13.3
Impact Factor
Theranostics 2021; 11(1):181-193. doi:10.7150/thno.48206 This issue Cite
Research Paper
Integrative analysis of long extracellular RNAs reveals a detection panel of noncoding RNAs for liver cancer
1. MOE Key Laboratory of Bioinformatics, Center for Synthetic and Systems Biology, School of Life Sciences, Tsinghua University, Beijing 100084, China.
2. Department of Maternal, Child and Adolescent Health, School of Public Health, Anhui Medical University, MOE Key Laboratory of Population Health Across Life Cycle, NHC Key Laboratory of Study on Abnormal Gametes and Reproductive Tract, Anhui Provincial Key Laboratory of Population Health and Aristogenics, No 81 Meishan Road, Hefei 230032, Anhui, China.
3. Department of Surgery, Eastern Hepatobiliary Surgery Hospital, Navy Medical University, Shanghai 200433, China.
4. Department of Epidemiology, Faculty of Navy Medicine, Navy Medical University, Shanghai 200433, China.
5. International Co-operation Laboratory on Signal Transduction, Eastern Hepatobiliary Surgery Institute, Second Military medical University, Shanghai 200438, China.
6. National Center for Liver Cancer, Shanghai 201805, China.
#Joint first-authors.
Abstract
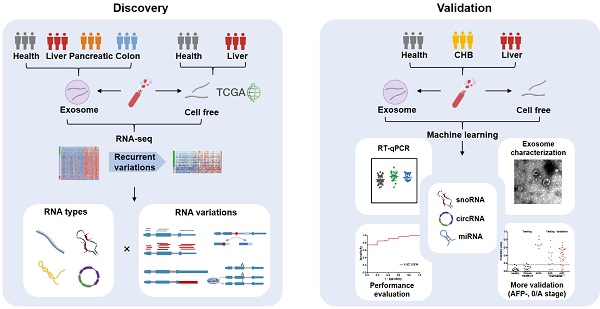
Rationale: Long extracellular RNAs (exRNAs) in plasma can be profiled by new sequencing technologies, even with low abundance. However, cancer-related exRNAs and their variations remain understudied.
Methods: We investigated different variations (i.e. differential expression, alternative splicing, alternative polyadenylation, and differential editing) in diverse long exRNA species (e.g. long noncoding RNAs and circular RNAs) using 79 plasma exosomal RNA-seq (exoRNA-seq) datasets of multiple cancer types. We then integrated 53 exoRNA-seq datasets and 65 self-profiled cell-free RNA-seq (cfRNA-seq) datasets to identify recurrent variations in liver cancer patients. We further combined TCGA tissue RNA-seq datasets and validated biomarker candidates by RT-qPCR in an individual cohort of more than 100 plasma samples. Finally, we used machine learning models to identify a signature of 3 noncoding RNAs for the detection of liver cancer.
Results: We found that different types of RNA variations identified from exoRNA-seq data were enriched in pathways related to tumorigenesis and metastasis, immune, and metabolism, suggesting that cancer signals can be detected from long exRNAs. Subsequently, we identified more than 100 recurrent variations in plasma from liver cancer patients by integrating exoRNA-seq and cfRNA-seq datasets. From these datasets, 5 significantly up-regulated long exRNAs were confirmed by TCGA data and validated by RT-qPCR in an independent cohort. When using machine learning models to combine two of these validated circular and structured RNAs (SNORD3B-1, circ-0080695) with a miRNA (miR-122) as a panel to classify liver cancer patients from healthy donors, the average AUROC of the cross-validation was 89.4%. The selected 3-RNA panel successfully detected 79.2% AFP-negative samples and 77.1% early-stage liver cancer samples in the testing and validation sets.
Conclusions: Our study revealed that different types of RNA variations related to cancer can be detected in plasma and identified a 3-RNA detection panel for liver cancer, especially for AFP-negative and early-stage patients.
Keywords: circular RNA, extracellular RNA, liquid biopsy, noncoding RNA, RNA biomarker, cancer
 Global reach, higher impact
Global reach, higher impact