13.3
Impact Factor
Theranostics 2020; 10(19):8880-8902. doi:10.7150/thno.47548 This issue Cite
Review
Long non-coding RNAs in gastric cancer: New emerging biological functions and therapeutic implications
1. State Key Laboratory of Biotherapy and Cancer Center, Department of Gastrointestinal Surgery, West China Hospital, Sichuan University, Chengdu 610041, China.
2. College of Traditional Chinese Materia Medica, Shenyang Pharmaceutical University, Shenyang 110064, China.
3. Department of Plastic Surgery, Peking University Third Hospital, Beijing 100191, China.
4. College of Stomatology, West China Hospital, Sichuan University, Chengdu 610041, China.
#These authors contributed equally to this work.
Received 2020-4-28; Accepted 2020-6-28; Published 2020-7-11
Abstract
Gastric cancer (GC) is currently the fourth most common malignancy and the third leading cause of cancer-related deaths worldwide. Long non-coding RNAs (lncRNAs), transcriptional products with more than 200 nucleotides, are not as well-characterized as protein-coding RNAs. Accumulating evidence has recently revealed that maladjustments of diverse lncRNAs may play key roles in multiple genetic and epigenetic phenomena in GC, affecting all aspects of cellular homeostasis, such as proliferation, migration, and stemness. However, the full extent of their functionality remains to be clarified. Considering the lack of viable biomarkers and therapeutic targets, future research should be focused on unravelling the intricate relationships between lncRNAs and GC that can be translated from bench to clinic. Here, we summarized the state-of-the-art advances in lncRNAs and their biological functions in GC, and we further discuss their potential diagnostic and therapeutic roles. We aim to shed light on the interrelationships between lncRNAs and GC with respect to their potential therapeutic applications. With better understanding of these relationships, the biological functions of lncRNAs in GC development will be exploitable, and promising new strategies developed for the prevention and treatment of GC.
Keywords: Long non-coding RNA (lncRNA), Gastric cancer (GC), Biological function, Biomarker, Therapeutic application
Introduction
Gastric cancer (GC), one of the most common malignant tumors, is the second leading cause of cancer-related deaths worldwide after lung cancer and is a major cause of cancer-related mortality in China [1]. GC is classified anatomically into gastric adenocarcinoma and gastro-esophageal borderline adenocarcinoma. Histologically, it can be divided into two types: diffuse and intestinal [2,3]. With the present surgical techniques and the implementation of traditional radiotherapy, chemotherapy, and neoadjuvant therapy, 5-year overall survival of patients with early GC can reach 95%. However, GC is seldom diagnosed early and therefore in most patients the disease has progressed to an advanced stage by the time of clinical manifestation. As a result of the malignant invasion and metastasis that is seen in later stages, the traditional treatments of surgery and chemoradiotherapy are not as effective as in early disease, and the 5-year survival rate is <20% [4]. Currently, the main treatment for advanced GC (AGC) includes combinations of chemoradiotherapy, molecular targeted therapy, and immunotherapy. In order to better understand the pathogenesis of GC and identify more effective biomarkers predicting prognosis and response to treatment, extensive research is needed on all aspects of the disease. Recently, compelling evidence has emerged for an important role of long non-coding RNAs (lncRNAs) in the pathogenesis and progression of GC through intricate molecular signaling networks.
The human genome contains a vast array of nucleotides, and these complex sequences can be transcribed and finally translated into more than 100,000 proteins. Of these, only 2% constitute transcripts that encode mature proteins; the remaining 98%, called non-coding RNAs (ncRNAs), do not encode proteins but still play essential roles in cell biology [5]. NcRNAs with more than 200 nucleotides are classified as lncRNAs; these account for more than 80% of ncRNAs, and are the new focus points in cancer research. Many lncRNAs have conserved secondary structures, shear forms, and subcellular localization; their conservation and specificity suggest that they are functional. However, their functions are more difficult to determine than those of microRNAs and proteins, because they cannot be inferred from their sequences or structures alone [6]. Based on their locations in the genome relative to the protein-coding genes, lncRNAs can be divided into five types: sense, antisense, bidirectional, intronic, and intergenic. Compared with the transcriptional orientation of the coding RNAs, the transcriptional orientation of lncRNAs can be antisense in addition to sense. Antisense lncRNAs transcripts are initiated inside or 3' of protein-coding genes, transcribed in the opposite direction to the protein-coding genes, and overlap by at least one exon. Bidirectional lncRNAs are initiated in different ways from the promoter of a protein-coding gene; the distance that constitutes bidirectionality is usually within hundreds of base pairs. Intron lncRNAs are initiated within introns of protein-coding genes and overlap entirely with these introns. Finally, intergenic lncRNAs are defined with different transcription units from protein-coding genes [7]. From another point of view, lncRNAs can be divided into nuclear lncRNAs and cytoplasmic lncRNAs, and this classification according to subcellular localization plays a key role in predicting their function. Most nuclear lncRNAs are involved in the regulation of epigenetic events and transcription, while lncRNAs in the cytoplasm play a major role at the post-transcriptional level. Thus, knowledge of such positional relationships is useful to infer the functions of lncRNAs. In addition, the promoter upstream transcripts (PROMPTs) and enhancer-associated RNAs (eRNAs) transcribed from promoters and enhancers, respectively, are similar to regulatory DNA elements in terms of their functional mechanisms [6]. Unlike other lncRNAs, PROMPTs share some regulatory elements with the promoters of adjacent protein-coding genes. The expression and function of PROMPTs is often associated with adjacent protein encoding transcripts. In mammals, PROMPTs are transcribed in the antisense orientation, about 0.5-2.5 kb from the transcription start sites (TSSs) of the protein-coding gene [8]. eRNAs are a group of lncRNAs transcribed from enhancers, which can promote enhancer function, and their regulation of gene expression is an emerging research field. eRNAs can recruit transcription factors, transcriptional co-activators, and chromatin remodelers in either cis or trans to regulate gene transcription and hence biological processes. Unlike common lncRNAs, eRNAs have similar transcription rates but produce fewer stable transcripts [9]. In addition, lncRNAs possess certain characteristics similar to those of mRNAs; for instance, lncRNAs are transcribed by RNA polymerase II, they contain a promoter, and their structure consists of multiple exons, equipped with 5' caps and 3' poly A tails that are spliced into mature transcripts by a typical mechanism [10]. Similar to the protein-coding genes, their genomic positions are marked by Histone H3 lysine K4 (H3K4) trimethylation at the transcription initiation site and H3K36 trimethylation enrichment throughout the genome; however, compared to the protein-coding transcripts, there are fewer exons in lncRNAs and the overall level of expression is lower [11]. Despite being initially regarded as “transcriptional noise,” accumulating evidence indicates that lncRNAs influence local or global gene expression via transcriptional, post-transcriptional, and epigenetic regulatory mechanisms [12]. Over the past decade, significant progress has been made in the study of GC-related lncRNAs, which are involved in a variety of tumor signaling pathways, such as Notch, mTOR, NF-κ B, and Wnt [13].
With the advent of high-throughput sequencing technology, novel lncRNAs, considered an important class of RNA, are being intensely investigated. Some lncRNAs have been found to have functional roles in the pathogenesis and development of cancer, and could be potential therapeutic targets or biomarkers for diagnosis and prognosis. Thus, these new findings will provide an insight into more new strategies for the diagnosis and treatment of lncRNAs in gastric cancer.
Multi-functional roles of lncRNAs in gastric cancer
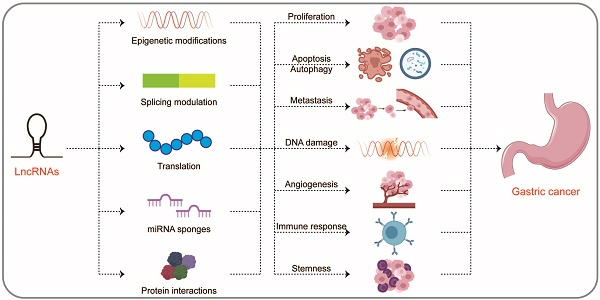
LncRNAs are involved in many epigenetic mechanisms, including histone modification, DNA methylation, hydroxymethylation, and chromatin remodeling, which control the expression of regulatory factors involved in the progression of GC, such as genes related to DNA repair, apoptosis, autophagy, and regulators of cell cycle, transcription, and signaling pathways [14]. In addition, lncRNAs can hybridize with pre-mRNAs to modulate their alternative splicing by blocking the recognition of splice sites by spliceosomes to produce alternative transcripts [15]. LncRNAs located in the cytoplasm most commonly act as molecular sponges of miRNAs, thereby regulating the expression of the downstream miRNAs [16]. Moreover, lncRNAs can bind directly to proteins, affecting protein activity, localization, and protein-protein interactions [17]. Therefore, here we specifically describe three levels of molecular mechanisms of lncRNAs in the progression of GC, namely at the DNA, RNA, and protein levels (Figure 1 and Table 1).
LncRNAs and epigenetic modifications
Histone modifications
Epigenetic modifiers are by far the most common protein partners of lncRNAs, and considerable attention has been lavished on their members, including histone methyltransferase and Polycomb Repressive Complex (PRC2) [18]. PRC2, a methyltransferase composed of EZH2, SUZ12, and EED, epigenetically regulates gene expression by catalyzing the methylation and trimethylation of lysine residue 27 of histone 3 (H3K27me3) [19].
Multi-functional roles of lncRNAs in gastric cancer
| Mode of mechanism | LncRNA | Expression | Molecular mechanism | Reference |
|---|---|---|---|---|
| Histone Modification | UCA1 | up | Mediates the trimethylation of H3K27 in promoters of p21 and SPRY1 by binding to EZH2. | [24] |
| MNX1-AS1 | up | Promotes GC progression through EZH2/BTG2 and miR-6785-5p/BCL2 aixs | [29] | |
| SNHG3 | up | Regulats neighboring MED18 gene methylation. | [30] | |
| ARHGAP27P1 | down | Epigenetically regulates p15 and p16 | [32] | |
| GCLNC1 | up | Acts as a modular scaffold of WDR5 and KAT2A complexes | [33] | |
| GCAWKR | up | Acts as a molecular scaffold of WDR5/KAT2A complexes | [34] | |
| LINC00673 | up | Acts as a scaffold for LSD1 and EZH2 | [35] | |
| FEZF1-AS1 | up | Represses the expression of P21 via binding with LSD1 | [21] | |
| HOTAIR | up | Recruits the PRC2 complex to silence target gene via H3K27me3 modification | [71] | |
| XIST | up | Interacts with EZH2 to suppress transcription of its potential target KLF2 | [72] | |
| TUG1 | up | Associates with PRC2 | [73] | |
| AGAP2-AS1 | up | Interacts with LSD1 and EZH2 and suppresses CDKN1A (P21) and E-cadherin transcription | [74] | |
| DNA methylation | SNHG1 | up | Promotes DNMT1 expression | [36] |
| MEG3 | down | DNA methylation affects the expression of MEG3 and its targeting miR-181a-5p | [39] | |
| HOTTIP | up | Increases HoxA13 expression by down-regulating DNA methylation at the E1 site | [75] | |
| LINC00673 | up | Suppresses KLF4 expression by interacting with EZH2 and DNMT1 | [76] | |
| Linc00441 | up | Recruits DNMT1 to the RB1 promoter and suppresses RB1 expression | [77] | |
| mRNA translation | MACC1-AS1 | up | Stabilizes MACC1 mRNA | [41] |
| GMAN | up | Promotes translation of ephrin A1 messenger RNA | [44] | |
| KRT7-AS | up | Stabilizes KRT7 mRNA | [78] | |
| CeRNA | KCNQ1OT1 | down | KCNQ1OT1/miR-9/LMX1A | [50] |
| LINC00682 | down | LINC00682/miR-9/LMX1A | [51] | |
| COL1A1-014 | up | COL1A1-014/miR-1273h-5p/CXCL12 | [52] | |
| MYOSLID | up | MYOSLID/miR-29c-3p/MCL-1 | [53] | |
| CTC-497E21.4 | up | CTC‑497E21.4/miR-22/NET1 | [57] | |
| LINC00346 | up | LINC00346/miR-34a-5p/CD44, NOTCH1, AXL | [79] | |
| UCA1 | up | UCA1/miR-26a/b, miR-193a, miR-214/PDL1 | [80] | |
| LINC01133 | down | LINC01133/miR-106a-3p/APC | [81] | |
| MT1JP | down | MT1JP/miR-92a-3p/FBXW7 | [82] | |
| LINC01939 | down | LINC01939/miR-17-5p/EGR2 | [83] | |
| H19 | up | H19/miR-22-3p/Snail | [84] | |
| UCA1 | up | UCA1/miR-203/ZEB2 | [85] | |
| HNF1A-AS1 | up | HNF1A-AS1/miR-661/CDC34 | [86] | |
| UFC1 | up | UFC1/miR-498/Lin28b | [87] | |
| GCRL1 | up | GCRL1/miR-885-3p/CDK4 | [88] | |
| SMARCC2 | down | SMARCC2/miR-551b-3p/TMPRSS4 | [89] | |
| BC032469 | up | BC032469/miR-1207-5p/hTERT | [90] | |
| LncRNAs act on proteins | MIR4435-2HG | up | Binds to DSP and inhibits its expression | [60] |
| KRT19P3 | down | Enhances COPS7A protein stability in GC cells | [62] | |
| LINC00707 | up | Interacts with mRNA stabilizing protein HuR | [65] | |
| HOXC-AS3 | up | Binds to YBX1 | [69] | |
| SNHG12 | up | Increases the phosphorylation of PI3K by directly binding to it | [91] | |
| SEMA3B-AS1 | down | Interacts with MLL4 | [92] | |
| pancEts-1 | up | Directly interacts with NONO to increase its interaction with ERG | [93] | |
| GALNT5 uaRNA | up | Binds with HSP90 | [94] | |
| HOTAIR | up | Interacts with Mex3b | [95] |
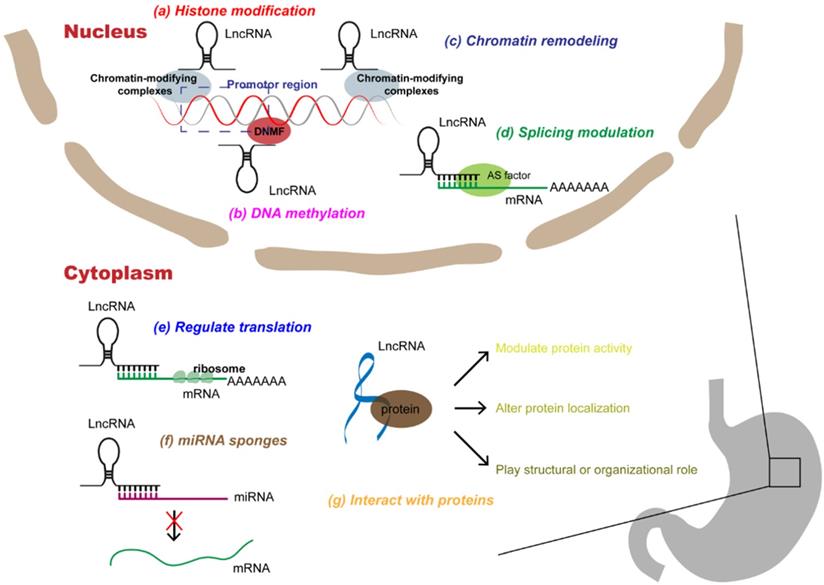
Multi-functional roles of lncRNAs in gastric cancer. In the nucleus, lncRNAs can participate in a variety of epigenetic mechanisms by recruiting chromatin modifiers, including (a) histone modifications, (b) DNA methylations, (c) chromatin remodeling; in addition, lncRNAs can participate in (d) splicing modulations of mRNAs. In the cytoplasm, lncRNAs can interact with RNAs, which can not only (e) rugulate translation of mRNAs, but also act as (f) miRNA sponges to alleviate miRNAs' inhibition of target genes. In addition, lncRNAs can directly (g) interact with proteins, affecting protein activity, localization, or as structural components.
About 20% of human lncRNAs are reported to associate physically with PRC2, suggesting that they may play a general role in recruiting PRC2 to its target genes [20]. Other epigenetic modifiers including LSD1, SMYD3, WDR5, SET1C, KMT2C, ING2, and PAQR3 may also be potential partners for lncRNAs [21-23]. One of the many lncRNAs acting on epigenetic modifiers is urothelial cancer-associated 1 (UCA1). RNA sequencing (RNA-seq) analysis showed that UCA1 knockout preferentially affects genes related to the cell cycle, proliferation, and migration. UCA1 promotes cell proliferation by binding to the histone methyltransferase EZH2 to inhibit the expression of Sprouty RTK signaling antagonist 1 (SPRY1) and P21, a cyclin-dependent kinase (CDK) inhibitor [24]. SPRY1, part of the mammalian sprouty gene family consisting of four members (SPRY1-4), has tumor suppressive activity and is downregulated in certain tumors such as prostate and breast cancer [25,26]. UCA1 was found to mediate the trimethylation of H3K27 in P21 and SPRY1 promoters [24]. Therefore, it can be concluded that UCA1, via its action on EZH2, can also influence GC progression, at least partially, through the epigenetic inhibition of P21 and SPYR1.
As an important member of the TEA domain (TEAD) family, TEAD4 is considered to be a key transcription factor involved in the development of various cancers including GC [27,28]. By binding to MNX1-AS1 (MNX1 antisense RNA 1) promoter, TEAD4 facilitates the transcription of MNX1-AS1, and acts as a carcinogen at the level of transcription. MNX1-AS1 can also act on EZH2 by recruiting EZH2 and H3K27me3 to the BTG2 (B-cell translocation gene 2) promoter and thus partially inhibiting BTG2, a tumor suppressor, thereby contributing to carcinogenicity in GC [29].
The expression of Small Nucleolar RNA Host Gene 3 (SNHG3) is elevated in GC, and high SNHG3 is associated with poor prognosis [30]. Because it is well-established that the mode of action of lncRNAs is by regulating adjacent genes, the relative expression of the MED18 (mediator subunit 18) gene adjacent to SNHG3 was analyzed [31]. MED18, a component of the Mediator complex and coactivator of DNA-binding factors that activate transcription by RNA polymerase II, is involved in regulation of lipid and peroxisome metabolism. SNHG3 knockdown weakened the enrichment of EZH2 in the MED18 promoter, which led to a decrease in methylation level and the upregulation of the MED18 promoter [30].
RNA immunoprecipitation (RIP) experiments showed that the lncRNA ARHGAP27P1 specifically bound only to the chromatin modifier Jumonji domain-containing 3 (JMJD3). Over-expression of ARHGAP27P1 enhanced the association between ARHGAP27P1 and JMJD3, resulting in a significant decrease in H3K27me3, thus eventually facilitating the transcription of P15, P16, and P57. Furthermore, the inhibitory effect of ARHGAP27P1 on cell proliferation and cell cycle progression can be reversed by knocking down JMJD3, P15, or P16 [32].
Studies have also revealed an involvement of some lncRNAs in GC through their action as modular scaffolds of WDR5 and KAT2A complexes, such as GCLNC1 and GCAWKR; some lncRNAs such as LINC00673 and FEZF1-AS1 act directly on LSD1, and thus regulate the expression of their downstream targets [21,33-35].
Increased lncRNA-AK058003 expression is frequently found in hypoxic GC. This regulates the expression of γ-synuclein (SNCG), a tumor metastasis-related gene involved in hypoxia-induced GC metastasis. AK058003 promotes hypoxia-induced GC metastasis and invasion by activating SNCG via the initiation of DNA demethylation of its CPG islands [37].
Studies on MEG3 in diabetic patients indicated that potential DNA methylation in the promoter region resulted in the reduction of target gene expression, thereby affecting disease development [38]. This was also observed in GC, where MEG3 levels are significantly lower and its methylation significantly greater in tumor cells than in para-cancerous tissues, as demonstrated in multiple GC cell lines [39]. Furthermore, DNA methylation affects the expression of MEG3 and its target miR-29 in HCC [40]. Similarly in GC, DNA methylation affects the expression of the MEG3 target miR-181a-5p, a tumor suppressor, thus downregulating the downstream target ATG4B (autophagy-associated 4B) [39].
LncRNAs act at the post-transcriptional level
mRNA translation
Some lncRNAs affect RNA translation by binding and stabilizing mRNAs. Recently, MACC1-AS1 (MACC1 antisense RNA 1), which is elevated under metabolic stress, was shown to promote the stability of MACC1 (metastasis-associated in colon cancer-1) mRNA by physically binding to it, as well as increasing its expression [41]. MACC1, a key regulator of the HGF/c-MET axis, is an important target for tumor therapy [42]. In addition, MACC1-AS1 has been shown to enhance metabolic plasticity by upregulating MACC1 and subsequently enhancing glycolysis and antioxidant capacity, possibly through the AMPK/Lin28 pathway [41]. AMPK and Lin28 are both important RNA-binding proteins (RBPs) that regulate the stability of mRNA.
Ephrin A1, which is either upregulated or downregulated in different cancers, is associated with tumor progression, malignancy, and prognosis [43]. GC cells exhibit high amounts of GMAN (gastric cancer metastasis associated long noncoding RNA), a sense lncRNA, which competitively binds to antisense GMAN RNA (GMAN-AS), resulting in polymer formation of the downstream Ephrin A1 mRNA and thus increasing the translation of Ephrin A1 mRNA. This promotes the invasive activity and metastasis of GC cells. Knocking out GMAN or Ephrin A1 in GC cell lines reduces their invasive activity and ability to form metastases in mice [44].
Competitive endogenous RNA (ceRNA)
It has become increasingly clear that multiple RNA transcripts, such as ncRNAs, lncRNAs, pseudogenes, and circular RNAs, can be act as ceRNAs, also known as “molecular sponges” of miRNAs, to regulate the expression of miRNA target genes through miRNA response elements (MREs) [45]. It is worth noting that a variety of lncRNAs involved in the progress of GC can be regulated in this manner, and lncRNA-miRNA-mRNA network alterations are considered indispensable mechanisms in GC. There are several examples of lncRNAs functioning as sponges and therefore oncogenes or tumor suppressor genes in GC.
KCNQ1OT1, an miR-504 sponge that targets CDK16 (cyclin‑dependent kinase 16) in HCC and CCNE2 (cyclin E2), and a miR-370 sponge, in glioma can also act as an ceRNA via neutralizing miR-9. This promotes the progression of GC by regulating the expression of Lim homeobox transcription factor 1 (LMX1A) [46,47]. LMX1A is a tumor suppressor and member of the LIM-homeodomain (LIM-HD) family [48], which is downregulated in cancers [49]. Moreover, in GC cells in which LMX1A is knocked out, over-expressing or silencing KCNQ1OT1 is completely ineffective, consistent with the notion that KCNQ1OT1 participates in GC progression by regulating the expression of miR-9-LMX1A [50]. Another lncRNA that can control the progression of GC by regulating the miR-9-LMX1A axis is LINC00682. Ectopic over-expression of LINC00682 can downregulate miR-9 and upregulate LMX1A, as well as decrease GC cell survival, proliferation, migration, and invasion [51].
The level of expression of chemokine (CXCmotif) ligand (CXCL12) was found to be significantly increased in GC tissues with high COL1A1-014 expression. Bioinformatics analysis revealed that the two genes have complementary sequences for miR-1273H-5P; COL1A1-014 might act as a ceRNA and compete with CXCL12 to bind with miR-1273h-5p, thus inhibiting the degradation of CXCL12 by miR-1273H-5P and promoting GC progression. Additionally, BGC-823 cells over-expressing COL1A1-014 also exhibited strong colony-forming ability, which may reflect the degree of malignancy and have important significance for prognosis [52].
MYOSLID, found to be elevated in GC, is mainly concentrated in the cytoplasm [53]. MCL-1 is a member of the anti-apoptotic BCL2 family that is over-expressed in many tumor types and protects tumor cells from invasion [54,55]. MYOSLID promotes cell proliferation and inhibits apoptosis by regulating the miR-29C-3P-MCL-1 axis and miR-29C-3P thus acts as a tumor inhibitor in GC [53].
Neuroepithelial cell transforming gene 1 (NET1) is a RHOA (Ras homolog gene family member A) specific guanine exchange factor (GEF); RHOA is one of the most widely studied members of the RHO GTPase family of proteins. The RHOA signaling pathway is involved in biological processes such as cell proliferation and apoptosis and is related to the invasion and metastasis of tumors [56]. CTC-497E21.4, involved in post-transcriptional regulation in the cytoplasm, competes with miR-22 and regulates the expression of NET1 through the RHOA signaling pathway [57]. Low miR-22 expression also promotes extracellular matrix (ECM) remodeling and epithelial-to-mesenchymal transition (EMT) by enhancing matrix metalloproteinase (MMP)14 and Snail expression in GC invasion and metastasis [58].
LncRNAs act on proteins
In addition to the above-mentioned complex regulations at the epigenetic and transcriptional levels, lncRNAs can also interact with functional proteins in the most direct manner. By regulating the activity and function of proteins, influencing the interactions between proteins, or changing the localization of proteins in cells, and even functioning as structural components, lncRNAs participate in the development of cancers.
Desmoplakin (DSP), the first discovered member of the Plakin family of proteins and a significant component of desmosomal plaques, is considered to play an important role in the early stages of tumor development [59]. There is a negative correlation between MIR4435-2HG and DSP, with the former inhibiting the latter, which stimulates the growth and metastasis of GC cells by inducing EMT and Wnt/β-catenin signaling. Further, the interaction between DSP and MIR4435-2HG has been verified by RNA pull-down test, and the positive rather than the negative MIR4435-2HG pull-down protein complex was detected [60].
KRT19P3 (Keratin 19 Pseudogene 3) can regulate the activity and enhance the stability of COPS7A in GC by binding directly to it. COPS7A belongs to the CSN complex and is composed of eight subunits designated CSN1-CSN8. Abnormal expression of CSN reportedly affects key processes in carcinogenesis such as signal transduction, cell cycling, and apoptosis. COPS7A enhances de-ubiquitination of IκBα (inhibitor of nuclear factor kappa-B (NF-κB)), by the CSN-associated de-ubiquitinase USP15; thus, KRT19P3 suppresses the NF-κB signaling pathway in a COPS7A-dependent manner [61,62]. As a consequence, decreased expression of KRT19P3 is associated with tumor size, TNM staging, Lauren grade, lymph node metastasis, and poor prognosis.
LINC00707, 3087 bp long, was previously reported to be an important oncogene in lung adenocarcinoma [63]. Recently, it has been found that LINC00707, which also functions as an oncogene in GC, can interact with HuR, which itself interacts with 3′UTR regions of mRNA after transcription and enhances its stability; the resulting LINC00707-HuR can further combine with VAV3/F11R mRNAs to increase their stability [64,65]. There is abundant evidence that VAV3 plays an important role in cell adhesion, angiogenesis, and cell differentiation, and F11R, also known as junctional adhesion molecule A (JAM-A), significantly influences epithelial cell morphology and migration [66,67].
In addition to HuR, RNA-binding protein YBX1 (Y-box-binding protein 1) also participates in the regulation of lncRNAs in GC. YBX1 is a protein with a common nucleic acid-binding domain in the gene promoter CCAA T-box, which influences many cell processes, including transcription and translation [68]. HOXC-AS3 potentially regulates a group of genes related to cell proliferation and migration of GC, including MMP7, WNT10B, HDAC5 and others, by binding to YBX1 [69]. Among these, HDAC5 is a key gene belonging to the histone deacetylase (HDAC) family. Because histone acetylation and deacetylation are essential for chromatin remodeling and gene expression, imbalances in these reactions lead to disorderly expression of many cancer-related genes [70]. In addition to HDAC5, numerous genes related to tumorigenesis are also regulated in a similar manner by HOXC-AS3 [69].
Relationships between lncRNAs and the hallmarks of gastric cancer
GC carcinogenesis is a multi-step process influenced by many different factors, such as heredity and environment. A variety of complex cancers share several common characteristics, as identified by Hanahan and Weinberg, including sustaining proliferation signals, avoidance of growth inhibitors, resistance to cell death, replicative immortality, induction of angiogenesis, activation of invasion, and metastasis [96]. Each factor contributes to the formation of large numbers of abnormal cells that grow unchecked. In recent years, the exploration and elucidation of the structures and functions of lncRNAs have shown these to be important molecular signal transduction media in cancer development in that they regulate the expression of specific genes and corresponding signal pathways. Below, we will discuss the regulatory role of some lncRNAs in GC (Table 2).
LncRNAs and proliferation
Sustained proliferation is a typical characteristic of cancer cells. Complex molecular pathways and dynamic crosstalk between tumor cells and their microenvironment maintain these characteristics. The ability of a cancer cell to sustain chronic proliferation is its most fundamental characteristic, achieved by the maintenance of growth signals via the autocrine and paracrine growth factor pathways [97]. Normal tissues strictly control the production and release of growth-promoting signals, which in turn control cell growth and division to ensure homeostasis and thus maintain normal structure and function [96]. Cancer cells regulate these signals to mediate tumor growth, and lncRNAs participate in this process by regulating the growth factors or receptors (Figure 2A).
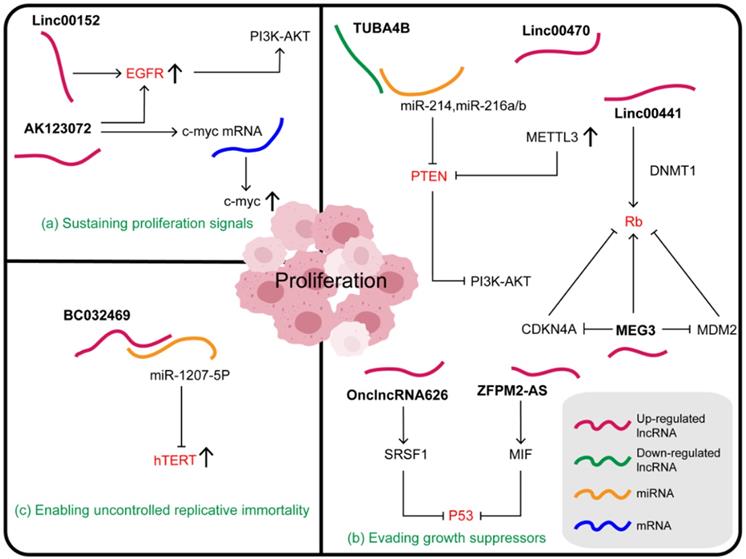
Relationships between lncRNAs and proliferation in gastric cancer. (a) Some lncRNAs mainly regulate the ability of tumors to grow by regulating growth factors or receptors. (b) Some lncRNAs affect the growth and proliferation of gastric cancer cells by directly or indirectly altering the expression of tumor suppressor factors. (c) Some lncRNAs play a role in the proliferation of gastric cancer cells by affecting telomerase activity.
The relationships between lncRNAs and the hallmarks of gastric cancer
| Function | LncRNA | Expression | Target | Reference |
|---|---|---|---|---|
| LncRNAs and proliferation | Linc00152 | up | EGFR | [99] |
| AK123072 | up | EGFR | [100] | |
| ZFPM2-AS1 | up | MIF/P53 | [104] | |
| OnclncRNA-626 | up | SRSF1/p53 | [107] | |
| GAS5 | up | eIF4A3/P53 | [147] | |
| PWRN1 | down | miR-425-5p/PTEN/Akt/MDM2/p53 | [148] | |
| AK023391 | up | PI3K/Akt/p53 | [149] | |
| TUBA4B | down | miR-214 and miR-216a/b /PTEN | [109] | |
| LINC00470 | up | METTL3/PTEN | [110] | |
| PCAT-1 | up | EZH2/PTEN | [150] | |
| Linc00441 | up | RB1 | [77] | |
| MEG3 | up | P53,RB | [151] | |
| BC032469 | up | BC032469/miR-1207-5p/hTERT | [90] | |
| LncRNAs and resistance to cell death | XIAP-AS1 | up | XIAP | [118] |
| TINCR | up | KLF2 | [121] | |
| LINC01419 | up | PI3K/Akt1/mTOR | [124] | |
| HAGLROS | up | miR-100-5p/mTOR ,mTORC | [152] | |
| LncRNAs and metastasis | MAGI2-AS3 | up | MAGI2-AS3/miR-141/200a/ZEB1 | [128] |
| DLX6-AS1 | up | miR-204-5p/OCT1 | [130] | |
| HULC | up | miR-9-5p/MYH9 | [153] | |
| DLX6-AS1 | up | FUS-regulated MAP4K1 | [154] | |
| SNHG16 | up | DKK3 | [155] | |
| ELIT-1 | up | TGFβ/Smad3 signaling | [156] | |
| LINC01133 | up | miR-106a-3p/APC | [81] | |
| MEG3 | down | E-cadherin,Vimentin | [157] | |
| LncRNA-ATB | up | MiR‐141‐3p/TGFβ2 | [158] | |
| LINC00675 | down | vimentin | [159] | |
| SNAI1 | up | CDH1 | [160] | |
| LncRNAs and DNA damage | PANDAR | up | NFYA, p53/CDKN1A | [134] |
| NORAD | up | miR-608 | [136] | |
| LncRNAs and angiogenesis | PVT1 | up | STAT3/VEGFA | [139] |
| LINC01314 | down | KLK4 | [140] | |
| ZEB2-AS1 | up | Wnt/β-catenin pathway | [161] | |
| MEG3 | down | MEG3/miR-21 | [162] | |
| LncRNAs and the immune response | UCA1 | up | miR-193a, miR-214/PDL1 | [80] |
| LncRNAs and the stemness of GC | LOXL1-AS1 | up | LOXL1-AS1/miR-708-5p/SOX2 | [145] |
| SPRY4-IT1 | up | SPRY4-IT1/miR-101-3p/AMPK | [146] | |
| MACC1-AS1 | up | MACC1-AS1/miR-145-5p | [163] | |
| MALAT1 | up | SOX2 mRNA | [164] |
Epidermal growth factor receptor (EGFR) belongs to the family of receptor tyrosine kinases (RTK), commonly over-expressed in tumors and considered as a driver and regulator of tumorigenesis [98]. Linc00152 in the cytoplasm can bind directly to EGFR, resulting in increased EGFR expression and activation of the EGFR signaling pathway. In addition, the inhibition of p-EGFR, p-AKT, and p-PI3K (phosphoinositide 3-kinase) in cells treated with Linc00152 shRNA suggested that Linc00152 might induce the constitutive activation of EGFR signaling and activate the downstream PI3K/AKT signaling pathway [99]. AK123072 is an intron antisense lncRNA that is frequently upregulated by hypoxia in GC. In hypoxia-induced AK123072 over-expression in GC cells, the CPG island methylation of the EGFR gene is significantly decreased, leading to its increased expression, thereby promoting the spread of cancer cells. Moreover, AK123072 can promote GC migration and invasion by increasing the stability and expression of c-Myc mRNA [100].
In addition to their remarkable ability to induce and maintain positive growth stimulating signals, cancer cells must also avoid the powerful mechanisms that negatively regulate cell proliferation. Several tumor suppressors, such as the classic p53, Rb, and PTEN [101,102], have been found to regulate the cell cycle and inhibit cell growth and proliferation in different ways. Characteristically, they are inactivated in cancer cells. Some lncRNAs affect the growth and proliferation of GC cells by directly or indirectly altering the expression of tumor suppressor factors (Figure 2B), for example, p53, a key gatekeeper of the cell. P53 is a stress-induced transcription factor, which can promote cell cycle arrest, apoptosis, and senescence [103]. The oncogene ZFPM2-AS1, which is an important upstream regulator of the MIF/P53 axis, promotes the proliferation and inhibits apoptosis of GC cells. The mechanism of ZFPM2-AS1 action involves regulating the expression and subcellular localization of p53 to inactivate the p53 signaling pathway through its physical interaction with MIF at the translational or post-translational level [104]. Previous studies have shown that MIF, a tumor promoter, decreases the stability of the p53 protein and prevents its nuclear translocation by physically associating with it [105]. Furthermore, it was found that the accumulation of MIF protein induced by ZFPM2-AS1 in GC cells was not due to increased protein synthesis, but was due to increased protein stability [104]. OnclncRNA-626 located on chromosome 11 can also affect p53 expression. It interacts specifically with serine- and arginine-rich splicing factor 1 (SRSF1), increasing its stability and upregulating it in GC tissues. RSF1, also known as SF2/ASF, which plays a regulatory role in many cancers, is a member of the SR family. It has two domains, RRM1 and RRM2, where RRM2 domain is the dominant binding site of OnclncRNA-626 [106,107]. Thus, OnclncRNA-626 may exert its biological function through the SRSF1/p53 pathway and may be a potential therapeutic target in GC [107].
Some lncRNAs also act on the tumor suppressor PTEN, whose frequent deletion in cancer suggests its physiological importance. PTEN governs a variety of biological processes by inhibiting the PI3K/AKT-mammalian target of rapamycin (mTOR) pathway through its lipid phosphatase activity; this includes maintaining genomic stability, cell survival, migration, proliferation, and metabolism [108]. TUBA4B, a ceRNA, was found to physically bind to and chelate miR-214 and miR-216a/b, thus increasing the expression of the common downstream target PTEN, decreasing the expression of p-PI3K and p-AKT, and thereby leading to the inactivation of the PI3K/AKT signaling pathway. Moreover, the weakened cell malignancy phenotype induced by TUBA4B was evidently rescued by PTEN silencing and AKT activation [109]. LINC00470, mainly distributed in the cytoplasm, can bind to PTEN mRNA and promote its degradation. This process, mediated by N6-methyladenosine (m6A) writer METTL3, enhanced its m6A methylation level resulting in high levels of m6A RNA and METTL3 in GC. It was also found that the LINC00470-METTL3-mediated degradation of PTEN mRNA was dependent on the human YTH domain family 2 (YTHDF2), an m6A reader protein [110]. LncRNAs that influence m6A modification play a key role in regulating cancer progression. For example, lncRNA GAS5-AS1 inhibits the growth of cervical cancer by interacting with RNA demethylase ALKBH5 to increase GAS5 stability and decrease GAS5 m6A modification [111]. It has also been reported that LINC00470 binds to fused in sarcoma (FUS) and AKT, which anchors FUS in the cytoplasm and activates AKT. Subsequently, the expression of ELFN2 is upregulated, thus affecting glycolysis, inhibiting autophagy, and promoting tumorigenesis [112,113]. Moreover, the mechanism of LINC00470 action on tumor suppressor PTEN also supports its influence on gastric cancer.
Finally, the key tumor suppressor Rb can also be regulated by lncRNAs. The Rb protein integrates signals from different extracellular and intracellular sources and determines whether the cell should continue its growth and division cycle [101]. When cells are stimulated by growth-promoting signals, cyclin D/CDK4 is induced to phosphorylate Rb1, so that the Rb-E2Fs complex releases E2Fs and cells can enter S-phase [114]. Linc00441, a carcinogenic lncRNA, recruits DNMT1 into the Rb1 promoter and inhibits the expression of RB1 in GC cells, leading to their proliferation [77]. In addition to the previously mentioned effects on the expression of ATG4B, MEG3 can inhibit MDM2, increase the active form of Rb, and then enhance the interaction between Rb and transcription factor E2Fs to block the transcription of its target gene. Additionally, MEG3 influences Rb via the regulation of RNA-protein binding or CDKN4A [115].
Telomerase, a DNA polymerase that adds telomeric repeats to the ends of telomeric DNA, maintains telomere length and protects the chromosome ends, crucial for the development and survival of tumors [96,116]. Telomerase reverse transcriptase (hTERT), a catalytic subunit of telomerase, plays an important role in maintaining telomere length, maintaining cell division, and replication [117]. A novel endogenous lncRNA BC032469, which is highly expressed in GC tissue, can combine directly with, and effectively act as a sponge for, miR-1207-5p to regulate hTERT expression and thus the cell cycle in the G0/G1 phase. Therefore, upregulation of BC032469 is associated with larger tumor volume, poor differentiation, and shorter survival time in patients with GC (Figure 2C) [90].
LncRNAs and resistance to cell death
Programmed cell death (PCD) refers to apoptosis, autophagy, and programmed necrosis, all of which are mediated by intracellular programming. The fate of tumor cells can be determined by the balance between cell death and survival mediated by PCD. Abnormal PCD processes can also result in faulty proliferation patterns of tumor cells. The cascade of events due to lncRNAs involved in tumorigenesis and malignant transformation are mostly related to the first two forms, namely apoptosis and autophagy (Figure 3). Another newly-described lncRNA, XIAP-AS1, mainly located in the nucleus, is transcribed from the first intron of the complementary chain of the XIAP (X-linked apoptosis inhibitor) gene. XIAP binds to the BIR2 and BIR3 domains of caspase-3 and caspase-9, respectively, to inhibit their activation and block apoptosis. Full-length XIAP-AS1 RNA interacts with SP1, which participates in the transcription of XIAP, and thus affects the apoptosis process mediated by XIAP. Furthermore, the downregulation of XIAP-AS1 promotes the apoptosis of gastric tumor cells induced by tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) [118,119]. TRAIL, a member of the TNF family of cytokines, induces apoptosis through both mitochondrial-dependent and death receptor pathways in tumor cells, preferentially killing tumor cells but not normal cells [120]. Therefore, XIAP-AS1 affects the apoptosis of GC cells by influencing the transcription of XIAP and acting as a potential target for TRAIL. Silencing the expression of TINCR inhibits cell proliferation, colony formation, tumorigenicity, and can promote cell apoptosis as reported for SGC7901 and BGC823 cell lines. TINCR binds to STAU1 protein, which can induce mRNA degradation, and the resulting complex affects the expression of Kruppel-like factor (KLF)2 through this STAU1-mediated mRNA degradation (SMD) [121]. SMD is a translation-dependent mechanism that occurs when STAU1, together with the nonsense-mediated mRNA decay factor UPF1, binds at a location downstream of the termination codon [122]. SMD is a translation-dependent mechanism that occurs when STAU1, together with the nonsense-mediated mRNA decay factor UPF1, binds at a location downstream of the termination codon [123]. When the expression of KLF2 is regulated by TINCR, KLF2 further regulates the transcription and expression of the CDK genes CDKN1A/P21 and CDKN2B/P15, thus affecting the proliferation and apoptosis of GC cells [121]. Further understanding of the function and clinical significance of TINCR and its target genes KLF2, CDKN1A/P21, and CDKN2B/P15 may be helpful for early diagnosis and treatment of GC. LINC01419 silencing inhibits the activation of the PI3K/AKT1/mTOR pathway, as demonstrated by reduced phosphorylation of AKT1 and mTOR, thus promoting autophagy of GC cells as well as inhibiting tumor growth and metastasis. These findings offer a new perspective in that the downstream PI3K/AKT1/MTOR axis mediated by LINC01419 may be a new target for GC therapy [124].
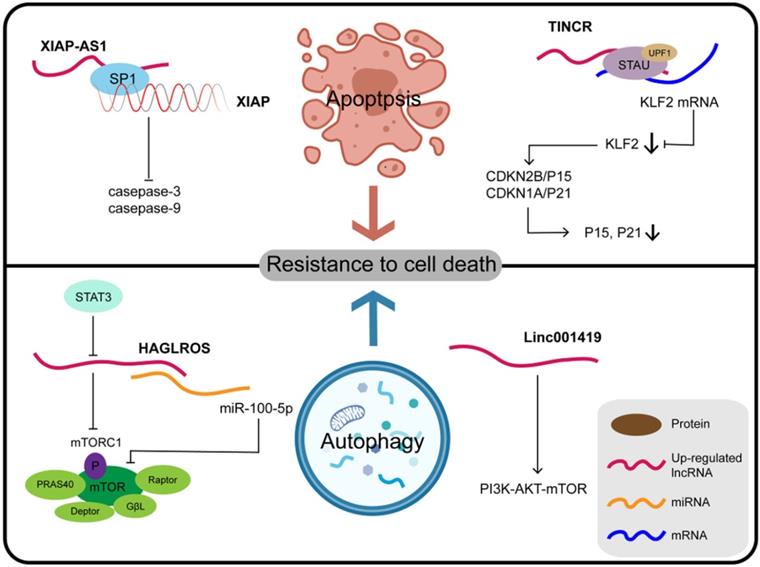
Some lncRNAs and their molecular partners or genomic targets play their key roles in resistance to cell death in gastric cancer.
Some lncRNAs play their important roles in the metastasis and invasion of gastric cancer by participating in the EMT process.
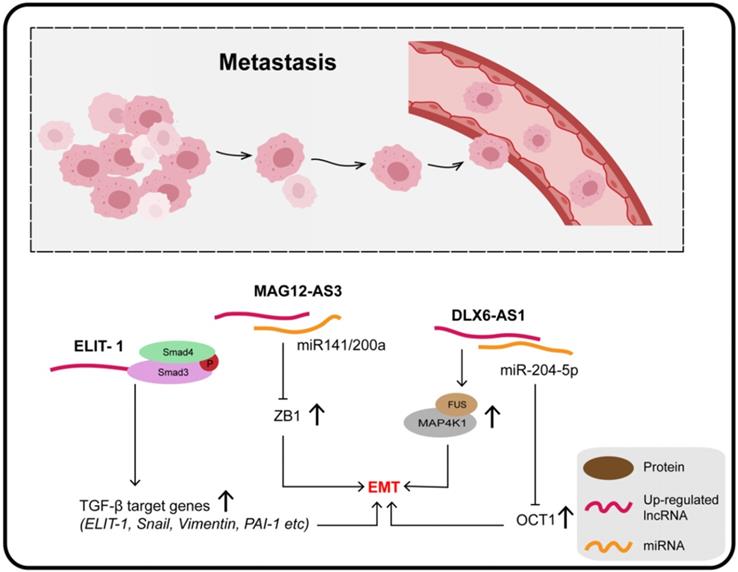
LncRNAs and metastasis
EMT, considered to be an important mechanism of tumor metastasis, is a cell program that is crucial for embryogenesis and wound healing, but also furthers malignant progression and can facilitate tumor initiation and metastatic potential of cancer cells. The process of EMT encompasses the disintegration of intercellular connections, reorganization of the actin cytoskeleton, and enhancement of cell viability and invasive capacity [125]. The destruction of tight junctions is related to the loss of the epithelial marker E-cadherin and the acquisition of mesenchymal markers N-cadherin and vimentin [126]. A variety of lncRNAs is involved in the development of GC by regulating EMT and metastasis (Figure 4).
Four molecular subtypes of GC can be defined on the basis of microsatellite stability (MSS) or instability (MSI) and p53 status, namely, MSS/TP53+, MSS/TP53-, MSI, and MSI/EMT [127]. Zhang et al. quantified lncRNA MAGI2-AS3 in these four subtypes of GC and reported that it was extremely high in the MSS/EMT subtype, suggesting that MAGI2-AS3 is associated with EMT. Moreover, miR-141 and miR-200a are known to be negative regulators of the ZEB1 gene and EMT pathway; MAGI2-AS3, mainly located in the cytoplasm, promotes tumor metastasis by binding miR-141/200a to maintain downstream ZEB1 over-expression, which is a well-accepted core factor regulating the EMT process [128,129].
It was confirmed that knockout of DLX6-AS1 inhibits the expression of N-cadherin, MMP9, and SLUG, as well as promoting the expression of E-cadherin. In addition, the distribution of the actin cytoskeleton and the presence of filamentous pseudopodia were significantly reduced when DLX6-AS1 was knocked out. The specific mechanism of its action involves DLX6-AS1 functioning as an endogenous cancer-promoting ceRNA that upregulates OCT1 by binding with miR-204-5p, which is downregulated in GC and inhibits tumor progression through various mechanisms. The 3-UTR region of OCT1 contains a potential binding site for miR-204-5p, and has been found to be a transcription factor contributing to carcinogenesis in GC and precancerous lesions. In addition, OCT1 was confirmed to enhance the expression of DLX6-AS1 by targeting the promoter region, revealing a positive feedback loop (OCT1/DLX6-AS1/miR-204-5p/OCT1) which promotes the progress of GC and EMT [130]. Together, DLX6-AS1 is upregulated in GC tissues and cell lines and is associated with distant metastasis, T3/T4 invasion, and poor clinical prognosis.
LncRNAs and DNA damage
An underlying feature of cancers is genomic instability, which is associated with a greater tendency to trigger DNA damage responses (DDR) [131]. The DDR which involves a complex regulatory network of lncRNAs may also participate in cancer progression (Figure 5A). For example, the NF-κB-HOTAIR axis links DDR to ovarian carcinogenesis [132], and LINC00261, an epigenetically regulated tumor suppressor, is critical for the activation of DDR in lung cancer [133].
In GC, PANDAR (promoter of CDKN1A antisense DNA damage-activated RNA), whose expression is induced by p53, is also a direct transcriptional target of the p53 protein. PANDAR is induced in response to DNA damage via inhibition of the function of the transcription factor nuclear transcription factor Y subunit alpha (NFYA). Upregulated HOTAIR inhibits the expression of p53 by enhancing the p53 promoter H3K27me3, while overexpression of PANDAR increases the stability of p53 in response to DNA damage [134].
Noncoding RNA activated by DNA damage (NORAD), a highly conserved lncRNA essential for mitotic cell division, plays a key role in DNA replication and DDR. NORAD responds to DNA damage by regulating the activity of the RNA-binding proteins PUM2 and PUM1 in order to maintain genomic integrity [135]. NORAD expression is upregulated in GC, which can also significantly increase FOXO6 abundance and promote tumor progression. Mechanistically, NORAD acts as a ceRNA sponge for miR-608 to upregulate the expression of FOXO6 that in turn enhances the tumorigenicity of GC cells by upregulating the oncogene c-Myc [136].
LncRNAs and angiogenesis
Angiogenesis is an essential feature of cancer, providing nutrients and oxygen for tumor growth. VEGFA, a major regulator of angiogenesis, can induce the proliferation and migration of endothelial cells to form new blood vessels [137] (Figure 5B).
(A) DNA damage involves a complex regulatory network and some lncRNAs can also participate in the regulation of DDR to affect cancer progression in gastric cancer. (B) Some lncRNAs have been shown to play a role in the development of gastric cancer by regulating angiogenesis.
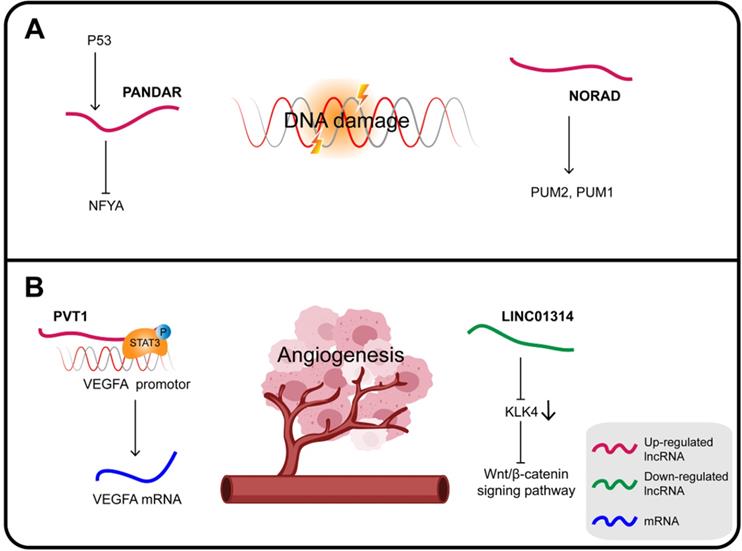
PVT1 is considered an oncogene interacting with different proteins and playing a variety of roles in carcinogenesis. In GC, PVT1 promotes cell proliferation by binding to EZH2 and also promotes migration and invasion by interacting with FOXM1 [138]. In addition, PVT1 can bind to nuclear p-STAT3 to form a complex that protects p-STAT3 from degradation in the polyubiquitin-proteasome, and then activates the STAT3/VEGFA axis to induce angiogenesis. Activated STAT3 in turn triggers PVT1 transcription, maintaining the oncogenicity of the PVT1/STAT3/VEGFA axis [139]. Therefore, the upregulation of lncRNA PVT1 is closely related to the high density of microvessels in GC and its poor prognosis; it can promote the proliferation of GC cells in vitro and tumor growth and angiogenesis in vivo.
The KLK4 gene is a direct target of LINC01314 that is poorly expressed in GC. KLK4 is a subunit of a serine endopeptidase found in a variety of cells and tissues, which is essential for diversified pathophysiological development, including angiogenesis in cancers. Over-expression of LINC01314 or KLK4 silencing suppresses angiogenesis in GC cells through negative regulation of the Wnt/β-catenin signaling pathway. At the same time, the tumor weight, the microvessel density of the transplanted tumor and, most importantly, the expression of both VEGF-C and VEGFR-3 were all decreased. In addition, LINC01314 over-expression can decrease N-cadherin and increase E-cadherin expression, therefore presumably it can also inhibit GC migration [140,141].
LncRNAs and the immune response
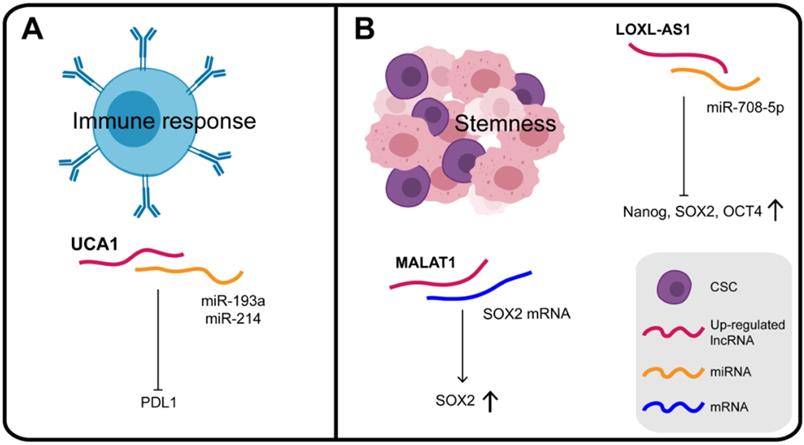
The differentiation and activation of innate and adaptive immune cells is highly dependent on a coordinated set of transcriptional and post-transcriptional events. LncRNAs are now emerging as important regulators of the differentiation and activation of immune cells. They do not directly encode innate or adaptive immune proteins but regulate the differentiation and function of immune cells, such as dendritic cell activity, T cell ratios and metabolism [142]. A dynamic equilibrium exists between the immune system and the malignant tumor, which can evade immune clearance and proliferate pathologically. Studies are increasingly revealing that lncRNAs have regulatory roles in the immune system, providing a reliable research basis for lncRNAs to participate in the immunotherapy of GC (Figure 6A). For instance, in addition to the effects of UCA1 on chromatin in GC, it can also act as a "miRNA sponge" for miR-193a and miR-214, reducing their targeting of the 3′ UTR of PDL1 and thus facilitating PDL1 expression [80]. PDL1 is a transmembrane protein, overexpressed in cancer cells, which suppresses the host's immune system by binding to PD1 on activated T cells, B cells, and bone marrow cells [143]. Therefore, UCA1, which protects PDL1 from the inhibitory effect of miRNAs and promotes immune escape of GC cells, may be a potential target, providing a new direction for immunotherapy of GC.
(A) Some lncRNAs play the regulatory roles in the immune system and participate in the balance between the immune system and gastric cancer cells. (B) Some lncRNAs play the key roles in maintaining the gastric cancer stem cell phenotype by influencing their expression.
In general, based on the theory of their binding with miRNAs and proteins, many types of lncRNAs found in exosomes could participate in the process of tumorigenesis as tumor antigens to activate anti-tumor immune response, and may thus be considered as targets for immunotherapy. However, the regulatory mechanism of existing RBPs such as lncRNA-miRNA-mRNA on immune regulation of GC is still unclear and requires further exploration.
LncRNAs and the stemness of gastric cancer
CSCs (cancer stem cells), recognized as tumor-initiating cells, are tumor cells with the capacity for self-renewal, metastasis, tumorigenicity, which exhibit chemotherapy resistance [144] (Figure 6B). Numerous studies have confirmed the genetic homology of lncRNA LOXL1 antisense RNA 1 (LOXL1-AS1) in various human cancers. Thus, it was confirmed that LOXL1-AS1 knockdown decreased the expression of stem cell factors such as Nanog, SOX2, and OCT4. Independently, MKN45 cells with lower expression of LOXL1-AS1 were found to be more sensitive to cisplatin therapy, which further supported the notion that LOXL1-AS1 promotes the maintenance of CSC features in GC [145]. In GC cells over-expressing SPRY4-IT1, CSC markers, including CD73, CD146, and ALDH1, were all significantly elevated. In contrast, inhibition of SPRY4-IT1 reduced the self-renewal ability of BGC823 cells and significantly reduced the expression of CSC markers [146]. These results consistently show that SPRY4-IT1 plays a role in maintaining the GC stem cell phenotype.
Potential clinical applications of lncRNAs in gastric cancer
LncRNAs as potential novel biomarkers
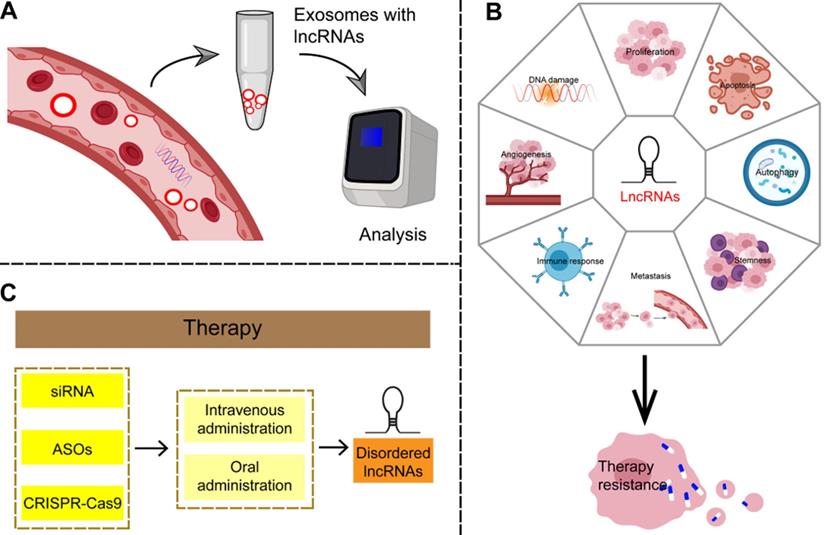
Because the majority of GC patients is diagnosed in the unsalvageable late stage, there is an urgent need to identify effective biomarkers for early disease. Some established circulating tumor-specific biomarkers, such as carcinoembryonic antigen (CEA), carbohydrate antigen (CA)242, CA724, and serum pepsinogen (SPG), are known to be of limited use in the diagnosis of GC [165]. LncRNAs can be detected in a highly stable form in plasma and are therefore considered to be non-invasive and promising biomarkers for cancer diagnosis (Figure 7A and Table 3). Studies have shown that cancer cells secrete exosomes, and lncRNAs are enriched and stabilized within these [166]. Exosomes, small vesicles with a diameter of about 30-150 nm that act as mediators of cell-to-cell communication, play an important role in the process of tumor proliferation and metastasis by delivering carcinogens [167]. As carriers of information between cells, exosomes represent a hitherto unknown biological communication system. However, the biological function of secretory lncRNAs in GC remains to be elucidated. LncRNAs such as ZFAS1, HOTTIP, and lncUEGC1 can be detected in plasma exosomes; significant differences between cancer patients and healthy controls were observed in their expression and area under the curve (AUC) values by receiver operating characteristic curve (ROC) analysis [168-170]. Therefore, a large number of lncRNAs has been found to be suitable as noninvasive biomarker candidates for GC.
Evidence indicates that plasma H19 levels can be used to distinguish patients with early GC from controls, with clinical results that are as satisfactory as traditional tumor biomarkers. After further analysis of 70 patients and 70 controls, it was concluded that the level of plasma H19 in patients with GC was significantly higher than in healthy controls (p < 0.0001); ROC analysis revealed that the AUC was 0.838 (p < 0.001; sensitivity, 82.9%; specificity, 72.9%). Furthermore, the expression of H19 distinguished early-stage GC from controls with an AUC of 0.877 (sensitivity, 85.5%; specificity, 80.1%). It has also been demonstrated that plasma H19 levels are stable during direct treatment with RNase and are therefore stable and detectable in plasma [171]. Therefore, plasma H19 may be a promising tumor marker for GC; however, many questions remain to be resolved before these findings can be translated into clinically useful, non-invasive screening strategies for GC patients.
In addition to the use of a single lncRNA in the diagnosis of GC, studies have shown that multiple plasma lncRNA combinations can provide more effective diagnostic power for GC diagnosis. Genome-wide plasma lncRNA profiles were screened to identify promising GC lncRNA biomarkers, with the result that plasma levels of FAM49B-AS, GUSBP11, and CTDHUT in GC patients were found to be significantly higher than in healthy controls after validation with data from 446 subjects (AUC of 0.818, 95% CI, 0.772-0.864). Moreover, the combination of these three lncRNAs with CA242 or CA724 was superior to the traditional CA242 or CA724 biomarkers alone for GC diagnosis; however, the AUC value of the combination (0.784) was lower than for the three lncRNAs. These three lncRNAs were stable and detectable in human plasma, and their amounts decreased significantly on the 10th day after surgical resection [172]. Therefore, the plasma lncRNAs FAM49B-AS, GUSBP11, and CTDHUT have great potential as non-invasive biomarkers for the diagnosis of GC.
Potential clinical applications of lncRNAs in gastric cancer. (A) LncRNAs are enriched and stable in exosomes, can be detected in plasma samples, and are considered as non-invasive and promising biomarkers for gastric cancer. (B) LncRNAs can be involved in the complex regulatory network of therapy resistance in a number of ways, including affecting the amplification or overexpression of oncogenes, suppression of tumor suppressor genes, immune escape, apoptosis, autophagy, EMT, angiogenesis, tumor stemness and so on. (C) Given the irreplaceable role of lncRNAs in the development of gastric cancer, targeting lncRNAs may produce reliable therapeutic results.
LncRNAs as diagnostic biomarkers in gastric cancer
| Clinical application | LncRNA | Expression | Diagnosis accuracy | Reference |
|---|---|---|---|---|
| Diagnostic biomarker | H19 | up | AUC was 0.838; p<0.001; sensitivity, 82.9%; specificity, 72.9% | [171] |
| FAM49B-AS, GUSBP11 and CTDHUT | up | The combined area under curve of these lncRNAs was 0.818 | [172] | |
| lnc-GNAQ-6:1 | down | AUC was 0.732 | [175] | |
| UCA1 | up | AUC was 0.759; The sensitivity and specificity were 93.20% and 78.63% | [176] | |
| B3GALT5-AS1 | up | AUC was 0.816 | [177] | |
| HOTAIR | up | AUC was 0.8416; The sensitivity and specificity were 66.67 and 87.04% | [178] | |
| lncUEGC1 | up | AUC was 0.8406 | [179] | |
| PANDAR, FOXD2-AS1, and SMARCC2 | up | The AUC of the combinative diagnostic value of these three lncRNAs was 0.839 | [180] | |
| TINCR, CCAT2, AOC4P, BANCR, and LINC00857 | up | The AUC of the combinative diagnostic value of these five lncRNAs was 0.91 | [181] |
LncRNAs as prognostic biomarkers in gastric cancer
| Clinical application | LncRNA | Expression | Clinical significance | Reference |
|---|---|---|---|---|
| Prognostic biomarker | CASC19 | up | Higher pathologic TNM stage, pathologic T stage, lymph node metastasis, and poor overall survival | [173] |
| SNHG6 | up | Invasion depth, lymph node metastasis, distant metastasis and TNM stage | [182] | |
| MIR100HG | up | TNM stage, tumor invasion, lymph node metastasis, and distant metastasis | [183] | |
| Sox2ot | up | TNM stage, tumor depth, lymph node metastasis, and distant metastasis | [184] | |
| MALAT1 | up | Distant metastasis | [185] | |
| NEAT1 | up | Clinical stage, histological type, lymph node metastasis, and distant metastasis | [174] | |
| SNHG8 | up | Predicts poor prognosis, shorter survival time | [186] | |
| CTD-2510F5.4 | Pathological grade, vascular or nerve invasion, AJCC TNM stage and OS, shorter MST | [187] |
LncRNAs and the resistance to therapy
| Clinical application | LncRNA | Expression | Pathway | Reference |
|---|---|---|---|---|
| chemoradiotherapy resistance | CRAL | down | miR-505/CYLD/ AKT | [188] |
| ARHGAP5-AS1 | up | ARHGAP5 | [189] | |
| PCAT1 | up | EZH2/PTEN | [192] | |
| MALAT1 | up | miR-23b-3p /ATG12 | [193] | |
| HULC | up | FoxM1 | [194] | |
| D63785 | up | miR-422a/MEF2D | [195] |
Evaluation of prognosis post-treatment is also essential for cancer patients. It can be used to assess the treatment status and provide evidence for adjusting treatment regimens. In recent years, together with tumor diagnostic biomarkers, the study of prognostic biomarkers related to lncRNAs has also made great progress (Table 4).
CASC19, also known as CARLo-6 or LINC01245, an intergenic lncRNA 324 bp in length, is encoded on chromosome 8q24.21. An innovative genomic analytical method has combined different lncRNA expression profiles with WGCNA, and resulting in the definition of CASC19 as a key lncRNA related to the development and progression of AGC. CASC19 was found to be upregulated in AGC tissue, and to correlate positively with abnormal clinicopathologic parameters. These included pathologic T stage, TNM stage, lymph node metastasis, and poorer prognosis in patients with AGC. Overall survival of patients with high CASC19 expression was significantly worse than patients with low CASC19 expression. In this cohort of 375 patients, CASC19 was therefore suggested to be an independent prognostic factor for AGC overall survival [173]. Thus, CASC19 is involved in the development of AGC through the regulation of MAPK, calcium, Wnt, and insulin signaling pathways and may be a new prognostic marker and potential therapeutic target for the disease.
The degree of over-expression of lncRNA NEAT1 in 140 GC specimens was found to be positively correlated with clinical stage, histological type, and distant or lymph node metastasis. Patients with low levels of NEAT1 had higher survival rates than those with high levels. Both univariate and multivariate analysis showed that overexpression of NEAT1 was an independent factor of poor prognosis in patients with GC. Knockout of NEAT1 significantly inhibited migration and invasion of GC cells in vitro and regulated the expression of EMT-related proteins. NEAT1 has also been shown to play a role in tumor development in esophageal cancer and glioma cells [174]. Thus, NEAT1 participates in the occurrence and development of GC and can be employed as a biomarker for its prognosis.
LncRNAs and resistance to therapy
Chemoradiotherapy (CRT) is the primary clinical treatment for multiple cancers including GC. Nevertheless, the development of acquired resistance to CRT greatly reduces its efficacy. Thus, resistance is the key mechanism of cancer escape from therapeutic attack and also the main obstacle for tumor therapy. Therefore, urgent exploration of the mechanisms of CRT resistance is required in order to improve therapeutic effects and reduce adverse reactions. Recent evidence suggests that lncRNAs may play vital roles in GC resistance to CRT (Figure 7B and Table 5).
LncRNA CRAL can regulate CYLD expression by binding to miR-505 and acting as an endogenous sponge, which can then reverse cisplatin resistance by increasing DNA damage and enhancing apoptosis. CYLD, a de-ubiquitinase, is considered a negative regulator of the PI3K/AKT/NF-κB signaling pathway. It was shown that in CRAL-deficient GC cells, CYLD expression was decreased and the activation of AKT increased. Blocking the PI3K/AKT signaling pathway reversed cisplatin resistance induced by CRAL deficiency and rendered GC cells sensitive to chemotherapeutic drugs. Thus, the suitability of AKT inhibitors for patients with GC could be determined based on the CRAL expression level. In summary, lncRNA CRAL may be a potential biomarker and therapeutic target for overcoming cisplatin resistance in GC by acting as a ceRNA via the miR-505/CYLD/AKT axis [188].
The autophagic adapter SQSTM1 (also known as the autophagic receptor) which recruits polyubiquitinated proteins to LC3-positive autophagosomes for degradation, can also recruit ARHGAP5-AS1 for autophagic degradation. However, in drug-resistant cancer cells with impaired autophagy, the amount of ARHGAP5-AS1 is upregulated and drug resistance increased. RBPs play an important role in regulating the stability of mRNA; it was also found that m6A modification can affect the stability of mRNA through RBPs such as HuR. ARHGAP5-AS1 modified ARHGAP5 mRNA by recruiting METTL3 for m6A modification and HuR binding, stimulated the transcription of ARHGAP5 in the nucleus, and also stabilized ARHGAP5 mRNA in the cytoplasm [189]. Therefore, targeting the ARHGAP5-AS1/ARHGAP5 axis may be a promising strategy for overcoming chemoresistance in GC.
Abnormal expression of PCAT1, an important regulator of multidrug resistance, is found in many cancers. For example, upregulation of PCAT1 in esophageal cancer tissue reduces the sensitivity of cancer cells to cisplatin in vitro [190]. Functional studies on Caco-2 and HT-29 cells showed that silencing PCAT1 increased the sensitivity of the cells to 5-fluorouracil by upregulating the expression of c-Myc [191]. PCAT1 was also significantly upregulated in CDDP-resistant GC tissues and cells, and caused cisplatin resistance in GC cells by binding to EZH2 to increase H3K27me3, that silenced PTEN [150]. In conclusion, PCAT1 is a positive regulator of cisplatin resistance in cancer cells and targeting PCAT1 may be an effective drug resistance strategy.
To sum up, a variety of lncRNAs has been implicated as regulators of downstream gene expression and/or function via their action as ceRNAs or via directly acting on proteins, thereby regulating drug resistance. They can participate in the complex regulatory network of CRT by affecting the amplification or overexpression of oncogenes, suppression of tumor suppressor genes, immune escape, apoptosis, autophagy, EMT, programmed death, tumor stemness and potentially by other mechanisms as well. Targeting these maladjusted lncRNAs may be a promising strategy to reverse CRT-induced resistance. In addition, targeted treatment of GC with lncRNAs in combination with traditional chemotherapeutic agents may be a promising alternative to reversing drug resistance and may help improve the prognosis of patients with AGC. This approach may ultimately open up new potential avenues for overcoming therapy resistance.
Therapeutic strategies targeting specific lncRNAs
Regulatory lncRNA networks can mediate anti-proliferative effects and reduce invasion of GC, thus mediating therapeutic effects, and making it possible to consider exploiting lncRNAs as therapeutic targets in GC. One advantage of targeting lncRNAs is that functional ablation of a lncRNA with regulatory function may have pleiotropic effects, resulting in simultaneous damage to multiple tumor-related pathways. Therefore, targeting lncRNAs may yield reliable therapeutic results that reduce drug resistance in tumors. Small interference RNAs (siRNAs) targeting abnormal GC-specific lncRNAs may be an effective strategy in this respect [196]. Antisense oligonucleotides (ASOs) can also correct aberrant lncRNA networks by inducing RNase H-dependent degradation or by spatially blocking lncRNA activity. In addition, specific chemical modification of lncRNA-targeted ASOs enhances their effectiveness and stability and allows free absorption in vivo without the need for transfer methods [197]. In addition, the regulation of lncRNA expression based on CRISPR-Cas9 has been demonstrated as a therapeutic tool for clinical cancer therapy [198]. Although there is no clear evidence that this approach can be useful in GC, it is worth further exploring this avenue based on these theories. Finally, because lncRNAs are emerging new targets but with an as-yet incomplete understanding of their mechanisms of action, it is important to use controls in gene silencing studies to ensure that the observed phenotypes are caused by the interaction of the targets. In addition, some challenges, such as missed target effects, modes of targeted delivery, and activation of the immune response, need to be resolved before these lncRNAs can be considered for clinical application [199]. Although most studies to date on the use of lncRNA-targeted drugs for GC are still in the preclinical phase, they nevertheless highlight the potential of this strategy (Figure 7C).
Conclusions and perspectives
In view of the high mortality and typically delayed diagnosis of gastric cancer (GC), a comprehensive understanding of the molecular pathogenesis and potential biomarkers of this disease is urgently needed to supplement existing treatment options. With the development of high throughput sequencing techniques such as microarrays or RNA-seq, important roles of various lncRNAs have been revealed. These have attracted the attention of investigators for their relevance also in the pathogenesis of GC. Large numbers of lncRNAs have been identified as being pertinent for pathology caused by imbalances in gene regulation and the resulting abnormalities in biological processes in GC, mainly due to the circumstantial evidence of their often-abnormal expression in GC. In recent decades, studies have shown that lncRNAs regulate gene expression at transcriptional, post-transcriptional, and epigenetic levels, and participate in the occurrence, development, and progression of GC.
Screening of lncRNAs revealed that most of them have a promoting effect on GC, for example, Linc00152 and PANDA promote proliferation and growth and MAGI2-AS3 promotes tumor invasion and metastasis; however, only a few lncRNAs inhibit GC in vitro and in vivo, including PWRN1 and TUBA4B that inhibit the proliferation of GC cells. Irrespective of their promoting or inhibiting action on tumor growth, the characteristics of lncRNAs involved in the regulation of GC makes them theoretically potential biomarkers for GC diagnosis, prognosis, prediction and recurrence, some of which have also progressed to clinical trials. For example, PC antigen 3 (PCA3) detection test has been developed to assess prostate cancer risk using urine samples; this indicates progress and provides encouragement for further investigations [200]. Compared with traditional biomarkers, lncRNAs have the following advantages: (1) They have higher specificity and sensitivity. (2) LncRNAs testing is a simple, non-invasive, and easy-to-accept method. (3) LncRNAs can be used as biomarkers independently, in combination with other lncRNAs, or as a consummate complement to classic cancer biomarkers. Moreover, because of the tissue- and disease-specific expression patterns of lncRNAs and their ability to control tumor micro-regulatory networks, they are of great significance not only as biomarkers but also as therapeutic targets. Thus, this is a promising field for molecular targeted therapy of GC. Strategies for targeting and regulating lncRNAs based on siRNAs, ASOs, CRISPR-Cas9, etc. are currently being explored, which will contribute to the development of more effective lncRNA-based GC treatment methods.
Moreover, some new types of lncRNAs have good prospects for applications and are worth exploring. Hitherto, some PROMPTs have been found to regulate cancer biology by interacting with adjacent proteins, and may also represent new biomarkers or therapeutic targets. Although there are few reports of PROMPTs specifically in the context of GC, they may provide new directions for future research on GC-related lncRNAs. Similar to PROMPTs, there is little data on eRNAs in GC, which may also gradually become a research hotspot.
Currently, there are numerous challenges for translating knowledge on lncRNAs to the clinic. First, their application must be based on knowledge of their mechanisms of action in vivo and in vitro, but only some of these have been well probed and elucidated. For example, how some lncRNAs affect DNA hydroxymethylation, chromatin remodeling, or alternative splicing of mRNAs in GC is still being explored. Owing to the complex nature of the tumor micro-environment and the involvement of multiple lncRNAs, we need to identify more lncRNAs affecting GC and construct a network of regulatory interactions between lncRNAs. Moreover, because lncRNAs function primarily through interactions with other biomolecules, elucidation of the secondary structure of their interaction domains is particularly necessary. The analysis of these lncRNA binding motifs may result in the identification of new RNA-based targets for disease prevention and treatment. At present, the study of lncRNAs in GC is mainly focused on their expression profiles, and the role of lncRNA mutants in gastric carcinogenesis needs further study. In addition, the low accessibility of lncRNAs is also a problem. Such problems can be tackled with the help of animal models and a new generation of technologies. The integration of functional lncRNAs and GC biology will further our understanding of the mechanisms involved in GC carcinogenesis. Although the application of specific lncRNAs to diagnosis, prognosis, or treatment of GC is still a distant goal, there is no doubt about their significance. A comprehensive exploration of this still-mysterious realm will be expected to provide more new strategies for the prevention and treatment of GC in the near future.
Abbreviations
AGC: advanced GC; ASO: antisense oligonucleotide; ATG4B: autophagy-associated 4B; AUC: area under the curve; BTG2: B-cell translocation gene 2; CA: carbohydrate antigen; CCNE2: cyclin E2; CDK: cyclin-dependent kinase; CDK16: cyclin‑dependent kinase 16; CEA: carcinoembryonic antigen; CRT: chemoradiotherapy; CSC: cancer stem cell; CXCL12: chemokine (CXCmotif) ligand; DDR: DNA damage response; DSP: desmoplakin; ECM: extracellular matrix; EGFR: epidermal growth factor receptor; EMT: epithelial-to-mesenchymal transition; eRNA: enhancer-associated RNA; FUS: fused in sarcoma; GC: gastric cancer; GEF: guanine exchange factor; GMAN: gastric cancer metastasis associated long noncoding RNA; GMAN-AS: antisense GMAN RNA; HDAC: histone deacetylase; hTERT: telomerase reverse transcriptase; H3K27me3: trimethylation of lysine residue 27 of histone 3; H3K4: histone H3 lysine K4; JAM-A: junctional adhesion molecule A; JMJD3: jumonji-domain containing 3; KLF: kruppel-like factor; KRT19P3: Keratin 19 Pseudogene 3; LIM-HD: LIM-homedomain; LMX1A: Lim homeobox transcription factor 1; lncRNA: long non-coding RNA; LOXL1-AS1: LOXL1 antisense RNA 1; MACC1: metastasis-associated in colon cancer-1; MACC1-AS1: MACC1 antisense RNA 1; MED18: mediator subunit 18; MMP: member matrix metalloproteinase; MNX1-AS1: MNX1 antisense RNA 1; MRE: miRNA response element; MSS: microsatellite stablity; mTOR: rapamycin; m6A: N6-methyladenosine; ncRNA: non-coding RNA; NET1: neuroepithelial cell transforming gene 1; NFYA: nuclear transcription factor Y subunit alpha; NF-κB: nuclear factor kappa-B inhibitor; NORAD: The noncoding RNA activated by DNA damage; PANDAR: promoter of CDKN1A antisense DNA damage activated RNA; PCA3: PC antigen 3; PCD: programmed cell death; p-PI3K: phosphoinositide 3-kinase; PRC2: polycomb repressive complex; PROMPT: promoter upstream transcript; RBP: RNA-binding protein; RHOA: Ras homolog gene family member A; RIP: RNA immunoprecipitation; ROC: receiver operating characteristic curve; RTK: receptor tyrosine kinases; siRNA: small interference RNA; SMD: STAU1 mediated mRNA degradation; SNCG: γ-synuclein; SNHG1: small nucleolar RNA host gene 1; SNHG3: small nucleolar RNA host gene 3; SPG: serum pepsinogen; SPRY1: sprouty RTK signaling antagonist 1; SRSF1: serine- and arginine-rich splicing factor 1; TEAD: TEA domain; TNF: tumor necrosis factor; TRAIL: tumor necrosis factor (TNF) related apoptosis inducing ligand; TSS: transcription start site; UCA1: urothelial cancer-associated 1; XIAP: X-linked apoptosis inhibitor; YBX1: Y-box binding protein 1; YTHDF2: YTH domain family 2.
Acknowledgements
We are grateful to Dr. Junjie Liu (UC Berkeley) and Prof. Yong Peng (Sichuan University) for their critical reviews on this manuscript. This work was supported in part by grants from National Science and Technology Major Project of the Ministry of Science and Technology of China (No. 2018ZX09735005), National Natural Science Foundation of China (Grant No. 81673455, Grant No. 81803365 and Grant No. 81873939), and Post-Doctorate Research Project (Grant No.2018M643510), Post-Doctorate Research Project of West China Hospital, Sichuan University (Grant No. 2018HXBH065).
Author Contributions
Y.C., Y.A., and B.L. conceived, extensively revised, formatted, and submitted the manuscript. H.T., S.Z., and J.Z. wrote the manuscript. L.Z., Y.C., and H.Y searched and archived the literature.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Correa P. Gastric cancer: overview. Gastroenterol Clin North Am. 2013;42:211-7
2. Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654-64
3. LAUREN P. THE TWO HISTOLOGICAL MAIN TYPES OF GASTRIC CARCINOMA: DIFFUSE AND SO-CALLED INTESTINAL-TYPE CARCINOMA. AN ATTEMPT AT A HISTO-CLINICAL CLASSIFICATION. Acta Pathol Microbiol Scand. 1965;64:31-49
4. Song Z, Wu Y, Yang J, Yang D, Fang X. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017;39:1393375038
5. Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126:2775-82
6. St LG, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239-51
7. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145-66
8. Preker P, Almvig K, Christensen MS, Valen E, Mapendano CK, Sandelin A. et al. PROMoter uPstream Transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Res. 2011;39:7179-93
9. Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD. et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310-25
10. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17:47-62
11. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915-27
12. Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298-307
13. Li H, Yu B, Li J, Su L, Yan M, Zhang J. et al. Characterization of differentially expressed genes involved in pathways associated with gastric cancer. Plos One. 2015;10:e125013
14. Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7:8601-12
15. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT. et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925-38
16. Bayoumi AS, Sayed A, Broskova Z, Teoh JP, Wilson J, Su H. et al. Crosstalk between Long Noncoding RNAs and MicroRNAs in Health and Disease. Int J Mol Sci. 2016;17:356
17. Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452-63
18. Hanly DJ, Esteller M, Berdasco M. Interplay between long non-coding RNAs and epigenetic machinery: emerging targets in cancer? Philos Trans R Soc Lond B Biol Sci. 2018 373
19. Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P. et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039-43
20. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea MD. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667-72
21. Liu YW, Xia R, Lu K, Xie M, Yang F, Sun M. et al. LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol Cancer. 2017;16:39
22. Zhang XD, Huang GW, Xie YH, He JZ, Guo JC, Xu XE. et al. The interaction of lncRNA EZR-AS1 with SMYD3 maintains overexpression of EZR in ESCC cells. Nucleic Acids Res. 2018;46:1793-809
23. Gu P, Chen X, Xie R, Han J, Xie W, Wang B. et al. lncRNA HOXD-AS1 Regulates Proliferation and Chemo-Resistance of Castration-Resistant Prostate Cancer via Recruiting WDR5. Mol Ther. 2017;25:1959-73
24. He X, Wang J, Chen J, Han L, Lu X, Miao D. et al. lncRNA UCA1 Predicts a Poor Prognosis and Regulates Cell Proliferation and Migration by Repressing p21 and SPRY1 Expression in GC. Mol Ther Nucleic Acids. 2019;18:605-16
25. Fritzsche S, Kenzelmann M, Hoffmann MJ, Muller M, Engers R, Grone HJ. et al. Concomitant down-regulation of SPRY1 and SPRY2 in prostate carcinoma. Endocr Relat Cancer. 2006;13:839-49
26. Lo TL, Yusoff P, Fong CW, Guo K, McCaw BJ, Phillips WA. et al. The ras/mitogen-activated protein kinase pathway inhibitor and likely tumor suppressor proteins, sprouty 1 and sprouty 2 are deregulated in breast cancer. Cancer Res. 2004;64:6127-36
27. He L, Yuan L, Sun Y, Wang P, Zhang H, Feng X. et al. Glucocorticoid Receptor Signaling Activates TEAD4 to Promote Breast Cancer Progression. Cancer Res. 2019;79:4399-411
28. Giraud J, Molina-Castro S, Seeneevassen L, Sifre E, Izotte J, Tiffon C. et al. Verteporfin targeting YAP1/TAZ-TEAD transcriptional activity inhibits the tumorigenic properties of gastric cancer stem cells. Int J Cancer. 2019
29. Shuai Y, Ma Z, Liu W, Yu T, Yan C, Jiang H. et al. TEAD4 modulated LncRNA MNX1-AS1 contributes to gastric cancer progression partly through suppressing BTG2 and activating BCL2. Mol Cancer. 2020;19:6
30. Xuan Y, Wang Y. Long non-coding RNA SNHG3 promotes progression of gastric cancer by regulating neighboring MED18 gene methylation. Cell Death Dis. 2019;10:694
31. Sun Y, Wei G, Luo H, Wu W, Skogerbo G, Luo J. et al. The long noncoding RNA SNHG1 promotes tumor growth through regulating transcription of both local and distal genes. Oncogene. 2017;36:6774-83
32. Zhang G, Xu Y, Zou C, Tang Y, Lu J, Gong Z. et al. Long noncoding RNA ARHGAP27P1 inhibits gastric cancer cell proliferation and cell cycle progression through epigenetically regulating p15 and p16. Aging (Albany NY). 2019;11:9090-110
33. Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC. et al. LncRNA GClnc1 Promotes Gastric Carcinogenesis and May Act as a Modular Scaffold of WDR5 and KAT2A Complexes to Specify the Histone Modification Pattern. Cancer Discov. 2016;6:784-801
34. Ma M, Zhang Y, Weng M, Hu Y, Xuan Y, Hu Y. et al. lncRNA GCAWKR Promotes Gastric Cancer Development by Scaffolding the Chromatin Modification Factors WDR5 and KAT2A. Mol Ther. 2018;26:2658-68
35. Huang M, Hou J, Wang Y, Xie M, Wei C, Nie F. et al. Long Noncoding RNA LINC00673 Is Activated by SP1 and Exerts Oncogenic Properties by Interacting with LSD1 and EZH2 in Gastric Cancer. Mol Ther. 2017;25:1014-26
36. Hu Y, Ma Z, He Y, Liu W, Su Y, Tang Z. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem Biophys Res Commun. 2017;491:926-31
37. Wang Y, Liu X, Zhang H, Sun L, Zhou Y, Jin H. et al. Hypoxia-inducible lncRNA-AK058003 promotes gastric cancer metastasis by targeting gamma-synuclein. Neoplasia. 2014;16:1094-106
38. Kameswaran V, Bramswig NC, McKenna LB, Penn M, Schug J, Hand NJ. et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014;19:135-45
39. Ding L, Tian Y, Wang L, Bi M, Teng D, Hong S. Hypermethylated long noncoding RNA MEG3 promotes the progression of gastric cancer. Aging (Albany NY). 2019;11:8139-55
40. Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S. et al. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750-6
41. Zhao Y, Liu Y, Lin L, Huang Q, He W, Zhang S. et al. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol Cancer. 2018;17:69
42. Stein U, Walther W, Arlt F, Schwabe H, Smith J, Fichtner I. et al. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat Med. 2009;15:59-67
43. Ieguchi K, Maru Y. Roles of EphA1/A2 and ephrin-A1 in cancer. Cancer Sci. 2019;110:841-8
44. Zhuo W, Liu Y, Li S, Guo D, Sun Q, Jin J. et al. Long Noncoding RNA GMAN, Up-regulated in Gastric Cancer Tissues, Is Associated With Metastasis in Patients and Promotes Translation of Ephrin A1 by Competitively Binding GMAN-AS. Gastroenterology. 2019;156:676-91
45. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353-8
46. Li C, Miao R, Zhang J, Qu K, Liu C. Long non-coding RNA KCNQ1OT1 mediates the growth of hepatocellular carcinoma by functioning as a competing endogenous RNA of miR-504. Int J Oncol. 2018
47. Gong W, Zheng J, Liu X, Liu Y, Guo J, Gao Y. et al. Knockdown of Long Non-Coding RNA KCNQ1OT1 Restrained Glioma Cells' Malignancy by Activating miR-370/CCNE2 Axis. Front Cell Neurosci. 2017;11:84
48. Doucet-Beaupre H, Ang SL, Levesque M. Cell fate determination, neuronal maintenance and disease state: The emerging role of transcription factors Lmx1a and Lmx1b. Febs Lett. 2015;589:3727-38
49. Dong W, Feng L, Xie Y, Zhang H, Wu Y. Hypermethylation-mediated reduction of LMX1A expression in gastric cancer. Cancer Sci. 2011;102:361-6
50. Feng L, Li H, Li F, Bei S, Zhang X. LncRNA KCNQ1OT1 regulates microRNA-9-LMX1A expression and inhibits gastric cancer cell progression. Aging (Albany NY). 2020;12:707-17
51. Zhang X, Li J, Li F, Zhao Z, Feng L. LINC00682 inhibits gastric cancer cell progression via targeting microRNA-9-LMX1A signaling axis. Aging (Albany NY). 2019;11:11358-68
52. Dong XZ, Zhao ZR, Hu Y, Lu YP, Liu P, Zhang L. LncRNA COL1A1-014 is involved in the progression of gastric cancer via regulating CXCL12-CXCR4 axis. Gastric Cancer. 2019
53. Han Y, Wu N, Jiang M, Chu Y, Wang Z, Liu H. et al. Long non-coding RNA MYOSLID functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-29c-3p in gastric cancer. Cell Prolif. 2019;52:e12678
54. Carrington EM, Tarlinton DM, Gray DH, Huntington ND, Zhan Y, Lew AM. The life and death of immune cell types: the role of BCL-2 anti-apoptotic molecules. Immunol Cell Biol. 2017;95:870-7
55. Chen G, Magis AT, Xu K, Park D, Yu DS, Owonikoko TK. et al. Targeting Mcl-1 enhances DNA replication stress sensitivity to cancer therapy. J Clin Invest. 2018;128:500-16
56. Murray D, Horgan G, Macmathuna P, Doran P. NET1-mediated RhoA activation facilitates lysophosphatidic acid-induced cell migration and invasion in gastric cancer. Br J Cancer. 2008;99:1322-9
57. Zong W, Feng W, Jiang Y, Cao Y, Ke Y, Shi X, et al. LncRNA CTC-497E21.4 promotes the progression of gastric cancer via modulating miR-22/NET1 axis through RhoA signaling pathway. Gastric Cancer. 2019
58. Zuo QF, Cao LY, Yu T, Gong L, Wang LN, Zhao YL. et al. MicroRNA-22 inhibits tumor growth and metastasis in gastric cancer by directly targeting MMP14 and Snail. Cell Death Dis. 2015;6:e2000
59. Chun MG, Hanahan D. Genetic deletion of the desmosomal component desmoplakin promotes tumor microinvasion in a mouse model of pancreatic neuroendocrine carcinogenesis. Plos Genet. 2010;6:e1001120
60. Wang H, Wu M, Lu Y, He K, Cai X, Yu X. et al. LncRNA MIR4435-2HG targets desmoplakin and promotes growth and metastasis of gastric cancer by activating Wnt/beta-catenin signaling. Aging (Albany NY). 2019;11:6657-73
61. Schweitzer K, Bozko PM, Dubiel W, Naumann M. CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. Embo J. 2007;26:1532-41
62. Zheng J, Zhang H, Ma R, Liu H, Gao P. Long non-coding RNA KRT19P3 suppresses proliferation and metastasis through COPS7A-mediated NF-kappaB pathway in gastric cancer. Oncogene. 2019;38:7073-88
63. Ma T, Ma H, Zou Z, He X, Liu Y, Shuai Y. et al. The Long Intergenic Noncoding RNA 00707 Promotes Lung Adenocarcinoma Cell Proliferation and Migration by Regulating Cdc42. Cell Physiol Biochem. 2018;45:1566-80
64. Grammatikakis I, Abdelmohsen K, Gorospe M. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA. 2017 8
65. Xie M, Ma T, Xue J, Ma H, Sun M, Zhang Z. et al. The long intergenic non-protein coding RNA 707 promotes proliferation and metastasis of gastric cancer by interacting with mRNA stabilizing protein HuR. Cancer Lett. 2019;443:67-79
66. Bustelo XR. Vav proteins, adaptors and cell signaling. Oncogene. 2001;20:6372-81
67. Severson EA, Parkos CA. Structural determinants of Junctional Adhesion Molecule A (JAM-A) function and mechanisms of intracellular signaling. Curr Opin Cell Biol. 2009;21:701-7
68. Kosnopfel C, Sinnberg T, Schittek B. Y-box binding protein 1-a prognostic marker and target in tumour therapy. Eur J Cell Biol. 2014;93:61-70
69. Zhang E, He X, Zhang C, Su J, Lu X, Si X. et al. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19:154
70. Peixoto P, Castronovo V, Matheus N, Polese C, Peulen O, Gonzalez A. et al. HDAC5 is required for maintenance of pericentric heterochromatin, and controls cell-cycle progression and survival of human cancer cells. Cell Death Differ. 2012;19:1239-52
71. Song Y, Wang R, Li LW, Liu X, Wang YF, Wang QX. et al. Long non-coding RNA HOTAIR mediates the switching of histone H3 lysine 27 acetylation to methylation to promote epithelial-to-mesenchymal transition in gastric cancer. Int J Oncol. 2019;54:77-86
72. Fang J, Sun CC, Gong C. Long noncoding RNA XIST acts as an oncogene in non-small cell lung cancer by epigenetically repressing KLF2 expression. Biochem Biophys Res Commun. 2016;478:811-7
73. Zhang E, He X, Yin D, Han L, Qiu M, Xu T. et al. Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109
74. Qi F, Liu X, Wu H, Yu X, Wei C, Huang X. et al. Long noncoding AGAP2-AS1 is activated by SP1 and promotes cell proliferation and invasion in gastric cancer. J Hematol Oncol. 2017;10:48
75. Wang SS, Wuputra K, Liu CJ, Lin YC, Chen YT, Chai CY. et al. Oncogenic function of the homeobox A13-long noncoding RNA HOTTIP-insulin growth factor-binding protein 3 axis in human gastric cancer. Oncotarget. 2016;7:36049-64
76. Ba MC, Long H, Cui SZ, Gong YF, Yan ZF, Wu YB. et al. Long noncoding RNA LINC00673 epigenetically suppresses KLF4 by interacting with EZH2 and DNMT1 in gastric cancer. Oncotarget. 2017;8:95542-53
77. Zhou J, Shi J, Fu X, Mao B, Wang W, Li W. et al. Linc00441 interacts with DNMT1 to regulate RB1 gene methylation and expression in gastric cancer. Oncotarget. 2018;9:37471-9
78. Huang B, Song JH, Cheng Y, Abraham JM, Ibrahim S, Sun Z. et al. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene. 2016;35:4927-36
79. Xu TP, Ma P, Wang WY, Shuai Y, Wang YF, Yu T. et al. KLF5 and MYC modulated LINC00346 contributes to gastric cancer progression through acting as a competing endogeous RNA and indicates poor outcome. Cell Death Differ. 2019;26:2179-93
80. Wang CJ, Zhu CC, Xu J, Wang M, Zhao WY, Liu Q. et al. Correction to: The lncRNA UCA1 promotes proliferation, migration, immune escape and inhibits apoptosis in gastric cancer by sponging anti-tumor miRNAs. Mol Cancer. 2019;18:129
81. Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z. et al. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/beta-catenin pathway. Mol Cancer. 2018;17:126
82. Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G. et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17:87
83. Chen M, Fan L, Zhang SM, Li Y, Chen P, Peng X. et al. LINC01939 inhibits the metastasis of gastric cancer by acting as a molecular sponge of miR-17-5p to regulate EGR2 expression. Cell Death Dis. 2019;10:70
84. Gan L, Lv L, Liao S. Long noncoding RNA H19 regulates cell growth and metastasis via the miR223p/Snail1 axis in gastric cancer. Int J Oncol. 2019;54:2157-68
85. Gong P, Qiao F, Wu H, Cui H, Li Y, Zheng Y. et al. LncRNA UCA1 promotes tumor metastasis by inducing miR-203/ZEB2 axis in gastric cancer. Cell Death Dis. 2018;9:1158
86. Liu HT, Liu S, Liu L, Ma RR, Gao P. EGR1-Mediated Transcription of lncRNA-HNF1A-AS1 Promotes Cell-Cycle Progression in Gastric Cancer. Cancer Res. 2018;78:5877-90
87. Zhang X, Liang W, Liu J, Zang X, Gu J, Pan L. et al. Long non-coding RNA UFC1 promotes gastric cancer progression by regulating miR-498/Lin28b. J Exp Clin Cancer Res. 2018;37:134
88. Lin Z, Zhou Z, Guo H, He Y, Pang X, Zhang X. et al. Long noncoding RNA gastric cancer-related lncRNA1 mediates gastric malignancy through miRNA-885-3p and cyclin-dependent kinase 4. Cell Death Dis. 2018;9:607
89. Yuan H, Chen Z, Bai S, Wei H, Wang Y, Ji R. et al. Molecular mechanisms of lncRNA SMARCC2/miR-551b-3p/TMPRSS4 axis in gastric cancer. Cancer Lett. 2018;418:84-96
90. Lu MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY. et al. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524-34
91. Zhang R, Liu Y, Liu H, Chen W, Fan HN, Zhang J. et al. The long non-coding RNA SNHG12 promotes gastric cancer by activating the phosphatidylinositol 3-kinase/AKT pathway. Aging (Albany NY). 2019;11:10902-22
92. Guo W, Liang X, Liu L, Guo Y, Shen S, Liang J. et al. MiR-6872 host gene SEMA3B and its antisense lncRNA SEMA3B-AS1 function synergistically to suppress gastric cardia adenocarcinoma progression. Gastric Cancer. 2019;22:705-22
93. Li D, Chen Y, Mei H, Jiao W, Song H, Ye L. et al. Ets-1 promoter-associated noncoding RNA regulates the NONO/ERG/Ets-1 axis to drive gastric cancer progression. Oncogene. 2018;37:4871-86
94. Guo H, Zhao L, Shi B, Bao J, Zheng D, Zhou B. et al. GALNT5 uaRNA promotes gastric cancer progression through its interaction with HSP90. Oncogene. 2018;37:4505-17
95. Xue M, Chen LY, Wang WJ, Su TT, Shi LH, Wang L. et al. HOTAIR induces the ubiquitination of Runx3 by interacting with Mex3b and enhances the invasion of gastric cancer cells. Gastric Cancer. 2018;21:756-64
96. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-74
97. Cheng N, Chytil A, Shyr Y, Joly A, Moses HL. Transforming growth factor-beta signaling-deficient fibroblasts enhance hepatocyte growth factor signaling in mammary carcinoma cells to promote scattering and invasion. Mol Cancer Res. 2008;6:1521-33
98. Weng X, Zhang H, Ye J, Kan M, Liu F, Wang T. et al. Hypermethylated Epidermal growth factor receptor (EGFR) promoter is associated with gastric cancer. Sci Rep. 2015;5:10154
99. Zhou J, Zhi X, Wang L, Wang W, Li Z, Tang J. et al. Linc00152 promotes proliferation in gastric cancer through the EGFR-dependent pathway. J Exp Clin Cancer Res. 2015;34:135
100. Yang Z, Wang R, Zhang T, Dong X. Hypoxia/lncRNA-AK123072/EGFR pathway induced metastasis and invasion in gastric cancer. Int J Clin Exp Med. 2015;8:19954-68
101. Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671-82
102. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. Rna Biol. 2012;9:703-19
103. Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749-58
104. Kong F, Deng X, Kong X, Du Y, Li L, Zhu H. et al. ZFPM2-AS1, a novel lncRNA, attenuates the p53 pathway and promotes gastric carcinogenesis by stabilizing MIF. Oncogene. 2018;37:5982-96
105. Fukaya R, Ohta S, Yaguchi T, Matsuzaki Y, Sugihara E, Okano H. et al. MIF Maintains the Tumorigenic Capacity of Brain Tumor-Initiating Cells by Directly Inhibiting p53. Cancer Res. 2016;76:2813-23
106. Shimoni-Sebag A, Lebenthal-Loinger I, Zender L, Karni R. RRM1 domain of the splicing oncoprotein SRSF1 is required for MEK1-MAPK-ERK activation and cellular transformation. Carcinogenesis. 2013;34:2498-504
107. Wu ZH, Liu CC, Zhou YQ, Hu LN, Guo WJ. OnclncRNA-626 promotes malignancy of gastric cancer via inactivated the p53 pathway through interacting with SRSF1. Am J Cancer Res. 2019;9:2249-63
108. Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283-96
109. Guo J, Li Y, Duan H, Yuan L. LncRNA TUBA4B functions as a competitive endogenous RNA to inhibit gastric cancer progression by elevating PTEN via sponging miR-214 and miR-216a/b. Cancer Cell Int. 2019;19:156
110. Yan J, Huang X, Zhang X, Chen Z, Ye C, Xiang W. et al. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun. 2020;521:887-93
111. Wang X, Zhang J, Wang Y. Long noncoding RNA GAS5-AS1 suppresses growth and metastasis of cervical cancer by increasing GAS5 stability. Am J Transl Res. 2019;11:4909-21
112. Liu C, Zhang Y, She X, Fan L, Li P, Feng J. et al. A cytoplasmic long noncoding RNA LINC00470 as a new AKT activator to mediate glioblastoma cell autophagy. J Hematol Oncol. 2018;11:77
113. Liu C, Fu H, Liu X, Lei Q, Zhang Y, She X. et al. LINC00470 Coordinates the Epigenetic Regulation of ELFN2 to Distract GBM Cell Autophagy. Mol Ther. 2018;26:2267-81
114. Kent LN, Bae S, Tsai SY, Tang X, Srivastava A, Koivisto C. et al. Dosage-dependent copy number gains in E2f1 and E2f3 drive hepatocellular carcinoma. J Clin Invest. 2017;127:830-42
115. He Y, Luo Y, Liang B, Ye L, Lu G, He W. Potential applications of MEG3 in cancer diagnosis and prognosis. Oncotarget. 2017;8:73282-95
116. Jafri MA, Ansari SA, Alqahtani MH, Shay JW. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016;8:69
117. Horikawa I, Barrett JC. Transcriptional regulation of the telomerase hTERT gene as a target for cellular and viral oncogenic mechanisms. Carcinogenesis. 2003;24:1167-76
118. Cai J, Wang D, Bai ZG, Yin J, Zhang J, Zhang ZT. The long noncoding RNA XIAP-AS1 promotes XIAP transcription by XIAP-AS1 interacting with Sp1 in gastric cancer cells. Plos One. 2017;12:e182433
119. Wilson TR, McEwan M, McLaughlin K, Le Clorennec C, Allen WL, Fennell DA. et al. Combined inhibition of FLIP and XIAP induces Bax-independent apoptosis in type II colorectal cancer cells. Oncogene. 2009;28:63-72
120. Suliman A, Lam A, Datta R, Srivastava RK. Intracellular mechanisms of TRAIL: apoptosis through mitochondrial-dependent and -independent pathways. Oncogene. 2001;20:2122-33
121. Xu TP, Liu XX, Xia R, Yin L, Kong R, Chen WM. et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648-61
122. Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120:195-208
123. Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4:206
124. Wang LL, Zhang L, Cui XF. Downregulation of long noncoding RNA LINC01419 inhibits cell migration, invasion, and tumor growth and promotes autophagy via inactivation of the PI3K/Akt1/mTOR pathway in gastric cancer. Ther Adv Med Oncol. 2019;11:432477605
125. Zhang H, Liu L, Wang Y, Zhao G, Xie R, Liu C. et al. KLF8 involves in TGF-beta-induced EMT and promotes invasion and migration in gastric cancer cells. J Cancer Res Clin Oncol. 2013;139:1033-42
126. Han Y, Ye J, Wu D, Wu P, Chen Z, Chen J. et al. LEIGC long non-coding RNA acts as a tumor suppressor in gastric carcinoma by inhibiting the epithelial-to-mesenchymal transition. Bmc Cancer. 2014;14:932
127. Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS. et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449-56
128. Li D, Wang J, Zhang M, Hu X, She J, Qiu X. et al. LncRNA MAGI2-AS3 Is Regulated by BRD4 and Promotes Gastric Cancer Progression via Maintaining ZEB1 Overexpression by Sponging miR-141/200a. Mol Ther Nucleic Acids. 2019;19:109-23
129. Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP. et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126:3219-35
130. Liang Y, Zhang CD, Zhang C, Dai DQ. DLX6-AS1/miR-204-5p/OCT1 positive feedback loop promotes tumor progression and epithelial-mesenchymal transition in gastric cancer. Gastric Cancer. 2019
131. O'Connor MJ. Targeting the DNA Damage Response in Cancer. Mol Cell. 2015;60:547-60
132. Ozes AR, Miller DF, Ozes ON, Fang F, Liu Y, Matei D. et al. NF-kappaB-HOTAIR axis links DNA damage response, chemoresistance and cellular senescence in ovarian cancer. Oncogene. 2016;35:5350-61
133. Shahabi S, Kumaran V, Castillo J, Cong Z, Nandagopal G, Mullen DJ. et al. LINC00261 Is an Epigenetically Regulated Tumor Suppressor Essential for Activation of the DNA Damage Response. Cancer Res. 2019;79:3050-62
134. Liu J, Ben Q, Lu E, He X, Yang X, Ma J. et al. Long noncoding RNA PANDAR blocks CDKN1A gene transcription by competitive interaction with p53 protein in gastric cancer. Cell Death Dis. 2018;9:168
135. Ventura A. NORAD: Defender of the Genome. Trends Genet. 2016;32:390-2
136. Miao Z, Guo X, Tian L. The long noncoding RNA NORAD promotes the growth of gastric cancer cells by sponging miR-608. Gene. 2019;687:116-24
137. Yu B, Wang S. Angio-LncRs: LncRNAs that regulate angiogenesis and vascular disease. Theranostics. 2018;8:3654-75
138. Xu MD, Wang Y, Weng W, Wei P, Qi P, Zhang Q. et al. A Positive Feedback Loop of lncRNA-PVT1 and FOXM1 Facilitates Gastric Cancer Growth and Invasion. Clin Cancer Res. 2017;23:2071-80
139. Zhao J, Du P, Cui P, Qin Y, Hu C, Wu J. et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37:4094-109
140. Tang L, Wen JB, Wen P, Li X, Gong M, Li Q. Long non-coding RNA LINC01314 represses cell migration, invasion, and angiogenesis in gastric cancer via the Wnt/beta-catenin signaling pathway by down-regulating KLK4. Cancer Cell Int. 2019;19:94
141. Kryza T, Achard C, Parent C, Marchand-Adam S, Guillon-Munos A, Iochmann S. et al. Angiogenesis stimulated by human kallikrein-related peptidase 12 acting via a platelet-derived growth factor B-dependent paracrine pathway. Faseb J. 2014;28:740-51
142. Denaro N, Merlano MC, Lo NC. Long noncoding RNAs as regulators of cancer immunity. Mol Oncol. 2019;13:61-73
143. Bersanelli M, Buti S. From targeting the tumor to targeting the immune system: Transversal challenges in oncology with the inhibition of the PD-1/PD-L1 axis. World J Clin Oncol. 2017;8:37-53
144. Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744-9
145. Sun Q, Li J, Li F, Li H, Bei S, Zhang X. et al. LncRNA LOXL1-AS1 facilitates the tumorigenesis and stemness of gastric carcinoma via regulation of miR-708-5p/USF1 pathway. Cell Prolif. 2019;52:e12687
146. Cao S, Lin L, Xia X, Wu H. lncRNA SPRY4-IT1 Regulates Cell Proliferation and Migration by Sponging miR-101-3p and Regulating AMPK Expression in Gastric Cancer. Mol Ther Nucleic Acids. 2019;17:455-64
147. Liu Y, Yin L, Chen C, Zhang X, Wang S. Long non-coding RNA GAS5 inhibits migration and invasion in gastric cancer via interacting with p53 protein. Dig Liver Dis. 2019
148. Chen Z, Ju H, Yu S, Zhao T, Jing X, Li P. et al. Prader-Willi region non-protein coding RNA 1 suppressed gastric cancer growth as a competing endogenous RNA of miR-425-5p. Clin Sci (Lond). 2018;132:1003-19
149. Huang Y, Zhang J, Hou L, Wang G, Liu H, Zhang R. et al. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J Exp Clin Cancer Res. 2017;36:194
150. Li H, Ma X, Yang D, Suo Z, Dai R, Liu C. PCAT-1 contributes to cisplatin resistance in gastric cancer through epigenetically silencing PTEN via recruiting EZH2. J Cell Biochem. 2020;121:1353-61
151. Al-Rugeebah A, Alanazi M, Parine NR. MEG3: an Oncogenic Long Non-coding RNA in Different Cancers. Pathol Oncol Res. 2019;25:859-74
152. Chen JF, Wu P, Xia R, Yang J, Huo XY, Gu DY. et al. STAT3-induced lncRNA HAGLROS overexpression contributes to the malignant progression of gastric cancer cells via mTOR signal-mediated inhibition of autophagy. Mol Cancer. 2018;17:6
153. Liu T, Liu Y, Wei C, Yang Z, Chang W, Zhang X. LncRNA HULC promotes the progression of gastric cancer by regulating miR-9-5p/MYH9 axis. Biomed Pharmacother. 2020;121:109607
154. Wu Q, Ma J, Meng W, Hui P. DLX6-AS1 promotes cell proliferation, migration and EMT of gastric cancer through FUS-regulated MAP4K1. Cancer Biol Ther. 2020;21:17-25
155. Zhou C, Zhao J, Liu J, Wei S, Xia Y, Xia W. et al. LncRNA SNHG16 promotes epithelial- mesenchymal transition via down-regulation of DKK3 in gastric cancer. Cancer Biomark. 2019;26:393-401
156. Sakai S, Ohhata T, Kitagawa K, Uchida C, Aoshima T, Niida H. et al. Long Noncoding RNA ELIT-1 Acts as a Smad3 Cofactor to Facilitate TGFbeta/Smad Signaling and Promote Epithelial-Mesenchymal Transition. Cancer Res. 2019;79:2821-38
157. Jiao J, Zhang S. Long noncoding RNA MEG3 suppresses gastric carcinoma cell growth, invasion and migration via EMT regulation. Mol Med Rep. 2019;20:2685-93
158. Li J, Li Z, Zheng W, Li X, Wang Z, Cui Y. et al. LncRNA-ATB: An indispensable cancer-related long noncoding RNA. Cell Prolif. 2017 50
159. Zeng S, Xie X, Xiao YF, Tang B, Hu CJ, Wang SM. et al. Long noncoding RNA LINC00675 enhances phosphorylation of vimentin on Ser83 to suppress gastric cancer progression. Cancer Lett. 2018;412:179-87
160. Wu H, Hu Y, Liu X, Song W, Gong P, Zhang K. et al. LncRNA TRERNA1 Function as an Enhancer of SNAI1 Promotes Gastric Cancer Metastasis by Regulating Epithelial-Mesenchymal Transition. Mol Ther Nucleic Acids. 2017;8:291-9
161. Wang F, Zhu W, Yang R, Xie W, Wang D. LncRNA ZEB2-AS1 contributes to the tumorigenesis of gastric cancer via activating the Wnt/beta-catenin pathway. Mol Cell Biochem. 2019;456:73-83
162. Xu G, Meng L, Yuan D, Li K, Zhang Y, Dang C. et al. MEG3/miR21 axis affects cell mobility by suppressing epithelialmesenchymal transition in gastric cancer. Oncol Rep. 2018;40:39-48
163. He W, Liang B, Wang C, Li S, Zhao Y, Huang Q. et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene. 2019;38:4637-54
164. Xiao Y, Pan J, Geng Q, Wang G. LncRNA MALAT1 increases the stemness of gastric cancer cells via enhancing SOX2 mRNA stability. Febs Open Bio. 2019;9:1212-22
165. Tian SB, Yu JC, Kang WM, Ma ZQ, Ye X, Cao ZJ. et al. Combined detection of CEA, CA 19-9, CA 242 and CA 50 in the diagnosis and prognosis of resectable gastric cancer. Asian Pac J Cancer Prev. 2014;15:6295-300
166. Bae S, Brumbaugh J, Bonavida B. Exosomes derived from cancerous and non-cancerous cells regulate the anti-tumor response in the tumor microenvironment. Genes Cancer. 2018;9:87-100
167. Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8:83
168. Pan L, Liang W, Fu M, Huang ZH, Li X, Zhang W. et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J Cancer Res Clin Oncol. 2017;143:991-1004
169. Zhao R, Zhang Y, Zhang X, Yang Y, Zheng X, Li X. et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer. 2018;17:68
170. Lin LY, Yang L, Zeng Q, Wang L, Chen ML, Zhao ZH. et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. 2018;17:84
171. Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516
172. Zheng R, Liang J, Lu J, Li S, Zhang G, Wang X. et al. Genome-wide long non-coding RNAs identified a panel of novel plasma biomarkers for gastric cancer diagnosis. Gastric Cancer. 2019;22:731-41
173. Wang WJ, Guo CA, Li R, Xu ZP, Yu JP, Ye Y. et al. Long non-coding RNA CASC19 is associated with the progression and prognosis of advanced gastric cancer. Aging (Albany NY). 2019;11:5829-47
174. Fu JW, Kong Y, Sun X. Long noncoding RNA NEAT1 is an unfavorable prognostic factor and regulates migration and invasion in gastric cancer. J Cancer Res Clin Oncol. 2016;142:1571-9
175. Li S, Zhang M, Zhang H, Hu K, Cai C, Wang J. et al. Exosomal long noncoding RNA lnc-GNAQ-6:1 may serve as a diagnostic marker for gastric cancer. Clin Chim Acta. 2020;501:252-7
176. Shan L, Liu C, Ma C. High Expression of Serum UCA1 may be a Potential Biomarker for Clinical Diagnosis of Gastric Cancer. Clin Lab. 2019 65
177. Feng W, Zong W, Li Y, Shen X, Cui X, Ju S. Abnormally expressed long noncoding RNA B3GALT5-AS1 may serve as a biomarker for the diagnostic and prognostic of gastric cancer. J Cell Biochem. 2020;121:557-65
178. Xu Z, Chen H, Yang B, Liu X, Zhou X, Kong H. The Association of HOTAIR with the Diagnosis and Prognosis of Gastric Cancer and Its Effect on the Proliferation of Gastric Cancer Cells. Can J Gastroenterol Hepatol. 2019;2019:3076345
179. Lin LY, Yang L, Zeng Q, Wang L, Chen ML, Zhao ZH. et al. Tumor-originated exosomal lncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer. 2018;17:84
180. Yang Z, Sun Y, Liu R, Shi Y, Ding S. Plasma long noncoding RNAs PANDAR, FOXD2-AS1, and SMARCC2 as potential novel diagnostic biomarkers for gastric cancer. Cancer Manag Res. 2019;11:6175-84
181. Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y. et al. Genome-Wide lncRNA Microarray Profiling Identifies Novel Circulating lncRNAs for Detection of Gastric Cancer. Theranostics. 2017;7:213-27
182. Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is Associated with Poor Prognosis of Gastric Cancer and Promotes Cell Proliferation and EMT through Epigenetically Silencing p27 and Sponging miR-101-3p. Cell Physiol Biochem. 2017;42:999-1012
183. Li J, Xu Q, Wang W, Sun S. MIR100HG: a credible prognostic biomarker and an oncogenic lncRNA in gastric cancer. Biosci Rep. 2019 39
184. Zhang Y, Yang R, Lian J, Xu H. LncRNA Sox2ot overexpression serves as a poor prognostic biomarker in gastric cancer. Am J Transl Res. 2016;8:5035-43
185. Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang Q. et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7:56209-18
186. Lin Y, Hu D, Zhou Q, Lin X, Lin J, Peng F. Correction: The fasting blood glucose and long non-coding RNA SNHG8 predict poor prognosis in patients with gastric carcinoma after radical gastrectomy. Aging (Albany NY). 2018;10:4294
187. Wang Z, Qin B. Prognostic and clinicopathological significance of long noncoding RNA CTD-2510F5.4 in gastric cancer. Gastric Cancer. 2019;22:692-704
188. Wang Z, Wang Q, Xu G, Meng N, Huang X, Jiang Z. et al. The long noncoding RNA CRAL reverses cisplatin resistance via the miR-505/CYLD/AKT axis in human gastric cancer cells. Rna Biol. 2020:1-14
189. Zhu L, Zhu Y, Han S, Chen M, Song P, Dai D. et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019;10:383
190. Zhen Q, Gao LN, Wang RF, Chu WW, Zhang YX, Zhao XJ. et al. LncRNA PCAT-1 promotes tumour growth and chemoresistance of oesophageal cancer to cisplatin. Cell Biochem Funct. 2018;36:27-33
191. Qiao L, Liu X, Tang Y, Zhao Z, Zhang J, Liu H. Knockdown of long non-coding RNA prostate cancer-associated ncRNA transcript 1 inhibits multidrug resistance and c-Myc-dependent aggressiveness in colorectal cancer Caco-2 and HT-29 cells. Mol Cell Biochem. 2018;441:99-108
192. Li H, Ma X, Yang D, Suo Z, Dai R, Liu C. PCAT-1 contributes to cisplatin resistance in gastric cancer through epigenetically silencing PTEN via recruiting EZH2. J Cell Biochem. 2020;121:1353-61
193. YiRen H, YingCong Y, Sunwu Y, Keqin L, Xiaochun T, Senrui C. et al. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol Cancer. 2017;16:174
194. Xin L, Zhou Q, Yuan YW, Zhou LQ, Liu L, Li SH. et al. METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J Cancer Res Clin Oncol. 2019;145:2507-17
195. Zhou Z, Lin Z, He Y, Pang X, Wang Y, Ponnusamy M. et al. The Long Noncoding RNA D63785 Regulates Chemotherapy Sensitivity in Human Gastric Cancer by Targeting miR-422a. Mol Ther Nucleic Acids. 2018;12:405-19
196. Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16:167-79
197. Arun G, Diermeier SD, Spector DL. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol Med. 2018;24:257-77
198. Yang J, Meng X, Pan J, Jiang N, Zhou C, Wu Z. et al. CRISPR/Cas9-mediated noncoding RNA editing in human cancers. Rna Biol. 2018;15:35-43
199. Lavorgna G, Vago R, Sarmini M, Montorsi F, Salonia A, Bellone M. Long non-coding RNAs as novel therapeutic targets in cancer. Pharmacol Res. 2016;110:131-8
200. Hessels D, Klein GJ, van Oort I, Karthaus HF, van Leenders GJ, van Balken B. et al. DD3(PCA3)-based molecular urine analysis for the diagnosis of prostate cancer. Eur Urol. 2003;44:8-15 15-6
Author contact
![]() Corresponding authors: Prof. Yi Chen, MD., Phone number: +86-28-85164063; E-mail: toddychancom; Prof. Yang An, MD., Phone number: +86-13264498382; E-mail: anyangdoctorcom; Prof. Bo Liu, Ph.D., phone number: +86-15708469925; E-mail: liubo2400com.
Corresponding authors: Prof. Yi Chen, MD., Phone number: +86-28-85164063; E-mail: toddychancom; Prof. Yang An, MD., Phone number: +86-13264498382; E-mail: anyangdoctorcom; Prof. Bo Liu, Ph.D., phone number: +86-15708469925; E-mail: liubo2400com.
 Global reach, higher impact
Global reach, higher impact