13.3
Impact Factor
Theranostics 2021; 11(6):2581-2593. doi:10.7150/thno.52366 This issue Cite
Review
N6-methyladenosine as a biological and clinical determinant in colorectal cancer: progression and future direction
1. Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Shanghai, China.
2. Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, China.
Received 2020-8-24; Accepted 2020-12-4; Published 2021-1-1
Abstract
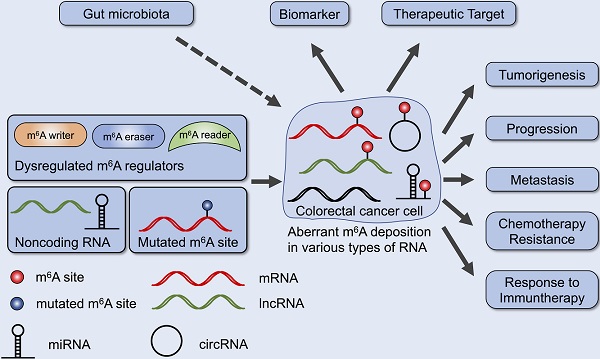
Colorectal cancer (CRC) is one of the most prevalent cancers and one of the leading causes of cancer death. Recent studies have provided evidence that N6-methyladenosine (m6A), the most abundant RNA modifications in eukaryote, performs many functions in RNA metabolism including translation, splicing, storage, trafficking and degradation. Aberrant regulation of m6A modification in mRNAs and noncoding RNAs found in CRC tissues is crucial for cancer formation, progression, invasion and metastasis. Further, m6A regulators and m6A-related RNAs may become promising biomarkers, prognosis predictors as well as therapeutic targets. Here, we review the biological and clinical roles of m6A modification in CRC, and discuss the potential of m6A in clinical translation.
Keywords: colorectal cancer, m6A, RNA modification, cancer progression, cancer treatment
Introduction
Colorectal cancer (CRC) is the third most prevalent cancer and the fourth leading causes of cancer death, with a rapidly increasing mobidity and mortality in developing countries and a stabilizing or decreasing trend in developed countries where the CRC burden remains among the highest worldwide [1]. The pathological mechanism of CRC development and progression includes chromosomal instability, microsatellite instability-high (MSI-H) and cytosine-phosphate-guanine (CpG) methylation, resulting in the mutations of oncogenes, tumor suppressor genes and genes related to mismatch repair [2]. Based on the novel understanding and advanced techniques, great improvement has been made to diagnose and treat CRC, which largely increases the overall survival. However, the prognosis of patients with advanced CRC is grim [3]. Therefore, the molecular mechanisms underlying CRC tumorigenesis and metastasis need to be further elucidated.
RNA modifications, including N6-methyladenosine (m6A), N1-methyladenosine (m1A), N7-methylguanidine (m7G), 5-methylcytidine (m5C), 2ʹ-O-methylation (Nm), pseudouridine (Ψ) and Inosine (I), are widely present in all types of RNAs, which mediate gene expression and participate in many biological processes, such as embryonic stem cell differentiation, circadian rhythms, temperature adaptation and meiotic progression [4]. The m6A is the most abundant epigenetic modification in eukaryotes occurring in various types of RNA, including message RNA (mRNA), micro RNA (miRNA), long noncoding RNA (lncRNA) and circular RNA (circRNA) [5-8]. As a reversible RNA methylation, m6A is installed by methyltransferase (also called writers), removed by demethylase (also called erasers), and recognized by some RNA-binding proteins (also called readers) (Figure 1). Typical consensus sequence of m6A modification sites is RRACH (R = G, A; H = A, C, U) [9,10]. The m6A readers recognize RNA methylation and perform different biological functions, including RNA translation, splicing, storage, trafficking and degradation [11].
m6A modification on RNAs. The m6A modification is installed by writers, multicomponent methyltransferase complex (composed of METTL3, METTL14, WTAP, RBM15/15B, KIAA1429, ZC3H13 and Flacc) or METTL16 alone. FTO or ALKBH5 are m6A erasers that remove m6A modifications. Readers are required to recognize m6A and exert post-transcriptional regulation.
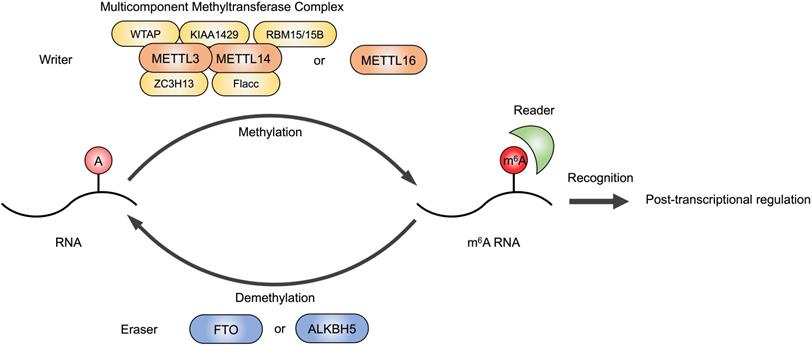
The m6A writer is a multicomponent methyltransferase complex in the nucleus, composed of a core protein heterodimer formed by methyltransferase like 3 (METTL3) and methyltransferase like 14 (METTL14), and other regulatory factors identified to interact with the METTL3/METTL14 complex to affect m6A deposition, including Wilms' tumor 1-associating protein (WTAP), Vir-like m6A methyltransferase-associated (KIAA1429/VIRMA), RNA-binding motif protein 15/15B (RBM15/15B), zinc finger CCCH domain-containing protein 13 (ZC3H13) and Fl(2)d-associated complex component (Flacc) [12-15]. METTL16 is also a methyltransferase that can function alone and catalyze m6A of mRNAs, lncRNAs and U6 small nuclear RNA (U6 snRNA) [16]. Fat mass and obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5) are the erasers that have been identified to have demethylation activity [17,18]. The dynamic regulation of m6A level in cellular RNAs is mediated by writers and erasers contributing to proper gene expression and protein production.
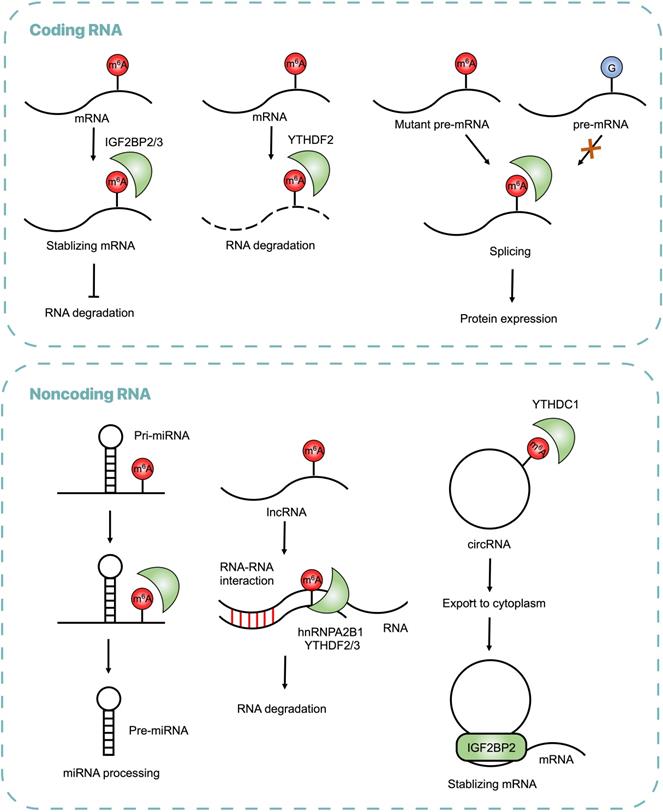
The well-studied m6A reader proteins are YT521-B homology (YTH) domain containing proteins, including YTHDC1-2 and YTHDF1-3, which bind RNA and recognize specific m6A sites to exert post-transcriptional function [19]. For example, YTHDC1 participated in pre-mRNA processing and changed the length of 3'UTR, contributing to mRNA polyadenylation and splicing [20]. YTHDF proteins altered translation efficiency and reduced stability of m6A modified RNAs [21]. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) are also identified as m6A readers, which promote the stability and storage of their target mRNAs [22].
In addition to mRNA processing, m6A modifications regulate metabolism of noncoding RNAs, including miRNA, lncRNA and circRNA. The miRNA is a short regulatory RNA (∼22 nucleotides, nt) encoded in introns of coding and non-coding pre-mRNAs, which represses its target mRNA [23]. Heterogeneous nuclear ribonucleoprotein A2B1 (HNRNPA2B1) is a reader protein that recruits the microprocessor complex to process primary miRNAs (pri-miRNAs) into mature miRNAs [24,25]. The lncRNA is the RNA transcript generally longer than 200nt that does not encode protein, is associated to many cellular functions, one of which is m6A-dependent [26]. lncRNA X-inactive specific transcript (XIST) mediated gene silence on the X chromosome via m6A installation and recognition [14]. The circRNA is a type of single-stranded non-coding RNA that forms a covalently closed loop, participating in pathological processes through a m6A-dependent manner [27].
Owing to the m6A sequencing techniques that allow the detection of m6A with high efficiency, numerous RNAs and proteins are found to be a part of m6A regulation in CRC tumorigenesis. Methylated RNA immunoprecipitation sequencing (MeRIP-seq or m6A-seq) is used widely to detect m6A despite of the resolution of near 100 nt [9,10]. New techniques, such as m6A individual-nucleotide-resolution cross-linking and immunoprecipitation (miCLIP) [28], m6A-sensitive RNA-endoribonuclease-facilitated sequencing (m6A-REF-seq or MAZTER-seq) [29,30], m6A-label-seq [31], deamination adjacent to RNA modification targets (DART-seq) [32], FTO-assisted m6A selective chemical labeling (m6A-SEAL) [33] are developed to map m6A at single-nucleotide resolution. Next generation sequencing methods on m6A epitranscriptome broaden our understanding of epigenomic marks in addition to traditional multi-omics analysis. Furthermore, the development of programmable RNA m6A editing by fusing CRISPR-Cas9 system with m6A writers or erasers without changing the primary sequence provides a powerful approach to uncover the mechanism under RNA modification during physiological and pathological processes [34].
It has been demonstrated that aberrant m6A deposition plays a critical role in various types of cancer [35]. In this review, we summarize the dysregulation of m6A in CRC as well as the mechanisms how m6A regulators and m6A-related RNAs participated in CRC pathogenesis. We also discuss the clinical potential of targeting m6A in CRC in future.
Aberrant Regulation of m6A in CRC
Cumulative evidence has revealed that m6A modification widely alters gene expression in CRC. Global abundance of m6A and its regulators, including writers, erasers and readers are found dysregulated in CRC, which exerts oncogenic and/or antitumor function in CRC through targeting different types of RNA and various signal pathways (Table 1). The mechanisms are shown (Figure 2). Transcriptome-wide m6A methylome showed global m6A modifications in CRC and found 1343 dysregulated m6A peaks in mRNA compared with adjacent normal tissues, among which 625 were upregulated and 718 were downregulated, crucial in regulating glucose metabolism, RNA metabolism and cancer stem cells [36]. In CRC, it was reported that m6A regulators, including METTL3, WTAP, FTO, YTHDF1, ZC3H13, HNRNPC, YTHDC, RBM15 and KIAA1429, were upregulated, while METTL14 and ALKBH5 were downregulated [37]. Some downstream pathways of m6A modification were identified, such as phosphatidylinositol-3-kinase (PI3K)/Akt and mammalian target of rapamycin (mTOR) signal pathway [38].
Dysregulation of m6A Writers
Aberrant expression of m6A writers in CRC leads to abnormal RNA metabolism, including mRNA stabilization, mRNA splicing, miRNA maturation and lncRNA degradation. Highly expressed METTL3 plays a critical role in CRC proliferation and progression by stabilizing target mRNAs through an m6A dependent manner. For instance, METTL3 raised m6A level of Cyclin E1 (CCNE1) mRNA in 3'UTR region and then increased CCNE1 mRNA stability in CRC cells [39]. Overexpression of METTL3 was also found in metastatic CRC tissues, with its downstream target SRY(sex determining region Y)-box 2 (SOX2) methylated in coding sequence (CDS) regions and subsequently recognized by reader protein IGF2BP2, which prevented SOX2 mRNA degradation [40]. Through interacting with the 5'/3'UTR regions of Hexokinase 2 (HK2) and the 3'UTR region of Glucose transporter 1 (GLUT1, also known as SLC2A1), METTL3 stabilized HK2 and GLUT1 transcripts to activate the glycolysis pathway and then promote CRC tumorigenesis [41].
Roles of m6A Regulators in CRC
| m6A Regulator | Role in CRC | Expression | m6A RNA | Mechanism | Ref. |
|---|---|---|---|---|---|
| Writer | |||||
| METTL3 | oncogenic | ↑ | mRNA SOX2 | enhance mRNA stability | [40] |
| oncogenic | ↑ | mRNA HK2 and SLC2A1 | enhance mRNA stability | [41] | |
| oncogenic | ↑ | mRNA CCNE1 | enhance mRNA stability | [39] | |
| oncogenic | ↑ | pre-mRNA p53 (273G>A) | promote pre-mRNA splicing | [91] | |
| oncogenic | ↑ | miR-1246 | promote pri-miRNA maturation | [42] | |
| oncogenic | mRNA Stat1 and Irf1 | promote mRNA degradation | [51] | ||
| oncogenic | ↑ | [37] | |||
| antitumor | ↓ | modulate p38/ERK pathways | [46] | ||
| METTL14 | antitumor | ↓ | lncRNA XIST | promote RNA degradation | [43] |
| antitumor | ↓ | miR-375 | promote pri-miRNA maturation | [45] | |
| antitumor | ↓ | [37] | |||
| antitumor | ↓ | mRNA SOX4 | promote mRNA degradation | [44] | |
| oncogenic | mRNA Stat1 and Irf1 | promote mRNA degradation | [51] | ||
| WTAP | oncogenic | ↓ | target the WTAP-WT1-TBL1 axis to suppress Wnt/β-catenin signalling | [55] | |
| oncogenic | ↑ | [37] | |||
| RBM15 | oncogenic | ↑ | [37] | ||
| ZC3H13 | oncogenic | ↑ | [37] | ||
| KIAA1429 | oncogenic | ↑ | [37] | ||
| ZCCHC4 | oncogenic | ↑ | [37] | ||
| Readers | |||||
| YTHDC1 | oncogenic | ↑ | circNSUN2 | increase cytoplastic export | [87] |
| oncogenic | ↑ | [37] | |||
| YTHDC2 | oncogenic | ↑ | promote translation of HIF-1α | [82] | |
| YTHDF1 | oncogenic | ↑ | transcriptionally upregulated by c-Myc | [84] | |
| oncogenic | ↑ | activate Wnt/β-catenin signaling pathway | [83] | ||
| oncogenic | ↓ | [37] | |||
| YTHDF3 | oncogenic | ↑ | lncRNA GAS5 | facilitate RNA degradation | [93] |
| IGF2BP2 | oncogenic | ↑ | mRNA HMGA2 | enhance mRNA stability | [87] |
| oncogenic | ↑ | mRNA SOX2 | enhance mRNA stability | [40] | |
| HNRNPC | oncogenic | ↑ | [37] | ||
| RBRP | oncogenic | ↑ | mRNA c-Myc | enhance mRNA stability | [86] |
| Erasers | |||||
| FTO | oncogenic | ↑ | [37] | ||
| ALKBH5 | oncogenic | ↓ | mRNA Mct4/Slc16a3 | promote pre-mRNA splicing and enhance mRNA stability | [81] |
| oncogenic | ↓ | [37] |
Regulatory Functions of m6A on RNAs in CRC. The m6A modifications recognized by reader proteins influence RNA metabolism, including RNA stabilization, splicing, processing, translocation and degradation.
In addition to the function of mRNA stabilization, the oncogenic role of METTL3 also affects noncoding RNA metabolism. Upregulated METTL3 markedly stabilized nascent lncRNA RP11 and increased its nuclear accumulation, which contributed to dissemination of CRC by escalating zinc finger E-Box binding homeobox 1 (Zeb1) [7]. The miRNA is also the target of METTL3. METTL3 enhanced the metastatic potential of CRC by promoting the maturation of pri-miR1246 in a DGCR8-dependent manner. The miR1246 negatively regulated anti-oncogene Sprouty Related EVH1 Domain Containing 2 (SPRED2) which prevented cancer cell migration and invasion through Raf/MEK/ERK pathway [42].
Different from the oncogenic role of METTL3, METTL14 acts as an antitumor gene that suppresses CRC proliferation and metastasis. Loss of METTL14 correlated with unfavorable prognosis of CRC patients. METTL14 induced methylation of lncRNA XIST as well as mRNA SOX4 to downregulate their expression via YTHDF2-mediated RNA degradation [43,44] METTL14 also suppressed CRC cell growth via miR-375/YAP1 pathway as well as inhibited CRC cell migration and invasion via miR-375/SP1 pathway [45].
In contrary to the studies listed above that supported the oncogenic role of METTL3 in CRC, Ru et al. showed an antitumor role of METTL3, whose expression was associated with better survival and suppressed cancer growth and metastasis via p38/ERK pathways [46]. Consistently, the controversial role of METTL3 has also been reported in other cancer types by different groups, such as glioblastoma [47,48]. Like the dual role of METTL3, it was also reported that METTL14 acts as an oncogene in acute myeloid leukemia (AML) and breast cancer [49,50]. The possible reason that may explain the controversial role of m6A writers is that the m6A sites and m6A modified RNAs reported in different studies are varied, which regulated different downstream targets and signal pathways, leading to the cancer heterogeneity. Hence, m6A and its regulators are promising biomarkers to distinguish cancer features.
It was reported that depletion of Mettl3 or Mettl14 in CRC cells enhanced the response of Patient-Derived Xenograft (PDX) mice to anti‐PD‐1 treatment by stabilizing the STAT1 and IRF1 mRNA and promoting IFN‐γ‐Stat1‐Irf1 signaling, which recruited cytotoxic tumor‐infiltrating CD8+ T cells in tumor microenvironment with escalated IFN‐γ, Cxcl9 and Cxcl10 [51]. In this case, METTL3 and METTL14 functioned synchronously to suppress anti-PD-1 treatment.
As a regulatory factor of m6A methyltransferase complex, the role of WTAP in CRC is far from understood. The association between WTAP expression and prognosis in patients with CRC varied from separate microarray databases [52,53], suggesting the multidimensional function of WTAP in tumor progression. Although some transcription factor binding sites have been identified in the promoter region of WTAP [54], the m6A related function of WTAP remains uncovered. Despite no direct evidence showing that WTAP could change m6A deposition in CRC cells, an interesting study demonstrated that the expression of WTAP was modulated by hypermethylation of CpG island, which might create a link between RNA modification and DNA modification. Transcriptional silence of CA4 in CRC tissues was mediated by hypermethylation with CpG sites within the CA4 promoter. CA4 interacted with WTAP and promoted its degradation through polyubiquitination and proteosome degradation, which in turn activated WT1 and increased the expression of downstream transcriptional factor Transducin β-Like Protein 1 (TBL1), resulting in the degradation of β-catenin and the inhibition of the Wnt signaling pathway [55]. This finding indirectly connected m6A regulator WTAP with CpG methylation, indicating that RNA modification and DNA modification might regulate each other.
Other than m6A, m5C is a common RNA modification identified in transfer RNA (tRNA) [56], mRNA [57], vault RNA (vtRNA) [58] and also mitochondrial tRNA (mt-tRNA) [59]. NOL1/NOP2/SUN domain family member 2 (NSUN2) is a RNA methyltransferase that introduces m5C into various RNAs to regulate RNA metabolism [56-59]. The research group of the scientist Wengong Wang reported that the m6A level of miR-125b was positively associated with the expression of NSUN2, suggesting a role of NSUN2 in repressing miRNA via m6A [60]. NSUN2, upregulated by proteinase-activated receptor 2 (PAR2) in CRC, was shown to interfere in the mature processing of miR-125b through a m6A dependent manner, thus regulating the expression of downstream gene Gab2, which contributed to CRC cell migration [61]. It was puzzled that m5C methyltransferase NSUN2 could alter m6A level in RNA and exert post-transcriptional regulation since there was no evidence that NSUN2 could catalyze m6A. Follow-up study from Wengong Wang's group reported a cooperative function of m6A and m5C, which explained the role of NSUN2 via m6A. The m5C methylation mediated by NSUN2 facilitated the m6A methylation by METTL3/METTL14 in p21 mRNA, and reciprocally METTL3/METTL14-mediated m6A methylation enhanced NSUN2-mediated m5C methylation, implicating that joint m6A and m5C modification of the same RNA may influence each other and therefore coordinately affected protein expression, added a new layer of post-transcriptional regulation by RNA modification [62].
Dysregulation of m6A Erasers
FTO has been considered essential for modulating fat mass and adipogenesis, whose single-nucleotide polymorphisms (SNPs) are related to the rising incidence of obesity and increasing risks of multiple cancer [63]. FTO mRNA showed high mutation rate in MSI-H CRC, and corresponding frameshift peptides were produced [64]. A number of studies presented different association between FTO SNPs and the risk of CRC based on different population and races [65-72]. Recently, FTO has been identified as m6A and m6Am demethylase of mRNA and small nuclear RNA (snRNA) in the cell nucleus and cytoplasm [73-75], which performs m6A-related function in many types of cancer [76-79]. FTO is also found upregulated in colorectal adenocarcinoma samples [34]. Whether FTO participates in the CRC tumorigenesis via m6A requires further study.
ALKBH5 is the second identified m6A demethylase that modulates RNA metabolism [80]. It was reported that ALKBH5 knockout (KO) in CRC cells enhanced efficacy of anti-PD-1 immunotherapy and improved mouse survival, indicating that ALKBH5 was a potential therapeutic target to improve immunotherapy outcome [81]. During anti-PD-1 treatment, ALKBH5 decreased m6A near splice sites and modulated splicing of MCT4/SLC16A3 mRNA, which regulated lactate accumulation and infiltered immune cell in tumor microenvironment [81].
Dysregulation of m6A Readers
The m6A readers are RNA binding proteins that recognize m6A modification at certain motif and control the modified RNA fate, so dysregulation of m6A readers may perturb RNA metabolism leading to pathological processes. For instance, upregulated YTHDC2 unwinded the 5′UTR of HIF-1α mRNA and promoted translation initiation, which contributed to colon cancer metastasis [82]. In colonospheres, overexpressed YTHDF1 regulated stem cell-like activity, thus promoting tumorigenicity and cell cycle progression through Wnt/β-catenin pathway [83]. In CRC, oncogenic transcription factor c-Myc may account for the amplified YTHDF1 [84].
Apart from YTH domain family proteins, IGF2BP proteins were also identified as m6A readers [85]. IGF2BP2 recognized m6A at CDS region of SOX2 mRNA maintaining its stability [40]. An additional regulatory subunit of m6A reader was first identified and named as RNA-binding regulatory peptide (RBRP), which was a 71-amino acid peptide encoded by a previously annotated lncRNA LINC00266-1 [86]. Through binding to IGF2BP1, RBRP strengthened m6A recognition by IGF2BP1 on c-Myc mRNA to increase the mRNA stability and expression, thereby promoting tumorigenesis [86].
Reader proteins also regulate non-coding RNA, such as circRNA. CircNSUN2 is a circRNA derived from the exons 4 and 5 regions within the NSUN2 locus. It was clinically reported that upregulated expressions of circNSUN2 and HMGA2 mRNA are more prevalent in liver metastasis tissues than in primary CRC tissues [87]. The cytoplasmic export of circNSUN2 was modulated by YTHDC1 at the GAACU motif. In the cytoplasm, circNSUN2 interacted with IGF2BP2 at the CAUCAU motif and then stabilized HMGA2 by forming a circNSUN2/IGF2BP2/HMGA2 RNA-protein ternary complex, which promoted liver metastasis [87]. In this case, two reader proteins, YTHDC1 and IGF2BP2, recognized the same RNA at different motifs and exerted different functions respectively.
Dysregulation of m6A regulators causes abnormal m6A modifications in various RNA and aberrantly regulates the expression of RNA and their downstream pathways, which plays a critical role in cancer.
Mutations in m6A sites in CRC
Generally, gene mutations are commonly found in cancer samples. However, little is known about the role of mutations in m6A sites in cancer. Mutated m6A sites of RNA may alter m6A deposition, which triggers aberrant post-transcriptional regulation and therefore leads to carcinogenesis. It was reported that he germline missense rs8100241 variant, located in the exon of Ankyrin Repeat and LEM Domain Containing 1 (ANKLE1) with a G>A change (Ala>Thr), was associated with decreased risk of CRC [88]. Less microsatellites was found in the ANKLE1 [A] than the ANKLE [G] allele, suggesting the ANKLE [A] could function as a potential tumor suppressor that inhibited cancer cell proliferation by maintaining genomic stability. Variant ANKLE1 [A] was methylated by METTL3 while ANKLE [G] could not be methylated, which facilitatd the stability of ANKLE1 mRNA via m6A and promoted the expression of ANKLE1 protein, resulting in the reduced risk of CRC [88].
Generally, p53 is the most frequently mutated gene in cancer [89]. In response to DNA damage stress and other oncogenic stresses, cells highly expressed p53 protein and upregulated its target genes, which triggered cell-cycle arrest, senescence and cell death by apoptosis or ferroptosis [90]. The point-mutated codon 273 (G>A) of p53 pre-mRNA promoted its splicing through methylation of METTL3, leading to the over production of p53 R273H mutant protein that contributed to multidrug resistance in CRC [91].
non-coding RNAs regulate m6A modification in CRC
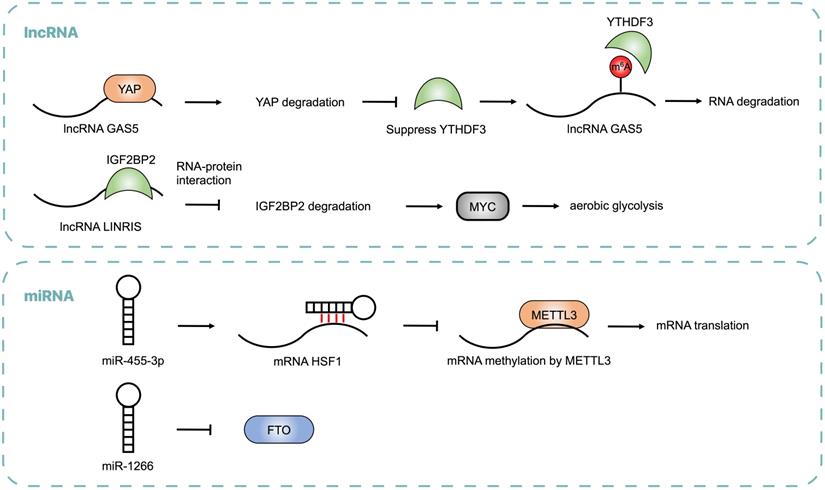
Emerging evidence showed that non-coding RNAs may regulate m6A modification by modulating the expression of m6A regulators and influence post-transcriptional gene expression in CRC (Table 2 and Figure 3). lncRNA LINRIS was upregulated in CRC patients with poor prognosis. LINRIS blocked K139 ubiquitination of IGF2BP2 and prevented its degradation through the autophagy-lysosome pathway (ALP), which promoted the MYC-mediated aerobic glycolysis in CRC cells [92]. The transcription of LINRIS was inhibited by GATA3 in CRC cells, thus suppressing the proliferation of tumors both in orthotopic models and PDX models. Another lncRNA GAS5 could upregulate YTHDF3 through a YAP-dependent manner, while YTHDF3 could also recognize m6A of GAS5 to promote its decay, which formed a negative function loop of GAS5-YAP-YTHDF3 that contributed to CRC progression [93]. Consistent with lncRNA, abnormal expression of miRNA was found in CRC. miR455-3p bound to 3'-UTR of HSF1 mRNA to block its interaction with METTL3 and repress its translation, thus inhibiting CRC progression. However, β-catenin suppressed the generation of miR455-3p and enhanced the expression of HSF1, which promoted glutaminolysis and activated mTOR in CRC [94]. It was reported that miR-1266 was lowly expressed in CRC tissues and negatively regulated the expression of FTO, leading to the proliferation of CRC [95]. The expression of miRNA-1266 was correlated to tumor size and TNM of CRC patients.
Noncoding RNAs, including lncRNA and miRNA, regulate m6A modification by modulating the expressions of m6A regulators, which is critical for CRC progression.
Roles of ncRNAs in CRC
| RNA type | Role in CRC | m6A enzyme | Mechanism | Ref |
|---|---|---|---|---|
| miRNA | ||||
| miR-1266 | antitumor | FTO | negatively regulate FTO | [95] |
| miR455-3p | antitumor | METTL3 | bind to the target mRNA of METTL3 to block their interaction and repress its translation | [94] |
| lncRNA | ||||
| GAS5 | antitumor | YTHDF3 | suppress YTHDF3 through YAP-mediated pathway by facilitating YAP translocation, phosphorylation and degradation | [93] |
| LINRIS | oncogenic | IGF2BP2 | prevent the degradation of IGF2BP2 from autophagy-lysosome pathway | [92] |
It has been found that non-coding RNAs are critical for cancer progression, but little is known about its function through an m6A-dependent manner. These results advance our understanding of non-coding RNA in cancer epigenetics.
m6A as clinical determinants in CRC
m6A as biomarkers of CRC
Most m6A regulators are dysregulated in CRC and their expression level was found correlated with clinical outcome of CRC patients, indicating the potential to become biomarkers for CRC [96]. For instance, the downregulation of METTL14 is closely related to malignant progression and poor recurrence-free survival and overall survival of patients, suggesting the potential role of METTL14 in predicting tumor metastasis and recurrence [45]. The expression of YTHDC2 was found to be positively correlated with the colon tumor stage, including metastasis [82]. CRC patients with high expression of RBRP have a poor prognosis [86].
However, the same m6A regulator may exert oncogenic or antitumor function according to different studies. Some studies identified METTL3 as an oncogene associated with poor prognosis [37,39-42,91], while another study suggested that positive expression of METTL3 is correlated with longer survival time [46]. Since the heterogeneity of m6A regulators made it difficult to detect cancer or predict prognosis, their target RNAs may be better biomarkers. Among various RNAs, circRNAs that can be detected in the blood are promising biomarkers. The m6A-modified circNSUN2 was found upregulated in serum and metastatic liver tissues of patients and was positively associated with CRC cell invasion, thus providing a novel diagnostic and prognostic predictor for colorectal liver metastasis [87]. Future studies should examine whether the serum concentration of m6A regulators and their target RNAs is correlated to CRC diagnosis or prognosis, rather than their expression level in tumor tissues.
Due to the rapid development of bioinformatics, many software tools have been developed to predict cancers. Bioinformatic tool RNAMethyPro, a novel gene expression signature that comprised of seven m6A regulators, is used to predict prognosis in multiple cancers [97]. Using comprehensive pan-cancer analysis, activated epithelial-mesenchymal transition (EMT) is identified as a highly conserved biological process across multiple cancer types, and further investigation on CRC revealed that high-risk patients were associated with the mesenchymal subtype, activated stromal infiltration and poor anti-EGFR therapeutic response [97].
m6A as therapeutic target of CRC
Developing inhibitors of oncogenic m6A regulator like METTL3 and agonist of antitumor m6A regulator like METTL14 is a promising therapeutic strategy to overcome cancer, improve immune responses and reduce drug resistance.
Although it remains unknown how FTO participates in CRC tumorigenesis, FTO inhibitors have been widely explored as anticancer drugs in other types of cancer. Meclofenamic acid (MA) is one of selective FTO inhibitors by competing with FTO binding sites [98]. Another inhibitor of FTO called FB23-2 has been developed to impair the proliferation and enhance differentiation of AML cells [99]. R-2-hydroxyglutarate (R-2HG) inhibits FTO demethylase activity and elevates m6A level in leukemia cells by enhancing the degradation of MYC and CEBPA, thus displaying anti-tumor activity [100]. Although some inhibitors have been applied to some types of cancer, there is still no application of these therapeutics in CRC patients.
The m6A modifications can regulate immune responses to anti‐PD‐1 therapy. As mentioned before, METTL3 or METTL14 KO as well as ALKBH5 KO in mice enhanced the efficacy of anti-PD-1 therapy [51,81]. Using ALKBH5-specific inhibitor could lead to the similar phenotype, indicating the future translational application [81]. Therefore, the inhibitors of m6A regulators can also be developed as an adjuvant therapy.
Mutations in m6A sites can induce multidrug resistance in CRC cells, so targeting m6A by regulating expression of m6A regulator is emerging as new therapeutics. For instance, mutant p53 proteins that promote drug resistance is a prospective therapeutic target. Either silencing METTL3 expression by using small interfering RNA or inhibiting RNA methylation with neplanocin A suppressed m6A formation in p53 pre-mRNA, and substantially increased its phosphorylation level which reduced p53 function in cells heterozygously carrying the R273H mutation, thereby re-sensitizing these cells to anticancer drugs [91].
Although targeting m6A seems a promising therapeutic strategy in addition to present treatments, the side-effect should not be ignored. RNA modification exists in all types of cells and exerts many fundamental function to maintain physiological function. Therefore, the application of inhibitors or agonist of m6A regulators may disturb its function in healthy organisms and lead to severe outcomes.
Potential of Gut microbiota to alter m6A in CRC
Gut microbiota has been an area of intense focus of biological and clinical research [101]. Gut microbiota comprises trillions of bacteria, viruses, fungi, archaea and protozoa, with their genome encoding numerous proteins that human cannot produce, which may have critical functions in physiological and pathological process [102]. Dysbiosis of gut microbiota may lead to abnormal composition of microorganisms and impact human health. Increasing diseases are being demonstrated relevant to host microbiota, including inflammatory bowel disease [103], diabetes [104], cardiovascular diseases [105], neurological diseases [106], and various types of cancer [107,108]. It was reported that gut microbiota might affect m6A deposition in the cecum and liver and influence pathways related to metabolism, inflammation and antimicrobial responses, which contributed to CRC tumorigenesis and metastasis [109]. For instance, compared with conventional mice, METTL16 was downregulated in germ-free mice with its target mRNA Mat2a less methylated [109]. Therefore, m6A modification represents a novel mechanism of interaction between host and commensal bacteria, setting the ground for future studies and promising therapies.
Increasing metabolites produced by gut microbiota have been demonstrated to influence CRC. Butyrate is a kind of short chain fatty acids as well as a classical intestinal microbial metabolite, which could modulate gut microbiota composition by increasing Firmicutes and Proteobacteria, and improve host immune response in mice with CRC liver metastasis [110]. Butyrate inhibited CRC development through an m6A dependent manner by downregulating METTL3 and related cyclin E1 [39]. Recently, aspirin has been discovered to reduce the risk of CRC in human and this function was confirmed in mice that gut microbes could affect the bioavailability and protective effects of aspirin to prevent colon tumor formation [111]. Further studies may examine whether oral administration of bacteria products or their metabolites could prevent cancer in human, minimizing the side effect of aspirin and maximizing its protective effect. Drugs other than aspirin can be tested to explore whether they exert the antitumor or oncogenic function through gut microbiota. Although accumulating evidence has been uncovering the relationship between gut bacteria and CRC tumorigenesis, there is little study on other types of microorganism, such as virus and fungi. Future studies should focus more on these species that colonize in intestinal tract which may also influence human health [112].
Conclusions and perspectives
The incidence and mortality for CRC has been largely reduced by regular screening with fecal occult blood test and colonoscopy, starting at age 50 years [113]. Patients diagnosed CRC at early stage benefit greatly from surgery and have a long overall survival. However, in the past few decades, there is an increasing trend of CRC patients diagnosed before age 50 years, also called early-onset CRC, with more evident for rectal cancer than colon cancer [114]. Coincidentally, the expression patterns of m6A regulators are different in colon cancer and rectal cancer, indicating different m6A features between colon cancer and rectal cancer [96]. Further, the overall survival of CRC patients at late stage is far from satisfactory, especially with liver metastasis. Therefore, the burden of CRC remains high in many countries.
The exploration of epigenetics is uncovering a new layer of cancer biology. As the most prevalent RNA modification in eukaryote, m6A has been detected in different types of RNAs to regulate post-transcriptional gene expression and participate in various biological processes. The m6A modification is raising increasingly broad concern due to cumulative evidence demonstrating that aberrant m6A level in RNAs may influence the occurrence and development of various types of cancers [115]. For example, m6A modification in gastric cancer promoted EMT and metastasis through METTL3/ZMYM1/E-cadherin signaling [116]. METTL3 promoted hepatocellular carcinoma progression through YTHDF2-dependent degradation of SOCS2 mRNA [117]. FTO, highly regulated in certain AML subtypes, abolished m6A of ASB2 and RARA mRNA, which enhanced leukemogenesis and inhibited all-trans-retinoic acid (ATRA)-induced AML cell differentiation [76]. Interestingly, it was reported that METTL3 was modified by small ubiquitin-like modifier (SUMO) in hepatocellular carcinoma, which promoted cancer progression through mediating Snail mRNA homeostasis [118]. Since dysregulated m6A modification is found in various types of cancer, systematic analysis of molecular features and clinical relevance of m6A regulators may further improve our understanding of cancer biology [119].
In addition to m6A, other RNA modifications have also been reported in CRC, such as m7G and Nm. The maturation of miRNA let-7e, mediated by m7G methylation of METTL1, interfered the translation of high mobility group AT-hook 2 (HMGA2) mRNA, which inhibited the progression of colon cancer [120]. lncRNA ZFAS1 stabilized multiple small nucleolar RNAs(snoRNA) and promoted Nm modification of rRNAs, thus regulating the RNA stability and translation of their downstream targets, leading to CRC initiation and maintenance [121]. The coordination of different RNA modifications, such as m6A and m5C, has been reported [62]. Moreover, the epigenetic modifications are not not isolated phenomena, but a complex regulatory network where multiple crosstalk of dysregulated modifications exists, which forms a comprehensive biological system [122,123].
There are two controversial issues on current researches of m6A.
Although both METTL3 and METTL14 are the core components of methyltransferase complex, METTL3 was upregulated while METTL14 was downregulated in CRC, which exerts oncogenic and antitumor effect respectively. The differences is partly because METTL3 is the catalytic unit while METTL14 is essential for stabilizing METTL3 conformation, substrates RNA binding and m6A sites deciding [124]. Constructing mouse model of METTL3 knockdown and knockout as well as METTL14 knockdown and knockout may help explain their different role in cancer. Interactions between methyltransferase complex and its regulatory factors may also contribute to their biological function, which requires further elucidation.
The other one is the contradiction that MELLT3, the m6A writer that increases the RNA m6A level, is an oncogene in CRC, while FTO or ALKBH5, the m6A eraser that decreases the RNA m6A level, is also an oncogene in CRC. We made a hypothesis that if the writer installs m6A at a specific site of a certain RNA, and the eraser uninstalls m6A in the same site of the same RNA, they may regulate the RNA conversely and represent opposite roles in cancer: one is oncogenic and the other is antitumor. It was reported that depletions of METTL3 and ALKBH5 resulted in substantially decreased and increased expression of a subset of small GTPase mRNAs and proteins [125], which supported our hypothesis to some extent. Therefore, the targets, including the RNAs, m6A sites and downstream pathways, of writers and erasers that reported in different studies are different, which reveals different mechanisms in cancer carcinogenesis. However, only a modest number of small GTPases were modulated by METTL3 and ALKBH5 [125], indicating other regulatory factors other than the writer and eraser. The m6A readers are crucial m6A regulators to exert post-transcriptional functions, which may account for the seemingly contradictory roles between the writer and eraser. Although some m6A methylases and demethylases have been identified, there are still potential m6A regulators undiscovered, which may participate in the regulation of m6A modification. Future studies could examine the effect of m6A writer and eraser on the same RNA simultaneously to explore whether there are other regulatory factors involved in m6A modification.
In conclusion, we summarized the current advances of m6A modification and its regulators in CRC, uncovering a novel dimension of cancer biology. Given that the m6A modification is one of numerous epigenetic modifications, it is essential to explore whether the m6A-mediated post-transcriptional regulation contributes more to the differences between the transcriptome and proteome. Despite the dual role of m6A regulators that exerted either oncogenic or antitumor role in various types of cancer, their downstream genes may become better biomarkers or therapeutic targets. The inhibitors of m6A regulators showed the potential of an adjuvant therapy to increase immune responses to anti-PD-1 therapy or reduce multidrug resistance. So far, there is no clinical application of therapeutic strategy targeting m6A in CRC. Therefore, more efforts should be made to elucidate the mechanism and develop novel treatments.
Abbreviations
ALKBH5: alkB homolog 5; ALP: autophagy-lysosome pathway; AML: acute myeloid leukemia; ANKLE1: Ankyrin Repeat and LEM Domain Containing 1; ATRA: all-trans-retinoic acid; CCNE1: Cyclin E1; CDS: coding sequence; circRNA: circular RNA; CpG: cytosine-phosphate-guanine; CRC: colorectal cancer; DART-seq: deamination adjacent to RNA modification targets; EMT: epithelial-mesenchymal transition; Flacc: Fl(2)d-associated complex component; FTO: Fat mass and obesity-associated protein; Gab2: GRB2 associated binding protein 2; GATA3: GATA binding protein 3; GLUT1: glucose transporter 1; HIF-1α: hypoxia-inducible factor 1α; HK2: hexokinase 2; HMGA2: high mobility group AT-hook 2; HNRNPA2B1: heterogeneous nuclear ribonucleoprotein A2B1; I: inosine; IFN: interferon; IGF2BPs: insulin-like growth factor 2 mRNA-binding proteins; IRF1: interferon regulatory factor 1; KO: knockout; lncRNA: long noncoding RNA; m1A: N1-methyladenosine; m5C: 5-methylcytidine; m6A-REF-seq: m6A-sensitive RNA-endoribonuclease-facilitated sequencing; m6A-SEAL: FTO-assisted m6A selective chemical labeling; m6A: N6 Methyladenosine; m6Am: N6,2′-O-dimethyladenosine; m7G: N7-methylguanidine; MA: meclofenamic acid; Mct4: monocarboxylate transporter 4; MeRIP-seq: methylated RNA immunoprecipitation sequencing; METTL14: methyltransferase like 14; METTL16: methyltransferase like 16; METTL3: methyltransferase like 3; miCLIP: m6A individual-nucleotide-resolution cross-linking and immunoprecipitation; miRNA: micro RNA; mRNA: message RNA; MSI-H: microsatellite instability-high; mt-tRNA: mitochondrial tRNA; mTOR: mammalian target of rapamycin; Nm: 2ʹ-O-methylation; NSUN2: NOL1/NOP2/SUN domain family member 2; PAR2: proteinase-activated receptor 2; PD1: programmed cell death protein 1; PDX: patient-derived xenograft; PI3K: phosphatidylinositol-3-kinase; pri-miRNAs: primary miRNAs; R-2HG: R-2-hydroxyglutarate; RBM15/15B: RNA-binding motif protein 15/15B; RBRP: RNA-binding regulatory peptide; SLC16A3: solute carrier family 16 member 3; snoRNA: small nucleolar RNAs; SNPs: single-nucleotide polymorphisms; snRNA: small nuclear RNA (U6); SOX: SRY (sex determining region Y)-box; SPRED2: Sprouty related EVH1 domain containing 2; STAT1: signal transducer and activator of transcription 1; TBL1: transducin β-like protein 1; tRNA: transfer RNA; UTR: untranslated region; vtRNA: vault RNA; WTAP: Wilms' tumor 1-associating protein; XIST: X-inactive specific transcript; YAP1: Yes-associated protein 1; YTH: YT521-B homology; YTHDC1-2: YTH domain-containing protein 1-2; YTHDF1-3: YTH domain family protein 1-3; ZC3H13: zinc finger CCCH domain-containing protein 13; Zeb1: zinc finger E-Box binding homeobox 1; Ψ: pseudouridine.
Acknowledgements
The authors take this opportunity to thank all of the participating patients and healthy volunteers for supporting this study by donating the precious samples used in this research.
Authors' contributions
JL and YM designed this study and JL drafted the manuscript. JL collected the data and conducted the picture processing. YM, YY and XL revised the paper and all authors read and approved the final manuscript.
Availability of supporting data
All data generated or analyzed during this study are included in this published article and its additional files.
Authors' information
Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, Shanghai, China; Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, China.
Jinming Li, Lei Liang, Yongzhi Yang, Xinxiang Li, Yanlei Ma.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No's. 81920108026, 81871964), the National Ten Thousand Plan Young Top Talents (for Dr. Yanlei Ma), the Shanghai Young Top Talents (for Dr. Yanlei Ma. No. QNBJ1701), the Shanghai Science and Technology Development Fund (No. 19410713300), the CSCO-Roche Tumor Research Fund (No. Y-2019Roche-079).
Competing Interests
The authors have declared that no competing interest exists.
References
1. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683-691
2. Mármol I, Sánchez-de-Diego C, Dieste AP, Cerrada E, Yoldi MJR. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017 18
3. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490-502
4. Haruehanroengra P, Zheng YY, Zhou Y, Huang Y, Sheng J. RNA modifications and cancer. RNA Biol. 2020;17:1560-75
5. Nicholas Dias, Yung Peng RK. M6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475-8
6. Sun Y, Zhao X, Zhou Y, Hu Y. MiR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol Rep. 2012;28:1346-52
7. Wu Y, Yang X, Chen Z, Tian L, Jiang G, Chen F. et al. M(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18:87
8. Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L. et al. N6-methyladenosine modification controls circular RNA immunity. Mol Cell. 2019;76:96-109
9. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201-6
10. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3' UTRs and near stop codons. Cell. 2012;149:1635-46
11. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15:293-306
12. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93-5
13. Schwartz S, Mumbach MR, Jovanovic M, Wang T, Maciag K, Bushkin GG. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5' sites. Cell Rep. 2014;8:284-96
14. Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M. et al. M(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369-73
15. Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH. et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32:415-29
16. Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Höbartner C. et al. Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18:2004-14
17. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang YY-G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885-7
18. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29
19. Liu S, Li G, Li Q, Zhang Q, Zhuo L, Chen X. et al. The roles and mechanisms of YTH domain-containing proteins in cancer development and progression. Am J Cancer Res. 2020;10:1068-84
20. Kasowitz SD, Ma J, Anderson SJ, Leu NA, Xu Y, Gregory BD. et al. Nuclear m6A reader YTHDC1 regulates alternative polyadenylation and splicing during mouse oocyte development. PLOS Genet. 2018;14:e1007412
21. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M. et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626
22. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-95
23. Bartel DP. Metazoan microRNAs. Cell. 2018;173:20-51
24. Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482-5
25. Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299-308
26. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47-62
27. Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D. et al. The role of N(6)-methyladenosine (m(6)A) modification in the regulation of circRNAs. Mol Cancer. 2020;19:105
28. Linder B, Grozhik A V, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12:767-72
29. Zhang Z, Chen L-Q, Zhao Y-L, Yang C-G, Roundtree IA, Zhang Z. et al. Single-base mapping of m(6)A by an antibody-independent method. Sci Adv. 2019;5:eaax0250
30. Garcia-Campos MA, Edelheit S, Toth U, Safra M, Shachar R, Viukov S. et al. Deciphering the “m(6)A Code” via antibody-independent quantitative profiling. Cell. 2019;178:731-747
31. Shu X, Cao J, Cheng M, Xiang S, Gao M, Li T. et al. A metabolic labeling method detects m(6)A transcriptome-wide at single base resolution. Nat Chem Biol. 2020;16:887-95
32. Meyer KD. DART-seq: an antibody-free method for global m(6)A detection. Nat Methods. 2019;16:1275-80
33. Wang Y, Xiao Y, Dong S, Yu Q, Jia G. Antibody-free enzyme-assisted chemical approach for detection of N(6)-methyladenosine. Nat Chem Biol. 2020;16:896-903
34. Liu XM, Zhou J, Mao Y, Ji Q, Qian SB. Programmable RNA N 6-methyladenosine editing by CRISPR-Cas9 conjugates. Nat Chem Biol. 2019;15:865-71
35. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176
36. Zhang Z, Wang Q, Zhang M, Zhang W, Zhao L, Yang C. et al. Comprehensive analysis of the transcriptome-wide m6A methylome in colorectal cancer by MeRIP sequencing. Epigenetics. 2020 p:1-11
37. Liu T, Li C, Jin L, Li C, Wang L. The Prognostic Value of m6A RNA Methylation Regulators in Colon Adenocarcinoma. Med Sci Monit. 2019;25:9435-45
38. Zhao Q, Zhao Y, Hu W, Zhang Y, Wu X, Lu J. et al. M(6)A RNA modification modulates PI3K/Akt/mTOR signal pathway in Gastrointestinal Cancer. Theranostics. 2020;10:9528-43
39. Zhu W, Si Y, Xu J, Lin Y, Wang J-Z, Cao M. et al. Methyltransferase like 3 promotes colorectal cancer proliferation by stabilizing CCNE1 mRNA in an m6A-dependent manner. J Cell Mol Med. 2020;24:3521-33
40. Li T, Hu P-S, Zuo Z, Lin J-F, Li X, Wu Q-N. et al. METTL3 facilitates tumor progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18:112
41. Shen C, Xuan B, Yan T, Ma Y, Xu P, Tian X. et al. M(6)A-dependent glycolysis enhances colorectal cancer progression. Mol Cancer. 2020;19:72
42. Peng W, Li J, Chen R, Gu Q, Yang P, Qian W. et al. Upregulated METTL3 promotes metastasis of colorectal cancer via miR-1246/SPRED2/MAPK signaling pathway. J Exp Clin Cancer Res. 2019;38:393
43. Yang X, Zhang S, He C, Xue P, Zhang L, He Z. et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer. 2020;19:46
44. Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B. et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19:106
45. Chen X, Xu M, Xu X, Zeng K, Liu X, Sun L. et al. METTL14 suppresses CRC progression via regulating N6-methyladenosine-dependent primary miR-375 Processing. Mol Ther. 2020;28:599-612
46. Deng R, Cheng Y, Ye S, Zhang J, Huang R, Li P. et al. M(6)A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco Targets Ther. 2019;12:4391-402
47. Li F, Yi Y, Miao Y, Long W, Long T, Chen S. et al. N6-methyladenosine modulates nonsense-mediated mRNA decay in human glioblastoma. Cancer Res. 2019;79:5785-98
48. Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G. et al. M6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622-34
49. Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L. et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m6A modification. Cell Stem Cell. 2018;22:191-205
50. Panneerdoss S, Eedunuri VK, Yadav P, Timilsina S, Rajamanickam S, Viswanadhapalli S. et al. Cross-talk among writers, readers, and erasers of m6A regulates cancer growth and progression. Sci Adv. 2018;4:eaar8263
51. Wang L, Hui H, Agrawal K, Kang Y, Li N, Tang R. et al. M(6)A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 2020;39:e104514
52. Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, Desai J. et al. Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15:7642-51
53. Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A. et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958-68
54. Wu L-S, Qian J-Y, Wang M, Yang H. Identifying the role of Wilms tumor 1 associated protein in cancer prediction using integrative genomic analyses. Mol Med Rep. 2016;14:2823-31
55. Zhang J, Tsoi H, Li X, Wang H, Gao J, Wang K. et al. Carbonic anhydrase IV inhibits colon cancer development by inhibiting the Wnt signalling pathway through targeting the WTAP-WT1-TBL1 axis. Gut. 2016;65:1482-93
56. Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S. et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900-5
57. Yang X, Yang Y, Sun B-F, Chen Y-S, Xu J-W, Lai W-Y. et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27:606-25
58. Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y. et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255-61
59. Shinoda S, Kitagawa S, Nakagawa S, Wei F-Y, Tomizawa K, Araki K. et al. Mammalian NSUN2 introduces 5-methylcytidines into mitochondrial tRNAs. Nucleic Acids Res. 2019;47:8734-45
60. Yuan S, Tang H, Xing J, Fan X, Cai X, Li Q. et al. Methylation by NSun2 represses the levels and function of microRNA 125b. Mol Cell Biol. 2014;34:3630-41
61. Yang L, Ma Y, Han W, Li W, Cui L, Zhao X. et al. Proteinase-activated receptor 2 promotes cancer cell migration through RNA methylation-mediated repression of miR-125b. J Biol Chem. 2015;290:26627-37
62. Li Q, Li X, Tang H, Jiang B, Dou Y, Gorospe M. et al. NSUN2-mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J Cell Biochem. 2017;118:2587-98
63. Deng X, Su R, Stanford S, Chen J. Critical Enzymatic Functions of FTO in Obesity and Cancer. Front Endocrinol (Lausanne). 2018;9:396
64. Linnebacher M, Wienck A, Boeck I, Klar E. Identification of an MSI-H tumor-specific cytotoxic T cell epitope generated by the (-1) frame of U79260(FTO). J Biomed Biotechnol. 2010;2010:841451
65. Zhao J, Huang X, Yang M, Li M, Zheng J. Association between the FTO rs8050136 polymorphism and cancer risk: a meta-analysis. Fam Cancer. 2016;15:145-53
66. Yamaji T, Iwasaki M, Sawada N, Shimazu T, Inoue M, Tsugane S. et al. Fat mass and obesity-associated gene polymorphisms, pre-diagnostic plasma adipokine levels and the risk of colorectal cancer: The Japan Public Health Center-based Prospective Study. PLoS One. 2020;15:e0229005
67. Yang B, Thrift AP, Figueiredo JC, Jenkins MA, Schumacher FR, Conti D V. et al. Common variants in the obesity-associated genes FTO and MC4R are not associated with risk of colorectal cancer. Cancer Epidemiol. 2016;44:1-4
68. Nock NL, Plummer SJ, Thompson CL, Casey G, Li L. FTO polymorphisms are associated with adult body mass index (BMI) and colorectal adenomas in African-Americans. Carcinogenesis. 2011;32:748-56
69. Huang X, Zhao J, Yang M, Li M, Zheng J. Association between FTO gene polymorphism (rs9939609 T/A) and cancer risk: a meta-analysis. Eur J Cancer Care. 2017 26
70. Lim U, Wilkens LR, Monroe KR, Caberto C, Tiirikainen M, Cheng I. et al. Susceptibility variants for obesity and colorectal cancer risk: the multiethnic cohort and PAGE studies. Int J cancer. 2012;131:E1038-43
71. Tarabra E, Actis GC, Fadda M, De Paolis P, Comandone A, Coda R. et al. The obesity gene and colorectal cancer risk: a population study in Northern Italy. Eur J Intern Med. 2012;23:65-9
72. Yamaji T, Iwasaki M, Sawada N, Shimazu T, Inoue M, Tsugane S. Fat mass and obesity-associated gene polymorphisms, pre-diagnostic plasma adipokine levels and the risk of colorectal cancer: The Japan Public Health Center-based Prospective Study. PLoS One. 2020;15:e0229005
73. Mauer J, Sindelar M, Despic V, Guez T, Hawley BR, Vasseur J-J. et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat Chem Biol. 2019;15:340-7
74. Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC. et al. Differential m(6)A, m(6)A(m), and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol Cell. 2018;71:973-85
75. Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik A V, Patil DP. et al. Reversible methylation of m(6)A(m) in the 5' cap controls mRNA stability. Nature. 2017;541:371-5
76. Li Z, Weng H, Su R, Weng X, Zuo Z, Li C. et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-methyladenosine RNA demethylase. Cancer Cell. 2017;31:127-41
77. Zhang C, Zhang M, Ge S, Huang W, Lin X, Gao J. et al. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019;8:4766-81
78. Li H-B, Tong J, Zhu S, Batista PJ, Duffy EE, Zhao J. et al. M6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338-42
79. Yang S, Wei J, Cui Y-H, Park G, Shah P, Deng Y. et al. M(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10:2782
80. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18-29
81. Li N, Kang Y, Wang L, Huff S, Tang R, Hui H. et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc Natl Acad Sci U S A. 2020;117:20159-70
82. Tanabe A, Tanikawa K, Tsunetomi M, Takai K, Ikeda H, Konno J. et al. RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett. 2016;376:34-42
83. Bai Y, Yang C, Wu R, Huang L, Song S, Li W. et al. YTHDF1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front Oncol. 2019;9:332
84. Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N. et al. Oncogene c-Myc promotes epitranscriptome m(6)A reader YTHDF1 expression in colorectal cancer. Oncotarget. 2018;9:7476-86
85. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285-95
86. Zhu S, Wang J-Z, Chen D, He Y-T, Meng N, Chen M. et al. An oncopeptide regulates m(6)A recognition by the m(6)A reader IGF2BP1 and tumorigenesis. Nat Commun. 2020;11:1685
87. Chen R-XX, Chen X, Xia L-PP, Zhang J-XX, Pan Z-ZZ, Ma X-DD. et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695
88. Tian J, Ying P, Ke J, Zhu Y, Yang Y, Gong Y. et al. ANKLE1 N(6)-methyladenosine-related variant is associated with colorectal cancer risk by maintaining the genomic stability. Int J cancer. 2020;146:3281-93
89. Hollstein M, Sidransky D, Vogelstein B, Harris CC. P53 mutations in human cancers. Science. 1991;253:49-53
90. Jiang L, Kon N, Li T, Wang S-J, Su T, Hibshoosh H. et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57-62
91. Uddin MB, Roy KR, Hosain SB, Khiste SK, Hill RA, Jois SD. et al. An N(6)-methyladenosine at the transited codon 273 of p53 pre-mRNA promotes the expression of R273H mutant protein and drug resistance of cancer cells. Biochem Pharmacol. 2019;160:134-45
92. Wang YY-N, Lu J-H, Wu Q-N, Jin Y, Wang D-S, Chen Y-X. et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol Cancer. 2019;18:174
93. Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A. et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18:143
94. Song P, Feng L, Li J, Dai D, Zhu L, Wang C. et al. β-catenin represses miR455-3p to stimulate m6A modification of HSF1 mRNA and promote its translation in colorectal cancer. Mol Cancer. 2020;19:129
95. Shen X-P, Ling X, Lu H, Zhou C-X, Zhang J-K, Yu Q. Low expression of microRNA-1266 promotes colorectal cancer progression via targeting FTO. Eur Rev Med Pharmacol Sci. 2019;22:8220-6
96. Liu X, Liu L, Dong Z, Li J, Yu Y, Chen X. et al. Expression patterns and prognostic value of m(6)A-related genes in colorectal cancer. Am J Transl Res. 2019;11:3972-91
97. Kandimalla R, Gao F, Li Y, Huang H, Ke J, Deng X. et al. RNAMethyPro: a biologically conserved signature of N6-methyladenosine regulators for predicting survival at pan-cancer level. NPJ Precis Oncol. 2019;3:13
98. Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H. et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373-84
99. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H. et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell. 2019;35:677-691
100. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y. et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell. 2018;172:90-105
101. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148:1258-70
102. Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:k2179
103. Nishida A, Inoue R, Inatomi O, Bamba S, Naito Y, Andoh A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018;11:1-10
104. Patterson E, Ryan PM, Cryan JF, Dinan TG, Ross RP, Fitzgerald GF. et al. Gut microbiota, obesity and diabetes. Postgrad Med J. 2016;92:286-300
105. Yamashita T, Emoto T, Sasaki N, Hirata K-I. Gut microbiota and coronary artery disease. Int Heart J. 2016;57:663-71
106. Quigley EMM. Microbiota-brain-gut axis and neurodegenerative diseases. Curr Neurol Neurosci Rep. 2017;17:94
107. Zhang X, Liu Q, Liao Q, Zhao Y. Pancreatic Cancer, Gut Microbiota, and Therapeutic Efficacy. J Cancer. 2020;11:2749-58
108. Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell. 2018;33:570-80
109. Jabs S, Biton A, Bécavin C, Nahori MA, Ghozlane A, Pagliuso A. et al. Impact of the gut microbiota on the m6A epitranscriptome of mouse cecum and liver. Nat Commun. 2020;11:1-16
110. Ma X, Zhou Z, Zhang X, Fan M, Hong Y, Feng Y. et al. Sodium butyrate modulates gut microbiota and immune response in colorectal cancer liver metastatic mice. Cell Biol Toxicol. 2020
111. Zhao R, Coker OO, Wu J, Zhou Y, Zhao L, Nakatsu G. et al. Aspirin Reduces Colorectal Tumor Development in Mice and Gut Microbes Reduce its Bioavailability and Chemopreventive Effects. Gastroenterology. 2020;159:969-83
112. Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. 2020;158:322-40
113. Helsingen LM, Vandvik PO, Jodal HC, Agoritsas T, Lytvyn L, Anderson JC. et al. Colorectal cancer screening with faecal immunochemical testing, sigmoidoscopy or colonoscopy: a clinical practice guideline. BMJ. 2019;367:l5515
114. Stoffel EM, Murphy CC. Epidemiology and mechanisms of the increasing incidence of colon and rectal cancers in young adults. Gastroenterology. 2020;158:341-53
115. Huang H, Weng H, Chen J. M(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270-88
116. Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z. et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18:142
117. Chen M, Wei L, Law C-T, Tsang FH-C, Shen J, Cheng CL-H. et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254-70
118. Xu H, Wang H, Zhao W, Fu S, Li Y, Ni W. et al. SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating Snail mRNA homeostasis in hepatocellular carcinoma. Theranostics. 2020;10:5671-86
119. Li Y, Xiao J, Bai J, Tian Y, Qu Y, Chen X. et al. Molecular characterization and clinical relevance of m(6)A regulators across 33 cancer types. Mol cancer. 2019;18:137
120. Liu Y, Zhang Y, Chi Q, Wang Z, Sun B. Methyltransferase-like 1 (METTL1) served as a tumor suppressor in colon cancer by activating 7-methyguanosine (m7G) regulated let-7e miRNA/HMGA2 axis. Life Sci. 2020;249:117480
121. Wu H, Qin W, Lu S, Wang X, Zhang J, Sun T. et al. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2'-O-methylation via NOP58 recruitment in colorectal cancer. Mol Cancer. 2020;19:95
122. Jung G, Hernández-Illán E, Moreira L, Balaguer F, Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol. 2020;17:111-30
123. Chen Y-T, Shen J-Y, Chen D-P, Wu C-F, Guo R, Zhang P-P. et al. Identification of cross-talk between m(6)A and 5mC regulators associated with onco-immunogenic features and prognosis across 33 cancer types. J Hematol Oncol. 2020;13:22
124. Huang H, Weng H, Deng X, Chen J. RNA modifications in cancer: functions, mechanisms, and therapeutic implications. Annu Rev Cancer Biol. 2020;4:221-40
125. Yang Y-Y, Yu K, Li L, Huang M, Wang Y. Proteome-wide interrogation of small GTPases regulated by N(6)-methyladenosine modulators. Anal Chem. 2020;92:10145-52
Author contact
![]() Corresponding author: Yanlei Ma, Department of Oncology, Shanghai Medical College of Fudan University; Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, No.270 Dongan' Road, Xuhui District, Shanghai 200032, China; E-mail: yanleimaedu.cn.
Corresponding author: Yanlei Ma, Department of Oncology, Shanghai Medical College of Fudan University; Department of Colorectal Surgery, Fudan University Shanghai Cancer Center, No.270 Dongan' Road, Xuhui District, Shanghai 200032, China; E-mail: yanleimaedu.cn.
 Global reach, higher impact
Global reach, higher impact