13.3
Impact Factor
Theranostics 2020; 10(13):5998-6010. doi:10.7150/thno.43427 This issue Cite
Research Paper
Investigation of the role and mechanism of ARHGAP5-mediated colorectal cancer metastasis
1. Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Guangzhou 510060, China
2. The Third Affiliated Hospital, Sun Yat-sen University, Guangzhou, 510630, China
*These authors contributed equally to this article.
Received 2019-12-27; Accepted 2020-4-18; Published 2020-5-1
Abstract
Background: Metastatic colorectal cancer (CRC) is a lethal disease; however, the underlying molecular mechanisms remain unclear and require further study.
Methods: RNA-Seq, PCR, Western blotting, immunohistochemistry, ChIP and RNAi assays were performed to investigate Rho GTPase-activating protein 5 (ARHGAP5, aslo known as p190RhoGAP-B, p190-B) expression and the clinical relevance, functional roles and regulatory mechanisms of this protein using human CRC cells and tissues. In vivo, two cell-based xenograft models were used to evaluate the roles of ARHGAP5 in CRC metastasis.
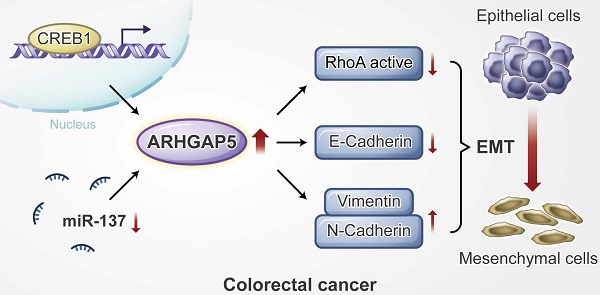
Results: Here, we report that ARHGAP5 expression is significantly increased in metastatic CRC tissues and is inversely associated with patient overall survival. The suppression of ARHGAP5 reduces CRC cell metastasis in vitro and in cell-based xenograft models. Furthermore, we show that ARHGAP5 promotes CRC cell epithelial-mesenchymal transition by negatively regulating RhoA activity. Mechanistically, cAMP response element-binding protein (CREB1) transcriptionally upregulates ARHGAP5 expression, and decreased miR-137 further contributes to ARHGAP5 mRNA stability in CRC.
Conclusions: Overall, our study highlights the crucial function of ARHGAP5 in CRC metastasis, thus suggesting novel prognostic biomarkers and hypothetical therapeutic targets.
Keywords: colorectal cancer, tumor metastasis, ARHGAP5, CREB1, miR-137
Introduction
Metastatic colorectal cancer (CRC) is a lethal disease that caused almost 861,000 deaths in 2018 worldwide [1, 2]. The liver is the most common organ for distant metastasis in CRC patients. Clinically, approximately 20-25% of CRC patients present with synchronous liver metastases at diagnosis, while approximately 50% of CRC patients develop metachronous liver metastases within 3 years of treatment [3-5]. Multidisciplinary team therapy is currently the main treatment mode for liver metastasis in CRC patients [4, 6]. However, the 5-year survival rate of these patients is only 20%, while the 5-year survival rate is 90% for early-stage CRC patients [1]. Therefore, investigations of the molecular mechanisms underlying CRC metastasis may help to develop novel prognostic biomarkers and therapeutic strategies for CRC patients.
RhoA, a member of the Rho family of small GTPases, is involved in regulating cell shape and movement through cytoskeletal remodeling, thus influencing cell migration and invasion [7]. Accordingly, the activity of this protein is tightly controlled by guanine nucleotide-exchange factors (GEFs) and GTPase-activating proteins (GAPs) that determine whether GTP or GDP is bound to this protein. RhoA is activated by GEFs, which catalyze the release of GDP and thus allow GTP to bind this protein [8, 9]. RhoA, in turn, is inactivated by GAPs, which bind to Rho proteins and induce them to hydrolyze their bound GTP to GDP. The GAPs of many small GTPases contain a conserved Arg residue that is essential for catalyzing nucleotide hydrolysis [10]. Rho GTPase-activating protein 5 (ARHGAP5, aslo known as p190RhoGAP-B, p190-B), a member of the RhoGAP family, negatively regulates RhoA activity [11-13]. Although previous studies suggested that ARHGAP5 may play an oncogenic role in tumor progression, its biological function and regulatory mechanisms in CRC are poorly understood.
In the present study, we aimed to discover the key molecules involved in CRC liver metastasis. We examined the clinical relevance, function and underlying regulatory mechanisms of ARHGAP5 in CRC metastasis. Additionally, our study suggests novel prognostic biomarkers and hypothetical therapeutic targets for metastatic CRC patients.
Materials and Methods
Patients and cells
Archived CRC tissue specimens (n=423) were collected after obtaining written informed consent, in accordance with our Institutional Review Board and the Declaration of Helsinki (Table S1). CRC, immortalized colon epithelial and 293T cell lines were purchased from ATCC (Manassas, VA, USA) and cultured under the conditions specified by the supplier. The cell lines SW480 derived from primary tumor and SW620 from a metastatic site in the same patient [14]. All cells were negative for mycoplasma contamination and were authenticated based on short tandem repeat DNA fingerprinting before use.
Immunoblotting and immunohistochemical (IHC) analysis
Immunoblotting and IHC analysis were conducted with standard procedures, as previously described [15, 16]. Blotting membranes were stripped and reprobed with anti-β-Actin antibody as a loading control. The degree of immunostaining of formalin-fixed, paraffin-embedded sections was reviewed and scored independently by two pathologists based on both the proportion of positively stained tumor cells and the intensity of the staining. The following antibodies were used for immunoblotting or IHC analysis: ARHGAP5 (#2562, WB, 1:1000), Vimentin (#5741; WB, 1:1000; IHC, 1:200), N-cadherin (#13116; WB, 1:1000; IHC, 1:100), E-cadherin (#14472; WB, 1:1000; IHC, 1:100), RhoA (#2171; WB, 1:1000), CREB1 (#9197; WB, 1:1000; IHC, 1:6000), β-Actin (#3700; WB, 1:3000) (all from Cell Signaling, Beverly, USA) and ARHGAP5 (#ab199160; IHC, 1:100) (Abcam, Cambridge, MA, USA).
Migration and invasion assays
The effects of ARHGAP5 on the migration and invasion of CRC cells were tested using transwell chambers, as previously reported [17, 18]. Briefly, CRC cells were harvested and suspended in 200 μL serum-free medium (2×105) and were plated in the top chamber with (migration) or without (invasion) a Matrigel-coated membrane (pore size, 8 µm)( BD Biosciences, San Jose, USA). The lower chambers were filled with serum as a chemoattractant. The cells were incubated for 24 h, and cells that did not migrate through the pores were removed with a cotton swab. Invading cells were fixed, stained with crystal violet (Sigma-Aldrich, St. Louis, USA) and counted in five random fields.
RhoA activity assay
Rho activity was assessed using an Active Rho Detection Kit (#8820, Cell Signaling, Beverly, USA). The measurement of GTPase activity was based on the ability of the GTP-bound (active) form to bind the Rhotekin-RBD fusion protein, which could then be immunoprecipitated with glutathione resin. Total RhoA levels were similarly analyzed using an aliquot of whole-cell lysate. Active RhoA and total RhoA were analyzed by immunoblotting analysis with an anti-RhoA antibody that was included in the kit.
Animal study
All animal experiments were performed in accordance with a protocol approved by our institutional Animal Care and Use Committee. Female BALB/c nude mice (4/5 weeks old) were obtained from the Animal Center of Guangdong Province (Guangzhou, China). To evaluate the effect of ARHGAP5 on CRC metastasis in vivo, two xenograft models were used. For liver metastasis, the mice were anesthetized, and CRC cells (2 × 106 per mouse) were injected into the distal tip of the spleen using a Hamilton syringe (8 mice/group). These mice were sacrificed six weeks postinjection. The spleen and liver were recovered, paraffin-embedded and stained with H&E. The micrometastases in the livers were examined and counted under a dissecting microscope. For mesenteric metastasis, mice were anesthetized, and CRC cells (2×106 per mouse) were orthotopically implanted into the cecum (8 mice/group). The mice were sacrificed, the intestines were removed, and the metastatic nodules in the intestines were counted after 6 weeks.
Statistical analysis
To identify the significant differences between two groups, a Student's t-test was used. Survival curves were plotted using the Kaplan-Meier method and compared by the log-rank test. For correlation analysis between two continuous variables, r values represent Pearson's correlation coefficients, and p-values were calculated by Pearson's correlation test. For the study of the association between the expressions of two genes, the Chi-square test was used. Statistical analyses were performed with GraphPad version 5.0. A P value of less than 0.05 was statistically significant. All statistical tests were two-sided.
The details of RNA extraction and qPCR analysis, lentiviral transduction, immunofluorescence analysis, chromatin immunoprecipitation (ChIP) and luciferase promoter assays are described in the Supplementary Materials and Methods.
Results
Increased ARHGAP5 expression is associated with CRC metastasis and poor prognosis
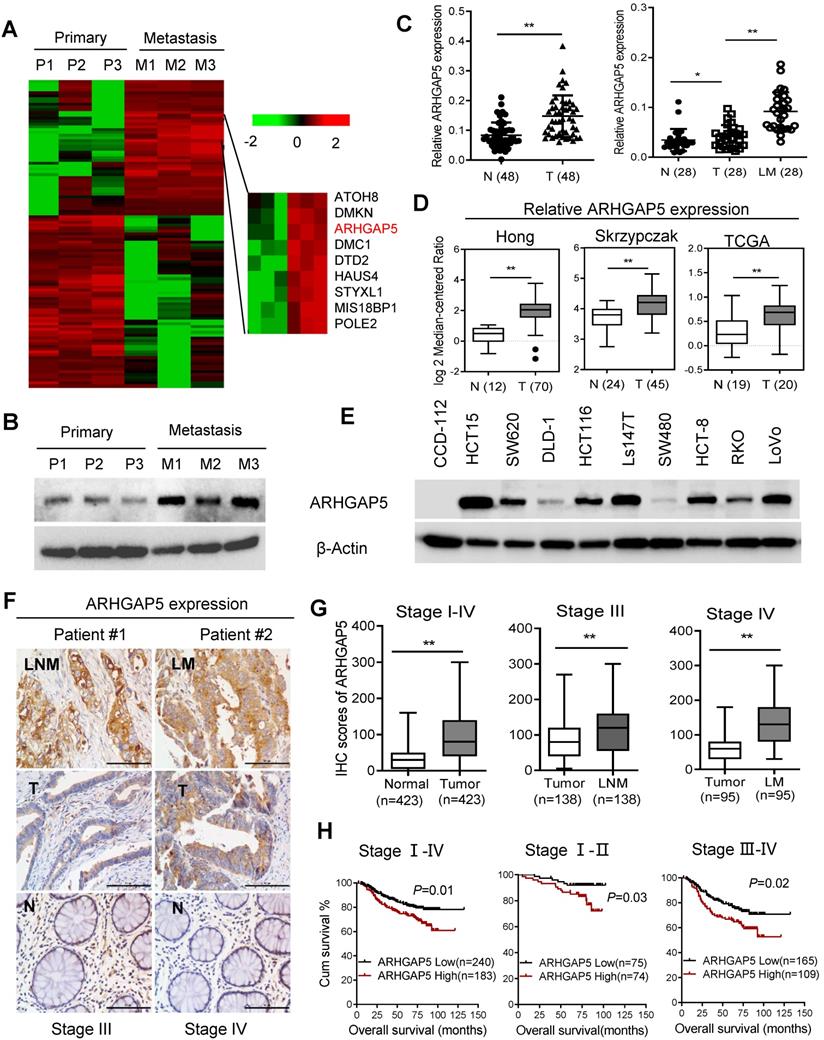
To explore the key molecules that modulate CRC hepatic metastasis, we performed RNA sequencing (RNA-Seq) analysis of three paired primary and liver metastasis CRC tissues. RNA-Seq analysis and validation with immunoblotting showed that ARHGAP5 was markedly overexpressed in liver metastatic tissues compared to matched primary tumor tissues (Figure 1A, 1B). ARHGAP5 and ARHGAP35 (p190RhoGAP-B, P190A) are known as the main negative regulator of RhoA [19], and ARHGAP35 degradation is implicated in metastatic CRC [20]. However, ARHGAP35 expression was not changed significantly in our RNA-seq analyses. PCR analysis also confirmed that ARHGAP5 was significantly upregulated in CRC liver metastatic tissues and primary tumor tissues compared to matched adjacent-normal tissues (Figure 1C). Furthermore, the overexpression of ARHGAP5 in CRC was also supported by the Oncomine database, including the Hong, Skrzypczak and TCGA datasets (Figure 1D). qPCR and immunoblotting analysis showed that the ARHGAP5 mRNA and protein levels were notably increased in CRC cells compared with colorectal epithelial cells (Figures 1E, S1). Consistently, the IHC analysis of tissue microarrays found that the ARHGAP5 expression levels were significantly upregulated in liver and lymph node metastatic tissues compared with paired primary tissues (Figure 1F-G). Strikingly, Kaplan-Meier survival analysis indicated that patients with high ARHGAP5 expression levels had a shorter overall survival (OS) and disease-free survival (DFS) (Figure 1H). Multivariate analysis also indicated that ARHGAP5 expression was an independent prognostic factor in CRC patients (Table S2). These results indicate that ARHGAP5 may serve as a potential prognostic biomarker and may contribute to CRC metastasis.
ARHGAP5 inhibition suppresses CRC metastasis in vitro and in vivo
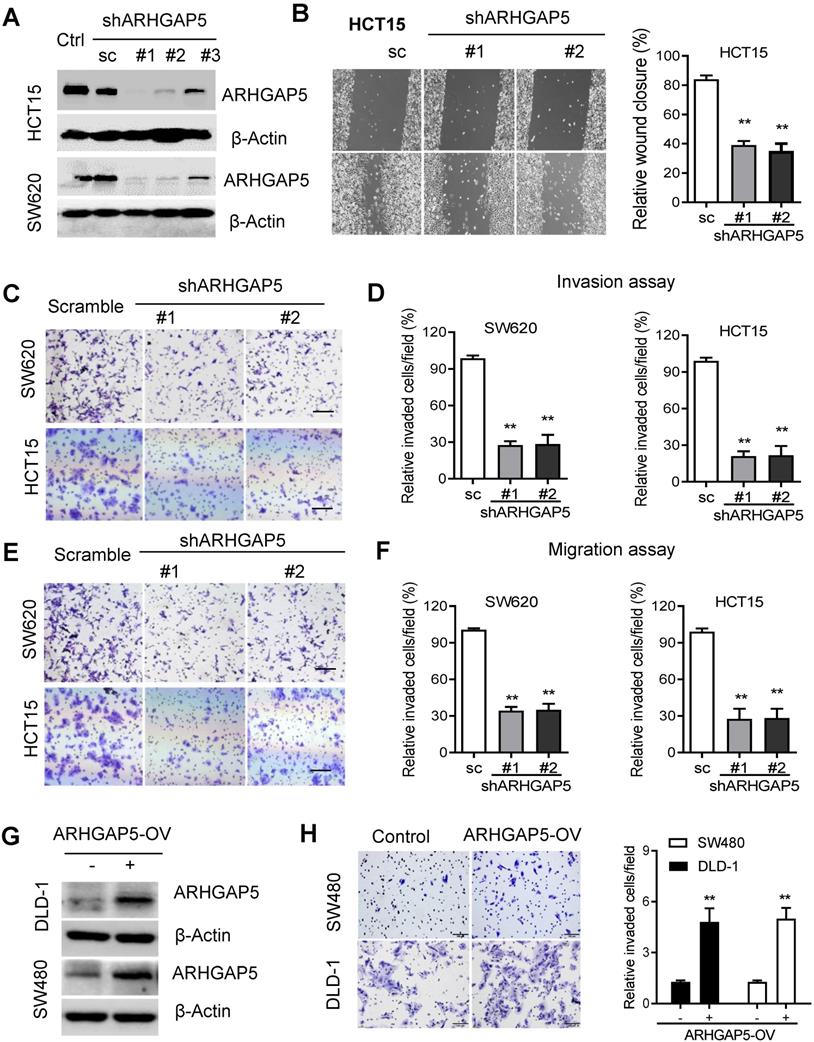
Given the prognostic value of ARHGAP5 and its correlation with metastasis in CRC, we next sought to determine whether ARHGAP5 contributes to metastasis by affecting cancer cell migration and/or proliferation, which are 2 critical determinants of metastasis. Then, we stably knocked down ARHGAP5 by the lentiviral infection of shRNA targeting ARHGAP5 in SW620 and HCT15 cells which expressed higher level of ARHGAP5 (Figures 1E, 2A, S2B). However, MTS assays and colony formation assays showed that ARHGAP5 knockdown had a small effect on CRC cell proliferation (Figure S1B). Interestingly, ARHGAP5 knockdown significantly inhibited CRC cell wound healing, migration and invasion (Figure 2B-F). On the other hand, the overexpression of ARHGAP5 significantly enhanced cell invasion in the DLD1 cell line and in SW480 cells which expressed lower level of ARHGAP5 (Figure 1E, 2G-H). These findings indicate that ARHGAP5 plays an important role in the enhancement of CRC metastatic ability in vitro.
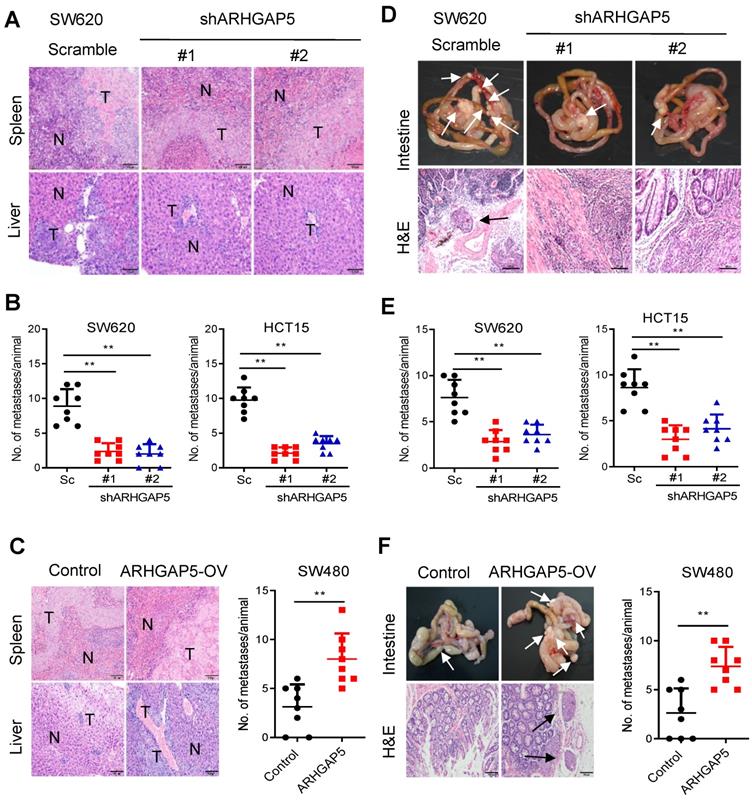
To further test whether ARHGAP5 contributes to CRC tumorigenesis in vivo, we performed cell-based xenograft experiments. For liver metastasis assays, the mice injected with control SW620 or HCT15 cells into the spleen had a heavy liver metastatic burden that was verified by histologic examination (H&E), whereas the knockdown of ARHGAP5 almost abolished this phenotype (Figure 3A, 3B). To further determine the effects of ARHGAP5 on the promotion of CRC metastasis, CRC cells were orthotopically implanted into the cecum of nude mice. The results indicated that ARHGAP5 suppression also reduced mesenteric metastatic nodules on the intestinal wall (Figure 3D, 3E). On the contrary, overexpressing ARHGAP5 in SW480 significantly enhanced the ability of these cells to form liver and mesenteric metastasis in vivo (Figure 3C, 3F). Overall, these results highlight the crucial roles of ARHGAP5 in promoting CRC metastasis in vivo.
Increased ARHGAP5 expression is associated with CRC metastasis and poor prognosis. (A) Heatmap profiling the gene expression of paired primary and liver metastasis CRC tissues (n=3), as analyzed by RNA-seq. (B) Immunoblotting analysis of ARHGAP5 expression in paired primary and liver metastasis CRC tissues. β-Actin was included as a loading control. (C) qPCR analysis of ARHGAP5 expression in 48 pairs of CRC tumor (T) and adjacent normal issues (N) and in 28 pairs of liver metastases (LM) and primary (T) tissues. (D) ARHGAP5 expression in multiple CRC microarray data sets available from the Oncomine database (www.oncomine.org). (E) Immunoblotting analysis of ARHGAP5 expression in CRC cells and epithelial colorectal cells (CCD112). (F-G) Representative IHC staining and quantification of ARHGAP5 in paired primary CRC tumor (n=423), lymph node metastatic (LNM, n=138) or liver metastatic tissues (LM, n=95). (H) Kaplan-Meier analysis of the overall survival (OS) or disease-free survival (DFS) of CRC patients based on ARHGAP5 expression (log-rank test). Data in C, D, and G are presented as the mean ± the SD. *P < 0.05, **P < 0.01 (Student's t-test).
ARHGAP5 inhibition suppresses CRC cell migration and invasion activity. (A) Immunoblotting evaluating the knockdown efficiency of ARHGAP5 with three unique shRNAs (#1, #2 and #3) in SW620 and HCT15 cells. Scrambled shRNA (sc) was used as a negative control, and β-Actin was included as a loading control. (B) Representative images and quantification showing the wound healing of HCT15 cells with or without ARHGAP5 knockdown. (C-F) Representative images and quantification showing the cell invasion and migration of the indicated CRC cells with or without ARHGAP5 knockdown. (G) Immunoblotting analysis of ARHGAP5 expression in DLD-1 and SW480 cells overexpressing ARHGAP5 (ARHGAP5-OV). (H) Representative images and quantification showing the cell invasion of the indicated CRC cells overexpressing ARHGAP5. Data in B, D, F and H are presented as the mean ± the SD (n=3). **P < 0.01 (Student's t-test).
Effects of ARHGAP5 knockdown or ARHGAP5 overexpression on CRC metastasis in vivo. (A-C) Representative H&E staining and statistical results of the micrometastatic nodules in the liver from mice injected with the indicated cells into the spleen for 45 days. Scale bar: 100 µm, N=8 per group. (D-F) Representative H&E staining and statistical results of metastatic tumors in the excised intestines of mice orthotopically implanted with the indicated cells for 60 days. Scale bar: 100 µm, N=8 per group. White arrows indicate the metastatic foci. Data in B, C, E and F are presented as the mean ± the SD. **P < 0.01 (Student's t-test).
ARHGAP5 promotes EMT by negatively regulating RhoA activity
Therefore, we further investigated the underlying mechanism of ARHGAP5 overexpression in CRC metastasis. Gene set enrichment analysis (GSEA) revealed that ARHGAP5 expression was positively corrected with the epithelial-mesenchymal transition (EMT) pathway in CRC (Figure 4A). Immunofluorescence analysis showed that the knockdown of ARHGAP5 significantly increased E-cadherin expression and decreased N-Cadherin expression in SW620 cells (Figure 4B), which was further verified by immunoblotting analysis (Figure 4C). As ARHGAP5 is a member of the RhoGAP family that negatively regulates RhoA activity [11, 12], we then detected total RhoA and active RhoA in SW620 and HCT15 cells with ARHGAP5 knockdown. The results indicated that the knockdown of ARHGAP5 obviously increased the amount of active RhoA (RhoA-GTP) pulled down by GS-RBD (Figure 4D). Furthermore, we found that knockdown of RhoA significantly restored CRC invasion capability that decreased due to ARHGAP5 depletion (Figure 4E). As expected, the overexpression of ARHGAP5 decreased active RhoA (RhoA-GTP) and E-cadherin expression but increased N-Cadherin and Vimentin expression compared with vector control SW480 cells (Figure 4F-G). These results also consistent with previous study that exogenous expression of constitutively active RhoA (RhoAQ63L) inhibited CRC and breast cancer cell invasion [21, 22]. Also, we constructed a mutant ARHGAP5 (Mu) with a point mutation in conserved Arg residue (R1297A, Arg to Ala) referred to previous studies [23, 24]. As shown, overexpression of mutant ARHGAP5 (Mu, R1297A) did not change RhoA activity and SW480 cell metastasis capability compared with vector control cells, further supporting that ARHGAP5 promotes CRC metastasis by suppressing RhoA activity (Figure 4H). Additionally, correlation studies in 423 CRC tissue specimens showed that ARHGAP5 expression was positively correlated with the expression of N-cadherin but was negatively correlated with the expression of E-cadherin (Figure 4I-J). Taken together, these results indicate that ARHGAP5 promotes EMT by negatively regulating RhoA activity in CRC.
CREB1 transcriptionally upregulates ARHGAP5 expression in CRC
To assess the molecular regulation of ARHGAP5, we first surveyed genetic alterations of this gene using the cBioPortal datasets and found that the ARHGAP5 locus is unamplified in CRC, indicating that ARHGAP5 may be transcriptionally regulated (Figure S2A). Bioinformatics analysis with the JASPAR and TCGA databases predicted that cAMP responsive element binding protein (CREB1) was a potential transcription factor of ARHGAP5, and there was a significant, positive correlation between CREB1 mRNA and ARHGAP5 mRNA expression (Figure 5A). qPCR analysis revealed that CREB1 expression was significantly upregulated in CRC liver metastatic tissues and primary tumor tissues compared to matched adjacent normal tissues (Figure 5B), and the expression of CREB1 was tightly correlated with the expression of ARHGAP5 in CRC samples available from SYSUCC (n=78) (Figure 5C). In addition, qPCR and immunoblotting analysis indicated that the depletion of CREB1 obviously decreased the expression of ARHGAP5 in HCT15 and SW620 cells (Figure 5D-E). As CREB1 activates target genes through cAMP (Adenosine 3',5'-cyclic monophosphate) response elements [25], we also treated SW480 and DLD-1 cells with addition of exogenous cAMP. The results shows that ARHGAP5 expression was significantly increased by adding exogenous cAMP (Figure S2B-C). Through bioinformatics analysis, we also identified two CREB1 DNA-binding sites in the human ARHGAP5 promoter region (Figure 5F). ChIP-PCR assays showed that CREB1 can bind to the promoter region of ARHGAP5 in HCT15 and SW620 cells (Figure 5G). A dual-luciferase reporter assay showed that the relative ARHGAP5 luciferase promoter activity increased or decreased with CREB1 overexpression or depletion, respectively, in the indicated cells (Figure 5H-I). Additionally, correlation studies in 423 CRC tissue specimens showed that ARHGAP5 expression was positively correlated with the expression levels of CREB1 (Figure 5J). In summary, these results indicate that CREB1 transcriptionally upregulates ARHGAP5 expression in CRC.
Decreased miR-137 contributes to ARHGAP5 overexpression in CRC
As microRNAs (miRNAs) are small, noncoding RNAs that act as the master regulators of gene expression in CRC cells [26, 27], we then investigated whether ARHGAP5 was regulated by specific miRNAs. Analysis using publicly available algorithms (TargetScan, miRanda and miRDB) showed that ARHGAP5 was the predicted target of miR-137, miR-486-5P and miR-107 (Figure 6A). Although qPCR analysis showed that these three miRNAs were significantly downregulated in CRC tumor tissues, only miR-137 was significantly negatively correlated with ARHGAP5 mRNA expression (Figures 6B-C, S3A-B). Further analysis using the TargetScan algorithm showed that the 3'UTR (from 1041 to 1048 bp) of ARHGAP5 was a predicted target of miR-137 (Figure S3C). Moreover, a luciferase reporter assay showed that the overexpression of miR-137 repressed the luciferase activity of ARHGAP5-3'UTR in HCT15 and SW620 cells with low miR-137 expression (Figure 6D, S3D). However, ectopically expressing miR-137 mutation mimics did not inhibit the ARHGAP5-3'UTR luciferase activity (Figure 6D). Accordingly, the ectopic expression of miR-137 significantly decreased both ARHGAP5 expression (Figure 6E) and inhibited cell invasion (Figure 6F) in HCT15 and SW620 cells. Clinically, qPCR analysis showed that the miR137 level was significantly lower in CRC liver metastatic tissues than in paired primary tumor tissues and was significantly lower in CRC patients with liver metastasis than in CRC patients without liver metastasis (Figure 6G). IHC staining and statistical analyses further revealed that miR-137 expression was positively correlated with the expression of N-cadherin but was negatively correlated with the expression of ARHGAP5 and E-cadherin (Figure 6H-I). These results clearly demonstrate that decreased miR-137 contributes to ARHGAP5 overexpression and enhances CRC metastasis.
ARHGAP5 negatively regulates RhoA activity. (A) Gene set enrichment analysis (GSEA) demonstrating the enrichment of EMT-related gene sets in the ranked gene list of ARHGAP5 up versus ARHGAP5 down available from the TCGA CRC database. (B) Representative images of immunofluorescence staining for E-cadherin and N-cadherin expression in SW620 cells with ARHGAP5 knockdown (scale bar: 10 µm). (C) Immunoblotting analysis of E-cadherin, Vimentin and N-cadherin expression in the indicated CRC cells with ARHGAP5 knockdown. (D) Immunoblotting analysis of active RhoA (RhoA-GTP) pulled down by GS-RBD and total RhoA from whole-cell lysates in indicated cells with ARHGAP5 knockdown. (E) Immunoblotting analysis of ARHGAP5 and RhoA expression in SW620 cells with ARHGAP5 and/or RhoA knockdown, and quantification of the cell invasion capability of these cells. (F) Immunoblotting analysis of active RhoA (RhoA-GTP) pulled down by GS-RBD, total RhoA, E-cadherin, Vimentin and N-cadherin expression in SW480 cells with ARHGAP5 overexpression. (G) The expression levels of E-cadherin, Vimentin and N-cadherin were quantified and normalized to the corresponding levels of β-Actin as a loading control. (H) Immunoblotting analysis of active RhoA, total RhoA and ARHGAP5 expression in SW480 cells with wide type (WT) or mutant (Mu, R1297A) ARHGAP5 overexpression, and quantification of the cell invasion capability of these cells. (I-J) Representative images and the percentage of samples showing low or high ARHGAP5 expression relative to E-cadherin and N-cadherin. Scale bar: 100 µm, *P < 0.05, **P < 0.01 (Chi-square test).
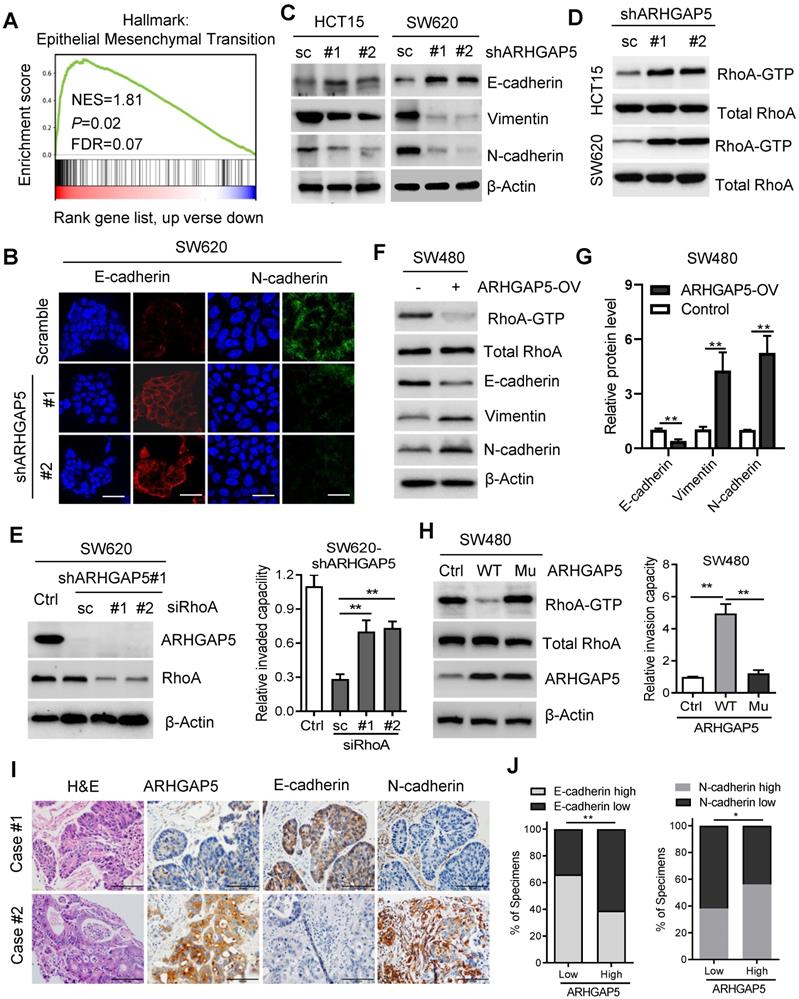
CREB1 transcriptionally upregulates ARHGAP5 expression. (A) Scatterplots of ARHGAP5 vs CREB1 mRNA expression in CRC samples available from the TCGA database (n=512). The Pearson correlation coefficient (r) and P value are displayed. (B) qPCR analysis of CREB1 mRNA levels in 28 pairs of CRC primary (T), liver metastasis (LM) and adjacent normal tissues (N). (C) Scatterplots of ARHGAP5 vs CREB1 mRNA expression in CRC samples available from SYSUCC (n=78). The Pearson correlation coefficient (r) and P value are displayed. (D-E) qPCR and immunoblotting analysis of ARHGAP5 expression in SW620 and HCT15 cells transfected with CREB1 siRNAs (#1 and #2). (F) Two CREB1 DNA-binding sites are present in the human ARHGAP5 promoter region. (G) ChIP-PCR analysis of CREB1 binding to the promoter region of ARHGAP5 in HCT15 and SW620 cells. (H-I) Relative ARHGAP5 luciferase promoter activity in the indicated cells with CREB1 overexpression or depletion. (J) Representative images and the percentage of samples showing low or high ARHGAP5 expression relative to CREB1. Scale bar: 100 µm, **P < 0.01 (Chi-square test). Data in B, D, G and H are presented as the mean ± the SD (n=3). **P < 0.01 (Student's t-test).
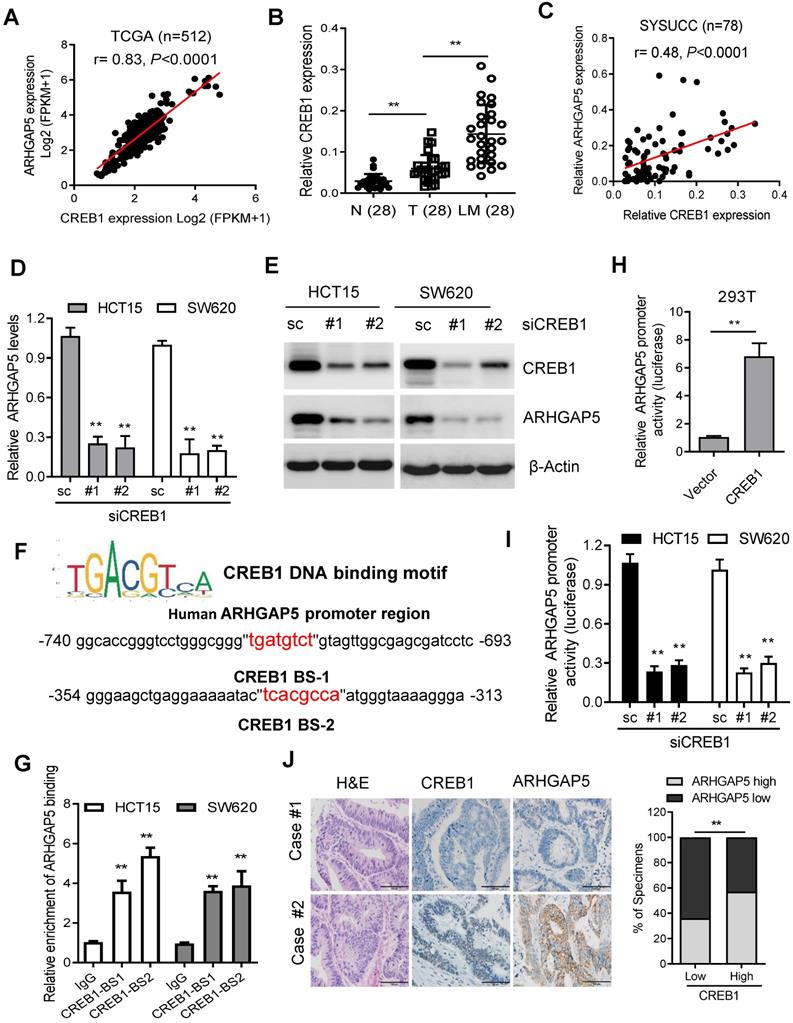
Decreased miR-137 contributes to ARHGAP5 overexpression. (A) Venn diagrams showing the number of potential miRNAs targeting the 3'UTR of ARHGAP5, as predicted by three databases. (B) qPCR analysis of miR-137 expression in paired CRC samples (n=48). (C) Scatterplots of ARHGAP5 vs miR-137 expression in CRC samples (n=48). The Pearson correlation coefficient (r) and P value are displayed. (D) Luciferase activity results of reporters containing the end of the 3'UTR of ARHGAP5 in cells transfected with a miR-137 mimic, miR-137 mutation mimics (miR-137-mut) and negative controls (NCs). (E) qPCR and immunoblotting analysis of ARHGAP5 expression in the indicated cells transfected with a miR-137 mimic. (F) Representative images and quantification of the effects of miR-137 on CRC cell invasion. Scale bar: 100 µm. (G) qPCR analysis of miR137 expression in 28 pairs of liver metastasis tissues and primary CRC tissues and in 48 pairs of CRC tissues with or without liver metastasis. (H-I) Representative images and the percentage of samples showing low or high miR137 expression relative to ARHGAP5, E-cadherin and N-cadherin. Scale bar: 100 µm, *P < 0.05, **P < 0.01 (Chi-square test). Data in D, E and F are presented as the mean ± SD (n=3). **P < 0.01 (Student's t-test).
Discussion
The distant metastasis of CRC patients is one of the most difficult challenges faced by clinicians. Progress has been made in the treatment of metastatic CRC in the past several decades due to the development of targeted drugs and immunotherapy [28]. Combining chemotherapeutic cytotoxic drugs with biologic monoclonal antibodies (cetuximab and bevacizumab) provides clinical benefits for metastatic CRC patients [4, 29]. The FDA recently approved immune checkpoint inhibitors (pembrolizumab and nivolumab) for the treatment of metastatic CRC patients with high microsatellite instability (MSI-H) [30, 31]. However, the prognosis of metastatic CRC patients is far from satisfactory. In particular, there was a lack of efficacy of these antibodies in the majority of CRC patients with KRAS mutant-type or microsatellites stable (MSS) disease [32]. Therefore, it is necessary to determine the underlying mechanisms, identify novel molecular biomarkers and develop appropriate therapies for metastatic CRC.
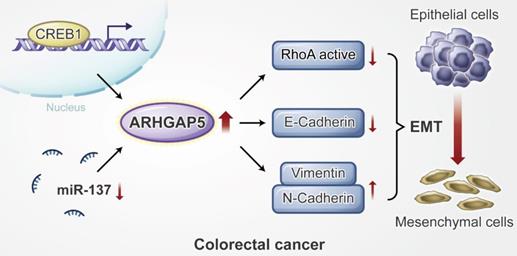
Proposed working model of this study. ARHGAP5 promotes CRC cell epithelial-mesenchymal transition by negatively regulating RhoA activity. ARHGAP5 is transcriptionally regulated by CREB1 and post-transcriptionally controlled by miR-137.
Epithelial to mesenchymal transition (EMT) is a developmental process of cell remodeling and is critical for embryogenesis, cell invasion and tumor metastasis [33, 34]. Cancer-associated EMT is driven by key transcription factors that are finely regulated by multiple signaling cues and cofactors [35]. In addition, the members of the Rho family of small GTPases, including RhoA, Rac1, Cdc42, etc., act as molecular switches that regulate cell shape, establish cell-cell junctional complexes and regulate the EMT process [36]. ARHGAP35 and ARHGAP5 are two main GAPs regulating the Rho family of small GTPases [19]. Previous studies have shown conflicting results that ARHGAP35 may be involved in CRC progression as an oncogene or tumor suppressor [20, 37, 38]. ARHGAP5 has been identified as an oncogene in lung cancer, hepatocellular carcinoma (HCC) and gastric cancer [39-41], and a tumor suppressor gene in invasive epithelial ovarian cancer [42]. In the current study, our results showed that ARHGAP5 also plays an important oncogenic role in the process of promoting CRC metastasis by both negatively regulating RhoA activity and promoting EMT. Although previous studies suggested that ARHGAP5 was associated with tumors, its underlying regulatory mechanisms in cancer cells are controversial and poorly understood. A previous study showed that ARHGAP5 was upregulated by the amplification of the chromosomal region 14q12 in a subgroup of hepatocellular carcinoma [40], by downregulation of miR-486-5p or miR-774 in lung cancer and nasopharyngeal carcinoma respectively [39, 43]. We report here that ARHGAP5 is transcriptionally regulated by CREB1 and is posttranscriptionally controlled by miR-137 in CRC. Although both CREB1 and miR-137 have been reported to regulate CRC aggression and tumorigenicity [44, 45], our findings show that these molecules also synergistically promote CRC metastasis by regulating ARHGAP5.
The substantial body of evidence that shows that Rho GTPases are related to cancer has made the key components of Rho GTPase signaling attractive therapeutic targets for cancer drug discovery [46]. A current promising approach is the development of small molecule inhibitors targeting protein kinase effectors upstream or downstream of Rho GTPase, and several inhibitors are currently in clinical trials for cancer treatment [47, 48]. These targeted inhibitors may offer efficacious treatment options for future precision cancer therapy, particularly in combination with chemotherapy or immunotherapy agents [48]. One of the remaining challenges is to better understand the detailed function and underlying regulatory mechanism of Rho GTPase signaling in the context of specific cancer types. Thus, this current study may not only provide a new understanding of CRC metastasis but also may enable the development of effective therapeutics for the treatment of metastatic CRC.
In conclusion, our findings reveal that ARHGAP5 is transcriptionally regulated by CREB1 and is post- transcriptionally controlled by miR- 137, and ARHGAP5 promotes CRC metastasis by negatively regulating RhoA activity. Additionally, our study suggests that ARHGAP5 might be used as a novel biomarker and therapeutic target for metastatic CRC patients.
Abbreviations
ARHGAP5: Rho GTPase-activating protein 5; CREB1: cAMP response element-binding protein; miR-137: mircro-RNA-137; CRC: colorectal cancer; GEF: guanine nucleotide-exchange factor; GAP: GTPase-activating protein; shRNAs: short hairpin RNAs; qPCR: quantitative real-time PCR; OS: overall survival; FISH: fluorescence in situ hybridization; ChIP: chromatin immunoprecipitation; RNAi: RNA interference; siRNAs: small interfering RNAs; H&E: hematoxylin and eosin; IHC: immunohistochemical; GSEA: Gene set enrichment analysis; RNA-seq: RNA sequencing; TCGA: The Cancer Genome Atlas; WT: wild-type; EMT: epithelial-mesenchymal transition.
Supplementary Material
Supplementary figures and tables.
Acknowledgements
This research was supported by the National Natural Science Foundation of China (81972569, 81672665, 81872267, 81772925, 81702761, 81972623), the Pearl River S&T Nova Program of Guangzhou (201806010002), the Guangdong Esophageal Cancer Institute Science and Technology Program (M201802, Q201702), the Natural Science Foundation of Guangdong Grant (2017A030313615, 2018A0303130282), and the Sci-Tech Project Foundation of Guangzhou City (201607020038).
Authors' Contributions
Conceptualization, WD, TT, XW and GL; Methodology, Investigation and Data Analysis, TT, ZC, ZZ, YL, QZ, YL, HQ, QL, MC, LL, FX, GL and XW; Writing-Original Draft, TT and ZC.; Writing - Review & Editing, TT, GL, XW and WD; Funding Acquisition, WD, MC, LL and FX; Resources, GL, XW and WD; Supervision, XW and WD.
Availability of data and materials
RNA-Seq data for CRC samples: Sequence Read Archive (SRA) PRJNA562113, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA562113.
The other datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors have declared that no competing interest exists.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7-34
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424
3. Siriwardena AK, Mason JM, Mullamitha S, Hancock HC, Jegatheeswaran S. Management of colorectal cancer presenting with synchronous liver metastases. Nat Rev Clin Oncol. 2014;11:446-59
4. Ju HQ, Zhao Q, Wang F, Lan P, Wang Z, Zuo ZX. et al. A circRNA signature predicts postoperative recurrence in stage II/III colon cancer. EMBO Mol Med. 2019: e10168.
5. Ju HQ, Lu YX, Chen DL, Zuo ZX, Liu ZX, Wu QN. et al. Modulation of Redox Homeostasis by Inhibition of MTHFD2 in Colorectal Cancer: Mechanisms and Therapeutic Implications. J Natl Cancer Inst. 2019;111:584-96
6. Sveen A, Kopetz S, Lothe RA. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat Rev Clin Oncol. 2020;17:11-32
7. Smithers CC, Overduin M. Structural Mechanisms and Drug Discovery Prospects of Rho GTPases. Cells. 2016 5
8. Chen P, Wang H, Zhang W, Chen Y, Lv Y, Wu D. et al. Loss of BAP1 Results in Growth Inhibition and Enhances Mesenchymal-Epithelial Transition in Kidney Tumor Cells. Mol Cell Proteomics. 2019;18:1320-9
9. Yu OM, Benitez JA, Plouffe SW, Ryback D, Klein A, Smith J. et al. YAP and MRTF-A, transcriptional co-activators of RhoA-mediated gene expression, are critical for glioblastoma tumorigenicity. Oncogene. 2018;37:5492-507
10. T B, B X, EJ D, G D, SB L, K N. et al. The structure of the GTPase-activating domain from p50rhoGAP. Nature. 1997;385:458-61
11. Zandvakili I, Lin Y, Morris JC, Zheng Y. Rho GTPases: Anti- or pro-neoplastic targets? Oncogene. 2017;36:3213-22
12. Ding Z, Dhruv H, Kwiatkowska-Piwowarczyk A, Ruggieri R, Kloss J, Symons M. et al. PDZ-RhoGEF Is a Signaling Effector for TROY-Induced Glioblastoma Cell Invasion and Survival. Neoplasia. 2018;20:1045-58
13. Stiegler AL, Boggon TJ. p190RhoGAP proteins contain pseudoGTPase domains. Nat Commun. 2017;8:506
14. Leibovitz A, Stinson JC, McCombs WB 3rd, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562-9
15. Ju HQ, Ying H, Tian T, Ling J, Fu J, Lu Y. et al. Mutant Kras- and p16-regulated NOX4 activation overcomes metabolic checkpoints in development of pancreatic ductal adenocarcinoma. Nat Commun. 2017;8:14437
16. Tian T, Li J, Li Y, Lu YX, Tang YL, Wang H. et al. Melatonin enhances sorafenib-induced cytotoxicity in FLT3-ITD acute myeloid leukemia cells by redox modification. Theranostics. 2019;9:3768-79
17. Ju HQ, Li H, Tian T, Lu YX, Bai L, Chen LZ. et al. Melatonin overcomes gemcitabine resistance in pancreatic ductal adenocarcinoma by abrogating nuclear factor-kappaB activation. J Pineal Res. 2016;60:27-38
18. Che Y, Li Y, Zheng F, Zou K, Li Z, Chen M. et al. TRIP4 promotes tumor growth and metastasis and regulates radiosensitivity of cervical cancer by activating MAPK, PI3K/AKT, and hTERT signaling. Cancer Lett. 2019;452:1-13
19. Heraud C, Pinault M, Lagree V, Moreau V. p190RhoGAPs, the ARHGAP35- and ARHGAP5-Encoded Proteins, in Health and Disease. Cells. 2019 8
20. Chen D, Li Y, Zhang X, Wu H, Wang Q, Cai J. et al. Ubiquitin ligase TRIM65 promotes colorectal cancer metastasis by targeting ARHGAP35 for protein degradation. Oncogene. 2019;38:6429-44
21. Y K, S M, Y K, K M, Y N, T I. et al. Cell cycle-dependent Rho GTPase activity dynamically regulates cancer cell motility and invasion in vivo. PLoS One. 2013;8:e83629
22. G K, C F, M Y, KC Y. Reduced RhoA expression enhances breast cancer metastasis with a concomitant increase in CCR5 and CXCR4 chemokines signaling. Scientific reports. 2019;9:16351
23. R L, B Z, Y Z. Structural determinants required for the interaction between Rho GTPase and the GTPase-activating domain of p190. The Journal of biological chemistry. 1997;272:32830-5
24. N T, DA L, IG M. The function of the p190 Rho GTPase-activating protein is controlled by its N-terminal GTP binding domain. The Journal of biological chemistry. 1998;273:34631-8
25. WA C, RS D, EJ N. The many faces of CREB. Trends in neurosciences. 2005;28:436-45
26. W S, J L, L Z, J H, R L, H Z. et al. The c-Myc/miR-27b-3p/ATG10 regulatory axis regulates chemoresistance in colorectal cancer. Theranostics. 2020;10:1981-96
27. L S, Y F, X W, Y H, F D, C L. et al. miR-302a Inhibits Metastasis and Cetuximab Resistance in Colorectal Cancer by Targeting NFIB and CD44. Theranostics. 2019;9:8409-25
28. Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol. 2017;14:235-46
29. Cremolini C, Loupakis F, Antoniotti C, Lupi C, Sensi E, Lonardi S. et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015;16:1306-15
30. Schrock AB, Ouyang C, Sandhu J, Sokol E, Jin D, Ross JS. et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann Oncol. 2019
31. Zhao P, Li L, Jiang X, Li Q. Mismatch repair deficiency/microsatellite instability-high as a predictor for anti-PD-1/PD-L1 immunotherapy efficacy. J Hematol Oncol. 2019;12:54
32. Venook AP, Niedzwiecki D, Lenz HJ, Innocenti F, Fruth B, Meyerhardt JA. et al. Effect of First-Line Chemotherapy Combined With Cetuximab or Bevacizumab on Overall Survival in Patients With KRAS Wild-Type Advanced or Metastatic Colorectal Cancer: A Randomized Clinical Trial. JAMA. 2017;317:2392-401
33. Liao TT, Yang MH. Revisiting epithelial-mesenchymal transition in cancer metastasis: the connection between epithelial plasticity and stemness. Mol Oncol. 2017;11:792-804
34. Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell. 2017;168:670-91
35. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178-96
36. Settleman J, Albright CF, Foster LC, Weinberg RA. Association between GTPase activators for Rho and Ras families. Nature. 1992;359:153-4
37. Li L, Li YM, Zhou P, Wang XS, Wang GY, Zhao XH. et al. Abnormal expression of p190RhoGAP in colorectal cancer patients with poor survival. Am J Transl Res. 2016;8:4405-14
38. Organ SL, Hai J, Radulovich N, Marshall CB, Leung L, Sasazuki T. et al. p120RasGAP is a mediator of rho pathway activation and tumorigenicity in the DLD1 colorectal cancer cell line. PLoS One. 2014;9:e86103
39. Wang J, Tian X, Han R, Zhang X, Wang X, Shen H. et al. Downregulation of miR-486-5p contributes to tumor progression and metastasis by targeting protumorigenic ARHGAP5 in lung cancer. Oncogene. 2014;33:1181-9
40. Gen Y, Yasui K, Zen K, Nakajima T, Tsuji K, Endo M. et al. A novel amplification target, ARHGAP5, promotes cell spreading and migration by negatively regulating RhoA in Huh-7 hepatocellular carcinoma cells. Cancer Lett. 2009;275:27-34
41. Dong G, Wang B, An Y, Li J, Wang X, Jia J. et al. SIRT1 suppresses the migration and invasion of gastric cancer by regulating ARHGAP5 expression. Cell Death Dis. 2018;9:977
42. Earp M, Tyrer JP, Winham SJ, Lin HY, Chornokur G, Dennis J. et al. Variants in genes encoding small GTPases and association with epithelial ovarian cancer susceptibility. PLoS One. 2018;13:e0197561
43. Fang Y, Zhu X, Wang J, Li N, Li D, Sakib N. et al. MiR-744 functions as a proto-oncogene in nasopharyngeal carcinoma progression and metastasis via transcriptional control of ARHGAP5. Oncotarget. 2015;6:13164-75
44. Liu Y, Lang T, Jin B, Chen F, Zhang Y, Beuerman RW. et al. Luteolin inhibits colorectal cancer cell epithelial-to-mesenchymal transition by suppressing CREB1 expression revealed by comparative proteomics study. J Proteomics. 2017;161:1-10
45. Sakaguchi M, Hisamori S, Oshima N, Sato F, Shimono Y, Sakai Y. miR-137 Regulates the Tumorigenicity of Colon Cancer Stem Cells through the Inhibition of DCLK1. Mol Cancer Res. 2016;14:354-62
46. Lin Y, Zheng Y. Approaches of targeting Rho GTPases in cancer drug discovery. Expert Opin Drug Discov. 2015;10:991-1010
47. Maldonado MDM, Dharmawardhane S. Targeting Rac and Cdc42 GTPases in Cancer. Cancer Res. 2018;78:3101-11
48. Humphries-Bickley T, Castillo-Pichardo L, Hernandez-O'Farrill E, Borrero-Garcia LD, Forestier-Roman I, Gerena Y. et al. Characterization of a Dual Rac/Cdc42 Inhibitor MBQ-167 in Metastatic Cancer. Mol Cancer Ther. 2017;16:805-18
Author contact
![]() Corresponding authors: Guangyu Luo, Sun Yat-Sen University Cancer Center, Guangzhou, China; E-mail: luogyorg.cn; or Xiaojun Wu, Sun Yat-Sen University Cancer Center, Guangzhou, China; E-mail: wuxjorg.cn; or Wuguo Deng, Sun Yat-Sen University Cancer Center, Guangzhou, China; E-mail: degnwgorg.cn.
Corresponding authors: Guangyu Luo, Sun Yat-Sen University Cancer Center, Guangzhou, China; E-mail: luogyorg.cn; or Xiaojun Wu, Sun Yat-Sen University Cancer Center, Guangzhou, China; E-mail: wuxjorg.cn; or Wuguo Deng, Sun Yat-Sen University Cancer Center, Guangzhou, China; E-mail: degnwgorg.cn.
 Global reach, higher impact
Global reach, higher impact