13.3
Impact Factor
Theranostics 2020; 10(16):7335-7350. doi:10.7150/thno.45971 This issue Cite
Research Paper
KRAS Mutation-Responsive miR-139-5p inhibits Colorectal Cancer Progression and is repressed by Wnt Signaling
1. State Key Laboratory of Cancer Biology and National Clinical Research Center for Digestive Diseases, Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Xi'an, Shaanxi 710032, China.
2. Department of Internal Medicine, The Hospital of the People's Liberation Army 63650 Corps, Malan, Xinjiang Uygur Autonomous Region 841700, China.
3. Department of Cardiovascular Medicine, First Affiliated Hospital of Medical School, Xi'an Jiaotong University, Xi'an, Shaanxi 710061, China.
4. Department of Biomedical Informatics and Center for Quantitative Sciences, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA.
5. International Cancer Center, Shenzhen University Health Science Center, Shenzhen, Guangdong 518060, China.
6. Comprehensive Cancer Center, The Ohio State University, Columbus, Ohio 43210, USA.
7. Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA.
8. State Key Laboratory of Translational Medicine and Innovative Drug Development, Jiangsu Simcere Diagnostics Co., Ltd., Nanjing, Jiangsu 210042, China.
9. National Institute of Biological Sciences, Beijing 102206, China.
#These authors contributed equally to this work.
Abstract
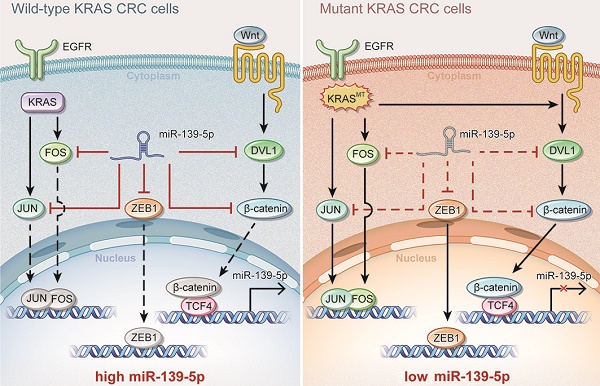
Introduction: Colorectal cancer (CRC) frequently harbors KRAS mutations that result in chemoresistance and metastasis. MicroRNAs (miRNAs) are usually dysregulated and play important regulatory roles in tumor progression. However, the KRAS mutation-responsive miRNA profile in CRC remains uninvestigated.
Methods: miR-139-5p was identified and evaluated by small RNA sequencing, qRT-PCR and in situ hybridization. The roles of miR-139-5p in CRC cells with and without KRAS mutation were determined by Cell Counting Kit-8 (CCK-8), colony formation, flow cytometry and transwell assays in vitro and by tumorigenesis and metastasis assays in vivo. Microarrays followed by bioinformatic analyses, luciferase reporter assays and Western blotting were applied for mechanistic studies.
Results: miR-139-5p was significantly downregulated in KRAS-mutated CRC cells and tissues compared with their wild-type counterparts. Low miR-139-5p expression was associated with aggressive phenotypes and poor prognosis in CRC patients. miR-139-5p overexpression inhibited CRC cell proliferation, migration and invasion in vitro, sensitized tumors to chemotherapy, and impaired tumor growth and metastasis in vivo. Transcriptomic profiling identified multiple modulators in the Ras (JUN and FOS) and Wnt (CTNNB1 and DVL1) signaling pathways and the epithelial-to-mesenchymal transition (EMT) process (ZEB1) as direct targets of miR-139-5p, and inverse correlations were confirmed in CRC clinical tissues. Aberrantly activated Wnt signaling in KRAS-mutant cells was demonstrated to transcriptionally repress miR-139-5p through TCF4, forming a miR-139-5p/Wnt signaling double-negative feedback loop.
Conclusions: We identified miR-139-5p as a KRAS-responsive miRNA and demonstrated its involvement in CRC progression. KRAS mutation disrupted the miR-139-5p/Wnt signaling reciprocal negative feedback mechanism, which might cause miR-139-5p downregulation and derepression of oncogenic signaling pathways and EMT. These results reveal a transcriptional regulatory mode of KRAS-driven malignant transformation and highlight miR-139-5p as a novel regulator of crosstalk between the Ras and Wnt signaling pathways in CRC.
Keywords: miR-139-5p, KRAS mutation, CRC, Ras signaling, Wnt/β-catenin signaling
 Global reach, higher impact
Global reach, higher impact