13.3
Impact Factor
Theranostics 2021; 11(2):649-664. doi:10.7150/thno.51479 This issue Cite
Research Paper
Rapid design and development of CRISPR-Cas13a targeting SARS-CoV-2 spike protein
1. Department of Neurosurgery, Tianjin Medical University General Hospital, Tianjin 300052, China;
2. Tianjin Neurological Institute, Key Laboratory of Post-neurotrauma Neuro-repair and Regeneration in Central Nervous System, Ministry of Education and Tianjin City, Tianjin 300052, China
* These authors contributed equally to this work.
Abstract
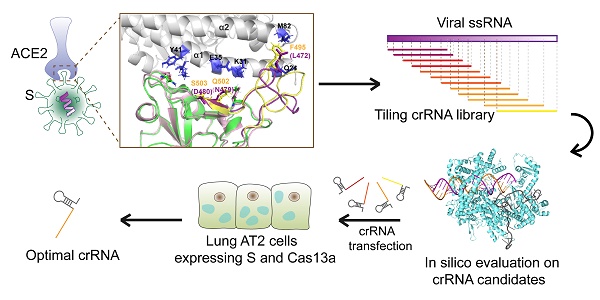
The novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a worldwide epidemic of the lethal respiratory coronavirus disease (COVID-19), necessitating urgent development of specific and effective therapeutic tools. Among several therapeutic targets of coronaviruses, the spike protein is of great significance due to its key role in host invasion. Here, we report a potential anti-SARS-CoV-2 strategy based on the CRISPR-Cas13a system.
Methods: A comprehensive set of bioinformatics methods, including sequence alignment, structural comparison, and molecular docking, was utilized to identify a SARS-CoV-2-spike(S)-specific segment. A tiling crRNA library targeting this specific RNA segment was designed, and optimal crRNA candidates were selected using in-silico methods. The efficiencies of the crRNA candidates were tested in human HepG2 and AT2 cells.
Results: The most effective crRNA sequence inducing a robust cleavage effect on S and a potent collateral cleavage effect were identified.
Conclusions: This study provides a rapid design pipeline for a CRISPR-Cas13a-based antiviral tool against SARS-CoV-2. Moreover, it offers a novel approach for anti-virus study even if the precise structures of viral proteins are indeterminate.
Keywords: CRISPR-Cas13a, coronavirus, SARS-CoV-2, bioinformatics, RNA-editing
 Global reach, higher impact
Global reach, higher impact