13.3
Impact Factor
Theranostics 2021; 11(1):461-469. doi:10.7150/thno.51963 This issue Cite
Review
tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections
1. Department of Biochemistry and Molecular Biology and Zhejiang Key Laboratory of Pathophysiology, Medical School of Ningbo University, Ningbo 315211, China.
2. Department of Gastrointestinal Surgery, Ningbo First Hospital, Ningbo University, Ningbo 315010, China.
Received 2020-8-14; Accepted 2020-10-2; Published 2021-1-1
Abstract
tRNA-derived fragments (tRFs) are a new category of regulatory noncoding RNAs with distinct biological functions in cancers and stress-induced diseases. Herein, we first summarize the classification and biogenesis of tRFs. tRFs are produced from pre-tRNAs or mature tRNAs. Based on the incision loci, tRFs are classified into several types: tRF-1, tRF-2, tRF-3, tRF-5, and i-tRF. Some tRFs participate in posttranscriptional regulation through microRNA-like actions or by displacing RNA binding proteins and regulating protein translation by promoting ribosome biogenesis or interfering with translation initiation. Other tRFs prevent cell apoptosis by binding to cytochrome c or promoting virus replication. More importantly, the dysregulation of tRFs has important clinical implications. They are potential diagnostic and prognostic biomarkers of gastric cancer, liver cancer, breast cancer, prostate cancer, and chronic lymphocytic leukemia. tRFs may become new therapeutic targets for the treatment of diseases such as hepatocellular carcinoma and respiratory syncytial virus infection. Finally, we point out the existing problems and future research directions associated with tRFs. In conclusion, the current progress in the research of tRFs reveals that they have important clinical implications and may constitute novel molecular therapeutic targets for modulating pathological processes.
Keywords: tRNA-derived fragment (tRF), mechanism, cancer, virus infection, therapeutic strategy
Introduction
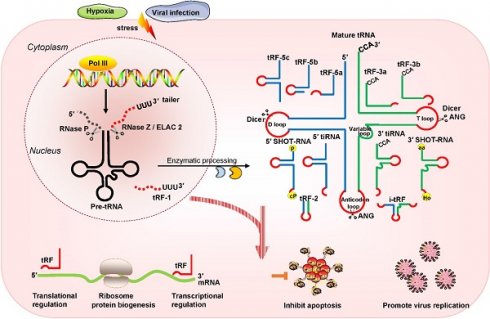
Recently, multiple studies have revealed that small noncoding RNAs (sncRNAs) exist extensively and have a diversity of functions in humans [1-3]. In the eukaryotic nucleus, RNA polymerase III (RNA Pol III) plays the role of transcribing tRNA genes into precursor tRNAs (pre-tRNAs) [4, 5]. During tRNA maturation, the typical cloverleaf structure of the pre-tRNA 5'-leader sequence as well as the 3'-tail ('poly-U') sequence are enzymatically digested by endoribonuclease P (RNase P) and endonuclease Z (RNase Z)/cytoplasmic homolog ribonuclease Z 2 (ELAC2), respectively; then, under the action of tRNA nucleotidyl transferase, the trinucleotide 'CCA' sequence is attached to the tail-free tRNAs at the 3' ends [4, 5]. The tRNA transcripts undergo enzymatic splicing, and chemical modification may yield new species of sncRNAs, such as tRNA-derived small RNAs (tsRNAs) [6, 7]. According to the cleavage loci and length, tsRNAs can be divided into two major categories: tRNA-derived stress-induced RNAs (tiRNAs) and tRNA-derived fragments (tRFs) (Figure 1). The generation of tiRNAs and tRFs takes place in multiple biological processes, which implies that these fragments are not random cleavage products [7].
tiRNAs are generated by ribonuclease angiogenin (ANG) incision at the middle site of the anticodons of mature tRNAs, so tiRNAs are also called tRNA halves [8, 9]. tiRNAs, with lengths of 31- 40 nucleotides (nt), have the 5′ hydroxyl but not the 5′ phosphate, which is different from the situation for microRNA (miRNA) 5′ phosphate ends, which are generated by Dicer enzymes [8, 9]. Based on whether the 5′ or 3′ sequencer of the anticodon cleavage position is included, tiRNAs are classified into two basic types: 5′ tiRNAs and 3′ tiRNAs [8, 9]. The 5′ tiRNAs encompass the 5′ end of the mature tRNA to the terminus of the anticodon loop, while the 3′ tiRNAs extend from the anticodon loop to the 3′ end of the mature tRNA (Figure 1). These tiRNAs are mainly generated by stress exposure, such as oxidative stress, hypoxia, and virus infection [8, 9].
Classification and biogenesis of tsRNAs. tRFs can be classified into five types: tRF-5, tRF-3, tRF-1, tRF-2, and i-tRF. tiRNAs are cleaved by ANG at the middle site of the anticodon of mature tRNAs. SHOT-RNAs are induced by sex hormones. The production of tRFs and tiRNAs is promoted by various stress conditions. They play roles in processes such as transcriptional regulation and the regulation of protein biogenesis. Abbreviations: tsRNAs, tRNA-derived small RNAs; Pol III, polymerase III; pre-tRNA, precursor tRNA; RNase Z, ribonuclease Z; ELAC2, cytoplasmic homolog ribonuclease Z 2; ANG, Angiogenin; SHOT-RNAs, sex hormone-dependent tRNA-derived RNAs; tRF, tRNA-derived fragments; tiRNA, tRNA-derived stress-induced RNA; P, phosphate; cP, cyclicphosphate; aa, amino acid; Ho, hydroxyl.
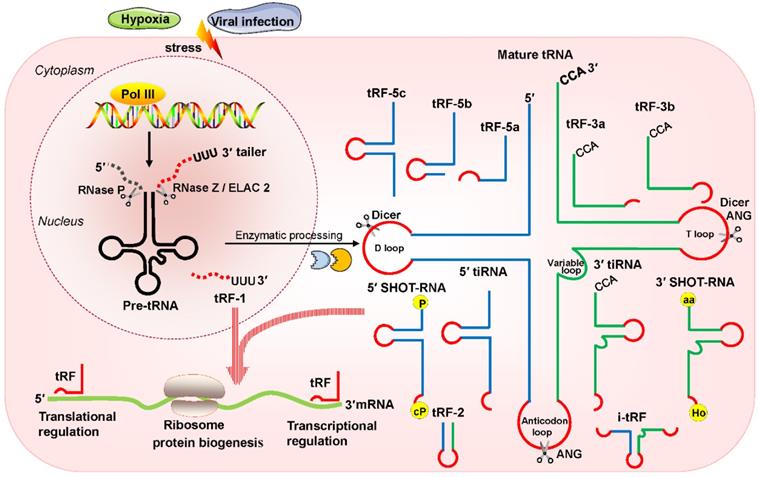
In addition, another type of tRNA half known as the sex hormone-dependent tRNA-derived RNA (SHOT-RNA) is induced by sex hormones and cleaved by ANG [10, 11]. SHOT-RNA, which is not induced by stress, is particularly highly expressed in androgen receptor (AR)-positive prostate cancer cells and estrogen receptor (ER)-positive breast cancer and is not expressed in other cancers at present. Specifically, 5′ SHOT-RNAs bear a phosphate at the 5′ end and a 2′,3′-cyclicphosphate at the 3′-end (Figure 1), while 3′ SHOT-RNAs are characterized by a hydroxyl at the 5′ end and an amino acid at the 3′ end, as they are derived from aminoacylated tRNAs [10, 12, 13].
High-throughput sequencing technology has revealed that there is more diversity in tsRNAs than is reflected in the existing classifications. Among tsRNAs, tRFs, with a length of 14 nt to 30 nt, have a greater number of distinct classes and function as key players in the regulation of gene expression at transcriptional as well as posttranscriptional levels, indicating that they are not merely byproducts of the random cleavage of tRNAs but are regulatory sncRNAs involved in physiological and pathological processes [1, 3].
Since tRFs are the major types of tsRNAs, in this review, we focused on tRFs. We present updated views focusing on the specific molecular mechanisms underlying tRF-mediated regulation of mRNA stability and translation, stress responses and viral infections and their potential roles as biomarkers or therapeutic targets in cancers and virus infections.
Classification and biogenesis of tRFs
tRFs are produced from pre-tRNAs or mature tRNAs [14, 15]. Notably, they are similar in size to miRNAs, with a 5′ phosphate and a 3′ hydroxyl [14]. According to the mapped locations, tRFs are largely categorized into two main classes: tRF-5 and tRF-3 [14, 15]. tRF-5s begin at the 5′ end of mature tRNAs and are cleaved by Dicer at the D-loop or the stem position between the D-loop and the anticodon loop [14, 15]. On the basis of the incision loci and lengths, tRF-5s are further classified into three subtypes: tRF-5a (14-16 nt), tRF-5b (22-24 nt) and tRF-5c (28-30 nt) [14, 15]. tRF-3s begin at the 3′ end (at the trinucleotide 'CCA' at the 3′ end) and are cleaved by Dicer and ANG at the T-loop of mature tRNAs. tRF-3s, which are approximately 18-22 nt in length, are further classified into two subgroups: tRF-3a and tRF-3b (Figure 1).
There are three additional groups of tRFs: tRF-1, tRF-2, and inter tRF (i-tRF) [15, 16]. tRF-1s are small fragments derived from the 3′ tails of pre-tRNsA (containing the 'poly-U' sequence at the 3′ ends) and are cleaved by RNase Z or ELAC2 [15]. tRF-2s comprise anticodon loop and stem sequences, excluding the 5′ end and 3′ end structures [15, 16]. However, the details of ribonuclease processing required for the production of tRF-2s and i-tRFs remain unclear [15, 16]. i-tRFs, originating from the internal body of mature tRNAs [17, 18], include the anticodon loop and segments of the D-loop and T-loop other than the 5′ terminal and 3′ terminal (Figure 1).
Regulation of mRNA stability
Studies have revealed that some tRFs target genic mRNAs and participate in posttranscriptional regulation in humans [19]. Here, we elaborate on tRFs targeting mRNA expression through miRNA-like actions and binding to RNA-binding proteins (RBPs) to control mRNA stability.
MiRNA-like actions
Preparatory bioinformatic analysis suggests that tRFs have sufficient sequence complementarity with endogenous mRNAs and thus may play a potential role in posttranscriptional regulation [19, 20]. As one kind of sncRNA with a length less than 30 nt, it seems logical to conclude that tRFs have functions similar to those of miRNAs [21, 22].
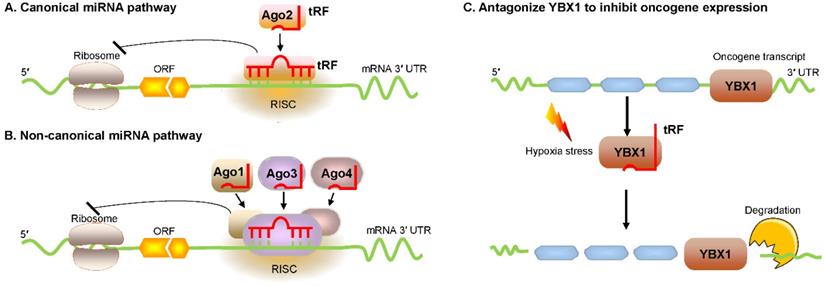
Huang et al. reported that tRF/miR-1280, which is derived from tRNALeu and pre-miRNA, inhibited colorectal cancer cell proliferation by inhibiting the Notch signaling pathway by directly interacting with the JAG2 mRNA 3′ untranslated region (UTR) [23]. In particular, these tRFs are physically related to Argonaute (Ago) proteins, which regulate gene expression [24, 25]. Recently, an increasing number of researchers have found that tRFs are loaded into Ago complexes [26, 27]. RNA-seq analysis proves that tRF-3009a guides Ago to inhibit the expression of targeted genes in a Dicer-independent manner posttranscriptionally [28]. Maute et al. verified in B cell lymphoma that tRF-3 derived from tRNAGly-GCC possessed an miRNA-like structure and functions by binding a complementary target site [29]. Using large-scale meta-analyses of available experimental data, researchers observed Ago1-loaded tRFs, and these tRFs interacted with target genes at the 3ʹ UTR [30]. Li et al. found that tRF-3s could guide Ago2 to cleave the target mRNA (Figure 2A) [31]. Kumar et al. intriguingly reported that tRF-5s and tRF-3s were preferentially associated with Ago1, Ago3, and Ago4 but not with Ago2 in human embryonic kidney 293 (HEK293) cells (Figure 2B) [32]. Hence, in conjunction with different Ago proteins, tRFs can regulate gene expression through either canonical or noncanonical miRNA-like actions [32-35]. Additionally, Ago-bound tRFs should be further explored because they have a propensity to target endogenous mRNA and to construct regulatory networks in humans [36, 37].
Regulation of mRNA stability by tRFs. (A, B) tRFs bind with different Ago proteins to inhibit targeted mRNA expression via pathways that exert either canonical or noncanonical miRNA-like activity. (C) tRFs cause oncogene transcript degradation by displacing YBX1 from the 3′ UTR of mRNA. Abbreviations: miRNA, microRNA; Ago, Argonaute; YBX1, Y-box-binding protein 1; 3′ UTR, 3′ untranslated region; RISC, RNA-induced silencing complex; ORF, open reading frame.
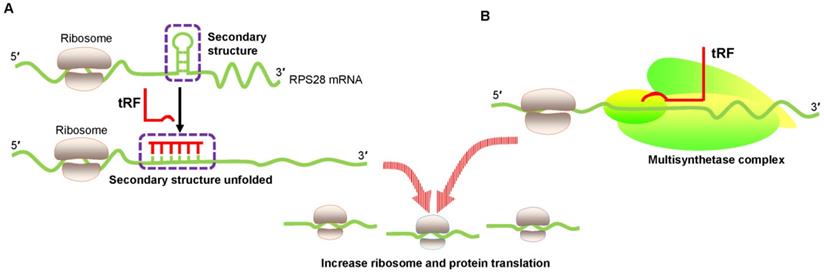
Translational activation by promoting ribosome biogenesis. (A) tRF increases ribosome biogenesis by unfolding the RPS28 mRNA secondary structure, thus enhancing translation. (B) tRF interacts with the human multisynthetase complex, thereby promoting RNA-binding protein translation. Abbreviations: RPS28, ribosomal protein S28.
Bind to RNA-binding proteins
tRFs may bind to RBPs and posttranscriptionally regulate gene expression [38, 39]. RBPs interact with targeted RNAs to control their stability [40].
In breast cancer cells, Goodarzi et al. reported that under hypoxic conditions, several tRFs were upregulated, which then suppressed oncogenic transcript stability by displacing the 3' UTR from the YBX1 protein [41]. These hypoxic stress-induced tRFs, which are mainly derived from tRNAAsp, tRNAGlu, tRNAGly, and tRNATyr, can competitively bind to YBX1 and block its interaction with oncogenic mRNAs [41]. YBX1 is an RBP with many biological roles. YBX1 maintains oncogene transcript stability and increases cell proliferation by binding with endogenous oncogenic mRNA [42]. YBX1 may also bind with several categories of regulatory RNAs, including tRFs [41]. Under hypoxic stress, cancer cells could produce more tRFs that can compete with oncogene transcripts and then bind to YBX1, thereby promoting mRNA degradation and eventually inhibiting the proliferation of cancer cells (Figure 2C). This posttranscriptional suppression depends on sequence complementary because tRFs have a motif that binds the sequence that YBX1 can recognize [41, 43].
In fact, YBX1 is not the only RBP that may be displaced by specific tRFs. In fact, a novel tRF derived from mature tRNAGlu has been found to be able to bind and displace the RBP nucleolin in breast cancer [44].
Regulation of protein translation
tRFs play additional biological roles by activating or inhibiting protein synthesis via different mechanisms [18].
Translational activation via promotion of ribosome biogenesis
An innovative study by Kim et al. proved that a 22 nt length tRF called 3′ tRFLeuCAG enhanced translation by facilitating ribosome protein biogenesis [45]. Ribosome gradient analysis showed that ribosomal protein S28 (RPS28) was needed for ribosomal RNA 18S rRNA biogenesis and was an integral part of the 40S ribosomal subunit [46]. The 3′ UTR target site of RPS28 mRNA forms a secondary structure that is a major region containing a translation initiation site. Several experiments involving target-site mutations demonstrated that 3′ tRFLeuCAG bound to duplexed secondary target sites in RPS28 mRNA and unwound the hairpin secondary structure to increase translation in human cancer cells [45] (Figure 3A). Thus, 3′ tRFLeuCAG plays a vital function in regulating the numbers of ribosomes. The greater the number of ribosomes, the more potential protein synthesis, eventually increasing cell growth and proliferation [45].
Keam et al. also convincingly showed the translational activation of tRFs [47]. 5′ tRFGln19 interacted with the human multisynthetase complex (MSC) and then promoted RBP translation (Figure 3B). However, the detailed mechanism needs to be determined.
Translational inhibition by interfering with translation initiation
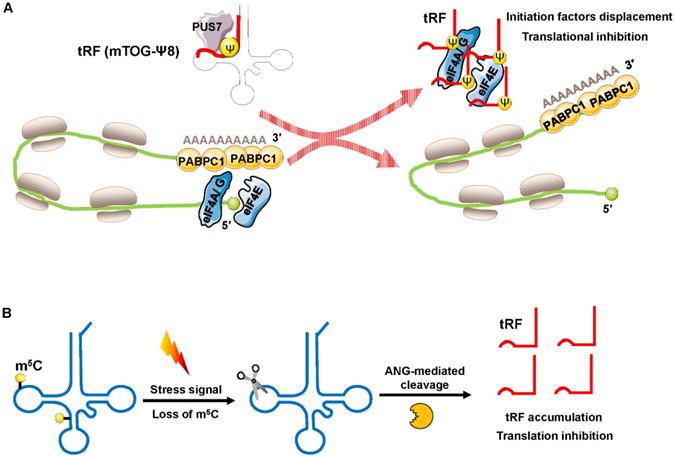
Recently, Guzzi et al. showed that in stem cells, pseudouridylation synthase 7 (PUS7) mediated pseudouridine (Ψ) disposition of particular tRFs (mTOG-Ψ8) to suppress translation (Figure 4A). They identified that PUS7 was enriched in embryonic and/or hematopoietic stem cells and then bound to diacritical tRNAs and modified uridine (U) into pseudouridine (Ψ) at the U8 position (Ψ8) [48]. PUS7-mediated “Ψ” controls the stem cell biogenesis of 5′ tRFs, which have a common oligoguanine motif at the terminus and are named mTOG-Ψ8 [48]. Under normal growth conditions, polysomes form a closed-loop translation complex (Figure 4A). In this structural model, translational initiation factors [eukaryotic initiation factor (eIF)-4 A/G, and E] interact with cytoplasmic poly(A) binding protein-1 (PABPC1) [49, 50]. In response to stress, PUS7 binds to diacritical tRNAs and governs biogenesis of mTOG-Ψ8; then, mTOG-Ψ8 preferentially binds to PABPC1, resulting in displacement of eIF-4 A/G and E from m7G-capped mRNAs (Figure 4A). PUS7 and mTOG-Ψ8 loss affects translation regulation, resulting in the increased biosynthesis of proteins and the impairment of hematopoietic stem cell commitment, potentially leading to myeloid malignancies [48].
Blanco et al. demonstrated that posttranscriptional methylation of tRNA at cytosine-5 (m5C) by the methyltransferase NSUN2 was an innovative mechanism to suppress global protein synthesis [51]. External stress stimulation represses NSUN2 activity, causing the loss of m5C; the dysregulation of m5C increases the affinity between tRNA and angiogenin, leading to the accumulation of tRF-5s, which then repress protein synthesis and promote squamous tumorigenesis (Figure 4B). Sobala et al. showed that tRF-5 derived from tRNAGln was able to inhibit translation and did not require complementary target sequences of mRNA [52]. Mechanistically, these inhibitory effects of tRFs on translation require a conserved 3′ “GG” dinucleotide [52].
Signs of cell stress and promotion of virus replication
The production of tRFs can be induced by stress conditions, such as high salinity, oxidative stress, and virus infection [53, 54]. Persistently activated stress responses result in inflammation and disease pathogenesis [54].
Signs of cell stress
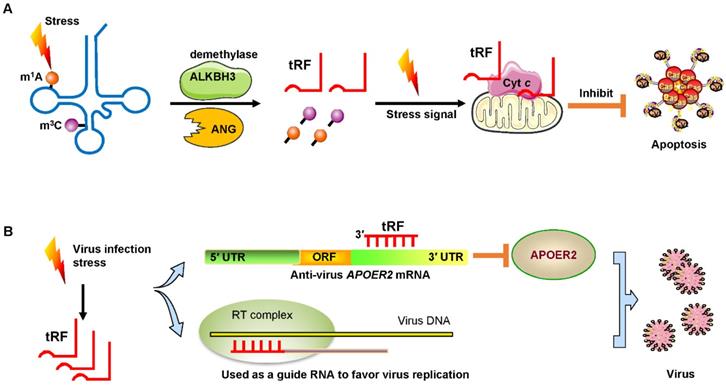
Under sodium arsenite stress, Chen et al. revealed that the demethylase α-KG-dependent alkB homolog 3 (ALKBH3) induced tRFs to interact with cytochrome c (Cyt c) to suppress cell apoptosis [55]. ALKBH3 catalyzes the demethylation of 1-methyladenosine (m1A) and 3-methylcytidine (m3C) in tRNAs. Demethylated tRNAs are more sensitive to the cleavage of ANG and easily generate tRF-5Gly-GCC [55, 56]. These tRFs bound to Cyt c that was released from the mitochondria and then eventually strengthened ribosome assembly and finally prevented apoptosis of cervical cancer cells (Figure 5A).
Translation inhibition by interfering with translation initiation. (A) Increased displacement of translational initiation factors from mRNA by the tRF mTOG-Ψ8 represses translation. Loss of PUS7 and mTOG-Ψ8 may contribute to human myeloid malignancies. (B) Loss of m5C increases the affinity between tRNA and ANG, leading to tRF-5 accumulation, which then decreases protein synthesis. Abbreviations: PUS7, pseudouridylation synthase 7; Ψ, pseudouridine; eIF4A/G, E, eukaryotic initiation factor 4 A/G, E; PABPC1, poly(A) binding protein-1; ANG, angiogenin.
Signs of stress and viral infection. (A) Under stress, the demethylase ALKBH3 induces tRFs to interact with Cyt c to suppress cancer cell apoptosis. (B) Virus infection-induced stress leads to the induction of specific tRFs. Some tRFs recognize the target site in the 3′ UTR of anti-virus protein APOER2 mRNA. The suppression of APOER2 will promote virus replication. Other tRFs are used as guide RNAs to initiate viral reverse transcription. Abbreviations: ALKBH3, α-KG-dependent alkB homolog 3; ANG, Angiogenin; Cyt c, cytochrome c; UTR, untranslated region; ORF, Open reading frame; APOER2, apolipoprotein E receptor 2; RT, reverse transcription.
Under oxidative stress, Gkatza et al. reported that the activity of the cytosine-5 RNA methyltransferase NSUN2 can be suppressed, leading to the reduction of tRNA methylation and then the intracellular biogenesis of tRFs related to the repression of protein synthesis [57].
Promotion of virus replication
Recent studies have demonstrated that infecting host cells with respiratory syncytial virus (RSV) can initiate a stress response by mediating ANG digestion of tRNAs to generate increases in tRFs [58, 59]. Viruses can exploit host tRFs as guide primers to improve their replication and promote the efficiency of infection [59, 60]. Deng et al. convincingly demonstrated that a specific tRF originating from the 5′ end of tRNAGluCTC (tRF-5GluCTC) was induced by RSV infection [61]. The 3′-portion of tRF-5GluCTC recognizes a target site in the 3′ UTR of apolipoprotein E receptor 2 (APOER2) mRNA (Figure 5B). APOER2 is a host anti-RSV protein whose inhibition favors RSV replication [61]. As is well-known, the 5′-end of miRNAs is critical for their gene silencing effect [62]. However, with regard to tRFs, the 3′-portion of tRF-5GluCTC is important for gene targeting [63]. Therefore, tRFs have different trans-silencing mechanisms than miRNAs. Zhou and his colleagues also found that RSV specifically led to the induction of two novel tRFs, tRF-5GlyCCC and tRF-5LysCTT [59]. These tRFs play a significant role in promoting RSV replication and impact RSV-induced cytokines/chemokines. Ruggero et al. proved that tRF-3019 in host cells is thoroughly complementary to primer binding site (PBS) in retroviral RNA from human T-cell leukemia virus type 1 (HTLV-1) [60]. Therefore, tRF-3019 could be utilized to guide RNA initiation of reverse transcription and increase virus amplification (Figure 5B).
Based on the above, we can see that tRFs play various roles in cancers and virus infection (Table 1).
Clinical value of tRFs
In recent years, tRFs have become rising stars in the regulation of biological processes, and their deregulation has important clinical implications [64]. Here, we describe the potential value of tRFs as diagnostic biomarkers and therapeutic targets (Table 2).
Potential as diagnostic and prognostic biomarkers
As high-throughput sequencing technology has spread, an increasing number of studies have demonstrated that aberrant expression of tRFs contributes to carcinogenesis and could represent new biomarkers for diagnosis [65]. tRFs carried by exosomes have been exploited as biomarkers and have been found to mediate communication between exosome-secreting cells and recipient cells [65]. Zhu et al. found that patients with liver cancer exhibited significantly higher levels of tRF-5GluCTC in plasma exosomes than healthy controls, indicating that exosomal tRFs in plasma can act as novel “liquid biopsy” biomarkers for the diagnosis of cancer [66]. Sun et al. also showed that patients with high expression of tRF-27-ZDXPHO53KSN and tRF-30-JZOYJE22RR33 in serum obtain less benefit from trastuzumab-based therapy, indicating that these two tRFs could be explored as potential intervention targets and biomarkers in trastuzumab-resistant breast cancer [67]. Biostatistical analysis revealed that i-tRFGlyCCC levels were significantly lower in peripheral blood mononuclear cells (PBMCs) from chronic lymphocytic leukemia (CLL) patients and could be considered a screening biomarker [68]. i-tRFGlyGCC and i-tRFPheGAA have been reported to have prognostic and diagnostic value in CLL, respectively [69, 70]. In addition, Olvedy et al. demonstrated that patients prostate cancer in which the expression of tRF-315/tRF-544 was increased in tissues had obviously poorer progression-free survival (PFS) [71]. These results indicated that abnormal tRFs could serve as fluid-based biomarkers for prospective screening research in the future.
Potential clinical therapeutic targets
Aberrant expression of tRFs not only plays a diagnostic and prognostic role in cancers but also indicates their potential usage as therapy targets for the treatment of disease [72]. For example, Kim et al. proved that tRF-3LeuCAG resolved the hairpin structure of RPS28 mRNA and facilitated ribosome protein biogenesis to promote hepatocellular carcinoma growth [45]. Using an antisense oligonucleotide to block tRF-3LeuCAG prevents it from binding RPS28 mRNA, resulting in diminished ribosome biogenesis and apoptosis of hepatocellular carcinoma cells [45]. This finding suggests the possibility that tRFs may be used as a therapeutic target in hepatocellular carcinoma therapy [45]. Wang et al. reported that the combined utilization of an anti-tRF oligonucleotide and a small interfering RNA (siRNA) could downregulate RSV-induced tRF-5GluCTC and eventually block RSV replication [63]. Since the dysregulation of tRFs is closely related to cancers, cellular stress responses and virus infection in humans [73], tRFs may become new therapeutic targets for the treatment of diseases.
Challenges and outlook
In this review, we mainly focused on the possible applications of tRFs in cancers and viral infections. However, tRFs also play roles in other types of diseases, including neurodegenerative and metabolic disorders [16, 57, 74, 75]. The study of tRFs remains at an early stage; there are many problems that still need to be resolved.
The mechanisms underlying the roles of tRFs in cancers and virus infections
| Function | Biological effect | Mechanism | tRF name/ID | Cancer type/Virus infection | References |
|---|---|---|---|---|---|
| Regulation of mRNA stability | MiRNA-like actions | Conjunction with different Ago proteins or direct interaction with mRNAs | tRFLeu-3a/miR-1280, | Colorectal cancer, | [23] |
| tRF-5Gln, | cervical carcinoma | [25] | |||
| tRF-3GlyGCC, | B lymphoma | [29] | |||
| tRF-3 (tRFHis-GTG, tRFLeu-CAG), | Chronic lymphocytic leukemia | [31] | |||
| tRF-3 (ts-3676, ts-4521) | Lung cancer | [35] | |||
| Binds to RNA-binding proteins | Binding to YBX-1 or nucleolin | tRFs (tRFAsp, tRFGlu, tRFGly, tRFTyr) | Breast cancer | [41, 44] | |
| Regulation of protein translation | Translational activation | Promotion of ribosome biogenesis | tRF-3LeuCAG | Hepatocellular carcinoma | [45] |
| Gln19 | Cervical carcinoma | [47] | |||
| Translational inhibition | Displacement of translational initiation factors from mRNA or tRF-5 accumulation for the loss of m5C | mTOG-Ψ8 | Myeloid malignancies | [48] | |
| tRF-5 | Squamous tumor | [51] | |||
| Signs of cell stress and promotion of virus replication | Suppression of apoptosis | Interaction with Cyt c to suppress stress-induced cell apoptosis | tRF-5GlyGCC | Cervical carcinoma | [55] |
| Promotion of virus replication | Targeting of anti-virus proteins or utilization as primers to favor virus replication | tRF-5GlyCCC, tRF-5LysCTT, tRF-5GluCTC, | Respiratory syncytial virus infection | [59, 61] | |
| tRF-3019 | T-cell leukemia virus type 1 infection | [60] |
Clinical value of tRFs in cancers and virus infections
| Function | Sample type | tRF name/ID | Cancer type/Virus infection | References |
|---|---|---|---|---|
| Potential diagnostic and prognostic predictive utility | Plasma (exosome) | tRF-5GluCTC | Liver cancer | [66] |
| Serum | tRF-30-JZOYJE22RR33, tRF-27-ZDXPHO53KSN | Breast cancer | [67] | |
| Blood (peripheral blood mononuclear cells) | i-tRF-GlyCCC, i-tRF-GlyGCC, i-tRF-PheGAA | Chronic lymphocytic leukemia | [68-70] | |
| Tissue | tRF-315, tRF-544 | Prostate cancer | [71] | |
| Potential clinical therapeutic targets | Cell, serum, patient-derived xenograft model | 3′tRF-LeuCAG | Hepatocellular carcinoma | [45] |
| Cell, virus | tRF-5GluCTC | Respiratory syncytial virus infection | [63] |
First, how many additional classes of tRFs exist? For restriction sequencing technology, the current nomenclature of tRFs is based on the different cleavage loci of tRNAs or their origins in tRNAs that transfer a specific amino acid. The nomenclature and classification of tRFs are still rudimentary and cannot provide clear basic information about tRFs. Second, which mechanisms are crucial for tRF roles? The distribution of tRFs may contribute to their biological roles. A recent report suggested that tRF-5s are located mostly in the nucleus, whereas tRF-3s and tRF-1s are mostly cytoplasmic [15]. The detailed mechanisms underlying the involvement of tRFs in cancers and stress may exceed our present knowledge and are worth more in-depth study in the future. Third, whether tRFs are safe and effective should be examined in the clinic. The applications of new bioinformatics techniques and additional experimental approaches, such as photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation (PAR-CLIP) and cross-linking, ligation and sequencing of hybrids (CLASH), to understand the exact molecular mechanisms and the establishment of clinical indicators are critically important for the therapeutic application of tRFs.
Taken together, the evidence suggests that the roles and applications of tRFs still require intensive study. We are confident that future in-depth efforts will contribute to elucidating the mechanisms of tRFs and developing them as novel diagnostic biomarkers and therapeutic targets.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (no. 81974316), the Scientific Innovation Team Project of Ningbo (no. 2017C110019), the Natural Science Foundation of Ningbo (2016A610157, 2018A610371), and the K.C. Wong Magna Fund in Ningbo University.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Shen Y, Yu X, Zhu L, Li T, Yan Z, Guo J. Transfer RNA-derived fragments and tRNA halves: biogenesis, biological functions and their roles in diseases. J Mol Med (Berl). 2018;96:1167-76
2. Li T, Zhu L, Xiao B, Gong Z, Liao Q, Guo J. CRISPR-Cpf1-mediated genome editing and gene regulation in human cells. Biotechnol Adv. 2019;37:21-7
3. Zhu L, Ge J, Li T, Shen Y, Guo J. tRNA-derived fragments and tRNA halves: The new players in cancers. Cancer Lett. 2019;452:31-7
4. Li S, Xu Z, Sheng J. tRNA-derived small RNA: A novel regulatory small non-coding RNA. Genes (Basel). 2018;9:246
5. Liapi E, van Bilsen M, Verjans R, Schroen B. tRNAs and tRNA fragments as modulators of cardiac and skeletal muscle function. Biochim Biophys Acta Mol Cell Res. 2020;1867:118465
6. Martinez G. tRNA-derived small RNAs: New players in genome protection against retrotransposons. RNA Biol. 2018;15:170-5
7. Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett. 2014;588:4297-304
8. Tao EW, Cheng WY, Li WL, Yu J, Gao QY. tiRNAs: A novel class of small noncoding RNAs that helps cells respond to stressors and plays roles in cancer progression. J Cell Physiol. 2020;235:683-90
9. Zhu L, Li T, Shen Y, Yu X, Xiao B, Guo J. Using tRNA halves as novel biomarkers for the diagnosis of gastric cancer. Cancer Biomark. 2019;25:169-76
10. Soares AR, Santos M. Discovery and function of transfer RNA-derived fragments and their role in disease. Wiley Interdiscip Rev RNA. 2017;8:e1423
11. Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I. et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. 2015;112:E3816-25
12. Xu WL, Yang Y, Wang YD, Qu LH, Zheng LL. Computational approaches to tRNA-derived small RNAs. Noncoding RNA. 2017;3:2
13. Zhu L, Liu X, Pu W, Peng Y. tRNA-derived small non-coding RNAs in human disease. Cancer Lett. 2018;419:1-7
14. Ma Z, Zhou J, Shao Y, Jafari FA, Qi P, Li Y. Biochemical properties and progress in cancers of tRNA-derived fragments. J Cell Biochem. 2020;121:2058-63
15. Kumar P, Kuscu C, Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs). Trends Biochem Sci. 2016;41:679-89
16. Qin C, Xu PP, Zhang X, Zhang C, Liu CB, Yang DG. et al. Pathological significance of tRNA-derived small RNAs in neurological disorders. Neural Regen Res. 2020;15:212-21
17. Park EJ, Kim TH. Fine-tuning of gene expression by tRNA-derived fragments during abiotic stress signal transduction. Int J Mol Sci. 2018;19:518
18. Kim HK. Transfer RNA-derived small non-coding RNA: Dual regulator of protein synthesis. Mol Cell. 2019;42:687-92
19. Boskovic A, Bing XY, Kaymak E, Rando OJ. Control of noncoding RNA production and histone levels by a 5' tRNA fragment. Genes Dev. 2020;34:118-31
20. Burroughs AM, Ando Y, de Hoon MJL, Tomaru Y, Suzuki H, Hayashizaki Y. et al. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011;8:158-77
21. Su Z, Frost EL, Lammert CR, Przanowska RK, Lukens JR, Dutta A. tRNA-derived fragments and microRNAs in the maternal-fetal interface of a mouse maternal-immune-activation autism model. RNA Biol. 2020 p: 1-13
22. Hasler D, Lehmann G, Murakawa Y, Klironomos F, Jakob L, Grässer FA. et al. The lupus autoantigen La prevents mis-channeling of tRNA fragments into the human microRNA pathway. Mol Cell. 2016;63:110-24
23. Huang B, Yang H, Cheng X, Wang D, Fu S, Shen W. et al. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res. 2017;77:3194-206
24. Loss-Morais G, Waterhouse PM, Margis R. Description of plant tRNA-derived RNA fragments (tRFs) associated with argonaute and identification of their putative targets. Biol Direct. 2013;8:6
25. Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JWS. et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147-60
26. Luo S, He F, Luo J, Dou S, Wang Y, Guo A. et al. Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res. 2018;46:5250-68
27. Tai H-C, Lim C. Gene silencing mechanisms revealed by dynamics of guide, target, and duplex binding to argonaute. J Chem Theory Comput. 2020;16:688-99
28. Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA. 2018;24:1093-105
29. Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K. et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1404-9
30. Guan L, Karaiskos S, Grigoriev A. Inferring targeting modes of Argonaute-loaded tRNA fragments. RNA Biol. 2019 p: 1-11
31. Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012;40:6787-99
32. Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78
33. Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673-95
34. Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23:2639-49
35. Pekarsky Y, Balatti V, Palamarchuk A, Rizzotto L, Veneziano D, Nigita G. et al. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc Natl Acad Sci U S A. 2016;113:5071-6
36. Bose M, Chatterjee S, Chakrabarty Y, Barman B, Bhattacharyya SN. Retrograde trafficking of Argonaute 2 acts as a rate-limiting step for de novo miRNP formation on endoplasmic reticulum-attached polysomes in mammalian cells. Life Sci Alliance. 2020;3:e201800161
37. Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22-32
38. Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393-406
39. Uchida Y, Chiba T, Kurimoto R, Asahara H. Post-transcriptional regulation of inflammation by RNA-binding proteins via cis-elements of mRNAs. J Biochem. 2019;166:375-82
40. Gallagher C, Ramos A. Joining the dots - protein-RNA interactions mediating local mRNA translation in neurons. FEBS Lett. 2018;592:2932-47
41. Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790-802
42. Blenkiron C, Hurley DG, Fitzgerald S, Print CG, Lasham A. Links between the oncoprotein YB-1 and small non-coding RNAs in breast cancer. PLoS One. 2013;8:e80171
43. Wurth L, Gebauer F. RNA-binding proteins, multifaceted translational regulators in cancer. Biochim Biophys Acta. 2015;1849:881-6
44. Falconi M, Giangrossi M, Zabaleta ME, Wang J, Gambini V, Tilio M. et al. A novel 3'-tRNA(Glu)-derived fragment acts as a tumor suppressor in breast cancer by targeting nucleolin. FASEB J. 2019;33:13228-40
45. Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H. et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57-62
46. Robledo S, Idol RA, Crimmins DL, Ladenson JH, Mason PJ, Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 2008;14:1918-29
47. Keam SP, Sobala A, Ten Have S, Hutvagner G. tRNA-derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation. J Proteome Res. 2017;16:413-20
48. Guzzi N, Cieśla M, Ngoc PCT, Lang S, Arora S, Dimitriou M. et al. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. 2018;173:1204-16.e26
49. Lyons SM, Achorn C, Kedersha NL, Anderson PJ, Ivanov P. YB-1 regulates tiRNA-induced stress granule formation but not translational repression. Nucleic Acids Res. 2016;44:6949-60
50. Evdokimova V, Ruzanov P, Imataka H, Raught B, Svitkin Y, Ovchinnikov LP. et al. The major mRNA-associated protein YB-1 is a potent 5' cap-dependent mRNA stabilizer. EMBO J. 2001;20:5491-502
51. Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J. et al. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016;534:335-40
52. Sobala A, Hutvagner G. Small RNAs derived from the 5' end of tRNA can inhibit protein translation in human cells. RNA Biol. 2013;10:553-63
53. Rashad S, Niizuma K, Tominaga T. tRNA cleavage: a new insight. Neural Regen Res. 2020;15:47-52
54. Elkordy A, Mishima E, Niizuma K, Akiyama Y, Fujimura M, Tominaga T. et al. Stress-induced tRNA cleavage and tiRNA generation in rat neuronal PC12 cells. J Neurochem. 2018;146:560-9
55. Chen Z, Qi M, Shen B, Luo G, Wu Y, Li J. et al. Transfer RNA demethylase ALKBH3 promotes cancer progression via induction of tRNA-derived small RNAs. Nucleic Acids Res. 2019;47:2533-45
56. Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J. et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell. 2016;167:816-28.e16
57. Gkatza NA, Castro C, Harvey RF, Heiß M, Popis MC, Blanco S. et al. Cytosine-5 RNA methylation links protein synthesis to cell metabolism. PLoS Biol. 2019;17:e3000297
58. Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185:35-42
59. Zhou J, Liu S, Chen Y, Fu Y, Silver AJ, Hill MS. et al. Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J Gen Virol. 2017;98:1600-10
60. Ruggero K, Guffanti A, Corradin A, Sharma VK, De Bellis G, Corti G. et al. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J Virol. 2014;88:3612-22
61. Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, Phan T. et al. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol Ther. 2015;23:1622-9
62. Haasnoot J, Berkhout B. RNAi and cellular miRNAs in infections by mammalian viruses. Methods Mol Biol. 2011;721:23-41
63. Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther. 2013;21:368-79
64. Feng W, Li Y, Chu J, Li J, Zhang Y, Ding X. et al. Identification of tRNA-derived small noncoding RNAs as potential biomarkers for prediction of recurrence in triple-negative breast cancer. Cancer Med. 2018;7:5130-44
65. Chiou NT, Kageyama R, Ansel KM. Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep. 2018;25:3356-70 e4
66. Zhu L, Li J, Gong Y, Wu Q, Tan S, Sun D. et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer. 2019;18:74
67. Sun C, Yang F, Zhang Y, Chu J, Wang J, Wang Y. et al. tRNA-derived fragments as novel predictive biomarkers for trastuzumab-resistant breast cancer. Cell Physiol Biochem. 2018;49:419-31
68. Katsaraki K, Artemaki PI, Papageorgiou SG, Pappa V, Scorilas A, Kontos CK. Identification of a novel, internal tRNA-derived RNA fragment as a new prognostic and screening biomarker in chronic lymphocytic leukemia, using an innovative quantitative real-time PCR assay. Leuk Res. 2019;87:106234
69. Karousi P, Katsaraki K, Papageorgiou SG, Pappa V, Scorilas A, Kontos CK. Identification of a novel tRNA-derived RNA fragment exhibiting high prognostic potential in chronic lymphocytic leukemia. Hematol Oncol. 2019;37:498-504
70. Karousi P, Adamopoulos PG, Papageorgiou SG, Pappa V, Scorilas A, Kontos CK. A novel, mitochondrial, internal tRNA-derived RNA fragment possesses clinical utility as a molecular prognostic biomarker in chronic lymphocytic leukemia. Clin Biochem. 2020; in press. doi: 10.1016/j.clinbiochem. 2020 07.005
71. Olvedy M, Scaravilli M, Hoogstrate Y, Visakorpi T, Jenster G, Martens-Uzunova ES. A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget. 2016;7:24766-77
72. Sun C, Fu Z, Wang S, Li J, Li Y, Zhang Y. et al. Roles of tRNA-derived fragments in human cancers. Cancer Lett. 2018;414:16-25
73. Balatti V, Pekarsky Y, Croce CM. Role of the tRNA-derived small RNAs in cancer: New potential biomarkers and target for therapy. Adv Cancer Res. 2017;135:173-87
74. Xie Y, Yao L, Yu X, Ruan Y, Li Z, Guo J. Action mechanisms and research methods of tRNA-derived small RNAs. Signal Transduct Target Ther. 2020;5:109
75. Zou G, Zhang X, Wang L, Li X, Xie T, Zhao J. et al. Herb-sourced emodin inhibits angiogenesis of breast cancer by targeting VEGFA transcription. Theranostics. 2020;10:6839-53
Author contact
![]() Corresponding authors: E-mail: xiaobingxiuedu.cn, or yanzhilongedu.cn; Tel: +86-574-87600758; Fax: +86-574-87608638.
Corresponding authors: E-mail: xiaobingxiuedu.cn, or yanzhilongedu.cn; Tel: +86-574-87600758; Fax: +86-574-87608638.
 Global reach, higher impact
Global reach, higher impact