the integrations of the machine learning approach with SERS can also help us to assign and analyze the multiple spectra. For example, the label-free detection of the gradients of multiple metabolites in the extracellular environment was achieved by the combination of machine learning and SERS optophysiology [79]. Artificial intelligence (AI) algorithm has many successful examples in medical imaging. In a recent Nature paper [153], they reported the X-ray data of a breast cancer developed jointly with Google and Deep Mind, which can reduce the false positive rate by 5.7% and false negative rate by 9.4%. Researchers used AI to diagnose the SRS images of brain tumor tissues [154]. The AI training model covered 12 common histologic categories and achieved a diagnosis accuracy rate of 94.6%. These developments have fully proved AI technology is powerful in image analysis. We can envision that the SERS united with other advanced algorithms will be a promising and hot trend in cellular and subcellular SERS researches.
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 21873039, 21573087 and 21573092) and the grant of China Scholarship Council. We acknowledge Prof. Chongyang Liang from Jilin University and Dr. Yuling Wang from Macquarie University for helpful discussions in the experimental aspects of cells and bio-SERS.
Abbreviations
NPs: nanoparticles; SERS: surface-enhanced Raman spectroscopy; ER: endoplasmic reticulum; ATP: adenosine triphosphate; ROS: reactive oxygen species; DNA: deoxyribonucleic acid; MNPs: metal nanoparticles; AuNRs: Au nanorods; GERTs: gap-enhanced Raman tags; MTT: 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; WST-1: 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2, 4-disulfophenyl)-2H, tetrazolium monosodium salt; PEG: polyethylene glycol; CTAB: cetyltrimethylammonium bromide; BSA: bovine serum albumin; APTMS: aminopropyltrimethoxysilane; SHINs: shell-isolated nanoparticles; HSC: human oral cancer cells; SV: simian virus; Edu: 5-ethynyl-2'-deoxyuridine; AuNPs: Au nanoparticles; PCA-LDA: principal component analysis-linear discriminant analysis; SRS: stimulated Raman scattering; CARS: coherent anti-Stokes Raman scattering; FA: folic acid; LHRH: luteinizing hormone-releasing hormone; SA: sialic acid; EpCAM: epithelial cell adhesion molecule; MPBA: 4-mercaptophenylboronic acid; AgNPs: Ag nanoparticles; NCL: nucleolin; Mpy: 4-mercaptopyridine; PTT: photothermal therapy; PDT: photodynamic therapy; Phe: phenylalanine; Dox: doxorubicin; AuNFs: Au nanoflowers; STORM: stochastic optical reconstruction; MBN: p-mercaptobenzonitrile; TEM: transmission electron microscopy.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Satori C, Henderson MM, Krautkramer EA, Kostal V, Distefano MM, Arriaga EA. Bioanalysis of eukaryotic organelles. Chem Rev. 2013;113:2733-811
2. Tani E, Nakano M, Itagaki T, Fukumori T. Cell membrane structure of human giant-celled glioblastoma. Acta Neuropathol. 1978;41:61-5
3. Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547-53
4. Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145-59
5. Castino R, Demoz M, Isidoro C. Destination 'Lysosome': a target organelle for tumour cell killing? J Mol Recognit. 2003;16:337-48
6. Schneider L, Zhang J. Lysosomal function in macromolecular homeostasis and bioenergetics in Parkinson's disease. Mol Neurodegener. 2010;5:14
7. Austin LA, Kang B, El-Sayed MA. A new nanotechnology technique for determining drug efficacy using targeted plasmonically enhanced single cell imaging spectroscopy. J Am Chem Soc. 2013;135:4688-91
8. Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787-95
9. Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet. 1992;2:324-9
10. Coutinho MF, Alves S. From rare to common and back again: 60 years of lysosomal dysfunction. Mol Genet Metab. 2016;117:53-65
11. Nixon RA, Yang D-S, Lee J-H. Neurodegenerative lysosomal disorders-A continuum from development to late age. Autophagy. 2008;4:590-9
12. Wishart DS. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15:473-84
13. Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338-50
14. Berners-Price SJ. Activating platinum anticancer complexes with visible light. Angew Chem Int Ed. 2011;50:804-5
15. Fan W, Huang P, Chen X. Overcoming the Achilles' heel of photodynamic therapy. Chem Soc Rev. 2016;45:6488-519
16. Bearer EL. Distribution of Xrel in the early Xenopus embryo: a cytoplasmic and nuclear gradient. Eur J Cell Biol. 1994;63:255-68
17. Nozaki T, Masutani M, Akagawa T, Sugimura T, Esumi H. Non-covalent interaction between poly(ADP-ribose) and cellular proteins: an application of a poly(ADP-ribose)-western blotting method to detect poly(ADP-ribose) binding on protein-blotted filter. Biochem Bioph Research Co. 1994;198:45-51
18. Tsitilou SG, Cox D, Mathias AP, Ridge D. The characterization of the non-histone chromosomal proteins of the main classes of nuclei from rat brain fractionated by zonal centrifugation. Biochem J. 1979;177:331-46
19. Bonner WM, West MH, Stedman JD. Two-dimensional gel analysis of histones in acid extracts of nuclei, cells, and tissues. Eur J Biochem. 1980;109:17-23
20. Wollscheid B, Bausch-Fluck D, Henderson C, O'Brien R, Bibel M, Schiess R. et al. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nat. Biotechnol. 2009;27:378-86
21. Barcena M, Koster AJ. Electron tomography in life science. Cell Dev. Biol. 2009;20:920-30
22. Liu H, Wen J, Xiao Y, Liu J, Hopyan S, Radisic M. et al. In situ mechanical characterization of the cell nucleus by atomic force microscopy. ACS Nano. 2014;8:3821-8
23. Guo Y, Li D, Zhang S, Yang Y, Liu J-J, Wang X. et al. Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales. Cell. 2018;175:1430-42
24. Kang B, Austin LA, El-Sayed MA. Real-time molecular imaging throughout the entire cell cycle by targeted plasmonic-enhanced Rayleigh/Raman spectroscopy. Nano Lett. 2012;12:5369-75
25. Fleischmann M, Hendra PJ, McQuillan AJ. Raman spectra of pyridine adsorbed at a silver electrode. Chem Phys Lett. 1974;26:163-6
26. Zong C, Xu M, Xu LJ, Wei T, Ma X, Zheng XS. et al. Surface-enhanced Raman spectroscopy for bioanalysis: reliability and challenges. Chem Rev. 2018;118:4946-80
27. Chen M, Zhang H, Ge Y, Yang S, Wang P, Fang Y. Surface-nanostructured single silver nanowire: A new one-dimensional microscale surface-enhanced Raman scattering interface. Langmuir. 2018;34:15160-5
28. Lane LA, Qian X, Nie S. SERS nanoparticles in medicine: from label-free detection to spectroscopic tagging. Chem Rev. 2015;115:10489-529
29. Qin X, Lyu M, Si Y, Yang J, Wu Z, Li J. Alkyne-based surface-enhanced Raman scattering nanoprobe for ratiometric imaging analysis of caspase 3 in live cells and tissues. Anal Chim Acta. 2018;1043:115-22
30. Guo X, Li J, Arabi M, Wang X, Wang Y, Chen L. Molecular-Imprinting-Based Surface-Enhanced Raman Scattering Sensors. ACS Sens. 2020;5:601-19
31. Cialla-May D, Zheng XS, Weber K, Popp J. Recent progress in surface-enhanced Raman spectroscopy for biological and biomedical applications: from cells to clinics. Chem Soc Rev. 2017;46:3945-61
32. Krafft C, Schmitt M, Schie IW, Cialla-May D, Matthäus C, Bocklitz T. et al. Label-free molecular imaging of biological cells and tissues by linear and nonlinear Raman spectroscopic approaches. Angew Chem Int Ed. 2017;56:4392-430
33. Cui L, Zhang D, Yang K, Zhang X, Zhu Y-G. Perspective on surface-enhanced Raman spectroscopic investigation of microbial world. Anal Chem. 2019;91:15345-54
34. Masango SS, Hackler RA, Large N, Henry A-I, McAnally MO, Schatz GC. et al. High-resolution distance dependence study of surface-enhanced Raman scattering enabled by atomic layer deposition. Nano Lett. 2016;16:4251-9
35. Kuku G, Altunbek M, Culha M. Surface-enhanced Raman scattering for label-free living single cell analysis. Anal Chem. 2017;89:11160-6
36. Li DW, Qu LL, Hu K, Long YT, Tian H. Monitoring of endogenous hydrogen sulfide in living cells using surface-enhanced Raman scattering. Angew Chem Int Ed. 2015;54:12758-61
37. Zhou H, Yang D, Ivleva NP, Mircescu NE, Niessner R, Haisch C. SERS detection of bacteria in water by in situ coating with Ag nanoparticles. Anal Chem. 2014;86:1525-33
38. Jarvis RM, Goodacre R. Characterisation and identification of bacteria using SERS. Chem Soc Rev. 2008;37:931-6
39. Kang B, Austin LA, El-Sayed MA. Observing real-time molecular event dynamics of apoptosis in living cancer cells using nuclear-targeted plasmonically enhanced Raman nanoprobes. ACS Nano. 2014;8:4883-92
40. Lussier F, Brule T, Vishwakarma M, Das T, Spatz JP, Masson JF. Dynamic-SERS optophysiology: A nanosensor for monitoring cell secretion events. Nano Lett. 2016;16:3866-71
41. Panikkanvalappil SR, Hira SM, El-Sayed MA. Elucidation of ultraviolet radiation-induced cell responses and intracellular biomolecular dynamics in mammalian cells using surface-enhanced Raman spectroscopy. Chem Sci. 2016;7:1133-41
42. Wang Y, Yan B, Chen L. SERS tags: novel optical nanoprobes for bioanalysis. Chem Rev. 2013;113:1391-428
43. Lenzi E, Jimenez de Aberasturi D, Liz-Marzán LM. Surface-enhanced Raman scattering tags for three-dimensional bioimaging and biomarker detection. ACS Sensors. 2019;4:1126-37
44. Liang L, Qu H, Zhang B, Zhang J, Deng R, Shen Y. et al. Tracing sialoglycans on cell membrane via surface-enhanced Raman scattering spectroscopy with a phenylboronic acid-based nanosensor in molecular recognition. Biosens Bioelectron. 2017;94:148-54
45. Zhou W, Li Q, Liu H, Yang J, Liu D. Building electromagnetic hot spots in living cells via target-triggered nanoparticle dimerization. ACS Nano. 2017;11:3532-41
46. Xu L, Yin H, Ma W, Kuang H, Wang L, Xu C. Ultrasensitive SERS detection of mercury based on the assembled gold nanochains. Biosens Bioelectron. 2015;67:472-6
47. Shen Y, Yue J, Shi W, Xu W, Xu S. Target-triggered hot spot dispersion for cellular biothiol detection via background-free surface-enhanced Raman scattering tags. Biosens Bioelectron. 2020;151:111957
48. Ma H, Liu S, Zheng N, Liu Y, Han XX, He C. et al. Frequency shifts in surface-enhanced Raman spectroscopy-based immunoassays: mechanistic insights and application in protein carbonylation detection. Anal Chem. 2019;91:9376-81
49. Wang W, Zhao F, Li M, Zhang C, Shao Y, Tian Y. A SERS optophysiological probe for the real-time mapping and simultaneous determination of the carbonate concentration and pH value in a live mouse brain. Angew Chem Int Ed. 2019;58:5256-60
50. Xie Y, Wang X, Han X, Song W, Ruan W, Liu J. et al. Selective SERS detection of each polycyclic aromatic hydrocarbon (PAH) in a mixture of five kinds of PAHs. J Raman Spectrosc. 2011;42:945-50
51. Qian X, Peng XH, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM. et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat. Biotechnol. 2008;26:83-90
52. Wang YW, Doerksen JD, Kang S, Walsh D, Yang Q, Hong D. et al. Multiplexed molecular imaging of fresh tissue surfaces enabled by convection-enhanced topical staining with SERS-coded nanoparticles. Small. 2016;12:5612-21
53. Chen Y, Bai X, Su L, Du Z, Shen A, Materny A. et al. Combined labelled and label-free SERS probes for triplex three-dimensional cellular imaging. Sci Rep. 2016 6
54. Huo S, Jin S, Ma X, Xue X, Yang K, Kumar A. et al. Ultrasmall gold nanoparticles as carriers for nucleus-based gene therapy due to size-dependent nuclear entry. ACS Nano. 2014;8:5852-62
55. Xu P, Van Kirk EA, Zhan Y, Murdoch WJ, Radosz M, Shen Y. Targeted charge-reversal nanoparticles for nuclear drug delivery. Angew Chem Int Ed. 2007;46:4999-5002
56. Gao G, Jiang YW, Yang J, Wu FG. Mitochondria-targetable carbon quantum dots for differentiating cancerous cells from normal cells. Nanoscale. 2017;9:18368-78
57. Zeng C, Hu F, Long R, Min W. A ratiometric Raman probe for live-cell imaging of hydrogen sulfide in mitochondria by stimulated Raman scattering. Analyst. 2018;143:4844-8
58. Busk M, Pytela R, Sheppard D. Characterization of the integrin alpha v beta 6 as a fibronectin-binding protein. J Biol Chem. 1992;267:5790-6
59. Takayama S, Hatori M, Kurihara Y, Kinugasa Y, Shirota T, Shintani S. Inhibition of TGF-beta1 suppresses motility and invasiveness of oral squamous cell carcinoma cell lines via modulation of integrins and down-regulation of matrix-metalloproteinases. Oncol Rep. 2009;21:205-10
60. Dehari H, Ito Y, Nakamura T, Kobune M, Sasaki K, Yonekura N. et al. Enhanced antitumor effect of RGD fiber-modified adenovirus for gene therapy of oral cancer. Cancer Gene Ther. 2003;10:75-85
61. Pan L, He Q, Liu J, Chen Y, Ma M, Zhang L. et al. Nuclear-targeted drug delivery of TAT peptide-conjugated monodisperse mesoporous silica nanoparticles. J Am Chem Soc. 2012;134:5722-5
62. Fan W, Shen B, Bu W, Zheng X, He Q, Cui Z. et al. Design of an intelligent sub-50 nm nuclear-targeting nanotheranostic system for imaging guided intranuclear radiosensitization. Chem Sci. 2015;6:1747-53
63. Fan W, Shen B, Bu W, Zheng X, He Q, Cui Z. et al. Intranuclear biophotonics by smart design of nuclear-targeting photo-/radio-sensitizers co-loaded upconversion nanoparticles. Biomaterials. 2015;69:89-98
64. Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677-90
65. Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83-96
66. Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695-701
67. Kawamura E, Yamada Y, Harashima H. Mitochondrial targeting functional peptides as potential devices for the mitochondrial delivery of a DF-MITO-Porter. Mitochondrion. 2013;13:610-4
68. Yamada Y, Akita H, Kamiya H, Kogure K, Yamamoto T, Shinohara Y. et al. MITO-Porter: A liposome-based carrier system for delivery of macromolecules into mitochondria via membrane fusion. Biochim Biophys Acta. 2008;1778:423-32
69. Ma Z, Han K, Dai X, Han H. Precisely striking tumors without adjacent normal tissue damage via mitochondria-templated accumulation. ACS Nano. 2018;12:6252-62
70. Zielonka J, Joseph J, Sikora A, Hardy M, Ouari O, Vasquez-Vivar J. et al. Mitochondria-targeted triphenylphosphonium-based compounds: syntheses, mechanisms of action, and therapeutic and diagnostic applications. Chem Rev. 2017;117:10043-120
71. Cai Y, Gui C, Samedov K, Su H, Gu X, Li S. et al. An acidic pH independent piperazine-TPE AIEgen as a unique bioprobe for lysosome tracing. Chem Sci. 2017;8:7593-603
72. Kuzmin AN, Pliss A, Lim CK, Heo J, Kim S, Rzhevskii A. et al. Resonance Raman probes for organelle-specific labeling in live cells. Sci Rep. 2016;6:28483
73. Vo-Dinh T, Kasili P. Fiber-Optic Nanosensors for single-cell monitoring. Anal Bioanal Chem. 2005;382:918-25
74. Vo-Dinh T, Scaffidi J, Gregas M, Zhang Y, Seewaldt V. Applications of fiber-optics-based nanosensors to drug discovery. Expert Opin Drug Discov. 2009;4:889-900
75. Niu JJ, Schrlau MG, Friedman G, Gogotsi Y. Carbon nanotube-tipped endoscope for in situ intracellular surface-enhanced Raman spectroscopy. Small. 2011;7:540-5
76. Vitol EA, Orynbayeva Z, Bouchard MJ, Azizkhan-Clifford J, Friedman G, Gogotsi Y. In situ intracellular spectroscopy with surface enhanced Raman spectroscopy (SERS)-enabled nanopipettes. ACS Nano. 2009;3:3529-36
77. Masson JF, Breault-Turcot J, Faid R, Poirier-Richard HP, Yockell-Lelièvre H, Lussier F, Spatz JP. Plasmonic nanopipette biosensor. Anal Chem. 2014;86:8998-9005
78. Vitol EA, Orynbayeva Z, Friedman G, Gogotsi Y. Nanoprobes for intracellular and single cell surface-enhanced Raman spectroscopy (SERS). J Raman Spectrosco. 2011;43:817-27
79. Lussier F, Missirlis D, Spatz JP, Masson JF. Machine-Learning-Driven Surface-Enhanced Raman Scattering Optophysiology Reveals Multiplexed Metabolite Gradients Near Cells. ACS Nano. 2019;13:1403-11
80. Wang J, Geng Y, Shen Y, Shi W, Xu W, Xu S. SERS-active fiber tip for intracellular and extracellular pH sensing in living single cells. Sensor Actuat B-Chem. 2019;290:527-34
81. Liang L, Huang D, Wang H, Li H, Xu S, Chang Y. et al. In situ surface-enhanced Raman scattering spectroscopy exploring molecular changes of drug-treated cancer cell nucleus. Anal Chem. 2015;87:2504-10
82. Pan X, Li L, Lin H, Tan J, Wang H, Liao M. et al. A graphene oxide-gold nanostar hybrid based-paper biosensor for label-free SERS detection of serum bilirubin for diagnosis of jaundice. Biosens Bioelectron. 2019;145:111713
83. Li JF, Zhang YJ, Ding SY, Panneerselvam R, Tian ZQ. Core-Shell Nanoparticle-Enhanced Raman Spectroscopy. Chem Rev. 2017;117:5002-69
84. Gu Y, Bi X, Ye J. Gap-enhanced resonance Raman tags for live-cell imaging. J Mater Chem B. 2020;8:6944-55
85. Khlebtsov NG, Lin L, Khlebtsov BN, Ye J. Gap-enhanced Raman tags: fabrication, optical properties, and theranostic applications. Theranostics. 2020;10:2067-94
86. Kang B, Mackey MA, El-Sayed MA. Nuclear targeting of gold nanoparticles in cancer cells induces DNA damage, causing cytokinesis arrest and apoptosis. J Am Chem Soc. 2010;132:1517-9
87. Doering WE, Nie S. Spectroscopic tags using dye-embedded nanoparticles and surface-enhanced Raman scattering. Anal. Chem. 2003;75:6171-6
88. Li MD, Cui Y, Gao MX, Luo J, Ren B, Tian ZQ. Clean substrates prepared by chemical adsorption of iodide followed by electrochemical oxidation for surface-enhanced Raman spectroscopic study of cell membrane. Anal Chem. 2008;80:5118-25
89. Kitahama Y, Tanaka Y, Itoh T, Ozaki Y. Time-resolved surface-enhanced resonance Raman scattering spectra of thiacyanine molecules in water. Chem. Lett. 2009;38:54-5
90. Lombardi JR, Birke RL, Haran G. Single molecule SERS spectral blinking and vibronic coupling. J Phys Chem C. 2011;115:4540-5
91. Gu Y, Zhang Y, Li Y, Jin X, Huang C, Ye J. Raman photostability of off-resonant gapenhanced Raman tags. RSC Adv. 2018;8:14434-44
92. Liu X, Knauer M, Ivleva NP, Niessner R, Haisch C. Synthesis of core-shell surface-enhanced Raman tags for bioimaging. Anal Chem. 2010;82:441-6
93. Li JF, Huang YF, Ding Y, Yang ZL, Li SB, Zhou XS. et al. Shell-isolated nanoparticle-enhanced Raman spectroscopy. Nature. 2010;464:392-5
94. Kang JW, So PT, Dasari RR, Lim DK. High resolution live cell Raman imaging using subcellular organelle-targeting SERS-sensitive gold nanoparticles with highly narrow intra-nanogap. Nano Lett. 2015;15:1766-72
95. Au L, Zheng D, Zhou F, Li ZY, Li X, Xia Y. A quantitative study on the photothermal effect of immuno gold nanocages targeted to breast cancer cells. ACS Nano. 2008;2:1645-52
96. Lin X-M, Cui Y, Xu Y-H, Ren B, Tian Z-Q. Surface-enhanced Raman spectroscopy: substrate-related issues. Anal. Bioanal. Chem. 2009;394:1729-45
97. Yan B, Reinhard BM. Identification of tumor cells through spectroscopic profiling of the cellular surface chemistry. J Phys Chem Lett. 2010;1:1595-8
98. Oyelere AK, Chen PC, Huang X, El-Sayed IH, El-Sayed MA, Peptide-conjugated gold nanorods for nuclear targeting. Bioconjugate Chem. 2007, 18: 1490-7.
99. Zhang J, Liang L, Guan X, Deng R, Qu H, Huang D. et al. In situ, accurate, surface-enhanced Raman scattering detection of cancer cell nucleus with synchronous location by an alkyne-labeled biomolecular probe. Anal Bioanal Chem. 2018;410:585-94
100. Karatas O F, Sezgin E, Aydin O, Culha M. Interaction of gold nanoparticles with mitochondria. Colloids Surf B Biointerfaces. 2009;71:315-8
101. Brazhe NA, Evlyukhin AB, Goodilin EA, Semenova AA, Novikov SM, Bozhevolnyi SI. et al. Probing cytochrome c in living mitochondria with surface-enhanced Raman spectroscopy. Sci Rep. 2015;5:13793
102. Guo X, Wei X, Chen Z, Zhang X, Yang G, Zhou S. Multifunctional nanoplatforms for subcellular delivery of drugs in cancer therapy. Prog Mater Sci. 2020;107:100599
103. Huefner A, Kuan WL, Muller KH, Skepper JN, Barker RA, Mahajan S. Characterization and visualization of vesicles in the endo-lysosomal pathway with surface-enhanced Raman spectroscopy and chemometrics. ACS Nano. 2016;10:307-16
104. Shen Y, Liang L, Zhang S, Huang D, Deng R, Zhang J. et al. Organelle-targeting gold nanorods for macromolecular profiling of subcellular organelles and enhanced cancer cell killing. ACS Appl Mater Interfaces. 2018;10:7910-8
105. Huefner A, Kuan WL, Barker RA, Mahajan S. Intracellular SERS nanoprobes for distinction of different neuronal cell types. Nano Lett. 2013;13:2463-70
106. Vitol EA, Orynbayeva Z, Bouchard MJ, Azizkhan-Clifford J, Friedman J, Gogotsi Y. In situ intracellular spectroscopy with surface enhanced Raman spectroscopy (SERS)-enabled nanopipettes. ACS Nano. 2009;3:3529-36
107. Evans CL, Xie XS. Coherent anti-stokes Raman scattering microscopy: chemical imaging for biology and medicine. Annu Rev Anal Chem. 2008;1:883-909
108. Lu FK, Basu S, Igras V, Hoang MP, Ji M, Fu D. et al. Label-free DNA imaging in vivo with stimulated Raman scattering microscopy. Proc Natl Acad Sci USA. 2015;112:11624-9
109. Chen Y, Ding L, Song W, Yang M, Ju H, Protein-specific Raman imaging of glycosylation on single cells with zone-controllable SERS effect. Chem Sci. 2016, 7: 569-74.
110. Chen Y, Ding L, Xu J, Song W, Yang M, Hu J. et al. Micro-competition system for Raman quantification of multiple glycans on intact cell surface. Chem Sci. 2015;6:3769-74
111. Cong L, Liang L, Cao F, Sun D, Yue J, Xu W. et al. Distinguishing cancer cell lines at a single living cell level via detection of sialic acid by dual-channel plasmonic imaging and by using a SERS-microfluidic droplet platform. Microchim Acta. 2019;367:186
112. Feng E, Zheng T, He X, Chen J, Tian Y. A novel ternary heterostructure with dramatic SERS activity for evaluation of PD-L1 expression at the single-cell level. Sci Adv. 2018;4:eaau3494
113. Nguyen T, Song M, Ly N, Lee S, Joo S. Nanostars on nanopipette tips: a Raman probe for quantifying oxygen levels in hypoxic single cells and tumours. Angew Chem Int Ed. 2019;58:2710-4
114. Shin H, Oh S, Kang D, Choi Y. Protein quantification and imaging by surface-enhanced Raman spectroscopy and similarity analysis. Adv. Sci. 2020;7:1903638
115. Jiang X, Tan Z, Lin L, He J, He C, Thackray BD. et al. Surface-enhanced Raman nanoprobes with embedded standards for quantitative cholesterol detection. Small Methods. 2018;2:1800182
116. Hanif S, Liu HL, Ahmed SA, Yang JM, Zhou Y, Pang J. et al. Nanopipette-based SERS aptasensor for subcellular localization of cancer biomarker in single Cells. Anal Chem. 2017;89:9911-7
117. Shen Y, Liang L, Zhang S, Huang D, Zhang J, Xu S. et al. Organelle-targeting surface-enhanced Raman scattering (SERS) nanosensors for subcellular pH sensing. Nanoscale. 2018;10:1622-30
118. Hanif S, Liu H, Chen M. et al. Organic cyanide decorated SERS active nanopipettes for quantitative detection of hemeproteins and Fe(3+) in single cells. Anal Chem. 2017;89:2522-30
119. Liu J, Yin D, Wang S, Chen H-Y, Liu Z. Probing low-copy-number proteins in a single living cell. Angew. Chem. Int. Ed. 2016;55:13215-8
120. Yue J, Shen Y, Liang L, Cong L, Guan X, Li Z. et al. In situ and ex situ surface-enhanced Raman spectroscopy (SERS) analysis of cell mitochondria. J Raman Spectrosc. 2020;51:602-10
121. Yue J, Shen Y, Liang L, Guan X, Zhang X, Xu S. et al. Tracing the molecular dynamics of living mitochondria under phototherapy via surface-enhanced Raman scattering spectroscopy. Analyst. 2019;144:5521-7
122. Sarycheva AS, Brazhe NA, Baizhumanov AA, Nikelshparg EI, Semenova AA, Garshev AV. et al. New nanocomposites for SERS studies of living cells and mitochondria. J Mater Chem B. 2016;4:539-46
123. Shen Y, Yang L, Liang L, Li Z, Zhang J, Shi W. et al. Ex situ and in situ surface-enhanced Raman spectroscopy for macromolecular profiles of cell nucleus. Anal Bioanal Chem. 2019;411:6021-9
124. Aioub M, El-Sayed MA. A Real-time surface enhanced Raman spectroscopy study of plasmonic photothermal cell death using targeted gold nanoparticles. J Am Chem Soc. 2016;138:1258-64
125. Ali MR, Wu Y, Han T, Zang X, Xiao H, Tang Y. et al. Simultaneous time-dependent surface-enhanced Raman spectroscopy, metabolomics, and proteomics reveal cancer cell death mechanisms associated with gold nanorod photothermal therapy. J Am Chem Soc. 2016;138:15434-42
126. Panikkanvalappil SR, James M, Hira SM, Mobley J, Jilling T, Ambalavanan N. et al. Hyperoxia induces intracellular acidification in neonatal mouse lung fibroblasts: real-time investigation using plasmonically enhanced Raman spectroscopy. J Am Chem Soc. 2016;138:3779-88
127. Austin LA, Kang B, Yen CW, El-Sayed MA. Nuclear targeted silver nanospheres perturb the cancer cell cycle differently than those of nanogold. Bioconjugate Chem. 2011;22:2324-31
128. Qi G, Zhang Y, Xu S, Li C, Wang D, Li H. et al. Nucleus and mitochondria targeting theranostic plasmonic surface-enhanced Raman spectroscopy nanoprobes as a means for revealing molecular stress response differences in hyperthermia cell death between cancerous and normal Cells. Anal Chem. 2018;90:13356-64
129. Yue J, Liang L, Shen Y, Guan X, Zhang J, Li Z. et al. Investigating Dynamic Molecular Events in Melanoma Cell Nucleus During Photodynamic Therapy by SERS. Front Chem. 2018;6:665
130. Qi G, Wang B, Zhang Y, Li H, Li C, Xu W. et al. Living-cell imaging of mitochondrial membrane potential oscillation and phenylalanine metabolism modulation during periodic electrostimulus. Anal Chem. 2019;91:9571-9
131. Deng R, Qu H, Liang L, Zhang J, Zhang B, Huang D. et al. Tracing the therapeutic process of targeted aptamer/drug conjugate on cancer cells by surface-enhanced Raman scattering spectroscopy. Anal Chem. 2017;89:2844-51
132. Luo R, Li Y, Zhou Q, Zheng J, Ma D, Tang P. et al. SERS monitoring the dynamics of local pH in lysosome of living cells during photothermal therapy. Analyst. 2016;141:3224-7
133. Yue J, Shen Y, Liang L, Cong L, Xu W, Shi W, Liang C, Xu S, Revealing mitochondrial microenvironmental evolution triggered by photodynamic therapy. Anal Chem. 2020; 92: 6081-6087.
134. Shen Y, Liang L, Zhang J, Li Z, Yue J, Wang J. et al. Interference-free surface-enhanced Raman scattering nanosensor for imaging and dynamic monitoring of reactive oxygen species in mitochondria during photothermal therapy. Sensor Actuat B-Chem. 2019;285:84-91
135. Chen P, Wang Z, Zong S, Zhu D, Chen H, Zhang Y. et al. pH-sensitive nanocarrier based on gold/silver core-shell nanoparticles decorated multi-walled carbon manotubes for tracing drug release in living cells. Biosens Bioelectron. 2016;75:446-51
136. Shao D, Zhang X, Liu W, Zhang F, Zheng X, Qiao P. et al. Janus silver-mesoporous silica nanocarriers for SERS traceable and pH-sensitive drug delivery in cancer therapy. ACS Appl Mater Interfaces. 2016;8:4303-8
137. Song C, Dou Y, Yuwen L, Sun Y, Dong C, Li F. et al. A gold nanoflower-based traceable drug delivery system for intracellular SERS imaging-guided targeted chemo-phototherapy. J Mater Chem B. 2018;6:3030-9
138. Sun D, Qi G, Ma K, Qu X, Xu W, Xu S. et al. Tumor microenvironment-activated degradable multifunctional nanoreactor for synergistic cancer therapy and glucose SERS feedback. iScience. 2020;23:101274
139. Zhao Y, Zhang Y, Meng J, Chen S, Li J, Tian Z. A facile method for the synthesis of large-size Ag nanoparticles as efficient SERS substrates. J. Raman Spectrosc. 2016;47:662-7
140. Gao P, Pan W, Li N, Tang B. Boosting cancer therapy with organelle-targeted nanomaterials. ACS Appl. Mater. Interfaces. 2019;11:26529-58
141. Frankel DJ, Pfeiffer JR, Surviladze Z, Johnson AE, Oliver JM, Wilson BS. et al. Revealing the topography of cellular membrane domains by combined atomic force microscopy/fluorescence imaging. Biophys J. 2006;90:2404-13
142. Park TH, Mirin N, Lassiter JB, Nehl CL, Halas NJ, Nordlander P. Optical properties of a nanosized hole in a thin metallic film. ACS Nano. 2008;2:25-32
143. Su JH, Zheng P, Kinrot SS, Bintu B, Zhuang X. Genome-scale imaging of the 3D organization and transcriptional activity of chromatin. Cell. 2020;182:1641-59
144. Li X, Duan X, Li L, Ye S, Tang B. An accurate and ultrasensitive SERS sensor with Au-Se interface for bioimaging and in situ quantitation. Chem Commun. 2020;56:9320-3
145. Zhang Y, Gu Y, Jing He, Thackray B, Ye J. Ultrabright gap-enhanced Raman tags for high-speed bioimaging. Nature Commun. 2019;10:3905
146. Khlebtsov B, Burov A, Bratashov D, Tumskiy R, Khlebtsov N. Petal-like gap-enhanced Raman tags with controllable structures for high-speed Raman imaging. Langmuir. 2020;36:5546-53
147. Lindquist N, Albuquerque C, Sobral-Filho R, Paci I, Brolo A . High-speed imaging of surface-enhanced Raman scattering fluctuations from individual nanoparticles. Nature Nanotechnol. 2019;14:981-7
148. Ayas S, Cinar G, Ozkan AD, Soran Z, Ekiz O, Kocaay D. et al. Label-free nanometer-resolution imaging of biological architectures through surface enhanced Raman scattering. Sci Rep. 2013;3:2624
149. Ertsgaard CT, McKoskey RM, Rich IS, Lindquist NC. Dynamic placement of plasmonic hotspots for super-resolution surface-enhanced Raman scattering. ACS Nano. 2014;8:10941-6
150. Olson AP, Spies KB, Browning AC, Soneral PAG, Lindquist NC. Chemically imaging bacteria with super-resolution SERS on ultra-thin silver substrates. Sci Rep. 2017;7:9135
151. Henry AI, Sharma B, Cardinal MF, Kurouski D, Van Duyne RP. Surface-enhanced Raman spectroscopy biosensing: In vivo diagnostics and multimodal imaging. Anal Chem. 2016;88:6638-47
152. Hou Y, Zhang W, Li S, Wang Z, Zhong H, Liu Z, Guo Z. Investigating the autophagy pathway in silver@gold core-shell nanoparticles-treated cells using surface-enhanced Raman scattering. Analyst. 2018;143:3677-85
153. McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H. et al. International evaluation of an AI system for breast cancer screening. Nature. 2020;577:89-94
154. Hollon TC, Pandian B, Adapa AR, Urias E, Save AV, Khalsa SSS. et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med. 2020;26:52-8
Author contact
![]() Corresponding author: E-mail: xuspedu.cn.
Corresponding author: E-mail: xuspedu.cn.
 Global reach, higher impact
Global reach, higher impact